Background: Cytochrome c maturation (Ccm) forms thioether bonds between heme b and c-type apocytochromes.
Results: CcmI chaperone exhibits a very high binding affinity for the class I c-type apocytochromes, which decreases drastically in the presence of heme.
Conclusion: CcmI holds the c-type apocytochromes tightly until heme coordination yields their b-type intermediates during Ccm.
Significance: Interactions between the c-type apocytochromes and chaperones are critical for the Ccm process.
Keywords: bacteria, chaperone, cytochrome, photosynthesis, protein-protein interaction, respiration, biolayer interferometry, Rhodobacter capsulatus, apocytochrome, cytochrome c maturation
Abstract
The c-type cytochromes are electron transfer proteins involved in energy transduction. They have heme-binding (CXXCH) sites that covalently ligate heme b via thioether bonds and are classified into different classes based on their protein folds and the locations and properties of their cofactors. Rhodobacter capsulatus produces various c-type cytochromes using the cytochrome c maturation (Ccm) System I, formed from the CcmABCDEFGHI proteins. CcmI, a component of the heme ligation complex CcmFHI, interacts with the heme-handling protein CcmE and chaperones apocytochrome c2 by binding its C-terminal helix. Whether CcmI also chaperones other c-type apocytochromes, and the effects of heme on these interactions were unknown previously. Here, we purified different classes of soluble and membrane-bound c-type apocytochromes (class I, c2 and c1, and class II c′) and investigated their interactions with CcmI and apoCcmE. We report that, in the absence of heme, CcmI and apoCcmE recognized different classes of c-type apocytochromes with different affinities (nm to μm KD values). When present, heme induced conformational changes in class I apocytochromes (e.g. c2) and decreased significantly their high affinity for CcmI. Knowing that CcmI does not interact with mature cytochrome c2 and that heme converts apocytochrome c2 into its b-type derivative, these findings indicate that CcmI holds the class I apocytochromes (e.g. c2) tightly until their noncovalent heme-containing b-type cytochrome-like intermediates are formed. We propose that these intermediates are subsequently converted into mature cytochromes following the covalent ligation of heme via the remaining components of the Ccm complex.
Introduction
The c-type cytochromes are ubiquitous electron transfer proteins involved in energy transduction in almost all living cells, and they also play critical roles in other cellular pathways (e.g. apoptosis in eukaryotes) (1–3). These proteins always contain at least one conserved heme-binding site (C1XXC2H), where heme b (protoporphyrin IX-Fe) is covalently ligated. The stereospecificity of the thioether bonds formed between the vinyl-2 and vinyl-4 of heme and the thiols of Cys1 and Cys2 of the heme-binding site, respectively, is universally conserved (4). The His residue of the heme-binding site, together with another Met or His residue, provides axial ligation to heme iron (5). Despite these common features, the c-type cytochromes are diverse in terms of size, three-dimensional structure, heme content, and physicochemical properties. Previously, Ambler (6) grouped the c-type cytochromes into four broad classes. Class I is a large group that includes small, globular, and soluble c-type cytochromes. They usually contain a single N-terminal heme-binding site with a Met residue as the sixth ligand, located near their C termini (e.g. mitochondrial cytochrome c). They are divided into subfamilies according to their structures, functions, and the properties of their cytochrome domains (1, 7). Class II c-type cytochromes include the high-spin cytochrome c′ with a C-terminally located heme-binding motif and a four-helical bundle fold. Class III c-type cytochromes comprise the low Em (redox potential) multi-heme proteins with generally bis-His coordination, and the c-type cytochromes with additional non-heme cofactors (e.g. flavins), such as flavocytochrome c3, are grouped in class IV.
Rhodobacter capsulatus produces a variety of c-type cytochromes under different growth conditions. These include the class I C-terminally membrane-bound cytochrome c1 subunit of the cytochrome bc1 complex (8) and the N-terminally membrane-attached cytochrome cp and co subunits of the cbb3-type oxygen reductase (9, 10), as well as the soluble cytochrome c2 and the N-terminally membrane-attached cytochrome cy as electron carriers (11, 12). The class II soluble high-spin cytochrome c′ is involved in NO detoxification (13), and the class III membrane-attached pentaheme c-type cytochrome DorC conveys electrons from the Q/QH2 pool to dimethyl sulfoxide (DMSO)3 reducing it to dimethylsulfide (14).
R. capsulatus and other α- and γ-proteobacteria, archaea, and mitochondria of plants and red algae carry out the process of covalent heme ligation to the c-type apocytochromes via a membrane complex, designated as cytochrome c maturation (Ccm) System I (15–18). The overall process relies on several cellular pathways, including post-translational modification and secretion of c-type apocytochromes and the folding and degradation of proteins, as well as maintenance of a suitable thioredox environment conducive to cofactor insertion. The Ccm complex involves nine membrane proteins (CcmABCDEFGHI) that are responsible for the chaperoning of c-type apocytochromes and heme as well as their covalent ligation (15).
CcmI is composed of two different domains and forms with CcmF and CcmH a multisubunit protein complex responsible for heme ligation (19–23). The N-terminal CcmI-1 domain is membrane-integral via two transmembrane helices and has a cytoplasmic loop with a leucine zipper-like motif. The large periplasmic C-terminal CcmI-2 domain contains three tetratricopeptide repeats (TPR) (20, 24–26). TPR domains are involved in protein-protein interactions and form two anti-parallel α-helices packed in tandem arrays as a superhelical structure with a convex and a concave surface where the target proteins bind (27). Genetic studies indicate that the CcmI-1 domain of R. capsulatus is required for the maturation of all c-type cytochromes, whereas some amount of C-terminally membrane-anchored cytochrome c1 is produced in the absence of CcmI-2 (20). Recently we (25) and others (28, 29) showed that CcmI binds as a chaperone the C-terminal helix of apocytochrome c2, primarily via its TPR-containing CcmI-2 domain.
CcmE is a heme-handling membrane protein with a β-barrel domain and a flexible C-terminal stretch (30–32). It binds covalently vinyl-2 of heme b through a surface-exposed His residue at its conserved HXXXY site (33–35). HoloCcmE formation and delivery of heme to c-type apocytochromes rely on a specific ABC-type transporter complex (CcmABCD). Once apoCcmE is heme-loaded, an ATP hydrolysis-dependent conformational change (36) renders it competent to deliver heme to c-type apocytochromes. Recently, we found that apoCcmE interacts with the N-terminal heme-binding region of apocytochrome c2 and forms a ternary complex together with CcmI in vitro (37). Moreover, in R. capsulatus membrane fractions, apoCcmE also interacts with both CcmI and CcmH (37). In addition, holoCcmE is known to form a complex with CcmF in Escherichia coli (38). Altogether, these findings indicate that the heme ligation complex, CcmFHI, contains CcmE and CcmG, possibly forming a large “maturase supercomplex” (15).
In this study, we investigated the binding interactions among CcmI, apoCcmE, and different c-type apocytochromes that are distinct from apocytochrome c2 in order to understand how R. capsulatus Ccm System I matures many structurally dissimilar c-type cytochromes. We also explored for the first time how the availability of heme affects these chaperone-apocytochrome interactions. We found that CcmI and apoCcmE bind different c-type apocytochromes with markedly different affinities (KD values) and that the strength of these interactions does not correlate with the distinct secondary structures. Remarkably, heme modulates these binding interactions significantly, suggesting that CcmI holds the c-type apocytochromes tightly until their intermediate b-type derivatives are formed. We propose that these intermediates are subsequently converted into mature c-type cytochromes upon completion of covalent heme ligation by the remaining components of the Ccm complex.
Experimental Procedures
Bacterial Strains and Growth Conditions
The bacterial strains and plasmids used in this work are described in Table 1. E. coli strains were grown aerobically at 37 °C in Luria-Bertani broth medium supplemented with ampicillin (100 μg/ml). Cultures were induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside (25). R. capsulatus strains were grown chemoheterotrophically (i.e. by respiration) at 35 °C on MPYE (mineral-peptone-yeast-extract) enriched medium supplemented with tetracycline or spectinomycin at 2.5 or 10 μg/ml, respectively (39).
TABLE 1.
Strains and plasmids used in this work
Res and Ps refer to respiratory and photosynthetic growth, respectively; Nadi refers to cytochrome c oxidase-dependent catalysis of α-naphtol to indophenol blue. R. capsulatus MT1131 is referred to as the wild type with respect to its c-type cytochrome profile and growth properties.
| Strains/Plasmids | Relevant characteristics | References |
|---|---|---|
| Bacteria | ||
| R. capsulatus | ||
| MT1131 | Wild type ctrD121crtD121; Rifr Res+ Nadi+ Ps+ | |
| MT-G4/S4 | Δ(cycA::kan); Res+ Nadi+ Ps+ | (58) |
| MTSRP1 | Δ(ccmI::kan); Res+ Nadi− Ps− | (19) |
| MD2 | Δ(ccmE::spec); Res+ Nadi− Ps− | (44 |
| E. coli | ||
| HB101 | F− Δ(gpt-proA)62 araC14 leuB6(Am) glnV44(AS) galK2(Oc) lacY1 Δ(mcrC-mrr) rpsL20(Strr) xylA5 mtl-1 thi-1 | Stratagene |
| Plasmids | ||
| pMAM1 | pCS1302 derivative, containing a truncated R. capsulatus petC gene cloned into the NdeI/BamHI sites, with a Strep-tag II fused at its 5′-end, without a signal sequence and missing the last 39 C-terminal residues, Ampr | This work |
| pMAM1ΔCys | pMAM1 derivative with Cys-34 and Cys-37 mutated to Ser, Ampr | This work |
| pAV6 | pCS1302 derivative, contains mature R. capsulatus cycP, cloned into the NdeI/BamHI sites without its native signal sequence and with a Strep-tag II fused at its 5′-end, Ampr | |
| pAV5 | pCS1302 derivative, contains mature H. thermophilus cyt c552 gene from pCS1208, cloned into the NdeI/BamHI without its signal sequence but with a Strep-tag II fused at its 5′-end, Ampr | This work |
| pAV5C13SC16S | pAV5 derivative with Cys-13 and Cys-16 mutated to Ser, Ampr | This work |
| pCS1302 | pET-3a derivative with a Strep-tag II sequence fused to GFP rendering GFP replaceable by cloning any gene of interest in-frame into NdeI and BamHI sites, Ampr | (23) |
| pCS1208 | pBSK derivative, contains full-length H. thermophilus cyt c552 gene carrying both 5′-end NdeI and 3′-end BamHI sites, cloned into the EcoRV site, Ampr | This work |
| pPET1-C144A/C167A/A181T | pPET1 derivative contains petABC operon (encoding R. capsulatus cyt bc1) with petC carrying the C144A, C167A, and A181T mutations, Ampr | (40) |
Molecular Genetic Techniques
Apocytochromes c1 and c′ and their derivatives were produced as done earlier for apocytochrome c2 (25). R. capsulatus native cytochrome c1 (petC) has four Cys residues (Cys-34 and Cys-37 of the C1XXC2H heme-binding site and Cys-144 and Cys-167, which form a disulfide bond) (39). Mutating the latter two Cys residues of cytochrome c1 does not affect its maturation but lowers its Em and renders it nonfunctional. An additional mutation, A181T, in the heme environment corrects this defect to yield a fully functional cytochrome c1 variant (40). This disulfide-less apocytochrome c1, chosen to avoid complications that could arise from the extra Cys residues, was considered the “wild type” for maturation purposes. Two different apocytochrome c1 derivatives (pMAM1 and pMAM2) were constructed by PCR amplification using the mutant petC allele on plasmid pPET1-C144A/C167A/A181T (40) as a template and the primers NdeI-Cytc1-Fw (inserting a NdeI restriction site at the 5′-end and removing the cytochrome c1 signal sequence) and Cytc1_BamHI-Rv or Cytc1t39_BamHI-Rv (inserting a BamHI restriction site either at the 3′-end of petC or 117 bp upstream of its stop codon, respectively) (Table 2). The PCR products were cloned into the same restriction sites in pCS1302 (23) to yield N-terminally Strep II-tagged signal sequence-less apocytochrome c1 derivatives with a Factor Xa cleavage site for tag removal. Plasmid pMAM1 encoded a soluble variant of apocytochrome c1 lacking its C-terminal 39-amino acid-long membrane anchor (apocytochrome c1t39), and pMAM2 encoded full-length apocytochrome c1 (Table 1). Similarly, a signal sequence-less and N-terminally Strep II-tagged apocytochrome c′ (RCC02682 corresponding to cycP) was obtained by PCR amplification using R. capsulatus chromosomal DNA as a template and the primers NdeI-c′-Fw and c′-BamHI-Rv containing the NdeI and BamHI restriction sites, yielding plasmid pAV6 after its cloning into pCS1302 (Tables 1 and 2). In addition, a mature form of Hydrogenobacter thermophilus cytochrome c552 was obtained by PCR amplification using plasmid pCS12084 (Table 1) as a template and primers Htssdel-Fw (inserting an NdeI site immediately downstream of its signal sequence) and CS46 (located 3′ of the BamHI restriction site) (Table 2). The PCR product was cloned into pCS1302 to yield pAV5, producing Ht-apocytochrome c552 lacking its signal sequence. Plasmid pAV5C13SC16S containing the double Cys to Ser substitutions at the heme-binding site of Ht-apocytochrome c552 was derived from pAV5 using a QuikChange site-directed mutagenesis kit (Invitrogen) and Htc552C13/C16-Fw and Htc552C13/C16-Rv primers (Tables 1 and 2) according to the supplier's recommendation. All constructs were analyzed by serial cloner 2.1 and confirmed by DNA sequencing.
TABLE 2.
Nucleotide sequences of the oligonucleotide primers used in this study
| Designation | Constructs | Nucleotide sequence |
|---|---|---|
| 5′ to 3′ | ||
| NdeI_Cytc1-Fw | pMAM1 and pMAM2 | GCCTTTGCGAACCTCCCATATGCCGGATCACGCCTTCAGC |
| Cytc1t39_BamHI-Rv | PMAM1 | CCCATCTGCTTGCGCGGATCCAGTTACGGTTCCGCGGCCC |
| Cytc1_BamHI-Rv | pMAM2 | CAGCTGTCCGGATCCTCTTAGGCCTTGTGGCC |
| Cytc1t39_C34SC37S-Fw | pMAM1Cys/Ser | CTACAACGAAGTCAGCTCGGCCAGCCACGGCATGAAG |
| Cytc1t39_C34SC37S-Rv | CTTCATGCCGTGGCTTGGCCGAGCTGACTTCGTTGTAG | |
| NdeI-c′-Fw | pAV6 | GGCTCGGCCGCCCATATGGCTGATACC |
| c′-BamHI-Rv | CCCCCGCCCCGGGATCCTTAGTCTTCTTCG | |
| Htssdel-Fw | pAV5 | GGCATACATATGGCCAATGAACAGCTTGCCAAGC |
| CS46 | GCTAGTTATTGCTCAGCGG | |
| Htc552C13/C16-Fw | pAV5C13SC16S | GCTTGCCAAGCAAAAGGGCGCTATGGCTGCCCACGATCTGAAAGCTAAG |
| Htc552C13/C16-Rv | CTTAGCTTTCAGATCGTGGGCAGCCATAGCGCCCTTTTGCTTGGCAAGC |
Protein Purification
The proteins His10-CcmI, His10-CcmI-2, His10-apoCcmE, and FLAG-CcmI were purified by affinity chromatography using Ni-Sepharose high performance (GE Healthcare) and anti-FLAG® M2 affinity (Sigma) resins, respectively (25, 37). Strep-tagged c-type apocytochromes were purified as described earlier (25). The Cys-less variant of H. thermophilus cytochrome c552, Ht-apocytochrome b-c552, was incubated overnight with 50 mm Tris-HCl, 50 mm NaCl, and 1 m imidazole for heme removal. The imidazole displaced the heme axial ligands, leading to precipitation of the heme, which was removed by centrifugation at 14,000 × g for 15 min, originating Ht-apocytochrome b-c552. Purified protein samples were checked by SDS-PAGE for purity (>95%), concentrated by ultrafiltration, and desalted using PD-10 columns (GE Healthcare). A synthetic peptide carrying a Strep II tag and Factor Xa cleavage site, corresponding to cytochrome c1 residues 222–241, was produced by Thermo Fisher Scientific.
Protein-Protein Interactions Monitored by Co-purification Assays
Direct interactions among His10-apoCcmE, FLAG-CcmI, and different Strep-tagged c-type apocytochromes were assayed as described previously (25). Briefly, equimolar amounts of Strep-tagged c-type apocytochromes (∼1 μm) were mixed with substoichiometric amounts of His10-apoCcmE or FLAG-CcmI (∼0.1 μm) in the assay buffer (50 mm Tris-HCl, 50 mm NaCl, pH 8.0, final volume of 400 μl) and incubated for 2 h at 25 °C with gentle shaking. The mixture was loaded onto a mini (200 μl volume) Strep-Tactin resin column equilibrated with the same buffer. The column was washed extensively with 2 ml of assay buffer (10 column volumes) and eluted with the same buffer containing 2.5 mm desthiobiotin. Flow-through and elution fractions were precipitated with methanol:acetone (7:2, v/v) overnight at −20 °C, and interacting partners were analyzed by SDS-PAGE. Binding assays using the synthetic peptides instead of the c-type apocytochromes followed a similar protocol. As appropriate, different amounts of hemin (i.e. heme chloride) (Frontier Scientific Inc.) dissolved in DMSO (determined using the extinction coefficient of 179 cm−1 mm−1 at 400 nm in 40% DMSO (41)) were added to the incubation mixtures.
Protein-Protein Interactions Monitored by Biolayer Interferometry
The binding kinetics of His10-CcmI and His10-apoCcmE to different Strep-tagged c-type apocytochromes was monitored quantitatively in real time by biolayer interferometry (BLI) using an Octet RED96 instrument (ForteBio). Purified c-type apocytochromes (i.e. ligands) were biotinylated using the EZ-LinkTM NHS-PEG4 biotinylation kit (Thermo Scientific) to immobilize them on streptavidin-coated biosensors (SA-sensors). SA-sensors were loaded with biotinylated c-type apocytochromes (Bt-apocytochromes) by soaking them in a buffer containing the desired Bt-apocytochrome at ∼400 nm, 50 mm Tris-HCl, pH 8, 100 mm NaCl, 0.01% n-dodecyl-β-d-maltoside, and 1% BSA at 30 °C and 1000 rpm with shaking. A reference sensor was dipped into a well containing the assay buffer and lacking the Bt-apocytochrome to assess nonspecific binding of the analyte (CcmI or apoCcmE) to the sensor. After the SA-sensors were washed with the same buffer, unoccupied residual streptavidin sites on the SA-sensors were blocked with biocytin (10 μg/ml), and following another washing step, a baseline signal was recorded. Different Bt-apocytochrome-loaded SA-sensors were incubated with increasing concentrations of analyte (i.e. CcmI from 4 nm to 30 μm or apoCcmE from 0.3 to 20 μm) (association step). Subsequent washing of the biosensors with the assay buffer released the analyte (CcmI or apoCcmE) from the immobilized ligand (dissociation step). An assay lacking the analyte was used as a negative control to confirm that the observed shifts were due to the ligand-analyte complexes. The collected data were used to determine the kinetic parameters. The range of concentrations used depended on the Bt-apocytochrome tested to obtain data under nonsaturating binding conditions. Higher concentrations of CcmI or apoCcmE were needed in the case of class II apocytochrome c′, which enhanced nonspecific binding to the sensors. To mitigate this problem, the n-dodecyl-β-d-maltoside concentration of the wash buffer was increased to 0.05%. The kinetics performed in the presence of heme used FLAG-CcmI instead of His10-CcmI as an analyte to avoid possible binding of heme to the His10 epitope. In addition, the standard assay buffer containing BSA (known to bind heme; see Ref. 42) was substituted with 50 mm Tris-HCl, pH 8, 150 mm NaCl, and 0.01% Tween-20. Under these conditions, hemin was used at concentrations ranging from 0.1 to 6.4 μm to monitor its binding to apocytochrome c2. To investigate the effect of heme on CcmI-apocytochrome c2 interactions, we first repeated full kinetic measurements using this buffer and FLAG-CcmI at concentrations ranging from 0.7 to 180 nm. Then, the assay buffer was supplemented with 2 μm hemin to yield a new baseline (accounting for binding of heme to apocytochrome c2). Increased FLAG-CcmI concentrations (from 0.07 to 2.4 μm) were used to account for decreased apparent association responses. The kon and koff rates of binding and the KD values for each interacting pair were determined by fitting the experimental data to 1:1 homogenous or 2:1 heterogeneous kinetic models describing bimolecular interactions according to the manufacturer's literature (ForteBio) (43). The quality of the fit between the experimental and calculated data was evaluated according to the following parameters: error values for kon and koff (at least an order of magnitude lower than the k values), residual values (<10% of the maximum response of the fitting curve), R2 (>0.95) and X2 (<3) (43).
SDS-PAGE and Immunoblot Analyses
SDS-PAGE under reducing conditions (5% β-mercaptoethanol) was performed using 15% gels according to Laemmli (44), and covalently bound heme-containing proteins were detected using tetramethylbenzidine (TMBZ) as described elsewhere (45). For apocytochrome c′ immunodetection, gel-resolved proteins were electroblotted onto Immobilon-P PVDF membranes (Millipore) and probed with rabbit polyclonal antibodies specific for R. capsulatus cytochrome c′ (a kind gift of Dr. R. Prince). Horseradish peroxidase-conjugated anti-rabbit IgG antibodies (GE Healthcare) were used as secondary antibodies, and detection was performed using SuperSignalTM West Pico chemiluminescent substrate from Thermo Scientific.
Circular Dichroism Spectroscopy
The far-UV circular dichroism (CD) spectra (195–240 nm) were recorded with a model 202 spectropolarimeter (AVIV® Biomedical, Inc) using a 2-mm-path length cuvette (Hellma, Inc.) as done previously (25). The CD spectra of proteins (15 μm) in 20 mm sodium phosphate buffer, pH 7.5, were recorded using a 3-nm bandwidth, a 2-nm step size, and a time constant of 10 s. The CD spectrum of the buffer was subtracted from the spectra of the proteins, and the absorbance values were converted to the mean residue ellipticity [θ]λ (deg cm2 dmol−1) at each wavelength using the relation [θ]λ = θλ/(10 × C × n × l), where θλ is the observed ellipticity in millidegrees at wavelength λ, C is the molar protein concentration, n is the number of amino acids of the protein, and l is the path-length of the cuvette in cm. The CD spectra monitoring the effect of hemin on apocytochrome c2 or CcmI or their interactions were recorded using 1-nm step size and 20 mm sodium phosphate buffer, pH 7.5, supplemented with 10 mm potassium cyanide to prevent hemin aggregation. Hemin stock solution was prepared in 100 mm NaOH, and the concentration was determined using the extinction coefficient of 5.84 × 104 cm−1 m−1 at 385 nm in the same solution (46). The spectra were recorded 2 h after hemin addition to ensure its complete binding to apocytochrome c2 and that full conformational changes had been induced under oxidizing conditions. The CD spectra of the buffers (with or without hemin) were subtracted from the spectra of the proteins, and the absorbance values were converted into the mean residue ellipticity [θ]λ (deg cm2 dmol−1) as described above. To determine the effect of heme on apocytochrome c2-CcmI interactions, the protein mixtures (molar ratio of apocytochrome c2 to CcmI, 2:1) were incubated for 2 h at room temperature without or with hemin (at 2–8-fold molar excess of apocytochrome c2), and their CD spectra were compared with the sum of the spectra of individual proteins obtained under the same conditions after subtraction of the spectral contributions of the corresponding buffers.
Reconstitution of b-Type Cytochrome Intermediates
A stoichiometric amount of hemin dissolved in DMSO was added slowly from a stock solution of 1 mm to 10 μm c-type apocytochrome in 50 mm Tris-HCl, 150 mm NaCl, pH 8. The sample was stirred for 5–10 min between each addition to reach equilibrium, and visible spectra between 380 and 650 nm were taken to monitor the binding of hemin to the c-type apocytochromes. Unbound hemin was removed by size exclusion chromatography (PD-10 column, GE Healthcare), and after concentration, the visible spectra of the newly formed b-type cytochromes were recorded as prepared (air-oxidized) and after dithionite reduction. The relative amounts of reconstituted b-type cytochrome derivatives of R. capsulatus c-type apocytochromes were determined by taking as 100% the amount of Ht-apocytochrome b-c552 reconstituted under the same conditions.
Chemicals
All chemicals and solvents were of high purity and HPLC spectral grades and were purchased from commercial sources.
Results
Overproduction and Purification of CcmI, apoCcmE, and c-Type Apocytochromes
We showed previously that R. capsulatus CcmI binds tightly to the C-terminal helix, whereas apoCcmE interacts with the N-terminal heme-binding region of apocytochrome c2 (25, 37). As this bacterium produces various c-type cytochromes, in the current work we inquired whether these interactions were exclusive to apocytochrome c2 or more general, including other c-type apocytochromes. Considering that maturation of cytochrome c′ in R. capsulatus had not been examined earlier, we first analyzed soluble extracts of R. capsulatus mutants lacking CcmI or CcmE (MT-SRP1 (20) or MD2 (47), respectively) using SDS-PAGE/TMBZ staining and immunodetection with cytochrome c′ antibodies. These mutants lacked cytochrome c′ (Fig. 1), confirming that CcmI and CcmE of Ccm System I were required for its maturation. Based on these findings, we chose in addition to apocytochrome c2 the class I membrane-anchored cytochrome c1, for which maturation is independent of the CcmI-2 domain of CcmI, and the class II-soluble cytochrome c′, which has a nonglobular three-dimensional structure (Fig. 2A).
FIGURE 1.
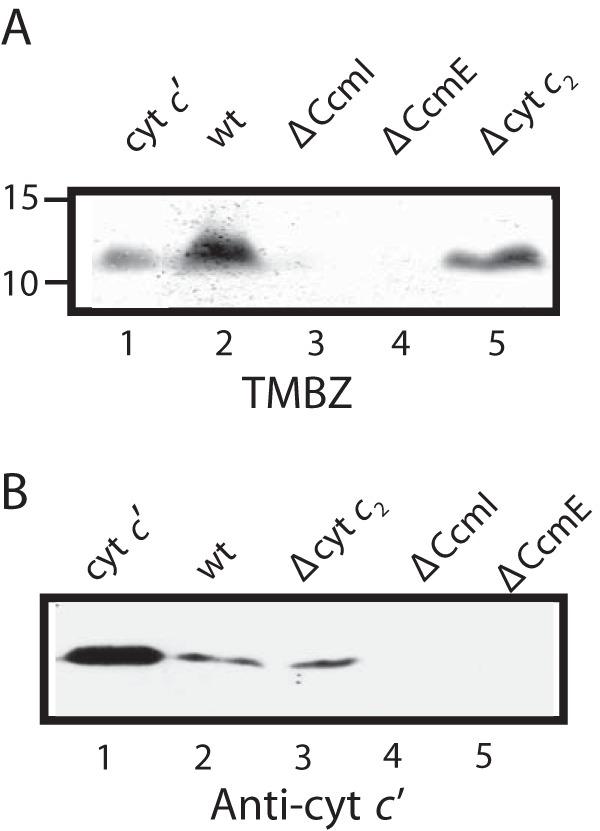
CcmI and CcmE are required for maturation of class II cytochrome c′. A, SDS-PAGE/TMBZ heme staining using 100 μg of total soluble protein extracts from the appropriate R. capsulatus strains: 2 μg of purified cytochrome c′ as a positive control for heme staining (lane 1), wild type MT1131 (lane 2), CcmI-null strain MTSRP1 (lane 3), CcmE-null strain MD2 (lane 4), and cytochrome c2-null strain MT-G4/S4 (lane 5). B, SDS-PAGE/immunoblot using anti-cytochrome c′ polyclonal antibodies with 100 μg of total soluble protein extracts from the appropriate R. capsulatus strains: 2 μg of purified cytochrome c′ as a positive control (lane 1), wild type MT1131 (lane 2), cytochrome c2-null strain MTG4/S4 (lane 3), CcmI-null strain MTSRP1 (lane 4), and CcmE-null strain MD2 (lane 5).
FIGURE 2.
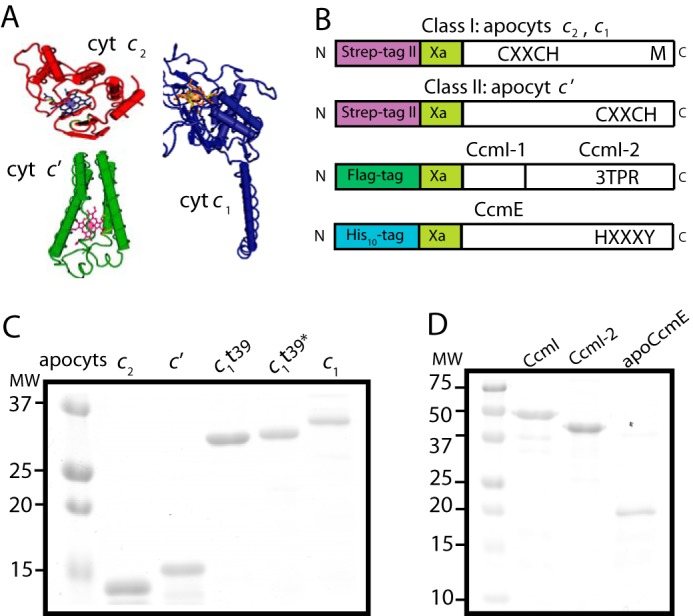
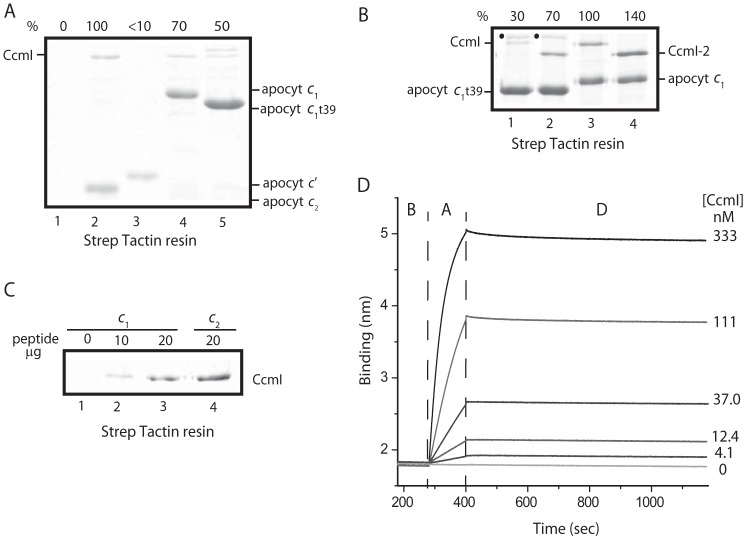
Purification of various R. capsulatus c-type apocytochromes, and CcmI and apoCcmE. A, three-dimensional structures of class I cytochrome c2 (Cyt c2; Protein Data Bank code: 1C2R), cytochrome c1 (Cyt c1; Protein Data Bank code: 1ZRT), and class II cytochrome c′ (Cyt c′; Protein Data Bank code: 1RCP). B, schematic representations of c-apocytochrome constructs with N-terminal Strep-tag followed by a Factor Xa cleavage site. The heme-binding site, C1XXC2H, is located close to the N and C termini in class I and class II c-type apocytochromes, respectively. The sixth axial ligand (M) in class I c-type apocytochromes is located at the C terminus, whereas in class II, apocytochrome c′ heme has no sixth axial ligand. CcmI, formed from the CcmI-1 and CcmI-2 (with its TPR motifs) domains, has an N-terminal FLAG tag followed by a Factor Xa cleavage site. ApoCcmE (with its HXXXY heme binding motif) has an N-terminal His10 tag fused at its membrane anchor. C, Coomassie Blue-stained SDS-PAGE containing 3 μg of Strep-Tactin Sepharose-purified Strep-apocytochrome c2 (lane 1), Strep-apocytochrome c′ (lane 2), Strep-apocytochrome c1t39 (lane 3), Cys-less derivative of Strep-apocytochrome c1t39* (lane 4), and Strep-apocytochrome c1 (lane 5). t39, refers to the truncation of 39 C-terminal amino acid residues encompassing the membrane anchor of cytochrome c1. D, Coomassie Blue-stained SDS-PAGE containing 3 μg of Anti-FLAG Sepharose-purified CcmI (lane 1), Ni-Sepharose-purified His10-CcmI-2 (lane 2), and apoCcmE (lane 3).
We overproduced in E. coli cytoplasm and purified Strep-tagged versions of the c-type apocytochromes and their derivatives, as done previously (25) (Fig. 2B). Strep-apocytochrome c′ (molecular mass of 15 kDa), produced at ∼1–2 mg/liter of culture, was prone to degradation, like the Strep-apocytochrome c2 (13.5 kDa) Fig. 2C, lanes 1 and 2). The full-length version (Strep-apocytochrome c1, 30 kDa), its C-terminal membrane-anchored (39 amino acid residues) truncated (Strep-apocytochrome c1t39, 27 kDa) soluble version, and also a Cys-less derivative of Strep-apocytochrome c1t39 were produced at large amounts (>10 mg/liter culture) (Fig. 2C). All Strep-Tactin affinity chromatography-purified c-type apocytochromes were devoid of heme, as confirmed by TMBZ staining and visible spectroscopy (data not shown). The truncated Strep-apocytochrome c1t39 readily formed intermolecular disulfide bonds to yield homodimers (∼55 kDa) even under reducing conditions. We also purified FLAG-CcmI (50 kDa), His10-CcmI (50 kDa), His10-CcmI-2 (42 kDa), and His-apoCcmE (18 kDa) proteins (37). The purity (>95%) of all proteins, confirmed by SDS-PAGE (Fig. 2, C and D) and immune detection with anti-CcmI and anti-CcmE antibodies (data not shown), was considered suitable for in vitro binding assays.
CcmI Discriminates among Different Classes of c-Type Apocytochromes
The chaperone activity of CcmI against the different classes of c-type apocytochromes was probed first using co-purification assays under the conditions established previously for apocytochrome c2 (25). SDS-PAGE analyses of elution fractions showed that different amounts of CcmI co-purified with different c-type apocytochromes (Fig. 3A). Semiquantitative image analyses of Coomassie-stained gels estimated that CcmI co-purified with apocytochrome c1 at about 70% (Fig. 3A, lane 4) of the amount of CcmI retained by apocytochrome c2 (lane 2). This decrease in CcmI retention was more obvious with the soluble apocytochrome c1t39 variant (Fig. 3A, lane 5). Remarkably, no detectable amount of CcmI co-purified with apocytochrome c′ (Fig. 3A, lane 3). We concluded that CcmI associates more readily with the class I than the class II c-type apocytochromes under the conditions used.
FIGURE 3.
CcmI recognizes differently class I and class II c-type apocytochromes. A, co-purification of FLAG-CcmI with different c-type apocytochromes. FLAG-CcmI does not bind to the Strep-Tactin resin (lane 1). Shown are co-purification of CcmI with apocytochrome c2 (lane 2), apocytochrome c′ (lane 3), apocytochrome c1 (lane 4), and apocytochrome c1t39 (lane 5). The amount of CcmI that co-purified with apocytochrome c2 was taken as 100% for semiquantitative estimation using ImageJ software and was compared with those seen with other c-type apocytochromes. Although the samples were reduced with β-mercaptoethanol, homodimers of apocytochrome c1t39 (marked as ·, above CcmI) were observed when its heme-binding site Cys residues were intact. B, co-purification of FLAG-CcmI and its derivative, His10-CcmI-2, with apocytochrome c1 and its truncated derivative, apocytochrome c1t39. The amount of CcmI that co-purified with apocytochrome c1 (lane 3) was taken as 100%, and that of the other c-type apocytochromes was determined as described above. C, co-purification of FLAG-CcmI with different amounts of a peptide corresponding to the C-terminal helix (see “Experimental Procedures”) following the membrane anchor of apocytochrome c1. FLAG-CcmI does not bind alone to the Strep-Tactin resin (lane 1). CcmI co-purified with 10 (lane 2) or 20 (lane 3) μg of this cytochrome c1 peptide as compared with 20 μg of the corresponding peptide from cytochrome c2 (i.e. its C-terminal helix) used as control. D, real-time protein-protein interactions between biotinylated Strep-apocytochrome c2 (Strep-Bt-apocytochrome c2) immobilized on a SA-biosensor (ligand) and His10-CcmI (analyte). The aligned sensorgram traces showing the baseline (B) followed by association (A) and dissociation (D) steps were obtained using 400 nm Bt-apocytochrome c2 and varying concentrations of FLAG-CcmI. The raw data were fitted with high accuracy to a homogeneous 1:1 bimolecular interaction model, and the kinetic parameters were determined (Table 3).
Using a full-length apocytochrome c1 and its membrane-anchorless variant, apocytochrome c1t39, we next probed the role of the different domains of CcmI in vitro. Co-purification assays conducted using intact CcmI or only its CcmI-2 domain with full-length or truncated apocytochrome c1 derivatives showed that the full-length apocytochrome c1 interacted better with CcmI than the truncated apocytochrome c1t39. Also, the amount of CcmI-2 that co-purified with either derivative of apocytochrome c1 was higher than CcmI (Fig. 3B). These findings suggested that neither the membrane anchor of apocytochrome c1 nor the CcmI-1 domain (i.e. the first transmembrane helix and the adjacent leucine zipper-containing cytoplasmic loop absent in the CcmI-2 derivative used) of CcmI is essential for these interactions in vitro. Lastly, using a synthetic peptide (NH2-WSHPQFEKIEGRTVDQMAQVDSAFLMWAAEPK-COOH) corresponding to the C-terminal helix of apocytochrome c1t39 (i.e. the C-terminal helix that interacts with the N-terminal heme-binding helix), we tested whether CcmI would recognize this helical sequence of apocytochrome c1, as observed elsewhere with that of apocytochrome c2 (25). Incubation of increasing amounts (10 and 20 μg) of this peptide with purified CcmI led to a concentration-dependent co-purification of the CcmI-peptide complex (Fig. 3C). The amount of CcmI co-purified was lower than that observed with the same amount of the apocytochrome c2 peptide used previously (25), paralleling the findings of the binding assays using apocytochromes c1 and c2 (Fig. 3A). The data indicated that in all class I c-type apocytochromes the C-terminal helix, which is orthogonal to the N-terminal heme binding site-containing helix, is sufficient to promote binding to CcmI.
ApoCcmE Recognizes Differently Class I and Class II c-Type Apocytochromes
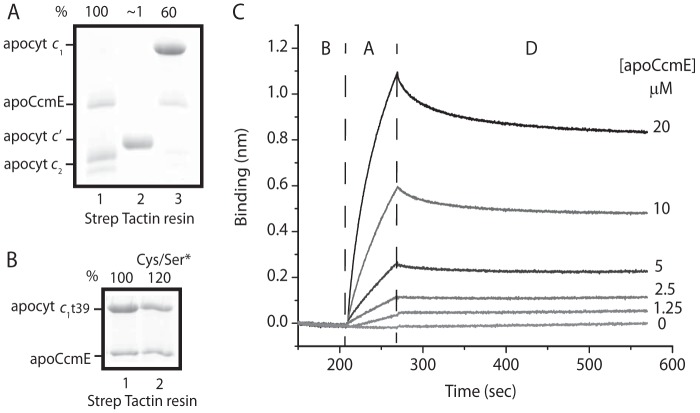
In this work, we extended the apoCcmE-apocytochrome c2 binding studies carried out earlier (37) to other c-type apocytochromes and found that apoCcmE, like CcmI, binds apocytochrome c1 but not apocytochrome c′ (Fig. 4A). Semiquantitative image analyses of Coomassie-stained gels revealed that the amounts of apoCcmE co-purifying with apocytochrome c1 decreased to 60% of that seen with apocytochrome c2 (Fig. 4A, lanes 1 and 3), whereas no detectable interaction was seen with apocytochrome c′ (lane 2). Moreover, a comparison of the truncated apocytochrome c1t39 with its Cys-less derivative (Cys to Ser substitutions at the heme-binding site) indicated that the occurrence of a disulfide bond at the heme-binding site had no effect on the apoCcmE-apocytochrome c1t39 interactions (Fig. 4B), unlike apocytochrome c2 (37).
FIGURE 4.
ApoCcmE differently recognizes class I and class II c-type apocytochromes. A, co-purification of His10-apoCcmE with stoichiometric amounts of different c-type apocytochromes. The amounts of apoCcmE co-purified with apocytochrome c2 (lane 1) was taken as 100% and compared with those seen with apocytochromes c′ and c1 (lanes 2 and 3, respectively). B, co-purification of His10-apoCcmE with apocytochrome c1t39 and with its Cys-less derivative, apocytochrome c1t39Cys/Ser*. The amounts of ApoCcmE co-purified with apocytochrome c1t39 (lane 1) and its Cys-less derivative (Cys/Ser*) were determined as described in the legend for Fig. 3. C, real-time protein-protein interactions between Strep-Bt-apocytochrome c2 immobilized on a SA-biosensor (ligand) and His10-apoCcmE (analyte). The aligned sensorgram traces showing baseline (B) followed by association (A) and dissociation (D) steps were obtained using 400 nm Bt-apocytochrome c2 and varying concentrations of apoCcmE. The raw data were fitted with high accuracy to a homogeneous 1:1 bimolecular interaction model, and the kinetic parameters were determined (Table 3).
Binding Kinetics of Various c-Type Apocytochromes to CcmI and apoCcmE
Using BLI, the binding affinities of CcmI and apoCcmE to c-type apocytochromes were quantified by real-time binding assays. The association (kon)/dissociation (koff) rates and the binding affinity (KD) constants of the appropriate protein couples were determined as described under “Experimental Procedures.” Negative controls lacking the analytes confirmed that the observed interference shifts originated from the ligand-analyte complexes. Using Octet data analysis software (ForteBio), experimental data (association and dissociation curves) were fit to a homogeneous 1:1 bimolecular protein-protein interaction model, and the binding parameters (kon, koff, and KD) of the ligand-analyte couple were determined (Table 3).
TABLE 3.
Kinetic analysis of protein-protein interactions between CcmI or apoCcmE and different c-type apocytochromes in the absence or presence of heme
Assay buffer A: 50 mm Tris-HCl, pH 8, 100 mm NaCl, 0.01% n-dodecyl-β-d-maltoside, and 1% BSA. Assay buffer B: 50 mm Tris-HCl, pH 8, 150 mm NaCl, and 0.01% Tween-20.
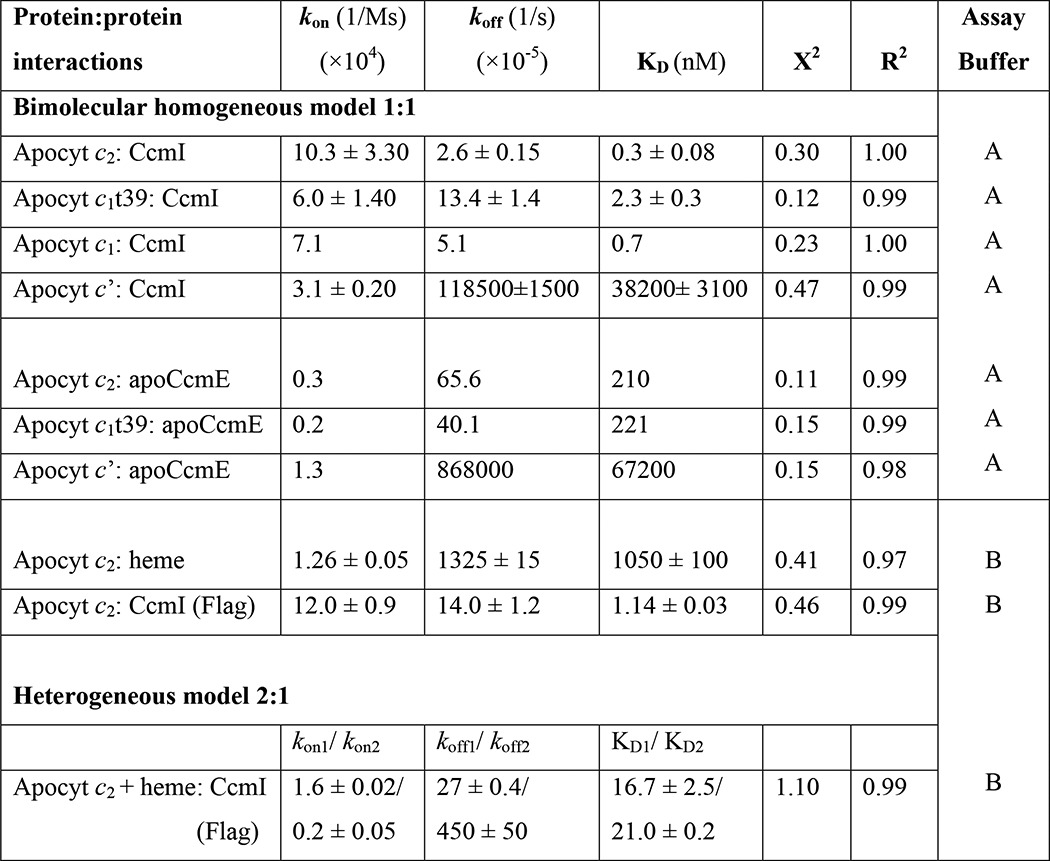
With the CcmI-Bt-apocytochrome c2 complexes, the association curves exhibited rapid increases until reaching equilibrium, and the dissociation curves followed slow decay kinetics (monitored for longer time periods for better data collection) (Fig. 3D). This behavior reflected fast binding of CcmI to apocytochrome c2 to form a stable complex. Similar experiments conducted with other c-type apocytochromes indicated that CcmI interacted strongly with the class I apocytochromes c1 and c1t39 tested (Table 3, KD values (nm range)). In contrast, although CcmI associated rapidly with apocytochrome c′, it also dissociated rapidly, indicating that it bound weakly to this class II c-type apocytochrome (Table 3, KD values (μm range)). Thus, the binding kinetics confirmed and quantified the findings of the co-purification assays (Fig. 3A). In all cases, the kon rates were comparable, indicating that the c-type apocytochromes bind rapidly to CcmI, but the koff rates were faster for unstable (i.e. apocytochrome c′) and slower for the stable (i.e. apocytochromes c2, c1, and c1t39) binary complexes.
Next, the binding of apoCcmE to c-type apocytochromes was examined using a similar approach (Fig. 4C). The kinetic data showed that apoCcmE associated with apocytochrome c2 or c1t39 at rates slower than those seen with CcmI and dissociated at faster rates, yielding KD values in the micromolar range (Table 3). Furthermore, even though apoCcmE bound apocytochrome c′ at rates similar to apocytochromes c2 or c1t39, it dissociated rapidly, showing ∼100-fold higher KD values. Overall the data established that both CcmI and apoCcmE recognized the class I c-type apocytochromes with higher affinities than their class II counterparts.
Probing the Secondary Structures of Various c-Type Apocytochromes Using CD Spectroscopy
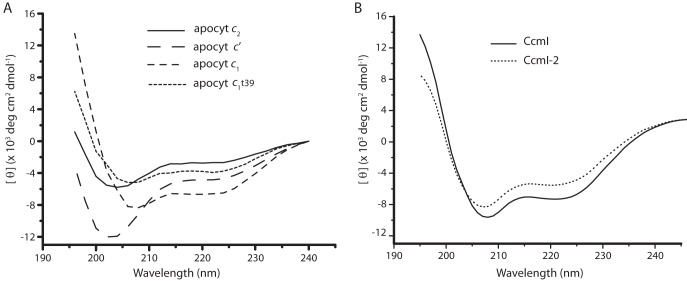
The secondary structures (globular class I cytochromes c2 and c1 versus four-helical bundle class II cytochrome c′) of different c-type apocytochromes were examined by CD spectroscopy in the far-UV region (Fig. 5A). The CD spectra of apocytochromes c2 and c′ exhibited a negative peak around 203 nm and low ellipticity below 215 nm, showing their random coil conformations, respectively. In contrast, both apocytochrome c1 and apocytochrome c1t39 exhibited CD spectra more characteristic of α-helical proteins, with two negative peaks at 208 and 223 nm. Interestingly, however, the two class I apocytochromes, c2 and c1, which have similar globular folds in their mature forms, exhibited different secondary structures in their apocytochrome forms (48, 49). In addition, the R. capsulatus class II apocytochrome c′ also differed from its E. coli homologue, Cyt b562, which forms a molten globule in the absence of heme (50), even though both holocytochromes have four-helical bundle structures. Finally, the CD spectra of CcmI and CcmI-2 showed the characteristics of α-helical proteins (Fig. 5B).
FIGURE 5.
CD spectra of various c-type apocytochromes and their interacting partners, CcmI and CcmI-2, in the absence of hemin. A, far-UV CD spectra between 195 and 240 nm of various c-type apocytochromes (15 μm) were recorded in the absence of hemin. Apocytochromes c2 and c′ exhibited CD spectra typical of disordered coils, whereas apocytochrome c1 and its truncated derivative, apocytochrome c1t39, showed characteristics of typical α-helical proteins. B, far-UV CD spectra between 195 and 240 nm of His10-CcmI and His10-CcmI-2 (1.5 μm) in the absence of hemin indicated their high α-helical contents. Both apocytochrome c1t39 and CcmI-2 are one helix shorter than apocytochrome c1 and CcmI, respectively, and the amplitudes of their ellipticity are comparatively lower.
Release of Apocytochrome c2 from CcmI Is Facilitated by the Presence of Hemin
The observed tight binding of CcmI to class I c-type apocytochromes was remarkable, leading us to probe whether heme affected these interactions. This was addressed by choosing cytochrome c2 as a prototype for class I c-type apocytochrome. We reasoned that if heme induces the formation of a b-type derivative of apocytochrome c2, with a molten globule-like structure (reminiscent of that of mature cytochrome c2), then this intermediate might not bind CcmI tightly, similar to what we observed earlier with a native form (25). To test this hypothesis, we first investigated the kinetics of heme binding to Bt-apocytochrome c2 using BLI. The data showed that heme bound to, and dissociated from, apocytochrome c2 rapidly (Fig. 6A). A 1:1 bimolecular kinetic model indicated that the apocytochrome c2-heme complex had an affinity constant (KD) in the order of 1 μm and was not very stable (Table 3). A similar low affinity has been reported for the horse heart apocytochrome c-heme complex under oxidizing conditions (46). Next, we examined the effects of heme on the CD spectra of apocytochrome c2 and CcmI. Upon the addition of hemin, the far-UV CD spectrum of apocytochrome c2 changed drastically, with increased helical content, paralleling increased amount of hemin (Fig. 6B). On the other hand, the CD spectrum of CcmI was hardly affected, with only minor spectral changes seen in the presence of excess (16-fold) hemin (Fig. 6C).
FIGURE 6.
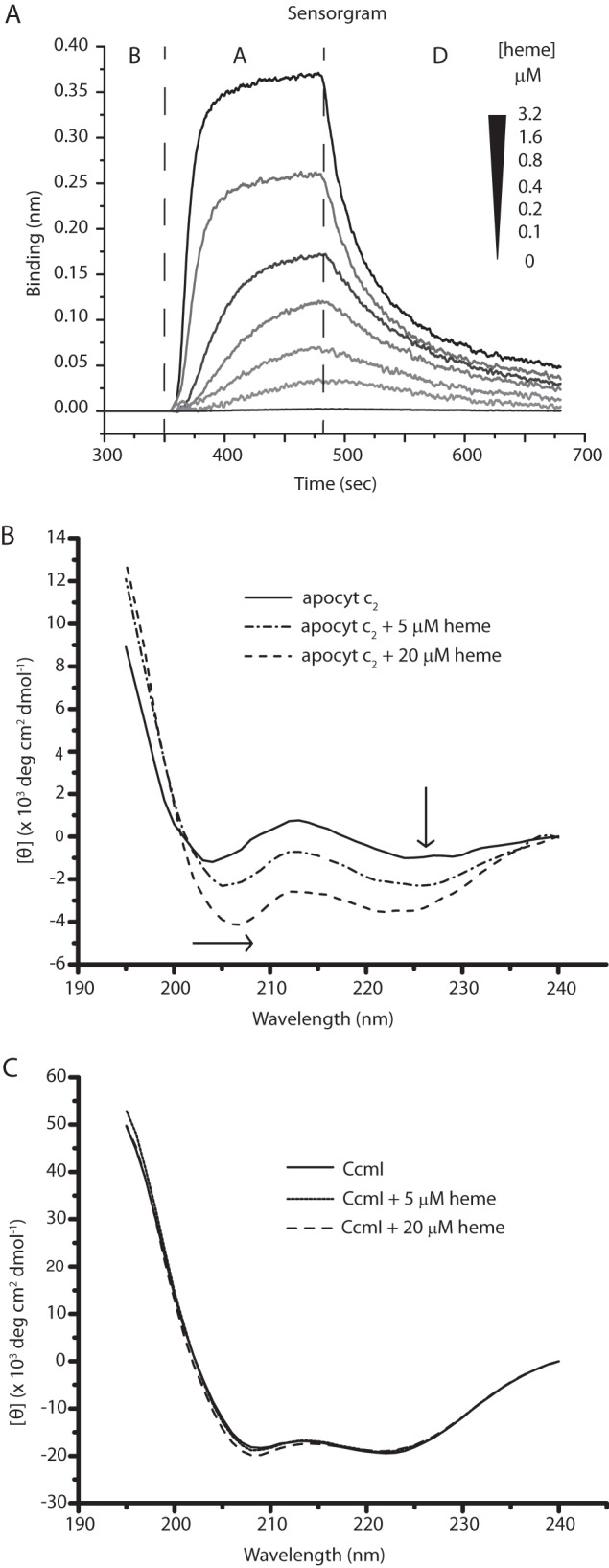
Binding of hemin to apocytochrome c2. A, real-time interactions between Strep-Bt-apocytochrome c2 immobilized on a SA-biosensor (ligand) and hemin (analyte). The aligned sensorgram traces showing baseline (B) followed by association (A) and dissociation (D) steps were obtained using 400 nm Bt-apocytochrome c2 and varying concentrations (0 to 3.2 μm) of hemin. The raw data were fitted with high accuracy to a homogeneous 1:1 bimolecular interaction model, and the kinetic parameters were determined (Table 3). B, far-UV CD spectra between 195 and 240 nm of Strep-apocytochrome c2 (2.5 μm) recorded in the absence or presence of 5 and 20 μm hemin. The arrows indicate the displacements of the spectra, reflecting the increased α-helical content of apocytochrome c2 in the presence of hemin. C, far-UV CD spectra between 195 and 240 nm of FLAG-CcmI (1.25 μm) in the absence or presence of 5 and 20 μm hemin. Note that hemin has no major effect on the α-helical content of CcmI.
For the effect of heme on CcmI-apocytochrome c2 interactions, we first tested the co-purification of CcmI with apocytochrome c2 in the presence of hemin (Fig. 7A). The addition of stoichiometric amounts of hemin (∼1 μm) decreased the amount of CcmI that co-purified with apocytochrome c2 by ∼25% (Fig. 7A). Next, the binding kinetics of CcmI to apocytochrome c2 were monitored by BLI in the absence of hemin but omitting BSA and replacing His10-CcmI with FLAG-CcmI to minimize spurious heme interference. As above, we observed KD values in the order of nanomolar for CcmI-apocytochrome c2 interactions for these derivatives (Table 3). However, when the assays were repeated in the presence of 2 μm hemin (∼5 fold molar excess of Bt-apocytochrome c2), the association and dissociation kinetics could not fit reliably to a 1:1 homogeneous model, suggesting the presence of a nonhomogeneous ligand population. In cases where “active” and “inactive” forms of ligands toward the analyte are expected, the use of a 2:1 heterogeneous model becomes appropriate. Indeed, upon the addition of heme, a fraction of apocytochrome c2 yielded a noncovalent heme-containing b-type cytochrome derivative (see below). When the kinetic data were fitted to a 2:1 heterogeneous model, two different KD values (with acceptable X2 and R2 values) for CcmI-apocytochrome c2 interactions were deduced (Table 3). These values were 15–20 times higher than the KD seen in the absence of hemin, clearly indicating that the CcmI-apocytochrome c2 interactions had weakened.
FIGURE 7.
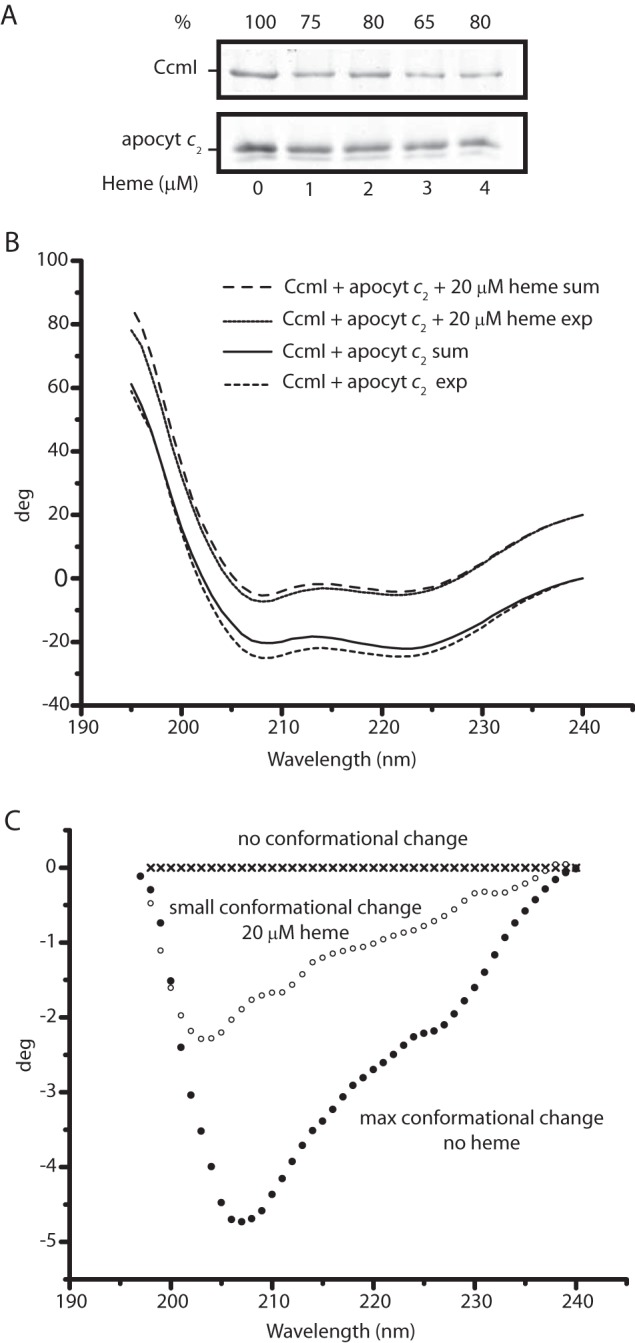
Effect of heme on CcmI-apocytochrome c2 interactions. A, co-purification of FLAG-CcmI with Strep-apocytochrome c2 in the absence (lane 1) or presence of 1 (lane 2), 2 (lane 3), 3 (lane 4), and 4 (lane 5) μm hemin. The amounts of the interacting partners were as described under “Materials and Methods,” and semiquantitative comparisons (as described in Fig. 3) showed that CcmI-apocytochrome c2 interactions were weakened, but not completely abolished, in the presence of hemin. B, effect of heme on the CD spectra of the CcmI-apocytochrome c2 mixture in the absence or presence of hemin (20 μm). Spectra were recorded after 2 h of incubation to ensure that all spectral changes were complete. exp and sum, refer to the experimental and calculated spectra, respectively. The calculated spectra were obtained by summing the spectra of each protein alone in the absence or presence of hemin (see Fig. 5, B and C). C, difference spectra between the experimental spectra of CcmI-apocytochrome c2 minus the calculated spectra shown in B in the absence or presence of hemin (20 μm). Decreased conformational changes were seen upon binding of apocytochrome c2 to CcmI in the presence of hemin, suggesting decreased binding interactions.
Previously, CD spectroscopy had shown that CcmI and apocytochrome c2 change their conformations upon binding to each other in the absence of hemin (25). This approach was used to further document the effect of heme on CcmI-apocytochrome c2 interactions. The secondary structure changes were monitored after incubating CcmI and apocytochrome c2 in the presence (molar excess) or absence of hemin, and the CD spectra obtained were compared with the sums of the spectra of the individual proteins recorded under the same experimental conditions (Fig. 7B). These comparisons showed that the conformational changes induced by the apocytochrome c2-CcmI interactions decreased markedly in the presence of hemin. This finding further supported the view that a fraction of apocytochrome c2 changed its secondary structure upon binding to heme and weakened its interactions with CcmI, lowering the CD-detected spectral changes (Fig. 7C).
The c-Type Apocytochromes Can Form b-Type Cytochrome Variants in the Presence of Hemin
Optical spectroscopy was used to monitor noncovalent binding of heme to apocytochrome c2 and to other selected c-type apocytochromes to assess whether indeed they form b-type cytochrome variants. As a control for formation of a b-type cytochrome variant from a c-type apocytochrome, a Cys-less derivative of H. thermophilus cytochrome c552 was used. When expressed in E. coli cytoplasm, native H. thermophilus cytochrome c552 contains covalently ligated heme (51) even in the absence of the Ccm System I and under aerobic growth conditions. Similarly, the Cys-less derivative of H. thermophilus cytochrome c552 produces a noncovalent heme-containing b-type cytochrome (called cytochrome b-c552) (52). Overnight incubation of purified cytochrome b-c552 in the presence of 1 m imidazole displaces its heme to yield apocytochrome b-c552 (52) (data not shown). We purified these variants of H. thermophilus cytochrome c552, and SDS-PAGE/TMBZ analyses confirmed the absence of covalently bound heme in both cytochrome b-c552 and its corresponding apocytochrome b-c552 (data not shown). The CD spectra of these proteins resembled those of typical α-helical proteins with the amounts of secondary structures being increased from apocytochrome b-c552 to cytochrome b-c552 to cytochrome c552 (data not shown). The binding of heme enhanced the secondary structure formation, even though apocytochrome b-c552 already had some secondary structure in the absence of heme as compared with the apocytochromes c2 and c′ (Fig. 5A). Thus, H. thermophilus cytochrome c552 and its derivatives provided valid controls for the formation of b-type cytochrome from the c-type apocytochromes.
The addition of stoichiometric amounts of hemin to R. capsulatus c-type apocytochromes resulted in changes in their visible spectra over time. After 30 min of incubation with hemin, apocytochromes c2 and c1, but not apocytochrome c′, exhibited spectral features that are typical of b-type cytochromes, with Soret and α-bands at 422 and 557 nm in apocytochrome c2 and at 425 and 559 nm in apocytochrome c1, respectively (Fig. 8). Under the conditions where full (100%) incorporation (as confirmed by comparison with similar amount of cytochrome b-c552 purified from E. coli) of heme to apocytochrome c552 occurred to yield cytochrome b-c552, ∼20% of the available heme was reconstituted into apocytochrome c2, ∼12% into apocytochrome c1, and no detectable amount into apocytochrome c′ (assuming similar extinction coefficients for all b-type cytochrome derivatives). These findings showed that a fraction of apocytochrome c2 was converted to its b-type cytochrome derivative in the presence of hemin. Moreover, apocytochromes c2 and c1 behaved in opposing ways with respect to the amounts of ellipticity they exhibited and the b-type cytochrome variants they yielded. Thus no direct correlation was seen between the helical content of a c-apocytochrome and its ability to bind heme to yield a b-type cytochrome variant in vitro.
FIGURE 8.
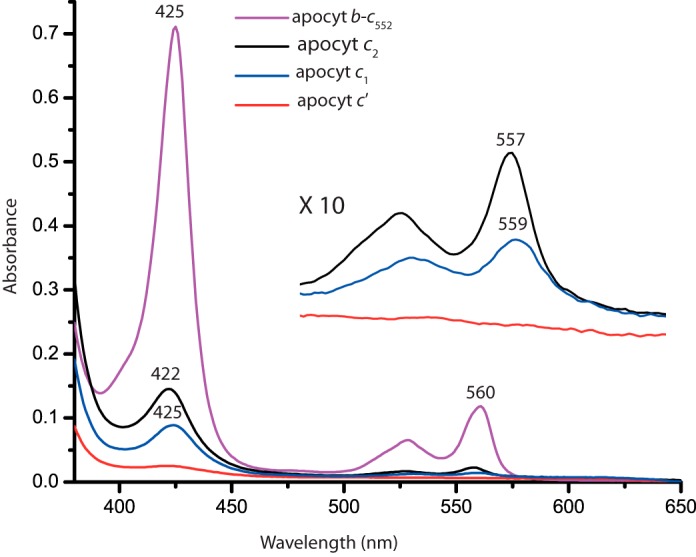
In vitro reconstitution of hemin to R. capsulatus c-type apocytochromes in the absence of Ccm System I components. Reduced minus oxidized optical difference spectra between 380 and 650 nm of 6.5 μm b-type cytochrome derivative formed after stoichiometric addition of hemin to 10 μm of H. thermophilus apocytochrome b-c552 (purple), which was used as a “heme reconstitution” control. Similar reconstitution experiments were repeated using the same amounts of R. capsulatus apocytochrome c2 (black), apocytochrome c1 (blue), and apocytochrome c′ (red) with stoichiometric amounts of hemin, and the obtained spectra were compared taking as 100% that of H. thermophilus apocytochrome b-c552. Inset, spectra depict the β- and α-bands of the b-type cytochrome derivatives of R. capsulatus apocytochromes c2, c1t39, and c′.
Discussion
Previously, we showed that the chaperone protein CcmI binds to the C-terminal helix of apocytochrome c2, whereas the heme-handling protein CcmE recognizes its heme-binding site (25, 37). In this work we addressed the next question, which is whether CcmI also chaperones other soluble and membrane-bound c-type apocytochromes in addition to apocytochrome c2. To this end, we chose the class I membrane-anchored apocytochrome c1 because the TPR motif-containing portion (CcmI-2) of CcmI is not essential for its maturation (20). We also investigated the soluble class II cytochrome c′, which has a different (four-helical bundle versus globular) structure than the class I members. The production in E. coli of cytochrome c′ from some species requires co-expression of the Ccm genes (53, 54), whereas that from others (e.g. Hydrogenophilus thermoluteolus cytochrome c′) does not (54). At the onset of this work, whether cytochrome c′ maturation in R. capsulatus relied on Ccm System I was unknown (20). Our data showed that cytochrome c′ maturation in R. capsulatus requires at least CcmE and CcmI and established it as a substrate for Ccm System I. Co-purification and real-time binding (BLI) assays demonstrated that both CcmI and apoCcmE distinguish different classes of c-type apocytochromes and that heme strongly affects these interactions.
Binding of CcmI and apoCcmE to Class I and Class II c-Type Apocytochromes in the Absence of Hemin
Real-time binding studies indicated for the first time that R. capsulatus class I c-type apocytochromes (c2, c1, and its anchor-less derivative, c1t39) had very high (∼nm range KD), whereas the class II apocytochrome c′ had much lower (∼μm range KD) binding affinities for CcmI (Table 3 and Fig. 9). How the c-type cytochromes interact with their chaperones has not yet been well studied. Only a single experiment, using the soluble portion of Pseudomonas aeruginosa CcmI and the class I apocytochrome c551 (or a dansylated peptide corresponding to its C-terminal end), has reported a low equilibrium binding KD (∼>100 μm) (29). However, a different experimental approach was used in those studies, which renders a direct data comparison difficult. Thus, the reason that different KD values were obtained here and in that study remains unclear.
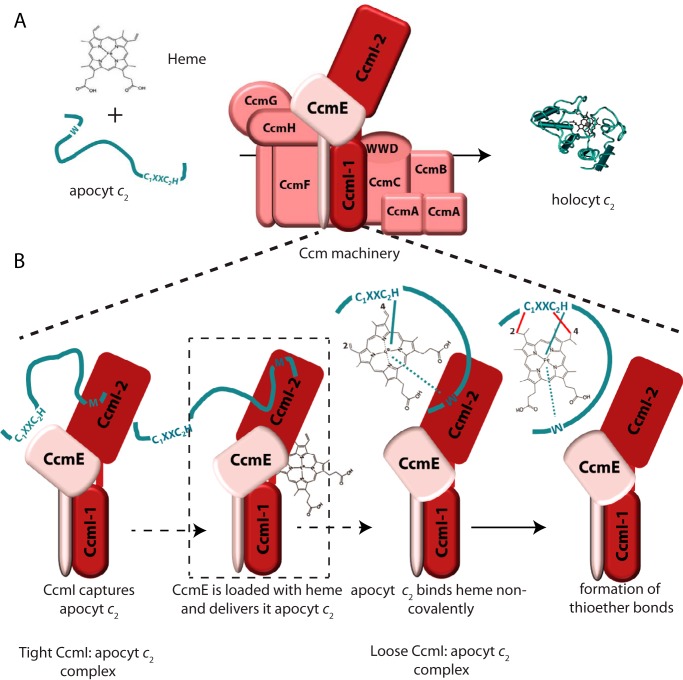
FIGURE 9.
CcmI-apocytochrome c2 interactions in the absence and presence of hemin during the Ccm process. A, shows the Ccm machinery and its two substrates, apocytochrome c2 and heme, forming holocytochrome c2. B, depicts the interactions between CcmI and apocytochrome c2 in the absence and presence of hemin (thought to be provided by CcmE). This hypothetical scheme takes into account all available data to show that the CcmI-2 portion of CcmI binds tightly the C-terminal helix of apocytochrome c2, bringing its heme-binding site near CcmE (left panel). Upon the availability of heme via CcmE, apocytochrome c2 binds heme noncovalently to form a b-type cytochrome intermediate that interacts less tightly with CcmI-2 (middle two panels). The subsequent formation of covalent thioether bonds between heme and apocytochrome c2, via currently unknown steps catalyzed by the remaining components of Ccm System I, yields holocytochrome c2 (right panel).
A major structural determinant of apocytochrome c2 for binding CcmI is its most C-terminal helix, the equivalent of which is conserved in most class I c-type cytochromes (55). This helix packs orthogonally against the heme-binding site containing N-terminal helix in mature c-type cytochromes. The presence of a hydrophobic molecule (e.g. a porphyrin ring) induces conversion of a random coiled c-type apocytochrome to a molten globular structure as a folding intermediate (56). This promotes interactions between its N- and C-terminal helices. In cytochrome c1, unlike the other R. capsulatus c-type cytochromes (i.e. c2, cy, co, and cp), this “C-terminal helix” precedes its most C-terminal membrane-anchoring helix but becomes the most C-terminal helix in the truncated apocytochrome c1t39 derivative. The availability of these two variants allowed us to probe the role of the anchoring and the C-terminal helix of cytochrome c1 on binding CcmI. Earlier genetic studies indicated that the CcmI-2 portion of CcmI is unnecessary for cytochrome c1 production, inferring that enough interactions occur during maturation between the Ccm System I and membrane-anchored apocytochrome c1 (20). The in vitro data obtained here further complemented these findings and showed that roughly similar KD values were observed for binding the native or truncated forms of apocytochrome c1 to CcmI (Table 3). Moreover, co-purification assays showed that CcmI-2 binds both derivatives of apocytochromes c1. Thus, the structural determinants recognized by CcmI-2 must also be present in the anchor-less apocytochrome c1t39. Indeed, similar to apocytochrome c2, a peptide corresponding to the C-terminal helix preceding the membrane anchor of cytochrome c1 is readily recognized by CcmI (Fig. 3C). Therefore, the overall data showed that apocytochrome c1 interacted via its C-terminal helix with CcmI-2 domain, but that it might also interact via its membrane anchor with CcmI-1 domain of CcmI. It remains to be seen whether the second transmembrane helix of CcmI-1 (present in the CcmI-2 variant used here) is also involved in the interactions between the apocytochrome c1 and CcmI.
How the class I c-type apocytochromes interact with the TPR motifs is not well known. The TPR-containing domain of E. coli NrfG (a functional homologue of R. capsulatus CcmI and E. coli CcmH), which is specific for the maturation of unusual c-type cytochromes (with a heme-binding sequence of C1XXC2K like the E. coli NrfA (28)) exhibits a KD of ∼10 μm toward a peptide mimicking the C terminus of NrfA. In this case, the TPR binding groove apparently recognizes a helix followed by a loop composed of six C-terminal residues (28). However, the TPR proteins are highly versatile with respect to the amino acid compositions of their TPR motifs, which modulate their ligand affinity. The three-dimensional structures of these proteins bound to their ligands show an intricate network of contacts, including electrostatic, hydrophobic, and Van der Waals interactions (27). For example, such a structure between a TPR protein and the Hsp70 or Hsp90 proteins define a conserved peptide (Met-Glu-Glu-Val-Asp) at the C termini of these proteins binding to the concave groove of the TPR protein (57). Yet, other TPR proteins present alternative interaction modes, including a binding site not located in the TPR groove but composed of the loops connecting the TPR domains (58). Clearly, much remains to be learned about the interactions of the c-type apocytochromes with their TPR-containing chaperones.
This study also showed that the heme-handling protein apoCcmE interacts not only with apocytochrome c2 (37) but also with other c-type apocytochromes. However, these interactions are weaker (μm versus nm KD values) than those seen with CcmI but are still discriminatory between the different classes of c-type apocytochromes, with about a 100-fold lower binding affinity for the class II apocytochrome c′. They are also unaffected by the redox state of the Cys residues at the heme-binding sites of apocytochromes.
Binding of Heme to c-Type Apocytochromes in the Absence of the Ccm Components
Heme triggers the folding of unstructured c-type apocytochromes, increasing their secondary structures (46). Several c-type apocytochromes, like the mitochondrial apocytochrome c (46, 59), Paracoccus denitrificans apocytochrome c550 (59) and R. capsulatus apocytochrome c2 are random coils that can form compact molten globular structures upon binding heme. However, they yield inefficiently b-type cytochrome derivatives. Conversely, some other c-type apocytochromes from thermophilic organisms like Hydrogenobacter thermophilus cytochrome c552 (51), Aquifex aeolicus cytochrome c555 (60), Thermus thermophilus cytochrome c552 (61), and H. thermoluteolus cytochrome c′ (62) exhibit secondary structures with high helical contents in the absence of heme (52, 62, 63) and readily yield b-type cytochrome derivatives upon heme availability in vitro (64). Even in exceptional cases, thermophilic molten globular c-type apocytochromes yield b-type cytochrome intermediates that are conducive to spontaneous covalent heme ligation (51). Hence, they are poorer models for cytochrome c maturation studies because of their structures that evolved to bind and ligate heme independently of the Ccm System I.
CD spectral data indicated that no direct correlation exists between the increased helical contents of c-type apocytochromes and their ability to yield b-type cytochrome derivatives upon addition of heme. For example, the class I apocytochrome c1 exhibited a high degree of secondary structure but was less efficient than apocytochrome c2 for binding heme (Fig. 8). On the other hand, R. capsulatus apocytochrome c′ had no secondary structure discernable by CD spectroscopy, like its homologue from mesophilic Allochromatium vinosum (62), and was unable to bind heme and yield a b-type cytochrome derivative. Conversely, an apocytochrome c-b562 derivative of E. coli cytochrome b562 (structural homologue of cytochrome c′) exhibits a fold that closely matches its final conformation and efficiently incorporates a stoichiometric amount of hemin independently of the Ccm machinery. So far, the only known mesophilic c-type apocytochrome that binds heme efficiently to form a b-type cytochrome derivative is horse cytochrome c (46, 59), but it is matured naturally by the cytochrome c biogenesis System III (65). Although a few c-type apocytochromes, because of their structure and intrinsic stability, can bind heme to yield either b-type cytochromes derivatives or mature c-type cytochromes in the absence of the Ccm components, apparently the “designed” four-helical bundles, which bind heme efficiently, still need the Ccm machinery to produce their c-type cytochrome derivatives (66). Thus, the high helical content of a c-type apocytochrome in the absence of hemin is not predictive of a form that is conducive to efficient heme binding. In addition to the interactions between the heme iron and its axial ligands, as well as those between the porphyrin ring and the polypeptide, the amino acid sequences of c-type apocytochromes might also contribute to providing a suitable environment for trapping heme. If this structural information is virtually absent in a c-type apocytochrome, then it relies heavily on the Ccm processes.
Effect of Hemin on the Interactions of Apocytochrome c2 with CcmI
A remarkable finding of this study is the very high binding affinity (∼nm) of CcmI for class I c-type apocytochromes (Fig. 9). Clearly, this high affinity is advantageous for the efficient capture of c-type apocytochromes by the Ccm complex following their translocation across the cytoplasmic membrane. However, it also raises an intriguing issue, which is their subsequent release upon maturation. Our earlier observation that CcmI does not bind holocytochrome c2 suggests that heme, and probably heme-mediated folding, might affect these interactions. Indeed, the addition of hemin to apocytochrome c2 promoted the formation of its b-type cytochrome-like derivative (Fig. 9). Co-purification assays in the presence of hemin indicated that the amount of CcmI that co-purified with apocytochrome c2 decreased. Interestingly, the decrease (∼25%) in the amount of CcmI that was retained by apocytochrome c2 coincided roughly with the amount (∼20%) of heme incorporated into apocytochrome c2 to yield its b-type derivative in vitro. This coincidence suggested that the b-type cytochrome derivative formed upon the addition of hemin interacted poorly with CcmI. This suggestion was also supported by decreased conformational changes seen by the CD spectra of CcmI-apocytochrome c2 complexes in the presence of hemin (Fig. 7). In agreement with these observations, the quantitative binding assays done in the presence of hemin documented higher KD values for the binding of apocytochrome c2 to CcmI (Table 3). It should be noted that thorough analyses of the tripartite interactions among hemin, apocytochrome c2, and CcmI are complicated due to the heterogeneity induced by binding of hemin to only a fraction of apocytochrome c2 to yield a b-type cytochrome-like derivative in the absence of any Ccm component in vitro. Nonetheless, the data analyses based on a 2:1 heterogeneous kinetic model supported a decrease in the affinity of CcmI for apocytochrome c2 (and possibly for other class I c-type apocytochromes) in the presence of hemin, possibly by promoting the formation of a partially folded apocytochrome c2 with coordinated heme moiety.
In summary, the detailed analyses of the binding process of CcmI to apocytochrome c2 summarized in Fig. 9 support our current notion that the release of a c-type apocytochrome from its chaperone during the Ccm process could occur upon the availability of heme (probably via CcmE). If so, then a b-type cytochrome derivative forms as an intermediate, which subsequently undergoes covalent heme ligation by the remaining components of the Ccm complex.
Acknowledgments
We thank Dr. C. Sanders for the plasmid pCS1208, Dr. R. Prince for the anti-cytochrome c′ antibody, Drs. W. Englander and W. Leland for help with CD spectroscopy, and members of ForteBio Inc. and Covance Development Services for assistance with biolayer interferometry data acquisitions and analyses.
This work was supported by U.S. Department of Energy Grant DOE DE-FG02-91ER20052 from the Division of Chemical Sciences, Geosciences and Biosciences, Office of Basic Energy Sciences. This work was also supported by National Institutes of Health Grant GM38237. The authors declare that they have no conflicts of interest with the contents of this article.
C. Sanders and F. Daldal, unpublished observations.
- DMSO
- dimethylsulfoxide
- Ccm
- cytochrome c maturation
- BLI
- biolayer interferometry
- Bt-
- biotinylated
- TPR
- tetratricopeptide repeat
- TMBZ
- tetramethylbenzidine
- SA
- streptavidin
- Ht
- H. thermophilus.
References
- 1. Moore G. R., Pettigrew G. W. (1990) Cytochromes c Evolutionary, Structural and Physicochemical Aspects, Springer-Verlag, New York [Google Scholar]
- 2. Bertini I., Cavallaro G., Rosato A. (2006) Cytochrome c: occurrence and functions. Chem. Rev. 106, 90–115 [DOI] [PubMed] [Google Scholar]
- 3. Jiang X., Wang X. (2004) Cytochrome c-mediated apoptosis. Annu. Rev. Biochem. 73, 87–106 [DOI] [PubMed] [Google Scholar]
- 4. Bowman S. E., Bren K. L. (2008) The chemistry and biochemistry of heme c: functional bases for covalent attachment. Nat. Prod. Rep. 25, 1118–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Allen J. W., Leach N., Ferguson S. J. (2005) The histidine of the c-type cytochrome CXXCH haem-binding motif is essential for haem attachment by the Escherichia coli cytochrome c maturation (Ccm) apparatus. Biochem. J. 389, 587–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ambler R. P. (1982) The structure and classification of cytochromes c, in Cyclotrons to Cytochromes (Robinson A., Kaplan N. eds) pp 263–280, Academic Press, New York [Google Scholar]
- 7. Ambler R. P. (1991) Sequence variability in bacterial cytochromes c. Biochim. Biophys. Acta 1058, 42–47 [DOI] [PubMed] [Google Scholar]
- 8. Daldal F., Davidson E., Cheng S. (1987) Isolation of the structural genes for the Rieske Fe-S protein, cytochrome b and cytochrome c1 all components of the ubiquinol:cytochrome c2 oxidoreductase complex of Rhodopseudomonas capsulata. J. Mol. Biol. 195, 1–12 [DOI] [PubMed] [Google Scholar]
- 9. Gray K. A., Grooms M., Myllykallio H., Moomaw C., Slaughter C., Daldal F. (1994) Rhodobacter capsulatus contains a novel cb-type cytochrome c oxidase without a CuA center. Biochemistry 33, 3120–3127 [DOI] [PubMed] [Google Scholar]
- 10. Koch H. G., Hwang O., Daldal F. (1998) Isolation and characterization of Rhodobacter capsulatus mutants affected in cytochrome cbb3 oxidase activity. J. Bacteriol. 180, 969–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jenney F. E., Jr., Daldal F. (1993) A novel membrane-associated c-type cytochrome, Cyt cy, can mediate the photosynthetic growth of Rhodobacter capsulatus and Rhodobacter sphaeroides. EMBO J. 12, 1283–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hochkoeppler A., Jenney F. E., Jr., Lang S. E., Zannoni D., Daldal F. (1995) Membrane-associated cytochrome cy of Rhodobacter capsulatus is an electron carrier from the cytochrome bc1 complex to the cytochrome c oxidase during respiration. J. Bacteriol. 177, 608–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cross R., Aish J., Paston S. J., Poole R. K., Moir J. W. (2000) Cytochrome c′ from Rhodobacter capsulatus confers increased resistance to nitric oxide. J. Bacteriol. 182, 1442–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shaw A. L., Hochkoeppler A., Bonora P., Zannoni D., Hanson G. R., McEwan A. G. (1999) Characterization of DorC from Rhodobacter capsulatus, a c-type cytochrome involved in electron transfer to dimethyl sulfoxide reductase. J. Biol. Chem. 274, 9911–9914 [DOI] [PubMed] [Google Scholar]
- 15. Verissimo A. F., Daldal F. (2014) Cytochrome c biogenesis system I: an intricate process catalyzed by a maturase supercomplex? Biochim. Biophys. Acta 1837, 989–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Travaglini-Allocatelli C. (2013) Protein machineries involved in the attachment of heme to cytochrome c: protein structures and molecular mechanisms. Scientifica 2013, 505714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kranz R. G., Richard-Fogal C., Taylor J. S., Frawley E. R. (2009) Cytochrome c biogenesis: mechanisms for covalent modifications and trafficking of heme and for heme-iron redox control. Microbiol. Mol. Biol. Rev. 73, 510–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ferguson S. J. (2012) New perspectives on assembling c-type cytochromes, particularly from sulphate reducing bacteria and mitochondria. Biochim. Biophys. Acta 1817, 1754–1758 [DOI] [PubMed] [Google Scholar]
- 19. Delgado M. J., Yeoman K. H., Wu G., Vargas C., Davies A. E., Poole R. K., Johnston A. W., Downie J. A. (1995) Characterization of the cycHJKL genes involved in cytochrome c biogenesis and symbiotic nitrogen fixation in Rhizobium leguminosarum. J. Bacteriol. 177, 4927–4934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lang S. E., Jenney F. E., Jr., Daldal F. (1996) Rhodobacter capsulatus CycH: a bipartite gene product with pleiotropic effects on the biogenesis of structurally different c-type cytochromes. J. Bacteriol. 178, 5279–5290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ritz D., Bott M., Hennecke H. (1993) Formation of several bacterial c-type cytochromes requires a novel membrane-anchored protein that faces the periplasm. Mol. Microbiol. 9, 729–740 [DOI] [PubMed] [Google Scholar]
- 22. Cinege G., Kereszt A., Kertész S., Balogh G., Dusha I. (2004) The roles of different regions of the CycH protein in c-type cytochrome biogenesis in Sinorhizobium meliloti. Mol. Genet. Genomics 271, 171–179 [DOI] [PubMed] [Google Scholar]
- 23. Sanders C., Turkarslan S., Lee D. W., Onder O., Kranz R. G., Daldal F. (2008) The cytochrome c maturation components CcmF, CcmH, and CcmI form a membrane-integral multisubunit heme ligation complex. J. Biol. Chem. 283, 29715–29722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sanders C., Boulay C., Daldal F. (2007) Membrane-spanning and periplasmic segments of CcmI have distinct functions during cytochrome c biogenesis in Rhodobacter capsulatus. J. Bacteriol. 189, 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Verissimo A. F., Yang H., Wu X., Sanders C., Daldal F. (2011) CcmI subunit of CcmFHI heme ligation complex functions as an apocytochrome c chaperone during c-type cytochrome maturation. J. Biol. Chem. 286, 40452–40463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. D'Andrea L. D., Regan L. (2003) TPR proteins: the versatile helix. Trends Biochem. Sci. 28, 655–662 [DOI] [PubMed] [Google Scholar]
- 27. Zeytuni N., Zarivach R. (2012) Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure 20, 397–405 [DOI] [PubMed] [Google Scholar]
- 28. Han D., Kim K., Oh J., Park J., Kim Y. (2008) TPR domain of NrfG mediates complex formation between heme lyase and formate-dependent nitrite reductase in Escherichia coli O157:H7. Proteins 70, 900–914 [DOI] [PubMed] [Google Scholar]
- 29. Di Silvio E., Di Matteo A., Malatesta F., Travaglini-Allocatelli C. (2013) Recognition and binding of apocytochrome c to P. aeruginosa CcmI, a component of cytochrome c maturation machinery. Biochim. Biophys. Acta 1834, 1554–1561 [DOI] [PubMed] [Google Scholar]
- 30. Arnesano F., Banci L., Barker P. D., Bertini I., Rosato A., Su X. C., Viezzoli M. S. (2002) Solution structure and characterization of the heme chaperone CcmE. Biochemistry 41, 13587–13594 [DOI] [PubMed] [Google Scholar]
- 31. Enggist E., Thöny-Meyer L., Güntert P., Pervushin K. (2002) NMR structure of the heme chaperone CcmE reveals a novel functional motif. Structure 10, 1551–1557 [DOI] [PubMed] [Google Scholar]
- 32. Aramini J. M., Hamilton K., Rossi P., Ertekin A., Lee H. W., Lemak A., Wang H., Xiao R., Acton T. B., Everett J. K., Montelione G. T. (2012) Solution NMR structure, backbone dynamics, and heme-binding properties of a novel cytochrome c maturation protein CcmE from Desulfovibrio vulgaris. Biochemistry 51, 3705–3707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schulz H., Hennecke H., Thöny-Meyer L. (1998) Prototype of a heme chaperone essential for cytochrome c maturation. Science 281, 1197–1200 [DOI] [PubMed] [Google Scholar]
- 34. Lee D., Pervushin K., Bischof D., Braun M., Thöny-Meyer L. (2005) Unusual heme-histidine bond in the active site of a chaperone. J. Am. Chem. Soc. 127, 3716–3717 [DOI] [PubMed] [Google Scholar]
- 35. Harvat E. M., Redfield C., Stevens J. M., Ferguson S. J. (2009) Probing the heme-binding site of the cytochrome c maturation protein CcmE. Biochemistry 48, 1820–1828 [DOI] [PubMed] [Google Scholar]
- 36. Feissner R. E., Richard-Fogal C. L., Frawley E. R., Kranz R. G. (2006) ABC transporter-mediated release of a haem chaperone allows cytochrome c biogenesis. Mol. Microbiol. 61, 219–231 [DOI] [PubMed] [Google Scholar]
- 37. Verissimo A. F., Mohtar M. A., Daldal F. (2013) The heme chaperone ApoCcmE forms a ternary complex with CcmI and apocytochrome c. J. Biol. Chem. 288, 6272–6283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. San Francisco B., Kranz R. G. (2014) Interaction of holoCcmE with CcmF in heme trafficking and cytochrome c biosynthesis. J. Mol. Biol. 426, 570–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Daldal F., Cheng S., Applebaum J., Davidson E., Prince R. C. (1986) Cytochrome c2 is not essential for photosynthetic growth of Rhodopseudomonas capsulata. Proc. Natl. Acad. Sci. U.S.A. 83, 2012–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Osyczka A., Dutton P. L., Moser C. C., Darrouzet E., Daldal F. (2001) Controlling the functionality of cytochrome c1 redox potentials in the Rhodobacter capsulatus bc1 complex through disulfide anchoring of a loop and a β-branched amino acid near the heme-ligating methionine. Biochemistry 40, 14547–14556 [DOI] [PubMed] [Google Scholar]
- 41. Collier G. S., Pratt J. M., De Wet C. R., Tshabalala C. F. (1979) Studies on haemin in dimethyl sulphoxide/water mixtures. Biochem. J. 179, 281–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Seery V. L., Muller-Eberhard U. (1973) Binding of porphyrins to rabbit hemopexin and albumin. J. Biol. Chem. 248, 3796–3800 [PubMed] [Google Scholar]
- 43. Tobias R., Kumaraswamy S. (2013) Biomolecular Binding Kinetics Assays on the Octet Platform, Application Note 14, ForteBio, Div. of Pall Life Sciences [Google Scholar]
- 44. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 45. Thomas P. E., Ryan D., Levin W. (1976) An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 75, 168–176 [DOI] [PubMed] [Google Scholar]
- 46. Dumont M. E., Corin A. F., Campbell G. A. (1994) Noncovalent binding of heme induces a compact apocytochrome c structure. Biochemistry 33, 7368–7378 [DOI] [PubMed] [Google Scholar]
- 47. Deshmukh M., Brasseur G., Daldal F. (2000) Novel Rhodobacter capsulatus genes required for the biogenesis of various c-type cytochromes. Mol. Microbiol. 35, 123–138 [DOI] [PubMed] [Google Scholar]
- 48. Fisher W. R., Taniuchi H., Anfinsen C. B. (1973) On the role of heme in the formation of the structure of cytochrome c. J. Biol. Chem. 248, 3188–3195 [PubMed] [Google Scholar]
- 49. Hamada D., Hoshino M., Kataoka M., Fink A. L., Goto Y. (1993) Intermediate conformational states of apocytochrome c. Biochemistry 32, 10351–10358 [DOI] [PubMed] [Google Scholar]
- 50. Feng Y. Q., Sligar S. G. (1991) Effect of heme binding on the structure and stability of Escherichia coli apocytochrome b562. Biochemistry 30, 10150–10155 [DOI] [PubMed] [Google Scholar]
- 51. Sambongi Y., Crooke H., Cole J. A., Ferguson S. J. (1994) A mutation blocking the formation of membrane or periplasmic endogenous and exogenous c-type cytochromes in Escherichia coli permits the cytoplasmic formation of Hydrogenobacter thermophilus holo cytochrome c552. FEBS Lett. 344, 207–210 [DOI] [PubMed] [Google Scholar]
- 52. Tomlinson E. J., Ferguson S. J. (2000) Conversion of a c-type cytochrome to a b-type that spontaneously forms in vitro from apo protein and heme: implications for c-type cytochrome biogenesis and folding. Proc. Natl. Acad. Sci. U.S.A. 97, 5156–5160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. McGuirl M. A., Lee J. C., Lyubovitsky J. G., Thanyakoop C., Richards J. H., Gray H. B., Winkler J. R. (2003) Cloning, heterologous expression, and characterization of recombinant class II cytochromes c from Rhodopseudomonas palustris. Biochim. Biophys. Acta 1619, 23–28 [DOI] [PubMed] [Google Scholar]
- 54. Inoue H., Wakai S., Nishihara H., Sambongi Y. (2011) Heterologous synthesis of cytochrome c′ by Escherichia coli is not dependent on the System I cytochrome c biogenesis machinery. FEBS J. 278, 2341–2348 [DOI] [PubMed] [Google Scholar]
- 55. Pielak G. J., Auld D. S., Beasley J. R., Betz S. F., Cohen D. S., Doyle D. F., Finger S. A., Fredericks Z. L., Hilgen-Willis S., Saunders A. J., Trojak S. K. (1995) Protein thermal denaturation, side-chain models, and evolution: amino acid substitutions at a conserved helix-helix interface. Biochemistry 34, 3268–3276 [DOI] [PubMed] [Google Scholar]
- 56. Colón W., Elöve G. A., Wakem L. P., Sherman F., Roder H. (1996) Side chain packing of the N- and C-terminal helices plays a critical role in the kinetics of cytochrome c folding. Biochemistry 35, 5538–5549 [DOI] [PubMed] [Google Scholar]
- 57. Cortajarena A. L., Wang J., Regan L. (2010) Crystal structure of a designed tetratricopeptide repeat module in complex with its peptide ligand. FEBS J. 277, 1058–1066 [DOI] [PubMed] [Google Scholar]
- 58. Lapouge K., Smith S. J., Walker P. A., Gamblin S. J., Smerdon S. J., Rittinger K. (2000) Structure of the TPR domain of p67phox in complex with Rac.GTP. Mol. Cell 6, 899–907 [DOI] [PubMed] [Google Scholar]
- 59. Daltrop O., Ferguson S. J. (2003) Cytochrome c maturation. The in vitro reactions of horse heart apocytochrome c and Paracoccus dentrificans apocytochrome c550 with heme. J. Biol. Chem. 278, 4404–4409 [DOI] [PubMed] [Google Scholar]
- 60. Kojima N., Yamanaka M., Ichiki S., Sambongi Y. (2005) Unexpected elevated production of Aquifex aeolicus cytochrome c555 in Escherichia coli cells lacking disulfide oxidoreductases. Biosci. Biotechnol. Biochem. 69, 1418–1421 [DOI] [PubMed] [Google Scholar]
- 61. Fee J. A., Todaro T. R., Luna E., Sanders D., Hunsicker-Wang L. M., Patel K. M., Bren K. L., Gomez-Moran E., Hill M. G., Ai J., Loehr T. M., Oertling W. A., Williams P. A., Stout C. D., McRee D., Pastuszyn A. (2004) Cytochrome rC552, formed during expression of the truncated Thermus thermophilus cytochrome c552 gene in the cytoplasm of Escherichia coli, reacts spontaneously to form protein-bound 2-formyl-4-vinyl (Spirographis) heme. Biochemistry 43, 12162–12176 [DOI] [PubMed] [Google Scholar]
- 62. Fujii S., Masanari M., Yamanaka M., Wakai S., Sambongi Y. (2014) High stability of apo-cytochrome c′ from thermophilic Hydrogenophilus thermoluteolus. Biosci. Biotechnol. Biochem. 78, 1191–1194 [DOI] [PubMed] [Google Scholar]
- 63. Yamanaka M., Mita H., Yamamoto Y., Sambongi Y. (2009) Heme is not required for Aquifex aeolicus cytochrome c555 polypeptide folding. Biosci. Biotechnol. Biochem. 73, 2022–2025 [DOI] [PubMed] [Google Scholar]
- 64. Daltrop O., Allen J. W., Willis A. C., Ferguson S. J. (2002) In vitro formation of a c-type cytochrome. Proc. Natl. Acad. Sci. U.S.A. 99, 7872–7876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kellogg J. A., Bren K. L. (2002) Characterization of recombinant horse cytochrome c synthesized with the assistance of Escherichia coli cytochrome c maturation factors. Biochim. Biophys. Acta 1601, 215–221 [DOI] [PubMed] [Google Scholar]
- 66. Anderson J. L., Armstrong C. T., Kodali G., Lichtenstein B. R., Watkins D. W., Mancini J. A., Boyle A. L., Farid T. A., Crump M. P., Moser C. C., Dutton P. L. (2014) Constructing a man-made c-type cytochrome maquette in vivo: electron transfer, oxygen transport, and conversion to a photoactive light harvesting maquette. Chem. Sci. 5, 507–514 [DOI] [PMC free article] [PubMed] [Google Scholar]