Background: Wnt5a signaling induces asymmetric localization of the melanoma cell adhesion molecules (MCAM).
Results: Wnt5a promotes MCAM depalmitoylation and point mutations in MCAM that block palmitoylation are sufficient to cause asymmetric MCAM localization.
Conclusion: Wnt5a induces polarized MCAM localization by promoting MCAM depalmitoylation.
Significance: These results reveal a mechanism for Wnt5a-induced polarized cell behavior.
Keywords: cell invasion, cell polarity, cell signaling, protein palmitoylation, Wnt signaling
Abstract
Wnt5a signaling regulates polarized cell behavior, but the downstream signaling events that promote cell polarity are not well understood. Our results show that Wnt5a promotes depalmitoylation of the melanoma cell adhesion molecule (MCAM) at cysteine 590. Mutation of Cys-590 to glycine is sufficient to polarize MCAM localization, similar to what is observed with Wnt5a stimulation. Inhibition of the depalmitoylating enzyme APT1 blocks Wnt5a-induced depalmitoylation, asymmetric MCAM localization, and cell invasion. Directly altering expression of the basal protein palmitoylation machinery is sufficient to promote cell invasion. Additionally, cancer mutations in palmitoyltransferases decrease MCAM palmitoylation and have impaired ability to suppress cell invasion. Our results provide evidence that Wnt5a induces protein depalmitoylation, which promotes polarized protein localization and cell invasion.
Introduction
The noncanonical Wnt pathway has been implicated in cancer progression and metastasis. Expression of the noncanonical Wnt ligand Wnt5a is increased in multiple cancers, including melanoma and gastric and colon cancer, which correlates with poor outcome (1–7).
Wnt5a signaling has been best studied in developmental systems, where it regulates polarized cell behavior and directional cell movement (8). In a cultured cell system, purified Wnt5a induces the formation of a dynamic polarized structure composed of Frizzled receptors, F-actin, and myosin II called the Wnt5a-induced receptor, actin, and myosin polarity structure (WRAMP4 structure) (9, 10). The localization of the WRAMP structure correlates with membrane retraction and nuclear movement and requires the cytosolic Wnt signaling protein Disheveled, protein kinase C, and the small GTPase RhoB for assembly (9, 10). This response also induces the polarized distribution of the melanoma cell adhesion molecule (MCAM), an immunoglobulin family cell adhesion molecule that is up-regulated in melanoma and promotes cell invasion (9–11). It is still unclear what downstream effectors promote Wnt5a-induced cell asymmetry.
Protein palmitoylation is a reversible lipid modification that plays a role in the trafficking, localization, and function of transmembrane proteins, including the canonical Wnt coreceptor LRP6, the cell adhesion molecule CD44, and integrin receptors (12–16). The transfer of palmitate to cysteine residues within target proteins is catalyzed by the DHHC family of four and six-pass transmembrane proteins (17). The removal of palmitate is mediated by the acyl-protein thioesterase enzymes APT1 and -2 (18). APT1 and -2 function to depalmitoylate proteins at the membrane, such as H-Ras and GAP-43 (19).
There are increasing examples of the critical role for protein palmitoylation in cell signaling. Protein palmitoylation plays a distinct role in determining receptor levels and localization in neurons and immune cells. The turnover of glutamate receptors at neuronal synapses is promoted by depalmitoylation of the postsynaptic density protein (PSD-95) in response to glutamate receptor activity (20). Similarly, immune T-cells require palmitoylation of T-cell co-receptors CD4 and CD8 for localization to lipid rafts during immune synapse formation (21). Aberrant palmitoylation is also implicated in human disease, particularly neurological disorders, including Huntington disease and X-linked intellectual disabilities (22). However, the function of protein palmitoylation in cancer is understudied. Alterations in palmitoyltransferases have been identified in cancer, but studies have mostly focused on H- and N-Ras membrane localization mediated by palmitoylation (23, 24). Our studies reveal a role for protein palmitoylation in regulating Wnt5a-induced asymmetric receptor localization and cell invasion.
Materials and Methods
Purification of Recombinant Wnt5a
Wnt5a was purified from mouse L-cells overexpressing human Wnt5a, as described previously (25).
Cell Culture and Transfection
HEK293T and WM239A cells were cultured in RPMI medium containing 10% FBS. MDA-MB-231 breast carcinoma cells were cultured in DMEM containing 10% FBS. All transfections were carried out using TransIT-LT1 (Mirus) according to the manufacturer's instructions. Plasmid-containing cells were selected with 6 μg/ml G418 or 100 μg/ml hygromycin. Palmostatin B (Calbiochem Millipore) was resuspended in DMSO and used at 1–5 μm final concentration. DMSO served as a vehicle control for palmostatin B. siRNAs (Dharmacon) were transfected with DMRIE-C (Life Technologies, Inc.) transfection reagent according to the manufacturer's instructions (control duplex, catalog no. D-001210-02; Disheveled-2 duplex, catalog no. D-004069-02).
Antibodies and Immunostaining
Anti-MCAM antibody (sc-18837) was from Santa Cruz Biotechnology. Anti-GFP (Ab290) antibody was from Abcam. Anti-FLAG M2 antibody was from Sigma. Anti-CD44 (catalog no. 3570) antibody was from Cell Signaling Technologies. Anti-ZDHHC20 antibody (catalog no. HPA014702) was from Sigma, and Ab110478 was from Abcam. Anti-caveolin 1 antibody (7C8) (catalog no. ab17052) was from Abcam.
Click Chemistry Palmitoylation Assay
WM239A melanoma cells and MDA-MB-231 breast cancer cells were grown in 6-cm dishes for 48 h and serum-starved for 15 h. Medium was switched to serum-free medium containing 100 μm palmitic acid azide (Life Technologies) and incubated for 4 h at 37 °C. HEK293T cells were not serum-starved and were labeled in RPMI containing 10% FBS. Control buffer or purified Wnt5a (150 ng/ml) was added and incubated for 30 min at 37 °C. Medium was removed, and cells were washed three times in PBS. Cells were lysed in 200 μl of lysis buffer (50 mm Tris, pH 7.5, 1% SDS, 1 μg/ml leupeptin, 1 μg/ml aprotinin, and 2 μg/ml pepstatin A). Lysates were sonicated and centrifuged at 15,000 rpm for 10 min. 50 μl of lysate was reacted with biotin alkyne (Life Technologies) using the Click-IT assay (Life Technologies) in a 200-μl final reaction volume as per the manufacturer's protocol. Biotinylated proteins were isolated using streptavidin-agarose (Pierce) and washed five times in wash buffer (50 mm Tris, pH 7.5, 0.1% SDS) and analyzed by SDS-PAGE and immunoblotting.
Acyl-Biotinyl Exchange (ABE) Assay
The ABE assay was performed as described (26).
Immunoprecipitation
HEK293T cells were seeded in 6-well plates. Each well of HEK293T cells was transfected with 2 μg of plasmids in total, including 1 μg of each plasmid as indicated. 48 h after transfection, cells were washed once with PBS and lysed by scraping in immunoprecipitation buffer (1% Triton X-100, 50 mm Tris, pH 7.5, 150 mm NaCl, and protease inhibitors). Lysates were incubated at 4 °C for 15 min while rotating and centrifuged at 17,000 × g for 10 min at 4 °C. For FLAG tag immunoprecipitation, the supernatant was added to 5 μl of prewashed anti-FLAG M2 magnetic beads (Sigma) and incubated at 4 °C overnight while rotating. For GFP tag immunoprecipitation, the supernatant was incubated with anti-GFP antibody overnight at 4 °C, followed by incubation with Protein A-Sepharose (GE Healthcare) for 2 h. Then the beads were washed four times and resuspended in 50 μl of SDS loading buffer and analyzed by SDS-PAGE and immunoblotting by standard methods.
Silencing of Human DHHC20
The oligonucleotides for shDHHC20 and shCtrl construct were synthesized (Integrated DNA Technologies) and inserted into the pLKO.1 vector. shCtrl encodes the non-targeting sequence of SHC002 (Sigma). The shRNA target sequence of human DHHC20 is 5′-GAGCTCTGCGTGTTTACTATT-3′ (shDHHC20-1). WM239A cells were transduced with lentivirus encoding shCtrl or shDHHC20 and selected by puromycin treatment (1 μg/ml). DHHC20 was rescued by expressing human DHHC20 containing a silent mutation in the shRNA 1 targeting sequence using the pRRL lentiviral vector or the vector alone as a control.
Collagen Invasion Assay
96-well plates were coated with 50 μl/well sterile 1.5% noble agar and solidified at room temperature for 10 min. 200 μl of 2.5 × 104 cells/ml cell suspension was added to each well. Spheroids formed at 37 °C and 4.0% CO2 for 24 h. Collagen matrix was prepared on ice using Advanced Biomatrix Pur Col purified bovine collagen, Hyclone RPMI 1640 (5×) with sodium bicarbonate diluted to 1× in total volume, and 10% FBS. Sterile NaOH was then added to correct collagen pH. 75 μl of collagen matrix was added to new wells and allowed to solidify at 37 °C for 1 h. Spheroids were resuspended in 125 μl of collagen matrix and transferred to wells containing 75 μl of collagen. After collagen solidified at 37 °C, 100 μl of fresh medium was added on top of collagen. Medium was changed after day 3. Images were taken every 24 h for 6–12 days. Wnt5a was added at a final concentration of 150 ng/ml and replaced in the medium every 48 h. Palmostatin B was added at a final concentration of 10 μm on day 3 and replaced in the medium every 48 h.
Live Cell and FRET Sensitized Emission Imaging
All live cell imaging was performed on a Leica AF6000 inverted microscope at 37 °C under 5% CO2. Relative CFP/YFP FRET efficiencies were determined using Leica LAS AF software Method 1: EA(i) = B − A × B − C × γ)/C, where EA is the apparent FRET efficiency; A, B, and C correspond to the intensities of the three signals (donor, FRET, and acceptor); and β and γ are calibration factors generated by acceptor only and donor only references. Cells were grown on Matek glass-bottomed dishes, and imaging was performed in Hanks' buffered saline solution with 1% FBS and 2 mm glutamine.
Xenograft Assay
Hairless CB17 mice (Charles River) were injected subcutaneously in the posterior flank with 1 × 106 cells mixed 1:1 with Matrigel (BD Biosciences). Tumors were measured by caliper when first palpable. After 3 weeks, tumors were removed and frozen in O.C.T medium and sectioned at −30 °C in 8-μm-thick sections. Sections were fixed in 4% paraformaldehyde, permeabilized in 0.1% Tween in TBS (TBST), and blocked with 5% BSA in TBST. Antibodies were diluted 1:200, and secondary antibody was diluted 1:1000. Sections were scored blind for straight borders between the tumor and the adjacent tissue or detached cells across the entire interface between the tumor and the adjacent tissue.
Results
Wnt5a Induces Protein Depalmitoylation via APT1
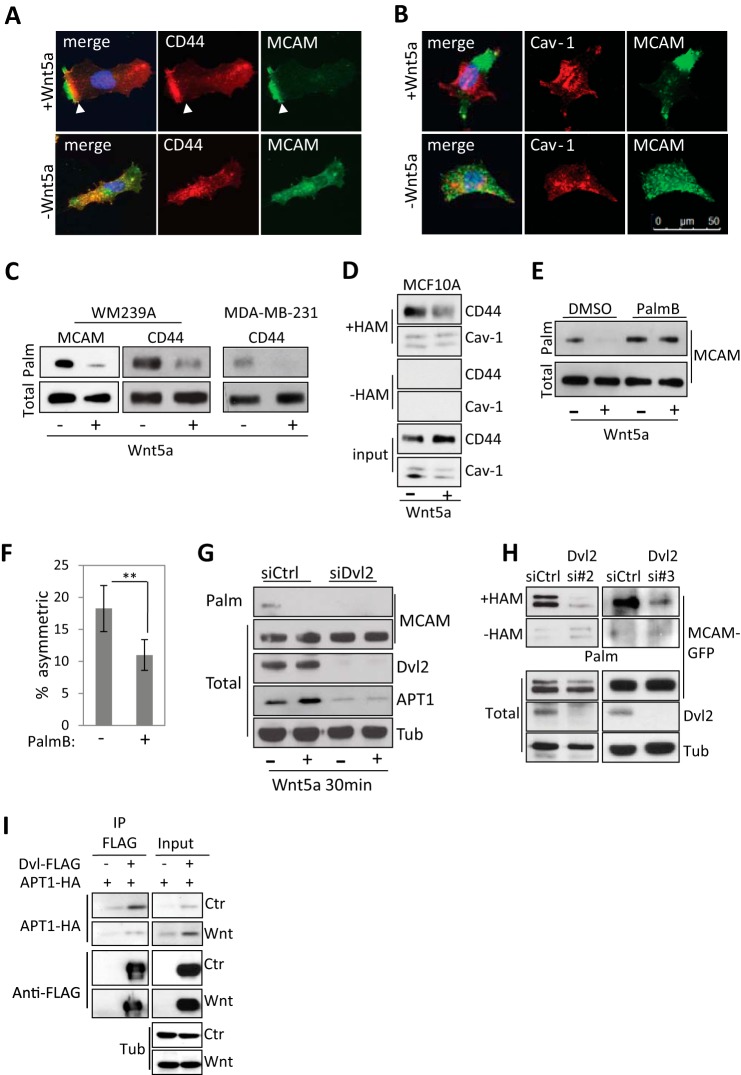
Wnt5a was previously shown to induce asymmetric localization of MCAM (9, 10). When we examined other membrane proteins, we found a similar asymmetric localization of the hyaluronic acid receptor CD44 in WM239A melanoma cells upon Wnt5a treatment (Fig. 1A). We also found that the lipid raft protein caveolin-1 (Cav-1) is excluded from the region of asymmetrically localized MCAM (Fig. 1B). It has been reported that palmitoylation of proteins can promote the association with lipid microdomains in the plasma membrane (27). CD44 and MCAM have both been reported to be palmitoylated, which may play a role in promoting the asymmetric localization (14, 28).
FIGURE 1.
Wnt5a promotes protein depalmitoylation. A, MCAM and CD44 localization in control- and Wnt5a-treated WM239A melanoma cells. Purification buffer (PBS + 1% CHAPS) serves as a vehicle control for Wnt5a. B, asymmetric MCAM is excluded from Cav-1-containing membrane domains in Wnt5a-treated cells. C, Wnt5a treatment decreases MCAM palmitoylation in WM239A melanoma cells and CD44 in WM239A cells and MDA-MB-231 breast cancer cells. Cells were metabolically labeled with palmitic acid azide. D, CD44 palmitoylation measured using the ABE in MCF10A breast epithelial cells decreases with Wnt5a treatment, whereas the levels of palmitoylated Cav-1 remain unchanged. Samples not treated with hydroxylamine (−HAM) serve as a negative control. E, Wnt5a-induced MCAM depalmitoylation is reduced with APT1 inhibition (palmostatin B, 1 μm) compared with DMSO control in WM239A melanoma cells. Error bars, S.E. F, palmostatin B (1 μm) inhibits MCAM asymmetry in Wnt5a-treated cells compared with DMSO control (Student's t test; *, p = 0.023; error bars, S.E.). G, levels of palmitoylated MCAM-GFP (Palm) in cells labeled with palmitic acid azide in control and Dvl2 siRNA-treated WM239A cells. Levels of APT1 decrease with Dvl2 siRNA treatment. H, levels of palmitoylated MCAM-GFP (Palm) determined using the acyl-biotinyl exchange (ABE) assay on WM239A cells expressing MCAM-GFP and transfected with siControl or siDvl2 oligonucleotides 2 and 3. Samples not treated with hydroxylamine (−HAM) serve as a negative control. I, immunoprecipitation (IP) of FLAG-tagged Dvl2 captures HA-tagged APT1.
To measure the levels of palmitoylated MCAM and CD44, cells were metabolically labeled with palmitic acid azide for 6 h, followed by conjugation to biotin using copper-catalyzed click chemistry and capture with streptavidin beads. Immunoblotting showed that the levels of palmitoylated MCAM and CD44 decreased in WM239A melanoma cells within 30 min of Wnt5a treatment, whereas the total levels remained unchanged (Fig. 1C). Wnt5a stimulation also decreased the levels of palmitoylated CD44 in the breast cancer cell line MDA-MB-231 and in WM239A cells (Fig. 1C). We finally asked if Wnt5a decreases CD44 palmitoylation in the non-transformed breast epithelial cell line MCF10A. In MCF10A cells, we could not detect palmitoylated CD44 by metabolic labeling, which is dependent on palmitoylation turnover. Instead an alternative assay was used to directly measure protein palmitoylation without metabolic labeling. The ABE assay uses hydroxylamine (HAM) to cleave palmitate from proteins exchanging it with biotin, allowing palmitoylated proteins to be isolated on streptavidin beads. Using the ABE assay, the level of palmitoylated CD44 decreased after 45 min of Wnt5a treatment. The level of palmitoylated Cav-1 was also monitored in the same samples. However, unlike CD44, the levels of palmitoylated Cav-1 remained unchanged. When hydroxylamine is left out of the preparation as a negative control (−HAM), the levels of CD44 and caveolin-1 are below detection (Fig. 1D).
The acyl-protein thioesterase 1 (APT1/LYPLA1) enzyme depalmitoylates proteins and is inhibited by the small molecule palmostatin B (29). We found that treatment with palmostatin B (1 μm) blocked the Wnt5a-induced decrease of palmitoylated MCAM (Fig. 1E). Treatment with Wnt5a stimulated cells with palmostatin B for 1 h also decreased the percentage of cells with asymmetrically localized MCAM (11.0 ± 2.4% (S.E.)) compared with cells pretreated with DMSO control (18.2 ± 3.6% (S.E.); Student's t test, p = 0.023) (Fig. 1F).
We next asked whether inhibition of the adaptor protein Dvl2 that is required for both canonical and noncanonical Wnt signaling pathways affects MCAM palmitoylation (30, 31). Inhibition of Dvl2 by siRNA decreased levels of palmitoylated MCAM in WM239A cells although total levels of APT1 were decreased. This indicates that the loss of MCAM palmitoylation with Dvl2 inhibition is not caused by increased APT1 protein levels (Fig. 1G). The decrease in APT1 observed with Dvl2 siRNA could indicate that APT1 is a transcriptional target of the canonical Wnt pathway, which also requires Dvl proteins. To confirm that the decrease in MCAM palmitoylation was not caused by a decrease in incorporation of labeled palmitate, the ABE assay was used to directly detect palmitoylated MCAM. In MCAM-GFP-expressing WM239A cells, we found that inhibition of Dvl2 with two separate siRNA duplexes reduced MCAM palmitoylation to levels similar to the negative control without hydroxylamine treatment (−HAM) (Fig. 1H).
To determine whether APT1 might be directly regulated by Wnt5a signaling, we examined possible APT1 interactions with Dvl2. FLAG-tagged Dvl2 was immunoprecipitated in HEK293T cells and was found in a complex with APT1-HA that decreased upon Wnt5a treatment compared with control-treated cells (Fig. 1I). These results suggest that Wnt5a may regulate APT1 function through an interaction between APT1 and Dvl2.
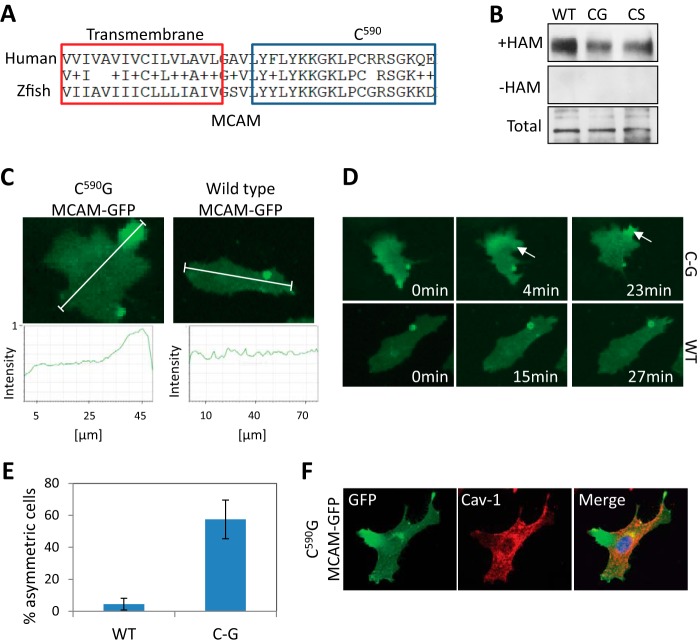
Mutation of Cysteine 590 Promotes Asymmetric MCAM Localization
The intracellular domain of MCAM contains one cysteine (Cys-590). Although human and zebrafish MCAM have only 32% identity, the cysteine residue is evolutionarily conserved (Fig. 2A). A single amino acid substitution of Cys-590 to glycine or serine measurably reduces palmitoylation of MCAM-GFP (Fig. 2B). We reasoned that blocking MCAM palmitoylation may also be sufficient to promote asymmetric MCAM localization. Time lapse imaging of cells expressing uniformly distributed C590G mutant MCAM-GFP showed relocalization to one end of the cell in 57% (n = 52) of recorded cells, compared with only 4% (n = 23) of WT MCAM-GFP-expressing cells (Fig. 2 (C–E) and supplemental Movies S1 and S2). The C590G mutant MCAM-GFP was also excluded from caveolin-containing regions of the plasma membrane, recapitulating what we observed in Wnt5a-treated cells (Fig. 2F).
FIGURE 2.
MCAM is palmitoylated at cysteine 590. A, sequence alignment of human and zebrafish MCAM transmembrane domain and juxtamembrane region containing a putative palmitoylated cysteine 590. B, mutation of cysteine 590 to glycine or serine reduces MCAM palmitoylation, as determined by the ABE assay. C, live cell imaging of C590G MCAM-GFP that translocates asymmetrically to one side of the cell in 57% (n = 33) of the cells compared with 4% (n = 22) in wild type-expressing cells without Wnt5a treatment. Cells were imaged for 1 h in 1% FBS after 1 h of serum starvation. White lines, regions of interest used for the line scans. D, still photographs of cells shown in Fig. 2C (arrows indicate a local increase in MCAM-GFP). E, quantification from live cell imaging (WT, n = 22; C590G, n = 33; Student's t test, p = 0.006; average of three experiments). F, asymmetrically localized C590G MCAM-GFP is excluded from caveolin-containing membrane domains. Error bars, S.E.
The C590G Mutation Promotes MCAM Self-association
We next asked whether the localization of C590G MCAM is distinct from WT MCAM. In Wnt5a-treated cells expressing both WT MCAM-YFP and C590G MCAM-CFP, we found both forms of MCAM were asymmetrically localized at one end of the cell, but there was a clear separation of the peak signal intensity between the WT MCAM-YFP from C590G MCAM-CFP (Fig. 3A and supplemental Movie S3). This is in contrast to WT MCAM-CFP and WT MCAM-YFP in which the positions of the peak intensities were closely aligned (Fig. 3B and supplemental Movie S4). The separation of the WT MCAM from the C590G MCAM may indicate a difference in affinity between MCAM molecules that could promote the accumulation of C590G MCAM to a spatially distinct membrane domain while excluding WT MCAM.
FIGURE 3.
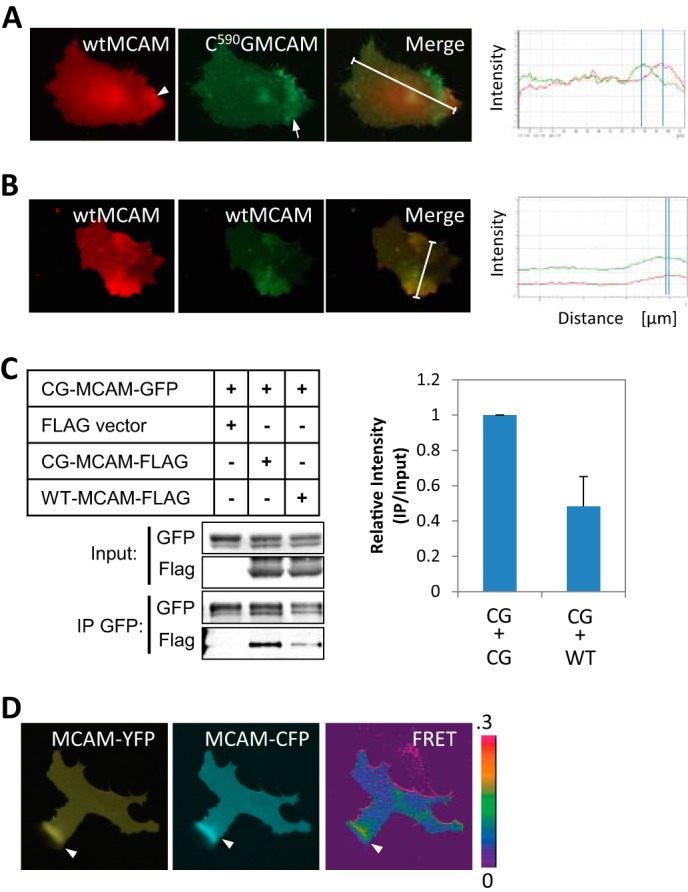
MCAM depalmitoylation promotes MCAM-MCAM interaction. A, wild type MCAM-YFP (red, arrowhead) localizes separately from palmitoylation-resistant C590G MCAM-CFP (green, arrow) in Wnt5a-treated melanoma cells. White lines, regions of interest used for the line scans. B, wild type MCAM-YFP (red) and wild type MCAM-CFP (green) co-localize asymmetrically in Wnt5a-treated WM239A melanoma cells. White lines, regions of interest used for the line scans. C, C590G MCAM-GFP forms a complex with C590G MCAM-FLAG with higher affinity than with wild type MCAM-FLAG in HEK293T cells (Student's t test, p = 0.006, average of three experiments). D, interaction between MCAM molecules detected by live CFP/YFP FRET imaging of melanoma cells expressing MCAM-YFP and MCAM-CFP. Increased FRET signal was detected at the area of asymmetrically localized MCAM (arrows). IP, immunoprecipitation. Error bars, S.E.
To address whether complex formation between C590G MCAM-GFP mutant receptors is preferential to formation with WT receptors, C590G MCAM-GFP was immunoprecipitated from HEK293T cells expressing either FLAG-tagged WT MCAM or C590G MCAM. We found the interaction between C590G MCAM-GFP and C590G MCAM-FLAG was 4-fold higher than with WT MCAM-FLAG (Fig. 3C).
Finally, live FRET imaging of WT MCAM-CFP and WT MCAM-YFP was performed in Wnt5a-stimulated cells to determine whether MCAM-MCAM interactions could be detected in a spatially distinct region. We found that the highest FRET emission was at regions of asymmetrically localized MCAM (Fig. 3D).
APT1 Is Necessary and Sufficient to Promote Cell Invasion
A three-dimensional collagen spheroid invasion assay was used to measure cell invasion of melanoma cells in the presence or absence of Wnt5a (32). A significant increase in collagen invasion was observed with Wnt5a added every 48 h compared with control treated cells (749.7 ± 50.8 versus 576.7 ± 74.9 μm (S.E.); Student's t test, p = 2.50 × 10−3) (Fig. 4A). Inhibition of APT1 with 10 μm palmostatin B blocked the Wnt5a-induced increase in invasion (516.0 ± 60.7 μm (S.E.); Student's t test, p = 9.27 × 10−5) (Fig. 4A). The modest effect of Wnt5a on collagen invasion might be caused by poor diffusion of the 35-kDa Wnt5a ligand through the collagen matrix. To determine whether blocking APT1 is sufficient to inhibit cell invasion in collagen, shRNA was used to reduce APT1 expression (Fig. 4B). Expression of APT1 shRNA decreased invasion of WM239A cells (54.1 ± 18.3 μm (S.D.)) compared with shRNA control-expressing cells (393.3 ± 44.3 μm (S.D.); Student's t test, p = 1.46 × 10−4) by day 12 (Fig. 4C). Conversely, overexpression of APT1-CFP-FLAG is sufficient to increase cell invasion (500.9 ± 54.1 μm (S.E.)) relative to control CFP-FLAG-expressing cells (178.7 ± 42.0 μm (S.E.); Student's t test, p = 2.05 × 10−9) by day 7 (Fig. 4, D and E).
FIGURE 4.
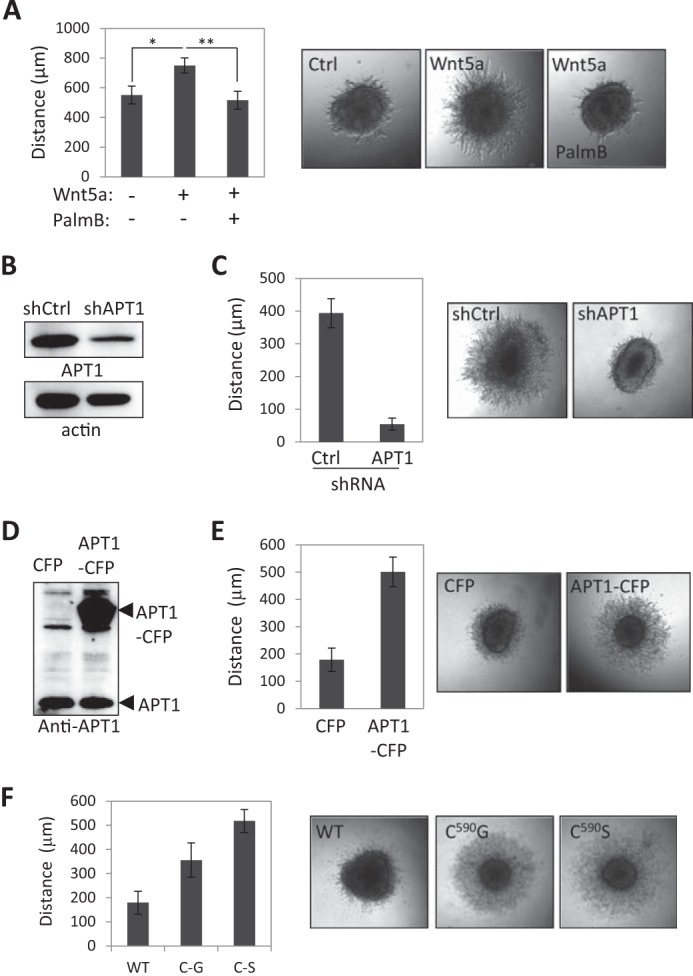
Protein depalmitoylation promotes cell invasion. A, palmostatin B inhibits Wnt5a-induced invasion in three-dimensional collagen measured on day 8 (n = 12 spheroids/condition; error bars, S.E.; Student's t test; *, p = 2.50 × 10−3; **, p = 9.27 × 10−5). DMSO serves as a vehicle control in conditions without palmostatin B. B, APT1 shRNA reduces APT1 protein expression. C, APT1 shRNA inhibits melanoma cell invasion compared with control shRNA measured on day 12 (shCtrl, n = 4; shAPT1, n = 8; Student's t test; p = 1.46 × 10−4; error bars, S.D.). D, expression level of ectopically expressed APT1-CFP detected with anti-APT1 antibody. E, cells expressing APT1-CFP are more invasive than control cells expressing CFP alone measured on day 7 (APT1-CFP, n = 16; control CFP, n = 14; Student's t test; p = 2.05 × 10−9; error bars, S.E.). F, cells expressing MCAM mutated at cysteine 590 to glycine or serine are more invasive in collagen than cells expressing WT MCAM measured on day 6 (WT, n = 16; C590G, n = 16; Student's t test; p < 0.0001). Error bars, S.E.
We next asked whether blocking MCAM palmitoylation specifically is sufficient to promote cell invasion. Expression of C590G or C590S MCAM-GFP increased collagen invasion (from 356 ± 71.2 to 518 ± 46.6 μm (S.D.)) compared with wild type MCAM-GFP-expressing cells (179 ± 47.8 μm (S.D.); Student's t test, p < 0.0001) (Fig. 4F).
C590G MCAM-GFP Expression Alters Local Cell Invasion in Vivo
To determine whether blocking MCAM palmitoylation affects melanoma cell invasion in vivo, a xenograft tumor model was used to measure local invasion by tumors expressing WT or C590G MCAM-GFP in CB17 SCID mice. Tumors grown from WM239A cells expressing palmitoylation-resistant C590G MCAM-GFP or WT MCAM-GFP stained positively for endogenous MCAM and contained regions of MCAM-GFP-expressing cells within the tumor (Fig. 5, A and B). The WT tumors had compact borders with very few areas where cells had separated from the primary tumor (Fig. 5A). In contrast to the WT tumors, the C590G MCAM-GFP-expressing tumors contained regions where melanoma cells appeared to detach from the primary tumor and invaded the adjacent tissue (Fig. 5B, arrows). Quantification of tumors performed blindly confirmed that mice with the C590G tumors had more MCAM-GFP-expressing cells in the adjacent tissue (78.0 ± 14.2% of fields imaged) compared with the WT MCAM-GFP-expressing tumors (41.6 ± 12.0% of fields imaged; Student's t test, p = 0.006) (Fig. 5C). This difference was not attributable to increased tumor growth because no appreciable difference in tumor volume between the C590G MCAM-GFP- and WT MCAM-GFP-expressing tumors was observed (406.9 ± 116.6 versus 489.5 ± 46.3 mm3 (S.D.); Student's t test, p = 0.84) (Fig. 5D). The growth rates of WT and C590G MCAM-GFP-expressing cells in two dimensions were also the same (Fig. 5E). Tumor F0 was removed from the data set because of the unusual shape and size relative to the other tumors (975.00 mm3, flat and multilobed). These results, together with the increased invasion in collagen, suggest that the palmitoylation state of MCAM promotes more invasive cell behavior.
FIGURE 5.
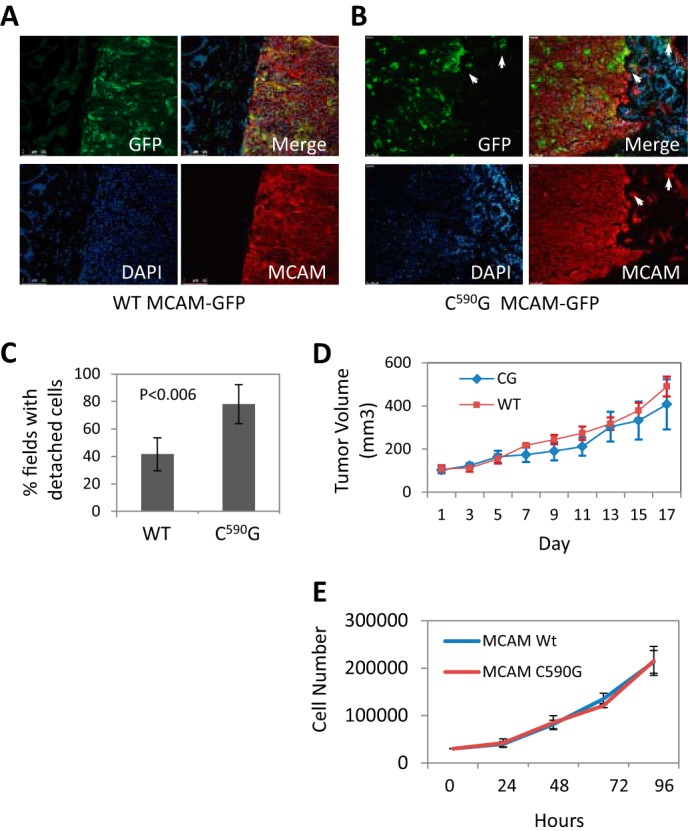
Blocking MCAM palmitoylation promotes melanoma invasion in vivo. A, xenograft tumors stained for endogenous MCAM (red) and WT MCAM-GFP (green). B, C590G MCAM-GFP-expressing tumors have irregular borders with C590G MCAM-GFP-expressing cells (green) detached (arrows) from the primary tumor residing in the adjacent mouse tissue (DAPI, blue). C, quantitation of detached tumor cells (average fields/tumor). Error bars, S.D. p value determined by Student's t test. D, xenograft tumor volumes, measured when first palpable (day 1). Error bars, S.D. E, growth rate of WT and C590G MCAM-GFP-expressing cells in two-dimensional culture.
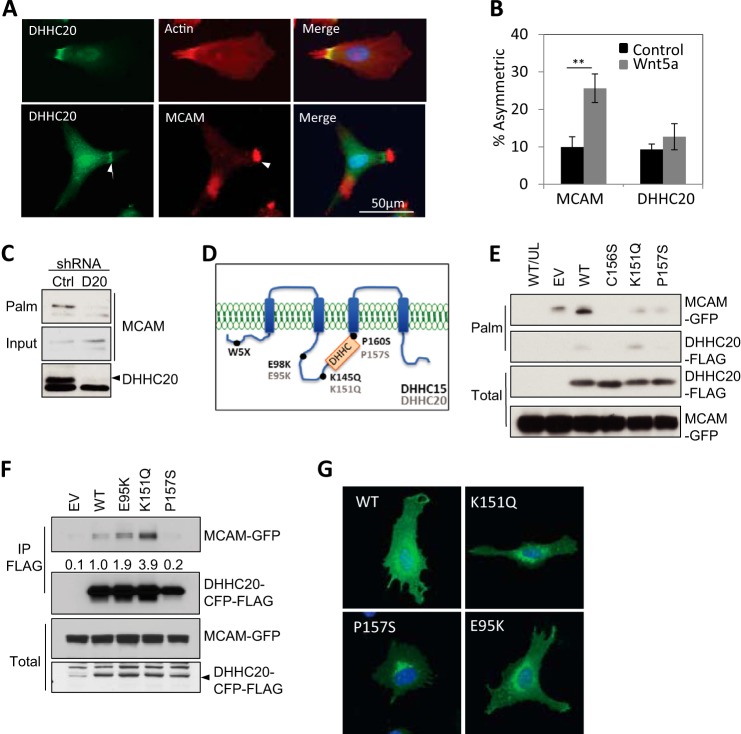
Palmitoyltransferase DHHC20 Is Asymmetrically Localized
A previous proteomic study of WM239A cells identified only two palmitoyltransferases, DHHC20 and DHHC5 (10). We chose to focus on DHHC20, because only one unique peptide was identified for DHHC5, whereas six peptides were identified for DHHC20. DHHC20 has also been shown to promote a transformation-like phenotype when exogenously expressed in NIH3T3 cells (33). Indirect immunofluorescence showed that endogenous DHHC20 protein is asymmetrically localized, overlapping with a filamentous actin (F-actin) structure in 9.3% of the cells (Fig. 6, A and B). This staining was abolished by expression of DHHC20 shRNA (data not shown). Cells were co-stained for both MCAM and DHHC20, and of the cells with asymmetric MCAM (9.9%), approximately half also display an adjacent but non-overlapping asymmetric pattern of DHHC20 (5.7%) (Fig. 6, A and B). In contrast to MCAM asymmetry, the asymmetric localization of DHHC20 is independent of Wnt5a treatment (Fig. 6, A and B).
FIGURE 6.
Cancer-associated mutations inhibit DHHC20 function. A, endogenous DHHC20, F-actin (arrows, top panels), and MCAM (bottom panels) localization. B, asymmetric DHHC20 localization is independent of Wnt5a (n > 300 cells/experimental condition; Student's t test; **, p < 0.005). Error bars, S.D. C, levels of palmitoylated MCAM decrease in WM239A cells expressing DHHC20 shRNA compared with control shRNA. D, positions of DHHC15 mutations (black) and analogous amino acids in DHHC20 (gray). E, HEK293T cells expressing DHHC20 shRNA were transfected with MCAM-GFP and wild type (WT) DHHC20 or mutants C156S, K151Q, and P157S that are shRNA-resistant. Cells were labeled with palmitic acid azide. Empty vector (EV) and unlabeled wild type cells (UL/WT). F, immunoprecipitation of wild type and mutant DHHC20-CFP-FLAG probed with anti-MCAM. G, representative cells expressing YFP-tagged DHHC20 wild type and point mutants in WM239A melanoma cells.
Loss of DHHC20 Reduces MCAM Palmitoylation
To determine whether DHHC20 palmitoylates MCAM, DHHC20 expression was inhibited by shRNA. Immunoblotting detected two bands at the approximate molecular weight of DHHC20, but the higher molecular weight band was effectively reduced by DHHC20 shRNA (Fig. 6C). When palmitoylated MCAM was measured in control shRNA- and DHHC20 shRNA-expressing cells, the level of palmitoylate MCAM was greatly reduced (Fig. 6C).
We next asked whether the protein palmitoylation pathway might be altered in cancer. Published exome sequencing data sets of human tumor samples were searched to identify mutations in MCAM or the DHHC family of palmitoyltransferases. Mutations in MCAM cysteine 590 were not identified in melanoma. However, mutations in a palmitoyltransferase DHHC15 were found in two of eight melanoma samples (K145Q and Trp-5 stop (W5X)) in one study and two additional mutations (E98K in melanoma and P160S in ovarian cancer) in two independent studies (Table 1) (34–36). Although DHHC15 mRNA was not detected by quantitative PCR in WM239A melanoma cells, DHHC20 is 55% identical to the DHHC15 and contains amino acids analogous to those mutated in DHHC15, allowing us to test the effect of the mutations on melanoma behavior (Fig. 6D and Table 1).
TABLE 1.
DHHC protein mutations identified in tumor samples
| Tumor | ZDHHC15 mutation | ZDHHC20 amino acid | Reference |
|---|---|---|---|
| Metastatic melanoma | E98K | E95K | Ref. 35 |
| Metastatic melanoma | K145Q | K151Q | Ref. 34 |
| Metastatic melanoma | W5Xa | NA | Ref. 34 |
| Ovarian carcinoma | P160S | P157S | Ref. 36 |
a Early stop mutation.
We first asked whether DHHC20 can palmitoylate MCAM. Ectopic expression of DHHC20 in HEK293T cells increased the levels of MCAM-GFP palmitoylation by 2-fold compared with cells transfected with the empty vector alone (Fig. 6E). Expression of DHHC20 harboring the mutations K151Q and P157S reduced MCAM palmitoylation below the levels in cells transfected with empty vector (Fig. 6E). However, the K151Q and P157S mutations did not reduce the levels of palmitoylated MCAM down to the levels of the catalytic mutation in the DHHC domain C156S required for enzymatic activity (Fig. 6E) (37).
Co-immunoprecipitation of the DHHC20-CFP-FLAG E95K and K151Q mutants showed increased interaction with MCAM-GFP by 2- and 4-fold, respectively, relative to WT DHHC20-CFP-FLAG (Fig. 6F). The increased interaction with MCAM could also contribute to inhibition of transferase activity by decreasing substrate release.
The expression of the DHHC20 mutations was examined to make sure the differences in function were not due to defects in expression. We found that the expression and localization of the DHHC20 mutant proteins were indistinguishable from those of the WT fusion protein (Fig. 6G). The expression is also consistent with the expression of a DHHC20-GFP fusion protein reported by others (38).
Palmitoyltransferase DHHC20 Inhibits Cell Invasion
If reduced protein palmitoylation promotes cell invasion, we would expect inhibition of DHHC20 to also increase cell invasion. Cells expressing DHHC20 shRNA invaded farther into collagen (339.4 ± 26.9 μm S.D.) compared with nonspecific shRNA control expressing cells (82.6 ± 22.5 μm (S.D.); one-way ANOVA with Tukey's multiple comparison test, p < 0.0001) (Fig. 7, A and B). Expression of shRNA-resistant DHHC20 partially rescued the increased invasion of the DHHC20 shRNA-expressing cells (211.0 ± 38.2 μm (S.D)) compared with rescue with an empty vector control (329.7 ± 70.5 μm (S.D.); one-way ANOVA with Tukey's multiple comparison test, p < 0.0001) (Fig. 7, A and B).
FIGURE 7.
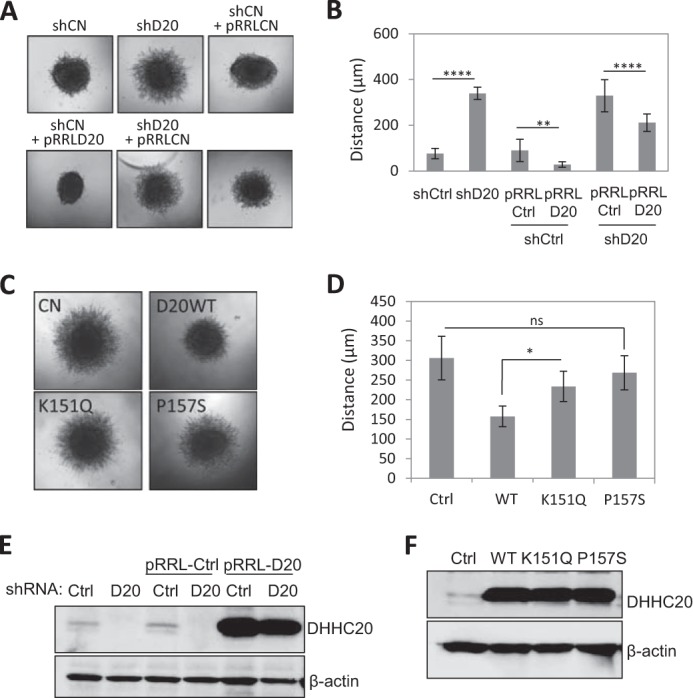
DHHC20 inhibits cell invasion. A, inhibition of DHHC20 increases cell invasion in three-dimensional collagen compared with control shRNA. Spheroid invasion was quantified 8 days following transfer into collagen. B, quantification of spheroids in A. Distance was measured from the edge of the core to the invasion front (shCtrl (n = 10), shDHHC20 (n = 11), shCtrl + pRRLCtrl (n = 12), shCtrl + pRRLD20 (n = 12), shDHHC20 + pRRLCtrl (n = 9), and shDHHC20 + pRRLD20 (n = 8)). Statistical analysis was performed using a one-way ANOVA with Tukey's multiple-comparison test (**, p < 0.01; ****, p < 0.0001). pRRLCtrl is a lentiviral vector expressing GFP, and pRRLD20 is a lentiviral vector expressing DHHC20 that is resistant to shRNA-mediated silencing. C, spheroid invasion assays of C590G MCAM-GFP-expressing cells expressing control pRRL-vector alone (Ctrl), WT, K151Q, or P157S mutant DHHC20 (D20). D, quantification of spheroids in C (*, p = 0.012; ns, p = 0.369). p values were determined using one-way ANOVA with Tukey's multiple-comparison test. Error bars, S.D. E, Western blot showing expression levels of DHHC20 in the cell lines used in A. F, Western blot showing expression levels of WT DHHC20 and the mutant forms of DHHC20 in the cell lines used in C.
Finally, we asked whether overexpression of DHHC20 is sufficient to suppress the increased invasion mediated by C590G MCAM-GFP expression. Expression of WT DHHC20 in C590G MCAM-GFP-expressing cells decreased invasion in collagen (157.6 ± 26.2 μm (S.D.)) compared with control vector-expressing cells (305.9 ± 55.2 μm (S.D.)) (Fig. 7, C and D). This indicates that there are probably other targets of DHHC20 that suppress cell invasion when palmitoylated in addition to MCAM. Expression of DHHC20 harboring the mutation P157S was unable to significantly decrease invasion compared with control cells (268.7 ± 43.4 versus 305.9 ± 55.2 μm (S.D.); one-way ANOVA with Tukey's multiple comparison test, p = 0.369). Similarly, cells expressing the K151Q mutation also display significantly higher levels of invasion than WT DHHC20-expressing cells (233.8 ± 38.5 versus 157.6 ± 26.2 μm (S.D.); one-way ANOVA with Tukey's multiple comparison test, p = 0.012) (Fig. 7, C and D). The levels of DHHC20 expression in the cell lines are shown (Fig. 7, E and F). These results demonstrate that the mutations found in human tumor samples that reduce enzyme function also impair the ability of DHHC20 to suppress cell invasion.
Discussion
Our results reveal that Wnt5a signaling rapidly induces depalmitoylation of cell adhesion molecules MCAM and CD44. Wnt5a induced MCAM asymmetry, and increased collagen invasion was blocked by the APT1 inhibitor, palmostatin B. Furthermore, a single point mutation that blocks MCAM palmitoylation is sufficient to phenocopy Wnt5a-induced asymmetric MCAM localization and promote invasive cell behavior in melanoma cells in vitro and in vivo. Although another function of cysteine 590 cannot be ruled out, the fact that inhibition of APT1 blocks asymmetric MCAM localization supports this interpretation. These results implicate the palmitoylation pathway in regulating invasive cell behavior.
Decreasing protein palmitoylation by overexpressing APT1 is sufficient to increase cell invasion, and conversely inhibition of APT1 by shRNA inhibits cell invasion. It is not known whether APT1 activity has specificity for certain protein substrates, and it is possible that overexpression of APT1 may function to decrease the global levels of palmitoylated proteins, including MCAM and CD44, causing either decreased cell adhesion or increased cell motility. Although the palmitoylation-resistant MCAM mutant C590G does increase melanoma cell invasion, overexpression of DHHC20 is sufficient to suppress this effect. This would suggest that depalmitoylation of MCAM alone is not necessarily sufficient to promote cell invasion. Therefore, it is not unexpected that a mutation in the palmitoylated cysteine in MCAM has not yet been identified in tumor samples.
Our studies reveal alternative mechanisms that could lead to decreased palmitoylation of MCAM, such as loss of function mutations in palmitoyltransferases found in human tumor samples. These mutations attenuate the enzyme's ability to palmitoylate MCAM and the ability to suppress invasion of the C590G MCAM-expressing cells. It should be noted that in cells expressing the mutant forms of DHHC20, the level of MCAM-GFP palmitoylation was lower than the levels of empty vector control cells. This could be caused by a dominant negative effect of DHHC proteins described previously by others (17, 39, 40). Palmitoyltransferases use a two-step catalytic process involving autopalmitoylation followed by transfer of palmitate to the substrate (41, 42). We observed a notable increase in DHHC20 palmitoylation in the K151Q mutation compared with WT that could be caused by a block in transfer of palmitate from DHHC20 to MCAM (Fig. 6E).
It will be important to understand how Wnt5a signals to the palmitoylation machinery. The downstream Wnt5a signaling events that increase protein depalmitoylation involve the Wnt adaptor protein Dvl2. Inhibition of Dvl2 by siRNA causes a marked reduction in palmitoylated MCAM in melanoma cells. These results suggest that Wnt5a may modulate APT1 function through the disruption of an inhibitory interaction with Dvl2. Although ectopic expression of APT1 is sufficient to increase cell invasion, it did not increase polarized MCAM localization (data not shown). Therefore, we hypothesize that Wnt5a signaling may regulate APT1 activity spatially through interaction with Dvl2, causing spatial changes in MCAM palmitoylation. Alternatively, Wnt5a could signal through DHHC20 and alter its activity. However, we have yet to see any obvious changes in DHHC20 protein level or interaction with MCAM in the presence or absence of Wnt5a. Future studies will examine changes in DHHC20 activity in response to Wnt5a.
Wnt5a treatment appears to promote localization of MCAM into membrane domains that are separate from caveolin-containing membrane regions. A similar localization was observed when MCAM palmitoylation was blocked with the cysteine mutation. This partitioning of MCAM into membrane subdomains could play a role in promoting the polarized MCAM localization. The increase in MCAM self-interaction induced by inhibiting MCAM palmitoylation could further facilitate the assembly of MCAM into asymmetric membrane domains.
Supplementary Material
Note Added in Proof
Supplemental Movie 4 showing asymmetric co-localization of wild type MCAM-YFP (red) and wild type MCAM-CFP (green) in Wnt5a-treated WM239A melanoma cells was inadvertently omitted from the version of this article that was published on May 5, 2015 as a Paper in Press. The supplemental movie is now available online.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-CA-181633-01A1 and the Melanoma Research Foundation. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains Movies S1–S4.
- WRAMP
- Wnt5a-induced receptor, actin, and myosin polarity
- MCAM
- melanoma cell adhesion molecule(s)
- ABE
- acyl-biotinyl exchange
- HAM
- hydroxylamine
- ANOVA
- analysis of variance
- Cav-1
- caveolin-1.
References
- 1. Bittner M., Meltzer P., Chen Y., Jiang Y., Seftor E., Hendrix M., Radmacher M., Simon R., Yakhini Z., Ben-Dor A., Sampas N., Dougherty E., Wang E., Marincola F., Gooden C., Lueders J., Glatfelter A., Pollock P., Carpten J., Gillanders E., Leja D., Dietrich K., Beaudry C., Berens M., Alberts D., Sondak V. (2000) Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature 406, 536–540 [DOI] [PubMed] [Google Scholar]
- 2. Weeraratna A. T., Jiang Y., Hostetter G., Rosenblatt K., Duray P., Bittner M., Trent J. M. (2002) Wnt5a signaling directly affects cell motility and invasion of metastatic melanoma. Cancer Cell 1, 279–288 [DOI] [PubMed] [Google Scholar]
- 3. Saitoh T., Mine T., Katoh M. (2002) Frequent up-regulation of WNT5A mRNA in primary gastric cancer. Int. J. Mol. Med. 9, 515–519 [PubMed] [Google Scholar]
- 4. Kurayoshi M., Oue N., Yamamoto H., Kishida M., Inoue A., Asahara T., Yasui W., Kikuchi A. (2006) Expression of Wnt-5a is correlated with aggressiveness of gastric cancer by stimulating cell migration and invasion. Cancer Res. 66, 10439–10448 [DOI] [PubMed] [Google Scholar]
- 5. Bakker E. R., Das A. M., Helvensteijn W., Franken P. F., Swagemakers S., van der Valk M. A., ten Hagen T. L., Kuipers E. J., van Veelen W., Smits R. (2013) Wnt5a promotes human colon cancer cell migration and invasion but does not augment intestinal tumorigenesis in Apc1638N mice. Carcinogenesis 34, 2629–2638 [DOI] [PubMed] [Google Scholar]
- 6. O'Connell M. P., Fiori J. L., Xu M., Carter A. D., Frank B. P., Camilli T. C., French A. D., Dissanayake S. K., Indig F. E., Bernier M., Taub D. D., Hewitt S. M., Weeraratna A. T. (2010) The orphan tyrosine kinase receptor, ROR2, mediates Wnt5A signaling in metastatic melanoma. Oncogene 29, 34–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nomachi A., Nishita M., Inaba D., Enomoto M., Hamasaki M., Minami Y. (2008) Receptor tyrosine kinase Ror2 mediates Wnt5a-induced polarized cell migration by activating c-Jun N-terminal kinase via actin-binding protein filamin A. J. Biol. Chem. 283, 27973–27981 [DOI] [PubMed] [Google Scholar]
- 8. Lin S., Baye L. M., Westfall T. A., Slusarski D. C. (2010) Wnt5b-Ryk pathway provides directional signals to regulate gastrulation movement. J. Cell Biol. 190, 263–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Witze E. S., Litman E. S., Argast G. M., Moon R. T., Ahn N. G. (2008) Wnt5a control of cell polarity and directional movement by polarized redistribution of adhesion receptors. Science 320, 365–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Witze E. S., Connacher M. K., Houel S., Schwartz M. P., Morphew M. K., Reid L., Sacks D. B., Anseth K. S., Ahn N. G. (2013) Wnt5a directs polarized calcium gradients by recruiting cortical endoplasmic reticulum to the cell trailing edge. Dev Cell. 26, 645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Satyamoorthy K., Muyrers J., Meier F., Patel D., Herlyn M. (2001) Mel-CAM-specific genetic suppressor elements inhibit melanoma growth and invasion through loss of gap junctional communication. Oncogene 20, 4676–4684 [DOI] [PubMed] [Google Scholar]
- 12. Conibear E., Davis N. G. (2010) Palmitoylation and depalmitoylation dynamics at a glance. J. Cell Sci. 123, 4007–4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Abrami L., Kunz B., Iacovache I., van der Goot F. G. (2008) Palmitoylation and ubiquitination regulate exit of the Wnt signaling protein LRP6 from the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 105, 5384–5389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guo Y. J., Lin S. C., Wang J. H., Bigby M., Sy M. S. (1994) Palmitoylation of CD44 interferes with CD3-mediated signaling in human T lymphocytes. Int. Immunol. 6, 213–221 [DOI] [PubMed] [Google Scholar]
- 15. Yang X., Kovalenko O. V., Tang W., Claas C., Stipp C. S., Hemler M. E. (2004) Palmitoylation supports assembly and function of integrin-tetraspanin complexes. J. Cell Biol. 167, 1231–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fukata M., Fukata Y., Adesnik H., Nicoll R. A., Bredt D. S. (2004) Identification of PSD-95 palmitoylating enzymes. Neuron 44, 987–996 [DOI] [PubMed] [Google Scholar]
- 17. Yeh D. C., Duncan J. A., Yamashita S., Michel T. (1999) Depalmitoylation of endothelial nitric-oxide synthase by acyl-protein thioesterase 1 is potentiated by Ca2+-calmodulin. J. Biol. Chem. 274, 33148–33154 [DOI] [PubMed] [Google Scholar]
- 18. Kong E., Peng S., Chandra G., Sarkar C., Zhang Z., Bagh M. B., Mukherjee A. B. (2013) Dynamic palmitoylation links cytosol-membrane shuttling of acyl-protein thioesterase-1 and acyl-protein thioesterase-2 with that of proto-oncogene H-ras product and growth-associated protein-43. J. Biol. Chem. 288, 9112–9125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. El-Husseini Ael-D, Schnell E., Dakoji S., Sweeney N., Zhou Q., Prange O., Gauthier-Campbell C., Aguilera-Moreno A., Nicoll R. A., Bredt D. S. (2002) Synaptic strength regulated by palmitate cycling on PSD-95. Cell 108, 849–863 [DOI] [PubMed] [Google Scholar]
- 20. Levental I., Grzybek M., Simons K. (2010) Greasing their way: lipid modifications determine protein association with membrane rafts. Biochemistry 49, 6305–6316 [DOI] [PubMed] [Google Scholar]
- 21. Hundt M., Tabata H., Jeon M. S., Hayashi K., Tanaka Y., Krishna R., De Giorgio L., Liu Y. C., Fukata M., Altman A. (2006) Impaired activation and localization of LAT in anergic T cells as a consequence of a selective palmitoylation defect. Immunity 24, 513–522 [DOI] [PubMed] [Google Scholar]
- 22. Young F. B., Butland S. L., Sanders S. S., Sutton L. M., Hayden M. R. (2012) Putting proteins in their place: palmitoylation in Huntington disease and other neuropsychiatric diseases. Prog. Neurobiol. 97, 220–238 [DOI] [PubMed] [Google Scholar]
- 23. Hancock J. F., Magee A. I., Childs J. E., Marshall C. J. (1989) All ras proteins are polyisoprenylated but only some are palmitoylated. Cell 57, 1167–1177 [DOI] [PubMed] [Google Scholar]
- 24. Eisenberg S., Laude A. J., Beckett A. J., Mageean C. J., Aran V., Hernandez-Valladares M., Henis Y. I., Prior I. A. (2013) The role of palmitoylation in regulating Ras localization and function. Biochem. Soc. Trans. 41, 79–83 [DOI] [PubMed] [Google Scholar]
- 25. Willert K., Brown J. D., Danenberg E., Duncan A. W., Weissman I. L., Reya T., Yates J. R., 3rd, Nusse R. (2003) Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature 423, 448–452 [DOI] [PubMed] [Google Scholar]
- 26. Wan J, Roth AF, Bailey AO, Davis NG. (2007) Palmitoylated proteins: purification and identification. Nat. Protoc. 2, 1573–1584 [DOI] [PubMed] [Google Scholar]
- 27. Shaul P. W., Smart E. J., Robinson L. J., German Z., Yuhanna I. S., Ying Y., Anderson R. G., Michel T. (1996) Acylation targets endothelial nitric-oxide synthase to plasmalemmal caveolae. J. Biol. Chem. 271, 6518–6522 [DOI] [PubMed] [Google Scholar]
- 28. Kang R., Wan J., Arstikaitis P., Takahashi H., Huang K., Bailey A. O., Thompson J. X., Roth A. F., Drisdel R. C., Mastro R., Green W. N., Yates J. R., 3rd, Davis N. G., El-Husseini A. (2008) Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature 456, 904–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dekker F. J., Rocks O., Vartak N., Menninger S., Hedberg C., Balamurugan R., Wetzel S., Renner S., Gerauer M., Schölermann B., Rusch M., Kramer J. W., Rauh D., Coates G. W., Brunsveld L., Bastiaens P. I., Waldmann H. (2010) Small-molecule inhibition of APT1 affects Ras localization and signaling. Nat. Chem. Biol. 6, 449–456 [DOI] [PubMed] [Google Scholar]
- 30. Gao C., Chen Y. G. (2010) Dishevelled: The hub of Wnt signaling. Cell. Signal. 22, 717–727 [DOI] [PubMed] [Google Scholar]
- 31. Ho H. Y., Susman M. W., Bikoff J. B., Ryu Y. K., Jonas A. M., Hu L., Kuruvilla R., Greenberg M. E. (2012) Wnt5a-Ror-Dishevelled signaling constitutes a core developmental pathway that controls tissue morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 109, 4044–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smalley K. S., Haass N. K., Brafford P. A., Lioni M., Flaherty K. T., Herlyn M. (2006) Multiple signaling pathways must be targeted to overcome drug resistance in cell lines derived from melanoma metastases. Mol. Cancer Ther. 5, 1136–1144 [DOI] [PubMed] [Google Scholar]
- 33. Draper J. M., Smith C. D. (2010) DHHC20: a human palmitoyl acyltransferase that causes cellular transformation. Mol. Membr. Biol. 27, 123–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stark M. S., Woods S. L., Gartside M. G., Bonazzi V. F., Dutton-Regester K., Aoude L. G., Chow D., Sereduk C., Niemi N. M., Tang N., Ellis J. J., Reid J., Zismann V., Tyagi S., Muzny D., Newsham I., Wu Y., Palmer J. M., Pollak T., Youngkin D., Brooks B. R., Lanagan C., Schmidt C. W., Kobe B., MacKeigan J. P., Yin H., Brown K. M., Gibbs R., Trent J., Hayward N. K. (2012) Frequent somatic MAP3K5 and MAP3K9 mutations in metastatic melanoma identified by exome sequencing. Nat. Genet. 44, 165–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Berger M. F., Hodis E., Heffernan T. P., Deribe Y. L., Lawrence M. S., Protopopov A., Ivanova E., Watson I. R., Nickerson E., Ghosh P., Zhang H., Zeid R., Ren X., Cibulskis K., Sivachenko A. Y., Wagle N., Sucker A., Sougnez C., Onofrio R., Ambrogio L., Auclair D., Fennell T., Carter S. L., Drier Y., Stojanov P., Singer M. A., Voet D., Jing R., Saksena G., Barretina J., Ramos A. H., Pugh T. J., Stransky N., Parkin M., Winckler W., Mahan S., Ardlie K., Baldwin J., Wargo J., Schadendorf D., Meyerson M., Gabriel S. B., Golub T. R., Wagner S. N., Lander E. S., Getz G., Chin L., Garraway L. A. (2012) Melanoma genome sequencing reveals frequent PREX2 mutations. Nature 485, 502–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cancer Genome Atlas Research Network (2011) Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Roth A. F., Feng Y., Chen L., Davis N. G. (2002) The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase J. Cell Biol. 159, 23–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ohno Y., Kihara A., Sano T., Igarashi Y. (2006) Intracellular localization and tissue-specific distribution of human and yeast DHHC cysteine-rich domain-containing proteins. Biochim. Biophys. Acta 176, 1474–1483 [DOI] [PubMed] [Google Scholar]
- 39. Fang C., Deng L., Keller C. A., Fukata M., Fukata Y., Chen G., Lüscher B. (2006) GODZ-mediated palmitoylation of GABA(A) receptors is required for normal assembly and function of GABAergic inhibitory synapses. J. Neurosci. 26, 12758–12768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Noritake J., Fukata Y., Iwanaga T., Hosomi N., Tsutsumi R., Matsuda N., Tani H., Iwanari H., Mochizuki Y., Kodama T., Matsuura Y., Bredt D. S., Hamakubo T., Fukata M. (2009) Mobile DHHC palmitoylating enzyme mediates activity-sensitive synaptic targeting of PSD-95. J. Cell Biol. 186, 147–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mitchell D. A., Mitchell G., Ling Y., Budde C., Deschenes R. J. (2010) Mutational analysis of Saccharomyces cerevisiae Erf2 reveals a two-step reaction mechanism for protein palmitoylation by DHHC enzymes. J. Biol. Chem. 285, 38104–38114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jennings B. C., Linder M. E. (2012) DHHC protein S-acyltransferases use similar ping-pong kinetic mechanisms but display different acyl-CoA specificities. J. Biol. Chem. 287, 7236–7245 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.