Summary
A variety of cancers depend on JAK2 signaling, including the high-risk subset of B-cell acute lymphoblastic leukemias (B-ALLs) with CRLF2 rearrangements. Type I JAK2 inhibitors induce paradoxical JAK2 hyperphosphorylation in these leukemias and have limited activity. To improve the efficacy of JAK2 inhibition in B-ALL, we developed the type II inhibitor CHZ868, which stabilizes JAK2 in an inactive conformation. CHZ868 potently suppressed the growth of CRLF2-rearranged human B-ALL cells, abrogated JAK2 signaling, and improved survival in mice with human or murine B-ALL. CHZ868 and dexamethasone synergistically induced apoptosis in JAK2-dependent B-ALLs and further improved in vivo survival compared to CHZ868 alone. These data support the testing of type II JAK2 inhibition in patients with JAK2-dependent leukemias and other disorders.
Introduction
Outcomes for most children with B-cell acute lymphoblastic leukemia (B-ALL) have drastically improved over the past thirty years through the iterative refinement of multi-agent chemotherapy regimens (Pui et al., 2004). However, the 15–30% of cases that harbor Philadelphia chromosome (Ph)-like transcriptional signatures but lack BCR-ABL rearrangements continue to have a poor prognosis (Mullighan et al., 2009b; Roberts et al., 2014; Roberts et al., 2012).
Recent studies have identified genetic alterations that activate kinase signaling in Ph-like BALLs (Roberts et al., 2014; Roberts et al., 2012), in some cases by directly fusing the kinase domains of ABL1, PDGFR, or JAK2 to N-terminal partners that may undergo homodimerization. This mimics the conformation of JAK2 at ligand-bound cytokine receptors such as EPOR and MPL, and results in signal activation that involves trans-phosphorylation between adjacent JAK2 molecules. Initial efforts to treat patients with these fusions using inhibitors of ABL (e.g. dasatinib) or JAK2 (e.g. ruxolitinib) have been highly promising (Roberts et al., 2014).
The most common rearrangements in Ph-like B-ALL, occurring in approximately 50% of cases, are translocations and intrachromosomal deletions that result in overexpression of the CRLF2 cytokine receptor (Hertzberg et al., 2010; Mullighan et al., 2009a; Russell et al., 2009b; Yoda et al., 2010). Unlike signaling downstream of EPOR or MPL, CRLF2 signaling is believed to involve heterodimerization of CRLF2 with the IL7Rα subunit and transduction through JAK2 (interacting with CRLF2) and JAK1 (interacting with IL7Rα) (Sessa et al., 2013; Wohlmann et al., 2010).
Overexpression of CRLF2 alone is not sufficient to constitutively activate downstream signaling in model systems. CRLF2-rearranged B-ALLs frequently harbor a second genetic alteration within the CRLF2 signaling cascade, most commonly in exon 16 of JAK2 (Hertzberg et al., 2010; Mesbah et al., 2012; Mullighan et al., 2009a; Russell et al., 2009a). Additional cases have alterations elsewhere in JAK2, in CRLF2 itself, JAK1, IL7R, SH2B3, or TSLP that activate JAK2/STAT5 signaling via CRLF2 (Roberts et al., 2014; Shochat et al., 2011; Shochat et al., 2014; Yoda et al., 2010).
We previously reported that our model systems of B-ALL cells dependent on JAK2 signaling downstream of CRLF2 are refractory to type I JAK2 inhibitors like ruxolitinib (Weigert et al., 2012), which target the ATP-binding pocket and stabilize JAK2 in the active confirmation. In these cells, type I inhibitors induce paradoxical JAK2 hyperphosphorylation. The Levine laboratory reported that type I JAK2 inhibitors can induce a state of persistent JAK2 signaling in EPOR- or MPL-expressing myeloid cells that involves heterodimerization and trans-phosphorylation of JAK2 by JAK1 or TYK2 (Koppikar et al., 2012). Importantly, persistent JAK2 signaling in myeloid cells was abrogated by treatment with the type II JAK2 inhibitor BBT594 (Koppikar et al., 2012), which stabilizes JAK2 in an inactive confirmation and blunts activation loop phosphorylation (Andraos et al., 2012). BBT594 (Figure 1A) was initially developed as an inhibitor of BCR-ABL T315I, but was found to also inhibit JAK2 by stabilizing the inactive conformation (Andraos et al., 2012). BBT594 has limitations in potency and selectivity for JAK2 as well as pharmacokinetic properties that preclude in vivo use. Thus, we developed another type II inhibitor to further explore the potential of type II JAK2 inhibition in B-ALL.
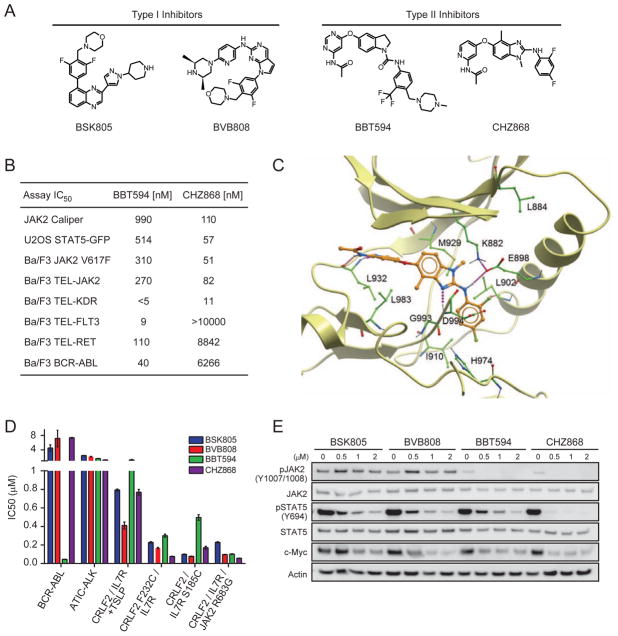
Figure 1. The type II JAK2 inhibitor NVP-CHZ868 blocks JAK2 signaling in vitro and in vivo.
(A) Chemical structures of type I and II JAK2 inhibitors. (B) IC50 values for CHZ868 and BBT594 in enzymatic and cell-based assays. (C) Binding mode model of CHZ868 to JAK2. Ribbon representation of the JAK2 kinase domain with CHZ868 illustrated as a stick model. Amino acid side chains interacting with the inhibitor are shown in green. Polar contacts between the protein and the inhibitor are highlighted with dotted purple lines. (D) IC50 values for type I and II JAK2 inhibitors in Ba/F3 cells expressing the indicated proteins in the absence of cytokines, except where “+TSLP” indicates 1 nM TSLP. Error bars represent SEM. (E) Immunoblotting against the indicated targets using lysates from Ba/F3-CRLF2/JAK2 R683G cells exposed to the indicated concentrations of JAK2 inhibitors for 2 hr. See also Figure S1 and Table S1.
Results
The type II JAK2 inhibitor CHZ868 blocks JAK2 signaling in vitro and in vivo
We launched a discovery program to identify type II JAK2 inhibitors with improved potency, selectivity and physicochemical properties. Mining of the Novartis database for compounds containing structural motifs canonical for type II kinase inhibition, followed by a cellular screening campaign using JAK2 V617F mutant SET-2 cells to identify compounds that suppress phosphorylation of both JAK2 and STAT5, revealed arylamino-benzimidazoles, originally described as RAF kinase inhibitors (Shiels et al., 2011), as an attractive starting point for an optimization program. Medicinal chemistry efforts, driven by property and protein structure based considerations, led to the discovery of CHZ868 (Figure 1A).
In terms of physicochemical and pharmacokinetic properties, CHZ868 is characterized by high passive permeability, good metabolic stability, and low water solubility, as well as by moderate blood clearance and good oral bioavailability (Table S1), making it suitable for in vivo use. Consistent with a type II binding mode, CHZ868 showed modest inhibitory activity in enzymatic assays with activated (phosphorylated) JAK2 kinase, but demonstrated excellent potency in JAK2-driven cellular assays (Figure 1B). Importantly, JAK2 inhibitory activity of CHZ868 is 3–9-fold improved over BBT594, whereas some of the BBT594 off-target activities, such as BCR-ABL, RET and FLT3 inhibition, are significantly reduced (Figure 1B).
Broader CHZ868 in vitro kinase selectivity was assessed in a panel consisting of over 400 kinases. At 100 nM CHZ868 had activity against 26 kinases, including JAK2 and TYK2 (Figures S1A and S1B). As the kinome panel is predominantly comprised of activated (i.e. phosphorylated) kinases, profiling of type II inhibitors may have limitations for selectivity predictions, given their preferential binding to inactive kinase conformations. Thus, CHZ868 kinome panel hits were followed-up in available cell-based kinase selectivity assays. In the cellular setting, low nanomolar activity against KIT, as well as PDGFR and VEGFR family kinases was substantiated, whereas activity against ABL, BRAF and RET was not corroborated (Figure S1B).
To determine the appropriate dose and schedule of CHZ868 in vivo, we utilized a murine model of polycythemia induced by exogenous erythropoietin (Baffert et al., 2010). Control mice receiving recombinant human erythropoietin (rhEpo) exhibited a marked induction of pSTAT5 in the spleen and bone marrow (Figures S1C and S1D). At an oral dose of 30 mg/kg/day CHZ868 completely blocked the induction of pSTAT5 by rhEpo in mice when administered 2 hr prior to sampling. At that timepoint, the concentration of CHZ868 within the spleen was 4.3 nmol/g (Figure S1C). By 6 hr after CHZ868 dosing, the drug level within the spleen was <1 nmol/g, consistent with the blood t1/2 in mice of 1 hr (Table S1). Despite the kinetics of CHZ868 elimination, the ability of rhEpo to induce pSTAT5 in the spleen and bone marrow remained attenuated for 48–72 hr (Figures S1C and S1D).
Next, we treated BALB/c mice with daily injections of either saline or rhEpo for four consecutive days. Mice also received either vehicle or CHZ868 and all animals were sacrificed on day 5. Treatment with CHZ868 at 30 mg/kg/day completely blocked splenomegaly induced by rhEpo (Figure S1E). The same dose of CHZ868 also blocked the increase in reticulocyte count induced by rhEpo but did not affect WBC count (Figure S1F). Importantly, treatment of wild-type mice with CHZ868 at 30 mg/kg/day for >6 weeks had no significant effects on body weight, peripheral blood cell counts, bone marrow cellularity, or the fractions of hematopoietic progenitors in the bone marrow (Figures S1G–S1M). Mice receiving CHZ868 had trends towards reduced spleen weights and elevated serum erythropoietin levels (Figures S1N and S1O), which are expected on-target effects. Taken together, these results show that CHZ868 30 mg/kg/day potently blocks JAK2 signaling with minimal toxicity in vivo.
Binding mode model of CHZ868 to JAK2
Our binding mode model of CHZ868 to JAK2 shares most of the characteristics described by the X-ray crystallographic structure of BBT594 binding to JAK2 (PDB entry 3UGC) (Andraos et al., 2012). CHZ868 is thought to engage with the hinge region of JAK2 through two H-bonds, formed between the amino-pyridine of CHZ868 and the backbone-NH/CO of L932, while the pyridine is occupying the adenine pocket of the ATP binding site (Figure 1C). Archetypal for a type II binding mode, the acceptor nitrogen of the 2-amino benzimidazole core is expected to interact with the backbone NH of D994, whereas the 2-amino benzimidazole NH binds to the side-chain of E898, which is also forming the usual salt bridge to the catalytic K882. The 2,4-difluorophenyl moiety is believed to fill the kinase back pocket, making close hydrophobic contacts to the C-helix, e.g. with L902. Importantly, the 4-methyl group of the benzimidazole was designed to interact in van der Waals distance with the side chain of L983 and the backbone of G993. G993 is the so-called pre-DFG residue, which is highly variable among protein kinases (Cesarman, 2013). Around 85% of all protein kinases have a residue sterically larger than glycine in the pre-DFG position. Therefore, the 4-methyl substituent of the benzimidazole is likely to be responsible for the improved kinase selectivity of CHZ868 (e.g. compared to BBT594) by causing steric clashes in the binding pocket of kinases with pre-DFG residues larger than glycine.
Activity of CHZ868 in Ba/F3 cell-based models of JAK2-dependent B-ALL
Next, we set out to characterize the type II inhibitors CHZ868 and BBT594, as compared to the type I JAK2 inhibitors BSK805 and BVB808, in Ba/F3 cell-based models of oncogenic signaling in B-ALL. All compounds potently inhibited the growth of Ba/F3-CRLF2/IL7R cells expressing the gain-of-function mutants CRLF2 F232C, IL7R S185C or JAK2 R683G (Figure 1D). CHZ868 was also highly active in Ba/F3 cells expressing TEL-JAK2 (Figure 1B). The growth of CRLF2 and IL7R expressing Ba/F3 cells cultured in the presence of TSLP (1 nM) was less potently inhibited by both type I and type II inhibitors. As we previously described (Weigert et al., 2012), treatment of Ba/F3-CRLF2/JAK2 R683G cells with type I JAK2 inhibitors induced hyperphosphorylation of JAK2. In contrast, CHZ868 completely abrogated pJAK2 and pSTAT5, with downregulation of the STAT5 target c-Myc (Figure 1E).
Discovery of a JAK2 L884P mutation that confers resistance to type II JAK2 inhibitors
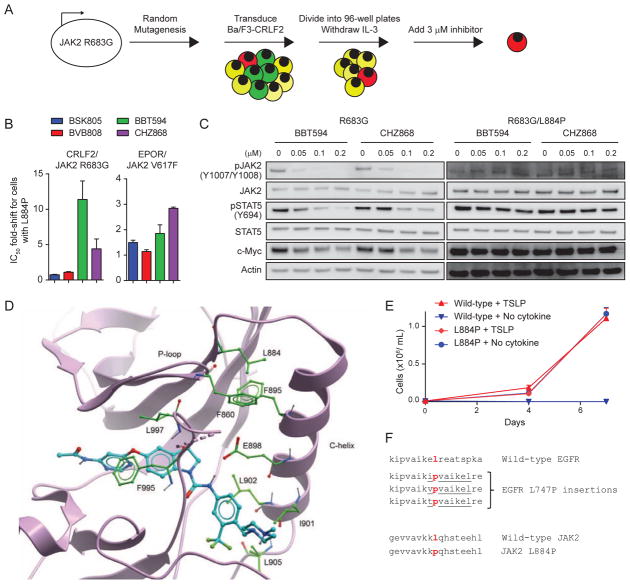
The ability of type II inhibitors to avoid JAK2 hyperphosphorylation raised the possibility that resistance to type II inhibitors will occur through genetic, rather than posttranslational, mechanisms. To identify mutations in JAK2 that confer resistance to type II inhibitors, Ba/F3-CRLF2 cells were transduced with a library of JAK2 R683G that was propagated in DNA repair-deficient bacteria. Clones were selected in 96-well plates in the absence of cytokines and the presence of either 3 μM BBT594 or 3 μM CHZ868 (Figure 2A). This approach allows for the parallel identification of multiple different alleles that confer resistance to each inhibitor. Over 30 individual clones were recovered after selection in the presence of BBT594 but none were recovered after selection with CHZ868. Remarkably, all BBT594-resistant clones harbored the same JAK2 L884P mutation. In comparison, a previous study using a mutagenized library of BCR-ABL in Ba/F3 cells identified over 100 different alleles that confer resistance to imatinib in vitro (Azam et al., 2003).
Figure 2. JAK2 L884P confers resistance to type II JAK2 inhibitors.
(A) Schematic of mutagenesis screen to identify mutations in JAK2 R683G that confer resistance to type II JAK2 inhibitors. Green cells are unable to proliferate in the absence of IL-3. Yellow cells can proliferate in the absence of IL-3 but not in the presence of JAK2 inhibitor. Red cells can proliferate both in the absence of IL-3 and in the presence of inhibitor. Cells are divided into multiple aliquots after transduction, allowing for the parallel selection of multiple clones that have independently acquired resistance. (B) Ba/F3-CRLF2 cells expressing JAK2 R683G or JAK2 R683G/L884P were treated with JAK2 inhibitor and cell proliferation was assessed after 48 hr. The same assay was performed on Ba/F3-EpoR cells expressing JAK2 V617F or JAK2 V617F/L884P. Data is presented as IC50 fold-shift for cells expressing L884P compared to cells without L884P. Results are from 8 replicates across one to two independent experiments. Error bars represent SEM. (C) Immunoblotting against the indicated targets using lysates from Ba/F3-CRLF2 cells expressing JAK2 R683G or R683G/L884P. Cells were exposed to varying concentrations of JAK2 inhibitor for 2 hr. (D) X-ray structure of JAK2 JH1 domain in complex with BBT594 (Andraos et al., 2012). Ribbon representation of the JAK2 kinase domain with BBT594 illustrated as a stick model. Amino acid side chains thought to be important for stabilizing the C-helix and the P-loop are shown in green. (E) Ba/F3-CRLF2/IL7Rα cells were transduced with either wild-type JAK2 or JAK2 L884P and then cultured in media with or without TSLP. Results are from 3 replicates from a representative experiment. Error bars represent SEM. (F) Homology of JAK2 L884P to EGFR L747P insertional mutations described in non-small cell lung cancer (He et al., 2012).
To confirm that JAK2 L884P is a bona fide resistance allele, we expressed JAK2 R683G/L884P in Ba/F3-CRLF2 cells. While the L884P allele had minimal effects on sensitivity to BSK805 or BVB808, the IC50 for cells expressing R683G with L884P was increased approximately 11-fold for BBT594 (Figure 2B). L884P induced only 4-fold resistance to CHZ868, potentially explaining the absence of CHZ868-resistant clones in the mutagenesis screen. The L884P mutation abrogated the suppressive effects of BBT594 and CHZ868 on pJAK2, pSTAT5, and c-Myc at concentrations up to 200 nM of each drug (Figure 2C). Interestingly, in the setting of JAK2 V617F, the L884P mutation led to only 2–3-fold increased IC50 values for BBT594 and CHZ868 (Figure 2B).
In the X-ray crystallographic structure of JAK2 in complex with BBT594 (Andraos et al., 2012), L884 is located at the end of the β3-strand in the N-terminal lobe and seems to play an important role in stabilizing the position of the C-helix, e.g. through interaction with F895 (Figure 2D). Substitution of leucine with proline may alter the conformation and flexibility of the loop linking the β3-strand to the C-helix. This effect and the loss of interaction with the side chain of F895 are likely to impact the position of the C-helix and thus the shape of the back pocket. Since hydrophobic contacts to the side chains of I901, L902, and L905 and the H-bond to the side chain of E898 are key interactions between the C-helix and BBT594 (Figure 2D), a different conformation of the C-helix is expected to negatively impact the binding of type II inhibitors.
In addition, the X-ray structure revealed a hydrophobic contact between the L884 and P-loop F860 side chains (Figure 2D). This interaction contributes to the stabilization of the P-loop when the F995 and L997 side chains are deeply buried in the ATP binding site, as generally observed in DFG-out or type II conformations. The loss of the favorable L884/F860 interaction through a L884P mutation would be predicted to destabilize the P-loop, favoring a DFG-in or type I conformation of the kinase. In fact, L884P was previously reported in two B-ALL cases that lack additional JAK2 mutations, suggesting that it is an activating allele (Torra et al., 2010). We confirmed that L884P is an activating allele, as Ba/F3-CRLF2/IL7Rα cells expressing JAK2 L884P proliferated in the absence of cytokines (Figure 2E). We also noted that JAK2 L884P is homologous to the activating EGFR L747P insertional mutations observed in non-small cell lung cancer (Figure 2F) (He et al., 2012).
CHZ868 targets JAK2 signaling in human B-ALL in vitro and in vivo
To expand on the Ba/F3 model findings, we assessed type I and type II JAK2 inhibitors using the human CRLF2-rearranged B-ALL cell line MHH-CALL4. In addition to an IGH@-CRLF2 fusion, MHH-CALL4 cells harbor a JAK2 I682F mutation and have constitutive phosphorylation of JAK2 and STAT5 (Russell et al., 2009b). We previously reported that MHH-CALL4 cells are insensitive to the type I JAK2 inhibitors BVB808, BSK805, JAK inhibitor I, ruxolitinib, and tofacitinib, with IC50 values exceeding 6–30 μM (Weigert et al., 2012).
Consistent with our prior studies, MHH-CALL4 cells were refractory to BVB808 and BSK805 with IC50 values exceeding 2 μM. In contrast to the type I inhibitors, BBT594 and CHZ868 both potently suppressed growth of MHH-CALL4 cells with IC50 values in the range of 100 nM (Figure 3A). Treatment of MHH-CALL4 cells with 1 μM of BVB808, BSK805 or ruxolitinib resulted in hyperphosphorylation of the JAK2 activation loop, while 1 μM of BBT594 or CHZ868 suppressed JAK2 phosphorylation (Figure 3B).
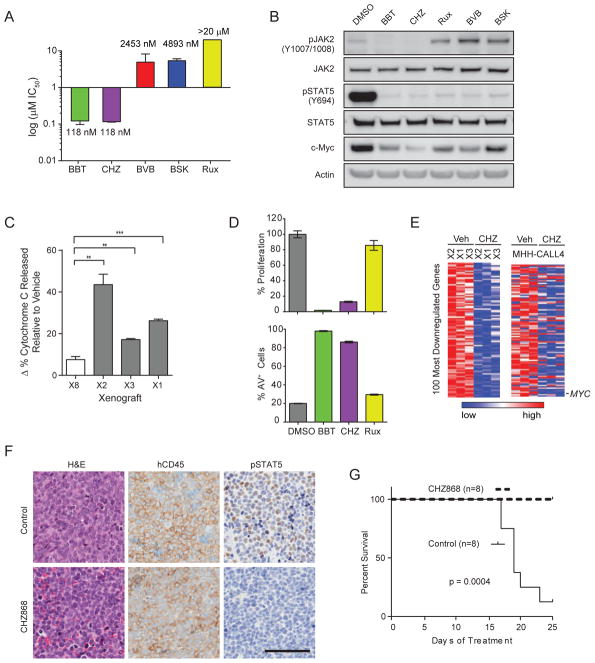
Figure 3. Type II JAK2 inhibitors potently inhibit JAK2-dependent B-ALL in vitro and in vivo.
(A) IC50 values for type I and II JAK2 inhibitors in MHH-CALL4 cells treated for 72 hr. Data represents 12 replicates over 3 experiments. (B) Immunoblotting against the indicated targets was performed on lysates from MHH-CALL4 cells exposed to vehicle (DMSO) or 1 μM of each inhibitor for 2 hr. (C) Splenocytes from three CRLF2-rearranged PDXs (B-ALL-X1 (X1), B-ALL-X2 (X2), and B-ALL-X3 (X3)) and an MLL-rearranged PDX (B-ALL-X8 (X8)) were treated ex vivo with vehicle or 300 nM CHZ868 for 16 hr and then exposed to varying concentrations of the pro-apoptotic peptide BIM. Change in % cytochrome C released for CHZ868-treated samples relative to vehicle-treated samples was quantified using flow cytometry. **p<0.01, ***p<0.001. (D) Proliferation relative to DMSO-treated cells and the percentage of Annexin V-positive (AV+) cells are shown for B-ALL-X2 cells treated for 48 hr. (E) Color-coded heatmap of the 100 most downregulated genes in splenocytes collected from mice injected with PDXs and treated with CHZ868 30 mg/kg/day for 3 days compared to vehicle-treated controls. The same 100 genes are displayed for MHH-CALL4 cells treated with CHZ868 compared to vehicle-treated controls. (F) Mice engrafted with PDXs received vehicle or CHZ868 30 mg/kg/day for 6 days. Representative examples of hematoxylin and eosin (H&E) staining and IHC from spleens of mice injected with B-ALL-X1 PDX are shown (scale bar, 100 μm). (G) Overall survival of mice combining all 4 PDXs (n = 2 mice/PDX/arm). All error bars in this figure indicate SEM. See also Figure S2 and Tables S2–S4.
To expand on the findings from MHH-CALL4 cells, we established a panel of CRLF2-rearranged B-ALL patient-derived xenograft (PDX) models in NOD.SCID.Il2rγ−/− (NSG) mice that included cases with JAK2 mutation, CRLF2 F232C, or no mutation in CRLF2, IL7R, JAK1 or JAK2 (Figure S2A, Table S2). First, we assessed the utility of “dynamic BH3 profiling” as a predictive biomarker for in vivo response to CHZ868 (Montero et al., 2015). To this end, leukemia cells isolated from animals engrafted with three different CRLF2-rearranged PDXs as well as an MLL-rearranged PDX were treated ex vivo with vehicle or CHZ868 for 16 hr and then exposed to varying concentrations of the pro-apoptotic peptide BIM. CHZ868 treatment of CRLF2-rearranged PDXs significantly increased cytochrome C release upon exposure to BIM compared to vehicle treatment of cells exposed to BIM (Figure 3C). These data suggest specificity of CHZ868 in targeting JAK2 dependence and predict that JAK2-dependent PDXs are selectively “primed” to undergo apoptosis through the mitochondrial pathway upon exposure to CHZ868.
Like in MHH-CALL4 cells, 1 μM CHZ868 or BBT594 consistently blocked proliferation and induced apoptosis ex vivo in CRLF2-rearranged PDXs that do (B-ALL-X2, B-ALL-X3) or do not (B-ALL-X4, B-ALL-X6) have activating JAK2 mutations (Figures 3D, S2B, and S2C). Again, ruxolitinib had little or no effect.
To gain insight into the transcriptional consequences of JAK2 inhibition by CHZ868 in vivo, we transplanted CRLF2-rearranged B-ALL PDXs into additional NSG recipients and randomized mice to receive either CHZ868 30 mg/kg/day or vehicle upon >2% peripheral blood involvement. Gene expression profiling of splenocytes isolated after 3 days of treatment identified the top 100 genes downregulated in the three PDXs after treatment with CHZ868 (Figure 3E, Table S3). This 100 gene signature was recapitulated in MHH-CALL4 cells treated with CHZ868 (Figure 3E), indicating that a similar pattern of expression changes is induced across models in vitro and in vivo. We noted multiple genes involved in cell cycle regulation, including MYC (third-most downregulated gene), in the signature. We queried multiple functionally-defined gene sets of either MYC or E2F1 using gene set enrichment analyses (Monti et al., 2012), which revealed that 4 of 4 E2F1 and 7 of 7 MYC functional gene sets were highly enriched among the genes downregulated by CHZ868 treatment of both the PDXs and MHH-CALL4 cells (Table S4, Figure S2D; FDR<0.001).
We then treated an expanded cohort of four CRLF2-rearranged B-ALL PDXs. Mice sacrificed after 6 days of treatment with CHZ868 had reduced splenomegaly and lower peripheral WBC counts compared to control animals (Figure S2E). Spleens from mice treated with CHZ868 for 6 days had persistent infiltration with human CD45 (hCD45)-positive B-ALL cells, but almost complete loss of pSTAT5 (Figure 3F). After 25 days of treatment, mice receiving CHZ868 had markedly improved overall survival compared to control animals (Figure 3G).
CHZ868 synergizes with dexamethasone in B-ALL cells in vitro and in vivo
To further improve on preclinical efficacy, we set out to identify CHZ868 combination partners that would act synergistically to kill CRLF2-rearranged B-ALLs. CHZ868 was screened in MHH-CALL4 cells for synergy with standard-of-care B-ALL chemotherapies, such as doxorubicin, cytarabine, vincristine, and dexamethasone. The most notable synergy was seen between dexamethasone and CHZ868 (Figure 4A), while the other drugs were either additive or synergistic (Figure S3A). Similar results were obtained in CRLF2-rearranged PDXs (Figures S3B and S3C). Thus, the combination of CHZ868 and dexamethasone may be an attractive therapeutic strategy, perhaps analogous to the finding that the vast majority of patients with BCR-ABL-rearranged B-ALL will achieve a complete remission upon treatment with the combination of a type II ABL inhibitor and a glucocorticoid such as dexamethasone (Foa et al., 2011).
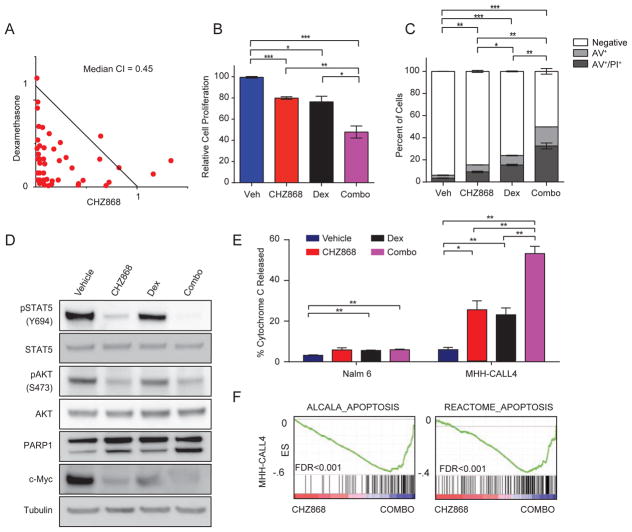
Figure 4. CHZ868 synergizes with dexamethasone in CRLF2-rearranged, JAK2-dependent human B-ALL cells.
(A) Isobologram analysis of CHZ868 and dexamethasone at multiple concentrations in MHH-CALL4 cells. Results are from a representative experiment performed in triplicate. CI: combination index. (B) MHH-CALL4 cells were exposed to vehicle, 300 nM CHZ868, 5 nM dexamethasone, or the combination (combo) for 48 hr. Relative cell proliferation was measured using CellTiterGlo. Results are from 18 replicates across 3 experiments. (C) Cells were treated as in (B) and the percentage of cells positive for Annexin V alone (AV+) or both Annexin V and Propidium Iodide (AV+/PI+) in each treatment arm was averaged across 18 replicates from 3 independent experiments. p values are comparing the percentage of negative cells. (D) Immunoblotting for the indicated targets performed on lysates from MHH-CALL4 cells treated with vehicle (DMSO), 300 nM CHZ868, 5 nM dexamethasone, or the combination for 24 hr. (E) Dynamic BH3 profiling was performed in Nalm 6 and MHH-CALL4 cells treated with vehicle, 300 nM CHZ868, 5 nM dexamethasone, or the combination for 16 hr, and then exposed to 0.1 μM BIM. (F) GSEA plots demonstrate enrichment of indicated apoptosis gene sets in MHH-CALL4 treated with the combination of dexamethasone and CHZ868 compared to single-agent CHZ868. In all panels of this figure, error bars represent SEM. *p<0.05, **p<0.01, ***p<0.001. See also Figure S3.
Treatment of MHH-CALL4 cells with CHZ868 (300 nM) and dexamethasone (5 nM) in combination reduced cell proliferation (Figure 4B) and increased the percentage of cells undergoing apoptosis (Figure 4C). Cells treated with the combination had further reductions in pSTAT5 and c-Myc compared to cells treated with CHZ868 alone and this was associated with increased PARP1 cleavage, a marker of apoptosis (Figure 4D). Dynamic BH3 profiling revealed that upon exposure to 0.1 μM BIM, MHH-CALL4 cells treated with CHZ868 and dexamethasone in combination showed a 2-fold increase in cytochrome C release relative to cells treated with either agent alone (Figure 4E). In contrast, the combination did not increase cytochrome C release in the JAK2-independent control cell line Nalm 6 over dexamethasone alone. Four independent apoptosis gene sets were highly enriched in the transcriptional signature of MHH-CALL4 cells treated with the combination of CHZ868 and dexamethasone compared to cells treated with CHZ866 alone (FDR<0.001; Figures 4F and S3D).
To study the in vivo effects of CHZ868 in an immunocompetent context, we generated transgenic mice with expression of human CRLF2 and mouse JAK2 R683G restricted to murine B cells. We previously reported that Eμ-CRLF2 transgenic mice crossed with Eμ-Jak2 R683G mice do not develop malignancies by 18 months of age (Lane et al., 2014). We crossed these mice or mice expressing the gain-of-function mutant CRLF2 F232C with mice carrying additional alleles known to contribute to B-ALL, including Pax5+/−, Cdkn2a−/−, and the Ts1Rhr model of trisomy 21. A subset of mice succumbed to leukemia involving the bone marrow and spleen between 131–378 days of age (Figures S4A–S4C). Each leukemia was confirmed to express CRLF2 and harbor a progenitor B-ALL immunophenotype (CD19+CD43+CD3−CD11b−) by flow cytometry (Figures S4A–S4C). All leukemias engrafted into secondary transplant recipients, satisfying the Bethesda criteria for B-ALL in mice (Morse et al., 2002).
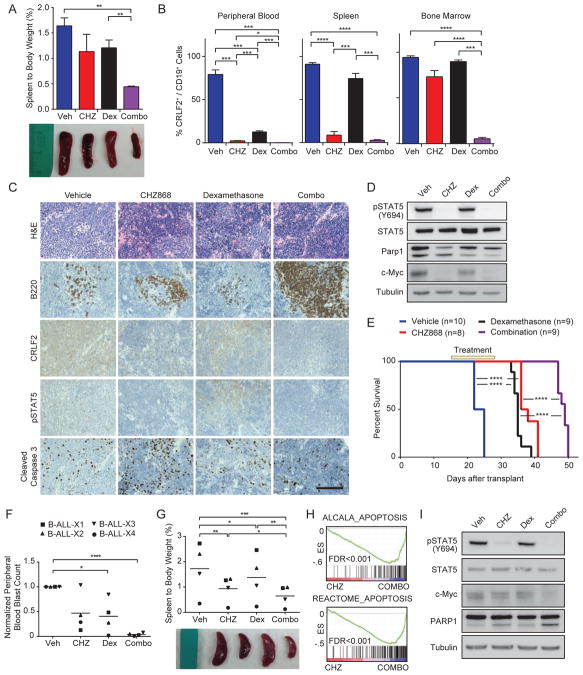
Mice were transplanted with Eμ-CRLF2/Eμ-Jak2 R683G/Ts1Rhr/Pax5+/− (line 2539p) B-ALL and randomized to treatment with vehicle, CHZ868 30 mg/kg/day by oral gavage, dexamethasone 1 mg/kg/day by intraperitoneal injection, or the combination beginning 15 days after transplant. After 5 days of treatment, mice receiving the combination had marked reductions in splenomegaly (Figure 5A), as well as decreased B-ALL in the peripheral blood, spleen, and bone marrow (Figure 5B). These findings were confirmed by IHC of the spleen, which showed restoration of murine B220+ cells in mice receiving the combination for only 5 days and clearance of cells expressing CRLF2 or pSTAT5 (Figure 5C). While cleaved caspase 3 was increased in spleens from mice treated with CHZ868 alone, it was not increased in the normal cells that occupied the spleens of mice treated with the combination (Figure 5C). Similarly, PARP1 cleavage was increased in the spleens of mice treated with CHZ868 alone but reduced in the spleens of mice treated with combination (Figure 5D), suggesting that the combination is not toxic to normal splenocytes.
Figure 5. CHZ868 with dexamethasone is highly active against CRLF2/JAK2-dependent leukemias in vivo.
Sublethally-irradiated wild-type mice were transplanted with Eμ-CRLF2/Eμ-Jak2 R683G/Pax5+/−/Ts1Rhr B-ALL. After 15 days, mice were treated with vehicle, CHZ868 (CHZ; 30 mg/kg/day by oral gavage), dexamethasone (dex; 1 mg/kg/day IP), or CHZ868 plus dexamethasone (combo). (A) Spleen weight as a percentage of total body weight after 5 days of treatment (n=3 mice/arm). Photograph of representative spleens from mice in each treatment arm are shown. (B) Percentage of human CRLF2+/mouse CD19+ cells in the peripheral blood, spleen, and bone marrow of mice after 5 days of treatment. (C, D) IHC (C, scale bar, 50 μm) and immunoblots (D) of spleens from mice in each treatment arm sacrificed after 5 days of treatment. (E) Survival of mice dosed with the indicated agents from days 15–28 after injection (yellow box). Survival was compared across arms using the log-rank test. (F) Upon engraftment of CRLF2-rearranged BALL PDXs (>2% peripheral blood human CD45+ cells), mice were treated for 5 days and then sacrificed. Normalized peripheral blood blast count (WBC count × %CD45+/CD19+ cells) relative to vehicle-treated controls. Horizontal lines denote the grand mean for each treatment condition. (G) Spleen weight as a percentage of total body weight is shown. Horizontal lines denote the grand mean for each treatment condition. (H) GSEA plots demonstrate enrichment of indicated apoptosis gene sets in PDXs treated with combination compared to CHZ868 alone for 3 days. (I) Representative immunoblotting of spleen lysates from B-ALL-X1 treated in vivo for 5 days with the indicated agents. In all panels of this figure, error bars represent SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. See also Figure S4.
We treated additional cohorts of mice transplanted with Eμ-CRLF2/Eμ-Jak2 R683G/Ts1Rhr/Pax5+/− B-ALL for 14 days beginning 15 days after transplant. Both dexamethasone and CHZ868 increased overall survival compared to vehicle treatment (Figure 5E). The combination further prolonged overall survival compared with CHZ868 or dexamethasone monotherapy, with an improvement in median survival from 23.5 days in vehicle-treated mice to 49 days in mice that received the combination for 14 days.
To confirm these findings in human B-ALL, we treated CRLF2-rearranged PDXs with vehicle, CHZ868 30 mg/kg/day, dexamethasone 1 mg/kg/day, or the combination for 5 days after engraftment (>2% peripheral blood human CD45+). Mice dosed with the combination also demonstrated marked reductions in peripheral blood leukemia compared with vehicle (Figure 5F). The combination treatment also reduced spleen involvement with B-ALL cells compared with vehicle, CHZ868, or dexamethasone monotherapy (Figure 5G). The transcriptional profiles of PDXs treated for 3 days in vivo with the combination compared to CHZ868 alone were highly enriched for all four apoptosis signatures (FDR<0.001; Figures 5H and S4D). Finally, the combination induced further loss of c-Myc and cleavage of PARP1 compared to either single agent (Figure 5I).
Discussion
Studies using human B-ALL cells with CRLF2 rearrangements reported that only a fraction are sensitive to type I JAK inhibitors, including ruxolitinib (Maude et al., 2012a; Tasian et al., 2012; Weigert et al., 2012). Treatment of CRLF2-rearranged B-ALL cells with type I inhibitors induces JAK2 hyperphosphorylation that may contribute to resistance to these agents (Weigert et al., 2012). This led us and others to pursue alternative strategies, such as HSP90 inhibitors (Weigert et al., 2012) or combinations of type I JAK2 inhibitors with additional agents (Maude et al., 2012b; Tasian et al., 2012; Waibel et al., 2013).
Approximately one-third of adult B-ALL cases harbor BCR-ABL rearrangements, which confer a very poor prognosis when treated with conventional chemotherapy regimens. The introduction of type II ABL kinase inhibitors like imatinib has dramatically changed the natural history of this disease (Foa et al., 2011). It has remained unclear whether patients with JAK2-dependent BALLs, including those with CRLF2-rearrangements, could achieve the same benefit through use of a type II JAK2 inhibitor.
Our drug discovery efforts led to the identification of CHZ868, a type II JAK2 inhibitor that potently blocks JAK2 signaling in vitro and in vivo. Broad kinome selectivity profiling suggested that CHZ868 interacts with a relatively small fraction of the kinome, including JAK2 and TYK2. However, we face the challenge that in vitro biochemical assays are predominantly based on phosphorylated/activated kinases. Thus, the selectivity profile obtained with type II inhibitors in biochemical assay formats may not reflect the selectivity in cells.
Recent studies using conditional deletions of Jak2 in mice have demonstrated defects in hematopoietic stem cells, as well as erythropoiesis and thrombopoiesis (Akada et al., 2014; Park et al., 2013). As a result, potent inhibition of JAK2 signaling could have significant hematopoietic toxicity and proper scheduling of JAK2 inhibition will be essential to achieve maximal anti-leukemic activity while maintaining tolerability. Of note, CHZ868 at a dose of 30 mg/kg/day was tolerated in NSG mice for up to 25 days and in immunocompetent mice for up to 44 days, with essentially no effects on peripheral blood counts. The trend towards elevated serum erythropoietin level in CHZ868-treated mice suggests that wild-type mice can return to homeostasis through physiological mechanisms. These data suggest that CHZ868 at doses comparable to 30 mg/kg/day in mice does not completely block JAK2 signaling in normal hematopoietic elements. This could provide a therapeutic index compared with JAK2-dependent leukemias that would not be present with complete loss of JAK2 function.
JAK1 associates with IL7Rα and JAK2 with CRLF2 in the context of the CRLF2/IL7Rα heterodimer (Sessa et al., 2013). Based on findings in myeloid cells (Koppikar et al., 2012), it is possible that TYK2 or other kinases could substitute for JAK1 and trans-phosphorylate JAK2 in B-ALL cells as well. This could help explain the relative insensitivity of CRLF2-rearranged B-ALL cells to agents with equipotent activity against JAK1 and JAK2 (ruxolitinib) or against all JAK family members (JAK inhibitor I) (Weigert et al., 2012). In contrast, type II inhibition stabilizes and locks JAK2 in the inactive conformation, preventing hyperphosphorylation and blocking downstream signaling (Andraos et al., 2012). Although we have not directly compared the in vivo efficacy of ruxolitinib to CHZ868, our results suggest that type II JAK2 inhibition may be a more effective strategy to target the broad genetic diversity of CRLF2-rearranged B-ALLs, including those with mutations in JAK2, CRLF2, IL7Rα, and SH2B3. This contrasts with B-ALLs that harbor JAK2 fusions, which appear to be highly sensitive to type I inhibitors (Roberts et al., 2014). Of note, Ba/F3 cells driven by TSLP are significantly less sensitive to both type I and type II inhibitors than cells dependent on mutant JAK2, CRLF2 or IL7Rα. Thus, future studies are needed to delineate the mechanisms that affect JAK2 inhibitor sensitivity in both TSLP-dependent and mutant JAK2-dependent contexts.
The mechanisms that mediate resistance to CHZ868 and other type II inhibitors in vivo were not explored in our study and remain unclear. Our in vitro resistance screen identified only one allele (L884P) that conferred resistance upon selection in BBT594 and none upon selection in CHZ868. This could have resulted from the relatively high ratio of inhibitor concentration to IC50 (approximately 100-fold), although our previous screens for resistance to type I inhibitors used similar ratios (Weigert et al., 2012). We failed to recover co-crystals of CHZ868 in complex with JAK2, but our binding model suggests that L884P confers resistance to type II inhibitors by changing the shape of the hydrophobic back pocket. Therefore, the failure to recover multiple different resistance alleles raises the intriguing possibility that few if any mutations within JAK2 are capable of altering this pocket without disrupting JAK2 function. Additional studies, including a comprehensive mutation analysis of all codons in the JAK2 kinase domain, will be necessary to confirm this possibility. The identification of JAK2 L884P as a de novo activating mutation (Torra et al., 2010) also indicates that patients who are candidates for type II JAK2 inhibitors will need to have their disease genotyped at this codon (and possibly others) to anticipate resistance.
Our data using PDX and transgenic mouse models, gene expression profiling, and BH3 profiling all suggest that the combination of CHZ868 with dexamethasone leads to greater apoptosis than either agent alone. Interrogation of the gene expression profiling data identified significant enrichment for apoptosis-associated gene sets but did not help clarify exactly how apoptosis may be initiated. Both JAK2 inhibition (Watanabe et al., 1996) and glucocorticoids (Ausserlechner et al., 2004; Rhee et al., 1995) can suppress MYC expression in lymphoid cells, as noted in our transgenic model. In addition, the Lock laboratory recently reported a glucocorticoid receptor binding site in a BIM intronic region that directly correlated with dexamethasone sensitivity in B-ALL PDX models (Jing et al., 2015). These results together further support the hypothesis that combinatorial effects of CHZ868 and dexamethasone involve the modulation of apoptotic networks.
In summary, CHZ868 abrogated JAK2 signaling in human CRLF2-rearranged B-ALL cells and transgenic CRLF2/JAK2-dependent leukemias, both in vitro and in vivo. We have also shown that the combination of CHZ868 and dexamethasone synergistically induced leukemia apoptosis in human CRLF2-rearranged cell lines and PDXs, and drastically improved overall survival in a murine B-ALL model. This combination is particularly attractive, based on the tolerability of dexamethasone, the ability of dexamethasone to promote apoptosis in B-ALL cells, and the precedent for combining tyrosine kinase inhibitors with glucocorticoids in patients with B-ALL. These data strongly argue for the translation of type II JAK2 inhibition in combination with dexamethasone into human trials.
Experimental procedures
All continuous variables were compared using a two-sided t-test or one-way ANOVA, followed by Dunnett’s test or Tukey’s test for multiple comparisons, and error bars in all figures represent the SEM. Survival curves were compared using the log-rank test with GraphPad software. BSK805, BVB808, BBT594, and CHZ868 were synthesized by Novartis. Dexamethasone, cytarabine, and doxorubicin were purchased from Sigma-Aldrich. Vincristine was purchased from Cayman Chemical. Ruxolitinib was purchased from SelleckChem.
Cell lines and random mutagenesis
Cell lines were maintained as previously described (Weigert et al., 2012). Ba/F3 (ATCC, USA) cells were stably transduced with CRLF2 (MSCVpuro), IL7Rα (MSCV-GFP), EpoR (MSCVpuro), or HA-hJAK2 (MSCVneo). JAK2 is either wild type or with the R683G, V617F, or L884P alteration. The mutagenesis screen was performed as previously described (Weigert et al., 2012), except that cells were selected in either 3 μM BBT594 or 3 μM CHZ868.
Immunoblotting
Immunoblotting was performed as previously described (Weigert et al., 2012), with pJAK2 (#3771), pSTAT5 (#4322), c-Myc (#9402), β-Actin (#4967), and JAK2 (#3230) antibodies from Cell Signaling Technologies, STAT5 antibody (sc-835) from Santa Cruz, and α-Tubulin antibody (T9026) from Sigma-Aldrich.
Flow cytometry
Splenocytes from transgenic murine leukemia models and primary human xenografts were prepared as previously described (Weigert et al., 2012). Cells were incubated with antibodies on ice for one hour before analysis. Human CD45-FITC (347463), CD19-PECy7 (557835), and CRLF2-PE (12-5499-42), mouse B220-PB (558108), CD3e-PB (55-8214), CD11b-FITC (11-0112-82), CD19-PerCPCy5.5 (45-0193-82), and CD43-APC (560663) antibodies were purchased from eBioscience. Apoptosis was quantified using Annexin V-FITC (Life Technologies, A13199) and Propidium Iodide (Roche Applied Science, 11348639001). All samples were analyzed on a BD FACSCanto II flow cytometer. Data were analyzed using FlowJo Version 9.5 (TreeStar).
In vitro inhibitor assays
Cell viability assays were performed as previously described (Weigert et al., 2012). Cells were plated at a density of 0.1 × 106 cells/mL (Ba/F3), 0.3 × 106 cells/mL (MHH-CALL4), or 0.6 × 106 cells/mL (PDXs) followed by addition of inhibitors or vehicle (DMSO). Media for PDX cells were supplemented with human cytokines (IL3, IL7, SCF, and Flt3L at 10 ng/mL, Peprotech), and 2-Mercaptoethanol (2 μL in 500 mL, Sigma-Aldrich). After 48 hr (Ba/F3 cells) or 72 hr (MHH-CALL4 and PDX cells), CellTiter-Glo® Luminescent Cell Viability Assay (Promega) was added (10 μL undiluted or 25 μL of a 1:2 dilution in each well) and plates were read by the 2104 EnVision® Multilabel Reader (PerkinElmer) or SpectraMax M3 Microplate Reader.
Each data point was quantified in triplicate or quadruplicate and cell line experiments were repeated at least twice. Analysis of pairwise dose-response data and isobologram plots was done according to the median-effect principle of Chou and Talalay (CalcuSyn Software). Dose response curves and plots were generated with GraphPad Prism software. Synergy experiments were performed in triplicate. Combination indices were calculated using CalcuSyn Software (Biosoft).
The JAK2 biochemical kinase assay was carried out in Caliper format with the predominantly activated (phosphorylated) kinase domain (amino acids 808-1132). Inhibition of erythropoietin-induced STAT5 nuclear translocation was assessed in U2OS cells that were stably transfected with the erythropoietin receptor and STAT5-GFP to enable imaging of cytoplasmic and nuclear levels of the transcription factor using the Cellomics automated imager. BH3 profiling was performed as previously described (Montero et al., 2015; Pan et al., 2014).
Murine experiments
De-identified human blood or bone marrow samples were obtained with informed consent under DFCI IRB-approved protocols #05-001 and #01-206. Xenografts were generated under DFCI IRB-approved protocol #13-351. All xenograft and transgenic animal studies were performed under DFCI IACUC approved protocol #13-034 and DFCI animal welfare assurance #A3023-01. Primary human B-ALLs (P0 or greater) were xenotransplanted into 6-week old NOD.SCID.IL2Rγ−/− (NSG) mice, as previously described (Weigert et al., 2012). Pharmacodynamic efficacy was assessed in mice 2 hr after the last dose. Immunohistochemistry was performed as previously described (Weigert et al., 2014; Baffert el al., 2010).
Gene expression profiling, marker selection and gene set enrichment analysis (GSEA)
MHH-CALL4 cells cultured at 1 × 106 cells/mL were treated with vehicle (DMSO), CHZ868 (500 nM), dexamethasone (250 nM), or the combination for 12 hr. The experiment was performed in triplicate. Primary human xenografts B-ALL-X1, -X2, and -X3 were treated in vivo with vehicle, CHZ868 (30 mg/kg/day PO), dexamethasone (1 mg/kg/day IP), or the combination for three days and then sacrificed 2 hr after the last dose. Total RNA was extracted from MHH-CALL4 cells and unselected splenocytes of the PDXs using Trizol followed by purification on an RNeasy column (Qiagen). RNA quality and concentration were confirmed by PicoGreen and profiling was performed on an Affymetrix human HTA-2_0 gene chip in the Dana-Farber Microarray Core (http://macf-web.dfci.harvard.edu).
Supplementary Material
Significance.
B-cell acute lymphoblastic leukemias (B-ALLs) with CRLF2 rearrangements have a poor outcome with currently available therapies. This has generated interest in targeting JAK2, which is downstream of CRLF2. Previous studies have demonstrated that type I JAK2 inhibitors have limited activity in cell lines and xenografts with CRLF2 rearrangements. In contrast, we show that CHZ868, a type II JAK2 inhibitor that stabilizes the kinase domain in an inactive conformation, is highly potent against these leukemias both in vitro and in vivo. CHZ868 and the glucocorticoid dexamethasone synergistically kill B-ALL cells dependent on JAK2, suggesting a promising combination strategy. Type II JAK2 inhibition should be pursued in patients with JAK2-dependent B-ALL as well as other disorders refractory to type I JAK2 inhibitors.
Acknowledgments
We thank Oliver Weigert for technical advice and previous constructs and Ilene Galinsky, Samia Ahmed, and Katharine Majewski for sample procurement and Shai Izraeli for thoughtful comments. We thank all the patients who contributed invaluable samples. We thank Aaron Thorner, Paul Van Hummelen, Matthew Ducar, and Bruce Wollison of the DFCI Center for Cancer Genome Discovery for assistance with genomic characterization. We thank Jerome Tamburini, Michael McKeown, and Elizabeth Townsend for their technical advice. This work was supported by the Dana-Farber/Novartis Drug Discovery Program (D.M.W.), the National Cancer Institute (R01 CA151898-01 (D.M.W.), T32 CA136432 (L.S.L.)), and the National Heart, Lung, and Blood Institute (T32 HL116324 (L.S.L.)).
Footnotes
Accession number
The Gene Expression Omnibus accession number for the gene expression data reported in this paper is GSE61696.
Additional experimental details are available in the Supplemental Material.
Author Contributions
S-C.W., L.S.L., N.K., T.R., C.G., and D.M.W. designed the experiments and interpreted the results. L.S.L., S-C.W., N.K., and D.M.W. wrote and edited the manuscript. S-C.W., L.S.L., N.K., J.M., J.Z., J.V.L., E.M., N.E., R.A., V.R., A.D., D.S., M.Z., A.L.C., H.L., A.C., and J.R. performed experiments and carried out analyses – in vitro and in vivo profiling of CHZ868 (E.M., N.E., R.A., V.R., A.D., D.S., and M.Z.), immunoblotting (S-C.W., L.S.L., and N.K.), mutagenesis screen (N.K. and J.V.L), in vitro inhibitor assays (L.S.L., S-C.W., N.K., A.Y., and J.Z.), mouse studies (E.M., N.E., F.B., S-C.W., L.S.L., A.L.C., H.L., and A.C.), genomic characterization of xenografts (N.K., L.S.L., and A.C.), BH3 profiling (L.S.L., J.M., and J.R.), and gene-expression profiling (L.S.L. and B.C.). S-C.W., A.L.C., D.V., T.T., and A.A.L. generated murine leukemia and PDX models. A.Y., D.J.D., R.M.S., D.S., M.W., M.H., L.B.S., and S.E.S. provided essential reagents and clinical samples. Immunohistochemistry was performed by M.M. and S.J.R. A.L. provided expertise in BH3 profiling. F.B., E.V., T.R., F.H., and C.G. developed NVP-CHZ868. E.V. and M.E. performed structural modeling.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute, National Heart, Lung, and Blood Institute, or the National Institutes of Health.
Declaration of Potential Conflicts of Interest
E.M., N.E., R.A., V.R., A.D., D.S., M.Z., M.M., F.H., F.B., E.V., T.R. and C.G. are or have been employees of Novartis Pharma AG. M.J.E. and D.M.W. received research funding from Novartis.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akada H, Akada S, Hutchison RE, Sakamoto K, Wagner KU, Mohi G. Critical role of Jak2 in the maintenance and function of adult hematopoietic stem cells. Stem Cells. 2014;32:1878–1889. doi: 10.1002/stem.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andraos R, Qian Z, Bonenfant D, Rubert J, Vangrevelinghe E, Scheufler C, Marque F, Regnier CH, De Pover A, Ryckelynck H, et al. Modulation of activation-loop phosphorylation by JAK inhibitors is binding mode dependent. Cancer discovery. 2012;2:512–523. doi: 10.1158/2159-8290.CD-11-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausserlechner MJ, Obexer P, Bock G, Geley S, Kofler R. Cyclin D3 and c-MYC control glucocorticoid-induced cell cycle arrest but not apoptosis in lymphoblastic leukemia cells. Cell Death Differ. 2004;11:165–174. doi: 10.1038/sj.cdd.4401328. [DOI] [PubMed] [Google Scholar]
- Azam M, Latek RR, Daley GQ. Mechanisms of autoinhibition and STI-571/imatinib resistance revealed by mutagenesis of BCR-ABL. Cell. 2003;112:831–843. doi: 10.1016/s0092-8674(03)00190-9. [DOI] [PubMed] [Google Scholar]
- Baffert F, Regnier CH, De Pover A, Pissot-Soldermann C, Tavares GA, Blasco F, Brueggen J, Chene P, Drueckes P, Erdmann D, et al. Potent and selective inhibition of polycythemia by the quinoxaline JAK2 inhibitor NVP-BSK805. Mol Cancer Ther. 2010;9:1945–1955. doi: 10.1158/1535-7163.MCT-10-0053. [DOI] [PubMed] [Google Scholar]
- Cesarman E. Pathology of lymphoma in HIV. Curr Opin Oncol. 2013;25:487–494. doi: 10.1097/01.cco.0000432525.70099.a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foa R, Vitale A, Vignetti M, Meloni G, Guarini A, De Propris MS, Elia L, Paoloni F, Fazi P, Cimino G, et al. Dasatinib as first-line treatment for adult patients with Philadelphia chromosome-positive acute lymphoblastic leukemia. Blood. 2011;118:6521–6528. doi: 10.1182/blood-2011-05-351403. [DOI] [PubMed] [Google Scholar]
- He M, Capelletti M, Nafa K, Yun CH, Arcila ME, Miller VA, Ginsberg MS, Zhao B, Kris MG, Eck MJ, et al. EGFR exon 19 insertions: a new family of sensitizing EGFR mutations in lung adenocarcinoma. Clin Cancer Res. 2012;18:1790–1797. doi: 10.1158/1078-0432.CCR-11-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzberg L, Vendramini E, Ganmore I, Cazzaniga G, Schmitz M, Chalker J, Shiloh R, Iacobucci I, Shochat C, Zeligson S, et al. Down syndrome acute lymphoblastic leukemia, a highly heterogeneous disease in which aberrant expression of CRLF2 is associated with mutated JAK2: a report from the International BFM Study Group. Blood. 2010;115:1006–1017. doi: 10.1182/blood-2009-08-235408. [DOI] [PubMed] [Google Scholar]
- Jing D, Bhadri VA, Beck D, Thoms JA, Yakob NA, Wong JW, Knezevic K, Pimanda JE, Lock RB. Opposing regulation of BIM and BCL2 controls glucocorticoid-induced apoptosis of pediatric acute lymphoblastic leukemia cells. Blood. 2015;125:273–283. doi: 10.1182/blood-2014-05-576470. [DOI] [PubMed] [Google Scholar]
- Koppikar P, Bhagwat N, Kilpivaara O, Manshouri T, Adli M, Hricik T, Liu F, Saunders LM, Mullally A, Abdel-Wahab O, et al. Heterodimeric JAK-STAT activation as a mechanism of persistence to JAK2 inhibitor therapy. Nature. 2012;489:155–159. doi: 10.1038/nature11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane AA, Chapuy B, Lin CY, Tivey T, Li H, Townsend EC, van Bodegom D, Day TA, Wu SC, Liu H, et al. Triplication of a 21q22 region contributes to B cell transformation through HMGN1 overexpression and loss of histone H3 Lys27 trimethylation. Nat Genet. 2014;46:618–623. doi: 10.1038/ng.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maude SL, Tasian SK, Vincent T, Hall JW, Sheen C, Roberts KG, Seif AE, Barrett DM, Chen IM, Collins JR, et al. Targeting JAK1/2 and mTOR in murine xenograft models of Ph-like acute lymphoblastic leukemia. Blood. 2012a;120:3510–3518. doi: 10.1182/blood-2012-03-415448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maude SL, Tasian SK, Vincent T, Hall JW, Sheen C, Roberts KG, Seif AE, Barrett DM, Chen IM, Collins JR, et al. Targeting JAK1/2 and mTOR in murine xenograft models of Ph-like acute lymphoblastic leukemia (ALL) Blood. 2012b doi: 10.1182/blood-2012-03-415448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesbah K, Rana MS, Francou A, van Duijvenboden K, Papaioannou VE, Moorman AF, Kelly RG, Christoffels VM. Identification of a Tbx1/Tbx2/Tbx3 genetic pathway governing pharyngeal and arterial pole morphogenesis. Human molecular genetics. 2012;21:1217–1229. doi: 10.1093/hmg/ddr553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero J, Sarosiek KA, DeAngelo JD, Maertens O, Ryan J, Ercan D, Piao H, Horowitz NS, Berkowitz RS, Matulonis U, et al. Drug-induced death signaling strategy rapidly predicts cancer response to chemotherapy. Cell. 2015;160:977–989. doi: 10.1016/j.cell.2015.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti S, Chapuy B, Takeyama K, Rodig SJ, Hao Y, Yeda KT, Inguilizian H, Mermel C, Currie T, Dogan A, et al. Integrative analysis reveals an outcome-associated and targetable pattern of p53 and cell cycle deregulation in diffuse large B cell lymphoma. Cancer Cell. 2012;22:359–372. doi: 10.1016/j.ccr.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse HC, 3rd, Anver MR, Fredrickson TN, Haines DC, Harris AW, Harris NL, Jaffe ES, Kogan SC, MacLennan IC, Pattengale PK, et al. Bethesda proposals for classification of lymphoid neoplasms in mice. Blood. 2002;100:246–258. doi: 10.1182/blood.v100.1.246. [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Collins-Underwood JR, Phillips LA, Loudin MG, Liu W, Zhang J, Ma J, Coustan-Smith E, Harvey RC, Willman CL, et al. Rearrangement of CRLF2 in B-progenitor- and Down syndrome-associated acute lymphoblastic leukemia. Nat Genet. 2009a;41:1243–1246. doi: 10.1038/ng.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Zhang J, Harvey RC, Collins-Underwood JR, Schulman BA, Phillips LA, Tasian SK, Loh ML, Su X, Liu W, et al. JAK mutations in high-risk childhood acute lymphoblastic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2009b;106:9414–9418. doi: 10.1073/pnas.0811761106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan R, Hogdal LJ, Benito JM, Bucci D, Han L, Borthakur G, Cortes J, DeAngelo DJ, Debose L, Mu H, et al. Selective BCL-2 inhibition by ABT-199 causes on-target cell death in acute myeloid leukemia. Cancer Discov. 2014;4:362–375. doi: 10.1158/2159-8290.CD-13-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SO, Wamsley HL, Bae K, Hu Z, Li X, Choe SW, Slayton WB, Oh SP, Wagner KU, Sayeski PP. Conditional deletion of Jak2 reveals an essential role in hematopoiesis throughout mouse ontogeny: implications for Jak2 inhibition in humans. PLoS One. 2013;8:e59675. doi: 10.1371/journal.pone.0059675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N Engl J Med. 2004;350:1535–1548. doi: 10.1056/NEJMra023001. [DOI] [PubMed] [Google Scholar]
- Rhee K, Bresnahan W, Hirai A, Hirai M, Thompson EA. c-Myc and cyclin D3 (CcnD3) genes are independent targets for glucocorticoid inhibition of lymphoid cell proliferation. Cancer research. 1995;55:4188–4195. [PubMed] [Google Scholar]
- Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang YL, Pei D, McCastlain K, Ding L, Lu C, Song G, et al. Targetable Kinase-Activating Lesions in Ph-like Acute Lymphoblastic Leukemia. N Engl J Med. 2014;371:1005–1015. doi: 10.1056/NEJMoa1403088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts KG, Morin RD, Zhang J, Hirst M, Zhao Y, Su X, Chen SC, Payne-Turner D, Churchman ML, Harvey RC, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22:153–166. doi: 10.1016/j.ccr.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell LJ, Capasso M, Vater I, Akasaka T, Bernard OA, Calasanz MJ, Chandrasekaran T, Chapiro E, Gesk S, Griffiths M, et al. Deregulated expression of cytokine receptor gene, CRLF2, is involved in lymphoid transformation in B-cell precursor acute lymphoblastic leukemia. Blood. 2009a;114:2688–2698. doi: 10.1182/blood-2009-03-208397. [DOI] [PubMed] [Google Scholar]
- Russell LJ, Capasso M, Vater I, Akasaka T, Bernard OA, Calasanz MJ, Chandrasekaran T, Chapiro E, Gesk S, Griffiths M, et al. Deregulated expression of cytokine receptor gene, CRLF2, is involved in lymphoid transformation in B cell precursor acute lymphoblastic leukemia. Blood. 2009b doi: 10.1182/blood-2009-03-208397. [DOI] [PubMed] [Google Scholar]
- Sessa C, Shapiro GI, Bhalla KN, Britten C, Jacks KS, Mita M, Papadimitrakopoulou V, Pluard T, Samuel TA, Akimov M, et al. First-in-human phase I dose-escalation study of the HSP90 inhibitor AUY922 in patients with advanced solid tumors. Clin Cancer Res. 2013;19:3671–3680. doi: 10.1158/1078-0432.CCR-12-3404. [DOI] [PubMed] [Google Scholar]
- Shiels MS, Pfeiffer RM, Gail MH, Hall HI, Li J, Chaturvedi AK, Bhatia K, Uldrick TS, Yarchoan R, Goedert JJ, Engels EA. Cancer burden in the HIV-infected population in the United States. Journal of the National Cancer Institute. 2011;103:753–762. doi: 10.1093/jnci/djr076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shochat C, Tal N, Bandapalli OR, Palmi C, Ganmore I, te Kronnie G, Cario G, Cazzaniga G, Kulozik AE, Stanulla M, et al. Gain-of-function mutations in interleukin-7 receptor-alpha (IL7R) in childhood acute lymphoblastic leukemias. J Exp Med. 2011;208:901–908. doi: 10.1084/jem.20110580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shochat C, Tal N, Gryshkova V, Birger Y, Bandapalli OR, Cazzaniga G, Gershman N, Kulozik AE, Biondi A, Mansour MR, et al. Novel activating mutations lacking cysteine in type I cytokine receptors in acute lymphoblastic leukemia. Blood. 2014;124:106–110. doi: 10.1182/blood-2013-10-529685. [DOI] [PubMed] [Google Scholar]
- Tasian SK, Doral MY, Borowitz MJ, Wood BL, Chen IM, Harvey RC, Gastier-Foster JM, Willman CL, Hunger SP, Mullighan CG, Loh ML. Aberrant STAT5 and PI3K/mTOR pathway signaling occurs in human CRLF2-rearranged B-precursor acute lymphoblastic leukemia. Blood. 2012;120:833–842. doi: 10.1182/blood-2011-12-389932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torra OS, Gundacker H, Pogosov G, Pogosova-Agadjanyan E, Forman SJ, O’Donnell M, Appelbaum FR, Willman C, Radich JP. Analysis of CRLF2/JAK Expression and Mutation Status In Adult ALL Patients. American Society of Hematology National Meeting; New Orleans, LA. 2010. p. Abstract 758. [Google Scholar]
- Waibel M, Solomon VS, Knight DA, Ralli RA, Kim SK, Banks KM, Vidacs E, Virely C, Sia KC, Bracken LS, et al. Combined targeting of JAK2 and Bcl-2/Bcl-xL to cure mutant JAK2-driven malignancies and overcome acquired resistance to JAK2 inhibitors. Cell reports. 2013;5:1047–1059. doi: 10.1016/j.celrep.2013.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Itoh T, Arai K. JAK2 is essential for activation of c-fos and c-myc promoters and cell proliferation through the human granulocyte-macrophage colony-stimulating factor receptor in BA/F3 cells. J Biol Chem. 1996;271:12681–12686. doi: 10.1074/jbc.271.21.12681. [DOI] [PubMed] [Google Scholar]
- Weigert O, Lane AA, Bird L, Kopp N, Chapuy B, van Bodegom D, Toms AV, Marubayashi S, Christie AL, McKeown M, et al. Genetic resistance to JAK2 enzymatic inhibitors is overcome by HSP90 inhibition. J Exp Med. 2012;209:259–273. doi: 10.1084/jem.20111694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlmann A, Sebastian K, Borowski A, Krause S, Friedrich K. Signal transduction by the atopy-associated human thymic stromal lymphopoietin (TSLP) receptor depends on Janus kinase function. Biological chemistry. 2010;391:181–186. doi: 10.1515/bc.2010.029. [DOI] [PubMed] [Google Scholar]
- Yoda A, Yoda Y, Chiaretti S, Bar-Natan M, Mani K, Rodig SJ, West N, Xiao Y, Brown JR, Mitsiades C, et al. Functional screening identifies CRLF2 in precursor B-cell acute lymphoblastic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:252–257. doi: 10.1073/pnas.0911726107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.