Background: EXTL2 belongs to the exostosin family of glycosyltransferases involved in heparan sulfate biosynthesis.
Results: We determined the effect of reduced or increased amounts of EXTL2 on heparan sulfate structure in a human cell line.
Conclusion: EXTL2 levels influence heparan sulfate chain elongation.
Significance: Knowledge about how EXTL2 influences heparan sulfate structure is crucial for understanding heparan sulfate dependent processes.
Keywords: chondroitin sulfate, glycosaminoglycan, glycosyltransferase, heparan sulfate, proteoglycan, EXTL2, Exostosin-like 2
Abstract
Heparan sulfate proteoglycans are ubiquitously located on cell surfaces and in the extracellular matrices. The negatively charged heparan sulfate chains interact with a multitude of different proteins, thereby influencing a variety of cellular and developmental processes, for example cell adhesion, migration, tissue morphogenesis, and differentiation. The human exostosin (EXT) family of genes contains five members: the heparan sulfate polymerizing enzymes, EXT1 and EXT2, and three EXT-like genes, EXTL1, EXTL2, and EXTL3. EXTL2 has been ascribed activities related to the initiation and termination of heparan sulfate chains. Here we further investigated the role of EXTL2 in heparan sulfate chain elongation by gene silencing and overexpression strategies. We found that siRNA-mediated knockdown of EXTL2 in human embryonic kidney 293 cells resulted in increased chain length, whereas overexpression of EXTL2 in the same cell line had little or no effect on heparan sulfate chain length. To study in more detail the role of EXTL2 in heparan sulfate chain elongation, we tested the ability of the overexpressed protein to catalyze the in vitro incorporation of N-acetylglucosamine and N-acetylgalactosamine to oligosaccharide acceptors resembling unmodified heparan sulfate and chondroitin sulfate precursor molecules. Analysis of the generated products revealed that recombinant EXTL2 showed weak ability to transfer N-acetylgalactosamine to heparan sulfate precursor molecules but also, that EXTL2 exhibited much stronger in vitro N-acetylglucosamine-transferase activity related to elongation of heparan sulfate chains.
Introduction
Most processes such as cell survival, division, migration, differentiation, and cancer development are largely determined by the concerted action of many signaling molecules that recognize and bind to membrane receptors to induce a signaling cascade that triggers a specific cell response. In addition to this ligand-receptor system, heparan sulfate proteoglycans are important modulators for optimizing many signaling pathways. The covalently attached heparan sulfate (HS)3 glycosaminoglycan chains play vital roles, through their interactions with a range of these ligand proteins and their receptors (1). Binding of HS to ligand proteins is influenced by HS chain length and depends on HS sulfation pattern, where the spacing and number of O-sulfate groups are of special importance.
HS chains are complex molecules created in a process that includes (a) assembly of the polysaccharide-protein linkage region; (b) generation of a polymer consisting of alternating glucuronic acid (GlcA) and GlcNAc residues; and (c) a series of modification reactions by which N- and O-sulfate groups are selectively introduced and some of the GlcA residues are C5-epimerized to iduronic acid units (2, 3). An HS chain is initiated by the formation of the so-called glycosaminoglycan-protein linkage region, which consists of the tetrasaccharide GlcA-galactose-galactose-xylose (GlcAβ1-3Galβ1-3Galβ1-4Xyl) where xylose is attached to a serine residue in the protein core (4). The addition of α-linked GlcNAc to the GlcA residue initiates HS formation. The addition of the first GlcNAc has been credited to be catalyzed by exostosin-like (EXTL)2 and/or EXTL3, both members of the exostosin (EXT) family of enzymes (5, 6). Once the first GlcNAc residue has been added, polymerization of HS, by alternating additions of GlcA and GlcNAc, is carried out by a hetero-complex of two other members of the EXT family, EXT1 and EXT2 (7–9).
There are five members of the EXT family in humans: EXT1, EXT2, EXTL1, EXTL2, and EXTL3. The genes encoding EXT1 and EXT2 were first identified as the genes defective in people with the disorder hereditary multiple osteochondromas, previously called hereditary multiple exostoses, an autosomal dominant disorder characterized by bone deformities and cartilage-capped bony outgrowths, called exostoses or osteochondromas, at the ends of the long bones (10, 11). The EXTL genes have not been linked to hereditary multiple osteochondromas; instead they belong to the EXT family based on amino acid sequence homology with EXT1 and EXT2. All members of the EXT family are suggested to be glycosyltransferases involved in HS biosynthesis (4).
EXTL2, the shortest member of the EXT family, is present in vertebrates, but not in invertebrates, such as Drosophila melanogaster and Caenorhabditis elegans, suggesting that EXTL2 may be necessary only for the production of vertebrate HS (12). Although several studies have established that EXT1, EXT2, and EXTL3 are involved in HS chain elongation, the function of EXTL2 in HS biosynthesis remains unclear. In vitro enzyme assays have demonstrated a soluble form of EXTL2 to have two glycosyltransferase activities, transfer of α-linked GlcNAc and α-linked GalNAc to an acceptor analog mimicking the tetrasaccharide linkage region (13). EXTL2 was also shown to transfer α-linked GalNAc, but not GlcNAc to an authentic tetrasaccharide linker substrate. The functional significance of the α-linked GalNAc transfer is not known because the product, GalNAcα1-4GlcAβ1-3Galβ1-3Galβ1-4Xyl, is not an acceptor for glycosyltransferases involved in glycosaminoglycan synthesis. However, the addition of the α-linked GalNAc may provide a stop signal that prevents glycosaminoglycan chain elongation (13). To assess the role of EXTL2 in mammalian HS chain elongation, we studied the effect on HS structure of reduced or up-regulated EXTL2 expression as well as EXTL2 in vitro enzyme activities in relation to HS chain elongation.
Experimental Procedures
siRNA-mediated Down-regulation of EXTL2 in HEK293 Cells
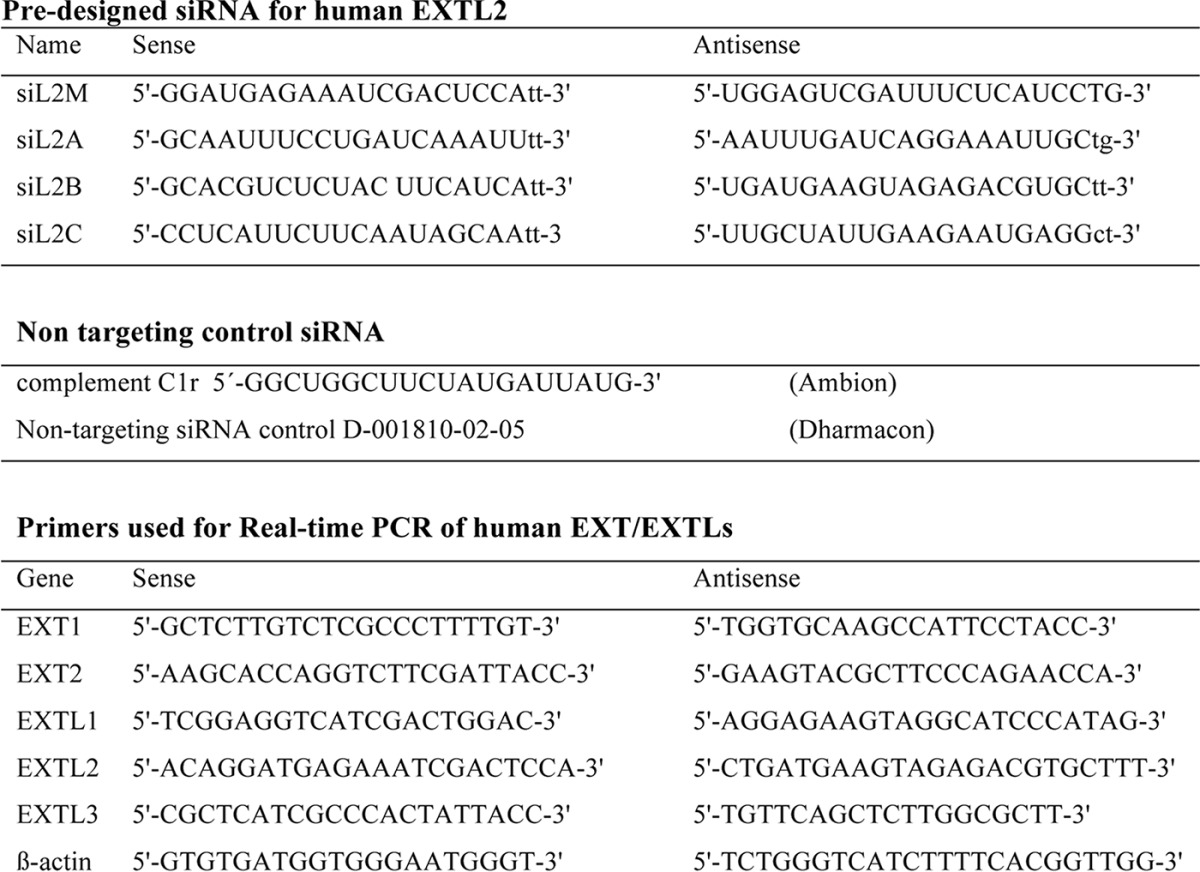
Four predesigned siRNAs directed against human EXTL2, siL2M, siL2A, siL2B, and siL2C, as well as complement C1r (non-targeting control siRNA), were all from Ambion. A second non-targeting control siRNA was from Dharmacon. Sequences of primers are listed in Table 1. In preliminary experiments, to determine which siRNA(s) was most effective in down-regulating EXTL2, HEK293 cells were transfected with 2, 5, 10, 20, 50, and 100 nm of different EXTL2 siRNAs. Down-regulation was evaluated by real-time PCR after 24 h. Based on these results, 50 nm was used in further experiments. HEK293 cells were transfected with the siRNAs (50 nm of each) using Lipofectamine 2000 according to the manufacturer's protocol (Invitrogen). Mock-transfected cells were treated with Lipofectamine 2000 only. Cells were cultivated 24 or 48 h before further experiments.
TABLE 1.
Primers used for siRNA and in real-time PCR
Construction of Expression Plasmid and Overexpression of EXTL2 in HEK293 Cells
Full-length human EXTL2 cDNA clone (I.M.A.G.E. Consortium Clone ID 5273246) (14), purchased from Geneservice Ltd., was amplified using sense primer, 5′-GGATCCATAAATCGGCTGGCCCTACT-3′, and antisense primer, 5′-GATATCTGGAAAACCAAACTGGGAAA-3′, and subcloned into pCR 2.1-TOPO vector (Invitrogen). EXTL2 was then excised using BamHI and EcoRV restriction sites (underlined in the primers) and subcloned into the corresponding site of pcDNA6/B Myc-His plasmid vector (Invitrogen). The insertions were confirmed by sequencing. Ligation into the expression vector resulted in a construct with EXTL2 in-frame with a C-terminal Myc/His tag (Myc-EXTL2). HEK293 cells were stably transfected with the EXTL2 plasmids or vector alone using Lipofectamine 2000 according to the manufacturer's protocol. Alternatively, HEK293 cells were transfected using the Nucleofector electroporation kit for adherent cells (Amaxa) with a C-terminal TurboGFP (tGFP)-tagged full-length human EXTL2 cDNA clone in the pCMV6-AC-GFP vector (OriGene). Selected cellular clones were maintained in DMEM (Invitrogen) complemented with 10% (v/v) fetal calf serum (Invitrogen), 1% penicillin G-streptomycin, and blasticidin (Fluka Analytical) (pcDNA6/B Myc-His) or Geneticin (G418 sulfate) (pCMV6-AC-GFP) at a concentration of 10 and 800 μg/ml, respectively. mRNA expression levels were determined by real-time PCR, and expression of recombinant proteins was examined by Western blotting. The tGFP-tagged construct was used in the majority of experiments, but the three cellular clones highly expressing the Myc-tagged EXTL2 were also analyzed for HS chain length, disaccharide composition, and glycosyltransferase assays with similar results as the tGFP-tagged EXTL2 construct.
Quantitative Real-time PCR (RT-PCR)
24 or 48 h after transfection, total RNA was isolated from HEK293 cells (overexpressing EXTL2, siRNA-treated, or mock-treated) using the RNeasy mini prep kit (Qiagen). Aliquots of 1 μg of total RNA were reverse-transcribed to cDNA using random primers (iScript cDNA synthesis kit, Bio-Rad) according to the manufacturer's instructions. Quantification of mRNA expression was conducted using iQ SYBR Green supermix (Bio-Rad) in a LightCycler 480 (Roche Applied Sciences). β-Actin was used as a reference gene for quantification. The primers used (Table 1) were selected using PrimerBank (15). Each primer/cDNA set was performed in duplicate or triplicate, and the expression level of EXTL2 mRNA was normalized to that of the β-actin mRNA level. The relative expression levels were calculated using the ΔΔCT method (16).
Western Blot Analysis
Protein samples from cell lysates were separated on a 10% SDS-PAGE gel and transferred to a nitrocellulose membrane (Amersham Biosciences) essentially as described (17). The membranes were incubated with the primary monoclonal anti-c-Myc antibody (1:1000, Abcam), 2H8 monoclonal anti-tGFP (1:2000, Origen), β-actin (1:1000, Sigma), or the polyclonal anti-EXTL2 (1:300, Sigma-Aldrich) for 2 h at room temperature. After washing in 0.5% TBS-T (25 mm Tris, 13.7 mm NaCl, 2.7 mm KCl, 0.1% Tween), bound antibodies were detected using HRP-conjugated goat anti-mouse and goat anti-rabbit IgG (1:5000, Santa Cruz Biotechnology) for 1 h at room temperature. Membranes were analyzed for chemiluminescence according to the manufacturer's instructions (ECL Plus detection kit; Pierce). Precision Plus Protein control (Bio-Rad) was used as molecular weight marker.
Metabolic Labeling, Isolation, and Analysis of HS Structure
Subconfluent cell cultures were incubated with 200 μCi/ml [35S]sulfate or 50 μCi/ml [6-3H]glucosamine/HCl (both from PerkinElmer) for 24 h. After incubation, the medium was saved, and the cell layers were washed twice with PBS and treated with 1 mg/ml trypsin for 5 min at 37 °C. Free glycosaminoglycan chains were isolated from the trypsin fraction (derived from cell surface/matrix proteoglycans) or from solubilized cell fractions as described in Ref. 5. Galactosaminoglycans were digested with chondroitinase ABC (Seikagaku) in 50 mm Tris-HCl, pH 8.0, 30 mm sodium acetate, and 0.1 mg/ml bovine serum albumin, and HS chain length was analyzed by gel chromatography on a Superose 6 HR10/30 column (Amersham Biosciences) eluted with 0.5 m NH4HCO3. For disaccharide analyses, labeled HS chains were depolymerized to disaccharides by treatment with nitrous acid at pH 1.5 (which cleaves the glucosaminidic linkage at GlcNS units) yielding disaccharides from contiguous N-sulfated domains followed by reduction with NaBH4. Disaccharides were isolated and then analyzed by anion-exchange HPLC using a Whatman Partisil 10-SAX column eluted with aqueous KH2PO4 of stepwise increasing concentration at a rate of 1 ml/min (5).
Purification of EXTL2 Protein
Cells overexpressing EXTL2 and mock-transfected cells were harvested, washed two to three times in PBS and solubilized in 1 ml each of solubilization buffer, 1% Triton X-100, 0.15 m NaCl, 10 mm Tris-HCl, pH 7.4, 1 mm MgCl2, 1 mm CaCl2 containing protease inhibitors (Complete Mini without EDTA protease inhibitor cocktail from Roche Diagnostics). Recombinant Myc-tagged proteins were captured on anti-Myc-agarose (Sigma-Aldrich) during overnight incubation at 4 °C, washed three times in PBS, and finally washed once with 0.15 m NaCl at 4 °C under vigorous shaking. After centrifugation at 13,000 rpm for 2 min, most of the supernatant was discarded, and the remaining 25 μl was mixed with the gel and used as an enzyme source in the glycosyltransferase assays. The tGFP-tagged EXTL2 proteins and proteins from mock-transfected cells were preincubated end-over-end for 2 h at 4 °C with 60 μl of a 50/50 slurry of protein G-Sepharose (DAKO) and 10 μl of serum. To the precleared supernatants, 60 μl of protein G-Sepharose (50/50 slurry) and 5 μg/ml anti-tGFP monoclonal antibody 2H8 were added followed by end-over-end incubation for 2 h at 4 °C. Next, the protein G-Sepharose beads were first washed three times with 1% Triton X-100, 0.5 m NaCl, 10 mm Tris-HCl, pH 7.4, 1 mm MgCl2, 1 mm CaCl2 and then washed three times in 0.1% Triton X-100, 0.15 m NaCl, 10 mm Tris-HCl, pH 7.4, 1 mm MgCl2, 1 mm CaCl2. After a final centrifugation at 16,000 × g, agarose beads were dissolved in 30 μl of PBS before for further analysis. Successful immunoprecipitation was verified by Western blotting before enzyme assays.
Glycosyltransferase Assay
Glycosyltransferase activities were measured essentially as described (5, 7). Briefly, anti-Myc- or protein G-agarose-bound enzyme protein preparations or crude cell extracts (40–80 μg of protein) were incubated overnight at 37 °C with radiolabeled UDP-GlcA and a GlcNAc[GlcA-GlcNAc]n oligosaccharide acceptor (measuring GlcA-transferase activity) or with radiolabeled UDP-GlcNAc or UDP-GalNAc and a [GlcA-GlcNAc]n acceptor, measuring GlcNAc-transferase and GalNAc-transferase activity, respectively, to heparan oligosaccharide acceptors. Oligosaccharide acceptors were generated from K5 polysaccharide, as described (18). Transfer of radiolabeled UDP-GalNAc to chondroitin oligosaccharides was done using oligosaccharide acceptors generated from defructosylated K4 polysaccharides (19). Labeled oligosaccharides were isolated by gel chromatography and quantified by scintillation counting.
Immunostaining
For immunocytochemical analysis, HEK293 cells, stably transfected with the Myc-His or tGFP expression plasmids, were cultured on coverslips. 24 h after transfection, the cells were washed twice with PBS, fixed with 4% paraformaldehyde in PBS for 10 min, washed three times with PBS, permeabilized in 0.1% Triton X-100 for 15 min in room temperature, and blocked with 10% bovine serum albumin in PBS. Cells were then incubated with the primary monoclonal anti-c-Myc antibody (Sigma) or the anti-tGFP monoclonal antibody 2H8 and the trans-Golgi marker rabbit anti-TGN46 polyclonal antibody (Abcam) at dilutions of 1:500, 1:200, and 1:1000, respectively, for 45 min at 37 °C. Cells were then rinsed three times with PBS and further incubated for 1 h at room temperature with secondary antibodies goat anti-mouse and goat anti-rabbit (both from Santa Cruz Biotechnology) at dilutions of 1:800 and 1:1000, respectively. Finally, cells were mounted in VECTASHIELD with DAPI (Vector Laboratories). The images were visualized under a Zeiss AxioScope microscope equipped with optics for observing fluorescence and captured using a digital AxioCam MRm camera.
Statistical Analysis
To identify differences between two groups, statistical analyses were performed with two-tailed unpaired Student's t test using GraphPad Prism v.6 (GraphPad software, Inc., La Jolla, CA). p values of <0.05 were considered significant.
Flow Cytometry
Cells were harvested using trypsin/EDTA and washed twice in cold PBS. An aliquot of the cells was resuspended in 1 ml of serum-free DMEM containing 20 milliunits of heparitinase I and II (Seikagaku Corp.) and incubated at room temperature for 2 h. After washing with PBS, heparitinase-treated and untreated cells were incubated with the primary monoclonal anti-HS antibody 10E4 (1:50) or 3G10 (1:50) (both from Seikagaku Corp.) at 4 °C for 30 min. After washing with PBS three times, the cells were incubated with the secondary antibodies, allophycocyanin-conjugated goat anti-mouse IgG for 10E4 and FITC-conjugated goat anti-mouse IgG + IgM (both from Jackson ImmunoResearch Laboratories, Inc.), for 3G10. Cells incubated with the secondary antibodies alone served as negative controls.
Analysis of FGF2-induced Signaling
Subconfluent cell cultures grown in a 6-well plate were washed with serum-deprived DMEM and starved for 18 h in the same medium. After changing to fresh starvation medium, FGF2 (R&D Systems) was added at 10 ng/ml final concentration and allowed to stimulate cultures for 10 min at 37 °C. Cell extracts were separated on SDS-10% polyacrylamide gels and electrotransferred onto a nitrocellulose membrane (Amersham Biosciences). For FGF2-mediated ERK phosphorylation, the membrane was incubated with mouse anti-phospho-ERK-1/2 (1:1000, Cell Signaling Technology) recognizing the activated, phosphorylated kinase, and after stripping, the membrane was incubated with mouse anti-total ERK-1/2 (1:1000, Cell Signaling Technology) recognizing both the unphosphorylated and the phosphorylated kinase and GAPDH (1:500, Santa Cruz Biotechnology). The membranes were developed using ECL reagent (Pierce), and the bands were visualized using ChemiDoc XRS device and the Quantity One 1-D Analysis Software (Bio-Rad) and quantified by using ImageJ software (National Institutes of Health, Bethesda, MD).
Results
EXTL2 has been described to have two in vitro glycosyltransferase activities, including transfer of GlcNAc and GalNAc to HS oligosaccharides representing the glycosaminoglycan protein linkage region and thus has been proposed to be an initiator and/or inhibitor of HS chain elongation (13). To assess the role of EXTL2 in HS chain elongation, we used an siRNA approach to reduce EXTL2 levels and an overexpression strategy to increase EXTL2 levels in HEK293 cells.
Effect of siRNA-mediated Silencing of EXTL2 on HS Structure
HEK293 cells were transiently transfected with different siRNAs targeting human EXTL2, non-targeting control siRNAs (Table 1), or mock-transfected. As determined by real-time PCR, EXTL2 mRNA expression levels in cells treated with siRNA against EXTL2 were reduced to ∼15–30% of that of the control siRNA- or mock-transfected cells (Fig. 1). To investigate the effect of EXTL2 reduction on HS synthesis, siRNA-treated cells were metabolically labeled for 24 h with [3H]glucosamine or [35S]sulfate. Proteoglycans were isolated from the cell surface/matrix and from the culture medium, and the glycosaminoglycan chains were released from the core proteins. For HS chain length analysis, only chains isolated from cell surface/matrix proteoglycans were used to ensure that the HS chains were intact and not processed by the lysosomal enzymes or derived from proteoglycans undergoing HS chain synthesis. Down-regulation of EXTL2 led to a consistent increased HS chain length (Fig. 2) and a small increase in labeled HS relative to chondroitin sulfate (Table 2), but no significant differences were noted in the yields of total labeled glycosaminoglycans (calculated as cpm/μg of cellular protein or as cpm/culture flask, data not shown).
FIGURE 1.
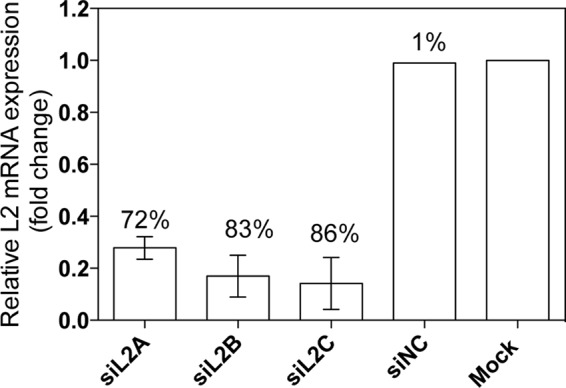
EXTL2 mRNA expression levels after siRNA-mediated knockdown of EXTL2 in HEK293 cells. HEK293 cells were transiently transfected with three different human siRNAs (siL2A, siL2B, and siL2C) targeting EXTL2, one negative non-targeting control siRNA (siNC), and transfection reagent only (Mock) as described under “Experimental Procedures.” 24 h after transfection, relative mRNA levels were determined by real-time PCR and normalized to those of β-actin. mRNA levels after siRNA treatments are expressed relative to the mock expression that was set to 1. The error bars represent the mean ± S.D. from at least four independent transfections. Each measurement was performed in duplicate or triplicate. The numbers above the bars show the percentage of down-regulation.
FIGURE 2.
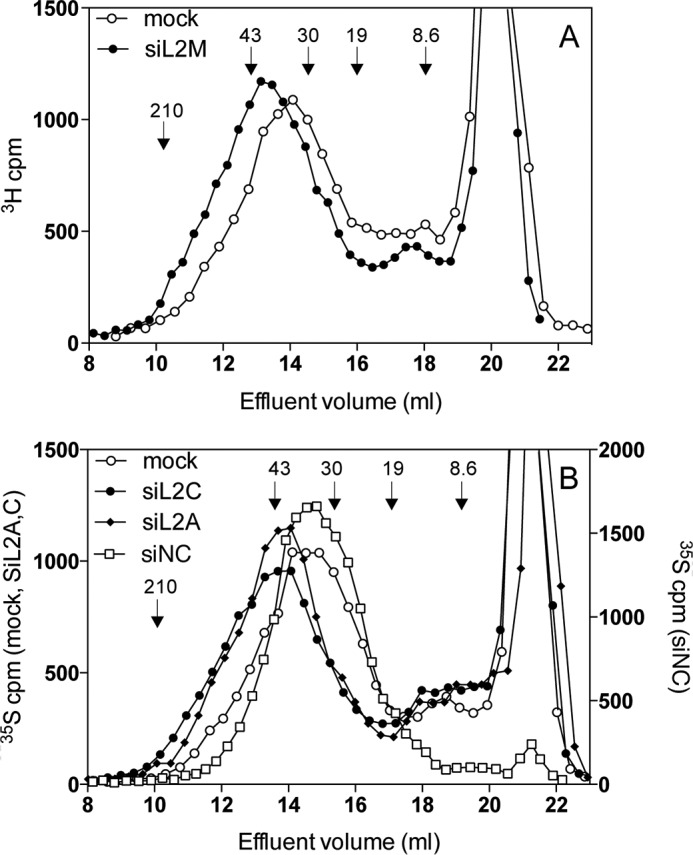
Effect of siRNA-mediated knockdown of EXTL2 on HS chain length. A and B, 3H-labeled (A) or 35S-labeled (B) HS was purified from the cell surface/extracellular matrix of siRNA (siL2M, siL2A, siL2C) and the non-targeting control siRNA-transfected (siNC) and mock-transfected (Mock) HEK293 cells 48 h after transfection as described under “Experimental Procedures.” Isolated glycosaminoglycans were digested with chondroitinase ABC and subjected to gel chromatography on a Superose 6 column. The retarded components eluting at ∼19–23 ml correspond to material that was degraded by chondroitinase ABC. Similar results were obtained from three independent transfections. The samples in A and B were run on two different Superose 6 columns calibrated using size-defined fragments of heparin and hyaluronan. The elution positions of molecular mass standards derived from heparin (8.6 kDa) and hyaluronan (19, 30, 43 and 210 kDa) are indicated by arrows.
TABLE 2.
Glycosaminoglycan synthesis and disaccharide composition of labeled HS isolated after mock or siRNA mediated knockdown of EXTL2 in HEK 293 cells
Labeled glycosaminoglycans were extracted from the cell surface and quantified. The percentage values of HS and CS are given from a single experiment (values within parentheses refer to a [3H[GlcN-labeled sample) or as means from three independent 35S labeling experiments ± mean deviation.35S-labeled samples were degraded by deamination at pH 1.5, followed by reduction of products with NaBH4. The resulting labeled disaccharides were analyzed by anion-exchange HPLC (see “Experimental Procedures”). The values for mock-treated and cells treated with siRNA against EXTL2 are given as means from two independent labelling experiments ± mean deviation. Mock, mock transfected cells; siNC, cells transfected with non-target control siRNA; siL2, cells transfected with siRNA against EXTL2.
| Cell type | Labeled glycosaminoglycans |
35S-labeled deamination productsa |
||||
|---|---|---|---|---|---|---|
| HSb | CSc | GlcA-aManR6Sd | IdoA-aManR6S | IdoA2S-aManR | IdoA2S-aManR6S | |
| % | % | |||||
| Mock | 51 ± 10 (42) | 49 ± 10 (52) | 18 ± 1.4 | 7 ± 1.4 | 56.5 ± 5.5 | 19 ± 4 |
| siNC | NDe | ND | 14 | 6 | 55 | 24 |
| siL2 | 57 ± 9 (57) | 44 ± 9 (40) | 17 ± 1 | 6.5 ± 0.5 | 58.5 ± 2.5 | 19 ± 4 |
a % of total O-sulfated disaccharides.
b Material resistant to chondroitinase ABC.
c Material susceptible to chondroitinase ABC.
d aManR, the 2,5-anhydromannitol deamination product of GlcNS residues.
e ND, not determined.
Detailed information regarding the distribution of sulfate groups was obtained by analysis of the disaccharides generated by low pH HNO2/NaBH4 treatment of 35S-labeled HS. The HS sulfation pattern occurs in a more or less block-wise fashion creating three structurally different domains (20, 21), contiguous N-sulfated regions, non-sulfated N-acetylated regions, and regions with short mixed sequences where N-acetylated disaccharides alternate with N-sulfated disaccharides. Depolymerization with nitrous acid at pH 1.5 and reduction with NaBH4 resulted in cleavage of HS chains at N-sulfated GlcN residues and conversion of GlcNS units to aManR. Under these conditions, N-acetylated units remain intact and consecutive N-sulfated disaccharides will be recovered as disaccharides. The resultant HS disaccharides were separated by anion-exchange HPLC. No significant difference in disaccharide composition was observed after siRNA treatment (Table 2). Thus, although EXTL2 down-regulation significantly increased HS chain length, it did not appear to significantly affect the sulfation pattern of HS. However, we cannot exclude that some variations in the sulfation pattern may have occurred in the N-acetylated units flanking the continuous N-sulfated domains.
Effect of EXTL2 Overexpression on HS Structure
To investigate whether overexpression of EXTL2 would affect HS chain elongation, we initially stably transfected HEK293 cells with a C-terminal Myc-His-tagged vector. Individual cell clones from EXTL2-transfected cells were selected for EXTL2 overexpression by Western blotting. However, immunofluorescence using an antibody against the Myc tag revealed that the level of overexpression was relatively low. Only ∼20–30% of the cells expressed the Myc-tagged EXTL2 (data not shown). We therefore stably transfected HEK293 cells with a tGFP-tagged EXTL2 construct and again analyzed clones for EXTL2 overexpression by Western blotting and immunofluorescence staining. Approximately 70–80% of the cells overexpressed the tGFP-tagged EXTL2. The fluorescence of the tagged EXTL2 protein was observed in the perinuclear region (Fig. 3), indicating an endoplasmic reticulum-Golgi localization. Real-time PCR (Fig. 4A) and Western blotting (Fig. 4B) confirmed a high level of EXTL2 overexpression. We were also interested to determine whether there was a possible co-regulation of EXT1, EXT2, EXTL1, EXTL3, and EXTL2 at the mRNA level. Therefore, we examined whether overexpression of EXTL2 would affect the transcript levels of the other EXT(L)s. We did not observe any statistically significant changes in EXT1, EXT2, or EXTL3 mRNA levels, although there was a small reproducible decrease in EXT2 mRNA levels for all clones tested (Fig. 4A, inset). Furthermore, real-time PCR revealed that HEK293 cells did not express EXTL1, which is in accordance with its known very restricted cell and tissue expression (12).
FIGURE 3.
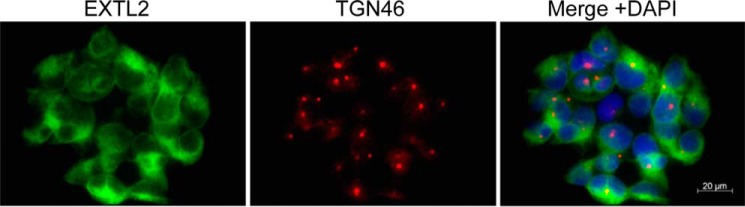
Subcellular localization of EXTL2 in HEK293 cells. HEK293 cells were stably transfected with full-length tGFP-EXTL2 expression plasmid. Transfectants were fixed and stained for EXTL2 (green) and the trans-Golgi marker TGN46 (red) as described under “Experimental Procedures.” The nuclei were counterstained with DAPI (blue).
FIGURE 4.
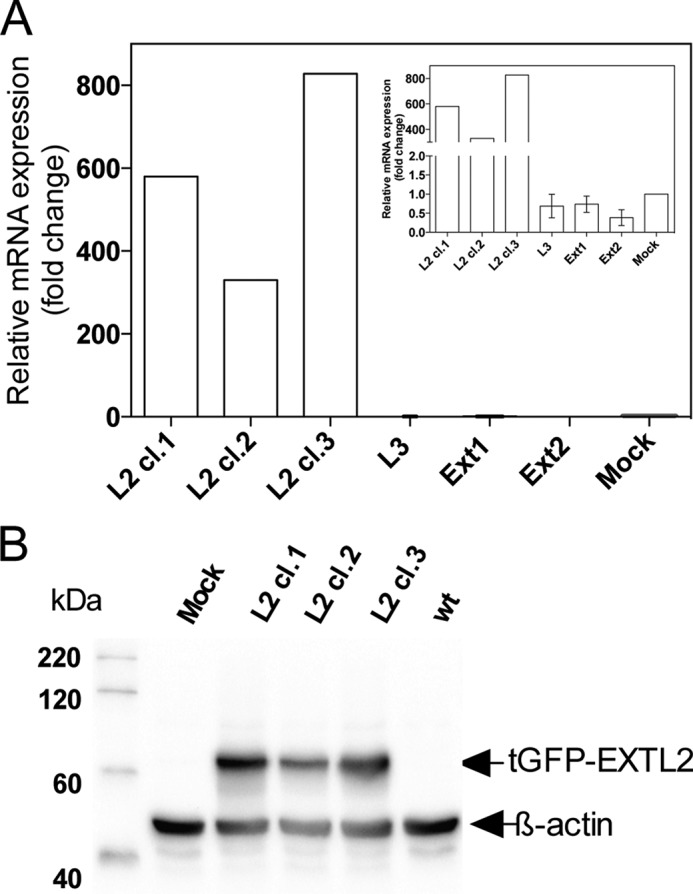
EXTL2 expression levels after overexpression of EXTL2 in HEK293 cells. HEK293 cells were stably transfected with the full-length tGFP-tagged EXTL2 and with vector only (Mock) as described under “Experimental Procedures.” A, mRNA levels for three clones stably overexpressing EXTL2 (L2cl.1, L2cl.2, and L2cl.3) were determined by real-time PCR and normalized to those of β-actin. The mRNA levels after overexpression are expressed as -fold changes relative to the mock expression that was set to 1. Each measurement was performed in triplicate. The mRNA levels of EXT1, EXT2, and EXTL3 (L3) were also determined for the three EXTL2-overexpressing clones (inset). The values for EXT1, EXT2, and EXTL3 are the average values across the three overexpressing clones from two independent experiments. The error bars represent mean values ± S.D. B, Western blot of the same three clones (L2cl.1, L2cl.2, and L2cl.3) stably overexpressing EXTL2 as in A. Cell extracts were separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane (“Experimental Procedures”). The blot was probed with antibodies against tGFP and β-actin and visualized by chemiluminescence.
Several EXTL2-overexpressing clones were selected and used to investigate the effect of increased EXTL2 expression on HS synthesis. Overexpression of EXTL2 did not significantly change the total amount of [3H]glucosamine- or [35S]sulfate-produced glycosaminoglycans nor the HS/chondroitin sulfate ratio as compared with mock-transfected cells (47 ± 7% HS and 46 ± 7% HS in mock- and EXTL2-transfected HEK293 cells, respectively), nor did the overexpression of EXTL2 significantly affect HS disaccharide composition (Table 3) or HS chain elongation, although some variations in chain length were observed between different labeling experiments (Fig. 5). The effect on HS chain length after EXTL2 overexpression was not due to the level of overexpression because similar results were obtained for the low overexpressing Myc-EXTL2 clones and the high expressing tGFP-EXTL2 clones. The relative amounts of cell surface HS expressed by HEK293 cells, mock-transfected cells, and the EXTL2-overexpressing cells were determined by flow cytometry using the 10E4 and 3G10 antibodies. The 10E4 antibody, specific for HS chains, recognizes sulfated regions within HS chains, whereas the 3G10 recognizes the unsaturated hexuronic acid-containing oligosaccharide stubs that remain associated with core proteins after heparitinase digestion (22); thus, 3G10 serves as a marker of the amount of HS chains. The binding of the 10E4 antibody to EXTL2-overexpressing cells indicated a larger, more diverse population of HS chains relative to wild-type and mock-transfected cells (Fig. 6A). No significant difference in the 3G10 antibody staining could be observed between the wild-type or mock-transfected cells and the EXTL2-expressing cells after digestion with a combination of heparitinase I and II (Fig. 6, B and C). Thus, our results indicate that the wild-type and EXTL2-overexpressing cells have similar numbers of HS chains exposed at the cell surface, but that these chains contain different binding epitopes for the 10E4 antibody.
TABLE 3.
Disaccharide composition of labeled HS isolated from mock or EXTL2 transfected HEK 293 cells.
Labeled samples were degraded by deamination at pH 1.5, followed by reduction of products with NaBH4. The resulting 35S- or 3H-labeled disaccharides were analyzed by anion-exchange HPLC (see “Experimental Procedures”). The values are given from single analyses or as means from metabolic labeling of two different EXTL2-overexpressing clones or mock-transfected clones ± mean deviation.
| Cell type |
35S-labeled deamination productsa |
3H-labeled deamination productsa |
||||||
|---|---|---|---|---|---|---|---|---|
| GlcA-aManR6Sb | IdoA-aManR6S | IdoA2S-aManR | IdoA2S-aManR6S | GlcA-aManR6S | IdoA-aManR6S | IdoA2S-aManR | IdoA2S-aManR6S | |
| % | % | |||||||
| HEK 293c | NDd | ND | ND | ND | 16 | 6 | 57 | 21 |
| Mock | 15 ± 0 | 6.5 ± 0.5 | 56.5 ± 3.5 | 22 ± 4 | 17.5 ± 0.5 | 6.5 ± 0.5 | 44.5 ± 1.5 | 31 ± 3 |
| EXTL2 | 17.5 ± 2.5 | 7 ± 1 | 60.5 ± 1.5 | 16 ± 6 | 16 ± 0 | 5.5 ± 0.5 | 55 ± 2 | 23.5 ± 2.5 |
a % of total O-sulfated disaccharides.
b Untransfected 293 cells.
c aManR, the 2,5- anhydromannitol deamination products of GlcNS residues.
d ND, not determined.
FIGURE 5.
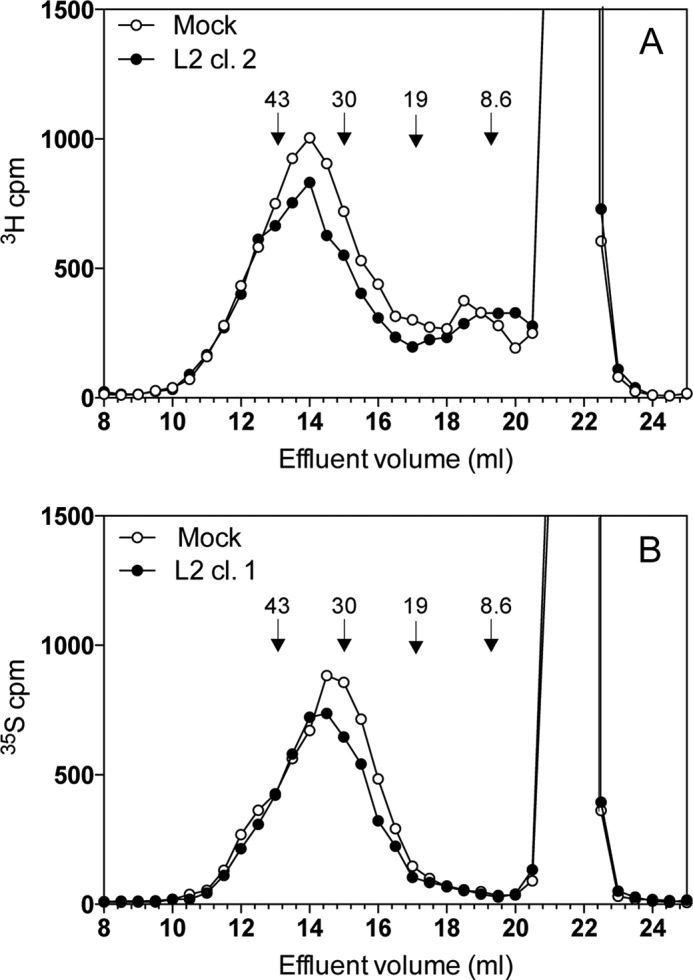
Effect of EXTL2 overexpression on HS chain length. A and B, 3H-labeled (A) or 35S-labeled (B) glycosaminoglycans were purified from the cell surface/extracellular matrix of HEK293 cells stably transfected with EXTL2, as described under “Experimental Procedures.” The resulting labeled polysaccharides were digested with chondroitinase ABC and subjected to chromatography on a Superose 6 column. The retarded component resulting at 20–23 ml corresponds to material that is degraded by chondroitinase ABC treatment. Similar results were obtained from four independent transfections with different overexpression levels. The elution positions of molecular mass standards derived from heparin (8.6 kDa) and hyaluronan (19, 30, and 43 kDa) are indicated by arrows.
FIGURE 6.
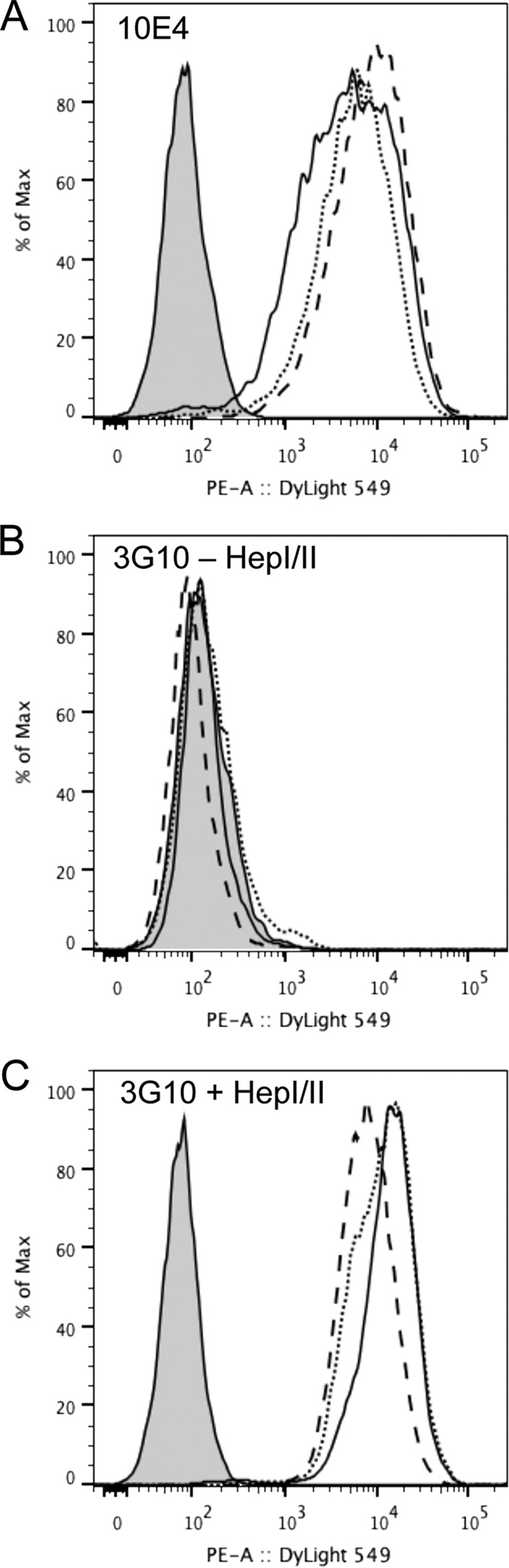
Cell surface expression of HS on HEK293 cells overexpressing EXTL2. A–C, EXTL2-overexpressing, mock-transfected, and wild-type cells were examined for cell surface expression of HS by flow cytometry using the 10E4 antibody (A) and the 3G10 antibody before (B) and after (C) heparitinase (Hep) digestion. The histograms show the intensity of the fluorescence (percentage of maximum (% of max)) and is representative of two independent experiments. Black line, EXTL2-overexpressing cells; dashed line, mock-transfected cells; dotted line, wild-type cells; filled gray profile, control (secondary antibody only). PE-A, Phycoerythrin-Area.
Glycosyltransferase Activities of Expressed EXTL2
As mentioned above, EXTL2 has been described to have two in vitro glycosyltransferase activities, including transfer GlcNAc and GalNAc to HS oligosaccharides representing the glycosaminoglycan protein linkage region, and thus has been proposed to be an initiator/terminator of HS synthesis (13). The addition of the GlcNAc residue to the linkage tetrasaccharide region is the key step to the initiation of HS polymerization. However, the reported GlcNAc-transferase activity of EXTL2 is weak and much lower than its α-GalNAc-transferase activity (13).
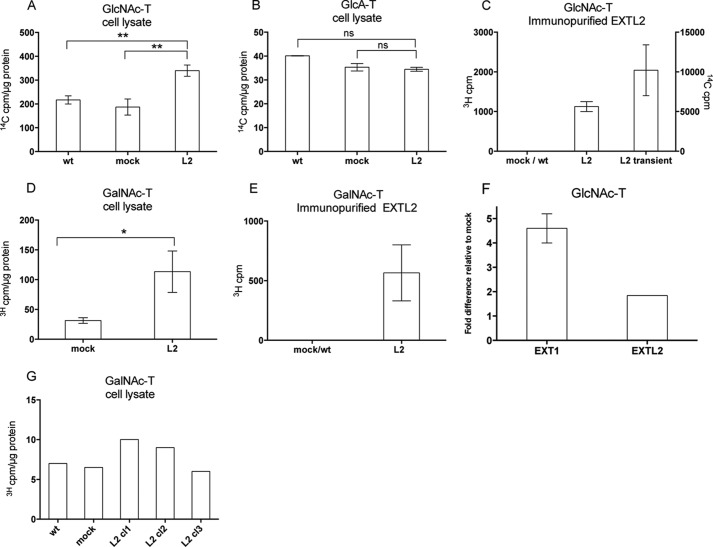
In view of these findings and the results on HS chain elongation after siRNA/overexpression treatment, we wanted to clarify whether EXTL2 also could transfer GlcNAc and GalNAc to the growing HS chains. Crude cell lysates of transfected cells were analyzed for GlcA-, GlcNAc-, and GalNAc-transferase activities using oligosaccharide preparations with nonreducing terminal GlcNAc or GlcA residues, respectively, as acceptors (see “Experimental Procedures”). These acceptor substrates mimic intermediates of the growing HS chain and are considered to measure enzyme activities involved in the elongation of HS. The analysis using crude cell lysates clearly demonstrated increased GlcNAc- and GalNAc-transferase activities in cells that had been transfected with EXTL2, whereas no significant differences were observed in the GlcA-transferase activities (Fig. 7, A, B, and D). Additionally, we did not observe any significant GalNAc transfer to chondroitin oligosaccharides (Fig. 7G). To relate the activity of EXTL2 to that of EXT1, with clearly demonstrated GlcNAc-transferase activities (7), the GlcNAc-transferase activities of EXTL2-overexpressing clones were compared with those of EXT1-overexpressing clones (5). The enzyme activities of the EXT1-expressing cells were ∼2.5-fold higher than the corresponding values for EXTL2-expressing cell clones (Fig. 7F). However, taking into account that the overexpression of EXTL2 was ∼30 times higher than that of EXT1, the actual transfer activities of EXTL2 appear much less than those of EXT1.
FIGURE 7.
Glycosyltransferase activities of HEK293 cells overexpressing EXTL2. Glycosyltransferase activities of cell extracts or immunopurified EXTL2, from untransfected (wt), empty vector-transfected (mock), EXTL2-transfected (L2), and EXT2-transfected HEK293 cells are shown as indicated. In A–F, oligosaccharides derived from Escherichia coli K5 capsular polysaccharide were used as acceptor substrates in transferase assays. The K5 polysaccharide has the same structure as the nonsulfated HS precursor molecule, and oligosaccharide derivatives thereof, with non-reducing terminal GlcA or GlcNAc residues served as acceptors in the GlcNAc- and GlcA-transferase reactions. C, shows the GlcNAc-transferase activity of immunopurified EXTL2 after transient transfection, whereas the other panels show enzyme activities after stable transfection. No GlcA-transferase activities were detected with immunopurified EXTL2 (data not shown). In G, oligosaccharides with a non-reducing GlcA residue, generated from defructosylated E. coli K4 capsular polysaccharide with the same structure as the nonsulfated chondroitin sulfate precursor molecule, were used as acceptor substrate in transferase assays. The error bars represent mean values ± S.D. from four (A) and two (B, D, and E) independent EXTL2-overexpressing clones and from one clone (F). G, representative results from one out of two independent analyses. Each clone was assayed at two different protein concentrations. *, p < 0.01, **, p < 0.001. ns, not significant.
To eliminate any effects of endogenous EXT proteins, the experiments above were repeated using immunopurified EXTL2. EXTL proteins were captured on Myc-agarose beads or on protein G-Sepharose (see “Experimental Procedures”) and analyzed for transferase activities. To ascertain that the EXTL2 protein was present on the agarose beads, a portion of the beads was analyzed by Western blotting with antibodies to Myc or to EXTL2. The immunopurified EXTL2 exhibited no detectable GlcA-transferase activity (data not shown), but exhibited highly reproducible GlcNAc and GalNAc transfer to HS oligosaccharides (Fig. 7, C and E). These results clearly demonstrated that, under our experimental conditions, EXTL2, in contrast to previous results (23), catalyzed the incorporation of GlcNAc and, although with much lower capacity, also catalyzed the incorporation of GalNAc to the growing HS chain in vitro.
Effect of EXTL2 Overexpression on HS Growth Factor Signaling
HS acts as an important co-receptor for several growth factors. The interaction of FGF2 with HS has received much attention because of the strong dependence on HS for FGF2-induced biological cellular responses. Therefore, to determine whether overexpression of EXTL2 had a biological effect, we examined FGF2-induced signaling in EXTL2-overexpressing, mock-transfected, and wild-type HEK293 cells. We observed no differences in signaling response between the different cell types (Fig. 8), indicating that EXTL2-overexpressing cells have HS chains with similar binding epitopes for FGF2.
FIGURE 8.
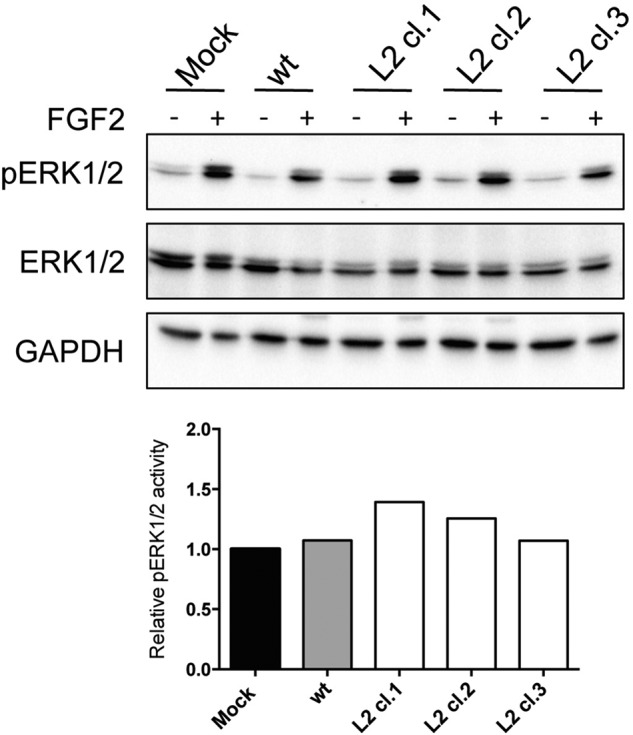
Effect of EXTL2 overexpression on FGF2-induced signaling. Three clones stably overexpressing EXTL2 (L2cl.1, L2cl.2, and L2cl.3) wild-type (wt) and mock-transfected (Mock) cells were starved in serum-free medium for 18 h and then stimulated with 10 ng/ml FGF2. After 10 min, cells were lysed, and the cell lysates were subjected to SDS-PAGE followed by Western blotting using phosphospecific ERK (pERK1/2), total ERK, and GAPDH antibodies. The lower panel shows the relative intensity of immunoreactive bands of phosphorylated versus total ERK normalized to the mock-transfected cells. The figures show representative results from one out of three independent experiments.
Discussion
Because the three EXTL proteins share amino acid sequence homology with EXT1 and EXT2 and have glycosyltransferase activities related to HS biosynthesis, it is expected that these proteins are involved in HS synthesis. However, except for EXTL3, which in contrast to the other two EXTLs is expressed in all investigated species from sea anemones to humans (24), and for which most published data suggest that it is an initiator of HS chains (12), the functions of the other EXTL proteins are unclear. In this study, we were particularly interested in studying the involvement of EXTL2 in human HS biosynthesis. Our data, based on siRNA-mediated down-regulation, clearly showed that the levels of EXTL2 influenced HS chain elongation.
EXTL2 has previously been shown to be able to transfer GalNAc and GlcNAc to artificial oligosaccharide acceptors resembling the polysaccharide protein linkage region but not to oligosaccharide substrates comparable with intermediates in HS polymerization (23). Consistent with previous results, recombinant EXTL2 did not show GlcA-transferase activity but, in contrast to previous studies (23), we found that EXTL2 exhibits significant GlcNAc-transferase activity toward oligosaccharide substrates. One possible explanation for this discrepancy may be that in the previous study, a truncated soluble form of EXTL2 was used as the enzyme source and thus did not contain full activity.
Our knowledge of how the synthesis of HS is regulated is surprisingly limited. The mechanism responsible for regulation of HS chain elongation is not known, and the exact role of each particular EXT and EXTL protein is still an enigma. EXTL2 was first suggested to be a terminator of HS and chondroitin sulfate chains, by transfer of GalNAc α-linked to the linkage tetrasaccharide GlcA residue (13). In a later publication, it was speculated that in cells lacking EXT1, HS chains were initiated by EXTL2 and polymerization was carried out by EXT2 (25). In a more recent publication by the same group, it was instead suggested that chain initiation by EXTL3 may be prevented by a FAM20B-catalyzed phosphorylation of the xylose residue followed by EXTL2 transfer of a GlcNAc residue to form the phosphorylated pentasaccharide, GlcNAc-GlcUA-Gal-Gal-Xyl(2-O-phosphate), which functions as a chain termination signal (26).
For structural studies, we focused on the cell surface/matrix-associated proteoglycans, and we found that, after siRNA treatment, cells produced longer HS chains and that the ratio of HS to chondroitin sulfate slightly increased without a significant change in the total amount of secreted glycosaminoglycans. Reducing the amounts of a chain terminator would naturally lead to longer HS chains in agreement with our results, but overexpression of EXTL2 did not significantly alter the HS chain length or the amounts of HS as would have been expected if EXTL2 were a terminator of HS chains. However, we cannot exclude that the structural changes induced by siRNA or the lack of changes after overexpression of EXTL2 could be specific for HEK293 cells and, possibly, distinctive from other cell types.
The observed lack of effect of EXTL2 overexpression was not due to the lack of FAM20B (Fig. 9) but could be due to the fact that the untransfected cells produce saturating levels of EXTL2, or that an unidentified partner is needed for EXTL2 to exhibit its function, or considering the proposed function of EXTL2 in regulating glycosaminoglycan synthesis (26), that FAM20B does not phosphorylate xylose in HEK293 cells.
FIGURE 9.
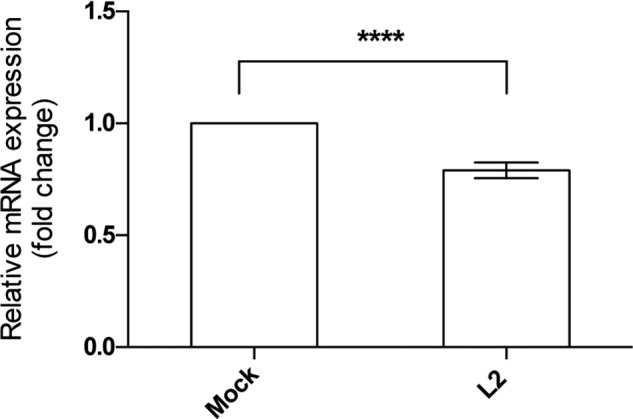
FAM20B mRNA expression level in HEK293 cells overexpressing EXTL2. FAM20B relative mRNA levels were determined by real-time PCR and normalized to those of β-actin. mRNA levels are expressed as fold changes relative to the levels of mock-transfected (Mock) cells that were set to 1. The error bars represent mean ± S.D. of values from four different EXTL2 (L2)-overexpressing clones measured in triplicates. ****, p < 0.0001.
In addition to FAM20B, other cellular components may influence EXTL2 function and HS biosynthesis. Although all the biosynthetic enzymes involved in HS biosynthesis have been cloned, we still know remarkably little about the organization of HS biosynthetic apparatus, the localization of the enzymes in the Golgi membrane, and their interaction with each other and with other proteins in the endoplasmic reticulum and in the Golgi apparatus. The GAGosome is one way to try to describe HS biosynthetic machinery and is defined as a complex of different HS enzyme/enzyme isoforms (27). In the GAGosome model, the relative amounts of each enzyme and the type of enzymes present in the GAGosome will have an impact on the degree of chain length and/or chain modification. However, whether the EXTL2 collaborates or forms complexes with other proteins involved in HS biosynthesis is still an open question. In an attempt to identify EXTL2-protein interactions, we performed co-immunoprecipitation-coupled mass spectroscopy. As of now for the most part, we have not been able to identify any EXTL2 interacting partners. The majority of the co-immunoprecipitated proteins were tubulin and heat shock proteins.4
Taken together, the findings regarding the activities of EXTL2 are contradictory and very confusing. Additional more refined experiments will be needed to resolve the function of EXTL2 and to determine whether it really plays any direct role in HS synthesis.
This work was supported by grants from the University of Bergen and the Norwegian Cancer Society (3292722-2012).
K. Katta, M. Grønning, and M. Kusche-Gullberg, unpublished results.
- HS
- heparan sulfate
- EXT
- exostosin
- EXTL
- exostosin-like
- GlcA
- d-glucuronic acid
- GlcNS
- N-sulfated d-glucosamine
- aManR
- 2,5-anhydro-d-mannitol (formed by reduction of terminal 2,5-anhydromannose residues with NaBH4)
- NS
- N-sulfate group
- 2S
- 2-O-sulfate group
- 6S
- 6-O-sulfate group
- tGFP
- TurboGFP.
References
- 1. Sarrazin S., Lamanna W. C., Esko J. D. (2011) Heparan sulfate proteoglycans. Cold Spring Harb. Perspect. Biol. 3, a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lindahl U., Li J. P. (2009) Interactions between heparan sulfate and proteins: design and functional implications. Int. Rev. Cell Mol. Biol. 276, 105–159 [DOI] [PubMed] [Google Scholar]
- 3. Whitelock J. M., Iozzo R. V. (2005) Heparan sulfate: a complex polymer charged with biological activity. Chem. Rev. 105, 2745–2764 [DOI] [PubMed] [Google Scholar]
- 4. Sugahara K., Kitagawa H. (2002) Heparin and heparan sulfate biosynthesis. IUBMB Life 54, 163–175 [DOI] [PubMed] [Google Scholar]
- 5. Busse M., Feta A., Presto J., Wilén M., Grønning M., Kjellén L., Kusche-Gullberg M. (2007) Contribution of EXT1, EXT2, and EXTL3 to heparan sulfate chain elongation. J. Biol. Chem. 282, 32802–32810 [DOI] [PubMed] [Google Scholar]
- 6. Kim B. T., Kitagawa H., Tamura J., Saito T., Kusche-Gullberg M., Lindahl U., Sugahara K. (2001) Human tumor suppressor EXT gene family members EXTL1 and EXTL3 encode α1,4-N-acetylglucosaminyltransferases that likely are involved in heparan sulfate/ heparin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 98, 7176–7181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Busse M., Kusche-Gullberg M. (2003) In vitro polymerization of heparan sulfate backbone by the EXT proteins. J. Biol. Chem. 278, 41333–41337 [DOI] [PubMed] [Google Scholar]
- 8. McCormick C., Duncan G., Goutsos K. T., Tufaro F. (2000) The putative tumor suppressors EXT1 and EXT2 form a stable complex that accumulates in the Golgi apparatus and catalyzes the synthesis of heparan sulfate. Proc. Natl. Acad. Sci. U.S.A. 97, 668–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Senay C., Lind T., Muguruma K., Tone Y., Kitagawa H., Sugahara K., Lidholt K., Lindahl U., Kusche-Gullberg M. (2000) The EXT1/EXT2 tumor suppressors: catalytic activities and role in heparan sulfate biosynthesis. EMBO Rep. 1, 282–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cook A., Raskind W., Blanton S. H., Pauli R. M., Gregg R. G., Francomano C. A., Puffenberger E., Conrad E. U., Schmale G., Schellenberg G., et al. (1993) Genetic heterogeneity in families with hereditary multiple exostoses. Am. J. Hum. Genet. 53, 71–79 [PMC free article] [PubMed] [Google Scholar]
- 11. Wu Y. Q., Heutink P., de Vries B. B., Sandkuijl L. A., van den Ouweland A. M., Niermeijer M. F., Galjaard H., Reyniers E., Willems P. J., Halley D. J. (1994) Assignment of a second locus for multiple exostoses to the pericentromeric region of chromosome 11. Hum. Mol. Genet. 3, 167–171 [DOI] [PubMed] [Google Scholar]
- 12. Busse-Wicher M., Wicher K. B., Kusche-Gullberg M. (2014) The exostosin family: proteins with many functions. Matrix Biol. 35, 25–33 [DOI] [PubMed] [Google Scholar]
- 13. Kitagawa H., Shimakawa H., Sugahara K. (1999) The tumor suppressor EXT-like gene EXTL2 encodes an α1,4-N-acetylhexosaminyltransferase that transfers N-acetylgalactosamine and N-acetylglucosamine to the common glycosaminoglycan-protein linkage region: the key enzyme for the chain initiation of heparan sulfate. J. Biol. Chem. 274, 13933–13937 [DOI] [PubMed] [Google Scholar]
- 14. Lennon G., Auffray C., Polymeropoulos M., Soares M. B. (1996) The I.M.A.G.E. Consortium: an integrated molecular analysis of genomes and their expression. Genomics 33, 151–152 [DOI] [PubMed] [Google Scholar]
- 15. Wang X., Seed B. (2003) A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 31, e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 17. Osterholm C., Barczyk M. M., Busse M., Grønning M., Reed R. K., Kusche-Gullberg M. (2009) Mutation in the heparan sulfate biosynthesis enzyme EXT1 influences growth factor signaling and fibroblast interactions with the extracellular matrix. J. Biol. Chem. 284, 34935–34943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yamada S., Busse M., Ueno M., Kelly O. G., Skarnes W. C., Sugahara K., Kusche-Gullberg M. (2004) Embryonic fibroblasts with a gene trap mutation in Ext1 produce short heparan sulfate chains. J. Biol. Chem. 279, 32134–32141 [DOI] [PubMed] [Google Scholar]
- 19. Lidholt K., Fjelstad M. (1997) Biosynthesis of the Escherichia coli K4 capsule polysaccharide: a parallel system for studies of glycosyltransferases in chondroitin formation. J. Biol. Chem. 272, 2682–2687 [DOI] [PubMed] [Google Scholar]
- 20. Lyon M., Gallagher J. T. (1998) Bio-specific sequences and domains in heparan sulphate and the regulation of cell growth and adhesion. Matrix Biol. 17, 485–493 [DOI] [PubMed] [Google Scholar]
- 21. Maccarana M., Sakura Y., Tawada A., Yoshida K., Lindahl U. (1996) Domain structure of heparan sulfates from bovine organs. J. Biol. Chem. 271, 17804–17810 [DOI] [PubMed] [Google Scholar]
- 22. David G., Bai X. M., Van der Schueren B., Cassiman J. J., Van den Berghe H. (1992) Developmental changes in heparan sulfate expression: in situ detection with mAbs. J. Cell Biol. 119, 961–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kitagawa H., Egusa N., Tamura J. I., Kusche-Gullberg M., Lindahl U., Sugahara K. (2001) rib-2, a Caenorhabditis elegans homolog of the human tumor suppressor EXT genes encodes a novel α1,4-N-acetylglucosaminyltransferase involved in the biosynthetic initiation and elongation of heparan sulfate. J. Biol. Chem. 276, 4834–4838 [DOI] [PubMed] [Google Scholar]
- 24. Feta A., Do A. T., Rentzsch F., Technau U., Kusche-Gullberg M. (2009) Molecular analysis of heparan sulfate biosynthetic enzyme machinery and characterization of heparan sulfate structure in Nematostella vectensis. Biochem. J. 419, 585–593 [DOI] [PubMed] [Google Scholar]
- 25. Okada M., Nadanaka S., Shoji N., Tamura J., Kitagawa H. (2010) Biosynthesis of heparan sulfate in EXT1-deficient cells. Biochem. J. 428, 463–471 [DOI] [PubMed] [Google Scholar]
- 26. Nadanaka S., Zhou S., Kagiyama S., Shoji N., Sugahara K., Sugihara K., Asano M., Kitagawa H. (2013) EXTL2, a member of the EXT family of tumor suppressors, controls glycosaminoglycan biosynthesis in a xylose kinase-dependent manner. J. Biol. Chem. 288, 9321–9333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Presto J., Thuveson M., Carlsson P., Busse M., Wilén M., Eriksson I., Kusche-Gullberg M., Kjellén L. (2008) Heparan sulfate biosynthesis enzymes EXT1 and EXT2 affect NDST1 expression and heparan sulfate sulfation. Proc. Natl. Acad. Sci. U.S.A. 105, 4751–4756 [DOI] [PMC free article] [PubMed] [Google Scholar]