Background: Streptococcus pneumoniae interacts with matricellular human thrombospondin-1 (hTSP-1), facilitating adhesion to and invasion into host cells.
Results: Pneumococcal surface proteins PavB and PspC bind hTSP-1 specifically.
Conclusion: PavB and PspC are important hTSP-1 adhesins of Gram-positive pneumococci.
Significance: This study demonstrates the importance of pneumococcal adhesins for hTSP-1-mediated host-pathogen interactions.
Keywords: adhesin, bacterial pathogenesis, extracellular matrix protein, platelet, thrombospondin, MSCRAMMs, pneumococci
Abstract
The human matricellular glycoprotein thrombospondin-1 (hTSP-1) is released by activated platelets and mediates adhesion of Gram-positive bacteria to various host cells. In staphylococci, the adhesins extracellular adherence protein (Eap) and autolysin (Atl), both surface-exposed proteins containing repeating structures, were shown to be involved in the acquisition of hTSP-1 to the bacterial surface. The interaction partner(s) on the pneumococcal surface was hitherto unknown. Here, we demonstrate for the first time that pneumococcal adherence and virulence factor B (PavB) and pneumococcal surface protein C (PspC) are key players for the interaction of Streptococcus pneumoniae with matricellular hTSP-1. PavB and PspC are pneumococcal surface-exposed adhesins and virulence factors exhibiting repetitive sequences in their core structure. Heterologously expressed fragments of PavB and PspC containing repetitive structures exhibit hTSP-1 binding activity as shown by ELISA and surface plasmon resonance studies. Binding of hTSP-1 is charge-dependent and inhibited by heparin. Importantly, the deficiency in PavB and PspC reduces the recruitment of soluble hTSP-1 by pneumococci and decreases hTSP-1-mediated pneumococcal adherence to human epithelial cells. Platelet activation assays suggested that PavB and PspC are not involved in the activation of purified human platelets by pneumococci. In conclusion, this study indicates a pivotal role of PavB and PspC for pneumococcal recruitment of soluble hTSP-1 to the bacterial surface and binding of pneumococci to host cell-bound hTSP-1 during adhesion.
Introduction
Streptococcus pneumoniae is a widespread Gram-positive bacterial commensal of the upper respiratory epithelia of humans. Colonization of mucosal surfaces with S. pneumoniae is often transient and asymptomatically. Especially children under the age of five, elderly, and immunocompromised individuals exhibit enhanced colonization rates (1, 2). Besides its role as a commensal, pneumococci are also major facultative human pathogens causing disease patterns ranging from mild local infections, such as otitis media or sinusitis, to more severe, life-threatening infections like pneumonia, meningitis, or sepsis (3, 4). The dissemination of pneumococci occurs via aerosols from person to person, as humans represent the main biological reservoir (5, 6). A prerequisite for a stable colonization is the capability to adhere to respiratory epithelial cells either directly or indirectly by targeting the extracellular matrix and the resistance against phagocytosis, antimicrobial substances, and mucus-mediated clearance (7). Here, the polysaccharide capsule of pneumococci represents the major barrier against the innate immunity (8). However, the capsule also represents a barrier for the interaction of bacterial surface proteins with receptors on the apical side of host cells or extracellular matrix (ECM),3 components essential for a stable colonization of the host. Therefore, S. pneumoniae undergoes phase variations in which the polysaccharide capsule is mainly degraded (8, 9). In this scenario the interaction of bacterial surface adhesins with soluble host proteins, host cellular receptors, or components of the ECM represents the major strategy essential to achieve a stable colonization (10, 11). The epithelial ECM comprises the interstitial matrix between cells and the basal lamina upon the epithelial layer, which is composed of proteoglycans and fibrous proteins (among others fibronectin, collagen, elastins, and laminins) as well as matricellular proteins like vitronectin (Vn) or hTSP-1 (12–16).
Human TSP-1 is a 420-kDa homotrimeric multidomain glycoprotein with multiple, often tissue-specific functions, which includes cell adhesion, signaling, proliferation, and angiogenesis of different cell types as well as immune regulation (17–20). Human TSP-1 monomers consist thereby of a globular N-terminal heparin binding domain, a von Willebrand factor binding domain, three properdin-like type I repeats, three epidermal growth factor-like type II repeats, eight calcium binding type III repeats, and a globular C-terminal domain (21, 22). The biological functions are mediated by the interaction of these hTSP-1 domains with a wide range of host molecules like cytokines, cell surface receptors, and components of the ECM (23). Human TSP-1 is stored in α-granules of platelets but is also synthesized and secreted by various other cell types like endothelial cells, smooth muscle cells, and macrophages (24–26). The plasma concentration of circulating hTSP-1 ranges usually between 20 and 300 ng/ml, which increases after activation-induced platelet α-granule release up to 30 μg/ml (27). In addition to the mentioned functions in eukaryotic organisms, Gram-positive bacteria were shown to recruit soluble hTSP-1 to their surface in a dose-dependent manner. This binding increases bacterial adhesion and invasion into various epithelial and endothelial cells, as shown for S. pneumoniae, Staphylococcus aureus, and other Gram-positive human pathogens but not for Gram-negative bacteria (28, 29). Binding of hTSP-1 to the eukaryotic cell surface and ECM components is well characterized, whereas the interaction partners on the bacterial site are largely unknown. In a recent study we were able to identify repeating structures of the main staphylococcal autolysin Atl in S. aureus and Staphylococcus epidermidis as a direct binding partner for hTSP-1 (29). Furthermore, the extracellular adherence protein (Eap) of S. aureus was shown to bind hTSP-1 (30).
For staphylococci, proteinaceous hTSP-1 interaction partners could be identified; however, the adhesins of other Gram-positive species are unknown. In previous experiments pneumococci showed a strong and dose-dependent acquisition of soluble hTSP-1 to their surface (29). Therefore, this study aimed to identify hTSP-1-binding proteins of S. pneumoniae. Interestingly, all yet identified staphylococcal hTSP-1-binding proteins contained repeating sequences that have been demonstrated as being essential for the interplay with hTSP-1. We, therefore, focused on pneumococcal surface adhesins containing repetitive motifs. The most promising candidates were pneumococcal surface protein C (PspC) and pneumococcal adherence and virulence factor B (PavB). Both proteins possess repeating structures and were previously shown to bind various human extracellular/matricellular matrix proteins such as fibronectin or Vn (11, 31), thereby mediating pneumococcal colonization and invasion of host cells (11, 32–36).
The family of pneumococcal PspC proteins (also termed CbpA or SpsA) is subdivided in 11 subtypes and two subgroups. The classical PspC (subtypes 1–6, subgroup one) exhibits a choline binding motif for a non-covalent anchoring to the choline moiety of cell wall-associated teichoic acids. The so called PspC-like proteins (subtypes 7–11, subgroup 2) contain a classical LPXTG motif for covalent linkage to the peptidoglycan via the sortase A enzyme. PspC binds to the secretory component of the human polymeric immunoglobulin receptor (pIgR) and the 67 kDa-laminin receptor, which leads to an internalization of pneumococci into host cells. Furthermore, PspC sequesters Factor H, Vn and the C4B-binding protein (C4BP), contributing to pneumococcal immune evasion (31, 37–41).
PavB, anchored covalently to the cell wall via the sortase, is an adhesin of pneumococci and composed of repeating sequences, which are known as streptococcal surface repeats (SSURE). Depending on the pneumococcal strain, PavB consists of 5–9 SSURE with an average of 150 amino acids per repeat (11). PavB was shown to be essential for pneumococcal adhesion to various epithelial cells as well as for the colonization and pathogenesis in the host as tested in different mouse models of infection and carriage. On the molecular level, PavB mainly interacts with fibronectin and plasminogen, both components of the ECM and serum (11, 42).
Besides binding to epithelial and endothelial cells, various bacteria are able to bind to human platelets leading to subsequent activation and aggregation (43). The interaction of bacteria or bacterial factors is either directly mediated by receptors of platelets or indirectly via recruited complement components or IgG (44, 45). Nevertheless, bacterial-platelet interaction can also be promoted by the utilization of plasma proteins, forming bridges between bacteria and platelet-activating receptors as shown for the fibronectin binding proteins of S. aureus (46). In addition, pneumococci were shown to interact with platelets, a process mainly being mediated by fibrin but also supported by hTSP-1 (47).
In this study, we investigated the interaction of hTSP-1 with the pneumococcal adhesins PspC and PavB and the molecular mechanisms underlying hTSP-1-mediated adherence of S. pneumoniae to human epithelial host cells. Therefore, we used pneumococcal deletion mutants and heterologously expressed PavB and PspC fragments. Furthermore, we investigated the role of PavB and PspC as human platelet stimulating factors.
Experimental Procedures
Bacterial Strains and Culture Conditions
S. pneumoniae wild-type and isogenic deletion mutants (Table 1) were cultivated in Todd-Hewitt broth (Roth) supplemented with 0.5% yeast extract (Roth) to mid-log phase (A600 = 0.35–0.40) at 37 °C and 5% CO2 or grown on Columbia blood agar plates (Oxoid) containing appropriate antibiotics (50 μg/ml kanamycin or 5 μg/ml erythromycin). Escherichia coli BL21 strains were cultured on solid Luria-Bertani (LB) medium plates or in liquid LB medium (Roth) in the presence of kanamycin (50 μg/ml) at 30 °C to mid-log phase (A600 = 0.8) on an environmental shaker.
TABLE 1.
Pneumococcal strains used in this study
| S. pneumoniae strain | Characteristics | Reference/Source |
|---|---|---|
| PN111 | D39Δcps (Δcps::kanr) | (28) |
| PN140 | D39ΔcpsΔpavB (ΔpavB::ermr) | (11) |
| PN194 | D39ΔcpsΔpspC (ΔpspC::ermr) | (31) |
| PN361 | D39ΔcpsΔpavBΔpspC (ΔpavB::ermr, ΔpspC::aad9) | This study |
| SP 37 | Serotype 35A (low encapsulated) | NCTC10319 |
| PN114 | Serotype 35AΔpspC (ΔpspC::ermr) | (38) |
| PN126 | Serotype 35AΔpavB (ΔpavB::ermr) | (11) |
| PN483 | Serotype 35AΔpavBΔpspC (ΔpavB::ermr, ΔpspC::aad9) | This study |
Isolation and Purification of Human TSP-1
Human thrombospondin-1 was purified from human platelets as described recently (29).
Fluorescein Isothiocyanate Labeling of Human TSP-1
Purified hTSP-1, dialyzed against 0.1 m carbonate buffer (pH 9.2), was labeled with FITC (AppliChem) using a standard protocol. In brief, hTSP-1 was incubated with FITC (dissolved in 0.1 m carbonate buffer (pH 9.2)) in a molecular ratio of 1:30 for 1.5 h at room temperature. Unbound FITC was removed by dialysis (12–14-kDa molecular weight cut off) against phosphate-buffered saline (PBS, pH 7.4) at 4 °C overnight.
Heterologous Expression and Purification of Recombinant Proteins
N-terminally His6-tagged PavB and PspC proteins used in this study have been described earlier (11, 31, 40, 48). Using Ni2+ affinity chromatography, His6-tagged proteins were purified from E. coli BL21 cell lysates after induction of protein expression (at A600 = 0.8) with 1 mm IPTG at 30 °C for 3 h. After using either 1-ml HisTrapTM FF crude columns (GE Healthcare) and ÄktaTM purifier system (GE Healthcare) or a prepacked Protino nickel column kit (Macherey-Nagel), proteins were dialyzed (12–14-kDa molecular weight cut off) against PBS (pH 7.4) at 4 °C overnight. The protein concentration was determined using Bradford reagent (Sigma) at A595. The stability and purity of expressed proteins was monitored by SDS-PAGE and Coomassie Brilliant Blue R-250 (CBB) staining.
Generation of Antibodies
Polyclonal antibodies against native PspC-SH2 and PavB-SSURE2+3 were generated by routine immunization of mice (CD-1, Charles River) using standard protocols. In brief, 20 μg of purified recombinant protein per mouse and incomplete Freund's adjuvant (50:50 v/v) (Sigma) were injected intraperitoneally. In a biweekly interval, mice were boosted twice with 20 μg of protein and incomplete Freund's adjuvant. After bleeding, purification of IgG was performed using Protein A-Sepharose® (Sigma) affinity chromatography. The polyclonal mouse anti-hTSP-1 IgG was generated as previously described (29). Polyclonal antibodies against pneumococci were generated by subcutaneous immunization of rabbits with a suspension of 1 × 109 heat inactivated bacteria in 0.15 m sodium chloride. The purification of IgG was performed as described above.
SDS-PAGE, Western Blot, and Immunological Detection
The purity of heterologously expressed PavB and PspC fragments was confirmed by SDS-PAGE and CBB R-250 staining. Furthermore, SDS-PAGE-separated proteins were transferred on a nitrocellulose membrane using a semi-dry blotting system (Bio-Rad). After blocking the membrane with 5% skim milk and 2% bovine serum albumin (BSA, Roth) in PBS overnight and washing 3 times with PBS, 0.05% Tween® 20, proteins were detected with polyclonal mouse anti-PspC-SH2 IgG (1:500 in PBS), polyclonal mouse anti-PavB-SSURE2+3 antibody (1:500 in PBS), or a monoclonal mouse anti-PentaHisTM IgG (Qiagen, 1:2000 in PBS) for 1 h at room temperature followed by incubation with a secondary alkaline phosphatase-coupled anti-mouse antibody (1:7500, Dianova) diluted in PBS for 1 h at room temperature. Subsequently, the nitrocellulose membrane was washed twice with PBS, 0.05% Tween® 20 and once with alkaline phosphatase buffer (0.1 m Tris, 0.1 m NaCl, 5 mm MgCl2·6H2O, pH 9.5). Protein bands were visualized using 5-bromo-4-chloro-3-indolyl-phosphate/nitro blue tetrazolium (0.25%/0.5%) in alkaline phosphatase buffer.
Surface Plasmon Resonance (SPR)
The direct protein-protein interactions between hTSP-1 and the His6-tagged pneumococcal protein fragments under native conditions were analyzed by SPR using a BIAcoreT100 optical biosensor (GE Healthcare). Human TSP-1 was immobilized as ligand on a carboxymethyl dextran (CM5) sensor chip using standard amine-coupling procedures as described (29). Binding analysis was performed with PBS, Hepes, and Hepes supplemented with 1 mm MgCl2 and 2 mm CaCl2 containing 0.05% Tween® 20 at 25 °C using a flow rate of 10 μl/min. Each interaction was measured at least three times. Data were analyzed using BIAcoreT100 Evaluation Software (Version 2.0.1.1).
Enzyme-linked Immunosorbent Assay (ELISA)
The binding between His6-tagged PavB and PspC derivates and purified hTSP-1 was analyzed by ELISA. Microtiter plates (96-well, MaxisorpTM, Nunc, Thermo Fisher Scientific) were coated with 0.1 μg/well hTSP-1 overnight at 4 °C. Cavities were washed 3 times with washing buffer (PBS, pH 7.4, 0.05% Tween® 20) and blocked with blocking buffer (PBS, 0.1% Tween® 20 supplemented with 1% BSA) for 1 h at room temperature. Next, wells were washed and incubated with increasing molecular ratios of pneumococcal PavB or PspC protein fragments, diluted in blocking buffer (1 h, room temperature). The protein-protein interactions were detected using a polyclonal antiserum (mouse anti-PavB-SSURE2+3 serum or polyclonal mouse anti-SH2 IgG, 1:500 in blocking buffer) and a secondary goat anti-mouse IgG coupled to horseradish peroxidase (HRP, 1:1000, Jackson ImmunoResearch Laboratories, Inc.), each incubated for 1 h at room temperature. 2,2′-Azino-di-[3-ethylbenzthiazoline sulfonate] diammonium salt (ABTS, Roche Applied Science) or O-phenylenediamine dihydrochloride (Dako) was used as a HRP substrate. Absorbance was measured at A405 or A492 using a FLUOstar Omega Fluoreader (BMG Labtech). In competition assays immobilized hTSP-1 was incubated with increasing concentrations of sodium chloride (0–1.0 m), heparin (0–5 mg/ml), or chondroitin sulfate A (Sigma) (0–5 mg/ml) and a constant molecular ratio (relating to hTSP-1) of the PavB or PspC proteins. To analyze the binding of soluble hTSP-1 to immobilized recombinant pneumococcal proteins, equimolar amounts of PavB or PspC fragments related to SSURE2 or SH3 (each 0.5 μg/well) were immobilized overnight at 4 °C in 96-well PolysorpTM microtiter plates (Thermo Scientific). After washing 3 times with washing buffer (PBS, pH 7.4), 0.01% Tween® 20) and subsequent blocking with PBS, 0.01% Tween® 20 supplemented with 1% BSA (blocking buffer), increasing concentrations of hTSP-1 (0–50 μg/ml) diluted in blocking buffer were incubated for 1 h at room temperature. Protein binding was detected with polyclonal mouse anti-hTSP-1 IgG (1:500 in blocking buffer) and a secondary goat anti-mouse IgG coupled to horseradish peroxidase using O-phenylenediamine dihydrochloride as substrate. In competition assays immobilized SH13 was incubated with 5 μg/ml hTSP-1 and increasing molar ratios of human Vn (relating to hTSP-1). Bound hTSP-1 was detected as described above. Vice versa, immobilized SH13 was also incubated with 2.5 μg/ml Vn and increasing molar ratios of hTSP-1 (relating to Vn). Bound Vn was detected using a polyclonal antiserum from rabbit (1:500 in blocking buffer, Complement Tec) and a secondary goat anti-rabbit IgG coupled to horseradish peroxidase (1:1000 in blocking buffer; Dianova) using O-phenylenediamine dihydrochloride as substrate.
Flow Cytometric Analysis of hTSP-1 Binding to S. pneumoniae and Epithelial Cells
S. pneumoniae D39Δcps and NCTC10319 (serotype 35A) were cultivated to mid-log phase, harvested at 3273 × g for 6 min, and washed with PBS (pH 7.4). A 50-μl suspension containing 2 × 108 bacteria was incubated with increasing concentrations of hTSP-1 (0–12.5 μg/ml) in 100 μl of PBS for 30 min at 37 °C and 5% CO2 in 96-well plates (U-bottom, Greiner Bio-One). After washing, hTSP-1 binding was detected using a polyclonal mouse anti-hTSP-1 IgG (1:500 in PBS, 0.5% FCS) and secondary AlexaFluor® 488-labeled goat anti-mouse IgG (Abcam) (1:500 in PBS/0.5% FCS), each incubated for 45 min at 4 °C. Finally, the bacteria were washed, fixed with 1% paraformaldehyde overnight at 4 °C, and fluorescence was determined using flow cytometry (FACS CaliburTM, BD Biosciences). Pneumococci were detected using log forward and log side scatter dot plot. To exclude debris and larger aggregates of bacteria, a gating region was set. 50,000 events were analyzed using log-scale amplification. The results of hTSP-1 binding to S. pneumoniae are shown as the geometric mean fluorescence intensity of the gated bacterial population multiplied by the percentage of labeled bacteria.
Human adenocarcinoma alveolar epithelial cells (A549, ATCC CCl-185; type II pneumocytes) were seeded in 24-well plates (Cellstar, Greiner) and cultivated to confluent monolayers with ∼2 × 105 cells in Dulbecco's modified Eagle's medium (DMEM, low glucose, PAA Laboratories) supplemented with 10% heat-inactivated fetal bovine serum (Gibco) and 2 mm l-glutamine (PAA Laboratories) for 48 h at 37 °C and 5% CO2. After washing with DMEM the cells were preincubated with 1 μm manganese chloride (MnCl2) for 1 h at 37 °C and 5% CO2. A549 cells were washed and incubated with FITC-labeled hTSP-1 (0–50 μg/ml in DMEM) for 30 min at 37 °C. To remove unbound hTSP-1, cells were washed twice with PBS, 0.5% FCS and subsequently fixed overnight at 4 °C with 1% paraformaldehyde. Binding of hTSP-1-FITC was assessed by flow cytometry using FACSCaliburTM (BD Biosciences). Detection of cells occurred within a log-forward and log-side scatter dot plot. For fluorescence analysis 25,000 events were counted using log-scale amplification. The results of hTSP-1-FITC binding to A549 cells are illustrated as histograms and geometric mean fluorescence intensity multiplied by the percentage of positive events. For data acquisition, CellQuestPro Software 6.0 (BD Biosciences) was used and data analysis was performed using Flowing Software 2.5.1 (by Perttu Terho, Turku Centre for Biotechnology).
Cell Culture Adherence Assays and Immunofluorescence Microscopy
Adherence of S. pneumoniae to A549 cells was calculated post infection by immune fluorescence microscopy. A549 cells were seeded on glass coverslips (diameter 12 mm) at a density of 5.2 × 104 cells per well and cultivated in DMEM (low glucose) supplemented with 10% fetal bovine serum and 2 mm l-glutamine (PAA Laboratories) to confluent monolayers with ∼2 × 105 cells per well. After washing twice with infection medium (DMEM, low glucose), preincubation with 1 μm MnCl2, and repeated washing, cells were incubated with hTSP-1 (0–50 μg/ml in DMEM) for 30 min at 37 °C. Pneumococci were cultivated to mid-log phase and washed with infection medium. The eukaryotic cell monolayer was infected with pneumococci in 500 μl of infection medium with a multiplicity of infection of 25. After spinfection (3 min, 120 × g) cells were incubated for 1.5 h at 37 °C and 5% CO2. Next, non-adherent bacteria were removed by rinsing with PBS, 0.5% FCS twice. Afterward, the infected cells were fixed on glass coverslips with 1% paraformaldehyde at 4 °C overnight. The coverslips were blocked with PBS, 10% FCS for 1 h at 37 °C. After washing with PBS, 0.5% FCS, cell-bound hTSP-1 and pneumococci were stained with polyclonal mouse anti-hTSP-1 antibody (1:500) and polyclonal rabbit anti-pneumococcal IgG (1:200) diluted in PBS, 0.5% FCS at 37 °C for 45 min followed by incubation with secondary AlexaFluor® 568-labeled goat anti-mouse IgG (Abcam) and secondary AlexaFluor® 488-labeled goat anti-rabbit IgG (Abcam), each 1:500 in PBS, 0.5% FCS for 45 min at 37 °C. Visualization of the cells was achieved by permeabilization with 0.1% Triton-X-100 (Serva) for 7 min and incubation with AlexaFluor® 488-coupled phalloidin (1:200 in PBS, 0.5% FCS; Invitrogen). The adherence of bacteria to 100 host cells was determined by counting attached bacteria using a fluorescence microscope LSM510 META (Zeiss) and VisiView 2.0.4 (Visitron Systems GmbH) software.
Platelet Isolation
The protocols for this study were reviewed and approved by the ethics board of the University Medicine of Greifswald. Human platelets were isolated from healthy volunteers as previously described (49–52). In brief, human blood, anticoagulated with acid-citrate-dextrose (ACD) (7 ml of ACD and 50 ml of blood) was centrifuged at room temperature for 20 min at 150 × g. Platelet-rich plasma was removed and centrifuged at 475 × g for 20 min in the presence of 3 μm prostaglandin E1 (PGE-1). The resulting platelet pellet was resuspended in 10 ml of PIPES-saline-glucose (pH 6.8) and supplemented with 3 μm PGE-1. Contaminating leukocytes were removed by CD45 microbead selection using autoMACS (Miltenyi Biotec). CD45-depleted platelets were centrifuged in the presence of 3 μm PGE-1 (475 × g, 20 min, room temperature) and resuspended in prewarmed serum-free medium M199 (Lonza) to 1 × 109 platelets per ml.
Platelet Activation with hTSP-1, Recombinant Pneumococcal Proteins, and S. pneumoniae
Platelets were incubated with PBS, 10 μg/ml PspC constructs SH13 or SH2, 25 μg/ml PavB fragment SSURE1–5, 25 μg/ml hTSP-1, or the pneumococcal proteins in combination with hTSP-1 (15 and 60 min, 37 °C, 5% CO2). Furthermore, platelets were incubated in suspension with nonencapsulated S. pneumoniae mutants at a bacterium-to-platelet ratio of 1:10. The pneumococci were preincubated either with PBS as negative control or 25 μg/ml hTSP-1 for 30 min at 37 °C and 5% CO2. Moreover, platelets were activated with 100 ng/ml convulxin (kindly provided by Dr K. J. Clemetson, University of Bern, Bern, Switzerland) as a positive control.
Measurement of Platelet Activation by Flow Cytometry
After incubation with either pneumococcal proteins, pneumococci, or controls, 4 × 106 platelets were incubated with mouse anti-human CD62P (P-selectin)-PE Cy5-conjugated antibody or the isotype-matched control or a mouse PAC-1 (anti-αIIbβIII)-FITC-labeled antibody (BD Biosciences) for 10 min at room temperature in the dark. Nonspecific PAC-1 antibody binding was controlled by preincubation of platelets with 1 mg/ml RGDS (Sigma; 10 min, room temperature). After fixation with Cellfix (BD Biosciences; 10 min, room temperature, in the dark) platelets were analyzed by flow cytometry (Cytomics FC 500; Beckman Coulter) using standard analysis protocols. Platelets were detected using log forward and log side scatter dot plots. A gating region was set to exclude debris and larger aggregates of platelets. 20,000 events were analyzed for fluorescence using log-scale amplification. The results of platelet activation are shown as percentage of labeled platelets.
Statistical Analysis
All data are reported as the mean ± S.D. Statistical analysis was performed with unpaired Student's t test. Platelet assays were analyzed with paired Student's t test. In all analyzes a p value < 0.05 was considered statistically significant.
Results
Binding of hTSP-1 Is Diminished in PavB- and PspC-deficient Pneumococci
To assess the impact of PavB and PspC for pneumococcal recruitment of soluble hTSP-1, wild-type pneumococci and isogenic mutants were incubated with increasing concentrations (0–12.5 μg/ml) of soluble hTSP-1, and bacterial bound hTSP-1 was analyzed by flow cytometry. Although all tested strains showed a dose-dependent binding of hTSP-1 (Fig. 1, A and B), binding of hTSP-1 to the single pavB and pspC deletion mutants was significantly decreased in comparison to the isogenic wild type. Human TSP-1 binding was even lower for the double mutants deficient for PavB and PspC, resulting in a decrease in hTSP-1 binding of 70% (nonencapsulated S. pneumoniae D39Δcps) or 85% (serotype 35A, NCTC10319), respectively. These results indicate the involvement of PavB and PspC in recruitment of soluble hTSP-1 to the bacterial surface of S. pneumoniae and exhibit an additive effect when both proteins were absent.
FIGURE 1.
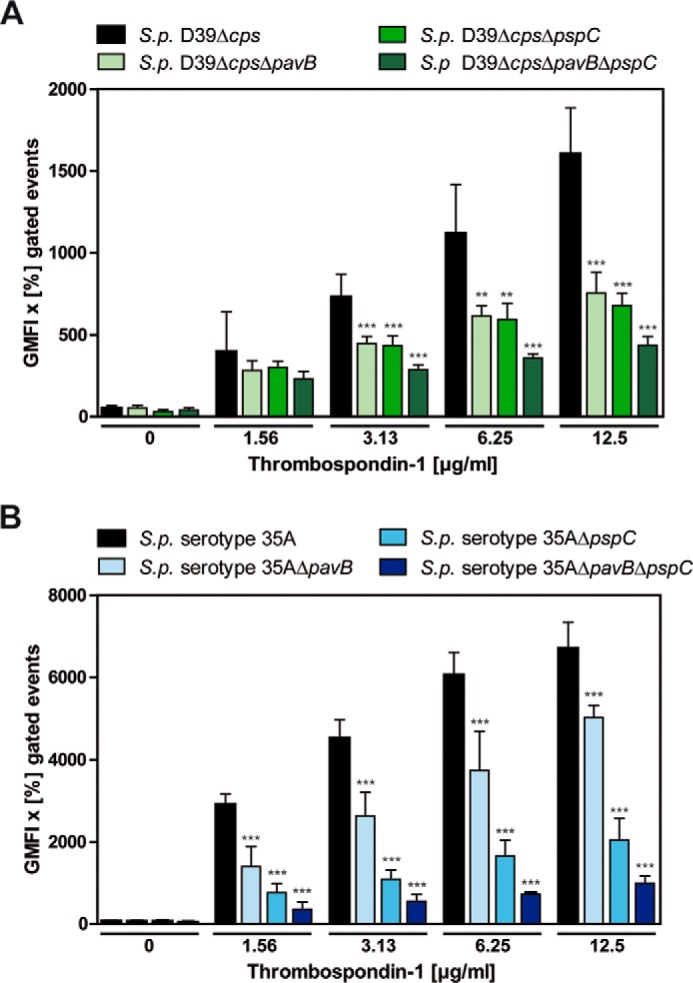
PavB and PspC of S. pneumoniae recruit soluble human thrombospondin-1 to the bacterial cell surface. A and B, concentration-dependent binding of soluble hTSP-1 to S. pneumoniae D39Δcps, S. pneumoniae serotype 35A, and isogenic mutants deficient for PavB, PspC or both. Bacteria (2 × 108) were incubated with increasing concentrations of hTSP-1 (0–12.5 μg/ml) in 100 μl of PBS. Binding of surface-associated hTSP-1 was measured by flow cytometry using a specific polyclonal mouse anti-hTSP-1 antibody and secondary AlexaFluor® 488-conjugated anti-mouse IgG. Binding of hTSP-1 is shown as the geometric mean fluorescence intensity (GMFI) multiplied by the percentage of gated events (GMFI × % gated events). The mean values of at least three independent experiments are shown with error bars corresponding to S.D. **, p < 0.01; ***, p < 0.001 versus S. pneumoniae (S.p.) D39Δcps or serotype 35A.
An additional deletion mutant of both pneumococcal strains lacking the LPXTG-anchored serine protease A (PrtA) served as the control, which shows no reduction in hTSP-1 binding. The acquisition of hTSP-1 to the cell envelope of the prtA deletion mutants was comparable with the isogenic wild types (data not shown). These data demonstrate a pivotal role for PavB and PspC in hTSP-1 recruitment to the bacterial surface and suggest that PavB and PspC are major hTSP-1-binding proteins of S. pneumoniae.
Heterologous Expression of PspC and PavB Fragments
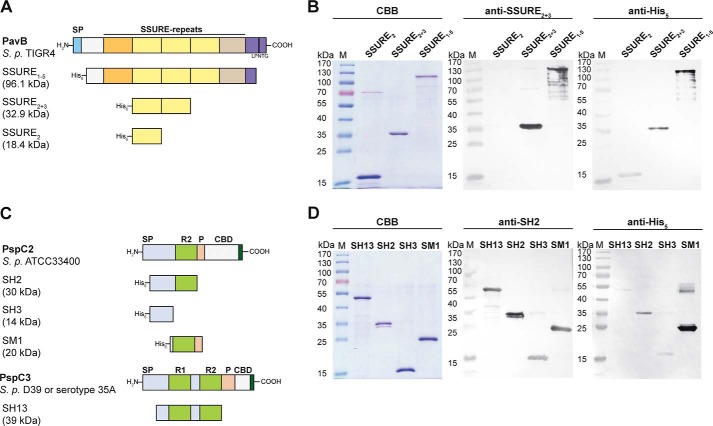
To examine a direct interaction between hTSP-1 and pneumococcal proteins PavB and PspC, respectively, different gene fragments encoding the mature PspC protein lacking the proline-rich sequence and choline binding domain, the mature PavB protein lacking the cell wall anchoring domain, or mature parts of the proteins were cloned into expression vectors and heterologously expressed in E. coli (11, 31, 40, 48). The PavB protein from S. pneumoniae TIGR4, containing the repeats SSURE 1–5, as well as fragments containing the repeats 2 + 3 or the single repeat 2 were expressed and purified as His6-tagged proteins (Fig. 2A). For PspC, two different PspC derivatives were used: PspC2 from S. pneumoniae ATCC33400 containing one repeating sequence and PspC3 originating from S. pneumoniae D39 or serotype 35A strains and exhibiting two repeating domains. Different PspC constructs containing one repetitive motif (SH2, SM1), two repeats (SH13), or a fragment without repetitive motifs (SH3) were heterologously expressed and purified as His6-tagged proteins (Fig. 2C). The purity of the various heterologously expressed proteins was verified using SDS-polyacrylamide gel electrophoresis and CBB staining as well as semidry blots (Fig. 2, B and D). The purified proteins were used for further binding studies by ELISA and SPR (Figs. 3–5).
FIGURE 2.
Heterologously expressed PavB and PspC fragments. A and C, schematic model of PavB of S. pneumoniae TIGR4 and PspC subtypes PspC2 (S. pneumoniae ATCC33400) and PspC3 (S. pneumoniae D39 or serotype 35A) and heterologously expressed His6-tagged fragments of both surface proteins. SP, signal peptide; SSURE, streptococcal surface repeats; LPNTG, sortase anchoring motif; R, repeat domain; P, proline-rich sequence; CBD, choline-binding domain. B, SDS-PAGE of heterologously expressed PavB fragments (SSURE2, SSURE2+3, SSURE1–5) stained with CBB R250 and corresponding immunoblots. Proteins were detected using a specific polyclonal mouse anti-SSURE2+3 antibody or a monoclonal mouse anti-penta-His antibody and an alkaline phosphatase-coupled secondary anti-mouse antibody. D, SDS-PAGE of heterologously expressed PspC2 (SH2, SH3, SM1) and PspC3 (SH13) fragments stained with CBB R250 and corresponding immunoblots. Proteins were detected using a specific polyclonal mouse anti-SH2 antibody or a monoclonal mouse anti-penta-His antibody and an alkaline phosphatase-coupled secondary anti-mouse antibody.
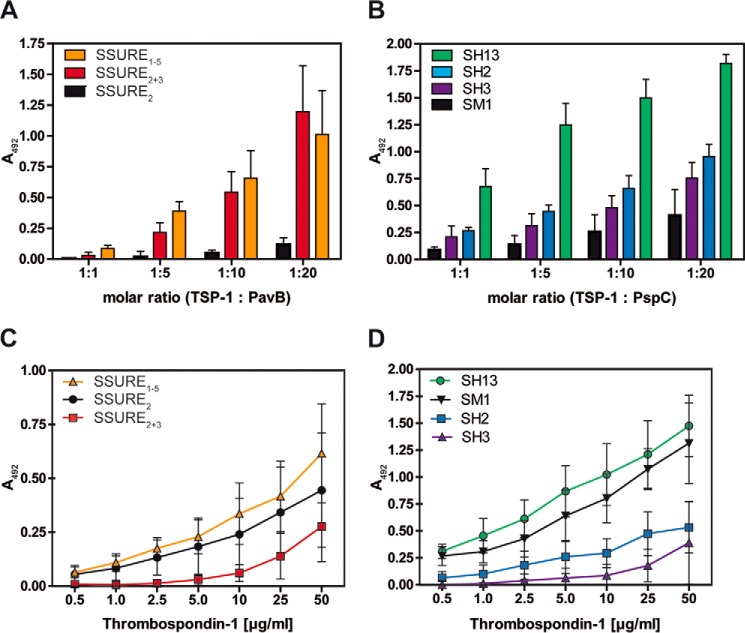
FIGURE 3.
Repetitive structures of PavB and PspC are involved in the interaction with hTSP-1. A and B, dose-dependent binding of soluble pneumococcal proteins PavB (A) or PspC (B) to immobilized hTSP-1. Human TSP-1 (0.1 μg in 100 μl/well) was coated on microtiter plates (MaxisorpTM, Nunc) and, after blocking, incubated with increasing molecular ratios of heterologously expressed PavB or PspC fragments. Bound pneumococcal proteins were detected using a polyclonal mouse anti-SSURE2+3 (PavB) antibody or a polyclonal mouse anti-SH2 (PspC) antibody and a peroxidase-coupled secondary anti-mouse antibody. Results are illustrated as mean values ± S.D. of at least three independent experiments. C and D, concentration-dependent binding of soluble hTSP-1 to immobilized, heterologously expressed PavB and PspC proteins. Pneumococcal proteins were immobilized on microtiter plates (PolysorpTM, Nunc) in equimolar amounts related to SSURE2 or SH3 (each 0.5 μg in 50 μl/well). Binding of hTSP-1 was detected after incubating the immobilized proteins with increasing concentrations of hTSP-1 (0–50 μg/ml) using a specific polyclonal mouse anti-hTSP-1 antibody and a peroxidase-coupled secondary anti-mouse antibody. Results are illustrated as mean values ± S.D. of at least four independent experiments.
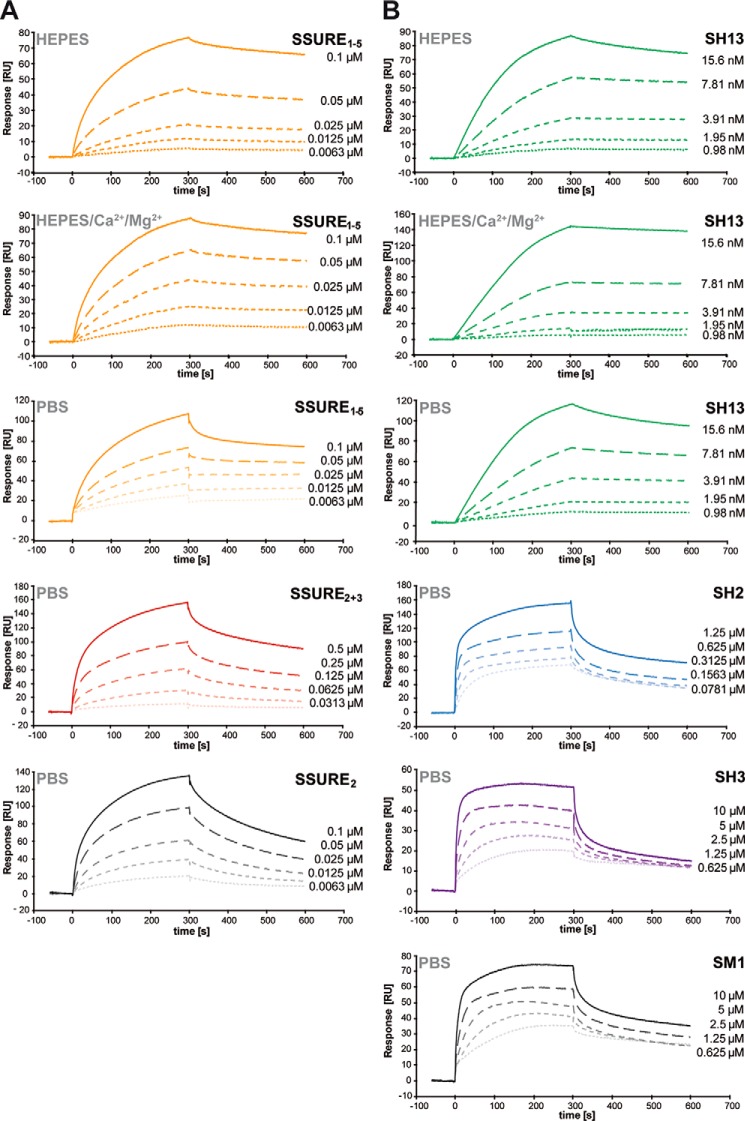
FIGURE 4.
Kinetics of the interaction between pneumococcal adhesins and hTSP-1. A and B, interactions of heterologously expressed pneumococcal proteins PavB and PspC with immobilized hTSP-1 were analyzed by surface plasmon resonance spectroscopy under different buffer conditions. Sensorgrams show the dose-dependent binding of the pneumococcal hTSP-1-binding proteins. A CM5-sensorchip was coated with native hTSP-1 (∼3000 response units), and the PavB and PspC proteins in PBS, Hepes, Hepes supplemented with 1 mm MgCl2 and 2 mm CaCl2 containing 0.05% Tween® 20 (pH 7.4) were used as analytes at a flow rate of 10 μl/min. The association and dissociation was observed, each for 300 s. Values of the control flow cells were subtracted from each sensorgram.
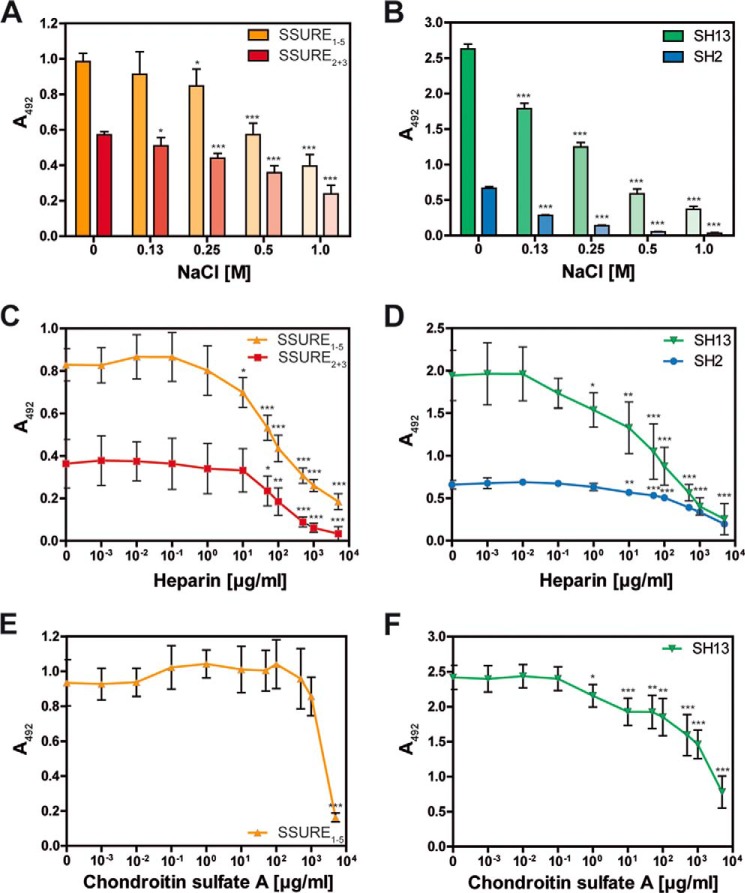
FIGURE 5.
Interaction between pneumococcal adhesins and hTSP-1 is charge-dependent and inhibited by heparin. A–F, human TSP-1 (0.1 μg in 100 μl/well) was immobilized on microtiter plates (MaxisorpTM, Nunc) and incubated with a constant molecular ratio of heterologously expressed PavB or PspC fragments (related to hTSP-1) in the presence of increasing concentrations of sodium chloride (0–1.0 m), heparin (0–5 mg/ml), or chondroitin sulfate A (0–5 mg/ml). Bound pneumococcal proteins were detected using a polyclonal mouse anti-SSURE2+3 (PavB) antibody or a polyclonal mouse anti-SH2 (PspC) antibody and a peroxidase-coupled secondary anti-mouse antibody. The mean values of at least three independent experiments are shown with error bars corresponding to S.D. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus buffer.
Repeating Structures of PavB and PspC Are Involved in the Interaction with hTSP-1
Protein-protein interactions with native proteins were analyzed under static conditions using an ELISA assay as well as under flow conditions using surface plasmon resonance spectroscopy. Using ELISA, PavB- and PspC proteins showed a dose-dependent binding to immobilized hTSP-1 (Fig. 3). The levels of SSURE1–5 and SSURE2+3 binding to immobilized hTSP-1 were comparable when used in similar molecular ratios (related to immobilized hTSP-1). In contrast, the SSURE2 protein, representing a single repeat of PavB, showed a very low level of hTSP-1 binding (Fig. 3A).
PspC SH13, containing two repetitive sequences termed R1 and R2 domain, exhibited a strong binding to immobilized hTSP-1, which is comparable to binding of SSURE2+3 (Fig. 2B). However, PspC proteins harboring only one R-domain or lacking the R-domain showed a reduced binding capacity to hTSP-1 when compared with PspC SH13 (Fig. 3B).
In a second series of experiments, PavB and PspC proteins were immobilized in equimolar ratios (related to SSURE2 and SH3, respectively) and soluble hTSP-1 was added in increasing concentrations. Binding of hTSP-1 to immobilized PavB and PspC proteins was dose-dependent. Similar to the experimental approach with immobilized hTSP-1, the highest binding capacity of soluble hTSP-1 was demonstrated to SSURE1–5 and PspC SH13 (Fig. 3, C and D). These data suggest that the binding efficiency between hTSP-1 and pneumococcal proteins PavB or PspC depends on the repetitive sequences of PavB and PspC. In addition, soluble hTSP-1 interacts with immobilized pneumococcal adhesins PavB and PspC, respectively, and vice versa, the soluble adhesins bind to the immobilized form of hTSP-1. However, PavB SSURE2 and PspC SM1 behave differently depending whether they are used in an immobilized or soluble form. We suppose that only one SSURE2 or SM1 molecule binds to one immobilized hTSP-1 molecule, whereas soluble hTSP-1 binds to more than one of the immobilized pneumococcal fragments resulting in a higher avidity interaction.
Kinetics of PavB and PspC Binding to hTSP-1 as Analyzed by Surface Plasmon Resonance
To analyze the association and dissociation between PavB or PspC and hTSP-1, SPR spectroscopy was performed. Human TSP-1 was immobilized on a CM5 sensor chip, whereas different PavB and PspC protein derivatives were used as soluble analytes (Fig. 4, A and B). Similar to the ELISA approach, all PavB-SSURE-proteins bound dose-dependently to immobilized hTSP-1 in a micromolar range. The association of SSURE1–5 and SSURE2 to hTSP-1 was comparable; however, a 5-fold higher concentration of SSURE2+3 had to be used to reach a similar level of binding as measured for SSURE2 (Fig. 4A). On the basis of a mathematical iteration (1:1 Langmuir binding model), SSURE1–5 showed the lowest dissociation constant (KD) followed by SSURE2 and SSURE2+3 (Table 2).
TABLE 2.
Data of surface plasmon resonance kinetics fitted to a 1:1 binding model
RU, response units.
| Fragment | ka | kd | KD | Rmax | χ2 |
|---|---|---|---|---|---|
| 1/ms | 1/s | m | RU | RU2 | |
| SSURE1–5_HEPES | 1.87E+4 | 3.63E−4 | 1.94E−8 | 175.7 | 4.22 |
| SSURE1–5_HEPES-Ca2+/Mg2+ | 8.35E+4 | 3.40E−4 | 4.07E−9 | 92.96 | 1.68 |
| SSURE1–5_PBS | 1.12E+5 | 6.20E−4 | 5.55E−9 | 89.56 | 8.69 |
| SSURE2 + 3_PBS | 8211 | 1.46E−3 | 1.78E−7 | 209.0 | 22.1 |
| SSURE2_PBS | 9.75E+4 | 2.72E−3 | 2.79E−8 | 152.8 | 8.80 |
| SH13_HEPES | 1.94E+6 | 5.43E−4 | 2.79E−10 | 86.80 | 4.22 |
| SH13_HEPES-Ca2+/Mg2+ | 2.04E+5 | 5.91E−3 | 2.90E−8 | 4207 | 1.68 |
| SH13_PBS | 7.88E+5 | 8.66E−4 | 1.10E−9 | 132.8 | 1.55 |
| SH2_PBS | 2.28E+4 | 2.15E−3 | 9.43E−8 | 103.0 | 91.6 |
| SH3_PBS | 1.06E+4 | 2.43E−3 | 2.29E−7 | 27.13 | 3.52 |
| SM1_PBS | 7914 | 1.70E−3 | 2.14E−7 | 47.79 | 12.8 |
The SPR experiments with PspC-proteins showed a dose-dependent binding when proteins were used in the nanomolar/micromolar range. But striking differences in their association to and dissociation from sensor-bound hTSP-1 were detected based on a 1:1 model. PspC SH13 containing two repeats bound more efficiently to hTSP-1 and dissociated only slowly from the biosensor suggesting a high binding affinity (Fig. 4B). The association of PspC SH2 containing one R domain was lower compared with SH13, and SM1 and SH3 bound even weaker than SH2 to hTSP-1. The lowest KD (highest binding affinity) was calculated for SH13 followed by SH2, SH3, and SM1, suggesting that the PspC protein binding affinity to immobilized hTSP-1 under dynamic conditions strictly depends on the number of repeats (Table 2). Binding experiments of PavB/PspC fragments to hTSP-1 were also carried out under various buffer conditions (Hepes and Hepes supplemented with Mg2+ and Ca2+) to assess an influence of conformational changes of hTSP-1 due to the presence of divalent cations (Fig. 4, A and B). Here, only minor differences could be observed, suggesting a negligible effect of divalent cations. Taken together, the results of the ELISA assays and SPR clearly demonstrate that the repetitive sequences of PavB and PspC are critical for the efficient interaction with soluble and immobilized hTSP-1.
PavB and PspC Binding to hTSP-1 Is Charge-dependent and Inhibited by Heparin
The influence of ionic forces on the interaction of PavB or PspC with hTSP-1 was analyzed by the addition of NaCl in increasing concentrations (Fig. 5, A and B). The results of the ELISA demonstrated that NaCl dose-dependently inhibited binding of PavB and PspC fragments to hTSP-1. Physiological concentrations of NaCl (0.13 m) had only a minor impact on binding of pneumococcal PavB fragments (11% reduction for SSURE2+3 and 7% reduction for SSURE1–5) and increases with higher salt concentrations (Fig. 5A). PspC fragments showed already at physiological concentrations a significant decrease in hTSP-1 binding (32% for SH13 and 57% for SH2), and increasing concentrations of salt led to concentration-dependent inhibition (Fig. 5B). These data confirmed the SPR data (Fig. 4), which suggested a low affinity interaction between hTSP-1 and pneumococcal adhesins containing only one R domain (PspC SH2) or less than five SSURE domains (PavB SSURE2+3).
To determine whether PavB or PspC compete with glycosaminoglycans for the binding to immobilized hTSP-1, heparin and chondroitin sulfate A were used in increasing concentrations to inhibit competitively binding of PavB SSURE1–5 or PspC SH13 to immobilized hTSP-1 (Fig. 5, C–F). The results of the competitive ELISA demonstrated that heparin dose-dependently inhibits binding of PavB and PspC proteins to immobilized hTSP-1. Interestingly, the competitor chondroitin sulfate A showed an inhibitory effect on the interaction between PspC and hTSP-1 but not on the interaction between PavB and hTSP-1. The binding of PavB SSURE1–5 was not diminished in a concentration range of 10−3-103 μg/ml chondroitin sulfate A (Fig. 5E). Only a concentration of 5 mg/ml significantly inhibited the SSURE1–5 binding to immobilized hTSP-1. The PspC SH13 binding was significantly decreased at a chondroitin sulfate A concentration of 1 μg/ml, comparable to the concentration of heparin (Fig. 5, D–F). These data suggest a charge-dependent binding of PavB and PspC to hTSP-1 and a participation of the heparin-binding site of hTSP-1 to these interactions. Furthermore, PavB and PspC seem to possess a different binding behavior toward hTSP-1.
PavB and PspC Contribute to hTSP-1-mediated Pneumococcal Adherence to Epithelial Cells
Cell-bound human TSP-1 was shown to act as a molecular bridge, and thereby promotes adherence of Gram-positive bacteria to epithelial cells (28). Here, we have assessed the contribution of PavB and PspC to pneumococcal adherence via hTSP-1. Binding of hTSP-1 was analyzed by flow cytometry (18). A549 cells bound hTSP-1 to their surface in a dose-dependent manner (Fig. 6, A and B).
FIGURE 6.
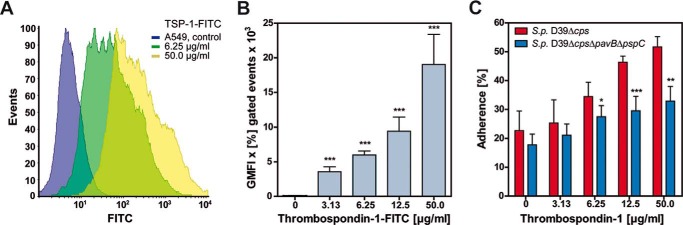
PavB and PspC contribute to pneumococcal adherence to A549 cells via hTSP-1. A and B, dose-dependent binding of hTSP-1-FITC to A549 cells was analyzed by flow cytometry. Approximately 2 × 105 cells were incubated with different concentrations of FITC labeled hTSP-1 (0–50 μg/ml) after preincubation with 1 μm MnCl2. Binding of hTSP-1 is shown as a histogram and geometric mean fluorescence intensity (GMFI) multiplied by the percentage of gated events (GMFI × % gated events). The mean values of three independent experiments are shown with error bars corresponding to S.D. *, p < 0.05; **, p < 0.01; ***, p < 0.001. C, A549 cells (2 × 105) were preincubated with 1 μm MnCl2 and increasing concentrations of hTSP-1 followed by an infection with S. pneumoniae D39Δcps or D39ΔcpsΔpavBΔpspC using a multiplicity of infection of 25 bacteria per epithelial cell. Adherent bacteria were detected using a polyclonal rabbit anti-pneumococci antibody followed by incubation with secondary AlexaFluor® 488-labeled goat anti-rabbit IgG. A549 cells were stained with AlexaFluor® 488-coupled phalloidin. Pneumococcal adherence of 100 cells was quantified 1.5 h post infection by immune fluorescence microscopy. Results are presented as percentage related to the multiplicity of infection for three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus S. pneumoniae D39Δcps.
For the adherence assays A549 cells were preincubated with Mn2+ followed by treatment with hTSP-1 and infection with either the parental strain S. pneumoniae D39Δcps or with its isogenic mutant S. pneumoniae D39ΔcpsΔpavBΔpspC (multiplicity of infection 25). The number of adherent bacteria was quantified by fluorescence microscopy. The results confirmed the function of host-cell-bound hTSP-1 as a molecular linker between pneumococci and epithelial cells (Fig. 6C). In the absence of hTSP-1 or at a low hTSP-1 concentration (3.13 μg/ml), adherence of pneumococcal strain D39Δcps and its isogenic mutant D39ΔcpsΔpavBΔpspC lacking the identified hTSP-1 adhesins was not significantly different (Fig. 6C).
However, at higher concentrations of hTSP-1 (6.25–50 μg/ml) a significant decrease in adherence to A549 cells compared with the parental strain was observed, indicating that these adhesins are involved in the hTSP-1 adherence mechanism (Fig. 6C). It is important to mention that the absence of PavB or PspC already influences adherence to host cells (11, 32). Moreover, A549 cells lack the polymeric immunoglobulin receptor, which was shown to interact with PspC (48). In conclusion, the surface-exposed PavB and PspC are hTSP-1-binding proteins that significantly contribute to the hTSP-1 host cell adherence mechanism of pneumococci.
Activation of Human Platelets Is Independent of Pneumococcal hTSP-1-binding Proteins PavB and PspC
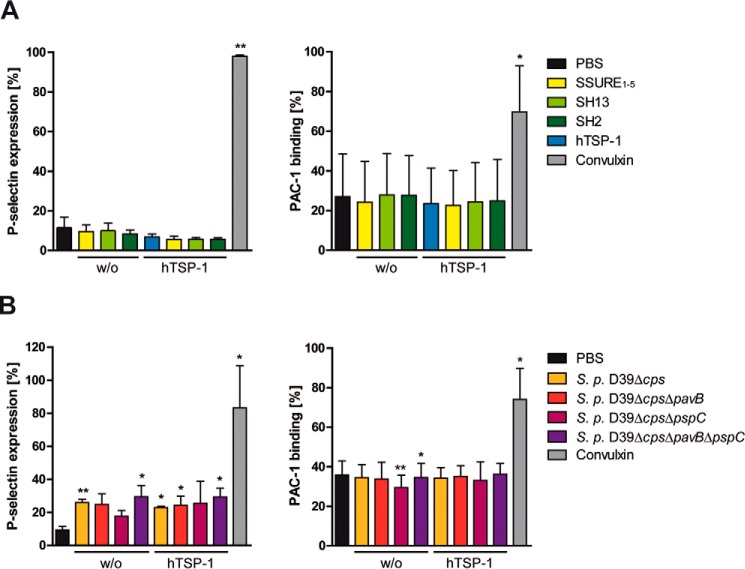
Human platelet activation by pneumococcal deletion mutants deficient in PavB, PspC, or both, and by heterologously expressed fragments of the adhesins was examined after two different incubation time points (15 and 60 min). The activation experiment was carried out in the absence or presence of exogenous hTSP-1. Flow cytometric analyses using heterologously expressed fragments of PavB or PspC revealed no activation of human platelets, whereas viable nonencapsulated pneumococci showed low levels of activation demonstrated by surface expression of P-selectin. However, no expression of the activated integrin αIIbβIII was observed as monitored by PAC-1 binding (Fig. 7, A and B). In conclusion, the measured pneumococci-induced activation of platelets was independent of surface-exposed PavB and PspC proteins. Furthermore, the presence of exogenously added hTSP-1 had no additional impact on platelet activation.
FIGURE 7.
Influence of S. pneumoniae and pneumococcal proteins on platelet activation. A and B, 1 × 109 platelets were incubated with the recombinant pneumococcal proteins PavB SSURE1–5 (25 μg/ml), PspC SH13 (10 μg/ml), PspC SH2 (10 μg/ml), or 1 × 108 pneumococcal deletion mutants in the absence or presence of exogenous hTSP-1 (25 μg/ml). Convulxin (100 ng/ml) was used as a positive control. Platelet activation was analyzed by flow cytometry using a specific mouse anti-human CD62P (P-selectin)-PE Cy5-conjugated antibody (left panels) or a mouse PAC-1 (anti-αIIbβIII)-FITC-labeled antibody (right panels) and expressed as percentage of labeled platelets. Results are presented as mean values ± S.D. of three independent experiments. *, p < 0.05; **, p < 0.01 versus PBS. S.p., S. pneumoniae.
Discussion
The initial adherence of bacteria to host cells is the obligatory step in an infection process. Therefore, bacteria exhibit an entire arsenal of surface structures to interact either directly with structures of various cell types or indirectly via components of the ECM.
In recent years it has been recognized that the ECM plays an important role in structure, cell proliferation, adhesion, and migration of eukaryotic cells (53). However, it has also been shown that the ECM represents an important target for various bacterial adhesins. In this study, we focused on the interplay between the human matricellular, multidomain glycoprotein hTSP-1 and proteinaceous surface adhesins of pneumococci, containing repetitive core sequences.
Previous studies demonstrated the ability of various Gram-positive bacteria, particularly S. pneumoniae, to recruit hTSP-1 to the bacterial surface (29) as well as the ability to exploit cell-bound hTSP-1 as a molecular bridge between bacterium and host cell (28). Whereas binding partners of hTSP-1 on eukaryotic cells have been identified, bacterial interaction partners are largely unknown and are only described for some staphylococcal species (29, 30).
In this study, we were able to demonstrate that the individual deletion of the adhesins pavB, pspC, or both in combination in different pneumococcal strains led to a dramatic decrease in the ability to acquire soluble hTSP-1 to the bacterial surface. Interestingly, the double deletion mutant D39ΔpavBΔpspC resulted in additive effects. Thus, the multifunctional adhesins PavB and PspC function as major hTSP-1-binding proteins of S. pneumoniae. However, a deletion of both genes did not cause a total loss of hTSP-1 binding activity, suggesting additional, yet unidentified pneumococcal hTSP-1-binding proteins.
To gain more insight into the molecular interaction of PavB and PspC from S. pneumoniae with hTSP-1, heterologously expressed fragments were purified and used in complementary binding assays. As shown for the staphylococcal hTSP-1-binding proteins Eap and Atl, repeating proteinaceous structures seem to be essential for a stable interaction between bacterial proteins and hTSP-1 (29, 30). Surprisingly, staphylococcal Eap and Atl as well as pneumococcal PspC and PavB share no amino acid sequence similarity or secondary structure homologies in repeating regions.
Using ELISA techniques, all tested PavB and PspC fragments showed a dose-dependent binding to immobilized hTSP-1, and vice versa, hTSP-1 showed dose-dependent binding to the immobilized fragments of PavB and PspC, respectively. In the case of PavB SSURE1–5, containing 5 repeats, binding to hTSP-1 was most efficient. Also PspC-SH13, containing two repeats, showed the most stable binding to hTSP-1.
Complementary to the ELISA assays, binding of various PavB and PspC fragments was analyzed under flow conditions using SPR and considered on a 1:1 binding iteration. Consistent with the ELISA, all analyzed proteins showed dose-dependent binding to immobilized hTSP-1.
Surprisingly, SSURE2 of PavB showed binding activities comparable to SSURE1–5. In summary, protein derivatives of PavB and PspC containing the largest number of repeating sequences (SSURE1–5 and SH13) showed the highest binding capacity with the lowest dissociation, suggesting an essential role of the repetitive structures of bacterial adhesins in the interaction with hTSP-1.
The binding efficiency of pneumococcal PavB and PspC proteins to hTSP-1 seems to increase with the number of repeating sequences, which was also shown for repeats R1abR2ab of AtlE from S. epidermidis and Eap of S. aureus (29, 30). Considering these data, one can hypothesize a zipper-like binding mechanism of these repeats with repeating domains within the hTSP-1 molecule.
To further characterize the type of chemical bond, binding of PavB and PspC constructs to hTSP-1 was analyzed in the presence of increasing concentrations of NaCl. Binding of bacterial adhesins was inhibited by NaCl in a dose-dependent manner, suggesting that this protein-protein interaction depends on ionic forces. Heparin and glycosaminoglycans were shown to bind to the N-terminal region and heparin also to type I repeats of hTSP-1 (54–56). Therefore, we tested the ability of heparin and chondroitin sulfate A to competitively inhibit binding of PavB SSURE1–5 or PspC SH13 (23, 57). Indeed, heparin was able to block the binding, suggesting that PavB and PspC compete with host heparin for binding to hTSP-1. In contrast, chondroitin sulfate A inhibited only the interaction of PspC, but not of PavB, indicating a different binding behavior of the pneumococcal proteins toward hTSP-1. Nevertheless, a sterical blocking of PspC binding through heparin cannot be excluded. Notably, a heparin-dependent inhibition of PspC binding to human Vn was recently demonstrated (31), further supporting the idea of the involvement of heparin binding domains in the interactions of PspC and PavB with hTSP-1 and Vn.
In a former study using staphylococcal Atl, hTSP-1 binding was shown to be inhibited by increasing molar ratios of human Vn (31). PspC has already been shown to interact with human Vn. Therefore, we analyzed the binding of hTSP-1 to immobilized PspC SH13 in the presence of increasing molar ratios of Vn and vice versa. Indeed, hTSP-1 binding was dose-dependently diminished by Vn and vice versa. However, lower molecular ratios of Vn seem to be necessary to inhibit hTSP-1 binding to SH13, suggesting a stronger binding affinity of Vn to SH13 compared with hTSP-1.
The in vitro relevance of the surface-exposed pneumococcal virulence factors PavB and PspC for the hTSP-1-mediated adherence to human alveolar epithelial cells (A549) was monitored by fluorescence microscopy. A previous study demonstrated a significantly enhanced pneumococcal adherence to and invasion into different human cells lines in the presence of host-cell bound hTSP-1 (28). We, therefore, infected A549 cells with S. pneumoniae D39Δcps and the isogenic PavB/PspC-deficient double mutant after preincubation with hTSP-1. The wild-type and the double mutant showed a concentration-dependent adherence. Importantly, a significant decrease in hTSP-1-mediated adherence was observed for PavB/PspC-deficient pneumococci, highlighting the role of these adhesins for the interaction with host cell-bound hTSP-1. However, based on the incomplete reduction of adherence observed for the mutant deficient for PavB and PspC, further bacterial surface structures may play a role during hTSP-1-mediated pneumococcal adherence to host cells.
We further speculate that hTSP-1 binding to the pneumococcal cell surface via PavB and PspC might play a pivotal role after bacteria breaking through the epithelial layer and circulating in the bloodstream. Human TSP-1 could be used by pneumococci to bind to circulating platelets, facilitating a hematogenous spreading within the host.
Bacterial bloodstream infections can also cause severe complications; among others, thrombocytopenia, which results in altered coagulation function (59–64). Gram-negative as well as Gram-positive pathogenic bacteria show the ability to bind to and activate human platelets (43). Recently, the extracellular adhesin protein Eap of S. aureus, interacting with various host ECM proteins, was shown to directly activate platelets (58).
We, therefore, analyzed the impact of the pneumococcal virulence factors PavB and PspC on human platelet activation using pneumococcal deletion mutants and recombinant expressed proteins. Additionally, exogenous hTSP-1 was added to analyze its supporting role in platelet activation.
The pneumococcal strain D39Δcps showed a moderate activation, exposing P-selectin after 15 min, which was also observed after 60 min (data not shown), but no activation of the integrin αIIbβIII could be detected. Moreover, P-selectin expression seemed to be independent of the presence of surface-associated PavB or PspC or exogenously added hTSP-1. The purified proteins PavB or PspC were not capable to activate platelets as demonstrated by measuring P-selectin expression and integrin αIIbβIII activation. These data suggest that platelet activation through S. pneumoniae is not triggered by its surface proteins PavB or PspC.
Taken together, in the present study we showed that various S. pneumoniae strains dose-dependently recruit human TSP-1 to their cell surface. Here, we identified the surface-displayed virulence factors PavB and PspC as novel pneumococcal hTSP-1-binding proteins. The number of repeating domains within these adhesins plays a crucial role for the interaction with hTSP-1. Moreover, these adhesins contribute to the hTSP-1-mediated pneumococcal adherence to human epithelial cells but did not participate in activation of human platelets.
Acknowledgments
We thank Sajida Kanwal for providing pneumococcal deletion mutant PN483 and Peggy Stremlow for excellent technical assistance.
This work was supported by Deutsche Forschungsgemeinschaft Grants SFB TRR 34, Project C10 (to S. H.) and DFG HA 3125/4-2.
- ECM
- extracellular matrix
- Vn
- vitronectin
- hTSP-1
- human matricellular glycoprotein thrombospondin-1
- Atl
- autolysin
- PspC
- pneumococcal surface protein C
- PavB
- pneumococcal adherence and virulence factor B
- SSURE
- streptococcal surface repeat
- CBB
- Coomassie Brilliant Blue
- SPR
- surface plasmon resonance
- Eap
- extracellular adherence protein.
References
- 1. Hussain M., Melegaro A., Pebody R. G., George R., Edmunds W. J., Talukdar R., Martin S. A., Efstratiou A., Miller E. (2005) A longitudinal household study of Streptococcus pneumoniae nasopharyngeal carriage in a UK setting. Epidemiol. Infect. 133, 891–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Garenne M., Ronsmans C., Campbell H. (1992) The magnitude of mortality from acute respiratory infections in children under 5 years in developing countries. World Health Stat. Q. 45, 180–191 [PubMed] [Google Scholar]
- 3. Bogaert D., De Groot R., Hermans P. W. (2004) Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect. Dis. 4, 144–154 [DOI] [PubMed] [Google Scholar]
- 4. Gamez G., Hammerschmidt S. (2012) Combat pneumococcal infections: adhesins as candidates for protein-based vaccine development. Curr. Drug Targets 13, 323–337 [DOI] [PubMed] [Google Scholar]
- 5. Gray B. M., Converse G. M., 3rd, Dillon H. C., Jr. (1980) Epidemiologic studies of Streptococcus pneumoniae in infants: acquisition, carriage, and infection during the first 24 months of life. J. Infect. Dis. 142, 923–933 [DOI] [PubMed] [Google Scholar]
- 6. Gwaltney J. M., Jr., Sande M. A., Austrian R., Hendley J. O. (1975) Spread of Streptococcus pneumoniae in families. II. Relation of transfer of S. pneumoniae to incidence of colds and serum antibody. J. Infect. Dis. 132, 62–68 [DOI] [PubMed] [Google Scholar]
- 7. Nelson A. L., Roche A. M., Gould J. M., Chim K., Ratner A. J., Weiser J. N. (2007) Capsule enhances pneumococcal colonization by limiting mucus-mediated clearance. Infect. Immun. 75, 83–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hammerschmidt S., Wolff S., Hocke A., Rosseau S., Müller E., Rohde M. (2005) Illustration of pneumococcal polysaccharide capsule during adherence and invasion of epithelial cells. Infect. Immun. 73, 4653–4667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weiser J. N., Austrian R., Sreenivasan P. K., Masure H. R. (1994) Phase variation in pneumococcal opacity: relationship between colonial morphology and nasopharyngeal colonization. Infect. Immun. 62, 2582–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Asmat T. M., Klingbeil K., Jensch I., Burchhardt G., Hammerschmidt S. (2012) Heterologous expression of pneumococcal virulence factor PspC on the surface of Lactococcus lactis confers adhesive properties. Microbiology 158, 771–780 [DOI] [PubMed] [Google Scholar]
- 11. Jensch I., Gámez G., Rothe M., Ebert S., Fulde M., Somplatzki D., Bergmann S., Petruschka L., Rohde M., Nau R., Hammerschmidt S. (2010) PavB is a surface-exposed adhesin of Streptococcus pneumoniae contributing to nasopharyngeal colonization and airways infections. Mol. Microbiol. 77, 22–43 [DOI] [PubMed] [Google Scholar]
- 12. Schaefer L., Schaefer R. M. (2010) Proteoglycans: from structural compounds to signaling molecules. Cell Tissue Res. 339, 237–246 [DOI] [PubMed] [Google Scholar]
- 13. Rozario T., DeSimone D. W. (2010) The extracellular matrix in development and morphogenesis: a dynamic view. Dev. Biol. 341, 126–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tan K., Lawler J. (2009) The interaction of Thrombospondins with extracellular matrix proteins. J. Cell Commun. Signal. 3, 177–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Preissner K. T. (1991) Structure and biological role of vitronectin. Annu. Rev. Cell Biol. 7, 275–310 [DOI] [PubMed] [Google Scholar]
- 16. Paterson G. K., Orihuela C. J. (2010) Pneumococcal microbial surface components recognizing adhesive matrix molecules targeting of the extracellular matrix. Mol. Microbiol. 77, 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tang M., Zhou F., Zhang W., Guo Z., Shang Y., Lu H., Lu R., Zhang Y., Chen Y., Zhong M. (2011) The role of thrombospondin-1-mediated TGF-β1 on collagen type III synthesis induced by high glucose. Mol. Cell Biochem. 346, 49–56 [DOI] [PubMed] [Google Scholar]
- 18. Calzada M. J., Sipes J. M., Krutzsch H. C., Yurchenco P. D., Annis D. S., Mosher D. F., Roberts D. D. (2003) Recognition of the N-terminal modules of thrombospondin-1 and thrombospondin-2 by α6β1 integrin. J. Biol. Chem. 278, 40679–40687 [DOI] [PubMed] [Google Scholar]
- 19. Isenberg J. S., Jia Y., Fukuyama J., Switzer C. H., Wink D. A., Roberts D. D. (2007) Thrombospondin-1 inhibits nitric oxide signaling via CD36 by inhibiting myristic acid uptake. J. Biol. Chem. 282, 15404–15415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Narizhneva N. V., Razorenova O. V., Podrez E. A., Chen J., Chandrasekharan U. M., DiCorleto P. E., Plow E. F., Topol E. J., Byzova T. V. (2005) Thrombospondin-1 up-regulates expression of cell adhesion molecules and promotes monocyte binding to endothelium. FASEB J. 19, 1158–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carlson C. B., Lawler J., Mosher D. F. (2008) Structures of thrombospondins. Cell. Mol. Life Sci. 65, 672–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bornstein P. (1995) Diversity of function is inherent in matricellular proteins: an appraisal of thrombospondin 1. J. Cell Biol. 130, 503–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Resovi A., Pinessi D., Chiorino G., Taraboletti G. (2014) Current understanding of the thrombospondin-1 interactome. Matrix Biol 37, 83–91 [DOI] [PubMed] [Google Scholar]
- 24. Raugi G. J., Lovett D. H. (1987) Thrombospondin secretion by cultured human glomerular mesangial cells. Am. J. Pathol. 129, 364–372 [PMC free article] [PubMed] [Google Scholar]
- 25. Asch A. S., Leung L. L., Shapiro J., Nachman R. L. (1986) Human brain glial cells synthesize thrombospondin. Proc. Natl. Acad. Sci. U.S.A. 83, 2904–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jaffe E. A., Ruggiero J. T., Falcone D. J. (1985) Monocytes and macrophages synthesize and secrete thrombospondin. Blood 65, 79–84 [PubMed] [Google Scholar]
- 27. Raugi G. J., Mumby S. M., Abbott-Brown D., Bornstein P. (1982) Thrombospondin: synthesis and secretion by cells in culture. J. Cell Biol. 95, 351–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rennemeier C., Hammerschmidt S., Niemann S., Inamura S., Zähringer U., Kehrel B. E. (2007) Thrombospondin-1 promotes cellular adherence of gram-positive pathogens via recognition of peptidoglycan. FASEB J. 21, 3118–3132 [DOI] [PubMed] [Google Scholar]
- 29. Kohler T. P., Gisch N., Binsker U., Schlag M., Darm K., Völker U., Zähringer U., Hammerschmidt S. (2014) Repeating structures of the major staphylococcal autolysin are essential for the interaction with human thrombospondin 1 and vitronectin. J. Biol. Chem. 289, 4070–4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hussain M., Haggar A., Peters G., Chhatwal G. S., Herrmann M., Flock J. I., Sinha B. (2008) More than one tandem repeat domain of the extracellular adherence protein of Staphylococcus aureus is required for aggregation, adherence, and host cell invasion but not for leukocyte activation. Infect. Immun. 76, 5615–5623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Voss S., Hallström T., Saleh M., Burchhardt G., Pribyl T., Singh B., Riesbeck K., Zipfel P. F., Hammerschmidt S. (2013) The choline-binding protein PspC of Streptococcus pneumoniae interacts with the C-terminal heparin-binding domain of vitronectin. J. Biol. Chem. 288, 15614–15627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rosenow C., Ryan P., Weiser J. N., Johnson S., Fontan P., Ortqvist A., Masure H. R. (1997) Contribution of novel choline-binding proteins to adherence, colonization and immunogenicity of Streptococcus pneumoniae. Mol. Microbiol. 25, 819–829 [DOI] [PubMed] [Google Scholar]
- 33. Hernani Mde L., Ferreira P. C., Ferreira D. M., Miyaji E. N., Ho P. L., Oliveira M. L. (2011) Nasal immunization of mice with Lactobacillus casei expressing the pneumococcal surface protein C primes the immune system and decreases pneumococcal nasopharyngeal colonization in mice. FEMS Immunol. Med. Microbiol. 62, 263–272 [DOI] [PubMed] [Google Scholar]
- 34. Ogunniyi A. D., LeMessurier K. S., Graham R. M., Watt J. M., Briles D. E., Stroeher U. H., Paton J. C. (2007) Contributions of pneumolysin, pneumococcal surface protein A (PspA), and PspC to pathogenicity of Streptococcus pneumoniae D39 in a mouse model. Infect. Immun. 75, 1843–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Balachandran P., Brooks-Walter A., Virolainen-Julkunen A., Hollingshead S. K., Briles D. E. (2002) Role of pneumococcal surface protein C in nasopharyngeal carriage and pneumonia and its ability to elicit protection against carriage of Streptococcus pneumoniae. Infect. Immun. 70, 2526–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Agarwal V., Hammerschmidt S. (2009) Cdc42 and the phosphatidylinositol 3-kinase-Akt pathway are essential for PspC-mediated internalization of pneumococci by respiratory epithelial cells. J. Biol. Chem. 284, 19427–19436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Voss S., Gámez G., Hammerschmidt S. (2012) Impact of pneumococcal microbial surface components recognizing adhesive matrix molecules on colonization. Mol. Oral Microbiol. 27, 246–256 [DOI] [PubMed] [Google Scholar]
- 38. Hammerschmidt S., Agarwal V., Kunert A., Haelbich S., Skerka C., Zipfel P. F. (2007) The host immune regulator factor H interacts via two contact sites with the PspC protein of Streptococcus pneumoniae and mediates adhesion to host epithelial cells. J. Immunol. 178, 5848–5858 [DOI] [PubMed] [Google Scholar]
- 39. Dieudonné-Vatran A., Krentz S., Blom A. M., Meri S., Henriques-Normark B., Riesbeck K., Albiger B. (2009) Clinical isolates of Streptococcus pneumoniae bind the complement inhibitor C4b-binding protein in a PspC allele-dependent fashion. J. Immunol. 182, 7865–7877 [DOI] [PubMed] [Google Scholar]
- 40. Hammerschmidt S., Tillig M. P., Wolff S., Vaerman J. P., Chhatwal G. S. (2000) Species-specific binding of human secretory component to SpsA protein of Streptococcus pneumoniae via a hexapeptide motif. Mol. Microbiol. 36, 726–736 [DOI] [PubMed] [Google Scholar]
- 41. Orihuela C. J., Mahdavi J., Thornton J., Mann B., Wooldridge K. G., Abouseada N., Oldfield N. J., Self T., Ala'Aldeen D. A., Tuomanen E. I. (2009) Laminin receptor initiates bacterial contact with the blood brain barrier in experimental meningitis models. J. Clin. Invest. 119, 1638–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bumbaca D., Littlejohn J. E., Nayakanti H., Rigden D. J., Galperin M. Y., Jedrzejas M. J. (2004) Sequence analysis and characterization of a novel fibronectin-binding repeat domain from the surface of Streptococcus pneumoniae. OMICS 8, 341–356 [DOI] [PubMed] [Google Scholar]
- 43. Fitzgerald J. R., Foster T. J., Cox D. (2006) The interaction of bacterial pathogens with platelets. Nat. Rev. Microbiol. 4, 445–457 [DOI] [PubMed] [Google Scholar]
- 44. Kerrigan S. W., Douglas I., Wray A., Heath J., Byrne M. F., Fitzgerald D., Cox D. (2002) A role for glycoprotein Ib in Streptococcus sanguis-induced platelet aggregation. Blood 100, 509–516 [DOI] [PubMed] [Google Scholar]
- 45. Byrne M. F., Kerrigan S. W., Corcoran P. A., Atherton J. C., Murray F. E., Fitzgerald D. J., Cox D. M. (2003) Helicobacter pylori binds von Willebrand factor and interacts with GPIb to induce platelet aggregation. Gastroenterology 124, 1846–1854 [DOI] [PubMed] [Google Scholar]
- 46. Fitzgerald J. R., Loughman A., Keane F., Brennan M., Knobel M., Higgins J., Visai L., Speziale P., Cox D., Foster T. J. (2006) Fibronectin-binding proteins of Staphylococcus aureus mediate activation of human platelets via fibrinogen and fibronectin bridges to integrin GPIIb/IIIa and IgG binding to the FcγRIIa receptor. Mol. Microbiol. 59, 212–230 [DOI] [PubMed] [Google Scholar]
- 47. Niemann S., Kehrel B. E., Heilmann C., Rennemeier C., Peters G., Hammerschmidt S. (2009) Pneumococcal association to platelets is mediated by soluble fibrin and supported by thrombospondin-1. Thromb. Haemost. 102, 735–742 [DOI] [PubMed] [Google Scholar]
- 48. Elm C., Braathen R., Bergmann S., Frank R., Vaerman J. P., Kaetzel C. S., Chhatwal G. S., Johansen F. E., Hammerschmidt S. (2004) Ectodomains 3 and 4 of human polymeric Immunoglobulin receptor (hpIgR) mediate invasion of Streptococcus pneumoniae into the epithelium. J. Biol. Chem. 279, 6296–6304 [DOI] [PubMed] [Google Scholar]
- 49. Rondina M. T., Schwertz H., Harris E. S., Kraemer B. F., Campbell R. A., Mackman N., Grissom C. K., Weyrich A. S., Zimmerman G. A. (2011) The septic milieu triggers expression of spliced tissue factor mRNA in human platelets. J. Thromb. Haemost 9, 748–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schwertz H., Tolley N. D., Foulks J. M., Denis M. M., Risenmay B. W., Buerke M., Tilley R. E., Rondina M. T., Harris E. M., Kraiss L. W., Mackman N., Zimmerman G. A., Weyrich A. S. (2006) Signal-dependent splicing of tissue factor pre-mRNA modulates the thrombogenicity of human platelets. J. Exp. Med. 203, 2433–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Denis M. M., Tolley N. D., Bunting M., Schwertz H., Jiang H., Lindemann S., Yost C. C., Rubner F. J., Albertine K. H., Swoboda K. J., Fratto C. M., Tolley E., Kraiss L. W., McIntyre T. M., Zimmerman G. A., Weyrich A. S. (2005) Escaping the nuclear confines: signal-dependent pre-mRNA splicing in anucleate platelets. Cell 122, 379–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Weyrich A. S., Denis M. M., Schwertz H., Tolley N. D., Foulks J., Spencer E., Kraiss L. W., Albertine K. H., McIntyre T. M., Zimmerman G. A. (2007) mTOR-dependent synthesis of Bcl-3 controls the retraction of fibrin clots by activated human platelets. Blood 109, 1975–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hynes R. O. (2009) The extracellular matrix: not just pretty fibrils. Science 326, 1216–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yu H., Tyrrell D., Cashel J., Guo N. H., Vogel T., Sipes J. M., Lam L., Fillit H. M., Hartman J., Mendelovitz S., Panel A., Roberts D. D. (2000) Specificities of heparin-binding sites from the amino-terminus and type 1 repeats of thrombospondin-1. Arch. Biochem. Biophys. 374, 13–23 [DOI] [PubMed] [Google Scholar]
- 55. Vogel T., Guo N. H., Krutzsch H. C., Blake D. A., Hartman J., Mendelovitz S., Panet A., Roberts D. D. (1993) Modulation of endothelial cell proliferation, adhesion, and motility by recombinant heparin-binding domain and synthetic peptides from the type I repeats of thrombospondin. J. Cell. Biochem. 53, 74–84 [DOI] [PubMed] [Google Scholar]
- 56. Panetti T. S., Kudryk B. J., Mosher D. F. (1999) Interaction of recombinant procollagen and properdin modules of thrombospondin-1 with heparin and fibrinogen/fibrin. J. Biol. Chem. 274, 430–437 [DOI] [PubMed] [Google Scholar]
- 57. Tan K., Duquette M., Liu J. H., Shanmugasundaram K., Joachimiak A., Gallagher J. T., Rigby A. C., Wang J. H., Lawler J. (2008) Heparin-induced cis- and trans-dimerization modes of the thrombospondin-1 N-terminal domain. J. Biol. Chem. 283, 3932–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bertling A., Niemann S., Hussain M., Holbrook L., Stanley R. G., Brodde M. F., Pohl S., Schifferdecker T., Roth J., Jurk K., Müller A., Lahav J., Peters G., Heilmann C., Gibbins J. M., Kehrel B. E. (2012) Staphylococcal extracellular adherence protein induces platelet activation by stimulation of thiol isomerases. Arterioscler. Thromb. Vasc. Biol. 32, 1979–1990 [DOI] [PubMed] [Google Scholar]
- 59. Sharma B., Sharma M., Majumder M., Steier W., Sangal A., Kalawar M. (2007) Thrombocytopenia in septic shock patients: a prospective observational study of incidence, risk factors and correlation with clinical outcome. Anaesth. Intensive Care 35, 874–880 [DOI] [PubMed] [Google Scholar]
- 60. Baughman R. P., Lower E. E., Flessa H. C., Tollerud D. J. (1993) Thrombocytopenia in the intensive care unit. Chest 104, 1243–1247 [DOI] [PubMed] [Google Scholar]
- 61. Stéphan F., Hollande J., Richard O., Cheffi A., Maier-Redelsperger M., Flahault A. (1999) Thrombocytopenia in a surgical ICU. Chest 115, 1363–1370 [DOI] [PubMed] [Google Scholar]
- 62. Vanderschueren S., De Weerdt A., Malbrain M., Vankersschaever D., Frans E., Wilmer A., Bobbaers H. (2000) Thrombocytopenia and prognosis in intensive care. Crit. Care Med 28, 1871–1876 [DOI] [PubMed] [Google Scholar]
- 63. Vincent J. L., Yagushi A., Pradier O. (2002) Platelet function in sepsis. Crit. Care Med. 30, S313–S317 [DOI] [PubMed] [Google Scholar]
- 64. Levi M. (2004) Platelets at a crossroad of pathogenic pathways in sepsis. J. Thromb. Haemost 2, 2094–2095 [DOI] [PubMed] [Google Scholar]