Background: H2A.Z is a histone variant that replaces H2A at strategic locations within the genome.
Results: Expression of non-acetylatable H2A.Z blocks myogenic differentiation.
Conclusion: H2A.Z acetylation is important for MyoD expression in myoblasts, which in turn is required for myogenesis.
Significance: These results demonstrate that expression of a mutant H2A.Z can lead to dysregulation and inhibition of cellular differentiation programs.
Keywords: chromatin, gene regulation, histone modification, myogenesis, transcription, H2A.Z, histone variant
Abstract
H2A.Z is a histone H2A variant that is essential for viability in Tetrahymena and Drosophila and also during embryonic development of mice. Although implicated in diverse cellular processes, including transcriptional regulation, chromosome segregation, and heterochromatin formation, its essential function in cells remains unknown. Cellular differentiation is part of the developmental process of multicellular organisms. To elucidate the roles of H2A.Z and H2A.Z acetylation in cellular differentiation, we examined the effects of expressing wild type (WT) or a non-acetylatable form of H2A.Z in the growth and differentiation of the myoblast C2C12 cell line. Ectopic expression of wild type or mutant H2A.Z resulted in distinct phenotypes in the differentiation of the C2C12 cells and the formation of myotubes. Most strikingly, expression of the H2A.Z non-acetylatable mutant (H2A.Z-Ac-mut) resulted in a complete block of myoblast differentiation. We determined that this phenotype is caused by a loss of MyoD expression in the Ac-mut-expressing cells prior to and after induction of differentiation. Moreover, chromatin accessibility assays showed that the promoter region of MyoD is less accessible in the differentiation-defective cells. Altogether, these new findings show that expression of the Ac-mut form of H2A.Z resulted in a dominant phenotype that blocked differentiation due to chromatin changes at the MyoD promoter.
Introduction
H2A.Z is a variant of the canonical histone H2A that is essential for viability in a number of organisms (1–3). At the amino acid level, the H2A.Z sequence is ∼90% conserved among vertebrate species, whereas H2A is only conserved at ∼60% (4, 5), suggesting that H2A.Z plays a critical function that is conserved throughout evolution. Although the precise nature of the essential function of H2A.Z is still unclear, a variety of cellular processes have been linked to H2A.Z, including transcriptional regulation, chromosome stabilization and segregation, establishment of boundaries to regulate heterochromatin formation, and regulation of cell cycle progression (6–13).
To date, the best studied function of H2A.Z is in transcriptional regulation. Evidence suggests that, depending on the organism and context, H2A.Z is associated with both positive and negative regulation of gene expression. For example, genome-wide studies in yeast have found that H2A.Z is localized at the transcription start sites of genes, and this enrichment is inversely correlated with transcription activity (7, 14–16). In mammalian cells, ChIP-sequencing studies in human T-cells found that H2A.Z is mostly associated with active genes (17), whereas H2A.Z is enriched at polycomb-silenced genes or bivalent domains in embryonic stem cells (18, 19). Consistent with its association with silenced genes, H2A.Z was found to function in heterochromatin formation during early development (9, 11, 13, 20). At present, how the function of H2A.Z links to both transcriptional activation and repression is still not clearly understood.
Differential post-translational modifications on H2A.Z, such as acetylation and monoubiquitylation, probably determine its specific roles in transcriptional regulation. For example, in mammalian cells, five lysines (Lys-4, -7, -11, -13, and -15) at the N terminus of H2A.Z are known to be acetylated, and three lysines at the C terminus (Lys-120, -121, and -125) are potential sites of ubiquitylation (21). Acetylated H2A.Z is most often associated with active gene transcription (7, 22, 23), whereas ubiquitylated H2A.Z is functionally linked to transcriptional repression and polycomb silencing (24, 25). More recently, the H2A.Z associated with bivalent (both H3K4- and H3K27-methylated)3 domains in embryonic stem cells was found to be both acetylated and ubiquitylated (19). These and other studies have led us to hypothesize that distinct modification patterns distinguish the functions of H2A.Z in activation versus repression (26).
Mammalian development is a highly complex program of events that requires precise coordination. As each cell type differentiates, different sets of genes are expressed in specific temporal order. Changes in the chromatin architecture can dictate gene expression by facilitating or restricting the binding of transcription factors to the target genes. However, how these different epigenetic states are established and maintained during the course of differentiation remains elusive. The transitions between epigenetic states probably involve alterations of the composition and modifications on chromatin. Several studies have suggested that H2A.Z is involved in the developmental process, especially because it is found at developmentally regulated genes (27). For example, knockdown of this variant results in dramatic developmental defects in Caenorhabditis elegans and Xenopus laevis (27–29). Most importantly, genetic deletion of H2A.Z is lethal in Tetrahymena and Drosophila and during mouse development (1–3); therefore, it must have an essential function during development. In mouse embryonic stem cells, H2A.Z has been found at the same developmentally related genes marked by a PcG protein, Suz12, a member of the PRC2 complex responsible for depositing the H3K27me3 mark (18). That study also showed that RNAi-mediated depletion of H2A.Z does not affect the growth of mouse embryonic stem cells, but this variant is required for neuronal differentiation. Therefore, the essential function of H2A.Z may be related to a role in regulating the changing gene expression program during differentiation.
Myogenesis is an excellent cellular model for studying control of cellular differentiation. This well defined process begins with the commitment of muscle precursor cells to the skeletal muscle lineage, and these committed myoblasts respond to external signals that activate expression of muscle-specific genes. In turn, the cascade of activated genes orchestrates the differentiation process, ultimately leading to the fusion of myoblasts to form multinucleated myotubes. MyoD, Myf5, myogenin, and MRF4 are a family of myogenic transcription factors that direct the temporal regulation of myogenesis. MyoD and Myf5 have roles in the commitment of cells to skeletal muscle cell fates, whereas myogenin and MRF4 are involved in terminal differentiation (30). Recent studies examining epigenetic changes during muscle differentiation have found that histone variants play an integral role in this process. For example, studies have found that the histone H3 variant, H3.3, is critical for myogenic fate determination and for the expression of muscle-specific genes, possibly through alteration of the chromatin environment at the MyoD locus (31, 32). In addition, Cuadrado et al. (33) reported that SRCAP-mediated deposition of H2A.Z at the myogenin promoter is important for the chromatin-modeling and subsequent activation of this gene during myogenic differentiation. Although the effects associated with loss of SRCAP suggest that H2A.Z is required for this process, the involvement of this variant in muscle differentiation has not been directly tested.
C2C12 mouse myoblast cells are commonly used to study the process of myogenesis. When the growth condition is switched from high-serum growth medium to low-serum differentiation medium, the normally mononuclear C2C12 myoblasts undergo myogenic differentiation and fuse to form multinucleated myotubes (34). To test whether H2A.Z and H2A.Z acetylation have specific roles in this process, we generated stable C2C12 cells that express wild type (WT) H2A.Z or a H2A.Z non-acetylatable mutant (H2A.Z-Ac-mut). We found that expression of ectopic WT or mutant H2A.Z had a dominant effect over the endogenous H2A.Z and resulted in distinct differentiation defects in the C2C12 myoblasts. Most strikingly, expression of H2A.Z-Ac-mut completely blocked differentiation of these cells. ChIP assays showed that RNA Pol II and histone modification marks associated with transcriptional activation were absent at the promoters of MyoD or myogenin in these cells. Further analyses showed that the apparent differentiation defect is due to a loss of MyoD expression under both normal growing and differentiation conditions and that ectopic expression of MyoD in the H2A.Z-Ac-mut-expressing cells could rescue the differentiation defect phenotype. Finally, chromatin accessibility assays showed that the MyoD promoter is less accessible in the H2A.Z-Ac-mut cells compared with control normal cells. Altogether, our findings suggest that expression of the non-acetylatable H2A.Z disrupted accessibility and activation of the MyoD gene, ultimately leading to an overall defect in the ability of C2C12 cells to differentiate.
Experimental Procedures
Cell Culture, Transfection, and Western Blotting
C2C12 mouse myoblasts were obtained from the American Tissue Culture Collection (ATCC). Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (growth medium (GM)). Differentiation was induced by replacing the GM with DMEM supplemented with 2% horse serum (differentiation medium) once the cells had reached 90% confluence. Cells were harvested 72 and 96 h after incubation in differentiation medium. Medium was changed every 2 days during differentiation. HEK293T cells for retroviral production were grown in DMEM supplemented with 10% fetal bovine serum. Transient transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. MyoD overexpression was performed by transient transfection of C2C12 cells with pEMSV-MyoD, a gift from Dr. John C. McDermott (York University). Total cell extracts were isolated, and protein levels were analyzed by immunoblotting on a 15% SDS-polyacrylamide gel and detected using the antibodies listed in Table 1. Quantification of proteins in Western blot analysis through chemiluminescence detection was performed using the Image Station 4000MM PRO (Carestream Molecular Imaging).
TABLE 1.
Primers
| Target | Forward primer | Reverse primer |
|---|---|---|
| Quantitative RT-PCR | ||
| MyoD | AGC ACT ACA GTG GCG | CAC TAT GCT GGA CAG |
| Myogenin | AGG CGC TGT GGG AGT | CAA CCA GGA GGA GCG |
| Myh3 | GCC AGG ATG GGA AAG | GGG CTC GTT CAG GTG |
| P21 | CCC TCT ATT TTG GAG | GTA CCC TGC ATA TAC |
| H2A.Z | GAG TTG GCA GGA AAT | GAT GAC ACC ACC ACC |
| Enhanced GFP | GCA GAA GAA CGG CAT | ACG AAC TCC AGC AGG |
| HPRT | GCC TAA GAT GAG CGC | TAC TAG GCA GAT GGC |
| ChIP-quantitative PCR | ||
| MyoD promoter (PRR) | CGC CCC CAG CCT CCC TTT CCA G | TGT CAG AGG AGT GGT GAA GAA A |
| MyoD upstream | GGA TGG GGC TCT CAA TGT CAG CG | TGA GTG TGC GTG CCT TCA CCA |
| Myogenin promoter | GAA TCA CAT GTA ATC CAC TGG A | ACA CCA ACT GCT GGG TGC CA |
| Myogenin upstream | GCC CAT CAC AGT TAG GAG CGG C | ACT GTG TTT CTC GGC AAC CCC A |
| CER | GGG ACA GCC GCC TCC AAA CG | TGA AGC GCC GGA CTC CAG GAA |
| −10kb | ACC CAG GGC ACC CCA AGT GT | CGG GCT GTC TTA GGG GAA GCC T |
| DRR | ATG GCG GCA GGA GAA CTG AGC | CAG CCA CCC CTT CTG GAG CG |
| β-Actin promoter | GCT TCT TTG CAG CTC CTT CGT TG | TTT GCA CAT GCC GGA GCC GTT GT |
| ChART-PCR | ||
| MyoD PRR | TAG ACA CTG GAG AGG CTT GGG CAG | CTG GGC TAT TTA TCC AGG GTA GCC |
| MyoD −7kb | GGC ATG GGA GGT TTA TAG CA | ATG CCA CTA TGC AAT CCA CA |
| GAPDH −1.5kb | GAC TCT GAA TCT GCC ATG CCT C | CCA GAG CCA AGG CTG TGT TAG |
Generation of H2A.Z Knockdown (KD) and H2A.Z-GFP Cells
Nineteen-base pair hairpin oligonucleotides for H2A.Z (5′-CA GCT GTC CAG TGT TGG TG-3′) were cloned into pSuper vector (Oligoengine). H2A.Z and H2A.Z mutants were tagged with GFP and cloned into pLNCX2 vector (Clontech) as described previously (24). Ecotropic retroviral supernatant were generated by transfection of a packaging vector and pSuper/pLNCX2 vector in a 1:1 ratio in HEK293T cells using Lipofectamine 2000 (Invitrogen). The viral supernatant was collected three times every 24 h and was used to infect C2C12 cells in 6-well tissue culture dishes with 4 μg/ml Polybrene (Sigma). Cells were infected for 4 h, viral supernatant was removed and replaced with fresh medium, and cells were allowed to recover for 24 h. Cells were then selected using puromycin or G418.
RT-Quantitative Polymerase Chain Reaction Analysis
Total RNA was isolated using an RNeasy kit (Qiagen) and eluted with 40 μl of RNase-free water according to the manufacturer's instructions. First strand cDNA was synthesized using 500 ng of total RNA in a reaction containing oligo(dT) random oligonucleotides and Superscript II (Invitrogen). Quantitative PCR was performed in triplicate using PerfeCta SYBR Green SuperMix (Quanta Biosciences) and transcript-specific primers according to the manufacturer's instructions. Reactions were run on an Applied Biosystems SDS7900HT thermal cycler in a 384-well format or the Bio-Rad Opticon 2 in a 96-well format. Gene expression was normalized to expression of the housekeeping gene HPRT. -Fold change of mRNA levels was calculated relative to the mRNA level in the H2A.Z-Ac-mut cells at the high-serum time point. Primer sequences are listed in Table 2.
TABLE 2.
Antibodies
| Antibodies | Source | Species | Catalog no. |
|---|---|---|---|
| Myosin heavy chain | DSHB | Mouse | MF-20 |
| Myogenin | DSHB | Mouse | F5D |
| MyoD | Santa Cruz Biotechnology | Rabbit | sc304 |
| RNA Pol II | Santa Cruz Biotechnology | Rabbit | sc899 |
| p21 | Santa Cruz Biotechnology | Rabbit | sc6246 |
| GFP | Santa Cruz Biotechnology | Mouse | sc9996 |
| H2A.Z | Active Motif | Rabbit | 39113 |
| H4ac | Millipore | Rabbit | 06-946 |
| H3K27me3 | Millipore | Rabbit | 07-449 |
| H3K27ac | Abcam | Rabbit | ab4729 |
| H3 | Abcam | Rabbit | ab1791 |
Chromatin Immunoprecipitation
Antibody-based ChIP assays were performed using our standard protocol (25), and GFP-ChIPs were done using the GFP-trap reagent (ChromoTek) as per the manufacturer's instructions. Samples were sonicated to generate DNA fragments of <500 bp. Chromatin fragments were immunoprecipitated using antibodies against H2A.Z (C-terminal), RNA Pol II, pan-acetyl-H4, H3K4me2/3, H3K27ac, H3K27me3, and H3 (Table 2). The precipitated DNA was amplified by real-time PCR, with the primer sets in Table 1. The data shown are representative of three or more independent experiments.
Immunofluorescence Analysis
C2C12 cells were seeded in 6-well tissue culture dishes containing sterile glass coverslips. Cells were fixed with 4% paraformaldehyde in PBS for 10 min at room temperature. Cells were permeabilized in PBS containing 0.2% Triton X-100. After blocking in 5% BSA in PBS, cells were incubated with primary antibody at 37 °C for 1 h. Cells were then washed in PBS and incubated with secondary antibody (1:800) conjugated to Alexa-Fluor-488 or Cy3 diluted in PBS-BSA for 1 h. Cell nuclei were counterstained with DAPI mounted in Prolong Gold antifade (Molecular Probes). Imaging was captured at ×10 magnification using a Zeiss AxioObserver microscope. The differentiation index (DI) is the percentage of myosin heavy chain (MHC)-positive cells divided by the total number of DAPI-stained nuclei. The fusion index (FI) was defined as the average number of nuclei in MHC-positive cells containing at least three nuclei. For each experimental condition and time point, a minimum of 10 fields were randomly imaged at identical magnification. The Click-iT proliferation assay was performed according to the manufacturer's protocol (Life Technologies, Inc.). MyoD fluorescence in transiently transfected cells was measured using ImageJ software for GFP-positive cells in C2C12 cells transfected with H2A.Z-Ac-mut-GFP, H2A.X-GFP, and NLS-GFP. The corrected total cell fluorescence is calculated as CTCF = (integrated density) − (area of selected cell × mean fluorescence of background), as described previously (35). Average MyoD fluorescence was also calculated for non-transfected C2C12 cells as a control.
Results
Reduced Expression of H2A.Z in C2C12 Cells Did Not Alter Their Ability to Differentiate
Although H2A.Z is required for development or viability in different organisms, whether it has a role in cellular differentiation of metazoans is not well understood. Moreover, whether acetylation of H2A.Z contributes to this process has also not been studied. Using mouse myoblast differentiation as a model system, we first investigated the requirement of H2A.Z during myogenesis. To that end, we generated stable H2A.Z knockdown C2C12 cell lines, stimulated them to differentiate, and then assessed their ability to form myotubes. Three different shRNAs targeting different regions of the 3′-UTR of H2A.Z mRNA were first tested in C2C12 cells by transient transfection (data not shown). The targeted region yielding the highest level of knockdown was used to generate stable KD cells using retroviral transduction (Fig. 1A). Following stimulation to differentiate, H2A.Z KD cells formed multinucleated myotubes in differentiation conditions (Fig. 1B). The formation of myotubes in H2A.Z KD cells was comparable with that of two control cells: normal C2C12 cells and C2C12 cells stably expressing a nonspecific, scrambled shRNA sequence. These findings suggest that continued expression of H2A.Z is not absolutely required for C2C12 differentiation. Alternatively, because residual levels of H2A.Z were still present in the KD cells (Fig. 1A), it is possible that low levels of H2A.Z were sufficient to support the differentiation of myoblasts to myotubes. Because the H2A.Z KD C2C12 cells were still able to differentiate, we next sought alternative means to examine the roles of H2A.Z in myogenesis. In particular, given that H2A.Z can be post-translationally modified by acetylation, we introduced WT or non-acetylatable mutant H2A.Z in C2C12 cells and tested their abilities to differentiate into myotubes.
FIGURE 1.
Characterization of H2A.Z knockdown and the expression of a H2A.Z non-acetylatable mutant in C2C12 cells. A, Western blot analysis of whole-cell lysates from non-infected C2C12 cells or C2C12 cells stably expressing either a scrambled shRNA (shScramble) control or a shRNA targeting H2A.Z mRNA (shH2A.Z) using an anti-H2A.Z antibody. H2A.Z levels were reduced in the H2A.Z knockdown cells compared with cells expressing scrambled shRNA or normal cells. Histone H3 expression was used as a loading control. B, bright field microscopy images showing C2C12 H2A.Z knockdown cells after differentiation and incubation in differentiation medium for 96 h. Knockdown of H2A.Z did not affect the differentiation of C2C12 myoblast cells into myotubes. C, schematic showing H2A.Z-GFP and H2A.Z-Ac-mut-GFP, where the five acetylatable lysines in the N terminus were mutated to arginine. Blue letters represent known sites of acetylation at the N terminus of H2A.Z. Red letters represent sites of lysine to arginine mutations in H2A.Z-Ac-mut. D, Western blot analysis using anti-GFP antibody detects the stable expression levels of NLS-GFP, H2A.X-GFP, H2A.Z-GFP, and H2A.Z-Ac-mut-GFP in C2C12 cell lines. The numbers represent quantification of the GFP-tagged proteins on the Western blot using chemiluminescence imaging. Histone H3 expression was used as a loading control. E, Western blot analysis using an anti-H2A.Z antibody shows that H2A.Z-GFP and H2A.Z-Ac-mut-GFP were not overexpressed as compared with endogenous H2A.Z levels. The numbers represent quantification of GFP-tagged H2A.Z and H2A.Z-Ac-mut as well as endogenous H2A.Z on the Western blot using chemiluminescence imaging. H2A.Z-Ac-mut represents ∼8% of endogenous H2A.Z at the protein level. The asterisk on the Western blot shows a nonspecific band detected by the antibody. F, real-time PCR amplification of mRNA isolated from C2C12 stable cell lines. Relative GFP mRNA levels detecting expression of exogenous histones and NLS-GFP were normalized to HPRT. G, immunofluorescence images showing the nuclear staining of GFP-tagged H2A.Z, H2A.Z-Ac-mut, and H2A.X in the C2C12 stable cell lines. E, GFP ChIP showed that H2A.Z-GFP as well as H2A.Z-Ac-mut-GFP is enriched at the MyoD promoter as compared with an upstream region and the promoter of β-actin. Experiments were performed in high-serum (HS) growth conditions.
Expression of H2A.Z-Ac-mut Blocks C2C12 Cell Differentiation
Our earlier work showed that GFP-tagged H2A.Z was incorporated into chromatin and monoubiquitylated in a similar manner as endogenous H2A.Z (24). Moreover, GFP tagging of H2A.Z significantly shifted its mobility on SDS gels, which facilitated distinction between the transfected and endogenous H2A.Z proteins. For these reasons, we generated a series of C2C12 cells stably expressing either WT H2A.Z-GFP or a non-acetylatable H2A.Z mutant (H2A.Z-Ac-mut), whereby all five potential acetylated lysines on H2A.Z-GFP (Lys-4, -7, -11, -13, and -15) were mutated to arginines (see Fig. 1C for details) for our studies. Additionally, we also generated C2C12 cell lines stably expressing GFP containing an added nuclear localization signal (NLS-GFP) to control for exogenous expression of a nuclear protein as well as H2A.X-GFP-expressing C2C12 cells as controls for exogenous expression of another H2A variant. All stable cell lines were generated and selected using the same methods and in parallel to minimize experimental variations.
Initial characterization of the stable C2C12 cells showed that they all grow similarly well under high-serum growth conditions. Western blot analyses showed that the control NLS-GFP and H2A.X-GFP are expressed at much higher levels than the WT and mutant H2A.Z-GFP in the respective stable cell lines (Fig. 1D). Western blots using an H2A.Z-specific antibody also showed that the tagged WT and H2A.Z-Ac-mut are expressed at significantly lower levels than endogenous H2A.Z (Fig. 1E). Consistent with previous reports, immunofluorescence analyses showed that all GFP-tagged histones are localized to the nuclei of expressing cells (Fig. 1G), and GFP-ChIP analyses confirmed that both WT- and H2A.Z-Ac-mut are incorporated into chromatin (specifically at the MyoD promoter for example, Fig. 1H). It is of interest to note that the mutant H2A.Z is expressed at about half the amount of the WT H2A.Z at the protein (Fig. 1D), steady-state mRNA (Fig. 1F), and chromatin-incorporated (Fig. 1H) levels. Altogether, these data show that although tagged H2A.X can be expressed at high levels in C2C12 cells, the overall levels of H2A.Z are more tightly regulated, and high levels of exogenous H2A.Z may not be tolerated.
To examine the effects of expressing WT and mutant H2A.Z on myogenic differentiation, we compared the phenotypes of the stable C2C12 cells under differentiation growth conditions. Upon serum withdrawal, all control cell lines (those expressing NLS-GFP, H2A-GFP, or H2A.X-GFP) formed long and parallel myotubes within 72–120 h (Fig. 2A, left). The myotubes formed by these control cells were morphologically identical to those formed by the normal differentiated C2C12 cells. In contrast, cells expressing either WT or mutant H2A.Z-GFP showed distinct and reproducible morphological changes compared with the control samples. More specifically, the myotubes formed from the WT H2A.Z-GFP-expressing cells were not aligned and were heterogeneous in diameter. Most strikingly, the cells expressing the H2A.Z-Ac-mut failed to form any myotubes during the same 120-h time frame.
FIGURE 2.
Expression of a non-acetylatable H2A.Z mutant results in defects in myoblast differentiation. A, bright field images and immunofluorescence images using MHC antibodies (red) examining myotube formation in C2C12 cells stably expressing NLS-GFP, H2A-GFP, H2A.X-GFP, H2A.Z-GFP, and H2A.Z-Ac-mut-GFP. Cells were counterstained with DAPI (blue) in the immunofluorescence images. Differentiation was induced by switching from high-serum (HS) growth medium to low-serum (LS) differentiation medium; images were then captured 120 h after switching of medium. Images show that H2A.Z-GFP cells formed irregular myotubes after differentiation, and H2A.Z-Ac-mut cells did not form any MHC-positive myotubes. B, differentiation index and Fusion index of the stable cell lines. Differentiation index measures the average percentage of myoblasts that commit to differentiation. Fusion index measures the average number of nuclei in a myotube containing at least three nuclei.
To better visualize the myotubes in these cell cultures, we used an antibody against MHC in immunofluorescence staining (Fig. 2A, right). MHC is expressed toward the end of the muscle differentiation process, and the MHC-positive cells comprise both differentiated single-nuclei myocytes and multinucleated myotubes. Consistent with the bright field microscopy results, the myotubes formed in the control NLS-GFP-, H2A-GFP-, or H2A.X-GFP-expressing cells were relatively uniform in size and well aligned. The H2A.Z-GFP-expressing cells showed a heterogeneous mixture of MHC-negative (undifferentiated), single-cell myocytes and large myotubes (Fig. 2A; see below for more quantitative measurements). Normally, the C2C12 differentiation and cell fusion processes are highly synchronized, leading to an organized layer of myotubes. The disorganized nature of the differentiated H2A.Z-GFP cells suggests that their differentiation is out of sync relative to one another. Finally, as with the bright field results, the majority of H2A.Z-Ac-mut cells were MHC-negative, and the rare MHC-positive cells were long and very thin. This confirmed that the majority of these cells failed to undergo the differentiation program.
To quantitatively analyze the extent of differentiation of these cells, we calculated the DI and FI of the samples as done in previous publications (36–38). The DI was calculated by dividing the number of nuclei (identified by DAPI staining) in MHC-positive cells (both differentiated myocytes and myotubes) by the total number of nuclei (including the undifferentiated MHC-negative cells) within the examined microscopy field. The average DI was calculated from three random fields and represents a measure of the proportion of cells that commit to differentiation compared with those that remain as reserve myoblasts within each culture. For example, in normal C2C12 cells, the average DI was 50%, suggesting that 50% of cells within each culture undergo the differentiation program to activate MHC expression. The DI of the WT H2A.Z-GFP- and H2A.X-GFP-expressing cells were roughly equivalent at 30%, whereas the DI of the H2A.Z-Ac-mut cells was only ∼4% (Fig. 2B). Consistent with the observation that the H2A.Z-Ac-mut cultures are defective in myotube formation, the DI analysis further showed that the majority of those cells failed to commit to the differentiation process.
To calculate the FI, we measured the number of nuclei per MHC-positive myotube that has at least three nuclei. This calculation only takes myotubes in the differentiated cultures into account, not the single-nucleated myocytes. We found that the average FI for the control H2A.X-GFP cells was ∼7, whereas the FI for the H2A.Z-GFP cells was closer to 16 (Fig. 2B). Insofar as the FI is a measure of the number of fusion events, the higher FI of the H2A.Z-GFP cell population is consistent with the formation of visibly thicker myotubes. Considering that the DI values of these two cultures were approximately the same, this also suggests that although the myotubes formed by the H2A.Z-GFP cells were bigger (approximately twice as many nuclei per myotube), there were fewer of them in number, and additional single nucleated myocytes (not counted in the FI calculations) were found in the H2A.Z-GFP cultures. Finally, the FI for the H2A.Z-Ac-mut culture was also very low because no myotubes were formed (Fig. 2B). Altogether, these findings suggest that ectopic expression of wild type H2A.Z as well as the non-acetylatable mutant of H2A.Z exerted dominant effects over the endogenous H2A.Z and resulted in distinct defects in the differentiation of the C2C12 cells.
Muscle-specific Genes Are Not Expressed in the Presence of Differentiation Cues in the H2A.Z-Ac-mut Cells
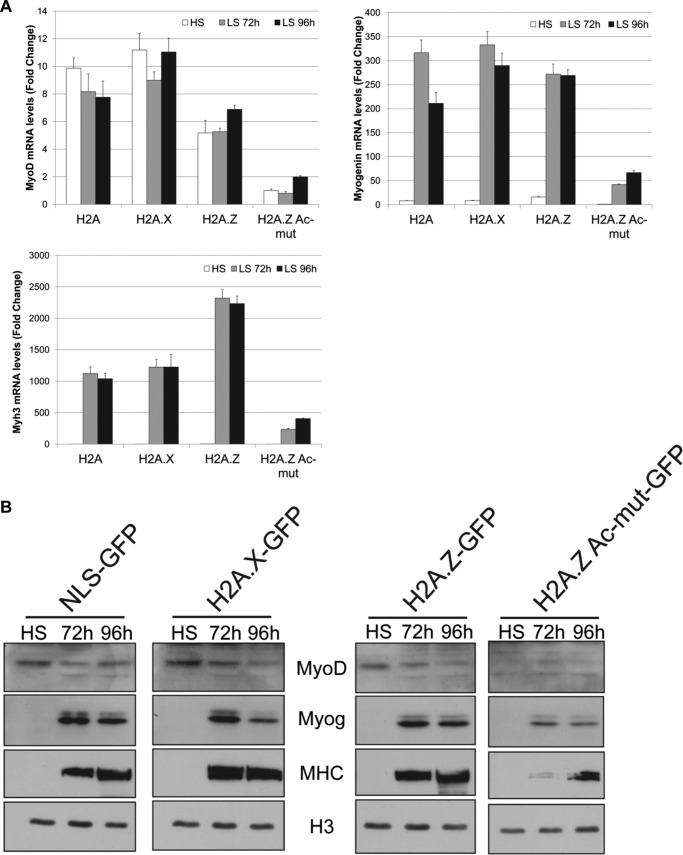
Our data so far suggest that the expression of H2A.Z-Ac-mut in C2C12 cells has rendered them unable to respond to external cues and differentiate into myotubes. To further characterize the phenotype and determine the possible cause, we examined the expression levels of the muscle differentiation genes after induction of differentiation. For example, we used RT-PCR to determine the mRNA levels of MyoD, myogenin, and Myh3 (an isoform of MHC) in cells under high-serum growth conditions and in cells grown in differentiation medium for 72 and 96 h (Fig. 3A). MyoD is normally expressed in myoblast cells grown under high-serum conditions, whereas myogenin and MHC are induced only after initiation of the differentiation process. We detected similar levels of MyoD transcripts in the two control cell lines, H2A-GFP- and H2A.X-GFP-expressing cells, at high- and low-serum conditions, respectively (Fig. 3A). H2A.Z-GFP cells expressed lower levels of MyoD transcripts at all stages of differentiation compared with control cells, whereas H2A.Z-Ac-mut cells showed little, if any, amount of MyoD expression. Because both myogenin and Myh3 are only induced upon differentiation, neither gene product was expressed in any of the tested cells under high-serum growth conditions. When cells were induced to differentiate, both myogenin and Myh3 were expressed at 72 and 96 h under low-serum conditions in H2A-GFP, H2A.X-GFP, and H2A.Z-GFP cells (Fig. 3A). The expression of Myh3 in H2A.Z-GFP-expressing cells was ∼2-fold higher than in the control cells, which may be associated with the morphological differences seen in these cells after differentiation. Consistent with the dramatic defects in myotube formation, myogenin and Myh3 were only expressed at low levels in the H2A.Z-Ac-mut even after the cells were grown in differentiation medium for 96 h (Fig. 3A). Complementing the RNA transcript results, Western blot analyses examining the protein levels of the same muscle cell-specific factors again showed comparable levels of MyoD, myogenin, and myosin heavy chain in control and H2A.Z-GFP cells, but their amounts were significantly diminished in H2A.Z-Ac-mut cells (Fig. 3B).
FIGURE 3.
H2A.Z-Ac-mut cells have impaired expression of genes critical for muscle differentiation. A, real-time PCR analysis of mRNA levels of critical muscle transcription factors, MyoD and myogenin, as well as a muscle structural gene, Myh3 (myosin heavy chain isoform 3) at high-serum (HS) growth conditions as well as at 72 and 96 h in low-serum (LS) differentiation conditions in H2A-GFP, H2A.X-GFP, H2A.Z-GFP, and H2A.Z-Ac-mut-GFP cells. H2A.Z-Ac-mut cells expressed MyoD, myogenin, and Myh3 at low levels even after incubation in low-serum differentiation medium as compared with the control cells. -Fold change of mRNA levels is calculated relative to the mRNA level in the H2A.Z-Ac-mut cells at the HS time point. B, Western blot analysis examining the protein levels of MyoD, myogenin, and MHC at high-serum growth conditions as well as at 72 and 96 h in low-serum differentiation conditions in H2A-GFP, H2A.X-GFP, H2A.Z-GFP, and H2A.Z-Ac-mut-GFP cells. Histone H3 expression was used as a loading control.
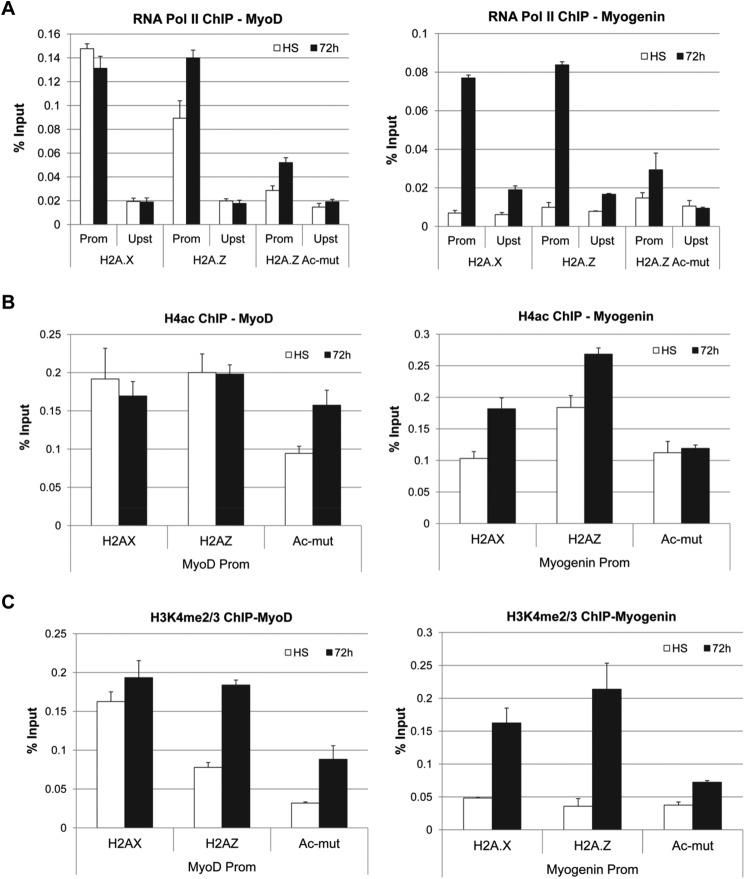
The reduced expression of the muscle cell-specific genes in the H2A.Z-Ac-mut cells is probably due to a block in transcription initiation. To test this, we performed ChIP assays to examine the localization of RNA Pol II and several histone modifications associated with transcriptional activation at the promoters of muscle differentiation genes (Fig. 4). As expected, RNA Pol II was present at the MyoD promoter during growth phase as well as differentiation phase in control cells and in H2A.Z-GFP cells (Fig. 4A). Also, consistent with the lack of MyoD expression in H2A.Z-Ac-mut cells, amounts of RNA Pol II at the MyoD promoter were consistently 2–3-fold lower in these cells compared with control cells. At the myogenin promoter, RNA Pol II was recruited after initiation of differentiation in H2A.X-GFP and H2A.Z-GFP cells (Fig. 4A) but not in H2A.Z-Ac-mut cells. The histone modification ChIP analyses showed that H4ac and H3K4me3 levels were also reduced at the MyoD promoter and at the myogenin promoter after differentiation in H2A.Z-Ac-mut cells as compared with cells expressing H2A.X-GFP or H2A.Z-GFP (Fig. 4, B and C). Altogether, these results demonstrate that loss of MyoD and myogenin expression in H2A.Z-Ac-mut cells may be due to defects in RNA Pol II recruitment and alterations of the H4ac and H3K4me3 marks at the respective promoters.
FIGURE 4.
Reduced RNA Pol II, H4ac, and H3K4me2/3 levels at MyoD and myogenin promoters in H2A.Z-Ac-mut cells. A, ChIP analysis of RNA Pol II enrichment at the promoter (Prom) and upstream (Upst) region of the MyoD and the myogenin gene in C2C12 cells stably expressing H2A.X-GFP, H2A.Z-GFP, and H2A.Z-Ac-mut-GFP in high-serum (HS) growth conditions and at 72 h in low-serum differentiation conditions. RNA Pol II is lost at the MyoD promoter and is not recruited to the myogenin promoter after differentiation induction in H2A.Z-Ac-mut cells. B, ChIP analysis of pan-acetylated H4 in H2A.Z-Ac-mut cells showed loss of H4ac at the MyoD promoter, and an increase in H4ac is not seen after induction of differentiation at the myogenin promoter as compared with control cells. C, H3K4me2/3 ChIP analysis showed reduction of this modification at the MyoD promoter, and an increase is not seen after induction of differentiation at the myogenin promoter as compared with the control cells.
Cell Cycle Withdrawal Defects in the H2A.Z-Ac-mut Cells
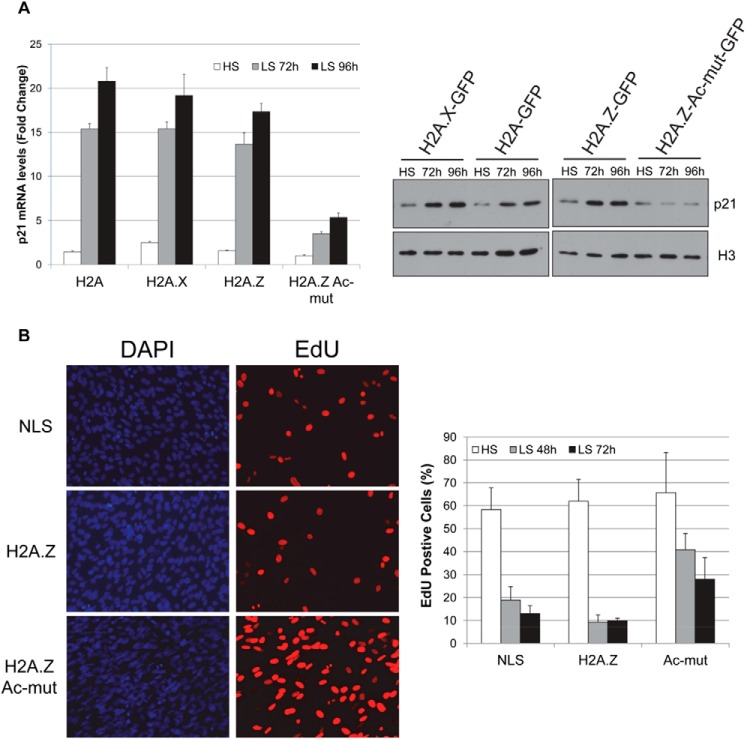
As part of any terminal differentiation, cells must exit the cell cycle and stop proliferation. In myogenic differentiation, MyoD activates expression of p21, which in turn binds and inhibits cyclin-dependent kinases, ultimately resulting in cell cycle withdrawal of the myoblast cells (39). Because H2A.Z-Ac-mut cells have a differentiation defect, we examined the expression of P21 at both mRNA and protein levels by RT-PCR and Western blot analysis (Fig. 5A). In the control (H2A-GFP- and H2A.X-GFP-expressing cells) and H2A.Z-GFP cells, P21 mRNA levels were greatly induced at 72 and 96 h after induction of differentiation (Fig. 5A). By comparison, 3–4-fold less P21 mRNA accumulated in H2A.Z-Ac-mut cells at the same differentiation time points compared with control cells. Western blot analysis confirmed that p21 protein levels did not increase after H2A.Z-Ac-mut cells were grown in low-serum medium (Fig. 5A). Consistent with the lack of p21 expression, assessment of the proliferation status using the Click-iT assay, which measures incorporation of Edu (a BrdU analog) into newly synthesized DNA, showed that 40% of H2A.Z-Ac-mut cells were still proliferating after switching to the differentiation conditions compared with 10–20% of H2A.Z-GFP cells and control NLS-GFP cells (Fig. 5B). Altogether, our data showed that the block in the differentiation of H2A.Z-Ac-mut cells is linked to the lack of p21 induction in those cells and their inability to initiate cell cycle withdrawal in response to normal differentiation signals.
FIGURE 5.
C2C12 cells stably expressing H2A.Z-Ac-mut did not exit cell cycle under differentiation conditions. A, real-time PCR analysis of P21 mRNA levels in H2A-GFP, H2A.X-GFP, H2A.Z-GFP, and H2A.Z-Ac-mut cells shows that expression of P21 in H2A.Z-Ac-mut cells was not induced to levels comparable with that in control cells upon differentiation: 72 and 96 h in low serum (LS) medium. Western blot analysis showed a defect in p21 induction when comparing p21 protein levels of H2A.Z-Ac-mut cells in high-serum (HS) growth medium and cells induced to differentiate for 72 or 96 h in low-serum (LS) medium. B, Click-iT proliferation assay allows EdU, a BrdU analog that is incorporated into the DNA during DNA synthesis, to be visualized by immunofluorescence. Cells that stain positively for EdU (red) have gone through cell division. Cells were counterstained with DAPI (blue). The percentage of EdU-positive cells was calculated relative to the total number of cells. This assay showed that 30–40% of cells continued to proliferate in the H2A.Z-Ac-mut cell population under differentiation conditions compared with 10% of H2A.Z-GFP cells.
Ectopic MyoD Overexpression Partially Rescues H2A.Z-Ac-mut Phenotype
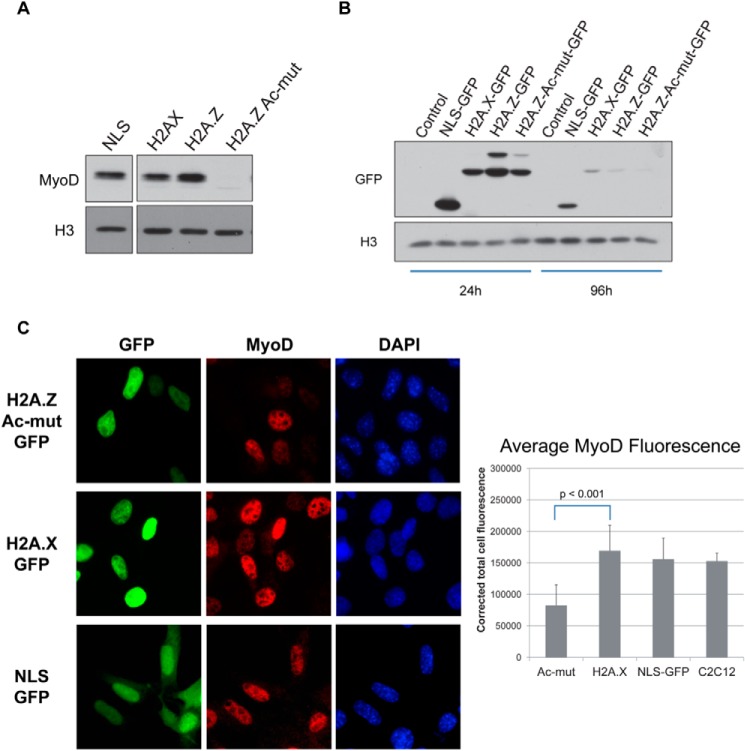
MyoD is the master regulator of the muscle gene expression program during differentiation and is responsible for activating p21 during the myoblast differentiation process. Indeed, analysis of MyoD protein levels in control and histone-expressing C2C12 cells showed a striking lack of MyoD in H2A.Z-Ac-mut cells even under the high-serum growth conditions (Figs. 3B and 6A). As a side note, we also tested the effects of H2A.Z-Ac-mut expression on endogenous MyoD expression in transiently transfected C2C12 cells. Compared with control cells expressing H2A.X-GFP or NLS-GFP, MyoD expression in the H2A.Z-Ac-mut-expressing cells (measured by quantifying the MyoD fluorescence in 50 random GFP-positive cells in each experimental condition by ImageJ) is consistently and significantly reduced (Fig. 6C). Because the reduction in MyoD expression was measured 24 h post-transfection, this finding suggests that the expression of H2A.Z-Ac-mut has an immediate and direct effect on the expression of the endogenous MyoD gene. However, because the expression of the transiently transfected histone genes was rapidly lost over the course of a few days (Fig. 6B), we have not used this transient transfection approach for further myotube differentiation studies.
FIGURE 6.
MyoD expression is reduced in C2C12 cells expressing H2A.Z-Ac-mut. A, Western blot analysis using anti-MyoD antibody shows that MyoD protein levels are undetectable in the stable H2A.Z-Ac-mut cells under growth conditions compared with cells expressing NLS-GFP, H2A.X-GFP, or H2A.Z-GFP. Histone H3 expression was used as a loading control. B, C2C12 cells were transiently transfected with NLS-GFP, H2A.X-GFP, H2A.Z-GFP, and H2A.Z-Ac-mut-GFP. Western blots showed strong expression of the transfected histone-GFP fusions 24 h post-transfection, but the expression levels of the transfected genes declined rapidly by 96 h post-transfection. C, MyoD immunofluorescence of C2C12 cells transiently transfected with H2A.Z-Ac-mut-GFP, H2A.X-GFP, and NLS-GFP was measured and quantified by ImageJ. The average MyoD fluorescence of H2A.Z-Ac-mut-GFP-transfected cells was reduced as compared with the H2A.X-GFP-transfected cells (p < 0.001, Student's t test). The average of the corrected total MyoD fluorescence calculated from 50 random GFP-positive cells per experimental conditions was plotted as shown. The average MyoD fluorescence of non-transfected C2C12 cells was also calculated as a control to determine the natural variations in MyoD expression/fluorescence in normal C2C12 cell cultures.
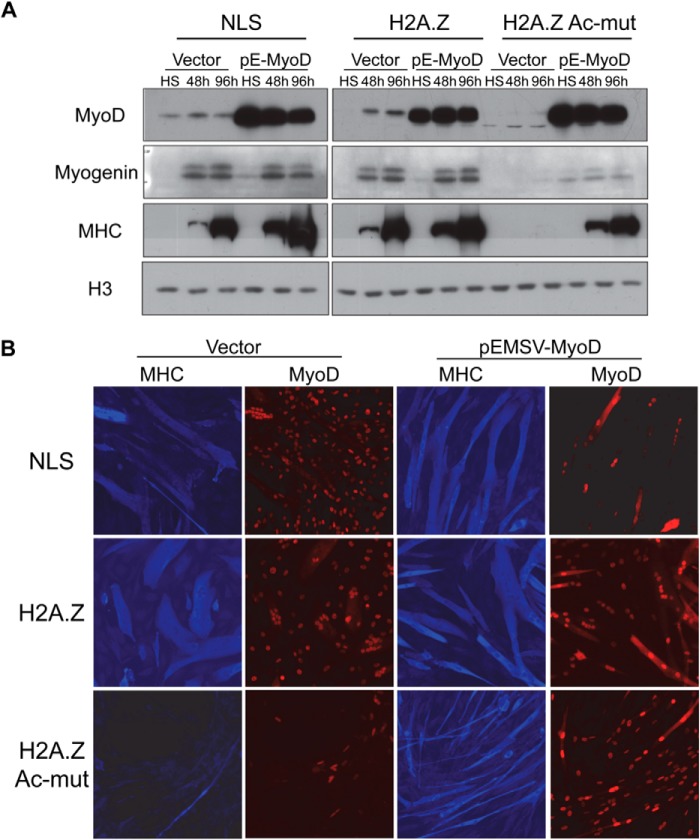
To test whether loss of MyoD expression is the main cause of the differentiation defect in the stable H2A.Z-Ac-mut cells, we asked whether re-expression of exogenous MyoD could rescue this differentiation defect phenotype. H2A.Z-Ac-mut cells as well as control cells were transfected with vector alone or with a MyoD-expressing construct. MyoD expression levels, as assessed by Western blots, increased dramatically in the cells transfected with the MyoD expression plasmid compared with cells transfected with the vector control (Fig. 7A). The high levels of MyoD expression in MyoD-transfected cells did not significantly alter expression of myogenin in differentiating NLS-GFP (control) and H2A.Z-GFP cells but consistently enhanced MHC expression after induction of differentiation. In H2A.Z-Ac-mut cells, expression of exogenous MyoD increased myogenin expression slightly but strongly enhanced MHC expression after induction of differentiation. Consistent with the Western blot results, immunofluorescence staining for MHC showed that there were significantly higher amounts of MHC in MyoD-transfected H2A.Z-Ac-mut cells. Most importantly, a significant portion of these cells now formed myotubes under differentiation conditions (Fig. 7B, bottom right). From the replicates of this experiment, we note that myotubes formed by MyoD-transfected H2A.Z-Ac-mut cells are consistently thinner than those formed by control cells (data not shown), which could be due to non-uniform expression levels of MyoD in the transfected culture. Nevertheless, the rescue of the ability to form myotubes strongly supports the interpretation that a defect in MyoD expression is the major cause of the differentiation defect in H2A.Z-Ac-mut cells.
FIGURE 7.
Ectopic expression of MyoD rescues the differentiation defect in the C2C12 H2A.Z-Ac-mut cells. A, pEMSV-MyoD (pE-MyoD) vector was transiently transfected into C2C12 cells stably expressing NLS-GFP control, H2A.Z-GFP, or H2A.Z-Ac-mut-GFP. A MyoD Western blot shows that MyoD is overexpressed in cells transfected with pEMSV-MyoD vector as compared with the cells transfected with the vector alone control. From Western blot analysis, MyoD, myogenin, and MHC expression was not detected in H2A.Z-Ac-mut cells expressing the vector alone. In the H2A.Z-Ac-mut cells overexpressing MyoD, increased levels of myogenin and MHC were seen upon differentiation: incubation in low-serum (LS) medium for 48 or 96 h as compared with cells growing under high-serum (HS) growth conditions. Histone H3 expression was used as a loading control. B, MHC immunofluorescence (blue) showed increased myotube formation upon differentiation induction in H2A.Z-Ac-mut cells overexpressing MyoD (red) as compared with cells expressing vector alone. Cells were differentiated for 96 h. in differentiation medium.
Expression of H2A.Z-Ac-mut Has Altered Chromatin Accessibility at the MyoD Locus
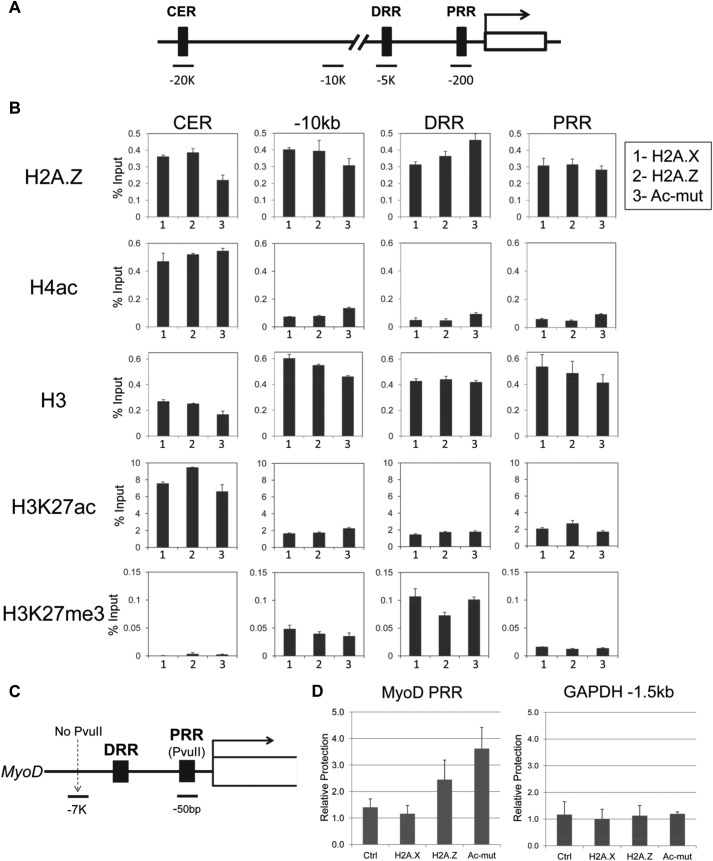
MyoD transcription is normally controlled by two enhancers: a distal enhancer (DRR) located ∼5 kb upstream and a core enhancer (CER) located ∼20 kb upstream of the MyoD gene (Fig. 8A) (40–45). To examine the enrichment of H2A.Z at these loci, we used an antibody that recognizes the C terminus of H2A.Z for ChIP analyses. Localization of H2A.Z was spread out over MyoD regulatory sequences in all cells (Fig. 8B, top row). However, at the MyoD core enhancer in H2A.Z-Ac-mut cells (Fig. 8B, top left), H2A.Z occupancy was ∼50% compared with that in control cells at the same region. This reduction was consistently seen with the replicates of the ChIP assay (data not shown), suggesting that expression of the non-acetylatable mutant of H2A.Z may interfere with the total amount of H2A.Z deposited at the MyoD core enhancer.
FIGURE 8.
Changes to the chromatin composition and architecture at the MyoD enhancer and promoter in H2A.Z-Ac-mut cells. A, schematic showing the MyoD gene and the different regions used for ChIP analysis, the CER, the −10 kb upstream control region (−10kb), DRR, and PRR. B, ChIP analysis of H2A.Z, H4ac, H3, H3K27ac, and H3K27me3 enrichment at the above regions in H2A.X-GFP (1), H2A.Z-GFP (2), and H2A.Z-Ac-mut-GFP (3) cells. The data shown are representative of three or more independent experiments. C, schematic showing the PvuII restriction enzyme cut site at the MyoD gene. This site is in the promoter regulatory region. D, intact nuclei were subjected to digestion by PvuII, and accessibility of this restriction enzyme site within the MyoD promoter (shown in Fig. 7C) was assayed by ChART-PCR. Relative protection was calculated by dividing the PCR product amount produced from primers that flank the PvuII site within the MyoD promoter by that produced from primers amplifying a −7 kb region upstream of the MyoD gene. This analysis showed that the promoter region of MyoD in H2A.Z-Ac-mut cells is more protected and less accessible as compared with the control cells. As a control, this assay was also performed at a region around the GAPDH promoter (GAPDH −1.5kb), which also contained a PvuII restriction enzyme cut site.
In mammalian cells, most enhancers are marked by monomethylation of H3K4 (H3K4me1) and methylation or acetylation of H3K27. In general, trimethylation of H3K27 (H3K27me3) marks a poised enhancer, whereas acetylation of this residue (H3K27ac) indicates an active enhancer (46, 47). Therefore, we also assessed the H3K27 modification status at MyoD regulatory sequences by ChIP analyses. High levels of H3K27ac enrichment were seen at the MyoD core enhancer region in all cell lines, suggesting that this enhancer is similarly active across the histone-expressing cells (Fig. 8B). H3K27me3 levels were low at the core enhancer but significantly higher at the distal enhancer region, suggesting that the distal enhancer may be poised but not active in these cell lines. Because the H3K27ac, H3K27me3, and H4ac patterns (Fig. 8B) are all similar for the different stable cell lines, this suggests that the epigenetic signatures marking the MyoD enhancer elements are not greatly altered in H2A.Z-Ac-mut cells.
Binding of transcription regulators at the MyoD core enhancer leads to structural changes at the MyoD promoter that render it more transcription-permissive (48). Therefore, we next examined the chromatin accessibility of the MyoD promoter using the chromatin accessibility by real-time PCR (ChART-PCR) assay established previously (31). This method assays the accessibility of the PvuII restriction enzyme site at the proximal regulatory region (PRR) of the MyoD promoter as a proxy to the chromatin compaction status of the region (Fig. 8C). Primers were generated flanking a PvuII cut site at the MyoD promoter such that, if the promoter were not accessible, PvuII would not cut at that site, and a PCR product could be amplified using these primers. Compared with control cells, there was a 3–4-fold increase in PvuII protection in H2A.Z-Ac-mut cells, suggesting that there is decreased access to the MyoD promoter in these cells (Fig. 8D). For comparison, we also tested the accessibility of the GAPDH promoter using the same ChART-PCR assay and found equal accessibility of the PvuII site within this locus in all stable cells (Fig. 8D), indicating that the changes in promoter accessibility in the H2A.Z-Ac-mut cells are specific to the MyoD promoter. It is of interest to note that chromatin accessibility of the MyoD promoter was also reduced in H2A.Z-GFP cells compared with controls but not as much as in H2A.Z-Ac-mut cells. This could be due to variations among individual cells within the H2A.Z-GFP population, possibly leading to the observed heterogeneous phenotype during differentiation (Fig. 2A). Overall, our data suggest that expression of exogenous H2A.Z-Ac-mut fusion protein in C2C12 cells reduces chromatin accessibility of the MyoD promoter and inhibits MyoD expression, which ultimately results in a block of the myogenic differentiation process.
Discussion
In this study, we used the C2C12 myoblast model to examine the effects of H2A.Z on cellular differentiation. We first found that shRNA knockdown of H2A.Z expression did not affect growth or differentiation of these cells. The lack of any phenotypic effects when H2A.Z expression is knocked down in these cells could indicate that the residual amounts of H2A.Z are sufficient to sustain growth and differentiation of C2C12 cells. In addition, recent studies showed that there are in fact two isoforms of H2A.Z encoded by distinct genes (49, 50). Because our shRNA specifically targets H2A.Z-1 but not the H2A.Z-2 isoform, it is a formal possibility that the H2A.Z-2 isoform in these cells is sufficient to compensate for the loss of H2A.Z-1 in these cells. Finally, it could be that the main function of H2A.Z is to set up the chromatin architecture of the genome to poise the promoters of various genes for activation, and once this architecture is established, sustained expression of H2A.Z is no longer required. In contrast to the knockdown results, we found that exogenous expression of WT H2A.Z as well as a H2A.Z non-acetylatable mutant resulted in distinct effects on differentiation of mouse C2C12 myoblast cells. Expression of the H2A.Z-Ac mutant in C2C12 cells did not alter growth of these cells under high-serum growth conditions; however, these cells were completely unable to differentiate into myotubes upon switching to growth in differentiation medium. Consistent with this phenotype, they do not express any of the differentiation markers, such as myogenin or myosin heavy chain, under differentiation conditions. Moreover, they lacked induction of p21 expression normally seen during myoblast differentiation and continued to proliferate. We further ascribed the cause of this phenotype to a loss of MyoD expression in these cells under both regular growth and differentiation conditions. As a validation of this conclusion, we found that expression of exogenous MyoD rescued their defects in differentiation and restored the ability to form myotubes in H2A.Z-Ac mutant-expressing cells.
MyoD is the master regulator transcription factor that drives myogenic differentiation in myoblast cells. Ectopic expression of MyoD in fibroblast or adipoblast cell lines is sufficient to convert and drive myogenesis in these cells (51, 52). Therefore, its expression in myoblast cells is paramount to the differentiation program. Prior to this study, a role of H2A.Z in MyoD expression has not been reported. MyoD expression is regulated through two different enhancers, the CER and the DRR. It is thought that the CER is important for directing the expression of MyoD during embryogenesis, whereas DRR activity is more important for expression in differentiated muscle (40, 43). Recently, Yang et al. (31) found that the histone H3 variant H3.3 is recruited to the CER and promoter upon differentiation and that loss of H3.3 or HIRA (the chaperone responsible for deposition of H3.3 into chromatin) blocks differentiation of C2C12 cells. H3.3 can partner with H2A.Z to form nucleosomes that mark nucleosome-free regions at active promoters (53), and our data suggest that expression of the H2A.Z-Ac-mut indeed altered the chromatin architecture of MyoD regulatory regions. For example, we found that total H2A.Z at the MyoD CER is reduced by ∼50% in H2A.Z-Ac-mut-expressing cells compared with control cells. Yang et al. (31) further used a restriction enzyme accessibility assay to determine that loss of H3.3 resulted in reduced accessibility of the MyoD promoter (proximal regulatory region). Using the same assay, we also observed partial loss of accessibility at the MyoD promoter, suggesting that expression of H2A.Z-Ac-mut affected chromatin compaction at the MyoD regulatory regions. At present, we do not know whether the changes in endogenous H2A.Z occupancy at the MyoD CER and the reduced accessibility of the MyoD promoter in the H2A.Z-Ac-mut cells are connected or independent events. For example, it is possible that loss of H2A.Z at the CER prevented the enhancer's ability to activate and decondense the chromatin at the MyoD promoter. Alternatively, incorporation of the H2A-Z-Ac-mut at the MyoD promoter may directly interfere with the chromatin state of that locus. In either case, the decreased chromatin accessibility of the MyoD promoter probably precluded recruitment of transcriptional machinery and ultimately blocked expression of the MyoD gene.
Finally, we note that expression of both WT and H2A.Z-Ac-mut are significantly lower than that of the endogenous H2A.Z in the stable cell lines, yet the mutant H2A.Z could drastically alter the differentiation potential of the C2C12 cells. This observation underscores the dominant effect of this mutant form of H2A.Z over the endogenous H2A.Z. Such a scenario is not without precedence. Recently, it has been shown that a significant percentage of pediatric glioblastoma patients harbor a point mutation in the H3.3 gene, resulting in the expression of a Lys-27 → Met mutant form of H3.3 (54). Moreover, expression of this H3.3 K27M mutant in 293T cells is sufficient to cause a global loss of Lys-27 methylation on all H3 isoforms although the H3.3 K27M mutant is expressed at much lower levels compared with the total wild type H3 in the cell. In that case, it was suggested that the mutant H3.3 binds to and inhibits the activity of the H3K27-methyltransferase, leading to a global loss of H3K27 methylation in those cells. By analogy, it is possible that H2A.Z-Ac-mut incorporated at the MyoD promoter exerts a local dominant negative effect by inhibiting a histone acetyltransferase that is normally recruited to acetylate the chromatin of this locus (see Fig. 9 for a model). Our earlier work showed that H2A.Z-nucleosomes are hyperacetylated at multiple histone components (55), and therefore, an inhibition of the locally recruited histone acetyltransferase could decrease acetylation of nearby wild type H2A.Z as well as other histones, leading to a loss of accessibility of the entire region. In addition, it is also possible that the chromatin-bound H2A.Z-Ac-mut at the CER enhancer inhibits or repels the SRCAP remodeling complex, leading to a reduction of H2A.Z occupancy at the CER and preventing the enhancer's ability to activate or decondense the MyoD promoter. Future studies on the biochemical effects of the non-acetylatable H2A.Z within the chromatin and nucleosome context could help to elucidate the mechanisms involved. Moreover, further examination of whether the inhibition of the differentiation phenotype is conserved in other systems, such as embryonic stem cells, could be of interest to both chromatin and stem cell biology fields.
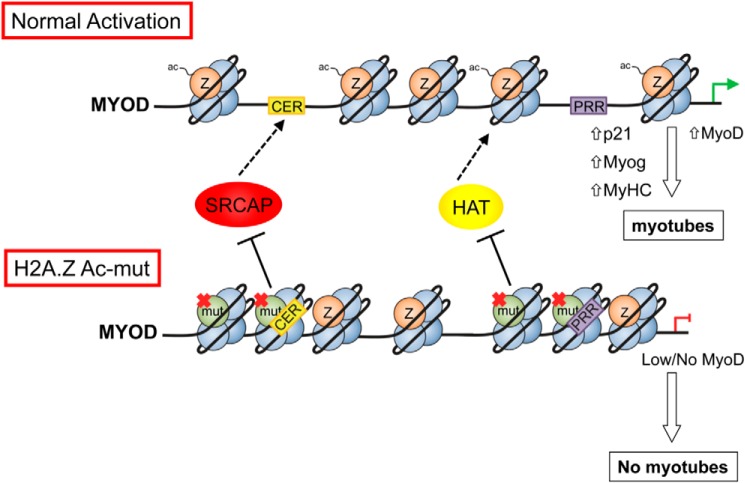
FIGURE 9.
Proposed model: H2A.Z deposition and acetylation are important for maintaining chromatin accessibility at the MyoD locus. For example, incorporation of H2A.Z at the CER maintains decondensation of the whole locus and permits activation of the downstream MyoD promoter (PRR) during differentiation into myotubes. In cells expressing the H2A.Z-Ac mutant, incorporation of the mutant at the CER could inhibit or repel deposition of wild type H2A.Z at the CER and thus impair the enhancer's ability to permit activation of the MyoD promoter. In addition, or alternatively, incorporation of the mutant H2A.Z at the PRR could also inhibit or repel histone acetyltransferases normally recruited to drive MyoD expression and permit differentiation.
Acknowledgments
We thank Drs. Scott Briggs, Sam Benchimol, and David Bazett-Jones for critical reading of the manuscript.
This work was supported in part by the Canadian Research Chair program and by a Canadian Cancer Society Research Institute operating grant (to P. C.).
- H3K4 and H3K27
- histone H3 Lys-4 and Lys-27, respectively
- ac
- me1, me2, and me3, acetylation, mono-methylation, dimethylation, and trimethylation, respectively
- H3 and H4
- histone H3 and H4, respectively
- H2A.Z-Ac-mut
- H2A.Z non-acetylatable mutant
- Pol
- polymerase
- GM
- growth medium
- DI
- differentiation index
- MHC
- myosin heavy chain
- FI
- fusion index
- NLS
- nuclear localization signal
- CER
- core enhancer region
- DRR
- distal regulatory region
- ChART-PCR
- chromatin accessibility by real-time PCR
- PRR
- proximal regulatory region
- EdU
- 5-ethynyl-2′-deoxyuridine.
References
- 1. Faast R., Thonglairoam V., Schulz T. C., Beall J., Wells J. R. E., Taylor H., Matthaei K., Rathjen P. D., Tremethick D. J., Lyons I. (2001) Histone variant H2A.Z is required for early mammalian development. Curr. Biol. 11, 1183–1187 [DOI] [PubMed] [Google Scholar]
- 2. Liu X., Li B., Gorovsky M. A. (1996) Essential and nonessential histone H2A variants in Tetrahymena thermophila. Mol. Cell. Biol. 16, 4305–4311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van Daal A., Elgin S. C. (1992) A histone variant, H2AvD, is essential in Drosophila melanogaster. Mol. Biol. Cell 3, 593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Thatcher T. H., Gorovsky M. A. (1994) Phylogenetic analysis of the core histones H2A, H2B, H3, and H4. Nucleic Acids Res. 22, 174–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jackson J. D., Gorovsky M. A. (2000) Histone H2A.Z has a conserved function that is distinct from that of the major H2A sequence variants. Nucleic Acids Res. 28, 3811–3816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Adam M., Robert F., Larochelle M., Gaudreau L. (2001) H2A.Z is required for global chromatin integrity and for recruitment of RNA polymerase II under specific conditions. Mol. Cell. Biol. 21, 6270–6279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wan Y., Saleem R. A., Ratushny A. V., Roda O., Smith J. J., Lin C. H., Chiang J. H., Aitchison J. D. (2009) Role of the histone variant H2A.Z/Htz1p in TBP recruitment, chromatin dynamics, and regulated expression of oleate-responsive genes. Mol. Cell. Biol. 29, 2346–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Larochelle M., Gaudreau L. (2003) H2A.Z has a function reminiscent of an activator required for preferential binding to intergenic DNA. EMBO J. 22, 4512–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rangasamy D., Berven L., Ridgway P., Tremethick D. J. (2003) Pericentric heterochromatin becomes enriched with H2A.Z during early mammalian development. EMBO J. 22, 1599–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rangasamy D., Greaves I., Tremethick D. J. (2004) RNA interference demonstrates a novel role for H2A.Z in chromosome segregation. Nat. Struct. Mol. Biol. 11, 650–655 [DOI] [PubMed] [Google Scholar]
- 11. Fan J. Y., Rangasamy D., Luger K., Tremethick D. J. (2004) H2A.Z alters the nucleosome surface to promote HP1α-mediated chromatin fiber folding. Mol. Cell 16, 655–661 [DOI] [PubMed] [Google Scholar]
- 12. Dhillon N., Oki M., Szyjka S. J., Aparicio O. M., Kamakaka R. T. (2006) H2A.Z functions to regulate progression through the cell cycle. Mol. Cell. Biol. 26, 489–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Greaves I. K., Rangasamy D., Devoy M., Marshall Graves J. A., Tremethick D. J. (2006) The X and Y chromosomes assemble into H2A.Z-containing facultative heterochromatin following meiosis. Mol. Cell Biol. 26, 5394–5405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guillemette B., Bataille A. R., Gévry N., Adam M., Blanchette M., Robert F., Gaudreau L. (2005) Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 3, e384–e384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li B., Pattenden S. G., Lee D., Gutiérrez J., Chen J., Seidel C., Gerton J., Workman J. L. (2005) Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc. Natl. Acad. Sci. U.S.A. 102, 18385–18390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zanton S. J., Pugh B. F. (2006) Full and partial genome-wide assembly and disassembly of the yeast transcription machinery in response to heat shock. Genes Dev. 20, 2250–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Barski A., Cuddapah S., Cui K., Roh T.-Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. (2007) High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 [DOI] [PubMed] [Google Scholar]
- 18. Creyghton M. P., Markoulaki S., Levine S. S., Hanna J., Lodato M. A., Sha K., Young R. A., Jaenisch R., Boyer L. A. (2008) H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell 135, 649–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ku M., Jaffe J. D., Koche R. P., Rheinbay E., Endoh M., Koseki H., Carr S. A., Bernstein B. E. (2012) H2A.Z landscapes and dual modifications in pluripotent and multipotent stem cells underlie complex genome regulatory functions. Genome Biol. 13, R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gévry N., Chan H. M., Laflamme L., Livingston D. M., Gaudreau L. (2007) p21 transcription is regulated by differential localization of histone H2A.Z. Genes Dev. 21, 1869–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ishibashi T., Dryhurst D., Rose K. L., Shabanowitz J., Hunt D. F., Ausió J. (2009) Acetylation of vertebrate H2A.Z and its effect on the structure of the nucleosome. Biochemistry 48, 5007–5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Millar C. B., Xu F., Zhang K., Grunstein M. (2006) Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 20, 711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bruce K., Myers F. A., Mantouvalou E., Lefevre P., Greaves I., Bonifer C., Tremethick D. J., Thorne A. W., Crane-Robinson C. (2005) The replacement histone H2A.Z in a hyperacetylated form is a feature of active genes in the chicken. Nucleic Acids Res. 33, 5633–5639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sarcinella E., Zuzarte P. C., Lau P. N. I., Draker R., Cheung P. (2007) Monoubiquitylation of H2A.Z distinguishes its association with euchromatin or facultative heterochromatin. Mol. Cell. Biol. 27, 6457–6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Draker R., Sarcinella E., Cheung P. (2011) USP10 deubiquitylates the histone variant H2A.Z, and both are required for androgen receptor-mediated gene activation. Nucleic Acids Res. 39, 3529–3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Draker R., Cheung P. (2009) Transcriptional and epigenetic functions of histone variant H2A.Z. Biochem. Cell Biol. 87, 19–25 [DOI] [PubMed] [Google Scholar]
- 27. Whittle C. M., McClinic K. N., Ercan S., Zhang X., Green R. D., Kelly W. G., Lieb J. D. (2008) The genomic distribution and function of histone variant HTZ-1 during C. elegans embryogenesis. PLoS Genet. 4, e1000187–e1000187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Updike D. L., Mango S. E. (2006) Temporal regulation of foregut development by HTZ-1/H2A.Z and PHA-4/FoxA. PLoS Genet. 2, e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ridgway P., Brown K. D., Rangasamy D., Svensson U., Tremethick D. J. (2004) Unique residues on the H2A.Z containing nucleosome surface are important for Xenopus laevis development. J. Biol. Chem. 279, 43815–43820 [DOI] [PubMed] [Google Scholar]
- 30. Berkes C. A., Tapscott S. J. (2005) MyoD and the transcriptional control of myogenesis. Semin. Cell Dev. Biol. 16, 585–595 [DOI] [PubMed] [Google Scholar]
- 31. Yang J. H., Song Y., Seol J. H., Park J. Y., Yang Y. J., Han J. W., Youn H. D., Cho E. J. (2011) Myogenic transcriptional activation of MyoD mediated by replication-independent histone deposition. Proc. Natl. Acad. Sci. U.S.A. 108, 85–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Harada A., Okada S., Konno D., Odawara J., Yoshimi T., Yoshimura S., Kumamaru H., Saiwai H., Tsubota T., Kurumizaka H., Akashi K., Tachibana T., Imbalzano A. N., Ohkawa Y. (2012) Chd2 interacts with H3.3 to determine myogenic cell fate. EMBO J. 31, 2994–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cuadrado A., Corrado N., Perdiguero E., Lafarga V., Muñoz-Canoves P., Nebreda A. R. (2010) Essential role of p18Hamlet/SRCAP-mediated histone H2A. Z chromatin incorporation in muscle differentiation. EMBO J. 29, 2014–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yaffe D., Saxel O. (1977) Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature 270, 725–727 [DOI] [PubMed] [Google Scholar]
- 35. Burgess A., Vigneron S., Brioudes E., Labbé J. C., Lorca T., Castro A. (2010) Loss of human Greatwall results in G2 arrest and multiple mitotic defects due to deregulation of the cyclin B-Cdc2/PP2A balance. Proc. Natl. Acad. Sci. U.S.A. 107, 12564–12569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bernacchioni C., Cencetti F., Blescia S., Donati C., Bruni P. (2012) Sphingosine kinase/sphingosine 1-phosphate axis: a new player for insulin-like growth factor-1-induced myoblast differentiation. Skelet. Muscle 2, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cottle D. L., McGrath M. J., Cowling B. S., Coghill I. D., Brown S., Mitchell C. A. (2007) FHL3 binds MyoD and negatively regulates myotube formation. J. Cell Sci. 120, 1423–1435 [DOI] [PubMed] [Google Scholar]
- 38. Filigheddu N., Gnocchi V. F., Coscia M., Cappelli M., Porporato P. E., Taulli R., Traini S., Baldanzi G., Chianale F., Cutrupi S., Arnoletti E., Ghè C., Fubini A., Surico N., Sinigaglia F., Ponzetto C., Muccioli G., Crepaldi T., Graziani A. (2007) Ghrelin and des-acyl ghrelin promote differentiation and fusion of C2C12 skeletal muscle cells. Mol. Biol. Cell 18, 986–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Guo K., Wang J., Andrés V., Smith R. C., Walsh K. (1995) MyoD-induced expression of p21 inhibits cyclin-dependent kinase activity upon myocyte terminal differentiation. Mol. Cell Biol. 15, 3823–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Goldhamer D. J., Faerman A., Shani M., Emerson C. P., Jr. (1992) Regulatory elements that control the lineage-specific expression of myoD. Science 256, 538–542 [DOI] [PubMed] [Google Scholar]
- 41. Brunk B. P., Goldhamer D. J., Emerson C. P., Jr. (1996) Regulated demethylation of the myoD distal enhancer during skeletal myogenesis. Dev. Biol. 177, 490–503 [DOI] [PubMed] [Google Scholar]
- 42. Chen J. C., Goldhamer D. J. (2003) Skeletal muscle stem cells. Reprod. Biol. Endocrinol. 1, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen J. C., Ramachandran R., Goldhamer D. J. (2002) Essential and redundant functions of the MyoD distal regulatory region revealed by targeted mutagenesis. Dev. Biol. 245, 213–223 [DOI] [PubMed] [Google Scholar]
- 44. Kucharczuk K. L., Love C. M., Dougherty N. M., Goldhamer D. J. (1999) Fine-scale transgenic mapping of the MyoD core enhancer: MyoD is regulated by distinct but overlapping mechanisms in myotomal and non-myotomal muscle lineages. Development 126, 1957–1965 [DOI] [PubMed] [Google Scholar]
- 45. L'honore A., Lamb N. J., Vandromme M., Turowski P., Carnac G., Fernandez A. (2003) MyoD distal regulatory region contains an SRF binding CArG element required for MyoD expression in skeletal myoblasts and during muscle regeneration. Mol. Biol. Cell 14, 2151–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Creyghton M. P., Cheng A. W., Welstead G. G., Kooistra T., Carey B. W., Steine E. J., Hanna J., Lodato M. A., Frampton G. M., Sharp P. A., Boyer L. A., Young R. A., Jaenisch R. (2010) Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 107, 21931–21936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rada-Iglesias A., Bajpai R., Swigut T., Brugmann S. A., Flynn R. A., Wysocka J. (2011) A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Taberlay P. C., Kelly T. K., Liu C. C., You J. S., De Carvalho D. D., Miranda T. B., Zhou X. J., Liang G., Jones P. A. (2011) Polycomb-repressed genes have permissive enhancers that initiate reprogramming. Cell 147, 1283–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dryhurst D., Ishibashi T., Rose K. L., Eirín-López J. M., McDonald D., Silva-Moreno B., Veldhoen N., Helbing C. C., Hendzel M. J., Shabanowitz J., Hunt D. F., Ausió J. (2009) Characterization of the histone H2A.Z-1 and H2A.Z-2 isoforms in vertebrates. BMC Biol. 7, 86–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Matsuda R., Hori T., Kitamura H., Takeuchi K., Fukagawa T., Harata M. (2010) Identification and characterization of the two isoforms of the vertebrate H2A.Z histone variant. Nucleic Acids Res. 38, 4263–4273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Weintraub H., Tapscott S. J., Davis R. L., Thayer M. J., Adam M. A., Lassar A. B., Miller A. D. (1989) Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc. Natl. Acad. Sci. U.S.A. 86, 5434–5438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lassar A. B., Paterson B. M., Weintraub H. (1986) Transfection of a DNA locus that mediates the conversion of 10T1/2 fibroblasts to myoblasts. Cell 47, 649–656 [DOI] [PubMed] [Google Scholar]
- 53. Jin C., Zang C., Wei G., Cui K., Peng W., Zhao K., Felsenfeld G. (2009) H3.3/H2A.Z double variant-containing nucleosomes mark “nucleosome-free regions” of active promoters and other regulatory regions. Nat. Genet. 41, 941–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kamakaka R. T., Biggins S. (2005) Histone variants: deviants? Genes Dev. 19, 295–310 [DOI] [PubMed] [Google Scholar]
- 55. Draker R., Ng M. K., Sarcinella E., Ignatchenko V., Kislinger T., Cheung P. (2012) A combination of H2A.Z and H4 acetylation recruits Brd2 to chromatin during transcriptional activation. PLoS Genet. 8, e1003047. [DOI] [PMC free article] [PubMed] [Google Scholar]