Abstract
Cytomegalovirus (CMV) is a beta-herpes virus present in a latent form in most people worldwide. In immunosuppressed individuals, CMV can reactivate and cause serious clinical complications, but the effect of the latent state on healthy people remains elusive. We undertook a systems approach to understand the differences between seropositive and negative subjects and measured hundreds of immune system components from blood samples including cytokines and chemokines, immune cell phenotyping, gene expression, ex vivo cell responses to cytokine stimuli and the antibody response to seasonal influenza vaccination. As expected, we found decreased responses to vaccination and an overall down-regulation of immune components in aged individuals regardless of CMV serostatus. In contrast, CMV-infected young adults exhibited an overall up-regulation of immune components including enhanced antibody responses to influenza vaccination, increased CD8+ T cell sensitivity, and elevated levels of circulating IFN-γ compared to uninfected individuals. Experiments with young mice infected with murine CMV also showed significant protection from an influenza virus challenge compared with uninfected animals, although this effect declined with time. These data show that CMV and its murine equivalent can have a beneficial effect on the immune response of young, healthy individuals, which may explain the continued coexistence of CMV and mammals throughout their evolution.
Introduction
Cytomegalovirus (CMV) is a common beta-herpes virus that infects most of the population worldwide. Primary infection often occurs during childhood and induces a strong immune response that, while neutralizing viral spread, does not prevent the virus from persisting in a latent form defined by a reversibly quiescent state in which viral genomes are maintained, but viral gene expression is highly restricted and no virus is produced (1). In immunocompromised individuals CMV can reactivate and cause substantial immunological changes in the host, resulting in serious clinical complications and death (2). In healthy individuals, CMV reactivation can also occur upon differentiation of myeloid cells, a reservoir for latent CMV (3), causing a state of chronic infection where the virus is persistently shed at low levels and for extended periods of time (4) In the immunocompetent host, this state of chronic infection is asymptomatic and is believed to occur intermittently, which appears to account for the substantial changes in the phenotype of T cells observed in the infected host (5) with characteristic shrinkages in naïve cells and increases in the memory T cell pool, with up to 10% of the latter being specific for CMV epitopes (5-9). Because this increase in both CD4+ and CD8+ memory T cells is also generally observed in aged individuals, it has been suggested that CMV promotes early immunological aging (10-13). However, the T cell compartment comprise only a fraction of the immune response and very little is known about the effect of CMV on other immune system components. Furthermore, the effect of these changes on immune function remains controversial (14, 15). For instance, in young humans and mice CMV may improve immune responses to unrelated antigens (16-19) whereas in older human cohorts, a number of association studies suggest that CMV seropositivity is linked to immune dysfunction and chronic inflammatory diseases including immunosenescence, cancer, cardiovascular disease, atherosclerosis, frailty and early mortality (11, 20-31). These observations suggest that the effect of CMV on the immune system may be highly dependent on an individuals’ age.
To study the influence of age and CMV in the immune system in an unbiased fashion, we undertook a systems biology approach. This type of approach enables comprehensive characterization of biological processes taking into consideration the diversity and interaction of the components involved and thus, it is often utilized to study complex systems. We obtained peripheral blood from 91 young and older individuals and measured the antibody responses to seasonal influenza vaccination, as well as hundreds of immune system components including serum cytokines and chemokines, immune cell phenotyping by multiparametric flow cytometry, gene expression and high-throughput analysis of cell function by determining cellular responses to multiple cytokine stimuli. As expected, we found a down-regulation of immune cell function with age including decreased responses to vaccination but no effect of CMV status in the older members of the cohort. In contrast, young CMV-infected adults exhibited an overall up-regulation of immune function including enhanced antibody responses to influenza vaccination, increased CD8+ T cell sensitivity, and elevated levels of circulating IFN-γ compared to uninfected individuals. These differences may be unique to CMV since we did not observe any significant changes in these parameters with Epstein-Barr virus (EBV) seropositivity, another lifelong herpes virus infection. In parallel experiments, mice infected with murine CMV (MCMV) showed improved T cell responses to influenza virus challenge and dramatically reduced influenza virus titers. This effect was IFN-γ-dependent and declined with time. These data indicate that CMV can boost the immune response in younger individuals and thus has features of a mutualistic agent, that is, one that confers benefits on the host (32).
Results
Positive contribution of CMV to immune function in young, but not older adults
To determine the influence of age and CMV in the immune system we undertook a systems analysis of peripheral blood from 91 young and older individuals from the Stanford-Ellison cohorts (Table S1) (33-35). We focused on immunological parameters representing different layers of the immune system (Fig. S1) including the level of soluble cytokines and chemokines, cell subset frequencies and function by measuring the phosphorylation levels of STAT proteins in response to multiple stimulations (Fig. S2), and genome-wide mRNA expression (gene modules, see www.cs.unc.edu/vjojic/fluy2-upd/). A gene module corresponds to a set of co-expressed genes sharing transcriptional regulatory programs (33, 36, 37).
We first determined the changes associated with CMV latency and aging across a total of 236 baseline immune parameters. To do so, we used a classifier to identify features that best separate each the following categories: young CMV-uninfected (yCMV−), young CMV-infected (yCMV+), older CMV-uninfected (oCMV−), and older CMV-infected (oCMV+) individuals. To minimize false positives and avoid overfitting, we conducted cross-validation, a machine learning procedure that allows for variable selection in an unbiased fashion. We identified a number of features that separate with good accuracy all combinations of groups of individuals, with the exception of oCMV− versus oCMV+ (63% model accuracy compared to 60% baseline accuracy, Table S2). This indicates that in older individuals the effect of CMV is negligible, since oCMV− cannot be distinguished from oCMV+ using the immunological parameters studied here. To isolate the contribution of age or CMV in young individuals, we focused on the comparisons of yCMV− vs yCMV+ individuals (CMV effect independent of age) and yCMV− vs oCMV− individuals (age effect independent of CMV). We also analyzed EBV-infected (EBV+) versus uninfected (EBV−) individuals, but the classification model was unable to distinguish EBV+ from EBV− subjects (the accuracy for the logistic regression model is lower that 50%) and multiple regression analysis (adjusted for age and sex) showed that the most significant EBV-related immune feature is only detected at a False Discovery Rate (FDR) of 75% (Q = 0.75) (Fig. S3a), suggesting no significant correlations with the immune biomarkers measured in this study.
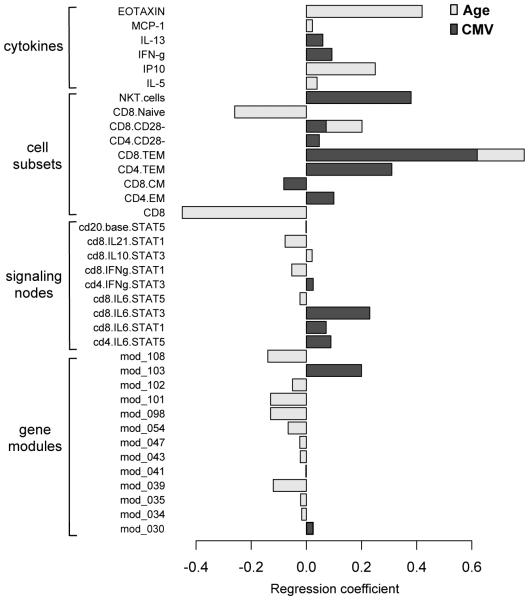
The accuracy of the computational models to distinguish yCMV− vs yCMV+ and yCMV− vs oCMV− were 79% and 91.7%, respectively (Table S2), indicating that aging has a more profound effect on the immune system than latent CMV. Strikingly, the effects of CMV and aging on the immunological variables measured here were almost entirely different (Fig. 1). The only exceptions to this were the CD8+ effector memory (TEM) and CD8+ CD28− cell frequencies, both being positively correlated with age and CMV. This suggests that, in general, the aging process and CMV infection have very different influences on the human immune system. Strikingly, while expression of most parameters (71%, 17/24) decreased with age, the majority (88%, 14/16) increased with CMV seropositivity in young individuals (Fig. 1 and Table S2), indicating an overall down-regulation of the immune response and associated parameters during aging and an up-regulation of several components of the immune system in young subjects with latent CMV.
Figure 1. Different immunological profiles in aging versus CMV seropositivity.
The contribution of age and CMV to immunological and gene expression profiles was estimated by a combination of nuclear norm and the Elastic net methods (see Methods). The magnitude of the regression coefficients used to separate the classes yCMV− and oCMV− (CMV-independent age effect) or yCMV− and yCMV+ (age-independent CMV effect) are shown in light and dark grey bars, respectively. Only two parameters, the frequency of CD8+ CD28− and CD8+ TEM cells, overlapped between these classification tasks. 14/16 (87.5%) of the parameters used to separate the yCMV− from yCMV+ classes were up-regulated in CMV, in contrast, the majority of parameters used to separate the yCMV− from oCMV− classes (16/23, 69.5%) were down-regulated in aging.
In particular, we found an elevation of circulating IL-13 and IFN-γ cytokines, and higher CD8+ pSTAT1 and pSTAT3 responses to IL-6 in CMV+ individuals in the younger cohort compared to the CMV− subjects (Fig. 1). This indicates that the former group has a generally activated immune system involving increased Th1 and Th2 cytokines, and also suggests that CMV improves the CD8+ response to IL-6 in young adults. It is interesting to note that compared to yCMV+ subjects, the oCMV+ individuals are defective in this pathway (Table S2, yCMV+ vs oCMV+), which suggests a degree of adaptation to chronic levels of inflammatory cytokines in older CMV-infected subjects. At the gene expression level, the CMV effect independent of age was an up-regulation of genes associated with immune activation. For example, expression of module 103 (antigen processing and presentation, P < 0.00001 and NK cell mediated cytotoxicity, P < 0.00001) was elevated in yCMV+ (Fig. 1) and this module includes several KIR2 and KIR3 genes as well as GZMH (see http://cs.unc.edu/~vjojic/fluy2-upd/mod103.html), genes typically highly expressed in NK cells and in CD4+ and CD8+ T lacking CD28 (38, 39).. The expression of the gene module 30 was also elevated in yCMV+ (Fig. 1) and this module is composed of HLA-DOA and –DOB genes , which clustered with APOD, LAMC1, MIR600 among other genes. Intriguingly, HLA-DOA and –DOB have been recently shown to confer susceptibility to hepatitis B virus infection and clearance [25528575]. The age effect independent of CMV (yCMV− vs oCMV−) revealed down-regulation of several gene modules (Fig. 1) including those associated with cell cycle (module 34 and 101, P = 0.019), protein synthesis (module 35 and 39, P = 0.0009 and P < 0.0001, respectively) amino-acid metabolism (module 43, P = 0.0023), cell-death (modules 47 and 54, P = 0.0021 and P < 0.0001, respectively), the ubiquitination pathway (module 98, P = 0.0092), HIF1α signaling (module 101, P < 0.0001), LXR/RXR activation, which is involved in cholesterol and lipid metabolism (module 102, P = 0.007) and the metabolism of carbohydrates (module 108, P = 0.00074). Many of these observations are consistent with a series of previous studies in diverse models of aging and in aged humans (40-42).
These results indicate a relatively restricted contribution of CMV to the expression of genes associated with immune activation and a broader contribution of age to critical aspects of cell function, such as cell cycle, protein synthesis and metabolism.
Negative contribution of age and positive contribution of CMV in the serological response to influenza vaccination
While aging typically has a negative effect on vaccination (33, 43), the influence of CMV infection on influenza vaccine responses has been controversial (14, 44, 45). To probe the immune system’s response in a standardized manner, we used seasonal trivalent inactivated influenza vaccination (IIV) and assayed antibody responses using the standard hemagglutinin-inhibition (HAI) assay at day 0 and 28±7 days after vaccination (Fig. S1) in two consecutive years, with 30 young individuals (20-30 years old) and 61 older individuals (60−>89 years old) in year 1; and 25 young and 52 older individuals in year 2. We computed the pre- and post-vaccine geometric mean titer (GMT) for all three strains in the vaccine and calculated a response score as the delta post- minus pre-GMT. As expected, a negative effect of age was observed in the antibody response to IIV (P < 0.0001) (Fig. 2a). Surprisingly, in the yCMV+ group the antibody response was higher than in yCMV− subjects (P = 0.03) (Fig. 2a). These CMV-related differences were not significant in the older cohort (Fig. 2a and b). A similar positive effect of CMV in the young (P = 0.04) but not the older cohort was observed in the 77 individuals who returned one year later during the 2009-2010 influenza season (Fig. 2b). In addition, these findings were validated in an independent cohort of 37 young individuals recruited during the 2010-2011 influenza season (P = 0.017) (Fig. 2c).
Figure 2. Young but not old CMV+ individuals have a better response to influenza vaccination.
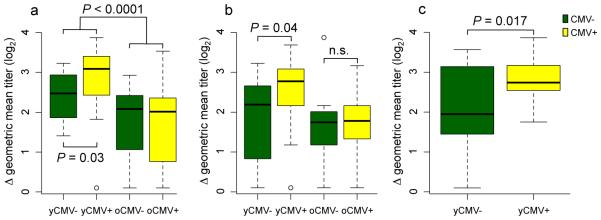
The geometric mean titer (GMT) for all three strains in the vaccine was calculated for each individual in the study and a standardized score (delta (Δ) post-pre GMT) for response was computed as described in Methods (y-axis). A higher response is observed in yCMV+ compared to yCMV− in the first (a) and second (b) year, as well as in an independent validation study conducted during the 2010-2011 influenza season (c). No significant differences were observed between oCMV− and oCMV+. Green bars = CMV-, yellow bars = CMV+. The age ranges for young and older individuals were 20-30 and 60->89 years, respectively (a); 22-32 (young) and 62->89 (older) years (b) and 19-44 years old (c).
These results demonstrate that in young individuals CMV infection may be beneficial since it improves the serological response to influenza vaccination and possibly other vaccines and infection.
Single nucleotide polymorphisms associated with CMV-related alteration in the CD4+ CD28− T cell pool
The alterations in the frequency of immune cell subsets, cytokines and other immune measurements observed in CMV+ subjects may contribute to improved response to vaccination. Indeed, yCMV+ subjects exhibited increased levels of IFN-γ, which was shown to mediate cross-protection in mice (16) (and see below). Notably, the CD4+ T cell response to CMV involves an amplification of cells lacking the CD28 receptor, which exhibit antigen-primed phenotypes and express cytolytic molecules including granzyme B and perforin. These cells emerge after cessation of the viral load; produce huge levels of IFN-γ and are observed only in CMV-infected persons (46). Thus, the variation in the CD4+ CD28− cell subset may be important for the immune status of the individuals in our study and likely contribute to the variation in other immune responses. To find genetic variations that may confer susceptibility to important immunological alterations in CMV+ subjects, we employed a single nucleotide polymorphism (SNP) genotyping approach (ImmunoChip) which analyzes ~200,000 immune-related SNPs (47). We restricted our analyses to SNPs with minor allele frequency greater than 5% (116,405 SNPs) (see Methods) and focused on the frequency of CD4+ CD28− cells, a hallmark of CMV infection (48) and the level of which is not significantly affected by age. Also, these cells often arise as a result of selective clonal expansions of a CMV-specific T cell response (46, 49) and in our data, they showed the lowest p-value for comparison of variance between CMV− and CMV+ individuals (P = 1.6 × 10−7, by one-tailed F-test). We identified 35 SNPs (on chromosomes 6, 9, 12, 14, 15, 17 and 19) associated with the frequency of CD4+ CD28− cells, at a false discovery rate (FDR) lower than 5% (P < 5 × 10−6, FDR Q < 0.05) (Fig. S4). Interestingly, one of the three SNPs on chromosome 6 (rs7744001 (6p21.32)) is located less than 3.5kb from HLA-DQB1, and 22kb and 140kb from HLA-DQA1/DRA, HLA-DRB5 and HLA-DRB1 (Fig. S5), which is in agreement with two recent studies showing multiple genomic loci at 6p21 in the HLA region associated with the total lymphocyte count (50), and with the levels of CD8+ CD28− cells (51). A cluster of 27 SNPs spanning a region in chromosome 9 from position 122,705,118 to 122,748,094 is located in the vicinity and within TRAF1 and 55kb centromeric from C5 (Fig. S6), both of which have been found to be associated with rheumatoid arthritis, where an elevated frequency of CD4+ CD28− T cells is a hallmark. These results indicate that polymorphisms in important immune-related genes could be responsible for the variation observed in the CD4+ T cell response to CMV.
Improved response to influenza challenge in mice previously infected with MCMV is IFN-γ dependent and declines with time
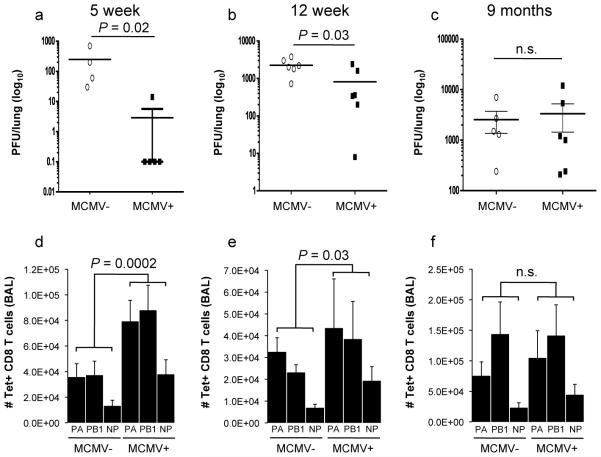
To determine whether the specific phenomenon observed in our human cohorts could be modeled in mice, we tested early and established latency with MCMV (5 and 12 weeks of MCMV infection, respectively) versus longstanding latency (9 months of MCMV infection) in C57BL/6 mice and then challenged them with the influenza A virus (IAV) strain ×31. Both in early and established latency, we observed better protection against influenza in the MCMV-infected (IAV+ MCMV+) versus the mock-infected (IAV+ MCMV−) mice at day 7 after IAV challenge as seen by the reduced influenza viral titers in the lungs (Fig. 3a and b). In contrast, in older mice exposed to long-standing MCMV infection these differences were lost (Fig. 3c). Consistent with this observation, the influenza-specific CD8+ T cell responses against the three major influenza epitopes (DbPA224--233 (PA), KbPB1703-711 (PB1), and DbNP366-374 (NP)) in BAL cells were higher in animals with early and established latency, but not after long-standing latency compared with the mock-infected group (Fig. 3d-f). Our previous results showing increased circulating levels of IFN-γ as well as better influenza vaccine responses in yCMV+ subjects, suggested that the observed CMV-mediated cross-protection might be mediated by IFN-γ. To test this, we monitored the influenza viral titer in the lungs and also the number of influenza-specific CD8+ T cells in IFN-γ knock-out (KO) or control mice challenged with IAV at 5 weeks after MCMV infection, as described earlier. Significantly higher viral titers and lower influenza-specific responses were observed in the IFN-γ KO compared to control mice (Fig 4a and b), demonstrating that IFN-γ is essential for the MCMV-induced cross-protection against influenza in young mice during early MCMV latency and probably also contributes to this effect directly through its antiviral activity.
Figure 3. Reduced viral titer and enhanced IAV-specific CD8+ T cell responses in early and established but not long-standing MCMV latency.
Groups of C57BL/6 mice were mock infected or infected with 4×104 PFU MCMV Smith strain i.p. and challenged with 106 EID50 of IAV ×31 i.n. 5 weeks (early latency) (a and d), 12 weeks (established latency) (b and e) or 9 months (long-standing latency) later (c and f). Seven days after IAV infection, influenza viral titer was determined (upper panel) and IAV-specific T cells were enumerated from the BAL of IAV+ MCMV− (MCMV−) or IAV+ MCMV+ (MCMV+) mice by tetramer staining for NP, PA and PB1-specific responses. Data are representative of three independent experiments with 4-6 mice per group in each experiment. Significance was determined by t-test for viral titer and using the Fisher’s combined probability test for comparison of specific T cell responses.
Figure 4. The effect of MCMV on cross-protection against influenza is IFN-γ-mediated.
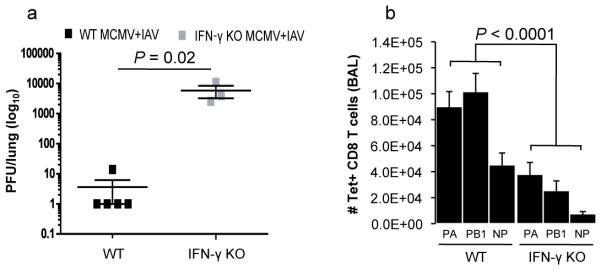
Groups of C57BL/6 mice or IFN-γ-deficient mice (on the C57BL/6 background) were mock infected or infected with 4×104 PFU MCMV Smith strain i.p. and challenged 5-6 weeks later with 106 EID50 of IAV x31 i.n. (a) IAV lung viral titers from control (wild-type, WT) or IFN-γ-deficient co-infected (IAV+ MCMV+) mice were determined at day 7 after IAV infection in early MCMV latency (>5 weeks). (b) IAV-specific CD8+ T cells were enumerated from the BAL of WT and IFN-γ-deficient MCMV and IAV co-infected mice by tetramer staining for NP, PA and PB1-specific responses, 7 days after IAV infection. Significance was determined by t-test. Data are representative of two independent experiments with 3-5 mice per group. Significance was determined by t-test for viral titer and using the Fisher’s combined probability test for comparison of specific T cell responses.
Discussion
In this study we identify major changes in the immune system of healthy individuals with age and CMV infection. Aging had a predominant and negative contribution on most immune parameters including the serological response to influenza vaccination, and CMV a positive contribution. This effect was not observed for another widespread member of the herpesviridae family, EBV. This observation contrasts with previous findings in mice showing that both murine gammaherpesvirus 68 and murine CMV, which are genetically similar to human EBV and CMV, can induce cross-protection against unrelated pathogens (16) but is consistent with studies in humans and monkeys showing that CMV induces much larger changes in immune cells than EBV, including the chronic activation of effector T cells (52). More specifically, since we show here that cross-protection in mice is IFN-γ-dependent, it is possible that these differences are due to the high frequencies of IFN-γ-producing CD4+ CD28− cells observed in persistent CMV but not EBV infection (53). Furthermore, increased percentages of circulating CD8+ CD45RA+ CD27− T cells, which also produce high levels of IFN-γ have been detected in CMV carriers but not after EBV or varicella-zoster virus infection nor after vaccination with the MMR (measles-mumps-rubella) vaccine (54). It is plausible that the differences between the CMV and EBV-associated changes in immune cells’ phenotype and function are due to the site of virus latency. For example, CMV reactivation occurs upon differentiation of myeloid precursors with systemic viral shedding and seemly far reaching effects, whereas latent EBV is found in differentiated memory B cells (55) with reactivation apparently occurring only locally upon recirculation of EBV-infected memory B cells into mucosa-associated lymphoid tissue (56). More studies are needed to address this possibility.
Young CMV+ subjects exhibited an increase in the circulating levels of Th1 and Th2 cytokines as well as a stronger CD8+ pSTAT1 and pSTAT3 responses to IL-6 compared to CMV− subjects. Interestingly, older CMV+ subjects were defective in this pathway. It is possible that the hyper-responsive T cells observed in young CMV+’s lose the ability to respond to further stimuli in the older CMV+ individuals, possibly the result of longterm in vivo exposure to inflammatory mediators and cell desensitization. In support of this hypothesis, CMV infection was found to induce high levels of IL-6 (57), which may affect T cell responses to cytokine stimuli in vivo in a manner dependent on the duration of infection. This has an implications for the immune response to infection and for vaccination strategies in humans of different ages, for example in prime-boost regimens where the transient elevation of inflammatory mediators and their receptors is required for vaccine efficacy (58).
At the gene level, aging was associated with the reduced expression of genes participating in critical aspects of cell function, such as cell cycle, protein synthesis, metabolism, autophagy and the stress response, which is consistent with a series of previous studies in diverse models of aging and in aged humans (40-42). In contrast, the higher expression of module 103 in yCMV+ versus yCMV− could be attributed to the known CMV-associated immune activation observed in infected individuals [20632887]. In agreement with the negative contribution of age and positive contribution of CMV to the immune components studied here, vaccine responses were reduced in older subjects and improved in young CMV-infected subjects. Whereas vaccine responses are known to be reduced in aging populations, the effect of CMV has been controversial, with some studies in older adults showing a negative effect of CMV while others showed no difference in CMV+ vs CMV− subjects (14, 44, 45). Of note, in young individuals, a negative association between the CMV antibody titer and the serological response to the influenza vaccine was found in one study; however this effect was weak (R = 0.16) and observed only for one out of four strains of the virus (60). In contrast, our analysis takes into account the response to all influenza strains in the vaccine for a given year and makes use of longitudinal and validation cohorts as well. Thus, our results indicate that CMV has no apparent effect on the serological response to influenza vaccination in older individuals and is consistent with a recent report analyzing more than 700 older subjects in long-term care facilities (44). Also in our studies CMV clearly boosts the immune responses of younger individuals, since yCMV+ subjects exhibited elevated serum levels of IFN-γ, stronger CD8+ T cell responses to cytokine stimuli and elevated antibody responses to the influenza vaccine. Consistent with these results, mice infected with MCMV showed a greater response to influenza challenge that is IFN-γ dependent and wanes with time in infected animals, much as older humans show no benefit. These results are consistent with longstanding experiments termed “heterologous immunity” in mice where infection with one pathogen can enhance the response to another (16, 61) and also has parallels with human work in which vaccination strategies directed against a specific pathogen has been shown to decrease mortality to different infectious diseases (62-64)
Other mechanisms can also be implicated in the CMV-mediated cross-protection. For example, Welsh and colleagues showed a degree of cross-reactivity of CD8+ T cell epitopes between influenza and CMV in mice and between influenza and EBV in humans (61). Furthermore, recently Su et al. (2013) have proposed that T cell cross-reactivity could be a factor based on the observation that healthy adults can have large numbers of memory phenotype CD4+ T cells specific for viral epitopes to which those individuals had never been exposed (65). However, we note that heterologous cross-reactivity has been directly examined for HCMV-specific responses (9) and was found to be rare, arguing against this mechanism in this situation. In addition, in this study, we did not find influenza-specific cells in the lung during the early stages of MCMV infection when IFN-γ-producing MCMV-specific cells were present suggesting that cross-protection in MCMV+ mice is not due to cross-reactivity in our system. Nonetheless, there may be other T cell epitopes or antibodies that are cross-reactive, which were not assessed here and that could contribute to the protective response.
Alternatively, the presence of high levels of circulating IFN-γ in young humans and mice infected with CMV strongly suggests that a by-stander effect is one mechanism of crossprotection, as the elimination of this gene shows that that this cytokine is essential for the protective effect. Also, in agreement with our observations in both humans and mice, in one study CMV-induced cross-protection to a bacterial infection in young mice that is lost in older animals (17).
In conclusion, these data demonstrate that the effects of CMV infection and aging on the immune system are almost entirely independent of each other, and offer no support for the hypothesis that this virus accelerates immunosenescence. In addition, and quite unexpectedly we found that CMV infection enhances the immune responses of younger adults, as it also in an inbred mouse model. The fact that it did not enhance the responses of older adults indicates that this effect requires a robust immune system, which is interesting in that it largely confines the benefits of infection to those of child-bearing age. Finally, while CMV is clearly a pathogen for immunodeficient individuals and some infants, the data presented here indicate that it is beneficial to a great many more people.
Materials and Methods
Study design, subjects and sample collection
Ninety-one healthy donors (ages 20 to >89) were enrolled in an influenza vaccine study at the Stanford-LPCH Vaccine Program during the fall of 2008, of which eighty-nine completed the study (33). The validation study consisted of the 77 individuals who returned during the fall of 2009 and an additional independent cohort of 37 individuals vaccinated in another study during the 2010 and 2011 influenza seasons. Given this sample size and 6 related tasks of sizes corresponding to classification tasks between 4 age/cmv combinations, an effect size exceeding 0.5 is detected with probability 73.03%. We synthesized data with randomly distributed weights across 6 tasks such that rank of weight matrix is 2. We generated 100 such synthetic datasets. We run our method 100 times on these datasets. We computed power of our method to detect an effect in excess of 0.5. One outlier for cytokine data was removed from the analysis. Since all the individuals were vaccinated, no randomization or blinding was done for this study. The protocol for this study was approved by the Institutional Review Board of the Research Compliance Office at Stanford University. Informed consent was obtained from all subjects. All individuals were ambulatory and generally healthy as determined by clinical assessment. Volunteers had no acute systemic or serious concurrent illness, no history of immunodeficiency, nor any known or suspected impairment of immunologic function, including clinically observed liver disease, diabetes mellitus treated with insulin, moderate to severe renal disease, blood pressure >150/95 at screening, chronic hepatitis B or C, recent or current use of immunosuppressive medication. In addition, none of the volunteers were recipients or donors of blood or blood products within the past 6 months and 6 weeks respectively nor showed any signs of febrile illness on day of enrollment and baseline blood draw. Peripheral blood samples were obtained at day 0 (pre-vaccine), and 28±7 days after receiving a single intramuscular dose of trivalent seasonal influenza vaccine Fluzone (Sanofi pasteur). Each dose of the vaccine contained 15 µg HA each of H1N1, H3N2 and B strains of the virus. Whole blood was used for gene expression analysis (below). Peripheral blood mononuclear cells (PBMC) were obtained by density gradient centrifugation (Ficoll-Paque) and frozen at −80°C for 24-48 hrs before transferring to LN2. Serum was separated by centrifugation of clotted blood, and stored at −80°C before use. Whole blood, PBMC or serum from the first visit (baseline - day 0) was processed and used for determination of gene expression, leukocyte subset frequency, signaling responses to stimulation, serum cytokine and chemokine levels, and CMV and EBV serostatus. Serum samples from day 0 and day ~28 were used for HAI titer determination.
Determination of CMV and EBV seropositivity
Determination of CMV and EBV seropositivity was conducted by ELISA using the cytomegalovirus (CMV) IgG ELISA kit (Calbiotech, cat. # CM027G). Serum samples were thawed at room temperature and dilutions were prepared according to the manufacturer’s recommendations. Calculation of results was done based on the controls provided by the vendor. Six individuals could not be classified because they exhibited an antibody index between 0.9 and 1.1 (“borderline positive” according to the manufacturer). The category (seropositive versus seronegative) for these unclassified individuals was imputed using the “Impute” package (R Bioconductor), which performs nearest neighbor averaging based on the entire cohort’s immune measurements and gene expression values.
Whole blood microarray analysis of gene expression
Total RNA was extracted from PAXgene blood RNA tubes (PreAnalytiX) using the QIAcube automation RNA extraction procedure according to the manufacturer’s protocol (Qiagene). The amount of total RNA, and A260/A280 and A260/A230 nm ratios were assessed using the NanoDrop 1000 (Thermo Fisher Scientific). RNA integrity was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies). For each sample, 750 ng of total RNA were hybridized to Beadchips (HumanHT-12v3 Expression Bead Chip, Illumina) that contains 48,771 probes for around 25,000 annotated genes. The hybridized Beadchips were scanned on an Illumina BeadScan confocal scanner and analyzed by Illumina's GenomeStudio software, version 2.0. After checking the quality of each individual array, the Feature Extraction Files were imported into R Bioconductor and analyzed using the Beadarray package for probe filtering, quantile normalization, replicate probe summarization and log2 transformation. The original microarray probelevel data files were entered into the GEO repository under accession number GSE41080.
Leukocyte subset frequency determination
PBMC were thawed in warm media, washed twice and stained with three separate antihuman antibody cocktails containing: (1) anti-CD3 AmCyan, CD4 Pacific Blue, CD8 APCH7, CD28 APC; (2) CD3 AmCyan, CD4 Pacific Blue, CD8 APCH7, CD27 PE, CD45RA PE-Cy5; (3) CD3 AmCyan, CD19 Alexa Fluor700, CD56 PE, CD33 PE-Cy7, TCR APC, all reagents from BD Biosciences. Additional information for these antibodies can be found on ImmPort (https://immport.niaid.nih.gov/) under accession number SDY212. Incubation with antibodies was performed for 40 min at 4ºC. Cells were washed with FACS buffer (PBS supplemented with 2% FBS and 0.1% Na Azide), and resuspended in 200 µL FACS buffer. Data was collected using DIVA software in an LRSII instrument (BD Biosciences). Analysis was performed using FlowJo 8.8.6 by gating on live cells based on forward vs side scatter profiles, then using double gating for singlet discrimination, followed by cell subset-specific gating.
Phosphorylation of intracellular proteins by phosphoflow analysis
Cells were thawed in warm media and rested at 37ºC in RPMI with 10% FBS. Cells were then distributed in 96-deep well blocks (2 ml) and stimulated with IFN-γ, IL-2, IL-6, IL-7, IL-10 or IL-21 at 50 ng/ml or with 104 U/ml IFN-α for 15 minutes. After stimulation, cells were immediately fixed with 1.5% PFA for 10 min at room temperature, washed with an excess of plain PBS and permeabilized with 95% ice-cold methanol for 20 min on ice. Different stimulus conditions were barcoded using a 3×3 matrix with Pacific Orange and Alexa Fluor 750 (Invitrogen Corp) at 0.03 and 0.04 µg/ml for low and 0.2 and 0.3 µg/ml for high staining, respectively. Incubation with barcoding dyes was performed at 4ºC for 30 min. After several washes with FACS buffer, stimulated and barcoded cells were pooled into single tubes and stained for 30 min at 4ºC with an antibody cocktail containing anti-pSTAT1 Alexa Fluor 488, pSTAT3 Alexa Fluor 647, pSTAT5 PE, CD3 Pacific Blue, CD4 PerCP-Cy5.5, CD20 PerCP-Cy5.5 and CD33 PE-Cy7 (all from BD Phosflow). Additional information for these antibodies can be found on ImmPort (https://immport.niaid.nih.gov/) under accession number SDY212. After washing, cells were resuspended in FACS buffer and acquisition was performed on an LSRII instrument (BD Biosciences). Data were collected using DIVA software. Data analysis was performed using FlowJo 8.8.6. by gating on live cells, then using double gating for singlet discrimination, followed by cell subset specific gating. Phosphorylation of STAT1, 3, and 5 proteins in B cells, CD4(+) or CD4(+) CD3(+) T cells or monocytes was analyzed by deconvolution of stimuli-specific gating. Baseline levels and fold increase between stimulated and unstimulated conditions were calculated using the 90th percentile fluorescence intensity of the pSTAT1, 3, or 5 signals.
Phosphorylation of Akt and PLC-γ was assessed in B cells by cross-linking of the B cell receptor. After resting PBMC samples at 37°C (as conducted for cytokine stimulations), cells were distributed in V-bottom 96-well plates at 0.5×106 cells per well, and incubated for 4 min at 37ºC in CO2 incubator with anti-IgM and anti-IgG both at 10 µg/ml (BD Biosciences) and 3% H2O2 for phosphatase inhibition. Cells were then fixed with 1.5% PFA for 10 min at room temperature. After washing twice with plain PBS, cells were permeabilized by 20 min incubation in 95% ice-cold methanol. Cells were then washed with FACS buffer and stained with an antibody cocktail containing: CD3 Pacific Blue, CD20 PerCp Cy5.5, CD27 PE Cy7, PLGγ2 (BD Biosciences) and pAkt-S473 Alexa Fluor 488 (Cell Signaling Technologies, Danvers MA). After 30 min incubation at 4ºC, cells were washed in FACS buffer and analyzed by flow cytometry (as for cytokine stimulation). Median fluorescence intensity was recorded and used for the calculation of baseline levels of phosphorylated proteins and fold increase after BCR stimulation. Additional information for all the antibodies used in this study can be found on ImmPort (https://immport.niaid.nih.gov/) under accession number SDY212.
Serum cytokine levels determination
Cytokines were measured using a Luminex system (Luminex Corp). 50-plex kits were purchased from Millipore and used according to manufacturer’s recommendations with modifications as described below. Briefly, serum samples were mixed with antibodylinked polystyrene beads on 96-well filter plates and incubated at room temperature for 2 h followed by overnight incubation at 4ºC. Plates were then vacuum filtered and washed twice prior to 2 h incubation with biotinylated detection antibody. Samples were filtered as above, washed twice and incubated with streptavidin-PE for 40 min, then filtered and washed twice again before resuspending in reading buffer. Each sample was measured in duplicate. Plates were read using a Luminex LabMap200 instrument with a lower bound of 100 beads per sample per measured cytokine. The Luminex LabMap200 outputs the fluorescence intensity of each bead measured for a given cytokine in a sample. For each well, we considered the median fluorescence intensity (MFI) of all beads measured for a given cytokine and averaged the MFI of the two replicates. Values were normalized to a control sample ran in each of the plates.
Hemagglutination inhibition assay
The HAI assay was performed on sera from day 0 and day 28 using a standard technique (66); serially diluted 25-µl aliquots of serum samples in PBS were mixed with 25-µl aliquots of virus matching the vaccine strain composition for that years, corresponding to 4 HA units, in V-bottom 96-well plates (Nunc) and incubated for 30 min at room temperature. At the end of the incubation, 50 µl of 0.5% chicken red blood cells was added and incubated for a minimum of 45 min before reading for HAI activity. The HAI titer of a given sample was defined as the reciprocal of the last serum dilution with no HA activity. A titer of 5 was assigned to all samples in which the first dilution (1:10) was negative. The geometric mean titer (GMT) to all three strains in the vaccine was computed for each individual. Wilcoxon rank-sum test was used to compare responses across age groups. To estimate vaccine response in young and older CMV− and CMV+, post-vaccination GMT was subtracted from pre-vaccination GMT (delta post-pre) and Wilcoxon rank-sum test was used to compare vaccine responses between age and CMV infection categories.
Mice and virus infections
Female 5- to 6-week-old control C57BL/6 and IFN-γ-knockout mice were obtained from Jackson laboratories. All mice were cared for under specific pathogen-free conditions in an approved animal facility at St Jude Children’s Research Hospital (SJCRH). All animal work was reviewed and approved by the appropriate institutional animal care and use committee at SJCRH (protocol #098) following guidelines established by the Institute of Laboratory Animal Resources and approved by the Governing Board of the U.S. National Research Council. The mouse-adapted influenza A virus HKx31, was grown in the allantoic fluid of 10-day-old embryonated chicken eggs (SPAFAS). The MCMV Smith strain (ATCC) was grown in mouse embryonic fibroblasts and passaged through BALB/c mice, where infectious virus was extracted from salivary glands at day 12 after infection. For MCMV virus infections, mice were infected intraperitoneally with 4×104 PFU of MCMV. For co-infections, at the indicated time after MCMV infection, avertin (2,2,2-tribromoethanol)-anesthesized animals were challenged intranasally with 1×106 EID50 of HKx31. Mice were considered in early latency at >5 weeks, established latency at 12 weeks, and long-standing latency at 9 months after MCMV infection.
Tissue Sampling
BAL fluid was recovered from infected animals challenged with 1×106 EID50 of HKx31 at the indicated time points. BAL samples were obtained by intra-tracheal HBSS wash in individual mice as described previously (67). Cells in the BAL were collected by centrifugation following standard procedures. For quantification of influenza in infected lungs, snap-frozen lungs were stored at −80°C until further processing.
Tetramer and Phenotypic Staining of CD8 T Cells
Influenza A peptides DbPA224-233 (SSLENFRAYV), KbPB1703-711 (SSYRRPVGI), and NP366–374 (ASNENMETM), and MCMV peptides M45985-993 (HGIRNASFI), m139419-426 (TVYGFCLL) and M38316-323 (SSPPMFRV) were synthesized by the Hartwell center, SJCRH. Class I monomers (H-2Db and H-2Kb MHC class I glycoprotein complexed with the influenza A virus (PA, PB1, NP) were synthesized through a collaboration with the Trudeau Institute and multimerized at SJCRH. Staining was done as described previously (68).
Quantification of IAV in infected lung tissue
Tissues were disrupted by chopping with scissors, homogenized and centrifuged at 10,000 rpm for 15 minutes. Lung homogenates were titered by plaque assay on Madin-Darby canine kidney (MDCK) cells. Near confluent 25-cm2 monolayers of MDCK cells were infected with 1 ml of homogenate or dilution of homogenate (in general, six tenfold dilutions of lungs were tested) for 1 h at 37°C. Cells were washed with PBS, 3 ml of MEM containing 1 mg/ml l-1-tosylamido-2-phenylethyl chloromethyl ketone-treated trypsin (Worthington Biochemical) 0.9% agarose was added, and cultures were incubated at 37°C with 5% CO2 for 72 h. Plaques were visualized with crystal violet.
Single-nucleotide polymorphism assay
All samples were genotyped using the custom designed ImmunoChip array (Illumina). The ImmunoChip array is focused on GWAS reported risk loci for immune-mediated diseases and includes 186 risk loci covered by 196,524 SNPs. Genotyping arrays were processed using 400ng of genomic DNA according to the manufactures protocols. Genotype calls were generated using the Gentrain2 Algorithm implemented in Illumina GenomeStudio software.
Statistical analysis of experiments conducted in mice
Data were analyzed using Prism 5.0 software (GraphPad). Experiments were repeated two to three times as indicated. We tested for homo vs heteroscedasticy (equal vs unequal variance) of each group of mice (CMV vs mock-infected) by the Breusch-Pagan test using the R package “lmtest” (http://cran.at.r-project.org/web/packages/lmtest/lmtest.pdf) and found that the groups had equal variance. The data presenting the differences between the groups were assessed using two-tailed unpaired Student t tests or by twoway ANOVA with Bonferroni post hoc settings. P < 0.05 indicates that the value of the test sample was significantly different from that of relevant controls. The Fisher’s combined probability test (69) was used for analysis of influenza-specific CD8+ T cell responses. To ensure reproducibility of our findings, we conducted three independent experiments with 4-6 mice that were randomized in each group for each experiment for the early versus long-standing latency experiments; and two independent experiments with 3-5 mice that were randomized in each group for the IFN-γ-KO versus wild-type mice experiments. No blinding was done for these experiments.
Supplementary Material
Acknowledgments
We thank all our volunteers for agreeing to participate in these studies. We also thank Garry Nolan for invaluable help with the phosphoflow assays, Yael Rosenberg-Hasson and Iris Herschmann for their expertise in cytokine level determination, and the Stanford-LPCH Vaccine Program staff: Sally Mackey MS, Sue Swope RN, Cynthia Walsh RN, Nancy Mastman RN, Ashima Goel, Kyrsten Spann, Thu Quan and Michele Ugur who enthusiastically enrolled subjects into the study and obtained blood samples from the participants. Funding: This work has been supported by grants from the Ellison Medical Foundation (AG-SS-1788), the Howard Hughes Medical Institute, and the National Institutes of Health (U19s AI057240 and AI090019 to M.M.D.). D.F. was supported by a fellowship from the Stanford Center on Longevity. C.J.L.A. as a Howard Hughes Medical Institute Medical Research Fellow. The study was also supported by an NIH/NCRR CTSA award (UL1 RR025744 to P.G.T.) and by a DP3 grant (DK085678) to S.O-G and P.C.
Footnotes
Author Contributions: D.F. performed pFlow experiments, CMV assays, antibody titer studies, and conducted data analysis; S.S. conducted infection and challenge experiments in mice; C.J.L.A. contributed to CMV assays and data analysis; H.T.M. coordinated gene expression, luminex and phenotyping experiments; S.O-G and P.C. provided support and assistance, and conducted SNP genotyping assays; D.F., V.J., B.K. and S.S-O. analyzed the data; C.L.D. coordinated, organized and conducted the clinical studies and contributed to study design; P.G.T. designed and supervised infection and challenge experiments in mice; D.F. and M.M.D. wrote the manuscript. All authors contributed to editing the manuscript.
Competing interests: All authors declare no conflicts of interest.
Data and materials availability: The original microarray probe-level data files were entered into the gene expression omnibus (GEO) repository under accession number GSE41080. The computer source code can be found at https://github.com/vjojic/CMVAge. Additional information for the materials used in this study found on ImmPort (https://immport.niaid.nih.gov/) under accession number SDY212.
References
- 1.Mocarski ES, Shenk T, Griffiths P, Pass RF. In: Fields Virology. Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, editors. Lippincott Williams & Wilkins; Philadelphia: 2013. pp. 1960–2014. [Google Scholar]
- 2.Richman DD, Whitley RJ, Hayden FG. In: Clinical Virology. Griffiths PD, Emery VC, editors. Churchill Livingstone; New York: 1997. pp. 445–470. [Google Scholar]
- 3.Reeves MB, MacAry PA, Lehner PJ, Sissons JG, Sinclair JH. Latency, chromatin remodeling, and reactivation of human cytomegalovirus in the dendritic cells of healthy carriers. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:4140–4145. doi: 10.1073/pnas.0408994102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goodrum F, Caviness K, Zagallo P. Human cytomegalovirus persistence. Cellular microbiology. 2012;14:644–655. doi: 10.1111/j.1462-5822.2012.01774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Hara GA, Welten SP, Klenerman P, Arens R. Memory T cell inflation: understanding cause and effect. Trends in immunology. 2012;33:84–90. doi: 10.1016/j.it.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 6.Chidrawar S, Khan N, Wei W, McLarnon A, Smith N, Nayak L, Moss P. Cytomegalovirus-seropositivity has a profound influence on the magnitude of major lymphoid subsets within healthy individuals. Clinical and experimental immunology. 2009;155:423–432. doi: 10.1111/j.1365-2249.2008.03785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moss P. The emerging role of cytomegalovirus in driving immune senescence: a novel therapeutic opportunity for improving health in the elderly. Current opinion in immunology. 2010;22:529–534. doi: 10.1016/j.coi.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 8.Snyder CM, Cho KS, Bonnett EL, Allan JE, Hill AB. Sustained CD8+ T cell memory inflation after infection with a single-cycle cytomegalovirus. PLoS pathogens. 2011;7:e1002295. doi: 10.1371/journal.ppat.1002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sylwester AW, Mitchell BL, Edgar JB, Taormina C, Pelte C, Ruchti F, Sleath PR, Grabstein KH, Hosken NA, Kern F, Nelson JA, Picker LJ. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. The Journal of experimental medicine. 2005;202:673–685. doi: 10.1084/jem.20050882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Looney RJ, Falsey A, Campbell D, Torres A, Kolassa J, Brower C, McCann R, Menegus M, McCormick K, Frampton M, Hall W, Abraham GN. Role of cytomegalovirus in the T cell changes seen in elderly individuals. Clinical immunology. 1999;90:213–219. doi: 10.1006/clim.1998.4638. [DOI] [PubMed] [Google Scholar]
- 11.Pawelec G, Derhovanessian E. Role of CMV in immune senescence. Virus research. 2011;157:175–179. doi: 10.1016/j.virusres.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 12.Pawelec G, Derhovanessian E, Larbi A, Strindhall J, Wikby A. Cytomegalovirus and human immunosenescence. Reviews in medical virology. 2009;19:47–56. doi: 10.1002/rmv.598. [DOI] [PubMed] [Google Scholar]
- 13.Sauce D, Larsen M, Fastenackels S, Duperrier A, Keller M, Grubeck-Loebenstein B, Ferrand C, Debre P, Sidi D, Appay V. Evidence of premature immune aging in patients thymectomized during early childhood. The Journal of clinical investigation. 2009;119:3070–3078. doi: 10.1172/JCI39269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Solana R, Tarazona R, Aiello AE, Akbar AN, Appay V, Beswick M, Bosch JA, Campos C, Cantisan S, Cicin-Sain L, Derhovanessian E, Ferrando-Martinez S, Frasca D, Fulop T, Govind S, Grubeck-Loebenstein B, Hill A, Hurme M, Kern F, Larbi A, Lopez-Botet M, Maier AB, McElhaney JE, Moss P, Naumova E, Nikolich-Zugich J, Pera A, Rector JL, Riddell N, Sanchez-Correa B, Sansoni P, Sauce D, van Lier R, Wang GC, Wills MR, Zielinski M, Pawelec G. CMV and Immunosenescence: from basics to clinics. Immun Ageing. 2012;9:23. doi: 10.1186/1742-4933-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sansoni P, Vescovini R, Fagnoni FF, Akbar A, Arens R, Chiu YL, Cicin-Sain L, Dechanet-Merville J, Derhovanessian E, Ferrando-Martinez S, Franceschi C, Frasca D, Fulop T, Furman D, Gkrania-Klotsas E, Goodrum F, Grubeck-Loebenstein B, Hurme M, Kern F, Lilleri D, Lopez-Botet M, Maier AB, Marandu T, Marchant A, Mathei C, Moss P, Muntasell A, Remmerswaal EB, Riddell NE, Rothe K, Sauce D, Shin EC, Simanek AM, Smithey MJ, Soderberg-Naucler C, Solana R, Thomas PG, van Lier R, Pawelec G, Nikolich-Zugich J. New advances in CMV and immunosenescence. Experimental gerontology. 2014;55:54–62. doi: 10.1016/j.exger.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 16.Barton ES, White DW, Cathelyn JS, Brett-McClellan KA, Engle M, Diamond MS, Miller VL, Virgin H. W. t. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature. 2007;447:326–329. doi: 10.1038/nature05762. [DOI] [PubMed] [Google Scholar]
- 17.Yager EJ, Szaba FM, Kummer LW, Lanzer KG, Burkum CE, Smiley ST, Blackman MA. gamma-Herpesvirus-induced protection against bacterial infection is transient. Viral immunology. 2009;22:67–72. doi: 10.1089/vim.2008.0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Terrazzini N, Bajwa M, Vita S, Thomas D, Smith H, Vescovini R, Sansoni P, Kern F. Cytomegalovirus infection modulates the phenotype and functional profile of the T-cell immune response to mycobacterial antigens in older life. Experimental gerontology. 2014;54:94–100. doi: 10.1016/j.exger.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pera A, Campos C, Corona A, Sanchez-Correa B, Tarazona R, Larbi A, Solana R. CMV latent infection improves CD8+ T response to SEB due to expansion of polyfunctional CD57+ cells in young individuals. PloS one. 2014;9:e88538. doi: 10.1371/journal.pone.0088538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michaelis M, Doerr HW, Cinatl J. The story of human cytomegalovirus and cancer: increasing evidence and open questions. Neoplasia. 2009;11:1–9. doi: 10.1593/neo.81178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Journal of the American College of Cardiology. 1999;34:1738–1743. doi: 10.1016/s0735-1097(99)00410-6. [DOI] [PubMed] [Google Scholar]
- 22.Wang GC, Kao WH, Murakami P, Xue QL, Chiou RB, Detrick B, McDyer JF, Semba RD, Casolaro V, Walston JD, Fried LP. Cytomegalovirus infection and the risk of mortality and frailty in older women: a prospective observational cohort study. American journal of epidemiology. 2010;171:1144–1152. doi: 10.1093/aje/kwq062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts ET, Haan MN, Dowd JB, Aiello AE. Cytomegalovirus antibody levels, inflammation, and mortality among elderly Latinos over 9 years of follow-up. American journal of epidemiology. 2010;172:363–371. doi: 10.1093/aje/kwq177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lau SK, Chen YY, Chen WG, Diamond DJ, Mamelak AN, Zaia JA, Weiss LM. Lack of association of cytomegalovirus with human brain tumors. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2005;18:838–843. doi: 10.1038/modpathol.3800352. [DOI] [PubMed] [Google Scholar]
- 25.Poltermann S, Schlehofer B, Steindorf K, Schnitzler P, Geletneky K, Schlehofer JR. Lack of association of herpesviruses with brain tumors. Journal of neurovirology. 2006;12:90–99. doi: 10.1080/13550280600654573. [DOI] [PubMed] [Google Scholar]
- 26.Huang TS, Lee JJ, Cheng SP. No evidence of association between human cytomegalovirus infection and papillary thyroid cancer. World journal of surgical oncology. 2014;12:41. doi: 10.1186/1477-7819-12-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathei C, Adriaensen W, Vaes B, Van Pottelbergh G, Wallemacq P, Degryse J. No relation between CMV infection and mortality in the oldest old: results from the Belfrail study. Age and ageing. 2014 doi: 10.1093/ageing/afu094. [DOI] [PubMed] [Google Scholar]
- 28.Vasto S, Colonna-Romano G, Larbi A, Wikby A, Caruso C, Pawelec G. Role of persistent CMV infection in configuring T cell immunity in the elderly. Immunity & ageing : I & A. 2007;4:2. doi: 10.1186/1742-4933-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simanek AM, Dowd JB, Pawelec G, Melzer D, Dutta A, Aiello AE. Seropositivity to cytomegalovirus, inflammation, all-cause and cardiovascular disease-related mortality in the United States. PloS one. 2011;6:e16103. doi: 10.1371/journal.pone.0016103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nieto FJ, Adam E, Sorlie P, Farzadegan H, Melnick JL, Comstock GW, Szklo M. Cohort study of cytomegalovirus infection as a risk factor for carotid intimal-medial thickening, a measure of subclinical atherosclerosis. Circulation. 1996;94:922–927. doi: 10.1161/01.cir.94.5.922. [DOI] [PubMed] [Google Scholar]
- 31.Muhlestein JB, Horne BD, Carlquist JF, Madsen TE, Bair TL, Pearson RR, Anderson JL. Cytomegalovirus seropositivity and C-reactive protein have independent and combined predictive value for mortality in patients with angiographically demonstrated coronary artery disease. Circulation. 2000;102:1917–1923. doi: 10.1161/01.cir.102.16.1917. [DOI] [PubMed] [Google Scholar]
- 32.Roossinck MJ. The good viruses: viral mutualistic symbioses. Nature reviews. Microbiology. 2011;9:99–108. doi: 10.1038/nrmicro2491. [DOI] [PubMed] [Google Scholar]
- 33.Furman D, Jojic V, Kidd B, Shen-Orr S, Price J, Jarrell J, Tse T, Huang H, Lund P, Maecker HT, Utz PJ, Dekker CL, Koller D, Davis MM. Apoptosis and other immune biomarkers predict influenza vaccine responsiveness. Mol Syst Biol. 2013;9:659. doi: 10.1038/msb.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Furman D, Hejblum BP, Simon N, Jojic V, Dekker CL, Thiebaut R, Tibshirani RJ, Davis MM. Systems analysis of sex differences reveals an immunosuppressive role for testosterone in the response to influenza vaccination. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:869–874. doi: 10.1073/pnas.1321060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang C, Liu Y, Xu LT, Jackson KJ, Roskin KM, Pham TD, Laserson J, Marshall EL, Seo K, Lee JY, Furman D, Koller D, Dekker CL, Davis MM, Fire AZ, Boyd SD. Effects of aging, cytomegalovirus infection, and EBV infection on human B cell repertoires. Journal of immunology. 2014;192:603–611. doi: 10.4049/jimmunol.1301384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Segal E, Shapira M, Regev A, Pe'er D, Botstein D, Koller D, Friedman N. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nature genetics. 2003;34:166–176. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- 37.Jojic V, Shay T, Sylvia K, Zuk O, Sun X, Kang J, Regev A, Koller D, C. Immunological Genome Project. Best AJ, Knell J, Goldrath A, Jojic V, Koller D, Shay T, Regev A, Cohen N, Brennan P, Brenner M, Kim F, Rao TN, Wagers A, Heng T, Ericson J, Rothamel K, Ortiz-Lopez A, Mathis D, Benoist C, Bezman NA, Sun JC, Min-Oo G, Kim CC, Lanier LL, Miller J, Brown B, Merad M, Gautier EL, Jakubzick C, Randolph GJ, Monach P, Blair DA, Dustin ML, Shinton SA, Hardy RR, Laidlaw D, Collins J, Gazit R, Rossi DJ, Malhotra N, Sylvia K, Kang J, Kreslavsky T, Fletcher A, Elpek K, Bellemare-Pelletier A, Malhotra D, Turley S. Identification of transcriptional regulators in the mouse immune system. Nature immunology. 2013;14:633–643. doi: 10.1038/ni.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snyder MR, Muegge LO, Offord C, O'Fallon WM, Bajzer Z, Weyand CM, Goronzy JJ. Formation of the killer Ig-like receptor repertoire on CD4+CD28null T cells. J Immunol. 2002;168:3839–3846. doi: 10.4049/jimmunol.168.8.3839. [DOI] [PubMed] [Google Scholar]
- 39.Xu J, Vallejo AN, Jiang Y, Weyand CM, Goronzy JJ. Distinct transcriptional control mechanisms of killer immunoglobulin-like receptors in natural killer (NK) and in T cells. J Biol Chem. 2005;280:24277–24285. doi: 10.1074/jbc.M500727200. [DOI] [PubMed] [Google Scholar]
- 40.Hsu HC, Mountz JD. Metabolic syndrome, hormones, and maintenance of T cells during aging. Curr Opin Immunol. 2010;22:541–548. doi: 10.1016/j.coi.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Magalhaes JP, Curado J, Church GM. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics. 2009;25:875–881. doi: 10.1093/bioinformatics/btp073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hasty P, Campisi J, Hoeijmakers J, van Steeg H, Vijg J. Aging and genome maintenance: lessons from the mouse? Science. 2003;299:1355–1359. doi: 10.1126/science.1079161. [DOI] [PubMed] [Google Scholar]
- 43.Goodwin K, Viboud C, Simonsen L. Antibody response to influenza vaccination in the elderly: a quantitative review. Vaccine. 2006;24:1159–1169. doi: 10.1016/j.vaccine.2005.08.105. [DOI] [PubMed] [Google Scholar]
- 44.den Elzen WP, Vossen AC, Cools HJ, Westendorp RG, Kroes AC, Gussekloo J. Cytomegalovirus infection and responsiveness to influenza vaccination in elderly residents of long-term care facilities. Vaccine. 2011;29:4869–4874. doi: 10.1016/j.vaccine.2011.03.086. [DOI] [PubMed] [Google Scholar]
- 45.Trzonkowski P, Mysliwska J, Szmit E, Wieckiewicz J, Lukaszuk K, Brydak LB, Machala M, Mysliwski A. Association between cytomegalovirus infection, enhanced proinflammatory response and low level of antihemagglutinins during the anti-influenza vaccination--an impact of immunosenescence. Vaccine. 2003;21:3826–3836. doi: 10.1016/s0264-410x(03)00309-8. [DOI] [PubMed] [Google Scholar]
- 46.van Leeuwen EM, Remmerswaal EB, Vossen MT, Rowshani AT, Wertheim-van Dillen PM, van Lier RA, ten Berge IJ. Emergence of a CD4+CD28- granzyme B+, cytomegalovirus-specific T cell subset after recovery of primary cytomegalovirus infection. Journal of immunology. 2004;173:1834–1841. doi: 10.4049/jimmunol.173.3.1834. [DOI] [PubMed] [Google Scholar]
- 47.Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis Res Ther. 2011;13:101. doi: 10.1186/ar3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Leeuwen EM, Remmerswaal EB, Heemskerk MH, ten Berge IJ, van Lier RA. Strong selection of virus-specific cytotoxic CD4+ T-cell clones during primary human cytomegalovirus infection. Blood. 2006;108:3121–3127. doi: 10.1182/blood-2006-03-006809. [DOI] [PubMed] [Google Scholar]
- 49.Pourgheysari B, Khan N, Best D, Bruton R, Nayak L, Moss PA. The cytomegalovirus-specific CD4+ T-cell response expands with age and markedly alters the CD4+ T-cell repertoire. Journal of virology. 2007;81:7759–7765. doi: 10.1128/JVI.01262-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nalls MA, Couper DJ, Tanaka T, van Rooij FJ, Chen MH, Smith AV, Toniolo D, Zakai NA, Yang Q, Greinacher A, Wood AR, Garcia M, Gasparini P, Liu Y, Lumley T, Folsom AR, Reiner AP, Gieger C, Lagou V, Felix JF, Volzke H, Gouskova NA, Biffi A, Doring A, Volker U, Chong S, Wiggins KL, Rendon A, Dehghan A, Moore M, Taylor K, Wilson JG, Lettre G, Hofman A, Bis JC, Pirastu N, Fox CS, Meisinger C, Sambrook J, Arepalli S, Nauck M, Prokisch H, Stephens J, Glazer NL, Cupples LA, Okada Y, Takahashi A, Kamatani Y, Matsuda K, Tsunoda T, Kubo M, Nakamura Y, Yamamoto K, Kamatani N, Stumvoll M, Tonjes A, Prokopenko I, Illig T, Patel KV, Garner SF, Kuhnel B, Mangino M, Oostra BA, Thein SL, Coresh J, Wichmann HE, Menzel S, Lin J, Pistis G, Uitterlinden AG, Spector TD, Teumer A, Eiriksdottir G, Gudnason V, Bandinelli S, Frayling TM, Chakravarti A, van Duijn CM, Melzer D, Ouwehand WH, Levy D, Boerwinkle E, Singleton AB, Hernandez DG, Longo DL, Soranzo N, Witteman JC, Psaty BM, Ferrucci L, Harris TB, O'Donnell CJ, Ganesh SK. Multiple loci are associated with white blood cell phenotypes. PLoS Genet. 2011;7:e1002113. doi: 10.1371/journal.pgen.1002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Orru V, Steri M, Sole G, Sidore C, Virdis F, Dei M, Lai S, Zoledziewska M, Busonero F, Mulas A, Floris M, Mentzen WI, Urru SA, Olla S, Marongiu M, Piras MG, Lobina M, Maschio A, Pitzalis M, Urru MF, Marcelli M, Cusano R, Deidda F, Serra V, Oppo M, Pilu R, Reinier F, Berutti R, Pireddu L, Zara I, Porcu E, Kwong A, Brennan C, Tarrier B, Lyons R, Kang HM, Uzzau S, Atzeni R, Valentini M, Firinu D, Leoni L, Rotta G, Naitza S, Angius A, Congia M, Whalen MB, Jones CM, Schlessinger D, Abecasis GR, Fiorillo E, Sanna S, Cucca F. Genetic variants regulating immune cell levels in health and disease. Cell. 2013;155:242–256. doi: 10.1016/j.cell.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vescovini R, Telera A, Fagnoni FF, Biasini C, Medici MC, Valcavi P, di Pede P, Lucchini G, Zanlari L, Passeri G, Zanni F, Chezzi C, Franceschi C, Sansoni P. Different contribution of EBV and CMV infections in very long-term carriers to age-related alterations of CD8+ T cells. Experimental gerontology. 2004;39:1233–1243. doi: 10.1016/j.exger.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 53.Amyes E, Hatton C, Montamat-Sicotte D, Gudgeon N, Rickinson AB, McMichael AJ, Callan MF. Characterization of the CD4+ T cell response to Epstein-Barr virus during primary and persistent infection. The Journal of experimental medicine. 2003;198:903–911. doi: 10.1084/jem.20022058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuijpers TW, Vossen MT, Gent MR, Davin JC, Roos MT, Wertheim-van Dillen PM, Weel JF, Baars PA, van Lier RA. Frequencies of circulating cytolytic, CD45RA+CD27-, CD8+ T lymphocytes depend on infection with CMV. Journal of immunology. 2003;170:4342–4348. doi: 10.4049/jimmunol.170.8.4342. [DOI] [PubMed] [Google Scholar]
- 55.Babcock GJ, Decker LL, Volk M, Thorley-Lawson DA. EBV persistence in memory B cells in vivo. Immunity. 1998;9:395–404. doi: 10.1016/s1074-7613(00)80622-6. [DOI] [PubMed] [Google Scholar]
- 56.Thorley-Lawson DA. Epstein-Barr virus: exploiting the immune system. Nature reviews. Immunology. 2001;1:75–82. doi: 10.1038/35095584. [DOI] [PubMed] [Google Scholar]
- 57.Botto S, Streblow DN, DeFilippis V, White L, Kreklywich CN, Smith PP, Caposio P. IL-6 in human cytomegalovirus secretome promotes angiogenesis and survival of endothelial cells through the stimulation of survivin. Blood. 2011;117:352–361. doi: 10.1182/blood-2010-06-291245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Badovinac VP, Messingham KA, Jabbari A, Haring JS, Harty JT. Accelerated CD8+ T-cell memory and prime-boost response after dendritic-cell vaccination. Nat Med. 2005;11:748–756. doi: 10.1038/nm1257. [DOI] [PubMed] [Google Scholar]
- 59.Denzin LK, Sant'Angelo DB, Hammond C, Surman MJ, Cresswell P. Negative regulation by HLA-DO of MHC class II-restricted antigen processing. Science. 1997;278:106–109. doi: 10.1126/science.278.5335.106. [DOI] [PubMed] [Google Scholar]
- 60.Turner JE, Campbell JP, Edwards KM, Howarth LJ, Pawelec G, Aldred S, Moss P, Drayson MT, Burns VE, Bosch JA. Rudimentary signs of immunosenescence in Cytomegalovirus-seropositive healthy young adults. Age. 2014;36:287–297. doi: 10.1007/s11357-013-9557-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Welsh RM, Che JW, Brehm MA, Selin LK. Heterologous immunity between viruses. Immunol Rev. 2010;235:244–266. doi: 10.1111/j.0105-2896.2010.00897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Veirum JE, Sodemann M, Biai S, Jakobsen M, Garly ML, Hedegaard K, Jensen H, Aaby P. Routine vaccinations associated with divergent effects on female and male mortality at the paediatric ward in Bissau, Guinea-Bissau. Vaccine. 2005;23:1197–1204. doi: 10.1016/j.vaccine.2004.02.053. [DOI] [PubMed] [Google Scholar]
- 63.Benn CS, Netea MG, Selin LK, Aaby P. A small jab - a big effect: nonspecific immunomodulation by vaccines. Trends in immunology. 2013;34:431–439. doi: 10.1016/j.it.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 64.Sorup S, Benn CS, Poulsen A, Krause TG, Aaby P, Ravn H. Live vaccine against measles, mumps, and rubella and the risk of hospital admissions for nontargeted infections. JAMA : the journal of the American Medical Association. 2014;311:826–835. doi: 10.1001/jama.2014.470. [DOI] [PubMed] [Google Scholar]
- 65.Su LF, Kidd BA, Han A, Kotzin JJ, Davis MM. Virus-specific CD4(+) memory-phenotype T cells are abundant in unexposed adults. Immunity. 2013;38:373–383. doi: 10.1016/j.immuni.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Webster R, Cox N, Stohr K. WHO Manual on Animal Influenza Diagnosis and Surveillance. World Health Organization/CDS/CSR/NCS/2002.5. 2002 [Google Scholar]
- 67.Nedrud JG, Liang XP, Hague N, Lamm ME. Combined oral/nasal immunization protects mice from Sendai virus infection. J Immunol. 1987;139:3484–3492. [PubMed] [Google Scholar]
- 68.Sharma S, Sundararajan A, Suryawanshi A, Kumar N, Veiga-Parga T, Kuchroo VK, Thomas PG, Sangster MY, Rouse BT. T cell immunoglobulin and mucin protein-3 (Tim-3)/Galectin-9 interaction regulates influenza A virus-specific humoral and CD8 T-cell responses. Proc Natl Acad Sci U S A. 2011;108:19001–19006. doi: 10.1073/pnas.1107087108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fisher RA. Statistical Methods for Research Workers. Oliver and Boyd; Edinburgh: 1925. [Google Scholar]
- 70.Akaike H. A new look at the statistical model identification. IEEE Transactions on Automatic Control. 1974;19:716–723. [Google Scholar]
- 71.Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 72.Koltchinskii V, Tsybakov AB, Lounici K. Nuclear norm penalization and optimal rates for noisy low rank matrix completion. Annals of Statistics. 2011;39:2302–2329. [Google Scholar]
- 73.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 1995;B-57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.