Background: Transcriptional repressor DAXX suppresses several tumor suppressor genes and is up-regulated in many cancers.
Results: We demonstrate that DAXX has potent growth-enhancing effects on primary prostatic malignancy through inhibition of autophagy.
Conclusion: In the early stages of tumorigenesis, autophagy suppresses prostate tumor formation.
Significance: This is the first study to link prostate cancer development to autophagy suppression by DAXX.
Keywords: autophagy, biomarker, cell death, prostate cancer, tumor cell biology, tumor promoter, tumor suppressor gene, DAXX
Abstract
The DAXX transcriptional repressor was originally associated with apoptotic cell death. However, recent evidence that DAXX represses several tumor suppressor genes, including the DAPK1 and DAPK3 protein kinases, and is up-regulated in many cancers argues that a pro-survival role may predominate in a cancer context. Here, we report that DAXX has potent growth-enhancing effects on primary prostatic malignancy through inhibition of autophagy. Through stable gene knockdown and mouse subcutaneous xenograft studies, we demonstrate that DAXX promotes tumorigenicity of human ALVA-31 and PC3 prostate cancer (PCa) cells in vivo. Importantly, DAXX represses expression of essential autophagy modulators DAPK3 and ULK1 in vivo, revealing autophagy suppression as a mechanism through which DAXX promotes PCa tumorigenicity. Furthermore, DAXX knockdown increases autophagic flux in cultured PCa cells. Finally, interrogation of the OncomineTM database suggests that DAXX overexpression is associated with malignant transformation in several human cancers, including prostate and pancreatic cancers. Thus, DAXX may represent a new cancer biomarker for the detection of aggressive disease, whose tissue-specific down-regulation can serve as an improved therapeutic modality. Our results establish DAXX as a pro-survival protein in PCa and reveal that, in the early stages of tumorigenesis, autophagy suppresses prostate tumor formation.
Introduction
The DAXX cell death modulator, originally identified as a pro-apoptotic protein (1), functions as a transcriptional repressor (2) and, together with ATRX, serves as a histone H3.3 chaperone (3). In its capacity as a transcriptional repressor, DAXX associates with RelB, a transcription factor of the NF-κB family that directly controls the expression of the DAPK1/3 tumor suppressor protein kinases (2), which are linked to autophagy (4, 5). Through its ability to repress tumor suppressors and autophagy regulators via a mechanism involving association with RelB and subsequent target promoter DNA methylation (2), DAXX would be expected to induce tumor growth or survival. However, its tumorigenic potential, if any, in prostate cancer (PCa)2 remains unknown.
Evidence suggests that in a cancer context, as in development (6), DAXX may have a pro-survival role. Strong positive DAXX staining is observed in PCa tissues, compared with benign prostatic hyperplasia tissues, suggesting that DAXX is overexpressed in PCa (7). Importantly, strong DAXX expression is linked to high Gleason score and increased cell proliferation index, suggesting that DAXX can serve as an independent prognosticator in PCa (8). Similarly to PCa, immunohistochemical analysis of DAXX in high grade urothelial carcinoma shows increased expression compared with normal urothelium (9). Likewise, increased DAXX expression in diffuse large B-cell lymphoma correlates with poor patient survival (10). Inactivating mutations in DAXX can have the opposite effect and are associated with improved pancreatic cancer patient survival (11). Alternative lengthening of telomeres, which is associated with loss of function of DAXX in pancreatic neuroendocrine tumors (11), is absent in adenocarcinomas of the prostate (12), implying that DAXX is functionally active in PCa.
These observations in multiple cancers suggest that the role of DAXX in cancer development is broadly relevant to tumor biology. The goal of this investigation was to determine the function of DAXX in PCa pathogenesis. We present evidence that, in the context of a mouse subcutaneous xenograft model of PCa, DAXX promotes tumorigenicity by suppressing autophagy.
Experimental Procedures
Cell Culture and Recombinant Lentivirus Transduction
Prostate cancer cell lines ALVA-31, DU145, LNCaP, and PC3, which were obtained from Scott Crist (Purdue University), were maintained in Roswell Park Memorial Institute-1640 (RPMI 1640) medium, containing 10% fetal bovine serum, FBS (HyClone), and 1% penicillin/streptomycin plus l-glutamine. The human prostatic epithelial cells, PWR-1E, derived from a normal prostate gland, were obtained from ATCC (catalogue no. CRL-11611) and grown in ATCC complete growth medium. For the generation of stable DAXX knockdown (K/D) PCa lines (ALVA-31, PC3, and DU145), recombinant lentiviruses targeting DAXX (constructed in the lentiviral backbone vector pLKO.1-puro) were purchased from Sigma (Clone ID NM_001350.x-2410s1c1; accession number NM_001350.3; region 3′-UTR). A nonspecific control virus was also purchased (SHC002V: MISSION® non-target shRNA control transduction particles). For the generation of ALVA-31 double knockdown cells (DAXX and ULK1 dK/D), a human ULK1 shRNA vector (TRCN0000000838), obtained from Reuben Shaw (Salk Institute), was used to transfect ALVA-31 DAXX K/D cells. When the cells reached 70–80% confluence, they were infected (MOI = 10) with the DAXX shRNA (ALVA-31, PC3, and DU145 cells), ULK1 shRNA (ALVA-31 DAXX K/D cells), or nonspecific control shRNA (ALVA-31 cells) virus vector. Hexadimethrine bromide (Polybrene, Sigma, catalog no. AL-118), at a concentration of 8 μg/ml, was added at the time of infection to enhance infection efficiency. After 24 h, the medium was changed and replaced with puromycin-containing medium (Sigma, catalog no. P9620; 2 μg/ml). Cells were cultured for ∼3 weeks in puromycin-containing medium before analyzing for DAXX or ULK1 expression and were subsequently used in subcutaneous xenograft studies.
qRT-PCR Analysis
ALVA-31, ALVA-31 DAXX K/D, PC3, and PC3 DAXX K/D cells were processed using the Power SYBR Green Cells-to-Ct kit (Ambion, catalog no. 4402953) to lyse cells, generate cDNA, and perform RT-PCR per the manufacturer's instructions. The sequences of the ULK1, LC3, p62, and control (GAPDH and CPH) qPCR primers are indicated below. For chromatin immunoprecipitation (ChIP)-qPCR experiments, ChIP assays were first performed using the ChIP-IT high sensitivity kit from Active Motif (catalog no. 53040). The resulting products were then subjected to qPCR analysis using ULK1 primers covering the five NF-κB binding sites shown below.
qPCR primers were as follows: ULK1 forward primer, 5′-GTG CAG TCG GCT GCC CTG GAC-3′; ULK1 reverse primer, 5′-TCA GGC ACA GAT GCC AGT CAG C-3′; LC3 forward primer, 5′-AAC AAA GAG TAG AAG ATG TCC GAC-3′; LC3 reverse primer, 5′-CTA ATT ATC TTG ATG AGC TCA CT-3′; p62 forward primer, 5′-CTA CAG ATG CCA GAA TCC GAA GGG-3′; p62 reverse primer, 5′-CAT CTG GGA GAG GGA CTC AAT-3′; GAPDH forward primer, 5′-ACA TCA AGA AGG TGG TGA AGC AGG-3′; GAPDH reverse primer, 5′-ACA AAG TGG TCG TTG AGG GCA ATG-3′; CPH forward primer, 5′-GAC CCA ACA CAA ATG GTT C-3′; CPH reverse primer, 5′-AGT CAG CAA TGG TGA TCT TC-3′.
For ChIP-qPCR experiments, the ULK1 primer sequences covering the five NFκB binding sites were as follows: ULK1 forward primer 1, 5′-CCG CAA GGA CCT GAT CGG CC-3′; ULK1 reverse primer 1, 5′-ACA GGC GGG GAA TCT CGG GG-3′; ULK1 forward primer 2, 5′-CAG GAT CCC CAC CCC GCG AC-3′; ULK1 reverse primer 2, 5′-GTT GCG GGG TGT CCC GGG GT-3′; ULK1 forward primer 3, 5′-GCG CGA TCC TCA ACC TGG CT-3′; ULK1 reverse primer 3, 5′-TGC ACT TGA CGG CGA CCT CC-3′; ULK1 forward primer 4, 5′-GTG CTG GGG GAG GGG GCG TG-3′; ULK1 reverse primer 4, 5′-CAG CAG ACC GCA GCC CAG AG-3′; ULK1 forward primer 5, 5′-TGC GTC ATG GCT CTG GGA GC-3′; ULK1 reverse primer 5, 5′-GGG GAG CCC TGG AGG GGA GC-3′.
Antibodies and Immunoblotting
Protein lysates were prepared as described previously (2). Aliquots of cell lysates, normalized for total protein content, were fractionated by SDS-PAGE and transferred to nitrocellulose blotting membranes (BA85 Protran, 0.45 μm, Whatman, catalogue no. 10401196). The following antibodies were used for immunoblotting: rabbit anti-DAXX (Novus Biologicals), rabbit anti-Atg1/ULK1 (Abcam); rabbit anti-ULK2 (Abcam), mouse anti-β-actin (Sigma), mouse anti-p62 (Sequestosome-1) (Millipore), and mouse anti-LC3 (MBL). Quantitative immunoblot detection was performed using the Odyssey Infrared Imaging System, version 3.0 (LI-COR Biosciences).
Deep Sequencing (ChIP-seq)
Active Motif's ChIP-IT high sensitivity kit (catalog no. 53040), was used, utilizing PC3 cells. Anti-DAXX (sc-7001) goat polyclonal antibody from Santa Cruz Biotechnology, Inc. was used. A two-step cross-linking procedure preceded ChIP as described (13). Deep sequencing (ChIP-seq) was performed using an Illumina HiSeq 2500 system. Genomic data analysis and visualization were done as described, using Bowtie2 and HOMER (13).
Confocal Microscopy
PC3 cells, wild type and DAXX knockdown, maintained in RPMI 1640 medium as described above, were plated onto Nunc Lab-Tek 4-well chamber slides (Sigma, Z691992), at a density of 50,000 cells/well. They were then transiently transfected with GFP-LC3 for 24 h. Following transfection, cells were left untreated, incubated in starvation medium (Earle's balanced salt solution, Life Technologies) for 2 h, or incubated in RPMI medium containing 50 nm bafilomycin A (BafA, Sigma) for 2 h. The cells were then fixed in 4% paraformaldehyde in PBS for 15 min, permeabilized in 0.2% Triton X-100 in PBS for 15 min, and then incubated with TO-PRO®-3 iodide (Life Technologies) for 15 min at room temperature in the dark to stain the nucleus, followed by extensive washing with PBS. Fluoromount-G (Southern Biotech) was used to mount the slides. Z-Stack confocal images of GFP-LC3-expressing cells were collected using the ×63 oil objective on a Zeiss LSM 710 microscope (Waitt Advanced Biophotonics Center, Salk Institute). 10–15 random fields were obtained and analyzed for each condition. The number of GFP-LC3 puncta in a cell was determined using IMARIS spot-counting software (Waitt Advanced Biophotonics Center, Salk Institute). Statistical analyses and graphs were constructed using the GraphPad Prism version 5.0 software package.
In Vivo Xenograft Studies
For subcutaneous xenograft studies, athymic nude male mice (8–10 weeks old) were purchased from the Jackson Laboratory and housed in the Salk Institute's vivarium. The subcutaneous xenograft study was performed three times, each time involving six mice per group. Ten million prostate cancer cells were injected subcutaneously into the right flank of the animals. Tumor size was measured biweekly using external calipers. Mice were monitored daily for any sign of illness. Tumors were harvested 4–7 weeks postinjections, and their respective weights (g) were determined immediately postexcision. Statistical analyses were performed using analysis of variance and Student's t test. Excised tumors were processed for 1) immunohistochemical (IHC) analyses and 2) immunoblotting. IHC analyses are described below. For immunoblotting, tumors were processed using a total protein extraction kit from Millipore (catalog no. 2140).
Immunohistochemistry
Tumor tissues were immediately excised from sacrificed mice (excision performed by Salk pathologist Mat LeBlanc), fixed in zinc-buffered formalin (Z-fix; Anatech, Ltd.), and embedded in paraffin (service performed by Pacific Pathology, Inc., San Diego, CA). Deparaffinized tissue sections were immunostained using rabbit polyclonal antibodies to DAXX (Santa Cruz Biotechnology), DAPK1 (Sigma-Aldrich), DAPK3 (Sigma-Aldrich), Ki67 (Thermo Scientific), and CD31 (Abcam) as well as mouse monoclonal antibodies to LC3 (NanoTools), p62 (Sequestosome-1) (Millipore), and multiubiquitin (MBL). Application of the primary antibody was followed by incubation with biotinylated secondary antibody (Vector Laboratories) and ImmPACT diaminobenzidine staining (Vector Laboratories), yielding a brown color. Mayer's hematoxylin solution (Sigma-Aldrich) was used as a nuclear counterstain, yielding a blue color.
Tissue Morphometry
Histological slides were scanned at a magnification of ×10–40 using a Zeiss Axio Observer VivaTome inverted fluorescence microscope (Waitt Advanced Biophotonics Center, Salk Institute). The ImageScope analysis software (version 9.1; Aperio Technologies, Inc.) was applied to quantify IHC stainings. The graphs were constructed using the GraphPad Prism version 5.0 statistical analysis software package.
Oncomine Database Interrogation
To assess expression levels of DAXX, DAPK1, and DAPK3 mRNA in normal prostate versus prostate cancer, the OncomineTM database (Compendia Bioscience) was interrogated using the Premium Edition package. The data were queried between December 3 and 6, 2012. Raw data in Excel format were provided. We collected the raw data to construct graphs using the GraphPad Prism version 5 software. The Oncomine interrogation included survival rates, metastases, drug resistance, and Gleason score analyses.
Results
DAXX Promotes Tumorigenicity of Human ALVA-31 and PC3 PCa Cells
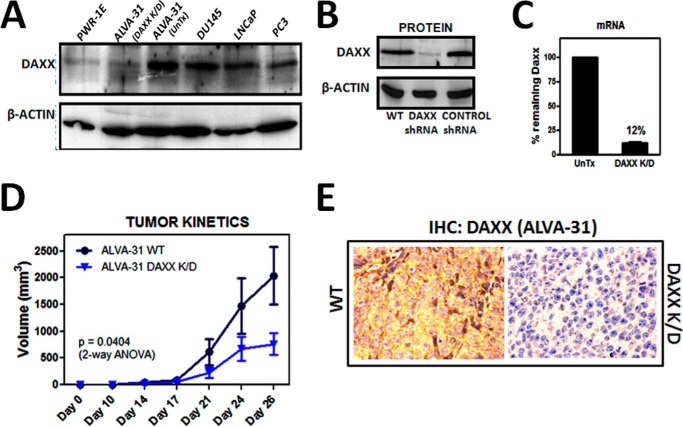
To explore the role of DAXX in PCa, we initially compared the endogenous DAXX levels in four human PCa cell lines (PC3, ALVA-31, LNCaP, and DU145) (Fig. 1A). DAXX levels were higher in all four PCa cell lines compared with non-tumorigenic PWR-1E human prostate epithelial cells (Fig. 1A), with the highest levels found in the hormone-refractory PCa line, ALVA-31 (Fig. 1A). We therefore selected ALVA-31, derived from a primary PCa (14), to initially analyze the consequences of knockdown of DAXX. Using high efficiency lentivirus-mediated gene delivery to introduce a DAXX targeting shRNA vector, we propagated multiple clones of puromycin-resistant ALVA-31 cells to produce (pooled) stable cell lines. DAXX knockdown was quantified via protein immunoblotting analysis for DAXX protein (Fig. 1B) and by reverse transcriptase-polymerase chain reaction (RT-PCR) for mRNA (Fig. 1C). We achieved ∼90% DAXX knockdown compared with untransduced cells (UnTx) (Fig. 1C). Therefore, the stable DAXX knockdown PCa cell line mimics non-tumorigenic prostate epithelial cells in its DAXX protein level (Fig. 1A).
FIGURE 1.
DAXX promotes tumorigenicity of ALVA-31 human PCa cells. A, DAXX levels across PCa cell lines were compared with normal PCa epithelia (PWR-1E), including ALVA-31 transfected with DAXX shRNA. B, DAXX shRNA transfection in ALVA-31 cells is shown. DAXX K/D was analyzed via immunoblotting, comparing DAXX protein levels with a loading control (β-actin). C, mRNA levels were determined via qRT-PCR. Respective quantifications of the degree of knockdown are shown, comparing DAXX shRNA-transfected samples (DAXX K/D) with the untransfected ones (UnTx). D, subcutaneous injection of 10 million ALVA-31 cells in nude mice (6 mice/group); ALVA-31 tumor growth kinetics, based on tumor volumes measured externally by calipers, shows that DAXX promotes tumorigenicity. The formula used to determine the tumor volume is L × W2/2 (where L is length and W is width). E, immunohistochemical analysis of DAXX was performed using excised tumor tissue corresponding to control WT (left) and DAXX K/D (right) tumors from ALVA-31 injections. Tissue sections were stained using an antibody specific for DAXX and hematoxylin. Representative ×40 images are shown. Error bars, S.E. ANOVA, analysis of variance.
To assess the role of DAXX in tumorigenesis in vivo, we utilized an athymic nude mouse model. Mice were injected subcutaneously in the flank with 10 million ALVA-31 DAXX K/D, ALVA-31 control shRNA, or untransfected ALVA-31 prostate cancer cells (WT). The tumors were allowed to grow for 4 weeks, tumor volume was monitored by external calipers twice a week, and the wet weights and final volumes of tumors were determined at the termination of the experiment. Mice injected with untransfected ALVA-31 (WT) or control shRNA-transfected ALVA-31 cells (data not shown) reproducibly developed tumors. In contrast, significantly fewer tumors formed in mice injected with DAXX K/D cells. Moreover, the tumors that arose in the DAXX shRNA group grew more slowly than tumors from the ALVA-31 WT group (Fig. 1D) and were smaller, as assessed by final volume and wet weight (data not shown). Immunohistochemistry analysis of tumor tissues confirmed reduced DAXX levels in DAXX K/D group, compared with the WT group (Fig. 1E). Thus, DAXX promotes the in vivo tumorigenicity of ALVA-31 cells in this subcutaneous tumor xenograft model.
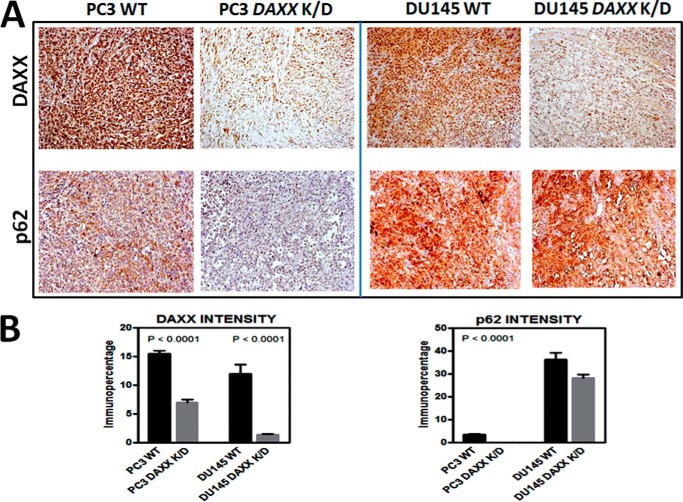
To exclude a cell type-specific effect, we extended ALVA-31 studies to other human PCa cell types by generating two additional DAXX K/D PCa lines, namely PC3 DAXX K/D and DU145 DAXX K/D (Fig. 2A). Although both of these lines share high tumorigenic potential and are commonly used in subcutaneous xenograft experiments (15), a recent study revealed that DU145 cells differ from other PCa cell lines, including PC3 cells, by being autophagy-defective as a result of ATG5 deficiency (16). Consistent with the ALVA-31 DAXX K/D xenograft data, the size of tumors elicited by injected PC3 DAXX K/D cells was dramatically reduced compared with PC3 WT cells, as determined by final tumor volumes and wet weights (Fig. 2, B and C) and tumor growth rate (Fig. 2D). In contrast, the autophagy-defective DU145 cells produced similar sized tumors regardless of DAXX status (Fig. 2, B–D). This effect was not due to differences in cell type viability or individual mouse health characteristics, because cells replated postinjection retained comparable viability (not shown), and mice from different groups were equally healthy (not shown).
FIGURE 2.
The tumorigenic potential of DAXX in human PCa cells is autophagy-dependent. A, PC3 and DU145 PCa cells were transfected with DAXX shRNA (left), followed by quantification of the degree of knockdown (protein; right). B, xenograft study consisted of subcutaneous injection of 10 million PC3 WT, PC3 DAXX K/D, DU145 WT, or DU145 DAXX K/D cells in nude mice (6 mice/group). Three representative images of tumor growth from each group at the end of the xenograft study are shown. Note the size difference among the tumors from two groups in the PC3 case and the lack of a difference in the size of tumors in the case of DU145. C, tumor volumes (left) and tumor wet weights (right) show that DAXX is required for tumor growth in PC3 cells, similarly to ALVA-31 cells. In contrast, in DU145 cells, which are autophagy-defective, DAXX does not significantly affect tumorigenicity. D, tumor growth kinetics in mice injected with PC3 cells (left) is contrasted to kinetics in mice injected with DU145 cells (right). Error bars, S.E.
DAXX Modulates Autophagy Markers in Vivo
With the observation that DAXX promotes ALVA-31 and PC3, but not DU145, tumor growth in mice, we performed histological and molecular analyses of tumors to examine markers of autophagy, proliferation, and angiogenesis in search of a potential explanation of the phenotype observed. Accordingly, fixed tumors were sectioned and evaluated by IHC analysis using antibodies to a variety of biomarkers, followed by quantitative image analysis using Aperio ImageScope. As expected, DAXX immunostaining was markedly reduced in tumor cells arising from PC3, DU145 (Fig. 3A), and ALVA-31 cells (Fig. 1C) transduced with DAXX shRNA. Immunohistochemical assessment of proliferation (Ki67) and angiogenesis markers (CD31) suggested that DAXX K/D tumors had slower division rates and were less vascularized (data not shown). Importantly, we noted differences in expression of autophagy marker proteins, including LC3 (not shown) and p62 (Fig. 3A), where immunostaining for p62 was decreased in PC3 DAXX K/D tumors (Fig. 3A). The change was statistically significant (Fig. 3B). The results were corroborated by immunoblotting of tumor tissue lysates (not shown).
FIGURE 3.
DAXX modulates autophagy markers in vivo. A, quantitative immunohistochemical analysis of protein markers was performed using excised tumor tissue corresponding to WT (n = 6) and DAXX K/D (n = 6) tumors from either PC3 or DU145 injections. Tissue sections were stained using antibodies specific for DAXX or p62. Representative ×10 images are shown. B, immunostaining was quantified and analyzed using Aperio ImageScope software. To construct graphs, GraphPad Prism version 5 software was used. Statistical significance was assessed by unpaired Student's t test. Note that DU145 cells, being autophagy-defective, show high levels of p62, regardless of the DAXX status. Error bars, S.E.
These initial results suggested a correlation between DAXX expression and decreased autophagy, because DAXX K/D tumors displayed elevated autophagy, as evidenced by decreased p62 in autophagy-intact PCa cells (PC3). During autophagy, cytosolic LC3-I is processed into an autophagosome-bound LC3-II form; a higher LC3-II/LC3-I ratio is taken as an indicator of increased autophagy (17). The p62 protein (Sequestosome 1) is a receptor that binds autophagosome-associated LC3-II and ubiquitin, allowing for specific elimination of ubiquitylated proteins via autophagy (18). p62 forms cytoplasmic aggregates in autophagic cells, with lower levels correlating with increased autophagic flux (19). It is the preferred marker for measuring autophagic flux, because it is degraded when the autophagosome fuses with the lysosome (17). The p62 IHC staining (Fig. 3, A and B) and p62 protein levels (not shown) of DU145 cells remained high regardless of DAXX presence, consistent with their defective autophagy status, providing a further clue that DAXX may affect tumorigenicity via the autophagic pathway.
DAXX Suppresses the Autophagy Machinery in Vivo
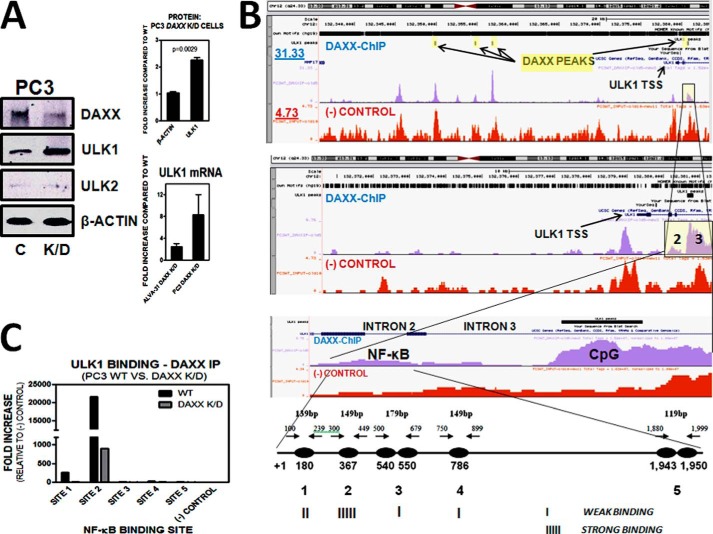
To determine whether autophagy is a mechanism by which DAXX affects tumorigenicity, we generated a stable dK/D PCa line, namely, ALVA-31 DAXX/ULK1 dK/D (Fig. 4A). The ULK1 protein kinase is an essential autophagy initiator protein (20). If autophagy were responsible for tumor growth suppression in DAXX K/D cells, then this effect would be reversed if ULK1 was depleted in these cells. Indeed, the growth of subcutaneous ALVA-31 DAXX K/D tumors was significantly enhanced when ULK1 was co-depleted in these cells, as determined by growth kinetics and tumor wet weights (Fig. 4B). Furthermore, the ALVA-31 DAXX/ULK1 dK/D tumors displayed significantly decreased levels of LC3 compared with ALVA-31 DAXX K/D tumors (Fig. 4C), suggesting that active autophagy is needed to suppress initial tumor growth of ALVA-31 cells. Of note, despite several attempts, we were unable to derive an ULK1 K/D ALVA-31 cell line to use as a control, because these cells were not viable. Through immunoblotting, ULK1 protein levels were found to be significantly increased in PC3 DAXX K/D cells (Fig. 5A), suggesting that DAXX normally suppresses expression of the autophagy initiator kinase ULK1, either directly or indirectly. Because ULK1 and its closely related family member ULK2 may have redundant functions, and, in some cells, one may compensate for the lack of the other (21), we checked the ULK2 protein levels in PC3 cells. The levels of ULK2 were negligible in these cells and were unaffected by DAXX knockdown (Fig. 5A), implying that ULK1, rather than ULK2, is the critical regulator of autophagy in PCa cells. Moreover, through qRT-PCR analysis, ULK1 mRNA levels in DAXX-deficient cells (ALVA-31 and PC3) were found to be increased 4–8-fold, respectively, compared with control counterparts (Fig. 5A), suggesting that the ULK1 promoter may be targeted by DAXX. In contrast, DAXX did not appear to affect the transcription of LC3, GAPDH (glyceraldehyde-3-phosphate dehydrogenase), or CPH (cyclophilin) (not shown).
FIGURE 4.
DAXX enhances tumorigenicity of PCa by specifically suppressing the autophagy machinery in vivo. A, ALVA-31 cells were stably transfected with control (C), DAXX, or DAXX and ULK1 (dK/D) shRNA (left), resulting in significant (∼93%) knockdown (right, protein levels). B, xenograft experiments were performed by injecting 10 million cells/mouse. There were six mice per group. Tumor kinetics (left) and wet weight (right) analyses demonstrate that when ULK1, an essential component of the autophagy machinery, is knocked down, tumor growth is restored in DAXX K/D cells. C, quantitative immunohistochemical analysis was performed using excised tumor tissue corresponding to CNTL (n = 6), DAXX K/D (n = 6), and DAXX/ULK1 dK/D (n = 6) tumors from ALVA-31 injections. Tissue sections were stained using an antibody specific for the autophagy marker LC3 (top row). A negative control (hematoxylin) was also included (bottom row). Immunostaining was quantified and analyzed using Aperio ImageScope software. Statistical significance was assessed by unpaired Student's t test. Representative ×40 images are shown. Error bars, S.E.
FIGURE 5.
DAXX binds to ULK1 and represses its expression. A, PC3 cells were stably transfected with control (C) or DAXX (K/D) shRNA. Lysates were analyzed for DAXX, ULK1, ULK2, and β-actin proteins levels via immunoblotting. ULK1 levels were significantly higher in DAXX K/D cells compared with control ones, as determined via ImageJ quantifications. In contrast, there was a negligible amount of ULK2 in PC3 cells, regardless of DAXX status. β-Actin served as a loading control. Q-PCR analysis was consistent with immunoblotting, revealing that ULK1 mRNA levels, like those of protein, were increased in DAXX K/D PC3 and ALVA-31 cells. B, to determine whether DAXX binds to the ULK1 gene, ChIP-seq and ChIP experiments were performed using an anti-DAXX antibody to immunoprecipitate the experimental sample (blue), whereas non-immunoprecipitated chromatin was used as a negative control (input, red). Several specific, prominent DAXX peaks were found in the ULK1 genomic region, which were not found in the negative control. These included a peak at +367 from the TSS in intron 2 and a peak in intron 3 +1,464 from the TSS close to a CpG island. To investigate the five predicted NF-κB binding sites in intron 2 (bottom) in more detail, primers were designed to encompass these sites, with a range of amplicon sizes between 119 and 179 bp. NF-κB binding sites were searched for using the DBTSS Web site. ChIP-seq experiments were performed using PC3 cells, WT and DAXX K/D. Note that the top panel highlights in yellow the four prominent DAXX ChIP-seq peaks in ULK1. ULK1 TSS is shown (black arrow). Also shown are the read counts (y axis) for the DAXX-ChIP (blue) and (−) control (red). The highlighted yellow region represents the regions that are enlarged in the second and third panels. Specifically, #2 represents intron 2 (predicted NF-κB binding site), and #3 represents intron 3 (CpG island). The schematic at the bottom represents the aforementioned NF-κB binding sites in intron 2, as described above. C, following DAXX immunoprecipitation, qRT-PCR revealed strongest binding of DAXX to the second (#2) NF-κB binding site in ULK1 in WT PC3 cells. The signal for DAXX K/D was weaker. In addition, site 1 displayed measurable binding affinity. Negative ChIP-PCR controls included the use of anti-TRAF6 and β-actin antibodies. Error bars, S.E.
We have previously shown that DAXX represses the expression of DAPK1 and DAPK3 by mediating NF-κB-dependent recruitment of the DNMT1 DNA methyltransferase to their promoters, resulting in increased local CpG methylation and decreased transcription (2). To determine whether DAXX associates with ULK1 in PC3 cells, we performed ChIP-seq analyses (Fig. 5B). ChIP-seq revealed that four prominent DAXX peaks were found in the genomic region encompassing the ULK1 gene (Fig. 5B, top). None of them was found in the negative control (input, non-immunoprecipitated chromatin). One of them, located at 1,464 bp downstream of the ULK1 transcription start site (TSS) in intron 3, corresponded to a CpG island, suggesting that perhaps DAXX utilizes DNA methylation to repress ULK1, in a manner similar to its suppression of DAPK1/3 (2). Interestingly, a DAXX ChIP-seq peak, located upstream of the CpG peak, in the second intron, was also detected (Fig. 5B, third panel). DBTSS (database of transcriptional start sites) revealed that there were five NF-κB binding sites in intron 2 of ULK1 (Fig. 5B, bottom). Chromatin immunoprecipitation (ChIP) analysis revealed that DAXX bound strongly to the second predicted NF-κB binding site in ULK1, 367 bp downstream of its TSS (Fig. 5C). The DAXX ChIP signal was lower in DAXX-deficient cells, as expected (Fig. 5C). A weak binding signal was detected for the first NF-κB site, and signals were negligible for the rest of the sites and negative controls (Fig. 5C). Therefore, we found that DAXX targets the essential autophagy gene ULK1, co-localizes with it, and represses its expression. The exact mechanism of repression remains to be elucidated.
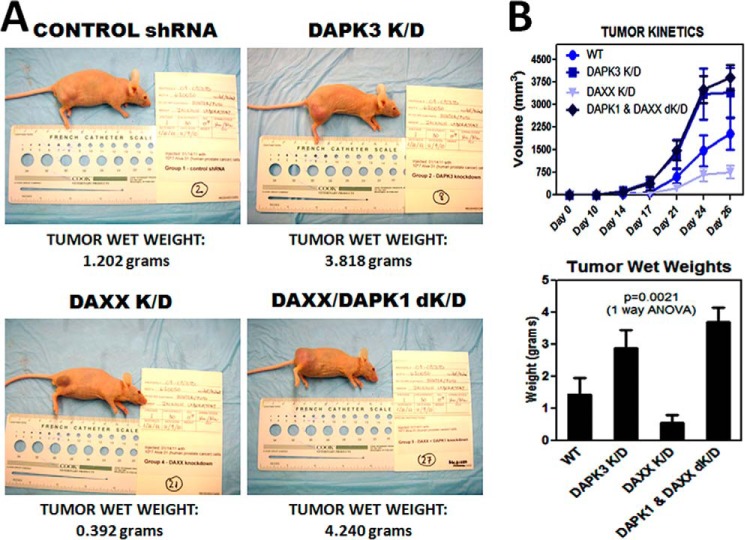
In addition to ULK1, DAXX may suppress the autophagic pathway by reducing expression of the DAPK3 and/or DAPK1 protein kinases. Not only are DAPK1 and -3 two of the transcriptional repression targets of DAXX (2), but they are also direct targets for ULK1 phosphorylation/activation (DAPK3 (5)) or upstream activators of Beclin1 (DAPK1 (4)), making them two additional components of the autophagic machinery. To determine whether DAXX affects tumor growth via DAPK3 suppression, DAPK3 was knocked down in ALVA-31 cells. Knocking down DAPK3 resulted in enhanced tumor growth rate and tumor size (Fig. 6, A and B) compared with the ALVA-31 control and DAXX K/D groups, suggesting that DAPK3 acts as a tumor suppressor, whereas DAXX acts as a tumor promoter. Furthermore, decreased tumor growth in ALVA-31 cells due to DAXX knockdown could be rescued by DAPK1 knockdown, suggesting that DAXX may also affect the DAPK1 pathway (Fig. 6, A and B). Of note, DAPK1 and DAPK3 are closely related serine/threonine kinases, with at least some functional redundancy between these two kinases (4). It is likely, therefore, that DAPK3 knockdown in ALVA-31 DAXX K/D cells, if generation of such a double knockdown line were successful, would give similar results.
FIGURE 6.
DAPKs act as tumor suppressors in the subcutaneous xenograft model. A, male athymic nude mice (6 mice/group) were injected in the right flank subcutaneously with 10 million cells from each of the cell groups indicated, and tumor volumes were measured over the course of 1 month using external calipers. Tumor appearance of a representative mouse per group at the conclusion of the experiment is shown. The respective tumor wet weights are shown at the bottom of each panel. B, tumor growth rate was faster (top), and tumor wet weights were greater (bottom) for DAPK3 K/D and DAXX/DAPK1 K/D groups, arguing that DAPKs are acting as tumor suppressors. Aliquots of various cell types used in injections were replated immediately following injections and determined to be fully viable (not shown). Error bars, S.E.
An inverse relationship between DAXX and DAPK3 was observed by IHC analysis of tumor tissues (Fig. 7, A–C), where tumors with depleted DAXX, which were significantly less vascularized (Fig. 7C), had significantly elevated levels of DAPK3 (Fig. 7A). This inverse relationship was also observed by immunoblotting of tumor tissue lysates (not shown). IHC staining for the LC3 and p62 autophagy markers confirmed significantly increased autophagy in tumor tissues arising from DAXX K/D group compared with all other groups (Fig. 7B), with increased LC3 (top) and decreased p62 (bottom). Thus, DAXX enhances tumor growth by affecting several components of the autophagy machinery in vivo.
FIGURE 7.
DAXX and DAPK3 are inversely correlated in vivo. A, quantitative immunohistochemical analyses were performed using excised tumor tissue corresponding to CNTL, DAXX K/D, and DAPK3 K/D tumors from ALVA-31 subcutaneous injections, as described above. Tissue sections were stained using an antibody specific for DAPK3. A negative control (hematoxylin) was also included. Representative ×10 images are shown. Immunostaining was quantified and analyzed using Aperio ImageScope software, and the graph was constructed using GraphPad Prism version 5 software. An inverse correlation between DAXX and DAPK3 is observed. B, immunostaining using LC3 or p62 antibodies shows increased autophagy in DAXX K/D tumors, based on LC3 increase and p62 decrease in these tumors. The pattern is reversed for DAPK3 K/D tumors. Representative ×40 images are shown. C, immunohistochemistry using CD31 marker for angiogenesis shows statistically significant increased proliferation in larger size tumors (DAPK3 K/D) compared with smaller ones (DAXX K/D). Quantification was done as in A above. Error bars, S.E.
DAXX Knockdown Modulates Autophagy Markers in Cultured PC3 Cells
To complement the in vivo studies, we analyzed the expression of autophagy markers in cultured PC3 cells (WT versus DAXX K/D). For this purpose, PC3 cells, transiently transfected with GFP-LC3, were analyzed for GFP-LC3 expression under basal (untreated) autophagy conditions (Fig. 8A) and serum starvation (autophagy induction) via Earle's balanced salt solution (not shown). Basal autophagy was significantly higher in DAXX K/D cells compared with WT counterparts, based on quantification of the number of LC3 puncta (Fig. 8A). Upon visual inspection, autophagy induction via serum starvation was also higher in the DAXX K/D group, based on the higher number of LC3 puncta (not shown), although quantification was complicated due to the fact that the size of the LC3 puncta was increased (in both groups), making it more difficult to discern and count individual puncta objectively. Inhibition of autophagy in PC3 cells using bafilomycin A (BafA), a potent and specific inhibitor of vacuolar H+ ATPase, in a time course experiment (Fig. 8B) resulted in increased p62 levels in both groups (Fig. 8B). The increase was more pronounced in the WT group, consistent with a lower autophagic flux when DAXX is overexpressed. Immunoblotting for DAXX confirmed the expected differences in its expression in WT versus DAXX K/D PC3 cells (Fig. 8B); as a control, no significant changes were observed in β-actin protein levels (Fig. 8B). Results similar to those for PC3 cells were obtained for p62 using ALVA-31 cells (not shown). In summary, using a range of assays that included measurement of endogenous p62, which reflects autophagic flux, we showed that reduction of DAXX expression had a stimulatory effect on autophagy (reduced p62 levels and increased LC3 puncta) in cultured PC3 cells (Fig. 8). We conclude that DAXX is a suppressor of autophagy in PCa.
FIGURE 8.
DAXX K/D increases autophagic flux in cultured PCa cells PC3. A, PC3 cells, control (WT) and DAXX K/D (K/D), transiently transfected with GFP-LC3 construct for 24 h, were cultured under normal (Basal) conditions as described under “Experimental Procedures.” The GFP-LC3 puncta were visualized using confocal microscopy (top two panels), and their numbers were assessed using IMARIS (bottom panel). At least 15 cells were analyzed. The number of LC3 puncta was increased in DAXX K/D, indicating higher basal autophagy in these cells. B, PC3 cells, control (WT) and DAXX K/D (K/D), were exposed to a time course of BafA (an inhibitor of autophagic flux) treatment (0–4 h). Cell lysates were subjected to immunoblotting with the indicated antibodies. DAXX and β-actin blots are included as controls. p62 levels were increased in WT cells upon autophagy inhibition, suggesting decreased autophagy. Similarly, whereas p62 levels were low in DAXX K/D under basal conditions (0 h), they were increased at later time points of autophagy inhibition but remained statistically lower than the WT group (bottom), pointing to increased overall autophagic flux in DAXX K/D compared with WT cells. ImageJ was used to quantify the blots, and GraphPad Prism version 5.0 was employed to construct the graph and perform statistical analyses. Error bars, S.E.
Human Malignancies Display Significant DAXX Up-regulation and an Inverse Correlation between DAXX and DAPK1/DAPK3
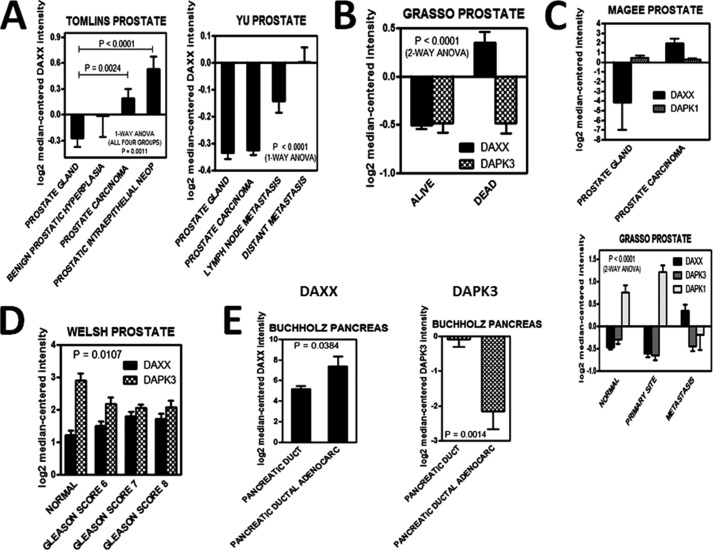
Based on our initial observation that DAXX levels are increased in PCa cell lines relative to a non-tumorigenic counterpart (Fig. 1A), we set out to assess whether expression levels of DAXX mRNA were similarly increased in primary prostate cancers. To that end, we interrogated the Oncomine database (Compendia Bioscience), using the Premium Edition package. DAXX mRNA levels were increased in metastatic PCa, whereas they were low in normal prostate glands (Fig. 9A). The opposite was true for two DAXX repression targets, the tumor suppressors DAPK1 and DAPK3 (Fig. 9C). Significantly, higher DAXX levels correlated with lower survival rates (Fig. 9B). Prostate tumors with a higher Gleason score are more aggressive and have a worse prognosis (22). Our Oncomine-derived analyses indicated that higher scores were associated with higher DAXX and lower DAPK3 levels (Fig. 9D). Several other cancers, including pancreatic cancer (Fig. 9E), displayed a similar trend, signaling an important role for DAXX in PCa development and progression/metastasis.
FIGURE 9.
Human prostatic and pancreatic malignancies display significant DAXX mRNA up-regulation and an inverse correlation between DAXX and DAPK1/3. The Oncomine Premium Edition database was interrogated in studies involving DAXX (A–E) and its repression targets, DAPK1/3 (B–E), in prostate (A–D) and pancreatic (E) cancers. The raw data were used to construct graphs using GraphPad Prism version 5 software. The data suggest that DAXX overexpression may be associated with malignant transformation (A), poor prognosis (B), and DAPK1/DAPK3 down-regulation (C). A high Gleason score correlated with an increase in DAXX and a decrease in DAPK3 levels (D). DAXX overexpression is also associated with malignant transformation in pancreatic cancer, whereas DAPK3 is down-regulated in this type of cancer (E). The analysis of variance statistical tests showed that the differences among various groups were statistically significant (p < 0.05). The title of each graph represents the name of the author that deposited the respective raw data in Oncomine (56–60). Error bars, S.E.
Discussion
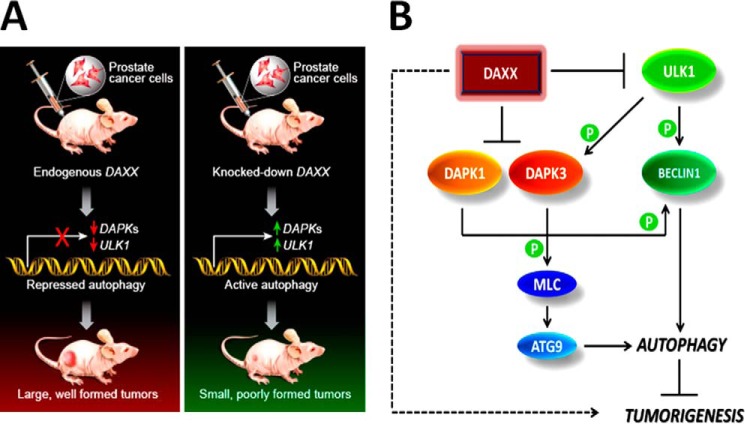
Using a subcutaneous xenograft model in nude mice, we have shown that the DAXX transcriptional repressor promotes prostate cancer development at least in part by inhibiting autophagy. DAXX has previously been associated with cell death, largely through studies in cell culture (1). In contrast, our in vivo studies with human PCa cell lines indicate that DAXX has a pro-survival function. Using several PCa cell lines, we found that DAXX promoted tumor growth and that there was a direct correlation between the level of DAXX and the rate of tumor growth. Specifically, ALVA-31 cells, with high DAXX levels (Fig. 1A), developed tumors faster than PC3 or DU145 cells, which have a lower level of DAXX. Our in vivo results with PCa cells are consistent with the recently reported pro-survival function of DAXX in ovarian cancer (23) and several reports of elevated DAXX expression in human cancer tissues compared with normal ones (8–10). Specifically, in ovarian cancer (23), DAXX overexpression increased cell proliferation and migration. Ovarian cancer cells that overexpressed DAXX displayed increased tumorigenesis in nude mice, whereas tumorigenesis was inhibited or reduced when DAXX was depleted (23), analogous to our findings. Yet, our data reveal autophagy suppression by DAXX as a mechanism for tumorigenesis promotion and thus provide, for the first time, mechanistic insights into the tumorigenic actions of DAXX. Our OncomineTM data analyses (Fig. 9) suggest that DAXX overexpression is associated with malignant transformation in several cancers, including prostate and pancreatic cancers. In contrast, two DAXX repression targets, the DAPK1/3 tumor suppressors and autophagy regulators, displayed reduced expression in various cancers, including PCa, suggesting their negative regulation by DAXX in cancer contexts. Our studies support the conclusion that DAXX acts to suppress autophagy in PCa tumorigenesis (in vivo) and in vitro settings. Protein and RNA analyses are consistent with DAXX suppression of key initiators of the autophagy machinery, including ULK1, resulting in tumorigenesis (illustrated in Fig. 10A). Both ULK1 (24) and DAPK1 (4) phosphorylate Beclin1 at distinct sites to increase its pro-autophagy function, an effect that is countered by increased DAXX protein (summarized in Fig. 10B).
FIGURE 10.
Model for DAXX-mediated induction of tumorigenesis. A, synopsis of experimental results outlines the effect of DAXX on subcutaneous tumor growth, emphasizing key autophagy effectors in the process. Green arrow, increased gene expression; red arrow, decreased gene expression. B, a mechanistic model of DAXX function in cancer. The dashed arrow on the left summarizes the overall effect of DAXX on tumorigenesis, whereas mechanistic details are depicted with solid lines. DAXX suppresses expression of DAPKs (2) and ULK1 (this investigation). ULK1 is the most upstream element in the autophagy pathway, without which phosphorylation-dependent activation of DAPK3 and Beclin1 and, ultimately, autophagy cannot proceed. DAXX may affect several components of the autophagy machinery simultaneously or sequentially, with the end result being increased tumorigenesis.
The function of autophagy in cancer is multilateral; depending on the system, autophagy can suppress or enhance tumorigenesis. For instance, autophagy reportedly suppresses angiogenesis, which is required for tumor growth (25) and invasion (26). Indeed, in our studies, smaller tumors arising from DAXX-depleted cells manifested increased autophagy and decreased vasculature, and vice versa. Immunohistochemical assessment of proliferation (Ki67; data not shown), angiogenesis markers (CD31), and autophagy markers (LC3 and p62) suggested that DAXX K/D tumors had slow division rates, were less vascularized, and displayed increased autophagy. Tumorigenicity of DAXX knockdown cells is, therefore, impaired, at least in part, due to increased autophagy, decreased angiogenesis, and reduced division rates.
Autophagy has been linked to tumor suppression in liver, breast, lung, and kidney cancers (19, 27–31). Similarly, we showed that prostate tumor growth is enhanced when autophagy is negatively regulated. Conversely, in other cases, autophagy has been shown to be required for tumor progression. Through the use of a genetically engineered mouse model of pancreatic ductal adenocarcinoma, autophagy was positively linked to advancement to malignancy (32). Additionally, a key role for autophagy in providing metabolites for mitochondrial function in V600-E BRAF mouse lung cancer was recently reported (33) (i.e. autophagy appears to be particularly important for tumors driven by Ras/Raf pathway activation).
Antagonistic functions of autophagy in tumorigenesis need not be irreconcilable. Specifically, autophagy can act as a tumor suppressor mechanism during the early stages of tumorigenesis by preventing genomic instability (34) and eliminating the p62 autophagy receptor (19), whereas it can protect tumor cells from oxidative damage during advanced stages of cancer (32). Indeed, ALVA-31 PCa cells used in our study were derived from an early stage tumor, established from a primary tumor biopsy specimen obtained during prostatectomy in a patient with well differentiated, stage B2 adenocarcinoma of the prostate (14). Consistent with our observation that smaller tumors displayed increased autophagy, autophagy appears to suppress early stage tumorigenesis, possibly through degradation of p62. In contrast to ALVA-31 cells, PC3 cells were isolated from a bone marrow metastasis (35), but the fact that they behaved similarly to ALVA-31 cells in their tumor growth kinetics and autophagy patterns suggests that the role of autophagy may predominantly be cancer type-specific rather than cancer stage-specific. In cancers such as lung (33) and pancreatic (32), autophagy may promote tumor progression, whereas in the prostate it may suppress it. In fact, metastatic PC3 PCa cells display relatively high basal levels of p62 RNA and protein (36), which is inversely correlated to autophagy. Metastatic PCa, therefore, does not appear to rely on autophagy for survival.
The tissue-specific mode of action of autophagy may stem from distinct molecular driving forces of tumorigenesis. Molecular drivers behind lung, pancreatic, colon, thyroid, and bladder cancers include the Ras oncoprotein family (37), where gain-of-function Ras mutations occur at high frequency. Ras-driven lung tumors in mouse models are dependent on autophagy for lipid catabolism and mitochondrial function (38). However, it is important to note that Ras-induced autophagy does not always promote tumor cell survival. For instance, tetracycline-inducible oncogenic H-Ras expression in human ovarian epithelial cells leads to autophagic cell death of tumor cells rather than survival (39). Similarly, Ras triggers autophagic cell death in neuroblastoma, causing tumor regression (40). Importantly, drug-induced autophagy causes K-Ras degradation and cell death in multiple tumors (41). Finally, p53 status influences the opposing effects of autophagy in pancreatic cancer, with p53 deficiency linked to a tumor-suppressive function of autophagy in K-Ras-driven pancreatic tumor development (42). In agreement with this role of p53 in pancreatic cancer, prostate cancers may also be affected by p53 status. Consistent with the pancreatic cancer findings, suppression of autophagy by DAXX resulted in accelerated tumor growth in ALVA-31 and PC3 PCa cells, which are both p53-deficient (43, 44), and this effect was reversed when DAXX levels were reduced. Notably, human PCa (45), including PC3 cells (46), does not generally harbor Ras mutations. Frequently mutated oncogenes in PCa include MYC (40% of primary tumors and 90% of metastases show increases in MYC copy number (47)), which is now generally accepted as an oncogenic factor in human PCa (48, 49). Importantly, MYC cooperates with the AKT kinase, part of the PI3K pathway (49). Generation of bigenic mice, where both activated human AKT1 and human MYC were expressed in the prostate, resulted in progression of PCa to advanced stages (49). Because AKT inhibits autophagy and enhances prostate tumorigenesis (50), these results agree with our finding that, in PCa, autophagy inhibition promotes tumor growth.
PCa initiation and progression does not depend only on oncogenes, such as MYC and AKT. Compelling data show that the PTEN tumor suppressor, which is deleted in 40% of localized PCa and mutated in 60% of metastases (51), has a vital role in human PCa (52). PTEN loss may account for increased AKT activation and decreased autophagy in PCa, because introducing PTEN in cancer cells that lack PTEN function negatively regulates AKT (50). Two of the three androgen-insensitive PCa lines that we utilized, namely, ALVA-31 and PC3, are PTEN-negative (at both the mRNA and protein levels) and phospho-AKT (activated AKT)-positive (53). Because the loss of PTEN (54) and increase in AKT (50) cause autophagy inhibition, ALVA-31- and PC3-originating tumors would not be expected to depend on autophagy for survival, because autophagy is inhibited in these tumors. Although we used cultured PCa cell lines and xenografts as model systems, we note that MYC and DAXX mRNA levels are similarly increased in human prostate cancers and are associated with poor prognosis (8, 47) (Fig. 9), whereas PTEN levels are decreased (51). Furthermore, therapy-induced autophagy enhances tumor cell death in a context-dependent manner (46), consistent with a tumor suppressor role for autophagy.
In conclusion, through the use of two stable DAXX knockdown PCa lines (ALVA-31 and PC3), depletion of essential autophagy genes (ULK1 and DAPKs), and the use of an autophagy-defective PCa line (DU145), we showed that DAXX exerts its tumor-promoting functions by reducing autophagy. The decrease in p62 immunostaining in DAXX K/D tumor tissue sections arising from PC3 cells compared with their wild-type counterparts (Fig. 3) suggests that DAXX knockdown indeed increases autophagy. The decrease in p62 was not due to decreased p62 transcription (not shown). Furthermore, a decrease in p62 was not observed using DU145 cells (ATG5-deficient) when DAXX was knocked down (Fig. 3), suggesting that DAXX cannot affect autophagy/tumorigenesis under autophagy-defective conditions. Corroboration of in vivo studies with cell culture analysis (Fig. 8), indicated that autophagic flux was increased in DAXX K/D cells.
DAXX suppresses expression of DAPK1/3 (2) and ULK1. ULK1 is the most upstream element in the autophagy pathway (Fig. 10B), without which activation of DAPK3 (5) and Beclin1 (24) and, thus, autophagy (Fig. 10B) cannot proceed. Although ULK1 has multiple targets in cell signaling and regulates other cellular processes besides autophagy (55), additional support that autophagy is indeed a mechanism by which DAXX affects tumorigenesis in PCa comes through our use of the ATG5-deficient cell line, DU145 (this study) and ChIP-seq/RNA-seq analyses (13). DAXX ChIP-seq peaks were found close to the TSSs of autophagy genes, and genes increased by DAXX K/D included those involved in autophagy. Furthermore, RNA-seq showed that DAXX was inversely correlated with the expression of positive regulators of autophagy (13).
Among many functions, DAXX is a regulator of NF-κB (2) and a histone H3.3 chaperone (3). Because DAXX is bound to the ULK1 promoter region in the vicinity of an NF-κB binding site, one could speculate that the repressive effect of DAXX on ULK1 expression is exerted directly, possibly through RelB/DAXX-mediated recruitment of DNMT1, leading to local repressive CpG methylation, as is the case for DAPK1 and DAPK3 (2) (data not shown). However, this could also involve DAXX/ATRX-mediated replacement of H3.3 at the DAPK1, DAPK3, or ULK1 promoters. Preliminary data have provided clues, but further work is required to determine the exact mechanistic details of repression. Indeed, DAXX can employ different repressive mechanisms for different genes. Overall, our current study serves as proof of concept unraveling the role of DAXX in tumorigenesis and its negative regulation of autophagy.
Acknowledgments
We are grateful to the following persons: Scott Crist (Purdue University) for providing ALVA-31, PC3, DU145, and LNCaP prostate cancer cells; Jill Meisenhelder and Amanda Chambers (Salk Institute) for help with animal care and tumor measurements; Mathias LeBlanc (Salk Institute) for carrying out tumor excisions and histologic preparation; Christopher Benner (Salk Institute) for helping to analyze the ChIP-seq data; and Deepti Wilkinson (Sanford-Burnham Medical Research Institute) for providing the GFP-LC3 construct.
This work was supported, in whole or in part, by National Institutes of Health, NCI, Grants CA14195 and CA082683 (to T. H.) and Fellowship 5T32CA009523-28 (to L. A. P.). This work was also supported by Department of Defense Prostate Cancer Research Program Fellowship DoD-W81XWH-08-1-0209 (to L. A. P.) and a grant from the Cancer Research UK Manchester Institute (to J. B.).
- PCa
- prostate cancer
- K/D
- knockdown
- qPCR
- quantitative PCR
- ChIP-seq
- ChIP sequencing
- RNA-seq
- RNA sequencing
- IHC
- immunohistochemical
- TSS
- transcription start site
- PTEN
- phosphatase and tensin homolog
- DAPK
- death-associated protein kinase.
References
- 1. Yang X., Khosravi-Far R., Chang H. Y., Baltimore D. (1997) Daxx, a novel Fas-binding protein that activates JNK and apoptosis. Cell 89, 1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Puto L. A., Reed J. C. (2008) Daxx represses RelB target promoters via DNA methyltransferase recruitment and DNA hypermethylation. Genes Dev. 22, 998–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lewis P. W., Elsaesser S. J., Noh K. M., Stadler S. C., Allis C. D. (2010) Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl. Acad. Sci. 107, 14075–14080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zalckvar E., Berissi H., Eisenstein M., Kimchi A. (2009) Phosphorylation of Beclin 1 by DAP-kinase promotes autophagy by weakening its interactions with Bcl-2 and Bcl-XL. Autophagy 5, 720–722 [DOI] [PubMed] [Google Scholar]
- 5. Tang H. W., Wang Y. B., Wang S. L., Wu M. H., Lin S. Y., Chen G. C. (2011) Atg1-mediated myosin II activation regulates autophagosome formation during starvation-induced autophagy. EMBO J. 30, 636–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Michaelson J. S., Bader D., Kuo F., Kozak C., Leder P. (1999) Loss of Daxx, a promiscuously interacting protein, results in extensive apoptosis in early mouse development. Genes Dev. 13, 1918–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kwan P. S., Lau C. C., Chiu Y. T., Man C., Liu J., Tang K. D., Wong Y. C., Ling M. T. (2013) Daxx regulates mitotic progression and prostate cancer predisposition. Carcinogenesis 34, 750–759 [DOI] [PubMed] [Google Scholar]
- 8. Tsourlakis M. C., Schoop M., Plass C., Huland H., Graefen M., Steuber T., Schlomm T., Simon R., Sauter G., Sirma H., Minner S. (2013) Overexpression of the chromatin remodeler death-domain-associated protein in prostate cancer is an independent predictor of early prostate-specific antigen recurrence. Hum. Pathol. 44, 1789–1796 [DOI] [PubMed] [Google Scholar]
- 9. Zizzi A., Montironi M. A., Mazzucchelli R., Scarpelli M., Lopez-Beltran A., Cheng L., Paone N., Castellini P., Montironi R. (2013) Immunohistochemical analysis of chromatin remodeler DAXX in high grade urothelial carcinoma. Diagn. Pathol. 8, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Horvilleur E., Sbarrato T., Hill K., Spriggs R. V., Screen M., Goodrem P. J., Sawicka K., Chaplin L. C., Touriol C., Packham G., Potter K. N., Dirnhofer S., Tzankov A., Dyer M. J., Bushell M., MacFarlane M., Willis A. E. (2014) A role for eukaryotic initiation factor 4B overexpression in the pathogenesis of diffuse large B-cell lymphoma. Leukemia 28, 1092–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jiao Y., Shi C., Edil B. H., de Wilde R. F., Klimstra D. S., Maitra A., Schulick R. D., Tang L. H., Wolfgang C. L., Choti M. A., Velculescu V. E., Diaz L. A., Jr., Vogelstein B., Kinzler K. W., Hruban R. H., Papadopoulos N. (2011) DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science 331, 1199–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Heaphy C. M., Subhawong A. P., Hong S. M., Goggins M. G., Montgomery E. A., Gabrielson E., Netto G. J., Epstein J. I., Lotan T. L., Westra W. H., Shih I. E. M., Iacobuzio-Donahue C. A., Maitra A., Li Q. K., Eberhart C. G., Taube J. M., Rakheja D., Kurman R. J., Wu T. C., Roden R. B., Argani P., De Marzo A. M., Terracciano L., Torbenson M., Meeker A. K. (2011) Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am. J. Pathol. 179, 1608–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Puto L. A., Benner C., Hunter T. (2015) The DAXX co-repressor is directly recruited to active regulatory elements genome-wide to regulate autophagy programs in a model of human prostate cancer. Oncoscience 2, 362–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Loop S. M., Rozanski T. A., Ostenson R. C. (1993) Human primary prostate tumor cell line, ALVA-31: a new model for studying the hormonal regulation of prostate tumor cell growth. Prostate 22, 93–108 [DOI] [PubMed] [Google Scholar]
- 15. Tsuji T., Du W., Nishioka T., Chen L., Yamamoto D., Chen C. Y. (2010) Phellinus linteus extract sensitizes advanced prostate cancer cells to apoptosis in athymic nude mice. PLoS One 5, e9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ouyang D. Y., Xu L. H., He X. H., Zhang Y. T., Zeng L. H., Cai J. Y., Ren S. (2013) Autophagy is differentially induced in prostate cancer LNCaP, DU145 and PC-3 cells via distinct splicing profiles of ATG5. Autophagy 9, 20–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mizushima N., Yoshimori T. (2007) How to interpret LC3 immunoblotting. Autophagy 3, 542–545 [DOI] [PubMed] [Google Scholar]
- 18. Puissant A., Fenouille N., Auberger P. (2012) When autophagy meets cancer through p62/SQSTM1. Am. J. Cancer Res. 2, 397–413 [PMC free article] [PubMed] [Google Scholar]
- 19. Mathew R., Karp C. M., Beaudoin B., Vuong N., Chen G., Chen H. Y., Bray K., Reddy A., Bhanot G., Gelinas C., Dipaola R. S., Karantza-Wadsworth V., White E. (2009) Autophagy suppresses tumorigenesis through elimination of p62. Cell 137, 1062–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mizushima N. (2010) The role of the Atg1/ULK1 complex in autophagy regulation. Curr. Opin. Cell Biol. 22, 132–139 [DOI] [PubMed] [Google Scholar]
- 21. Lee E. J., Tournier C. (2011) The requirement of uncoordinated 51-like kinase 1 (ULK1) and ULK2 in the regulation of autophagy. Autophagy 7, 689–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lowe B. A., Listrom M. B. (1988) Incidental carcinoma of the prostate: an analysis of the predictors of progression. J. Urol. 140, 1340–1344 [DOI] [PubMed] [Google Scholar]
- 23. Pan W. W., Zhou J. J., Liu X. M., Xu Y., Guo L. J., Yu C., Shi Q. H., Fan H. Y. (2013) Death domain-associated protein DAXX promotes ovarian cancer development and chemoresistance. J. Biol. Chem. 288, 13620–13630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Russell R. C., Tian Y., Yuan H., Park H. W., Chang Y. Y., Kim J., Kim H., Neufeld T. P., Dillin A., Guan K. L. (2013) ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 15, 741–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ramakrishnan S., Nguyen T. M., Subramanian I. V., Kelekar A. (2007) Autophagy and angiogenesis inhibition. Autophagy 3, 512–515 [DOI] [PubMed] [Google Scholar]
- 26. Sakurai T., Okumura H., Matsumoto M., Uchikado Y., Setoyama T., Omoto I., Owaki T., Maemura K., Ishigami S., Natsugoe S. (2013) The expression of LC-3 is related to tumor suppression through angiogenesis in esophageal cancer. Med. Oncol. 30, 701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liang X. H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. (1999) Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402, 672–676 [DOI] [PubMed] [Google Scholar]
- 28. Takamura A., Komatsu M., Hara T., Sakamoto A., Kishi C., Waguri S., Eishi Y., Hino O., Tanaka K., Mizushima N. (2011) Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 25, 795–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang R. C., Wei Y., An Z., Zou Z., Xiao G., Bhagat G., White M., Reichelt J., Levine B. (2012) Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science 338, 956–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu Z., Chen P., Gao H., Gu Y., Yang J., Peng H., Xu X., Wang H., Yang M., Liu X., Fan L., Chen S., Zhou J., Sun Y., Ruan K., Cheng S., Komatsu M., White E., Li L., Ji H., Finley D., Hu R. (2014) Ubiquitylation of autophagy receptor optineurin by HACE1 activates selective autophagy for tumor suppression. Cancer Cell 26, 106–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu X. D., Yao J., Tripathi D. N., Ding Z., Xu Y., Sun M., Zhang J., Bai S., German P., Hoang A., Zhou L., Jonasch D., Zhang X., Conti C. J., Efstathiou E., Tannir N. M., Eissa N. T., Mills G. B., Walker C. L., Jonasch E. (2014) Autophagy mediates HIF2α degradation and suppresses renal tumorigenesis. Oncogene 10.1038/onc.2014.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang S., Wang X., Contino G., Liesa M., Sahin E., Ying H., Bause A., Li Y., Stommel J. M., Dell'antonio G., Mautner J., Tonon G., Haigis M., Shirihai O. S., Doglioni C., Bardeesy N., Kimmelman A. C. (2011) Pancreatic cancers require autophagy for tumor growth. Genes Dev. 25, 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Strohecker A. M., Guo J. Y., Karsli-Uzunbas G., Price S. M., Chen G. J., Mathew R., McMahon M., White E. (2013) Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer Discov. 3, 1272–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Karantza-Wadsworth V., Patel S., Kravchuk O., Chen G., Mathew R., Jin S., White E. (2007) Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 21, 1621–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kaighn M. E., Narayan K. S., Ohnuki Y., Lechner J. F., Jones L. W. (1979) Establishment and characterization of a human prostatic carcinoma cell line (PC-3). Invest. Urol. 17, 16–23 [PubMed] [Google Scholar]
- 36. Chang M. A., Morgado M., Warren C. R., Hinton C. V., Farach-Carson M. C., Delk N. A. (2014) p62/SQSTM1 is required for cell survival of apoptosis-resistant bone metastatic prostate cancer cell lines. Prostate 74, 149–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fernández-Medarde A., Santos E. (2011) Ras in cancer and developmental diseases. Genes Cancer 2, 344–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guo J. Y., Karsli-Uzunbas G., Mathew R., Aisner S. C., Kamphorst J. J., Strohecker A. M., Chen G., Price S., Lu W., Teng X., Snyder E., Santanam U., Dipaola R. S., Jacks T., Rabinowitz J. D., White E. (2013) Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 27, 1447–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Elgendy M., Sheridan C., Brumatti G., Martin S. J. (2011) Oncogenic Ras-induced expression of Noxa and Beclin-1 promotes autophagic cell death and limits clonogenic survival. Mol. Cell 42, 23–35 [DOI] [PubMed] [Google Scholar]
- 40. Kitanaka C., Kato K., Ijiri R., Sakurada K., Tomiyama A., Noguchi K., Nagashima Y., Nakagawara A., Momoi T., Toyoda Y., Kigasawa H., Nishi T., Shirouzu M., Yokoyama S., Tanaka Y., Kuchino Y. (2002) Increased Ras expression and caspase-independent neuroblastoma cell death: possible mechanism of spontaneous neuroblastoma regression. J. Natl. Cancer Inst. 94, 358–368 [DOI] [PubMed] [Google Scholar]
- 41. Kohli L., Kaza N., Carroll S. L., Roth K. A. (2013) Protector turns predator: autophagic death via selective degradation of KRAS. Autophagy 9, 1438–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rosenfeldt M. T., O'Prey J., Morton J. P., Nixon C., MacKay G., Mrowinska A., Au A., Rai T. S., Zheng L., Ridgway R., Adams P. D., Anderson K. I., Gottlieb E., Sansom O. J., Ryan K. M. (2013) p53 status determines the role of autophagy in pancreatic tumour development. Nature 504, 296–300 [DOI] [PubMed] [Google Scholar]
- 43. van Bokhoven A., Varella-Garcia M., Korch C., Hessels D., Miller G. J. (2001) Widely used prostate carcinoma cell lines share common origins. Prostate 47, 36–51 [DOI] [PubMed] [Google Scholar]
- 44. Hostanska K., Nisslein T., Freudenstein J., Reichling J., Saller R. (2005) Apoptosis of human prostate androgen-dependent and -independent carcinoma cells induced by an isopropanolic extract of black cohosh involves degradation of cytokeratin (CK) 18. Anticancer Res. 25, 139–147 [PubMed] [Google Scholar]
- 45. Moul J. W., Friedrichs P. A., Lance R. S., Theune S. M., Chang E. H. (1992) Infrequent RAS oncogene mutations in human prostate cancer. Prostate 20, 327–338 [DOI] [PubMed] [Google Scholar]
- 46. Guo J., Zhu T., Chen L., Nishioka T., Tsuji T., Xiao Z. X., Chen C. Y. (2010) Differential sensitization of different prostate cancer cells to apoptosis. Genes Cancer 1, 836–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jenkins R. B., Qian J., Lieber M. M., Bostwick D. G. (1997) Detection of c-myc oncogene amplification and chromosomal anomalies in metastatic prostatic carcinoma by fluorescence in situ hybridization. Cancer Res. 57, 524–531 [PubMed] [Google Scholar]
- 48. Dong J. T. (2006) Prevalent mutations in prostate cancer. J. Cell. Biochem. 97, 433–447 [DOI] [PubMed] [Google Scholar]
- 49. Clegg N. J., Couto S. S., Wongvipat J., Hieronymus H., Carver B. S., Taylor B. S., Ellwood-Yen K., Gerald W. L., Sander C., Sawyers C. L. (2011) MYC cooperates with AKT in prostate tumorigenesis and alters sensitivity to mTOR inhibitors. PLoS One 6, e17449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Degtyarev M., De Mazière A., Orr C., Lin J., Lee B. B., Tien J. Y., Prior W. W., van Dijk S., Wu H., Gray D. C., Davis D. P., Stern H. M., Murray L. J., Hoeflich K. P., Klumperman J., Friedman L. S., Lin K. (2008) Akt inhibition promotes autophagy and sensitizes PTEN-null tumors to lysosomotropic agents. J. Cell Biol. 183, 101–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Phin S., Moore M. W., Cotter P. D. (2013) Genomic Rearrangements of PTEN in Prostate Cancer. Front. Oncol. 3, 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hollander M. C., Blumenthal G. M., Dennis P. A. (2011) PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat. Rev. Cancer 11, 289–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Festuccia C., Muzi P., Millimaggi D., Biordi L., Gravina G. L., Speca S., Angelucci A., Dolo V., Vicentini C., Bologna M. (2005) Molecular aspects of gefitinib antiproliferative and pro-apoptotic effects in PTEN-positive and PTEN-negative prostate cancer cell lines. Endocr. Relat. Cancer 12, 983–998 [DOI] [PubMed] [Google Scholar]
- 54. Ueno T., Sato W., Horie Y., Komatsu M., Tanida I., Yoshida M., Ohshima S., Mak T. W., Watanabe S., Kominami E. (2008) Loss of Pten, a tumor suppressor, causes the strong inhibition of autophagy without affecting LC3 lipidation. Autophagy 4, 692–700 [DOI] [PubMed] [Google Scholar]
- 55. Roach P. J. (2011) AMPK → ULK1 → autophagy. Mol. Cell. Biol. 31, 3082–3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Magee J. A., Araki T., Patil S., Ehrig T., True L., Humphrey P. A., Catalona W. J., Watson M. A., Milbrandt J. (2001) Expression profiling reveals hepsin overexpression in prostate cancer. Cancer Res. 61, 5692–5696 [PubMed] [Google Scholar]
- 57. Tomlins S. A., Mehra R., Rhodes D. R., Cao X., Wang L., Dhanasekaran S. M., Kalyana-Sundaram S., Wei J. T., Rubin M. A., Pienta K. J., Shah R. B., Chinnaiyan A. M. (2007) Integrative molecular concept modeling of prostate cancer progression. Nat. Genet. 39, 41–51 [DOI] [PubMed] [Google Scholar]
- 58. Grasso C. S., Wu Y. M., Robinson D. R., Cao X., Dhanasekaran S. M., Khan A. P., Quist M. J., Jing X., Lonigro R. J., Brenner J. C., Asangani I. A., Ateeq B., Chun S. Y., Siddiqui J., Sam L., Anstett M., Mehra R., Prensner J. R., Palanisamy N., Ryslik G. A., Vandin F., Raphael B. J., Kunju L. P., Rhodes D. R., Pienta K. J., Chinnaiyan A. M., Tomlins S. A. (2012) The mutational landscape of lethal castration-resistant prostate cancer. Nature 487, 239–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Welsh J. B., Sapinoso L. M., Su A. I., Kern S. G., Wang-Rodriguez J., Moskaluk C. A., Frierson H. F., Jr., Hampton G. M. (2001) Analysis of gene expression identifies candidate markers and pharmacological targets in prostate cancer. Cancer Res. 61, 5974–5978 [PubMed] [Google Scholar]
- 60. Wang X. D., Reeves K., Luo F. R., Xu L. A., Lee F., Clark E., Huang F. (2007) Identification of candidate predictive and surrogate molecular markers for dasatinib in prostate cancer: rationale for patient selection and efficacy monitoring. Genome Biol. 8, R255. [DOI] [PMC free article] [PubMed] [Google Scholar]