Background: SNX-PXB proteins SNX20 and SNX21 are poorly characterized members of the sorting nexin (SNX) family.
Results: We find the SNX-PXB proteins are localized to endosomes, and that the PXB domain has a tetratricopeptide repeat (TPR)-fold.
Conclusion: The PXB domain has an atypical TPR-fold with conserved surfaces likely to mediate protein-protein interactions.
Significance: SNX-PXB proteins are endosome-associated scaffolds that mediate membrane and protein interactions.
Keywords: endosome, membrane trafficking, phosphoinositide, small-angle x-ray scattering (SAXS), X-ray crystallography, sorting nexin, tetratricopeptide repeat
Abstract
Sorting nexins (SNX) orchestrate membrane trafficking and signaling events required for the proper distribution of proteins within the endosomal network. Their phox homology (PX) domain acts as a phosphoinositide (PI) recognition module that targets them to specific endocytic membrane domains. The modularity of SNX proteins confers a wide variety of functions from signaling to membrane deformation and cargo binding, and many SNXs are crucial modulators of endosome dynamics and are involved in a myriad of physiological and pathological processes such as neurodegenerative diseases, cancer, and inflammation. Here, we have studied the poorly characterized SNX20 and its paralogue SNX21, which contain an N-terminal PX domain and a C-terminal PX-associated B (PXB) domain of unknown function. The two proteins share similar PI-binding properties and are recruited to early endosomal compartments by their PX domain. The crystal structure of the SNX21 PXB domain reveals a tetratricopeptide repeat (TPR)-fold, a module that typically binds short peptide motifs, with three TPR α-helical repeats. However, the C-terminal capping helix adopts a highly unusual and potentially self-inhibitory topology. SAXS solution structures of SNX20 and SNX21 show that these proteins adopt a compact globular architecture, and membrane interaction analyses indicate the presence of overlapping PI-binding sites that may regulate their intracellular localization. This study provides the first structural analysis of this poorly characterized subfamily of SNX proteins, highlighting a likely role as endosome-associated scaffolds.
Introduction
The trafficking of proteins in eukaryotic cells is a tightly regulated process. A complex array of signaling proteins, molecular transporters, and lipids target transmembrane proteins to their appropriate location in the cell (1). Acting as a central hub in this process, the endosome is a multifunctional organelle regulating the sorting and delivery of biological regulators such as receptors, adhesion molecules, and ion channels (2), as well as acting as specialized platforms for cell signaling (3). One of the major protein families regulating endosomal function and organization are the sorting nexins (SNXs).4 All SNX proteins possess a phox homology (PX) domain that binds to phosphoinositide (PI) lipids contained in the membranes of different organelles, thus mediating their attachment to the cytoplasmic leaflets of endosomal compartments (4). Most members of the SNX family also incorporate a variety of distinct structural and functional domains, which are then targeted by the PX module to appropriate membranes within the endosomal network (5, 6). The SNX proteins can thus be classified into different subfamilies based on the structural arrangements of these different scaffolding, enzymatic, and regulatory domains (4, 5, 7). As the SNX proteins are established regulators of endosomal sorting and signaling, they have also been shown to play an important role in pathological states such as chronic and neurodegenerative diseases, pathogen invasion, and inflammation (reviewed in Ref. 5). Although the roles of many of the SNXs have been partly deciphered and attributed to particular cellular functions, many other members of the SNX family still remain poorly characterized. Given the increasing appreciation of their importance in maintaining cell homeostasis and driving pathogenesis, it is crucial to gain new molecular insights into the functions of the SNX family protein members that remain unexplored.
Although not all SNX proteins play a direct role in membrane trafficking, it is clear that the association of SNXs with membranes of the endocytic system is often coupled to protein-protein interactions that regulate recruitment of trafficking cargoes and signaling proteins. As just one example, the SNX-4.1/ezrin/radixin/moesin subfamily composed of the members SNX17, SNX27, and SNX31, possess both PSD95-discs-large-zonula occludens and 4.1/ezrin/radixin/moesin domains that associate with early endosomes through their PX domain, and couple transmembrane proteins with specific peptide motifs to the retromer complex to mediate endosomal trafficking (8–12). Synergistic action of the PX, PSD95-discs-large-zonula occludens, and 4.1/ezrin/radixin/moesin domains can reinforce binding avidity to the membrane microenvironment through coincidence detection (13).
One of the most poorly characterized SNX subfamilies are the so-called SNX-PXB proteins SNX20 and SNX21 (also called SNX-L) (5), where PXB stands for “PX-associated domain B.” SNX20 appears to be mainly expressed in macrophages and immune cells, whereas SNX21 appears to be expressed more broadly but with particularly high levels in fetal liver tissue (14, 15). A single study of the SNX20 molecule identified it as an endosomal protein that could associate with the cytosolic domain of the transmembrane adhesion receptor P-selectin glycoprotein ligand 1 (PSGL-1) (16). Overexpression of SNX20 caused a redistribution of PSGL-1 from the cell surface into endosomal organelles, suggesting a role in PSGL-1 trafficking. However, SNX20 knock-out mice showed no overt phenotypes related to endothelial cell adhesion, and therefore its precise function is still not clear. Down-regulation of SNX21 by siRNA also has a modest effect on the endocytic degradation of the epidermal growth factor receptor (17), further suggesting a role for these proteins in endosomal transport, although the mechanism underlying this effect has also not been characterized in detail.
In this study, we report the crystal structure of the domain-swapped C-terminal PXB domain of SNX21, demonstrating that it possesses a tetratricopeptide repeat (TPR)-fold and suggesting that this SNX subfamily is likely to be involved in protein-protein interactions. The topology of the SNX21 TPR domain is highly unusual, suggesting a potential autoinhibitory conformation and bearing some structural similarity to the TPR domain of the functionally unrelated yeast protein Fis1p. This provides structural clues that allow us to speculate on the function and regulation of this domain in SNX21. The solution structures of the SNX-PXB proteins determined by small angle x-ray scattering (SAXS) reveal a compact globular architecture, which may facilitate coincidence detection of PIs and potential transmembrane protein cargoes. Mutagenesis and liposome pelleting assays demonstrate the ability of these proteins to bind the endosomal PI lipid phosphatidylinositol 3-phosphate (PtdIns3P) required for endosomal recruitment. However, they also unexpectedly bind PtdIns(4,5)P2 via an overlapping PI binding pocket although the functional significance of this is still unclear. Overall, our data provides new insights into the structural organization of the SNX-PXB proteins that will underpin future functional studies.
Experimental Procedures
Antibodies
The following primary antibodies were used: mouse monoclonal anti-Myc tag (9B11) antibody (Cell Signaling Technology, 2272); mouse anti-EEA1 antibody (BD Transduction Laboratories, 610457), mouse anti-human CD107a/LAMP-1 antibody (BD Pharmingen, 555798), mouse monoclonal anti-human CD8a purified clone OKT8 antibody (eBioscience, 14-0086-80), rabbit polyclonal c-Myc antibody (Novus Biologicals, NB600-336), and anti-CD8 (18). Donkey anti-mouse coupled to Alexa Fluor® 488 or 647 and donkey anti-rabbit coupled to Alexa Fluor® 488 or 647 (Life Technologies) were used as secondary antibodies for immunofluorescence. The IRDye®800CW donkey anti-rabbit IgG and IRDye®680LT goat anti-mouse IgG were purchased from LI-COR Biosciences.
Mammalian Expression Plasmids
pcDNA-3.1-n-myc-SNX20 was generated by inserting amplified murine SNX20 cDNA using primers BamHI, forward CGC GGATCC GCA AGT CCA GAG CAT CCT and Xba, reverse CCC TCTAGA TCA GGA CAG ATA CTC CCG into the BamHI/XbaI sites of pcDNA-3.1-n-myc backbone. pcDNA-3.1-n-myc-SNX21 was generated by inserting amplified murine SNX21 cDNA using PCR primers infusion_1, AGAAGACCTG GGATCC GCC TCG CGG CTC CTA CAC CGG and infusion_2, GCCC TCTAGA CTCGAG TTA GTC CAG CAC CTC TTT GAT into the BamHI/XbaI sites of pcDNA-3.1-n-myc backbone. pcDNA-3.1-n-myc-SNX20-R313Q and pcDNA-3.1-n-myc-SNX21-R331Q were generated by site-directed mutagenesis using a QuikChange II kit (Stratagene) in accordance with the manufacturer's instructions. CD8-P-selectin and CD8-PSGL-1 were cloned by ligating the P-selectin and PSGL-1-amplified cDNA flanked with restriction sites corresponding to BamHI (5′) and XhoI (3′) enzymes into a BamHI/XhoI-digested pcDNA-3.1-n-GFP-CD8 expression vector.
Recombinant Protein Expression and Purification
cDNAs corresponding to residues 1–313 of SNX20 (mouse), 93–363 of SNX21 (mouse), and 231–363 of SNX21 (mouse) were cloned using circular polymerase extension cloning. The SNX-PXB genes were inserted into a pHUE vector yielding a N terminally His6-ubiquitin-tagged protein containing a ubiquitin-protease cleavage site. Site-directed mutagenesis was carried out using the QuikChange II kit (Stratagene) with 100 ng of parental DNA for all experiments. Proteins were expressed in the Escherichia coli BL21(DE3) strain overnight at 20 °C and purified using a two-step procedure involving affinity chromatography followed by gel filtration. Proteins were first purified on a 5-ml nickel-nitrilotriacetic acid gravity column and eluted with 300 mm imidazole in a buffer containing 25 mm Tris (pH 8.0), 500 mm NaCl, 20 mm imidazole, and 10% (v/v) glycerol. The ubiquitin tag was cleaved off for 12 h by dialysis into an imidazole-free buffer in the presence of ubiquitin protease, and the His6-ubiquitin was separated from the proteins by elution through a nickel-nitrilotriacetic acid gravity column. As a second step, proteins were gel filtered in a buffer containing 25 mm Tris (pH 8.0), 500 mm NaCl, and 1 mm DTT using a Sepharose S200 16/60 column attached to an AKTA system (GE Healthcare).
Protein Crystallization, Data Collection, and Structure Determination
SNX21-(231–363) (PXB) fractions were pooled and concentrated to ∼20 mg/ml. Eight 96-well crystallization hanging-drop screens were set-up using a Mosquito Liquid Handling robot (TTP LabTech) at 20 °C. Diffraction quality crystals were obtained using the sitting drop vapor diffusion method and a buffer containing 10% (w/v) PEG 8000, 20% (v/v) ethylene glycol, 0.1 m MES/imidazole (pH 6.5), 0.02 m sodium formate, 0.02 m ammonium acetate, 0.02 m trisodium citrate, 0.02 m sodium potassium l-tartrate, and 0.02 m sodium oxamate. The derivative crystal was produced by soaking for 1 h in the crystallization solution supplemented with 10 mm ethyl-HgCl2. Crystals were then transferred into a cryoprotectant solution composed of mother liquor supplemented with 20% glycerol and cooled to 100 K prior to data collection.
Native data were collected at the UQ ROCX diffraction facility on a Rigaku FR-E Superbright generator with Osmic Vari-Max HF optics and Rigaku Saturn 944 CCD detector. Derivative data were collected at 100 K at beamline MX-2 (UQ ROCX and Australian Synchrotron, Table 1) and integrated and scaled with MOSFLM (19) and SCALA (20). The SNX21 PXB domain structure was solved by single isomorphous replacement with anomalous scattering (SIRAS) using CRUNCH2 and BP3 for heavy atom location, SOLOMON (21) for density modification and phase improvement, and BUCCANNEER for automated model building as implemented within the CRANK automated platform in CCP4i (22). After phase determination, BUCCANNEER was able to automatically build 271 residues of a total of 288. A model was built using COOT (23) followed by repeated refinement and model building with PHENIX.REFINE and COOT. All structural figures were generated using PyMOL.
TABLE 1.
Data collection, phasing and refinement statistics for SNX21 PXB domain
Highest resolution shell is shown in parentheses.
| Native | Mercury derivative | |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 1.5418 | 1.5418 |
| Space group | P212121 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 100.9, 100.9, 63.6 | 100.6, 100.6, 64.3 |
| a, b, g (°) | 90, 90, 120 | 90, 90, 120 |
| Resolution (Å) | 21.5-2.40 (2.53-2.40) | 21.55-2.63 (2.78-2.63) |
| Rmerge | 0.088 (0.330) | 0.141 (0.613) |
| 〈I〉/σI | 10.9 (2.8) | 9.8 (2.8) |
| Total number reflections | 60,610 | 97,578 |
| Total unique reflections | 13,890 | 11,386 |
| Completeness (%) | 93.0 (66.4) | 99.8 (100.0) |
| Multiplicity | 4.4 (2.9) | 8.6 (7.8) |
| Wilson B factor | 47.6 | 59.1 |
| Phasing (SIRAS) | ||
| Number Hg sites found | 4 | |
| Figure of merit | 0.42 | |
| Figure of merit after density modification | 0.73 | |
| Number of residues automatically built | 271 of 288 | |
| Refinement | ||
| Resolution (Å) | 21.5-2.4 (2.59-2.40) | |
| No. reflections/No. Rfree | 13873 (1932)/692 (123) | |
| Rwork/Rfree | 0.230 (0.295)/0.267 (0.332) | |
| No. atoms | ||
| Protein | 2,048 | |
| Solvent | 21 | |
| Average B-factor (Å2) | 48.3 | |
| Root mean square deviations | ||
| Bond lengths (Å) | 0.008 | |
| Bond angles (°) | 1.37 | |
Multi-angle Laser Light Scattering
Samples (20–50 μl) of purified truncated SNX20 and SNX21 proteins were injected onto either a Superdex 200 HR 10/30 size-exclusion column (GE Healthcare), with a flow rate of 0.5 ml/min, or Wyatt WTC-030N5 SEC protein silica column (5 μm 300 Å 4.6 × 300 mm), with a flow rate of 0.2 ml/min. The chromatography system was coupled to a three-angle light scattering detector (mini-DAWN TRISTAR) and a refractive index detector (Optilab DSP, Wyatt Technology). Data were collected every 0.5 s. Data analysis was carried out using ASTRA software.
Small-angle X-ray Scattering
The molar masses and monodispersity of homogeneous solutions of truncated SNX20 and 21 proteins were confirmed using MALLS. SAXS data were collected at the SAXS/WAXS beamline at the Australian Synchrotron from a concentration series of each protein between 2.5 and 7.5 mg/ml. Scattering data were measured in a q range between 0.035 and 0.6 Å-1 at 12 keV with a 1.6-m camera length. Data were measured on a Pilatus 1 M camera (Dectris) and absolute scaled using distilled water. For each concentration, 20 1-s exposures were background-corrected, averaged, and scaled using ScatterBrain software. All further processing was carried out using the ATSAS program suite (24). PRIMUS was used for initial data evaluation, with Kratky plots were used to assess the folded state of the proteins. The radius of gyration (Rg) values listed in Table 2 were determined using the AutoRg function, with data quality of ≥90%. The consistency of Rg values with increasing concentration, together with inspection of Guinier plots for each data set, confirmed the absence of aggregation or interparticle interference. Similar Rg values were obtained by GNOM, the program used to calculate the pair distance distribution function (P(r)) and maximum diameter (Dmax) of the SNX20 and SNX21 proteins. Normalized scattering intensity (I(0)) values were estimated using extrapolations of the Guinier plot in the linear region qmax × Rg ≤ 1.3. Molecular masses were determined by evaluating the scattering data on an absolute scale and also by comparison with a secondary protein standard (glucose isomerase).
TABLE 2.
Summary of MALLS and SAXS analyses of the SNX-PXB proteins
| SNX21 PXB/TPR | SNX20Δ50 | SNX21Δ93 | |
|---|---|---|---|
| Structural parametersa | |||
| I(0) (cm−1) (Guinier/P(r)) | 0.031 | 0.050 | 0.041 |
| Rg (Å) (Guinier) | 21.78 ± 0.28 | 24.37 ± 0.24 | 25.78 ± 0.08 |
| Rg (Å) (P(r)) | 21.57 ± 0.25 | 24.41 ± 0.16 | 25.34 ± 0.07 |
| Dmax (Å) | 67.0 ± 2.0 | 74 ± 2.0 | 84 ± 2.0 |
| Molecular mass (Mr) determination | |||
| Sequence (kDa) | 29.6 (dimer) | 31.0 | 40.8a |
| MALLS | 29.3 | 28.9 | 40.6 |
| SAXS | 30.6 | 33.2 | 42.1 |
| Ab initio analysis | |||
| χ (GASBOR) | 0.36 | 0.38 | 0.70 |
| Rigid-body modeling/validation | |||
| χ2 | 0.435 | 0.794 | 1.21 |
| CRYSOL | BUNCH | SASREF | |
a Including a 10-kDa ubiquitin tag.
Ab initio free atom modeling was performed using the program GASBOR with a total of 40 independent simulations carried out for each protein. The accuracy of each model was assessed by the normalized residual coefficient, χ, which provides a value for the goodness of fit. Low values (close to 1.0) indicate good agreement of the models with the experimental data. The 40 individual GASBOR models were averaged using DAMAVER and filtered based on occupancy and volume to generate restored ab initio shapes of each protein. Rigid body modeling of the SNX 20 and 21 proteins was performed using either SASREF (SNX20Δ50) or BUNCH (SNX21Δ93RQ) with partial scattering amplitudes computed by the program CRYSOL. Models for SNX20 and SNX21 PX domains were generated using the structure of kinesin16B (PDB code 2V14) and cytokine-independent survival kinase (PDB code 1XTN) as templates, respectively (using Swiss-model), and used in combination with the monomeric domain of the SNX21TPR crystal structure (PDB code 4YMR). CRYSOL was used to evaluate the fitting of the theoretical curve of SNX21TPR to the experimental scattering data. The atomic models generated were docked into the SAXS-based volumes using the program SUPCOMB20.
Liposome Pelleting Assays
PIs were protonated using HCl and mixed with POPC (1-palmitoyl-2-oleoylphosphatidylcholine) lipids (POPC-phosphatidylethanolamine:PI mixed as 70–30:10%) or Folch lipids (Folch:PI mixed as 90:10%). Liposomes were prepared using a 100-nm membrane for extrusion to obtain small unilamellar vesicles. The assay was conducted using 20 μg of protein diluted in a 1 mg/ml of liposome solution in assay buffer (25 mm HEPES (pH 7), 150 mm NaCl, 1 mm DTT) to a total volume of 100 μl. After a 20-min incubation at room temperature, the liposomes were pelleted by ultracentrifugation at 100,000 × g using a benchtop ultracentrifuge (Optima TL centrifuge, rotor TLA100.3). The supernatant was collected and the pellet was resuspended in 50 μl of assay buffer to be analyzed by SDS-PAGE and Coomassie staining.
Isothermal Titration Calorimetry
Isothermal titration calorimetry experiments were performed on a microcal iTC200 instrument in 50 mm Tris (pH 8.0), 100 mm NaCl. Phospholipids (diC8-PtdIns3P, diC8-PtdIns(3,4)P2, diC8-PtdIns(3,5)P2, diC8-PtdIns(4,5)P2, diC8-PtdIns(3,4,5)P3, diC8-PtdIns4P, diC8-PtdIns) were dissolved in the respective buffers at 500 μm and titrated into 15 μm SNX20 protein solutions in 13 3.1-μl aliquots at 25 °C. Data were processed using ORIGIN to extract the thermodynamic parameters ΔH, Ka (1/Kd), and the stoichiometry n. ΔG and ΔS were derived from the relationship: ΔG = −RTln Ka and ΔG = ΔH − TΔS.
Cell Culture and Transfection
Human epithelial HeLa (CCL-2) cells were maintained in complete Dulbecco's modified Eagle's medium (DMEM) (Life Technologies) supplemented with 10% (v/v) fetal bovine serum (FBS) and 2 mm l-glutamine. Cells were incubated in humidified air/atmosphere (5% CO2) at 37 °C. Cells were transfected with Lipofectamine 2000 as per the manufacturer's instructions (Life Technologies). 0.8 μg of DNA and 2 μl of Lipofectamine 2000 was used per well of a 24-well plate.
Immunofluorescence Imaging
Cells were grown on glass coverslips, fixed with 4% formaldehyde in 250 mm HEPES, and permeabilized with Triton X-100 (0.25% in PBS) before incubation with antibodies. Cells on coverslips were incubated with primary antibodies in 1% BSA in PBS for 1 h, followed by wash in PBS and a 1-h incubation with secondary antibodies with or without DAPI (all in 1% BSA in PBS). After washing in PBS, coverslips were rinsed in water and mounted on glass slides using Dako Fluorescent Mounting Medium. Fluorescent images were acquired using Zeiss LSM 710 with Plan-Apochromatic ×63/1.4 Oil DIC objective confocal microscope, operated by ZEN2009 acquisition software.
Cell Lysis and Immunoprecipitation
HeLa cells on 10-cm dishes were transfected using Lipofectamine 2000 (Life Technologies). 16 h post-transfection the cells were washed twice with cold PBS and lysed in TK lysis buffer containing 50 mm HEPES, 150 mm NaCl, 10 mm Na4P2O7, 30 mm NaF, 2 mm Na3VO4, 10 mm EDTA, 0.5% Nonidet P-40, 0.5 mm 4-benzenesulfonyl fluoride hydrochloride and protease inhibitor mixture. Lysates were centrifuged at 13,300 rpm for 10 min at 4 °C, and the resulting supernatant was subjected to BCA assay to determine protein concentrations. For co-immunoprecipitation, equivalent amounts of protein samples were pre-cleared with 30 μl of protein G-conjugated agarose beads (50% slurry in PBS) for 1 h at 4 °C. The cleared supernatants were then subjected to CD8 or Myc immunoprecipitation by using mouse monoclonal anti-CD8 or anti-myc antibody coupling with Protein G-conjugated agarose beads overnight at 4 °C. The beads were then washed three times with lysis buffer, before bound proteins were eluted by boiling the beads for 5 min in SDS-PAGE protein loading buffer.
SDS-PAGE and Western Blotting
Equal amounts of cell lysates or immunoprecipitation samples resuspended in SDS-PAGE protein loading buffer were resolved on SDS-PAGE gels and transferred onto a PVDF membrane (Immobilon-FL, Millipore) using a semi-dry transfer apparatus (Bio-Rad) according to the manufacturer's instructions. After blocking in diluted Odyssey blocker (1:1 ratio in PBS) at room temperature for 1 h, membranes primary antibodies in diluted Odyssey blocker with 0.1% Tween 20 overnight at 4 °C. Membranes were washed in PBST three times for 5 min each wash, followed by the incubation of membranes with IRDye® secondary antibodies for 1 h at room temperature. Fluorescence intensities were detected by LI-COR Odyssey Infrared Imaging System (LI-COR Biosciences). For the detection of less sensitive signals, the enhanced chemiluminescence (ECL) detection kit (Super Signal ECL substrate, Thermo Scientific) was used.
Results
SNX20 and SNX21 Share Conserved Domain Architectures
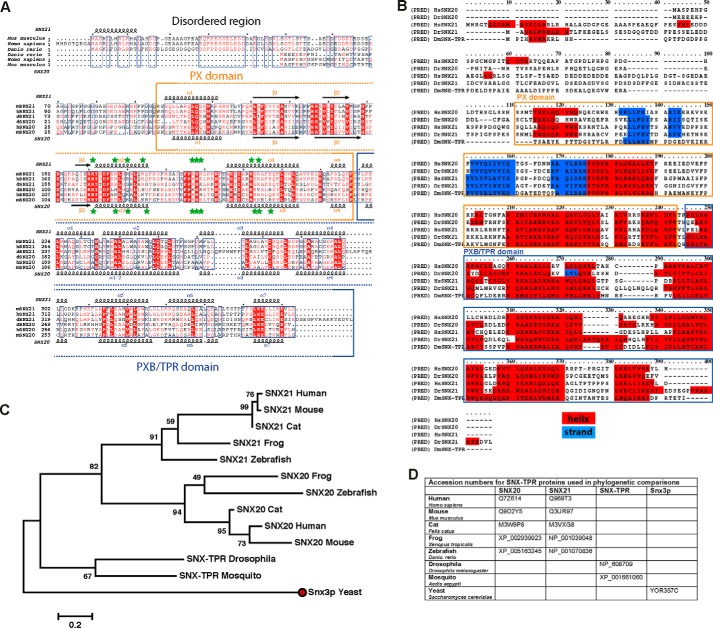
Based on sequence alignment and secondary structure prediction analysis, the SNX20 and SNX21 proteins have a similar domain architecture and share ∼40% similarity in their protein sequence. Both SNX20 and SNX21 have a predicted N-terminal disordered region, followed by a PX domain closely linked by a short sequence to the C-terminal PXB domain we defined previously (5) (Fig. 1, A and B). The helical repeats of the latter are weakly predicted to form a TPR-fold, a domain typically involved in protein-protein interactions (25). Interestingly, the very N terminus of SNX21 is predicted to include a short conserved stretch of ∼10 residues with α-helical secondary structure that is absent in SNX20, and its conservation across various species suggests a potentially unique role in SNX21 (Fig. 1B). As almost all other structural elements in both SNX20 and SNX21 align perfectly, it is clear these two proteins will share a similar tertiary structure and likely have related and perhaps redundant cellular functions. In phylogenetic analyses of the SNX20 and SNX21 proteins we were not able to trace the ancestry of these proteins further back than the Insecta class where only a single homologous protein is found (Fig. 1, C and D). This suggests the PXB domain was likely acquired late in eukaryotic evolution, and subsequently this ancestral PXB domain-containing protein underwent gene duplication and neofunctionalization that created two paralogues in chordates. SNX21 shows a slightly higher similarity to the ancestral Drosophila melanogaster protein compared with SNX20, although the difference is not significant enough to be confidant of stating whether SNX20 or SNX21 is the closest functional relative of the insect protein.
FIGURE 1.
SNX20 and SNX21 share conserved PX and PXB/TPR domains. A, sequence alignment and domain boundaries of mouse, human, and zebrafish SNX20 and SNX21. Secondary structure predictions were calculated with JPRED for the entire SNX20 sequence and the SNX21 PX domain, or determined from the crystal structure of the SNX21 PXB/TPR domain. These are indicated for SNX21 (above) and SNX20 (below). Green stars indicate point mutants generated for biochemical studies. B, SNX20 and SNX21 sequence alignment based on secondary structure predictions. Chordates and insect sequences were aligned using PRALINE (35). Residues predicted to form helical or extended structures are highlighted in red and blue, respectively. C, maximum likelihood phylogenetic tree for the SNX20 and SNX21 proteins, constructed in MEGA 5.1 with 1000 bootstrap test iterations (36). The tree was built with 10 chordates and 2 insects sequences, and was rooted using yeast Snx3p as an ancestral PX protein (red dot). The scale bar represents the genetic distance of protein members in a subtree from the corresponding node.
The SNX21 C-terminal PXB Domain Is a Tetratricopeptide Repeat (TPR)-fold
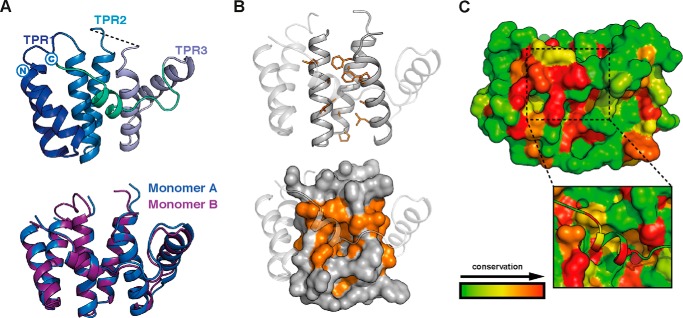
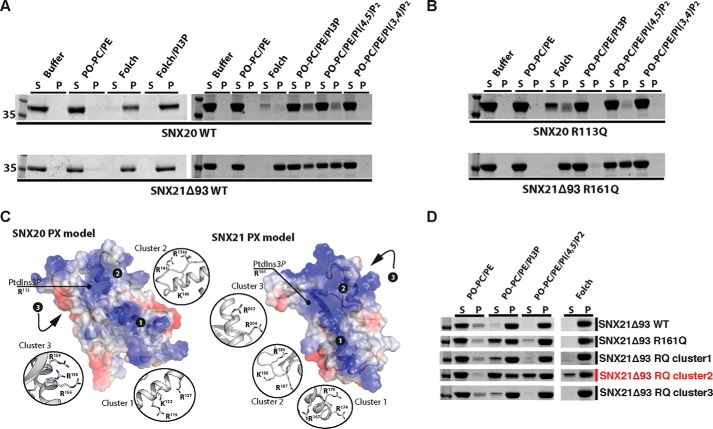
As SNXs for the most part couple the PX domain with other modules, they possess a wide range of functions including phosphoinositide kinase activity, phospholipase activity, and protein-protein interactions among others (4, 5, 7). Based on our structural predictions, we accordingly designed a number of full-length and truncated constructs that were used in subsequent structural and biochemical experiments (Fig. 2A). Multi-angle laser light scattering (MALLS) studies of these recombinant proteins indicated that SNX20Δ50 and SNX21Δ93 are both monomeric in solution, whereas the SNX21 PXB domain (SNX21Δ230) appears to form a stable homodimer. To gain insight into the possible functions of the SNX20 and SNX21 proteins, we crystallized and determined the structure of the C-terminal SNX21 PXB domain by single isomorphous replacement and anomalous scattering (SIRAS) at a resolution of 2.4 Å (Fig. 2B; Table 1). The structure shows the PXB domain does indeed consist of TPR α-helical repeats. Clear electron density reveals that the crystallized protein forms a domain-swapped dimer, linked between helices α4 and α5, stabilizing the two molecules in the asymmetric unit (Fig. 2B). This unconventional domain-swapped conformation correlates with the MALLS data that showed the SNX21 PXB/TPR domain to be dimeric in solution (Fig. 2A). In addition, the domain-swapped SNX21 PXB/TPR crystal structure showed excellent agreement with SAXS data, indicating that the domain-swapped organization of this protein is not due to its crystalline arrangement but represents a low energy, stable assembly adopted by the recombinant production in solution (Fig. 2C; Table 2).
FIGURE 2.
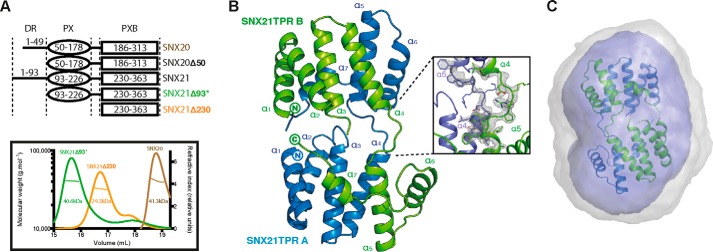
The SNX21 PXB domain crystallizes as a domain-swapped dimer. A, constructs used in this study (top) and analysis of their oligomeric state by MALLS (bottom). The center line crossing the scattering profile represents the size distribution over the peak. Asterisk indicates the presence of an N-terminal ubiquitin tag. B, top, crystal structure of the SNX21 TPR domain. The separate protein chains are colored in green and blue. The SNX21 TPR dimer reveals a domain swapping supported by the loops linking helices α4 and α5 of each chain. Refined 2Fo − Fc electron density contoured at 2.0 σ for the helices supporting the domain swapping is displayed in gray (dashed ellipse). C, the ab initio SAXS model calculated by GASBOR overlaid with the SNX21 PXB/TPR crystal structure. Averaged and filtered envelopes are shown in gray and blue, respectively. Comparison of the theoretical scattering for the SNX21 PXB/TPR domain-swapped dimer gives an excellent agreement to the experimental scattering data with a χ2 of 0.44 using CRYSOL (Table 2).
Although the SNX21 PXB domain forms a domain-swapped dimer, because both full-length SNX20 and SNX21Δ93 are monomeric, PXB dimerization is unlikely to be biologically relevant. We believe this low-energy pairing of the PXB domain forms during recombinant expression in E. coli, due to the lack of the N-terminal PX domain. Notably, neither the PXB domain of SNX20, nor the isolated PX domains of either SNX20 or SNX21 are soluble when expressed in E. coli, suggesting an intimate stabilizing association between the PX and PXB domain in the full-length proteins (further confirmed by SAXS studies described below). Therefore we based our further analyses on the extrapolated SNX21 TPR domain monomer (Fig. 3A). The SNX21 TPR domain exhibits the typical helix-turn-helix motif common to TPR proteins. The first six α-helices (α1-α6) correspond to three typical TPR repeats, whereas the seventh and final C-terminal α-helix (α7) folds in an atypical conformation in both protomers, traversing back onto the core structure (Fig. 3, A and B). The exposed surface where a α7 capping helix would normally pack against helices α5 and α6 is predominantly hydrophilic (including exposed Arg, Glu, and Asn side chains), and does not show the expected hydrophobic nature required for typical α7 packing. We deduce that the unusual α7 conformation is therefore a natural result of small sequence alterations in SNX21 protein precluding this normal arrangement.
FIGURE 3.
The SNX21 PXB domain has an atypical TPR structure. A, the monomeric SNX21 PXB domain was defined by linking the loop between α4 and α5 at the point where domain swapping occurs in the crystal structure. The monomer is composed of three TPR repeats. Alignment of both monomers shows an identical structural architecture. B, ribbon and surface representation of a patch of hydrophobic residues (orange) occluded by the C-terminal helix α7, which could potentially form an interaction surface exposed on the inner concave side of the SNX21 TPR core. C, ribbon and surface representation of the conservation of SNX21 residues among 80 relatives. The conserved atypical helix α7 (dashed line) hides another conserved patch of residues possibly implicated in cargo binding (black square).
Each TPR repeat consists of an inner helix and an outer helix that forms a concave structure, which has almost exclusively been shown to promote protein-protein interactions (25). The α7 helix itself is highly conserved, and the amino acids lining the concave groove beneath the helix also form a hydrophobic surface that is conserved in evolution among SNX21 proteins (Fig. 3C). This suggests that this region of the protein may function in establishing hydrophobic contacts with protein partners. However, the presence of the α7-helix completely obscures this hydrophobic surface. Although highly speculative, the exposed loop between α6 and α7 contains serine, threonine, and proline residues, suggesting some degree of flexibility and a potential for phosphorylation-induced regulation.
The SNX21 PXB/TPR Domain Shares Structural Similarities to the Yeast Protein Fis1p
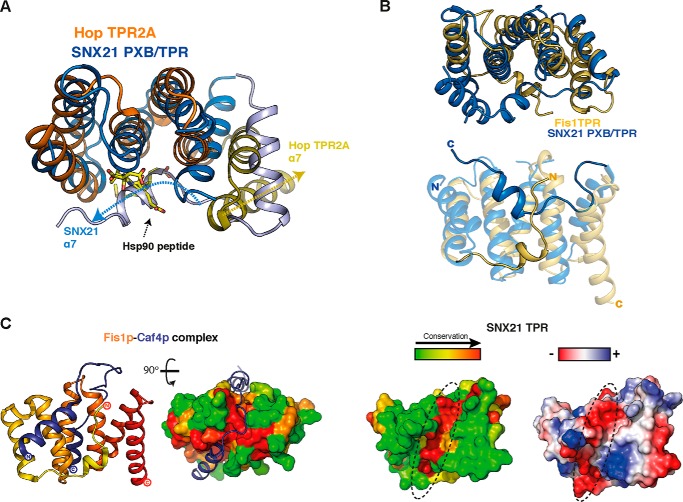
In almost all TPR domains, the extended surface shaped by the TPR repeats forms an interface for controlling interactions with various protein partners (25). For example, the concave surface of the TPR domain often associates with short linear peptide motifs in their protein partners, as seen for the Hop TPR2A domain in complex with an Hsp90 peptide (26) (PDB code 1ELR) (Fig. 4A). All TPR domains possess an odd number of α-helices, where the last helix forms a final capping structure at the C terminus of the protein. In SNX21, however, the final odd helix α7 instead adopts a unique conformation that in fact folds back across the surface of the TPR repeats, obscuring the putative concave region of the TPR domain likely to promote interactions. For example, aligning the SNX21 and Hop/Hsp90 structures highlights the fact that the α7 helix of SNX21 occludes this typical ligand-binding interface (Fig. 4A). In Hop TPR2A, the Hsp90 motif is able to bind in the groove formed by the three core TPR repeats, because the C-terminal helix (corresponding to α7 in SNX21) is folded in the typical fashion parallel to and capping the last TPR repeat.
FIGURE 4.
Potential ligand binding sites in the SNX21 PXB/TPR domain. A, the SNX21 PXB/TPR domain (blue) is aligned with the Hop TPR2A domain-Hsp90 peptide complex (Hop TPR2A, orange; Hsp90 peptide, yellow sticks). The cores of the two TPR domains superpose closely, whereas the atypical SNX21 C-terminal α7 helix lies across the canonical binding interface that mediates Hsp90 binding in Hop TPR2A. The directions of the Hop TPR2A and SNX21 α7 helices are indicated by dashed arrows to highlight the different orientations. B, structural alignment of SNX21 (blue) and yeast Fis1p TPR domains (yellow; PDB code 2PQR) highlights the structural conservation of the TPR cores, whereas the C-terminal α7 of SNX21 and the N-terminal α0 of Fis1p-fold against the concave surface from opposite directions. C, left, the Fis1p-Caf4p complex structure shows the binding region of Caf4p forms two α-helices that associate with opposite faces of the TPR domain, whereas the linker regions bind within a conserved groove at the top of Fis1p TPR domain highlighted in surface view. Right, the SNX21 TPR also possesses a similar conserved groove at the same position (dashed ellipse). This conserved surface groove on the SNX21 TPR domain is acidic, as represented by electrostatic charges plotted on its surface (dashed ellipse).
The SNX21 conformation suggests a potential self-regulatory interaction by the C-terminal α7-helix. Interestingly, we noticed a similar occlusion of the concave TPR surface in only one other TPR-containing protein: the yeast mitochondrial fission protein Fis1p. Fis1p is a mitochondrial membrane protein with a single-pass C-terminal transmembrane sequence, and a cytoplasmic N terminus recruiting the dynamin-related Dnm1p protein that mediates fission at the mitochondrial division site via adaptors Mdv1p and Caf4p (27). This cytosolic domain is a TPR-fold with an N-terminal extension shaped in a short α-helix, which traces back across the concave surface of the TPR domain, in a similar location to the SNX21 α7 helix (28, 29) (Fig. 4B). Fis1p expression in FIS1−/− yeast strains rescues mitochondrial fission-deficient phenotypes, whereas complementation with Fis1pΔN (lacking the N-terminal α-helix) does not (28). This indicates that the N-terminal helix plays a crucial role in Fis1p function in yeast, and structural studies have provided a mechanistic explanation. Yeast Fis1p effectors Mdv1 and Caf4 both associate in a hairpin fashion by clamping two interaction areas on each side of Fis1p (29) (Fig. 4C, left). This interaction is stabilized by two structural elements: a helix from Caf4p stacks against the N-terminal extension of Fis1p, and a conserved groove on the top part of the Fis1p TPR domain accommodates the stretch of sequence linking the two Caf4p helices. This binding groove is conserved in Fis1p, and most interestingly a similarly located and highly conserved groove is also seen in SNX21 (Fig. 4C). In SNX21 this groove is acidic in nature suggesting the potential to bind positively charged sequences (Fig. 4C).
SNX20 and SNX21 Adopt a Globular and Compact Architecture
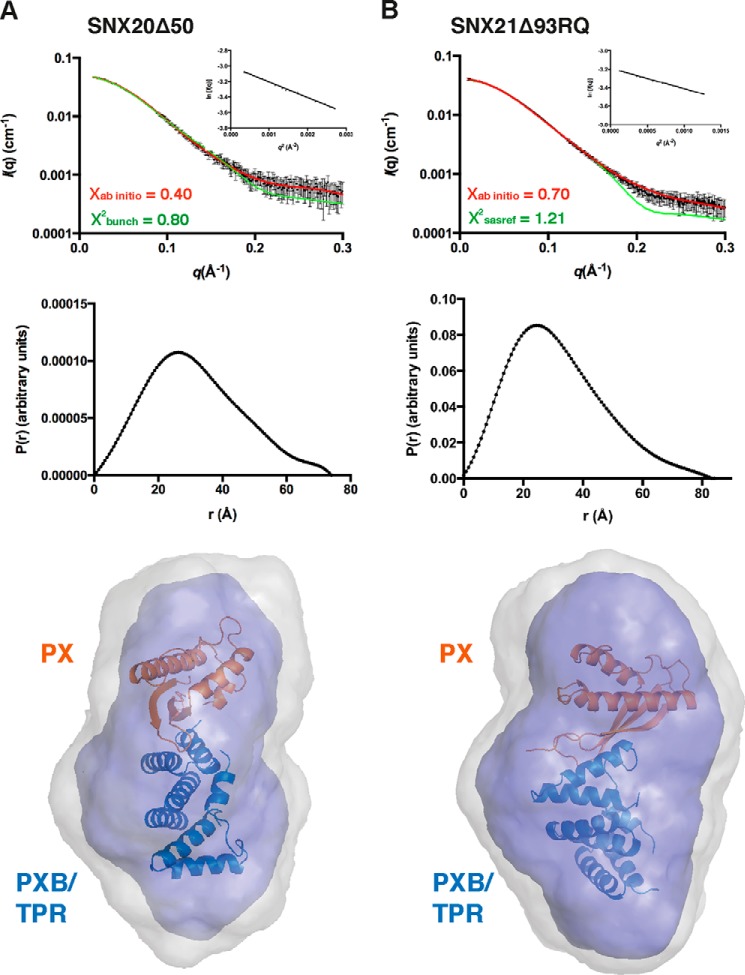
To define the overall architecture of the SNX-PXB proteins in solution, we used SAXS to examine both SNX20 and SNX21 proteins with N-terminal deletions to remove the predicted disordered regions (Fig. 5; Table 2). The scattering curves and pair distribution functions (Fig. 5A) indicate that SNX20Δ50 and SNX21Δ93 possess a relatively compact globular conformation in solution. Models for SNX20 and SNX21 PX domains were generated using the structure of kinesin16B (30) (PDB code 2V14) and cytokine-independent survival kinase (31) (PDB code 1XTN) as templates, respectively (using Swiss model), and in combination with the monomeric domain of the SNX21 TPR crystal structure were used by the program BUNCH to perform rigid body modeling. Combining these two structural modules by rigid body refinement allowed us to obtain a full-length solution structure that was compared with the SNX20Δ50 and SNX21Δ93 SAXS envelopes. The modeled domains were refined against the scattering data with excellent χ2 scores, indicating that their architecture and orientation in solution reflects their true conformation (Fig. 5B, Table 2). This derived hybrid structure overlays well with the ab initio models calculated by the program GASBOR, and reveal that the PX and PXB/TPR domains of SNX20 and SNX21 are closely packed to form a compact tertiary structure. Altogether, this data suggests that the PX and PXB/TPR domains of the SNX-PXB proteins are tightly packed against each other, and are likely to use this rigid architecture to simultaneously bind PIs and protein partners by increasing binding avidity.
FIGURE 5.
SAXS solution structures of the SNX-PXB proteins. SAXS data for the SNX20Δ50 (A) and SNX21Δ93RQ (B) proteins. Top, experimental SAXS data (black) overlaid with the theoretical profiles calculated from rigid body modeling by CRYSOL (green), or the ab initio model determined by the program GASBOR (red). Middle, distance distribution functions P(r) calculated from the scattering data of SNX20Δ50 and SNX21Δ93RQ. Bottom, molecular models of the SNX-PXB proteins show their globular compact arrangement in solution. The SNX21 TPR domain crystal structure (blue) was combined with a model of the SNX20 PX domain (generated by Swiss-model, orange) using BUNCH, and overlaid with the ab initio structures of SNX20Δ50 and SNX21Δ93RQ.
The SNX-PXB Proteins Localize to Early Endosomes at Steady State
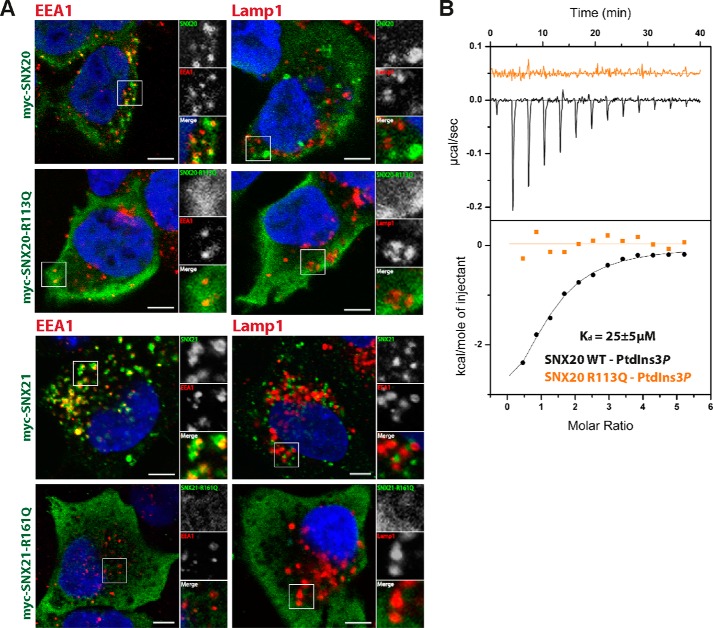
The localization of the SNX proteins to specific regions of the endocytic system is critical to their function. Previous studies of SNX20 indicated an endosomal localization of the protein in COS and CHO cells (16). Here we compared the localization of both SNX20 and SNX21 in HeLa cells using N-terminal myc-tagged myc-SNX20 and myc-SNX21 constructs (Fig. 6A). These experiments confirm the localization of SNX20 on early endosomal EEA1-positive punctate structures, but not late endosomal LAMP1-positive compartments indicating a predominantly early endosomal distribution (Fig. 7A). SNX21 shows an almost identical distribution indicating the two paralogues are both early endosome-associated SNX family members.
FIGURE 6.
The SNX-PXB proteins are localized to early endosomes via PtdIns3P interaction. A, HeLa cells were grown on coverslips and transfected with myc-SNX20, myc-SNX20-R113Q, or myc-SNX21, myc-SNX21-R161Q. Cells were co-labeled with anti-myc antibody with anti-rabbit Alexa 488 (green) and anti-EEA1 antibody or anti-Lamp1 antibody with anti-mouse Alexa 647 (red). Scale bars = 5 μm. B, native and mutant SNX20 were tested for binding against PtdIns3P by isothermal titration calorimetry and affinity measures are presented with standard deviation over three experiments. Raw data are at the top; integrated normalized data at the bottom.
FIGURE 7.
The SNX-PXB proteins possess overlapping binding sites for PtdIns3P and PtdIns(4,5)P2in vitro. A, membrane binding of SNX20 and SNX21Δ93 to liposomes containing different PI combinations was assessed by incubation following ultracentrifugation and subsequent protein content analysis of supernatant (S) and pellet (P) fractions. B, mutation of SNX20 and SNX21Δ93 at the key PtdIns3P-binding arginine side chain prevents binding to PtdIns3P but still allows interaction with negatively charged Folch membranes, and liposomes containing PtdIns(4,5)P2. C, SNX20 and SNX21 PX domain models reveal three distinct conserved basic clusters based on homology modeling. D, arginine and lysine residues forming these three clusters were mutated to glutamine to test for PI binding disruption.
The membranes of the early endosomal compartment are naturally enriched in PtdIns3P at steady state, and the structural basis for canonical PtdIns3P recognition by PX domains is now relatively well understood, involving a key arginine residue that contacts the 3-phosphate group (Arg113 in SNX20 and Arg161 in SNX21) (5). Using isothermal titration calorimetry we find that SNX20 binds PtdIns3P headgroups with a Kd ∼ 25 μm, whereas a mutation in the canonical PtdIns3P-binding pocket (R113Q) abolishes this interaction (Fig. 6B). As expected, when SNX20 and SNX21 protein constructs are mutated at the 3-phosphate binding arginine residues of their PX domains they show a diffuse cytoplasmic localization in HeLa cells and are no longer recruited to endosomal membranes (Fig. 6A).
The SNX-PXB Proteins Interact with PtdIns3P and PtdIns(4,5)P2-containing Membranes in Vitro via Overlapping Pockets
Although cellular experiments confirm the binding of SNX20 and SNX21 to PtdIns3P-enriched early endosomes in cells we also tested their qualitative interactions with membranes in vitro using liposome-pelleting assays with full-length SNX20 and SNX21Δ93 (Fig. 7A). Both SNX20 and SNX21Δ93 display quite similar binding profiles to PI-containing liposomes. SNX20 and SNX21 were both found to bind PtdIns3P-containing liposomes as predicted from their sequences and endosomal localization (5). PtdIns3P-enriched liposomes pelleted slightly higher levels of SNX21, suggesting a tighter interaction than with SNX20. Surprisingly, however, a substantial amount of protein was also found in the pellet fraction of liposomes containing PtdIns(4,5)P2. This appeared to be selective as proteins were not bound to the similar isoform PtdIns(3,4)P2. This data also correlates with the fact that the SNX-PXB proteins bind very strongly to Folch lipids, a brain-extracted lipid mixture containing multiple species of PIs but only relatively low levels of PtdIns3P (Fig. 7A). In our experience, proteins that bind PtdIns3P alone are not fully pelleted by Folch liposomes in these assays. In liposome pelleting assays using SNX20 and SNX21 proteins with mutations in the canonical PtdIns3P-binding site (R113Q and R161Q, respectively), we see complete loss of binding to PtdIns3P-containing liposomes, however, binding to Folch lipids and PtdIns(4,5)P2 remains relatively unaffected (Fig. 7B). This suggests that the canonical binding pocket of the SNX20 and SNX21 PX domains is able to associate with PtdIns3P normally, but that a second PtdIns(4,5)P2 binding site exists somewhere within the proteins. Notably, we were not able to detect binding to the soluble PtdIns(4,5)P2 headgroup by isothermal titration calorimetry (not shown) suggesting this interaction occurs preferentially within a membrane environment.
Surface representations of the SNX20 and SNX21 PX domain homology models (generated for our SAXS experiments) suggest the presence of additional basic, positively charged putative phospholipid binding sites, some of which overlap to some extent with the canonical PtdIns3P-binding site (Fig. 7C). We next mutated these clusters of positively charged residues in SNX21Δ93 to glutamine to test their potential role in PI interactions. As described above SNX21Δ93 (R161Q) was perturbed in binding PtdIns3P but was still pelleted by Folch liposomes. In fact we found that all mutant proteins were somewhat perturbed in binding to PtdIns3P in this system, however, the greatest effects were seen for the canonical R161Q site, and also for cluster 2 (residues Arg187-Lys188-Arg189) (Fig. 7D). Similarly this cluster 2 mutant, and R161Q to a lesser extent, also reduced binding to both Folch and PtdIns(4,5)P2. Cluster 2 residues lie in the extended loop connecting helices α1 and α2 of the PX domain. Typically this loop is highly flexible, and adjusts its conformation to embrace the bound canonical PtdIns3P headgroup (5). Our data therefore suggests that the PI-binding pocket is competent for binding to PtdIns3P in the usual manner, but can also accommodate PtdIns(4,5)P2, although the precise orientation of PtdIns(4,5)P2 in the binding pocket is unknown and will require further high-resolution structural analyses to define. Altogether, our data indicates that PtdIns(4,5)P2 binding is mediated by an overlapping binding pocket to the canonical PtdIns3P cavity on the PX domain of the SNX-PXB proteins.
No Evidence for Stable Association of SNX20 or SNX21 with PSGL-1
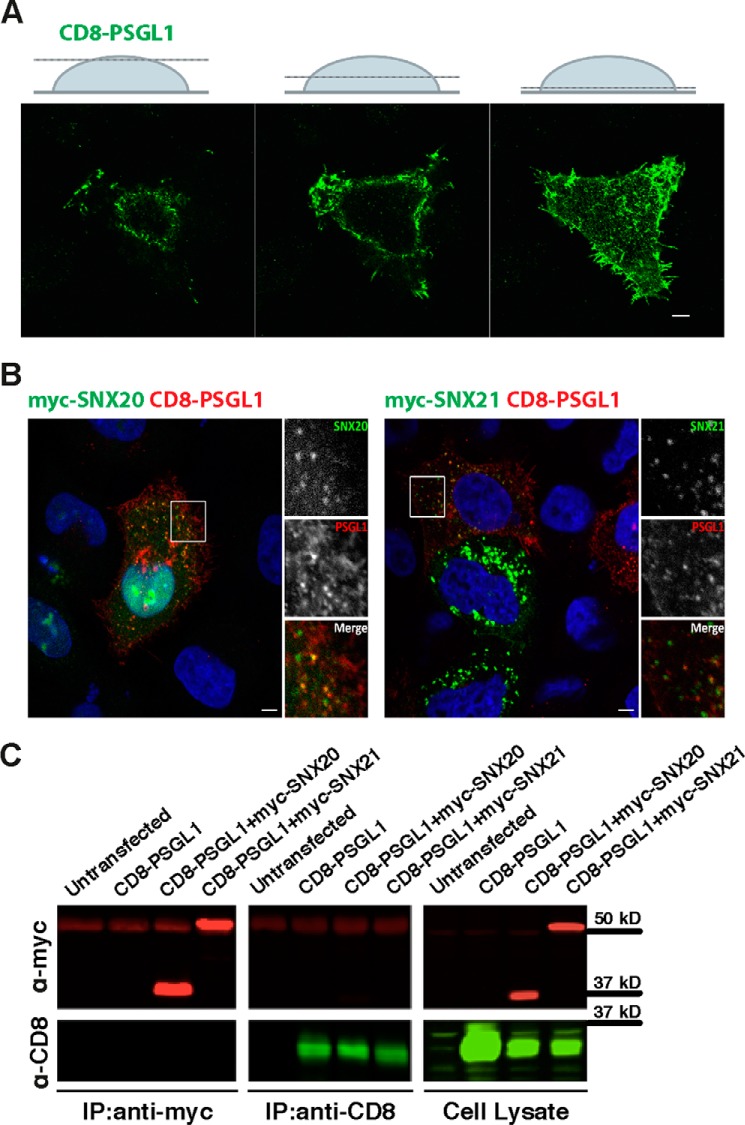
The major question regarding the SNX-PXB proteins is what is the function(s) of these apparently conserved endosomal scaffolds? Previously it was reported that SNX20 could interact with the cytoplasmic domain of the PSGL-1 receptor, and it was observed that overexpression of SNX20 caused a redistribution of PSGL-1 from a predominantly cell-surface location to SNX20-decorated endosomal compartments (16). We attempted to confirm these findings using a CD8-PSGL-1 fusion reporter, where the cytoplasmic tail of PSGL-1 is fused to the lumenal and transmembrane domains of CD8. In immunofluorescence experiments, we observe that CD8-PSGL-1 is localized at the plasma membrane of HeLa cells and also decorates intracellular membrane compartments (Fig. 8A). When the CD8-PSGL-1 reporter is co-expressed with myc-SNX20 or myc-SNX21 we do observe colocalization of the proteins at early endosomal compartments, however, we do not see any change in CD8-PSGL-1 cellular distribution relative to control experiments as was reported for SNX20 previously (16) (Fig. 8B). In conjunction with this we also do not find stable association with CD8-PSGL-1 using either immunoprecipitations (Fig. 8C) or GST-tagged PSGL-1 pulldowns with purified SNX-PXB proteins (not shown). Therefore, whereas our data shows that both SNX20 and SNX21 share overlapping early endosomal localization with PSGL-1, we do not see evidence for a stable interaction with PSGL-1 in this context.
FIGURE 8.
SNX20 and SNX21 do not stably associate with the putative cargo PSGL-1 in HeLa cells. A, HeLa cells were grown on coverslips and transfected with CD8-PSGL-1. 24 h post-transfection, cells were fixed and permeabilized and indirect immunofluorescence was performed using antibody with rabbit anti-CD8 antibodies and anti-rabbit Alexa 488 (green). Z-stack sections depict highlight of plasma CD8-PSGL-1 membrane localization at steady state. Scale bars = 5 μm. B, CD8-PSGL1 was co-expressed with myc-SNX20 (left) or myc-SNX21 (right) and their co-localization determined by indirect immunofluorescence. Scale bars = 5 μm. C, co-immunoprecipitation (IP) from HeLa cells transiently expressing the indicated constructs was performed using anti-myc or anti-CD8 antibodies, respectively. Calculated relative molecular mass of each protein is indicated on the gels.
Discussion
Although the functions of many of the mammalian SNX proteins have been studied, revealing roles in a variety of signaling and cellular trafficking processes (4, 5, 7), a large number still remain very poorly characterized. Two of these proteins, SNX20 and SNX21, belong to the SNX-PXB subfamily, and to gain insights into the possible functions of these SNX proteins we have examined both their structure and membrane binding properties in vitro and in situ. Overall our data indicates that SNX-PXB proteins will possess a compact modular structure, which will likely be employed to engage multiple membrane-associated ligands including lipid headgroups and transmembrane proteins at the endosomal compartment.
The PX domain is generally employed by SNX family proteins to bind specifically to the PI lipid PtdIns3P, leading to the large majority of SNX proteins being localized at the PtdIns3P-enriched early endosomal compartment (5, 32). Although we find that both SNX20 and SNX21 are localized to endosomal compartments, as was also shown previously for SNX20 (16), we discovered an unexpected ability to recognize both PtdIns3P and PtdIns(4,5)P2. This is promoted by a large positively charged binding pocket, with overlapping specificity for these two phospholipids. It should be noted that our results differ somewhat from previous experiments conducted by Schaff et al. (16) where lipid-strip assays suggested SNX20 could bind to numerous PI species other than the two we identified. The presence of two distinct binding pockets in the same PX domain has previously been described, for example, in phospholipase D1 (33). In this case a second distinct lipid binding pocket is formed by a cluster of cationic residues that bind with moderate affinity to anionic lipids and the secondary messenger phosphatidic acid. Simultaneous association with both distinct binding pockets increases the efficiency of phospholipase D1 PX domain-mediated membrane targeting. The mechanistic basis and more importantly the functional significance of the binding of SNX-PXB proteins to PtdIns(4,5)P2 is still unclear, especially given the obvious lack of plasma membrane recruitment of SNX20 and SNX21 where PtdIns(4,5)P2 levels are highest. However, it suggests a potential for regulating the localization of SNX-PXB proteins to different membrane domains, or following stimulation-induced changes in PI levels.
Our findings confirmed that the previously defined C-terminal “PXB” domain of the SNX20 and SNX21 proteins possesses a TPR-fold, containing three TPR α-helical repeats. TPR scaffolds are composed of a sequential array of paired α-helices that form inherently curved structures, with concave surfaces typically employed to bind associated ligands. More than 100 TPR structures deposited in the PDB show that these domains exhibit different surface properties, promoting selectivity for targets. The amino acid insertions that modify the topology of the loops and length and angles of the helices give rise to impressive variations in structure, and thus make this module a highly versatile binding scaffold (reviewed in Ref. 34). The usual topology of TPR proteins is supported by an odd number of helices, the final one capping the hydrophobic core of the last TPR repeat for solubility. In our case, the three SNX21 TPR repeats are not capped in the usual fashion; instead the atypical helix α7-folds back to cover the hydrophobic concave groove of the TPR domain. In this conformation, helix α7 covers the region normally involved in heterotypic protein-protein interactions. Although Fis1p and SNX20/SNX21 are functionally unrelated, their similar topologies lead us to speculate that the mechanistic properties of Fis1p may provide some clues to the potential binding regions of the SNX20/SNX21 proteins. By comparison with the yeast TPR protein Fis1p (28), we suggest one possibility is that membrane-targeted SNX20/SNX21 proteins may engage transmembrane ligands through an unusual binding surface similarly to the binding of the Caf4p adaptor to Fis1p (Figs. 4C and 9). A contrasting alternative model is that the atypical α7 helix could play a potential regulatory role, whereby ligand binding or perhaps signal-induced conformational changes such as phosphorylation of the α6-α7 linker region cause a movement in α7 allowing ligand association with the conserved hydrophobic pocket buried beneath.
FIGURE 9.
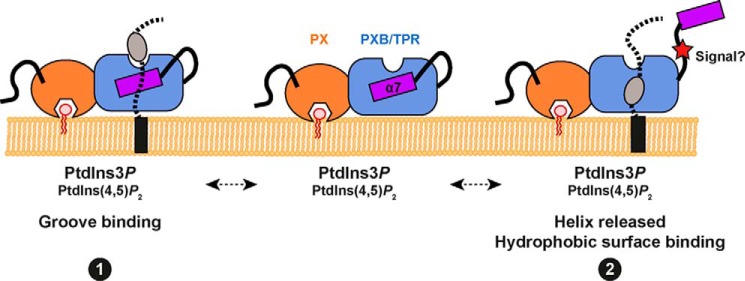
A model for membrane and ligand engagement by the SNX-PXB proteins. We propose two alternatives for coincident lipid and protein binding to the SNX-PXB proteins. The PX domain can bind the endosomal lipid PtdIns3P, as well as the lipid PtdIns(4,5)P2 although the functional significance of this is still unclear. In conjunction with membrane targeting the TPR domain of the SNX-PXB proteins may then engage ligands through an unusual binding groove similarly to the binding of the Caf4p adaptor to Fis1p (see Fig. 4C) (binding mode 1). Alternatively the atypical α7 helix could play a regulatory role, whereby ligand binding or perhaps signal-induced conformational changes such as phosphorylation of the α6-α7 linker region cause a movement in α7 allowing ligand association with the conserved hydrophobic pocket buried beneath (binding mode 2).
This speculation regarding ligand binding is predicated on the assumption that the TPR domains of SNX-PXB proteins are involved in modulating protein-protein interactions (as is typically the case), either with soluble effectors or transmembrane cargo proteins. However, defining the mechanism of ligand attachment to SNX20 or SNX21 will first require identification of SNX20- and SNX21-associated proteins. Schaff et al. (16) proposed a role for SNX20 in trafficking of the inflammatory adhesion receptor PSGL-1 after identifying an interaction with PSGL-1 in a yeast two-hybrid screen. Unfortunately we were not able to replicate these reported interactions between PSGL-1 and SNX-PXB proteins, although we do observe an overlapping cellular distribution of the proteins. We note that a confusing aspect to this previous study is that the region of SNX20 binding to PSGL-1 identified in two-hybrid screens was a C-terminal fragment of the PX domain plus the PXB domain, however, peptide array studies suggested the PX domain alone was responsible for binding. Thus the molecular basis for PSGL-1 interaction with SNX20 remains to be firmly established, and whether SNX20 or SNX21 contribute to PSGL-1 trafficking remains unclear.
In conclusion, we show that the SNX20 and SNX21 constitute a subfamily of SNX proteins possessing both a PX domain and C-terminal PXB/TPR domain. These proteins show the clear hallmarks of being membrane-associated scaffolding proteins, with a potential to mediate interactions with ligands including transmembrane proteins that require transport within the endosomal system, cytosolic effectors, or proteins regulating SNX-PXB function. Our work provides the first molecular insights into these proteins, and a foundation for understanding the mechanisms by which they influence endosomal function.
Acknowledgments
We acknowledge support from the staff and facilities of the University of Queensland Remote Operation Crystallization and x-ray (UQ ROCX) facility, and the Australian Synchrotron. We thank in particular Simon Williams for assistance with multi-angle laser light scattering experiments and Oleksiy Kovtun for providing the expression vector and cloning materials used in this study.
This work is supported in part by Australian Research Council ARC Grant DP0985029 and National Health and Medical Research Council (NHMRC) Grants APP1042082 and APP1058734.
The atomic coordinates and structure factors (code 4YMR) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- SNX
- sorting nexins
- PI
- phosphoinositide
- PX
- phox homology
- PSGL-1
- P-selectin glycoprotein ligand 1
- TRP
- tetratricopeptide repeat
- SAXS
- small angle x-ray scattering
- MALLS
- multi-angle laser light scattering
- PDB
- Protein Data Bank
- PXB
- PX-associated domain B.
References
- 1. Scita G., Di Fiore P. P. (2010) The endocytic matrix. Nature 463, 464–473 [DOI] [PubMed] [Google Scholar]
- 2. Hsu V. W., Bai M., Li J. (2012) Getting active: protein sorting in endocytic recycling. Nat. Rev. Mol. Cell Biol. 13, 323–328 [DOI] [PubMed] [Google Scholar]
- 3. Sorkin A., von Zastrow M. (2009) Endocytosis and signalling: intertwining molecular networks. Nat. Rev. Mol. Cell Biol. 10, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cullen P. J. (2008) Endosomal sorting and signalling: an emerging role for sorting nexins. Nat. Rev. Mol. Cell Biol. 9, 574–582 [DOI] [PubMed] [Google Scholar]
- 5. Teasdale R. D., Collins B. M. (2012) Insights into the PX (phox-homology) domain and SNX (sorting nexin) protein families: structures, functions and roles in disease. Biochem. J. 441, 39–59 [DOI] [PubMed] [Google Scholar]
- 6. Carlton J., Bujny M., Rutherford A., Cullen P. (2005) Sorting nexins: unifying trends and new perspectives. Traffic 6, 75–82 [DOI] [PubMed] [Google Scholar]
- 7. Seet L. F., Hong W. (2006) The Phox (PX) domain proteins and membrane traffic. Biochim. Biophys. Acta 1761, 878–896 [DOI] [PubMed] [Google Scholar]
- 8. Ghai R., Mobli M., Norwood S. J., Bugarcic A., Teasdale R. D., King G. F., Collins B. M. (2011) Phox homology band 4.1/ezrin/radixin/moesin-like proteins function as molecular scaffolds that interact with cargo receptors and Ras GTPases. Proc. Natl. Acad. Sci. U.S.A. 108, 7763–7768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ghai R., Bugarcic A., Liu H., Norwood S. J., Skeldal S., Coulson E. J., Li S. S., Teasdale R. D., Collins B. M. (2013) Structural basis for endosomal trafficking of diverse transmembrane cargos by PX-FERM proteins. Proc. Natl. Acad. Sci. U.S.A. 110, E643–E652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gallon M., Clairfeuille T., Steinberg F., Mas C., Ghai R., Sessions R. B., Teasdale R. D., Collins B. M., Cullen P. J. (2014) A unique PDZ domain and arrestin-like fold interaction reveals mechanistic details of endocytic recycling by SNX27-retromer. Proc. Natl. Acad. Sci. U.S.A. 111, E3604–E3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ghai R., Tello-Lafoz M., Norwood S. J., Yang Z., Clairfeuille T., Teasdale R. D., Merida I., Collins B. M. (2014) Phosphoinositide binding by the SNX27 FERM domain regulates localisation at the immune synapse of activated T-cells. J. Cell Sci. 28, 553–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Steinberg F., Gallon M., Winfield M., Thomas E. C., Bell A. J., Heesom K. J., Tavaré J. M., Cullen P. J. (2013) A global analysis of SNX27-retromer assembly and cargo specificity reveals a function in glucose and metal ion transport. Nat. Cell. Biol. 15, 461–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lemmon M. A. (2008) Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9, 99–111 [DOI] [PubMed] [Google Scholar]
- 14. Wu C., Orozco C., Boyer J., Leglise M., Goodale J., Batalov S., Hodge C. L., Haase J., Janes J., Huss J. W., 3rd, Su A. I. (2009) BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 10, R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zeng W., Yuan W., Wang Y., Jiao W., Zhu Y., Huang C., Li D., Li Y., Zhu C., Wu X., Liu M. (2002) Expression of a novel member of sorting nexin gene family, SNX-L, in human liver development. Biochem. Biophys. Res. Commun. 299, 542–548 [DOI] [PubMed] [Google Scholar]
- 16. Schaff U. Y., Shih H. H., Lorenz M., Sako D., Kriz R., Milarski K., Bates B., Tchernychev B., Shaw G. D., Simon S. I. (2008) SLIC-1/sorting nexin 20: a novel sorting nexin that directs subcellular distribution of PSGL-1. Eur. J. Immunol. 38, 550–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Danson C., Brown E., Hemmings O. J., McGough I. J., Yarwood S., Heesom K. J., Carlton J. G., Martin-Serrano J., May M. T., Verkade P., Cullen P. J. (2013) SNX15 links clathrin endocytosis to the PtdIns3P early endosome independently of the APPL1 endosome. J. Cell Sci. 126, 4885–4899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lieu Z. Z., Derby M. C., Teasdale R. D., Hart C., Gunn P., Gleeson P. A. (2007) The golgin GCC88 is required for efficient retrograde transport of cargo from the early endosomes to the trans-Golgi network. Mol. Biol. Cell 18, 4979–4991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Leslie A. G. (2006) The integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 62, 48–57 [DOI] [PubMed] [Google Scholar]
- 20. Collaborative Computational Project, Number 4. (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 21. Abrahams J. P., Leslie A. G. (1996) Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D Biol. Crystallogr. 52, 30–42 [DOI] [PubMed] [Google Scholar]
- 22. Potterton E., Briggs P., Turkenburg M., Dodson E. (2003) A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 59, 1131–1137 [DOI] [PubMed] [Google Scholar]
- 23. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Petoukhov M. V., Franke D., Shkumatov A. V., Tria G., Kikhney A. G., Gajda M., Gorba C., Mertens H. D., Konarev P. V., Svergun D. I. (2012) New developments in the program package for small-angle scattering data analysis. J. Appl. Crystallogr. 45, 342–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zeytuni N., Zarivach R. (2012) Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure 20, 397–405 [DOI] [PubMed] [Google Scholar]
- 26. Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F. U., Moarefi I. (2000) Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell 101, 199–210 [DOI] [PubMed] [Google Scholar]
- 27. Mozdy A. D., McCaffery J. M., Shaw J. M. (2000) Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 151, 367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Suzuki M., Neutzner A., Tjandra N., Youle R. J. (2005) Novel structure of the N terminus in yeast Fis1 correlates with a specialized function in mitochondrial fission. J. Biol. Chem. 280, 21444–21452 [DOI] [PubMed] [Google Scholar]
- 29. Zhang Y., Chan D. C. (2007) Structural basis for recruitment of mitochondrial fission complexes by Fis1. Proc. Natl. Acad. Sci. U.S.A. 104, 18526–18530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Blatner N. R., Wilson M. I., Lei C., Hong W., Murray D., Williams R. L., Cho W. (2007) The structural basis of novel endosome anchoring activity of KIF16B kinesin. EMBO J. 26, 3709–3719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xing Y., Liu D., Zhang R., Joachimiak A., Songyang Z., Xu W. (2004) Structural basis of membrane targeting by the Phox homology domain of cytokine-independent survival kinase (CISK-PX). J. Biol. Chem. 279, 30662–30669 [DOI] [PubMed] [Google Scholar]
- 32. Yu J. W., Lemmon M. A. (2001) All phox homology (PX) domains from Saccharomyces cerevisiae specifically recognize phosphatidylinositol 3-phosphate. J. Biol. Chem. 276, 44179–44184 [DOI] [PubMed] [Google Scholar]
- 33. Stahelin R. V., Ananthanarayanan B., Blatner N. R., Singh S., Bruzik K. S., Murray D., Cho W. (2004) Mechanism of membrane binding of the phospholipase D1 PX domain. J. Biol. Chem. 279, 54918–54926 [DOI] [PubMed] [Google Scholar]
- 34. Allan R. K., Ratajczak T. (2011) Versatile TPR domains accommodate different modes of target protein recognition and function. Cell Stress Chaperones 16, 353–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bawono P., Heringa J. (2014) PRALINE: a versatile multiple sequence alignment toolkit. Methods Mol. Biol. 1079, 245–262 [DOI] [PubMed] [Google Scholar]
- 36. Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]