Background: Base excision repair (BER) is the major pathway for repair of single oxidized nucleobases.
Results: Bifunctional DNA glycosylases and AP endonucleases are able to remove cross-linked guanine in guanine(C8)-thymine(N3) intrastrand cross-links.
Conclusion: BER pathways can repair the intrastrand cross-links.
Significance: Oxidatively generated intrastrand cross-linked DNA lesions can be repaired in HeLa cell extracts not only by nucleotide excision repair, but also by multiple BER pathways.
Keywords: base excision repair (BER), DNA damage, DNA endonuclease, DNA repair, nucleotide excision repair
Abstract
Oxidatively generated guanine radical cations in DNA can undergo various nucleophilic reactions including the formation of C8-guanine cross-links with adjacent or nearby N3-thymines in DNA in the presence of O2. The G*[C8-N3]T* lesions have been identified in the DNA of human cells exposed to oxidative stress, and are most likely genotoxic if not removed by cellular defense mechanisms. It has been shown that the G*[C8-N3]T* lesions are substrates of nucleotide excision repair in human cell extracts. Cleavage at the sites of the lesions was also observed but not further investigated (Ding et al. (2012) Nucleic Acids Res. 40, 2506–2517). Using a panel of eukaryotic and prokaryotic bifunctional DNA glycosylases/lyases (NEIL1, Nei, Fpg, Nth, and NTH1) and apurinic/apyrimidinic (AP) endonucleases (Apn1, APE1, and Nfo), the analysis of cleavage fragments by PAGE and MALDI-TOF/MS show that the G*[C8-N3]T* lesions in 17-mer duplexes are incised on either side of G*, that none of the recovered cleavage fragments contain G*, and that T* is converted to a normal T in the 3′-fragment cleavage products. The abilities of the DNA glycosylases to incise the DNA strand adjacent to G*, while this base is initially cross-linked with T*, is a surprising observation and an indication of the versatility of these base excision repair proteins.
Introduction
Reactions of reactive intermediates such as free radicals and oxidizing agents with DNA can give rise to interstrand and intrastrand cross-linked DNA lesions. Interstrand cross-linked DNA lesions (ICL)6 that result from reactions of a variety of bifunctional agents such as cisplatin (1) with DNA, have received much attention because they are difficult to remove by DNA repair mechanisms and are therefore highly genotoxic (2, 3). While ICL lesions arise from the covalent coupling of two nucleotides on opposite DNA strands, the coupling of two nucleotides on the same strand gives rise to intrastrand cross-linked (IntraCL) lesions. Well known examples are the UV photo-induced cyclobutanepyrimidine dimers (CPD) and 6–4 photoproducts (4). Oxidatively generated IntraCL lesions include cross-links between the C8-atom of guanine and a 3′-adjacent 5-methyl group of thymine (G[8–5m]T) (5), and the analogous G[8–5]C (6), and G[8–5m]C (7) intrastrand tandem lesions. Other forms of intrastrand lesions are the intranucleotide 8,5′-cyclo-2′-deoxypurine lesions that are characterized by covalent linkages between the C5′ deoxyribose and C8 carbon atoms of adenine (cdA) or guanine (cdG); these types of lesions were first discovered in dilute DNA solutions exposed to γ-radiation (8, 9).
Recently, the novel guanine(C8)-thymine(N3) tandem lesions (G*[C8-N3]T*) were identified in vitro (10) and detected in human HeLa cells by isotope dilution LC-MS/MS methods (11). In these IntraCLs, guanine and thymine bases are either adjacent to one another (G*T*) or separated by one intervening cytosine (G*CT*) linked by a covalent bond between the C8(G) and N3(T) atoms (Fig. 1A). Like the cdG and cdA lesions embedded in double-stranded DNA (12–16), the G*T* and G*CT* lesions are moderate to good substrates, respectively, of the human nucleotide excision repair (NER) system in extracts from HeLa cells (17). Furthermore, evidence was presented that both types of IntraCL lesions are also incised at the sites of the lesions. The origins of these incisions were not further investigated (17), but suggested that a base excision repair (BER) pathway could have been responsible for these incisions. However, BER repair enzymes are not known to incise intrastrand DNA cross-links. Here, we considered whether conventional BER pathways (18–20) and/or the nucleotide incision repair (NIR) pathway (21, 22) could account for the non-NER incisions reported earlier by Ding et al. (17).
FIGURE 1.
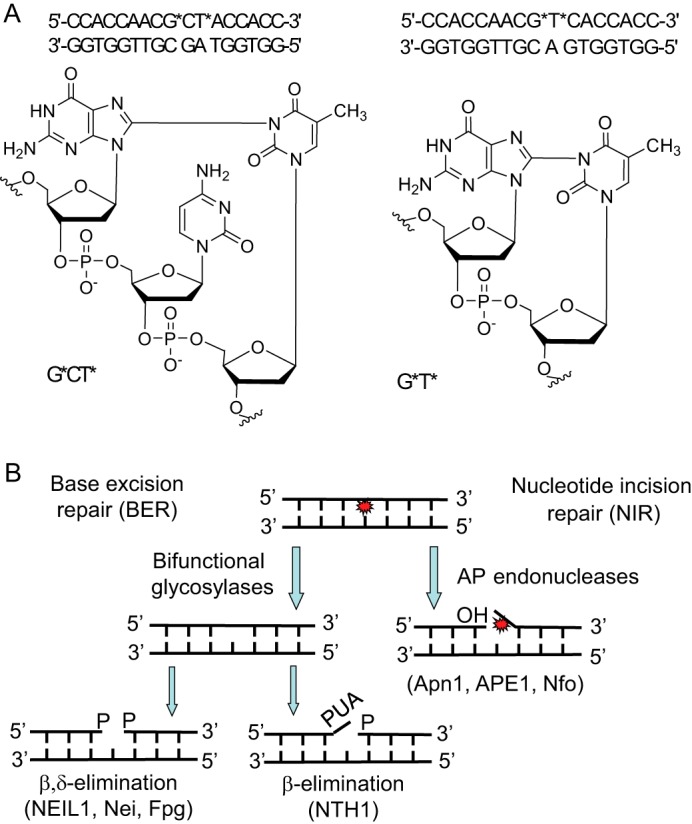
A, structures of the G*[C8-N3]T* intrastrand cross-links and the sequences of the DNA duplexes used in the experimental studies. The starred bases denote the modified sites. B, BER (18) and NIR (21) pathways of removal of oxidatively generated DNA lesions.
To gain insights into these mechanisms of incision of the G*CT* and G*T* IntraCLs embedded in double-stranded DNA, we incubated the 17-mer double-stranded constructs with different Escherichia coli and human DNA glycosylases/AP lyases Nei and NEIL1 (endonuclease VIII and oxidized pyrimidine-specific DNA glycosylase), Nth and NTH1 (endonuclease III and thymine glycol-DNA glycosylase), respectively, E. coli Fpg (8-oxoguanine DNA glycosylase), and E. coli Nfo, Saccharomyces cerevisiae Apn1 and human APE1 proteins, and monitored the formation of incision products. We demonstrate here that the bacterial, yeast, and human bifunctional DNA glycosylases and AP endonucleases cleave the strands adjacent to the G*CT* and G*T* IntraCL (Fig. 1A) embedded in site-specifically modified oligonucleotide duplexes. Analysis of the cleavage products by denaturing polyacrylamide gel electrophoresis (PAGE) and MALDI-TOF/MS methods showed that the DNA glycosylases/AP lyases excise the cross-linked guanine and cleave the resulting abasic sites via β- and β,δ-elimination mechanisms. In turn, the AP endonucleases of E. coli Nfo, yeast Apn1 and human APE1 cleave the duplex DNA containing G*[C8-N3]T* lesions 5′ next to cross-linked guanine initiating the nucleotide incision repair (NIR) pathway (22).
Experimental Procedures
G*T* and G*CT* Duplexes and Proteins
The 17-mer oligonucleotides containing the G*T* or G*CT* lesions were synthesized as described by Crean et al. (10). Before the enzymatic assays, oligonucleotides were either 5′-end-labeled by T4 polynucleotide kinase (New England Biolabs) in the presence of [γ-32P]ATP (3,000 Ci/mmol, PerkinElmer-Life Science Research), or 3′-end-labeled by terminal deoxynucleotidyl transferase (TdT, New England Biolabs) in the presence of 3′-[α-32P]dATP (cordycepin 5′-triphosphate, 5,000 Ci/mmol) employing the standard protocols. The radioactively labeled oligonucleotides were desalted on a Sephadex G-25 column equilibrated in water and then annealed to the required complementary strand for 3 min at 65 °C in a buffer containing 20 mm Hepes-KOH (pH 7.6) and 50 mm KCl, and then slowly cooled to room temperature. The purified E. coli uracil-DNA glycosylase (Ung), Fpg, Nth, and human NTH1 and APE1 proteins were from laboratory stock and prepared as described (23). Purifications of the E. coli Nfo and S. cerevisiae Apn1 proteins were performed as described (22, 24). The expression vectors phNEIL1 (25) and pET24b-EndoVIII (26) were generously provided by Drs. Hiroshi Ide (Hiroshima University, Japan) and Dmitry Zharkov (ICBFM, Novosibirsk, Russia), respectively. The E. coli Nei and full-length native NEIL1 proteins were purified as described (25, 26).
Generation of DNA Size Markers
The 5′-[32P]-labeled 17-mer oligonucleotide duplexes d(CCACCAACUCTACCACC) containing single uracil residues at position 9 were incubated with Ung to generate an AP site. Subsequently, the DNA was treated either with the Nfo protein, or heated in 10% (v/v) aqueous piperidine at 37 °C, or at 90 °C for 15 min to generate 5′- 32P-labeled 8-mer cleavage fragments containing 3′-hydroxyl (3′-OH), or 3′-phosphoaldehyde (3′-PUA), or 3′-phosphate (3′-P) ends, respectively.
Repair Assays
The standard reaction mixtures (20 μl) contained 5 nm concentrations of the 32P-labeled G*T* or G*CT* duplexes and 50 nm of the purified repair protein, were incubated for 30 min at 37 °C unless stated otherwise. The DNA repair activities of APE1 protein were tested either in the NIR buffer, which is optimal for the nucleotide incision activity and contained 50 mm KCl, 20 mm Hepes-KOH (pH 6.9), 0.1 mg/ml BSA, 1 mm DTT, and 0.1 mm MgCl2, or in the BER+Mg2+ buffer, which is optimal for the AP endonuclease activity, containing 50 mm KCl, 20 mm Hepes-KOH (pH 7.6), 0.1 mg/ml BSA, 1 mm DTT, and 5 mm MgCl2. The same buffer was used for the S. cerevisiae Apn1 protein whereas, for the E. coli Nfo protein, MgCl2 was omitted. The incision activities were determined from the amount of cleaved oligonucleotide substrates. The DNA glycosylase activity was measured in the BER+EDTA buffer containing 5 nm of an oligonucleotide duplex, 50 mm KCl, 20 mm Hepes-KOH (pH 7.6), 0.1 mg/ml BSA, 1 mm DTT, 1 mm EDTA, and 50 nm of the required purified protein for 30 min at 37 °C unless stated otherwise. All reactions were terminated by adding 10 μl of a stop solution containing 0.5% SDS and 20 mm EDTA, and then desalted by hand-made spin-down columns filled with Sephadex G25 (Amersham Biosciences) equilibrated in 7.5 m urea. The purified reaction products were separated by electrophoresis in denaturing 20% (w/v) polyacrylamide gel (7.5 m urea, 0.5× TBE, 42 °C). The gels were exposed to a Fuji FLA-3000 Phosphor Screen, then scanned with Fuji FLA-3000 or FLA-9500, and analyzed using Image Gauge V4.0 software.
MALDI-TOF/MS Analysis of the Cleavage Products
Mass spectrometry measurements were performed as described previously (45). Typically, 10 pmol of the non-labeled lesion-containing oligonucleotide duplexes (in 100 μl) were incubated with repair proteins (100 nm) in the relevant buffer solution at 37 °C either for 1 h with APE1 or for 15 min with NEIL1 and NTH1. The reaction products were precipitated with 2% lithium perchlorate in acetone, desalted, and then dissolved in water prior to analysis by MALDI-TOF/MS. The mass spectra were obtained in the negative mode on a time-of-flight Microflex mass spectrometer (Bruker), equipped with a 337-nm nitrogen laser and pulsed delay source extraction. The matrix was prepared by dissolving 3-hydroxypicolinic acid in 10 mm ammonium citrate buffer. Prior to MALDI-TOF mass analysis, oligonucleotide solutions were purified and concenterd on ZipTip pipette tips (Millipore). A mixture of purified DNA sample (10 pmol; 1 μl) was added to matrix (1 μl) and spotted on a polished stainless target plate using the dried droplet method. Spectra were calibrated using reference oligonucleotides of known masses.
Results
Characterization of Duplexes with G*[C8-N3]T* Cross-links
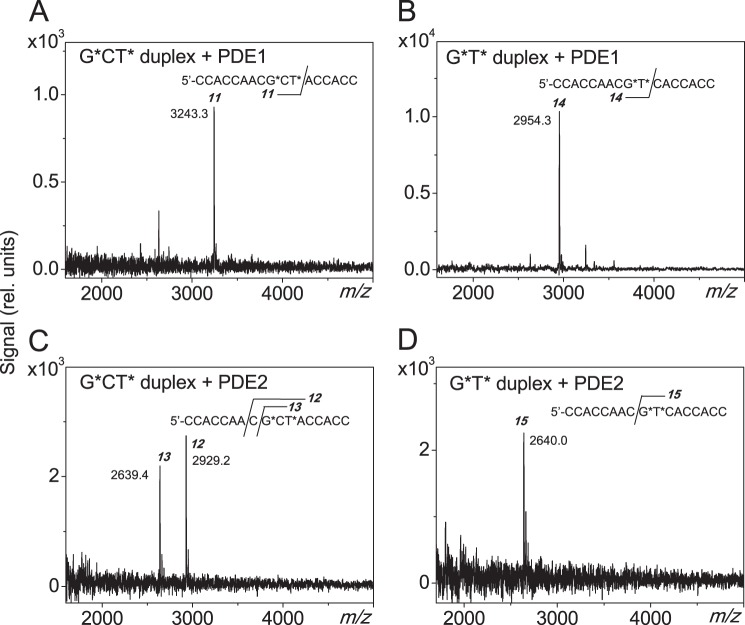
Such duplexes are referred to as G*CT* and G*T*, depending on the nature of the cross-linked lesion (Fig. 1A). The 17-mer sequences used are defined in Fig. 1A. The differences in molar mass between the G*CT* and G*T* duplexes and the normal duplexes without these cross-links are only 2 Da. Since the absorption spectra of these cross-linked and normal duplexes are identical, it is not straightforward to verify the presence of these lesions (10). While mass spectrometry combined with two-dimensional 13C NMR yields definitive characterization (10), a more simple approach is to monitor the stepwise degradation of the single-stranded oligonucleotides using exonucleases that stall at the sites of the cross-linked nucleotides (27). The MALDI-TOF/MS analysis of the digestion products of G*CT* and G*T* duplexes generated by snake venom phosphodiesterase 1 (digestion from the 3′-end) and calf spleen phosphodiesterase 2 (digestion from the 5′-end) clearly demonstrated the presence and integrity of the intrastrand cross-links in these duplexes (Fig. 2).
FIGURE 2.
MALDI-TOF/MS analysis of the mixture of oligonucleotides arising from the incubation of the 17-mer G*CT* and G*T* duplexes with phosphodiesterases 1 and 2. Typically, 40 pmol of the lesion containing oligonucleotide duplexes were incubated with either 100 nm of PDE1 or PDE2 in the appropriate reaction buffer (100 μl) at 37 °C for 5 h (PDE1) or overnight (PDE2). The products were desalted on a MicroSpin G-25 column, prior subjection to the MALDI-TOF/MS measurements. The experimental m/z values are agreed with those calculated from monoisotopic masses provided below in brackets. A, treatment of G*CT* duplex with PDE1; peak 11: 5′-CCACCAACG*CT* (m/z 3241.6). B, treatment of G*T* duplex with PDE1; peak 14: 5′-CCACCAACG*T* (m/z 2952.5). C, treatment of G*CT* duplex with PDE2 for overnight; peak 12: 5′-CG*CT*ACCACC (m/z 2928.5), peak 13: 5′-G*CT*ACCACC (m/z 2639.5). D, treatment of G*T* duplex with PDE2; peak 15: 5′-G*T*CACCACC (m/z 2639.5).
Overall Approach
In this work, the 17-mer duplexes containing the G*CT* and G*T* lesions were exposed to either bifunctional DNA glycosylases (BER) or AP endonucleases that are involved in the NIR pathway. In the case of BER, DNA glycosylases recognize the lesions and then cleave the N-glycosyl bond releasing the damaged base, thus producing abasic sites (18) (Fig. 1B). Bifunctional DNA glycosylases, in addition to base excision, also exhibit AP lyase activity and cleave the abasic sites formed, leaving single-strand breaks flanked by 5′-phosphates and either 3′-phosphate groups (generated by β,δ-elimination), or a 3′-α,β-unsaturated aldehyde (PUA) group (β-elimination). In the NIR pathway, AP endonucleases incise the phosphodiester bond adjacent to and on the 5′-side of the damaged bases in a DNA glycosylase-independent manner, leaving single-strand breaks flanked by proper 3′-OH termini for subsequent primer extension, and 5′-damaged nucleotides in the 3′-downstream cleavage fragments (21) (Fig. 1B). These cleavage products, generated by BER or NIR mechanisms, were distinguished by denaturing PAGE and identified by MALDI-TOF/MS methods (18, 28).
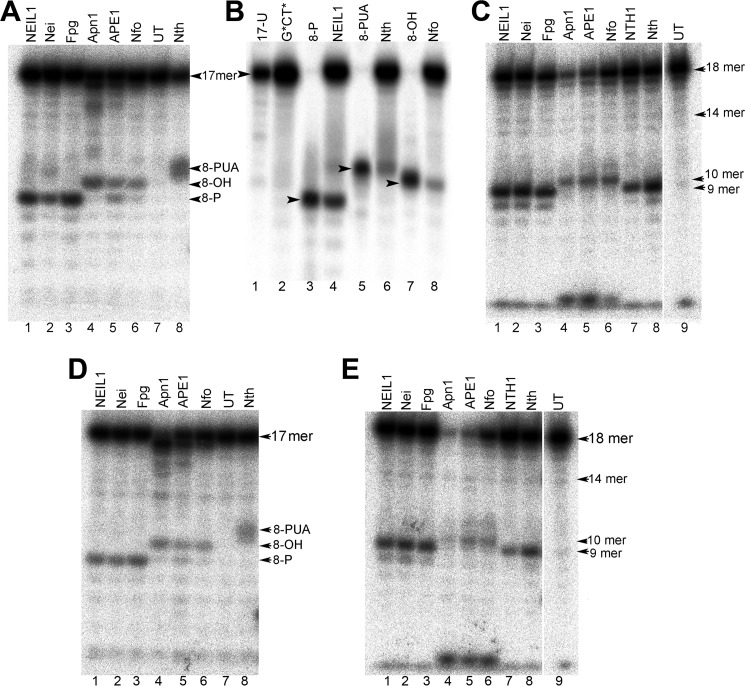
DNA duplexes with either 32P-labeled 5′-strands (17-mers) or 32P-labeled 3′-strands (18-mers) containing G*CT* (Fig. 3, A–C) or G*T* lesions (Fig. 3, D and E) were incubated with either (i) E. coli or the human DNA glycosylases Nei and NEIL1 (endonuclease VIII or oxidized pyrimidine-specific DNA glycosylase), (ii) Nth or NTH1 (endonuclease III or thymine glycol-DNA glycosylase), (iii) E. coli Fpg (8-oxoguanine DNA glycosylase), (iv) E. coli Nfo, (v) S. cerevisiae Apn1, or (vi) human APE1.
FIGURE 3.
Denaturing PAGE analysis of the cleavage patterns generated by DNA glycosylases/AP lyases (BER) and NIR-AP endonucleases (NIR) in duplexes containing G*CT* and G*T* lesions. A and D, duplexes constructed from either 5′-32P-labeled 17 mer G*CT* (A), or G*T* (D) strands hybridized with their natural complementary strands. B, size marker 8-mer oligonucleotide standards with 5′-[32P]-labeled 8-mer containing 3′-hydroxyl (3′-OH), or 3′-phosphoaldehyde (3′-PUA), or 3′-phosphate (3′-P) ends are shown in lanes 3, 5, and 7, respectively. These standards were derived from the parent 17-mer strands containing uracil as described under “Experimental Procedures.” C and E, duplexes constructed from cordycepin 3′-32P-endlabeled 18 mer G*CT* (C) or G*T* (E) strands hybridized with their complementary 17-mer complementary strands. DNA glycosylase activities were performed in BER+EDTA buffer containing 5 nm of 5′- or 3′-32P-labeled duplex and 50 nm of pure enzyme. Apn1 activity was measured in BER buffer containing 5 mm MgCl2. The NIR activity of APE1 protein was performed in NIR buffer containing 0.1 mm MgCl2. After 30 min of incubation at 37 °C, the enzymatic reactions were stopped, and the DNA was purified before denaturing PAGE analysis. UT indicates untreated oligonucleotide. For details see “Experimental Procedures.”
G*CT* Oligonucleotide Duplexes
The bifunctional DNA glycosylases NEIL1, Nei, and Fpg cleave the 5′-32P-labeled DNA strand, thus giving rise to fast migrating ∼8-mers with a 3′-phosphate end (Fig. 3A, lanes 1–3), as expected for the bifunctional DNA glycosylases that exhibit β,δ-elimination activity as depicted in Fig. 1B. On the other hand, APE1, Apn1, and Nfo generate slower migrating 8-mer cleavage fragments with 3′-OH ends (Fig. 3A, lanes 4–6), which are expected from the hydrolytic mechanism of action of AP endonucleases (29). Finally, the bifunctional DNA glycosylase Nth that gives rise to β-elimination (Fig. 1B) generates the most slowly migrating 8-mer fragments containing the 3′-α,β-unsaturated aldehyde group (Fig. 3A, lane 8). The latter is unstable and typically exists in the hydrated form (PUAH) (30, 31). These cleavage fragments were therefore identified as the 5′-CCACCAAC-p-PUAH sequence, which was confirmed by MALDI-TOF/MS methods (see below). Lane 7 in Fig. 3A depicts a control experiment (untreated G*CT* duplex). To corroborate the chemical nature of the 3′-terminal residue of the cleavage products of G*CT* duplexes, the electrophoretic mobilities of the reaction products were compared with standard 8-mer oligonucleotide markers with 3′-P, 3′-PUA and 3′-OH ends. As expected, the cleavage products of NEIL1, Nth and Nfo acting upon 5′-32P-labeled G*CT* duplexes co-migrated with 8-mer 3′-P, 8-mer 3′-PUA, and 8-mer 3′-OH size markers, respectively (Fig. 3B, lanes 3–8).
In the case of the 3′-labeled G*CT* duplexes, the modified strands are longer by one nucleotide because of the 3′-end-labling method. The bifunctional DNA glycosylases NEIL1, Nei, and Fpg yield fast-migrating ∼9-mer fragments with a 5′-phosphate end that are consistent with cleavage of the G* site (Fig. 3C, lanes 1–3). The NTH1 and Nth glycosylases also yield the same fast-migrating 3′-end-labeled cleavage fragments (Fig. 3B, lanes 7 and 8) since the bulky PUA groups remain on the 5′-unlabeled fragment rather than on the 3′-end-labeled one (29) according to the cleavage mechanism (Fig. 1B). As expected from the established mechanism of action of NIR-AP endonucleases, Apn1, APE1, and Nfo generate slower migrating DNA cleavage fragments (Fig. 3C, lanes 4–6). Based on their mobilities, these fragments could be identified either as 10-mers with a phosphorylated damaged guanine residue G* at its 5′-end (21), or as slower migrating 9-mer fragments with a 5′-OH end, which is more consistent with the MALDI-TOF/MS results (see below). It should be noted that all three NIR-AP endonucleases exhibit nonspecific 3′→5′ exonuclease activity (32, 33), which degrades 5′- and 3′-end-labeled 17-mer and 18-mer oligonucleotides, respectively (Fig. 3, A and C). This activity accounts for the dark bands on the bottom of the gels in lanes 4–6 (Fig. 3, C and E) that are due to cordycepin monophosphate 3′-dAM32P, which are products of the 3′→5′ exonuclease activity of AP endonucleases. Similarly, some degradation of the 5′-end-labeled 17-mers is observable in Fig. 3, A and D (lanes 4 and 5).
G*T* Oligonucleotide Duplexes
Incubation of the G*T* 17-mer duplexes (Fig. 3, D and E) with the same enzymes as the G*CT* 17-mer duplexes (Fig. 3, A and C) yields similar results. Bifunctional DNA glycosylases excise the G* residue and the resulting AP sites thus formed are subsequently cleaved on their 3′-sides generating a 3′-deoxyribose-phosphate group at the 5′-cleavage fragment. The AP endonucleases on the other hand, incise the damaged strand on the 5′-side and adjacent to G*.
MALDI-TOF/MS Analysis of Cleavage Fragments
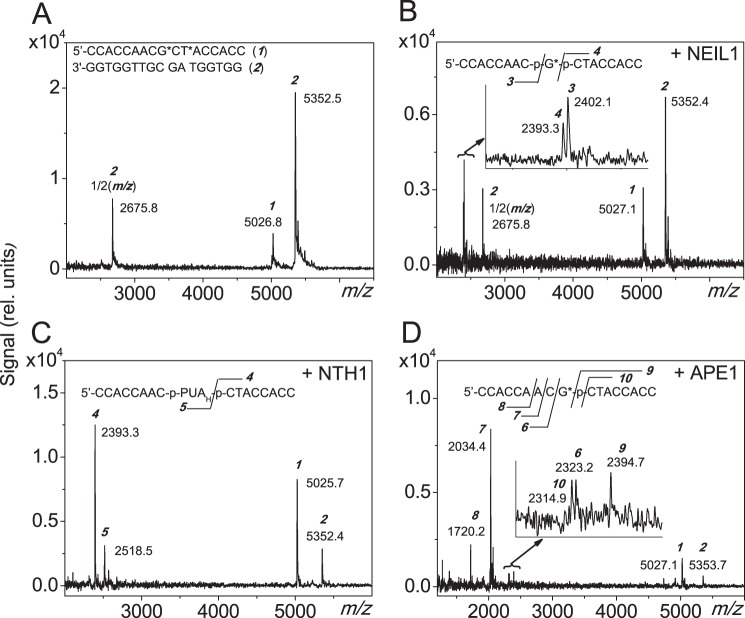
Further insights into the identities of the cleavage fragments generated by BER and NIR pathways were obtained using negative mode mass spectrometric methods. Here we focus on the results obtained with G*CT* duplexes (Fig. 4), since the G*T* and G*CT* duplexes yield similar cleavage products (Fig. 3). Results obtained with untreated G*CT* 17-mer duplexes are depicted in Fig. 4A. The two peaks shown are due to the G*CT* IntraCL-containing strand and the complementary strand, and the lack of other smaller molecular mass products indicates that the starting material is not degraded (Fig. 4A). On the other hand, after treatment with NEIL1, two additional, closely spaced fragments are observed at m/z 2402.1 and 2393.3 (Fig. 4B). These peaks are identified as the 5′-CCACCAAC-p and p-CTACCACC-3′ 8-mer fragments that are the expected cleavage products induced by bifunctional glycosylase (Fig. 1B). These results are also consistent with the gel electrophoresis results (Fig. 3, A and C, lane 1) and the conclusion that the cross-linked base G* is excised.
FIGURE 4.
Negative MALDI-TOF mass spectra of the cleavage fragments generated by DNA repair proteins in the 17-mer G*CT* duplexes incubated with NEIL1, NTH1, and APE1 proteins. Typically, 40 pmol of the lesion-containing oligonucleotide duplexes were incubated with either 100 nm NEIL1 and NTH1 or 20 nm APE1 in the appropriate reaction buffer (100 μl) at 37 °C for 60 min. The products were desalted prior to the MALDI-TOF/MS measurements. The experimental m/z values of the molecular ions, [M-H]− are consistent with those calculated from monoisotopic masses provided below in brackets; the doubly charged ions, [M-2H]2− are marked as ½(m/z). A, control non-treated duplex; peak 1: 5′-CCACCAACG*CT*ACCACC (m/z 5023.9), peak 2: 5′-GGTGGTAGCGTTGGTGG (m/z 5349.9). B, duplex treated with NEIL1; peak 3: 5′-CCACCAAC-p (m/z 2401.4), peak 4: 5′-p-CTACCACC (m/z 2392.4). C, duplex treated with NTH1; peak 5: 5′-CCACCAAC-3′-p-PUAH (hydrated form, m/z 2518.6). D, duplex treated with APE1; peak 6: 5′-CCACCAAC (m/z 2321.5); peak 7: 5′-CCACCAA (m/z 2032.4), peak 8: 5′-CCACCA (m/z 1720.2), peak 9: 5′-p-CTACCACC (m/z 2392.4), peak 10: 5′-CTACCACC (m/z 2313.6). For details see “Experimental Procedures.”
After treatment with NTH1, one of the cleavage fragments is p-CTACCACC-3′ (m/z 2393.3 (Fig. 4C) which is also observed in the case of NEIL1 (Fig. 4B). The other one at m/z 2518.5 is the fragment 5′-CCACCAAC-3′-p-PUAH (Fig. 4C), as expected for NTH1 (Fig. 3A, lane 8). In this fragment the PUAH is the hydrated form (-CH2CHOHCHOHCH2CHO, M = 117.0 Da) of the aldehyde since its mass is 18 Da higher than the mass of the α,β-unsaturated aldehyde (-CH2CHOHCH = CHCHO, M = 99.0 Da) (30, 31). Again the G* base is missing from all of these oligonucleotide fragments; furthermore the former T* base is present in its intact form T in all 3′-downstream cleavage fragments.
The negative mass spectra of the products of APE1-catalyzed incision of G*CT* duplexes exhibits a series of molecular ions (Fig. 4D): 1) The cleavage fragment at m/z 2323.2 is 5′-CCACCAAC generated by the cleavage adjacent to and on the 5′-side of G*, as expected from Fig. 3A (lane 5). Two other fragments at m/z 2034.4 and 1720.2, corresponding to the 7-mer oligonucleotides 5′-CCACCAA-3′ and the 6-mer 5′-CCACCA-3′, respectively, are attributed to degradation catalyzed by the known 3′→5′ exonuclease activity of APE1 (32, 33). 2) The APE1-cleavage fragments at m/z 2394.7 and 2314.9, assigned to the 8-mer oligonucleotide 5′-p-CTACCACC-3′ and its dephosphorylated form 5′-CTACCACC-3′, respectively, both released from the cleavage of the strand on the 3′-side of G* in the G*CT* duplexes. As in the case of 5′-upstream cleavage fragments, these are shorter by one nucleotide than a 9-mer with a G (or G*) at the 5′-end of the 3′-downstream cleavage fragment (21, 22).
Taken together, these results suggest that all the DNA glycosylases tested excise the guanine G* in G*CT* and G*T* cross-links and then cleave the DNA strands on the 3′-side of the apurinic site generated by the removal of G*, either via β or β,δ-elimination mechanisms (Fig. 1B). The AP endonucleases tested, cleave the modified strands on the 5′-side of G* of the G*T* and G*CT* cross-links and induce further degradation of the cross-link with the release of the G* base that is missing from the 3′-side cleavage fragments analyzed by mass spectrometric methods.
Discussion
The primary DNA base target of oxidizing agents, that principally function via one electron transfer mechanisms, is guanine (34), the most easily oxidizable nucleic acid base (35). Oxidatively modified guanines include a diverse group of different single guanine base oxidation products and tandem lesions such as intra- and interstrand crosslinks; the latter are more genotoxic than single oxidized bases (2). The tandem lesions detected in cells include 8,5′-cyclo-2′-deoxypurine (36–38), guanine(C8)-thymine(5-methyl) (39), and guanine(C8)-cytosine(C5) (40) IntraCLs. The repair of such oxidatively generated lesions is a crucial factor in maintaining genomic stability during oxidative stress (20).
It is well established that the 8,5′-cyclo-2′-deoxypurine lesions are not repaired by DNA glycosylase-mediated BER mechanisms, but are excellent substrates of mammalian NER (12, 13, 16, 41, 42) and prokaryotic NER repair pathways (43). However, a limited amount of information is available about IntraCLs that involve covalent bonds between two different nucleotides on the same strand. It has been shown that the G[8–5m]C, G[8–5mT] and G[8–5m]C IntraCLs are substrates of the prokaryotic UvrABC system (44, 45). Based on differences of levels of these lesions in NER-deficient and proficient mammalian cells and tissues, it was concluded that the G[8–5m]T IntraCL is a substrate of NER in vivo (15). The G*CT* and G*T* IntraCLs embedded in 135-mer DNA were found to be excellent-to-modest NER substrates, respectively, in human cell extracts as shown in Fig. 5 (17). In the same cell extract incubation experiments, substantial amounts of 67-mer cleavage fragments, corresponding to cleavage at the sites of the lesions, were also noted, but not further investigated (17).
FIGURE 5.
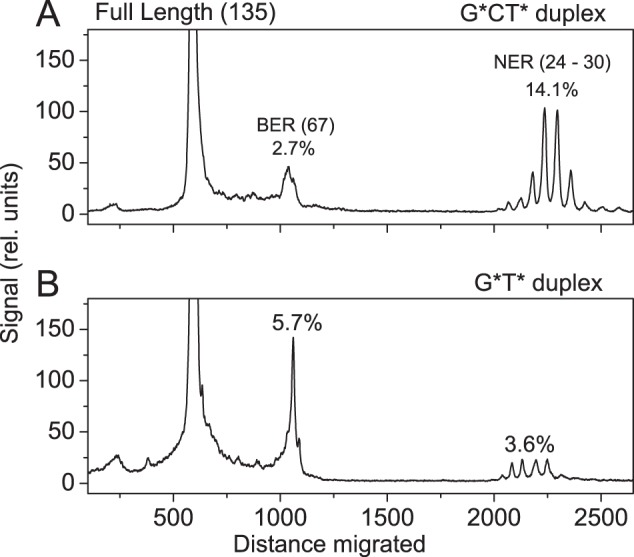
Examples of histograms derived from denaturing gel autoradiographs depicting the relative distributions of BER and NER excision products in the 135-mer G*CT* (A) and G*T* (B) duplexes after incubation in HeLa cell extracts for 40 min (17). The 17-mer sequences with either G*CT*, or G*T* lesions were incorporated into the 135-mer duplexes. The exact modified strand containing the 17-mer G*T* sequence (underlined) is: 5′-d(GACCTGAACACGTACGGAATTCGATATCCTCGAGCC AGATCTGCGCCAGCTGGCCACCC[32P]CCACCAACG*T*CACCACCCGCCAAGCTTGGGCTGCAGCAGGTCGACTCTAGAGGATCCCGGGCGAGCTCGAATTCGC). The duplex with the G*CT* sequence is the same except that it substitutes the G*T*C sequence in the 135-mer strand shown. The position of the 32P-labeled phosphodiester bond is indicated by [32P]. The NER reaction gives rise to a series of dual incision products 24–30 nucleotides in lengths containing the lesions. Typical NER results for the G*T* and G*CT* lesions were published earlier (17), but the quantitative analysis of the BER reaction products was omitted in that publication.
Our in vitro biochemical studies suggest that all the DNA glycosylases/AP lyases tested excise the guanine G* in G*CT* and G*T* cross-links and then cleave the DNA strands on the 3′-side of the apurinic site resulting from the removal of G*, via either β or β,δ-elimination mechanisms (Fig. 1B). All of the NIR-specific AP endonucleases tested, cleave the damaged strands on the 5′-side of G* and appear to generate cordycepin-labeled ∼9-mers with either phosphate or OH-groups at the 5′-ends, the latter exhibiting a mobility similar to that of a 10-mer with a phosphate residue at its 5′-end. However, the MALDI-TOF/MS results are consistent with the 9-mer interpretation. Nevertheless, incision at the 3′-side of a lesion by NIR mechanisms is not supported by previous studies (21) (Fig. 1B), and we cannot exclude that the initially formed 3′-fragment does contain a G* residue at its 5′-end and that AP endonucleases induce further degradation of DNA with the loss of the G* nucleotide to yield the observed 5′-p-CTACCCCACC and 5′-CTACCACC fragments (Fig. 4D, peaks 9 and 10), under conditions used to prepare samples for MALDI-TOF/MS analysis.
It is interesting to note that the bifunctional DNA glycosylases and AP endonucleases studied are capable of excising the cross-linked G* in the G*T* and G*CT* duplexes, but not the originally cross-linked T* according to the denaturing gel electrophoresis results (Fig. 3) and MALDI-TOF/MS analysis (Fig. 4). In contrast, denaturing gel electrophoresis showed that the standard hot piperidine treatment cleaves 5′-CCATCG*CT*ACC at G* and T* sites with the release of 5′-CCATCp and 5′-pACC fragments according to MALDI-TOF/MS analysis (27). This is clear evidence that hot alkali is unable to hydrolyze the G*[C8-N3]T* bond to form the intact T that is generated by the bifunctional DNA glycosylases and AP endonucleases (Figs. 3 and 4). The G*[C8-N3]T* bond is also resistant to nuclease P1, which generates cross-linked dinucleotide d(G*-T*) and d(G*pT*) fragments after complete hydrolysis of the single-stranded DNA containing G*CT* and G*T* IntraCL, respectively (10, 27). The phosphodiesterases 1 and 2 do not cleave the phosphodiester bonds between the cross-linked nucleotides (Fig. 2), and the combined action of these enzymes generates only d(G*pCpT*) and d(G*pT*) fragments (10, 27).
The cross-linked guanine has two covalent N-C bonds, one is the normal G*[N7-C1′] glycosydic bond, and the second involves the C8 atom on the same imidazole ring linked to N3 of thymine in G*T* or to the T on the 3′-side of C in G*CT* duplexes (Fig. 1A). Based on our data we propose that the bifunctional DNA glycosylases and AP endonucleases are able to cleave both the G*[C8-N3]T* and the G*[N7-C1′] bonds.
To summarize, we have shown that the cleavage observed in cell extracts at the sites of the G*T* and G*CT* intrastrand lesions in double-stranded DNA (17) can be attributed to BER mechanisms. It should be noted that in the same experiments both G*CT* and G*T* lesions are also removed by NER mechanisms as shown in Fig. 5 (17). It is noteworthy that the higher yield of NER products in the case of G*CT* is accompanied by a lower yield of BER products (Fig. 5A). By contrast, the yield of NER products is ∼5 times smaller in the case of G*T* while the BER yield is significantly higher (Fig. 5B). This inverse correlation between BER and NER product yields suggests that these two processes may be competing with one another. The origins of these effects are presently under investigation. It is remarkable that both BER and NER can function in parallel in incising the G*T* and G*CT* intrastrand cross-links in DNA in the same human cell extract experiments (17). These observations suggest that these two pathways may complement one another in cellular environments.
Acknowledgments
Components of this work were conducted in the Shared Instrumentation Facility at NYU that was constructed with support from a Research Facilities Improvement Grant (C06 RR-16572) from the National Center for Research Resources, National Institutes of Health. The acquisition of the MALDI-TOF mass spectrometer at NYU was supported by the National Science Foundation (CHE-0958457).
This work was supported by the National Institute of Environmental Health Sciences Grant R01 ES 011589 (to V. S.), and Grants from Electricité de France RB 2014-26 (to M. K. S. and D. G.).
- ICL
- interstrand cross-link
- BER
- base excision repair
- NIR
- nucleotide incision repair
- NER
- nucleotide excision repair
- IntraCL
- intrastrand cross-link
- G*[C8-N3]T*
- guanine-thymine cross-link with a covalent bond between the C8 (G*) and thymine N3 (T*) atoms
- G*T*
- IntraCl between adjacent G and T bases
- G*CT*
- IntraCl between G and T separated by one C base
- AP
- apurinic/apyrimidinic.
References
- 1. Jung Y., Lippard S. J. (2007) Direct cellular responses to platinum-induced DNA damage. Chem. Rev. 107, 1387–1407 [DOI] [PubMed] [Google Scholar]
- 2. Cadet J., Ravanat J. L., TavernaPorro M., Menoni H., Angelov D. (2012) Oxidatively generated complex DNA damage: tandem and clustered lesions. Cancer Lett. 327, 5–15 [DOI] [PubMed] [Google Scholar]
- 3. Clauson C., Scharer O. D., Niedernhofer L. (2013) Advances in understanding the complex mechanisms of DNA interstrand cross-link repair. Cold Spring Harb. Perspect. Biol. 5, a012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Friedberg E. C. (2003) DNA damage and repair. Nature 421, 436–440 [DOI] [PubMed] [Google Scholar]
- 5. Box H. C., Budzinski E. E., Dawidzik J. D., Wallace J. C., Evans M. S., Gobey J. S. (1996) Radiation-induced formation of a crosslink between base moieties of deoxyguanosine and thymidine in deoxygenated solutions of d(CpGpTpA). Radiat. Res. 145, 641–643 [PubMed] [Google Scholar]
- 6. Box H. C., Dawidzik J. B., Budzinski E. E. (2001) Free radical-induced double lesions in DNA. Free Radic. Biol. Med. 31, 856–868 [DOI] [PubMed] [Google Scholar]
- 7. Zhang Q., Wang Y. (2003) Independent generation of 5-(2′-deoxycytidinyl)methyl radical and the formation of a novel cross-link lesion between 5-methylcytosine and guanine. J. Am. Chem. Soc. 125, 12795–12802 [DOI] [PubMed] [Google Scholar]
- 8. Dizdaroglu M. (1986) Free-radical-induced formation of an 8,5′-cyclo-2′-deoxyguanosine moiety in deoxyribonucleic acid. Biochem. J. 238, 247–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dirksen M. L., Blakely W. F., Holwitt E., Dizdaroglu M. (1988) Effect of DNA conformation on the hydroxyl radical-induced formation of 8,5′-cyclopurine 2′-deoxyribonucleoside residues in DNA. Int. J. Radiat. Biol. 54, 195–204 [DOI] [PubMed] [Google Scholar]
- 10. Crean C., Uvaydov Y., Geacintov N. E., Shafirovich V. (2008) Oxidation of single-stranded oligonucleotides by carbonate radical anions: generating intrastrand cross-links between guanine and thymine bases separated by cytosines. Nucleic Acids Res. 36, 742–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Madugundu G. S., Wagner J. R., Cadet J., Kropachev K., Yun B. H., Geacintov N. E., Shafirovich V. (2013) Generation of guanine-thymine cross-links in human cells by one-electron oxidation mechanisms. Chem. Res. Toxicol. 26, 1031–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kuraoka I., Bender C., Romieu A., Cadet J., Wood R. D., Lindahl T. (2000) Removal of oxygen free-radical-induced 5′,8-purine cyclodeoxynucleosides from DNA by the nucleotide excision-repair pathway in human cells. Proc. Natl. Acad. Sci. U.S.A. 97, 3832–3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brooks P. J. (2008) The 8,5′-cyclopurine-2′-deoxynucleosides: candidate neurodegenerative DNA lesions in xeroderma pigmentosum, and unique probes of transcription and nucleotide excision repair. DNA Repair 7, 1168–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. You C., Dai X., Yuan B., Wang J., Wang J., Brooks P. J., Niedernhofer L. J., Wang Y. (2012) A quantitative assay for assessing the effects of DNA lesions on transcription. Nat. Chem. Biol. 8, 817–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang J., Cao H., You C., Yuan B., Bahde R., Gupta S., Nishigori C., Niedernhofer L. J., Brooks P. J., Wang Y. (2012) Endogenous formation and repair of oxidatively induced G[8–5 m]T intrastrand cross-link lesion. Nucleic Acids Res. 40, 7368–7374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pande P., Das R. S., Sheppard C., Kow Y. W., Basu A. K. (2012) Repair efficiency of (5'S)-8,5′-cyclo-2′-deoxyguanosine and (5'S)-8,5′-cyclo-2′-deoxyadenosine depends on the complementary base. DNA Repair 11, 926–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ding S., Kropachev K., Cai Y., Kolbanovskiy M., Durandina S. A., Liu Z., Shafirovich V., Broyde S., Geacintov N. E. (2012) Structural, energetic and dynamic properties of guanine(C8)-thymine(N3) cross-links in DNA provide insights on susceptibility to nucleotide excision repair. Nucleic Acids Res. 40, 2506–2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wallace S. S., Murphy D. L., Sweasy J. B. (2012) Base excision repair and cancer. Cancer Lett. 327, 73–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krokan H. E., Bjøras M. (2013) Base excision repair. Cold Spring Harb. Perspect. Biol. 5, a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Scott T. L., Rangaswamy S., Wicker C. A., Izumi T. (2014) Repair of oxidative DNA damage and cancer: recent progress in DNA base excision repair. Antioxid. Redox Signal. 20, 708–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ischenko A. A., Saparbaev M. K. (2002) Alternative nucleotide incision repair pathway for oxidative DNA damage. Nature 415, 183–187 [DOI] [PubMed] [Google Scholar]
- 22. Ishchenko A. A., Deprez E., Maksimenko A., Brochon J. C., Tauc P., Saparbaev M. K. (2006) Uncoupling of the base excision and nucleotide incision repair pathways reveals their respective biological roles. Proc. Natl. Acad. Sci. U.S.A. 103, 2564–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Faure V., Saparbaev M., Dumy P., Constant J. F. (2005) Action of multiple base excision repair enzymes on the 2′-deoxyribonolactone. Biochem. Biophys. Res. Commun. 328, 1188–1195 [DOI] [PubMed] [Google Scholar]
- 24. Ishchenko A. A., Ide H., Ramotar D., Nevinsky G., Saparbaev M. (2004) Alpha-anomeric deoxynucleotides, anoxic products of ionizing radiation, are substrates for the endonuclease IV-type AP endonucleases. Biochemistry 43, 15210–15216 [DOI] [PubMed] [Google Scholar]
- 25. Katafuchi A., Nakano T., Masaoka A., Terato H., Iwai S., Hanaoka F., Ide H. (2004) Differential specificity of human and Escherichia coli endonuclease III and VIII homologues for oxidative base lesions. J. Biol. Chem. 279, 14464–14471 [DOI] [PubMed] [Google Scholar]
- 26. Zharkov D. O., Golan G., Gilboa R., Fernandes A. S., Gerchman S. E., Kycia J. H., Rieger R. A., Grollman A. P., Shoham G. (2002) Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. EMBO J. 21, 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Crean C., Lee Y. A., Yun B. H., Geacintov N. E., Shafirovich V. (2008) Oxidation of guanine by carbonate radicals derived from photolysis of carbonatotetramminecobalt(III) complexes and the pH dependence of intrastrand DNA cross-links mediated by guanine radical reactions. ChemBioChem. 9, 1985–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Prorok P., Alili D., Saint-Pierre C., Gasparutto D., Zharkov D. O., Ishchenko A. A., Tudek B., Saparbaev M. K. (2013) Uracil in duplex DNA is a substrate for the nucleotide incision repair pathway in human cells. Proc. Natl. Acad. Sci. U.S.A. 110, E3695–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hegde M. L., Hazra T. K., Mitra S. (2008) Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 18, 27–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bourdat A. G., Gasparutto D., Cadet J. (1999) Synthesis and enzymatic processing of oligodeoxynucleotides containing tandem base damage. Nucleic Acids Res. 27, 1015–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Darwanto A., Farrel A., Rogstad D. K., Sowers L. C. (2009) Characterization of DNA glycosylase activity by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Biochem. 394, 13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mazouzi A., Vigouroux A., Aikeshev B., Brooks P. J., Saparbaev M. K., Morera S., Ishchenko A. A. (2013) Insight into mechanisms of 3′-5′ exonuclease activity and removal of bulky 8,5′-cyclopurine adducts by apurinic/apyrimidinic endonucleases. Proc. Natl. Acad. Sci. U.S.A. 110, E3071–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wong D., DeMott M. S., Demple B. (2003) Modulation of the 3′–>5′-exonuclease activity of human apurinic endonuclease (Ape1) by its 5′-incised Abasic DNA product. J. Biol. Chem. 278, 36242–36249 [DOI] [PubMed] [Google Scholar]
- 34. Cadet J., Douki T., Ravanat J. L. (2008) Oxidatively generated damage to the guanine moiety of DNA: mechanistic aspects and formation in cells. Acc. Chem. Res. 41, 1075–1083 [DOI] [PubMed] [Google Scholar]
- 35. Steenken S., Jovanovic S. V. (1997) How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J. Am. Chem. Soc. 119, 617–618 [Google Scholar]
- 36. Jaruga P., Dizdaroglu M. (2008) 8,5′-Cyclopurine-2′-deoxynucleosides in DNA: mechanisms of formation, measurement, repair and biological effects. DNA Repair 7, 1413–1425 [DOI] [PubMed] [Google Scholar]
- 37. Wang Y. (2008) Bulky DNA lesions induced by reactive oxygen species. Chem. Res. Toxicol. 21, 276–281 [DOI] [PubMed] [Google Scholar]
- 38. Wang J., Clauson C. L., Robbins P. D., Niedernhofer L. J., Wang Y. (2012) The oxidative DNA lesions 8,5′-cyclopurines accumulate with aging in a tissue-specific manner. Aging Cell 11, 714–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jiang Y., Hong H., Cao H., Wang Y. (2007) In vivo formation and in vivo replication of guanine-thymine intrastrand cross-link lesion. Biochemistry 46, 12757–12763 [DOI] [PubMed] [Google Scholar]
- 40. Hong H., Cao H., Wang Y. (2007) Formation and genotoxicity of a guanine-cytosine intrastrand cross-link lesion in vivo. Nucleic Acids Res. 35, 7118–7127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kropachev K., Ding S., Terzidis M. A., Masi A., Liu Z., Cai Y., Kolbanovskiy M., Chatgilialoglu C., Broyde S., Geacintov N. E., Shafirovich V. (2014) Structural basis for the recognition of diastereomeric 5′,8-cyclo-2′-deoxypurine lesions by the human nucleotide excision repair system. Nucleic Acids Res. 42, 5020–5032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brooks P. J., Wise D. S., Berry D. A., Kosmoski J. V., Smerdon M. J., Somers R. L., Mackie H., Spoonde A. Y., Ackerman E. J., Coleman K., Tarone R. E., Robbins J. H. (2000) The oxidative DNA lesion 8,5′-(S)-cyclo-2′-deoxyadenosine is repaired by the nucleotide excision repair pathway and blocks gene expression in mammalian cells. J. Biol. Chem. 275, 22355–22362 [DOI] [PubMed] [Google Scholar]
- 43. Jasti V. P., Das R. S., Hilton B. A., Weerasooriya S., Zou Y., Basu A. K. (2011) (5'S)-8,5′-cyclo-2′-deoxyguanosine is a strong block to replication, a potent pol V-dependent mutagenic lesion, and is inefficiently repaired in Escherichia coli. Biochemistry 50, 3862–3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gu C., Zhang Q., Yang Z., Wang Y., Zou Y., Wang Y. (2006) Recognition and incision of oxidative intrastrand cross-link lesions by UvrABC nuclease. Biochemistry 45, 10739–10746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yang Z., Colis L. C., Basu A. K., Zou Y. (2005) Recognition and incision of g-radiation-induced cross-linked guanine-thymine tandem lesion G[8,5-Me]T by UvrABC nuclease. Chem. Res. Toxicol. 18, 1339–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]