Background: RUNX1 is indispensable for endothelial to hematopoietic transition involving generation of hematopoietic stem cells from hemogenic endothelium.
Results: Histone chaperone HIRA interacts with RUNX1, incorporates H3.3 variant within enhancer element, and regulates downstream targets of RUNX1 implicated in definitive hematopoiesis.
Conclusion: HIRA regulates RUNX1.
Significance: A histone chaperone-transcription factor complex is implicated in hemogenic to hematopoietic transition.
Keywords: hematopoiesis, hematopoietic stem cells, histone, histone chaperone, transcription factor, H3.3 variant, hematopoietic precursor
Abstract
RUNX1 (Runt-related transcription factor 1) is indispensable for the generation of hemogenic endothelium. However, the regulation of RUNX1 during this developmental process is poorly understood. We investigated the role of the histone chaperone HIRA (histone cell cycle regulation-defective homolog A) from this perspective and report that HIRA significantly contributes toward the regulation of RUNX1 in the transition of differentiating mouse embryonic stem cells from hemogenic to hematopoietic stage. Direct interaction of HIRA and RUNX1 activates the downstream targets of RUNX1 implicated in generation of hematopoietic stem cells. At the molecular level, HIRA-mediated incorporation of histone H3.3 variant within the Runx1 +24 mouse conserved noncoding element is essential for the expression of Runx1 during endothelial to hematopoietic transition. An inactive chromatin at the intronic enhancer of Runx1 in absence of HIRA significantly repressed the transition of cells from hemogenic to hematopoietic fate. We expect that the HIRA-RUNX1 axis might open up a novel approach in understanding leukemogenesis in future.
Introduction
Multipotent hematopoietic stem cells (HSCs)5 arise during development and gradually colonize within liver, spleen, and finally bone marrow. Embryonic hematopoiesis encompasses two distinct processes of primitive and definitive hematopoiesis. Primitive hematopoiesis originates in the yolk sac at E7.25. In the case of definitive hematopoiesis, erythroid and myeloid progenitors are generated at E8.25 (1), and at E10.5 HSCs are detected in the aorta gonad mesonephros (AGM) region. Erythroid and myeloid progenitors are generated from a special subset of endothelial cells scattered within blood vessels called hemogenic endothelium (HE) (2, 3). HE undergoes endothelial to hematopoietic transition (EndHT) (4) to generate HSCs. EndHT is predominantly regulated by RUNX1 that belongs to the Runt-related transcription factor (RUNX) family of proteins. The RUNX1 knock-out embryos displayed a lack of fetal liver hematopoiesis (5) and died between E11.5 and E12.5 because of hemorrhaging in the central nervous system (6). Interestingly, RUNX1 can potentially modify the epigenetic status at the loci involved in hemogenic to hematopoietic transition (7). It can also physically interact with the histone methyltransferase to regulate hematopoiesis (8). However, the regulation of RUNX1 itself at the epigenetic level is poorly understood. Earlier, we demonstrated that HIRA-dependent histone H3.3 acetylation at lysine 56 residues could regulate the expression of endothelial specific genes (9). Functionally, HIRA acts to incorporate histone variant H3.3 into chromatin in a DNA replication-independent manner (10). The importance of HIRA during mammalian development is evident from the targeted mutagenesis study in mouse (11). HIRA null mice die between E10 and E11 and display a wide range of phenotypes secondary to defective mesendodermal development (11). Because RUNX1 is essential for the endothelial to hematopoietic transition, we were interested to investigate the potential role of histone chaperone HIRA in regulation of RUNX1 in endothelial to hematopoietic transition to HSCs.
Materials and Methods
Mouse Yolk Sac Study
E9.5 C57BL/6J mouse embryos were isolated along with the yolk sac. Yolk sacs were cleaned from the debris, fixed in 3.7% formaldehyde, and treated as described for the immunofluorescence study. Six yolk sac samples from two different pregnant mice were studied.
Culture of ES Cells
Mouse W9.5 (control) and Hira−/− ES cells (12) were cultured on a mitomycin C treated feeder layer for initial passages. For experiments, the cells were cultured in feeder-free conditions in ES medium composed of Iscove's modified Dulbecco's medium (Invitrogen; 12440053), 15% ES qualified serum (Invitrogen; 10439024), 1% antimycotic/antibiotic (Invitrogen; 15240062), 0.0124% 1-thioglycerol (Sigma; M6145), and 1% penicillin and streptomycin (Invitrogen; 10378016). We maintained them in undifferentiated condition by addition of 105 units of LIF/ml (Millipore; ESG1106) (13).
Embryoid Body (EB) Generation
3 × 105 control and Hira−/− ES cells were grown in ES medium for 2 days in feeder-free conditions before differentiation. To generate EBs, ES cells were grown in the absence of LIF in ES differentiation medium of Iscove's modified Dulbecco's medium containing 15% FCS selected for endothelial cell differentiation (Stem Cell Technologies; 06907), 1% ascorbic acid (5 mg/ml in 100 ml; Stem Cell Technologies; 07157), and 0.0124% 1-thioglycerol for 5 days (13).
ESC Differentiation to Hemogenic Endothelium and Hematopoietic Precursors
For the differentiation of HE from undifferentiated ES cells, we followed the protocol described by Chiang and Wong (14) with few modifications. Briefly, control and Hira−/− ES cells were cultured in serum- and feeder-free N2B27 medium (15) for 48 h. At day 2, 5 ng/ml BMP4 (R&D Systems; 314-BP-050), 4 ng/ml activin A (R&D Systems; 338-AC-010), 12.5 ng/ml FGF2 (R&D Systems; 233-FB-025), and 3 μm CHIR99021 (Stemgent; 130-095-555) were supplemented in the medium. At day 4, cells cultured in the presence of mesodermal inducer (mentioned above) were dissociated, sorted for FLK1+ cells, and replated on 12-well plate at a concentration of 5 × 104 cells/well in N2B27 medium supplemented with 20 ng/ml BMP4, 12.5 ng/ml FGF2, 20 ng/ml VEGF165, 0.25 mm BrcAMP (Sigma; B5386), and 4 μm ALKi (Merck; 616461). At day 6, adherent cells and floating fractions were collected by trypsinization and trituration, respectively, for further study (14).
Culture of Kasumi-1 Cells
Kasumi-1 cells were cultured in RPMI medium (Himedia; AL1621) supplemented with 20% FBS, 1% antimycotic/antibiotic, and 1% penicillin and streptomycin.
Ectopic Expression of HIRA
Mouse Hira cDNA (accession number BC156807; MGC premier ORF clone TOM6004) purchased from TransOMICS Technologies was PCR-amplified and subcloned into pEGFP-C1 (Clontech; catalog number 6084-1) at XhoI and SalI sites.
Transfection
2 × 105 control and Hira−/− ES cells were seeded in Opti-MEM (Invitrogen; 31985088) on 12-well plates. Cells were transfected with Lipofectamine 2000 (Invitrogen; 11668019) and 2 μg of H3.3-Flag-tagged plasmid (Gift from Prof. James J. Bieker) and selected for 72 h with G418 (Sigma; G8168). In Hira−/− ES cells, HIRA was ectopically expressed by transfection of pEGFP-C1/Hira cDNA plasmid with Lipofectamine 2000. Transfected cells were observed for the expression of GFP and selected with G418 for 72 h.
Fluorescence-associated Cell Sorting
Control and Hira−/− ES cells were trypsinized and centrifuged at 1200 rpm for 5 min. Pellet was washed with FACS buffer (Dulbecco's phosphate-buffered saline with 0.8% (v/v) FBS and 0.263 mm EDTA (Sigma; E9884)) twice at 2000 rpm for 5 min. The cells were suspended in the FACS buffer and incubated with 0.5 μg of fluorescent-conjugated monoclonal antibody for 30 min in dark on ice. Following incubation, cells were centrifuged and resuspended in 500 μl of FACS buffer, filtered by cell strainer, and subsequently sorted by BD FACS AviaTM II.
Quantitative RT-PCR
Total RNA from cells were extracted with TRIzol reagent (Invitrogen; 15596018); cDNA preparation was accomplished by a high capacity cDNA reverse transcription kit (ABI; 4368814), and the expression of genes were analyzed by qRT-PCR using Sybr Green master mix (ABI, #4367659). Primer sequences are listed in Table 1.
TABLE 1.
Primers for qRT-PCR analysis
| Gene | Forward (5′ → 3′) | Reverse (3′ → 5′) |
|---|---|---|
| Hira | CAGGAGGATGACGAGAAGGA | ACTGTTTGACCACCGCACAC |
| Runx1 | CCATAGAGCCATCAAAATCAC | GCTTGGTCTGATCATCTAGTTTC |
| Cmyb | GTCCTCTGTCTTCCCACAGG | TGTCCTCAAAGCCTTTACCG |
| Tie2 | CTTAGTGACCATTCTCCCTCC | TGTCCAAGAAACCATAGCTG |
| Sox17 | ACGCTAGCTCAGCGGTCTACTATT | AGGGATTTCCTTAGCGCTTCCAGG |
| Gata4 | GAAAACGGAAGCCCAAGAAC | GGGAGGGTCTCACCAGCA |
| Nestin | TGTCCCTTAGTCTGGAAGTGG | GGTGTCTGCAAGCGAGAGTT |
| Gfi1 | TCCGAGTTCGAGGACTTTTG | ACTGCCGATAGCTCTGCAC |
| Gata2 | AAAGGGGCTGAATGTTTCG | GCGTGGGTAGGATGTGTC |
| Pu.1 | GAGTTTGAGAACTTCCCTGAG | TGGTAGGTCATCTTCTTGCGG |
| Flk1 | AATGGAGAAACAGAGCCACA | ATCCATAGGCGAGATCAAGG |
| Gfi1b | ATGGGGAATCACCACTCTCTG | GGGGTCTGTGTGTAGCTGT |
| Gapdh | TGCCCCCATGTTTGTGATG | TGTGGTCATGAGCCCTTCC |
| Runx1 (human) | CTGCTCCGTGCTGCCTAC | AGCCATCACAGTGACCAGAGT |
| Gata1 (human) | TGCTCTGGTGTCCTCCACAC | TGGGAGAGGAATAGGCTGCT |
| β-Actin (human) | CCAGCTCACCATGGATGATG | ATGCCGGAGCCGTTGTC |
Western Blot
Protein concentrations were determined by the Bradford reagent (Bio-Rad) and resolved by 8 or 10% SDS-PAGE. Antibodies have been listed in Table 2.
TABLE 2.
Antibodies used in the study
IF, immunofluorescence; WB, Western blotting; BRG1, Brahma-related gene-1; PE, phycoerythrin.
| Company | Catalog number | |
|---|---|---|
| Primary antibody | ||
| RUNX1(WB; IP; IF) | Santa Cruz | sc-365644 |
| RUNX1 (ChIP; IF) | Abcam | ab23980 |
| HIRA (WB; IF) | Abcam | ab20655 |
| β-Actin (EB) | Sigma | A5441 |
| BRG1 (WB) | Santa Cruz | sc-10768 |
| GATA2 | Abcam | ab22849 |
| FLAG (DDDDK) | Abcam | ab1162 |
| FLK1 | Cell Signaling | 2479S |
| Histone H3 | Cell Signaling | 9715S |
| Secondary antibody | ||
| Anti-mouse | Santa Cruz | sc-2005 |
| Anti-rabbit | Santa Cruz | sc-2004 |
| FACS antibody | ||
| PE rat anti-mouse Flk1 | BD Pharmingen | 555308 |
Immunofluorescence and Confocal Microscopy
Immunostaining to detect expression of RUNX1 and HIRA in E9.5 yolk sacs, undifferentiated ES cells, and HE cells was performed using standard protocols. ESCs were cultured on coverslips under different culture conditions. Briefly, cells or yolk sacs were fixed with 3.7% paraformaldehyde (Sigma; P6148) and permeabilized with 0.2% Tween 20 (Sigma; P1379) in Dulbecco's phosphate-buffered saline (Invitrogen; 14190235). Nonspecific binding was blocked with 10% goat serum (Jackson Immunoresearch; 005-00-121), and cells were incubated overnight with primary antibody at a dilution of 1:200. Fluorescent conjugated secondary antibodies (Alexa Fluor 568; Invitrogen; A11004 and Alexa Flour 488; Invitrogen; A11008) were used at a 1:200 dilution. Coverslips were mounted on slides and observed using a confocal microscope. We used Hoechst dye (Sigma; #B1155, 1 μg/ml) for nuclear staining. For hematopoietic precursors, slides were coated with 0.1% poly-l-lysine (Sigma; 9813) for 30 min. Drops of cell culture suspension were added onto the slides and left for 4 h of incubation in a CO2 incubator at 37 °C. Slides were further processed as described previously.
RNA Interference
Mouse Hira shRNA validated from our earlier study (9) was used to knockdown Hira in ES cells. Human Hira shRNAs were generated using iRNAi software. Lentiviral vectors containing shRNA targeting human Hira was cloned in the Plko.1 (Addgene) vector. Lentiviral supernatant was produced in HEK293T cells by transient transfection using calcium chloride (9). Briefly HEK293T cells were grown to 80% confluency. pRRE (gag/pol), pMD2G (VSVG), pRSV (Rev), and Hira shRNA plasmids were mixed in 0.25 m CaCl2 and equilibrated with an equal volume of 2× HEPES-buffered saline. Lentiviral particle containing supernatants were collected after 24 h of incubation. ES cells or Kasumi-1 cells were grown to 70% confluence and transfected with lentiviral soups. Transfected cells were selected by the addition of 1 μg/ml of puromycin (Sigma, #P8833). After 3 days, RNA and protein were extracted for analysis. Quantitative RT-PCR and Western blotting confirmed the knockdown. We screened three different sets of shRNA, where sh247 (CCACAATGGCAAGCCGATT) worked best for the knockdown of Hira in Kasumi-1 cells.
Co-immunoprecipitation
Protein lysates were prepared in radio immunoprecipitation assay buffer (10 mm Tris-HCl, pH 7.6, 1% Triton X-100, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS, 150 mm NaCl, 5 mm EDTA, 1 mm sodium orthovanadate, 1 mm PMSF, 10 mg/ml aprotinin) (16). Protein concentrations were determined by Bradford reagent (Bio-Rad). Lysates were immunoprecipitated with primary antibody. Immune complexes were adsorbed to protein A-Sepharose (Sigma; P3391) and resolved on 8% or 10% PAGE.
ChIP Analysis
Real time PCR-based quantitative ChIP analysis was performed as described previously (9, 13). Briefly, cells were trypsinized, cross-linked with formaldehyde (1%), and sonicated to generate chromatin fragments. Antibodies were used to immunoprecipitate protein-DNA cross-linked fragments. Precipitated complexes were eluted and reverse cross-linked. Enrichment of chromatin fragments was measured by qRT-PCR using Sybr Green fluorescence relative to a standard curve of input chromatin. Primer sequences are listed in the Tables 3 and 4.
TABLE 3.
Primers for q-ChIP analysis
q-ChIP, quantitative ChIP.
| Locus | Forward (5′ → 3′) | Reverse (3′ → 5′) |
|---|---|---|
| (−)14-kb Pu.1 | CCTGGTTTCAGTCACTCCTCTGCT | TACTTCCTGCCTCCTCCCAGAAATC |
| (−)35-kb Gfi1 | CCACATGACCTCATGAATGC | CCACAAACAGAACAGCTGGA |
| CNE3 | CCAGCATGACACAGCAAATC | CCCAGGGCTTATCTGCAC |
| CNE2 | GCTATTTCTGCCAAGGGTGA | GGGGTCTGAGGACCATGATA |
TABLE 4.
Primers of q-ChIP at Runx1 +24/+25 mCNE locus
q-ChIP, quantitative ChIP.
| Locus | Forward (5′ → 3′) | Reverse (3′ → 5′) |
|---|---|---|
| +24mCNE- L2 | AAGTTCAGGCCACAGTGCTAGG | CTTCCATGCGGATCTTCACAG |
| +24mCNE- L2A | GCTTCAAACTCCCGTTCCTTCT | GGATGGGAGAGTGTGTGTGTGA |
| +24mCNE- L1A | CCACTGCACCTGCTAGGTTCTC | TCCCAGGCTCTTTGAGAAGAAA |
| +24mCNE″ | GGGGCCCTCACTACCTCTTTTC | GGCTTCAACTGCCGGTTTATTT |
| +24mCNE′ | ATGGTGTGAGGAGGAGACAGGA | ATGGTGTGAGGAGGAGACAGGA |
| +24mCNE- R1a | GCAGAGCTGATCAGAGGGTAGC | CAAGACCAGGGTCTGCAAGC |
| +24mCNE- R2d | CAGCAAATGGCATCTAAGACGTG | AACTTAGGTGTCAGGATCCAGAAGG |
Statistical Analysis
Student's two-tailed, unpaired t test was used to determine statistical significance. p values less than 0.05 were considered to be significant.
Results
HIRA Influence RUNX1 during Development
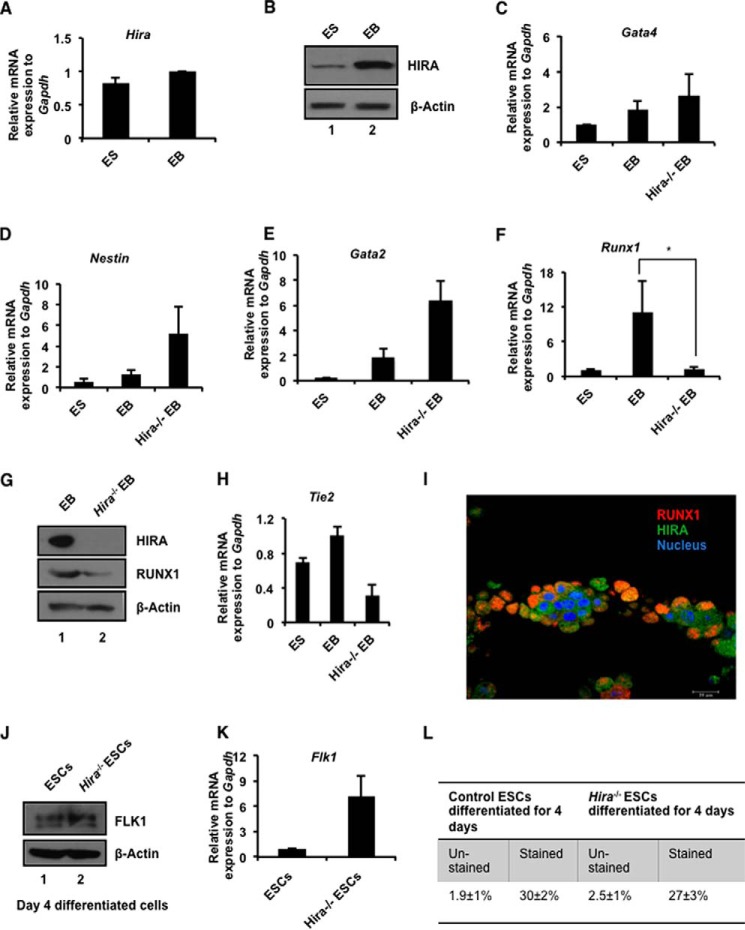
Establishment of hemogenic endothelium forms an integral part of development. At E9.5, mouse yolk sac expresses the key hemogenic marker RUNX1 in the circulation (17). To explore the function of HIRA in development, we utilized the classical embryonic stem cell (ESC) differentiation model for the formation of EBs. During differentiation, there is no significant change of Hira at the mRNA level (Fig. 1A), whereas Western blot analysis showed that protein expression was significantly increased (Fig. 1B). To assess the requirement for HIRA during differentiation, we generated EBs from Hira−/− ESCs. In concordance with an earlier report (18), we observed that the expression of the developmental genes Gata4, Nestin, and Gata2 (Fig. 1, C–E) were induced in absence of HIRA. However, RUNX1 expression was significantly abrogated in absence of HIRA (Fig. 1F). This was confirmed by the down-regulation of RUNX1 protein in Hira−/− EBs (Fig. 1G). An analogous trend was observed in the Tie2 mRNA level in EBs deprived of HIRA (Fig. 1H). Next, we examined the correlation between HIRA and RUNX1 protein expression during development in mouse, specifically within the yolk sac. Immunofluorescence study revealed that RUNX1 and HIRA co-localize in E9.5 mouse yolk sac (Fig. 1I).
FIGURE 1.
HIRA influence RUNX1 in development. A, murine ES cells were differentiated into EBs in LIF free EB-specific medium and was subjected to RNA and protein isolation. Quantitative RT-PCR analysis of Hira in the ES-EB system (means ± S.E. for three independent experiments). B, the same sets of cells used in A were analyzed for the protein expression of HIRA by Western blot. C–F, control and Hira−/− ESCs were differentiated to EB formation. Quantitative RT-PCR analyses for different germ layer markers in ES/EB system (means ± S.E. for three independent experiments). F, the plot shows (p < 0.05) significant loss in expression of Runx1 in Hira−/−EB. G, Western blots for the same set of samples analyzed in C–F. H, quantitative RT-PCR analysis of Tie2 in ES-EB system (means ± S.E. for three independent experiments). I, mouse yolk sac was isolated at E9.5 and immunostained for RUNX1 and HIRA expression. Magnification was 63× under oil immersion. J, control and Hira−/− ESCs were differentiated toward mesodermal lineage by mesoderm inducer growth factors for the generation of HE. Cells at day 4 were analyzed for the expression of FLK1 at protein level. K, same set of cells used in J was analyzed for the expression of Flk1 at the mRNA level (means ± S.E. for three or more independent experiments). L, cells analyzed in J and K were sorted for FLK1 by FACS. The figure represents the fraction of FLK1+ control and Hira−/− cells. Averages of three independent experiments have been presented.
HIRA Regulates RUNX1 in Hemogenic to Hematopoietic Transition
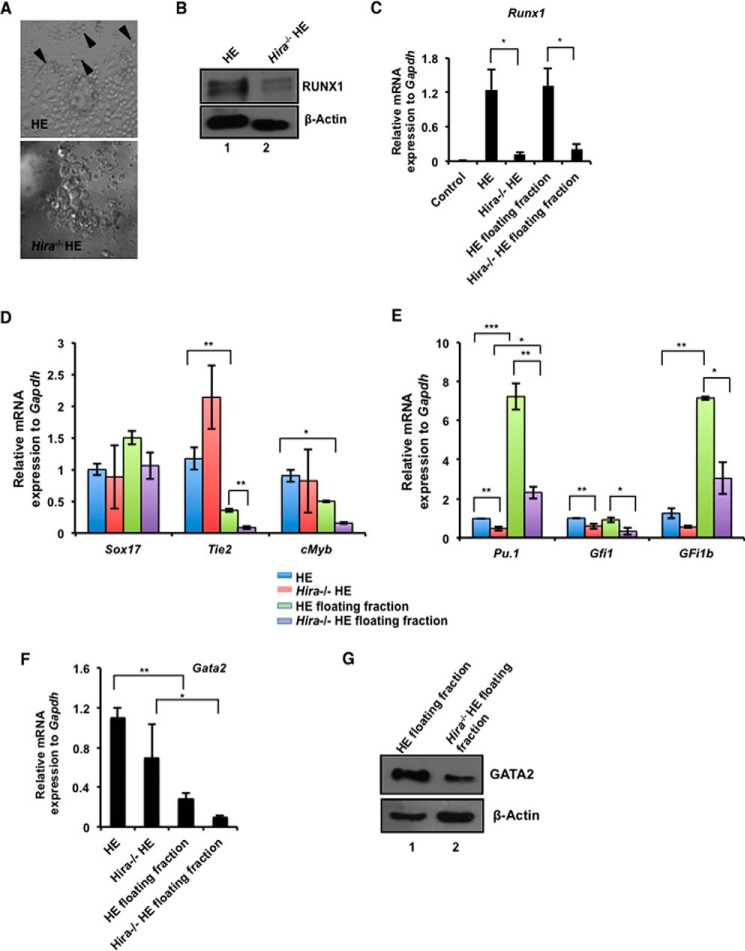
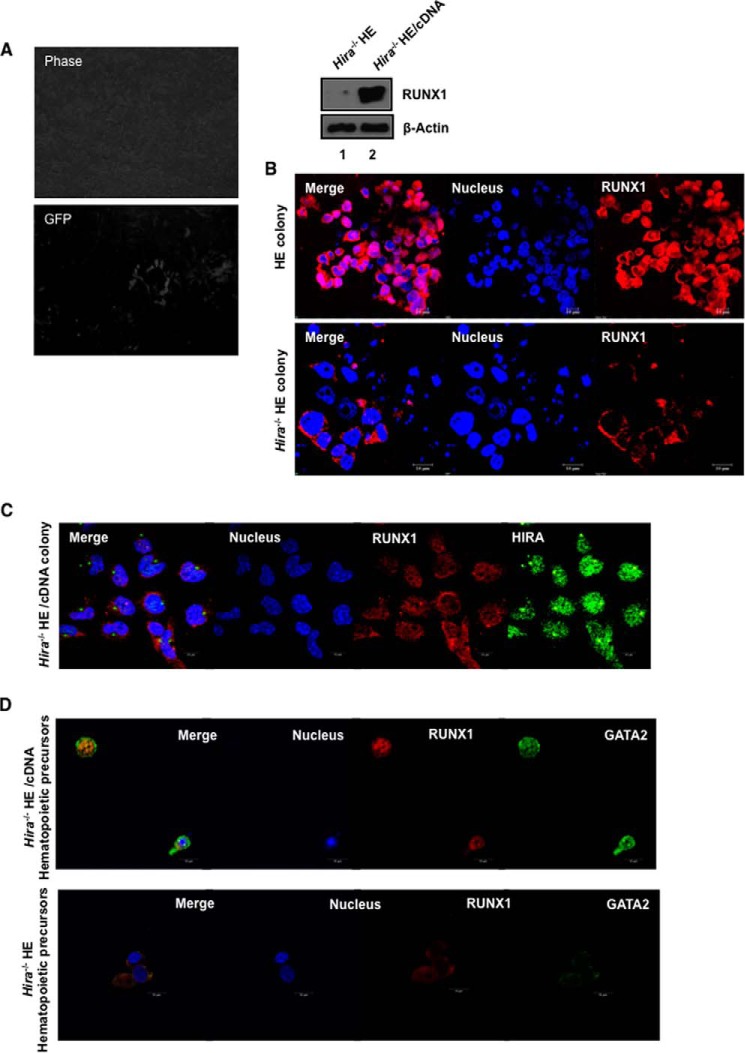
RUNX1 drives the generation of hemogenic endothelium and is required until the emergence of HSCs (4); on commitment to hematopoiesis, the role of RUNX1 becomes redundant. Because we demonstrated that RUNX1 expression is modulated by HIRA, we further investigated this relationship with regard to the hemogenic to hematopoietic transition. We generated HE from control and Hira−/− ESCs following the protocol described by Chiang and Wong (14). First, we questioned whether HIRA could influence the initial stages of differentiation of ESCs to HE. The differentiation in presence of mesodermal inducer and BMP4 induce the ESCs in endothelial lineage (14). Therefore, at day 4, we analyzed the expression of FLK1 (fetal liver kinase-1) in control and Hira−/− ES differentiated cells. FLK1+ cells are common progenitors for endothelial and hematopoietic lineage. We observed that Hira−/− cells did not demonstrate any significant alteration in the expression of Flk1 (Fig. 1, J and K). FACS analysis revealed a comparable number of FLK1+ cells arising from both control and Hira−/− ESCs (Fig. 1L). Effectively, absence of HIRA could not inhibit or influence the differentiation of ESCs till the mesoderm stage. In the next stage, during hemogenic to hematopoietic transition, the phenotype of the cells undergoes a shift first from flattened to spherical cells and then from loosely bound cells to floating cells. Control ESCs conformed to this pattern of HE colony development (Fig. 2A, upper panel), whereas Hira−/− ESCs failed to generate colonies with an analogous morphology (Fig. 2A, lower panel). The black arrows in Fig. 2A (upper panel) indicate the small clusters of cells undergoing the classical EndHT. RUNX1 expression was significantly down-regulated at the protein level in hemogenic endothelial cells derived from Hira−/− ESCs (Fig. 2B). During the differentiation of ESCs toward HE, HSC precursors repopulate the floating fraction of the culture (14). Thus, we analyzed both the adherent cells and floating cells of the HE culture. In absence of HIRA, Runx1 expression was significantly reduced in both the floating and adherent cells of the culture (Fig. 2C). We also analyzed the status of other genes associated with HE within the same samples. Sox17 has been implicated in HE formation (19), and we observed that in HIRA depleted cells, there was no significant difference in its expression between the mutant and wild type HE cells (Fig. 2D). However, expression of c-Myb, a partner of RUNX1 (20) induced in hematopoietic differentiation (21), is significantly down-regulated within the Hira−/− HE (Fig. 2D). Endothelial marker Tie2 was significantly down-regulated within the floating fraction of cells in the absence of HIRA (Fig. 2D). RUNX1 is a positive modulator of definitive hematopoiesis. Along with other transcription factors, RUNX1 regulates the dynamic pattern of genes implicated in hematopoiesis. Pu.1, Gfi1, and Gfi1B are reported to be direct downstream targets of RUNX1 (22, 23). Interestingly, the expression of the transcription factors Pu.1, Gfi1, and Gfi1b (Fig. 2E) were significantly down-regulated both in HE cells and hematopoietic precursors in the absence of HIRA. We conclude that HIRA is specifically involved in the regulation of RUNX1 during hemogenic program in the hemogenic-defined circuitry. Genes that are essential for hematopoietic progenitors are also affected as evident from the expression of Gata2 (Fig. 2, F and G) or c-myb (Fig. 2E). On ectopic expression of HIRA (tagged with GFP) in Hira−/− ESCs (Fig. 3A, left panels), the expression of RUNX1 was rescued in differentiating HE cells (Fig. 3A, right panel).
FIGURE 2.
HIRA regulates RUNX1 in endothelial to hematopoietic transition. A, ES cells were differentiated toward hemogenic endothelium. Cells were cultured in serum-free medium for 48 h followed by culturing in the presence of BMP4, FGF2, GSKi, and Activin for the subsequent 48 h. Next, cells were allowed to differentiate in presence of VEGF, BrcAMP, BMP4, FGF2, or ALKi for 48 h to induce them toward HE. A phase contrast image of HE colony formed from control (upper panel) and Hira−/− ES cells (lower panel). The black arrowheads indicate the formation of typical round-shaped clusters of cells in control ES cells. These cells have undergone the EndHT. The representative clusters are absent in cells differentiated from Hira−/− ES cells. B, the above set of samples was analyzed for the presence of RUNX1 by Western blot. C, qRT-PCR analyses for the expression of Runx1 in the same set of samples including the floating fractions representing hematopoietic precursors (means ± S.E. for three independent experiments). D, quantitative RT-PCR analyses of other hemogenic and endothelial markers in the same samples analyzed in C. Statistical analyses indicate the significant change in the Tie2 expression in adherent and floating fractions of control cells and within floating fractions of control and Hira−/− cells. Significant change was observed in c-myb expression in adherent and floating cells of control set. E, qRT-PCR analysis of RUNX1 target genes in hematopoiesis were analyzed in HE cells and hematopoietic precursors generated from control and Hira−/− ES cells (means ± S.E. for three independent experiments). Significant down-regulation of all the hematopoietic targets (Pu.1, Gfi1, and Gfi1b) of Runx1 is observed in the floating fractions of Hira−/− cells. F, quantitative RT-PCR analysis of Gata2 in hematopoietic precursors and HE cells (means ± S.E. for three independent experiments). Significant down-regulation in Gata2 expression within Hira−/− ESCs is observed. Statistical analyses were performed using Student's t test function. *, p < 0.05; **, p < 0.01; ***, p < 0.001. G, Western blot analysis for GATA2 expression in samples used in F.
FIGURE 3.
HIRA rescues RUNX1 expression. A, Hira (tagged to GFP) was ectopically expressed in Hira−/− ESCs (left panel) and differentiated to HE. Western blot analysis demonstrates the rescue of RUNX1 in the presence of HIRA (right panel). B, confocal study for the expression of RUNX1 within HE colonies generated from control and Hira−/− ESCs. C, localization of RUNX1 in Hira−/− HE cells with ectopic expression of HIRA. D, confocal study for the expression of RUNX1 and GATA2 in hematopoietic precursors of same set of cells analyzed in C.
To further understand the role of HIRA in the context of RUNX1 regulation, we studied the expression of RUNX1 in control and Hira−/− ESCs by immunofluorescence. Intriguingly, in absence of HIRA, we observed a lower intensity and altered localization of RUNX1 (Fig. 3B, lower panel) within the Hira−/− HE colony (Fig. 3B, lower panel) than within the control HE colony (Fig. 3B, upper panel). Interestingly, upon rescue by ectopic expression of HIRA, RUNX1 expression was induced with distinct localization within the nucleus (Fig. 3C). The hematopoietic precursors demonstrated intense RUNX1 and GATA2 co-localization in these rescued cells (Fig. 3D, upper panel), whereas the Hira−/− hematopoietic precursors displayed lowered and altered localization of RUNX1 (Fig. 3D, lower panel).
HIRA Level Linked to Nuclear Expression of RUNX1
What triggers the altered localization of RUNX1 in the hemogenic or hematopoietic clusters in absence of HIRA? To understand the mechanism for this altered or lesser nuclear expression of RUNX1 in the absence of HIRA, we generated a model of ESCs wherein we knocked down HIRA by shRNA expression (Fig. 4A). Interestingly, in a similar manner, EBs generated from Hira-kd ESCs showed significant down-regulation of Runx1 (Fig. 4B) as observed in Hira−/− EBs (Fig. 1F). Immunofluorescence studies demonstrated that the intensity of RUNX1 was lowered in HE cells generated from Hira-kd ESCs (Fig. 4C). Effectively, few cells (indicated with white arrows in Fig. 4C, lower panel) appeared to have altered RUNX1 expression. Therefore, we infer that the reduced HIRA expression might be responsible for the inhibition of nuclear entry of the RUNX1, and hence an altered localization of RUNX1 in HE cells is observed.
FIGURE 4.
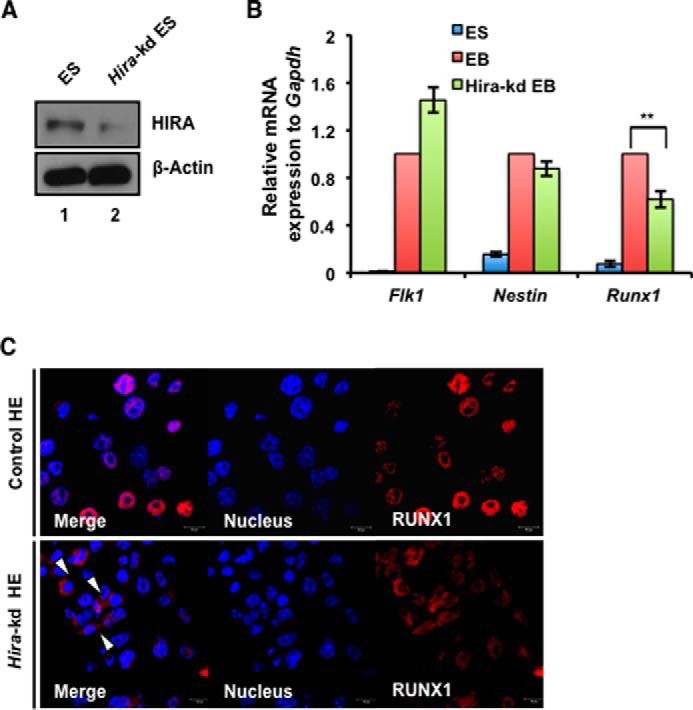
Reduced level of HIRA associated with decreased and hence altered localization of RUNX1. A, Hira was knocked down in control ESCs by shRNA expression. Western blot analysis demonstrates the knockdown of HIRA. B, EBs were generated from control and Hira-kd ESCs and analyzed for the expression of Flk1, Runx1, and Nestin. The expression of Runx1 was significantly down-regulated in Hira-kd EBs. C, confocal study for the expression of RUNX1 in control and Hira-kd HE cells demonstrates differential localization of RUNX1. The arrows indicate cells showing altered RUNX1 localization. Statistical analyses were performed using Student's t test function. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
HIRA Acts through RUNX1 to Modulate Definitive Hematopoiesis
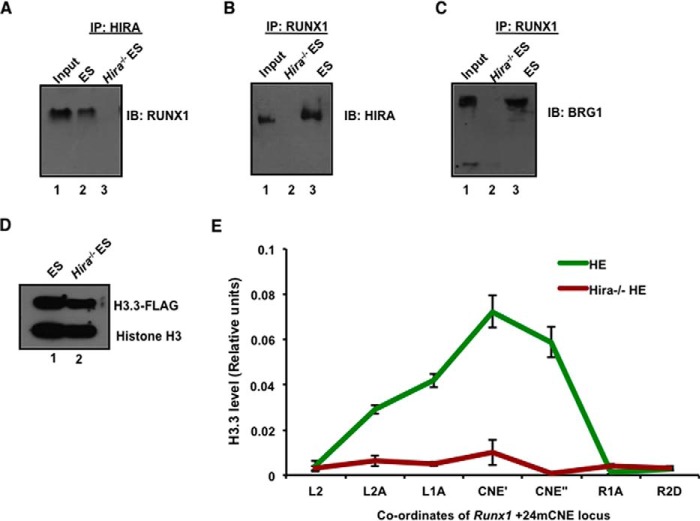
Thus, we postulated that if depletion of HIRA reduces nuclear entry of RUNX1, then RUNX1 will be unable to bind to its targets and hematopoiesis will fail to initiate. We demonstrated that the expression of Runx1 and its targets gets significantly down-regulated especially within the hematopoietic precursors (Fig. 2E). Further, RUNX1 binding to highly conserved and transcription factor rich binding regions at target loci is integral to the initiation of hematopoiesis. We therefore analyzed the region around (−)14 kb Pu.1, (−)35 kb Gfi1, and the enhancer elements CNE2 and CNE3 of Gfi1b by ChIP (7). RUNX1 has been reported to bind these regions and reshape the chromatin such that it becomes poised for hematopoiesis (7). Interestingly, ChIP analyses in HE cells showed that, in the absence of HIRA, RUNX1 is not recruited at either the Pu.1 and Gfi1b upstream regions or the enhancer elements of Gfi1b (Fig. 5, A–D). Analyses using EBs demonstrated a similar pattern of RUNX1 recruitment to target loci in the presence and absence of HIRA (Fig. 5, A–D). These data indicate HIRA could effectively regulate the RUNX1 targets in the context of HE and HSC generation. At the molecular level, HIRA and its associated complex (comprising ASF1, CAIN, and UBN1) physically interact with transcription factors, a chromatin insulator, and an ATP-dependent chromatin-remodeling complex (24). Therefore, we investigated whether HIRA and RUNX1 interacted within a protein complex. Co-immunoprecipitation experiments showed that RUNX1 physically associated with HIRA and vice versa (Fig. 6, A and B). The BRG1 component of the SWI-SNF chromatin remodeling complex has been implicated in regulating RUNX1 in the setting of leukemia and in the binding of the HIRA-complex to transcription factors (24, 25). Immunoprecipitation analyses demonstrated that BRG1 and RUNX1 interacted only in the presence of HIRA (Fig. 6C). Hence, the protein complex of HIRA, BRG1, and RUNX1 might subsequently modulate the expression of other hematopoietic genes targeted by RUNX1 within the nucleus.
FIGURE 5.
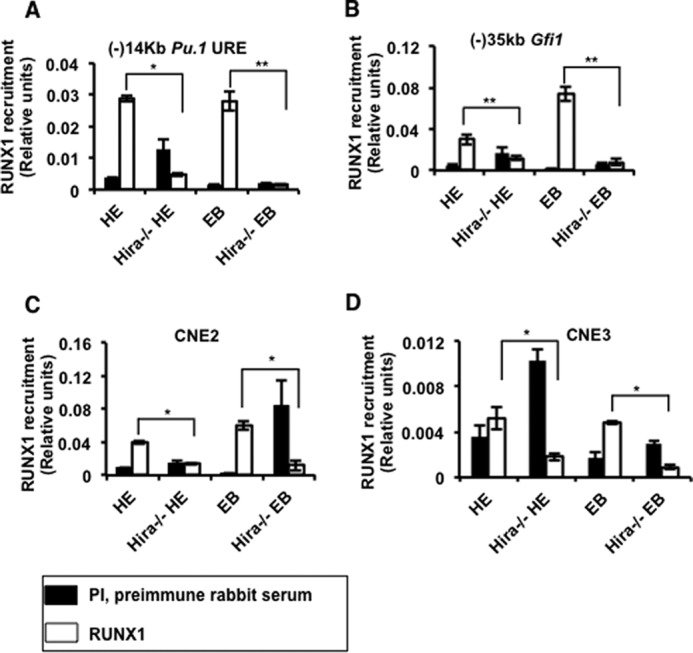
HIRA in regulation of definitive hematopoiesis. A–D, HE cells and EBs were generated from control and Hira−/− ES cells, cross-linked with 1% formaldehyde, and subjected to ChIP. The plots represent the recruitment of RUNX1 at the enhancer rich regions of Pu.1 and Gfi1 and at the enhancer elements (CNE3 and CNE2) of Gfi1b (means ± S.E. for three independent experiments). Preimmune rabbit serum (PI) is used as the negative control. Statistical analyses were performed using Student's t test function. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
FIGURE 6.
HIRA-mediated incorporation of histone H3.3 variant within Runx1 intronic enhancer drives hemogenic to hematopoietic transition. A and B, co-immunoprecipitation of HIRA and RUNX1 in control and Hira−/− ESCs. Shown are results of immunoprecipitation (IP) with HIRA and immunoblot (IB) with RUNX1 and vice versa, wherein input is 14%. C, co-immunoprecipitation of RUNX1 and BRG1 in control and Hira−/− ESCs. Shown are results of immunoprecipitation with RUNX1 and immunoblot with BRG1, wherein input is 14%. D, H3.3-Flag-tagged plasmid was transfected in ESCs by Lipofectamine and selected with G418 for 72 h. Western blot for FLAG-H3.3 in control and Hira−/− ESCs. E, HE cells were generated from transfected control and Hira−/− ESCs, cross-linked with 1% formaldehyde, and subjected to ChIP. A 3.6-kb locus flanking +24mCNE (conserved noncoding elements) region was analyzed to determine the extent of H3.3 incorporation. The plot shows the reduced or loss in incorporation of H3.3 in Hira−/− HE cells within +24mouse conserved noncoding elements (CNE) implicated in hemogenic to hematopoietic transition. PI is preimmune rabbit serum, used as negative control. Statistical analyses were performed using Student's t test function. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
HIRA-mediated H3.3 Enrichment within Runx1 Locus in Hemogenic to Hematopoietic Transition
Next, we asked how HIRA could regulate the expression of RUNX1 at the molecular level? Histone chaperone HIRA is responsible for the incorporation of Histone H3.3 variant in a replication-independent manner within the chromatin (10). Earlier reports indicate that a regulatory intronic enhancer termed as Runx1 +24 mouse conserved noncoding elements (m CNE) is open and active for the expression of Runx1 in cells undergoing hemogenic to hematopoietic transition (26). We determined the incorporation of Histone H3.3 variant within the enhancer region. We transfected control and Hira−/− ESCs with FLAG-tagged H3.3 construct (Fig. 6D) and differentiated them toward HE. We observed that H3.3 incorporation was significantly reduced in absence of HIRA within the enhancer region (Fig. 6E). We infer that HIRA-mediated histone H3.3 incorporation is required for the expression of Runx1 during the hemogenic to hematopoietic transition.
Discussion
Histone chaperone HIRA mediated H3.3 acetylation at residue Lys-56 could regulate in vitro and in vivo model of angiogenesis by endothelial cells (9). Endothelial specific genes were down-regulated in HIRA knocked down yolk sac endothelial cells. Targeted mutagenesis of Hira led to abnormal mesendoderm development and disturbed gastrulation resulting in death of embryo in utero (11). To replicate a similar development pattern in vitro, we differentiated control and Hira−/− ES cells to form EBs. Expression of endodermal or ectodermal markers or genes implicated in early hematopoiesis were either induced or remained unaltered in EBs in absence of HIRA (Fig. 1). Earlier studies suggested that the presence of unbound histones in Hira−/− EBs induces the expression of different germ layer markers (18). However, our data indicated that HIRA might have a differential regulatory role that could modulate the expression of RUNX1 during development.
These observations shaped the basis of determining the role of Hira in regulation of endothelial to hematopoietic transition. RUNX1 is essential for this particular cellular transition. Hence, we analyzed the HE colonies generated from control and Hira−/− cells and their subsequent transition to hematopoietic precursors. However, HIRA did not regulate the differentiation of ESCs toward the mesoderm because FLK1 expression remained unchanged in Hira−/− cells. It can be recalled here that up-regulation of FLK1 has been associated with down-regulation of RUNX1 in endothelial cells (27). Also, in our earlier studies we showed that expression of Flk1 in response to angiogenic signals was independent of HIRA in endothelial cells (9). In the differentiation protocol adopted in this study, inclusion of BMP4 in the culture media and the sequential differentiation from ES cells eventually allow a higher yield of ∼80% for both endothelial cells and blood or hematopoietic precursors (14) without the use of sorting processes between differentiation steps. We observed that on down-regulation of RUNX1 expression, the formation of specific morphological pattern of colonies of cells experiencing the EndHT was eventually distorted (Fig. 2A). Intriguingly, the absence of HIRA altered the subcellular localization of RUNX1 in HE cells (Fig. 3B). However, we demonstrated that the lower expression of Hira eventually led to the nonaccumulation or localization of nuclear RUNX1 (Fig. 4).
Understandably, nonavailability of RUNX1 within the nucleus affected its downstream targets especially those involved in definitive hematopoiesis. Other nontargets genes like c-myb were also affected in the hematopoietic precursors. We reasoned that a decrease in the level of hematopoietic specific markers is quite natural when the transition to hematopoiesis is hindered. Recently, Soni et al. (28) reported that Hira is critical for β-globin expression and few erythropoietic regulators but not the early hematopoietic marker Gata2 in EBs. Our analyses in EBs demonstrated a similar result in respect to Gata2 (Fig. 1E), but in the context of generation of hematopoietic precursors from Hira−/− HE cells, Gata2 was significantly down-regulated in the hematopoietic precursors (Fig. 2F) and in HE cells at the mRNA level (Fig. 2F). We inferred that HIRA could regulate RUNX1 and its targets during EndHT. To understand the transcription factor and histone chaperone interaction, it should be noted that the HIRA complex of UBN1, CAIN, and ASF1 with chromatin modulators BRG1/INI1 could regulate the chromatin landscape on the basis of gene transcription (24). Interaction studies indicated that Hira is essential for the formation of the protein complex formed of RUNX1 and BRG1 to bind to other downstream targets of RUNX1.
EndHT followed by hematopoietic stem cell differentiation are structured by different sets of transcription factors including RUNX1. Gfi1, Gfi1b, and Pu.1 have been implicated as direct targets of RUNX1 to establish the downstream hierarchy of hematopoiesis (23, 7, 29). The upstream region of myeloid master regulator Pu.1 has more than one functionally essential RUNX1 binding sites (29). Similarly, upstream region and enhancer elements of Gfi1 and Gfi1b have putative RUNX1 binding sites associated with definitive hematopoiesis (23). We demonstrated that reduced expression of RUNX1 in the nucleus within the cells deprived of HIRA could neither induce nor bind to the targets and hence could not establish the global reorganization at the chromatin level needed for the proper initiation of hematopoiesis (7), but how HIRA could influence the RUNX1 expression is the major concern of the study. Therefore, we explored the cis-regulatory elements of Runx1, essential for its expression during the transition. Ng et al. (26) reported that the Runx1 +24 mCNE was specifically active in HE cells and sites associated with HSC generation. We observed that the region spanning ∼3.6 kb has significant enrichment of H3.3 in control HE cells. We concluded that H3.3 incorporation by HIRA is essential to retain an active state of the enhancer element of Runx1.
Considering the fact that even in the complete absence of HIRA, there is always a constitutive expression of RUNX1, as evident from figures included in the study. There are few factors that might contribute to this level of expression of RUNX1. First, histone chaperone DAXX can occasionally complement the role of HIRA (30). Second, the incorporation of H3.3 is not completely abolished at the intronic Runx1 enhancer (Fig. 6E). Therefore, we predict that few other aspects might contribute toward the regulation of RUNX1.
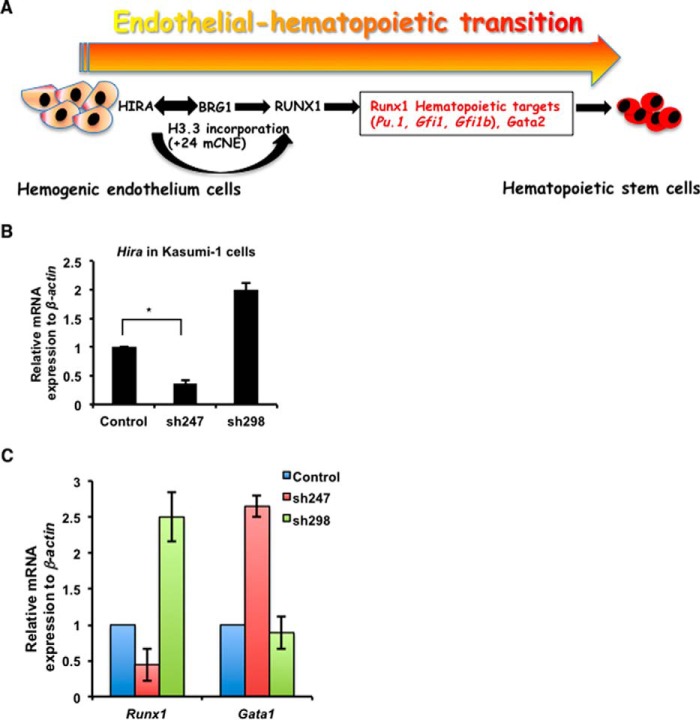
The transcription factor RUNX1 is also known as acute myeloid leukemia 1 protein or AML1. RUNX1 has been implicated in different types of leukemia in the context of chromosomal translocation or point mutation within this gene (31). Recent findings revealed that altered subnuclear localization of RUNX1 can switch cancer cells between proliferation and differentiation (32). Other histone chaperones including Asf1b and Caf1p60 have been implicated in cancer (33–35), but how Hira could modulate different genes in the context of cancer has never been addressed. Based on our initial observation of HIRA-mediated regulation of RUNX1 in hematopoiesis, we considered whether this phenomenon could be relevant in leukemia cells. Our preliminary findings in leukemia cell line harboring AML1/ETO fusion revealed that on knockdown of HIRA in Kasumi cells (gift from Dr. O. Williams; Fig. 7B), a reduced expression of RUNX1 was observed (Fig. 7C). Interestingly, a significant gain in the expression of Gata1 was detected at the RNA level (Fig. 7C). Gata1 and other Gata factors are repressed under the influence of AML1/ETO and hence are responsible for the blockade of cells toward erythroid and megakaryocytic lineage (36). We anticipate that HIRA-mediated regulation of RUNX1 followed by induction in GATA1 level might shift the balance from proliferative to differentiation of leukemia cells. In summary, we report for the first time that the regulation of the transcription factor RUNX1 by histone chaperone HIRA operates within the murine hemogenic endothelium, and this pathway subsequently regulates genes expressed in HSCs and thus regulates the endothelial to hematopoietic transition (Fig. 7).
FIGURE 7.
HIRA regulates RUNX1. A, summary of the proposed mechanism represent that HIRA interacting with the chromatin remodeler BRG1 regulates RUNX1 in EndHT and further contributes toward the regulation of hematopoietic targets of RUNX1 in generation of HSCs. B, Kasumi-1 cells harboring the t(8;21) translocation were analyzed for the HIRA-RUNX1 axis in leukemia. Quantitative RT-PCR analysis of Hira knockdown in Kasumi-1 cells. C, the same cells were analyzed for the expression of Runx1 and Gata1 by qRT-PCR. Control is unmanipulated Kasumi-1 cells. Statistical analyses were performed using Student's t test function. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Acknowledgments
We thank Prof. James Bieker, Dr. Owen Williams, and Dr. Arumugam Rajavelu for sharing reagents. We acknowledge the technical facilities at the host institute. The corresponding author also acknowledges the support of Dr. Ananda Mukherjee for providing critical comments on the manuscript.
Note Added in Proof
Khaja Mohieddin Syed's name was listed incorrectly in the version of the article that was published as a Paper in Press on April 6, 2015.
This work was supported by Grant SB/FT/LS-158/2012 from the Department of Science & Technology, India, and by intramural funding from the Department of Biotechnology, India.
- HSC
- hematopoietic stem cell
- ESC
- embryonic stem cell
- qRT-PCR
- quantitative RT-PCR
- EB
- embryoid body
- En
- embryonic day n
- HE
- hemogenic endothelium
- EndHT
- endothelial to hematopoietic transition.
References
- 1. Palis J., Robertson S., Kennedy M., Wall C., Keller G. (1999) Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development 126, 5073–5084 [DOI] [PubMed] [Google Scholar]
- 2. Bertrand J. Y., Chi N. C., Santoso B., Teng S., Stainier D. Y., Traver D. (2010) Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 464, 108–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boisset J. C., van Cappellen W., Andrieu-Soler C., Galjart N., Dzierzak E., Robin C. (2010) In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 464, 116–120 [DOI] [PubMed] [Google Scholar]
- 4. Chen M. J., Yokomizo T., Zeigler B. M., Dzierzak E., Speck N. A. (2009) Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature 457, 887–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Okuda T., van Deursen J., Hiebert S. W., Grosveld G., Downing J. R. (1996) AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84, 321–330 [DOI] [PubMed] [Google Scholar]
- 6. Wang Q., Stacy T., Binder M., Marin-Padilla M., Sharpe A. H., Speck N. A. (1996) Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl. Acad. Sci. 93, 3444–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lichtinger M., Ingram R., Hannah R., Müller D., Clarke D., Assi S. A., Lie-A-Ling M., Noailles L., Vijayabaskar M. S., Wu M., Tenen D. G., Westhead D. R., Kouskoff V., Lacaud G., Göttgens B., Bonifer C. (2012)) RUNX1 reshapes the epigenetic landscape at the onset of haematopoiesis. EMBO J. 31, 4318–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Koh C. P., Wang C. Q., Ng C. E., Ito Y., Araki M., Tergaonkar V., Huang G., Osato M. (2013) RUNX1 meets MLL: epigenetic regulation of hematopoiesis by two leukemia genes. Leukemia 27, 1793–1802 [DOI] [PubMed] [Google Scholar]
- 9. Dutta D., Ray S., Home P., Saha B., Wang S., Sheibani N., Tawfik O., Cheng N., Paul S. (2010) Regulation of angiogenesis by histone chaperone HIRA-mediated incorporation of lysine 56-acetylated histone H3.3 at chromatin domains of endothelial genes. J. Biol. Chem. 285, 41567–41577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Green E. M., Antczak A. J., Bailey A. O., Franco A. A., Wu K. J., Yates J. R., 3rd, Kaufman P. D. (2005) Replication-independent histone deposition by the HIR complex and Asf1. Curr. Biol. 15, 2044–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Roberts C., Sutherland H. F., Farmer H., Kimber W., Halford S., Carey A., Brickman J. M., Wynshaw-Boris A., Scambler P. J. (2002) Targeted mutagenesis of the Hira gene results in gastrulation defects and patterning abnormalities of mesoendodermal derivatives prior to early embryonic lethality. Mol. Cell. Biol. 22, 2318–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goldberg A. D., Banaszynski L. A., Noh K. M., Lewis P. W., Elsaesser S. J., Stadler S., Dewell S., Law M., Guo X., Li X., Wen D., Chapgier A., DeKelver R. C., Miller J. C., Lee Y. L., Boydston E. A., Holmes M. C., Gregory P. D., Greally J. M., Rafii S., Yang C., Scambler P. J., Garrick D., Gibbons R. J., Higgs D. R., Cristea I. M., Urnov F. D., Zheng D., Allis C. D. (2010) Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140, 678–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dutta D., Ray S., Home P., Larson M., Wolfe M. W., Paul S. (2011) Self-renewal versus lineage commitment of embryonic stem cells: protein kinase C signaling shifts the balance. Stem Cells 29, 618–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chiang P. M., Wong P. C. (2011) Differentiation of an embryonic stem cell to hemogenic endothelium by defined factors essential role of bone morphogenetic protein 4. Development 138, 2833–2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ying Q. L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. (2008) The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ray S., Dutta D., Rumi M. A., Kent L. N., Soares M. J., Paul S. (2009) Context-dependent function of regulatory elements and a switch in chromatin occupancy between GATA3 and GATA2 regulate Gata2 transcription during trophoblast differentiation. J. Biol. Chem. 284, 4978–4988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Samokhvalov I. M., Samokhvalova N. I., Nishikawa S. (2007) Cell tracing shows the contribution of the yolk sac to adult haematopoiesis. Nature 446, 1056–1061 [DOI] [PubMed] [Google Scholar]
- 18. Meshorer E., Yellajoshula D., George E., Scambler P. J., Brown D. T., Misteli T. (2006) Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell 10, 105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Clarke R. L., Yzaguirre A. D., Yashiro-Ohtani Y., Bondue A., Blanpain C., Pear W. S., Speck N. A., Keller G. (2013) The expression of Sox17 identifies and regulates haemogenic endothelium. Nat. Cell Biol. 15, 502–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hernandez-Munain C., Krangel M. S. (1994) Regulation of the T-cell receptor delta enhancer by functional cooperation between c-Myb and core-binding factors. Mol. Cell. Biol. 14, 473–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Soza-Ried C., Hess I., Netuschil N., Schorpp M., Boehm T. (2010) Essential role of c-myb in definitive hematopoiesis is evolutionarily conserved. Proc. Natl. Acad. Sci. U.S.A. 107, 17304–17308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jin H., Li L., Xu J., Zhen F., Zhu L., Liu P. P., Zhang M., Zhang W., Wen Z. (2012) Runx1 regulates embryonic myeloid fate choice in zebrafish through a negative feedback loop inhibiting Pu.1expression. Blood 119, 5239–5249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lancrin C., Mazan M., Stefanska M., Patel R., Lichtinger M., Costa G., Vargel O., Wilson N. K., Möröy T., Bonifer C., Göttgens B., Kouskoff V., Lacaud G. (2012) GFI1 and GFI1B control the loss of endothelial identity of hemogenic endothelium during hematopoietic commitment. Blood 120, 314–322 [DOI] [PubMed] [Google Scholar]
- 24. Pchelintsev N. A., McBryan T., Rai T. S., van Tuyn J., Ray-Gallet D., Almouzni G., Adams P. D. (2013) Placing the HIRA histone chaperone complex in the chromatin landscape. Cell Rep. 3, 1012–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bakshi R., Hassan M. Q., Pratap J., Lian J. B., Montecino M. A., van Wijnen A. J., Stein J. L., Imbalzano A. N., Stein G. S. (2010) The human SWI/SNF complex associates with RUNX1 to control transcription of hematopoietic target genes. J. Cell. Physiol. 225, 569–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ng C. E., Yokomizo T., Yamashita N., Cirovic B., Jin H., Wen Z., Ito Y., Osato M. (2010) A Runx1 intronic enhancer marks hemogenic endothelial cells and hematopoietic stem cells. Stem Cells 28, 1869–1881 [DOI] [PubMed] [Google Scholar]
- 27. Hirai H., Samokhvalov I. M., Fujimoto T., Nishikawa S., Imanishi J., Nishikawa S. (2005) Involvement of Runx1 in the down-regulation of fetal liver kinase-1 expression during transition of endothelial cells to hematopoietic cells. Blood 106, 1948–1955 [DOI] [PubMed] [Google Scholar]
- 28. Soni S., Pchelintsev N., Adams P. D., Bieker J. J. (2014) Transcription factor EKLF (KLF1) recruitment of the histone chaperone HIRA is essential for β-globin gene expression. Proc. Natl. Acad. Sci. U.S.A. 111, 13337–13342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hoogenkamp M., Krysinska H., Ingram R., Huang G., Barlow R., Clarke D., Ebralidze A., Zhang P., Tagoh H., Cockerill P. N., Tenen D. G., Bonifer C. (2007) The Pu.1 locus is differentially regulated at the level of chromatin structure and noncoding transcription by alternate mechanisms at distinct developmental stages of hematopoiesis. Mol. Cell. Biol. 27, 7425–7438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Drané P., Ouararhni K., Depaux A., Shuaib M., Hamiche A. (2010) The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 24, 1253–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Osato M. (2004) Point mutations in the RUNX1/AML1 gene: another actor in RUNX leukemia. Oncogene 23, 4284–4296 [DOI] [PubMed] [Google Scholar]
- 32. Zaidi S. K., Dowdy C. R., van Wijnen A. J., Lian J. B., Raza A., Stein J. L., Croce C. M., Stein G. S. (2009) Altered Runx1 subnuclear targeting enhances myeloid cell proliferation and blocks differentiation by activating a miR-24/MKP-7/MAP kinase network. Cancer Res. 69, 8249–8255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Corpet A., De Koning L., Toedling J., Savignoni A., Berger F., Lemaître C., O'Sullivan R. J., Karlseder J., Barillot E., Asselain B., Sastre-Garau X., Almouzni G. (2011) Asf1b, the necessary Asf1 isoform for proliferation, is predictive of outcome in breast cancer. EMBO J. 30, 480–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Staibano S., Mascolo M., Mancini F. P., Kisslinger A., Salvatore G., Di Benedetto M., Chieffi P., Altieri V., Prezioso D., Ilardi G., De Rosa G., Tramontano D. (2009) Overexpression of chromatin assembly factor-1 (CAF-1) p60 is predictive of adverse behaviour of prostatic cancer. Histopathology 54, 580–589 [DOI] [PubMed] [Google Scholar]
- 35. Burgess R. J., Zhang Z. (2013) Histone chaperones in nucleosome assembly and human disease. Nat. Struct. Mol. Biol. 20, 14–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Elagib K. E., Racke F. K., Mogass M., Khetawat R., Delehanty L. L., Goldfarb A. N. (2003) RUNX1 and GATA-1 coexpression and cooperation in megakaryocytic differentiation. Blood 101, 4333–4341 [DOI] [PubMed] [Google Scholar]