Background: TREM2 is a DAP12-coupled receptor associated with neurodegenerative diseases.
Results: Co-expression of DAP12 increased the level of TREM2 C-terminal fragment (TREM2-CTF) which suppressed the release of pro-inflammatory cytokines.
Conclusion: A major function of DAP12 is to stabilize TREM2-CTF, which regulates inflammatory responses in microglia.
Significance: Our studies unraveled a novel function of DAP12 and provided new link between TREM2/DAP12 complexes and neuroinflammation.
Keywords: Alzheimer disease, cytokine, inflammation, lipopolysaccharide (LPS), microglia, γ-secretase, DAP12, TREM-CTF, TREM2
Abstract
Triggering receptor expressed on myeloid cells 2 (TREM2) is a DAP12-associated receptor expressed in microglia, macrophages, and other myeloid-derived cells. Previous studies have suggested that TREM2/DAP12 signaling pathway reduces inflammatory responses and promotes phagocytosis of apoptotic neurons. Recently, TREM2 has been identified as a risk gene for Alzheimer disease (AD). Here, we show that DAP12 stabilizes the C-terminal fragment of TREM2 (TREM2-CTF), a substrate for γ-secretase. Co-expression of DAP12 with TREM2 selectively increased the level of TREM2-CTF with little effects on that of full-length TREM2. The interaction between DAP12 and TREM2 is essential for TREM2-CTF stabilization as a mutant form of DAP12 with disrupted interaction with TREM2 failed to exhibit such an effect. Silencing of either Trem2 or Dap12 gene significantly exacerbated pro-inflammatory responses induced by lipopolysaccharides (LPS). Importantly, overexpression of either full-length TREM2 or TREM2-CTF reduced LPS-induced inflammatory responses. Taken together, our results support a role of DAP12 in stabilizing TREM2-CTF, thereby protecting against excessive pro-inflammatory responses.
Introduction
Triggering receptor expressed on myeloid cells 2 (TREM2)4 is an innate immune receptor expressed on the surface of a subset of myeloid cells, including macrophages, dendritic cells, osteoclasts and microglia (1–3). Human TREM2 consists of an ectodomain, a transmembrane region and a short cytoplasmic tail lacking any signaling motifs. It requires a signaling adaptor protein DNAX-activating protein of 12 kDa (DAP12, also known as KARAP or TYROBP), bearing an immune-receptor tyrosine activation motif (ITAM), to initiate the signaling pathways (4). The endogenous ligand for TREM2 has yet to be identified and its signal transducing mechanisms are largely unknown. Homozygous loss-of-function mutations in the TREM2 gene result in Nasu-Hakola disease (NHD), an autosomal recessive disorder, or polycystic lipomembranous osteodysplasia with sclerosing leukoencephalopathy (PLOSL). NHD is characterized by bone cysts, bone fractures, late-onset dementia, demyelination, and cerebral atrophy with wide-spread activation of microglia (5). Recently, genetic screenings have also identified heterozygous missense mutations in the TREM2 gene as risk factors for Alzheimer disease (AD), Parkinson disease (PD), frontotemporal dementia (FTD), and amyotrophic lateral sclerosis (ALS) (6–9). Together these studies indicate that TREM2 is likely a general risk gene for multiple neurodegenerative diseases.
TREM2 is a type I transmembrane protein mainly expressed in microglia cells within the brain. In association with another transmembrane protein DAP12, TREM2 regulates critical functions of microglia including inhibition of pro-inflammatory responses and stimulation of phagocytosis of apoptotic neurons (3, 10–12). In addition to TREM2, several other receptors of the TREM receptor family, including TREM1, also associate with DAP12 (13). Given TREM1, in contrast to TREM2, serves as a critical amplifier of inflammatory signaling (14, 15), DAP12 could modulate either a pro-inflammatory or anti-inflammatory response depending on the upstream receptor partner. Importantly, DAP12 has been identified as a master regulator of the molecular networks that are perturbed in late-onset AD patients (16).
Neuroinflammation induced by microglia activation is an important pathological feature and an early event in the pathogenesis of AD (17–20). Genome-wide association studies also provide evidence of the role of inflammation in AD (21–23). Mounting evidence now implicates both TREM2 and DAP12 as key regulators of immune-responses within microglia. However, it remains largely unknown how DAP12 and TREM2 coordinate to execute their important physiological and pathophysiological functions. Recent studies have shown that TREM2 undergoes a two-step proteolytic cleavage similar to that of the amyloid precursor protein (APP) (24). The APP intracellular domain (AICD), generated from γ-secretase cleavage of APP C-terminal fragments, has been reported to regulate the expression of various genes involved in regulating neuronal or disease-related pathways (25). Whether the TREM2 fragments function similarly to APP fragments and whether DAP12 affects the level and function of TREM2 fragments remain unknown.
Herein, we confirmed that TREM2-CTF, which was generated through extracellular shedding of TREM2, serves as a substrate for the γ-secretase. Intriguingly, we observed that the level of TREM2-CTF, but not that of full-length TREM2, was significantly increased upon co-expression of DAP12 in both HEK293T and microglial BV2 cells. In the presence of DAP12, the turnover rate of TREM2-CTF was significantly reduced. However, the DAP12 D50A mutation, which abolishes its physical interaction with TREM2, failed to stabilize TREM2-CTF. Silencing of either Trem2 or Dap12 gene exacerbated lipopolysaccharides (LPS)-stimulated inflammation in microglial BV2 cells. In contrast, overexpression of full-length TREM2 or a TREM2-CTF variant reduced the LPS-induced expression of pro-inflammatory cytokines. Our studies illuminate a novel function of DAP12 in stabilizing TREM2-CTF, which protects against the deleterious pro-inflammatory responses induced by LPS, and provide a functional link between TREM2/DAP12 signaling and neuroinflammation.
Experimental Procedures
Reagents and Antibodies
γ-Secretase inhibitor DAPT as well as protein synthesis inhibitor cycloheximide were purchased from Sigma. γ-Secretase inhibitor Compound E was purchased from Millipore. Primers for cloning and quantitative RT-PCR were synthesized by Life Technologies. Restriction enzymes were purchased from Fermentas. SYBR Green for quantitative RT-PCR was purchased from Roche. Antibodies used in this study are as follows: mouse anti-c-Myc (9E10), rabbit anti-HA, mouse IgG, and rabbit IgG (Santa Cruz Biotechnology); rabbit anti-GFP (Proteintech); goat anti-mouse IgG, and goat anti-rabbit IgG antibody conjugated with horseradish peroxidase (Thermo).
cDNA Constructs
The following vectors were used in this study: TREM2-Myc, TREM2-CTF-Myc, DAP12-Myc, TREM2-GFP, DAP12-GFP, DAP12 D50A-GFP. For cloning these constructs, both TREM2 and DAP12 specific primers containing each tag sequence as needed were designed and used to amplify the TREM2 or DAP12 cDNA. The PCR products were digested with BamHI and XhoI and ligated into pcDNA3.1 or pEGFP-N1 vector. The HA-TREM2-Myc plasmid contains a HA tag at the N terminus immediately following the signal peptide and a Myc tag at the extreme C terminus. All constructs were verified by sequencing.
Cell Culture
Human embryonic kidney cell line (HEK293T) and the immortalized murine microglial cell line BV2 cells were grown in Dulbecco's modified Eagle's medium (DMEM) (Hyclone) supplemented with 10% heat-inactivated fetal bovine serum (Gibco) and 1% penicillin streptomycin solution (100 units/ml penicillin, 100 μg/ml streptomycin; Invitrogen) at 37 °C in a humidified atmosphere containing 5% CO2.
Cell Transfection
Cells were grown to 80% confluence and transfected with Turbofect Transfection Reagent (Thermo) according to the manufacturer's instruction. After 4 h, cells were transfected with another plasmid for co-expression experiments. Medium was changed 4 h after the final transfection. For BV2 cells, plasmids were transfected using Amaxa® Cell Line Nucleofector® Kit T (VCA-1002, Lonza).
Western Blotting
Cells were harvested with Nonidet P-40 lysis buffer (1% Nonidet P-40, 50 mm Tris-HCl, pH 8.0, 150 mm sodium chloride supplemented with protease inhibitors mixture) 24 h post-transfection. Protein concentrations were determined using the BCA protein assay kit according to the manufacturer's instruction (Thermo). Equal amounts of total proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to PVDF membrane (Millipore). Proteins were visualized using ECL Western blotting detection reagents (Millipore). Immunoreactive bands were quantified using ImageJ software.
Cell Surface Biotinylation
A previously described protocol was used for the cell surface biotinylation assay (26). Briefly, cells were washed twice with ice-cold phosphate-buffered saline containing 1 mm CaCl2 and 1 mm MgCl2 and then incubated twice with 0.5 mg/ml NHS-SS-biotin (Pierce) for 20 min at 4 °C. Cell lysates were prepared in Nonidet P-40 lysis buffer (1% Nonidet P-40, 50 mm Tris-HCl, pH 8.0, 150 mm sodium chloride supplemented with protease inhibitors mixture). After affinity precipitation with streptavidin beads (Pierce), biotinylated proteins were eluted with sample buffer and subjected to SDS-PAGE, followed by Western blot analysis.
Co-immunoprecipitation
HEK293T cells were co-transfected with TREM2-Myc and wild-type (DAP12 WT-GFP) or mutant (DAP12 D50A-GFP) DAP12 plasmids. Twenty-four hours after transfection, cells were lysed in Nonidet P-40 lysis buffer (0.5% Nonidet P-40, 50 mm Tris-HCl, pH 8.0, 150 mm sodium chloride supplemented with protease inhibitors). Lysates were immunoprecipitated using antibodies against mouse IgG, mouse anti-Myc antibody, rabbit IgG, or rabbit anti-GFP antibody, followed by immunoblotting with antibody against the Myc or GFP epitope.
Immunofluorescence Staining
Cells were transfected with indicated plasmids. Twenty-four hours after transfection, cells were washed three times with PBS and fixed for 15 min with 4% paraformaldehyde and then permeabilized with 0.2% Triton X-100 for 5 min. All solutions were made in PBS. After 3 rinses with PBS, cells were blocked in 5% bovine serum albumin (BSA) for 30 min at room temperature and incubated with appropriate primary antibodies for 1 h. Cells were then washed twice in 0.1% Tween-20 (0.1% PBST) for 10 min and incubated for 30 min with secondary antibodies conjugated to Alexa Fluor-488 or Alexa Fluor-594 (Life Technologies). Fluorophore-labeled cells were further stained with DAPI for 1 min and then washed twice in 0.1% PBST for 15 min. Coverslips were mounted on glass slides using antifade reagent (Life Technologies) and observed using OLYMPUS FV10-ASW confocal microscope.
Quantitative RT-PCR
Total RNAs were extracted from BV2 cells using TRIzol reagent (Invitrogen). One microgram RNA was reverse-transcribed into first-strand cDNA using ReverTra Ace qPCR RT Master Mix (TOYOBO) according to the manufacturer's protocol. Quantitative PCR was performed using the FastStart Universal SYBR Green Master (Roche). The primer sequences for Trem2, Dap12, Il-1β, Il-6, and β-actin were as followed: Trem2-Forward: 5′-TGCTGGCAAAGGAAAGGTG-3′; Trem2-Reverse: 5′-GTTGAGGGCTTGGGACAGG-3′; Dap12-Forward: 5′-GTTGACTCTGCTGATTGCCCT-3′; Dap12-Reverse: 5′-CCCTTCCGCTGTCCCTTGA-3′; Il-1β-Forward: 5′-CAGGCAGGCAGTATCACTCATTG-3′; Il-1β-Reverse: 5′-GCTTTTTTGTTGTTCATCTCGGA-3′; Il-6-Forward: 5′-CAATGGCAATTCTGATTGTATG-3′; Il-6-Reverse: 5′-AGGACTCTGGCTTTGTCTTTC-3′; β-actin-Forward: 5′-AGTGTGACGTTGACATCCGTA-3′; β-actin-Reverse: 5′-GCCAGAGCAGTAATCTCCTTC-3′.
RNA Interference
siRNA at a concentration of 300 nm was transfected into BV2 cells using Amaxa® Cell Line Nucleofector® Kit T (VCA-1002, Lonza). Cells were harvested 48 h post-transfection followed by RNA extraction and quantitative RT-PCR analysis. The siRNA sequences for Trem2 and Dap12 were as followed: Trem2 siRNA1: 5′-CCAGUCCUUGAGGGUGUCAUGUACU-3′; Trem2 siRNA2: 5′-ACCCUUGCUGGAACCGUCACCAUCA-3′; Dap12 siRNA1: 5′-CCAAGAUGCGACUGUUCUU-3′; Dap12 siRNA2: 5′-GGUGUUGACUCUGCUGAUU-3′.
Statistical Analyses
Statistical analyses were performed with GraphPad Prism. Unpaired t test, one-way ANOVA test or two-way ANOVA test was used. Data are presented as mean values from at least three independent experiments (±S.E.). To classify and indicate significant values, the following p values were used: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Results
Co-overexpression of TREM2 and DAP12 Increases the Level of TREM2-CTF
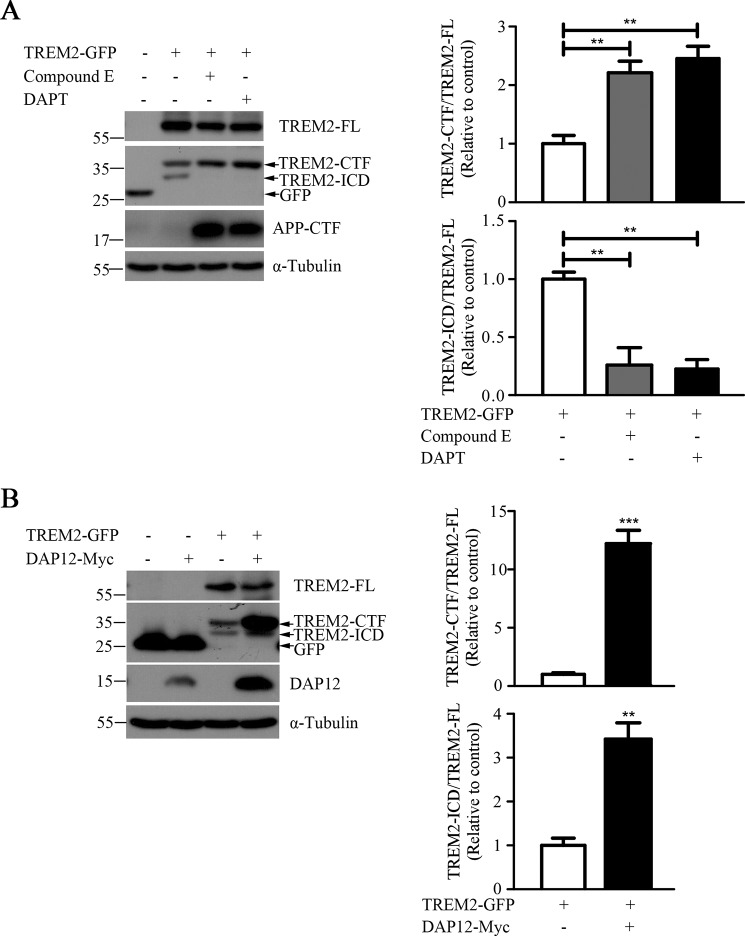
To determine how DAP12 regulates TREM2 expression and processing, we first co-expressed TREM2-Myc with DAP12-GFP in HEK293T cells. We detected a band for the full-length TREM2 (TREM2-FL) protein at about 38 kDa and a lower molecular mass species at about 15 kDa, which is consistent with the size of the C-terminal fragment of TREM2 (TREM2-CTF) (Fig. 1A). Intriguingly, we observed significantly elevated levels of TREM2-CTF but not TREM2-FL when TREM2-Myc was co-expressed with DAP12-GFP (Fig. 1A). Furthermore, biotinylation experiments demonstrated a selective accumulation of TREM2-CTF at the cell surface when DAP12 was co-expressed in HEK293T cells (Fig. 1B). Given TREM2 is predominantly expressed in microglia in the brain (27), we sought to further confirm our findings in the immortalized murine microglial cell line BV2, a model frequently used for in vitro studies of microglial functions (28, 29). Consistent with our findings in HEK293T cells, the level of TREM2-CTF was significantly elevated in the presence of DAP12 co-expression in BV2 cells (Fig. 1C). TREM2 localization in the presence or absence of DAP12 was further visualized with a specific antibody against the C-terminal Myc tag. TREM2 was localized mainly in punctate structures in the absence of DAP12. Upon DAP12 co-expression, the majority of TREM2 or its CTFs were co-localized with DAP12 leading to foci formation (Fig. 2A). Consistent with previous findings that TREM2-CTF is a substrate for the γ-secretase (24), we observed an increased amount of TREM2-CTF after treatment with γ-secretase inhibitor Compound E or DAPT (Fig. 2B). To examine potential presence of TREM2-ICD, we also tested an alternative TREM2 construct with a larger C-terminal tag, TREM2-GFP. Indeed, we observed a band at about 30 kDa, consistent with the putative size of the TREM2 intracellular domain (TREM2-ICD) fused to GFP (Fig. 3A). Consistent with a γ-secretase processing product, the level of this putative TREM2-ICD was significantly decreased upon treatments with γ-secretase inhibitor Compound E or DAPT. Interestingly, the level of this putative TREM2-ICD was increased upon DAP12 co-expression, likely reflecting a result of TREM2-CTF stabilization by DAP12 (Fig. 3B). Together, these results in multiple cell types suggest that DAP12 stabilizes TREM2-CTF and its related TREM2 processing products.
FIGURE 1.
Co-expression of DAP12 with TREM2 increases the level of TREM2-CTF. A, HEK293T cells were transfected with TREM2-Myc in combination with vector control or DAP12-GFP. Cells were harvested 24 h after transfection and the lysates were analyzed by Western blotting. The levels of TREM2 and DAP12 were detected by antibodies against the Myc or GFP epitope, respectively. Blots with α-Tubulin in this and subsequent figures serve as loading controls. Bar graph to the right shows the quantification of Western blots as ratios of TREM2-CTF/FL (n = 4). **, p < 0.01. B, HEK293T cells were transfected with TREM2-GFP in combination with vector control or DAP12-Myc. Cell surface proteins were biotinylated with sulfo-NHS-biotin followed by precipitation with streptavidin beads. Biotinylated proteins were eluted with SDS-PAGE sample buffer and subjected to Western blotting. Bar graphs to the right show the quantification of Western blots as either relative levels of TREM2-FL or TREM2-CTF/TREM2-FL ratios on the cell surface (n = 3). ns, not significant; ***, p < 0.001. C, BV2 cells were transfected with TREM2-Myc in combination with vector control or DAP12-GFP. Cell lysates were collected at 24 h post-transfection. The levels of TREM2 and DPAP12 were detected by antibodies against Myc or GFP epitope, respectively. Bar graph to the right shows the quantification of Western blots as ratios of TREM2-CTF/TREM2-FL (n = 3). **, p < 0.01.
FIGURE 2.
TREM2-CTF is a substrate for γ-secretase. A, HEK293T cells were transfected with TREM2-Myc in combination with DAP12-GFP or vector control for 24 h. Following immunostaining with antibody to Myc, cells were observed by fluorescence confocal microscopy with representative staining shown. Scale bar: 10 μm. Quantification shows the relative intensity of TREM2 fluorescence (n = 3). **, p < 0.01. B, HEK293T cells were transiently transfected with TREM2-Myc plasmids. After 8 h, cells were treated with 500 nm Compound E, 10 μm DAPT or vehicle control for 16 h. As a positive control, APP-CTF was detected by Western blotting using the 369 antibody (45). Bar graph to the right shows the quantification of Western blots as ratios of TREM2-CTF/TREM2-FL (n = 3). *, p < 0.05.
FIGURE 3.
Co-expression of DAP12 with TREM2 increases the level of TREM2-ICD. A, HEK293T cells were transfected with TREM2-GFP then incubated for 16 h with 500 nm Compound E, 10 μm DAPT or vehicle control. Bar graphs to the right show the quantification of Western blots as ratios of TREM2-CTF/TREM2-FL or TREM2-ICD/TREM2-FL (n = 3). **, p < 0.01. B, BV2 cells were transfected with TREM2-GFP in combination with DAP12-Myc or vector control. Cells were harvested 24 h post-transfection, followed by Western blotting with antibodies against GFP (TREM2-FL, TREM2-CTF and TREM2-ICD), Myc (DAP12) and α-Tubulin. Bar graphs to the right show the quantification of Western blots as ratios of TREM2-CTF/TREM2-FL or TREM2-ICD/TREM2-FL (n = 3). **, p < 0.01; ***, p < 0.001.
Stabilization of TREM2-CTF by DAP12 Requires Their Interaction
The interaction between TREM2 and its signaling adaptor protein DAP12 is mediated by charged amino acid residues within their respective transmembrane domains (13, 30). An aspartic acid in the transmembrane domain of DAP12 allows its association with TREM2 via an electrostatic interaction. As such, we sought to determine whether interaction between TREM2 and DAP12 is required for DAP12-mediated increases in the level of TREM2-CTF. We mutated the aspartic acid residue at position 50 of DAP12 to an alanine, thus eliminating its interaction with TREM2 (Fig. 4A). While co-expression of TREM2 and wild-type DAP12 in HEK293T cells increased the level of TREM2-CTF, the DAP12 D50A mutant failed to exhibit such an effect (Fig. 4B). These results suggest that the interaction between TREM2 and DAP12 is required for the function of DAP12 in stabilizing TREM2-CTF.
FIGURE 4.
Effects of DAP12 on TREM2-CTF require their interaction. A, HEK293T cells were co-transfected with TREM2-Myc and wild-type (DAP12 WT-GFP) or mutant (DAP12 D50A-GFP) DAP12 plasmids. Lysates were immunoprecipitated using antibodies against mouse IgG (M IgG), mouse anti-Myc antibody (Myc), rabbit IgG (R IgG), or rabbit anti-GFP antibody (GFP), followed by immunoblotting with antibody against the Myc or GFP epitope. Five percentage of total protein was loaded as an input control. Arrowheads indicate each specific band recognized by the antibodies or the IgG heavy or light chain. B, HEK293T cells were co-transfected with TREM2-Myc and wild-type (DAP12 WT-GFP) or mutant (DAP12 D50A-GFP) DAP12 plasmids. Cells were harvested 24 h after transfection, and the lysates were analyzed by Western blotting. Bar graph to the right shows the quantification of Western blots as ratios of TREM2-CTF/TREM2-FL (n = 3). **, p < 0.01.
DAP12 Stabilizes TREM2-CTF by Suppressing Its Turnover
Given TREM2-CTF arises from the proteolytic shedding of the ectodomain and undergoes subsequent processing by the γ-secretase, the increased levels of TREM2-CTF by DAP12 could be explained by two alternative pathways: either a DAP12-mediated increase in TREM2-CTF protein stability or an enhanced ectodomain processing of the full-length TREM2/decreased cleavage of TREM2-CTF by the γ-secretase. To further explore the mechanism, we generated a TREM2 construct containing a HA tag at the N terminus immediately following the signal peptide and a Myc tag at the extreme C terminus. This allows for the detection of both soluble TREM2 (sTREM2) and TREM2-CTF derived from ectodomain processing of the full-length TREM2. As shown in Fig. 5A, the level of sTREM2 did not differ in the absence or presence of DAP12 co-expression. The γ-secretase inhibitor Compound E blocked TREM2-CTF processing to similar extent in both the absence and presence of DAP12 (Fig. 5B). Together, these data suggest that the increased level of TREM2-CTF in the presence of DAP12 is unlikely due to enhanced ectodomain processing of the full-length TREM2 or decreased cleavage of TREM2-CTF by the γ-secretase. To examine the effect of DAP12 on the stability of TREM2-CTF protein, we performed protein turnover assays by treating cells for different periods of time in the presence of a protein synthesis inhibitor CHX in HEK293T cells. DAP12 co-expression had minimal effect on the half-life of the full-length TREM2; however, DAP12 significantly increased the half-life of TREM2-CTF (Fig. 5C). This effect of DAP12 requires an interaction with TREM2 as co-expression of the mutant DAP12 (D50A) did not affect the level or half-life of TREM2-CTF (Fig. 5C). Together, these results indicate that co-expression of DAP12 increases the level of TREM2-CTF by reducing its turnover rate.
FIGURE 5.
DAP12 stabilizes TREM2-CTF by suppressing its degradation. A, HEK293T cells stably expressing HA-TREM2-Myc (scheme) were transfected with wild-type (DAP12 WT-GFP) or mutant (DAP12 D50A-GFP) DAP12 plasmids. Conditioned medium was collected 24 h post-transfection and precipitated with trichloroacetic acid (TCA). Cells were harvested and the lysates were analyzed by Western blotting. sTREM2 in the conditioned medium was detected by Western blotting with an anti-HA antibody. The levels of TREM2 and DPAP12 in the cell lysates were detected by antibody against the Myc or GFP epitope, respectively. Bar graph to the right shows the relative protein level of sTREM2 in the medium (n = 3). ns, not significant. B, HEK293T cells were transfected with TREM2-Myc in combination with vector control or DAP12-GFP. After 16 h, cells were incubated with 500 nm Compound E or vehicle control. Bar graph to the right shows the quantification of Western blots as ratios of TREM2-CTF/TREM2-FL, which was calculated by subtracting the TREM2-CTF/TREM2-FL ratio in the absence of Compound E from the TREM2-CTF/TREM2-FL ratio in the presence of Compound E (n = 3). ns, not significant. C, HEK293T cells were co-transfected with TREM2-Myc and wild-type (DAP12 WT-GFP) or mutant (DAP12 D50A-GFP) DAP12 plasmids. Cells were treated with 500 nm CHX for indicated time periods (hour, hr). Bar graphs to the right show the relative levels of TREM2-FL or TREM2-CTF (n = 3). The relative signal intensities of TREM2-FL and TREM2-CTF at various time points were normalized to the “0” time point. ***, p < 0.001.
TREM2-CTF Suppresses the Pro-inflammatory Responses in Microglial Cells
As we observed a function of DAP12 in stabilizing TREM2-CTF, we sought to explore their potential physiological functions in microglial cells. Previous studies have demonstrated that TREM2 negatively regulates Toll-like receptor-induced inflammatory cytokine production (11, 12, 31). DAP12 signaling was reported to play either an inhibitory (32) or a stimulatory role (33) in the macrophage response to microbial products. To clarify their functions, we first examined the effects of Trem2 or Dap12 silencing on cytokine production. We found that knockdown of either Trem2 or Dap12 gene in microglial BV2 cells significantly increased the mRNA levels of pro-inflammatory cytokines IL-1β and IL-6 in the presence of LPS (Fig. 6), suggesting that TREM2 and DAP12 inhibit pro-inflammatory responses upon LPS stimulation in microglia cells. To address the physiological function of TREM2-CTF, we generated a TREM2 construct lacking the TREM2 ectodomain (Δ21–163). Expression of this TREM2 construct revealed a band on SDS-PAGE at ∼15 kDa, consistent with the predicted size of the TREM2-CTF fused to a C-terminal Myc tag (Fig. 7A). Immunofluorescence localization experiment showed that both full-length and this C-terminal fragment of TREM2 co-localized extensively with Golgi markers, including giantin and TGN46 (Fig. 7B), in line with the previous reports of TREM2 subcellular localization (34). Similar to the effect of full-length TREM2, overexpression of TREM2-CTF in BV2 cells significantly decreased the mRNA levels of pro-inflammatory cytokines IL-1β and IL-6 in the presence of LPS (Fig. 7C). These results suggest that TREM2-CTF stabilized by DAP12 inhibits LPS-induced inflammation in microglia cells.
FIGURE 6.
Knockdown of Trem2 or Dap12 gene exacerbates LPS-stimulated pro-inflammatory cytokine production. A, BV2 cells were transiently transfected with non-targeting siRNA (NT) or Trem2-specific siRNAs for 48 h. The relative mRNA levels of Trem2 were determined by quantitative RT-PCR and shown as bar graph (n = 3). β-Actin was used as an internal control. **, p < 0.01; ***, p < 0.001. B, Cells from panel A were treated with 500 ng/ml LPS or vehicle control for 6 h. RNA was extracted and the relative mRNA levels of IL-1β and IL-6 shown as bar graph were determined by quantitative RT-PCR (n = 3). β-Actin was used as an internal control. ***, p < 0.001. C, BV2 cells were transiently transfected with non-targeting siRNA (NT) or Dap12-specific siRNAs for 48 h. The relative mRNA levels of Dap12 were determined by quantitative RT-PCR and shown as bar graph (n = 3). β-Actin was used as an internal control. ***, p < 0.001. D, cells from panel C were treated with 500 ng/ml LPS or vehicle control for 6 h. RNA was extracted and the relative mRNA levels of IL-1β and IL-6 shown as a bar graph were determined by quantitative RT-PCR (n = 3). β-Actin was used as an internal control. **, p < 0.01; ***, p < 0.001.
FIGURE 7.
TREM2-CTF reduces LPS-induced pro-inflammatory cytokine production. A, BV2 cells were transiently transfected with TREM2-FL-Myc or TREM2-CTF-Myc and harvested for protein analysis 24 h post transfection. B, BV2 cells were transiently transfected with TREM2-FL-Myc and TREM2-CTF-Myc for 24 h. Following immunostaining with antibodies to TGN46, Myc or Giantin, cells were observed by fluorescence confocal microscopy with representative staining shown. Scale bar: 10 μm. C, cells from panel A were treated with 500 ng/ml LPS or vehicle control for 6 h. RNA was extracted and the relative mRNA levels of IL-1β and IL-6 shown as a bar graph were determined by quantitative RT-PCR (n = 4). β-Actin was used as an internal control. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Discussion
In this study, we provide strong evidence that DAP12 stabilizes TREM2-CTF by interacting with TREM2 and reducing its turnover rate. To our knowledge, this is the first report indicating an alternative role of DAP12 beyond its known function in mediating TREM2 signaling. As both the full-length TREM2 and the TREM2-CTF exhibit inhibitory functions in pro-inflammatory responses in microglial cells, our studies have strong implications for the molecular mechanisms by which DAP12 partners with TREM2.
Within the brain, TREM2 and DAP12 are predominantly expressed in microglia, the tissue-resident macrophage in the central nervous system (3, 27). TREM2 lacks cytoplasmic signaling elements but contains charged residue(s) in the transmembrane domain that facilitates its association with DAP12 for signal transduction. It has been demonstrated that DAP12-coupled TREM2 signaling promotes the phagocytosis of apoptotic neurons (3, 35) and inhibits the production of pro-inflammatory cytokines (10, 11). However, it remains largely unknown how DAP12 and TREM2 coordinate to execute their important physiological functions. As TREM2 is readily shed by ADAM10 to generate sTREM2 and TREM2-CTF (24, 36), it is unknown whether DAP12 continues to associate with TREM2 upon its shedding. Our current studies provide strong evidence that DAP12 stabilizes TREM2-CTF, suggesting that TREM2 retains its ability to partner with DAP12 even upon shedding to carry out its downstream signaling function.
Reliable TREM2 antibodies that specifically detect endogenous TREM2 are lacking; therefore, our current study on TREM2 relied primarily on overexpression systems. Nevertheless, our data provide a mechanistic insight by showing that DAP12 specifically stabilizes the C-terminal fragment of TREM2 (Figs. 1 and 5C). The presence of DAP12 has minimal effects on the level of soluble TREM2 or the activity of the γ-secretase (Fig. 5, A and B). Therefore, the increased level of TREM2-CTF by DAP12 could not be explained by either the enhanced ectodomain processing of the full-length TREM2 or the decreased γ-secretase cleavage of TREM2-CTF. Instead, DAP12 dramatically extended the half-life of TREM2-CTF by slowing its degradation (Fig. 5C). The increased level of TREM2-CTF by DAP12 reduced the production of pro-inflammatory cytokines, in line with the important functions of TREM2/DAP12 signaling (Fig. 7C). As shown in Fig. 2B, the presence of γ-secretase inhibitor increased the amount of TREM2-CTF. It would be interesting to examine if the increased level of TREM2-CTF by γ-secretase inhibitor could restrain the production of pro-inflammatory cytokines. However, such studies have potential caveats as γ-secretase has multiple substrates including Notch, APP, and TREM2; therefore, potential modulatory effects of γ-secretase inhibitor on inflammation might be difficult to interpret. As TREM2-CTF plays important roles in regulating inflammatory responses within microglia, it would be intriguing to further study the cellular regulation of TREM2-CTF by defining its trafficking pathways and identifying proteases involved in its degradation. Although beyond the scope of the current report, such studies could help to identify novel therapeutic targets to treat potentially harmful effects of neuroinflammation in neurodegenerative diseases.
It remains unknown how the full-length and the C-terminal fragments of TREM2 regulate the transcription of inflammation-related genes. Previous reports have shown that multiple type I transmembrane proteins, including APP and Notch, exhibit regulatory functions on gene expression after sequential proteolytic processing (37). Specifically, following cleavage by sheddases, namely α-secretase, β-secretase, or furin, their C-terminal fragments undergo subsequent cleavage by the γ-secretase to release their intracellular domains (ICDs), which can often translocate into the nucleus to regulate the expression of target genes (38–40). In future studies, it would be intriguing to investigate whether the full-length TREM2, TREM2-CTF, and/or TREM2-ICD regulate the expression of inflammatory genes through a related mechanism.
Recently, rare heterozygous mutations in TREM2 have been identified by genome-wide association studies that increase the risk of late-onset AD (LOAD) (6, 7). Similar associations have been reported in other AD cohorts (41–43) and in other neurodegenerative diseases, including FTD, PD, and ALS (6–9), demonstrating that TREM2 is a general neurodegenerative disease risk gene. Although DAP12 has been identified as a master regulator of the molecular networks that are perturbed in LOAD patients, there are yet reports on the potential association of DAP12 mutations with AD (16). In our study, we provided evidence that a mutation (D50A) that abolishes the electrostatic interaction between TREM2 and DAP12 compromises the ability of DAP12 to stabilize TREM2-CTF. Therefore, it would be of interest to study whether pathogenic mutations within the TREM2 and DAP12 genes that are associated with various neurodegenerative diseases abolish the interaction between TREM2 and DAP12. In this regard, it is interesting to note that a mutation within the transmembrane domain of TREM2 (K186N), which potentially disrupts the interaction between TREM2 and DAP12, is associated with Nasu-Hakola disease (44).
Taken together, data from our current study demonstrate a novel function of DAP12 in stabilizing the C-terminal fragment of TREM2 which protects against the deleterious inflammatory response induced by pro-inflammatory stimuli. We herein provide a new mechanistic insight into the functional link between TREM2/DAP12 receptor complex and neuroinflammation. Our results likely have implications for the molecular mechanism by which DAP12 regulates and mediates the functions of TREM family members that interact with DAP12. Finally, our studies also provide insights into mechanism-based therapies for neurodegenerative diseases.
Acknowledgment
We thank Caroline T. Stetler for critical reading of the manuscript.
This work was supported by National Institutes of Health Grants R01AG027924, R01AG035355, R01AG046205 (to G. B.) and Grants (81370459, 31400914) from the National Natural Science Foundation of China (to X. C.). The authors declare that they have no conflicts of interest with the contents of this article.
- TREM2
- triggering receptor expressed on myeloid cells 2
- AD
- Alzheimer disease
- DAP12
- DNAX-activating protein of 12 kDa
- CTF
- C-terminal fragment
- ICD
- intracellular domain
- CHX
- cycloheximide.
References
- 1. Schmid C. D., Sautkulis L. N., Danielson P. E., Cooper J., Hasel K. W., Hilbush B. S., Sutcliffe J. G., Carson M. J. (2002) Heterogeneous expression of the triggering receptor expressed on myeloid cells-2 on adult murine microglia. J. Neurochem. 83, 1309–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Colonna M. (2003) TREMs in the immune system and beyond. Nat. Rev. Immunol. 3, 445–453 [DOI] [PubMed] [Google Scholar]
- 3. Takahashi K., Rochford C. D., Neumann H. (2005) Clearance of apoptotic neurons without inflammation by microglial triggering receptor expressed on myeloid cells-2. J. Exp. Med. 201, 647–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lanier L. L., Corliss B. C., Wu J., Leong C., Phillips J. H. (1998) Immunoreceptor DAP12 bearing a tyrosine-based activation motif is involved in activating NK cells. Nature 391, 703–707 [DOI] [PubMed] [Google Scholar]
- 5. Paloneva J., Autti T., Raininko R., Partanen J., Salonen O., Puranen M., Hakola P., Haltia M. (2001) CNS manifestations of Nasu-Hakola disease: a frontal dementia with bone cysts. Neurology 56, 1552–1558 [DOI] [PubMed] [Google Scholar]
- 6. Guerreiro R., Wojtas A., Bras J., Carrasquillo M., Rogaeva E., Majounie E., Cruchaga C., Sassi C., Kauwe J. S., Younkin S., Hazrati L., Collinge J., Pocock J., Lashley T., Williams J., Lambert J. C., Amouyel P., Goate A., Rademakers R., Morgan K., Powell J., St George-Hyslop P., Singleton A., Hardy J., and Alzheimer Genetic Analysis, G. (2013) TREM2 variants in Alzheimer's disease. N. Engl. J. Med. 368, 117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jonsson T., Stefansson H., Steinberg S., Jonsdottir I., Jonsson P. V., Snaedal J., Bjornsson S., Huttenlocher J., Levey A. I., Lah J. J., Rujescu D., Hampel H., Giegling I., Andreassen O. A., Engedal K., Ulstein I., Djurovic S., Ibrahim-Verbaas C., Hofman A., Ikram M. A., van Duijn C. M., Thorsteinsdottir U., Kong A., Stefansson K. (2013) Variant of TREM2 associated with the risk of Alzheimer's disease. N. Engl. J. Med. 368, 107–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rayaprolu S., Mullen B., Baker M., Lynch T., Finger E., Seeley W. W., Hatanpaa K. J., Lomen-Hoerth C., Kertesz A., Bigio E. H., Lippa C., Josephs K. A., Knopman D. S., White C. L., 3rd, Caselli R., Mackenzie I. R., Miller B. L., Boczarska-Jedynak M., Opala G., Krygowska-Wajs A., Barcikowska M., Younkin S. G., Petersen R. C., Ertekin-Taner N., Uitti R. J., Meschia J. F., Boylan K. B., Boeve B. F., Graff-Radford N. R., Wszolek Z. K., Dickson D. W., Rademakers R., Ross O. A. (2013) TREM2 in neurodegeneration: evidence for association of the p.R47H variant with frontotemporal dementia and Parkinson's disease. Molecular Neurodegeneration 8, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cady J., Koval E. D., Benitez B. A., Zaidman C., Jockel-Balsarotti J., Allred P., Baloh R. H., Ravits J., Simpson E., Appel S. H., Pestronk A., Goate A. M., Miller T. M., Cruchaga C., Harms M. B. (2014) TREM2 variant p.R47H as a risk factor for sporadic amyotrophic lateral sclerosis. JAMA Neurol. 71, 449–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hamerman J. A., Jarjoura J. R., Humphrey M. B., Nakamura M. C., Seaman W. E., Lanier L. L. (2006) Cutting edge: inhibition of TLR and FcR responses in macrophages by triggering receptor expressed on myeloid cells (TREM)-2 and DAP12. J. Immunol. 177, 2051–2055 [DOI] [PubMed] [Google Scholar]
- 11. Turnbull I. R., Gilfillan S., Cella M., Aoshi T., Miller M., Piccio L., Hernandez M., Colonna M. (2006) Cutting edge: TREM-2 attenuates macrophage activation. J. Immunol. 177, 3520–3524 [DOI] [PubMed] [Google Scholar]
- 12. Takahashi K., Prinz M., Stagi M., Chechneva O., Neumann H. (2007) TREM2-transduced myeloid precursors mediate nervous tissue debris clearance and facilitate recovery in an animal model of multiple sclerosis. PLoS Med. 4, e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lanier L. L. (2009) DAP10- and DAP12-associated receptors in innate immunity. Immunol. Rev. 227, 150–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bouchon A., Dietrich J., Colonna M. (2000) Cutting edge: inflammatory responses can be triggered by TREM-1, a novel receptor expressed on neutrophils and monocytes. J. Immunol. 164, 4991–4995 [DOI] [PubMed] [Google Scholar]
- 15. Bouchon A., Facchetti F., Weigand M. A., Colonna M. (2001) TREM-1 amplifies inflammation and is a crucial mediator of septic shock. Nature 410, 1103–1107 [DOI] [PubMed] [Google Scholar]
- 16. Zhang B., Gaiteri C., Bodea L. G., Wang Z., McElwee J., Podtelezhnikov A. A., Zhang C., Xie T., Tran L., Dobrin R., Fluder E., Clurman B., Melquist S., Narayanan M., Suver C., Shah H., Mahajan M., Gillis T., Mysore J., MacDonald M. E., Lamb J. R., Bennett D. A., Molony C., Stone D. J., Gudnason V., Myers A. J., Schadt E. E., Neumann H., Zhu J., Emilsson V. (2013) Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer's disease. Cell 153, 707–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Akiyama H., Barger S., Barnum S., Bradt B., Bauer J., Cole G. M., Cooper N. R., Eikelenboom P., Emmerling M., Fiebich B. L., Finch C. E., Frautschy S., Griffin W. S., Hampel H., Hull M., Landreth G., Lue L., Mrak R., Mackenzie I. R., McGeer P. L., O'Banion M. K., Pachter J., Pasinetti G., Plata-Salaman C., Rogers J., Rydel R., Shen Y., Streit W., Strohmeyer R., Tooyoma I., Van Muiswinkel F. L., Veerhuis R., Walker D., Webster S., Wegrzyniak B., Wenk G., Wyss-Coray T. (2000) Inflammation and Alzheimer's disease. Neurobiol. Aging 21, 383–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eikelenboom P., van Exel E., Hoozemans J. J., Veerhuis R., Rozemuller A. J., van Gool W. A. (2010) Neuroinflammation - an early event in both the history and pathogenesis of Alzheimer's disease. Neurodegener Dis. 7, 38–41 [DOI] [PubMed] [Google Scholar]
- 19. Lemere C. A. (2013) Immunotherapy for Alzheimer's disease: hoops and hurdles. Molecular Neurodegeneration 8, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Landel V., Baranger K., Virard I., Loriod B., Khrestchatisky M., Rivera S., Benech P., Féron F. (2014) Temporal gene profiling of the 5XFAD transgenic mouse model highlights the importance of microglial activation in Alzheimer's disease. Molecular Neurodegeneration 9, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lambert J. C., Heath S., Even G., Campion D., Sleegers K., Hiltunen M., Combarros O., Zelenika D., Bullido M. J., Tavernier B., Letenneur L., Bettens K., Berr C., Pasquier F., Fiévet N., Barberger-Gateau P., Engelborghs S., De Deyn P., Mateo I., Franck A., Helisalmi S., Porcellini E., Hanon O., European Alzheimer's Disease Initiative Investigators, de Pancorbo M. M., Lendon C., Dufouil C., Jaillard C., Leveillard T., Alvarez V., Bosco P., Mancuso M., Panza F., Nacmias B., Bossù P., Piccardi P., Annoni G., Seripa D., Galimberti D., Hannequin D., Licastro F., Soininen H., Ritchie K., Blanché H., Dartigues J. F., Tzourio C., Gut I., Van Broeckhoven C., Alpérovitch A., Lathrop M., Amouyel P. (2009) Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat. Genet. 41, 1094–1099 [DOI] [PubMed] [Google Scholar]
- 22. Hollingworth P., Harold D., Sims R., Gerrish A., Lambert J. C., Carrasquillo M. M., Abraham R., Hamshere M. L., Pahwa J. S., Moskvina V., Dowzell K., Jones N., Stretton A., Thomas C., Richards A., Ivanov D., Widdowson C., Chapman J., Lovestone S., Powell J., Proitsi P., Lupton M. K., Brayne C., Rubinsztein D. C., Gill M., Lawlor B., Lynch A., Brown K. S., Passmore P. A., Craig D., McGuinness B., Todd S., Holmes C., Mann D., Smith A. D., Beaumont H., Warden D., Wilcock G., Love S., Kehoe P. G., Hooper N. M., Vardy E. R., Hardy J., Mead S., Fox N. C., Rossor M., Collinge J., Maier W., Jessen F., Rüther E., Schürmann B., Heun R., Kolsch H., van den Bussche H., Heuser I., Kornhuber J., Wiltfang J., Dichgans M., Frolich L., Hampel H., Gallacher J., Hull M., Rujescu D., Giegling I., Goate A. M., Kauwe J. S., Cruchaga C., Nowotny P., Morris J. C., Mayo K., Sleegers K., Bettens K., Engelborghs S., De Deyn P. P., Van Broeckhoven C., Livingston G., Bass N. J., Gurling H., McQuillin A., Gwilliam R., Deloukas P., Al-Chalabi A., Shaw C. E., Tsolaki M., Singleton A. B., Guerreiro R., Muhleisen T. W., Nothen M. M., Moebus S., Jockel K. H., Klopp N., Wichmann H. E., Pankratz V. S., Sando S. B., Aasly J. O., Barcikowska M., Wszolek Z. K., Dickson D. W., Graff-Radford N. R., Petersen R. C., Alzheimer's Disease Neuroimaging, I., van Duijn C. M., Breteler M. M., Ikram M. A., DeStefano A. L., Fitzpatrick A. L., Lopez O., Launer L. J., Seshadri S., consortium C., Berr C., Campion D., Epelbaum J., Dartigues J. F., Tzourio C., Alperovitch A., Lathrop M., consortium E., Feulner T. M., Friedrich P., Riehle C., Krawczak M., Schreiber S., Mayhaus M., Nicolhaus S., Wagenpfeil S., Steinberg S., Stefansson H., Stefansson K., Snaedal J., Bjornsson S., Jonsson P. V., Chouraki V., Genier-Boley B., Hiltunen M., Soininen H., Combarros O., Zelenika D., Delepine M., Bullido M. J., Pasquier F., Mateo I., Frank-Garcia A., Porcellini E., Hanon O., Coto E., Alvarez V., Bosco P., Siciliano G., Mancuso M., Panza F., Solfrizzi V., Nacmias B., Sorbi S., Bossu P., Piccardi P., Arosio B., Annoni G., Seripa D., Pilotto A., Scarpini E., Galimberti D., Brice A., Hannequin D., Licastro F., Jones L., Holmans P. A., Jonsson T., Riemenschneider M., Morgan K., Younkin S. G., Owen M. J., O'Donovan M., Amouyel P., Williams J. (2011) Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat. Genet. 43, 429–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bertram L., Lange C., Mullin K., Parkinson M., Hsiao M., Hogan M. F., Schjeide B. M., Hooli B., Divito J., Ionita I., Jiang H., Laird N., Moscarillo T., Ohlsen K. L., Elliott K., Wang X., Hu-Lince D., Ryder M., Murphy A., Wagner S. L., Blacker D., Becker K. D., Tanzi R. E. (2008) Genome-wide association analysis reveals putative Alzheimer's disease susceptibility loci in addition to APOE. Am. J. Hum. Genet. 83, 623–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wunderlich P., Glebov K., Kemmerling N., Tien N. T., Neumann H., Walter J. (2013) Sequential proteolytic processing of the triggering receptor expressed on myeloid cells-2 (TREM2) protein by ectodomain shedding and γ-secretase-dependent intramembranous cleavage. J. Biol. Chem. 288, 33027–33036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Müller T., Meyer H. E., Egensperger R., Marcus K. (2008) The amyloid precursor protein intracellular domain (AICD) as modulator of gene expression, apoptosis, and cytoskeletal dynamics-relevance for Alzheimer's disease. Prog. Neurobiol. 85, 393–406 [DOI] [PubMed] [Google Scholar]
- 26. Wang H., Luo W. J., Zhang Y. W., Li Y. M., Thinakaran G., Greengard P., Xu H. (2004) Presenilins and γ-secretase inhibitors affect intracellular trafficking and cell surface localization of the γ-secretase complex components. J. Biol. Chem. 279, 40560–40566 [DOI] [PubMed] [Google Scholar]
- 27. Sessa G., Podini P., Mariani M., Meroni A., Spreafico R., Sinigaglia F., Colonna M., Panina P., Meldolesi J. (2004) Distribution and signaling of TREM2/DAP12, the receptor system mutated in human polycystic lipomembraneous osteodysplasia with sclerosing leukoencephalopathy dementia. Eur. J. Neurosci. 20, 2617–2628 [DOI] [PubMed] [Google Scholar]
- 28. Bocchini V., Mazzolla R., Barluzzi R., Blasi E., Sick P., Kettenmann H. (1992) An immortalized cell line expresses properties of activated microglial cells. J. Neurosci. Res. 31, 616–621 [DOI] [PubMed] [Google Scholar]
- 29. Lucin K. M., O'Brien C. E., Bieri G., Czirr E., Mosher K. I., Abbey R. J., Mastroeni D. F., Rogers J., Spencer B., Masliah E., Wyss-Coray T. (2013) Microglial beclin 1 regulates retromer trafficking and phagocytosis and is impaired in Alzheimer's disease. Neuron 79, 873–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lanier L. L., Bakker A. B. (2000) The ITAM-bearing transmembrane adaptor DAP12 in lymphoid and myeloid cell function. Immunol. Today 21, 611–614 [DOI] [PubMed] [Google Scholar]
- 31. Ito H., Hamerman J. A. (2012) TREM-2, triggering receptor expressed on myeloid cell-2, negatively regulates TLR responses in dendritic cells. Eur. J. Immunol. 42, 176–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hamerman J. A., Tchao N. K., Lowell C. A., Lanier L. L. (2005) Enhanced Toll-like receptor responses in the absence of signaling adaptor DAP12. Nat. Immunol. 6, 579–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Turnbull I. R., McDunn J. E., Takai T., Townsend R. R., Cobb J. P., Colonna M. (2005) DAP12 (KARAP) amplifies inflammation and increases mortality from endotoxemia and septic peritonitis. J. Exp. Med. 202, 363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Prada I., Ongania G. N., Buonsanti C., Panina-Bordignon P., Meldolesi J. (2006) Triggering receptor expressed in myeloid cells 2 (TREM2) trafficking in microglial cells: continuous shuttling to and from the plasma membrane regulated by cell stimulation. Neuroscience 140, 1139–1148 [DOI] [PubMed] [Google Scholar]
- 35. Hsieh C. L., Koike M., Spusta S. C., Niemi E. C., Yenari M., Nakamura M. C., Seaman W. E. (2009) A role for TREM2 ligands in the phagocytosis of apoptotic neuronal cells by microglia. J. Neurochem. 109, 1144–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kleinberger G., Yamanishi Y., Suarez-Calvet M., Czirr E., Lohmann E., Cuyvers E., Struyfs H., Pettkus N., Wenninger-Weinzierl A., Mazaheri F., Tahirovic S., Lleo A., Alcolea D., Fortea J., Willem M., Lammich S., Molinuevo J. L., Sanchez-Valle R., Antonell A., Ramirez A., Heneka M. T., Sleegers K., van der Zee J., Martin J. J., Engelborghs S., Demirtas-Tatlidede A., Zetterberg H., Van Broeckhoven C., Gurvit H., Wyss-Coray T., Hardy J., Colonna M., Haass C. (2014) TREM2 mutations implicated in neurodegeneration impair cell surface transport and phagocytosis. Science Translational Medicine 6, 243ra286. [DOI] [PubMed] [Google Scholar]
- 37. Nakayama K., Nagase H., Koh C. S., Ohkawara T. (2011) gamma-Secretase-regulated mechanisms similar to notch signaling may play a role in signaling events, including APP signaling, which leads to Alzheimer's disease. Cell Mol. Neurobiol. 31, 887–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang Y. W., Wang R., Liu Q., Zhang H., Liao F. F., Xu H. (2007) Presenilin/gamma-secretase-dependent processing of β-amyloid precursor protein regulates EGF receptor expression. Proc. Natl. Acad. Sci. U.S.A. 104, 10613–10618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu Q., Zerbinatti C. V., Zhang J., Hoe H. S., Wang B., Cole S. L., Herz J., Muglia L., Bu G. (2007) Amyloid precursor protein regulates brain apolipoprotein E and cholesterol metabolism through lipoprotein receptor LRP1. Neuron 56, 66–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schroeter E. H., Kisslinger J. A., Kopan R. (1998) Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393, 382–386 [DOI] [PubMed] [Google Scholar]
- 41. Benitez B. A., Cooper B., Pastor P., Jin S. C., Lorenzo E., Cervantes S., Cruchaga C. (2013) TREM2 is associated with the risk of Alzheimer's disease in Spanish population. Neurobiol. Aging 34, 1711 e1715-e1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pottier C., Wallon D., Rousseau S., Rovelet-Lecrux A., Richard A. C., Rollin-Sillaire A., Frebourg T., Campion D., Hannequin D. (2013) TREM2 R47H variant as a risk factor for early-onset Alzheimer's disease. J. Alzheimers Dis. 35, 45–49 [DOI] [PubMed] [Google Scholar]
- 43. Cuyvers E., Bettens K., Philtjens S., Van Langenhove T., Gijselinck I., van der Zee J., Engelborghs S., Vandenbulcke M., Van Dongen J., Geerts N., Maes G., Mattheijssens M., Peeters K., Cras P., Vandenberghe R., De Deyn P. P., Van Broeckhoven C., Cruts M., Sleegers K., and BELNEU consortium (2014) Investigating the role of rare heterozygous TREM2 variants in Alzheimer's disease and frontotemporal dementia. Neurobiol. Aging 35, 726 e11-e9 [DOI] [PubMed] [Google Scholar]
- 44. Paloneva J., Manninen T., Christman G., Hovanes K., Mandelin J., Adolfsson R., Bianchin M., Bird T., Miranda R., Salmaggi A., Tranebjaerg L., Konttinen Y., Peltonen L. (2002) Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am. J. Hum. Genet. 71, 656–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Buxbaum J. D., Gandy S. E., Cicchetti P., Ehrlich M. E., Czernik A. J., Fracasso R. P., Ramabhadran T. V., Unterbeck A. J., Greengard P. (1990) Processing of Alzheimer beta/A4 amyloid precursor protein: modulation by agents that regulate protein phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 87, 6003–6006 [DOI] [PMC free article] [PubMed] [Google Scholar]