Background: Gata3 directly transactivates the Il5 gene.
Results: The methylation-mimicking Gata3 mutant at Arg-261 had a selective defect in the induction of IL-5 production via recruitment of Hsp60 to prevent transactivation.
Conclusion: Arginine methylation on Gata3 may play a critical role in the organization and function of the Gata3 transcriptional complex.
Significance: A new regulatory mechanism of Gata3-mediated Il5 transactivation is revealed.
Keywords: GATA transcription factor, post-translational modification (PTM), protein methylation, T helper cells, transcription factor, Gata3, Hsp60, IL-5, Th2, transcription factor
Abstract
Gata3 acts as a master regulator for T helper 2 (Th2) cell differentiation by inducing chromatin remodeling of the Th2 cytokine loci, accelerating Th2 cell proliferation, and repressing Th1 cell differentiation. Gata3 also directly transactivates the interleukin-5 (Il5) gene via additional mechanisms that have not been fully elucidated. We herein identified a mechanism whereby the methylation of Gata3 at Arg-261 regulates the transcriptional activation of the Il5 gene in Th2 cells. Although the methylation-mimicking Gata3 mutant retained the ability to induce IL-4 and repress IFNγ production, the IL-5 production was selectively impaired. We also demonstrated that heat shock protein (Hsp) 60 strongly associates with the methylation-mimicking Gata3 mutant and negatively regulates elongation of the Il5 transcript by RNA polymerase II. Thus, arginine methylation appears to play a pivotal role in the organization of Gata3 complexes and the target gene specificity of Gata3.
Introduction
The GATA family of transcription factors (Gata1–6) selectively binds GATA sites in vertebrate genomes to regulate specific gene expression programs (1). Each GATA family member possesses two highly conserved type IV zinc fingers, referred to as the N-terminal zinc finger (N-finger) and C-terminal zinc finger (C-finger), both of which are involved in DNA binding and protein-protein interactions (2). Among the six vertebrate homologues, Gata1–3 play key roles in the development and maintenance of hematopoietic and immune cells. Gata3 plays a critical role in T cell development in the thymus; it has roles in the CD4 versus CD8 lineage choice and at the β-selection checkpoint, and in Th2 2 cell differentiation through the activation and repression of transcription (3–5).
After antigenic stimulation in a particular cytokine milieu, naive CD4 T cells differentiate into one of the several T helper cell subsets, including Th1, Th2, and Th17 cells (6–8).The differentiation of Th2 cells requires IL-4 stimulation, which leads to Stat6 phosphorylation and the up-regulation of Gata3 transcription (9–11). In addition, the Ras-ERK MAPK cascade controls Gata3 stability through the ubiquitin/proteasome-dependent pathway (12–14). The deletion of Gata3 in peripheral CD4 T cells prevents their differentiation into the Th2 lineage, causing cells to differentiate toward a Th1 phenotype in the absence of polarizing cytokines (15). Conversely, the overexpression of Gata3 in Th1 cells switches their polarity to a Th2 phenotype (16). Gata3 forms functionally distinct complexes and controls the differentiation of naive CD4 T cells into Th2 cells by the induction of chromatin remodeling of the Th2 cytokine loci, facilitation of Th2 cell proliferation, and inhibition of Ifng (5, 17, 18). Gata3 also directly transactivates the Il5 gene via additional mechanisms that are not well understood (19–21).
IL-5 is one of the key cytokines produced by effector Th2 cells, which are involved in the regulation of eosinophilic inflammation (22). In addition, we recently identified pathogenic IL-5-producing memory Th2 cells that play a critical role in the development of eosinophilic airway inflammation (23, 24). IL-5 has important roles in the activation of eosinophils and their migration into the asthmatic lung (25). Activated eosinophils secrete a series of inflammatory cytokines and chemokines and are a potent source of the chemical mediator, leukotriene C4 (26). Gata3 binds to the proximal promoter region and mediates the transactivation of the Il5 gene (19–21). Although Gata3 cooperates with AP-1 and Ets1 to mediate the transactivation of the Il5 gene (27), the molecular mechanisms underlying the Gata3-mediated induction of Il5 expression have not been fully elucidated.
Transcription factors are regulated by means of several different posttranslational modifications, including phosphorylation, acetylation, ubiquitination, sumoylation, and methylation (28). Arginine methyltransferases are a major regulator of gene expression by both the direct methylation of transcription factors, including p53, Stat1, and Nip45, and indirectly via histone modifications (29). For example, arginine methylation is regulated during the p53 responses and affects the target gene specificity of p53, and arginine methylation of Stat1 and Nip45 modulates their interaction with cofactors (30–32). Although the phosphorylation of Gata3 in a human T cell line has been reported (33), no definitive analysis has yet been reported regarding the arginine methylation of Gata3 and its roles in the functions of Gata3.
In this study, we identified novel arginine methylations in the N-finger of Gata3 as a key mechanism regulating the Il5 gene expression in Th2 cells. Interestingly, although the methylation-mimicking Gata3 mutant retained the ability to induce IL-4 and repress IFNγ expression, the IL-5 production was selectively impaired. A methylation-mimicking Gata3 mutant strongly associated with Hsp60 in Th2 cells and was not able to transactivate the Il5 promoter. Moreover, the defect in the transactivation of the Il5 gene in the Gata3 mutant was rescued by knockdown of Hsp60, indicating that Hsp60 is a key molecule involved in the arginine methylation-mediated regulation of Il5 expression. Therefore, arginine methylation appears to play a pivotal role in the organization of Gata3 complexes and the target gene specificity of Gata3.
Experimental Procedures
Mice
C57BL/6 mice were purchased from CLEA Co. (Tokyo, Japan). All mice were maintained under specific pathogen-free conditions and were used at 6–8 weeks of age. All animal care was conducted in accordance with the guidelines of Chiba University.
Identification of the Methylation Sites of Gata3 in the Th2 Cell Clone, D10G4.1
FLAG-tagged Gata3 proteins were purified from D10G4.1 as described previously (18). The proteins were digested with trypsin. After adding 0.1% formic acid to the supernatant, the peptides were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) with an LTQ MASS SPECTROMETER (Thermo scientific). The resulting MS/MS data were analyzed using the Mascot search engine (Matrix Science).
The Generation of Th1 and Th2 Cells
Th1 and Th2 cells were generated as described previously (34). In brief, CD4 T cells with a naive phenotype (CD44low) from C57BL/6 mice were purified using a FACSAria instrument (BD Biosciences), yielding a purity of >98%, and were stimulated with 3 μg/ml of immobilized anti-TCRβ mAb plus 1 μg/ml anti-CD28 mAb under the Th1 or Th2 conditions in vitro. The Th1 conditions were as follows: 25 units/ml IL-2, 10 units/ml IL-12, and an anti-IL-4 mAb. The Th2 conditions were: 25 units/ml IL-2, 10 units/ml IL-4, and anti-IL12 and anti-IFNγ mAbs.
Retroviral Vectors and Infection
The retrovirus vector, pMXs-IRES-hNGFR, was provided by Dr. T. Kitamura (The University of Tokyo, Tokyo, Japan). The methods used to generate the virus supernatant and for infection were described previously (35). Infected cells were collected 6 days after stimulation and were subjected to a quantitative RT-PCR (RT-qPCR) analysis, intracellular staining, and a ChIP assay.
RT-qPCR
Total RNA was isolated using the TRIzol reagent (Invitrogen). Reverse transcription was performed using SuperScript II (Invitrogen). For the quantitative real-time PCR, a TaqMan universal PCR master mix was used for all reactions (Applied Biosystems) with an ABI Prism 7500 sequence detection system. The primers and probes used to detect Gata3, Tbx21, Il4, Il5, Il13, Ifng, Hsp60, and Hprt were described previously (34). The data are shown as the relative expression levels normalized to the Hprt signal.
Luciferase Reporter Assay
A single copy of an Il5 promoter (−1200 bp) and a single copy of an Il13 promoter (−254 bp) in the luciferase reporter plasmid, pGL3 Basic (Promega), were used. M12 cells were used for transfection by electroporation. In addition, 5 ng of a Renilla luciferase reporter vector with the HSV thymidine kinase promoter (pRL-TK; Promega) was added into each transfection as an internal control for the transfection efficiency. Two days after electroporation, the cell extracts were prepared and subjected to a luciferase assay using the manufacturer's instructions for the Dual-luciferase reporter (Promega).
Pulldown Assay
Cell lysates from 293T cells were incubated with the indicated biotinylated oligonucleotides. Bound protein was eluted and separated on an SDS-polyacrylamide gel and then subjected to immunoblotting with an anti-FLAG mAb. The oligonucleotide probe used for the pulldown assay for the Il5 promoter was: 5′-CCTCTATCTGATTGTTAGCA-3′
Chromatin Immunoprecipitation Assay
ChIP assays were performed using anti-Gata3 (mixed 1:1, Santa Cruz Biotechnology; HG3-31, R&D Systems; 634913), anti-JunB (Santa Cruz Biotechnology; N-17), anti-Ets (Santa Cruz Biotechnology; C-20), anti-trimethyl histone H3-K4 (Millipore), anti-acetyl histone H3-K9 (Millipore), anti-RNA polymerase II (Abcam), anti-RNA polymerase II phospho-CTD Ser-5 (Abcam), anti-RNA polymerase II phospho-CTD Ser-2 (Active Motif), and anti-FLAG (M2) antibodies as described previously (36). The quantitative-PCR analyses were performed on an ABI prism 7500 real-time PCR machine with probes from the Roche Diagnostics Universal Probe Library System. The specific primers and TaqMan probes used in this study were described previously (24).
Immunoprecipitation and Immunoblotting
Immunoprecipitation was performed as described previously (18). After electrophoresis, the proteins were subjected to immunoblotting as described previously (37). The antibodies used for the immunoblot analyses were anti-FLAG (M2), anti-Myc (My3), and anti-Hsp60 (Santa Cruz Biotechnology; N-20).
Expression Plasmids
FLAG-tagged Gata3 (pFLAG-CMV2-Gata3) mutants were generated by a PCR-based method. Expression plasmids were transfected into 293T cells using the TransIT-LT1 transfection reagent (Mirus) according to the manufacturer's protocol.
Knockdown Assay
M12 cells were infected with a lentivirus vector containing control (pLKO.1-puro-shControl) or Hsp60 shRNA (pLKO.1-puro-shHsp60) (Sigma-Aldrich) and were cultured in the presence of puromycin (10 μg/ml) for 2 weeks, and resistant cells were subjected to further analyses.
Results
Methylation at Arg-256 and Arg-261 in the N-terminal Zinc Finger of Gata3 Regulates IL-5 Production from Th2 Cells
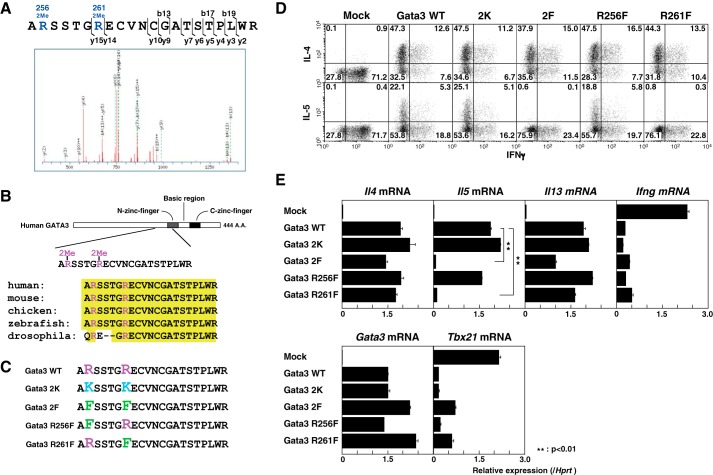
To elucidate whether posttranslational modifications of Gata3 play a role in Th2 cell differentiation, we first performed a LC-MS/MS analysis to identify Gata3 posttranslational modifications using immunopurified FLAG-Gata3 derived from the Th2 cell clone, D10G4.1. We found that Arg-256 and Arg-261 in the N-finger motif of Gata3 were methylated concurrently (Fig. 1A). These two Gata3 arginine residues are conserved from Drosophila to humans (Fig. 1B).
FIGURE 1.
Identification of Gata3 methylation and its role in IL-5 production. A, D10G4.1 cells were infected with a lentivirus encoding FLAG-Gata3, and then the immunopurified Gata3 was subjected to an LC-MS/MS analysis to assess the posttranslational modifications as described under “Experimental Procedures.” The mass spectrometry profile of Gata3 residues 255–275 is shown. B, the methylated residues of Gata3 in the N-terminal zinc finger are highly conserved from Drosophila to humans. A.A., amino acids. C, schematic representations of the WT and mutant Gata3 (Gata3 2K, methylation-impaired; 2F, R256F,R261F, methylation-mimic) are shown. D and E, naive CD4 T cells from C57BL/6 mice were stimulated under Th1 conditions and then infected with a retrovirus vector carrying WT or mutant Gata3 cDNAs. Four days later, the cells were restimulated and subjected to IL-4, IL-5, and IFNγ staining, followed by a FACS analysis (D). The retrovirus-infected human nerve growth factor receptor (hNGFR)-expressing cells were purified, and the mRNA levels of Tbx21 and Gata3 were measured by RT-qPCR. To induce cytokine production, the cells were stimulated with an immobilized anti-TCRβ mAb for another 4 h, and then the extracted RNA was subjected to RT-qPCR for Il4, Il5, Il13, and Infg mRNA expression. The relative expression (/Hprt) is shown with standard deviations (E). Mock, mock-transfected. **, p < 0.01 by Student's t test.
Next, we generated point mutants of Gata3 in which these Arg residues were substituted to Lys (Gata3 2K, positively charged, Arg methylation-deficient mutant) or Phe (Gata3 2F, bulky hydrophobic, Arg methylation-mimicking mutant) (Fig. 1C) (38). To characterize the functions of the Gata3 mutants in Gata3-mediated Th2 cell differentiation, we expressed Gata3 WT and mutant proteins in Th1 cells using a retrovirus system and examined the production of IL-4, IL-5, and IFNγ. Interestingly, although WT and mutant Gata3 retained the ability to induce IL-4 and repress IFNγ production, the IL-5 production was selectively impaired in the methylation-mimicking Gata3 2F mutant (Fig. 1D).
To evaluate the precise contribution of each methylation site, we also generated single point mutants at Arg-256 and Arg-261 (Fig. 1C). Notably, the IL-5 induction was significantly more impaired in the Gata3 R261F mutant than in the R256F mutant (Fig. 1D). Furthermore, an RT-qPCR analysis revealed that methylation-mimicking Gata3 mutants (2F and R261F) had defects in the induction of Il5 expression, without altering the Il4 and Ifng expression, whereas the levels of Gata3 introduced into the cells were comparable (Fig. 1E). The Gata3 2F and R261F mutants showed a tendency to have slightly decreased expression of IL-13 and less repression of Tbx21 expression (Fig. 1E).
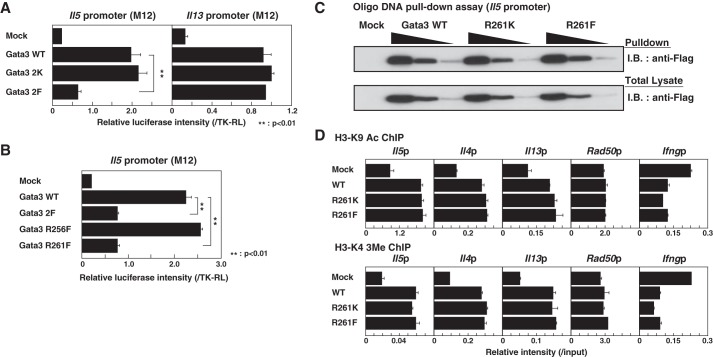
The Gata3 R261F Mutant Fails to Transactivate the Il5 Gene
We next assessed whether the Gata3 mutants were able to induce the transcription of the Il5 and the Il13 promoters. The introduction of Gata3 WT or 2K mutants into an M12 B cell line resulted in the induction of reporter activity of the Il5 promoter, whereas that of the 2F mutant failed to induce Il5 transcription (Fig. 2A). We found that there were no significant differences in the transcriptional activities of the Il13 promoter among the WT, 2K, and 2F mutants, suggesting that the 2F mutant has a selective defect in transactivating the Il5 promoter (Fig. 2A). In addition, in a comparison between the two methylation sites, the transactivation of the Il5 promoter was more significantly impaired in the Gata3 R261F mutant than in the cells with the Gata3 R256F mutant (Fig. 2B). When we examined the DNA binding activity of the Gata3 WT and mutants by an oligonucleotide DNA pulldown assay, the binding of the Gata3 R261F mutant to the GATA consensus sequence in the Il5 promoter was not impaired (Fig. 2C). Furthermore, we investigated the histone modifications at the Th2 cytokine gene loci in Gata3 WT- and mutant-introduced Th1 cells, particularly histone H3-K9 acetylation (H3-K9 Ac) and H3-K4 trimethylation (H3-K4 3Me), which are typically associated with the transcriptionally active state of chromatin. The introduction of WT Gata3 modestly enhanced the H3-K9 Ac and H3-K4 3Me at the Th2 cytokine promoter regions, including the Il5 promoter (Fig. 2D). However, there was no significant difference in the acetylation and trimethylation among the WT, R261K, and R261F mutants in Th1 cells, suggesting that the R261F mutant is capable of binding to the Il5 promoter (Fig. 2C) and inducing chromatin remodeling of the Il5 promoter to the same extent as WT Gata3 (Fig. 2D).
FIGURE 2.
The transactivational activity for the Il5 promoter is impaired by methylated Gata3. A and B, reporter assays with the Il5 promoter and the Il13 promoter were performed using the M12 cell line that expresses no Gata3 family proteins. WT or mutant Gata3 cDNAs were introduced, and the luciferase activity was measured. The data indicate the mean results of three independent experiments with standard deviations. Mock, mock-transfected. **, p < 0.01 by Student's t test. C, 293T cells were transfected with an expression plasmid encoding FLAG-tagged Gata3 WT or one of the mutants, and total extracts were subjected a pulldown assay using Il5 promoter oligonucleotides (Oligo) as described under “Experimental Procedures.” The results of immunoblotting of the total lysates are also shown. D, naive CD4 T cells were stimulated under Th1 conditions and then infected with a retrovirus vector carrying WT or mutant Gata3 cDNAs. Four days later, the status of H3-K9 Ac and H3-K4 3Me in the Th2 cytokines and the Ifng promoters was determined by a ChIP assay with a qPCR analysis. The relative intensities (/input) are shown with standard deviations.
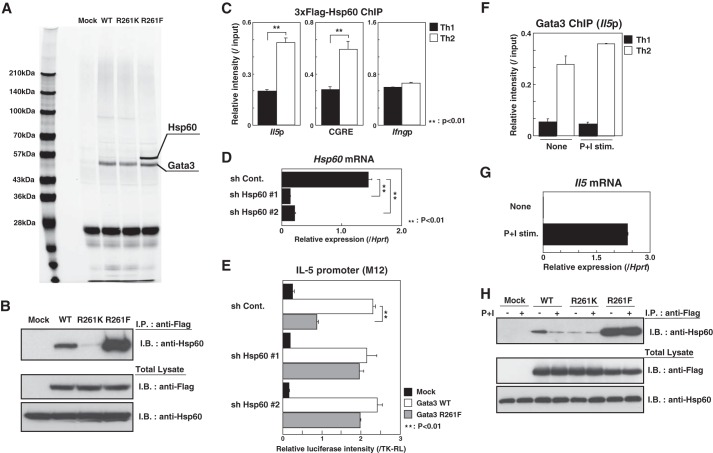
Hsp60 Represses the Transactivation of the Il5 Promoter by Methylated Gata3
Next, to address whether the Gata3 R261F mutant organizes a functional transcription activation complex like WT Gata3, D10G4.1 cells expressing FLAG-tagged WT or mutant Gata3 were subjected to two-step affinity purification using anti-FLAG and anti-Gata3 mAbs, followed by SDS-PAGE and silver staining. We found that the binding of molecules around 60 kDa was increased in the Gata3 R261F mutant when compared with the cells with WT and R261K Gata3 (Fig. 3A). A mass spectrometry analysis identified several polypeptides, including Hsp60, from the band around 60 kDa. To confirm the association of Gata3 with Hsp60, cell extracts from FLAG-Gata3-expressing D10G4.1 cells were subjected to I.P. with an anti-FLAG mAb, and increased binding of endogenous Hsp60 in the R261F mutant was detected, whereas R261K mutant had decreased binding activity to Hsp60 (Fig. 3B). To determine whether Hsp60 binds to the Il5 promoter in primary Th2 cells, 3×FLAG-tagged Hsp60 proteins were introduced into developing Th1 and Th2 cells, and the lysates were subjected to a ChIP assay using an anti-FLAG mAb. When compared with Th1 cells, higher binding of Hsp60 to the Th2 cytokine loci, including the Il5 promoter and conserved Gata3 response element (CGRE) regions, was observed in Th2 cells, indicating that Hsp60 can be recruited to the Il5 promoter (Fig. 3C).
FIGURE 3.
Hsp60 represses the transactivation of the Il5 promoter by methylated Gata3. A, total extracts from FLAG-Gata3 (WT or mutants)-expressing D10G4.1 cells were subjected to two-step affinity purification using a FLAG mAb and a Gata3 mAb, followed by SDS-PAGE and silver staining. The specific polypeptides were identified by mass spectrometry. Mock, mock-transfected. B, the FLAG-Gata3 (WT or mutants)-expressing D10G4.1 cells were subjected to an I.P. assay using a FLAG mAb, followed by immunoblotting (I.B.) with an Hsp60 Ab (upper). The total lysates were also subjected to I.B. in parallel (lower). C, naive CD4 T cells were stimulated under Th1 or Th2 conditions for 2 days and then infected with a retroviral vector carrying 3×FLAG-Hsp60 cDNA. Four days later, the binding of Hsp60 to the Il5 and Ifng promoters and conserved Gata3 response element (CGRE) region was determined by a ChIP assay using an anti-FLAG mAb. **, p < 0.01 by Student's t test. D, M12 cells were infected with a lentivirus encoding control (sh Cont.) or shHsp60 bicistronically with a puromycin resistance gene. The mRNA expression levels of Hsp60 were determined by RT-qPCR. **, p < 0.01 by Student's t test. E, reporter assays with the Il5 promoter were performed using Hsp60 knockdown M12 cells as in Fig. 2A. **, p < 0.01 by Student's t test. F, developing Th1 and Th2 cells were stimulated with or without phorbol 12-myristate 13-acetate (10 ng/ml) and ionomycin (500 nm) (P+I stim.) for 24 h. Then, the binding of Gata3 to the Il5 promoter was determined by a ChIP assay. G, the expression levels of Il5 mRNA were determined by RT-qPCR using a portion of the same cultured cells used in panel F. H, the FLAG-Gata3-expressing D10G4.1 cells were stimulated with or without phorbol 12-myristate 13-acetate (50 ng/ml) and ionomycin (500 nm) for 24 h. The amount of endogenous Hsp60 associated with the FLAG-tagged Gata3 was assessed by I.P., followed by I.B. (upper panel). The total lysates were also subjected to I.B. in parallel (lower panel).
To clarify the role of Hsp60 in the transactivation of the Il5 promoter, we investigated the effects of Hsp60 knockdown on the transcriptional activity of the Gata3 R261F mutant for the Il5 promoter. To accomplish this, we engineered two Hsp60 knockdown M12 cell lines (#1 and #2) stably expressing an shRNA against Hsp60 via a lentivirus system (Fig. 3D) and subjected the cells to a reporter assay using the Il5 promoter. Interestingly, we found that the impairment of the transcriptional activity of Gata3 R261F in control M12 cells was restored to a level similar to that in WT cells by the introduction of shHsp60 (Fig. 3E). These results indicate that Hsp60 strongly associates with the methylated Gata3 and represses its ability to transactivate the Il5 gene.
The Dissociation of Hsp60 from Gata3 Is Induced in Activated Th2 Cells
Next, to examine whether Hsp60 is involved in the induction of the transcription of the Il5 gene by endogenous Gata3 in Th2 cells, we first performed a ChIP assay for Gata3 using Th1 and Th2 cells with and without restimulation. Although Il5 expression was induced after restimulation, the binding of Gata3 at the Il5 promoter was not dramatically changed (Fig. 3, F and G). These results prompted us to examine the levels of Hsp60 binding to Gata3 in restimulated Th2 cells, and we found that the activation of Th2 cells induces a substantial dissociation of Hsp60 from Gata3 WT, whereas there was no difference in the amount of Hsp60 binding in R261K and R261F mutants (Fig. 3H). Taken together, these results indicate that the dissociation of Hsp60 from Gata3 is induced after restimulation and that this results in the activation of the Il5 gene in Th2 cells. Therefore, Hsp60 appears to be involved in regulating the Il5 transcription by Gata3 in primary Th2 cells.
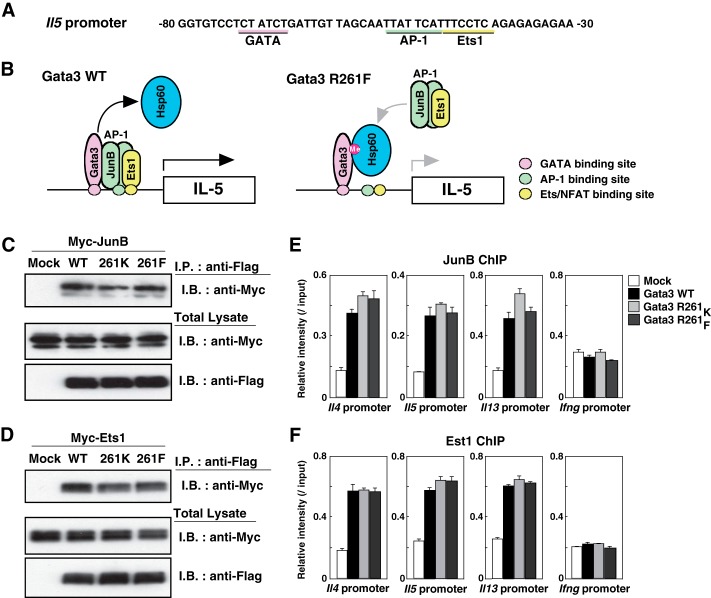
Methylated Gata3 Is Able to Recruit JunB and Ets1 to the Il5 Promoter
It has been reported that the proximal GATA site, along with AP-1 and Ets/NFAT (nuclear factor of activated T-cells) sites, has an important role in the transactivation of the Il5 gene, and Gata3 cooperates with AP-1 and Ets1 to mediate the transcription of Il5 (Fig. 4A) (27). The AP-1 transcription factor consists of a variety of dimers composed of members of the Fos, Jun, and activating transcription factor (ATF) families of proteins (39). Among the Jun family proteins, we previously found that JunB was a Gata3-interacting molecule in Th2 cells (18). Moreover, JunB was identified as one of the components of the transcriptional activation complex at the Il5 proximal promoter (40). Therefore, Gata3 may organize a transcriptional activation complex including JunB and Ets1 to transactivate the Il5 gene in Th2 cells, and Hsp60 may block the association of Gata3 with JunB and Ets1 and their subsequent recruitment to the Il5 promoter (Fig. 4B).
FIGURE 4.
Methylated Gata3 is able to recruit JunB and Ets1 to the Il5 promoter. A, the DNA sequences of the proximal Il5 promoter region are shown. The GATA, AP-1, and Ets1 consensus binding sites are underlined. The numbers indicate positions relative to the transcriptional start site of the Il5 gene. NFAT, nuclear factor of activated T-cells. B, schematic representations of the transcriptional activation Gata3 complex bound to the Il5 promoter. C and D, 293T cells were transfected with expression plasmids encoding FLAG-tagged Gata3 and Myc-tagged JunB (C) or Ets1 (D). Two days later, the extracts were immunoprecipitated with a FLAG mAb, followed by I.B. with a Myc mAb (upper panel). The total lysates were also subjected to I.B. in parallel (lower panel). Mock, mock-transfected. E and F, naive CD4 T cells were stimulated under Th1 conditions and then infected with a retrovirus vector carrying WT or mutant Gata3 cDNAs. Four days later, the cells were restimulated with phorbol 12-myristate 13-acetate and ionomycin for 16 h, and the binding of JunB and Est1 at the indicated regions was determined by a ChIP assay with a qPCR analysis. The relative intensities (/input) are shown with standard deviations.
To assess this possibility, FLAG-tagged Gata3 and Myc-tagged JunB or Ets1 were ectopically co-expressed in 293T cells, and then immunoprecipitation was performed with an anti-FLAG mAb. Specific complexes containing Gata3 and JunB or Ets1 were easily detected, and the Gata3 R261F mutant retained its JunB and Ets1 binding (Fig. 4, C and D). In addition, we found that there was no difference in the recruitment of JunB and Ets1 to the Th2 cytokine gene loci, including the Il5 promoter, in Th1 cells transfected with the WT, R261K mutant, or R261F mutant Gata3 (Fig. 4, E and F). These results indicate that the methylated Gata3 is able to organize a transactivation complex with AP-1 and Ets1 and recruit them to the Il5 promoter.
Methylated Gata3 Is Not Able to Induce the Elongation of the Il5 Transcript by RNA Polymerase II
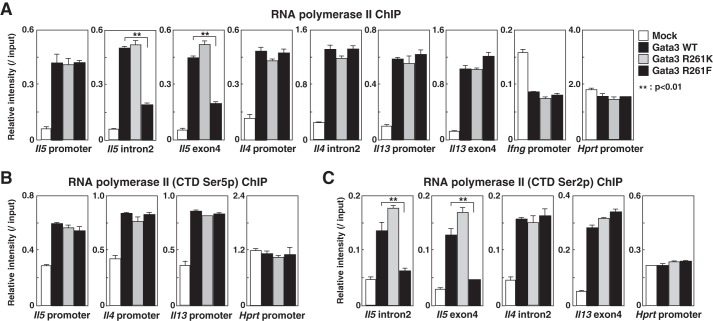
A recent study indicated that Hsp90, the sister chaperone of Hsp60, is a transcriptional regulator required for RNA polymerase II pausing that functions by stabilizing the negative elongation factor complex (41). Thus, we examined whether methylated Gata3-associated Hsp60 maintains the pausing property of RNA polymerase II at the Il5 locus after restimulation in Th2 cells. We expressed WT and R261 mutants in Th1 cells using a retrovirus system and assessed the recruitment of RNA polymerase II to the Th2 cytokine loci. Interestingly, although the recruitment of RNA polymerase II to the Th2 cytokine promoters and the coding region of the Il4 and Il13 genes was unaffected, RNA polymerase II recruitment to the coding region of the Il5 gene (intron1 and exon4) was significantly compromised in R261F-expressing cells (Fig. 5A).
FIGURE 5.
Methylated Gata3 is unable to induce transcriptional elongation by RNA polymerase II at the Il5 locus. A–C, naive CD4 T cells were stimulated under Th1 conditions and then infected with a retrovirus vector carrying WT or mutant Gata3 cDNAs. Four days later, the binding of RNA polymerase II at the indicated regions was determined by a ChIP assay. The relative intensities (/input) are shown with standard deviations. Ser5p, phospho-Ser-5; Ser2p, phospho-Ser-2.
Previous studies have shown that productive transcription requires the phosphorylation of the C-terminal domain (CTD) of RNA polymerase II. Subsequent to the phosphorylation of CTD Ser-5 at the promoter region, CTD Ser-2 phosphorylation concurs with the entry of RNA polymerase II in the productive elongation phase of transcription (42). By a ChIP analysis, we found that there was a significant decrease in the amount of the phospho-Ser-2 form of RNA polymerase II in the coding region of the Il5 gene, whereas there was no difference in the amount of the phospho-Ser-5 at the Il5 promoter in R261F mutant-introduced cells (Fig. 5, B and C). These results indicate that methylated Gata3-associated Hsp60 blocks the transition of the phosphorylated form of RNA polymerase II from initiation to elongation at the Il5 locus.
Discussion
We have herein demonstrated that Gata3 is methylated in Th2 cells and that methylated Gata3 had a selective defect in the induction of IL-5 production. Hsp60 strongly associates with methylated Gata3 and represses the transcriptional elongation of the Il5 gene by RNA polymerase II.
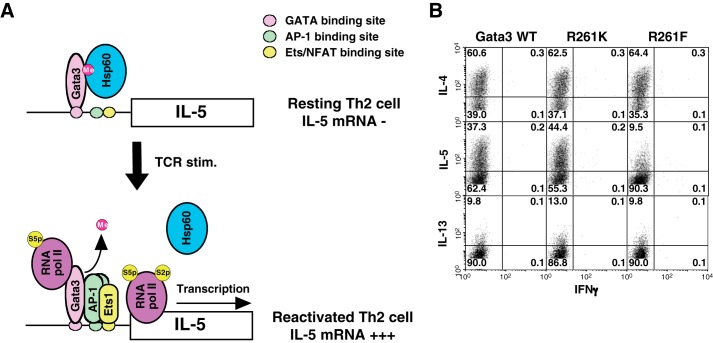
This finding highlights a novel molecular mechanism that controls the IL-5 production from Th2 cells. The transcription of the Il5 gene is strictly regulated by several steps, including Gata3 binding to the Il5 promoter, AP-1 and Ets1 activation and recruitment to the Il5 promoter after TCR restimulation, and the demethylation of Gata3 followed by dissociation of Hsp60 from the Gata3 complex (Fig. 6 A). This strict regulation of Il5 transcription may serve to prevent the induction of harmful IL-5-mediated eosinophilic inflammation. A similar inhibitory mechanism of Il-5 expression through Eomesodermin was reported in memory Th2 cells (24).
FIGURE 6.
Methylated Gata3 has a dominant-negative effect on the Gata3-mediated transactivation of the Il5 gene. A, schematic representation of Gata3-mediated transactivation of the Il5 gene in resting and TCR-reactivated Th2 cells. In resting Th2 cells, methylated Gata3 binds to the Il5 promoter with Hsp60 (upper). After TCR restimulation (TCR stim.), AP-1 and Ets1 are activated and organize the complex with Gata3 at the Il5 locus. Demethylation of Gata3 can be induced by unknown mechanisms resulting in the dissociation of Hsp60 from Gata3 complex, leading to the Il5 transcription (lower). NFAT, nuclear factor of activated T-cells; RNA pol II, RNA polymerase II; S5p, phospho-Ser-5; S2p, phospho-Ser-2. B, naive CD4 T cells from C57BL/6 mice were stimulated under Th2 conditions and then infected with a retrovirus vector carrying WT or mutant Gata3 cDNAs. Four days later, the cells were restimulated and subjected to IL-4, IL-5, IL-13, and IFNγ staining, followed by a FACS analysis.
We have shown that a methylation-mimicking Gata3 mutant had a defect in the induction of IL-5 production, without altering the IL-4 and IFNγ expression. The introduction of the methylation-mimicking Gata3 mutant selectively reduced the production of IL-5 from Th2 cells (Fig. 6B). It is therefore likely that methylated Gata3 has a dominant-negative effect on the Gata3-mediated transactivation of the Il5 gene. Further detailed analyses of the methylation of Gata3 and methylated Gata3 complex may lead to the discovery of novel therapeutic targets for IL-5-mediated inflammatory disorders, including allergic asthma and eosinophilic chronic rhinosinusitis (43).
It is also known that Gata3 expression is observed in the developing parathyroid glands, inner ear, and kidneys in humans and mice. HDR syndrome is an autosomal dominant disorder characterized by hypoparathyroidism, sensorineural deafness, and renal dysplasia (44). Loss-of-function mutations of Gata3 have been reported to be associated with HDR syndrome. Importantly, a mutation in Arg-261 was found in patients with familial hypoparathyroidism and deafness syndrome (45). Thus, arginine methylation has a pivotal role in regulating the expression of Gata3 target genes in the parathyroid glands and inner ear, together with Th2 cells. This finding indicates that methylation at Arg-261 in the N-terminal zinc finger of Gata3 plays an important pathophysiological role in humans.
In summary, we have herein demonstrated that methylation of arginine on Gata3 plays a critical role in the organization and function of the Gata3 transcriptional complex, as well as expression of a specific Gata3 target gene.
Acknowledgments
We thank Kaoru Sugaya, Hikari Kato, Yoko Ozawa, Setsuko Miyano, Mizuho Oda, Emiko Koba, Yuki Matsuzaki, Eishika Shalini Dissanayake, and Toshihiro Ito for expert technical assistance. This work was partly performed in the Cooperative Research Project Program of the Medical Institute of Bioregulation, Kyushu University.
This work was supported by Grants-in-aid for Scientific Research S 26221305, C 24592083, and 26460569 and Exploratory Research and Young Scientists B Grants 24790461 and 23790522; the Takeda Science Foundation; the Astellas Foundation for Research on Metabolic Disorders; the Uehara Memorial Foundation; the Manpei Suzuki Diabetes Foundation; and the Kanae Foundation for the Promotion of Medical Science.
- Th2
- T helper 2
- Hsp
- heat shock protein
- RT-qPCR
- quantitative RT-PCR
- CTD
- C-terminal domain
- TCR
- T-cell receptor
- I.P.
- immunoprecipitation
- I.B.
- immunoblotting.
References
- 1. Bresnick E. H., Martowicz M. L., Pal S., Johnson K. D. (2005) Developmental control via GATA factor interplay at chromatin domains. J. Cell. Physiol. 205, 1–9 [DOI] [PubMed] [Google Scholar]
- 2. Chen Y., Bates D. L., Dey R., Chen P. H., Machado A. C., Laird-Offringa I. A., Rohs R., Chen L. (2012) DNA binding by GATA transcription factor suggests mechanisms of DNA looping and long-range gene regulation. Cell Rep. 2, 1197–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rothenberg E. V., Scripture-Adams D. D. (2008) Competition and collaboration: GATA-3, PU.1, and Notch signaling in early T-cell fate determination. Semin. Immunol. 20, 236–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hosoya T., Maillard I., Engel J. D. (2010) From the cradle to the grave: activities of GATA-3 throughout T-cell development and differentiation. Immunol. Rev. 238, 110–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ho I. C., Tai T. S., Pai S. Y. (2009) GATA3 and the T-cell lineage: essential functions before and after T-helper-2-cell differentiation. Nat. Rev. Immunol. 9, 125–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Reiner S. L. (2007) Development in motion: helper T cells at work. Cell 129, 33–36 [DOI] [PubMed] [Google Scholar]
- 7. Zhu J., Yamane H., Paul W. E. (2010) Differentiation of effector CD4 T cell populations. Annu. Rev. Immunol. 28, 445–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ansel K. M., Djuretic I., Tanasa B., Rao A. (2006) Regulation of Th2 differentiation and Il4 locus accessibility. Annu. Rev. Immunol. 24, 607–656 [DOI] [PubMed] [Google Scholar]
- 9. Onodera A., Yamashita M., Endo Y., Kuwahara M., Tofukuji S., Hosokawa H., Kanai A., Suzuki Y., Nakayama T. (2010) STAT6-mediated displacement of polycomb by trithorax complex establishes long-term maintenance of GATA3 expression in T helper type 2 cells. J. Exp. Med. 207, 2493–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nakayama T., Yamashita M. (2008) Initiation and maintenance of Th2 cell identity. Curr. Opin. Immunol. 20, 265–271 [DOI] [PubMed] [Google Scholar]
- 11. Kurata H., Lee H. J., O'Garra A., Arai N. (1999) Ectopic expression of activated Stat6 induces the expression of Th2-specific cytokines and transcription factors in developing Th1 cells. Immunity 11, 677–688 [DOI] [PubMed] [Google Scholar]
- 12. Yamashita M., Shinnakasu R., Asou H., Kimura M., Hasegawa A., Hashimoto K., Hatano N., Ogata M., Nakayama T. (2005) Ras-ERK MAPK cascade regulates GATA3 stability and Th2 differentiation through ubiquitin-proteasome pathway. J. Biol. Chem. 280, 29409–29419 [DOI] [PubMed] [Google Scholar]
- 13. Shinnakasu R., Yamashita M., Kuwahara M., Hosokawa H., Hasegawa A., Motohashi S., Nakayama T. (2008) Gfi1-mediated stabilization of GATA3 protein is required for Th2 cell differentiation. J. Biol. Chem. 283, 28216–28225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hosokawa H., Kimura M. Y., Shinnakasu R., Suzuki A., Miki T., Koseki H., van Lohuizen M., Yamashita M., Nakayama T. (2006) Regulation of Th2 cell development by Polycomb group gene bmi-1 through the stabilization of GATA3. J. Immunol. 177, 7656–7664 [DOI] [PubMed] [Google Scholar]
- 15. Zhu J., Min B., Hu-Li J., Watson C. J., Grinberg A., Wang Q., Killeen N., Urban J. F., Jr., Guo L., Paul W. E. (2004) Conditional deletion of Gata3 shows its essential function in TH1-TH2 responses. Nat. Immunol. 5, 1157–1165 [DOI] [PubMed] [Google Scholar]
- 16. Zheng W., Flavell R. A. (1997) The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 89, 587–596 [DOI] [PubMed] [Google Scholar]
- 17. Hosokawa H., Tanaka T., Kato M., Shinoda K., Tohyama H., Hanazawa A., Tamaki Y., Hirahara K., Yagi R., Sakikawa I., Morita A., Nagira M., Poyurovsky M. V., Suzuki Y., Motohashi S., Nakayama T. (2013) Gata3/Ruvbl2 complex regulates T helper 2 cell proliferation via repression of Cdkn2c expression. Proc. Natl. Acad. Sci. U.S.A. 110, 18626–18631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hosokawa H., Tanaka T., Suzuki Y., Iwamura C., Ohkubo S., Endoh K., Kato M., Endo Y., Onodera A., Tumes D. J., Kanai A., Sugano S., Nakayama T. (2013) Functionally distinct Gata3/Chd4 complexes coordinately establish T helper 2 (Th2) cell identity. Proc. Natl. Acad. Sci. U.S.A. 110, 4691–4696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang D. H., Cohn L., Ray P., Bottomly K., Ray A. (1997) Transcription factor GATA-3 is differentially expressed in murine Th1 and Th2 cells and controls Th2-specific expression of the interleukin-5 gene. J. Biol. Chem. 272, 21597–21603 [DOI] [PubMed] [Google Scholar]
- 20. Inami M., Yamashita M., Tenda Y., Hasegawa A., Kimura M., Hashimoto K., Seki N., Taniguchi M., Nakayama T. (2004) CD28 costimulation controls histone hyperacetylation of the interleukin 5 gene locus in developing Th2 cells. J. Biol. Chem. 279, 23123–23133 [DOI] [PubMed] [Google Scholar]
- 21. Siegel M. D., Zhang D. H., Ray P., Ray A. (1995) Activation of the interleukin-5 promoter by cAMP in murine EL-4 cells requires the GATA-3 and CLE0 elements. J. Biol. Chem. 270, 24548–24555 [DOI] [PubMed] [Google Scholar]
- 22. Rothenberg M. E., Hogan S. P. (2006) The eosinophil. Annu. Rev. Immunol. 24, 147–174 [DOI] [PubMed] [Google Scholar]
- 23. Endo Y., Hirahara K., Yagi R., Tumes D. J., Nakayama T. (2014) Pathogenic memory type Th2 cells in allergic inflammation. Trends Immunol. 35, 69–78 [DOI] [PubMed] [Google Scholar]
- 24. Endo Y., Iwamura C., Kuwahara M., Suzuki A., Sugaya K., Tumes D. J., Tokoyoda K., Hosokawa H., Yamashita M., Nakayama T. (2011) Eomesodermin controls interleukin-5 production in memory T helper 2 cells through inhibition of activity of the transcription factor GATA3. Immunity 35, 733–745 [DOI] [PubMed] [Google Scholar]
- 25. Takatsu K., Nakajima H. (2008) IL-5 and eosinophilia. Curr. Opin. Immunol. 20, 288–294 [DOI] [PubMed] [Google Scholar]
- 26. Rosenberg H. F., Dyer K. D., Foster P. S. (2013) Eosinophils: changing perspectives in health and disease. Nat. Rev. Immunol. 13, 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang J., Shannon M. F., Young I. G. (2006) A role for Ets1, synergizing with AP-1 and GATA-3 in the regulation of IL-5 transcription in mouse Th2 lymphocytes. Int. Immunol. 18, 313–323 [DOI] [PubMed] [Google Scholar]
- 28. Filtz T. M., Vogel W. K., Leid M. (2014) Regulation of transcription factor activity by interconnected post-translational modifications. Trends Pharmacol. Sci. 35, 76–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bedford M. T., Clarke S. G. (2009) Protein arginine methylation in mammals: who, what, and why. Mol. Cell 33, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jansson M., Durant S. T., Cho E. C., Sheahan S., Edelmann M., Kessler B., La Thangue N. B. (2008) Arginine methylation regulates the p53 response. Nat. Cell Biol. 10, 1431–1439 [DOI] [PubMed] [Google Scholar]
- 31. Mowen K. A., Schurter B. T., Fathman J. W., David M., Glimcher L. H. (2004) Arginine methylation of NIP45 modulates cytokine gene expression in effector T lymphocytes. Mol. Cell 15, 559–571 [DOI] [PubMed] [Google Scholar]
- 32. Mowen K. A., Tang J., Zhu W., Schurter B. T., Shuai K., Herschman H. R., David M. (2001) Arginine methylation of STAT1 modulates IFNα/β-induced transcription. Cell 104, 731–741 [DOI] [PubMed] [Google Scholar]
- 33. Maneechotesuwan K., Xin Y., Ito K., Jazrawi E., Lee K. Y., Usmani O. S., Barnes P. J., Adcock I. M. (2007) Regulation of Th2 cytokine genes by p38 MAPK-mediated phosphorylation of GATA-3. J. Immunol. 178, 2491–2498 [DOI] [PubMed] [Google Scholar]
- 34. Yamashita M., Hirahara K., Shinnakasu R., Hosokawa H., Norikane S., Kimura M. Y., Hasegawa A., Nakayama T. (2006) Crucial role of MLL for the maintenance of memory T helper type 2 cell responses. Immunity 24, 611–622 [DOI] [PubMed] [Google Scholar]
- 35. Kimura M., Koseki Y., Yamashita M., Watanabe N., Shimizu C., Katsumoto T., Kitamura T., Taniguchi M., Koseki H., Nakayama T. (2001) Regulation of Th2 cell differentiation by mel-18, a mammalian polycomb group gene. Immunity 15, 275–287 [DOI] [PubMed] [Google Scholar]
- 36. Yamashita M., Ukai-Tadenuma M., Kimura M., Omori M., Inami M., Taniguchi M., Nakayama T. (2002) Identification of a conserved GATA3 response element upstream proximal from the interleukin-13 gene locus. J. Biol. Chem. 277, 42399–42408 [DOI] [PubMed] [Google Scholar]
- 37. Omori M., Yamashita M., Inami M., Ukai-Tadenuma M., Kimura M., Nigo Y., Hosokawa H., Hasegawa A., Taniguchi M., Nakayama T. (2003) CD8 T cell-specific downregulation of histone hyperacetylation and gene activation of the IL-4 gene locus by ROG, repressor of GATA. Immunity 19, 281–294 [DOI] [PubMed] [Google Scholar]
- 38. Campbell M., Chang P. C., Huerta S., Izumiya C., Davis R., Tepper C. G., Kim K. Y., Shevchenko B., Wang D. H., Jung J. U., Luciw P. A., Kung H. J., Izumiya Y. (2012) Protein arginine methyltransferase 1-directed methylation of Kaposi sarcoma-associated herpesvirus latency-associated nuclear antigen. J. Biol. Chem. 287, 5806–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jochum W., Passegué E., Wagner E. F. (2001) AP-1 in mouse development and tumorigenesis. Oncogene 20, 2401–2412 [DOI] [PubMed] [Google Scholar]
- 40. Karlen S., D'Ercole M., Sanderson C. J. (1996) Two pathways can activate the interleukin-5 gene and induce binding to the conserved lymphokine element 0. Blood 88, 211–221 [PubMed] [Google Scholar]
- 41. Sawarkar R., Sievers C., Paro R. (2012) Hsp90 globally targets paused RNA polymerase to regulate gene expression in response to environmental stimuli. Cell 149, 807–818 [DOI] [PubMed] [Google Scholar]
- 42. Heidemann M., Hintermair C., Voß K., Eick D. (2013) Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim. Biophys. Acta 1829, 55–62 [DOI] [PubMed] [Google Scholar]
- 43. Ortega H. G., Liu M. C., Pavord I. D., Brusselle G. G., FitzGerald J. M., Chetta A., Humbert M., Katz L. E., Keene O. N., Yancey S. W., Chanez P. (2014) Mepolizumab treatment in patients with severe eosinophilic asthma. N. Engl. J. Med. 371, 1198–1207 [DOI] [PubMed] [Google Scholar]
- 44. Van Esch H., Groenen P., Nesbit M. A., Schuffenhauer S., Lichtner P., Vanderlinden G., Harding B., Beetz R., Bilous R. W., Holdaway I., Shaw N. J., Fryns J. P., Van de Ven W., Thakker R. V., Devriendt K. (2000) GATA3 haplo-insufficiency causes human HDR syndrome. Nature 406, 419–422 [DOI] [PubMed] [Google Scholar]
- 45. Nakamura A., Fujiwara F., Hasegawa Y., Ishizu K., Mabe A., Nakagawa H., Nagasaki K., Jo W., Tajima T. (2011) Molecular analysis of the GATA3 gene in five Japanese patients with HDR syndrome. Endocr. J. 58, 123–130 [DOI] [PubMed] [Google Scholar]