Background: Cohesin-dockerin interactions support the binding of multienzyme complexes (cellulosomes) onto the cell surface.
Results: The structures of novel Coh-Doc complexes reveal a dual binding mode.
Conclusion: A dual binding mode is present in Coh-Doc interactions supporting cell-surface attachment.
Significance: The dual binding mode introduces flexibility into highly populated multienzyme complexes attached to the cell surface.
Keywords: cell surface, cellulase, cellulose, cellulosome, protein-protein interaction, cell-surface attachment
Abstract
Protein-protein interactions play a pivotal role in the assembly of the cellulosome, one of nature's most intricate nanomachines dedicated to the depolymerization of complex carbohydrates. The integration of cellulosomal components usually occurs through the binding of type I dockerin modules located at the C terminus of the enzymes to cohesin modules located in the primary scaffoldin subunit. Cellulosomes are typically recruited to the cell surface via type II cohesin-dockerin interactions established between primary and cell-surface anchoring scaffoldin subunits. In contrast with type II interactions, type I dockerins usually display a dual binding mode that may allow increased conformational flexibility during cellulosome assembly. Acetivibrio cellulolyticus produces a highly complex cellulosome comprising an unusual adaptor scaffoldin, ScaB, which mediates the interaction between the primary scaffoldin, ScaA, through type II cohesin-dockerin interactions and the anchoring scaffoldin, ScaC, via type I cohesin-dockerin interactions. Here, we report the crystal structure of the type I ScaB dockerin in complex with a type I ScaC cohesin in two distinct orientations. The data show that the ScaB dockerin displays structural symmetry, reflected by the presence of two essentially identical binding surfaces. The complex interface is more extensive than those observed in other type I complexes, which results in an ultra-high affinity interaction (Ka ∼1012 m). A subset of ScaB dockerin residues was also identified as modulating the specificity of type I cohesin-dockerin interactions in A. cellulolyticus. This report reveals that recruitment of cellulosomes onto the cell surface may involve dockerins presenting a dual binding mode to incorporate additional flexibility into the quaternary structure of highly populated multienzyme complexes.
Introduction
The dynamic nature of plant cell walls is of growing environmental and industrial significance as the demand for renewable sources for energy and novel molecules for the chemical industry increases. Plant cell wall polysaccharides, primarily cellulose and hemicelluloses, are a major reservoir of carbon and energy (1). The deconstruction of the plant cell wall requires an extensive array of hydrolytic enzymes to attack the heterogeneous, insoluble, and highly recalcitrant substrate, which requires the catalytic entities to act in synergy to degrade this composite structure (2). Anaerobes have adopted an elegant alternative strategy for degrading structural plant carbohydrates, through the organization of enzymes into multiprotein complexes termed cellulosomes. The cellulosomal organization of Clostridium thermocellum is the most well defined and characterized system, and it is therefore used as a blueprint for typical cellulosome assembly. In this system, assembly occurs through the binding of type I cohesin modules found in a primary scaffoldin subunit to enzyme-borne type I dockerin modules. Primary scaffoldins are large noncatalytic cohesin-containing proteins, which characteristically include a carbohydrate-binding module that directs the cellulosome and therefore the cellulosomal enzymes to its cellulosic substrate. A sequence-divergent type II dockerin, located at the C terminus of some primary scaffoldins, tethers the cellulosome to the peptidoglycan layer of the bacterial cell envelope through an interaction with type II cohesin modules located in cell-surface anchoring scaffoldins (1, 3). The organization and structural architecture of cellulosomes are therefore orchestrated by the specificity of the different cohesin and dockerin modules (4). Cohesin-dockerin (Coh-Doc)3 pairs exhibit one of the strongest protein-protein affinities found in nature, and their interaction is crucial for both cellulosome assembly and cell-surface attachment (5, 6).
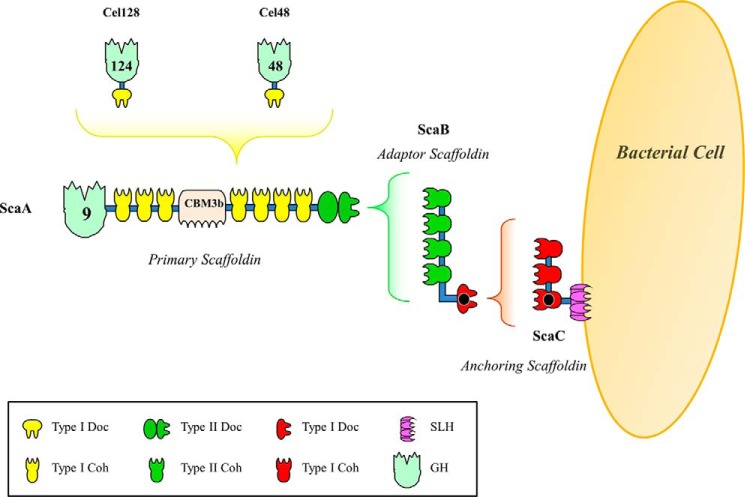
The mesophilic anaerobic bacterium Acetivibrio cellulolyticus produces a highly efficient cellulosome capable of hydrolyzing a range of cellulosic materials, including crystalline cellulose, with a higher activity than that of Aspergillus niger and Trichoderma viride systems (7, 8). Initial sequencing of an A. cellulolyticus gene cluster identified four tandem scaffoldin genes (scaA, scaB, scaC, and scaD (9, 10)). The primary scaffoldin (where the enzymes of the cellulosome are recruited), ScaA, contains an internal carbohydrate-binding module, bordered by seven type I cohesin modules and a single X module that provides structural stability to the neighboring C-terminal type II dockerin domain. Intriguingly, ScaA also contains an N-terminal family 9 glycoside hydrolase, a unique property within primary scaffoldins (11), although in the corresponding fungal cellulosomes, it has been proposed that the dockerins of the catalytic subunits bind to a GH3 enzyme, which acts as a scaffoldin (12). Downstream of scaA are genes encoding an adaptor scaffoldin and an anchoring protein, ScaB and ScaC, respectively. ScaB contains four type II cohesin modules, which interact with the C-terminal type II dockerin of ScaA, and a divergent C-terminal type I dockerin, which specifically interacts with the type I cohesin modules found in ScaC. ScaC, in turn, acts as an anchoring scaffoldin by virtue of its C-terminal SLH module (Fig. 1) (9). The recent sequencing of the A. cellulolyticus CD2 genome identified numerous additional cellulosomal components, gene regulatory elements, and cell anchoring modules (identified by the presence of dockerins or cohesins), suggestive of a much more elaborate and sophisticated cellulosome system than originally observed (13). Analysis of the genome uncovered 41 putative cohesin modules distributed among 16 scaffoldins (including ScaA, ScaB, ScaC, and ScaD), some of which have both cohesins and dockerins in the same polypeptide chain. All of the identified scaffoldins, with the exception of ScaI, appear to contain a signal peptide, suggesting that these proteins are secreted (13). The genome of A. cellulolyticus encodes 143 dockerin-containing proteins, which is about twice the number of such proteins in clostridial bacteria, but fewer than the 220 cellulosomal proteins encoded by the Ruminococcus flavefaciens FD-1 genome (13, 14).
FIGURE 1.
Schematic representation of the A. cellulolyticus cellulosome. Type I dockerin modules located in A. cellulolyticus cellulosomal enzymes bind reiterated cohesin modules located in the primary scaffoldin ScaA. ScaB plays the role of an adaptor scaffoldin with its cohesins binding the type II dockerin of ScaA. ScaB also contains a type I dockerin that specifically binds the complementary type I cohesins of the anchoring scaffoldin ScaC. The entire complex is tethered to the bacterial cell surface via the resident SLH module of ScaC. ScaA also contains a carbohydrate-binding module and a GH9 catalytic module. The structures of the ScaB dockerin in complex with the third cohesin of ScaC, solved here, are highlighted with a black dot.
Although structurally related, there is no cross-specificity between type I and type II Coh-Doc partners, which allows for the efficient assembly and cell-surface attachment of bacterial cellulosomes (15, 16). Structural studies on type I Coh-Doc complexes of C. thermocellum (6, 17) and Clostridium cellulolyticum (18) provided insights into the molecular determinants of the interaction responsible for cellulosome assembly. It is now known that the sequence duplication displayed by type I dockerins, from a variety of organisms beyond C. thermocellum, supports a dual binding mode within Coh-Doc complexes (5, 18). The sequence and structural symmetry within the type I dockerin module from C. thermocellum support a dual binding mode with the main Coh-contacting residues at positions 11 and 12 in the N-terminal (Ser-11 and Thr-12) or C-terminal (Ser-45 and Thr-46) helix of the protein domain (17). This symmetry is evident in the enzyme-borne dockerins of A. cellulolyticus that interact with ScaA, where the specificity residues in the two helices are Ser-Ile (or Ser-Leu). In the type I ScaB A. cellulolyticus dockerin, the specificity residues at positions 11 and 12 of the two duplicated segments consist of a conserved Ile-Asn motif. Thus, a lack of cross-specificity between the type I-Doc interactions that modulate the binding of ScaB into ScaC or the cellulosomal enzymes into ScaA (9, 19) may be related to differences at these key residues.
Nevertheless, the striking symmetry of the duplicated segments does not directly constitute evidence that both modes of the wild-type dockerin will bind the cohesin. In this context, mutagenesis studies, combined with structural and affinity-based binding data, serve to verify the dual mode of binding between a given cohesin-dockerin pair. Here, we investigated the molecular determinants for type I Coh-Doc interactions in the A. cellulolyticus cellulosome. We describe structures comprising the third type I cohesin from the anchoring ScaC scaffoldin in complex with the type I dockerin of the adaptor ScaB scaffoldin in which the dockerin is in two different orientations. The data reveal that the type I Coh-Doc interaction between adaptor scaffoldins and anchoring proteins in A. cellulolyticus cellulosome displays a dual binding mode.
Experimental Procedures
Gene Synthesis and DNA Cloning
To promote stability, the A. cellulolyticus ScaB type I dockerin module (DocScaB) was co-expressed in vivo with the third type I A. cellulolyticus ScaC cohesin module (CohScaC). The genes encoding the two proteins were synthesized (NZYTech Ltd., Lisboa, Portugal) and cloned into the same plasmid under the control of separate T7 promoters. Additionally, a cloning strategy was developed to engineer hexahistidine tags either at the N terminus of the dockerin module or the C terminus of the cohesin module. In total, three DNA fragments were synthesized comprising the following sequences (from the 5′ to the 3′ end): dockerin DNA, T7 terminator, T7 promoter, and cohesin DNA. Genes were designed to encode DocScaB (residues 868–942) and CohScaC (residues 326–467) with codon usage optimized for expression in Escherichia coli. The three constructs encode wild-type CohScaC for co-expression with wild-type DocScaB (Coh_DocScaB), DocScaB mutated at positions I15S/N16I (Coh_DocI15S/N16I), and DocScaB mutated at positions I51S/N52I (Coh_DocI51S/N52I). The three DNA constructs contained 5′- and 3′-engineered restriction sites for cloning into pET28a in two different forms. The first had an engineered hexahistidine tag located at the N terminus of the dockerin (NheI-SalI), and the second had the hexahistidine tag positioned at the C terminus of the cohesin (NcoI-XhoI). In total, six recombinant pET28a derivatives were generated from the three DNA constructs synthesized.
To produce recombinant A. cellulolyticus type I cohesin and dockerin modules individually, the following approaches were utilized. The gene encoding CohScaC was isolated in pET28a by excising the DocScaB sequence in a BglII DNA fragment from a plasmid containing the two genes, followed by re-ligation. CohScaC contained an engineered C-terminal hexahistidine tag. DocScaB constructs were expressed fused with thioredoxin to improve solubility and stability The genes encoding DocScaB, DocI15S/N16I, and DocI51S/N52I were amplified from the respective plasmids encoding the protein complexes described above by PCR using NZYProof polymerase (NZYTech Ltd.) and primers described in Table 1. DocScaB genes were subsequently ligated into EcoRI-XhoI-digested pET32a (Novagen). Site-directed mutagenesis was used to create DocI15S/N16I/I51S/N52I with putative recognition residues at positions 11 and 12 of the two duplicated segments changed from IN to SI (Tables 1 and 2).
TABLE 1.
Primers used to obtain the genes encoding the cohesin and the dockerins used in this study
Engineered restriction sites are shown in bold and mutations are underlined.
| Clone | Sequence (5′ → 3′) | Direction |
|---|---|---|
| DocScaB, DocI15S/N16I and DocI51S/N52I | CTCGAATTCAAGTTTATTTATGGCGATG | Forward |
| DocScaB, DocI15S/N16I and DocI51S/N52I | CACCTCGAGTAATTCCTTTTCTTCCACC | Reverse |
| DocI15S/N16I/I51S/N52I | GGCTCGGTTCGTTCGATTGATGCTGTGCTGATTCGTG | Forward |
| DocI15S/N16I/I51S/N52I | CACGAATCAGCACAGCATCAATCGAACGAACCGAGCC | Reverse |
| CohScaA | CACACCATGGCAAACGGGCTTTAATCTGAGC | Forward |
| CohScaA | CACACTCGAGATTGACAGCACCGTTGGTAAC | Reverse |
TABLE 2.
Protein sequences of A. cellulolyticus type I enzyme dockerin (DocGH5), ScaB dockerin (DocScaB), and ScaB mutant dockerin derivatives
Amino acid changes are shown as bold and underlined.

To identify the dockerin residues that modulate type I Coh-Doc specificity in A. cellulolyticus, various dockerin derivatives were designed. The type I GH5 dockerin (residues 502–573, ZP_09464781; DocGH5) and six mutant derivatives of DocScaB (DocR14N/K50N, DocI15S/I51S, DocN16I/N52I, DocA18F/A54F, DocL20Y/L56Y, and DocD23Q/D59Q) were synthesized (NZYTech Ltd.) with codon usage optimized for expression in E. coli (Table 2). The genes contained engineered EcoRI and XhoI recognition sequences at the 5′ and 3′ ends, respectively, and were subcloned into the pET32a vector (Novagen). The gene region encoding the sixth cohesin of the primary scaffoldin ScaA (residues 1472–1611; CohScaA) was also isolated through PCR using the primers described in Table 1 and cloned into the NcoI-XhoI sites of pET28a.
Expression and Purification of Recombinant Proteins
Preliminary expression screens revealed that higher levels of CohScaC-DocScaB complexes were obtained when the hexahistidine tag was located at the C terminus of CohScaC (data not shown). The respective pET28a derivatives were used to transform E. coli Tuner cells, which were grown at 37 °C to an A600 of 0.5. Recombinant protein expression was induced by the addition of isopropyl β-d-1-thiogalactopyranoside to a final concentration of 0.2 mm followed by induction at 19 °C with shaking for 16 h. Cells were centrifuged for 15 min at 5000 rpm at 4 °C and resuspended in 20 ml of binding buffer (50 mm HEPES, pH 7.5, 10 mm imidazole, 1 m NaCl, 5 mm CaCl2). Cells were disrupted by sonication, and cell-free extract was recovered by centrifugation at 15,000 × g. Prior to crystallization trials, SDS-PAGE analysis (data not shown) indicated that expression levels of free CohScaC were higher than those of DocScaB, suggesting that the majority of CohScaC was in complex with DocScaB, with an excess of unbound CohScaC. CohScaC-DocScaB complexes and unbound CohScaC were initially purified by immobilized metal-ion affinity chromatography using Sepharose columns charged with nickel (HisTrapTM) following conventional protocols and a 0–25 mm imidazole gradient. Fractions containing the CohScaC-DocScaB complex and unbound CohScaC were buffer-exchanged into 20 mm Tris-HCl, pH 8.0, 5 mm CaCl2 using a PD-10 Sephadex G-25 M size exclusion column (Amersham Biosciences). A further anion-exchange chromatography purification step was performed to separate the CohScaC-DocScaB complexes from free CohScaC using a gradient elution with 0–1 m NaCl (Amersham Biosciences). Fractions containing the purified CohScaC-DocScaB complex were pooled and concentrated using Amicon Ultra-15 centrifugal devices with a 10-kDa cutoff membrane (Millipore) and washed three times with 1 mm CaCl2. The final protein concentration was adjusted to 42 mg/ml (Coh-DocI15S/N16I) and 30 mg/ml (Coh-DocI51S/N52I), in a storage buffer consisting of H2O (Sigma) and 0.5 mm CaCl2. Recombinant DocScaB, DocI15S/N16I, DocI51S/N52I, DocI15S/N16I/I51S/N52I, DocGH5, and the mutant derivatives of DocScaB were expressed in E. coli Origami cells. Recombinant CohScaA and CohScaC were expressed in E. coli Tuner cells. Growth was performed at 37 °C to an A600 of 0.5 in Luria broth. Recombinant protein expression was induced with 1 mm (Origami) or 0.2 mm (Tuner) isopropyl β-d-1-thiogalactopyranoside and incubated for 16 h at 19 °C. The individual cohesin and dockerin constructs were purified using immobilized metal-ion affinity chromatography as described above. Unless otherwise stated, the recombinant cohesin and dockerin constructs were buffer-exchanged to 50 mm HEPES, pH 7.5, 2 mm CaCl2, and 0.5 mm tris(2-carboxyethyl)phosphine hydrochloride.
X-ray Crystallography, Structural Determination, and Refinement
Crystallization conditions were screened by the sitting-drop vapor diffusion method using an Oryx8 robotic nanodrop dispensing system (Douglas Instruments) for the three protein complexes (CohScaC-DocScaB, CohScaC-DocI15S/N16I, and CohScaC-DocI51S/N52I). Crystals of CohScaC-DocI15S/N16I (42 mg/ml) grew over a period of ∼21 days at 19 °C in 0.5 m ammonium sulfate, 0.1 m HEPES, pH 7.5, 30% (v/v) 2-methyl-2,4-pentanediol and were cryoprotected with 30% (v/v) glycerol (20). Crystals of the Coh-DocI51S/N52I complex (30 mg/ml) grew over a period of ∼21 days at 19 °C in 1 m sodium citrate, 0.1 m MES, pH 6.5, and were cryoprotected with 30% (v/v) glycerol. Crystals were harvested in rayon fiber loops and flash-frozen in liquid nitrogen. Data were collected for both constructs using single crystals at the Diamond Light Source (beamline IO4) to a resolution of 1.49 Å for the CohScaC-DocI15S/N16I crystals and 1.81 Å for the Coh-DocI51S/N52I. A summary of the data processing statistics is given in Table 3. BALBES was used for the CohScaC-DocI15S/N16I data to carry out molecular replacement (21). In total, 42 structures with a sequence identity greater than 15% for either cohesin or dockerin or both modules were found. The best solution was found using the mutant cohesin-dockerin complex from C. thermocellum (Protein Data Bank code 2ccl (17)). Two copies of the heterodimer Coh-Doc complex are present in the asymmetric unit in the space group P41212, with final Rfactor/Rfree of 39.8/41.2 and Q-factor (22) of 0.706 after REFMAC5 at the end of the BALBES run. This model was adjusted and refined using REFMAC5 (23) interspersed with model adjustment in COOT (24) to give the final model (Protein Data Bank code 4uyp; Table 3). The final round of refinement was performed using the TLS/restrained refinement procedure using each module as a single group. The molecular replacement program PHASER (25) and the atomic coordinates of CohScaC-DocI15S/N16I (Protein Data Bank code 4uyp) were used as a search model against the highest resolution data (1.81 Å) obtained for a single complex of Coh-DocI51S/N52I. A successful solution was obtained in space group P43 with a TFZ score of 34.2 and LLG of 3308. The structure was refined as above. The root mean square deviation of the bond lengths, bond angles, torsion angles, and other indicators was continuously monitored using the validation tools in COOT (24) and MOLPROBITY (26). A summary of the refinement statistics for both structures is shown in Table 3.
TABLE 3.
Data collection and refinement statistics for the final models of the A. cellulolyticus type I cohesin-dockerin complexes
Values in parentheses are for the highest resolution shell.
| Data Quality | Coh_DocI15S/N16I | Coh_DocI51S/N52I |
|---|---|---|
| X-ray source | Diamond Light Source (104) | Diamond Light Source (104) |
| Wavelength (Å) | 0.9795 | 0.9762 |
| Unit cell parameters | ||
| a (Å) | 107.75 | 68.17 |
| b (Å) | 107.75 | 68.17 |
| c (Å) | 100.81 | 57.14 |
| Space group | P 41 21 2 | P 43 |
| Resolution range (Å) | 76.19–1.49 (1.543–1.49) | 68.17–1.81 (1.879–1.814) |
| Total reflections | 641814 (43638) | 52759 (2311) |
| Unique reflections | 96788 (9512) | 23100 (2183) |
| Multiplicity | 6.6 (6.2) | 2.3 (2.0) |
| Completeness (%) | 99.85 (99.17) | 96.89 (93.05) |
| Mean I/σ (I) | 15.92 (2.92) | 5.35 (2.01) |
| Wilson B-factor | 13.99 | 17.92 |
| Rmergea | 6.6 (58.7) | 11.2 (41.8) |
| Rpimb | 3.1 (27.6) | 8.8 (36.4) |
| CC½c | 0.998 (0.808) | 0.981 (0.687) |
| Average mosaicity | 0.21 | 0.85 |
| R-work | 0.1555 (0.2164) | 0.1497 (0.2404) |
| R-free | 0.1854 (0.2424) | 0.1950 (0.2080) |
| No. of non-hydrogen atoms | 4397 | 2004 |
| Macromolecules | 3580 | 1682 |
| Ligands | 177 | 3 |
| Water | 640 | 319 |
| Protein residues | 440 | 219 |
| Root mean square(bonds) | 0.027 | 0.023 |
| RMS (angles) | 2.85 | 2.16 |
| Ramachandran favored (%) | 100 | 100 |
| Ramachandran outliers (%) | 0.21 | 0 |
| Clash score | 29 | 11.66 |
| Average B-factor | 19.1 | 24.7 |
| Macromolecules | 16.4 | 22.5 |
| Ligands | 32.9 | 25.3 |
| Solvent | 30.4 | 36.2 |
| PDB entry | 4uyp | 4uyq |
a Rmerge = Σh Σi|I(h,i) − 〈I(j)〉|/Σh Σi I(h,i), where I(h,i) is the intensity of the measurements of reflection h and 〈I(h)〉 is the mean value of I(h,i) for all i measurements.
b Rpim = (Σhkl √1/(n − 1) Σj = 1n|Ihkl, j − 〈Ihkl〉|)/ΣhklΣjIhkl, j), where Ihklj is the average of symmetry-related observations of a unique reflection.
c CC½ is the half-data set correlation coefficient.
Nondenaturing Gel Electrophoresis (NGE)
For NGE experiments, dockerin constructs (DocScaB, DocI15S/N16I, DocI51S/N52I, DocGH5, DocR14N/K50N, DocI15S/I51S, DocN16I/N52I, DocA18F/A54F, DocL20Y/L56Y, DocD23Q/D59Q, and DocI15S/N16I/I51S/N52I) at a concentration of 25 μm were incubated in the absence and presence of 25 μm CohScaC for 1 h at room temperature and separated on a 10% native gel. Electrophoresis was carried out at room temperature. Gels were stained with Coomassie Blue, and protein complexes were detected by the presence of an additional band displaying a lower electrophoretic mobility.
Isothermal Titration Calorimetry
Isothermal titration calorimetry (ITC) experiments were carried out as described previously (17, 18), with the exception that the titrations were performed at 35 °C. During titrations, the dockerin constructs (25 μm) were stirred at 307 revolutions/min in the reaction cell and titrated with 27 successive 10-μl injections of CohScaC (180 μm for dual binding experiments and 120 μm for specificity experiments) at 220-s intervals. Integrated heat effects, after correction for heats of dilution, were analyzed by nonlinear regression using a single site-binding model (Microcal Software, ORIGIN, Version 7.0).
Surface Plasmon Resonance (SPR)
The interactions between DocScaB, DocI15S/N16I/I51S/N52I, DocGH5, and mutant derivatives of DocScaB and CohScaC were analyzed using a BIAcore X100 (GE Healthcare). CohScaC was diluted to 10 μg/ml in 10 mm sodium acetate, pH 4.5, and immobilized (∼150 resonance units) on a CM5 sensor chip (GE Healthcare) containing a dextran matrix with free carboxylic groups. The chip surface was prepared using conventional carbodiimide coupling chemistry and subsequent deactivation of excess active esters with ethanolamine (EDC/NHS coupling kit, GE Healthcare). Immobilization followed the procedure guidelines provided by the vendor (BIAcoreTM, GE Healthcare). The dockerin constructs were diluted in running buffer (10 mm HEPES, pH 7.5, 0.15 mm NaCl, 3.4 mm CaCl2, 0.05% Tween 20) and allowed to interact with the sensor chip surface during a 180-s injection. In all experiments, five different concentrations (0.05, 0.1, 1, 5, and 15 nm) of the ligand were injected at a flow rate of 30 μl/min. The sensor chip surface was regenerated with 10 mm glycine, pH 1.8. The association rate constant (kon) and dissociation rate constant (koff) were each calculated using the BIAcore X100 evaluation software, version 2.0.1. The dissociation constant (KD) was determined as koff/kon. The data were interpreted on the basis of a 1:1 binding model.
Results and Discussion
Expression and Crystallization of A. cellulolyticus Coh-Doc Complexes
Because the two different type I cohesin-dockerin interactions in A. cellulolyticus are not cross-reactive, DocScaB mutations were designed to replace the putative recognition residues in positions 11 and 12 (Ile-Asn) with those of the enzyme-borne dockerins (Ser-Ile), rather than the commonly applied alanine substitution.
Preliminary experiments thus evaluated the levels of expression of the following three Coh-Doc complexes: the wild-type Coh_DocScaB, Coh_DocI15S/N16I, and Coh_DocI51S/N52I mutants, wherein the hexahistidine tag was located at either the N terminus of the dockerin or the C terminus of CohScaC. The data (not shown) revealed that the levels of expression of the protein complexes were higher when the affinity tag was located at the C terminus of CohScaC. The recombinant plasmids encoding complexes with this latter cohesin construct were therefore selected to produce highly pure Coh-Doc complexes for crystallization. Initial attempts to crystallize the A. cellulolyticus type I CohScaC-DocScaB complex failed. Analysis of the DocScaB sequence revealed a high degree of internal symmetry, which suggested that this dockerin contained two identical cohesin binding interfaces. Failure to crystallize the complex may have reflected the lack of conformational homogeneity resulting from the two potential binding modes of DocScaB to CohScaC. It is well established that in type I dockerins residues at positions 11 and 12 of each of the two duplicated segments dominate cohesin recognition. In ScaB dockerins, Ile and Asn occupy these positions, which is in contrast with enzyme-borne dockerins of this bacterium that contain a Ser-Ile (or Ser-Leu) pair and specifically bind the ScaA cohesins. To generate proteins where only one of the cohesin-binding sites in DocScaB was functional, two variants of the protein complex, Coh-DocI15S/N16I and Coh-DocI51S/N52I, were generated to enforce conformational homogeneity within the complex. Replacement of one of the Ile-Asn pairs with Ser-Ile would result in a single conformation, therefore facilitating crystal formation. Diffracting crystals of CohScaC in complex with either DocScaB mutant derivative were obtained.
Structure of the Type I Coh-Doc A. cellulolyticus Complex
The structures of the Coh-DocI15S/N16I and Coh-DocI51S/N52I complexes were solved by molecular replacement using the crystal structure of C. thermocellum type I Coh-Doc complex (17, 20) as a search model for Coh_DocI15S/N16I and for Coh_DocI51S/N52I to resolutions of 1.49 and 1.81 Å, respectively (Fig. 2). The Coh-DocI15S/N16I model included two molecules of the heterodimer with each dockerin coordinating two calcium ions and a total of 640 water molecules. The two complexes in the asymmetric unit overlay with an r.m.s.d of 0.7 Å for 176 Cα atoms. Although the Coh-DocI15S/N16I complex behaved as a monomer in solution (i.e. no oligomerization occurs, as determined by size exclusion), in crystal it is a dimer resulting from interactions established between two CohScaC modules, although the biological relevance of these interactions, if any, is unclear. In contrast, the Coh-DocI51S/N52I crystal structure consisted of one heterodimer in the asymmetric unit together with two calcium atoms, bound to the dockerin, and 319 water molecules.
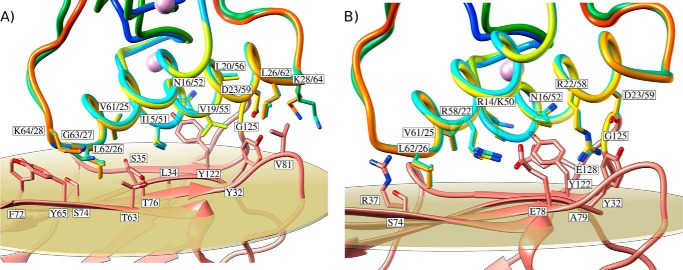
FIGURE 2.
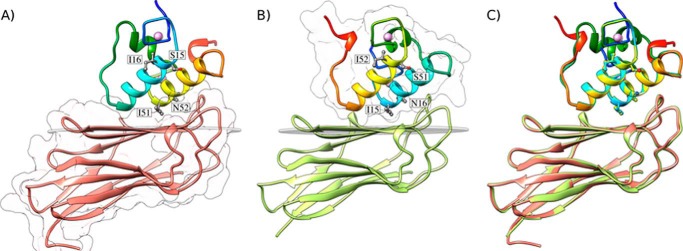
Structures of the A. cellulolyticus cohesin-dockerin complexes. A, structure of Coh_DocI15S/N16I with the dockerin color-ramped from N terminus (blue) to C terminus (red) and the cohesin in salmon. Ile-51 and Asn-52 that dominate cohesin recognition and engineered residues Ser-15 and Ile-16, to force a single binding mode, are labeled and shown as ball-and-stick configuration. Ca2+ ions are depicted as purple spheres. B, structure of Coh_DocI51S/N52I with the dockerin color-ramped from N terminus (blue) to C terminus (red) and the cohesin in green. Ile-15 and Asn-16 that dominate cohesin recognition and engineered residues Ser-51 and Ile-52, to force a single binding mode, are again labeled and shown as ball-and-stick representations. C, overlay of the two binding modes showing the high degree of overall similarity reflecting the internal 2-fold symmetry of the dockerin module. The transparent gray disk in A and B marks the plane defined by β-sheet B, and the β-strands form a distinctive dockerin interacting plateau. A and B also depict a representation of the molecular surface contour of the cohesin and dockerin, respectively.
Structure of A. cellulolyticus ScaC Type I Cohesin in Complex with Its Cognate Dockerin
ScaC type I cohesin in complex with its cognate dockerin displayed an elliptical structure comprising two β-sheets aligned in an elongated β-sandwich with a classical jellyroll fold (Fig. 2). The two sheets included β-strands 9, 1, 2, 7, and 4 on one face and β-strands 5, 6, 3, and 8 on the other face. β-Strand 8 is predominantly aligned with β-strand 3 but, at its N terminus, also makes interactions with β-strand 9. There is a disorganization of the secondary structure of β-strand 8 between Asn-115 and Ser-120, which leads to the above-mentioned capacity of β-strand 8 to contact both β-sheets. Strands 1 and 9 are aligned parallel to each other, whereas the other β-strands are antiparallel. As described above, the structure of CohScaC is essentially identical in the two protein complexes. Significantly, the A. cellulolyticus CohScaC also displays striking structural similarities to the type I cohesin modules of C. thermocellum (r.m.s.d. 1.41 Å) and C. cellulolyticum (r.m.s.d. 1.31 Å).
Structure of A. cellulolyticus ScaB Type I Dockerin
In both complexes, the ScaB dockerin displays an identical structure that comprises two helices arranged in an antiparallel orientation comprising residues Ile-15 to Gly-27 (helix-1) and Ile-51 to Gly-63 (helix-3), respectively, whereas the loop connecting these secondary structures contains a six-residue α-helix extending from Glu-35 to Asp-42 (helix-2) (Fig. 2). The overall structure of A. cellulolyticus DocScaB is very similar to both C. thermocellum (r.m.s.d. 1.2 Å) and C. cellulolyticum (r.m.s.d. 1.1 Å) type I dockerin modules. In both complexes, DocScaB contains two Ca2+ ions coordinated by several amino acid residues, similar to canonical EF-hand loop motifs. The Ca2+ ion located at the dockerin N terminus is coordinated by the side chains of residues Asp-6, Asp-8, Asn-10, and Asp-17 (both the Oδ1 and Oδ2), the latter belonging to the N-terminal α-helix (helix-1) of this module. The octahedral geometry of the coordination of this Ca2+ ion is fulfilled by the main chain carbonyl of Ser-12 and by a water molecule, which hydrogen bonds to Asn-16. The second Ca2+ site stabilizes the loop connecting the internal and C-terminal α-helix (helix-3) of the dockerin module. This Ca2+ ion is coordinated by the side chains of residues Asp-42, Asp-44, Asn-46, and Asp-53 (both the Oδ1 and Oδ2), as well as by the carbonyl of Ser-48, and by a water molecule, which is hydrogen-bonded to Asn-52. Thus, both Ca2+ sites show coordination to residues n, n + 2, n + 4, n + 6 (main-chain O atom), and n + 11, with a water molecule bridging to residue n + 10.
A. cellulolyticus Type I Coh-DocI15S/N16I and Coh-DocI51S/N52I Interfaces
DocScaB interacted with the 8–3-5–6 sheet of the CohScaC β-sandwich, which presents a predominantly flat surface. The structures of the Coh-DocI15S/N16I and Coh-DocI51S/N52I complexes were found to be very similar to each other, with a backbone r.m.s.d. of 0.7 Å (Fig. 2C). In addition, helix-1 and helix-3 of DocI15S/N16I overlapped almost perfectly with helix-3 and helix-1, respectively, of DocI51S/N52I as a result of a 180° rotation on dockerin DocI51S/N52I upon binding (Fig. 2C). This internal structural conservation within DocScaB has been previously observed in other type I dockerins and results from the near-perfect 2-fold dyad symmetry of the dockerin module (6, 17). In contrast, helix-2 that links the two duplicated segments exhibited a different spatial position when the two complexes were overlaid. Thus, the internal structural symmetry presented by the dockerins enables cohesin recognition through two highly similar protein ligand-binding surfaces, complexes that have a buried surface area of ∼820 Å2 and that are described below.
Hydrophobic interactions play a key role in Coh-DocI15S/N16I and Coh-DocI51S/N52I complex assembly and are displayed in Fig. 3A. The intermolecular interface also includes 10 direct polar interactions, including two salt bridges (Table 4 and Fig. 3B). The number of polar and apolar interactions identified at the complex interface is at least 1.25 times greater than that of other type I Coh-Doc complexes (6, 17, 18). The DocScaB residues at the complex interface are located in both helices 1 and 3 and remain unchanged upon the 180° rotation of the dockerin module on the CohScaC surface, reflecting the internal symmetry of the ScaB dockerin. However, the two dockerin helices provide different contributions for cohesin recognition. Although only the C-terminal region of DocScaB helix-3 interacts with the CohScaC, the entire length of the dockerin helix-1 contacts its protein partner in Coh-DocI51S/N52I and vice versa in Coh-DocI15S/N16I. Henceforth, the helix interacting through its entire length with CohScaC is defined as helix-A, and the helix only making interactions with the cohesin exclusively at one of its termini is termed helix-B (see below and Fig. 4). Cα atoms of the DocScaB helix contacting CohScaC along its entire length (helix-A) are 6–7 Å away from the Cα atoms of the respective complementary contact residues located at the cohesin surface. In contrast, Cα atoms of the dockerin helix only contacting the cohesin at one of its termini (helix-B) are distant from their corresponding binding partners by 5 Å at the region in closer proximity with CohScaC and more than 14 Å from the distal helix region that faces away from the cohesin module.
FIGURE 3.
Cohesin-dockerin interface in the two A. cellulolyticus cohesin-dockerin complexes. A, shows the hydrophobic interactions at the complex interface. B, shows the polar interactions at the complex interface. Residues at the interface are shown as sticks. The symmetry-related dockerins are overlaid and displayed color-ramped. ScaC cohesin is depicted in salmon. In both panels the residue labeling of the dockerins indicates the residue number(s) in equivalent positions on one or the other binding orientation.
TABLE 4.
Main hydrogen-bound contact residues at the cohesin-dockerin interfaces of complexes Coh_DocI15S/N16I and Coh_DocI51S/N52I
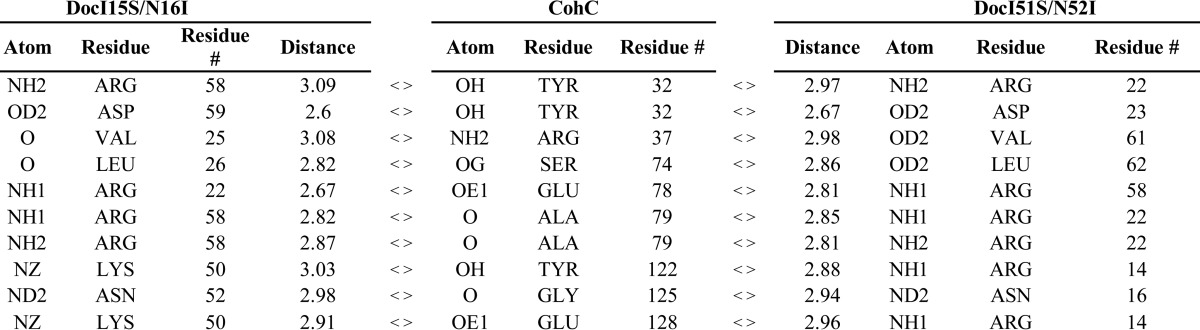
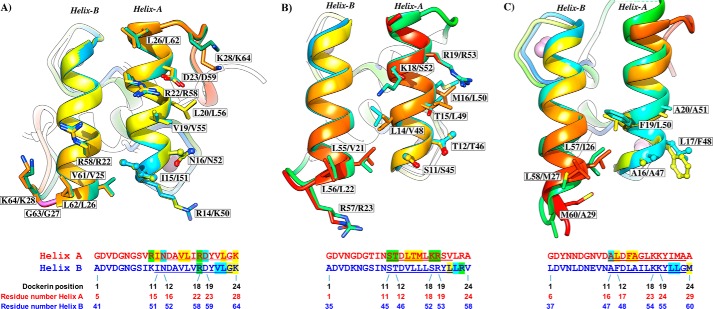
FIGURE 4.
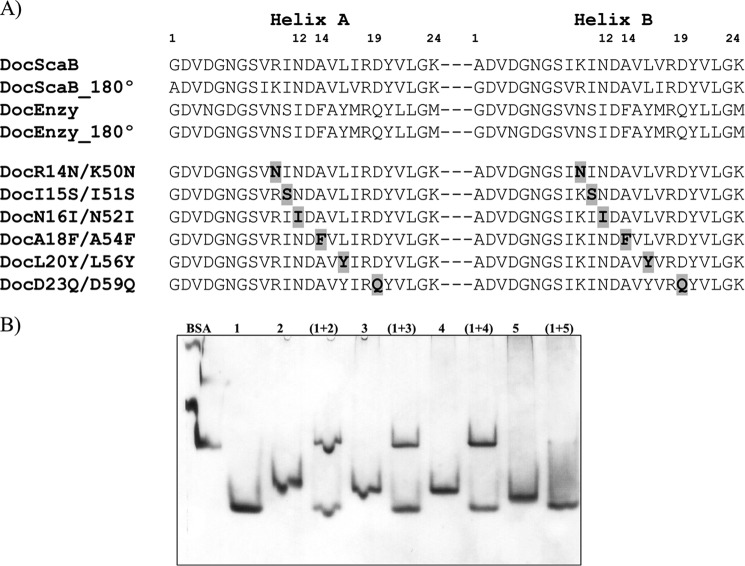
Two identical cohesin-binding faces support the dual binding mode of type I dockerins from A. cellulolyticus (A), C. thermocellum (B), and C. cellulolyticum (C). Dockerins are color-ramped and were overlaid with its 180° rotated alternative binding mode structure. The most important cohesin contact residues are highlighted and displayed as ball-and-stick or sticks. Below the representation of the two cohesin-interacting faces, the primary sequence alignment of the respective dockerin is provided (dockerin position number and residue number indicated below each alignment). Residues in green are involved in polar contacts, yellow in hydrophobic contacts, and blue in both. Helix-A defines the helix that dominates cohesin recognition. Helix-B only contacts the cohesin at one of its termini (see text for details).
The interactions between helix-A of DocScaB and CohScaC are dominated by the Arg-14/Lys-50, Ile-15/Ile-51, and Asn-16/Asn-52 of DocScaB and Thr-76 and Tyr-122 of CohScaC (Fig. 3). Nη1 of Arg-14/Lys-50 contributes an important salt bridge with Oδ1 of Glu-128 located in β-strand 8 of CohScaC. The Ile-15/Ile-51 side chain occupies the hydrophobic pocket formed by Leu-34 and Ala-33. The more distal helix-A Arg-22/Arg-58 and Asp-23/Asp-59 pair contributes an important hydrogen bond network with the CohScaC residues Ala-79 and Tyr-32, whereas the aliphatic side chains of Asp-23/Asp-59 make comprehensive hydrophobic contacts with Val-81. In helix-B, the contacts are dominated by the important salt bridge established between Nη1 of Arg-58/Arg-22 and Oδ1 of Glu-78 of CohScaC. In addition, Leu-62/Leu-26 located at the extremity of helix-B, which is in closer proximity with CohScaC, makes nonpolar contacts with Thr-63, Thr-64, Tyr-65, Ser-74, and Thr-76. Val-61/Val-25 forms van der Waals interactions with the side chain of CohScaC Tyr-118. The carbonyl groups of these two dockerin residues (of each of the rotated modes) also form hydrogen bonds with the side-chain groups of Arg-37 and Ser-74 in CohSacC. Collectively, these observations suggest that DocScaB presents two cohesin-binding interfaces that span through helix-A and -B and are revealed by the 180° rotation of the dockerin module on the 8–3-5–6 face of CohScaC. Dockerin residues at the interface of both structures are conserved, suggesting that affinities would be similar, if not identical (see below). Thus, these structural data strongly support the presumed dual binding mode of DocScaB to CohScaC.
Exploration of the A. cellulolyticus proteome revealed that there are no homologues of DocScaB within the bacterium. However, Blast analysis revealed that the cellulosome of Clostridium clariflavum contains an adaptor scaffoldin in which the C-terminal type I dockerin shares 85% identity with DocScaB (27). The dockerin residues described above, which mediate cohesin recognition, are conserved in the two orthologous proteins suggesting that the two dockerins share identical ligand specificities. In contrast, CohScaC is highly homologous with the other two cohesin modules found in this anchoring scaffoldin. The residues that participate in dockerin recognition are mostly invariant in the three cohesins (data not shown), suggesting that ScaC can accommodate three ScaB proteins via the recognition of the C-terminal dockerin module. In addition, a second A. cellulolyticus protein termed ScaJ, accession number WP_010243813, contains a single N-terminal cohesin module that shares extensive similarity with the ScaC cohesins, primarily at the residues important for protein recognition (13, 19). ScaJ also contains a C-terminal SLH domain, which suggests that, like ScaC, it constitutes an anchoring scaffoldin with the ability to recognize a single ScaB molecule.
Comparison of A. cellulolyticus with C. cellulolyticum and C. thermocellum Type I Coh-Doc Complexes
It was previously observed that the A. cellulolyticus type I DocScaB specifically recognizes type I cohesins of ScaC and will not bind type I cohesins of ScaA or type I cohesins from C. thermocellum or C. cellulolyticum (19). This lack of cross-specificity between Coh-Doc pairs of different species may assist accurate and efficient assembly of a coherent set of enzymes into each cellulosome. The Coh-Doc interactions in A. cellulolyticus, C. thermocellum, and C. cellulolyticum provide insight into the molecular basis for the distinct specificities displayed by these protein module pairs. When compared, A. cellulolyticus, C. cellulolyticum, and C. thermocellum type I Coh-Doc complexes reveal significant structural conservation with an overall r.m.s.d. of 1.5 Å (for both cohesin and dockerin). The location of the interface between the two protomers within the heterodimers is also conserved, despite a lack of cross-specificity between the three complexes (6, 17, 18). Therefore, subtle yet distinct residue-specific changes at the cohesin and dockerin surfaces modulate Coh-Doc specificity (Fig. 4). In contrast to what was previously observed in C. thermocellum and C. cellulolyticum complexes, the residue that precedes helix-A in the A. cellulolyticus dockerin (Arg-14/Lys-50) participates in cohesin recognition through the establishment of two important polar interactions with cohesin residues Tyr-122 and Glu-128. In Clostridia, this dockerin position is usually occupied by an asparagine, in which the side chain is exclusively involved in the coordination of the Ca2+ ions. The position occupied by Tyr-122 in the A. cellulolyticus cohesin is replaced in C. thermocellum and C. cellulolyticum by Ala and Gly residues, respectively, where the side chains would be unable to produce a salt bridge with A. cellulolyticus Arg-14/Lys-50. This major change at the Coh-Doc interface may explain, in part, why A. cellulolyticus DocScaB is unable to interact with Clostridia type I cohesins. Inspection of the Coh-Doc intermolecular interfaces provided further evidence for the species-restricted protein-protein specificity. The Ala-(Leu/Phe)-(Phe/Leu) triplet (dockerin positions 11–12-14) that dominates cohesin recognition by helix-A of the C. cellulolyticum dockerin is accommodated by a hydrophobic platform that is absent in A. cellulolyticus CohScaC. In addition, the bulky Phe/Leu side chains at position 12 in C. cellulolyticum dockerins would cause a steric clash with Tyr-122 of the A. cellulolyticus cohesin, further supporting the lack of cross-species recognition between these protein partners. In addition, the Ser-Thr pair that dominates cohesin recognition in helix-A of C. thermocellum through the provision of a hydrogen binding network, primarily with the side chains of the cohesin residues Asn-37 and Asp-39, will be unable to bind the A. cellulolyticus cohesin modules as the equivalent residues, Ala-33 and Ser-35, do not contribute sufficient hydrogen-binding sites. As stated above, in A. cellulolyticus DocSacB the specificity determinants at position 11 and 12 of helix-A, Ile-15/51 and Asn-+16/52, occupy a hydrophobic pocket and make a single hydrogen bond with CohSacC, respectively. Finally, an arginine located in the loop preceding helix B of the C. thermocellum dockerin (a glycine in the A. cellulolyticus dockerin), which forms a hydrogen bond with the C. thermocellum cohesin, would cause a steric clash with A. cellulolyticus cohesin residues Arg-37 and Tyr-118. Detailed analysis of the other complexes suggests that polar contacts are scarce in the C. cellulolyticum complex, where the interactions are predominantly hydrophobic, and in C. thermocellum the Coh-Doc interface is largely dominated by hydrogen bonds. In contrast, the number of both polar and hydrophobic interactions in the A. cellulolyticus protein complexes are much more extensive, in helix-A and -B, when compared with their Clostridia counterparts. Therefore, although different from type II complexes, where both dockerin helices (A and B) contact the cohesin surface over its entire length, the number of polar and apolar contacts identified in the protein complex formed between DocScaB and CohScaC is intriguingly more comparable with a type II interaction. Because of the higher number of contacts observed in the A. cellulolyticus Coh-Doc interface, the affinity between the two modules is predicted to be significantly higher than that observed in other type I complexes as further discussed below.
Type I Coh-Doc Complexes That Anchor A. cellulolyticus Cellulosomes into the Cell Surface Present a Dual Binding Mode
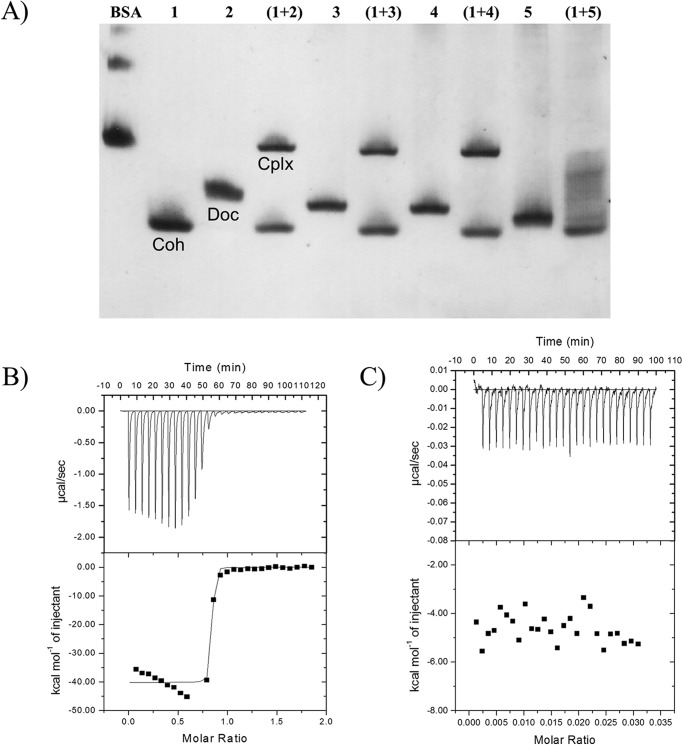
NGE and SPR showed that DocSacB, and the mutant derivatives DocI15S/N16I and DocI51S/N52I bound to CohScaC (Fig. 5A and Table 5) with very similar affinities. In contrast, NGE, SPR, and ITC showed that DocI15S/N16I/I51S/N52I was unable to interact with CohScaC (Fig. 5, A and C, and Table 5) suggesting that DocScaB does, in fact, contain two cohesin-binding interfaces and that complete abrogation of cohesin recognition requires the inactivation of both of these binding faces. Overlaying the structure of DocSacB in protein complexes DocI15S/N16I and DocI51S/N52I indicates that two cohesin molecules would be unable to bind to a single dockerin molecule simultaneously because residues in the cohesin loop extending from Ile-56 to Phe-62 will make steric clashes. Titrations of wild-type, DocI15S/N16I, and DocI51S/N52I DocScaB constructs with the CohScaC, at 25 °C, revealed a 1:1 stoichiometry and ΔH values ranging between −25 and −35 kcal mol−1. Because of the very high affinity (Ka >109 m−1) of DocSacB for CohSacC, an accurate Ka value could not be determined by ITC (see below). These data suggest that substitution of the helix-A Ile/Asn residues for Ser/Ile disrupts cohesin recognition, which is consistent with the structural data showing that CohSacC recognition is dominated by the Ile/Asn pair. DocScaB of A. cellulolyticus is involved in the attachment of the cellulosome to the bacterial cell surface through its interaction with the type I cohesin modules of the ScaC anchoring scaffoldin. This is in contrast to the majority of other type I Coh-Doc interactions, which are usually involved in the binding of enzymes into cellulosomes. It is believed that the dual binding mode displayed by type I Coh-Doc complexes may introduce quaternary flexibility into cellulosomes thus promoting substrate targeting and enzyme synergism. Considering this, the dual binding mode displayed by the A. cellulolyticus type I complex may further contribute to overall cellulosome architecture by reducing steric constraints that are undoubtedly imposed in the assembling of a large number of cellulosomes onto the bacterial cell surface.
FIGURE 5.
ScaB dockerin displays a dual binding mode. A, CohScaC(lane 1), DocScaB (lane 2), DocI15S/N16I (lane 3), DocI51S/N52I (lane 4), and DocI15S/N16I/I51S/N52I (lane 5), with a slight excess of cohesin, were mixed and separated by NGE. Each dockerin was incubated with CohScaC (lane 1) for 1 h, after which complex formation was evaluated. Cohesin, dockerin, and complex bands are indicated as Coh, Doc, and Cplx, respectively, in the 1st 3 lanes of the figure. The interaction of ScaB dockerin (B) or its mutant derivative DocI15S/N16I/I51S/N52I (C) with ScaC cohesin was evaluated through ITC analysis at 35 °C. The upper parts of each result show the raw heats of binding, and the lower parts are the integrated heats after correction for heats of dilution. The curve in B represents the best fit to a single-site model.
TABLE 5.
Identification of the binding of dockerin polypeptides to CohScaC cohesin as evaluated by native gel electrophoresis and isothermal titration calorimetry corroborated with the association and dissociation constants for the binding of DocScaB and mutant derivatives to immobilized CohScaC evaluated by surface plasmon resonance using a BIAcore system
KD was determined as koff/kon. The following symbols are used: +, binding; NB, no binding; ND, experiment not done.
| Dockerin | Native gel | ITC | SPR |
||
|---|---|---|---|---|---|
| kon | koff | KD | |||
| m−1 s−1 | s−1 | m | |||
| DocScaB | + | + | 2.8 × 106 | 3.5 × 10−5 | 1.3 × 10−11 |
| DocI15S/N16I | + | + | 2.1 × 106 | 4.2 × 10−5 | 2.0 × 10−11 |
| DocI51S/N52I | + | + | 2.0 × 106 | 2.9 × 10−5 | 1.4 × 10−11 |
| DocI15S/N16I/I51S/N52I | NB | NB | NB | NB | NB |
| DocR14N/K50N | + | + | 4.6 × 106 | 2.9 × 10−4 | 6.4 × 10−11 |
| DocI15S/I51S | + | + | 5.6 × 105 | 3.1 × 10−4 | 5.6 × 10−10 |
| DocN16I/N52I | + | + | 4.0 × 106 | 5.4 × 10−2 | 1.3 × 10−8 |
| DocA18F/A54F | + | + | 1.2 × 106 | 6.7 × 10−2 | 5.4 × 10−8 |
| DocL20Y/L56Y | + | + | 9.1 × 106 | 2.7 × 10−4 | 3.0 × 10−11 |
| DocD23Q/D59Q | − | + | 1.7 × 106 | 6.4 × 10−2 | 3.8 × 10−8 |
| DocGH5 | + | + | ND | ND | ND |
Structural Determinants of Type I Coh-Doc Specificity in A. cellulolyticus
The A. cellulolyticus cellulosome has evolved two different co-existing type I Coh-Doc specificities as follows: the first one is responsible for assembling dockerin-bearing enzymes onto ScaA, and the second one, described here at the structural level, is responsible for the attachment of cellulosomes and poly-cellulosomes to the cell surface (Fig. 1). These two different type I subclasses do not cross-react (19, 28). To identify the residues that modulate type I Coh-Doc specificity in A. cellulolyticus, a mutagenesis study, based on the crystal structure of DocScaB and the alignment of this dockerin with the primary consensus sequence of type I dockerins of A. cellulolyticus cellulosomal enzymes (∼100 primary sequences of A. cellulolyticus enzyme dockerins were used), was initiated (Fig. 6A). The analysis suggests that there are six main amino acid differences that may explain the observed specificity as follows: R14N/K50N, I15S/I51S, N16I/N52I, A18F/A54F, L20Y/L56Y, and D23Q/D59Q (ScaB dockerin versus the enzyme dockerin that targets ScaA). These amino acid residues are all located in helix-A of DocScaB, as helix-A seems to be remarkably conserved in the two type I dockerin subclasses. To probe the importance of these residues for type I specificity, the DocScaB backbone was altered at the above-mentioned positions to reflect the sequence of enzyme dockerins. Because a dual binding mode is likely to operate in both A. cellulolyticus Coh-Doc type I subclasses, dockerin residues were changed at both helices (Fig. 6A). The affinity of the resulting variants for CohScaC was evaluated initially by NGE. The qualitative data revealed that all six mutated DocScaB derivatives efficiently recognized CohScaC (Fig. 6B). Although NGE may detect Coh-Doc interactions with Ka ≥105 m−1, the data suggest that the changes introduced into helix-A of DocScaB have a marginal effect on the capacity of the dockerin to recognize its cognate cohesin. To quantify the effect of the amino acid changes on the affinity of the different dockerins to CohScaC, Coh-Doc interactions were evaluated by SPR. The data revealed a calculated dissociation constant of the DocScaB-CohScaC interaction of 1.3 × 10−11 m (Table 5; Fig. 7), which is significantly higher than the affinities reported for clostridial type I Coh-Doc interactions that were found to display KD values of >10−9 m. With respect to the DocScaB mutants, a marginal decrease in affinity was observed with DocR14N/K50N when the basic Arg/Lys residues were replaced by Asn, the residue at the first position before helix-A in the enzyme-borne dockerin modules. This may reflect the capacity of Asn to participate in the hydrogen bond network established by Arg/Lys. Similarly, DocL20Y/L56Y displayed an affinity similar to wild-type DocScaB suggesting that the tyrosine residue identified in several enzyme-borne dockerins may contribute to the hydrophobic nature of the interaction. A more pronounced reduction in affinity, ∼100-fold, was observed with DocI15S/I51S, suggesting that nonpolar properties of Ile in helix-A make a significant contribution to the affinity of the interaction. However, ∼1000-fold lower affinity relative to the wild-type DocScaB was observed for dockerins DocN16I/N52I, DocA18F/A54F, and DocD23Q/D59Q. Thus, the important hydrogen bonds contributed by DocScaB Asn-16/Asn-52 are removed by the Ile replacement. In addition, introduction of the bulky side chains (i.e. Phe) within helix-A causes steric clashing with those residues on the dockerin-binding surface of CohScaC. Finally, when the Asp residue located in helix-A is replaced by a Gln, the critical hydrogen bonds established by this residue will be compromised. Overall, the data suggest that residues Asn/Ile, Ala/Phe, and Asp/Gln (ScaB/Enzyme dockerin, respectively) located at dockerin positions 12, 14, and 19 in helix-A (DocScaB residues 883, 885, and 890, respectively) are the most important modulators of type I Coh-Doc specificity in A. cellulolyticus cellulosome.
FIGURE 6.
Residue determinants of type I cohesin-dockerin specificity in A. cellulolyticus. A, identifies the residues that may modulate type I Coh-Doc specificity in A. cellulolyticus (DocScaB) aligned with the primary consensus sequence of a type I enzyme dockerin (Doc_Enzy) of A. cellulolyticus. Key residues located in either helix-A or helix-B, are highlighted in bold, within these sequences. The two cohesin-binding faces were revealed by rotating each dockerin by 180°, demonstrating the highly symmetrical nature of dockerin sequences. The importance of the proposed residues of type I specificity have been probed by producing DocScaB mutant derivatives (DocR14N/K50N to DocD23Q/D59Q) with single amino acid changes at both helices. The changes (highlighted in bold and shaded gray) were made to reflect those of the type I enzyme dockerin (Doc_Enzy). B, shows an example of the affinity of the engineered dockerins DocR14N/K50N (lane 2), DocI15S/I51S (lane 3), DocN16I/N52I (lane 4), and the type I enzyme dockerin, DocGH5, as a negative control (lane 5) for CohScaC (lane 1), evaluated qualitatively by NGE.
FIGURE 7.
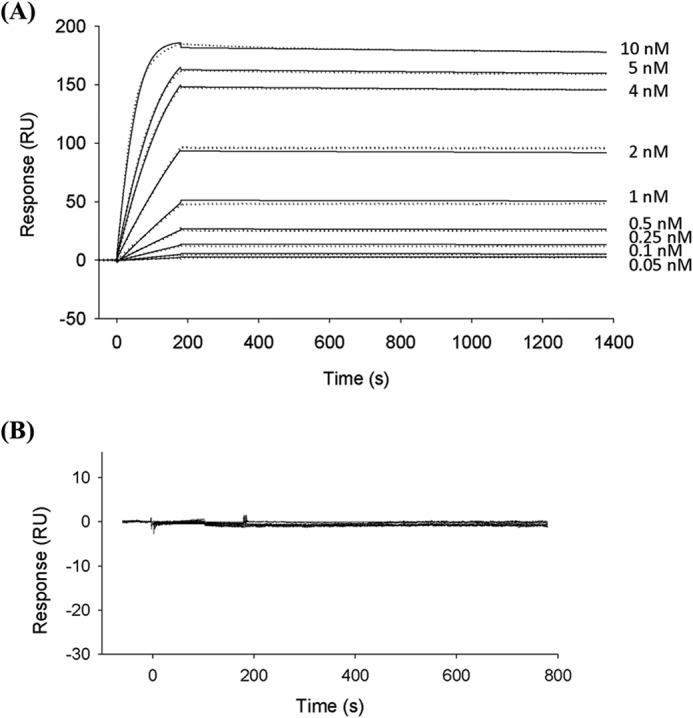
Sensorgrams showing the interactions between immobilized CohScaC to DocScaB (A) and DocI15S/N16I/I51S/N52I (B), monitored by SPR. CohScaC was immobilized on a CM5 sensor chip as described under “Experimental Procedures.” The sensor chip surface was subsequently exposed to different concentrations (shown in A) of wild-type Doc ScaB. The SPR sensorgrams are shown as dotted lines with kinetic fits of binding superimposed with solid lines. B, exemplifies a no binding result obtained with the DocI15S/N16I/I51S/N52I mutant. Both were fitted with a 1:1 Langmuir binding model using BIAevaluation software (Version 2.0.1). The affinity constant was calculated as koff/kon.
Conclusions
In nature, type I Coh-Doc interactions are essential for the assembly of cellulosomal enzymes onto a primary scaffoldin, which in turn attaches to the cell surface via a type II Coh-Doc pair. Generally, type I Coh-Doc interactions are remarkably flexible as exemplified by the capacity of type I dockerins to present a dual binding mode resulting from the incidence of two identical cohesin-binding faces. However, in contrast, cell-surface attachment does not appear to require this increased flexibility, as type II dockerins were shown to display a single binding mode. The structure of DocScaB revealed an internal symmetry that supports the presence of two virtually identical cohesin-binding faces. Thus, in conjunction with previous studies of the C. thermocellum and C. cellulolyticum cellulosome, this report shows that flexibility in cohesin recognition seems to be a general feature of type I dockerin modules, including those that recruit cellulosomes into the cell surface. Although flexibility resulting from a dual binding mode may not be universal to all cellulosomes, in A. cellulolyticus it seems to be essential not only for enzyme incorporation, but also when the multienzyme complexes are targeted to the cell surface. In this context, the three ScaC cohesins are positioned at the N terminus of the scaffoldin without any detectable linker segment between them. The dual mode of binding may therefore provide the conformational plasticity required for sufficient attachment of three ScaB adaptor subunits, together with its complement of enzyme-laden ScaA complexes. All of these components must fit en masse into the spatial confines of the particularly elaborate cellulosome architecture that characterizes the surface of this bacterial species.
Acknowledgments
We thank Drs. Pierpaulo Romano, Juan Sanchez-Weatherby, Teresa Santos Silva, and Catarina Coelho for help with data collection at Diamond Light Source and Cecilia Bonifacio for help with crystallization.
This work was supported in part by Fundacao para a Ciencia e a Tecnologia (Lisbon, Portugal) Grants PTDC/BIA-PRO/103980/2008 and EXPL/BIA-MIC/1176/2012, European Union Seventh Framework Programme FP7 2007-2013 under the WallTraC Project Grant 263916, and BioStruct-X Grant 283570.
The atomic coordinates and structure factors (codes 4uyp and 4uyq) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- Coh-Doc
- cohesin-dockerin
- NGE
- nondenaturing gel electrophoresis
- SPR
- surface plasmon resonance
- ITC
- isothermal titration calorimetry
- r.m.s.d.
- root mean square deviation.
References
- 1. Fontes C. M., Gilbert H. J. (2010) Cellulosomes: highly efficient nanomachines designed to deconstruct plant cell wall complex carbohydrates. Annu. Rev. Biochem. 79, 655–681 [DOI] [PubMed] [Google Scholar]
- 2. Gilbert H. J. (2007) Cellulosomes: microbial nanomachines that display plasticity in quaternary structure: cohesin dockerin recognition. Mol. Microbiol. 63, 1568–1576 [DOI] [PubMed] [Google Scholar]
- 3. Leibovitz E., Béguin P. (1996) A new type of cohesin domain that specifically binds the dockerin domain of the Clostridium thermocellum cellulosome-integrating protein CipA. J. Bacteriol. 178, 3077–3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ding S. Y., Bayer E. A., Steiner D., Shoham Y., Lamed R. (2000) A scaffoldin of the Bacteroides cellulosolvens cellulosome that contains 11 type II cohesins. J. Bacteriol. 182, 4915–4925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bayer E. A., Belaich J. P., Shoham Y., Lamed R. (2004) The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 58, 521–554 [DOI] [PubMed] [Google Scholar]
- 6. Carvalho A. L., Dias F. M., Prates J. A., Nagy T., Gilbert H. J., Davies G. J., Ferreira L. M, Romão M. J., Fontes C. M. (2003) Cellulosome assembly revealed by the crystal structure of the cohesin-dockerin complex. Proc. Natl. Acad. Sci. U.S.A. 100, 13809–13814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Khan A. W. (1980) Cellulolytic enzyme system of Acetivibrio cellulolyticus, a newly isolated anaerobe. Microbiology 121, 499–502 [DOI] [PubMed] [Google Scholar]
- 8. Lamed R., Naimark J., Morgenstern E., Bayer E. A. (1987) Specialized cell surface structures in cellulolytic bacteria. J. Bacteriol. 169, 3792–3800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu Q., Gao W., Ding S. Y., Kenig R., Shoham Y., Bayer E. A., Lamed R. (2003) The cellulosome system of Acetivibrio cellulolyticus includes a novel type of adaptor protein and a cell surface anchoring protein. J. Bacteriol. 185, 4548–4557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xu Q., Barak Y., Kenig R., Shoham Y., Bayer E. A., Lamed R. (2004) A novel Acetivibrio cellulolyticus anchoring scaffoldin that bears divergent cohesins. J. Bacteriol. 186, 5782–5789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ding S. Y., Bayer E. A., Steiner D., Shoham Y., Lamed R. (1999) A novel cellulosomal scaffoldin from Acetivibrio cellulolyticus that contains a family 9 glycosyl hydrolase. J. Bacteriol. 181, 6720–6729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nagy T., Tunnicliffe R. B., Higgins L. D., Walters C., Gilbert H. J., Williamson M. P. (2007) Characterization of a double dockerin from the cellulosome of the anaerobic fungus Piromyces equi. J. Mol. Biol. 373, 612–622 [DOI] [PubMed] [Google Scholar]
- 13. Dassa B., Borovok I., Lamed R., Henrissat B., Coutinho P., Hemme C. L., Huang Y., Zhou J., Bayer E. A. (2012) Genome-wide analysis of Acetivibrio cellulolyticus provides a blueprint of an elaborate cellulosome system. BMC Genomics 13, 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Berg Miller M. E., Antonopoulos D. A., Rincon M. T., Band M., Bari A., Akraiko T., Hernandez A., Thimmapuram J., Henrissat B., Coutinho P. M., Borovok I., Jindou S., Lamed R., Flint H. J., Bayer E. A., White B. A. (2009) Diversity and strain specificity of plant cell wall degrading enzymes revealed by the draft genome of Ruminococcus flavefaciens FD-1. PLoS One 4, e6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miras I., Schaeffer F., Béguin P., Alzari P. M. (2002) Mapping by site-directed mutagenesis of the region responsible for cohesin-dockerin interaction on the surface of the seventh cohesin domain of Clostridium thermocellum CipA. Biochemistry 41, 2115–2119 [DOI] [PubMed] [Google Scholar]
- 16. Schaeffer F., Matuschek M., Guglielmi G., Miras I., Alzari P. M., Béguin P. (2002) Duplicated dockerin subdomains of Clostridium thermocellum endoglucanase CelD bind to a cohesin domain of the scaffolding protein CipA with distinct thermodynamic parameters and a negative cooperativity. Biochemistry 41, 2106–2114 [DOI] [PubMed] [Google Scholar]
- 17. Carvalho A. L., Dias F. M., Nagy T., Prates J. A., Proctor M. R., Smith N., Bayer E. A., Davies G. J., Ferreira L. M., Romão M. J., Fontes C. M., Gilbert H. J. (2007) Evidence for a dual binding mode of dockerin modules to cohesins. Proc. Natl. Acad. Sci. U.S.A. 104, 3089–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pinheiro B. A., Proctor M. R., Martinez-Fleites C., Prates J. A., Money V. A., Davies G. J., Bayer E. A., Fontesm C. M., Fierobe H. P., Gilbert H. J. (2008) The Clostridium cellulolyticum dockerin displays a dual binding mode for its cohesin partner. J. Biol. Chem. 283, 18422–18430 [DOI] [PubMed] [Google Scholar]
- 19. Hamberg Y., Ruimy-Israeli V., Dassa B., Barak Y., Lamed R., Cameron K., Fontes C. M., Bayer E. A., Fried D. B. (2014) Elaborate cellulosome architecture of Acetivibrio cellulolyticus revealed by selective screening of cohesin-dockerin interactions. Peer J. 2, e636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cameron K., Alves V. D., Bule P., Ferreira L. M., Fontes C. M., Najmudin S. (2012) Purification, crystallization and preliminary x-ray characterization of the Acetivibrio cellulolyticus type I cohesin ScaC in complex with the ScaB dockerin. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 68, 1030–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Long F., Vagin A. A., Young P., Murshudov G. N. (2008) BALBES: a molecular-replacement pipeline. Acta Crystallogr. D Biol. Crystallogr. 64, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Krissinel E., Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 23. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A. (2011) REFMAC 5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 25. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Artzi L., Dassa B., Borovok I., Shamshoum M., Lamed R., Bayer E. A. (2014) Cellulosomics of the cellulolytic thermophile Clostridium clariflavum. Biotechnol. Biofuels 7, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haimovitz R., Barak Y., Morag E., Voronov-Goldman M., Shoham Y., Lamed R., Bayer E. A. (2008) Cohesin-dockerin microarray: diverse specificities between two complementary families of interacting protein modules. Proteomics 8, 968–979 [DOI] [PubMed] [Google Scholar]