Background: 3′ repair exonuclease 1 (TREX1) is a DNase involved in autoimmune disorders and the antiviral response.
Results: TREX1 also degrades single-stranded RNA or RNA in a RNA/DNA hybrid molecule.
Conclusion: TREX1 is a human homolog of Escherichia coli RNase T.
Significance: The novel RNase activity of TREX1 is crucial for understanding its physiological role.
Keywords: deoxyribonuclease (DNase), DNA, ribonuclease, RNA, RNA degradation, trex1
Abstract
3′ repair exonuclease 1 (TREX1) is a known DNA exonuclease involved in autoimmune disorders and the antiviral response. In this work, we show that TREX1 is also a RNA exonuclease. Purified TREX1 displays robust exoribonuclease activity that degrades single-stranded, but not double-stranded, RNA. TREX1-D200N, an Aicardi-Goutieres syndrome disease-causing mutant, is defective in degrading RNA. TREX1 activity is strongly inhibited by a stretch of pyrimidine residues as is a bacterial homolog, RNase T. Kinetic measurements indicate that the apparent Km of TREX1 for RNA is higher than that for DNA. Like RNase T, human TREX1 is active in degrading native tRNA substrates. Previously reported TREX1 crystal structures have revealed that the substrate binding sites are open enough to accommodate the extra hydroxyl group in RNA, further supporting our conclusion that TREX1 acts on RNA. These findings indicate that its RNase activity needs to be taken into account when evaluating the physiological role of TREX1.
Introduction
3′ repair exonuclease 1 (TREX1)3 is the most abundant 3′ to 5′ DNA exonuclease in mammalian cells (1–3). Mutations in TREX1 cause autoimmune disorders, including Aicardi-Goutieres syndrome (AGS) type 1, systemic lupus erythematosus, chilblain lupus, and retinal vasculopathy with cerebral leukodystrophy (4–8). Failure to degrade cytosolic DNA was believed to be the principal cause of TREX1-mediated autoimmune diseases (9–11). Additionally, recent data showed that defects in TREX1 also conferred resistance to RNA viruses including human immunodeficiency virus (HIV), vesicular stomatitis virus, influenza virus, West Nile virus, and Sendai virus (12, 13).
Intriguingly, TREX1 resembles bacterial RNase T, a founding member of the DEDD superfamily of exoribonucleases, not only in its primary sequence (1, 14), but also in that both TREX1 and RNase T are active as homodimers (1, 15, 16). Inasmuch as RNase T plays an important role in Escherichia coli RNA metabolism including tRNA end turnover and 3′ maturation of tRNAs and rRNAs, and also exhibits high affinity and robust exonuclease activity on DNA like all other members of the DEDD superfamily (14, 17), we speculated that TREX1 also might have exoribonuclease activity. This was of particular interest because of the involvement of TREX1 in the autoimmune response to cellular nucleic acids as well as its immune response to RNA viruses (9, 10, 12, 13). However, in earlier work exoribonuclease activity was not detected for TREX1 (1).
Nevertheless, we thought it would be worthwhile to reevaluate the biochemical activity of TREX1. To do this, we designed a novel protocol to overexpress and purify full-length human TREX1 from insect cells. In the previous work, TREX1 was expressed in the bacterium, E. coli, and was not the full-length protein (1, 18). Using the insect-cell purified protein, we demonstrate here that TREX1 has 3′ to 5′ exoribonuclease activity on single-stranded RNA (ssRNA) and on DNA/RNA hybrids, but not on double-stranded RNA (dsRNA). Interestingly, human TREX1 and E. coli RNase T share strikingly similar catalytic properties, and based on our observations, we believe they are homologs. Possible roles for the exoribonuclease activity of TREX1 in cellular RNA metabolism and in immunity to RNA viruses are discussed.
Experimental Procedures
Protein Expression, Fractionation, and Purification
The identity of human TREX1 cDNA was confirmed by sequencing and found to match exactly the open reading frame of NCBI Reference Sequence NM_033629 (isoform b) (2). The TREX1-D200N mutant was created by using the QuikChange site-directed mutagenesis kit (Agilent Technologies). Overexpression of non-tagged TREX1 was carried out in insect High Five cells using the Bac-to-Bac expression system (Invitrogen). Expression of TREX1 was confirmed by Western blot analysis using a Pierce ECL kit (Pierce). Antibody against TREX1 was purchased from Santa Cruz Biotechnology (Dallas, TX) (SC-271870).
Upon expression of human TREX1, the insect cells were resuspended in a hypotonic buffer (20 mm Hepes-KOH, pH 7.5, 0.5 mm MgCl2, 5 mm KCl, 5 mm β-mercaptoethanol, and a mixture of protease inhibitors (19)). Nuclei were separated from the cytoplasm of insect cells by homogenization on ice using 10 strokes of a Dounce homogenizer followed by centrifugation at 4,200 rpm (Beckman Coulter rotor JLA-10.500) at 4 °C for 6 min. The nuclear pellet was re-suspended in a cold hypertonic buffer (20 mm Hepes-KOH, pH 7.5, 10% sucrose, 500 mm NaCl, 5 mm β-mercaptoethanol, and the mixture of protease inhibitors), incubated on ice for 10 min, and centrifuged at 12,000 rpm (Beckman Coulter rotor JA-25.50) at 4 °C for 30 min. The precleaned nuclei were then resuspended in the hypertonic buffer containing 0.5% Nonidet P-40, incubated on ice for 10 min, and centrifuged at 16,000 rpm (Beckman Coulter rotor JA-25.50) at 4 °C for 1 h.
The supernatant fraction containing TREX1 was desalted to 100 mm KCl in buffer Q (50 mm KPO4, pH 7.4, 0.5 mm EDTA, 0.01% Nonidet P-40, 10% glycerol, 2 mm DTT and protease inhibitors) and loaded onto a 5-ml Q-Sepharose column. The flow-through from the Q-Sepharose column was directly loaded onto a 1-ml Mono S column (GE Healthcare) in buffer Q. The Mono S column was then resolved using a 30-ml 100 to 500 mm KCl gradient. TREX1 was eluted at ∼280 mm KCl. The pooled TREX1 was concentrated by ultracentrifugation and loaded on a Superdex 200 column (GE Healthcare) in buffer Q with 160 mm KCl. Depending on the purity of TREX1 after Superdex chromatography, it can be further purified on a ssDNA cellulose column using 330 mm KCl for elution from the column. TREX1 protein was followed by SDS-PAGE and Western blot analysis throughout the purification process. Mutant TREX1-D200N was purified using the same protocol as for the wild-type protein. Protein concentration was determined by the Coomassie (Bradford) Protein Assay Reagent (Pierce). The purified protein was stored in aliquots at −80 °C.
Oligonucleotide Substrates and Exonuclease Assays
RNA and DNA oligos were synthesized by Integrated DNA Technology and Sigma, respectively. The sequence of the 20-nucleotide single-stranded RNA (ssRNA) is 5′-GACGCUGCCGAAUUCUACCA-3′. The sequence of the 20-nucleotide ssDNA is 5′-GACGCTGCCGAATTCTACCA-3′. We also used poly(A)17, poly(U)17, poly(C)17, and poly(A)4 RNA oligos (20) and poly(dA)17, poly(dT)17, and poly(dC)17 DNA oligos. All oligos were 32P-labeled at their 5′ end using phage T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP. For nuclease assays, a specified amount of RNA or DNA substrate was incubated with an indicated amount of protein in a 5-μl reaction mixture containing 25 mm Hepes-KOH, pH 7.6, 1 mm DTT, 3 mm MgCl2, 6.5% glycerol, 120 μg/ml of BSA, and 100 mm KCl. After incubation for the specified time at 37 °C, the reaction was terminated by adding 5 μl of sequencing dye with 0.2% SDS and denaturing at 90 °C for 3 min, followed by resolution using a 20% denaturing sequencing gel and analyzed by autoradiography.
Determination of Kinetic Parameters
To measure kinetic parameters, the analyses were repeated three times using increasing amounts of the 20-nt ssRNA (1 nm, 10 nm, 100 nm, 1 μm, and 10 μm) or ssDNA substrate (1, 3, 9, 27, 81, and 243 nm). In the presence of TREX1 (16 nm for ssRNA and 0.4 nm for ssDNA), the reaction was carried out for 20 min at 37 °C. Data were fitted to the Michaelis-Menten equation: v = Vmax[S]/(Km + [S]), where v is the reaction rate and [S] is the concentration of substrate. Km and Vmax were determined by plotting v against [S] using Origin software through nonlinear curve fitting. kcat was calculated by Vmax/[E], where [E] is the enzyme concentration. Quantitation of the nucleic acid excision was carried out using Image J.
Nuclease Assay on tRNA
The reactions were carried out with the addition of 4 nm TREX1 protein and 9.3 μm of a mixture of tRNA-CC[3H]As (21). The 30-μl reaction mixtures were incubated at 37 °C for 30 min. The reaction was stopped by the addition of 150 μl of 0.5% (w/v) yeast RNA and 200 μl of 10% trichloroacetic acid. The sample was placed in ice for 30 min and then centrifuged at 16,000 × g for 15 min at 4 °C. The radioactivity in 200 μl of the supernatant fraction was determined by liquid scintillation counting using a LS 6500 multipurpose scintillation counter (Beckman Coulter, Inc.).
Results
Purification of Non-tagged Human TREX1 Protein
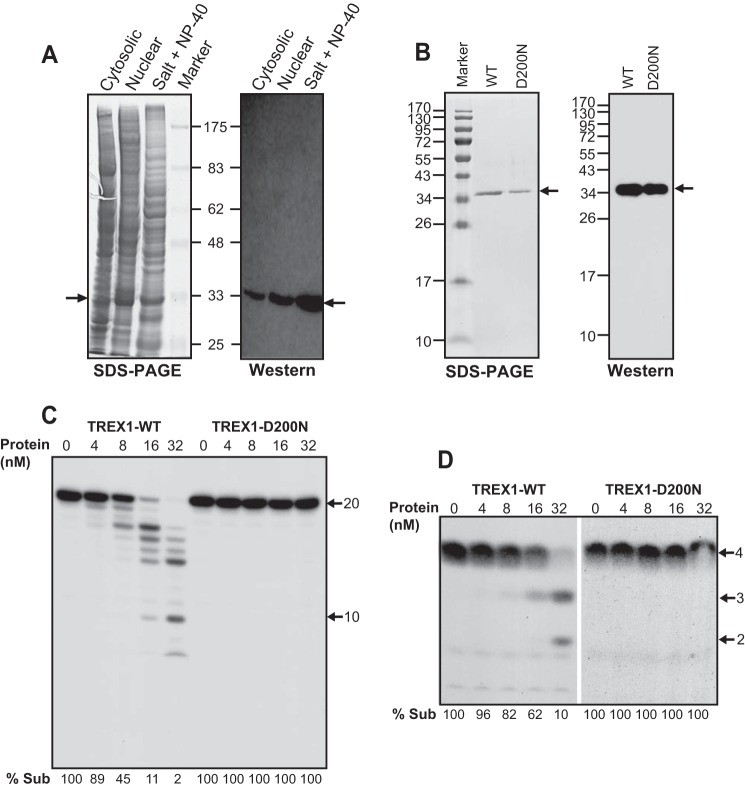
To examine the nuclease activity of native human TREX1, we developed a simple protocol for enriching and purifying the non-tagged, full-length protein from insect cells in which it was overexpressed (Fig. 1A). Isolated nuclei, which contain a large amount of TREX1 (Fig. 1A), were treated with a high salt concentration (500 mm NaCl), which results in nuclear shrinkage and forces most nuclear matrix proteins out of the nuclei. The “shrunken” nuclei were then treated with 0.5% Nonidet P-40 and 500 mm salt for release of nuclear membrane-associated and chromatin-associated proteins. This procedure greatly enriches the TREX1 protein sample before column purification (Fig. 1A, compare salt + Nonidet P-40 lanes with nuclear lanes). The non-tagged TREX1-WT and TREX1-D200N mutant proteins were then purified to homogeneity using Q-Sepharose, Mono S, Superdex 200, and ssDNA columns. Western analysis confirmed that both homogeneous proteins were indeed TREX1 (Fig. 1B).
FIGURE 1.
Purified human TREX1 displays exoribonuclease activity on ssRNA. A, SDS-PAGE and Western blots of fractionated TREX1-containing extracts (40 μg each lane). Salt + Nonidet P-40 indicates the TREX1 protein samples after osmotic and detergent treatments. Arrows point to TREX1. Protein markers in kilodaltons are indicated. B, SDS-PAGE and Western analysis of the purified WT and D200N TREX1 proteins. The PAGE gel was stained with Coomassie Brilliant Blue R-250. Arrows point to the purified TREX1. Protein markers in kilodaltons are indicated. C, titration of TREX1-WT and TREX1-D200N proteins on a 20-nucleotide ssRNA substrate (2 nm) labeled at its 5′ end by [γ-32P]ATP using T4 polynucleotide kinase. Reaction time is 10 min. Arrows point to positions of the substrate and a 10-nt standard. % Sub indicates the percentage of the remaining substrate. D, activity of TREX1-WT and D200N on a 4-nucleotide poly(A) RNA substrate (2 nm) labeled at its 5′ end. Reaction time is 10 min. Arrows point to position of the 4-nt substrate and positions of the 2- and 3-nt standards.
Human TREX1 Has Exoribonuclease Activity
After verifying the DNA exonuclease activity of the purified protein (see examples in Figs. 3, 4A, and 5A), we investigated whether TREX1 also has ribonuclease activity by incubating the protein with a 5′-end labeled 20-nt long RNA (see “Experimental Procedures” for sequence). As shown in Fig. 1C, TREX1 showed distributive exoribonuclease activity on this RNA substrate that increased with increasing concentrations of enzyme. At the highest levels of TREX1 protein (16 and 32 nm), almost all of the RNA oligo was converted to shorter fragments due to removal of residues from its 3′ end. In contrast, the D200N mutant of TREX1, which lacks DNA exonuclease activity and leads to AGS disease (9, 10, 22), is completely defective in shortening the ssRNA substrate (Fig. 1C). Inasmuch as the WT and mutant proteins were purified using the same procedure, these data also indicate that the RNase activity is a property of TREX1 and not a contaminating protein. Additionally, an N-terminal hexahistidine-tagged version of TREX1 that was purified through a different protocol (nickel and Mono S columns) shows similar RNase activity (data not shown).
FIGURE 3.
Effect of metal ions on the exonuclease activity of TREX1. The experiments were done using 16 nm purified human TREX1 for RNA and 1 nm for DNA, and the indicated oligo substrates (2 nm). The cations were added to the reaction mixture at a final concentration of 3 mm. Incubation time is shown on the top. % Sub indicates the percentage of the remaining substrate. P indicates the 32P labeling.
FIGURE 4.
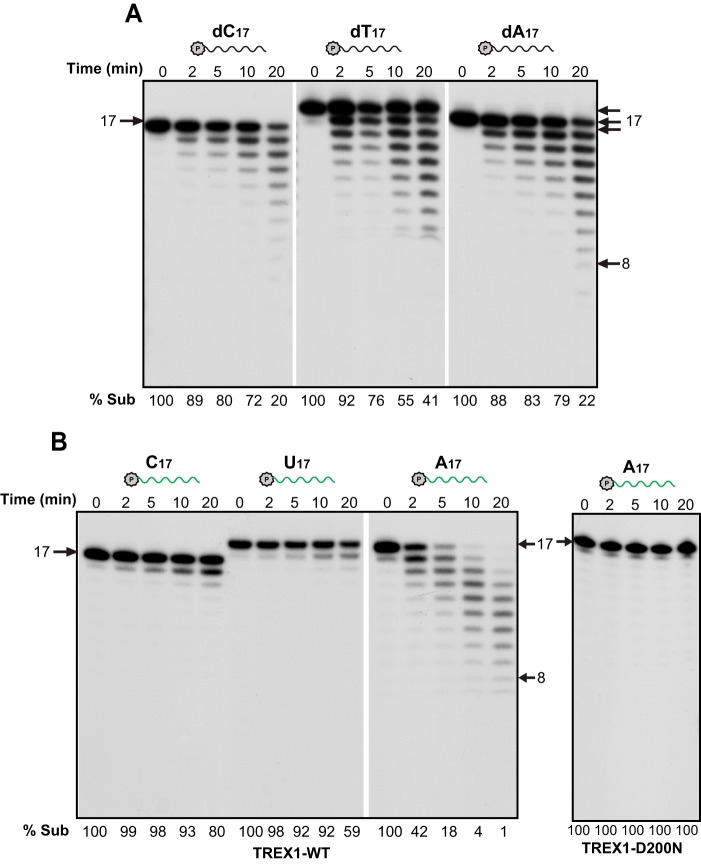
TREX1 discriminates against pyrimidines in RNA. Time course experiment using 16 nm purified human TREX1 and TREX1-D200N (for A17 only) and the indicated DNA (A) and RNA (B) substrates (2 nm, green, RNA; black, DNA).
FIGURE 5.
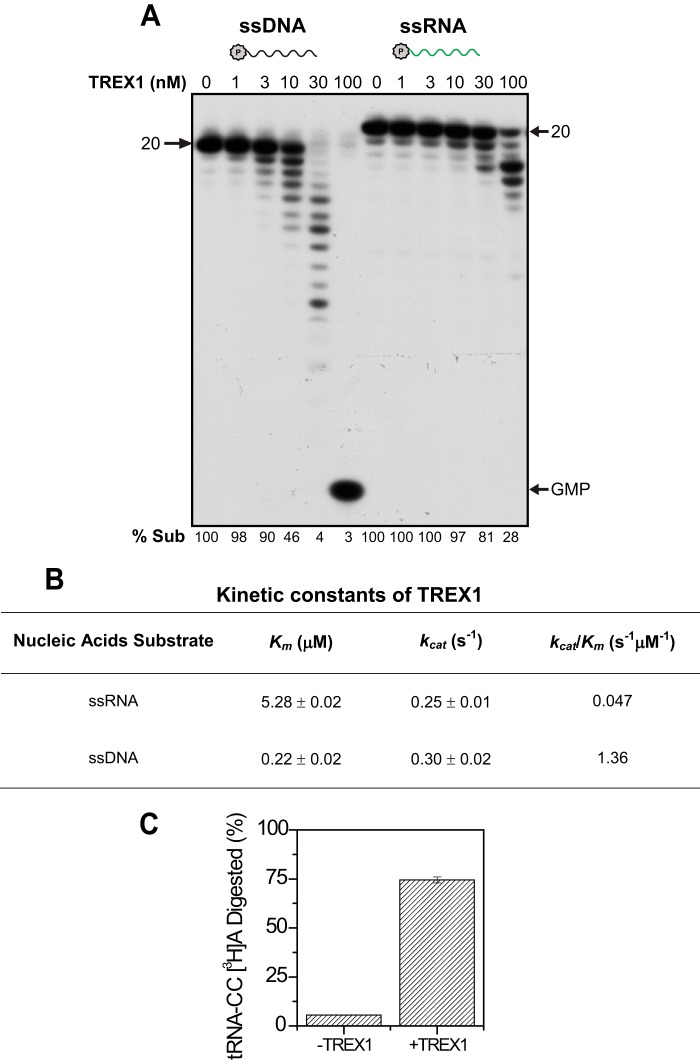
Kinetic analysis of TREX1 on ssRNA and ssDNA and comparison of its DNA and RNA exonuclease activity. A, TREX1 activity on 20-nucleotide ssDNA (black) or ssRNA (green) substrate (2.4 μm). The reaction time is 10 min. Arrows point to substrate position and position of GMP. % Sub indicates the percentage of remaining substrate. P in a circle indicates the 32P labeling. B, kinetic measurement of TREX1. In the presence of 16 nm TREX1, ssDNA, or ssRNA were added at varying concentrations (“Experimental Procedures”) in a 20-min reaction. Data were fitted to the Michaelis-Menten equation and Km and kcat were calculated as described under “Experimental Procedures.“ C, activity of TREX1 on tRNA. 4 nm TREX1 protein was incubated with 9.3 μm tRNA-CC[3H]A at 37 °C for 30 min. The radioactivity in 200 μl of the supernatant fraction was determined by liquid scintillation counting. Error bars, standard deviation.
Under the incubation conditions used in Fig. 1C, most of the degradation products derived from the 20-nt ssRNA substrate were longer than 10 nucleotides. To examine whether TREX1 also can degrade a small RNA substrate, we incubated TREX1 with a 4-nucleotide oligo, A4 (Fig. 1D). Although TREX1 is about 2–3-fold less active on the 4-nucleotide substrate than the 20-nucleotide RNA (compare Fig. 1, C with D), it effectively shortens A4 to A3 and A2, indicating that TREX1 can digest even very small RNAs. Again, the D200N mutant is completely defective in shortening the A4 substrate (Fig. 1D), confirming that the activity on A4 is due to TREX1.
TREX1 Cleaves the RNA in a RNA/DNA hybrid, but Is Inactive on Double-stranded RNA
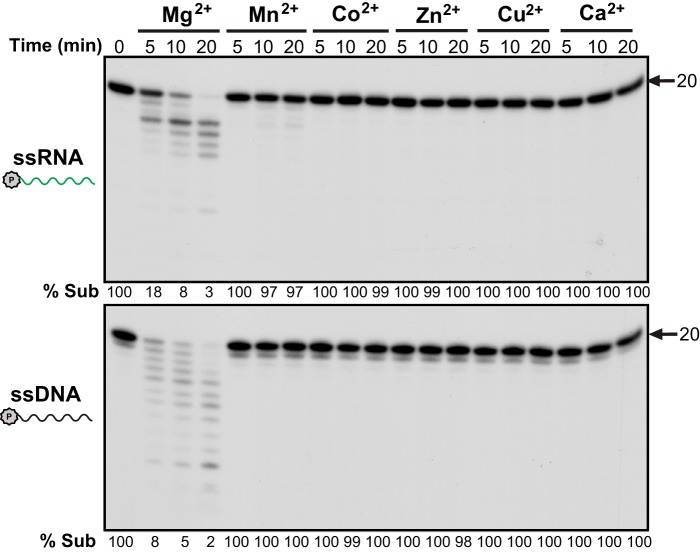
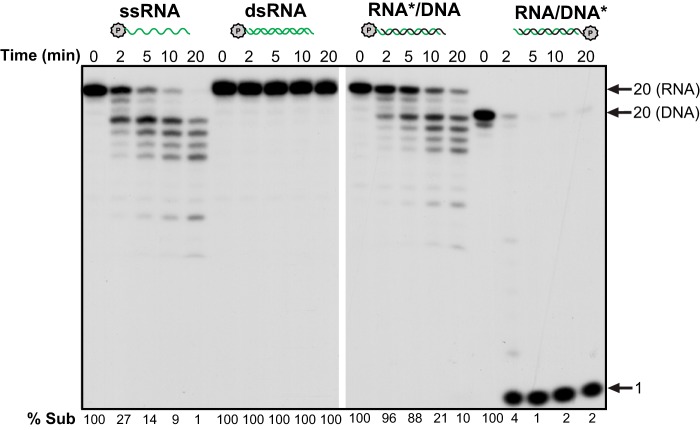
In early work, TREX1 was shown to have strong exonuclease activity on dsDNA (18). To determine whether TREX1 also can degrade dsRNA, we examined its activity on a variety of substrates using 16 nm TREX1. Although TREX1 rapidly degrades ssRNA in a time-dependent manner, it is inactive on the dsRNA counterpart with no loss of substrate even after 20 min of incubation (Fig. 2). This observation is similar to what was reported for E. coli RNase T (23), another exoribonuclease of the DEDD family. In contrast, TREX1 is able to degrade the RNA strand in an RNA/DNA hybrid molecule, although the activity is lower than that on ssRNA. It is likely that RNA degradation in the DNA-RNA hybrid is due to release of ssRNA following rapid degradation of the DNA strand, because, as shown in Fig. 2, the DNA strand is completely degraded in 2 min, whereas degradation of the RNA strand is just beginning at that time.
FIGURE 2.
TREX1 action on RNA and RNA/DNA hybrid substrates. The experiments were done using 16 nm purified human TREX1 for RNA, the indicated oligo substrates (2 nm), and an increasing incubation time. Arrows point to substrate position and 1-nt standard. % Sub indicates the percentage of remaining substrate. P in a circle and stars indicate the 32P labeling. Green, RNA; black, DNA.
As previously observed for its DNA exonuclease activity (Fig. 3, bottom panel, and Ref. 18), TREX1 requires Mg2+ for optimal RNase activity (Fig. 3, top panel). Other cations, Mn2+, Co2+, Zn2+, Cu2+, and Ca2+ are unable to stimulate activity when present at 3 mm.
The Exoribonuclease Activity of TREX1 Displays a Dramatic Base Specificity
E. coli RNase T is known to display strong base specificity and to discriminate against pyrimidine nucleotides (23). Consequently, we investigated the action of TREX1 on ssRNA oligos with different base compositions. In contrast to its DNA exonuclease activity, which acts almost equally on dC17, dT17, and dA17 (Fig. 4A), the RNase activity of TREX1 is very sequence dependent. As shown in Fig. 4B, TREX1 effectively degrades an A17 substrate, but exhibits very limited activity with either C17 or U17 substrates, removing, at most, only a few residues. Again, the D200N mutant is completely incapable of degrading A17 (Fig. 4B, right panel). These findings indicate that the similarity between TREX1 and E. coli RNase T extends to its base specificity (23).
TREX1 Has Lower Affinity for ssRNA Than for ssDNA
Inasmuch as TREX1 is active on both ssDNA and ssRNA, we directly compared these two activities. As shown in Fig. 5A, TREX1 degrades both ssDNA and ssRNA in a distributive manner. However, TREX1 appears to show stronger activity on ssDNA, being ∼10-fold more active on DNA under the specific experimental conditions (compare 10 and 100 nm lanes on DNA and RNA (2.4 μm), respectively, in Fig. 5A).
To determine the catalytic property, we measured the kinetic constants of TREX1 on ssRNA and ssDNA. The apparent Km of TREX1 for ssRNA is 5.28 μm (Fig. 5B), comparable with that of E. coli RNase T (10 μm) (24). This is considerably higher than the Km of TREX1 for ssDNA (0.22 μm, Fig. 5B, and 19 nm in Ref. 18), and again resembles E. coli RNase T in terms of affinity for RNA and DNA substrates (17). Interestingly, the kcat of TREX1 for ssRNA is very similar to that for ssDNA (Fig. 5B). Thus, the resulting overall enzyme efficiency of TREX1 for ssDNA is ∼30-fold higher than for ssRNA (Fig. 5B, kcat/Km). These kinetic parameters indicate that the DNase activity of TREX1 is not dramatically stronger than its RNase activity (similar kcat), but it is more efficient on ssDNA due to its higher substrate affinity to DNA (lower Km).
TREX1 Acts on tRNA
RNase T plays an important role in E. coli tRNA end turnover and 3′ maturation of tRNAs and rRNAs (14, 17). To determine whether TREX1 also acts on natural RNA substrates, we incubated purified TREX1 with a native tRNA sample that had been 3′ end labeled with [3H]A, an assay commonly used for RNase T (21). As shown in Fig. 5C, TREX1 efficiently removes the [3H]AMP residue from tRNA, amounting to 75% of the tRNA ends in 30 min. Essentially no AMP is removed in the absence of added enzyme. This indicates that human TREX1 is active on native tRNA, again showing that its catalytic properties are similar to those of E. coli RNase T (17).
Discussion
In this report, we demonstrate that the DNA exonuclease TREX1, like all other members of the DEDD superfamily of nucleases, is also an exoribonuclease. We suggest that TREX1 is a human homolog of E. coli RNase T based on the following observations: 1) TREX1 and RNase T share sequence similarity (Fig. 7) (1, 2); 2) both TREX1 and RNase T need to form a homodimer to exhibit nuclease activity (1, 15, 16, 24); 3) TREX1 and RNase T both display exoribonuclease and DNA exonuclease activity (this study and Ref. 17 for RNase T); 4) both TREX1 and RNase T display a higher Km for ssRNA than for ssDNA (this study and Refs. 17 and 18 for RNase T); and 5) both TREX1 and RNase T discriminate against pyrimidine nucleotides for their exoribonuclease activities (this study and Ref. 23 for RNase T). Thus, TREX1 might be involved in RNA metabolism in human cells just as RNase T is in E. coli (14, 17). Indeed, our data show that human TREX1, like RNase T, is competent in trimming tRNA ends (Fig. 5C). It would be of considerable interest to further examine TREX1 for possible roles in RNA metabolism.
FIGURE 7.
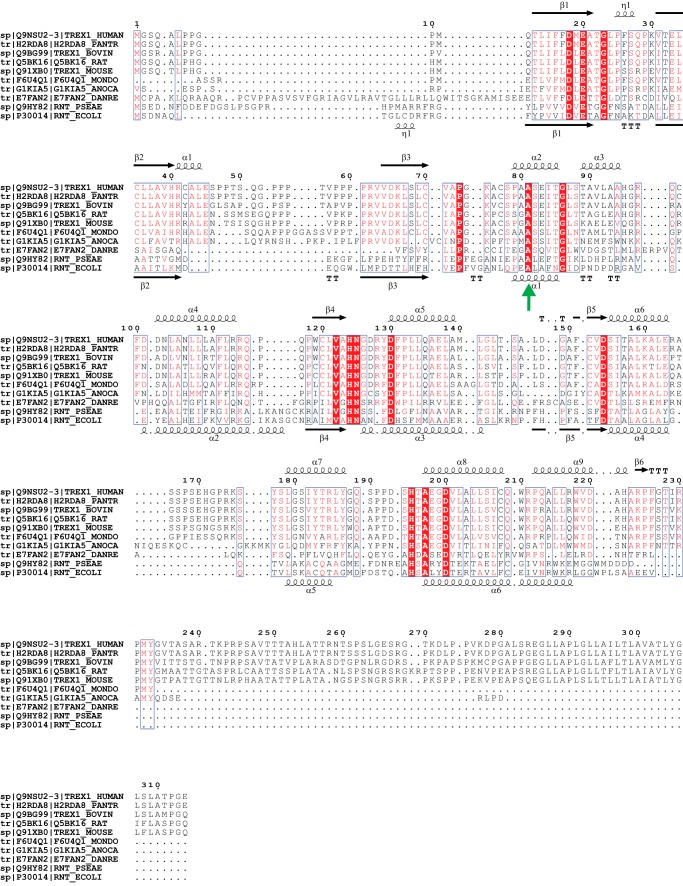
Multiple sequence alignment of selected TREX1 and RNase T homologs. Human TREX1 (UniProt accession number Q9NSU2, isoform b) was aligned with several putative TREX1 homologs as well as two RNase T sequences. Structure guided multiple sequence alignment was carried out using T-Coffee and Expresso (32), with minor adjustments based on RNase T structures (33). Homologs were chosen to include sequence diversity, and are H2RDA8 from Pan troglodytes, Q9BG99 from Bos taurus, Q5BK16 from Rattus norvegicus, Q91XB0 from Mus musculus, F6U4Q1 from Monodelphis domestica, G1KIA5 from Anolis carolinensis, and the more distant (26% identity) from Danio rerio (zebrafish). RNase T sequences are from Pseudomonas aeruginosa (Q9HY82) and E. coli (P30014). Sequence similarity is highlighted using ESPRIPT (34), and the secondary structure schematics shown are based on the mouse TREX1 structure (top; Protein Data Bank code 3U6F, Ref. 9) and the E. coli RNase T structure (bottom; Protein Data Bank code 2IS3, Ref. 33). Residue 81 in human TREX1, closest to the 2′ position on the 3′ nucleotide at the cleavage site, is conserved as an alanine in TREX1 homologs (position marked with a green arrow).
Analysis of the available TREX1 crystal structures reveals that the substrate binding sites are open enough to accommodate the 2′ hydroxyls in an RNA substrate (9) (Fig. 6). The closest approach between the enzyme and the 2′ positions on the nucleic acid substrate is at the 3′ end in a co-crystal structure of mouse TREX1 and a DNA substrate (9). The residue closest to the 2′ position at the 3′ end of the substrate has a short side chain (Ala-81 in mouse TREX1) and can easily accommodate a hydroxyl group. This position (residue 81) in human TREX1 is also an alanine, and sequence alignment of TREX1 homologs suggests that this may be conserved, presumably to maintain sufficient space for binding RNA substrates (Fig. 7).
FIGURE 6.
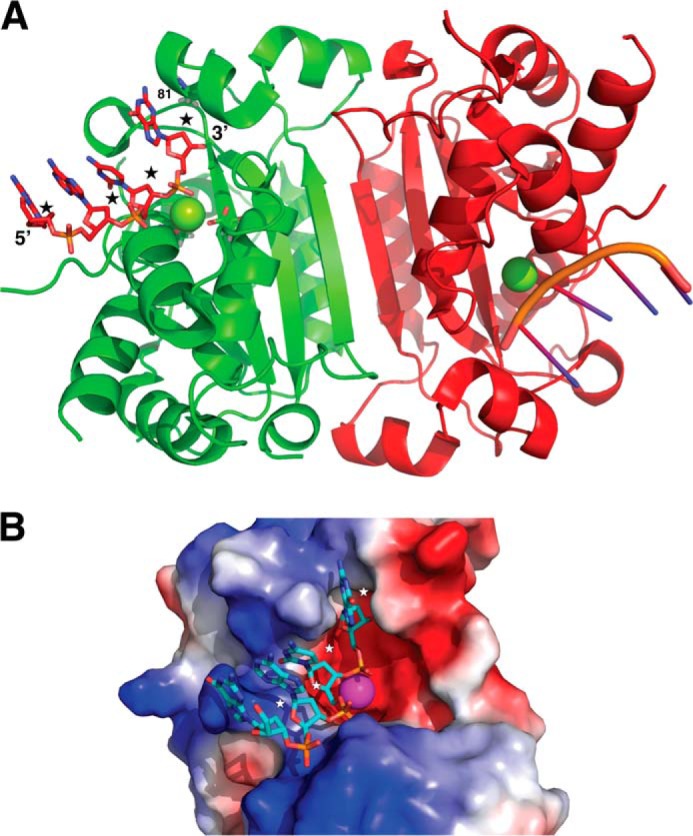
Mouse TREX1 structure shows that this enzyme can bind RNA substrates. A, ribbon structure of the mouse TREX1 (TREX1-D200N; Protein Data Bank code 3U6F (9)) shows a dimeric protein that binds nucleic acid substrates on two oppositely positioned solvent-exposed clefts (DNA substrate from this structure is shown in a stick representation). The binding sites are open enough to accommodate the 2′ OH in an RNA substrate (approximate expected positions of the 2′ oxygen atoms are marked by stars; Mg at the active site is shown as a sphere, and an active site Asp is shown as sticks). The closest approach between the enzyme and the 2′ positions on the nucleic acid substrate is at the 3′ end. This position (residue 81 in human TREX1) is always a short side chain (alanine), which provides enough space to accommodate a 2′ hydroxyl. B, a molecular surface view showing one of the substrate binding clefts of TREX1. The molecular surface is colored by electrostatic charge using PyMol (31).
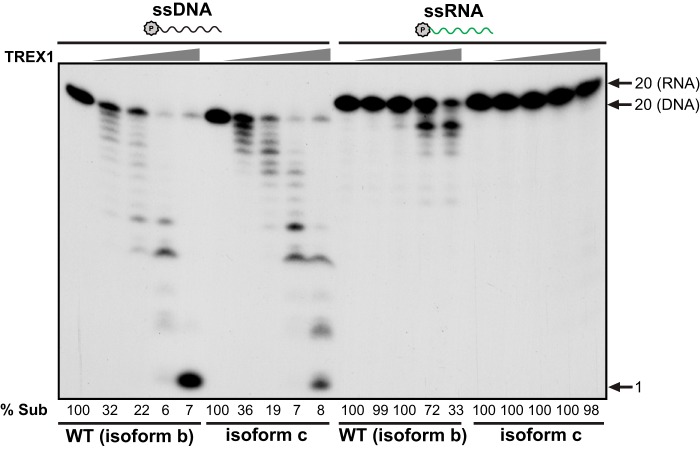
Human TREX1 has three transcript variants with different lengths of extra residues at the N terminus. The highest expressed TREX1 variant in human cells is isoform b that was used in this study (2). Earlier work failed to detect the RNase activity of TREX1 (1). One likely possibility to explain this result is that the TREX1 protein expressed in the earlier work was the shortest isoform (isoform c), lacking 10 amino acids at its N terminus compared with isoform b (1, 2, 18). Such a change in TREX1 structure could affect its substrate affinity or catalytic activity considering that the active site residues (Asp-8 and Glu-10 in isoform c or Asp-18 and Glu-20 in isoform b) of TREX1 are located in the N-terminal region (9, 25). To test this possibility, we purified isoform c and compared its DNase and RNase activities with isoform b (referred as WT in this study). Under conditions in which both isoform b (WT) and isoform c show similar DNase activity, isoform c fails to degrade the ssRNA substrate, in contrast to isoform b (Fig. 8). These data explain the previous observation (1) and suggest that the structural difference between isoform b and isoform c affects primarily the RNase activity. In this regard, it will be interesting to examine the DNase and RNase activities of the longest TREX1 isoform a (2). Nevertheless, based on our data, it is clear that TREX1 isoform b possesses RNase activity, and that TREX1 is closely related to E. coli RNase T, another nuclease that possesses both DNase and RNase activity (17).
FIGURE 8.
The N-terminal 1–10 truncation present in isoform c of TREX1 diminishes RNase activity. The protein concentrations of WT (isoform b, NCBI Ref. sequence NP_338599.1) and isoform c (NCBI Ref. sequence NP_009179.2) of TREX1 were adjusted so that their activities against ssDNA (2 nm) were identical. Under these same conditions their respective activities against ssRNA (2 nm) were determined. Assays were carried out at 37 °C for 20 min. Arrows point to positions of the substrate and a 1-nt standard. % Sub indicates the percentage of remaining substrate.
Mutations in TREX1 are known to cause AGS1 (7, 26). Moreover, the heterotrimeric endoribonuclease, RNase H2, interacts with TREX1. Defects in the RNase H2 subunits, namely RNASEH2B, RNASEH2C, and RNASEH2A, are known to cause AGS2, AGS3, and AGS4 types of the disease, respectively (26–29). Although it is tempting to speculate that the TREX1-RNase H2 complex functions as a holo-RNase enzyme in removing RNA debris that may cause the autoimmune response, it is possible that the endoribonuclease activity of RNase H2 first cuts RNA into smaller fragments and then the exoribonuclease activity of TREX1 further degrades the RNA fragments to nucleotides (Fig. 1, C and D). It is therefore conceivable that defects in RNA degradation may contribute to accumulation of RNA species and thus induce other aberrant autoimmune responses such as systemic lupus erythematosus, chilblain lupus, and retinal vasculopathy with cerebral leukodystrophy. It would be interesting to investigate whether and how the exoribonuclease activity of TREX1 and the endoribonuclease activity of RNase H2 coordinate with each other to achieve efficient RNA removal.
The DNA exonuclease activity of TREX1 was shown to be responsible for preventing HIV replication by degrading the reverse-transcribed ssDNA and hiding the virus from detection by the immune system (12, 30). Beyond the proposed DNA removal of the HIV virus in suppressing the immune response (12, 30), it is also possible, based on our observations, that TREX1 may directly degrade the single-stranded HIV RNA virus or the RNA/DNA hybrid (Fig. 2A) and contribute to suppression of HIV replication. Although it has been shown that a knock-out of TREX1 causes aberrant lysosomal biogenesis, which elicits innate immune responses and inhibits replication of RNA viruses (12, 13), it is also likely that the RNase activity of TREX1 is directly involved in degrading RNA virus and preventing it from being duplicated.
In addition to the discovery of the exoribonuclease activity of TREX1, we also describe an efficient protocol for enriching and precleaning proteins that are associated with chromatin and membranes. This protocol not only makes it possible to isolate and purify high quality non-tagged human TREX1 protein for this study, but it also provides a universal strategy for isolating non-tagged membrane-bound and chromatin-bound proteins.
Acknowledgments
We are grateful for insightful discussions with Dr. Guo-Min Li at the University of Kentucky College of Medicine. Thanks to Dr. Fred W. Perrino at the Wake Forest School of Medicine for providing the cDNA for human TREX1, Dr. Thomas Hollis at the Wake Forest School of Medicine for inspiring us to initiate this research, and Dr. Dan Stetson at the University of Washington for providing the TREX1 knock-out MEFs.
This work was supported, in whole or in part, by National Institutes of Health Grants HL105631 (to Y. Z.) and GM16317 (to M. D.).
- TREX1
- 3′ repair exonuclease 1
- AGS
- Aicardi-Goutieres syndrome
- ssRNA
- single-stranded RNA
- ssDNA
- single-stranded DNA
- dsRNA
- double-stranded RNA
- nt
- nucleotide.
References
- 1. Mazur D. J., Perrino F. W. (1999) Identification and expression of the TREX1 and TREX2 cDNA sequences encoding mammalian 3′ → 5′ exonucleases. J. Biol. Chem. 274, 19655–19660 [DOI] [PubMed] [Google Scholar]
- 2. Mazur D. J., Perrino F. W. (2001) Structure and expression of the TREX1 and TREX2 3′ → 5′ exonuclease genes. J. Biol. Chem. 276, 14718–14727 [DOI] [PubMed] [Google Scholar]
- 3. Höss M., Robins P., Naven T. J., Pappin D. J., Sgouros J., Lindahl T. (1999) A human DNA editing enzyme homologous to the Escherichia coli DnaQ/MutD protein. EMBO J. 18, 3868–3875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Richards A., van den Maagdenberg A. M., Jen J. C., Kavanagh D., Bertram P., Spitzer D., Liszewski M. K., Barilla-Labarca M. L., Terwindt G. M., Kasai Y., et al. (2007) C-terminal truncations in human 3′-5′ DNA exonuclease TREX1 cause autosomal dominant retinal vasculopathy with cerebral leukodystrophy. Nat. Genet. 39, 1068–1070 [DOI] [PubMed] [Google Scholar]
- 5. Lee-Kirsch M. A., Gong M., Chowdhury D., Senenko L., Engel K., Lee Y. A., de Silva U., Bailey S. L., Witte T., Vyse T. J., et al. (2007) Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat. Genet. 39, 1065–1067 [DOI] [PubMed] [Google Scholar]
- 6. Lee-Kirsch M. A., Chowdhury D., Harvey S., Gong M., Senenko L., Engel K., Pfeiffer C., Hollis T., Gahr M., Perrino F. W., Lieberman J., Hubner N. (2007) A mutation in TREX1 that impairs susceptibility to granzyme A-mediated cell death underlies familial chilblain lupus. J. Mol. Med. 85, 531–537 [DOI] [PubMed] [Google Scholar]
- 7. Crow Y. J., Hayward B. E., Parmar R., Robins P., Leitch A., Ali M., Black D. N., van Bokhoven H., Brunner H. G., Hamel B. C., et al. (2006) Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat. Genet. 38, 917–920 [DOI] [PubMed] [Google Scholar]
- 8. Kavanagh D., Spitzer D., Kothari P. H., Shaikh A., Liszewski M. K., Richards A., Atkinson J. P. (2008) New roles for the major human 3′-5′ exonuclease TREX1 in human disease. Cell Cycle 7, 1718–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bailey S. L., Harvey S., Perrino F. W., Hollis T. (2012) Defects in DNA degradation revealed in crystal structures of TREX1 exonuclease mutations linked to autoimmune disease. DNA Repair 11, 65–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fye J. M., Orebaugh C. D., Coffin S. R., Hollis T., Perrino F. W. (2011) Dominant mutation of the TREX1 exonuclease gene in lupus and Aicardi-Goutieres syndrome. J. Biol. Chem. 286, 32373–32382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barber G. N. (2011) Cytoplasmic DNA innate immune pathways. Immunol. Rev. 243, 99–108 [DOI] [PubMed] [Google Scholar]
- 12. Yan N., Regalado-Magdos A. D., Stiggelbout B., Lee-Kirsch M. A., Lieberman J. (2010) The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat. Immunol. 11, 1005–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hasan M., Koch J., Rakheja D., Pattnaik A. K., Brugarolas J., Dozmorov I., Levine B., Wakeland E. K., Lee-Kirsch M. A., Yan N. (2013) Trex1 regulates lysosomal biogenesis and interferon-independent activation of antiviral genes. Nat. Immunol. 14, 61–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zuo Y., Deutscher M. P. (2001) Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 29, 1017–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Orebaugh C. D., Fye J. M., Harvey S., Hollis T., Perrino F. W. (2011) The TREX1 exonuclease R114H mutation in Aicardi-Goutieres syndrome and lupus reveals dimeric structure requirements for DNA degradation activity. J. Biol. Chem. 286, 40246–40254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li Z., Zhan L., Deutscher M. P. (1996) Escherichia coli RNase T functions in vivo as a dimer dependent on cysteine 168. J. Biol. Chem. 271, 1133–1137 [DOI] [PubMed] [Google Scholar]
- 17. Zuo Y., Deutscher M. P. (1999) The DNase activity of RNase T and its application to DNA cloning. Nucleic Acids Res. 27, 4077–4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mazur D. J., Perrino F. W. (2001) Excision of 3′ termini by the Trex1 and TREX2 3′ → 5′ exonucleases: characterization of the recombinant proteins. J. Biol. Chem. 276, 17022–17029 [DOI] [PubMed] [Google Scholar]
- 19. Benitez A., Yuan F., Nakajima S., Wei L., Qian L., Myers R., Hu J. J., Lan L., Zhang Y. (2014) Damage-dependent regulation of MUS81-EME1 by Fanconi anemia complementation group A protein. Nucleic Acids Res. 42, 1671–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dutta T., Deutscher M. P. (2009) Catalytic properties of RNase BN/RNase Z from Escherichia coli: RNase BN is both an exo- and endoribonuclease. J. Biol. Chem. 284, 15425–15431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Deutscher M. P., Ghosh R. K. (1978) Preparation of synthetic tRNA precursors with tRNA nucleotidyltransferase. Nucleic Acids Res. 5, 3821–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lehtinen D. A., Harvey S., Mulcahy M. J., Hollis T., Perrino F. W. (2008) The TREX1 double-stranded DNA degradation activity is defective in dominant mutations associated with autoimmune disease. J. Biol. Chem. 283, 31649–31656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zuo Y., Deutscher M. P. (2002) The physiological role of RNase T can be explained by its unusual substrate specificity. J. Biol. Chem. 277, 29654–29661 [DOI] [PubMed] [Google Scholar]
- 24. Zuo Y., Deutscher M. P. (2002) Mechanism of action of RNase T: I. identification of residues required for catalysis, substrate binding, and dimerization. J. Biol. Chem. 277, 50155–50159 [DOI] [PubMed] [Google Scholar]
- 25. de Silva U., Choudhury S., Bailey S. L., Harvey S., Perrino F. W., Hollis T. (2007) The crystal structure of TREX1 explains the 3′ nucleotide specificity and reveals a polyproline II helix for protein partnering. J. Biol. Chem. 282, 10537–10543 [DOI] [PubMed] [Google Scholar]
- 26. Rice G., Newman W. G., Dean J., Patrick T., Parmar R., Flintoff K., Robins P., Harvey S., Hollis T., O'Hara A., et al. (2007) Heterozygous mutations in TREX1 cause familial chilblain lupus and dominant Aicardi-Goutieres syndrome. Am. J. Hum. Genet. 80, 811–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. O'Driscoll M. (2008) TREX1 DNA exonuclease deficiency, accumulation of single stranded DNA and complex human genetic disorders. DNA Repair 7, 997–1003 [DOI] [PubMed] [Google Scholar]
- 28. Rigby R. E., Leitch A., Jackson A. P. (2008) Nucleic acid-mediated inflammatory diseases. Bioessays 30, 833–842 [DOI] [PubMed] [Google Scholar]
- 29. Reijns M. A., Jackson A. P. (2014) Ribonuclease H2 in health and disease. Biochem. Soc. Trans. 42, 717–725 [DOI] [PubMed] [Google Scholar]
- 30. Geijtenbeek T. B. (2010) Host DNase TREX1 hides HIV from DNA sensors. Nat. Immunol. 11, 979–980 [DOI] [PubMed] [Google Scholar]
- 31. Delano W. L. (2002) The PyMOL Molecular Graphics System, version 1.4, Schrödinger, LLC, New York [Google Scholar]
- 32. Notredame C., Higgins D. G., Heringa J. (2000) T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217 [DOI] [PubMed] [Google Scholar]
- 33. Zuo Y., Zheng H., Wang Y., Chruszcz M., Cymborowski M., Skarina T., Savchenko A., Malhotra A., Minor W. (2007) Crystal structure of RNase T, an exoribonuclease involved in tRNA maturation and end turnover. Structure 15, 417–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Robert X., Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 [DOI] [PMC free article] [PubMed] [Google Scholar]