Background: Severe inflammatory reactions delay wound healing of bone.
Results: Tumor necrosis factor α (TNFα) inhibition of osteoblast differentiation is associated with increased cAMP response element-binding protein H (CREBH) and Smurf1 expression.
Conclusion: CREBH mediates the inhibitory actions of TNFα in bone regeneration.
Significance: CREBH is identified as a new mediator of inflammation-dependent bone degradation and a potential therapeutic target.
Keywords: cAMP response element-binding protein (CREB), endoplasmic reticulum stress (ER stress), osteoblast, SMAD transcription factor, tumor necrosis factor (TNF)
Abstract
Endoplasmic reticulum (ER) stress transducers, such as old astrocyte specifically induced substance (OASIS) and activating transcription factor 6 (ATF6), which are induced by bone morphogenetic protein 2 (BMP2), regulate bone formation and osteoblast differentiation. Here, we examined the role of cAMP response element-binding protein H (CREBH), a member of the same family of ER membrane-bound basic leucine zipper (bZIP) transcription factors as OASIS and ATF6, in osteoblast differentiation and bone formation. Proinflammatory cytokine TNFα increased CREBH expression by up-regulating the nuclear factor-κB (NF-κB) signaling pathway in osteoblasts, increased the level of N-terminal fragment of CREBH in the nucleus, and inhibited BMP2 induction of osteoblast specific gene expression. Overexpression of CREBH suppressed BMP2-induced up-regulation of the osteogenic markers runt-related transcription factor 2 (Runx2), alkaline phosphatase (ALP), and osteocalcin (OC) in MC3T3-E1 cells and primary osteoblasts, as well as BMP2-induced ALP activity and OC protein production. In contrast, knockdown of CREBH attenuated the inhibitory effect of TNFα on BMP2-induced osteoblast differentiation. Mechanistic studies revealed that CREBH increased the expression of Smad ubiquitination regulatory factor 1 (Smurf1), leading to ubiquitin-dependent degradation of Smad1, whereas knockdown of CREBH inhibited TNFα-mediated degradation of Smad1 by Smurf1. Consistent with these in vitro findings, administration of Ad-CREBH inhibited BMP2-induced ectopic and orthotopic bone formation in vivo. Taken together, these results suggest that CREBH is a novel negative regulator of osteoblast differentiation and bone formation.
Introduction
BMP25 is an important regulator of osteoblast differentiation, bone development, and the repair of bone defects (1, 2). Recently, it was reported that the BMP2 signaling pathway activates unfolded protein response molecules during osteogenesis. ER stress is caused by the accumulation of excessive amounts of unfolded proteins in the ER (3, 4). The physiological significance of ER stress has been definitively associated with the activation of three major unfolded protein response transducers: protein kinase RNA-like endoplasmic reticulum kinase (PERK), inositol-requiring kinase 1 (IRE-1), and ATF6 (5–8). BMP2 stimulates osteoblast differentiation and bone formation by activating several unfolded protein response transducers, including OASIS and ATF6, which induce osteoblast-specific gene expression (9, 10). However, the ability of BMPs to induce bone formation in clinical settings is limited, and the repair of bone defects by BMPs is negatively regulated by other factors (11).
OASIS and ATF6 are structurally similar to CREBH; these three proteins belong to the ER membrane-bound bZIP transcription factor family and are collectively referred to as OASIS family members (12). However, the mechanisms by which CREBH and ATF6 sense unfolded proteins and translocate from the ER to the Golgi differ from those of other OASIS family members (13). In addition, OASIS family members have distinct expression patterns, suggesting that these transcription factors may be associated with cell- or tissue-specific physiological responses. CREBH is induced and activated by the proinflammatory cytokines TNFα, IL-6, and LPS (14). Hence, it integrates the proinflammatory response and regulation of ER stress, thereby underlining its importance to inflammatory responses. Although these transcriptional factors have a structural similarity, they can produce an opposite effect on biological systems. In glucose metabolism, CREBH promotes gluconeogenic activity in a CRTC2-independent manner (15), and ATF6 inhibits hepatic glucose output by competing with CREB for interaction with CRTC2 (16).
Inflammatory cytokines inhibit BMP-induced osteogenesis and bone formation (17, 18); in fact, TNFα is a major inflammatory mediator responsible for bone loss in a number of bone-related inflammatory diseases (19, 20). TNFα inhibits BMP signaling by interfering with the DNA-binding ability of Smads via activation of the nuclear factor-κB (NF-κB) pathway, regulating Runx2 expression, and inhibiting BMP-induced osteoblast differentiation (18, 21, 22). In addition, TNFα induces the expression of Smad7 and Msx2, which also inhibit BMP signaling and related osteogenesis (23, 24). Activation of ERK by TNFα results in inhibition of the transcription factor osterix, and TNFα-mediated induction of Smad ubiquitination regulatory factor 1 (Smurf1) and Smurf2 accelerates the degradation of Runx2 protein through the proteasomal degradation pathway (25). Despite these findings, the molecular mechanisms underlying inflammatory actions in osteoblast differentiation are not fully understood.
Here, we examined the effect of CREBH on osteoblast differentiation in vitro and ectopic and orthotopic bone formation in vivo. The results indicate that CREBH functions as a modulator of TNFα-mediated inhibition of osteogenesis, and that this action is mediated mainly by Smurf1-induced degradation of Smad1. Overall, we describe a novel signaling pathway that encourages further analyses of the relationship between ER stress and bone formation.
Experimental Procedures
Reagents and Antibodies
Recombinant human BMP2 was obtained from Cowellmedi Co. (Busan, Korea) and TNFα was obtained from R&D Systems (Minneapolis, MN). BAY11-7082, an inhibitor of κB kinase (IKK), was purchased from Biomol (Plymouth, PA). Antibodies against Runx2 and β-actin were from Santa Cruz Biotechnology (Santa Cruz, CA). The anti-CREBH antibody was described previously (12).
Plasmids and Adenoviruses
The luciferase reporter driven by the osteocalcin promoter (OG2-Luc) was a kind gift from Dr. R. T. Franceschi (University of Michigan). The Smurf1-Luc reporter plasmid and the Smurf1, -2, and dnIκBα (S32A/S36A) expression constructs were a kind gift from Dr. J. H. Baek (Seoul National University, Korea). The CREBH-Luc reporter plasmid, CREBH expression construct (pcDNA-CREBH), and adenoviruses containing the nuclear form of CREBH (Ad-CREBH-N), green fluorescence protein (GFP) transcribed from the CMV promoter (Ad-GFP), the CREBH-specific short hairpin (sh) RNA (Ad-CREBHi), or an unspecific shRNA (Ad-USi), have been described previously (12, 26).
Cell Culture, Transient Transfection, and Viral Infection
The pre-osteoblast MC3T3-E1 cells and primary osteoblasts have been described previously (27). The cells were cultured in α-minimal essential medium (Gibco-BRL, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS; GIBCO-BRL) and antibiotics, and incubated at 37 °C in a humidified atmosphere containing 5% CO2. Transient transfections were performed as described previously (28). For viral infection, the cells were treated with the indicated viruses at the designated multiplicity of infection (m.o.i.) under serum-free conditions. After 4 h, the medium was replaced with an equivalent volume of medium containing 10% FBS, and the cells were incubated for an additional 24–48 h.
qRT-PCR Analysis
Total RNA was isolated from the cultured cells using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. RT-PCR was performed using 1 μg of total RNA. For quantification of gene transcription, cDNA was generated with the Maxime RT PreMix Kit (iNtRon, Sungnam, Korea), and then amplified on the StepOnePlus Real-time PCR System (ABI, Abilene, TX) using the QuantiTect SYBR PCR Kit (Qiagen, Valencia, CA) and specific primers. Cycling conditions were 95 °C for 10 min, followed by 40 cycles of 95 °C for 30 s, 58 °C for 1 min, and 72 °C for 30 s. Post-run samples were analyzed using ABI software and the relative expression was quantified using the 2−ΔΔCt method with endogenous β-actin levels. The primer sequences have been described previously (29).
Western Blotting
Total cells or nuclear fractions were harvested in lysis buffer (Cell Signaling Technology, Cambridge, MA) and centrifuged at 12,000 × g for 15 min at 4 °C. The nuclear and cytoplasmic fractions were prepared using the NE-PER Nuclear and Cytoplasmic Extraction Kit (Pierce Biotechnology), according to the manufacturer's instructions. Quantification of total protein was performed using the BCA Protein Assay Kit (Bio-Rad). Proteins were resolved by 10% SDS-PAGE and transferred to a PVDF membrane. After blocking using Tris-buffered saline (TBS) containing 0.1% Tween 20 and 5% milk, the membrane was incubated with specific primary antibodies. Signals were detected using an enhanced chemiluminescence reagent (Santa Cruz Biotechnology) according to the manufacturer's instructions. Densitometric analysis of the membrane was performed using a LAS-4000 lumino-image analyzer (Fujifilm, Tokyo, Japan).
ChIP Assay
MC3T3-E1 cells were treated with TNFα for the designated times and ChIP assays were performed as described previously (30). The DNA samples were quantified by qPCR using two pairs of primers. The primer sequences for the p50 and p65 binding regions of the CREBH promoter were 5′-CCAACTCTCAAGAATCAGTCAGC-3′ (forward) and 5′-GCTTTGCATCTGTGACAGGATG-3′ (reverse). The control primer sequences were 5′-GTTCTTGCATAGACCAGGCCA-3′ (forward) and 5′-TGGCCTGGTCTATGCAAGAAC-3′ (reverse). For quantitative comparison with qPCR, the ΔCT method was applied. A ΔCT value was calculated by subtracting the CT value of the input from that of the immunoprecipitated sample. A ΔΔCT value was then obtained by subtracting the ΔCT value of the sample immunoprecipitated with p65 or p50 antiserum from that of the corresponding control sample with normal rabbit IgG. Fold-differences were determined by raising 2 to the ΔΔCT power.
Alkaline Phosphatase Staining and Osteocalcin Production Assay
For detection of alkaline phosphatase (ALP), the cultured cells were fixed with 70% ethanol, rinsed three times with deionized water, and treated with BCIP®/nitro blue tetrazolium solution (Sigma) for 15 min. The stained cultures were then documented on an Epson Perfection V700 photo scanner (Seiko Epson, Nagano, Japan). For quantitative comparison, color intensities were measured from scanned images using Image J software and normalized to the value of the untreated control group. The level of osteocalcin (OC) secreted into the culture medium was determined using a mouse osteocalcin ELISA kit (Biomedical Technologies, Stoughton, MA), according to the manufacturer's instructions.
Animals and Surgical Procedure
The study was performed in accordance with the guidelines of the Chonnam National University Animal Care and Use Committee. C57BL/6 mice were purchased from Daehan Biolink (Eumsung, Korea) and 6-week-old male mice were randomly assigned to each experimental group. The animals were anesthetized by an intraperitoneal injection of a mixture of Zoletil (30 mg/kg; Virbac, Carros Cedex, France) and Rompun (10 mg/kg; Bayer Korea, Seoul, Korea). For ectopic bone formation, a sagittal incision (0.8–1.0 cm) was made on the back of each mouse and the subcutaneous pocket was formed by blunt dissecting. Absorbable collagen sponges (Colladerm, Bioland, Ochang, Korea) containing Ad-BMP2 and Ad-CREBH were implanted into the pocket. For an orthotopic model, a sagittal incision was made on the scalp, and the calvarium was exposed by blunt dissection. A critical-sized defect was created by means of a 5-mm diameter trephine bur (Fine Science Tools, Foster City, CA) under low speed drilling and copious phosphate-buffered saline irrigation. Ad-BMP2 and Ad-CREBH were administered into the defect with absorbable collagen sponges. The total amount of viruses was adjusted by adding Ad-GFP control virus. Three weeks after the implantation, bone formation was evaluated using a three-dimensional micro-computed tomography (μCT) system (model 1172, Skyscan, Aartselaar, Belgium). For the μCT analysis, the scanned images were collected at 50 kV and 200 μA and were reconstructed using the NRecon and CT analyzer software (Skyscan). For histology study, the implant or calvarial specimens were harvested, fixed in 10% neutral-buffered formalin, decalcified in Calci-Clear Rapid (National Diagnostics, Atlanta, GA), embedded in paraffin, and then cut into sections of 4 μm thickness. The sections were stained with hematoxylin/eosin, and evaluated for general tissue response and bone formation.
Statistical Analyses
All experiments were repeated at least three times and statistical analyses were performed using a Student's t test or analysis of variance followed by Duncan's multiple comparison test. p < 0.05 was considered significant. The results are expressed as the mean ± S.E. of triplicate independent experiments.
Results
TNFα Increases the Expression and Activation of CREBH in Osteoblasts
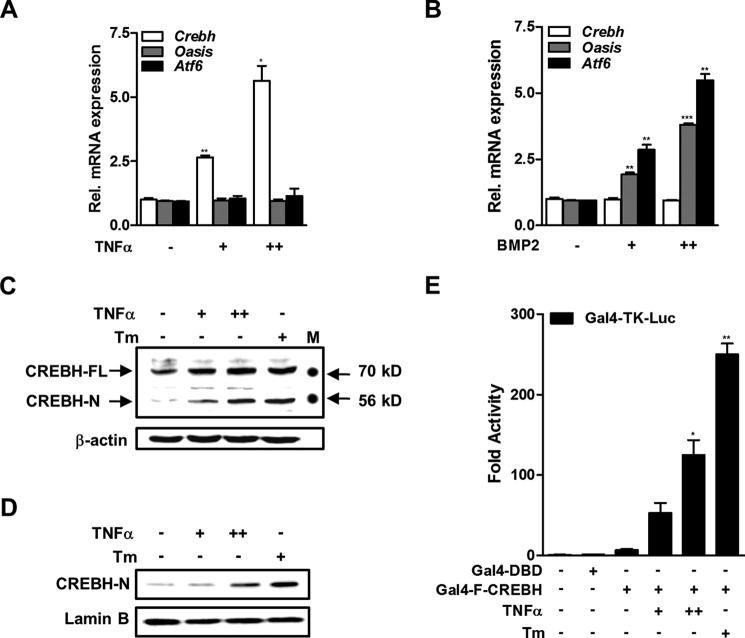
The proinflammatory cytokines TNFα and IL-6 increase Crebh mRNA expression in hepatocytes (31). Therefore, we examined whether CREBH expression is also regulated by TNFα in osteoblasts. Treatment of MC3T3-E1 cells with TNFα increased Crebh mRNA expression significantly, but did not affect the expression of the Atf6 and Oasis mRNAs (Fig. 1A). In contrast, the bone-forming cytokine BMP2 induced Atf6 and Oasis mRNA, as described in other studies (9, 32), but did not alter Crebh expression (Fig. 1B). Western blot analyses using total or nuclear protein extracts showed that TNFα also increased the CREBH protein level in a dose-dependent manner. Notably, both TNFα and tunicamycin, a strong ER stress inducer, increased the amount of the cleaved (nuclear) form of CREBH protein (Fig. 1, C and D).
FIGURE 1.
TNFα induces CREBH expression and cleavage of the N-terminal fragment in osteoblasts. A and B, real-time qRT-PCR measurements of the expression levels of the Crebh, Oasis, and Atf6 mRNAs. MC3T3-E1 cells were treated with or without TNFα (+, 5 ng/ml; ++, 20 ng/ml) for 6 h (A) or BMP2 (+, 200 ng/ml; ++, 500 ng/ml) for 24 h (B). *, p < 0.05; **, p < 0.01; and ***, p < 0.001 versus the untreated group. C and D, Western blot analyses of cellular (C) and nuclear (D) levels of CREBH. MC3T3-E1 cells were treated with or without TNFα (+, 5 ng/ml; ++, 20 ng/ml) and with or without tunicamycin (Tm; 1 μg/ml) as a positive control for 12 h. FL, full-length; N, nuclear; M, molecular size marker. E, the effects of TNFα and tunicamycin on the luciferase activity of Gal4-fused full-length CREBH. MC3T3-E1 cells were co-transfected with Gal4-TK-Luc (200 ng) and Gal4-FL-CREBH (100 ng) or Gal4-DBD (100 ng) as a negative control. At 12 h post-transfection, cells were exposed to TNFα for 24 h. The results are expressed as the luciferase activity relative to that of the control. *, p < 0.05 and **, p < 0.01 versus the Gal4-FL-CREBH transfected group.
In response to ER stress, membrane-bound transcription factors, such as ATF6 and OASIS, are cleaved and a part of the cytosolic component is translocated to the nucleus to function as a transcription factor. Therefore, we determined whether TNFα can promote the translocation of the cleaved (active) form of CREBH protein to the nucleus using a GAL4-based luciferase reporter assay. Exposure of TNFα increased the luciferase activity of GAL4-fused full-length CREBH in a dose-dependent manner (Fig. 1E). Taken together, these results suggest that TNFα increases CREBH expression and may enhance its transcriptional activity in the nucleus.
NF-κB Signaling Pathway Is Involved in TNFα-induced CREBH Expression
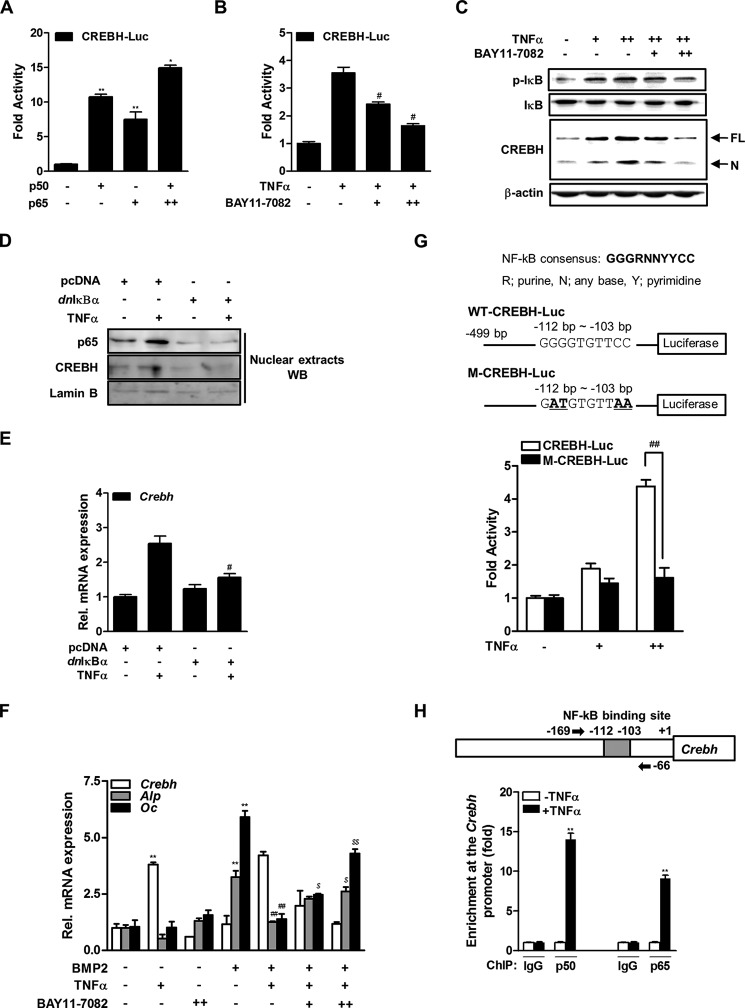
TNFα regulates the expression of several genes related to inflammation, by activation of the NF-κB pathway (33). To determine whether this pathway is involved in TNFα-induced CREBH expression in osteoblasts, MC3T3-E1 cells were co-transfected with a Crebh reporter construct (CREBH-Luc), and a plasmid expressing the p50 and/or p65 subunits of NF-κB. The overexpression of p50 and p65 increased the luciferase activity of CREBH-Luc (Fig. 2A). Treatment of BAY11-7082, a chemical inhibitor of the IKK/NF-κB pathway, decreased the TNFα-induced Crebh promoter activity in a dose-dependent manner (Fig. 2B). The compound also abrogated the TNFα induction of CREBH protein expression with the reduced phosphorylation of IκB (Fig. 2C).
FIGURE 2.
NF-κB signaling pathway is involved in TNFα induction of CREBH expression. A, the effects of p50 and p65 overexpression on CREBH-Luc activity in MC3T3-E1 cells. Luciferase activity was measured 48 h after transfection. β-Galactosidase plasmid was used as an internal control. The results are expressed as the luciferase activity relative to that of the control. *, p < 0.05 and **, p < 0.01 versus the untransfected control. B, the effects of TNFα and BAY11-7082 on CREBH-Luc activity. At 24 h post-transfection of CREBH-Luc, MC3T3-E1 cells were treated with TNFα (20 ng/ml) and/or BAY11-7082 (+, 0.1 μm; ++, 0.5 μm) for 6 h, and then luciferase activity was measured as in A. #, p < 0.05 versus the TNFα-treated group. C, the cells were treated with TNFα (20 ng/ml) and BAY11-7082 (0.1 or 0.5 μm) for 6 h. After total protein was extracted, and Western blot analysis was performed with the indicated antibodies. p-IκB, phosphorylated IκB; FL, full-length; N, nuclear CREBH. D and E, effects of dnIκBα overexpression on TNFα-induced CREBH expression. The cells were transiently transfected with pcDNA or dnIκBα expression construct and incubated in the presence of TNFα (20 ng/ml) for 24 h. D, nuclear fractions were prepared and Western blotting was performed with the designated antibodies. E, expression of CREBH mRNA was determined by qRT-PCR analysis. The expressions were normalized to those of β-actin. #, p < 0.05 versus the TNFα-treated group. F, effects of BMP2, TNFα, and BAY11-7082 on Crebh, Alp, and Oc mRNA levels. The cells were cultured with BMP2 (200 ng/ml) for 2 days and then treated with TNFα and/or BAY11-7082 for 6 h. The expression levels were normalized to those of β-actin. **, p < 0.01 versus the untreated control; ##, p < 0.01 versus the BMP2-treated group; $, p < 0.05 and $$, p < 0.01 versus the BMP2 with TNFα-treated group. G, effects of TNFα on activities of WT-CREBH-Luc or M-CREBH-Luc plasmid. The upper panel shows the consensus NF-κB binding site identified in the Crebh promoter. The lower panel shows the effects of TNFα on the luciferase activities of WT-CREBH-Luc and M-CREBH-Luc, the latter of which contained two point mutations of the NF-κB binding site (underlined in the upper panel). The cells were transfected with the plasmids for 24 h, and then treated with TNFα (20 ng/ml) for 6 h prior to measuring luciferase activity. H, ChIP-qPCR analysis for p50 or p65 binding to Crebh gene. The upper panel shows a schematic representation of the Crebh promoter region depicting the NF-κB binding motif and the qPCR primer binding sites (arrow). The lower panel shows relative binding of p50 or p65 to Crebh promoter gene. The cells were cultured with TNFα (20 ng/ml) for 12 h and then immunoprecipitated with anti-p50 or anti-p65 antibody or IgG as a negative control. Precipitated DNA was subjected to qPCR analysis with the primer pairs described in the upper panel. Fold-differences were calculated by the ΔCT method. All data represent the mean ± S.E. from three independent experiments. **, p < 0.01 versus the TNFα-untreated control.
To more confirm the involvement of specific IKK/IκBα/NF-κB pathway in the TNFα-induced CREBH expression, we examined the effects of the dnIκBα (S32A/S36A) mutant, which is unable to be phosphorylated and proteolytically degraded (23). Transfection of dnIκBα effectively blocked TNFα-increased p65 and CREBH protein levels in nucleus and Crebh mRNA expression in the cells (Fig. 2, D and E). On the other hand, TNFα inhibited BMP2-induced Alp and Oc mRNA expressions and treatment of BAY-11-7082 was partially rescued the TNFα action (Fig. 2F). An in silico analysis revealed a consensus NF-κB binding site at nucleotides −112 to −103 relative to the transcription initiation site of the Crebh gene. TNFα failed to activate the luciferase activity of a mutant form of the Crebh promoter (M-CREBH-Luc) in which four nucleotides in the NF-κB binding site were substituted, compared with the WT-CREBH-Luc activity (Fig. 2G). In addition, ChIP-qPCR analysis showed treatment of TNFα enhanced the binding of NF-κB (p50 or p65) on the Crebh promoter (Fig. 2H). Taken together, these results suggest that TNFα stimulates CREBH expression by activating the NF-κB pathway.
TNFα Suppresses Osteoblast Differentiation by Stimulating CREBH Expression
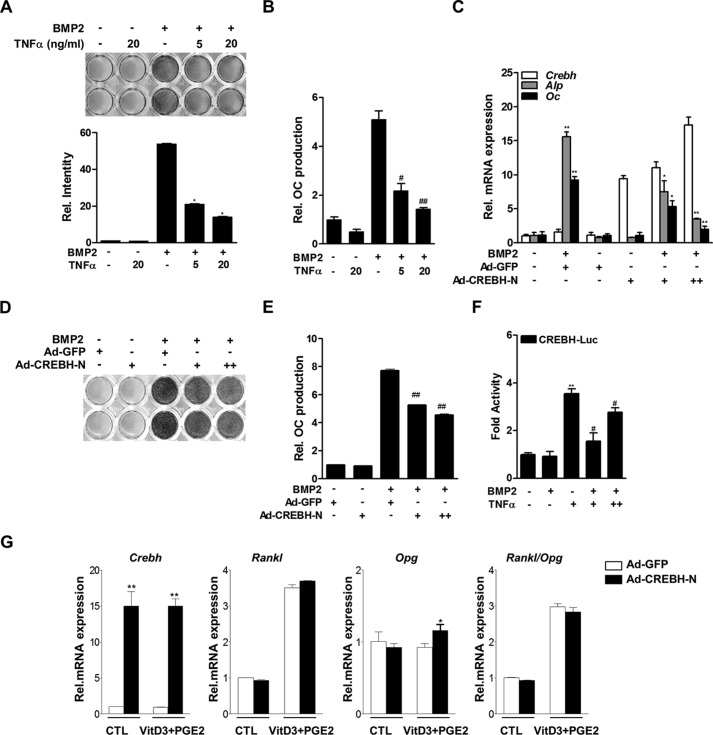
TNFα suppresses osteoblast differentiation by inhibiting the BMP signaling pathway (21). Therefore, we determined whether CREBH is involved in this process using gain- or loss-of-function experiments. TNFα inhibited BMP2-induced ALP activity and OC protein production (Figs. 3, A and B). In this condition, overexpression of CREBH using adenovirus encoding the nuclear form of CREBH (Ad-CREBH-N) dramatically inhibited BMP2 induction of ALP activity and OC production, as well as expression of osteogenic markers in a dose-dependent manner (Fig. 3, C–E). TNFα still stimulated the activity of the Crebh promoter to some extent in the presence of BMP2 (Fig. 3F). However, overexpression of CREBH did not affect the expression of the mRNAs encoding osteoprotegerin (Opg) and receptor activator of NF-κB ligand (Rankl) in primary osteoblasts, indicating that the expression of CREBH in osteoblasts might not be directly related to osteoclastogenesis (Fig. 3G).
FIGURE 3.
TNFα inhibits BMP2-induced osteoblast differentiation by stimulating CREBH expression. A and B, the effects of TNFα on BMP2-induced ALP activity in primary calvarial osteoblasts (A) and OC protein production in osteoblasts (B). Cells were cultured with BMP2 (200 ng/ml) for 3 days and then treated with TNFα for 24 h. Cells were stained with a BCIP®/nitro blue tetrazolium solution to determine ALP activity (A), and the level of OC protein in the culture medium was measured using an OC-specific ELISA kit (B). C–E, the effects of BMP2 and/or infection with an Ad-CREBH-N on the expression levels of the Crebh, Alp, and Oc mRNAs (C), ALP activity (D), and OC production (E) in MC3T3-E1 cells (C) or primary osteoblasts (D and E). Cells were infected with Ad-CREBH-N (+, 50 m.o.i.; ++, 100 m.o.i.) for 24 h and then treated with BMP2 (200 ng/ml) for 3 days. The expression levels of the target mRNAs were measured by qRT-PCR and normalized to those of β-actin. *, p < 0.05; **, p < 0.01; #, p < 0.05; and ##, p < 0.01 versus the BMP2-treated group. F, the effects of BMP2 and TNFα on CREBH-Luc activity. Twelve hours after transfection, cells were treated with BMP2 (200 ng/ml) for 48 h and/or TNFα (+, 5 ng/ml; ++, 20 ng/ml) for 6 h, and then luciferase activity was measured. **, p < 0.01 versus the untreated control; #, p < 0.01 versus the BMP2-treated group. G, the effects of a 3-day exposure to vitamin D3 (VitD3, 10 nm) and prostaglandin E2 (PGE2, 1 μm) on the expression levels of the Crebh, Rankl, and Opg mRNAs in primary osteoblasts infected with Ad-CREBH-N. The expression levels of the mRNAs were evaluated by qRT-PCR and normalized to those of β-actin. CTL, control.
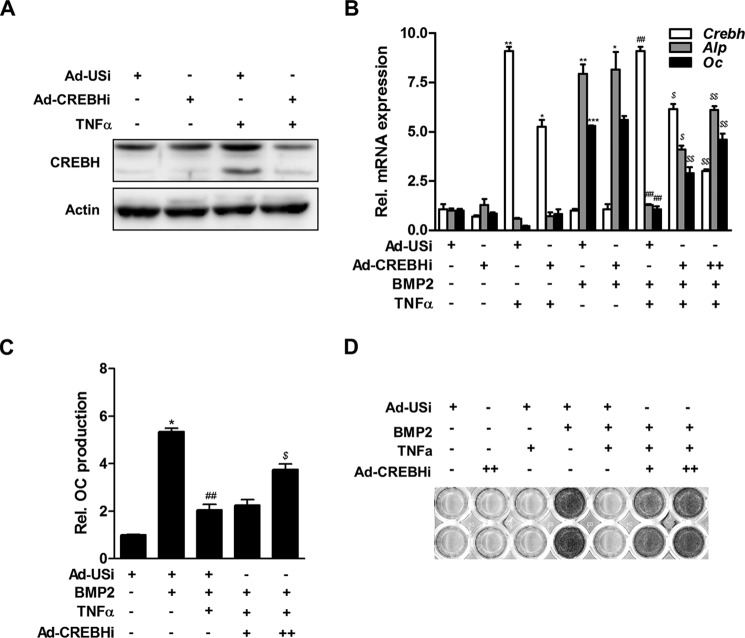
Next, we examined the effect of knockdown of CREBH using adenoviruses encoding an unspecific control shRNA (Ad-USi) or a CREBH-specific shRNA (Ad-CREBHi) on BMP2-induced osteoblast differentiation. Western blot analysis confirmed that treatment of Ad-CREBHi decreased the level of CREBH protein in MC3T3-E1 cells (Fig. 4A). Knockdown of CREBH attenuated the TNFα-mediated suppression of BMP2-induced expression of the Alp and Oc mRNAs (Fig. 4B), as well as OC protein production (Fig. 4C) and ALP activity (Fig. 4D). These results indicate that CREBH negatively regulates BMP2-induced osteoblast differentiation.
FIGURE 4.
Inhibition of CREBH attenuates TNFα-mediated suppression of BMP2-induced osteoblast differentiation. A, Western blot analysis for efficiency of Ad-CREBHi. MC3T3-E1 cells were infected with adenoviruses encoding an unspecific siRNA (Ad-USi) as a control virus or CREBH-specific shRNA (Ad-CREBHi) for 24 h, and then treated with TNFα (20 ng/ml) for 12 h. Western blotting was performed with CREBH and β-actin antibodies. B, the effect of knockdown of CREBH using adenoviruses encoding a CREBH-specific shRNA (Ad-CREBHi) on the expression levels of the Crebh, Alp, and Oc mRNAs in BMP2- and/or TNFα-treated MC3T3-E1 cells. The cells were infected with Ad-CREBHi (+, 50 m.o.i.; ++, 100 m.o.i.) or Ad-USi (100 m.o.i.). Ad-USi for unspecific shRNA was used as a control. Twenty-four hours after infection, the cells were treated with BMP2 (200 ng/ml) for 3 days and then treated with TNFα (20 ng/ml). qRT-PCR analyses were used to examine the expression levels of the Alp, Oc, and Crebh mRNAs. The mRNA expression levels were normalized to those of β-actin. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus the untreated control; ##, p < 0.01 versus the BMP2-treated group; and $, p < 0.05 and $$, p < 0.01 versus the BMP2 with TNFα-treated group. C and D, the effect of Ad-CREBHi on TNFα-mediated suppression of osteoblast differentiation. MC3T3-E1 cells (C) or primary osteoblasts (D) were cultured as described in B. The level of OC protein (C) and ALP activity (D) were measured as described in the legend to Fig. 3. *, p < 0.05 versus the BMP2-untreated control; ##, p < 0.01 versus the BMP2-treated group; and $, p < 0.05 versus the BMP2 with TNFα treated group.
CREBH Promotes Smad1 Degradation by Inducing Smurf1 Expression
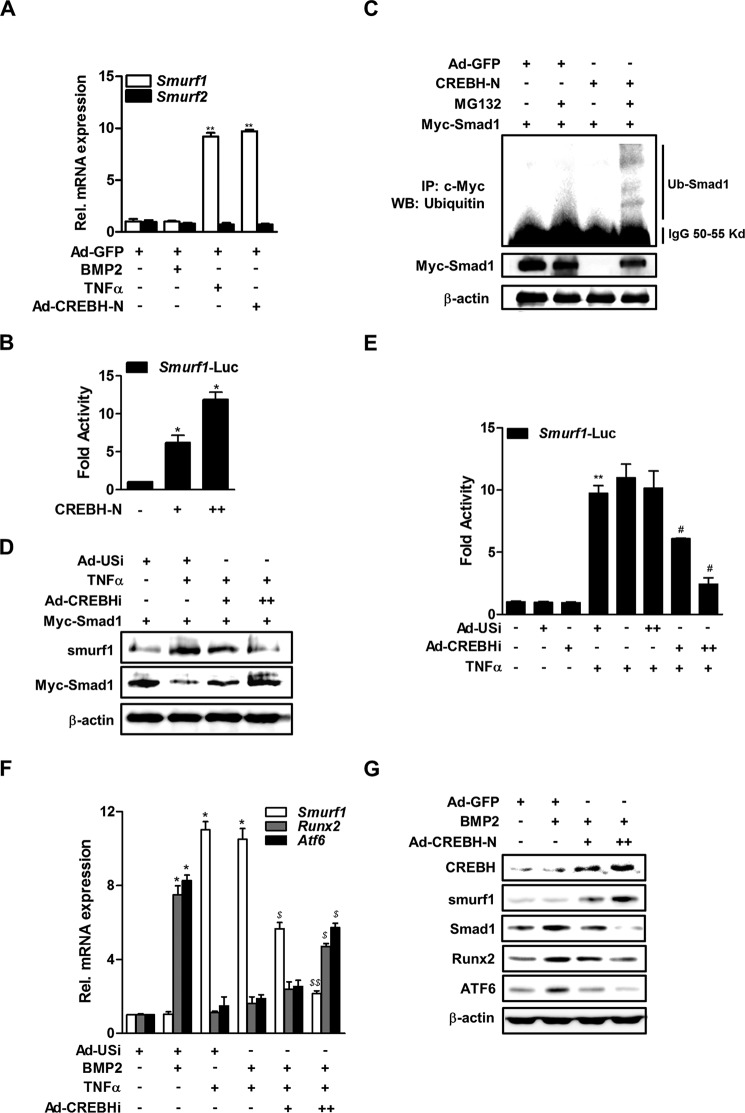
Smurf1 is a negative regulator of BMP signaling that suppresses osteoblast function by promoting Smad1 degradation via a proteosomal-dependent mechanism (34). To determine whether TNFα or CREBH affects Smurf1 expression, MC3T3-E1 cells were treated with TNFα or infected with Ad-CREBH-N, and then Smurf1 mRNA expression was measured by qRT-PCR. TNFα and Ad-CREBH-N increased the expression of Smurf1 mRNA significantly, whereas infection with a control adenovirus (Ad-GFP) did not (Fig. 5A). However, TNFα and Ad-CREBH-N did not affect the expression of Smurf2 mRNA (Fig. 5A). In addition, introduction of CREBH-N increased the Smurf1 promoter activity in a dose-dependent manner (Fig. 5B).
FIGURE 5.
CREBH promotes Smad1 degradation by inducing Smurf1 expression. A, the effects of TNFα and Ad-CREBH-N on Smurf1 and Smurf2 mRNA levels. MC3T3-E1 cells were infected with Ad-CREBH-N or Ad-GFP as a control virus (100 m.o.i.). Twenty-four hours after infection, the cells were treated with BMP2 (200 ng/ml) for 3 days and then with TNFα (20 ng/ml) for 6 h. The mRNA levels were determined by qRT-PCR and normalized to those of β-actin. **, p < 0.01 versus the untreated control. B, the effect of CREBH overexpression on Smurf1 promoter activity. Cells were transfected with the Smurf1-Luc and CREBH-N expression plasmids (+, 50 ng; ++, 200 ng) and luciferase activity was measured 24 h later. *, p < 0.05 versus the untransfected control. C, the effects of CREBH-N overexpression and proteasome inhibition on Smad1 ubiquitination. Cells were transfected with a Myc-Smad1 expression plasmid with or without a CREBH-N expression plasmid for 24 h, and then exposed to 10 μm MG132 (lanes 2 and 4) for 12 h. Immunoprecipitation (IP) and Western blotting (WB) analyses were performed with an anti-Myc or anti-ubiquitin antibody, respectively. D, the effects of inhibition of CREBH on TNFα-mediated changes in Smad1 and Smurf1 protein levels. Cells were transfected with a Myc-Smad1 with or without an Ad-USi or a CREBH-specific shRNA (+, 50 m.o.i.; ++, 100 m.o.i.). Twenty-four hours after transfection, the cells were treated with or without TNFα (20 ng/ml) for 6 h. Western blotting was performed with the designated antibodies. E, the effects of inhibition of CREBH on TNFα-mediated activation of the Smurf1 promoter. Cells were transfected with Smurf1-Luc and Ad-CREBHi or Ad-USi, and then exposed to TNFα (20 ng/ml) for 12 h. Luciferase activity was measured 24 h later. **, p < 0.01 versus the untreated control and #, p < 0.05 versus the TNFα treated group. F, the effects of inhibition of CREBH on TNFα-mediated changes in Smurf1, Runx2, and Atf6 mRNA levels. Cells were transfected with Ad-USi or Ad-CREBHi for 12 h and then exposed to TNFα (20 ng/ml) for 6 h prior to treatment with BMP2 (200 ng/ml) for 2 days. Total viral titer was held constant at 100 m.o.i. by addition of the appropriate amount of Ad-USi, a control Ad-shRNA. The levels of the mRNAs were determined by qRT-PCR and normalized to those of β-actin. *, p < 0.05 versus the untreated control, and $, p < 0.05 and $$, p < 0.01 versus the BMP2/TNFα-treated group. G, Western blot analyses of the effects of overexpression of CREBH on BMP2-mediated changes in Smad1, Runx2, ATF6, and CREBH protein levels. Cells were treated with BMP2 (200 ng/ml) for 2 days and infected with Ad-CREBH-N (+, 50 m.o.i.; ++, 100 m.o.i.) or Ad-GFP (50 m.o.i.) for an additional 24 h.
Next, we determined whether CREBH affects the expression level of Smad1 protein by regulating Smurf1. Western blot analyses of MC3T3-E1 cells that were transfected with an expression vector harboring Myc-Smad1 revealed that exposure to the potent proteasome inhibitor MG132 resulted in the production of small amounts of ubiquitinated Smad1. However, the level of ubiquitinated Smad1 was increased markedly by overexpression of Ad-CREBH-N (Fig. 5C), indicating that CREBH induces ubiquitination of Smad1, leading to its breakdown through proteasomal degradation. Moreover, TNFα decreased the level of Smad1 protein with the increased Smurf1, whereas inhibition of CREBH with Ad-CREBHi blocked TNFα-mediated Smad1 degradation and reduced Smurf1 expression (Fig. 5D). In addition, a luciferase reporter assay revealed that inhibition of CREBH reduced TNFα-mediated Smurf1 promoter activation in a dose-dependent manner (Fig. 5E).
In the study, we also examined the effects of CREBH on the expression levels of the Smad1 downstream factors Runx2 and ATF6, which are required for osteoblast differentiation. Inhibition of CREBH attenuated TNFα-mediated suppression of Runx2 and Atf6 mRNA levels, as well as the TNFα-mediated increase in Smurf1 expression (Fig. 5F). In contrast, overexpression of CREBH reduced BMP2-mediated induction of Smad1, Runx2, and ATF6 protein levels, and increased Smurf1 protein expression (Fig. 5G), suggesting that CREBH is involved in TNFα-mediated suppression of BMP2 downstream signals. Overall, these results indicate that CREBH negatively regulates BMP2-induced osteoblast differentiation by inducing Smurf1-related degradation of Smad1.
CREBH Suppresses BMP2-induced Bone Formation in Vivo
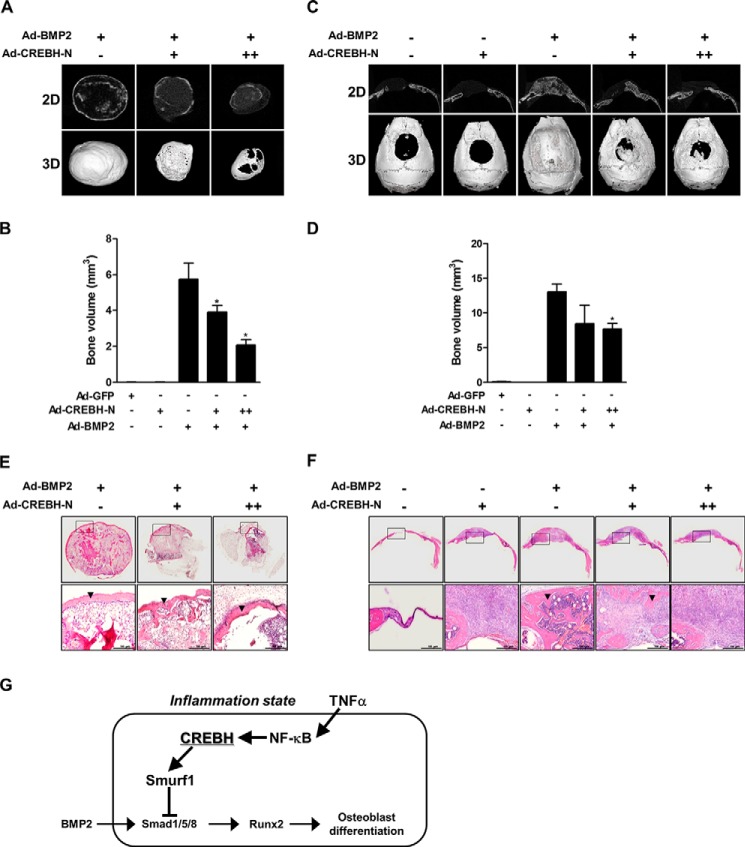
The results described above suggested that CREBH suppresses BMP2-induced osteoblast differentiation. Therefore, we investigated the role of CREBH in BMP2-induced bone formation in vivo. Radiographic analyses showed that administration of BMP2 strongly induced ectopic and orthotopic bone formation, and co-administration with CREBH significantly reduced BMP2-induced bone formation at both ectopic subcutaneous and orthotopic calvarial defect models (Fig. 6, A and C). A quantitative μCT analysis also confirmed that co-administration of CREBH decreased the BMP2 action (Fig. 6, B and D). However, administration of Ad-CREBH-N or the control Ad-GFP did not produce any significant changes (Fig. 6, B and D). Histology analysis consistently showed that BMP2 regenerated newly mineralized tissues in the administered regions, whereas co-administration of BMP2 and CREBH produced less or immature mineralized tissues (Fig. 6, E and F). Taken together, these findings suggest that CREBH has an inhibitory role in BMP2-induced bone formation in vivo.
FIGURE 6.
CREBH suppresses BMP2-induced ectopic and orthotopic bone formation. Ad-GFP (5 × 1010 particle number, PN), Ad-BMP2 (5 × 1010 PN), and/or Ad-CREBH-N (5 × 1010 or 10 × 1010 PN) with absorbable collagen sponges were subcutaneously implanted into the backs or critical sized cranial defects of mice. The total amount of implanted virus was adjusted by adding Ad-GFP virus. Three weeks after implantation, the implants were harvested and ectopic or orthotopic bone formation was evaluated. A and B, two- and three-dimensional μCT reconstructions (A) and volume (B) of subcutaneously formed ectopic bones. *, p < 0.05 versus the Ad–MP2treated group. C and D, μCT reconstructions (C) and volume (D) of newly formed orthotopic bones in critical-sized calvarial defects. *, p < 0.05 versus the Ad-BMP2-treated group. E and F, histology of ectopic (E) or orthotopic (F) regenerated bones. Lower panels are magnified images of the squared areas in upper panel, respectively. Black arrowheads indicate newly formed mineralized tissues. Representative data are shown (n = 4). G, an overview of the role of CREBH in BMP2-induced osteoblast differentiation. Under normal conditions, BMP2 stimulates osteoblast differentiation via Smad1/5/8-dependent intracellular signaling. In the presence of severe inflammation, TNFα induces the expression of CREBH and Smurf1 by NF-κB pathway, leading to the suppression of BMP2-induced osteoblast differentiation through Smurf1-dependent degradation of Smad1.
Discussion
Inflammation is triggered by inflammatory cytokines, such as TNFα and LPS, which suppress BMP2-induced osteoblast differentiation in vitro and contribute to bone loss in inflammatory bone diseases, such as rheumatoid arthritis (17, 35). Therefore, BMP2 and inflammatory cytokines have opposing roles in osteoblast differentiation. However, other cytokines, such as IL-6, IL-1β, and nitric oxide, are also secreted into inflammatory environments and may play different roles in osteogenesis (36–39). Moreover, local inhibitors of inflammation such as triptolide and BMP-binding peptide, can enhance the osteoinductive efficacy of BMP-2 in vivo (40, 41). However, direct evidence supporting an inhibitory effect of inflammation on BMP2-induced osteoblast differentiation is currently lacking. This study examined the effect of the inflammatory mediator CREBH on BMP2-induced osteoblast differentiation.
ER membrane-bound bZIP transcription factors ATF6, OASIS, and CREBH can be cleaved by cellular stresses, such as ER stress, to provide important signals for regulating cellular physiology (42). OASIS−/− mice exhibit severe osteopenia with reduced levels of collagen type αI in the bone matrix and reduced activity of osteoblasts (9). In our previous study, ATF6 stimulated osteoblast differentiation by regulating osteocalcin gene expression directly (32). However, the mechanisms involved in the sensing of unfolded proteins and translocation from the ER to the Golgi differ between OASIS family members. In addition, OASIS family members have unique cell- or tissue-specific expression patterns, suggesting that these transcription factors may be activated by, and associated with, distinct physiological responses that are dependent on particular environments (31). The results presented here demonstrate that the expression properties of CREBH differ from those of OASIS and ATF6; specifically, Crebh mRNA expression in osteoblasts was not affected by BMP2 but was increased in response to TNFα, whereas the expression levels of Oasis and Atf6 mRNAs were up-regulated by BMP2 but were not affected by TNFα (Fig. 1, A and B). Although CREBH belongs to the same family of transcription factors as OASIS and ATF6, it seems to play a different role in osteoblast differentiation due to differences in its activating stimuli, tissue distribution, and response element binding.
Here, we established that overexpression and knockdown of CREBH enhance and inhibit TNFα-mediated inhibition of BMP2-induced osteoblast differentiation, respectively. Furthermore, we sought to clarify the molecular mechanism by which TNFα regulates CREBH expression. Luciferase reporter assays and Western blot analysis showed that TNFα increased the CREBH promoter activity and the protein levels. Treatment of BAY-11-7082 or dnIκBα (S32A/S36A) consistently inhibited the TNFα-induced CREBH expression with the decreased phosphorylation of IκB or level of p65 subunit. In addition, one particular NF-κB binding site was crucial for TNFα-mediated stimulation of Crebh promoter activity (Fig. 2G). NF-κB subunits p50 and p65 bound to a consensus NF-κB site in the Crebh gene promoter and may utilize this cis-acting element to regulate promoter activity (Fig. 2H). In this study, we demonstrate for the first time in osteoblasts that TNFα may regulate CREBH expression via the IKK/IκBα/NF-κB signaling pathway.
TNFα suppresses BMP2-induced osteoblast differentiation and increases the expression of Smurf1, leading to the subsequent degradation of Smad1 and Runx2, which are critical mediators of BMP2 signaling (25, 43). Smurf1 interacts directly with Smad1 and Runx2, and stimulates the degradation of these proteins in ubiquitin- and proteasome-dependent manners (44). However, the factors downstream of TNFα that regulate Smurf expression were previously unknown. The results presented here suggest that CREBH mediates TNFα-induced Smurf1 expression in osteoblasts. Treatment of cells with TNFα or overexpression of CREBH stimulated ubiquitin-mediated degradation of Smad1. In addition, BMP2 increased the levels of the Smad1, Runx2, and ATF6 proteins in osteoblasts (Fig. 5G), as described previously (32, 45), and overexpression of CREBH attenuated these BMP2 effects with the increased Smurf1 level. These indicate that TNFα may suppress BMP2-induced osteoblast differentiation through the CREBH/Smurf/Smad1 regulatory system.
We also examined the in vivo effects of CREBH overexpression on BMP2-induced bone formation, using a subcutaneous ectopic model and a critical-sized calvarial defect model in mice. Consistent with the results of in vitro cell experiments, BMP2 strongly regenerated new bones within defected calvariae and subcutaneous spaces, and overexpression of CREBH significantly reduced the BMP2 effects. These in vivo findings firmly support that CREBH has an inhibitory role in BMP2-induced bone formation.
Overall, our findings reveal a novel mechanism by which TNFα inhibits BMP2-induced osteogenesis, namely the up-regulation of CREBH and subsequent stimulation of the Smurf1 E3 ligase to promote Smad1 degradation. Fig. 6G summarizes our proposed model of the inhibitory role of CREBH in osteoblast differentiation. Our results suggest that this interplay network might regulate several biological and pathological processes and provide valuable insights into why the repair of bone defects associated with severe inflammation is delayed.
This work was supported in part by grants from the National Research Foundation of Korea (NRF) funded by Korea government Grant MSIP 2011–0030121 (to J. T. K.) and National Creative Research Initiatives Grant 20110018305 from the NRF (funded by Ministry of Science, ICT & Future Planning of the Korean government) (to H. S. C.).
- BMP2
- bone morphogenetic protein 2
- CREBH
- cAMP response element-binding protein H
- ER
- endoplasmic reticulum
- IKK
- inhibitor of κB kinase
- OASIS
- old astrocyte specifically induced substance
- bZIP
- basic leucine zipper
- SMURF1
- Smad ubiquitination regulatory factor 1
- m.o.i.
- multiplicity of infection
- ALP
- alkaline phosphatase
- OC
- osteocalcin
- Ad
- adenovirus.
References
- 1. Urist M. R. (1965) Bone: formation by autoinduction. Science 150, 893–899 [DOI] [PubMed] [Google Scholar]
- 2. Wozney J. M., Rosen V., Celeste A. J., Mitsock L. M., Whitters M. J., Kriz R. W., Hewick R. M., Wang E. A. (1988) Novel regulators of bone formation: molecular clones and activities. Science 242, 1528–1534 [DOI] [PubMed] [Google Scholar]
- 3. Ron D., Walter P. (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 4. Malhotra J. D., Kaufman R. J. (2007) The endoplasmic reticulum and the unfolded protein response. Semin. Cell Dev. Biol. 18, 716–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Harding H. P., Zhang Y., Ron D. (1999) Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397, 271–274 [DOI] [PubMed] [Google Scholar]
- 6. Korennykh A. V., Egea P. F., Korostelev A. A., Finer-Moore J., Zhang C., Shokat K. M., Stroud R. M., Walter P. (2009) The unfolded protein response signals through high-order assembly of Ire1. Nature 457, 687–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Okada T., Yoshida H., Akazawa R., Negishi M., Mori K. (2002) Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem. J. 366, 585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yoshida H., Matsui T., Hosokawa N., Kaufman R. J., Nagata K., Mori K. (2003) A time-dependent phase shift in the mammalian unfolded protein response. Dev. Cell 4, 265–271 [DOI] [PubMed] [Google Scholar]
- 9. Murakami T., Saito A., Hino S., Kondo S., Kanemoto S., Chihara K., Sekiya H., Tsumagari K., Ochiai K., Yoshinaga K., Saitoh M., Nishimura R., Yoneda T., Kou I., Furuichi T., Ikegawa S., Ikawa M., Okabe M., Wanaka A., Imaizumi K. (2009) Signalling mediated by the endoplasmic reticulum stress transducer OASIS is involved in bone formation. Nat. Cell Biol. 11, 1205–1211 [DOI] [PubMed] [Google Scholar]
- 10. Saito A., Ochiai K., Kondo S., Tsumagari K., Murakami T., Cavener D. R., Imaizumi K. (2011) Endoplasmic reticulum stress response mediated by the PERK-eIF2α-ATF4 pathway is involved in osteoblast differentiation induced by BMP2. J. Biol. Chem. 286, 4809–4818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kusumoto K., Bessho K., Fujimura K., Akioka J., Okubo Y., Wang Y., Iizuka T., Ogawa Y. (2002) Osteoinduction by recombinant human bone morphogenetic protein-2 in muscles of non-human primates. J. Int. Med. Res. 30, 251–259 [DOI] [PubMed] [Google Scholar]
- 12. Lee M. W., Chanda D., Yang J., Oh H., Kim S. S., Yoon Y. S., Hong S., Park K. G., Lee I. K., Choi C. S., Hanson R. W., Choi H. S., Koo S. H. (2010) Regulation of hepatic gluconeogenesis by an ER-bound transcription factor, CREBH. Cell Metab. 11, 331–339 [DOI] [PubMed] [Google Scholar]
- 13. Saito A. (2014) Physiological functions of endoplasmic reticulum stress transducer OASIS in central nervous system. Anat. Sci. Int. 89, 11–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gentile C. L., Wang D., Pfaffenbach K. T., Cox R., Wei Y., Pagliassotti M. J. (2010) Fatty acids regulate CREBh via transcriptional mechanisms that are dependent on proteasome activity and insulin. Mol. Cell. Biochem. 344, 99–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee A. H. (2012) The role of CREB-H transcription factor in triglyceride metabolism. Curr. Opin. Lipidol. 23, 141–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang Y., Vera L., Fischer W. H., Montminy M. (2009) The CREB coactivator CRTC2 links hepatic ER stress and fasting gluconeogenesis. Nature 460, 534–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gilbert L., He X., Farmer P., Boden S., Kozlowski M., Rubin J., Nanes M. S. (2000) Inhibition of osteoblast differentiation by tumor necrosis factor-α. Endocrinology 141, 3956–3964 [DOI] [PubMed] [Google Scholar]
- 18. Nanes M. S. (2003) Tumor necrosis factor-α: molecular and cellular mechanisms in skeletal pathology. Gene 321, 1–15 [DOI] [PubMed] [Google Scholar]
- 19. Choy E. H., Panayi G. S. (2001) Cytokine pathways and joint inflammation in rheumatoid arthritis. N. Engl. J. Med. 344, 907–916 [DOI] [PubMed] [Google Scholar]
- 20. Beklen A., Ainola M., Hukkanen M., Gürgan C., Sorsa T., Konttinen Y. T. (2007) MMPs, IL-1, and TNF are regulated by IL-17 in periodontitis. J. Dent. Res. 86, 347–351 [DOI] [PubMed] [Google Scholar]
- 21. Yamazaki M., Fukushima H., Shin M., Katagiri T., Doi T., Takahashi T., Jimi E. (2009) Tumor necrosis factor alpha represses bone morphogenetic protein (BMP) signaling by interfering with the DNA binding of Smads through the activation of NF-κB. J. Biol. Chem. 284, 35987–35995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gilbert L., He X., Farmer P., Rubin J., Drissi H., van Wijnen A. J., Lian J. B., Stein G. S., Nanes M. S. (2002) Expression of the osteoblast differentiation factor RUNX2 (Cbfa1/AML3/Pebp2α A) is inhibited by tumor necrosis factor-α. J. Biol. Chem. 277, 2695–2701 [DOI] [PubMed] [Google Scholar]
- 23. Lee H. L., Yi T., Woo K. M., Ryoo H. M., Kim G. S., Baek J. H. (2010) Msx2 mediates the inhibitory action of TNF-α on osteoblast differentiation. Exp. Mol. Med. 42, 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yan X., Chen Y. G. (2011) Smad7: not only a regulator, but also a cross-talk mediator of TGF-β signalling. Biochem. J. 434, 1–10 [DOI] [PubMed] [Google Scholar]
- 25. Kaneki H., Guo R., Chen D., Yao Z., Schwarz E. M., Zhang Y. E., Boyce B. F., Xing L. (2006) Tumor necrosis factor promotes Runx2 degradation through up-regulation of Smurf1 and Smurf2 in osteoblasts. J. Biol. Chem. 281, 4326–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chanda D., Kim Y. H., Kim D. K., Lee M. W., Lee S. Y., Park T. S., Koo S. H., Lee C. H., Choi H. S. (2012) Activation of cannabinoid receptor type 1 (Cb1r) disrupts hepatic insulin receptor signaling via cyclic AMP-response element-binding protein H (Crebh)-mediated induction of Lipin1 gene. J. Biol. Chem. 287, 38041–38049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jeong B. C., Lee Y. S., Bae I. H., Lee C. H., Shin H. I., Ha H. J., Franceschi R. T., Choi H. S., Koh J. T. (2010) The orphan nuclear receptor SHP is a positive regulator of osteoblastic bone formation. J. Bone Miner. Res. 25, 262–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jang W. G., Kim E. J., Lee K. N., Son H. J., Koh J. T. (2011) AMP-activated protein kinase (AMPK) positively regulates osteoblast differentiation via induction of Dlx5-dependent Runx2 expression in MC3T3E1 cells. Biochem. Biophys. Res. Commun. 404, 1004–1009 [DOI] [PubMed] [Google Scholar]
- 29. Jang W. G., Kim E. J., Koh J. T. (2011) Tunicamycin negatively regulates BMP2-induced osteoblast differentiation through CREBH expression in MC3T3E1 cells. BMB Rep. 44, 735–740 [DOI] [PubMed] [Google Scholar]
- 30. Lee Y. S., Kim D. K., Kim Y. D., Park K. C., Shong M., Seong H. A., Ha H. J., Choi H. S. (2008) Orphan nuclear receptor SHP interacts with and represses hepatocyte nuclear factor-6 (HNF-6) transactivation. Biochem. J. 413, 559–569 [DOI] [PubMed] [Google Scholar]
- 31. Zhang K., Shen X., Wu J., Sakaki K., Saunders T., Rutkowski D. T., Back S. H., Kaufman R. J. (2006) Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell 124, 587–599 [DOI] [PubMed] [Google Scholar]
- 32. Jang W. G., Kim E. J., Kim D. K., Ryoo H. M., Lee K. B., Kim S. H., Choi H. S., Koh J. T. (2012) BMP2 protein regulates osteocalcin expression via Runx2-mediated Atf6 gene transcription. J. Biol. Chem. 287, 905–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ghosh S., Hayden M. S. (2008) New regulators of NF-κB in inflammation. Nat. Rev. Immunol. 8, 837–848 [DOI] [PubMed] [Google Scholar]
- 34. Zhu H., Kavsak P., Abdollah S., Wrana J. L., Thomsen G. H. (1999) A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature 400, 687–693 [DOI] [PubMed] [Google Scholar]
- 35. Huang R. L., Yuan Y., Zou G. M., Liu G., Tu J., Li Q. (2014) LPS-stimulated inflammatory environment inhibits BMP-2-induced osteoblastic differentiation through crosstalk between TLR4/MyD88/NF-κB and BMP/Smad signaling. Stem Cells Dev. 23, 277–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mukai T., Otsuka F., Otani H., Yamashita M., Takasugi K., Inagaki K., Yamamura M., Makino H. (2007) TNF-α inhibits BMP-induced osteoblast differentiation through activating SAPK/JNK signaling. Biochem. Biophys. Res. Commun. 356, 1004–1010 [DOI] [PubMed] [Google Scholar]
- 37. Yamashita M., Otsuka F., Mukai T., Otani H., Inagaki K., Miyoshi T., Goto J., Yamamura M., Makino H. (2008) Simvastatin antagonizes tumor necrosis factor-α inhibition of bone morphogenetic proteins-2-induced osteoblast differentiation by regulating Smad signaling and Ras/Rho-mitogen-activated protein kinase pathway. J. Endocrinol. 196, 601–613 [DOI] [PubMed] [Google Scholar]
- 38. Hess K., Ushmorov A., Fiedler J., Brenner R. E., Wirth T. (2009) TNF-α promotes osteogenic differentiation of human mesenchymal stem cells by triggering the NF-κB signaling pathway. Bone 45, 367–376 [DOI] [PubMed] [Google Scholar]
- 39. Matsumoto Y., Otsuka F., Takano M., Mukai T., Yamanaka R., Takeda M., Miyoshi T., Inagaki K., Sada K. E., Makino H. (2010) Estrogen and glucocorticoid regulate osteoblast differentiation through the interaction of bone morphogenetic protein-2 and tumor necrosis factor-α in C2C12 cells. Mol. Cell. Endocrinol. 325, 118–127 [DOI] [PubMed] [Google Scholar]
- 40. Lee K. B., Murray S. S., Taghavi C. E., Song K. J., Brochmann E. J., Johnson J. S., Keorochana G., Liao J. C., Wang J. C. (2011) Bone morphogenetic protein-binding peptide reduces the inflammatory response to recombinant human bone morphogenetic protein-2 and recombinant human bone morphogenetic protein-7 in a rodent model of soft-tissue inflammation. Spine J. 11, 568–576 [DOI] [PubMed] [Google Scholar]
- 41. Ratanavaraporn J., Furuya H., Tabata Y. (2012) Local suppression of pro-inflammatory cytokines and the effects in BMP-2-induced bone regeneration. Biomaterials 33, 304–316 [DOI] [PubMed] [Google Scholar]
- 42. Asada R., Kanemoto S., Kondo S., Saito A., Imaizumi K. (2011) The signalling from endoplasmic reticulum-resident bZIP transcription factors involved in diverse cellular physiology. J. Biochem. 149, 507–518 [DOI] [PubMed] [Google Scholar]
- 43. Guo R., Yamashita M., Zhang Q., Zhou Q., Chen D., Reynolds D. G., Awad H. A., Yanoso L., Zhao L., Schwarz E. M., Zhang Y. E., Boyce B. F., Xing L. (2008) Ubiquitin ligase Smurf1 mediates tumor necrosis factor-induced systemic bone loss by promoting proteasomal degradation of bone morphogenetic signaling proteins. J. Biol. Chem. 283, 23084–23092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhao M., Qiao M., Oyajobi B. O., Mundy G. R., Chen D. (2003) E3 ubiquitin ligase Smurf1 mediates core-binding factor α1/Runx2 degradation and plays a specific role in osteoblast differentiation. J. Biol. Chem. 278, 27939–27944 [DOI] [PubMed] [Google Scholar]
- 45. Upton P. D., Long L., Trembath R. C., Morrell N. W. (2008) Functional characterization of bone morphogenetic protein binding sites and Smad1/5 activation in human vascular cells. Mol. Pharmacol. 73, 539–552 [DOI] [PubMed] [Google Scholar]