Background: Anillin is an evolutionarily conserved protein essential for cytokinesis.
Results: Importin β2 targets anillin to the nucleus during interphase.
Conclusion: Anillin is spatio-temporally regulated during the cell cycle to prevent disruption of the interphase cellular architecture.
Significance: Sequestering proteins with key mitotic functions in the nucleus is a vital part of their cell cycle regulation.
Keywords: cell cycle, cell division, nuclear transport, nucleus, protein targeting
Abstract
The compartmentalization of cell cycle regulators is a common mechanism to ensure the precise temporal control of key cell cycle events. For instance, many mitotic spindle assembly factors are known to be sequestered in the nucleus prior to mitotic onset. Similarly, the essential cytokinetic factor anillin, which functions at the cell membrane to promote the physical separation of daughter cells at the end of mitosis, is sequestered in the nucleus during interphase. To address the mechanism and role of anillin targeting to the nucleus in interphase, we identified the nuclear targeting motif. Here, we show that anillin is targeted to the nucleus by importin β2 in a Ran-dependent manner through an atypical basic patch PY nuclear localization signal motif. We show that although importin β2 binding does not regulate anillin's function in mitosis, it is required to prevent the cytosolic accumulation of anillin, which disrupts cellular architecture during interphase. The nuclear sequestration of anillin during interphase serves to restrict anillin's function at the cell membrane to mitosis and allows anillin to be rapidly available when the nuclear envelope breaks down to remodel the cellular architecture necessary for successful cell division.
Introduction
The spatio-temporal localization of proteins within a cell determines where and when a particular biochemical reaction or pathway is activated. Such mechanisms are evident throughout evolution, the most fundamental being between the cytoplasm and the nucleus. The nucleus is separated from the cytosol of eukaryotic cells by the nuclear envelope, a double membrane studded with pores that allow the trafficking of proteins and RNA into and out of the nucleus (1). Proteins are targeted to the nucleus through distinct motifs, nuclear localization signals (NLS)2 that bind to karyopherins, a family of nuclear transport receptors (NTRs). Classical NLSs contain basic patches that are either monopartite (K(K/R)X(K/R)) or bipartite (KRX10–12KRRK) where the basic clusters are separated by 10–12 amino acids (2–4). The best studied NTRs that bind to these basic patch NLS motifs are a complex of importin α and importin β1 (3, 4). However, the karyopherin importin β2 binds to a different motif, called a PY-NLS as they almost always contain a proline-tyrosine (PY) dipeptide at the C-terminal end of the NLS (5, 6). PY-NLS motifs are often much larger than classical NLSs, up to 100 amino acids (7). The PY is preceded by a basic residue such that the C-terminal portion of the PY-NLS motif consists of the consensus sequence RX2–5PY. The N-terminal region of the PY-NLS falls into one of two categories, a basic patch (subclass bPY-NLS) or a hydrophobic patch (subclass hPY-NLS) (6).
Targeting to the nucleus can be dynamic; some proteins cycle in and out of the nucleus (8), whereas the targeting of other proteins to the nucleus is regulated by signal transduction pathways (9). The nucleus itself is a dynamic organelle, most markedly in eukaryotes that undergo an open mitosis that involves nuclear envelope breakdown. Interestingly, many proteins with “cytoplasmic” functions during mitosis are targeted to the nucleus during interphase prior to nuclear envelope breakdown, for example anillin (10), TPX2 (11), and MgcRacGAP (12). As these proteins are expressed during interphase, it has been proposed that their nuclear sequestration during interphase could spatially restrict function prior to nuclear envelope breakdown. For instance, the nuclear targeting of MgcRacGAP is required for the functional incorporation of CENP-A at the centromere (12), whereas the nuclear targeting of TPX2 may serve a dual purpose as follows: to allow TPX2 to respond to DNA damage (13, 14) and to prevent the accumulation of TPX2 in the cytosol where it could re-organize microtubules, a function of TPX2 when released into the cytosol during the early stages of apoptosis (15).
Anillin is an essential multidomain protein found at the plasma membrane during cytokinesis (16–19). Anillin is linked to the plasma membrane (17) and to different cytoskeletal elements, including actin (20), myosin (21), septins (22), and microtubules during cytokinesis (23). During interphase, anillin is targeted to the nucleus, suggesting that it may either have novel (and as yet unknown) nuclear functions and/or it is sequestered there prior to nuclear envelope breakdown to restrict its activity to mitosis. In this study, we determined how and why anillin is targeted to the nucleus during interphase.
Experimental Procedures
Plasmids
cDNA fragments of human anillin (17), importin β2 (IMAGE clone 30059046), CD2AP (IMAGE clone 7262392), and mDia2 (Addgene plasmid 25407) (24) were generated by PCR using the Pfx polymerase (Life Technologies, Inc.) and oligonucleotide primers (IDT) (data not shown). The cDNA fragments were purified, A-tailed using TaqDNA polymerase (Fermentas), and subcloned into the plasmid vectors pCR8/GW/TOPO (Life Technologies, Inc.). To generate recombinant protein fragments of anillin, the cDNA fragments subcloned in pCR8/GW/TOPO were recombined into the vector pDEST17 (Life Technologies, Inc.) to generate His6-tagged proteins, pDEST15 (Life Technologies, Inc.) to generate GST fusion proteins, or pKM596 (Addgene plasmid 8837) (25) to generate MBP fusion proteins. To generate GFP-anillin fusion proteins to be expressed in HeLa cells, the anillin cDNA fragments in pCR8/GW/TOPO were recombined into pcDNA6.2/N-EmGFP-DEST (Life Technologies, Inc.). To generate mutations within the anillin cDNA, complementary primers incorporating the mutation (data not shown) were used to make partial cDNA fragments that were purified and stitched together to make the desired region of anillin that contained the mutation. These cDNA fragments were then cloned as described above. All constructs and mutations were verified by sequencing. The Myc-tagged MBP M9M and Myc-tagged MBP containing mammalian expression vectors were gifts from Y. M. Chook (7).
To generate stable cells lines that expressed GFP-anillin(K68A/K69A/R70A) fusion proteins under the control of the tetracycline repressor, constructs were generated using the Flp-In system from Invitrogen as described previously (19). Briefly, a full-length anillin(K68A/K69A/R70A) fusion protein was generated, as described above containing a ligation-independent cloning sequence at the 5′ and 3′ end. Large single-stranded “sticky ends” were created by incubating the cDNA fragment with T4 DNA polymerase (Life Technologies, Inc.) in the presence of 2.5 mm dTTP. A pcDNA5/FRT/TO/eGFP-N vector previously modified to contain complementary LIC sequences (19) was linearized with FseI (New England Biolabs) and then large single-stranded sticky ends were generated by incubating with T4 DNA polymerase in the presence of 2.5 mm dATP. Plasmid and cDNA were annealed by heating to 95 °C then cooled slowly to room temperature prior to transforming into TOP10 cells.
Protein Purification
Recombinant proteins were purified from BL21 Escherichia coli transfected with plasmids containing His6 or GST fusion proteins or ER2523 E. coli (New England Biolabs) with plasmids containing MBP fusion proteins generated as described above. BL21 cells were grown in LB media at 37 °C to an optical density of 0.6 at A600. Recombinant fusion protein expression was induced by the addition of 1 mm isopropyl β-d-1-thiogalactopyranoside (IPTG) with further incubation at 16 °C overnight. Cells were harvested by centrifugation, and the cell pellet was stored at −80 °C until protein purification.
For the purification of MBP fusion protein, E. coli cells were resuspended in 25 mm HEPES, pH 7.5, 500 mm NaCl, 100 mm KCl, 0.5 mm β-mercaptoethanol, 1 mm PMSF and lysed by sonication. Cell lysates were cleared by centrifugation at 10,000 × g for 30 min at 4 °C and then applied to amylose resin. The resin was washed with 10 column volumes of column buffer (25 mm HEPES, pH 7.5, 500 mm NaCl, 100 mm KCl, 0.5 mm β-mercaptoethanol, 1 mm PMSF, 0.1% (v/v) Triton X-100), and the MBP fusion proteins were eluted in column buffer containing 10 mm maltose.
For purification of His6-tagged proteins, cell pellets were resuspended in 25 mm HEPES, pH 7.5, 500 mm NaCl, 5% (v/v) glycerol, 5 mm imidazole, 0.5 mm β-mercaptoethanol, 1 mm PMSF and lysed by sonication. Cell lysates were cleared by centrifugation at 10,000 × g for 30 min at 4 °C and then applied to nickel-Sepharose beads (Amersham Biosciences). The resin was then washed with 10 volumes of column buffer containing 20 mm imidazole, and the His6-tagged fusion proteins were then eluted in column buffer containing 500 mm imidazole.
Eluted proteins were dialyzed into 10 mm HEPES, pH 7.6, 100 mm KCl, 2 mm MgCl2, 50 mm sucrose at 4 °C overnight and then concentrated using Millipore Ultrafree spin columns with a 10-kDa cutoff. Proteins were then aliquoted, flash-frozen in N2(l), and stored at −80 °C.
Binding Assays
0.05 nmol of MBP-anillin fusion protein was diluted in 100 μl of MBP-binding buffer (50 mm HEPES, pH 7.5, 1 mm DTT, 1 mm EDTA, 5 mm MgCl2, 0.3% (v/v) Triton X-100, 1 mm β-mercaptoethanol) prebound to 20 μl of amylose resin at 4 °C for 1 h. The resin was washed three times in binding buffer and then incubated for 4 h at 4 °C with 100 μl of binding buffer containing 0.02 nmol of either His6 importin β1, His6 importin β2, or GST-RanGTP. The resin was then washed three times with binding buffer and resuspended in a final volume of 100 μl of binding buffer, boiled in SDS sample buffer, and analyzed by immunoblotting for the presence of co-purifying His6-tagged fusion proteins using an anti-His6 antibody (MP Biomedicals). Alternatively, 0.05 nmol of His6-tagged fusion proteins was diluted in 100 μl of His-binding buffer (25 mm HEPES, pH 7.5, 120 mm NaCl, 1 mm EGTA, 0.3% (v/v) Triton X-100, 1 mm β-mercaptoethanol) and bound to 10 μl of nickel-Sepharose. The binding reaction containing 0.02 mmol of MBP fusion protein was carried out as described above with co-purifying MBP fusion detected using an anti-MBP antibody (New England Biolabs).
Cell Culture and Transfection
HeLa cells were cultured in DMEM3 (Sigma) supplemented with 10% fetal bovine serum (Life Technologies, Inc.) in a 5% CO2 atmosphere at 37 °C. Plasmids expressing emGFP-anillin fusion proteins were transfected into HeLa cells grown on glass coverslips using Lipofectamine 2000 (Life Technologies, Inc.) following the manufacturer's directions. 48 h after transfection, cells were fixed in 3.7% paraformaldehyde in PBS for 15 min at room temperature and then permeabilized in 0.1% Triton X-100 in PBS for 15 min at room temperature.
Immunostaining and Microscopy and Quantitation
Fixed cells were then stained with 4′,6-diamidino-2-phenylindole (DAPI) to visualize DNA and a mouse anti-α-tubulin, clone DM1A (Sigma), followed by a secondary goat anti-mouse antibody conjugated to Alexa 594 (Life Technologies) to visualize the cytosolic microtubule network. Alternatively, to visualize the actin cytoskeleton, fixed cells were stained with DAPI and rhodamine-labeled phalloidin (Life Technologies). Coverslips were mounted on glass slides using Mowiol (polyvinyl alcohol 4–88, Fluka). Cells were visualized using a Nikon TE8000 microscope with a ×60/1.4 NA oil-immersion objective lens and 1.515 immersion oil (Nikon) at room temperature. Images were acquired using Metamorph software (Molecular Devices) driving an electron multiplying CCD camera (ImagEM, Hamamatsu). Z sections (0.2 μm apart) were acquired to produce a stack that was then imported into Autoquant X2 (Media Cybernetics) for deconvolution (10 iterations). Maximum projections and cross-sections were done using Metamorph. Images were overlaid in Photoshop (Adobe), involving adjustments in brightness and contrast of images.
To analyze changes in cell shape, the ratio of cell width to cell length was determined using the calibrate distances function in Metamorph (Molecular Devices) to measure the cell width and the length. The ratio was calculated from over 200 cells in each experiment.
To quantify the relative proportion of different GFP-anillin constructs in the nucleus compared with the cytosol, the ratio of fluorescence intensity between the nucleus and cytoplasm was determined. A region of common size was selected in the nucleus and the cytosol, and the fluorescence intensity within each region was determined using the “calibrate gray levels” function of the Metamorph. The ratio of fluorescence intensity between the nucleus and cytoplasm of a cell was then calculated and averages were over at least 100 cells per experiment.
Results
Nuclear Targeting Element in the Anillin N Terminus
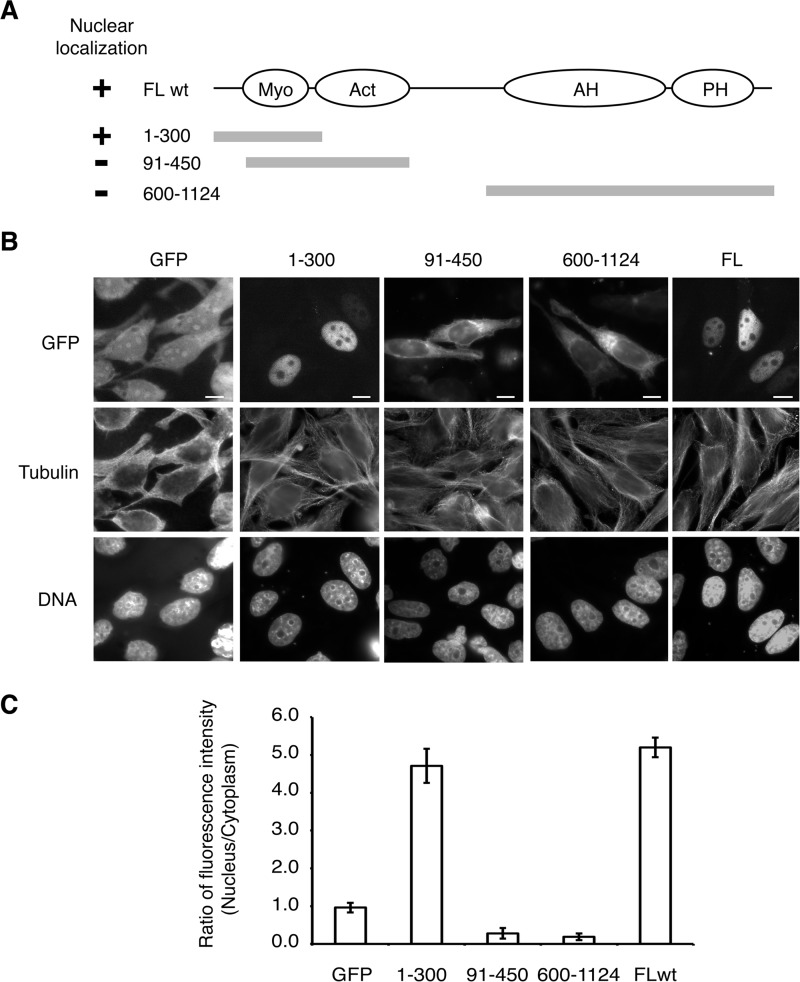
To determine how anillin is targeted to the nucleus in metazoan cells, we fused different fragments of anillin to GFP to identify which regions of anillin contained the NLS (Fig. 1A). An anillin fragment containing the C-terminal half of the protein, amino acids 600–1124, did not localize to the nucleus (Fig. 1, B and C). In contrast, an anillin fragment containing amino acids 1–300 localized to the nucleus in interphase cells, suggesting that the NLS lies in the N terminus of anillin, consistent with previous observations (10). Further deletion of the first 91 amino acids disrupted the nuclear localization (Fig. 1, B and C), mapping the NLS to within the first 91 amino acids of anillin.
FIGURE 1.
Nuclear localization sequence of anillin lies in the first 91 amino acids. A, schematic of the domain organization of anillin and the GFP-anillin fusion proteins expressed in HeLa cells shown in B. FL, full-length anillin sequence; wt, wild type sequence. B, micrographs of HeLa cells transiently expressing different GFP-anillin fusion proteins described in A. C, quantitation of the ratio of GFP fluorescence between the nuclear and cytoplasmic compartments for HeLa cells expressing different GFP-anillin fusion proteins described in A and B. Bar, 5 μm. Myo, myosin; Act, actin; AH, anillin homology domain; PH, pleckstrin homology domain.
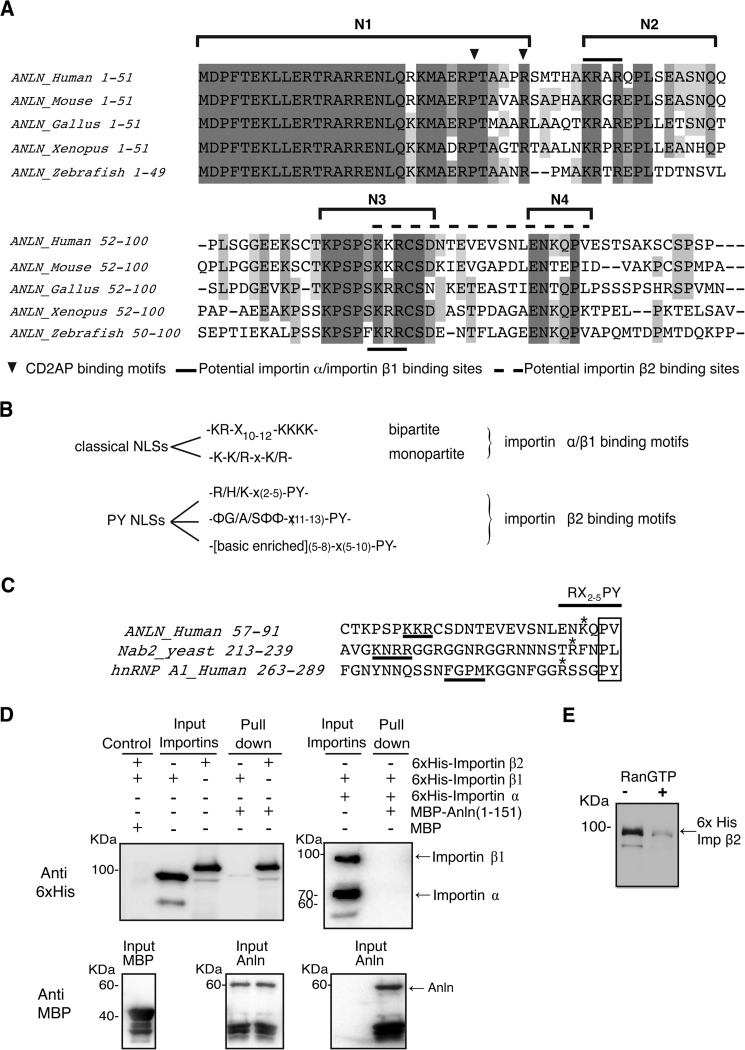
Sequence alignments of the first 91 amino acids of human anillin with other vertebrate species revealed four highly conserved regions that we termed N1, N2, N3, and N4 (Fig. 2A). CD2AP, an actin regulator, is known to bind to amino acids within N1 (26), but the function of the other regions is unknown. Putative NLS consensus motifs, K(K/R)X(K/R), exist in region N2 (KRAR, amino acids 38–41) and in region N3 (KKR, amino acids 68–70) (Fig. 2, A and B).
FIGURE 2.
N terminus of anillin has distinct conserved regions across different vertebrate species and binds to importin β2 in a Ran-dependent manner. A, alignment of the first 100 amino acids of anillin in different vertebrate species. B, consensus motifs for different classes of nuclear localization sequences. C, comparison of the anillin bPY motif to other bPY motifs. D, recombinant His6-tagged importin α and β1 or β2 were incubated with a MBP-anillin(1–151) fusion protein. The MBP-anillin fusion protein was re-isolated on amylose beads, and co-purifying His6-tagged proteins were detected by SDS-PAGE and Western blotting. E, His6 importin β2-MBP-anillin binding reaction described in A was repeated in the presence and absence of recombinant GST-RanGTP. Alignment of the anillin importin β2-binding motif with other known importin β2-binding motifs.
Anillin Binds to Importin β2
To determine the mechanism of anillin targeting to the nucleus, we analyzed the direct binding of recombinant anillin fragments to NTRs. Typically, basic patch NLSs are associated with binding to a complex of importin α and importin β1 (4). Surprisingly, an MBP-anillin(1–151) fragment did not bind to the importin α/importin β1 (Fig. 2C), suggesting that another NTR must be involved in targeting anillin to the nucleus. Further analysis of the anillin sequence revealed a conserved proline (Pro-87) in the N4 region that is preceded by basic residues (Fig. 2, A, B and E). Although the proline is not followed by a tyrosine as observed in a typical PY-NLS, the combination of conserved regions N3 and N4 is reminiscent of a basic patch PY motif known to bind to importin β2 and target factors to the nucleus (Fig. 2E) (5). To test this model, recombinant importin β2 was purified and assessed for its ability to bind to anillin. MBP-anillin bound to His6-tagged importin β2 (Fig. 2C), an interaction that was inhibited by the presence of RanGTP (Fig. 2D). These data suggest that the anillin-importin β2 interaction reflects a physiological NTR-cargo interaction where high concentrations of RanGTP in the nucleus prompt the release of cargo from NTRs and suggest that anillin is targeted to the nucleus in an importin β2-dependent manner.
Importin β2 Targets Anillin to the Nucleus
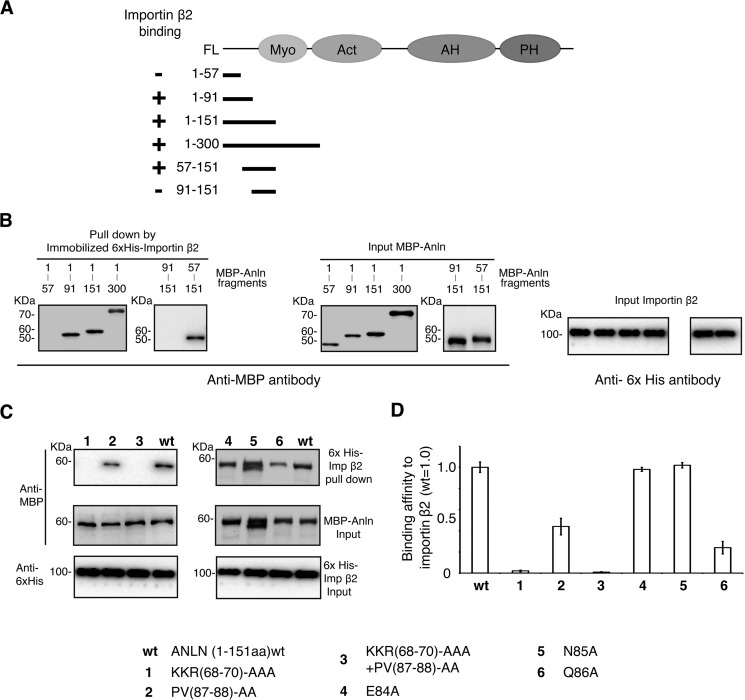
To better define the importin β2-binding site within anillin, we generated a series of recombinant anillin fragments fused to MBP and performed in vitro binding assays. Anillin fragments containing amino acid residues 57–91 bound to importin β2 (Fig. 3, A and B), consistent with regions N3 and N4 being involved in importin β2 binding. We next made a series of point mutations of amino acids within the putative importin β2-binding site that are conserved across species to assess the contributions of different amino acids in regions N3 and N4 (Fig. 2A). Mutation of the basic patch residues 68KKR70 to alanine reduced the binding to importin β2 by 93 ± 1.5% (Fig. 3, C and D). In contrast, mutation of 87PV88 each to alanine reduced anillin binding by 52 ± 7.5%. Combining both mutants (68AAA70 and 87AA88) reduced anillin binding to importin β2 by 97 ± 1% (Fig. 3, C and D). Further examination of the cross-species sequence alignments identified three residues directly upstream of Pro-87 that were conserved across vertebrates, Glu-83, Asn-84, and Gln-86 (Fig. 2A). Mutation of these residues to alanine revealed that Gln-86 reduced anillin's binding to importin β2 by 66 ± 5% (Fig. 3, C and D). These data demonstrate that both the basic patch KKR and the downstream QPV motif are involved in binding to importin β2.
FIGURE 3.
Mapping the importin β2-binding site in anillin. A, schematic outlining the MBP-anillin fusion proteins used in the importin β2 binding assay in relation to the domain organization of anillin. B, recombinant MBP-anillin fragments were incubated with His6 importin β2 and then re-isolated on His6 beads. Co-purifying MBP-tagged anillin fragments were detected by SDS-PAGE and Western blotting using an anti-MBP antibody. C, recombinant MBP-anillin(1–151) carrying different mutations was incubated with His6 importin β2 and then re-isolated on His6 beads. Co-purifying MBP-tagged anillin fragments were detected by SDS-PAGE and Western blotting using an anti-MBP antibody. D, quantitation of the amount of the different MBP-anillin mutants co-purifying with importin β2 normalized to the amount of importin β2 co-purifying to a wild type MBP-anillin fusion protein. Blots from three different experiments were compared and quantitated. Error bars indicate ± S.E.
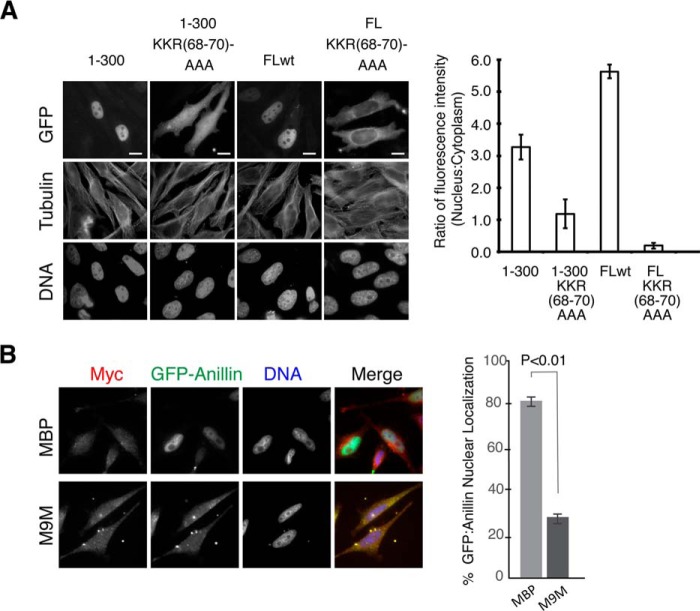
We next determined whether importin β2 binding was required for anillin targeting to the nucleus. Anillin fragments containing wild type or the 68AAA70 mutant sequence were fused to GFP and expressed in HeLa cells. GFP-anillin with wild type sequence localized to the nucleus. In contrast, full-length anillin or an N-terminal fragment of anillin(1–300) containing the 68AAA70 mutation failed to localize to the nucleus (Fig. 4). These data demonstrate that the KKR motif is necessary to target anillin to the nucleus during interphase.
FIGURE 4.
Basic patch of the atypical bPY NLS of anillin is required for targeting anillin to the nucleus. A, micrographs showing the localization of different GFP-anillin fusion proteins in HeLa cells. B, quantitation of the ratio of GFP fluorescence between the nuclear and cytoplasmic compartments for HeLa cells expressing different GFP-anillin fusion proteins depicted in A. Bar, 5 μm.
We next asked whether importin β2 is required for the nuclear import of anillin. We specifically blocked importin β2-mediated nuclear import by expressing a competitive peptide inhibitor of importin β2 binding. The M9M peptide incorporates two importin β2-binding sites, one from heterogeneous nuclear ribonucleoprotein-A1 and another from heterogeneous nuclear ribonucleoprotein-M that has a high affinity for importin β2, thereby allowing it to specifically out-compete cargo from importin β2 (27, 28). When expressed in cells as an MBP fusion protein, M9M-MBP significantly reduced the level of GFP-anillin localized to the nucleus compared with expressing MBP alone (Fig. 4B). Therefore, both in vitro and in vivo data indicate that importin β2 targets anillin to the nucleus.
Importin β2 Binds to a Unique Site on Anillin
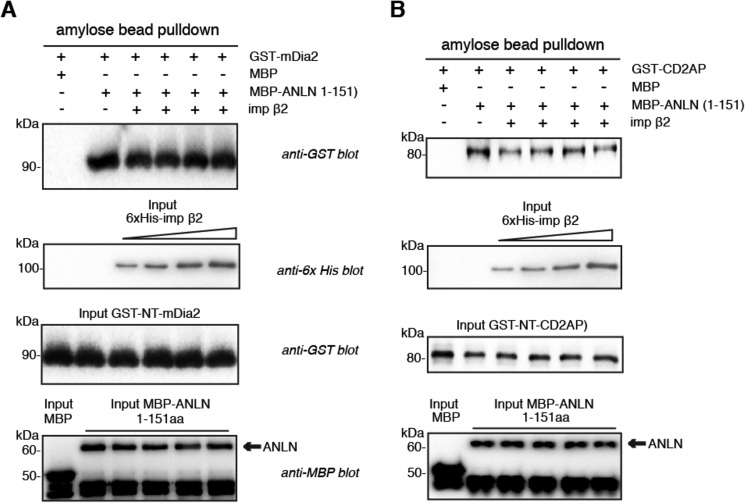
The binding of NTRs to mitotic proteins can regulate the function of a mitotic protein, for instance by displacing downstream targets (29–32), and can also serve to spatially constrain a protein's activity within the cell (29, 30, 33, 34). Because the N-terminal region of anillin mediates its interaction with CD2AP (26) and mDia2 (35), we tested whether importin β2 competed with either protein for binding to anillin in vitro. Increasing concentrations of importin β2 had no effect on the binding of CD2AP or mDia2 to anillin (Fig. 5, A and B), suggesting that importin β2, CD2AP, and mDia2 have distinct and nonoverlapping anillin-binding sites.
FIGURE 5.
Importin β2-binding site on anillin does not overlap with the CD2AP- and mDia2-binding sites. A, recombinant MBP-anillin(1–151) was incubated with GST-mDia2(89–533) in the presence of increasing concentrations of His6-importin β2. MBP-anillin was re-isolated on amylose beads, and co-purified GST-mDia2 was detected by SDS-PAGE and Western blotting. B, recombinant MBP-anillin(1–151) was incubated with GST-CD2AP(1–350) in the presence of increasing concentrations of His6-importin β2. MBP-anillin was re-isolated on amylose beads, and co-purified GST-CD2AP was detected by SDS-PAGE and Western blotting.
Importin β2 Binding Is Not Required for Anillin's Role in Cytokinesis
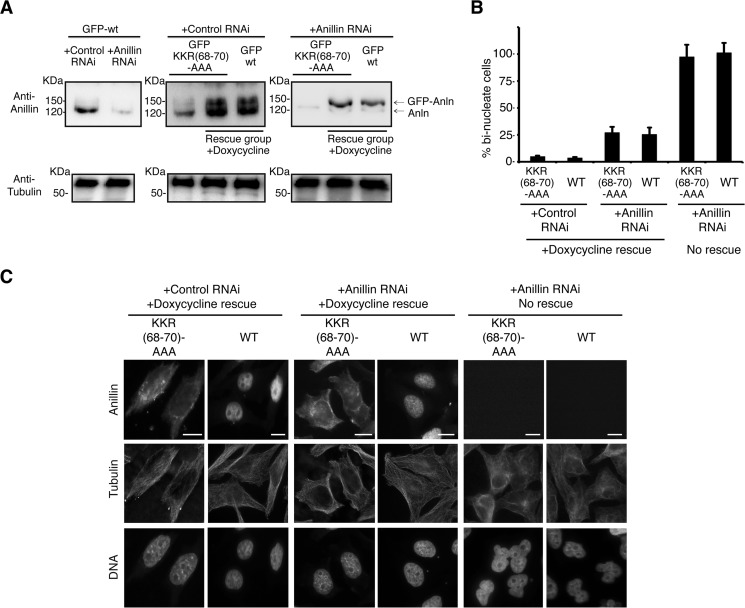
We next assessed whether importin β2 binding could regulate anillin's function in cytokinesis by monitoring cytokinesis in HeLa cells expressing anillin(K68A/K69A/R70A). A stable HeLa cell line expressing anillin(K68A/K69A/R70A) under the control of a tet repressor was generated, and culture conditions developed to induce the anillin(K68A/K69A/R70A) transgene expression at a similar level to endogenous anillin (Fig. 6A). Endogenous anillin expression was then suppressed by siRNA targeted to the 3′UTR of the endogenous anillin mRNA. In the absence of transgene expression, depletion of anillin resulted in the accumulation of bi-nucleate cells (81 ± 7.1% of the total cells, Fig. 6, B and C), a phenotype characteristic of cytokinetic failure and consistent with previous results (17, 19, 36). The bi-nucleate phenotype was rescued upon the expression of either GFP-anillin or the non-importin β2-binding mutant, GFP-anillin(K68A/K69A/R70A) (Fig. 6, B and C), indicating that importin β2 binding does not regulate anillin's cytokinetic function.
FIGURE 6.
Importin β2 binding to anillin does not regulate anillin function during cytokinesis. A, GFP-anillin and GFP-mutant anillin stably expressing HeLa cells lines where GFP-anillin expression was under the control of the tetracycline repressor. Expression conditions were developed to induce GFP-anillin at near endogenous anillin levels. A siRNA strategy was developed to prevent the expression of endogenous anillin, although GFP-anillin expression continued. B, quantitation of the number of bi-nucleate cells observed upon expression of different GFP-anillins in the presence and absence of siRNA to deplete endogenous anillin protein levels. Error bars are ± S.E. C, micrographs showing nuclear morphology in HeLa cells expressing different GFP-anillins in the presence and absence of endogenous anillin. Bar, 5 μm.
Cytosolic Accumulation of Anillin Disrupts Cell Shape
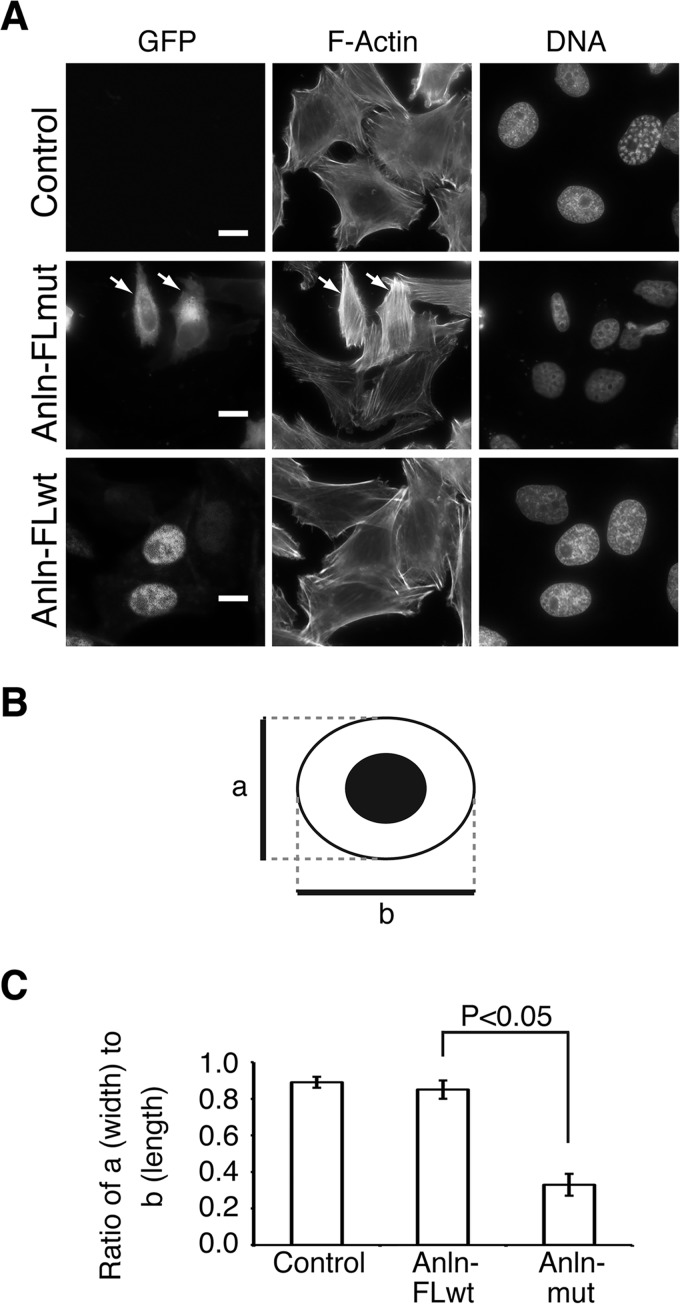
Successful cell division requires that cytokinesis takes place at the end of mitosis, after the completion of chromosome segregation. Given anillin's crucial role in linking the cell membrane with the actin and septin cytoskeletons at the cytokinetic furrow, we postulated that the presence of anillin in the cytoplasm during interphase would disrupt the cellular cytoskeleton, thus necessitating that anillin be kept inactive by sequestration in the nucleus during interphase. To test this model, we transiently overexpressed GFP-anillin or GFP-anillin(K68A/K69A/R70A) in HeLa cells. When overexpressed, GFP-anillin still localized to the nucleus, and the cytosolic actin organization and cell shape were similar to cells not expressing GFP-anillin (Fig. 7A). In contrast, in 93 ± 3.5% of cells overexpressing GFP-anillin(K68A/K69A/R70A), a dramatic alteration in cell shape was observed, with cells being notably longer and narrower (Fig. 7C). This change in cell shape correlated with an increase in the intensity of phalloidin staining, indicating an increase in actin filament assembly in the cytosol (Fig. 7A). Furthermore, these data suggest that the anillin NLS prevents disruption of the actin cytoskeleton during interphase by targeting anillin to the nucleus.
FIGURE 7.
Elevated cytoplasmic levels of anillin cause change in cell shape. A, microgrpahs of HeLa cells transiently expressing different GFP-anillins and staining with phalloidin to show actin organization and cell shape. White arrows indicate transfected cells. B, schematic outlining the different cell parameters measured to define changes in cell shape. a, cell width; b, cell length. C, quantitation of the ratio of cell width to cell length, outlined in B, in HeLa cells expressing different GFP-anillins. Error bars indicate ± S.E.
Discussion
The specific targeting of proteins to different subcellular locations is essential for localized biochemical activities within a cell. Here, we describe how anillin, an essential cytokinetic protein, is sequestered to the nucleus during interphase through a novel importin β2-binding motif. We determined that anillin binds to importin β2 rather than a complex of importin α and importin β1. Importin β2-binding motifs show a greater diversity than classical basic path NLS motifs that bind to a complex of importin α and importin β1. Importin β2 motifs fall broadly into two classes as follows: 1) a hydrophobic patch followed downstream by a PY motif (hPY) or 2) a basic patch followed downstream by a PY (bPY). Our data extend this concept by demonstrating that the PY motif is not absolutely conserved in metazoans. For instance, in many vertebrate species, the tyrosine in the PY motif of anillin is replaced by an aliphatic amino acid, whereas in Xenopus laevis, a lysine is present. Crystallographic studies of the Saccharomyces cerevisiae importin β2-Nab2 complex found that 238PL239, the equivalent of the PY motif, interacted primarily through hydrophobic interactions with side chain residues of importin β2 (Fig. 2E) (37). We would thus predict anillin forms similar interactions as Nab2 with importin β2.
Anillin is a crucial regulator of cytokinesis, modulating the organization and function of the actomyosin and septin cytoskeletons at the cytokinetic furrow during mitosis. Like many mitotic proteins, anillin is targeted to the nucleus in interphase. Although our study found no evidence of a nuclear function for anillin, we cannot rigorously rule out that anillin may have as yet unknown interphase functions. We do, however, find that deleting the anillin NLS leads to re-organization of the actin cytoskeleton and subsequently changes in cell shape. Our data suggest that the sequestration of anillin into the nucleus during interphase prevents anillin from prematurely re-organizing the plasma membrane and actomyosin cytoskeleton, and thus anillin availability serves to restrict fundamental aspects of cytokinesis to mitosis, specifically post-nuclear envelope breakdown.
There is precedent for the use of nuclear localization as a mechanism to regulate proteins with mitotic functions. TPX2, a microtubule-associated spindle assembly factor, is targeted to nuclei prior to mitosis, facilitating a nuclear function (13, 14) and also preventing a deleterious cytosolic function (15, 29). By using nuclear targeting to control anillin's localization in the cell, the availability of anillin can be coordinated with cell cycle progression to constrain its plasma membrane remodeling activity until post-nuclear envelope breakdown, yet without the lag time of waiting for protein levels to increase as would be required for mitotic expression. Such a mechanism could be applied to a broad spectrum of mitotic proteins that remodel cellular structures in ways that would hinder cellular function in interphase. It is noteworthy that although anillin is released into the cytoplasm in late prophase, its cytokinetic function occurs post-chromosome segregation. Mitotic proteins that localize to the nucleus during interphase by NTRs are further regulated by NTRs in mitosis (33, 34). In these instances, the binding of importin α/β1 inhibits the function of TPX2, NuMA, Kid, and XCTK2 in mitotic spindle assembly, an inhibition reversed in the presence of RanGTP (29–31). As RanGTP generated on chromosomes spatially regulates importin α/β1 binding around the spindle, these mitotic proteins are most active in regions of the cell where RanGTP is at it highest concentration. We tested whether a similar mechanism could regulate anillin function; however, cells expressing an anillin mutant refractory to importin β2 binding exhibited no cytokinesis defects, indicating that anillin activity is primarily determined by its intracellular localization and concentration at the plasma membrane. Additional regulatory pathways are likely to impinge on orchestrating the correct timing of anillin function, as anillin is released into the cytoplasm following nuclear envelope breakdown, but it is required through telophase, once the daughter nuclei have reformed (19). Indeed, anillin released into the cytoplasm in late prophase is not initially targeted to the future site of furrow ingression but rather interacts diffusely with the cell membrane, suggesting that the onset of membrane remodeling requires additional factors to either activate anillin and/or mark the site where anillin will accumulate and function, such as RhoA (36) and phosphatidylinositol 4,5-bisphosphate (17).
It is also possible that the anillin-importin β2 interaction is subject to further cell cycle regulation. Anillin has essential cytokinetic functions during telophase (19). After nuclear envelope reformation, anillin remains enriched at the intercellular bridge. Whether this is achieved through post-translational modification, protein-protein interactions, and/or the blocking of importin, β2 binding remains to be determined. Further suggestion that anillin's importin β2 interaction may be actively regulated comes from observations that Drosophila anillin is not always in the nucleus during interphase. In syncytial nuclear cycles of Drosophila embryos, anillin does not enter the nucleus, rather it is either cytosolic or targeted to the pseudo-cleavage furrow (32, 38). However, anillin enters the nucleus promptly after cellularization, on a time scale of minutes. Similarly, anillin is targeted to cell junctions of differentiated epithelial cells of X. laevis (39); however, whether this stems from modifications to anillin or importin β2 is uncertain. Combined, these observations demonstrate that a critical aspect of coordinating anillin function with cell cycle progression is the regulation of anillin subcellular localization, which is at least in part achieved through targeting anillin to the nucleus by binding to importin β2.
Acknowledgments
We thank Dr. Pawson (Tanenbaum Lunenfeld Research Institute, Toronto, Canada) for cDNA clones.
Footnotes
- NLS
- nuclear localization signal
- NTR
- nuclear transport receptor
- MBP
- maltose-binding protein
- b
- basic
- h
- hydrophobic.
References
- 1. Görlich D., Kutay U. (1999) Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15, 607–660 [DOI] [PubMed] [Google Scholar]
- 2. Xu D., Farmer A., Chook Y. M. (2010) Recognition of nuclear targeting signals by karyopherin-β proteins. Curr. Opin. Struct. Biol. 20, 782–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Marfori M., Mynott A., Ellis J. J., Mehdi A. M., Saunders N. F., Curmi P. M., Forwood J. K., Bodén M., Kobe B. (2011) Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim. Biophys. Acta 1813, 1562–1577 [DOI] [PubMed] [Google Scholar]
- 4. Chook Y. M., Süel K. E. (2011) Nuclear import by karyopherin-βs: recognition and inhibition. Biochim. Biophys. Acta 1813, 1593–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee B. J., Cansizoglu A. E., Süel K. E., Louis T. H., Zhang Z., Chook Y. M. (2006) Rules for nuclear localization sequence recognition by karyopherin β2. Cell 126, 543–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Süel K. E., Gu H., Chook Y. M. (2008) Modular organization and combinatorial energetics of proline-tyrosine nuclear localization signals. PLoS Biol. 6, e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang Z. C., Satterly N., Fontoura B. M., Chook Y. M. (2011) Evolutionary development of redundant nuclear localization signals in the mRNA export factor NXF1. Mol. Biol. Cell 22, 4657–4668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gama-Carvalho M., Carmo-Fonseca M. (2001) The rules and roles of nucleocytoplasmic shuttling proteins. FEBS Lett. 498, 157–163 [DOI] [PubMed] [Google Scholar]
- 9. O'Neill E. M., Kaffman A., Jolly E. R., O'Shea E. K. (1996) Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science 271, 209–212 [DOI] [PubMed] [Google Scholar]
- 10. Oegema K., Savoian M. S., Mitchison T. J., Field C. M. (2000) Functional analysis of a human homologue of the Drosophila actin binding protein anillin suggests a role in cytokinesis. J. Cell Biol. 150, 539–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wittmann T., Wilm M., Karsenti E., Vernos I. (2000) TPX2, A novel xenopus MAP involved in spindle pole organization. J. Cell Biol. 149, 1405–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lagana A., Dorn J. F., De Rop V., Ladouceur A. M., Maddox A. S., Maddox P. S. (2010) A small GTPase molecular switch regulates epigenetic centromere maintenance by stabilizing newly incorporated CENP-A. Nat. Cell Biol. 12, 1186–1193 [DOI] [PubMed] [Google Scholar]
- 13. Neumayer G., Belzil C., Gruss O. J., Nguyen M. D. (2014) TPX2: of spindle assembly, DNA damage response, and cancer. Cell. Mol. Life Sci. 71, 3027–3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Neumayer G., Nguyen M. D. (2014) TPX2 impacts acetylation of histone H4 at lysine 16: implications for DNA damage response. PLoS One 9, e110994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moss D. K., Wilde A., Lane J. D. (2009) Dynamic release of nuclear RanGTP triggers TPX2-dependent microtubule assembly during the apoptotic execution phase. J. Cell Sci. 122, 644–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Piekny A., Maddox A. (2010) The myriad roles of anillin during cytokinesis. Semin. Cell Dev. Biol. 9, 881–891 [DOI] [PubMed] [Google Scholar]
- 17. Liu J., Fairn G. D., Ceccarelli D. F., Sicheri F., Wilde A. (2012) Cleavage furrow organization requires PIP2-mediated recruitment of anillin. Curr. Biol. 22, 64–69 [DOI] [PubMed] [Google Scholar]
- 18. Kechad A., Jananji S., Ruella Y., Hickson G. R. (2012) Anillin acts as a bifunctional linker coordinating midbody ring biogenesis during cytokinesis. Curr. Biol. 22, 197–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Renshaw M. J., Liu J., Lavoie B. D., Wilde A. (2014) Anillin-dependent organization of septin filaments promotes intercellular bridge elongation and Chmp4B targeting to the abscission site. Open Biol. 4, 130190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Field C. M., Alberts B. (1995) Anillin, a contractile ring protein that cycles from the nucleus to the cell cortex. J. Cell Biol. 131, 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Straight A. F., Field C. M., Mitchison T. J. (2005) Anillin binds nonmuscle myosin II and regulates the contractile ring. Mol. Biol. Cell 16, 193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kinoshita M., Field C. M., Coughlin M. L., Straight A. F., Mitchison T. J. (2002) Self- and actin-templated assembly of mammalian septins. Dev. Cell 3, 791–802 [DOI] [PubMed] [Google Scholar]
- 23. van Oostende Triplet C., Jaramillo Garcia M., Haji Bik H., Beaudet D., Piekny A. (2014) Anillin interacts with microtubules and is part of the astral pathway that defines cortical domains. J. Cell Sci. 127, 3699–3710 [DOI] [PubMed] [Google Scholar]
- 24. Tominaga T., Sahai E., Chardin P., McCormick F., Courtneidge S. A., Alberts A. S. (2000) Diaphanous-related formins bridge Rho GTPase and Src tyrosine kinase signaling. Mol. Cell 5, 13–25 [DOI] [PubMed] [Google Scholar]
- 25. Fox J. D., Routzahn K. M., Bucher M. H., Waugh D. S. (2003) Maltodextrin-binding proteins from diverse bacteria and archaea are potent solubility enhancers. FEBS Lett. 537, 53–57 [DOI] [PubMed] [Google Scholar]
- 26. Monzo P., Gauthier N. C., Keslair F., Loubat A., Field C. M., Le Marchand-Brustel Y., Cormont M. (2005) Clues to CD2-associated protein involvement in cytokinesis. Mol. Biol. Cell 16, 2891–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cansizoglu A. E., Lee B. J., Zhang Z. C., Fontoura B. M., Chook Y. M. (2007) Structure-based design of a pathway-specific nuclear import inhibitor. Nat. Struct. Mol. Biol. 14, 452–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Desmond C. R., Atwal R. S., Xia J., Truant R. (2012) Identification of a karyopherin β1/β2 proline-tyrosine nuclear localization signal in huntingtin protein. J. Biol. Chem. 287, 39626–39633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Trieselmann N., Armstrong S., Rauw J., Wilde A. (2003) Ran modulates spindle assembly by regulating a subset of TPX2 and Kid activities including Aurora A activation. J. Cell Sci. 116, 4791–4798 [DOI] [PubMed] [Google Scholar]
- 30. Tsai M.-Y., Wiese C., Cao K., Martin O., Donovan P., Ruderman J., Prigent C., Zheng Y. (2003) A Ran signalling pathway mediated by the mitotic kinase Aurora A in spindle assembly. Nat. Cell Biol. 5, 242–248 [DOI] [PubMed] [Google Scholar]
- 31. Ems-McClung S. C., Zheng Y., Walczak C. E. (2004) Importin α/β and Ran-GTP regulate XCTK2 microtubule binding through a bipartite nuclear localization signal. Mol. Biol. Cell 15, 46–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Silverman-Gavrila R. V., Hales K. G., Wilde A. (2008) Anillin-mediated targeting of peanut to pseudocleavage furrows is regulated by the GTPase Ran. Mol. Biol. Cell 19, 3735–3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wiese C., Wilde A., Moore M. S., Adam S. A., Merdes A., Zheng Y. (2001) Role of Importin-β in coupling Ran to downstream targets in microtubule assembly. Science 291, 635–656 [DOI] [PubMed] [Google Scholar]
- 34. Nachury M. V., Maresca T. J., Salmon W. C., Waterman-Storer C. M., Heald R., Weis K. (2001) Importin b is a mitotic target of the small GTPase Ran in spindle assembly. Cell 104, 95–106 [DOI] [PubMed] [Google Scholar]
- 35. Watanabe S., Okawa K., Miki T., Sakamoto S., Morinaga T., Segawa K., Arakawa T., Kinoshita M., Ishizaki T., Narumiya S. (2010) Rho and anillin-dependent control of mDia2 localization and function in cytokinesis. Mol. Biol. Cell 21, 3193–3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Piekny A. J., Glotzer M. (2008) Anillin is a scaffold protein that links RhoA, actin, and myosin during cytokinesis. Curr. Biol. 18, 30–36 [DOI] [PubMed] [Google Scholar]
- 37. Soniat M., Sampathkumar P., Collett G., Gizzi A. S., Banu R. N., Bhosle R. C., Chamala S., Chowdhury S., Fiser A., Glenn A. S., Hammonds J., Hillerich B., Khafizov K., Love J. D., Matikainen B., et al. (2013) Crystal structure of human Karyopherin β2 bound to the PY-NLS of Saccharomyces cerevisiae Nab2. J. Struct. Funct. Genomics 14, 31–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Field C. M., Coughlin M., Doberstein S., Marty T., Sullivan W. (2005) Characterization of anillin mutants reveals essential roles in septin localization and plasma membrane integrity. Development 132, 2849–2860 [DOI] [PubMed] [Google Scholar]
- 39. Carazo-Salas R. E., Guarguaglini G., Gruss O. J., Segref A., Karsenti E., Mattaj I. W. (1999) Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature 400, 178–181 [DOI] [PubMed] [Google Scholar]