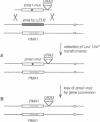
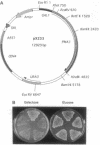
Abstract
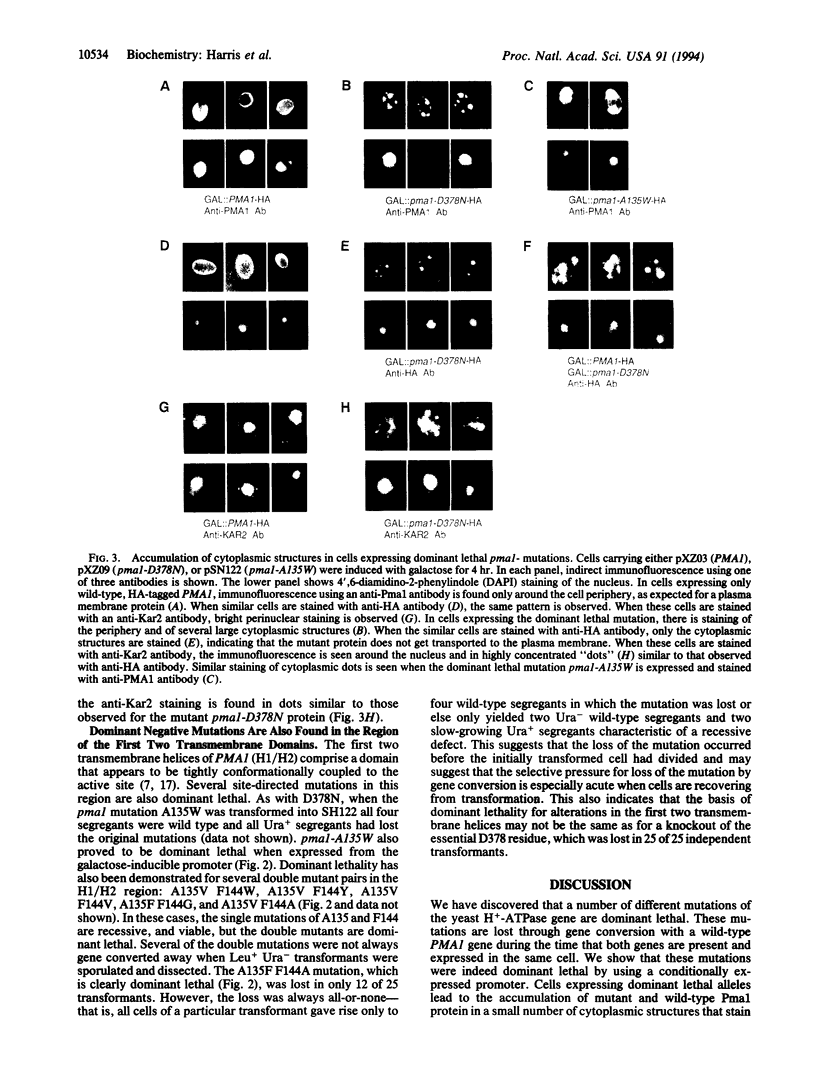
The plasma membrane H(+)-ATPase of Saccharomyces cerevisiae is an essential protein that is required to establish cellular membrane potential and maintain a normal internal pH. An Asp-378 to Asn substitution at the residue phosphorylated during catalysis is dominant lethal when the pma1-D378N mutation is expressed along with a wild-type plasma membrane H(+)-ATPase (PMA1) gene. Several mutations in the first two putative transmembrane domains are also dominant lethal. However, these dominant lethal mutants often appear to be innocuous, because they are frequently lost by gene conversion to the wild-type sequence during the process of introducing the mutant sequence and subsequently removing the wild-type gene. Loss of the mutation by gene conversion does not occur while introducing recessive lethal mutations. Cells carrying the wild-type PMA1 gene on the chromosome and a dominant lethal mutation under the control of a GAL1 promoter on a centromere-containing plasmid exhibit a galactose-dependent lethality. Indirect immunofluorescence staining using anti-Pma1 antibodies shows that induction of dominant lethal PMA1 mutations leads to the accumulation of a number of intensely staining cytoplasmic structures that are not coincident with the nucleus and its immediately surrounding endoplasmic reticulum. These structures also accumulate the endoplasmic reticulum protein Kar2. Expression of the dominant lethal protein also prevents transport of the wild-type ATPase to the plasma membrane.
Full text
PDF
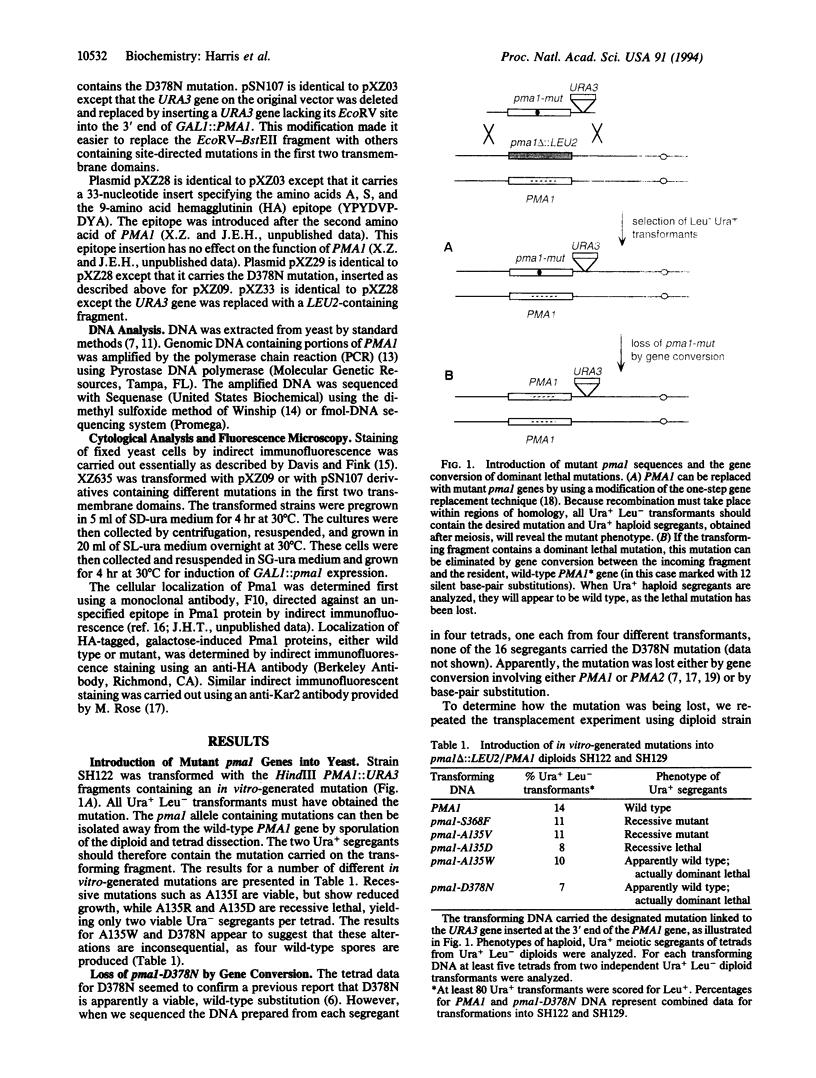
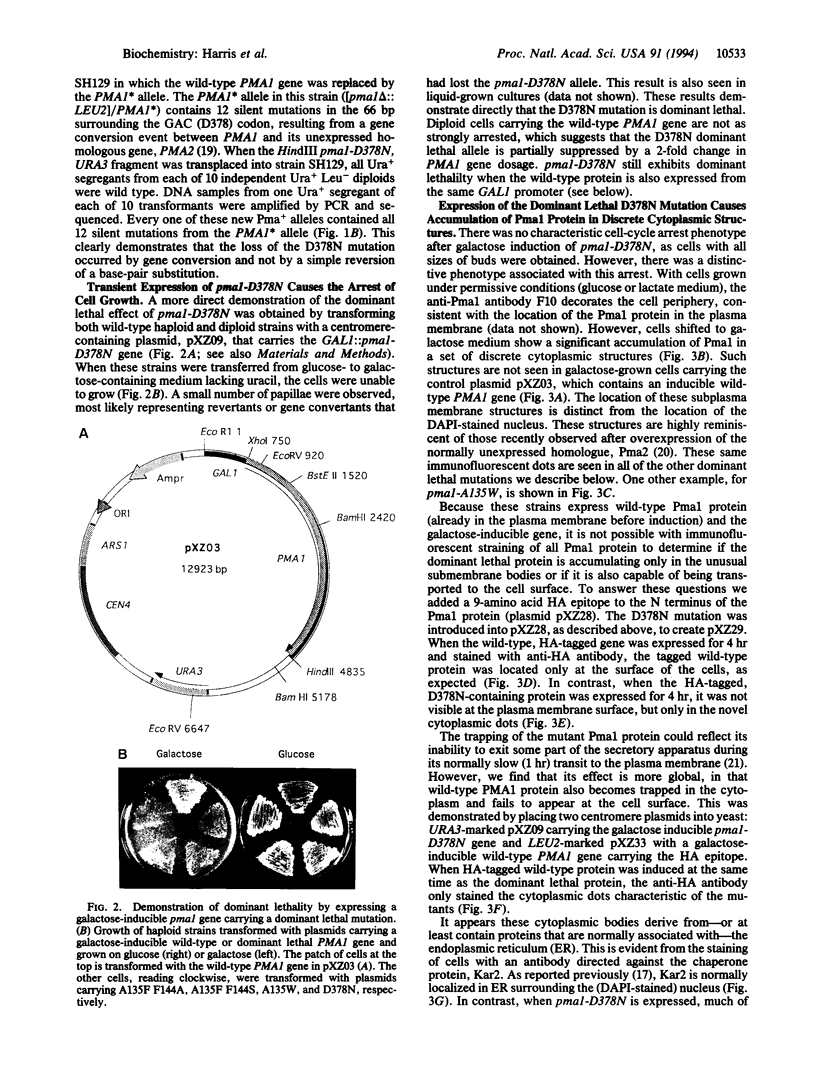

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amory A., Gofffeau A. Characterization of the beta-aspartyl phosphate intermediate formed by the H+-translocating ATPase from the yeast Schizosaccharomyces pombe. J Biol Chem. 1982 May 10;257(9):4723–4730. [PubMed] [Google Scholar]
- Boeke J. D., Trueheart J., Natsoulis G., Fink G. R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Chang A., Slayman C. W. Maturation of the yeast plasma membrane [H+]ATPase involves phosphorylation during intracellular transport. J Cell Biol. 1991 Oct;115(2):289–295. doi: 10.1083/jcb.115.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dame J. B., Scarborough G. A. Identification of the phosphorylated intermediate of the Neurospora plasma membrane H+-ATPase as beta-aspartyl phosphate. J Biol Chem. 1981 Oct 25;256(20):10724–10730. [PubMed] [Google Scholar]
- Davis L. I., Fink G. R. The NUP1 gene encodes an essential component of the yeast nuclear pore complex. Cell. 1990 Jun 15;61(6):965–978. doi: 10.1016/0092-8674(90)90062-j. [DOI] [PubMed] [Google Scholar]
- Harris S. L., Perlin D. S., Seto-Young D., Haber J. E. Evidence for coupling between membrane and cytoplasmic domains of the yeast plasma membrane H(+)-ATPase. An analysis of intragenic revertants of pma1-105. J Biol Chem. 1991 Dec 25;266(36):24439–24445. [PubMed] [Google Scholar]
- Harris S., Rudnicki K. S., Haber J. E. Gene conversions and crossing over during homologous and homeologous ectopic recombination in Saccharomyces cerevisiae. Genetics. 1993 Sep;135(1):5–16. doi: 10.1093/genetics/135.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziej P. A., Young R. A. Epitope tagging and protein surveillance. Methods Enzymol. 1991;194:508–519. doi: 10.1016/0076-6879(91)94038-e. [DOI] [PubMed] [Google Scholar]
- Kruse C., Johnson S. P., Warner J. R. Phosphorylation of the yeast equivalent of ribosomal protein S6 is not essential for growth. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7515–7519. doi: 10.1073/pnas.82.22.7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungdahl P. O., Gimeno C. J., Styles C. A., Fink G. R. SHR3: a novel component of the secretory pathway specifically required for localization of amino acid permeases in yeast. Cell. 1992 Oct 30;71(3):463–478. doi: 10.1016/0092-8674(92)90515-e. [DOI] [PubMed] [Google Scholar]
- Mann C., Buhler J. M., Treich I., Sentenac A. RPC40, a unique gene for a subunit shared between yeast RNA polymerases A and C. Cell. 1987 Feb 27;48(4):627–637. doi: 10.1016/0092-8674(87)90241-8. [DOI] [PubMed] [Google Scholar]
- Maruyama K., MacLennan D. H. Mutation of aspartic acid-351, lysine-352, and lysine-515 alters the Ca2+ transport activity of the Ca2+-ATPase expressed in COS-1 cells. Proc Natl Acad Sci U S A. 1988 May;85(10):3314–3318. doi: 10.1073/pnas.85.10.3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker J. H., Perlin D. S., Haber J. E. Pleiotropic plasma membrane ATPase mutations of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Nov;7(11):4082–4088. doi: 10.1128/mcb.7.11.4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na S., Perlin D. S., Seto-Young D., Wang G., Haber J. E. Characterization of yeast plasma membrane H(+)-ATPase mutant pma1-A135V and its revertants. J Biol Chem. 1993 Jun 5;268(16):11792–11797. [PubMed] [Google Scholar]
- Portillo F., Serrano R. Dissection of functional domains of the yeast proton-pumping ATPase by directed mutagenesis. EMBO J. 1988 Jun;7(6):1793–1798. doi: 10.1002/j.1460-2075.1988.tb03010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao R., Slayman C. W. Mutagenesis of conserved residues in the phosphorylation domain of the yeast plasma membrane H(+)-ATPase. Effects on structure and function. J Biol Chem. 1993 Mar 25;268(9):6708–6713. [PubMed] [Google Scholar]
- Rose M. D., Misra L. M., Vogel J. P. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989 Jun 30;57(7):1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Schiestl R. H., Gietz R. D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989 Dec;16(5-6):339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- Serrano R., Portillo F. Catalytic and regulatory sites of yeast plasma membrane H(+)-ATPase studied by directed mutagenesis. Biochim Biophys Acta. 1990 Jul 25;1018(2-3):195–199. doi: 10.1016/0005-2728(90)90247-2. [DOI] [PubMed] [Google Scholar]
- Supply P., Wach A., Thinès-Sempoux D., Goffeau A. Proliferation of intracellular structures upon overexpression of the PMA2 ATPase in Saccharomyces cerevisiae. J Biol Chem. 1993 Sep 15;268(26):19744–19752. [PubMed] [Google Scholar]
- Villalba J. M., Palmgren M. G., Berberián G. E., Ferguson C., Serrano R. Functional expression of plant plasma membrane H(+)-ATPase in yeast endoplasmic reticulum. J Biol Chem. 1992 Jun 15;267(17):12341–12349. [PubMed] [Google Scholar]
- Wach A., Schlesser A., Goffeau A. An alignment of 17 deduced protein sequences from plant, fungi, and ciliate H(+)-ATPase genes. J Bioenerg Biomembr. 1992 Jun;24(3):309–317. doi: 10.1007/BF00768851. [DOI] [PubMed] [Google Scholar]
- Winship P. R. An improved method for directly sequencing PCR amplified material using dimethyl sulphoxide. Nucleic Acids Res. 1989 Feb 11;17(3):1266–1266. doi: 10.1093/nar/17.3.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]