Background: Diaminopimelic acid (DAP) is mainly found in amidated form in cell wall peptidoglycan of Corynebacteriales.
Results: Corynebacterium glutamicum ltsA gene product is a glutamine amidotransferase that specifically amidates peptidoglycan lipid intermediates.
Conclusion: LtsA accounts for the peptidoglycan structural modification by DAP amidation observed in Corynebacteriales.
Significance: Loss of peptidoglycan DAP amidation results in hyper-susceptibility of bacterial cells to lysozyme and β-lactam antibiotics.
Keywords: antibiotics, bacterial metabolism, cell wall, enzyme, gene knockout, glutaminase, peptidoglycan, Corynebacteriales, DAP amidation, lysozyme
Abstract
A gene named ltsA was earlier identified in Rhodococcus and Corynebacterium species while screening for mutations leading to increased cell susceptibility to lysozyme. The encoded protein belonged to a huge family of glutamine amidotransferases whose members catalyze amide nitrogen transfer from glutamine to various specific acceptor substrates. We here describe detailed physiological and biochemical investigations demonstrating the specific role of LtsA protein from Corynebacterium glutamicum (LtsACg) in the modification by amidation of cell wall peptidoglycan diaminopimelic acid (DAP) residues. A morphologically altered but viable ΔltsA mutant was generated, which displays a high susceptibility to lysozyme and β-lactam antibiotics. Analysis of its peptidoglycan structure revealed a total loss of DAP amidation, a modification that was found in 80% of DAP residues in the wild-type polymer. The cell peptidoglycan content and cross-linking were otherwise not modified in the mutant. Heterologous expression of LtsACg in Escherichia coli yielded a massive and toxic incorporation of amidated DAP into the peptidoglycan that ultimately led to cell lysis. In vitro assays confirmed the amidotransferase activity of LtsACg and showed that this enzyme used the peptidoglycan lipid intermediates I and II but not, or only marginally, the UDP-MurNAc pentapeptide nucleotide precursor as acceptor substrates. As is generally the case for glutamine amidotransferases, either glutamine or NH4+ could serve as the donor substrate for LtsACg. The enzyme did not amidate tripeptide- and tetrapeptide-truncated versions of lipid I, indicating a strict specificity for a pentapeptide chain length.
Introduction
Bacteria belonging to Corynebacteriales, an order of Actinobacteria, possess an atypic, complex multilayered envelope (1, 2). In contrast to other bacteria, the peptidoglycan of these species is covalently bound to arabinogalactan, a huge polysaccharidic polymer mainly composed of arabinose and galactose, which itself is covalently linked to mycolic acids (3). This complex, named the mycoloyl-arabinogalactan-peptidoglycan complex (MAPc),3 constitutes with other lipids the cell wall that contains the outer membrane (mycomembrane) (1, 2, 4). Arabinogalactan is bound to peptidoglycan by its galactan domain via a conserved disaccharide linker (α-l-rhamnose-(1→3)-α-d-N-acetylglucosamine-(1→P)) that is attached to the C-6 position of some of the peptidoglycan N-acetylmuramic acid residues (5). The peptidoglycan structure of different genera of Corynebacteriales, such as Mycobacterium, Corynebacterium, and Rhodococcus, shows similarities with that of Escherichia coli (6–8). In all cases, the glycan strands are composed of alternating β-1→4-linked N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) units and are cross-linked via short MurNAc-linked peptides that are initially synthesized as l-Ala-γ-d-Glu-meso-DAP-d-Ala-d-Ala (DAP, diaminopimelic acid). This peptidoglycan belongs to the A1γ-type (6), in which meso-DAP and d-Ala (at position 4) from adjacent stems are directly cross-linked. In fact, recent detailed analyses of these cross-links in Mycobacterium tuberculosis, Mycobacterium abscessus, and Corynebacterium jeikeium showed that they occurred either between meso-DAP and d-Ala (4→3 cross-links) or between two meso-DAP (3→3 cross-links) (9–13), demonstrating the participation of both d,d- and l,d-transpeptidase activities, respectively, in this process. Cross-links of the 3→3 type predominated in M. tuberculosis and M. abscessus (70–80%) but not in C. jeikeium (38%). They were comparatively much less abundant (<5%) in the E. coli polymer. Some other modifications of the peptidoglycan structure were identified in Corynebacteriales, i.e. glycine residues bound to the ϵ-amine of meso-DAP as peptide linker in M. tuberculosis (11), or the glycolylation of MurNAc residues in M. tuberculosis and Mycobacterium smegmatis (11, 14). Also, in all mycobacterial species analyzed to date, as well as in C. jeikeium, the α-carboxyl group of d-Glu and the ϵ-carboxyl group of meso-DAP are amidated (8, 11–13, 15).
Amidation of DAP has been observed in other bacteria (16–18). The enzyme responsible for this modification has been recently identified in Lactobacillus plantarum (16) as the product of the asnB1 gene, which is part of the asnB1-thrA1-murE locus. The asnB1 gene might be essential in this species because a mutant partially deficient in meso-DAP amidation showed major growth defects (16). A search for asnB1 homologues in the Corynebacterium glutamicum genome retrieved one ORF (cg2410) corresponding to a gene named ltsA that had been previously identified for its ability to confer resistance to lysozyme in C. glutamicum and Rhodococcus erythropolis (19, 20). In contrast to what was observed in L. plantarum, ltsA was not essential in the latter two species, and its disruption did not induce a filamentous phenotype. LtsA, as AsnB1, was expected to belong to the large family of glutamine-dependent asparagine synthases (EC 6.3.5.4) that contains some extensively characterized members, such as the E. coli AsnB protein (AsnBEc) (21, 22). Lysozyme sensitivity of the R. erythropolis ltsA mutant could be complemented by ltsA homologues from C. glutamicum, M. tuberculosis, and Bacillus subtilis but not by the asnB gene from E. coli, suggesting that, although they belonged to the same family, LtsA and AsnBEc might have different activities or substrate specificities (20). The observation that ltsA from R. erythropolis and C. glutamicum could not complement an asparagine-requiring mutant of E. coli (in which the two genes encoding asparagine synthases AsnA (the l-aspartate:ammonia ligase) and AsnB were inactivated) further supported this assumption (19, 20). Moreover, although both LtsARe and AsnBEc displayed ATP-dependent glutaminase activity in vitro, the subsequent step of transfer of the amino group from glutamine to aspartic acid was only observed for AsnBEc. By contrast, LtsARe displayed a synthetase activity when lysozyme-treated cell wall extracts were added to the incubation mixture, suggesting the presence of a glutamine amino group acceptor in the cell wall of this species, which was not further identified (20). It should be noted that the M. smegmatis LtsA homologue (AsnBMs) was also identified while screening a transposon insertion library of mutants for antibiotic hyper-susceptibility (23). This M. smegmatis asnB mutant exhibited sensitivity to several hydrophobic drugs, suggesting a cell wall permeability defect, but no other phenotypical difference with the wild-type strain was reported.
Although the data obtained with the ltsA mutants strongly suggested that a cell wall formation process was affected by these mutations, no obvious cell envelope defect was associated, and the exact function of the LtsA proteins in these bacteria remained to be elucidated. In light of the recent results obtained by Bernard et al. (16) on L. plantarum AsnB1, we reinvestigated the function of LtsA in C. glutamicum. We here demonstrate that this protein is responsible for the amidation of DAP residues present in the C. glutamicum peptidoglycan and that this modification confers a high lysozyme resistance level to this bacterial species. It is also shown that the heterologous expression of LtsACg in E. coli results in a massive and toxic incorporation of amidated DAP residues (DAPNH2) in the peptidoglycan of this host. Finally, in vitro enzymatic assays were developed that clearly demonstrated the glutamine amidotransferase activity of LtsACg and further specified its substrate specificity.
Experimental Procedures
Strains, Plasmids, and Growth Conditions
The E. coli strain DH5α (supE44 ΔlacU169 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 φ80 dlacZ ΔM1) (Invitrogen) was used as the host for plasmids, and strain C43(DE3)(pLysS) (F− ompT hsdSB rB− mB− gal dcm DE3) (Novagen) was used for protein production and physiological studies. C. glutamicum strain RES167, a restriction-less derivative of ATCC 13032 (24), and its ΔltsA derivative were cultured in brain-heart infusion (BHI) medium (Difco) at 30 °C. E. coli cells were grown in 2YT medium (25) or Luria Bertani (LB) medium (Difco) at 37 °C. Antibiotics were added when required, at final concentrations of 100 μg/ml for ampicillin, 25 μg/ml for kanamycin (Km), 30 μg/ml (E. coli), and 6 μg/ml (C. glutamicum) for chloramphenicol. Growth was monitored at 600 nm.
Molecular Biology Techniques
Polymerase chain reaction (PCR) amplification of genes was performed using Expand High Fidelity Polymerase (Roche Applied Science) or Phusion High Fidelity Polymerase (Thermo Scientific). Standard procedures for DNA digestion, ligation, and agarose gel electrophoresis were used (26). E. coli cells were transformed with plasmid DNA as described by Dagert and Ehrlich (27) or by electroporation. C. glutamicum cells were transformed by electroporation (28).
Construction of an ltsA Deleted Strain
Deletion of the ltsA gene (cg2410) was done using a strategy described previously (29). In brief, two DNA fragments overlapping the ltsA gene at its 5′ and 3′ extremities were amplified by PCR from C. glutamicum total DNA using primers 5′-TTGAAGATCTTCGTGGGTTTCG-3′/5′-ATACCCGCGGATTGAAAAATCCTCC-3′ and 5′-ATCCCCGCGGTCTTAAAGCCTAAAC-3′/5′-TATGCTCGAGCTAAGGCACTCATC-3′, respectively. These fragments were purified and inserted flanking a kanamycin resistance (aphIII gene) cassette into plasmid pMCS5 (MoBiTec, Göttingen, Germany). The resulting plasmid (pMCS5-ΔltsA) was transferred into C. glutamicum RES167 by electroporation, and transformants were selected on Km-containing plates. Transformants in which allelic replacement had occurred were selected by PCR analysis using combinations of primers localized upstream and downstream of ltsA and in the aphIII sequence. After sequencing of the PCR products, one strain, ΔltsA, was selected for further studies.
Construction of Expression Plasmids
The pET2160 plasmid (pET21d derivative) used for expression of proteins with a C-terminal six-histidine tag (His6) has been previously described (30). Plasmids allowing high level expression of the C. glutamicum LtsA protein were constructed as follows. PCR primers 5′-CGCGACATGTGCGGCCTTCTTGGCATATTGACTGC-3′ and 5′-CGCGAAGCTTAAAGCTCGACTGGGTAGGAGCGGTCCTC-3′ were used to amplify the 1,920-bp gene from the strain ATCC 13032 chromosome, and the resulting material was treated with BspLU11I and HindIII and ligated between the compatible sites NcoI and HindIII of pET21d vector, generating the pMLD288 plasmid. For expression of the protein with a C-terminal His6 tag, the gene was similarly amplified using this time as reverse primer 5′-CGCGGGATCCAAGCTCGACTGGGTAGGAGCGGTCCTC-3′, and the PCR product was cleaved by BspLU11I and BamHI and cloned between the compatible NcoI and BglII sites of pET2160 vector, yielding pMLD290. Sequences of cloned inserts were controlled by DNA sequencing.
Microscopy Analyses
Optical Microscopy
Bacteria pelleted and resuspended in PBS buffer were visualized using a DMIRE2 optical microscope (Leica) equipped with a CCD camera (CoolSNAP HQ2, Roper Scientific).
Cryo-transmission Electron Microscopy
Bacteria were pelleted and resuspended in PBS buffer. A 4-μl suspension was adsorbed onto a glow-discharged holey carbon-coated grid (Quantifoil, Germany), blotted with Whatman filter paper, and vitrified into liquid ethane at −178 °C using a Vitrobot (FEI Co., The Netherlands). Frozen grids were transferred onto a Philips CM200-FEG electron microscope using a Gatan 626 cryo-holder. Electron micrographs were recorded at an accelerating voltage of 200 kV and a nominal magnification of ×50,000, using a low dose system (10 e−/Å2) and keeping the sample at −175 °C. Defocus values were −3 μm. Micrographs were recorded with a 4K × 4K CMOS camera (TVIPS, Germany).
Peptidoglycan Purification and Structure Analysis
E. coli C43(DE3)(pLysS) cells (0.8-liter cultures) carrying either the pMLD288 plasmid or the empty vector pET21d were grown exponentially in 2YT medium. At an OD600 of 0.4, 1 mm IPTG was added, and incubation was continued until a decrease in the growth rate was detected about 90 min later, when the absorbance of the culture of ltsA-overexpressing cells reached a plateau at about 0.9–1.0. Cultures (0.5-liter) of wild-type and ΔltsA C. glutamicum strains were arrested at an OD600 of 5 (exponential phase). In all cases, bacteria were harvested in the cold, washed with a cold 0.85% NaCl solution, and then rapidly suspended under vigorous stirring in 40 ml of a hot (95 to 100 °C) aqueous 4% SDS solution for 1 h. After standing overnight at room temperature, the suspensions were centrifuged for 30 min at 200,000 × g, and the pellets were washed several times with water. After final resuspension in 10 ml of water, aliquots were hydrolyzed (16 h at 95 °C in 6 m HCl) and analyzed with a Hitachi model 8800 amino acid analyzer (ScienceTec). The peptidoglycan content was expressed in terms of its characteristic and specific constituents, muramic acid and diaminopimelic acid (31, 32). The peptidoglycan material was further purified by treatment with proteases to remove contaminating proteins, as described earlier (31). The C. glutamicum preparation was also specifically treated with 48% hydrofluoric acid to remove covalently linked polysaccharides (33). The purity of the resulting material was confirmed by analysis of its amino acid and amino sugar composition, as described above.
The structure of the peptidoglycan was determined by using the classical procedure of Glauner (34). The purified peptidoglycan preparations were treated with a mixture of lysozyme and mutanolysin, and the released fragments (muropeptides) were reduced with sodium borohydride and separated by HPLC on a 3-μm ODS-Hypersil column (4.6 × 250 mm, Thermo Scientific), using a gradient of methanol (from 0 to 25% in 120 min) in 50 mm sodium phosphate buffer, pH 4.5, at a flow rate of 0.5 ml/min. Peaks were detected at 207 nm, and the collected muropeptides were desalted on the same column, this time using 0.05% trifluoroacetic acid and a 0–25% methanol gradient for elution. They were identified by amino acid and amino sugar analyses and by MALDI-TOF mass spectrometry.
Quantitation of Peptidoglycan Nucleotide Precursors
C43(DE3)(pLysS) cells (0.8-liter cultures) carrying either the pMLD288 plasmid or the empty vector pET21d were grown at 37 °C in 2YT medium, and expression of ltsACg was induced for 90 min with 1 mm IPTG, as described above. Cultures were rapidly chilled to 0 °C, and cells were harvested in the cold. The conditions used for the extraction of the peptidoglycan nucleotide precursors as well as the analytical procedure used for their separation and quantification were described previously (31, 35). The final separation step of the different UDP-MurNAc peptides was performed as follows: aliquots were applied onto a μ-Bondapak C18 HPLC column (7.8 × 300 mm), and elution was done at a flow rate of 3 ml/min with 50 mm ammonium formate for 15 min at pH 3.35, followed by a gradient of pH, from 3.35 to 4.75, applied between 15 and 50 min. Nucleotide precursors were identified on the basis of their retention time, as compared with authentic standards. Their identity was confirmed by analysis of their amino acid and amino sugar composition, as well as by MALDI-TOF mass spectrometry.
Mass Spectrometry Analyses
MALDI-TOF Mass Spectrometry
Positive and negative spectra were recorded in the reflectron mode with delayed extraction on a PerSeptive Voyager-DE STR instrument (Applied Biosystems) equipped with a 337-nm laser. Compounds (peptidoglycan precursors and muropeptides; 1 μl at 50–100 pmol/μl in water) were deposited on the plate, followed by 2,5-dihydroxybenzoic acid (1 μl at 10 mg/ml in 0.1 m citric acid). After evaporation of water, spectra were recorded at an acceleration voltage of ± 20 kV and an extraction delay time of 200 ns. External calibration was performed using the calibration mixture 1 of the SequazimeTM peptide mass standards kit (Applied Biosystem) in the positive mode or a mixture of UDP-MurNAc, UDP-MurNAc-dipeptide, and UDP-MurNAc pentapeptide in the negative mode.
Tandem Mass Spectrometry
The structure of the purified reduced muropeptides was determined with an electrospray time-of-flight mass spectrometer operating in the positive mode (Qstar Pulsar I, Applied Biosystems), as described previously (36). Briefly, [M + H]+ ions were selected on the basis of the m/z value in the high resolution mode, and fragmentation was performed with nitrogen as the collision gas. The collision energy was typically of 36–40 eV.
Production and Purification of LtsACg
C43(DE3)(pLysS) cells harboring the pMLD290 plasmid were grown exponentially at 37 °C in 2YT/ampicillin medium (1-liter cultures). Expression of the LtsA-encoding gene was induced with 1 mm IPTG when the OD600 of the culture reached 0.9. Cells were harvested 4 h later and were washed with 40 ml of cold 20 mm potassium phosphate buffer, pH 7.4, containing 0.1% 2-mercaptoethanol (buffer A). The wet cell pellet was suspended in 10 ml of the same buffer and disrupted by sonication (Bioblock Vibracell sonicator, model 72412) for 10 min with cooling. The resulting suspension was centrifuged at 4 °C for 30 min at 200,000 × g in a Beckman TL100 centrifuge, and both the pellet (membrane fraction) and the supernatant (soluble fraction) were kept and stored at −20 °C. The membrane fraction was washed several times with buffer A before performing LtsA activity assays. SDS-PAGE analysis of proteins was performed as described previously using 8% polyacrylamide gels (37), and protein concentrations were determined by the method of Bradford, using bovine serum albumin as a standard (38).
Enzymatic Assays, Coupled MraY-LtsA Activity Assay
The MraY exchange assay, in which the exchange of [14C]UMP with the unlabeled UMP moiety of UDP-MurNAc pentapeptide is followed (39), was performed in a reaction mixture containing, in a final volume of 40 μl, 25 mm Tris-HCl, pH 7.4, 12.5 mm MgCl2, 5 mm KCl, 5 μm [14C]UMP (16.6 GBq/mmol), 0.2 mm UDP-MurNAc pentapeptide, and membranes (40 μg of protein). To concomitantly assay the LtsA amidotransferase activity present in these membranes, the reaction mixture in addition contained ATP (5 mm) and either glutamine, asparagine, or ammonium sulfate (amine donor, 5 mm). Some assays aimed at determining the substrate specificity of LtsA were also performed, with UDP-MurNAc tetrapeptide or UDP-MurNAc tripeptide instead of UDP-MurNAc pentapeptide. Assays aimed at identifying the acceptor substrate (nucleotide precursor or lipid I) were performed in reaction mixtures containing 25 mm Tris-HCl, pH 7.4, 12.5 mm MgCl2, 5 mm KCl, 20 μm UDP-MurNAc-14C-labeled pentapeptide (500 Bq), 5 mm ATP, 5 mm glutamine, and membranes from LtsACg-expressing cells (40 μg of protein). Tunicamycin (10 μm) was added in some of these assays to totally inhibit the MraY activity. In all cases, after 30–120 min of incubation at 37 °C, the reaction was stopped by addition of 110 μl of 50 mm ammonium formate buffer, at a pH of 3.9 or 4.2, depending on the acceptor substrate used, and 135 μl were injected onto a 5-μm Nucleosil 100 C18 column (4.6 × 150 mm; Alltech-France) using the same buffer at 0.6 ml/min as the mobile phase. Detection was performed with a radioactive flow detector (model LB506-C1, Berthold) using the Quicksafe Flow 2 scintillator (Zinsser Analytic) at 0.6 ml/min. Quantitation was carried out with the Winflow software (Berthold).
Glutaminase Assay
The glutaminase activity of LtsA was assayed in a reaction mixture containing, in a final volume of 40 μl, 25 mm Tris-HCl, pH 7.4, 5 mm ATP, 12.5 mm MgCl2, 5 mm KCl, 5 mm glutamine, and membranes (40 μg of protein). To test whether this activity could be stimulated in the presence of an amine group acceptor, MraY-dependent synthesis of lipid I was induced in some assays by addition of UDP-MurNAc pentapeptide at a concentration of 1 mm. Mixtures were incubated for 20 min at 37 °C, and reactions were stopped by addition of 160 μl of 67 mm trisodium citrate/HCl buffer, pH 2.2. Aliquots were injected in the Hitachi amino acid analyzer, allowing the HPLC separation and quantification of the reaction substrate (glutamine) and product (glutamic acid).
Sensitivity to Lysozyme and Antibiotics of C. glutamicum Strains
Dose effects of lysozyme and antibiotics were determined on exponentially growing cells. Overnight cultures of wild-type and ΔltsA strains were inoculated into fresh BHI medium to an OD600 of 0.2 and grown until the OD600 reached 3. Cultures were then dispatched, and drugs were added at different concentrations. Growth was monitored until the stationary phase was achieved. The percentage of growth inhibition was calculated by reference to the OD600 of the culture without drug in the stationary phase. IC50 values were determined from the curves obtained from these measurements (percentage of growth inhibition as a function of drug concentration) and correspond to the drug concentration for which the growth inhibition is 50%.
Chemicals
Unlabeled and 14C-radiolabeled forms of UDP-MurNAc peptides were prepared and purified as described earlier (40, 41). DAPNH2-containing UDP-MurNAc tripeptide was enzymatically produced from UDP-MurNAc-l-Ala-d-Glu and DAPNH2 using purified E. coli MurE ligase (17), and DAPNH2-containing UDP-MurNAc pentapeptide was isolated from vancomycin-treated Clostridium difficile cells, as described earlier (42). [14C]UMP was obtained from Hartmann Analytic, and IPTG from Eurogentec. Proteases, lysozyme, mutanolysin, tunicamycin, antibiotics, amino acids, and reagents were from Sigma. DNA ligase and restriction enzymes were obtained from New England Biolabs or Fermentas, and DNA purification kits were from Promega and Macherey-Nagel. Synthesis of oligonucleotides and DNA sequencing was performed by Eurofins-MWG or IDT.
Results
Mutant Construction and Phenotypical Analysis
A BLAST search analysis in the C. glutamicum ATCC 13032 genome identified LtsA (cg2410) as a potential L. plantarum AsnB1 homologue. Similar homologues were also found in genomes of all other Corynebacteriales species, and alignment analyses confirmed important sequence conservation between these proteins (Fig. 1). Sequence homology was comparatively significantly lower with the E. coli asparagine synthase AsnBEc, especially in the C-terminal domain.
FIGURE 1.
Multiple sequence alignment of LtsA homologues from different bacterial species (ClustalW). Protein sequences are as follows: AsnB1 from L. plantarum WCFS1 (GI:28377797); LtsA from C. glutamicum ATCC 13032 (GI:62391036); LtsA from R. erythropolis PR4 (GI:226307090); and AsnB from M. tuberculosis H37RV (GI:15609338). Identical and similar amino acid residues are highlighted in blue and green, respectively. * indicates positions that have a single and fully conserved residue; : indicates conservation between groups of strongly similar properties, and . indicates conservation between groups of weakly similar properties.
To investigate the role of LtsA in Corynebacteriales, we attempted to delete the corresponding gene in C. glutamicum. For this purpose, a kanamycin resistance cassette flanked by sequences identical to the 5′ (408 bp upstream of the ATG) and the 3′ (434 bp downstream of the stop codon) parts of ltsA was cloned in a vector unable to replicate in corynebacteria. This construct was transferred into the ATCC 13032 strain, and several KmR transformants were analyzed by PCR using appropriate combinations of primers. One clone giving the amplification pattern expected from an allelic exchange between the wild-type copy of ltsA and the km cassette was chosen and subsequently analyzed (ΔltsA strain).
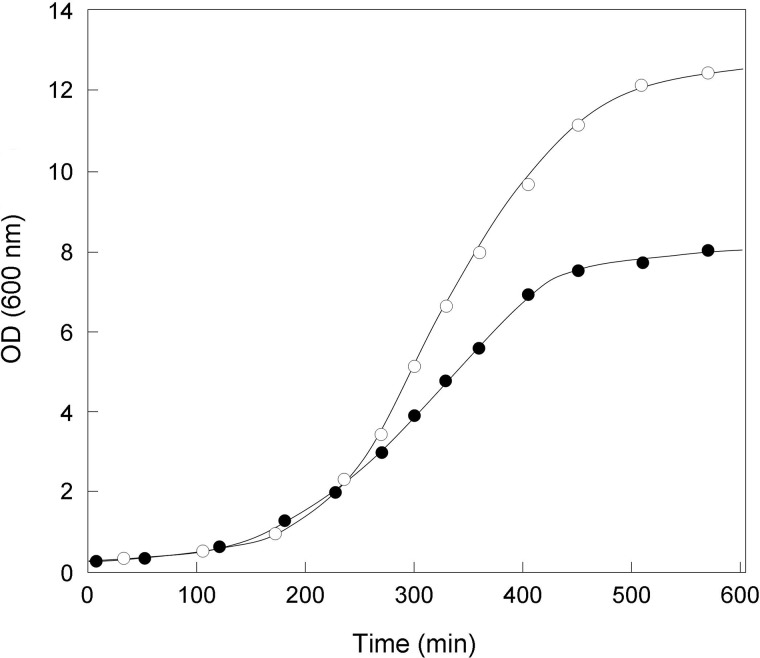
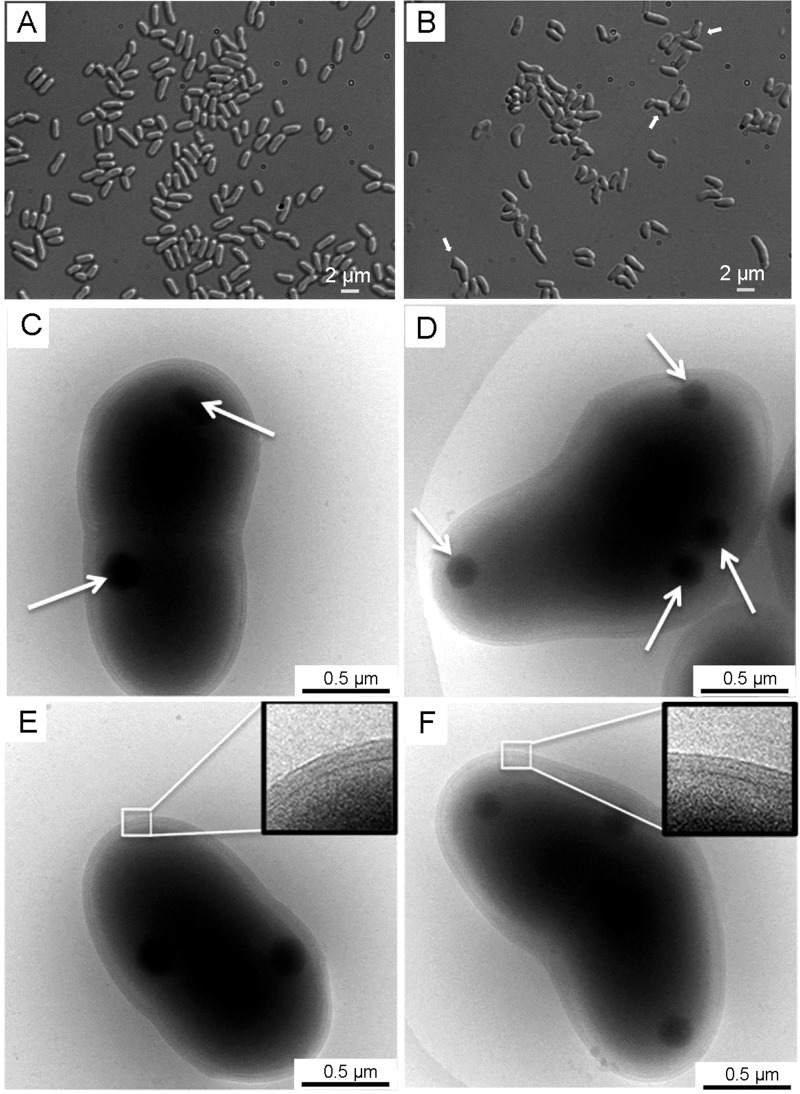
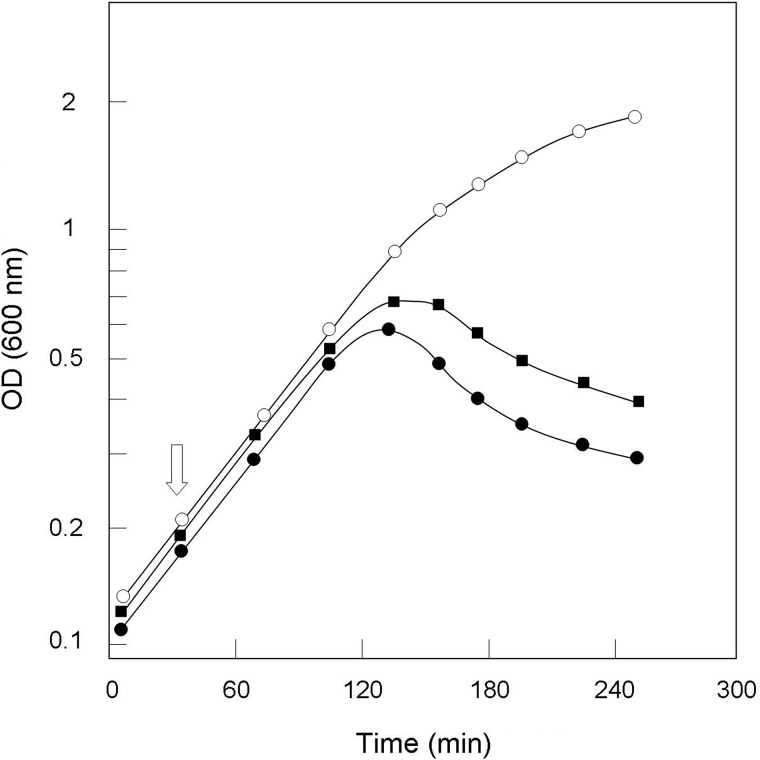
As compared with the wild-type strain, the ΔltsA mutant strongly aggregated in liquid culture, grew more slowly (generation time of 100 min versus 60 min for the wild-type), and entered stationary phase at a lower absorbance value (Fig. 2). Enumeration of bacteria from growing cultures revealed a 2-fold lower number of colony-forming units for this mutant at a given OD600, suggesting that LtsA was required for optimal growth and that its inactivation might have induced cell morphological alterations. Optical microscopy observation of ΔltsA and wild-type cells in exponential or stationary growth phases showed that a majority of mutant cells presented rod- and club-shaped morphologies that were quite similar to that of the parental cells. However, about a quarter of them exhibited abnormal shapes (Fig. 3, A and B). Cell filamentation was not detected in our ΔltsA C. glutamicum mutant, in contrast to the L. plantarum mutant deficient in DAP amidation (16). A transmission electron microscopy analysis of wild-type and ΔltsA cells was performed to better visualize these morphological changes. As shown in Fig. 3, C–F, although the parental cells presented a normal shape with electron-dense granules (volutin granules of poly(P)) on either side of the dividing bacteria, many of the ΔltsA mutant cells exhibited an asymmetrical shape with a larger volume and a large number of electron-dense granules randomly distributed in the cytoplasm. However, no significant difference was observed between the structures of the cell envelopes in these two strains (Fig. 3, E and F). This was consistent with an absence of difference in their corynomycolic acid profiles (data not shown).
FIGURE 2.
Growth curves of the wild-type (○) and ΔltsA (●) C. glutamicum strains in BHI medium at 30 °C.
FIGURE 3.
Morphologies of C. glutamicum strains. A and B, optical micrographs of exponentially growing C. glutamicum ATCC 13032 (A) and ΔltsA mutant (B) cells. Arrows indicate examples of cells exhibiting abnormal shapes. C–F, electron micrographs of frozen hydrated C. glutamicum ATCC 13032 cells (C and E) and of ΔltsA mutant cells exhibiting an irregular shape (D and F). The inset in E and F is an enlargement of the cell envelope. Arrows in C and D show the electron-dense granules.
Effect of the ltsA Mutation on the C. glutamicum Peptidoglycan Structure
MAPc was extracted from exponentially growing cells of wild-type and ΔltsA mutant strains of C. glutamicum (ATCC 13032) by treatment with 4% SDS at 100 °C. Analysis of the amino acid and amino sugar contents of these extracts indicated that the peptidoglycan content did not vary between the two strains (∼12.5 μmol of DAP/liter of culture at OD600 = 1, i.e. 30 μmol per g of cell dry weight). The overall compositions of these crude MAPc preparations were quite similar, and the only difference observed on the profile of the amino acid analyzer was the ratio between two peaks corresponding to glucosamine (GlcN), originating from peptidoglycan GlcNAc, and galactosamine (GalN), originating from arabinogalactan (43). The GlcN/GalN ratio was 1.8 and 2.6 in the wild-type and ΔltsA mutant strains, respectively, suggesting that the arabinogalactan content could be decreased in the mutant strain. As expected, GalN was not detected any longer in the insoluble peptidoglycan fraction when the MAPc preparations were preliminarily treated with 48% hydrofluoric acid, a treatment known to release polysaccharides that are covalently linked to peptidoglycan (33).
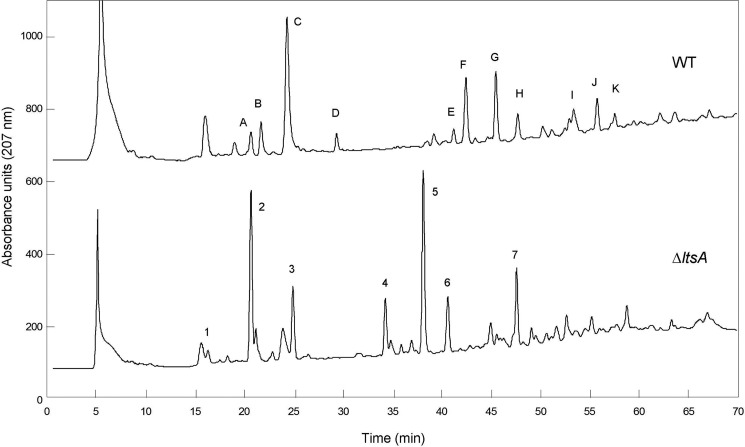
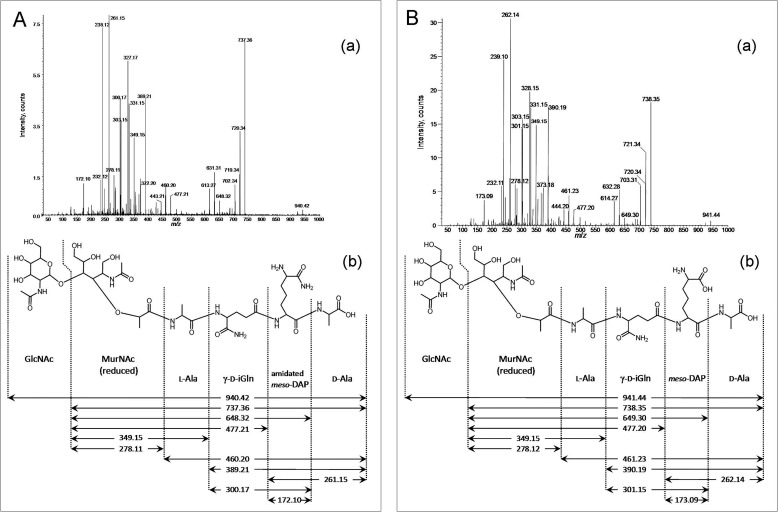
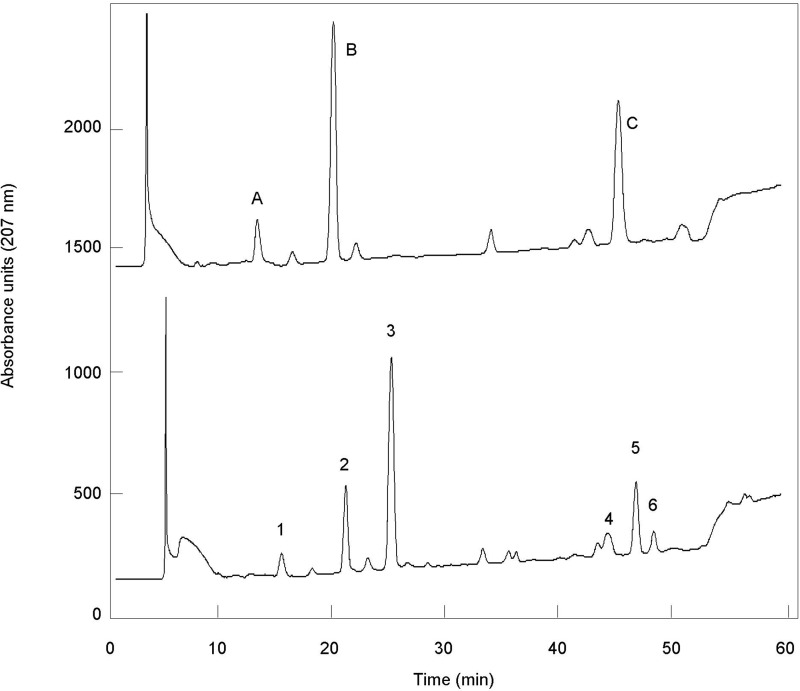
To further analyze the fine structure of the peptidoglycan from these two strains, the MAPc preparations were digested by specific muramidases, and the resulting muropeptides were separated by HPLC. The elution profiles appeared completely different between the wild-type and ΔltsA strains and revealed a characteristic general displacement toward the lower retention times of many of the peaks for the ΔltsA mutant, as compared with the parental strain, consistent with a loss of amidation in the peptidoglycan peptide chains (Fig. 4). The main muropeptides were collected, further purified, and desalted by HPLC, and their composition (amino acids and amino sugars) and molecular mass were determined with the Hitachi amino acid analyzer and the MALDI-TOF mass spectrometer, respectively. The data reported in Table 1 showed that muropeptides from the parental strain mainly contained two amide groups per stem peptide, i.e. two and four in monomers and dimers, respectively. Thus, the two potential amidation sites existing in peptidoglycan, namely the α- and ϵ-carboxyl groups of d-Glu and meso-DAP residues, respectively, were both amidated in C. glutamicum peptidoglycan, as observed previously in C. jeikeium (12). In contrast, only a single amidation per stem peptide was detected in muropeptides from the ΔltsA mutant (Table 1). Tandem mass spectrometry performed on muropeptides isolated from the parental and ΔltsA strains showed that although both the α- and ϵ-carboxyl groups of d-Glu and meso-DAP were amidated in the wild-type strain, only that of d-Glu was amidated in the mutant strain (Fig. 5). These results demonstrated that LtsA was required for amidation of meso-DAP in the peptidoglycan of C. glutamicum.
FIGURE 4.
HPLC analysis of peptidoglycan fragments (muropeptides) generated by digestion of peptidoglycan from wild-type (WT) and ΔltsA C. glutamicum strains with muramidases (lysozyme and mutanolysin). Muropeptides were reduced by sodium borohydride and separated by HPLC on a 3-μm ODS-Hypersil column (4.6 × 250 mm), using a gradient of methanol (from 0 to 25% in 120 min) in 50 mm sodium phosphate buffer, pH 4.5, at a flow rate of 0.5 ml/min. mAU, absorbance unit × 103 at 207 nm. Their identity is indicated in Table 1.
TABLE 1.
Mass spectrometry analysis of C. glutamicum peptidoglycan muropeptides
Muropeptides resulting from the digestion of peptidoglycan purified from the wild-type or ΔltsA C. glutamicum strain were reduced with sodium borohydride, purified by HPLC (Fig. 4) and analyzed by MALDI-TOF mass spectrometry as described in the “Experimental Procedures.”
| Peak no. (see Fig. 4) | Muropeptides | MALDI-TOF m/z [M + H]+ | Calculated monoisotopic molecular mass |
|---|---|---|---|
| Wild-type C. glutamicum strain | |||
| Monomers | |||
| A | Tri (2 NH2) | 869.3 | 868.4 |
| B | Tetra(1 NH2) | 941.6 | 940.4 |
| C | Tetra(2 NH2) | 940.6 | 939.4 |
| D | Penta(2 NH2) | 1011.5 | 1010.5 |
| Dimersa | |||
| E | Tetra-tri(4 NH2) | 1791.1 | 1789.8 |
| F | Tetra-tetra(3 NH2) | 1863.2 | 1861.9 |
| G | Tetra-tetra(4 NH2) | 1862.0 | 1860.9 |
| H | Tetra-penta(4 NH2) | 1933.4 | 1931.9 |
| Trimersa | |||
| I | Tetra-tetra-tetra(5 NH2) | 2784.4 | 2783.3 |
| J | Tetra-tetra-tetra(6 NH2) | 2783.8 | 2782.3 |
| K | Tetra-tetra-penta(6 NH2) | 2854.0 | 2853.3 |
| ΔltsA C. glutamicum strain | |||
| Monomers | |||
| 1 | Tri(1 NH2) | 870.5 | 869.4 |
| 2 | Tetra(1 NH2) | 941.6 | 940.4 |
| 3 | Penta(1 NH2) | 1012.6 | 1011.5 |
| Dimersa | |||
| 4 | Tetra-tri(2 NH2) | 1793.1 | 1791.8 |
| 5 | Tetra-tetra(2 NH2) | 1863.9 | 1862.9 |
| 6 | Tetra-penta(2 NH2) | 1935.1 | 1933.9 |
| Trimersa | |||
| 7 | Tetra-tetra-tetra(3 NH2) | 2786.6 | 2785.2 |
a The “donor” or “acceptor” nature of peptide chains (tri, tetra, or penta) participating in the cross-links within these different dimers and trimers was not determined. The total number of amidated residues (×NH2) present in these muropeptides is indicated in parentheses.
FIGURE 5.
MS-MS analysis of the main monomer muropeptides from wild-type and ΔltsA C. glutamicum strains. A, fragmentation of a reduced disaccharide tetrapeptide containing an amidated DAP residue. This muropeptide was isolated from wild-type C. glutamicum strain ATCC 13032 (peak C in Fig. 4). Panel a, fragmentation of the ion at m/z 940.6; panel b, inferred structure. B, fragmentation of a reduced disaccharide tetrapeptide containing a nonamidated DAP residue. This muropeptide was isolated from ΔltsA C. glutamicum strain (peak 2 in Fig. 4). Panel a, fragmentation of the ion at m/z 941.6; panel b, inferred structure.
Integration of peaks from the HPLC profiles shown in Fig. 4 indicated that ∼80% of DAP residues were amidated in the peptidoglycan of the wild-type strain (Table 2). Mass spectrometry analyses of the main monomers, dimers, and trimers derived from the ΔltsA peptidoglycan showed that they contained one, two, and three amidations, respectively. Although the presence of minor muropeptides totally lacking amidation cannot be excluded, this result showed that a major part (if not all) of the residues of d-Glu were amidated in the peptidoglycan of C. glutamicum. A calculation of the ratios between the main peaks of monomers and dimers observed on the HPLC profiles (Fig. 4) showed that the extent of peptide chain cross-linking was quite similar in the peptidoglycan from wild-type and ΔltsA strains (Table 2). This indicated that the peptidoglycan polymerases that catalyze this transpeptidation reaction accepted nonamidated DAP-containing peptide stems as substrates.
TABLE 2.
Effect of the expression of LtsACg on DAP amidation and cross-linking of the peptidoglycan in C. glutamicum and E. coli cells
Effects of LtsAcg Expression in E. coli
The C. glutamicum ltsA gene was cloned in pET vectors allowing expression of the protein in either the wild-type (pMLD288) or His6-tagged (pMLD290) form. When the expression of LtsA from these plasmids was induced in exponentially growing E. coli C43(DE3)(pLysS) cells by addition of 1 mm IPTG, an arrest of growth was observed ∼90 min later, followed by cell lysis (Fig. 6). This phenotype, suggesting a loss of cell wall integrity, was likely due to an interference with peptidoglycan biosynthesis. To confirm this assumption, the peptidoglycan content and structure were analyzed in these cells. The amounts of this polymer extracted from IPTG-induced C43 cells, carrying either the empty vector or the pMLD288 plasmid (LtsA-expressing cells were harvested when the first effects on cell growth were observed), were similar, i.e. 5.8 and 5.3 μmol of DAP/liter of culture at OD600 = 1, respectively. The HPLC profiles of muropeptides obtained by digestion of the purified peptidoglycans by muramidases revealed significant differences between the ltsA-expressing and parental E. coli strains (Fig. 7). In particular, production of LtsA led to additional peaks corresponding to new monomers and dimers with increased retention times, as expected for an increased hydrophobicity resulting from amidation of carboxyl groups. The main muropeptides were purified, and their composition and molecular mass were determined. The results shown in Table 3 revealed a decrease in the mass of muropeptides by 1 dalton (monomers) or 1 or 2 daltons (dimers) lower as compared with the classical ones, as expected for the amidation of one carboxyl group per stem peptide. The amino acid and amino sugar compositions of these muropeptides were otherwise identical.
FIGURE 6.
Bacteriolytic effect of expression of LtsA enzyme from C. glutamicum in E. coli cells. C43(DE3)(pLysS) cells carrying either the plasmid vector pET2160 (○), the pMLD288 plasmid expressing wild-type LtsACg (●), or the pMLD290 plasmid expressing His-tagged LtsACg (■) were grown at 37 °C in 2YT medium. When the OD600 reached 0.2, expression of the LtsA protein was induced by addition of 1 mm IPTG. An arrest of growth followed by cell lysis was observed about 90–120 min later.
FIGURE 7.
HPLC analysis of muropeptides released by digestion of peptidoglycan from wild-type and LtsACg-expressing E. coli cells with muramidases (lysozyme and mutanolysin). See the legend of Fig. 4 for details on HPLC conditions. mAU, absorbance unit × 103 at 207 nm. The identity of the muropeptides is indicated in Table 3.
TABLE 3.
Mass spectrometry analysis of the main E. coli peptidoglycan muropeptides
Muropeptides resulting from the digestion of peptidoglycan from E. coli cells expressing or not the LtsACg protein were reduced with sodium borohydride, purified by HPLC (Fig. 7), and analyzed by MALDI-TOF mass spectrometry as described under “Experimental Procedures.” The total number of amidated residues (×NH2) present in these muropeptides is indicated in parentheses.
| Peak no. (see Fig. 7) | Muropeptides | MALDI-TOF m/z [M + H]+ | Calculated monoisotopic molecular mass |
|---|---|---|---|
| Wild-type E. coli strain | |||
| A | Tri | 871.5 | 870.4 |
| B | Tetra | 942.5 | 941.4 |
| C | Tetra-tetra | 1866.0 | 1864.9 |
| LtsA-expressing E. coli strain | |||
| 1 | Tri | 871.4 | 870.4 |
| 2 | Tetra | 942.6 | 941.4 |
| 3 | Tetra(1 NH2) | 941.7 | 940.4 |
| 4 | Tetra-tetra | 1866.3 | 1864.9 |
| 5 | Tetra-tetra(1 NH2) | 1865.3 | 1863.9 |
| 6 | Tetra-tetra(2 NH2) | 1864.0 | 1862.9 |
Integration of peaks from the HPLC profiles showed that incorporation of amidated DAP had massively occurred in the E. coli cell wall polymer following LtsACg expression. It was estimated at 70 and 45% in the main monomer (tetra) and dimer (tetra-tetra), respectively (Table 2). These results confirmed the essential role of LtsA in DAP amidation and showed that implementation of this peptidoglycan modification in E. coli cells was tolerated to some extent, but it rapidly turned out to be toxic.
Analysis of Peptidoglycan Nucleotide Precursors
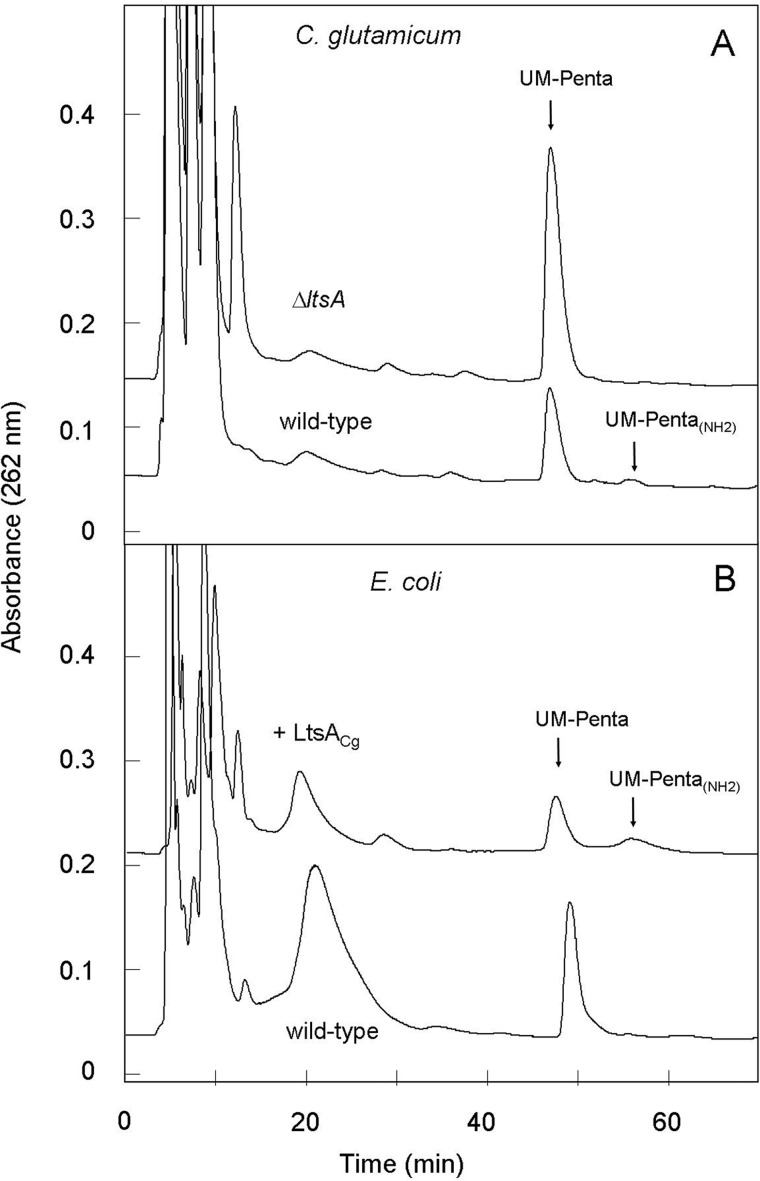
To localize the step of the peptidoglycan pathway where the LtsA-dependent DAP amidation reaction occurs, peptidoglycan nucleotide precursors from both E. coli and C. glutamicum cells, expressing or not LtsACg, were extracted and analyzed using previously described procedures (31, 42). In all cases, the HPLC profiles revealed the presence of a main peak corresponding to the classical UDP-MurNAc pentapeptide precursor (UDP-MurNAc-l-Ala-γ-d-Glu-meso-DAP-d-Ala-d-Ala) that does not contain any amidated residue, as confirmed by mass spectrometry (Fig. 8). The pool of this precursor did not change significantly following induction of LtsA expression in E. coli (Fig. 8B), and quite similar levels were observed in the wild-type and ΔltsA mutant C. glutamicum strains (Fig. 8A). Interestingly, a very small peak of compound eluted at a higher retention time, which could correspond to an amidated form of this precursor, was detected in only two of our extracts as follows: those prepared from LtsACg-expressing E. coli cells and from wild-type C. glutamicum cells (Fig. 8). Its identification as authentic amidated UDP-MurNAc pentapeptide was confirmed by mass spectrometry. Its very low pool level, as compared with the nonamidated precursor, strongly suggested that the LtsACg-dependent amidation reaction should mainly occur downstream in the pathway, most likely at the level of the peptidoglycan lipid intermediates.
FIGURE 8.
Analysis of the pools of amidated and nonamidated forms of the peptidoglycan UDP-MurNAc pentapeptide nucleotide precursor in C. glutamicum (A) and E. coli (B) strains. Nucleotide precursors were extracted from exponentially growing cells as described under “Experimental Procedures.” Aliquots were analyzed by HPLC on a column of μ-Bondapak C18 (7.8 × 300 mm). Elution at 3 ml/min was with 50 mm ammonium formate for 15 min at pH 3.35, followed by a gradient of pH, from 3.35 to 4.75, applied between 15 and 50 min.
In Vitro Enzymatic Activity of LtsA
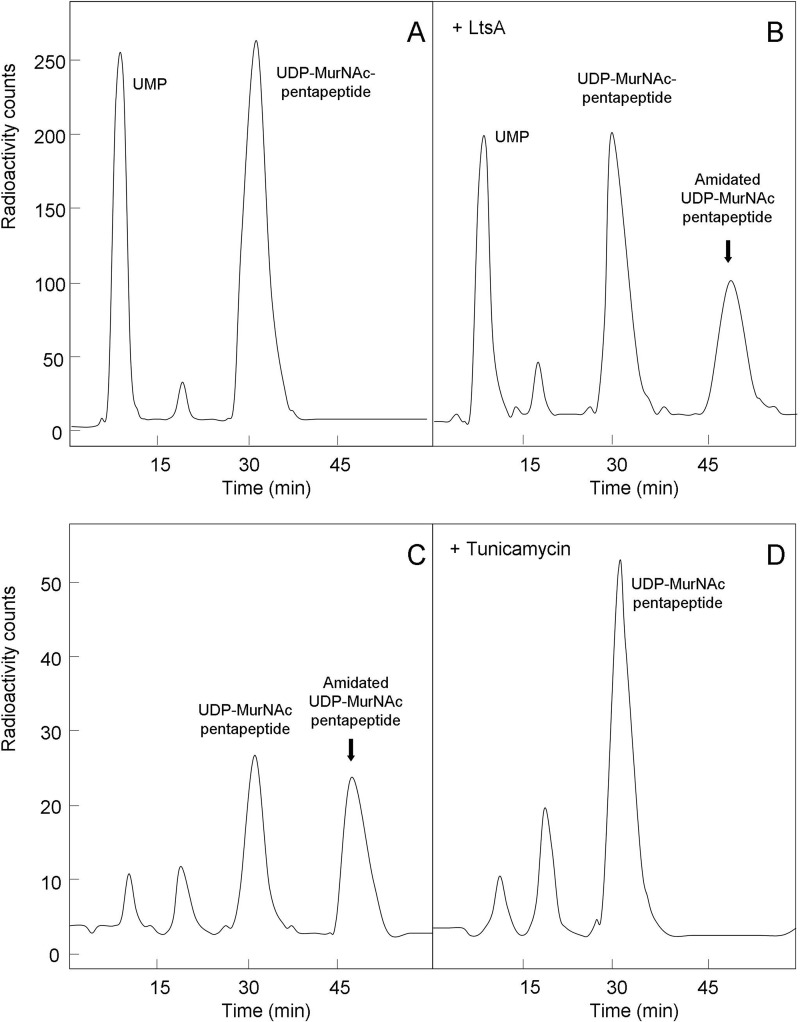
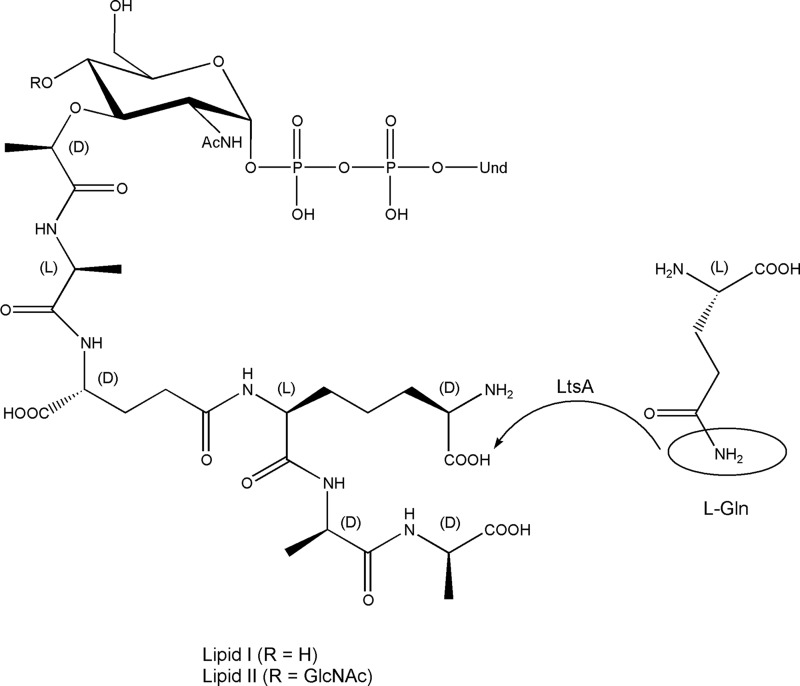
Attempts to purify the His6-tagged LtsACg protein from E. coli cell extracts were made. As the expression of this protein was toxic and led to cell lysis, C43(pLysS)(pMLD290) cells were induced with IPTG when entering the stationary phase. Whatever the induction conditions (3 h, overnight) and temperature (15–37 °C) used for expression, no significant overproduction of a protein species was detectable in cell extracts by SDS-PAGE. Only a faint band that could correspond to LtsA (predicted molecular mass of 72.4 kDa) was observed in both the membrane and soluble fractions. As our previous results suggested that LtsA should preferentially use membrane-associated peptidoglycan lipid intermediates, rather than nucleotide precursors as substrates, enzymatic assays were performed with the membrane fraction. The latter fraction indeed contains the MraY and MurG enzymes and free undecaprenyl-phosphate (C55-P) that can be used to generate these lipid intermediates, namely C55-PP-MurNAc pentapeptide (lipid I) and C55-PP-MurNAc(pentapeptide)-GlcNAc (lipid II), to analyze their possible modification by LtsA. Interestingly, the reaction catalyzed by MraY, i.e. the formation of lipid I from C55-P and UDP-MurNAc pentapeptide, is reversible as follows: C55-P + UDP-MurNAc pentapeptide 〈=〉 C55-PP-MurNAc pentapeptide + UMP.
This property has been earlier used to develop an “exchange assay” for MraY, in which a radiolabeled UMP is introduced into UDP-MurNAc pentapeptide (39, 44). As shown in Fig. 9A, incubation of membranes from control E. coli cells with UDP-MurNAc pentapeptide and [14C]UMP indeed resulted in the appearance of [14C]UDP-MurNAc pentapeptide. When this experiment was performed with membranes prepared from LtsA-expressing cells, the same exchange reaction occurred, but an additional peak of radiolabeled compound was observed on the HPLC profile (Fig. 9B), which was then identified as amidated UDP-MurNAc pentapeptide (co-migration with an authentic standard, mass spectrometry analysis). To identify the donor of amine used in this amidation reaction, membranes were extensively washed, and the exchange assay was performed in the presence of glutamine, asparagine, or ammonium sulfate. Although the exchange reaction similarly occurred in all conditions, the coupled amidation reaction was only observed when either glutamine or ammonium sulfate was present (data not shown). These in vitro results clearly confirmed that LtsA exhibited an amidotransferase activity. They also indicated that at least part of this protein has remained associated to the E. coli cell membranes following extraction and washing procedures.
FIGURE 9.
In vitro LtsA glutamine amidotransferase activity assays. A and B, MraY-catalyzed reaction of exchange between [14C]UMP and the UMP moiety of UDP-MurNAc pentapeptide was assayed in membrane fractions prepared from control (A) or LtsACg-expressing (B) E. coli cells. Reaction mixtures containing as substrates C55-P (provided by membranes), 14C-radiolabeled UMP, UDP-MurNAc pentapeptide, ATP, and glutamine were incubated with membrane extracts for 30 min at 37 °C. Amidated UDP-MurNAc pentapeptide was formed only when LtsA enzyme was present. C and D, radiolabeled UDP-MurNAc pentapeptide was incubated with the membrane extract from LtsACg-expressing E. coli cells, ATP and glutamine, in the absence (C) or presence (D) of tunicamycin (an MraY inhibitor). Amidated UDP-MurNAc pentapeptide was formed only when MraY was functional, demonstrating that LtsA accepts lipid I but not UDP-MurNAc pentapeptide as a substrate. In all cases, the radiolabeled substrate and products were separated by HPLC as described under “Experimental Procedures.”
As amidated UDP-MurNAc pentapeptide detected in the MraY-LtsA assay may originate either from UDP-MurNAc pentapeptide or from lipid I (or both), the identity of the acceptor substrate(s) used by LtsA in vivo needed to be determined. To test whether the nucleotide precursor was a substrate or not of the enzyme, radiolabeled UDP-MurNAc pentapeptide was incubated with the LtsA-containing membrane extract, ATP and glutamine, in the presence or absence of tunicamycin, an MraY inhibitor. As shown in Fig. 9, C and D, amidated UDP-MurNAc pentapeptide was formed only when the exchange reaction of MraY was allowed to proceed, i.e. in the absence of tunicamycin. It was thus clear that the amidated nucleotide precursor originated exclusively from amidated lipid I, thereby demonstrating that the LtsACg enzyme essentially worked on lipid intermediates and did not accept UDP-MurNAc pentapeptide as a substrate. This LtsA-catalyzed amidotransferase reaction is depicted in Fig. 10.
FIGURE 10.
LtsA-catalyzed amidotransferase reaction. LtsA catalyzes the transfer of an NH2 group between l-glutamine (donor) and lipid I or II (acceptor). The NH2 group is transferred to the carboxyl function linked to the d-carbon of the meso-DAP residue, thereby resulting in the formation of an amidated meso-DAP residue in the peptide stem. Ammonium sulfate can also act as the donor. Und, undecaprenyl.
Other assays were then developed that were aimed at determining the substrate specificity of LtsA for the peptide chain length. DAP-containing UDP-MurNAc tripeptide and UDP-MurNAc tetrapeptide were tested in the exchange assay, in place of the pentapeptide nucleotide precursor. Although these two compounds were substrates of MraY and thus became progressively radiolabeled in the exchange reaction, no peaks corresponding to amidated forms of these UDP-MurNAc peptides could be detected on the HPLC profiles (data not shown). This demonstrated that the LtsACg amidotransferase was specific for peptidoglycan precursors carrying a pentapeptide moiety.
The glutaminase activity was also assayed in these membrane preparations. Hydrolysis of glutamine into glutamic acid was observed with membranes prepared from LtsACg-expressing E. coli cells but not with membranes from control cells. The glutaminase activity we detected in this crude membrane extract was around 100 nmol/min/mg of protein. It did not require ATP and was slightly increased (by ∼20%) when 1 mm UDP-MurNAc pentapeptide was added to the reaction mixture. This result suggested that the presence of an amine acceptor, i.e. the lipid I that is generated by MraY in the presence of this nucleotide precursor, was not required for the glutaminase activity of LtsA but stimulated it to some extent. As the amount of LtsA protein present in this membrane fraction is not known, specific activity values could not be more precisely determined.
Effect of DAP Amidation on Lysozyme Sensitivity of C. glutamicum Cells and Purified Peptidoglycan
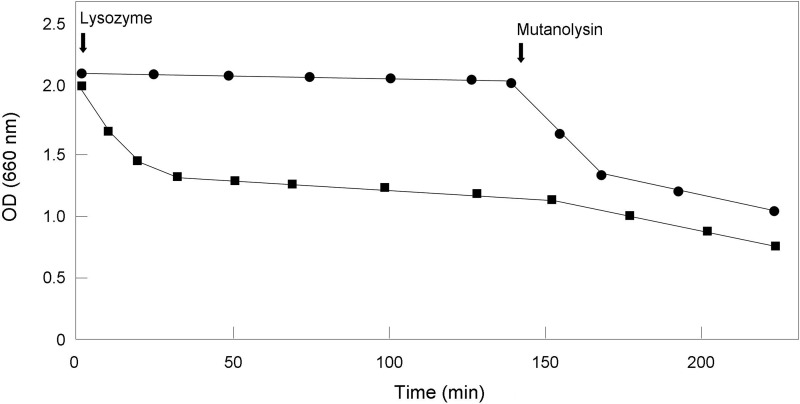
C. glutamicum is known to be highly resistant to lysozyme. Indeed, a 200 μg/ml concentration was necessary to totally inhibit the growth of the wild-type strain in liquid culture. In comparison, as little as 0.2 μg/ml lysozyme inhibited growth of the ΔltsA mutant in the same conditions. The increased sensitivity of the ΔltsA mutant could be the result of a dramatic increase of the cell wall permeability to lysozyme or more likely to the modification of the peptidoglycan structure we observed, i.e. the absence of DAP amidation. We thus directly tested the effect of lysozyme on purified peptidoglycan (after treatment of the MAPc complex with hydrofluoric acid, as described above). As shown in Fig. 11, the peptidoglycan of the parental strain was digested readily by mutanolysin but was not hydrolyzed, or only very slowly, by lysozyme, whereas both enzymes hydrolyzed the peptidoglycan from the ΔltsA strain quite efficiently. These results showed that the presence of DAPNH2 residues in the peptidoglycan inhibited in some way the hydrolase activity of lysozyme, a finding perfectly consistent with the higher resistance of the C. glutamicum wild-type cells toward this enzyme.
FIGURE 11.
Lysozyme sensitivity of peptidoglycan purified from wild-type and ΔltsA mutant C. glutamicum strains. Purified peptidoglycan (∼300 μg) from wild-type (circles) and mutant (squares) strains was incubated at 37 °C in 2 ml of 25 mm potassium phosphate buffer, pH 7.8. Lysozyme (75 μg/ml, final concentration) was added at t = 0 and peptidoglycan digestion was followed by measuring the decrease of absorbance at 660 nm. Mutanolysin (50 units) was subsequently added at t = 150 min.
Antibiotic Sensibility of the ΔltsA Mutant Strain
The in vitro experiments described above clearly established that the hyper-susceptibility to lysozyme of the ΔltsA mutant was related to the lack of DAP amidation in its peptidoglycan. However, it could be associated in vivo with an enhanced permeability of the outer membrane to lysozyme. To determine whether the cell wall permeability of the mutant strain was different from that of the wild-type strain, the effects of different antibiotics were tested on the growth of these two strains. The sensitivity of the ΔltsA mutant to novobiocin, erythromycin, d-cycloserine, vancomycin, and bacitracin was not significantly different from that of the wild-type strain, and only a slightly lower IC50 value was observed for chloramphenicol. Surprisingly, more striking differences were observed when antibiotics of the β-lactam family were tested; the mutant strain exhibited hyper-sensitivity to all members of this family that we tested, namely ampicillin, carbenicillin, ceftazidim, and imipenem (Table 4). Because the sensitivity of the mutant strain toward the other classes of antibiotics we tested was not modified (in particular vancomycin, bacitracin, and d-cycloserine, which also target peptidoglycan biosynthesis), this hyper-sensitivity to β-lactam antibiotics could therefore be considered as a specific and direct effect of the peptidoglycan structure modification (loss of DAP amidation) rather than a manifestation of a global cell wall permeability alteration.
TABLE 4.
IC50 (μg/ml) of various antibiotics for C. glutamicum wild-type and ΔltsA strains
The IC50 value corresponds to the drug concentration for which 50% inhibition of bacterial cell growth is observed. Each value is the mean of at least three independent experiments and is given ± S.D.
| Wild type (ATCC 13032) | ΔltsA | |
|---|---|---|
| Chloramphenicol | 2.6 ± 0.4 | 1.4 ± 0.4 |
| Ampicillin | 6.0 ± 0.1 | 0.24 ± 0.04 |
| Carbenicillin | 2.3 ± 0.2 | 0.23 ± 0.06 |
| Ceftazidim | 27 ± 3.6 | 0.10 ± 0.03 |
| Imipenem | 0.038 ± 0.001 | 0.0080 ± 0.0001 |
Discussion
In this study, we describe the characterization of the function of LtsA in C. glutamicum and the consequences of its inactivation on the physiology of this bacterial species. LtsA had been identified years ago for its ability to confer resistance to lysozyme to Corynebacteriales, and it was suspected to amidate a cell wall component (20). Its function, however, remained to be elucidated. Very recently, we found the ltsA gene within a transposon insertion library that was screened for mutants impaired in cell wall integrity.4 Based on the recent results of Bernard et al. (16) who showed that AsnB1, an LtsA orthologue, was responsible for DAP amidation in the L. plantarum peptidoglycan, we decided to reinvestigate the role of LtsA in C. glutamicum cell wall biogenesis. For that purpose, we inactivated the ltsA gene in this bacterial species. Unlike asnB1, which in L. plantarum is essential and co-transcribed with murE (encoding the peptidoglycan UDP-MurNAc-l-Ala-d-Glu:meso-DAP ligase) and thrA1 (encoding a putative aspartokinase) (16), ltsA from C. glutamicum is predicted to be monocistronic and could be replaced by allelic exchange without any difficulty. As compared with the wild-type strain, the resulting mutant showed growth defects and an altered morphology for some bacteria but none of the important septation defects that the L. plantarum asnB1 conditional mutant exhibited (16). Cell wall structural analyses showed that d-Glu and meso-DAP were both amidated in the peptidoglycan of the wild-type C. glutamicum strain, at nearly 100 and 80%, respectively. Only d-Glu was found to be amidated in the peptidoglycan of the ΔltsA mutant, which demonstrated the specific role of LtsA in DAP amidation. The extent of d-Glu amidation was unchanged in the mutant (almost 100%), indicating that DAP amidation was not a prerequisite for the modification of this other peptide stem residue.
Most (80%) but not all of the DAP residues were found amidated in the C. glutamicum peptidoglycan, raising the question of a potential role of the nonamidated DAP residues in peptidoglycan biosynthesis or bacterial physiology. Almost 100% of the DAP residues are amidated in the peptidoglycan of C. jeikeium (12). Amidation of DAP was also found to be extensive in Mycobacterium species, but not total, and this extent may slightly differ in the function of the growth phase (8, 11, 13). Interestingly, in the M. tuberculosis polymer, only nonamidated DAP residues were found to be linked via their ϵ-amino group to glycine residues (11), a modification whose significance remains to be established. Amidation of DAP was also shown to be a major feature of B. subtilis peptidoglycan (45). Only a few minor nonamidated muropeptides were detected in this species, whose role in spore peptidoglycan formation has been suggested (46). Only the latter nonamidated fragments were detected by the intracellular Nod1 innate immunity recognition protein (17). The presence of DAPNH2 residues was also reported recently in the peptidoglycan of C. difficile (42). Unsuspected until now, this peptidoglycan modification was fortuitously discovered and shown to occur only in very specific conditions, i.e. following induction by vancomycin of the expression of a cryptic vanG cluster present in the C. difficile genome (42).
Although it contributes to a global change of peptidoglycan chemical properties, the physiological role of peptidoglycan amidation remains unclear. It neutralizes the free acidic carboxyl groups present in the peptide chains and therefore reduces the charge density in the cell wall. Lack of DAP amidation in C. glutamicum resulted in a reduced cell resistance toward lysozyme and β-lactam antibiotics. The same increase of cell sensitivity toward these molecules and some defensins was observed when d-Glu amidation was inhibited in Staphylococcus aureus (47, 48). In Lactococcus lactis, amidation of interpeptide d-Asp residues was shown to increase cell resistance against the activities of endogenous autolysins, lysozyme and nisin (49). As shown in this study, not only the whole ΔltsA cells but also the peptidoglycan polymer purified from them exhibited a decreased resistance toward lysozyme, demonstrating that DAP amidation directly impacted the muramidase activity of lysozyme and therefore constituted a key factor mediating lysozyme resistance in Corynebacteriales. As we did not observe any other modification in the composition of the ΔltsA peptidoglycan nor in its cross-linking extent, this difference in sensitivity could be the result of a change in the tridimensional structure of this polymer arising from the lack of amidation that could have facilitated the lysozyme accessibility to its target (i.e. the β-1→4 glycosidic bond between the C-1 carbon of MurNAc and the C-4 carbon of GlcNAc). It also could result from a change in the ratio between the two types of peptidoglycan cross-links (4→3 and 3→3), which had been proposed to influence the flexibility/rigidity of the polymer (50). A similar increase in lysozyme sensitivity was observed with peptidoglycan purified from S. aureus strains in which d-Glu amidation was conditionally expressed and modulated (48). The modification of the peptide charge resulting from the amidation of either of these residues could have a repulsive effect on lysozyme and inhibit the peptidoglycan binding and/or peptidoglycan hydrolyzing activity of this enzyme. As no three-dimensional structure of lysozyme in complex with a peptidoglycan fragment has been solved to date, whether and how peptide stems of this cell wall polymer interact with the lysozyme active site remains to be determined. Other cell wall modifications encountered mainly in Gram-positive species are known to be implicated in lysozyme resistance mechanisms, namely the O-acetylation at C-6 position of MurNAc (51, 52), de-N-acetylation of amino sugars (52, 53), and the presence of peptidoglycan covalently linked polysaccharides, such as teichoic acids (54, 55).
In C. glutamicum, the peptidoglycan polymer is attached to another polysaccharidic polymer, the arabinogalactan. The decrease of the galactosamine to glucosamine ratio we observed in MAP complex extracted from the ΔltsA C. glutamicum strain suggested that the absence of DAP amidation might have reduced the amount of peptidoglycan-bound arabinogalactan in this mutant. Further work is required to validate this hypothesis.
The impact of the ltsA mutation on C. glutamicum growth rate and morphology suggested that amidated peptidoglycan precursors may provide better substrates for proteins that catalyze peptidoglycan biosynthesis and cell division. The lack of the amide group may indeed create an unbalance between the synthetic and the hydrolytic machineries of the cell. Moreover, as mentioned above, some changes in the types of cross-links could also occur in the ΔltsA peptidoglycan. Indeed, both d,d-transpeptidases (PBP) and l,d-transpeptidases (Ldt), which specifically form 4→3 and 3→3 cross-links, respectively, are known to participate in peptidoglycan polymerization in this and other Corynebacteriales species (11–13). The increased susceptibility of the ltsA mutant toward β-lactams is interesting in this respect, as these antibiotics are known to more specifically inhibit PBPs than Ldts. Further work is needed to determine the real impact of DAP amidation on the activity of these transpeptidases. Heterologous expression of LtsACg in E. coli cells provided some information on the specificity of the host transpeptidases. Indeed, the detection of a tetra(NH2)-tetra(NH2) muropeptide dimer having its two DAP residues amidated suggested that some of the E. coli PBPs could accept, at least to some extent, DAPNH2-containing peptides both as acceptor and donor chains for the formation of cross-links. However, the lower incorporation in dimers as compared with monomers reflected a reduced efficiency of these enzymes for amidated peptide chains. The decrease of overall peptide cross-linking observed in the peptidoglycan of LtsACg-expressing E. coli cells, 22 versus 29% in control cells, was consistent with this assumption. As LtsA expression resulted in an arrest of growth and cell lysis, this massive but not total incorporation of DAPNH2 in the E. coli cell peptidoglycan (70% in monomers and 45% in dimers) likely corresponded to the maximal level compatible with cell viability. A total replacement was therefore expected to be lethal for E. coli cells.
LtsA proteins exhibit significant sequence similarity with members of the glutamine amidotransferase family. The LtsA protein from R. erythropolis was earlier shown to display a glutaminase activity that was stimulated in the presence of a lysozyme-treated cell wall extract, suggesting that some cell wall component(s) may act as amide acceptor(s). It was also shown not to function as an asparagine synthase (20). LtsA from C. glutamicum is demonstrated here to catalyze amidotransfer from glutamine to the ϵ-carboxyl group of DAP present in peptidoglycan precursors, thereby mediating the incorporation of DAPNH2 into this cell wall polymer. In vitro assays showed that LtsACg preferentially used the lipid intermediates as substrates and displayed a high specificity for a pentapeptide chain-containing precursor, i.e. did not amidate tripeptide and tetrapeptide derivatives of lipid I. This was consistent with the observation that only trace amounts of amidated UDP-MurNAc peptide precursors could be detected in LtsACg-expressing E. coli cells, although huge amounts of DAPNH2 were incorporated into the polymer in these conditions. The low pool of amidated UDP-MurNAc pentapeptide detected in LtsA-expressing C. glutamicum and E. coli cells could either result from a very low activity of LtsA on the nucleotide precursor or be due to the reversibility of the MraY-catalyzed reaction. Whether the same specificity (lipid intermediates versus nucleotide precursors) is shared by LtsA homologues from other bacterial species is not known. Mitani et al. (20) showed earlier that ltsA genes from M. tuberculosis and B. subtilis could complement the lysozyme-sensitive phenotype of their C. glutamicum ltsA mutant. We also observed that expression of ltsA genes from B. subtilis and Enterococcus faecium in E. coli cells resulted in an arrest of growth and cell lysis, as observed here with the ltsACg gene,5 suggesting that the latter LtsA orthologues all have the same function. Further work is now required to characterize this enzyme and its kinetic properties in more detail. Unfortunately, expression of LtsACg was very poor in E. coli, and amounts of protein that could be purified from the soluble fraction were quite low and did not show significant in vitro activity. Mitani et al. (20) mentioned that LtsA protein from R. erythropolis was extremely labile and should be tested immediately after purification (20), as also observed previously for AsnBEc (56). Although LtsA is not predicted to be a membrane protein, we here took advantage that part of the LtsACg protein remained associated to the E. coli cell membranes to conveniently assay its activity and main properties.
Once the last nucleotide precursor of this pathway (UDP-MurNAc pentapeptide) and the lipid intermediates I and II have been synthesized, several additional structural modifications may occur in bacteria before lipid II is translocated to the periplasmic side of the membrane where peptidoglycan polymerization reactions take place. These are the amidation of peptide chain residues (DAP, d-Glu, and d-Asp), the addition of supplementary amino acid residues (interpeptide bridge) by Fem transferases, and the N-glycolylation of muramic acid residues (52, 57). Depending on the bacterial species and the enzyme concerned, the latter modifications are introduced either on UDP-MurNAc pentapeptide, on the lipid intermediates I and II, or on both. Most if not all of these peptidoglycan structural modifications result in bacterial resistance to lysozyme, a major bacterial killing factor of the host innate immune system.
Very recently, genes and enzymes involved in peptidoglycan d-Glu amidation were identified in S. aureus (47, 48). Two proteins acting in concert and forming a physically stable complex were shown to be required for the latter modification, a glutamine amidotransferase-like protein (GatD) and a Mur ligase homologue (MurT). As shown in this study, only a single protein species, LtsA, was needed for the modification of DAP residues in C. glutamicum. Indeed, heterologous expression in E. coli of the sole LtsACg protein was sufficient to provoke a massive incorporation of DAPNH2 into the peptidoglycan of this species. Two genes encoding GatD and MurT homologues (cg0299 and cg0300, respectively) were detected in the C. glutamicum genome, which should therefore be responsible for peptidoglycan d-Glu amidation in this species. It will be interesting to determine whether this modification is essential for growth in C. glutamicum, as observed in S. aureus, or whether it is dispensable and could be totally abolished, alone or together with the DAP modification.
Acknowledgments
We thank Ahmed Bouhss and Nicolas Bayan for helpful discussions; Muriel Masi, Luis Augusto, and Martine Caroff for help in peptidoglycan purification procedures; and Magali Prigent from the microscopy platform of IGM for help in optical microscopy analyses.
This work was supported by grants from CNRS (UMR 8619 and UMR 8621), the University of Paris-Sud, and the Agence Nationale de la Recherche (Bactoprenyl Project ANR-11-BSV3-002).
C. de Sousa-d'Auria and C. Houssin, unpublished data.
D. Mengin-Lecreulx, unpublished data.
- MAPc
- mycoloyl-arabinogalactan-peptidoglycan complex
- DAP
- meso-2,6-diaminopimelic acid
- DAPNH2
- meso-DAP whose carboxyl group linked to the D carbon is amidated
- LtsACg
- LtsA from C. glutamicum
- Km
- kanamycin
- IPTG
- isopropyl β-d-thiogalactopyranoside.
References
- 1. Hoffmann C., Leis A., Niederweis M., Plitzko J. M., Engelhardt H. (2008) Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc. Natl. Acad. Sci. U.S.A. 105, 3963–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zuber B., Chami M., Houssin C., Dubochet J., Griffiths G., Daffé M. (2008) Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J. Bacteriol. 190, 5672–5680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Crick D. C., Mahapatra S., Brennan P. J. (2001) Biosynthesis of the arabinogalactan-peptidoglycan complex of Mycobacterium tuberculosis. Glycobiology 11, 107R–118R [DOI] [PubMed] [Google Scholar]
- 4. Puech V., Chami M., Lemassu A., Lanéelle M. A., Schiffler B., Gounon P., Bayan N., Benz R., Daffé M. (2001) Structure of the cell envelope of corynebacteria: importance of the non-covalently bound lipids in the formation of the cell wall permeability barrier and fracture plane. Microbiology 147, 1365–1382 [DOI] [PubMed] [Google Scholar]
- 5. McNeil M., Daffé M., Brennan P. J. (1990) Evidence for the nature of the link between the arabinogalactan and peptidoglycan of mycobacterial cell walls. J. Biol. Chem. 265, 18200–18206 [PubMed] [Google Scholar]
- 6. Schleifer K. H., Kandler O. (1972) Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36, 407–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Petit J. F., Adam A., Wietzerbin-Falszpan J., Lederer E., Ghuysen J. M. (1969) Chemical structure of the cell wall of Mycobacterium smegmatis. I. Isolation and partial characterization of the peptidoglycan. Biochem. Biophys. Res. Commun. 35, 478–485 [DOI] [PubMed] [Google Scholar]
- 8. Mahapatra S., Crick D. C., McNeil M. R., Brennan P. J. (2008) Unique structural features of the peptidoglycan of Mycobacterium leprae. J. Bacteriol. 190, 655–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gupta R., Lavollay M., Mainardi J. L., Arthur M., Bishai W. R., Lamichhane G. (2010) The Mycobacterium tuberculosis protein LdtMt2 is a nonclassical transpeptidase required for virulence and resistance to amoxicillin. Nat. Med. 16, 466–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kumar P., Arora K., Lloyd J. R., Lee I. Y., Nair V., Fischer E., Boshoff H. I., Barry C. E., 3rd (2012) Meropenem inhibits dd-carboxypeptidase activity in Mycobacterium tuberculosis. Mol. Microbiol. 86, 367–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lavollay M., Arthur M., Fourgeaud M., Dubost L., Marie A., Veziris N., Blanot D., Gutmann L., Mainardi J. L. (2008) The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by l,d-transpeptidation. J. Bacteriol. 190, 4360–4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lavollay M., Arthur M., Fourgeaud M., Dubost L., Marie A., Riegel P., Gutmann L., Mainardi J. L. (2009) The β-lactam-sensitive d,d-carboxypeptidase activity of Pbp4 controls the l,d- and d,d-transpeptidation pathways in Corynebacterium jeikeium. Mol. Microbiol. 74, 650–661 [DOI] [PubMed] [Google Scholar]
- 13. Lavollay M., Fourgeaud M., Herrmann J. L., Dubost L., Marie A., Gutmann L., Arthur M., Mainardi J. L. (2011) The peptidoglycan of Mycobacterium abscessus is predominantly cross-linked by l,d-transpeptidases. J. Bacteriol. 193, 778–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mahapatra S., Scherman H., Brennan P. J., Crick D. C. (2005) N Glycolylation of the nucleotide precursors of peptidoglycan biosynthesis of Mycobacterium spp. is altered by drug treatment. J. Bacteriol. 187, 2341–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mahapatra S., Yagi T., Belisle J. T., Espinosa B. J., Hill P. J., McNeil M. R., Brennan P. J., Crick D. C. (2005) Mycobacterial lipid II is composed of a complex mixture of modified muramyl and peptide moieties linked to decaprenyl phosphate. J. Bacteriol. 187, 2747–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bernard E., Rolain T., Courtin P., Hols P., Chapot-Chartier M. P. (2011) Identification of the amidotransferase AsnB1 as being responsible for meso-diaminopimelic acid amidation in Lactobacillus plantarum peptidoglycan. J. Bacteriol. 193, 6323–6330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Girardin S. E., Travassos L. H., Hervé M., Blanot D., Boneca I. G., Philpott D. J., Sansonetti P. J., Mengin-Lecreulx D. (2003) Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2. J. Biol. Chem. 278, 41702–41708 [DOI] [PubMed] [Google Scholar]
- 18. Slamti L., de Pedro M. A., Guichet E., Picardeau M. (2011) Deciphering morphological determinants of the helix-shaped Leptospira. J. Bacteriol. 193, 6266–6275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hirasawa T., Wachi M., Nagai K. (2000) A mutation in the Corynebacterium glutamicum ltsA gene causes susceptibility to lysozyme, temperature-sensitive growth, and l-glutamate production. J. Bacteriol. 182, 2696–2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mitani Y., Meng X., Kamagata Y., Tamura T. (2005) Characterization of LtsA from Rhodococcus erythropolis, an enzyme with glutamine amidotransferase activity. J. Bacteriol. 187, 2582–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Larsen T. M., Boehlein S. K., Schuster S. M., Richards N. G., Thoden J. B., Holden H. M., Rayment I. (1999) Three-dimensional structure of Escherichia coli asparagine synthetase B: a short journey from substrate to product. Biochemistry 38, 16146–16157 [DOI] [PubMed] [Google Scholar]
- 22. Boehlein S. K., Richards N. G., Schuster S. M. (1994) Glutamine-dependent nitrogen transfer in Escherichia coli asparagine synthetase B. Searching for the catalytic triad. J. Biol. Chem. 269, 7450–7457 [PubMed] [Google Scholar]
- 23. Ren H., Liu J. (2006) AsnB is involved in natural resistance of Mycobacterium smegmatis to multiple drugs. Antimicrob. Agents Chemother. 50, 250–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dusch N., Pühler A., Kalinowski J. (1999) Expression of the Corynebacterium glutamicum panD gene encoding l-aspartate-α-decarboxylase leads to pantothenate overproduction in Escherichia coli. Appl. Environ. Microbiol. 65, 1530–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Miller J. H. (1972) Experiments in Molecular Genetics, pp. 431–435, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 26. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 27. Dagert M., Ehrlich S. D. (1979) Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene 6, 23–28 [DOI] [PubMed] [Google Scholar]
- 28. Bonamy C., Guyonvarch A., Reyes O., David F., Leblon G. (1990) Interspecies electro-transformation in Corynebacteria. FEMS Microbiol. Lett. 54, 263–269 [DOI] [PubMed] [Google Scholar]
- 29. Portevin D., De Sousa-D'Auria C., Houssin C., Grimaldi C., Chami M., Daffé M., Guilhot C. (2004) A polyketide synthase catalyzes the last condensation step of mycolic acid biosynthesis in mycobacteria and related organisms. Proc. Natl. Acad. Sci. U.S.A. 101, 314–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Barreteau H., Bouhss A., Fourgeaud M., Mainardi J. L., Touzé T., Gérard F., Blanot D., Arthur M., Mengin-Lecreulx D. (2009) Human- and plant-pathogenic Pseudomonas species produce bacteriocins exhibiting colicin M-like hydrolase activity towards peptidoglycan precursors. J. Bacteriol. 191, 3657–3664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mengin-Lecreulx D., Flouret B., van Heijenoort J. (1982) Cytoplasmic steps of peptidoglycan synthesis in Escherichia coli. J. Bacteriol. 151, 1109–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mengin-Lecreulx D., van Heijenoort J. (1985) Effect of growth conditions on peptidoglycan content and cytoplasmic steps of its biosynthesis in Escherichia coli. J. Bacteriol. 163, 208–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jayatissa P. M., Rose A. H. (1976) Role of wall phosphomannan in flocculation of Saccharomyces cerevisiae. J. Gen. Microbiol. 96, 165–174 [DOI] [PubMed] [Google Scholar]
- 34. Glauner B. (1988) Separation and quantification of muropeptides with high-performance liquid chromatography. Anal. Biochem. 172, 451–464 [DOI] [PubMed] [Google Scholar]
- 35. Mengin-Lecreulx D., Flouret B., van Heijenoort J. (1983) Pool levels of UDP N-acetylglucosamine and UDP N-acetylglucosamine-enolpyruvate in Escherichia coli and correlation with peptidoglycan synthesis. J. Bacteriol. 154, 1284–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Arbeloa A., Hugonnet J. E., Sentilhes A. C., Josseaume N., Dubost L., Monsempes C., Blanot D., Brouard J. P., Arthur M. (2004) Synthesis of mosaic peptidoglycan cross-bridges by hybrid peptidoglycan assembly pathways in Gram-positive bacteria. J. Biol. Chem. 279, 41546–41556 [DOI] [PubMed] [Google Scholar]
- 37. Laemmli U. K., Favre M. (1973) Maturation of the head of bacteriophage T4. I. DNA packaging events. J. Mol. Biol. 80, 575–599 [DOI] [PubMed] [Google Scholar]
- 38. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 39. Mengin-Lecreulx D., Texier L., Rousseau M., van Heijenoort J. (1991) The murG gene of Escherichia coli codes for the UDP-N-acetylglucosamine:N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase involved in the membrane steps of peptidoglycan synthesis. J. Bacteriol. 173, 4625–4636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bouhss A., Crouvoisier M., Blanot D., Mengin-Lecreulx D. (2004) Purification and characterization of the bacterial MraY translocase catalyzing the first membrane step of peptidoglycan biosynthesis. J. Biol. Chem. 279, 29974–29980 [DOI] [PubMed] [Google Scholar]
- 41. El Ghachi M., Bouhss A., Barreteau H., Touzé T., Auger G., Blanot D., Mengin-Lecreulx D. (2006) Colicin M exerts its bacteriolytic effect via enzymatic degradation of undecaprenyl phosphate-linked peptidoglycan precursors. J. Biol. Chem. 281, 22761–22772 [DOI] [PubMed] [Google Scholar]
- 42. Ammam F., Meziane-Cherif D., Mengin-Lecreulx D., Blanot D., Patin D., Boneca I. G., Courvalin P., Lambert T., Candela T. (2013) The functional vanGCd cluster of Clostridium difficile does not confer vancomycin resistance. Mol. Microbiol. 89, 612–625 [DOI] [PubMed] [Google Scholar]
- 43. Marchand C. H., Salmeron C., Bou Raad R., Méniche X., Chami M., Masi M., Blanot D., Daffé M., Tropis M., Huc E., Le Maréchal P., Decottignies P., Bayan N. (2012) Biochemical disclosure of the mycolate outer membrane of Corynebacterium glutamicum. J. Bacteriol. 194, 587–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Geis A., Plapp R. (1978) Phospho-N-acetylmuramoyl pentapeptide-transferase of Escherichia coli K12. Properties of the membrane-bound and the extracted and partially purified enzyme. Biochim. Biophys. Acta 527, 414–424 [DOI] [PubMed] [Google Scholar]
- 45. Atrih A., Bacher G., Allmaier G., Williamson M. P., Foster S. J. (1999) Analysis of peptidoglycan structure from vegetative cells of Bacillus subtilis 168 and role of PBP 5 in peptidoglycan maturation. J. Bacteriol. 181, 3956–3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Atrih A., Zöllner P., Allmaier G., Williamson M. P., Foster S. J. (1998) Peptidoglycan structural dynamics during germination of Bacillus subtilis 168 endospores. J. Bacteriol. 180, 4603–4612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Münch D., Roemer T., Lee S. H., Engeser M., Sahl H. G., Schneider T. (2012) Identification and in vitro analysis of the GatD/MurT enzyme-complex catalyzing lipid II amidation in Staphylococcus aureus. PLoS Pathog. 8, e1002509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Figueiredo T. A., Sobral R. G., Ludovice A. M., Almeida J. M., Bui N. K., Vollmer W., de Lencastre H., Tomasz A. (2012) Identification of genetic determinants and enzymes involved with the amidation of glutamic acid residues in the peptidoglycan of Staphylococcus aureus. PLoS Pathog. 8, e1002508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Veiga P., Erkelenz M., Bernard E., Courtin P., Kulakauskas S., Chapot-Chartier M. P. (2009) Identification of the asparagine synthase responsible for d-Asp amidation in the Lactococcus lactis peptidoglycan interpeptide crossbridge. J. Bacteriol. 191, 3752–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Desmarais S. M., De Pedro M. A., Cava F., Huang K. C. (2013) Peptidoglycan at its peaks: how chromatographic analyses can reveal bacterial cell wall structure and assembly. Mol. Microbiol. 89, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bera A., Herbert S., Jakob A., Vollmer W., Götz F. (2005) Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol. Microbiol. 55, 778–787 [DOI] [PubMed] [Google Scholar]
- 52. Vollmer W. (2008) Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol Rev. 32, 287–306 [DOI] [PubMed] [Google Scholar]
- 53. Zipperle G. F., Jr., Ezzell J. W., Jr., Doyle R. J. (1984) Glucosamine substitution and muramidase susceptibility in Bacillus anthracis. Can. J. Microbiol. 30, 553–559 [DOI] [PubMed] [Google Scholar]
- 54. Bera A., Biswas R., Herbert S., Kulauzovic E., Weidenmaier C., Peschel A., Götz F. (2007) Influence of wall teichoic acid on lysozyme resistance in Staphylococcus aureus. J. Bacteriol. 189, 280–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mauck J., Glaser L. (1972) On the mode of in vivo assembly of the cell wall of Bacillus subtilis. J. Biol. Chem. 247, 1180–1187 [PubMed] [Google Scholar]
- 56. Humbert R., Simoni R. D. (1980) Genetic and biomedical studies demonstrating a second gene coding for asparagine synthetase in Escherichia coli. J. Bacteriol. 142, 212–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vollmer W., Blanot D., de Pedro M. A. (2008) Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32, 149–167 [DOI] [PubMed] [Google Scholar]