Background: Translational regulation might underlie the high expression levels of the protease cathepsin L (CTSL) associated with poor breast cancer prognosis.
Results: Translation of CTSL mRNA is highly stress-resistant and promotes metastasis of murine breast cancer.
Conclusion: CTSL mRNA circumvents translational shutdown in cancer-associated stress conditions.
Significance: High expression of a metastasis promoting protease is maintained by translational regulation.
Keywords: alternative splicing, breast cancer, cysteine protease, metastasis, translation control
Abstract
The cysteine protease cathepsin L (CTSL) is often thought to act as a tumor promoter by enhancing tumor progression and metastasis. This goes along with increased CTSL activity in various tumor entities; however, the mechanisms leading to high CTSL levels are incompletely understood. With the help of the polyoma middle T oncogene driven breast cancer mouse model expressing a human CTSL genomic transgene, we show that CTSL indeed promotes breast cancer metastasis to the lung. During tumor formation and progression high expression levels of CTSL are maintained by enduring translation of CTSL mRNA. Interestingly, human breast cancer specimens expressed the same pattern of 5′ untranslated region (UTR) splice variants as the transgenic mice and the human cancer cell line MDA-MB 321. By polyribosome profiling of tumor tissues and human breast cancer cells, we observe an intrinsic resistance of CTSL to stress-induced shutdown of translation. This ability can be attributed to all 5′ UTR variants of CTSL and is not dependent on a previously described internal ribosomal entry site motif. In conclusion, we provide in vivo functional evidence for overexpressed CTSL as a promoter of lung metastasis, whereas high CTSL levels are maintained during tumor progression due to stress-resistant mRNA translation.
Introduction
One hallmark of tumor progression is the switch from benign hyperplasia to invasive growth and subsequent formation of metastases. Several steps to invasive tumor growth are facilitated by proteolysis (1). Among other protease families lysosomal cysteine cathepsins, especially cathepsin B and L, are often found to be highly expressed in aggressive tumors (2–4). Tumor mouse models for pancreatic islet carcinoma showed retarded tumor growth upon cathepsin B or L deficiency (5). Furthermore, high activity of these proteases positively correlates with poor clinical outcome in several cancer entities. Especially the prognostic value of high cathepsin L (CTSL)2 activity in breast cancer is appreciated (6–9).
Gain of CTSL activity might be explained on several levels of regulation. First, CTSL mRNA can be elevated in tumor cells either by enhanced transcription as observed in ErbB2 positive breast cancer (10) or by epigenetic regulation (11, 12). Second, CTSL protein stability and activity may be modulated by protein turnover and expression of endogenous cathepsin inhibitors (13). Interestingly, regulation on the level of mRNA translation into protein has been brought up as another aspect of CTSL regulation. The CTSL transcript is produced in different 5′ UTR variants and it has been proposed that these variants differ in their efficiency to be translated into protein. Most of them vary in their length of the 5′ UTR caused by different splice acceptor sites of exon 1 that are joined to the 5′ end of exon 2 as depicted in Fig. 3A (14, 15). The CTSL open reading frame starts in exon 2 so all splice variants encode for the same functional protein. In previous studies contradictory findings about translation efficiencies have been reported. Some reports assign the highest translation efficiency to the shortest variant (15), whereas others state that the longest variant is favored (16). Only some of the previous reports take into account that translation has to be assessed upon conditions that prevail within the cancer tissue.
FIGURE 3.
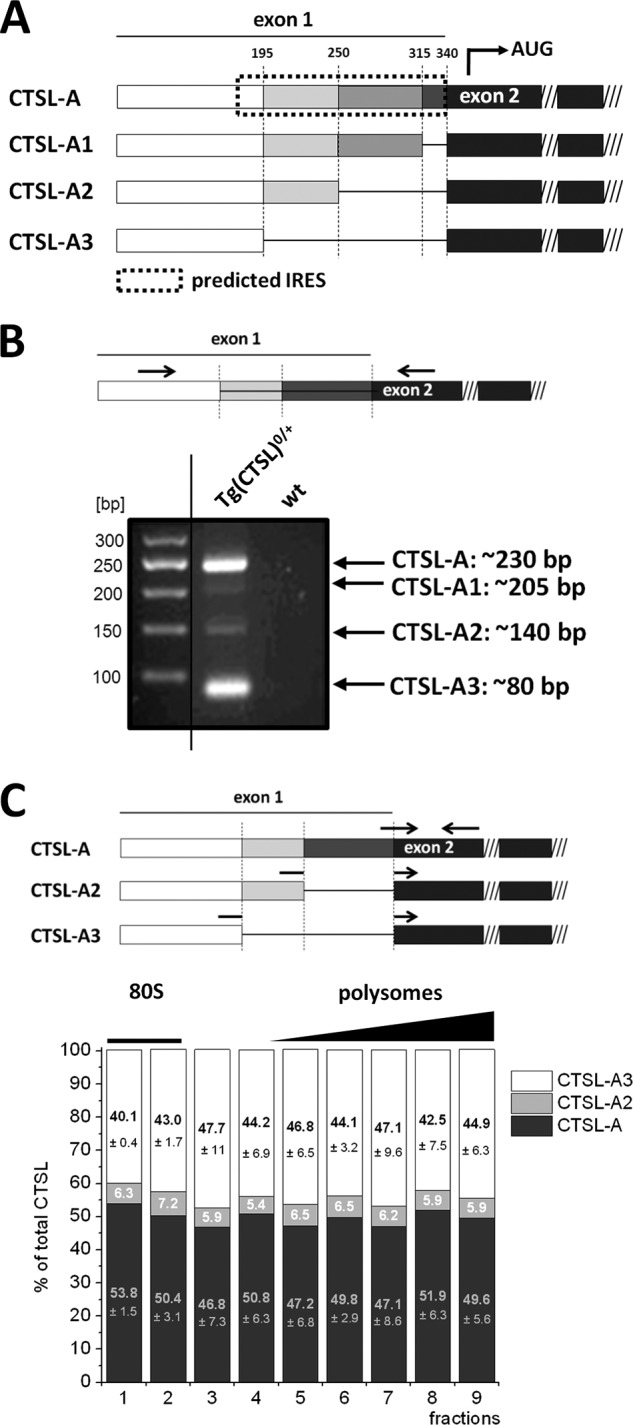
Polyribosome profiling of CTSL 5′ UTR splice variants in murine breast cancer. A, schematic overview of the 5′ UTR splice variants of CTSL. The complete predicted IRES motif is only present in the longest variant A. B, qualitative detection of CTSL splice variants in tumor tissue of PyMT-Tg(CTSL)0/+ mice. RT-PCR with primers that span exons 1 and 2 (shown in scheme) to detect the 5′ UTR splice variants of CTSL. Analysis of RT-PCR shows amplification of CTSL mRNA splice variants of the expected length. C, distribution of 5′ UTR splice variants on the polyribosome gradient. Splice site-specific primers allow for quantification of 5′ UTR splice variants (shown in scheme). Ratios of the different splice variants in fractions of the polyribosome gradient are shown (n = 3, mean ± S.E.).
Tumor-associated stress conditions coin the state of translation as described in the following. Especially in rather large solid tumors, e.g. tumors of the mammary gland, cellular stress due to reduced oxygen and nutrient supply is common. It is known that such conditions cause a general decline in translation of mRNAs into protein (17). Translation is enabled and regulated by at least 12 eukaryotic translation initiation factors (eIFs) (18). Under stress conditions a general shutdown of translation is mediated by reduced phosphorylation of eIF2α, which abrogates formation of the pre-initiation complex of the 40S ribosomal subunit, the initiating methionyl tRNA, and eIFs. Under normal conditions this complex is recruited to the 5′ cap of the mRNA. Stress signaling interferes in this process by activation of 4E-BP, a factor that hinders cap recognition. The key pathway to mediate translational shutdown is the mTOR pathway (19). Active mTOR inactivates 4E-BP by phosphorylation and keeps up activity of other eIFs to maintain cap-dependent translation. Consequently, pharmacological inhibition of mTOR by rapamycin or Torin-1 is a way to induce translational shutdown. Under such conditions mechanisms of cap-independent translation come into play. This can be facilitated by the use of internal ribosomal entry sites (IRES), a concept known from viral polycistronic mRNAs. Several eukaryotic mRNAs encoding for proteins that are essential for survival of the cell contain potential IRES domains in their 5′ UTR (20). The longest CTSL splice variant is thought to form an IRES structure that enables favored translation under stress conditions (21). The basic functionality of the IRES structure has been shown by experiments with bicistronic reporter vectors (16, 21). However, the functionality and actual impact of IRES structures on cellular mRNAs is still under debate (22–24).
In this study we address if one of the CTSL splice variants does indeed represent a stress-resistant source for CTSL in tumor tissue. Similar to previous reports we observed a discrepancy between CTSL mRNA and protein levels. However, to investigate whether this phenomenon is due to increased CTSL translation we choose a different approach than previous studies. Polyribosome profiling allowed us to analyze efficiency of translation of single splice variants transcribed from the genuine gene locus. We observed that all CTSL splice variants were recruited to the polyribosome with high efficiency in a stress-resistant manner. This stress resistance was further confirmed by expression of single splice variants under hypoxia as well as mTOR inhibition. The circumvention of translational shutdown might be due to escape from translationally silent mRNA accumulations like stress granules or P-bodies rather than the predominant use of an IRES structure. Furthermore, expression of a human genomic CTSL transgene in the MMTV-PyMT mouse model of metastasizing breast cancer revealed increased metastasis, which might be fostered by the stress resistance of CTSL biosynthesis.
Experimental Procedures
Mice
FVB/N mice harboring the genomic human cathepsin L construct (Tg(CTSL)+/0) (25) were crossed with the transgenic mouse strain FVB/N-TgN(MMTV-PyMT)634-Mul/J (MMTV-PyMT) (26). Mouse work in this study was performed in accordance to the German law for animal protection (Tierschutzgesetz) as published on May 25, 1998 (ethics approval G-07/26 regional council Freiburg).
Patient Material
Patient material was obtained and worked with according to guidelines set by the Ethics Committee Freiburg (ethics approval 324/09_120807).
Tumor Phenotype
MMTV-PyMT mice were sacrificed at 10 or 14 weeks, and tumor tissue was resected from all mammary glands to measure the total tumor weight. Histopathological grading of hematoxylin and eosin (HE)-stained tumor sections of left thoracic mammary glands was performed in a blinded setting by an experienced histopathologist as described previously (27). For histomorphometric analysis, the area and number of metastatic foci on lung sections was measured using Axiovision LE 4.4 software (Zeiss, Oberkochen, Germany). For analysis of hypoxic areas, 1.5 mg of pimonidazole-HCl (Hypoxyprobe®) (Hypoxyprobe, Inc., Burlington, MA) was injected intraperitoneally 30 min prior to sacrifice of tumor bearing mice. Binding of pimonidazole was visualized on paraffin sections using a FITC-labeled α-hypoxyprobe antibody and subsequent usage of a POD-labeled α-FITC antibody (catalog number MAB045P; Millipore, Darmstadt, Germany). Peroxidase activity was detected by incubation with diaminobenzidine (Sigma). Adobe Photoshop software was used for moderate contrast enhancement.
Preparation of Protein Lysate
Fresh tissue samples (∼100 mg) were lysed in 1 ml of homogenization buffer (100 mm Na-acetate, 5 mm EDTA, 1 mm dithiothreitol, 0.05% Brij, pH 5.5, and protease inhibitor (Complete® inhibitor mixture, Roche, Basel, Switzerland)) using an Ultra-Turrax and debris was pellet at 1000 × g for 15 min at 4 °C. Whole cell lysates were prepared by on-plate lysis. Cells were washed with PBS and lysed by adding lysis buffer containing 50 mm Tris-HCl, pH 8, 250 mm NaCl, 2.5 mm EDTA, 2% Nonidet P-40, 0.1% SDS, 0.5% sodium deoxycholate and protease inhibitors (Complete® inhibitor mixture). Protein concentrations were determined by BCA assay (Thermo Scientific, Waltham, MA).
Immunoblotting
10–40 μg of lysate was loaded onto 12% SDS-polyacrylamide gels. After electrophoretic separation, proteins were transferred on polyvinylidene fluoride membranes by semidry blotting (Bio-Rad). After blocking with 3% milk powder in PBS with 0.1% Tween, the membranes were exposed to the primary antibodies (biotinylated polyclonal goat α-human cathepsin L, 1:500 (catalog number BAF952; R&D Systems, Minneapolis, MN), polyclonal goat α-human/mouse/rat hypoxia inducible factor-1α, 1:500 (catalog number AF1935; R&D Systems), mouse α-mouse/human tubulin, 1:1000 (catalog number T6199; Sigma), mouse α-mouse actin, 1:1000 (catalog number 691001; MP Biomedicals, Solon, OH), polyclonal rabbit α-mouse histone H3, 1:1000 (catalog number ab1791; Abcam, Cambridge, UK), polyclonal rabbit α-human phospho-eIF2α (pSer52) (catalog number 44-728G; Invitrogen, Paisley, UK), monoclonal mouse α-human/mouse eIF2α (catalog number 2103; Cell Signaling, Leiden, The Netherlands) overnight at 4 °C. Membranes were washed and incubated for 2 h with the respective secondary antibody (goat α-mouse IgG-POD (catalog number A4416; Sigma) or goat α-rabbit IgG-POD (catalog number 5196-2504; Bio-Rad) or with a streptavidin-POD conjugate (Roche Applied Science). Membranes were washed and developed with the West Pico Chemiluminescent substrate (Pierce, Rockford, IL). Light emission was detected using the Fusion® SL Detection System (Peqlab, Erlangen, Germany).
Cell Culture and Stress Treatments
MDA-MB 231 cells (catalog number 300275; CLS, Eppelheim, Germany) and BT-474 cells (ATCC, Manassas, VA) were cultured in Dulbecco's modified Eagle's medium (Gibco/Invitrogen, Paisley, UK) supplemented with 10% fetal calf serum (PAN, Aidenbach, Germany), 1% penicillin/streptomycin, and 2 mm l-glutamine (both from Gibco/Invitrogen) in humidified air containing 5% CO2. For stress treatments 1 × 106 cells were seeded in a 100-mm dish and left overnight for cells to attach. For hypoxia cells were incubated in Anaerocult A® bags (Merck, Darmstadt, Germany) for 16 h. In these sealed bags a hypoxic milieu of <0.5% is created within the first 90 min of incubation. During the experiment maintenance of the hypoxic milieu was monitored using Anaerotest® strips (Merck). For mTOR inhibition equivalent cell numbers were treated with 250 nm Torin-1 (Tocris, Bristol, UK). For polyribosome gradient preparations ∼6 × 106 cells were seeded. In the case of cells carrying an inducible expression vector, CTSL expression was induced by treatment with 2 μg/ml of doxycycline (Sigma). For blockage of stress granules formation of cells were treated with 10 μm nocodazole (Sigma).
Cloning of Inducible Expression Vectors
The different CTSL transcript variants (CTSL-A: catalog number SC124186; CTSL-A3: SC100267; Origene, Rockville, MD) were amplified by PCR with mutagenesis primers, generating two new restriction sites at the ends of the PCR product. The purified PCR product was cloned into a lentiviral pTRIPZ shRNAmir vector (Thermo Fisher Scientific) via AgeI and MluI restriction sites. For cloning of the CTSL-CDS variant, primers for amplification of the insert were flanking the coding sequence only.
Preparation of PyMT Cells with Inducible CTSL Expression
Primary cells were prepared from tumors of MMTV-PyMT mice by mechanical disruption and enzymatic digestion of the tissue as previously described (27). Cells were spontaneously immortalized by repeated passaging for at least 25 passages. For the expression of different CTSL cDNAs cells were stably transduced by lentiviral transduction, employing the pMISSION® system (Sigma) as previously described (28).
RT-PCR and qPCR Analysis
RNA was isolated from tissues and cells using the RNeasy Mini Kit® (Qiagen, Hamburg, Germany) and transcribed to cDNA using the iSCRIPT® cDNA synthesis system (Bio-Rad). Primer used for RT-PCR to visualize production of splice variants had the following sequence: CTSL-ex1 fw, 5′-GAC AGG GAC TGG AAG AGA GGA C-3′; CTSL-ex2 rev, 5′-AAA GGC AGC AAG GAT GAG TGT AGG ATT CAT-3′. Primer sequences for quantitative real-time PCR were as follows: mβ-actin fw, 5′-ACC CAG GCA TTG CTG ACA GG-3′; mβ-actin rev, 5′-GGA CAG TGA GGC CAG GAT GG-3′; hGAPDH fw, 5′-CGA CCA CCT TGT CAA GCT CA-3′; hGAPDH rev, 5′-AGG GGT CTA CAT GGC AAC TG-3′; CTSL-A fw, 5′-GGG TGG ACA CAG GTT TTA AAA-3′; CTSL-A2 fw, 5′-TTG AGC GGG CAG GTT TTA AA-3′; CTSL-A3 fw, 5′-CGC GGT CGA GTA GGT TTT AAA-3′; CTSL-ex2 rev, 5′-TGG TGC ATC GCC TTC CAC T-3′; CTSL total fw, 5′-GAA TCC TAC ACT CAT CCT TGC TGC C-3′; CTSL total rev, 5′-ACA CTG CTC TCC TCC ATC CTT CTT C-3′; heEF2 fw, 5′-CCT CTA TGC CAG TGT GCT GA-3′; heEF2 rev, 5′-TCC TGT TCA AAA CCC CGT AG-3′; meEF2 fw, 5′-GCG AGG ACA AAG ACA AGG AG-3′; meEF2 rev, 5′-GGG ATG GTA AGT GGA TGG TG-3′. qRT-PCR was performed using Platinum® SYBR® Green qPCR SuperMix-UDG (Life Technologies, Darmstadt, Germany) and PCR was run in the CFX96® real-time PCR machine (Bio-Rad). RT-PCR products were analyzed on a sodium/borate-buffered 1% agarose gel.
Polyribosome Profiles
Ten to 50% sucrose gradients containing 40 mm Tris, pH 8, 20 mm KCl, 10 mm MgCl2, 100 μg/ml of cycloheximide were prepared in 12-ml polyallomer ultracentrifuge tubes (Beckman Coulter). Cells were incubated for 5 min with 1× PBS, 100 μg/ml of cycloheximide and detached from the cell culture plate by trypsination. Cells were pelleted and washed one time with PBS/cycloheximide. The cell pellet was resuspended in 500 μl of hypotonic polyribosome extraction buffer (5 mm Tris-HCl, pH 7.5, 0.5% (w/v) sodium deoxycholate, 2.5 mm MgCl2, 1.5 mm KCl, 100 μg/ml of cycloheximide, 2 mm DTT, 0.5% (v/v) Triton X-100). Lysates were passed through 27-gauge needles twice to ensure break up of cell membranes. For dissociation of tissue samples ∼300 mg of frozen tumor tissue was ground into fine powder in liquid nitrogen. Only after addition of 600 μl of hypotonic polysome extraction buffer were samples allowed to thaw on ice. Lysates from cells or tissues were centrifuged at 13,000 × g for 7 min to pellet cellular membranes. Optical density values at 260 nm (A260) were determined and 10 A260 units were loaded onto sucrose gradients. The gradients were centrifuged at 275,000 × g (TH-641 rotor (Sorvall), Thermo Scientific) for 2 h and 30 min at 4 °C. Twenty fractions with each 600 μl were collected while measuring the absorbance at 254 nm using a density gradient fractionator system (Teledyne ISCO, Lincoln, NE). The polyribosome profile obtained shows an ribonucleoprotein peak, the 40S and 60S subunits, and a single assembled ribosome appearing as the 80S peak followed by peaks representing mRNA with increasing numbers of assembled ribosomes. Profiles are presented under “Results” with the 80S peak, 0.5 m lithium chloride and 3 volumes of 100% ethanol were added to gradient fractions (both solutions were pre-cooled). RNA was precipitated for 2 h and 30 min at −80 °C before tubes were centrifuged for 40 min at 16,500 rpm at 4 °C. Supernatants were removed and pellets were briefly dried before adding 300 μl of RNA Lysis buffer T. RNA was then purified using the “RNeasy Mini Kit®” columns and buffers according to the manufacturer's instructions (Qiagen).
Results
Transgenic Expression of Human CTSL in a Murine Breast Cancer Model
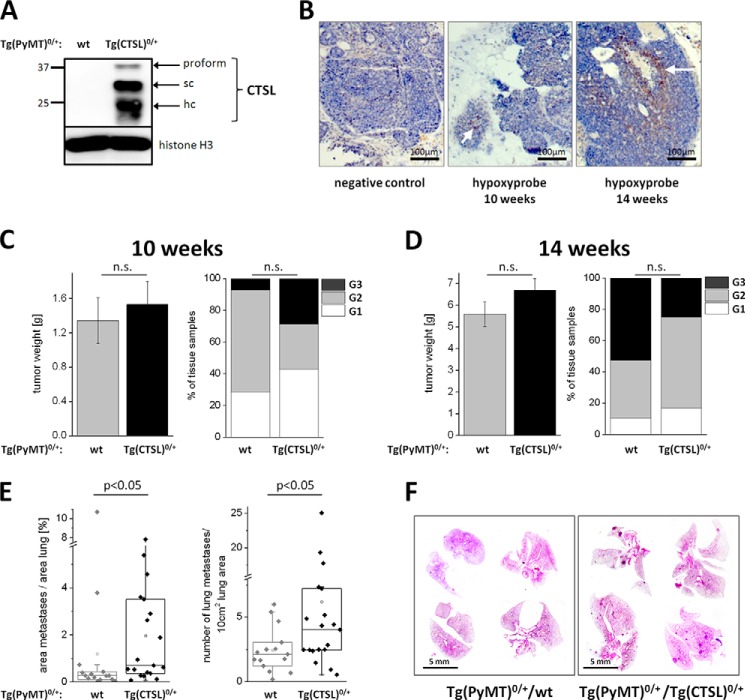
To examine the influence of human CTSL expression on breast cancer progression we generated a breast cancer mouse model (expressing the PyMT oncogene under the control of the MMTV long terminal repeat/promoter (26, 29)) with an additional genomic transgene for CTSL (25, 30). Importantly, the CTSL transgene consists of a genomic construct, which requires RNA splicing and enables transcriptional control via the genuine human CTSL promoter allowing for experimental in vivo studies of human CTSL regulation (25). Expression of CTSL in tumor cells prepared from breast tumors out of this model was shown by immunoblotting (Fig. 1A). The CTSL protein presents in three distinct bands: a 37-kDa inactive proform that is further processed in the endolysosomal compartment into the active single chain form of 27 kDa and the active single chain form of which the heavy chain of 21 kDa is visible. The expression of the CTSL transgene induces about a 4-fold increase of CTSL proteolytic activity as compared with wild type mice (30). Tumor growth in the MMTV-PyMT model starts around 4 weeks along with progressing puberty in the mice (29). At around 10 weeks tumors are palpable in most of the 10 mammary glands and first micrometastases can be found in the lungs. When tissue sections of the primary tumor in this stage are stained for hypoxic conditions using a pimonidazol hydrochloride-based probe only a few areas stain positive as expected for rather small tumor volume (marked by arrow, Fig. 1B). At 14 weeks single tumors reach sizes around 1 cm3 and most animals show metastases in the lung. In this stage tumors often show necrosis and extended areas of hypoxia (marked by arrow, Fig. 1B). Furthermore, tumor and metastasis burden were measured at these two time points. Transgenic expression of CTSL did not influence the weight and grading of the primary tumors formed at 10 or 14 weeks of age (Fig. 1, C and D). When the lungs of 14-week-old MMTV-PyMT mice were analyzed, animals carrying the CTSL transgene showed a higher metastatic burden compared with human CTSL negative tumor mice (Fig. 1E). Representative pictures of lungs from 14-week-old MMTV-PyMT mice with and without CTSL transgene are shown in Fig. 1F.
FIGURE 1.
Transgenic expression of human CTSL in MMTV-PyMT breast cancer mice. A, expression of human CTSL in primary murine breast cancer cells isolated from MMTV-PyMT mice. Immunoblot of cell lysates from tumor cells derived from PyMT mice (Tg(PyMT)0/+) without any additional transgene (wt) and mice that carry the genomic construct of human CTSL in addition to the oncogene (Tg(CTSL)0/+). The immunoblot was stained with an antibody specific for the human form of CTSL. sc, single chain form; hc, heavy chain of the two-chain form. B, hypoxia in tumors in the MMTV-PyMT model. Paraffin sections of tumors from 10- and 14-week-old mice after injection and staining of hypoxyprobe. Hypoxic areas are marked by the arrows. C, development of the primary tumor in wt versus Tg(CTSL)0/+ mice. Mass of the primary tumor in all 10 mammary glands in wt and Tg(CTSL)0/+ PyMT mice at 10 weeks (wt n = 14; Tg(CTSL)0/+, n = 21; mean ± S.E., p > 0.05, unpaired t test). Tumor grading in wt and Tg(CTSL)0/+ mice at 10 weeks was: grade 1 (G1), well differentiated; grade 2 (G2), moderately differentiated; grade 3 (G3), poorly differentiated (wt n = 14; Tg(CTSL)0/+ n = 21; p > 0.05, χ2 test). D, development of the primary tumor with 14 weeks (wt n = 19; Tg(CTSL)0/+ n = 24; mean ± S.E., p > 0.05, unpaired t test for tumor weight, χ2 test for grading). E, metastasis in wt versus Tg(CTSL)0/+ mice. Percentage of lung area that is covered by metastases and number of metastases per 10 cm2 lung area in wt versus Tg(CTSL)0/+ mice with 14 weeks (wt n = 15; Tg(CTSL)0/+, n = 19; p < 0.05, Mann-Whitney). F, representative pictures of lungs from wt versus Tg(CTSL)0/+ mice with 14 weeks in H&E staining. Lung metastases appear as dense spots within the lung parenchyma.
Translation of CTSL mRNA in Murine Breast Cancer Tissue
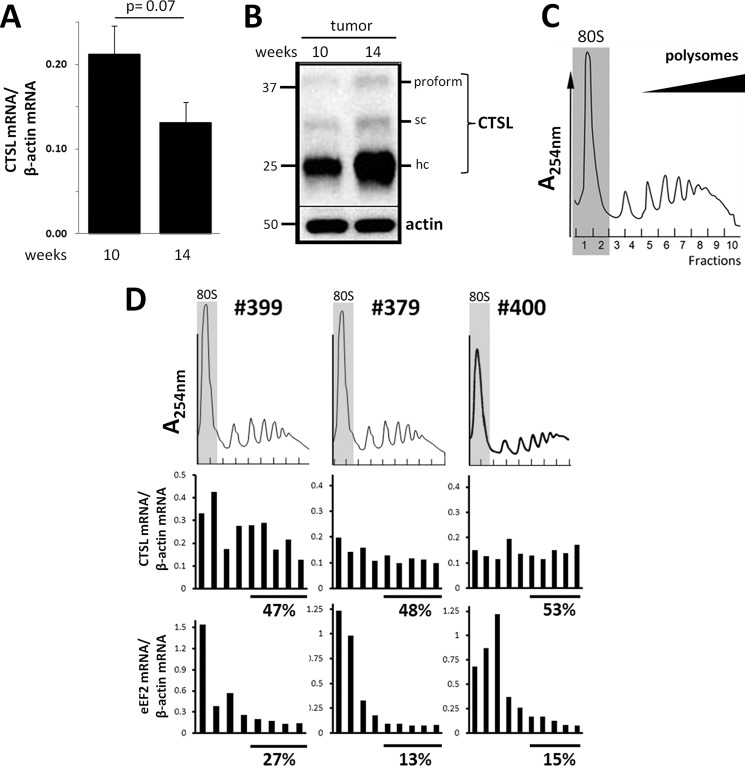
High expression of CTSL in the tumor increased the ability of metastases formation. This is in line with clinical findings of high CTSL levels as a poor prognostic factor (6). However, how tumor cells maintain high levels of CTSL activity needs to be further elucidated. In the tumor tissue of MMTV-PyMT mice, abundance of the CTSL mRNA derived from the genomic transgene was not increased with progression of the tumor from 10 to 14 weeks (Fig. 2A). In contrast, the abundance of CTSL protein in tumor tissue lysates increased over this period of time (Fig. 2B). We wondered if this discrepancy in CTSL mRNA and protein levels might be due to high efficiency of CTSL mRNA translation under tumor-associated stress conditions like hypoxia. To analyze translation in vivo, polyribosome profiles of tissue lysates from 14-week-old mice were prepared and fractions were analyzed by qRT-PCR. In these profiles the 80S peak is followed by peaks representing mRNAs with increasing numbers of associated ribosomes, i.e. the polyribosomes (example in Fig. 2C). mRNAs found in the fractions with high numbers of ribosomes are thought to be strongly translated (31). The translation status of CTSL was assessed in tumor samples of three different mice (Fig. 2D). The presence of CTSL mRNA in different fractions of the polyribosome gradient was normalized to β-actin. In addition the distribution of eukaryotic elongation factor 2 (eEF2) mRNA over the gradient was measured to assure that distribution in the profiles is not due to changes in β-actin ribosome association. Around 50% of CTSL mRNAs found in ribosome fractions range within high polyribosomes (shown by the bars below the graphs in Fig. 2D). The eEF2 mRNA is less efficiently translated as it can be found mainly in the 80S peaks and within the single ribosome fractions. In conclusion, high CTSL protein levels in the late stage tumors of MMTV-PyMT mice might indeed be caused by high efficiency in mRNA translation.
FIGURE 2.
Translation of CTSL in mammary cancer tissue. A, abundance of CTSL mRNA in tumors of 10- and 14-week-old PyMT-Tg(CTSL)0/+ mice. Normalized to β-actin expression (n (10 weeks) = 7; n (14 weeks) = 5, mean ± S.E., unpaired t test). B, immunoblot of CTSL in tumors of 10- and 14-week-old PyMT-Tg(CTSL)0/+ mice. C, polyribosome profile obtained from a primary tumor of a 14-week-old PyMT mouse. D, polyribosome association of CTSL mRNA in 3 samples of primary tumors of PyMT-Tg(CTSL)0/+ mice with 14 weeks. The percentage below the graph states the ratio of mRNA found in the high polyribosome fractions. Polyribosome association of CTSL and eukaryotic translation elongation factor 2 (eEF2) were normalized to β-actin mRNA.
Polyribosome Profiling of CTSL 5′ UTR Splice Variants in Murine Breast Cancer
Next, we addressed if high translation efficiency of CTSL can be assigned to one of the 5′ UTR splice variants in particular. The CTSL splice variants vary in their length of exon 1 caused by different splice acceptor sites that are joined to exon 2 (Fig. 3A). The longest variant with an exon 1 of 340 bp named CTSL-A (NCBI entry NM_001912.4) has been proposed to build a three-dimensional structure that functions as an internal ribosomal entry site (predicted IRES is marked by the dotted line box in Fig. 3A (21)). The segment in position 250 to 340 bp has been described as critical for the formation of the IRES structure (21). The shorter variants CTSL-A1 (NCBI entry NM_125791.1), CTSL-A2 (described in Ref. 15), and CTSL-A3 (NCBI entry NM_145918.2) are lacking gradually longer parts of the 3′ part of exon 1. The size of exon 1 in these transcripts is 315 bp for CTSL-A1, 250 bp for CTSL-A2, and 195 bp for CTSL-A3. If primers span from the constant 5′ part of exon 1 to exon 2 a ladder of PCR products can be amplified (scheme in Fig. 3B). In tumor samples of 14-week-old mice all 5′ UTR splice variants were produced from the CTSL transgene, although the A1 variant was detectable only in very small abundance and, therefore, not further analyzed (Fig. 3B). Splice site-specific primers were used for quantification of splice variants by qRT-PCR (scheme in Fig. 3C). The CTSL-A, -A2, and -A3 variants were quantified by taking the specific efficiencies of the different PCRs into account. Ratios were calculated by defining the sum of the three variants as 100%. In tumor samples of 14-week-old MMTV-PyMT mice the CTSL-A variant represents about 49%, CTSL-A3 accounts for ∼45%, and the CTSL-A2 mRNA makes up to ∼6% of the total CTSL mRNA. The percentage distribution of splice variants over the polyribosome profile remained fairly constant without increased polyribosome association of individual CTSL splice variants (Fig. 3C). In summary, our in vivo analysis of polyribosome-associated 5′ UTR CTSL splice variants did not provide evidence for one of the mRNA variants being superior to the others in terms of translation initiation.
Expression of CTSL Splice Variants in Clinical Samples
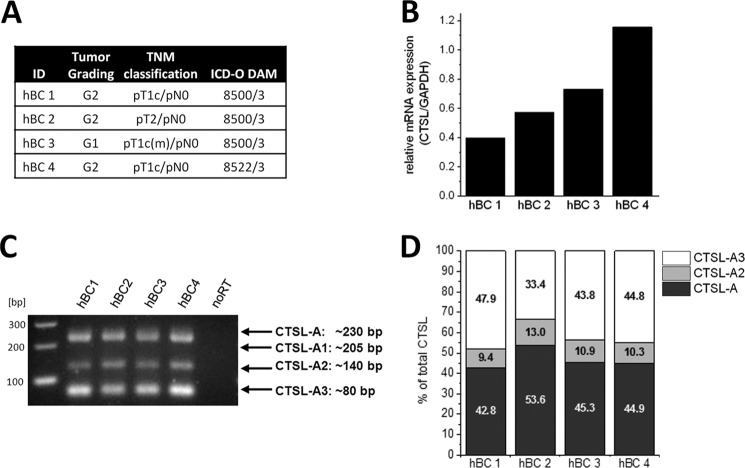
Although our mouse tumor samples do carry the human genomic construct of CTSL, its transcript is expressed in murine cells. Translational co-factors might be species specific and thereby translational regulation of human CTSL may differ in murine cells. To ensure that the distribution of splice variants is comparable in MMTV-PyMT mouse cancers and clinical breast cancer samples, we analyzed four different mammary cancer biopsies all classified as ductal carcinomas in the premetastatic stage (overview in Fig. 4A). To first assess abundance of total CTSL mRNA in these samples qRT-PCRs were performed. The mRNA abundance varied by about 50% (Fig. 4B). However, in this rather small cohort we did not see a correlation of grading or classification to the quantity of CTSL mRNA. The CTSL-A, -A2, and -A3 splice variants were found by RT-PCR in the biopsy samples, whereas the CTSL-A1 variant was not detectable (Fig. 4C). Corroborating the results in the MMTV-PyMT mouse tumor samples, CTSL-A and -A3 mRNAs are the major transcripts among the splice variants each accounting for ∼45% of the total transcripts, whereas the remaining 10% are represented by the CTSL-A2 variant (Fig. 4D). The percentage distribution of CTSL splice variants is similar between the four cancer biopsies, only the sample hBC 2 showed somewhat increased CTSL-A mainly on the cost of reduced CTSL-A3 levels. However, due to the lack of biopsy material, the correlation of this splice variant distribution to the CTSL protein could not be addressed. Therefore, we sought to analyze translation of CTSL splice variants in human breast cancer cells in the next step.
FIGURE 4.
Cathepsin L 5′ UTR splice variants in human breast cancer samples. A, overview of breast cancer biopsies employed with grading, TNM classification, and subtype according to the St Gallen molecular subtype classification. B, abundance of total CTSL mRNA in breast cancer tissues. C, detection of 5′ UTR splice variants of CTSL by RT-PCR. D, distribution of 5′ UTR splice variants of CTSL in clinical breast cancer samples.
Stress-resistant Translation of CTSL-mRNA in Human Breast Cancer Cells
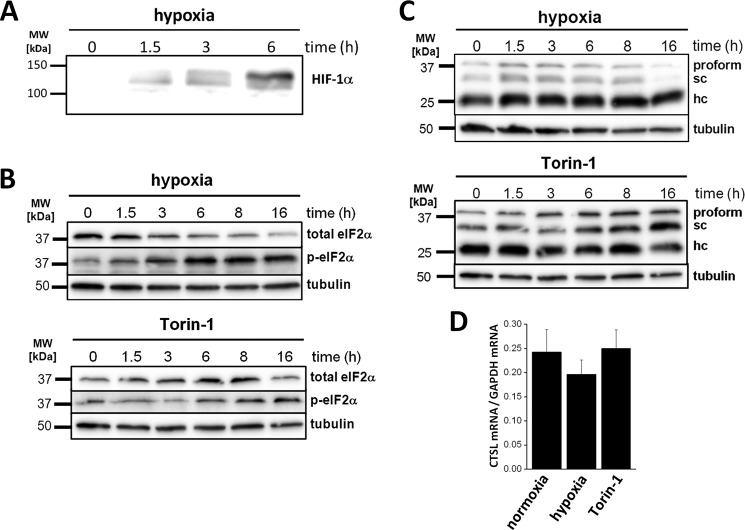
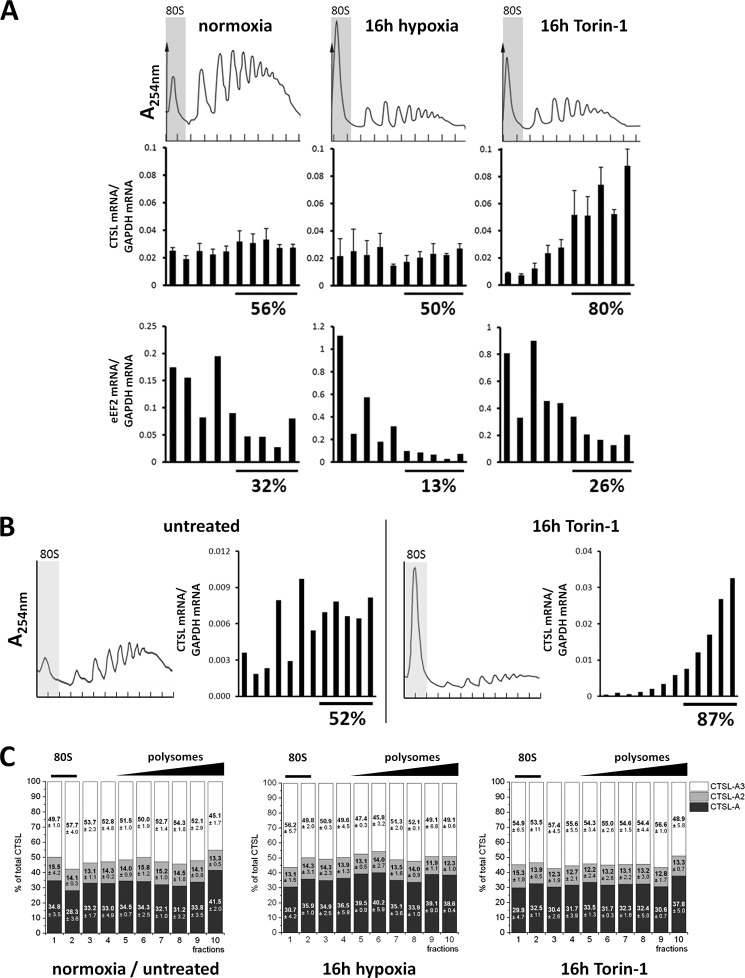
To address translation of CTSL mRNA in human breast cancer cells under defined stress conditions, MDA-MB231 cells were exposed to hypoxia or chemical mTOR inhibition by Torin-1 for 16 h. Hypoxia was monitored by accumulation of hypoxia inducible factor-1α in whole cell lysates (Fig. 5A). Chemical inhibition of mTORC1 is a more defined way to suppress translation (19) and might give us further insight into stress-resistant translation of CTSL. Torin-1 is superior over rapamycin regarding inhibition of mTORC1 especially with regard to 4E-BP1 regulation (32). Hypoxia as well as Torin-1 treatment led to a general decline in translation indicated by the increase of the 80S peak and the lowering of the high polyribosome peaks (Fig. 6A, upper panel). Additionally, stress was monitored by an increase in phosphorylation of eIF2A (Fig. 5B). Levels of total eIF2A decreased upon hypoxic conditions (Fig. 5B), whereas protein levels of CTSL were not influenced by hypoxia and even increased during Torin-1 treatment (Fig. 5C). CTSL mRNA abundance remained stable under hypoxic or Torin-1 stress (Fig. 5D). In line with the protein levels, abundance of CTSL mRNA in high polyribosome fractions stayed constant during hypoxia, whereas eEF2 mRNA shifted into the 80S or single ribosome fractions (Fig. 6A). Notably, the percentage of CTSL mRNA in high polyribosome fractions increased to ∼80% upon mTOR inhibition (Fig. 6A), which is in line with the higher abundance of CTSL protein under these conditions (Fig. 5C). The shift of eEF2 mRNA toward low polyribosome fractions was not that pronounced upon Torin-1 treatment compared with hypoxia (Fig. 6A). MDA-MB 231 cells have been described previously to express high CTSL protein levels, which correspond with high invasiveness (33). They are derived from pleural effusion and kept their potential to form metastases in the lung (34). Their molecular subtype corresponds to basal-like breast cancers, which differ from the more luminal B-like subtype of tumors formed in the MMTV-PyMT model. To test if the observed stress resistance is dependent on the breast cancer subtype, we analyzed the polyribosome association of CTSL under Torin-1 treatment in BT-474 cells, classified as luminal B mammary carcinomas (Fig. 6B). In BT-474 cells, Torin-1 treatment increased the ratio of CTSL in high polyribosome fractions comparable with the results in MDA-MB 231 cells.
FIGURE 5.
CTSL expression in human breast cancer cells upon cell stress. A, immunoblot of hypoxia inducible factor-1α (HIF-1α) in cell lysates of MDA-MB 231 cultivated under hypoxic conditions. B, levels of phosphorylated eukaryotic initiation factor 2 (eIF2) in hypoxia and Torin-1-treated cells. Immunoblot for total and phosphorylated eIF2 in cell lysates of stress-treated MDA-MB 231. C, CTSL protein levels in stress-treated MDA-MB 231. D, abundance of total CTSL mRNA in stress-treated MDA-MB 231 (n = 3, mean ± S.E.).
FIGURE 6.
Polyribosome association of CTSL mRNA in human breast cancer cells. A, polyribosome association of CTSL mRNA in MDA-MB 231 under stress conditions. One representative polyribosome profile is shown. Association of CTSL and eEF2 mRNA to polyribosomes normalized to GAPDH mRNA (n = 3, mean ± S.E.). B, polyribosome association of CTSL mRNA in BT-474 cells under Torin-1 treatment. C, distribution of 5′ UTR splice variants on the polyribosome gradient in MDA-MB 231 under stress conditions.
Furthermore, the MDA-MB 231 cells were used to assess translation of the 5′ UTR splice variants by analysis of polyribosome association under defined stress conditions. The ratios of splice variants did not vary dependent on localization in the polyribosome gradient (Fig. 6C). Here, we corroborate the finding from the mouse tumors that none of the splice variants is superior in translation over the others. In summary, we observe that translation of CTSL mRNA is stress-resistant especially upon direct inhibition of mTOR. However, all CTSL splice variants are translated with comparable efficiencies.
Stress-resistant Translation Is Mediated by the CTSL UTRs Independently of the CTSL-A “IRES” Element
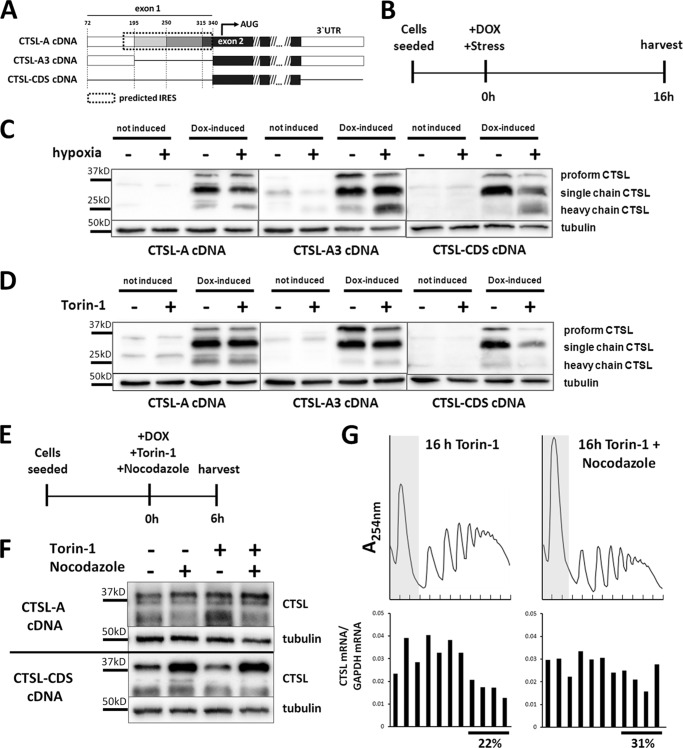
To further prove the translational advantage of CTSL mRNAs, we expressed CTSL transcript variants by stable transduction of murine breast cancer cells with doxycyline-inducible cDNA expression vectors (overview in Fig. 7A). Doxycycline treatment and exposure to stress conditions was started simultaneously to minimize the bias by effects of protein turnover (scheme in Fig. 7B). When the CTSL-A variant was expressed under hypoxia or Torin-1 treatment, stress did not affect biosynthesis of the CTSL protein (Fig. 7, C and D, left panel). The same outcome was observed when we expressed the shortest splice variant, i.e. CTSL-A3, which does not contain the predicted IRES element (Fig. 7, C and D, middle panel). In contrast, translation of a CTSL transcript without flanking UTRs (only the coding sequence (CDS) of CTSL) is strongly impaired by stress-induced shutdown of translation (Fig. 7, C and D, right panel). Apparently, the 5′ UTRs of all investigated CTSL splice variants are able to mediate stress-resistant translation, although the 5′ UTR of the CTSL-A3 variant does not contain a potential IRES or alternative regulatory motifs that may enhance translation initiation. This has been checked by scanning the sequence using RegRNA 2.0 (35) (data not shown). Therefore, we thought about alternative ways of how mRNA translation is regulated. mRNAs can be silenced by deposition in translationally inactive mRNA/protein accumulations, called stress granules or P-bodies (36). The inhibition of microtubule association by nocodazole is a well established method to block formation of stress granules (37). Cells that express either the CTSL-A transcript or the CTSL coding sequence devoid of UTRs were treated with stress and nocodazole from the time point of doxycycline induction (see Fig. 7E). As long exposure with nocodazole is cytotoxic, protein levels were analyzed after 6 h by immunoblotting. Due to this short period of doxycycline induction the cathepsin L protein presents mainly as the still unprocessed proform of 37 kDa. Nocodazole treatment did not affect CTSL protein production in cells expressing the UTR-bearing full-length cDNA (Fig. 7F). Importantly, nocodazole treatment fully rescued biosynthesis of CTSL protein under stress conditions in cells expressing the UTR-free cDNA. In line when Torin-1-stressed cells were treated with nocodazole, the UTR-free mRNA variant partially shifted from the 80S and low polyribosome fractions into the high polyribosome fraction (Fig. 7G). Thus, nocodazole prevented dislocation of UTR-free CTSL mRNA from polyribosomes, whereas CTSL biosynthesis from the full-length mRNA was not affected (Fig. 7F). Taken together, this series of experiments suggests that full-length CTSL mRNA intrinsically avoids its targeting to translationally inactive cytoplasmic mRNA deposits thereby enabling its persistent polyribosome association and CTSL protein biosynthesis upon cancer-associated cell stress conditions.
FIGURE 7.
Stress resistance is mediated by the UTRs but is not dependent on the IRES element. A, inducible expression of single CTSL cDNA variants in murine PyMT cells. Overview of expressed variants. The variant CTSL-A contains the complete described IRES motif. CTSL-A3 corresponds to the shortest described splice variant. CTSL-CDS does not contain any UTR. B, treatment scheme to assess newly synthesized CTSL under stress conditions. C, production of CTSL protein under hypoxia upon expression of different cDNA variants. D, production of CTSL protein under Torin-1 upon expression of different cDNA variants. E, treatment scheme to assess newly synthesized CTSL under Torin-1 and nocodazole treatment. F, production of cathepsin L protein under Torin-1 and nocodazole upon expression of different cDNA variants. G, polyribosome association of CTSL-CDS mRNA under Torin-1 treatment with/without addition of nocodazole.
Discussion
Often the up-regulation of cancer-associated genes can be directly attributed to the level of gene transcription. The cysteine protease CTSL has been frequently reported to be highly abundant and prognostic in tumors, however, the mechanisms leading to these high CTSL levels are incompletely understood. Here, we report an intrinsic resistance of CTSL to stress-induced shutdown of translation in tumor tissues and provide in vivo functional evidence for overexpressed CTSL as promoter of metastasis.
In the tumor context the stable translation yielding constantly high levels of proteolysis by CTSL is very likely to contribute to the progression and metastasis of carcinomas. In our analysis, the transgenic expression of human CTSL in mouse mammary cancer led to an increase in lung metastatic burden in agreement with clinical studies that established positive correlations of CTSL with metastasis and poor prognosis in mammary cancer patients (6, 7, 9). It needs to be further explored at which stage of tumor progression high CTSL levels are especially critical. However, cancer cells are repeatedly exposed to stress conditions during the complex metastatic cascade from primary tumor to colonization of a secondary organ. Thus maintaining high CTSL levels during cell stress might actively promote several steps of metastasis (38).
We assessed CTSL translation by monitoring the association of multiple ribosomes to mRNA, a method called polyribosome profiling, which represents the gold standard for translation analysis (31). It allowed for the first time to analyze translation of intrinsically expressed human CTSL mRNA either directly from cancer tissue or under distinct stress conditions. With help of this method we established that association of CTSL mRNA to the ribosome is consistently strong in tumor tissues with prevalent hypoxia when compared with mRNAs of housekeeping genes. Even the intense exposure of tumor cells to hypoxia, which led to a general decline in polyribosomes, did not reduce recruitment of CTSL mRNA to the remaining polyribosomes. Interestingly, inhibition of mTORC1 even led to a further shift of CTSL mRNA to high polyribosome fractions. This might imply that upon complete mTORC1 inhibition the translation of housekeeping mRNAs might be reduced to a state allowing for a more pronounced CTSL translation. Inhibition of mTOR is a common tool to induce autophagy in cells. The advantage of CTSL mRNA translation under such conditions might mirror its functional role in autophagy as it is needed for the turnover of autophagolysosomes (39).
A major starting point of this study was the question if certain 5′ UTR variants of CTSL serve as preferred sources for CTSL protein upon stress. The longest splice variant, variant CTSL-A, is able to form a secondary mRNA structure that functioned as an IRES element in bicistronic reporter assays (21). This suggested that CTSL-A would be preferentially translated upon stress-induced shutdown of cap-dependent translation. However, when we analyzed the translation of different CTSL splice variants, the ratios of the different transcripts in total cell and tissue lysates reflected the ratios found in the polyribosome fractions. This means all 5′ UTR splice variants of CTSL are recruited to ribosomes with the same efficiency. Indeed, the biological relevance of IRES-mediated translation on cellular mRNAs has recently been doubted for some classical cellular “IRES genes” like hypoxia inducible factor and vascular endothelial growth factor (24, 40). Thus, although IRES sequences proved to function in bicistronic assays, they might play a minor role in translational regulation in cancers or upon cell stress, at least in case of CTSL.
The above notion gained further support by expressing individual CTSL splice variants under stress conditions and quantification of the protein produced from these variants. Here, we found that the CTSL-A variant as well as the CTSL-A3 variant, which does not contain the putative IRES sequence, are both able to maintain efficient CTSL translation under stress. At the same time a CTSL variant devoid of UTRs was sensitive to stress-induced translational shutdown. This sensitivity was suspended when microtubule association was blocked by nocodazole treatment showing that this regulation is dependent on trafficking of mRNA within the cell. Nocodazole treatment blocks the formation of translationally inactive stress granules. However, the exact fate of the stress-sensitive CTSL mRNA variant still needs to be further defined. At the same time nocodazole treatment had no influence on the translation of the UTR-bearing CTSL variants supporting that the UTRs mediate the escape from stress-induced deposit in translationally inactive granules. How this is facilitated has to be further explored, especially as there are no annotated features proposing any translational regulation in the mRNA of the CTSL-A3 splice variant. The escape of certain mRNAs from stress granules has been observed before (41). For instance, mRNAs translated at the endoplasmic reticulum have been suggested to be protected from deposit in stress granules (42, 43). However, the CTSL variant without UTRs does contain an endoplasmic reticulum-import signal and should thereby be normally recruited to the endoplasmic reticulum. Mechanisms of selective translational control of distinct mRNAs are emerging as regulators of gene expression. These are based on RNA-binding proteins in combination with micro-RNAs binding mainly to UTRs thereby regulating the initiation step. As examples, selective regulations serve diminished translation of mRNAs with 5′ terminal oligopyrimidine tracts (44), the usage of iron responsive elements (45), and more recently described the function of cytosolic metadherin as a regulatory RNA-binding protein (46). Especially the regulation by metadherin might be interesting for our observation as it is highly expressed in solid tumors and even further up-regulated by hypoxia (47). The analysis of different breast cancer cell lines revealed that the observed resistance of CTSL mRNA to translational shutdown is not dependent on the molecular subtype, suggesting a more general mechanism. However, the critical factors that mediate persistent translation of CTSL need to be further defined.
In summary, breast cancer cells that face stress conditions during tumor progression maintain or even increase CTSL levels unaffected by the stress-induced general shutdown of protein biosynthesis. This stress resistance of CTSL translation is mediated by its UTR that might enable the CTSL mRNA to escape from deposit in translationally inactive stress granules. This posttranscriptional mechanism is likely to promote CTSL-mediated tumor progression and metastasis.
Acknowledgments
We thank S. Dollwet-Mack and U. Reif for excellent technical assistance. BT-474 cells were kindly provided by T. Brummer, Freiburg.
Note Added in Proof
Fig. 3B was not formatted correctly in the version of this article that was published on May 8, 2015 as a Paper in Press. Specifically, Fig. 3B did not show the borders between images of separate gel sections. Fig. 3B has been corrected to conform with JBC polices regarding figures assembled from separate images. These corrections do not affect the interpretation of the results or the conclusions.
This work was supported by Deutsche Forschungsgemeinschaft Grants SFB850 project B7 and RE1584/6-1 (to T. R.) and SFB850 project C8 (to E. S.). The authors declare that they have no conflicts of interest with the contents of this article.
- CTSL
- cathepsin L
- Tg
- transgene
- PyMT
- polyoma middle T oncogene
- MMTV
- mouse mammary tumor virus
- IRES
- internal ribosome entry site
- mTOR
- mammalian target of rapamycin
- 4E-BP
- eukaryotic initiation factor 4E binding protein-1
- qRT
- quantitative RT.
References
- 1. Mason S. D., Joyce J. A. (2011) Proteolytic networks in cancer. Trends Cell Biol. 21, 228–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nouh M. A., Mohamed M. M., El-Shinawi M., Shaalan M. A., Cavallo-Medved D., Khaled H. M., Sloane B. F. (2011) Cathepsin B: a potential prognostic marker for inflammatory breast cancer. J. Transl. Med. 9, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Miyamoto K., Iwadate M., Yanagisawa Y., Ito E., Imai J., Yamamoto M., Sawada N., Saito M., Suzuki S., Nakamura I., Ohki S., Saze Z., Kogure M., Gotoh M., Omicronbara K., Ohira H., Tasaki K., Abe M., Goshima N., Watanabe S., Waguri S., Takenoshita S. (2011) Cathepsin L is highly expressed in gastrointestinal stromal tumors. Int. J. Oncol. 39, 1109–1115 [DOI] [PubMed] [Google Scholar]
- 4. Skrzypczak M., Springwald A., Lattrich C., Häring J., Schüler S., Ortmann O., Treeck O. (2012) Expression of cysteine protease cathepsin L is increased in endometrial cancer and correlates with expression of growth regulatory genes. Cancer Invest. 30, 398–403 [DOI] [PubMed] [Google Scholar]
- 5. Gocheva V., Zeng W., Ke D., Klimstra D., Reinheckel T., Peters C., Hanahan D., Joyce J. A. (2006) Distinct roles for cysteine cathepsin genes in multistage tumorigenesis. Genes Dev. 20, 543–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harbeck N., Alt U., Berger U., Krüger A., Thomssen C., Jänicke F., Höfler H., Kates R. E., Schmitt M. (2001) Prognostic impact of proteolytic factors (urokinase-type plasminogen activator, plasminogen activator inhibitor 1, and cathepsins B, D, and L) in primary breast cancer reflects effects of adjuvant systemic therapy. Clin. Cancer Res. 7, 2757–2764 [PubMed] [Google Scholar]
- 7. Foekens J. A., Kos J., Peters H. A., Krasovec M., Look M. P., Cimerman N., Meijer-van Gelder M. E., Henzen-Logmans S. C., van Putten W. L., Klijn J. G. (1998) Prognostic significance of cathepsins B and L in primary human breast cancer. J. Clin. Oncol. 16, 1013–1021 [DOI] [PubMed] [Google Scholar]
- 8. Levicar N., Kos J., Blejec A., Golouh R., Vrhovec I., Frkovic-Grazio S., Lah T. T. (2002) Comparison of potential biological markers cathepsin B, cathepsin L, stefin A and stefin B with urokinase and plasminogen activator inhibitor-1 and clinicopathological data of breast carcinoma patients. Cancer Detect. Prev. 26, 42–49 [DOI] [PubMed] [Google Scholar]
- 9. Thomssen C., Schmitt M., Goretzki L., Oppelt P., Pache L., Dettmar P., Jänicke F., Graeff H. (1995) Prognostic value of the cysteine proteases cathepsins B and cathepsin L in human breast cancer. Clin. Cancer Res. 1, 741–746 [PubMed] [Google Scholar]
- 10. Rafn B., Nielsen C. F., Andersen S. H., Szyniarowski P., Corcelle-Termeau E., Valo E., Fehrenbacher N., Olsen C. J., Daugaard M., Egebjerg C., Bøttzauw T., Kohonen P., Nylandsted J., Hautaniemi S., Moreira J., Jäättelä M., Kallunki T. (2012) ErbB2-driven breast cancer cell invasion depends on a complex signaling network activating myeloid zinc finger-1-dependent cathepsin B expression. Mol. Cell 45, 764–776 [DOI] [PubMed] [Google Scholar]
- 11. Jean D., Rousselet N., Frade R. (2006) Expression of cathepsin L in human tumor cells is under the control of distinct regulatory mechanisms. Oncogene 25, 1474–1484 [DOI] [PubMed] [Google Scholar]
- 12. Samaiya M., Bakhshi S., Shukla A. A., Kumar L., Chauhan S. S. (2011) Epigenetic regulation of cathepsin L expression in chronic myeloid leukaemia. J. Cell Mol. Med. 15, 2189–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zajc I., Sever N., Bervar A., Lah T. T. (2002) Expression of cysteine peptidase cathepsin L and its inhibitors stefins A and B in relation to tumorigenicity of breast cancer cell lines. Cancer Lett. 187, 185–190 [DOI] [PubMed] [Google Scholar]
- 14. Rescheleit D. K., Rommerskirch W. J., Wiederanders B. (1996) Sequence analysis and distribution of two new human cathepsin L splice variants. FEBS Lett. 394, 345–348 [DOI] [PubMed] [Google Scholar]
- 15. Abudula A., Rommerskirch W., Weber E., Günther D., Wiederanders B. (2001) Splice variants of human cathepsin L mRNA show different expression rates. Biol. Chem. 382, 1583–1591 [DOI] [PubMed] [Google Scholar]
- 16. Mittal S., Mir R. A., Chauhan S. S. (2011) Post-transcriptional regulation of human cathepsin L expression. Biol. Chem. 392, 405–413 [DOI] [PubMed] [Google Scholar]
- 17. Spriggs K. A., Bushell M., Willis A. E. (2010) Translational regulation of gene expression during conditions of cell stress. Mol. Cell 40, 228–237 [DOI] [PubMed] [Google Scholar]
- 18. Silvera D., Formenti S. C., Schneider R. J. (2010) Translational control in cancer. Nat. Rev. Cancer 10, 254–266 [DOI] [PubMed] [Google Scholar]
- 19. Mamane Y., Petroulakis E., LeBacquer O., Sonenberg N. (2006) mTOR, translation initiation and cancer. Oncogene 25, 6416–6422 [DOI] [PubMed] [Google Scholar]
- 20. Spriggs K. A., Stoneley M., Bushell M., Willis A. E. (2008) Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol. Cell 100, 27–38 [DOI] [PubMed] [Google Scholar]
- 21. Jean D., Rousselet N., Frade R. (2008) Cathepsin L expression is up-regulated by hypoxia in human melanoma cells: role of its 5′-untranslated region. Biochem. J. 413, 125–134 [DOI] [PubMed] [Google Scholar]
- 22. Gilbert W. V. (2010) Alternative ways to think about cellular internal ribosome entry. J. Biol. Chem. 285, 29033–29038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kozak M. (2005) A second look at cellular mRNA sequences said to function as internal ribosome entry sites. Nucleic Acids Res. 33, 6593–6602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shatsky I. N., Dmitriev S. E., Terenin I. M., Andreev D. E. (2010) Cap- and IRES-independent scanning mechanism of translation initiation as an alternative to the concept of cellular IRESs. Mol. Cells 30, 285–293 [DOI] [PubMed] [Google Scholar]
- 25. Houseweart M. K., Pennacchio L. A., Vilaythong A., Peters C., Noebels J. L., Myers R. M. (2003) Cathepsin B but not cathepsins L or S contributes to the pathogenesis of Unverricht-Lundborg progressive myoclonus epilepsy (EPM1). J. Neurobiol. 56, 315–327 [DOI] [PubMed] [Google Scholar]
- 26. Guy C. T., Cardiff R. D., Muller W. J. (1992) Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol. Cell Biol. 12, 954–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vasiljeva O., Korovin M., Gajda M., Brodoefel H., Bojic L., Krüger A., Schurigt U., Sevenich L., Turk B., Peters C., Reinheckel T. (2008) Reduced tumour cell proliferation and delayed development of high-grade mammary carcinomas in cathepsin B-deficient mice. Oncogene 27, 4191–4199 [DOI] [PubMed] [Google Scholar]
- 28. Bengsch F., Buck A., Günther S. C., Seiz J. R., Tacke M., Pfeifer D., von Elverfeldt D., Sevenich L., Hillebrand L. E., Kern U., Sameni M., Peters C., Sloane B. F., Reinheckel T. (2014) Cell type-dependent pathogenic functions of overexpressed human cathepsin B in murine breast cancer progression. Oncogene 33, 4474–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lin E. Y., Jones J. G., Li P., Zhu L., Whitney K. D., Muller W. J., Pollard J. W. (2003) Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am. J. Pathol. 163, 2113–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sevenich L., Pennacchio L. A., Peters C., Reinheckel T. (2006) Human cathepsin L rescues the neurodegeneration and lethality in cathepsin B/L double-deficient mice. Biol. Chem. 387, 885–891 [DOI] [PubMed] [Google Scholar]
- 31. Esposito A. M., Mateyak M., He D., Lewis M., Sasikumar A. N., Hutton J., Copeland P. R., Kinzy T. G. (2010) Eukaryotic polyribosome profile analysis. J. Vis. Exp. 40, 1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thoreen C. C., Sabatini D. M. (2009) Rapamycin inhibits mTORC1, but not completely. Autophagy 5, 725–726 [DOI] [PubMed] [Google Scholar]
- 33. Caserman S., Kenig S., Sloane B. F., Lah T. T. (2006) Cathepsin L splice variants in human breast cell lines. Biol. Chem. 387, 629–634 [DOI] [PubMed] [Google Scholar]
- 34. Minn A. J., Gupta G. P., Siegel P. M., Bos P. D., Shu W., Giri D. D., Viale A., Olshen A. B., Gerald W. L., Massagué J. (2005) Genes that mediate breast cancer metastasis to lung. Nature 436, 518–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chang T. H., Huang H. Y., Hsu J. B., Weng S. L., Horng J. T., Huang H. D. (2013) An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinformatics 14, S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Buchan J. R., Parker R. (2009) Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36, 932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ivanov P. A., Chudinova E. M., Nadezhdina E. S. (2003) Disruption of microtubules inhibits cytoplasmic ribonucleoprotein stress granule formation. Exp. Cell Res. 290, 227–233 [DOI] [PubMed] [Google Scholar]
- 38. Mohamed M. M., Sloane B. F. (2006) Cysteine cathepsins: multifunctional enzymes in cancer. Nat. Rev. Cancer 6, 764–775 [DOI] [PubMed] [Google Scholar]
- 39. Dennemärker J., Lohmüller T., Müller S., Aguilar S. V., Tobin D. J., Peters C., Reinheckel T. (2010) Impaired turnover of autophagolysosomes in cathepsin L deficiency. Biol. Chem. 391, 913–922 [DOI] [PubMed] [Google Scholar]
- 40. Young R. M., Wang S. J., Gordan J. D., Ji X., Liebhaber S. A., Simon M. C. (2008) Hypoxia-mediated selective mRNA translation by an internal ribosome entry site-independent mechanism. J. Biol. Chem. 283, 16309–16319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Piccirillo C. A., Bjur E., Topisirovic I., Sonenberg N., Larsson O. (2014) Translational control of immune responses: from transcripts to translatomes. Nat. Immunol. 15, 503–511 [DOI] [PubMed] [Google Scholar]
- 42. Unsworth H., Raguz S., Edwards H. J., Higgins C. F., Yagüe E. (2010) mRNA escape from stress granule sequestration is dictated by localization to the endoplasmic reticulum. FASEB J. 24, 3370–3380 [DOI] [PubMed] [Google Scholar]
- 43. Reid D. W., Nicchitta C. V. (2012) Primary role for endoplasmic reticulum-bound ribosomes in cellular translation identified by ribosome profiling. J. Biol. Chem. 287, 5518–5527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ivanov P., Kedersha N., Anderson P. (2011) Stress puts TIA on TOP. Genes Dev. 25, 2119–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wilkinson N., Pantopoulos K. (2014) The IRP/IRE system in vivo: insights from mouse models. Front. Pharmacol. 5, 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Meng X., Zhu D., Yang S., Wang X., Xiong Z., Zhang Y., Brachova P., Leslie K. K. (2012) Cytoplasmic Metadherin (MTDH) provides survival advantage under conditions of stress by acting as RNA-binding protein. J. Biol. Chem. 287, 4485–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Noch E., Bookland M., Khalili K. (2011) Astrocyte-elevated gene-1 (AEG-1) induction by hypoxia and glucose deprivation in glioblastoma. Cancer Biol. Ther. 11, 32–39 [DOI] [PMC free article] [PubMed] [Google Scholar]