Abstract
The p21-activated kinases (PAKs) are a family of six serine/threonine kinases that act as key effectors of RHO family GTPases in mammalian cells. PAKs are subdivided into two groups: type I PAKs (PAK1, PAK2, and PAK3) and type II PAKs (PAK4, PAK5, and PAK6). Although these groups are involved in common signaling pathways, recent work indicates that the two groups have distinct modes of regulation and have both unique and common substrates. Here, we review recent insights into the molecular level details that govern regulation of type II PAK signaling. We also consider mechanisms by which signal transduction is regulated at the level of substrate specificity. Finally, we discuss the implications of these studies for clinical targeting of these kinases.
Keywords: CDC42, protein structure, Ras-related C3 botulinum toxin substrate 1 (Rac1), serine/threonine protein kinase, signal transduction
Introduction
The type II PAKs2 are members of the Sterile 20 (Ste20) family of STE (homologs of yeast Sterile 7, Sterile 11, and Sterile 20) group serine/threonine kinases. They are important for signaling from small GTPases of the RHO family, particularly the CDC42 (cell division cycle 42) and to a lesser extent RAC (Ras-related C3 botulinum toxin substrate) isoforms, to the actin cytoskeleton and to growth and survival pathways. Type II PAKs share significant sequence similarity with the well studied type I PAKs (PAK1, PAK2, and PAK3), which are extensively reviewed elsewhere (1–5), but the roles of the type I and type II PAKs in GTPase signaling pathways, and their mechanisms of regulation, differ considerably. In the sections below, we consider the function of the type II PAK serine/threonine kinases in small GTPase pathways, the structural and biochemical basis for their regulation, mechanisms of their targeting to substrates, and some of the current strategies for targeted small molecule inhibition of these enzymes.
Type II PAKs in Signal Transduction
The type II PAKs are binding partners of RHO family small GTPases. Their dominant role in signal transduction is as serine/threonine kinases, where transfer of the γ-phosphate from an ATP molecule to a substrate protein results in consequent regulation of downstream effector pathways. The activation of type II PAK kinase signaling is associated with small GTPase binding, but as discussed below, direct activation does not seem to occur upon small GTPase binding. Rather, GTPases are thought to primarily regulate type II PAKs by controlling their subcellular localization. Type II PAKs also have reported kinase-independent roles as scaffolding proteins, with functions that are still emerging.
The type II PAKs are placed at critical nodal points in multiple signaling pathways that are associated with cell growth, cytoskeletal dynamics, cell polarity, survival, and development (6–8). They are located directly downstream of RHO family GTPase activation. Numerous substrates of type II PAKs have been identified that are thought to act as downstream effectors in distinct cellular processes. For example, direct phosphorylation of LIM kinases (9, 10), p210/β-catenin (11, 12), slingshot phosphatase (SSH) (13), GEF-H1 (8, 14), and PDZ-RHOGEF (15) has been implicated in transducing type II PAK signaling to the actin cytoskeleton, and phosphorylation of Par6B (16), integrin β5 (17–19), and paxillin (20) regulates cell adhesion. Likewise, growth and survival signals emanating from type II PAKs may involve direct phosphorylation of Raf (21, 22), MDM2 (23), Bcl2-associated agonist of cell death (BAD) (24–27), and RAN (28). From studies in animal models, only PAK4 has been found to be essential for life; Pak4 knock-out mice display severe defects in angiogenesis, in the cardiovascular system, and in neuronal development (29, 30). Pak5 and Pak6 knock-out mice are viable and show few phenotypes; their double knock-out, however, is associated with cognition and locomotive defects (31, 32), which are potentially associated with disruption of the interaction between two PAK substrates, Pacsin-1 and Synaptojanin-1, that normally control synaptic vesicle trafficking (33). There are clear differences between the type II PAKs in tissue expression profile (3, 8, 34), with PAK4 widely expressed and most abundant in prostate, testis, and colon (35), PAK5 predominantly found in the brain and pancreas (32, 36), and PAK6 found largely in testis, prostate, and brain, and possibly the kidneys and placenta (34, 37, 38). Subcellular localization also differs between the family members (3), as discussed in the next section, which may contribute to selective targeting of some substrates. For example, so far androgen receptor has only been found to be a substrate of PAK6 (38). These enzymes therefore play important roles in signal transduction within multiple pathways.
Specificity and Impact of PAKs as Binders and Effectors of RHO Family Small GTPases
Small GTPases are elegant switches that adopt two basic structural conformations, the GDP-loaded “inactive” conformation and the GTP-loaded “active” conformation. The GTP-loaded active conformation is associated with binding to effector molecules and consequent activation of downstream signaling pathways (39). When the effector protein is a protein kinase, GTPase binding is often associated with activation of catalytic activity, and this is the case for the PAK serine/threonine kinases. Signaling from the PAKs is controlled by their binding to small GTPases; however, the mechanism of control differs between the type I and type II subgroups (Table 1).
TABLE 1.
Similarities and differences between the type I and type II PAKs
| Type I PAKs (PAK1, PAK2, PAK3) | Type II PAKs (PAK4, PAK5, PAK6) | |
|---|---|---|
| GTPase binding | Yes, via CRIB domain | Yes, via CRIB domain |
| GTPase specificity | RAC ≈ CDC42 | CDC42 > RAC |
| Direct activation by GTPase | Yes | Noa |
| Autoinhibition mechanism | Trans-autoinhibited homodimer | Pseudosubstrate autoinhibition |
| Autoinhibited stoichiometry in solution | Homodimeric | Monomeric |
| Activation loop phosphorylation state (autoinhibited) | Unphosphorylated | Phosphorylated |
| Activation mechanism | GTPase binding-associated conformational change; activation loop phosphorylation | Pseudosubstrate release |
| Phosphoacceptor residue specificity | Ser > Thr | Ser > Thr |
a Most studies indicate that the type II PAKs are not directly activated by small GTPase binding.
All PAKs harbor a GTPase-binding region termed the CRIB (CDC42/RAC interactive binding) domain (Fig. 1A). CRIB domains are highly similar among the six family members with identities of over 50% between the type I and type II PAKs. CRIB domains interact specifically with RHO family small GTPases when in the GTP-loaded state. Structurally, the CRIB domains of type I and II PAKs interact with small GTPases in a similar fashion (Protein Data Bank (PDB) IDs: 2QME, 2OV2, and 2ODB) (40, 41) (Fig. 1B), and NMR studies indicate that the CRIB domain of type II PAKs is unfolded in the absence of small GTPase (42). Despite their similarity, CRIB domains from type I and type II PAKs appear to specifically target different GTPases, with type I PAKs acting downstream of RAC isoforms and CDC42 and the type II PAKs preferring CDC42 (35, 37, 38). RhoA, RhoB, and RhoC have not been implicated as PAK-binding small GTPases (3), but PAK5 has been shown to additionally interact with RhoD and RhoH (43) and PAK6 to interact with RhoV (Wrch-2) (44) and RhoJ (Tc10-like GTP-binding protein (TCL)) (45). Systematic comprehensive analysis of PAK specificity has yet to be performed and could highlight more intricate differences as GTPase effectors.
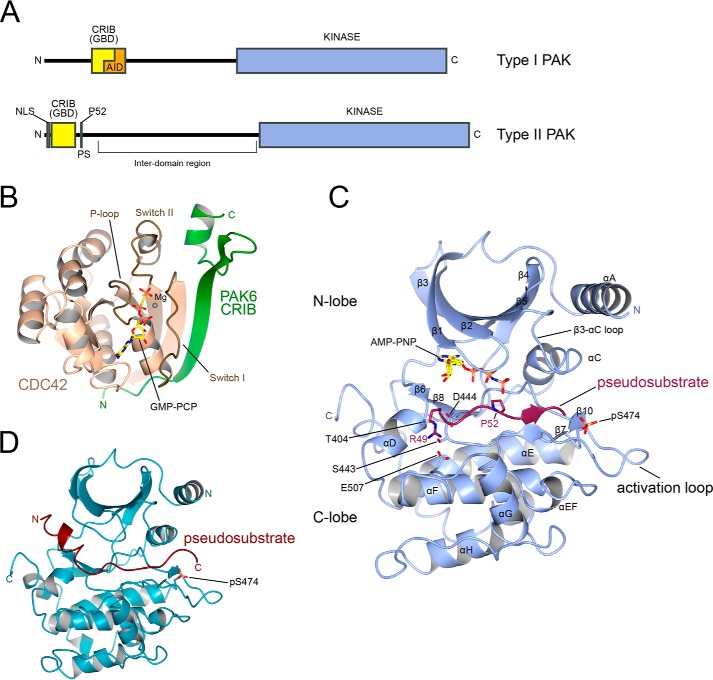
FIGURE 1.
Type II PAK domains and structure. A, schematic diagram showing domain assignments of the type I and II PAKs. NLS indicates nuclear localization sequence, GBD indicates GTPase-binding domain, and PS indicates pseudosubstrate. The location of residue Pro52 is indicated. B, the crystal structure of CDC42 in complex with the CRIB domain of PAK6 shown in graphic format. CDC42 is shown in tan, and PAK6 is shown in green. Switch I, Switch II, and P-loop regions of CDC42 are indicated. GTP analogue GMP-PCP is shown in stick format. Mg2+ is shown as a sphere. N and C termini of PAK6 are indicated. PDB ID: 2ODB. C, the crystal structure of PAK4 catalytic domain in complex with its N-terminal pseudosubstrate shown in graphic format. PAK4 catalytic domain is shown in blue, and PAK4 pseudosubstrate is shown in maroon. Secondary structure elements are labeled. Residues discussed in the text and ATP analogue AMP-PNP are shown in stick format and labeled. Activation loop phosphoserine Ser(P)474 (pS474) is indicated. N and C termini of PAK4 catalytic domain are indicated. PDB ID: 4FIF (46). D, the crystal structure of PAK4 catalytic domain in complex with a longer fragment of its N-terminal pseudosubstrate. A helical region of the pseudosubstrate is clearly visible. PDB ID: 4L67 (42).
For the type I PAKs, the interaction with small GTPases is associated with activation of kinase activity (1, 4) and is further discussed below. For the type II PAKs, however, most studies find that GTPase binding does not directly activate kinase activity (35–38, 46) (Table 1), although PAK4 isolated from mammalian cells has reportedly higher activity when associated with CDC42 (47). In contrast, interaction of the type II PAKs with small GTPase is thought to be important for regulating subcellular localization (35, 38, 48–50), The different type II PAKs localize differently, with PAK4 targeted to the Golgi upon CDC42 binding (35) and found close to the cell surface (17, 51), PAK5 to the mitochondria, nucleus (25, 43), and filopodia (43), and PAK6 to the mitochondria (25), cytoplasm (52), nucleus (38), and cell-cell junctions (53). Taken together these studies suggest that small GTPase binding to the type II PAKs does not directly impact catalytic activity, but instead targets these proteins to specific locations in the cell to prime the kinase for activation by another signal and/or place the kinase in proximity to its substrate.
The PAK Interdomain Region
Between the N-terminal CRIB domain and the C-terminal kinase domain of the type II PAKs, there is a non-conserved region of between 160 and 310 amino acids in length depending on the isoform. Although poorly conserved among the type II PAKs, in each isoform, this region contains multiple proline-rich motifs, and this region can interact with protein-binding partners (14). It is therefore possible that PAK isoform-specific functions could be driven through binding to this interdomain region. The interdomain region may well turn out to be critically important for isoform-specific interplay with signaling pathways.
Autoregulation of PAKs
Since their discovery, the mechanism of autoregulation for the type II PAKs has been unclear. High sequence similarity between the type I and type II PAKs is observed for the kinase and CRIB domains, but outside of those domains, there is little to suggest conservation of fold or binding partners. Importantly, the AID (autoinhibitory domain) that is highly conserved among the three type I PAK family members is not present in the type II PAKs. The absence of this region in the type II PAKs was both informative (it suggested differences in the regulation mechanisms of the two PAK subgroups) and a paradox (how might these proteins be regulated?), so it has presented a challenge to the community. A mechanism of autoregulation unique to PAK5, involving residues 119–123 that are not conserved in PAK4 or PAK6, was observed (49), but how this might function has remained elusive. The system therefore required detailed biochemical and structural analysis to investigate its mechanisms of regulation.
Biochemical analysis of full-length PAK4 confirms a difference between the type I and type II PAKs. The type I PAKs utilize both their CRIB domains and their AID domains to generate a trans-autoinhibited homodimer (1, 54, 55). In this state, the AIDs of each molecule bind to the substrate recognition groove of the other, thus preventing catalysis. This dimeric autoinhibited conformation is stabilized by the CRIB domains, which form the majority of the dimerization interface. Binding of small GTPase to the type I PAK CRIB domain is incompatible with the trans-autoinhibited homodimer, so it can readily impact the equilibrium between catalytically inactive and active states (55). The type I PAK trans-autoinhibited homodimer is a biochemically stable species that can be purified as a dimer by size exclusion chromatography (54). In contrast, size exclusion chromatography of purified full-length PAK4 yields an exclusively monomeric species (46, 47), implying that the type II PAKs do not form a similar stable trans-autoinhibited dimer in solution.
Early studies indicated that the catalytic domain of PAK4 had greatly increased kinase activity when compared with the full-length protein when isolated from mammalian cells (35). More recently, a biochemical study showed that the N-terminal 68 residues of PAK4 were sufficient to suppress kinase activity and that a fusion of these N-terminal residues and the catalytic domain, but lacking interdomain residues 69–285, also suppressed kinase activity, suggesting that autoinhibition of PAK4 was controlled by this N-terminal region (47). A parallel study showed through a series of structure-directed functional assays that suppression of PAK4 catalytic activity was controlled by a portion of the N terminus centered around residue Pro52 (46). X-ray crystallographic data of full-length (isoform 2) PAK4, sequence conservation analysis, and kinase activity assays focused studies on a portion of the N terminus encompassing residues Arg49-Pro50-Lys51-Pro52-Leu53-Val54, which could suppress catalytic activity of the PAK4 kinase domain, and when mutated in the context of the full-length protein, this region resulted in a significant increase in catalytic activity. When this region was co-crystallized with the PAK4 catalytic domain to yield a high-resolution structure, it became clear that this region suppresses catalytic activity by adopting a pseudosubstrate mode of binding to the catalytic domain (Fig. 1C) (46). The peptide occupied the substrate-binding cleft of the kinase domain, with Pro52 sitting precisely in the site normally occupied by the phosphoacceptor residue. Additionally, salt bridges were formed between Arg49 and acidic residues in the substrate cleft, which are discussed under “Specificity of PAKs toward Downstream Substrates” below. In all three crystal structures of PAK4 in complex with its pseudosubstrate, the kinase was found to be in an active state (46), with the DFG motif in the “DFG-in” conformation and Ser474 in the activation loop-phosphorylated (Ser(P)474). Because of the high conservation of this pseudosubstrate region across the type II PAKs, its ability to bind active-state kinase, and increases in catalytic activity upon its mutation in full-length protein in vitro and in cells (discussed below), the pseudosubstrate was proposed to be the general mode of autoregulation for the type II PAKs (46).
Supporting the pseudosubstrate mechanism of type II PAK regulation, double mutagenesis of Arg48/Arg49 results in an increase in PAK4 kinase activity (47). Further, PAK6 is autoregulated by the pseudosubstrate region and can be activated in cells by mutation of Pro52 (56). Another study also used x-ray crystallography to observe an identical pseudosubstrate conformation for the interaction of a longer peptide fragment with the PAK4 kinase domain (42). Interestingly, this later study also discovered that 10 residues N-terminal of the pseudosubstrate (Trp39–Arg48) may fold into an α-helical conformation that appears to block nucleotide access to the ATP-binding site (Fig. 1D) (42). NMR titrations suggested conformational changes in this region of the N terminus upon interaction with the catalytic domain (42). It is therefore an intriguing possibility that autoinhibition of type II PAK activity may be further stabilized by an additional “clamp” mechanism involving sequences N-terminal to the pseudosubstrate region.
Phosphorylation of the activation loop is a mechanism for regulation of the catalytic activity common to many protein kinases (57, 58). In most protein kinases, the autoinhibited or inactive conformation of the kinase is strongly disfavored once the activation loop is phosphorylated. Activation loop phosphorylation is also associated with an extended conformation (57), with the active DFG-in state of the DFG motif (59), and with fully formed active-state hydrophobic spines within the catalytic domain (60). Thus activation loop phosphorylation is often used as a biochemical surrogate to determine the activation state of a protein kinase. For the type I PAKs, significant increases in phosphorylation in cells are observed on GTPase binding, indicating that kinase activation is driven by the interaction with small GTPase (61). In contrast, the type II PAKs do not follow this paradigm and are observed to be constitutively phosphorylated in cells (35, 47). Type II PAKs are consistently activation loop-phosphorylated when produced in Escherichia coli for structural studies (42, 46, 56, 62), and although PAK4 activation loop phosphorylation has been used as a marker for activation in cell-based assays and as a histological marker (19, 63–66), the validity of this readout has been recently questioned (47). Therefore, the role of modulating phosphorylation in the activation loop of the type II PAKs as a regulatory mechanism is not completely clear. Autoregulation by the pseudosubstrate is consistent with constitutive phosphorylation of the kinase, but further work may be required to investigate whether the type II PAKs access an inactive conformation that is non-phosphorylated on the activation loop, and whether this is functionally important.
The conformational flexibility of type II PAKs has also been probed at a structural level. When the initial structures of the catalytic domains of PAK4, PAK5, and PAK6 were determined, unusual conformational flexibility was observed between these kinase domains, although all were observed to be phosphorylated on their activation loops (1, 62, 63, 67, 68). Three prominent features from these structures included an open-closed hinging of the kinase N-lobe, an open-closed hinging of the glycine-rich P-loop, and a conformational change in the β3-αC loop and αC helix (Fig. 1C). The gross hinging of the N-lobe and the opening of the P-loop are conformational movements that are associated with binding and release of nucleotide to the active site and are also observed in other kinases (57). In contrast, the conformational changes in the β3-αC loop are perhaps more specific to the type II PAKs, and not observed, for example, in the type I PAKs. In these type II PAK structures, the αC helix can vary in length by an additional turn that replaces some of the flexible β3-αC loop. The longer αC helix is associated with a shorter β3-αC loop and a closed conformation active-like state. The differences in αC helix length are also associated with different conformational states of an arginine residue (Arg359, Arg487, and Arg445 for PAK4, PAK5, and PAK6) that is sometimes observed to be part of the β3-αC loop and sometimes part of the first turn of the αC helix. In the array of fully active structures of PAK4 in complex with substrate or pseudosubstrate peptides, this arginine invariably forms a salt bridge with the activation loop phosphoserine,3 and the glutamine residue immediately N-terminal of it displays a propensity to hydrogen-bond to substrate peptide backbone. This structural evidence therefore seems to suggest that for cycling of nucleotide, concerted conformational changes of the kinase domain are required to open the cleft for ADP release and to close the cleft to form the optimal ATP-bound catalytic conformation. It is also possible that substrate or pseudosubstrate binding can stabilize the active state by forming hydrogen bonds with the β3-αC loop and helping stabilize the arginine-Ser(P) interaction by stacking against aliphatic parts of the extended arginine. This would consolidate autoinhibition by the pseudosubtrate, and/or aid phosphotransfer by helping hold the substrate in place until phosphotransfer is complete.
The recent structural studies on the type II PAKs have therefore provided insight into the autoinhibition mechanisms of these kinases, and also into the conformational flexibility accessible to the active state. These studies also suggest intriguing possibilities for understanding the mechanism of activation for these proteins. Because of the presence of proline-rich motifs in and around the pseudosubstrate, SH3-domain proteins or other protein scaffolding domains were proposed to play a role in stabilizing an active state, where the pseudosubstrate is sequestered away from the kinase domain to allow catalytic activity (46). Studies to investigate this possibility are continuing.
Specificity of PAKs toward Downstream Substrates
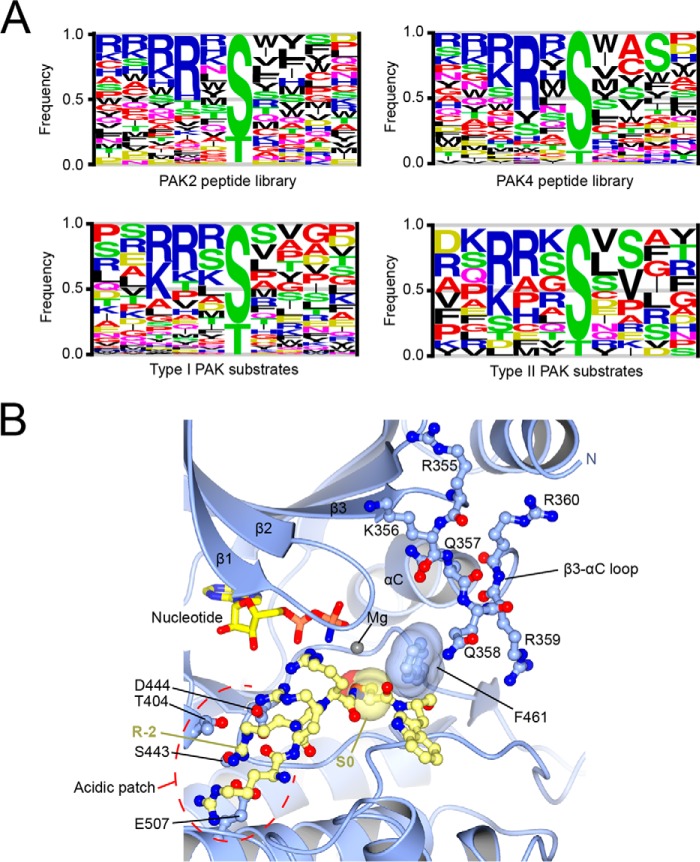
Although type I and type II PAKs have some common substrates, recently a number of type II PAK-specific substrates have been identified (6). Specificity of PAKs for downstream substrates is likely to be controlled at many levels, including subcellular localization as well as direct physical interactions with substrates. Early studies with synthetic peptide substrates defined a consensus phosphorylation site motif for PAK2 (69), including an essential Arg residue at the −2 position in the substrate and a strong preference for basic residues (Lys/Arg) at the −3 position. Peptide library analysis of both type I and type II PAKs has confirmed these studies and provided additional detail, including distinct sequence preferences for type I and type II PAKs (70). For example, these studies revealed that other residues, such as Tyr and His, could substitute for the Arg at the −2 position and that all PAKs prefer to have a hydrophobic residue at the +1 position. Further, all PAKs prefer Ser over Thr as the phosphoacceptor residue. In addition to these common features, type I and type II PAKs differ with respect to residues preferred downstream of the phosphorylation site, including a relative preference for small residues at the +2 position by type II PAKs when compared with larger hydrophobic residues preferred by type I PAKs (Fig. 2A).
FIGURE 2.
Type II PAK substrate specificity and its determinants. A, the sequence logos showing specificity of type I and type II PAKs on peptide and protein substrates. Logos were generated using either positional scanning peptide library data (top) or data from all reported sites of phosphorylation in protein substrates (bottom) as compiled on the PhosphoSitePlus database (98). Sites of phosphorylation for the reported type II PAK substrates androgen receptor, LIMK1, Pacsin-1, Synaptojanin-1, MDM2, and PAR6B, which are not included in PhosphoSitePlus, were taken from the literature (9, 16, 23, 33, 38). Logos were generated using the program EnoLOGOS (99) and are scaled such that the height of the letter is proportional to the frequency of the residue at the indicated position. B, close-up view showing the interaction of PAK4 catalytic domain with an optimized PAKtide substrate peptide. PAK4 is shown in graphic format in blue, and amino acids discussed in the text are shown in stick format. PAKtide is shown in stick format in yellow. The DFG+1 residue Phe461 and the phosphoacceptor residue Ser0 are indicated. The acidic patch and arginine −2 residue are indicated. Nucleotide is shown in stick format. PDB ID: (4JDI) (71).
Structural studies of PAK4-peptide complexes and biochemical analysis of PAK mutants have pointed to key features of the kinase catalytic domains that dictate common and unique phosphorylation site preferences (71). Selectivity at the −2 position for example appears to involve direct interactions between the guanidino head group of the substrate Arg residue with two basic residues in the kinase catalytic cleft (Asp444 and Glu507) (72). This −2 specificity pocket is fully analogous to the site in PKA that binds to the −2 Arg residue in its substrates (73). Selectivity for Ser versus Thr as the phosphoacceptor residue for PAKs, and indeed for Ser-Thr kinases in general, was shown to depend on the identity of the so-called DFG+1 residue, located immediately downstream of the conserved Asp-Phe-Gly sequence that begins the kinase activation loop. Kinases with β-branched amino acids in this position have a propensity to phosphorylate threonine as the substrate residue, but a bulky amino acid in the DFG+1 position results in a propensity to phosphorylate serine (Fig. 2B). All PAKs have a Phe as the DFG+1 residue, and mutation to Val converted PAK4 to a Thr-preferring kinase (71). Features unique to type I versus type II PAKs at positions downstream of the phosphorylation site are strongly influenced by the β3-αC loop sequence that as discussed above can differ in conformation between the two groups. Indeed, mutants in which two residues in this region were exchanged between PAK2 and PAK4 (PAK2-P286Q,K287R and PAK4-Q358P,R359K) could partly interconvert their peptide sequence specificities (70).
Known sites of phosphorylation mapped on protein substrates are enriched for features common to the type I and type II PAK consensus motifs (Fig. 2A). Notably, not all PAK sites conform closely to the peptide consensus motif; for example, several substrates are phosphorylated on Thr residues rather than the preferred Ser. By analogy with other kinases, phosphorylation of non-preferred sequences may facilitate modulation of the level of phosphorylation in response to changes in PAK activity in a manner that may depend on competition by the kinase for more optimal phosphorylation sites (74, 75). Sites phosphorylated by type I and type II PAKs appear to be similar in sequence, suggesting that differences in phosphorylation site motifs may play little role in controlling which specific PAKs are responsible for phosphorylation of a given substrate. However, it was shown that specific phosphorylation of Pacsin-1 in vitro by type II PAKs is strongly influenced by the phosphorylation site sequence. Although wild-type PAK2 poorly phosphorylated Pacsin-1 in vitro, the PAK2 P286Q,K287R exchange mutant could phosphorylate Pacsin-1 with comparable efficiency to PAK5. These observations suggest that for at least some substrates, PAK phosphorylation sites are specifically tuned to optimize phosphorylation by either type I or type II PAKs.
Alteration of Type II PAK Signaling in Cancer
Multiple studies have found genomic amplification, protein overexpression, or point mutation of type II PAKs in cancer (6, 76). Gene amplification has been most frequently observed for PAK4, specifically in pancreatic, ovarian, and colon cancers (65, 77–79) as well as multiple tumor cell lines (6, 76, 80). Amplifications of PAK5 in colorectal (81, 82) and PAK6 in prostate cancer (83) have also been observed. The assumption for these tumors is that catalytic activity for the type II PAKs is up-regulated and potentially plays a role in driving the malignancy. These notions are corroborated by cell-based assays showing, for example, that silencing expression type II PAKs by RNAi can retard growth (84, 85). The type II PAKs are also thought to have a role in invasion and metastasis (6, 67, 76), and PAK4 may also play a role in proliferation of BRAF- or KRAS-driven cancers (85). These discoveries have provided a framework to drive efforts to achieve ATP-competitive small molecule inhibitors of the type II PAKs (6, 76, 86), with PF-3758309 the most prominent example (63).
In addition to amplifications at the genomic or protein level, somatic point mutations have also been discovered for the type II PAKs. In lung and colon cancers, PAK5 is observed to be particularly susceptible to acquisition of in-frame missense mutations (87, 88), although PAK4 somatic mutation has also been found to be associated with cancer (84, 89). Although these mutations do not cluster to specific regions within the proteins, one tumor-associated PAK5 mutant was demonstrated to have increased signaling, presumably due to increased kinase activity. Interestingly, the mutation (T538N) maps to the catalytic cleft, suggesting release of pseudosubstrate inhibition as a potential mechanism (87). A recurrent PAK6 mutation at the central Pro residue in the pseudosubstrate region (P52L) found in melanomas (90, 91) resulted in increased catalytic activity (56). Further studies will be required to investigate whether, or how, other cancer-associated mutations impact catalytic activity or other PAK functions.
ATP-competitive small molecule kinase inhibitors are often classified by the structural conformation of the protein kinase that they bind (92). Type I inhibitors bind to the active, DFG-in state, and type II inhibitors bind to the inactive, DFG-out, conformation. Following the failure of PF-3758309 in phase I trials, the clinical potential of type I PAK4 inhibitors is not currently clear (63, 93), although LCH-7749944 in this class is still in development (94, 95). Interestingly, structural flexibility of the back pocket of the ATP-binding site close to the gatekeeper residue may provide an additional mechanism to obtain type II or type I 1/2 inhibitors (86, 96) that could have increased specificity. Structural biology is therefore aiding the progress toward PAK-specific drug discovery. The reader is also directed to further recent in-depth discussions on the roles and targeting of PAKs in disease (6, 67, 76, 86, 97).
Conclusions
The type II PAKs are emerging as important components in RHO family signaling cascades. Although first described over 15 years ago, the molecular level mechanisms by which these proteins are regulated are still being described, and recent studies have significantly improved our understanding.
This work was partially supported by National Institutes of Health Grant R01GM102262 (to B. E. T. and T. J. B.) and by a grant from Gilead Sciences.
In structure 4JDH, a steric clash prevents the salt bridge from forming.
- PAK
- p21-activated kinase
- RAC
- Ras-related C3 botulinum toxin substrate
- CRIB
- CDC42/RAC interactive binding
- AID
- autoinhibitory domain
- AMP-PNP
- 5′-adenylyl-β,γ-imidodiphosphate.
References
- 1. Eswaran J., Soundararajan M., Kumar R., Knapp S. (2008) UnPAKing the class differences among p21-activated kinases. Trends Biochem. Sci. 33, 394–403 [DOI] [PubMed] [Google Scholar]
- 2. Bokoch G. M. (2003) Biology of the p21-activated kinases. Annu. Rev. Biochem. 72, 743–781 [DOI] [PubMed] [Google Scholar]
- 3. Arias-Romero L. E., Chernoff J. (2008) A tale of two Paks. Biol. Cell 100, 97–108 [DOI] [PubMed] [Google Scholar]
- 4. Hofmann C., Shepelev M., Chernoff J. (2004) The genetics of Pak. J. Cell Sci. 117, 4343–4354 [DOI] [PubMed] [Google Scholar]
- 5. Rane C. K., Minden A. (2014) p21 activated kinases: structure, regulation, and functions. Small GTPases 5, e28003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Radu M., Semenova G., Kosoff R., Chernoff J. (2014) PAK signalling during the development and progression of cancer. Nat. Rev. Cancer 14, 13–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dart A. E., Wells C. M. (2013) p21-activated kinase 4: not just one of the PAK. Eur. J. Cell Biol. 92, 129–138 [DOI] [PubMed] [Google Scholar]
- 8. Wells C. M., Jones G. E. (2010) The emerging importance of group II PAKs. Biochem. J. 425, 465–473 [DOI] [PubMed] [Google Scholar]
- 9. Dan C., Kelly A., Bernard O., Minden A. (2001) Cytoskeletal changes regulated by the PAK4 serine/threonine kinase are mediated by LIM kinase 1 and cofilin. J. Biol. Chem. 276, 32115–32121 [DOI] [PubMed] [Google Scholar]
- 10. Ahmed T., Shea K., Masters J. R., Jones G. E., Wells C. M. (2008) A PAK4-LIMK1 pathway drives prostate cancer cell migration downstream of HGF. Cell. Signal. 20, 1320–1328 [DOI] [PubMed] [Google Scholar]
- 11. Wong L. E., Reynolds A. B., Dissanayaka N. T., Minden A. (2010) p120-catenin is a binding partner and substrate for Group B Pak kinases. J. Cell. Biochem. 110, 1244–1254 [DOI] [PubMed] [Google Scholar]
- 12. Li Y., Shao Y., Tong Y., Shen T., Zhang J., Li Y., Gu H., Li F. (2012) Nucleo-cytoplasmic shuttling of PAK4 modulates β-catenin intracellular translocation and signaling. Biochim. Biophys. Acta 1823, 465–475 [DOI] [PubMed] [Google Scholar]
- 13. Soosairajah J., Maiti S., Wiggan O., Sarmiere P., Moussi N., Sarcevic B., Sampath R., Bamburg J. R., Bernard O. (2005) Interplay between components of a novel LIM kinase-slingshot phosphatase complex regulates cofilin. EMBO J. 24, 473–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Callow M. G., Zozulya S., Gishizky M. L., Jallal B., Smeal T. (2005) PAK4 mediates morphological changes through the regulation of GEF-H1. J. Cell Sci. 118, 1861–1872 [DOI] [PubMed] [Google Scholar]
- 15. Barac A., Basile J., Vázquez-Prado J., Gao Y., Zheng Y., Gutkind J. S. (2004) Direct interaction of p21-activated kinase 4 with PDZ-RhoGEF, a G protein-linked Rho guanine exchange factor. J. Biol. Chem. 279, 6182–6189 [DOI] [PubMed] [Google Scholar]
- 16. Jin D., Durgan J., Hall A. (2015) Functional cross-talk between Cdc42 and two downstream targets, Par6B and PAK4. Biochem. J. 467, 293–302 [DOI] [PubMed] [Google Scholar]
- 17. Zhang H., Li Z., Viklund E. K., Strömblad S. (2002) p21-activated kinase 4 interacts with integrin αvβ5 and regulates αvβ5-mediated cell migration. J. Cell Biol. 158, 1287–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Z., Zhang H., Lundin L., Thullberg M., Liu Y., Wang Y., Claesson-Welsh L., Strömblad S. (2010) p21-activated kinase 4 phosphorylation of integrin β5 Ser-759 and Ser-762 regulates cell migration. J. Biol. Chem. 285, 23699–23710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li Z., Lock J. G., Olofsson H., Kowalewski J. M., Teller S., Liu Y., Zhang H., Strömblad S. (2010) Integrin-mediated cell attachment induces a PAK4-dependent feedback loop regulating cell adhesion through modified integrin αvβ5 clustering and turnover. Mol. Biol. Cell 21, 3317–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wells C. M., Whale A. D., Parsons M., Masters J. R., Jones G. E. (2010) PAK4: a pluripotent kinase that regulates prostate cancer cell adhesion. J. Cell Sci. 123, 1663–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu X., Carr H. S., Dan I., Ruvolo P. P., Frost J. A. (2008) p21 activated kinase 5 activates Raf-1 and targets it to mitochondria. J. Cell. Biochem. 105, 167–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cammarano M. S., Nekrasova T., Noel B., Minden A. (2005) Pak4 induces premature senescence via a pathway requiring p16INK4/p19ARF and mitogen-activated protein kinase signaling. Mol. Cell Biol. 25, 9532–9542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu T., Li Y., Gu H., Zhu G., Li J., Cao L., Li F. (2013) p21-activated kinase 6 (PAK6) inhibits prostate cancer growth via phosphorylation of androgen receptor and tumorigenic E3 ligase murine double minute-2 (Mdm2). J. Biol. Chem. 288, 3359–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gnesutta N., Qu J., Minden A. (2001) The serine/threonine kinase PAK4 prevents caspase activation and protects cells from apoptosis. J. Biol. Chem. 276, 14414–14419 [DOI] [PubMed] [Google Scholar]
- 25. Cotteret S., Jaffer Z. M., Beeser A., Chernoff J. (2003) p21-activated kinase 5 (Pak5) localizes to mitochondria and inhibits apoptosis by phosphorylating BAD. Mol. Cell. Biol. 23, 5526–5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang M., Siedow M., Saia G., Chakravarti A. (2010) Inhibition of p21-activated kinase 6 (PAK6) increases radiosensitivity of prostate cancer cells. Prostate 70, 807–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gnesutta N., Minden A. (2003) Death receptor-induced activation of initiator caspase 8 is antagonized by serine/threonine kinase PAK4. Mol. Cell. Biol. 23, 7838–7848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bompard G., Rabeharivelo G., Frank M., Cau J., Delsert C., Morin N. (2010) Subgroup II PAK-mediated phosphorylation regulates Ran activity during mitosis. J. Cell Biol. 190, 807–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qu J., Li X., Novitch B. G., Zheng Y., Kohn M., Xie J. M., Kozinn S., Bronson R., Beg A. A., Minden A. (2003) PAK4 kinase is essential for embryonic viability and for proper neuronal development. Mol. Cell. Biol. 23, 7122–7133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tian Y., Lei L., Cammarano M., Nekrasova T., Minden A. (2009) Essential role for the Pak4 protein kinase in extraembryonic tissue development and vessel formation. Mech. Dev. 126, 710–720 [DOI] [PubMed] [Google Scholar]
- 31. Nekrasova T., Jobes M. L., Ting J. H., Wagner G. C., Minden A. (2008) Targeted disruption of the Pak5 and Pak6 genes in mice leads to deficits in learning and locomotion. Dev. Biol. 322, 95–108 [DOI] [PubMed] [Google Scholar]
- 32. Li X., Minden A. (2003) Targeted disruption of the gene for the PAK5 kinase in mice. Mol. Cell. Biol. 23, 7134–7142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Strochlic T. I., Concilio S., Viaud J., Eberwine R. A., Wong L. E., Minden A., Turk B. E., Plomann M., Peterson J. R. (2012) Identification of neuronal substrates implicates Pak5 in synaptic vesicle trafficking. Proc. Natl. Acad. Sci. U.S.A. 109, 4116–4121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jaffer Z. M., Chernoff J. (2002) p21-activated kinases: three more join the Pak. Int. J. Biochem. Cell Biol. 34, 713–717 [DOI] [PubMed] [Google Scholar]
- 35. Abo A., Qu J., Cammarano M. S., Dan C., Fritsch A., Baud V., Belisle B., Minden A. (1998) PAK4, a novel effector for Cdc42Hs, is implicated in the reorganization of the actin cytoskeleton and in the formation of filopodia. EMBO J. 17, 6527–6540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dan C., Nath N., Liberto M., Minden A. (2002) PAK5, a new brain-specific kinase, promotes neurite outgrowth in N1E-115 cells. Mol. Cell. Biol. 22, 567–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pandey A., Dan I., Kristiansen T. Z., Watanabe N. M., Voldby J., Kajikawa E., Khosravi-Far R., Blagoev B., Mann M. (2002) Cloning and characterization of PAK5, a novel member of mammalian p21-activated kinase-II subfamily that is predominantly expressed in brain. Oncogene 21, 3939–3948 [DOI] [PubMed] [Google Scholar]
- 38. Yang F., Li X., Sharma M., Zarnegar M., Lim B., Sun Z. (2001) Androgen receptor specifically interacts with a novel p21-activated kinase, PAK6. J. Biol. Chem. 276, 15345–15353 [DOI] [PubMed] [Google Scholar]
- 39. Bishop A. L., Hall A. (2000) Rho GTPases and their effector proteins. Biochem. J. 348, 241–255 [PMC free article] [PubMed] [Google Scholar]
- 40. Morreale A., Venkatesan M., Mott H. R., Owen D., Nietlispach D., Lowe P. N., Laue E. D. (2000) Structure of Cdc42 bound to the GTPase binding domain of PAK. Nat. Struct. Biol. 7, 384–388 [DOI] [PubMed] [Google Scholar]
- 41. Gizachew D., Guo W., Chohan K. K., Sutcliffe M. J., Oswald R. E. (2000) Structure of the complex of Cdc42Hs with a peptide derived from p-21 activated kinase. Biochemistry 39, 3963–3971 [DOI] [PubMed] [Google Scholar]
- 42. Wang W., Lim L., Baskaran Y., Manser E., Song J. (2013) NMR binding and crystal structure reveal that intrinsically-unstructured regulatory domain auto-inhibits PAK4 by a mechanism different from that of PAK1. Biochem. Biophys. Res. Commun. 438, 169–174 [DOI] [PubMed] [Google Scholar]
- 43. Wu X., Frost J. A. (2006) Multiple Rho proteins regulate the subcellular targeting of PAK5. Biochem. Biophys. Res. Commun. 351, 328–335 [DOI] [PubMed] [Google Scholar]
- 44. Shepelev M. V., Korobko I. V. (2012) Pak6 protein kinase is a novel effector of an atypical Rho family GTPase Chp/RhoV. Biochemistry 77, 26–32 [DOI] [PubMed] [Google Scholar]
- 45. Rolland T., Taşan M., Charloteaux B., Pevzner S. J., Zhong Q., Sahni N., Yi S., Lemmens I., Fontanillo C., Mosca R., Kamburov A., Ghiassian S. D., Yang X., Ghamsari L., Balcha D., Begg B. E., Braun P., Brehme M., Broly M. P., Carvunis A. R., Convery-Zupan D., Corominas R., Coulombe-Huntington J., Dann E., Dreze M., Dricot A., Fan C., Franzosa E., Gebreab F., Gutierrez B. J., Hardy M. F., Jin M., Kang S., Kiros R., Lin G. N., Luck K., MacWilliams A., Menche J., Murray R. R., Palagi A., Poulin M. M., Rambout X., Rasla J., Reichert P., Romero V., Ruyssinck E., Sahalie J. M., Scholz A., Shah A. A., Sharma A., Shen Y., Spirohn K., Tam S., Tejeda A. O., Trigg S. A., Twizere J. C., Vega K., Walsh J., Cusick M. E., Xia Y., Barabási A. L., Iakoucheva L. M., Aloy P., De Las Rivas J., Tavernier J., Calderwood M. A., Hill D. E., Hao T., Roth F. P., Vidal M. (2014) A proteome-scale map of the human interactome network. Cell 159, 1212–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ha B. H., Davis M. J., Chen C., Lou H. J., Gao J., Zhang R., Krauthammer M., Halaban R., Schlessinger J., Turk B. E., Boggon T. J. (2012) Type II p21-activated kinases (PAKs) are regulated by an autoinhibitory pseudosubstrate. Proc. Natl. Acad. Sci. U.S.A. 109, 16107–16112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Baskaran Y., Ng Y. W., Selamat W., Ling F. T., Manser E. (2012) Group I and II mammalian PAKs have different modes of activation by Cdc42. EMBO Rep. 13, 653–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Callow M. G., Clairvoyant F., Zhu S., Schryver B., Whyte D. B., Bischoff J. R., Jallal B., Smeal T. (2002) Requirement for PAK4 in the anchorage-independent growth of human cancer cell lines. J. Biol. Chem. 277, 550–558 [DOI] [PubMed] [Google Scholar]
- 49. Ching Y. P., Leong V. Y., Wong C. M., Kung H. F. (2003) Identification of an autoinhibitory domain of p21-activated protein kinase 5. J. Biol. Chem. 278, 33621–33624 [DOI] [PubMed] [Google Scholar]
- 50. Baldassa S., Calogero A. M., Colombo G., Zippel R., Gnesutta N. (2010) N-terminal interaction domain implicates PAK4 in translational regulation and reveals novel cellular localization signals. J. Cell. Physiol. 224, 722–733 [DOI] [PubMed] [Google Scholar]
- 51. Wells C. M., Abo A., Ridley A. J. (2002) PAK4 is activated via PI3K in HGF-stimulated epithelial cells. J. Cell Sci. 115, 3947–3956 [DOI] [PubMed] [Google Scholar]
- 52. Lee S. R., Ramos S. M., Ko A., Masiello D., Swanson K. D., Lu M. L., Balk S. P. (2002) AR and ER interaction with a p21-activated kinase (PAK6). Mol. Endocrinol. 16, 85–99 [DOI] [PubMed] [Google Scholar]
- 53. Fram S., King H., Sacks D. B., Wells C. M. (2014) A PAK6-IQGAP1 complex promotes disassembly of cell-cell adhesions. Cell. Mol. Life Sci. 71, 2759–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lei M., Lu W., Meng W., Parrini M. C., Eck M. J., Mayer B. J., Harrison S. C. (2000) Structure of PAK1 in an autoinhibited conformation reveals a multistage activation switch. Cell 102, 387–397 [DOI] [PubMed] [Google Scholar]
- 55. Parrini M. C., Lei M., Harrison S. C., Mayer B. J. (2002) Pak1 kinase homodimers are autoinhibited in trans and dissociated upon activation by Cdc42 and Rac1. Mol. Cell 9, 73–83 [DOI] [PubMed] [Google Scholar]
- 56. Gao J., Ha B. H., Lou H. J., Morse E. M., Zhang R., Calderwood D. A., Turk B. E., Boggon T. J. (2013) Substrate and inhibitor specificity of the type II p21-activated kinase, PAK6. PLoS One 8, e77818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Huse M., Kuriyan J. (2002) The conformational plasticity of protein kinases. Cell 109, 275–282 [DOI] [PubMed] [Google Scholar]
- 58. Nolen B., Taylor S., Ghosh G. (2004) Regulation of protein kinases; controlling activity through activation segment conformation. Mol. Cell 15, 661–675 [DOI] [PubMed] [Google Scholar]
- 59. Levinson N. M., Kuchment O., Shen K., Young M. A., Koldobskiy M., Karplus M., Cole P. A., Kuriyan J. (2006) A Src-like inactive conformation in the Abl tyrosine kinase domain. PLoS Biol. 4, e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. McClendon C. L., Kornev A. P., Gilson M. K., Taylor S. S. (2014) Dynamic architecture of a protein kinase. Proc. Natl. Acad. Sci. U.S.A. 111, E4623–E4631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chong C., Tan L., Lim L., Manser E. (2001) The mechanism of PAK activation: autophosphorylation events in both regulatory and kinase domains control activity. J. Biol. Chem. 276, 17347–17353 [DOI] [PubMed] [Google Scholar]
- 62. Eswaran J., Lee W. H., Debreczeni J. E., Filippakopoulos P., Turnbull A., Fedorov O., Deacon S. W., Peterson J. R., Knapp S. (2007) Crystal structures of the p21-activated kinases PAK4, PAK5, and PAK6 reveal catalytic domain plasticity of active group II PAKs. Structure 15, 201–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Murray B. W., Guo C., Piraino J., Westwick J. K., Zhang C., Lamerdin J., Dagostino E., Knighton D., Loi C. M., Zager M., Kraynov E., Popoff I., Christensen J. G., Martinez R., Kephart S. E., Marakovits J., Karlicek S., Bergqvist S., Smeal T. (2010) Small-molecule p21-activated kinase inhibitor PF-3758309 is a potent inhibitor of oncogenic signaling and tumor growth. Proc. Natl. Acad. Sci. U.S.A. 107, 9446–9451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mak G. W., Chan M. M., Leong V. Y., Lee J. M., Yau T. O., Ng I. O., Ching Y. P. (2011) Overexpression of a novel activator of PAK4, the CDK5 kinase-associated protein CDK5RAP3, promotes hepatocellular carcinoma metastasis. Cancer Res. 71, 2949–2958 [DOI] [PubMed] [Google Scholar]
- 65. Siu M. K., Chan H. Y., Kong D. S., Wong E. S., Wong O. G., Ngan H. Y., Tam K. F., Zhang H., Li Z., Chan Q. K., Tsao S. W., Strömblad S., Cheung A. N. (2010) p21-activated kinase 4 regulates ovarian cancer cell proliferation, migration, and invasion and contributes to poor prognosis in patients. Proc. Natl. Acad. Sci. U.S.A. 107, 18622–18627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Paliouras G. N., Naujokas M. A., Park M. (2009) Pak4, a novel Gab1 binding partner, modulates cell migration and invasion by the Met receptor. Mol. Cell. Biol. 29, 3018–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Eswaran J., Soundararajan M., Knapp S. (2009) Targeting group II PAKs in cancer and metastasis. Cancer Metastasis Rev. 28, 209–217 [DOI] [PubMed] [Google Scholar]
- 68. Knapp S. (2013) 3D structure and physiological regulation of PAKs. in PAKs, RAC/CDC42 (p21)-activated Kinases: Towards the Cure of Cancer and Other PAK-dependent Diseases (Maruta H., ed), pp. 137–148, Elsevier Science Bv, Amsterdam [Google Scholar]
- 69. Tuazon P. T., Spanos W. C., Gump E. L., Monnig C. A., Traugh J. A. (1997) Determinants for substrate phosphorylation by p21-activated protein kinase (γ-PAK). Biochemistry 36, 16059–16064 [DOI] [PubMed] [Google Scholar]
- 70. Rennefahrt U. E., Deacon S. W., Parker S. A., Devarajan K., Beeser A., Chernoff J., Knapp S., Turk B. E., Peterson J. R. (2007) Specificity profiling of Pak kinases allows identification of novel phosphorylation sites. J. Biol. Chem. 282, 15667–15678 [DOI] [PubMed] [Google Scholar]
- 71. Chen C., Ha B. H., Thévenin A. F., Lou H. J., Zhang R., Yip K. Y., Peterson J. R., Gerstein M., Kim P. M., Filippakopoulos P., Knapp S., Boggon T. J., Turk B. E. (2014) Identification of a major determinant for serine-threonine kinase phosphoacceptor specificity. Mol. Cell 53, 140–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhu G., Fujii K., Liu Y., Codrea V., Herrero J., Shaw S. (2005) A single pair of acidic residues in the kinase major groove mediates strong substrate preference for P-2 or P-5 arginine in the AGC, CAMK, and STE kinase families. J. Biol. Chem. 280, 36372–36379 [DOI] [PubMed] [Google Scholar]
- 73. Knighton D. R., Zheng J. H., Ten Eyck L. F., Xuong N. H., Taylor S. S., Sowadski J. M. (1991) Structure of a peptide inhibitor bound to the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253, 414–420 [DOI] [PubMed] [Google Scholar]
- 74. Kang S. A., Pacold M. E., Cervantes C. L., Lim D., Lou H. J., Ottina K., Gray N. S., Turk B. E., Yaffe M. B., Sabatini D. M. (2013) mTORC1 phosphorylation sites encode their sensitivity to starvation and rapamycin. Science 341, 1236566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kim S. Y., Ferrell J. E., Jr. (2007) Substrate competition as a source of ultrasensitivity in the inactivation of Wee1. Cell 128, 1133–1145 [DOI] [PubMed] [Google Scholar]
- 76. King H., Nicholas N. S., Wells C. M. (2014) Role of p-21-activated kinases in cancer progression. Int. Rev. Cell Mol. Biol. 309, 347–387 [DOI] [PubMed] [Google Scholar]
- 77. Chen S., Auletta T., Dovirak O., Hutter C., Kuntz K., El-ftesi S., Kendall J., Han H., Von Hoff D. D., Ashfaq R., Maitra A., Iacobuzio-Donahue C. A., Hruban R. H., Lucito R. (2008) Copy number alterations in pancreatic cancer identify recurrent PAK4 amplification. Cancer Biol. Ther. 7, 1793–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kimmelman A. C., Hezel A. F., Aguirre A. J., Zheng H., Paik J. H., Ying H., Chu G. C., Zhang J. X., Sahin E., Yeo G., Ponugoti A., Nabioullin R., Deroo S., Yang S., Wang X., McGrath J. P., Protopopova M., Ivanova E., Zhang J., Feng B., Tsao M. S., Redston M., Protopopov A., Xiao Y., Futreal P. A., Hahn W. C., Klimstra D. S., Chin L., DePinho R. A. (2008) Genomic alterations link Rho family of GTPases to the highly invasive phenotype of pancreas cancer. Proc. Natl. Acad. Sci. U.S.A. 105, 19372–19377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Begum A., Imoto I., Kozaki K., Tsuda H., Suzuki E., Amagasa T., Inazawa J. (2009) Identification of PAK4 as a putative target gene for amplification within 19q13.12-q13.2 in oral squamous-cell carcinoma. Cancer Sci. 100, 1908–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Tyagi N., Bhardwaj A., Singh A. P., McClellan S., Carter J. E., Singh S. (2014) p-21 activated kinase 4 promotes proliferation and survival of pancreatic cancer cells through AKT- and ERK-dependent activation of NF-κB pathway. Oncotarget 5, 8778–8789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Gong W., An Z., Wang Y., Pan X., Fang W., Jiang B., Zhang H. (2009) p21-activated kinase 5 is overexpressed during colorectal cancer progression and regulates colorectal carcinoma cell adhesion and migration. Int. J. Cancer 125, 548–555 [DOI] [PubMed] [Google Scholar]
- 82. Wang X., Gong W., Qing H., Geng Y., Wang X., Zhang Y., Peng L., Zhang H., Jiang B. (2010) p21-activated kinase 5 inhibits camptothecin-induced apoptosis in colorectal carcinoma cells. Tumour. Biol. 31, 575–582 [DOI] [PubMed] [Google Scholar]
- 83. Kaur R., Yuan X., Lu M. L., Balk S. P. (2008) Increased PAK6 expression in prostate cancer and identification of PAK6 associated proteins. Prostate 68, 1510–1516 [DOI] [PubMed] [Google Scholar]
- 84. Whale A. D., Dart A., Holt M., Jones G. E., Wells C. M. (2013) PAK4 kinase activity and somatic mutation promote carcinoma cell motility and influence inhibitor sensitivity. Oncogene 32, 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tabusa H., Brooks T., Massey A. J. (2013) Knockdown of PAK4 or PAK1 inhibits the proliferation of mutant KRAS colon cancer cells independently of RAF/MEK/ERK and PI3K/AKT signaling. Mol. Cancer Res. 11, 109–121 [DOI] [PubMed] [Google Scholar]
- 86. Rudolph J., Crawford J. J., Hoeflich K. P., Wang W. (2015) Inhibitors of p21-activated kinases (PAKs). J. Med. Chem. 58, 111–129 [DOI] [PubMed] [Google Scholar]
- 87. Fawdar S., Trotter E. W., Li Y., Stephenson N. L., Hanke F., Marusiak A. A., Edwards Z. C., Ientile S., Waszkowycz B., Miller C. J., Brognard J. (2013) Targeted genetic dependency screen facilitates identification of actionable mutations in FGFR4, MAP3K9, and PAK5 in lung cancer. Proc. Natl. Acad. Sci. U.S.A. 110, 12426–12431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Greenman C., Stephens P., Smith R., Dalgliesh G. L., Hunter C., Bignell G., Davies H., Teague J., Butler A., Stevens C., Edkins S., O'Meara S., Vastrik I., Schmidt E. E., Avis T., Barthorpe S., Bhamra G., Buck G., Choudhury B., Clements J., Cole J., Dicks E., Forbes S., Gray K., Halliday K., Harrison R., Hills K., Hinton J., Jenkinson A., Jones D., Menzies A., Mironenko T., Perry J., Raine K., Richardson D., Shepherd R., Small A., Tofts C., Varian J., Webb T., West S., Widaa S., Yates A., Cahill D. P., Louis D. N., Goldstraw P., Nicholson A. G., Brasseur F., Looijenga L., Weber B. L., Chiew Y. E., DeFazio A., Greaves M. F., Green A. R., Campbell P., Birney E., Easton D. F., Chenevix-Trench G., Tan M. H., Khoo S. K., Teh B. T., Yuen S. T., Leung S. Y., Wooster R., Futreal P. A., Stratton M. R. (2007) Patterns of somatic mutation in human cancer genomes. Nature 446, 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Parsons D. W., Wang T. L., Samuels Y., Bardelli A., Cummins J. M., DeLong L., Silliman N., Ptak J., Szabo S., Willson J. K., Markowitz S., Kinzler K. W., Vogelstein B., Lengauer C., Velculescu V. E. (2005) Colorectal cancer: mutations in a signalling pathway. Nature 436, 792. [DOI] [PubMed] [Google Scholar]
- 90. Krauthammer M., Kong Y., Ha B. H., Evans P., Bacchiocchi A., McCusker J. P., Cheng E., Davis M. J., Goh G., Choi M., Ariyan S., Narayan D., Dutton-Regester K., Capatana A., Holman E. C., Bosenberg M., Sznol M., Kluger H. M., Brash D. E., Stern D. F., Materin M. A., Lo R. S., Mane S., Ma S., Kidd K. K., Hayward N. K., Lifton R. P., Schlessinger J., Boggon T. J., Halaban R. (2012) Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat. Genet. 44, 1006–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wei X., Walia V., Lin J. C., Teer J. K., Prickett T. D., Gartner J., Davis S., NISC Comparative Sequencing Program, Stemke-Hale K., Davies M. A., Gershenwald J. E., Robinson W., Robinson S., Rosenberg S. A., Samuels Y. (2011) Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat. Genet. 43, 442–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zhao Z., Wu H., Wang L., Liu Y., Knapp S., Liu Q., Gray N. S. (2014) Exploration of type II binding mode: a privileged approach for kinase inhibitor focused drug discovery? ACS Chem. Biol. 9, 1230–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Bradshaw-Pierce E. L., Pitts T. M., Tan A. C., McPhillips K., West M., Gustafson D. L., Halsey C., Nguyen L., Lee N. V., Kan J. L., Murray B. W., Eckhardt S. G. (2013) Tumor P-glycoprotein correlates with efficacy of PF-3758309 in in vitro and in vivo models of colorectal cancer. Front. Pharmacol. 4, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Guo Q., Su N., Zhang J., Li X., Miao Z., Wang G., Cheng M., Xu H., Cao L., Li F. (2014) PAK4 kinase-mediated SCG10 phosphorylation involved in gastric cancer metastasis. Oncogene 33, 3277–3287 [DOI] [PubMed] [Google Scholar]
- 95. Zhang J., Wang J., Guo Q., Wang Y., Zhou Y., Peng H., Cheng M., Zhao D., Li F. (2012) LCH-7749944, a novel and potent p21-activated kinase 4 inhibitor, suppresses proliferation and invasion in human gastric cancer cells. Cancer Lett. 317, 24–32 [DOI] [PubMed] [Google Scholar]
- 96. Staben S. T., Feng J. A., Lyle K., Belvin M., Boggs J., Burch J. D., Chua C. C., Cui H., DiPasquale A. G., Friedman L. S., Heise C., Koeppen H., Kotey A., Mintzer R., Oh A., Roberts D. A., Rouge L., Rudolph J., Tam C., Wang W., Xiao Y., Young A., Zhang Y., Hoeflich K. P. (2014) Back pocket flexibility provides group II p21-activated kinase (PAK) selectivity for type I 1/2 kinase inhibitors. J. Med. Chem. 57, 1033–1045 [DOI] [PubMed] [Google Scholar]
- 97. Dummler B., Ohshiro K., Kumar R., Field J. (2009) Pak protein kinases and their role in cancer. Cancer Metastasis Rev. 28, 51–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Hornbeck P. V., Zhang B., Murray B., Kornhauser J. M., Latham V., Skrzypek E. (2015) PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Workman C. T., Yin Y., Corcoran D. L., Ideker T., Stormo G. D., Benos P. V. (2005) enoLOGOS: a versatile web tool for energy normalized sequence logos. Nucleic Acids Res. 33, W389–W392 [DOI] [PMC free article] [PubMed] [Google Scholar]