Background: Spinal muscular atrophy (SMA), a neurodegenerative disease, is caused by low levels of expression of the survival motor neuron (SMN) protein.
Results: Gemin5 binds to and regulates translation of the mature SMN mRNA.
Conclusion: Gemin5 functions in the regulation of the SMN mRNA.
Significance: Investigation of the regulation of SMN expression is critical for understanding SMA and developing therapeutics.
Keywords: mRNA, neurodegenerative disease, post-transcriptional regulation, RNA-binding protein, translation, Gemin5, SMA, SMN, spinal muscular atrophy, survival motor neuron
Abstract
Reduced expression of SMN causes spinal muscular atrophy, a severe neurodegenerative disease. Despite the importance of maintaining SMN levels, relatively little is known about the mechanisms by which SMN levels are regulated. We show here that Gemin5, the snRNA-binding protein of the SMN complex, binds directly to the SMN mRNA and regulates SMN expression. Gemin5 binds with high specificity, both in vitro and in vivo, to sequence and structural elements in the SMN mRNA 3′-untranslated region that are reminiscent of the snRNP code to which Gemin5 binds on snRNAs. Reduction of Gemin5 redistributes the SMN mRNA from heavy polysomes to lighter polysomes and monosomes, suggesting that Gemin5 functions as an activator of SMN translation. SMN protein is not stoichiometrically present on the SMN mRNA with Gemin5, but the mRNA-binding activity of Gemin5 is dependent on SMN levels, providing a feedback mechanism for SMN to regulate its own expression via Gemin5. This work both reveals a new autoregulatory pathway governing SMN expression, and identifies a new mechanism through which SMN can modulate specific mRNA expression via Gemin5.
Introduction
The survival motor neuron (SMN)2 complex functions in the assembly of small nuclear ribonucleoproteins (snRNPs), the RNP components of the spliceosome that perform pre-mRNA splicing (1–7). The SMN complex itself is comprised of the SMN protein, seven proteins termed Gemins 2–8, and unrip. In the cytoplasm, the SMN complex binds to seven Sm proteins (Sm B/B′, D1, D2, D3, E, F, and G) and assembles them into a heptameric ring on each of seven small-nuclear RNAs (U1, U2, U4, U5, U11, U12, and U4atac snRNAs) (8–10). The snRNAs are delivered to the SMN complex by Gemin5 (11, 12). Gemin5 binds directly to snRNAs by recognizing a set of sequence and structural features found in the snRNAs (11, 13). Gemin5 specifically binds to an AU5–6 sequence within the highly conserved Sm site flanked by a short stem loop (the snRNP code) (11, 14). In this way, Gemin5 recognizes and differentiates the snRNAs from all other cellular RNAs, and targets them for assembly into spliceosomal snRNPs.
Several lines of evidence suggest that, in addition to binding snRNAs, Gemin5 may also function directly with mRNAs (15–17). For example, Gemin5 was recently identified in a screen for mRNA-binding proteins (15). Additionally, Gemin5 has been shown to interact with the eukaryotic translation initiation machinery (16–18). In the best-characterized system to date, Gemin5 has been shown to bind to internal ribosome entry sites (IRESs) on foot-and-mouth disease (FMDV) and hepatitis C virus (HCV) mRNAs (17). In these systems, Gemin5 modulates IRES-dependent translation of viral mRNAs (17).
Reduced expression of SMN resulting from mutation of the SMN1 gene causes the neurodegenerative disease, spinal muscular atrophy (SMA) (19). Mutation and deletion of the SMN gene results in low levels of SMN expression, which leads to reduced snRNP assembly (7, 20, 21). It is unclear how reduced snRNP assembly capacity leads to a motor neuron-specific disease, as snRNPs are required for splicing in all cell types. Most therapeutics for SMA have focused on up-regulating SMN expression or correction of mis-splicing that occurs with an SMN2 gene copy, also in an effort to increase levels (22). However, relatively little is known about the 3′ processing, localization, stability and translation of the SMN mRNA. Understanding the regulation of the SMN mRNA could not only provide clues as to the etiology of the disease, but also lead to further targets for SMA therapeutics.
We recently identified Gemin5 in a proteomic screen for proteins that specifically bind to the SMN 3′-UTR (23). Here, we show that Gemin5 associates with the SMN mRNA in vitro as well as in both HeLa and motor-neuron-derived MN1 cells. Gemin5 directly binds to the mature SMN 3′-UTR immediately upstream of the poly(A) tail. The SMN complex is not stoichiometrically present with Gemin5 on the SMN mRNA, and Gemin5 is the sole component of the SMN complex that binds directly to the mRNA. Gemin5 binds to a stem-loop and U-rich single-stranded sequence of the SMN mRNA 3′-UTR that is remarkably similar to its binding site on snRNAs. Gemin5 binding to the SMN 3′-UTR activates expression of SMN protein by increasing translation of the SMN mRNA. Furthermore, the mRNA binding activity of Gemin5 is directly dependent on SMN levels, providing a feedback mechanism whereby SMN regulates its own expression through Gemin5. This work identifies both a new function for the SMN complex in the direct regulation of mRNAs via Gemin5, and a new mechanism regulating the expression of the SMN protein itself. These findings have broad implications for better understanding the pathology of SMN reduction and optimizing treatments for SMA.
Experimental Procedures
Modified PAR-CLIP
Procedure was as described in Hafner, 2010, but modified to allow for PCR detection of transcripts (24). HeLa or mouse MN1 cells were grown and treated with 100 μm 4-thiouridine (Sigma) for 18 h. After treatment, the cells were irradiated with 365 nm UV and harvested. Cells were lysed in 1× RSB-100 (10 mm Tris, pH 7.4, 100 mm NaCl, 2.5 mm MgCl2) + 1% Empigen and immunoprecipitated with antibodies immobilized on Dynabeads protein G (Invitrogen). After washing in 1× RSB-100 + 1% Empigen buffer, immunoprecipitates were digested with proteinase K (1.2 mg/ml), and RNA was extracted with TRIzol (Invitrogen) as per manufacturer's specifications. cDNA was made with oligo dT primers (Invitrogen) using SMART MMLV reverse transcriptase (Clontech) following manufacturer's protocol. PCR was performed with the SMN or GAPDH primers using Titanium Taq (Clontech). Primer sequences were SMN Forward ACAGATCTGGAATGTGAAGCGTTA, SMN Reverse AAGAGTTACCCATTCCACTTCCTTT, GAPDH Forward GAAGGTGAAGGTCGGAGTC, GAPDH Reverse GAAGATGGTGATGGGATTTC. The PCR conditions were 95 °C 30 s, 60 °C C 1 min, 72 °C 2 min for 40 cycles (SMN) or 20 cycles (GAPDH) on a Bio-Rad T100 thermal cycler. PCR samples were then visualized on agarose gels stained with ethidium bromide. Gels were visualized on a Bio-Rad GelDoc XR+ System with ImageQuant Software.
Biotinylated RNA Pulldowns
Streptavidin pulldowns were performed as described in Workman, 2014 (23). Deletion of the 3′ end of the SMN 3′-UTR fragment was performed by PCR and inserted into pUC19 with T7 promoter sequences for in vitro RNA transcription. A 60 nucleotide nonspecific control RNA was transcribed from the T7 promoter of a pSP72 plasmid linearized with AccI.
Direct RNA Binding Assays
SMN 3′-UTR constructs were cloned into pUC19 along with T7 promoter sequences. Deletions of helical regions were constructed by PCR and site-directed mutagenesis was performed to create the point mutations. RNAs were in vitro transcribed and radiolabeled. The control RNA was transcribed from pSP72 as stated above for biotinylation. Gemin5, Gemin3, SMN, or control antibodies were immobilized on protein G-Sepharose beads (Invitrogen) and used to immunopurify the specific proteins from HeLa cell extract. The beads were washed extensively in 1× RSB-100 (10 mm Tris, pH 7.4, 100 mm NaCl, 2.5 mm MgCl2) containing 1% Empigen BB. Purification of each protein was confirmed by Western blotting and silver staining with the SilverQuest kit (Invitrogen) as per manufacturer's instructions. The immunopurified proteins were then incubated with in vitro transcribed and radiolabeled (32P-UTP) RNA. 100,000 CPM RNA was incubated with the purified proteins for 30 min at room temperature in the presence of binding buffer (1× RSB-100 with 0.01% Nonidet P-40). The beads were washed extensively with the same buffer before extracting the RNA with TRIzol (Invitrogen) according to the manufacturer's specifications. The RNA was then resuspended in formamide load buffer and loaded onto 6% native polyacrylamide gels. The gels were then exposed to autoradiography film.
Antibodies
Antibodies used in Western blots were mouse monoclonal anti-Gemin5 (Millipore), mouse monoclonal anti-beta tubulin (Sigma), mouse monoclonal anti-SMN (BD Biosciences), mouse monoclonal anti-Gemin4 (Millipore), mouse monoclonal anti-Gemin3 (Abcam). The Li-cor secondary antibody used was goat anti-mouse IRDye 800 (Rockland). The following antibodies were used in immunoprecipitations: mouse monoclonal anti-Gemin5 (Millipore), mouse monoclonal anti-SMN (Millipore), mouse monoclonal anti-Gemin3 (BD Biosciences) or mouse monoclonal isotype control (BioLegend).
Dual Luciferase Assay
SMN 3′-UTR sequences were cloned into the pmirGLO (Promega) vector 3′ of the luciferase gene in PmeI to XbaI sites. This assay was performed essentially as described in Workman et al. (23). Briefly, luciferase plasmids were transfected into HeLa cell lines stably or transiently expressing Gemin5 or control siRNA using Lipofectamine2000 (Life Technologies) or TransIT-LT1 (Mirus) according to the manufacturer's protocol. Cells were incubated for 24 h before harvesting and assaying for luciferase activity using the Dual Luciferase Reporter Assay (Promega) according to the manufacturer's protocol. The reporter assay data were collected using the Tecan Infinite F200.
Cell Lines
pLKO.1 plasmids used to express non-targeting GFP control or Gemin5-directed shRNA driven by the U6 promoter were obtained from Open Biosystems. Lentivirus packaging plasmids were obtained from Addgene. Lentivirus particles containing the pLKO.1 plasmids were produced and used for infection of target cells as per The RNA Consortium Cloning Vector Protocol available online (Broad Institute). Stable cell lines were created by infecting HeLa cells with lentivirus containing the shRNA constructs and selecting with puromycin. Transient RNAi was also performed on HeLa cells. Briefly, a non-targeting siRNA control sequence (Ambion Ctrl #2), Gemin5 SMARTpool (Dharmacon) or SMN SMARTpool (Dharmacon) were transfected into HeLa cells using Dharmafect 1 (Dharmacon) as per manufacturer's protocol. Cells were incubated for 48 h with siRNA and assayed for protein or RNA expression.
Real-time PCR
Cells expressing Gemin5 siRNA or GFP (control) cells were harvested with TRIzol (Invitrogen), and the RNA was extracted. cDNA was made from the RNA using SMART MMLV Reverse Transcriptase (Clontech) according to the manufacturer's protocol using an oligo dT (Invitrogen) or random hexamer primer (GE Healthcare). Real-time PCR reactions were set up using SYBR Green PCR Master Mix (Applied Biosystems) according to the manufacturer's protocol. PCR conditions were 95°C for 10 min followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The real-time PCR was performed on the Applied Biosystems 7300 and quantitated using the comparative Ct method. Samples were normalized to 18S rRNA. Primers were 18S Forward CCTTTAACGAGGATCCATTGGA, 18S Reverse CGAGCTTTTTAACTGCAGCAACT, SMN Forward ACGGTTGCATTTACCCAGCTA and SMN Reverse CAGATTTTGCTCCTCTCTATTTCCA.
RNA Decay
HeLa cells were grown and after 24 h, 200 ng/μl actinomycin D was administered to the cells. Cells were harvested at 0, 1, 2, 4, 6, and 8 h after actinomycin D was added. RNA was extracted from the cells with TRIzol (Invitrogen) as above and subjected to real-time PCR as above. The same primers as above were used to detect SMN and 18S. Data were plotted as the log of transcript remaining and the half-life was determined by the equation of the line.
Polysome Fractionation and RNA Quantification
Performed essentially as described in Mukherjee et al. (25). 0.2 ng of β-globin RNA transcript was used to spike each RNA fraction and normalize fractions for RNA content. RNA was isolated and cDNA was made as described above. The same primers as above were used to detect SMN. Control primers used to normalize fractions were β-globin Forward ACATTTGCTTCTGACACAACT and β-globin Reverse ACAGGGCAGTAACGGCAGA.
Statistics
All data are presented as the means of the standard error from at least three independent experiments. Statistical significance between two groups was determined by using the two-tailed Student's t test, and resulting p values less than 0.05 were considered significant.
Results
Gemin5 Is Directly Bound to SMN mRNA in Vivo
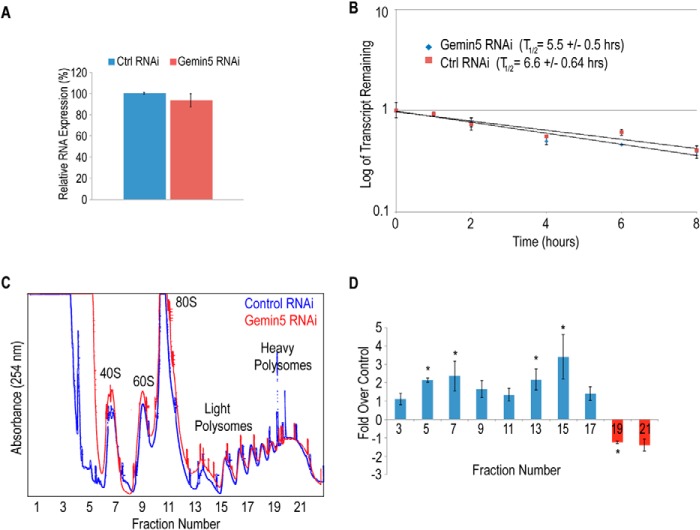
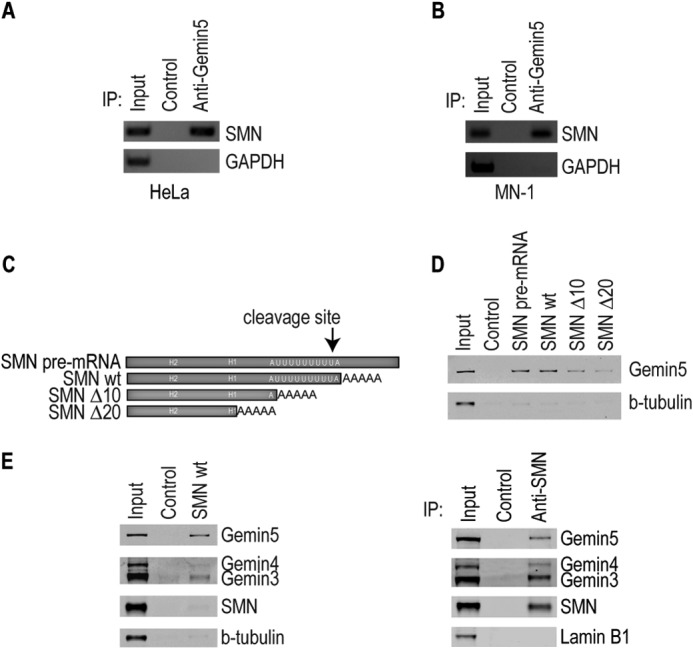
Previous work identified Gemin5 associated with a fragment of the SMN mRNA 3′-UTR when the fragment was biotinylated and added to cytoplasmic extract (23). While that experiment showed that Gemin5 has the capability to associate with the SMN 3′-UTR, it left unanswered whether Gemin5 associates with the SMN mRNA in living cells, and whether that association is direct or mediated by other proteins. To investigate this, we performed a modified PAR-CLIP experiment based on the method of Hafner, 2010 (24, 26). HeLa or MN1 cells were incubated with 4SU, a photo-activatable uridine analog that is incorporated into cellular RNA. The cells were then irradiated with UV light, allowing zero-distance, direct covalent RNA-protein crosslinks to form (24, 26). Gemin5 was immunoprecipitated under stringent conditions that still allowed the covalent RNA-protein crosslinks to be maintained. After immunoprecipitation, the proteins were removed by proteolysis and the crosslinked SMN mRNA was extracted and detected by RT-PCR. Fig. 1, A and B show that Gemin5 protein is directly and specifically bound to the SMN mRNA in cells. Further, we found that Gemin5 is bound to the SMN mRNA in both HeLa cells (Fig. 1A) and mouse motor neuron-derived MN-1 cells (Fig. 1B).
FIGURE 1.
Gemin5 binds to the mature SMN mRNA in vivo and in vitro. RT-PCR of modified PAR-CLIP to detect SMN mRNA bound to Gemin5 in both (A) HeLa and (B) MN1 cell extract. Control is an isotype-matched IgG antibody. C, diagram of RNA transcripts used in D and E. The SMN pre-mRNA construct is 192 nucleotides in length and 118 nucleotides for the mature mRNA exclusive of the 5 nucleotide pA tail. The regions deleted in subsequent experiments are indicated by H2, H1, as well as the U-rich region (white text). Further 10 or 20 nucleotide deletions were made to the 3′-end as indicated. D, Western blot of mutant and wild type SMN mRNA fragments in streptavidin pulldown assays. Control RNA used is a 60-nucleotide nonspecific RNA transcribed from empty pSP72. E, (left) Western blot of mutant and wild type SMN probing for proteins of the SMN complex in a streptavidin pulldown assay. Right, Western blot of SMN immunoprecipitation showing expected ratios of complex proteins. Control is the same as in D. For all, input represents extract from cell lines. IP, immunoprecipitation; Δ10 and Δ20, deletion of 10 or 20 nucleotides; wt, wild type; H2, helical region 2; H1, helical region 1.
Gemin5 Associates with the 3′-End of the Mature SMN mRNA in Extract
We have previously shown that Gemin5 associates with the 3′-UTR of the SMN pre-mRNA (23). The construct used for those experiments included sequences of the mature SMN 3′-UTR upstream of the polyadenylation site, as well as sequences of the SMN pre-mRNA downstream of the cleavage and polyadenylation site (Fig. 1C). To determine if Gemin5 associates with the mature SMN 3′-UTR, we created a construct that ends at the polyadenylation site and includes a short poly(A) tail (Fig. 1C). These mRNAs were biotinylated, incubated in cell extract, and then pulled down with streptavidin beads. Western blot analysis of the pulldowns showed that Gemin5 binds to the mature, polyadenylated SMN 3′-UTR construct as well as it did to the pre-mRNA construct, while no binding was observed to a control RNA (Fig. 1D). This suggests that the Gemin5 binding site is somewhere in the mature mRNA upstream of the polyadenylation site. In order to better define the region of the SMN mRNA bound by Gemin5, we created new constructs that deleted 10 or 20 nucleotides off the end of the polyadenylated mRNA (Fig. 1C). Deletion of the first 10 or 20 nucleotides of the mature mRNA resulted in decreased binding of Gemin5, showing that Gemin5 associates with a sequence at the 3′-end of the mature mRNA immediately upstream of the polyadenylation site (Fig. 1D).
Gemin5 Binds to the SMN mRNA as Part of an SMN-depleted Complex
Roughly half of Gemin5 is stably associated with the SMN complex at any given time, whereas the other half is found in SMN-depleted complexes (27). The SMN-free pool of Gemin5 binds to snRNAs and delivers them to SMN for snRNP assembly (11, 27). To determine whether Gemin5 binds to snRNAs as part of the SMN complex, we performed streptavidin pulldowns as described above and probed for other proteins in the SMN complex (Fig. 1E). We found that although Gemin5 is readily detectable bound to the SMN mRNA, Gemin3 and Gemin4 were only faintly detectable by Western analysis, and SMN was not detectable. To compare the ratios of Gemin5 to SMN and Gemins 3 and 4 as would be expected in the SMN complex, we immunoprecipitated SMN, isolating canonical, intact SMN complexes (Fig. 1E, right). Comparison of the ratios of Gemin5 to SMN in Fig. 1E, left (mRNA complex) and right (SMN complex) shows that the proteins associated with the SMN mRNA are strongly depleted in SMN, Gemin3 and Gemin4 (left) compared with the intact SMN complexes (SMN IP) (right). This suggests that it is the SMN-free pool of Gemin5, possibly associated in a previously reported complex with Gemin3 and Gemin4, that binds to the SMN mRNA (27). However, it remains possible that the SMN complex transiently associates with the SMN mRNA but is not captured in this assay.
Gemin5 Binds Directly and Specifically to the SMN 3′-UTR in Vitro
To investigate whether Gemin5 binds directly to the SMN mRNA, we immunopurified Gemin5 and the other components of the SMN complex under previously published stringent conditions (1% Empigen-BB) where the SMN complex is disrupted, and Gemin5 is effectively purified from other cellular proteins (11, 12). We then performed direct RNA-binding experiments by immobilizing the immunopurified proteins on magnetic beads and incubating them with in vitro transcribed, radiolabeled SMN mRNA, as well as a nonspecific control RNA. As shown in Fig. 2A, Gemin5 specifically binds to the SMN 3′ UTR but does not bind the control RNA. We detected no direct binding of purified SMN or Gemin3 to the SMN 3′-UTR. Further, deletion of 10 or 20 nucleotides from the 3′-end of the SMN mRNA resulted in complete loss of Gemin5 binding, confirming that Gemin5 directly binds to the extreme 3′-end of the SMN mRNA immediately upstream of the poly(A) tail (Fig. 2B).
FIGURE 2.
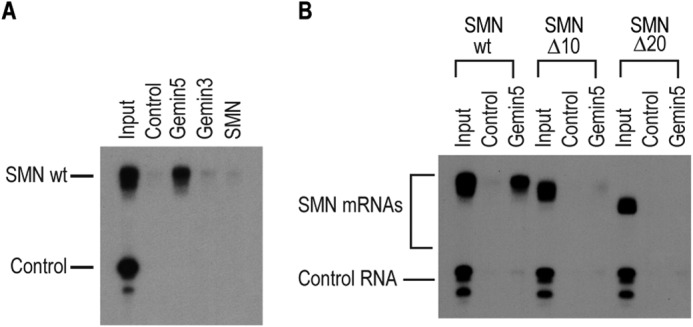
Gemin5 directly binds to the SMN mRNA. A, direct RNA binding assay performed with radiolabeled wild type SMN 3′-UTR or a control RNA sequence (as described in Fig. 1D). The RNA was bound to immunopurified Gemin5, Gemin3 or SMN protein or to a nonspecific isotype matched control IgG antibody. B, direct RNA binding assay as in A performed with 10 or 20 nucleotide truncations of the SMN 3′-UTR. For both, input represents 1% or 1,000 CPM of RNA used in each binding assay.
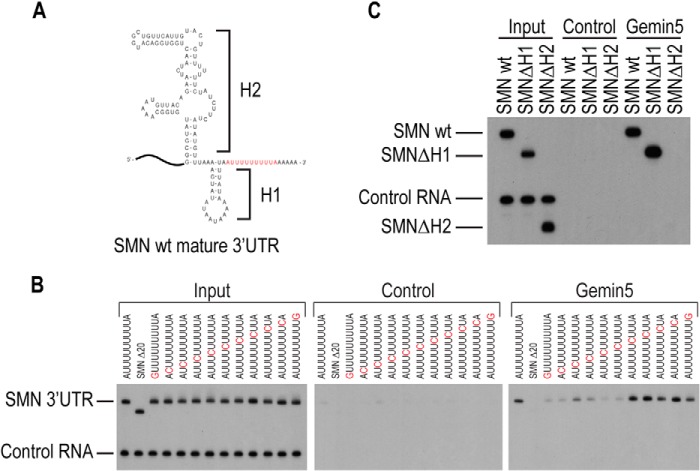
Gemin5 has been shown to directly bind snRNAs by recognizing a short single-stranded sequence of an A followed by 5 U residues, flanked at either the 5′- or 3′-ends by a stem-loop of varied length and composition. RNA structure prediction using the mfold structure prediction program (28) revealed that the 3′-end of the SMN mRNA folds into two helical regions that may coaxially stack, which we have termed H1 and H2 (Fig. 3A). These helical regions are followed by a predicted single-stranded U-rich sequence that matches the primary Gemin5 binding sites on snRNAs. To determine whether Gemin5 binds to the SMN mRNA in a mode similar to its recognition of snRNAs, we changed each nucleotide of the putative Gemin5 sequence, 5′-AUUUUUUUUUA-3′, at the 3′-end of the SMN 3′-UTR (Fig. 3B). Adenosine nucleotides were replaced with guanine and uridines with cytosine in a sequential manner. Direct binding assays showed that mutation of the initial adenosine as well as the first, fourth, and fifth uridines strongly reduced Gemin5 binding. In addition, mutation of the second and third uridines resulted in reduced binding. Mutation of the remaining uridine positions had no effect on the ability of Gemin5 to bind in vitro, nor did mutation of the final adenosine. These data are consistent with previous work showing that Gemin5 binds to an adenosine followed by 5 uridines in snRNAs (11).
FIGURE 3.
Gemin5 binds to specific sequence and structure elements of the SMN 3′-UTR. A, mfold structure prediction for the 3′ fragment of the SMN 3′-UTR. B, RNA binding assay performed with radiolabeled SMN RNA that contain point mutations in the AU sequence as indicated above each lane. C, RNA binding assay performed with stem-loop deleted SMN RNA as indicated in A. Input and control are the same as described in Fig. 2. H1, helical region 1; H2, helical region 2; wt, wild type.
In addition to the single-stranded AU5 Sm-site sequence, Gemin5 binding to snRNAs requires a flanking stem-loop structure. To test whether either of the upstream helical regions is required for Gemin5 binding to the SMN mRNA, we deleted each of the regions and tested them for direct binding to Gemin5. We found that the deletion of the H2 region caused a loss of Gemin5 binding, demonstrating that the H2 region performs a similar role as the flanking stem loop in snRNAs (Fig. 3C). This indicates that Gemin5 requires a stem loop structure in addition to AU sequence to bind mRNA in the same manner it binds to snRNAs.
Gemin5 Knockdown Reduces Protein Expression from Endogenous and Reporter SMN
To investigate the function of Gemin5 binding to the SMN mRNA, we created stable knockdown cell lines expressing shRNA directed against Gemin5 or a control shRNA targeting GFP. Using this system, we were able to significantly reduce Gemin5 expression in these cells by more than 95% (Fig. 4A and quantitated in B). We found that reduction of Gemin5 causes a greater than 40% reduction of endogenous SMN protein. To confirm that the effect of Gemin5 reduction is mediated by the SMN 3′-UTR we transfected the GFP or Gemin5 knockdown cell lines with luciferase constructs containing either the SMN 3′-UTR or a control 3′-UTR. We found that Gemin5 reduction caused a specific and significant reduction of luciferase expression from the reporter bearing the SMN 3′-UTR (Fig. 4C).
FIGURE 4.
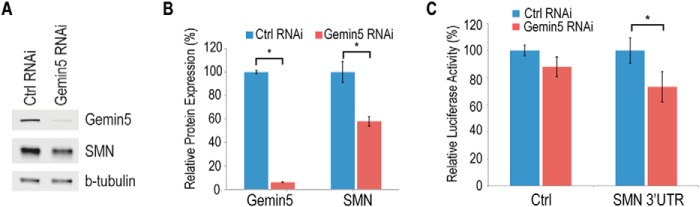
Gemin5 regulates expression of endogenous and reporter SMN. A, Western blot of protein from HeLa cells stably expressing shRNA targeting GFP (control) or Gemin5. B, quantitation of protein in A using ImageStudio for Li-Cor and normalizing signals to the loading control tubulin. The signal is expressed as a percentage of the signal in the control RNAi cells. C, dual luciferase reporter assay with a control or SMN 3′-UTR in the presence of transiently transfected control (nonspecific nontargeting siRNA) or Gemin5 siRNA. The control on the x-axis represents a luciferase expressed without an inserted 3′-UTR sequence. Ctrl, control; *, p < 0.05.
Gemin5 could affect SMN expression by altering SMN transcription, mRNA stability, or translation of the SMN mRNA. We measured the overall expression levels of the SMN mRNA by qRT-PCR and found that the RNA levels are unchanged as a result of Gemin5 RNAi (Fig. 5A). Further, we performed mRNA turnover experiments in the Gemin5 or control RNAi cells and found no significant changes in the stability of the SMN mRNA (Fig. 5B). Since SMN mRNA levels and stability are unaffected by Gemin5 reduction, and Gemin5 has previously been shown to interact with the translation apparatus to regulate translation of viral IRES-containing mRNAs, we assessed whether Gemin5 plays a role in the translation of the SMN mRNA. We loaded cytoplasmic extracts from the control or Gemin5 RNAi cells on a sucrose density gradient to separate mRNPs, monosomes, and polysomes. As shown in Fig. 5C, reduction of Gemin5 does not produce a general effect on mRNA translation. To determine if translation of the SMN mRNA was affected, we isolated mRNA from each fraction and determined the amount of SMN mRNA in each fraction by qRT-PCR. Following reduction of Gemin5 by RNAi, we observed a significant loss of SMN mRNA from the heavier polysomes, and a shifting of the SMN mRNA to lighter polysomes, monosomes, and mRNP fractions (Fig. 5D). These results suggest that reduction of Gemin5 results in a reduction in the translation of the SMN mRNA.
FIGURE 5.
Knockdown of Gemin5 does not affect SMN RNA stability or turnover, but does affect translation. A, real-time quantitative PCR of SMN in RNA extracts from control or Gemin5 RNAi cells. B, real-time quantitation of SMN RNA remaining after treatment with actinomycin D in cells with or without Gemin5 knockdown. C, UV absorbance trace of polysome fractions obtained from a sucrose gradient of cell lysates from control or Gemin5 knockdown cells. D, real-time PCR quantitation of SMN RNA in odd fractions of polysome gradient shown in C. For all, control RNAi represents a nonspecific nontargeting siRNA. Ctrl, control; *, p < 0.05.
SMN Autoregulates Its Own Expression through Gemin5
SMA results from a reduction in the SMN protein. Under normal conditions, Gemin5 is in equilibrium between SMN-bound and SMN-free complexes. When SMN is reduced, Gemin5 is freed from the SMN complex and accumulates in SMN-free complexes bound to snRNA (12, 27). To determine whether SMN reduction as observed in SMA affects the ability of Gemin5 to bind the SMN mRNA, we incubated biotinylated SMN 3′-UTR RNA fragments in cytoplasmic extract from either SMN RNAi cells or cells treated with a non-targeting siRNA. Following incubation, the biotinylated RNAs were isolated using streptavidin beads, and associated proteins were detected by Western blot. As shown in Fig. 6A, reduction of SMN results in an increase of Gemin5 binding to the SMN 3′-UTR, consistent with what has been observed for Gemin5 binding to snRNAs.
FIGURE 6.
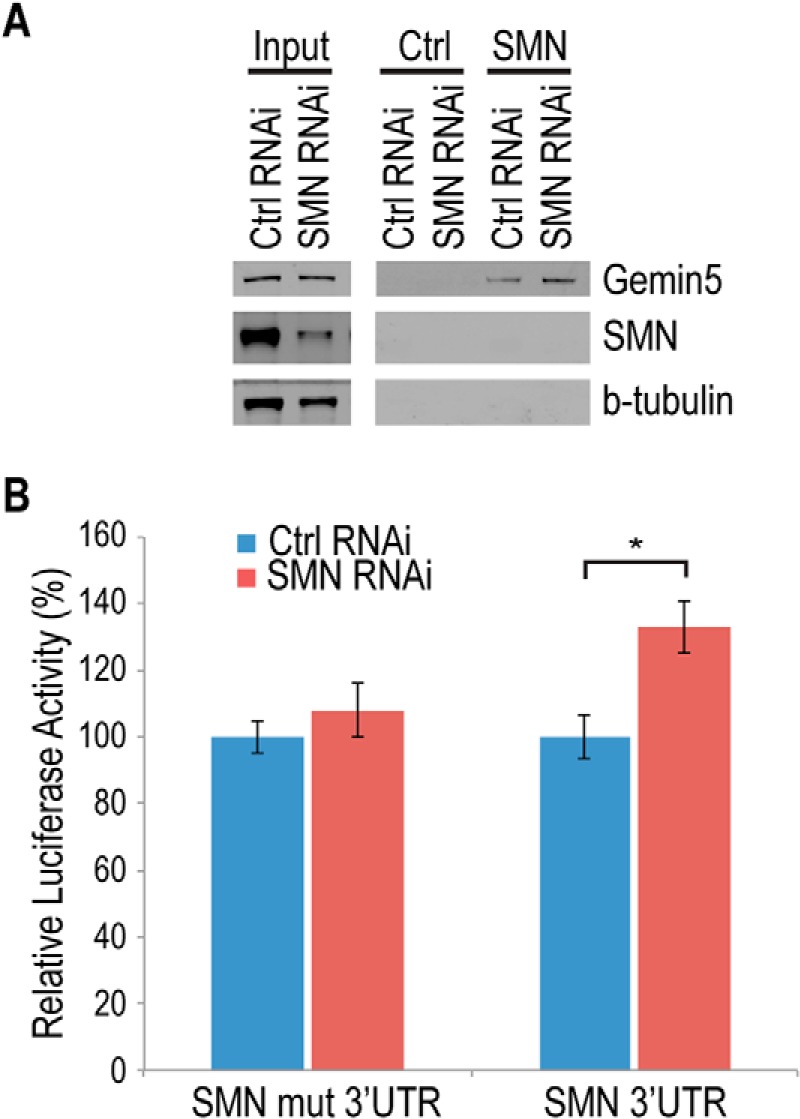
Knockdown of SMN causes Gemin5-dependent up-regulation of SMN reporter. A, Western blot of streptavidin pulldown assay using cell extracts from control (nontargeting siRNA) or SMN knockdown cells. Control RNA used is nonspecific RNA transcribed from an empty pSP72 vector. Input is 10 μg of total protein from respective cell lines. B, dual luciferase reporter assay performed with a wild type SMN RNA (SMN 3′-UTR) or a Gemin5 binding sequence-deleted mutant of SMN (SMN mut 3′-UTR) expressed in cells also transfected with control or SMN RNAi. Ctrl, control; mut, mutant, *, p < 0.05.
To determine if the increase in the mRNA-binding activity of Gemin5 results in an increase in SMN expression, we transfected luciferase reporters tagged with either the SMN 3′-UTR or an SMN 3′-UTR mutated in the Gemin5 binding site into either the SMN RNAi cells or cells targeted with a non-targeting siRNA. As shown in Fig. 6B, SMN reduction causes an increase in expression of the reporter bearing the wild-type SMN 3′-UTR, but not the reporter with the identical sequence containing a mutation of the Gemin5 binding site. Taken together, these data are consistent with a classic autoregulatory feedback mechanism in which reduction of SMN increases the Gemin5 binding to the SMN mRNA, in turn increasing SMN expression.
Discussion
We show here that Gemin5, the snRNA-binding protein of the SMN complex, binds to the SMN mRNA and modulates its expression by controlling translation of the SMN mRNA. The mRNA-binding activity of Gemin5 is dependent on levels of SMN protein, creating an autoregulatory feedback mechanism that allows SMN to regulate its own expression via Gemin5. This discovery identifies both a new function for an SMN complex protein in the direct regulation of cellular mRNA, and a new mechanism regulating expression of SMN.
Gemin5 binds to the SMN mRNA both in vitro and in vivo. Gemin5 binds directly to specific sequence and structural elements in the SMN 3′-UTR reminiscent of elements required for it to bind to snRNAs. Within snRNAs, Gemin5 binds to a bipartite site containing the single-stranded Sm site as well as an adjacent stem loop (11, 14). In the Sm site, Gemin5 specifically recognizes a sequence of an adenosine followed by 5 uridines (11). The requirements for the adjacent stem loop are less well understood. While the presence of an adjacent stem loop is absolutely required, the sequence and size of the stem loop are not conserved among snRNAs, and the stem-loop can be either 5′ or 3′ of the Sm site (11, 14). With the SMN mRNA, Gemin5 likewise binds with high sequence specificity to a single-stranded adenosine followed by 5 uridines adjacent to a required stem-loop structure. In this case, the stem-loop is 5′ of the Sm-like element, in a configuration similar to that found in the U4atac snRNA. Gemin5 has also been reported binding to viral mRNAs through an IRES element; however, that sequence differs from the specificity seen for snRNAs and for SMN mRNA (29). The viral IRES binding site for Gemin5 contains a stem loop, but lacks the canonical AU5–6 sequence motif (29). Therefore, the Gemin5 binding site on the SMN mRNA more closely resembles the well-characterized binding site on snRNAs, rather than the binding site found in viral IRES structures.
Although Gemin5 is an integral component of the SMN complex, the Gemin5 complexes isolated with the SMN mRNA are depleted in SMN protein. This is in contrast to either purified SMN complexes or complexes isolated with snRNAs where Gemin5 and SMN are present together in stoichiometric amounts (11, 30, 31). The data here are consistent with several possible models. Since Gemin5 exists in the cytoplasm in both SMN-bound and SMN-free complexes, the simplest explanation is that it is the SMN-free pool of Gemin5 that binds to the SMN mRNA, perhaps as part of a previously reported complex with Gemin3 and Gemin4 (27). However, although we do not detect stoichiometric amounts of SMN, the possibility remains that Gemin5 initially binds the SMN mRNA as part of the SMN complex, but unlike the situation with snRNAs, SMN is selectively released following binding, or associates only at a particular step, such as axonal transport.
SMN and Gemin5 have been found to localize to axons (32) and SMN has been associated with mRNA transport complexes in neuronal cells (33–35). For instance SMN has been shown to associate with β-actin mRNA in axons (35). It has also been shown to interact with alpha-COP vesicles and HuD, both of which are involved in axonal transport complexes (36–39). SMN's involvement with mRNA transport coupled with Gemin5's capacity to bind to mRNA and regulate translation could suggest that these proteins may function together to control specific axonal gene expression. This is supported by the report that Gemin5 is the most highly associated of the Gemin proteins with SMN within axons (32). Further work will determine whether SMN and Gemin5 function together with the same mRNAs, perhaps sequentially, or whether they function with different mRNAs.
Reduction of Gemin5 results in a parallel reduction in SMN expression. While overall SMN mRNA levels and stability remain unchanged, Gemin5 reduction results in selective reduction of SMN mRNA associated with heavier polysomes, indicating that Gemin5 acts as a modulator of SMN translation. These data are consistent with a model in which Gemin5 is directly or indirectly acting as an activator of SMN translation. Attempts to simply overexpress Gemin5 in either normal or SMN-reduced cells did not result in an increase of SMN protein levels (data not shown), suggesting that the mechanism is likely more complex than direct translational activation, and that perhaps Gemin5 binding displaces an inhibitor of SMN translation. In contrast, when Gemin5 binds to viral IRES-containing mRNAs, it results in down-regulation of translation (17). In that context, Gemin5 functions as an inhibitor of translation. These two systems together support the idea that Gemin5 may not directly function with the translation machinery, but depending on the local circumstance, can displace either positive or negative modulators of translation. Further work will be necessary to understand the mechanism of Gemin5's ability to affect translation.
The binding of Gemin5 to the SMN mRNA is dependent on SMN protein levels in that reduction of SMN results in an increase in the mRNA binding activity of Gemin5. This is similar to what has been reported for the snRNA binding activity of Gemin5. In that work, reduction of SMN was shown to shift Gemin5 from the SMN-associated to the SMN-free pool, and this SMN-free Gemin5 accumulates bound to RNAs (12, 27). Our new data suggest a classic feedback mechanism in which, when SMN is low, Gemin5 is freed from the SMN protein to bind to the SMN mRNA in an attempt to restore SMN expression to proper levels. In this way, SMN modulates its own expression levels via Gemin5.
Understanding the regulation of SMN expression is fundamental to developing effective treatment for SMA. Much research has focused on up-regulating the levels of SMN in the hopes of ameliorating the disease. Further understanding of how SMN levels are mechanistically maintained in the cell can provide additional targets for therapeutic intervention as well as aid in understanding the effectiveness of therapies currently being investigated. Here, we have identified a new pathway controlling SMN expression levels through regulated translation of the SMN transcript. This pathway is at least transiently activated in cell lines in which SMN is reduced. This work allows investigation of whether this pathway is active in SMA, and whether other mRNAs are targets for Gemin5.
Acknowledgment
We thank Dr. Daniel Kiss for technical assistance performing polysome gradients. The authors declare that they have no conflicts of interest with the contents of this article.
This work was supported by National Institutes of Health Grant R01NS077010 (to D. B.).
- SMN
- survival motor neuron
- SMA
- spinal muscular atrophy
- snRNP
- small nuclear ribonucleoprotein.
References
- 1. Eggert C., Chari A., Laggerbauer B., Fischer U. (2006) Spinal muscular atrophy: the RNP connection. Trends Mol. Med. 12, 113–121 [DOI] [PubMed] [Google Scholar]
- 2. Fischer U., Englbrecht C., Chari A. (2011) Biogenesis of spliceosomal small nuclear ribonucleoproteins. Wiley Interdiscip. Rev. RNA 2, 718–731 [DOI] [PubMed] [Google Scholar]
- 3. Liu Q., Fischer U., Wang F., Dreyfuss G. (1997) The Spinal Muscular Atrophy Disease Gene Product, SMN, and Its Associated Protein SIP1 Are in a Complex with Spliceosomal snRNP Proteins. Cell 90, 1013–1021 [DOI] [PubMed] [Google Scholar]
- 4. Meister G., Eggert C., Fischer U. (2002) SMN-mediated assembly of RNPs: a complex story. Trends Cell Biol. 12, 472–478 [DOI] [PubMed] [Google Scholar]
- 5. Paushkin S., Gubitz A. K., Massenet S., Dreyfuss G. (2002) The SMN complex, an assemblyosome of ribonucleoproteins. Curr. Opin. Cell Biol. 14, 305–312 [DOI] [PubMed] [Google Scholar]
- 6. Simic G. (2008) Pathogenesis of proximal autosomal recessive spinal muscular atrophy. Acta Neuropathol. 116, 223–234 [DOI] [PubMed] [Google Scholar]
- 7. Wan L., Battle D. J., Yong J., Gubitz A. K., Kolb S. J., Wang J., Dreyfuss G. (2005) The Survival of Motor Neurons Protein Determines the Capacity for snRNP Assembly: Biochemical Deficiency in Spinal Muscular Atrophy. Mol. Cell Biol. 25, 5543–5551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Achsel T., Stark H., Luhrmann R. (2001) The Sm domain is an ancient RNA-binding motif with oligo(U) specificity. Proc. Natl. Acad. Sci. U.S.A. 98, 3685–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kambach C., Walke S., Nagai K. (1999) Structure and assembly of the spliceosomal small nuclear ribonucleoprotein particles. Curr. Opin. Struct. Biol. 9, 222–230 [DOI] [PubMed] [Google Scholar]
- 10. Stark H., Dube P., Luhrmann R., Kastner B. (2001) Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature 409, 539–542 [DOI] [PubMed] [Google Scholar]
- 11. Battle D. J., Lau C. K., Wan L., Deng H., Lotti F., Dreyfuss G. (2006) The Gemin5 protein of the SMN complex identifies snRNAs. Mol. Cell 23, 273–279 [DOI] [PubMed] [Google Scholar]
- 12. Yong J., Kasim M., Bachorik J. L., Wan L., Dreyfuss G. (2010) Gemin5 Delivers snRNA Precursors to the SMN Complex for snRNP Biogenesis. Mol. Cell 38, 551–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lau C. K., Bachorik J. L., Dreyfuss G. (2009) Gemin5-snRNA interaction reveals an RNA binding function for WD repeat domains. Nat. Struct. Mol. Biol. 16, 486–491 [DOI] [PubMed] [Google Scholar]
- 14. Golembe T. J., Yong J., Dreyfuss G. (2005) Specific Sequence Features, Recognized by the SMN Complex, Identify snRNAs and Determine Their Fate as snRNPs. Mol. Cell Biol. 25, 10989–11004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B. M., Strein C., Davey N. E., Humphreys D. T., Preiss T., Steinmetz L. M., Krijgsveld J., Hentze M. W. (2012) Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406 [DOI] [PubMed] [Google Scholar]
- 16. Fierro-Monti I., Mohammed S., Matthiesen R., Santoro R., Burns J. S., Williams D. J., Proud C. G., Kassem M., Jensen O. N., Roepstorff P. (2006) Quantitative proteomics identifies Gemin5, a scaffolding protein involved in ribonucleoprotein assembly, as a novel partner for eukaryotic initiation factor 4E. J. Proteome Res. 5, 1367–1378 [DOI] [PubMed] [Google Scholar]
- 17. Pacheco A., López de Quinto S., Ramajo J., Fernández N., Martínez-Salas E. (2009) A novel role for Gemin5 in mRNA translation. Nucleic Acids Res. 37, 582–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bradrick S. S., Gromeier M. (2009) Identification of gemin5 as a novel 7-methylguanosine cap-binding protein. PLoS ONE 4, e7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lefebvre S., Bürglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. (1995) Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80, 155–165 [DOI] [PubMed] [Google Scholar]
- 20. Gabanella F., Carissimi C., Usiello A., Pellizzoni L. (2005) The activity of the spinal muscular atrophy protein is regulated during development and cellular differentiation. Hum. Mol. Genet. 14, 3629–364216236758 [Google Scholar]
- 21. Winkler C., Eggert C., Gradl D., Meister G., Giegerich M., Wedlich D., Laggerbauer B., Fischer U. (2005) Reduced U snRNP assembly causes motor axon degeneration in an animal model for spinal muscular atrophy. Genes Dev. 19, 2320–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arnold W. D., Burghes A. H. (2013) Spinal muscular atrophy: Development and implementation of potential treatments. Ann. Neurol. 74, 348–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Workman E., Veith A., Battle D. J. (2014) U1A regulates 3′ processing of the survival motor neuron mRNA. J. Biol. Chem. 289, 3703–3712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M., Jungkamp A. C., Munschauer M., Ulrich A., Wardle G. S., Dewell S., Zavolan M., Tuschl T. (2010) PAR-CliP–a method to identify transcriptome-wide the binding sites of RNA binding proteins. J. Vis. Exp. 41, e2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mukherjee C., Patil D. P., Kennedy B. A., Bakthavachalu B., Bundschuh R., Schoenberg D. R. (2012) Identification of cytoplasmic capping targets reveals a role for cap homeostasis in translation and mRNA stability. Cell Rep. 2, 674–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M., Jr., Jungkamp A. C., Munschauer M., Ulrich A., Wardle G. S., Dewell S., Zavolan M., Tuschl T. (2010) Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Battle D. J., Kasim M., Wang J., Dreyfuss G. (2007) SMN-independent subunits of the SMN complex. Identification of a small nuclear ribonucleoprotein assembly intermediate. J. Biol. Chem. 282, 27953–27959 [DOI] [PubMed] [Google Scholar]
- 28. Zuker M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Piñeiro D., Fernández N., Ramajo J., Martínez-Salas E. (2013) Gemin5 promotes IRES interaction and translation control through its C-terminal region. Nucleic Acids Res. 41, 1017–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gubitz A. K., Mourelatos Z., Abel L., Rappsilber J., Mann M., Dreyfuss G. (2002) Gemin5, a novel WD repeat protein component of the SMN complex that binds Sm proteins. J. Biol. Chem. 277, 5631–5636 [DOI] [PubMed] [Google Scholar]
- 31. Feng W., Gubitz A. K., Wan L., Battle D. J., Dostie J., Golembe T. J., Dreyfuss G. (2005) Gemins modulate the expression and activity of the SMN complex. Hum. Mol. Genet. 14, 1605–1611 [DOI] [PubMed] [Google Scholar]
- 32. Todd A. G., Morse R., Shaw D. J., Stebbings H., Young P. J. (2010) Analysis of SMN-neurite granules: Core Cajal body components are absent from SMN-cytoplasmic complexes. Biochem. Biophys. Res. Commun. 397, 479–485 [DOI] [PubMed] [Google Scholar]
- 33. Jablonka S., Sendtner M. (2003) Molecular and cellular basis of spinal muscular atrophy. Amyotroph. Lateral Scler Other Motor Neuron Disord. 4, 144–149 [DOI] [PubMed] [Google Scholar]
- 34. Zhang H. L., Pan F., Hong D., Shenoy S. M., Singer R. H., Bassell G. J. (2003) Active Transport of the Survival Motor Neuron Protein and the Role of Exon-7 in Cytoplasmic Localization. J. Neurosci. 23, 6627–6637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rossoll W., Jablonka S., Andreassi C., Kröning A. K., Karle K., Monani U. R., Sendtner M. (2003) Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons. J. Cell Biol. 163, 801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hubers L., Valderrama-Carvajal H., Laframboise J., Timbers J., Sanchez G., Côté J. (2011) HuD interacts with survival motor neuron protein and can rescue spinal muscular atrophy-like neuronal defects. Hum. Mol. Genet. 20, 553–579 [DOI] [PubMed] [Google Scholar]
- 37. Fallini C., Zhang H., Su Y., Silani V., Singer R. H., Rossoll W., Bassell G. J. (2011) The survival of motor neuron (SMN) protein interacts with the mRNA-binding protein HuD and regulates localization of poly(A) mRNA in primary motor neuron axons. J. Neurosci. 31, 3914–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Peter C. J., Evans M., Thayanithy V., Taniguchi-Ishigaki N., Bach I., Kolpak A., Bassell G. J., Rossoll W., Lorson C. L., Bao Z. Z., Androphy E. J. (2011) The COPI vesicle complex binds and moves with survival motor neuron within axons. Hum. Mol. Genet. 20, 1701–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Todd A. G., Lin H., Ebert A. D., Liu Y., Androphy E. J. (2013) COPI transport complexes bind to specific RNAs in neuronal cells. Hum. Mol. Genet. 22, 729–736 [DOI] [PMC free article] [PubMed] [Google Scholar]