ABSTRACT
The interferon (IFN) response is the earliest host immune response dedicated to combating viral infection. As such, viruses have evolved strategies to subvert this potent antiviral response. Two closely related gammaherpesviruses, Kaposi's sarcoma-associated herpesvirus (KSHV) and rhesus macaque rhadinovirus (RRV), are unique in that they express viral homologues to cellular interferon regulatory factors (IRFs), termed viral IRFs (vIRFs). Cellular IRFs are a family of transcription factors that are particularly important for the transcription of type I IFNs. Here, we demonstrate a strategy employed by RRV to ensure rapid inhibition of virus-induced type I IFN induction. We found that RRV vIRF R6, when expressed ectopically, interacts with a transcriptional coactivator, CREB-binding protein (CBP), in the nucleus. As a result, phosphorylated IRF3, an important transcriptional regulator in beta interferon (IFN-β) transcription, fails to effectively bind to the IFN-β promoter, thus inhibiting the activation of IFN-β genes. In addition, we found R6 within RRV virion particles via immunoelectron microscopy and, furthermore, that virion-associated R6 is capable of inhibiting the type I IFN response by preventing efficient binding of IRF3/CBP complexes to the IFN-β promoter in the context of infection. The work shown here is the first example of a vIRF being associated with either the KSHV or RRV virion. The presence of this immunomodulatory protein in the RRV virion provides the virus with an immediate mechanism to evade the host IFN response, thus enabling the virus to effectively establish an infection within the host.
IMPORTANCE Kaposi's sarcoma-associated herpesvirus (KSHV) and the closely related rhesus macaque rhadinovirus (RRV) are the only viruses known to encode viral homologues to cellular interferon regulatory factors (IRFs), known as vIRFs. In KSHV, these proteins have been shown to play major roles in a variety of cellular processes and are particularly important in the evasion of the host type I interferon (IFN) response. In this study, we delineate the immunomodulatory mechanism of an RRV vIRF and its ability to assist the virus in rapid immune evasion by being prepackaged within the virion, thus providing evidence, for the first time, of a virion-associated vIRF. This work further contributes to our understanding of the mechanisms behind immunomodulation by the RRV vIRFs during infection.
INTRODUCTION
Activation of type I interferons (IFNs) is a fundamental component of a host's antiviral response. Type I IFNs (alpha and beta interferon [IFN-α and -β]) are produced by virus-infected cells and are responsible for initiating the primary response to viral infection. IFN-α is produced largely by plasmacytoid dendritic cells (pDCs) (1), whereas IFN-β is produced by a variety of cell types, including epithelial and endothelial cells, fibroblasts, monocytes/macrophages, and B cells (2, 3). The induction of IFNs is a tightly regulated process controlled by IFN regulatory factors (IRFs). Within the broad family of IRFs, IRF3 and -7 are specifically important for the induction of type I IFNs (4, 5). IRF3 is constitutively expressed in all cell types, while IRF7 is induced by type I IFNs. As a consequence of viral infection or treatment with double-stranded RNA (dsRNA), inactive cytoplasmic IRF3 is phosphorylated, after which it forms a homodimer and is translocated to the nucleus (2, 5). Phosphorylated IRF3 (pIRF3) then interacts with the transcriptional coactivator CREB-binding protein (CBP)/p300 and binds the positive regulatory domain I (PRDI)-PRDIII element of the IFN-β promoter, thus inducing transcription. To effectively establish infection within a host, viruses have evolved a variety of mechanisms that specifically target cellular IRFs, induction of IFNs, and downstream signaling pathways induced by IFNs (6–8).
Kaposi's sarcoma (KS)-associated herpesvirus (KSHV) and rhesus rhadinovirus (RRV) are two closely related gamma-2 herpesviruses that are capable of inducing lymphoproliferative disorders in their respective immunocompromised hosts. Both KSHV and RRV encode multiple viral homologues to cellular proteins that play various roles in cellular processes, such as apoptosis, cellular growth and differentiation, and immune signaling. These viral proteins have the ability to manipulate the host environment in a manner that maximizes conditions for efficient viral replication. In particular, these viral homologues, namely, viral CD200 (vCD200), viral interleukin 6 (vIL-6), viral G protein-coupled receptor (vGPCR), and the viral interferon regulatory factors (vIRFs), are critical modulators of the immune response and cell growth and also major players in disease pathogenesis (6). Although it is a common attribute of many viruses to carry genes that subvert the immune system, KSHV and RRV are unique in that they are the only two viruses known to encode vIRFs, which are homologous to cellular IRFs (9–12). RRV encodes a cluster of 8 vIRFs, with genes localized in the same genomic region as the genes encoding 4 vIRFs found in KSHV (9, 12). The unique sequence homology shared between the vIRFs and cellular IRFs suggests a possible role for vIRFs in evasion of the IFN response. In fact, it has been shown that 3 of the KSHV vIRFs (vIRF1, -2, and -3) disrupt functions of cellular IRFs, ultimately altering IFN induction, as well as downstream signaling of IFN (13–25). Interestingly, each of these vIRFs has distinct functions in targeting particular elements of the IFN response. For example, KSHV vIRF1, the best studied of the IRFs, blocks transcription of type I IFNs in a variety of ways. KSHV vIRF1 binds the transcriptional coactivator proteins CBP and p300, thus interfering with their binding and function (14, 18, 26, 27). Also, the interaction between vIRF1 and p300 displaces CBP/p300-asssociated factor (pCAF), a protein that possesses histone acetyltransferase (HAT) activity, effectively inhibiting the HAT activity of p300, which is important for altering chromatin structure and making DNA available for transcription (26). Furthermore, vIRF1 can displace IRF3 from CBP/p300, thus inhibiting the transcriptional activity of IRF3 (18). KSHV vIRF2 blocks both NF-κB- and IRF1-dependent IFN-β transcription (15) and has recently been found to target IFN-stimulated gene (ISG) factor 3 (ISGF3) as a means of inhibiting type I IFN signaling (25). Work on KSHV vIRF3 has been less clear, as some studies have reported an increase in IFN-α transcription and expression in the presence of vIRF3 (17) and others have shown inhibition of IRF7-mediated signal transduction (22) and decreased IFN-α expression (16). Not only do KSHV vIRFs function to limit the IFN response, but they also play a significant role in modulation of the cell cycle and apoptosis. When expressed in NIH 3T3 cells, vIRF1 displayed oncogenic properties, and this was further corroborated when vIRF1-expressing cells formed tumors after injection into nude mice (21). Furthermore, KSHV vIRF1, -3, and -4 all act as negative modulators of the p53 pathway, a vital pathway involved in the induction of cell cycle arrest or apoptosis and thus important in tumor suppression (27–31). Additionally, KSHV vIRF2 downregulates CD95L expression in activated T cells, thus inhibiting activation-induced cell death (32) and providing evidence for yet another immunomodulatory function of the KSHV vIRFs. The above-mentioned studies have provided a vast amount of insight into the molecular mechanisms of the KSHV vIRFs, albeit the majority of the work has been done in the absence of de novo KSHV infection. This is mainly due to the fact that in vitro replication of KSHV is particularly inefficient. RRV, however, displays robust lytic replication in vitro, and with its 8 vIRFs that share homology to the KSHV vIRFs, RRV provides a suitable model for the study of the function of vIRFs early in de novo infection.
Recent studies have tested whether the RRV vIRFs are capable of antagonizing the function of cellular IRFs and IRF-mediated induction of IFN (33, 34). This was achieved via the construction of a recombinant RRV lacking all 8 vIRF open reading frames (ORFs) (vIRF-ko RRV) (33), the backbone of which is derived from a bacterial artificial chromosome (BAC) clone of wild-type (WT) RRV17757 (WTBACRRV) (35). Infection of rhesus macaques with vIRF-ko RRV resulted in lower viral loads and increased levels of plasma IFN-α. Interestingly, although persistence was established in the absence of the RRV vIRFs, decreased levels of persistent virus were detected in B cells, along with decreased development of B cell hyperplasia (34). The decrease in B cell hyperplasia, however, could be due to smaller amounts of circulating virus within these animals. In vitro, the vIRFs prevented early nuclear accumulation of pIRF3, the activated state of the transcription factor that prompts the induction of type I IFNs (33). No change was observed, however, in total IRF3 levels or in IRF3 dimerization, indicating that the mechanism of action of the RRV vIRFs occurs after IRF3 phosphorylation, potentially by interfering with nuclear accumulation of pIRF3 itself (33). Examination of individual RRV vIRFs in the absence of infection revealed a potential interaction between R6 and IRF3 and also identified R6 as an important inhibitor of IRF3-mediated transcription (33).
To further dissect the potential immunomodulatory role of R6, we examined its effect on the induction of IFN and the signaling cascade leading up to IFN-β transcription. Here, we show a delay in IRF3-mediated transcription, in accordance with previous data (33), along with a decrease in the transcription of IFN-β. In terms of the mechanism, we found that R6 competes with IRF3 for binding to CBP, resulting in a decrease of IRF3/CBP complexes present on the IFN-β promoter. Remarkably, we detected R6 in purified virus preparations, as well as in the context of infection without protein translation. Our results strongly suggest that R6 is a virion-associated protein that functions to inhibit early type I IFN responses in virus-infected cells.
MATERIALS AND METHODS
Cells, virus, plasmids, and drugs.
Primary rhesus fibroblasts (RFs), telomerized rhesus fibroblasts (tRFs), tRF–interferon-stimulated response elements (ISREs) (36), and HEK 293T/17 cells were grown in Dulbecco's modified Eagle's medium (DMEM) (Mediatech, Herndon, VA) supplemented with 10% fetal bovine serum (FBS) (HyClone, Ogden, UT). All cell culture incubations were carried out at 37°C in a humidified atmosphere containing 5% CO2. The viruses used in these studies included WT BAC-derived RRV17577 (WTBACRRV) (35), as well as vIRF-ko RRV17577 (vIRF-ko RRV) (33) and a WTBACRRV with a hemagglutinin (HA) tag at the C terminus of the R6 ORF (R6HA RRV). All virus stocks were purified through a 30% sorbitol cushion and resuspended in phosphate-buffered saline (PBS) unless otherwise indicated, and titers in RFs were determined using a standard plaque assay. Construction of plasmids pcDNA3.1-R6HA and pcDNA3.1-R7HA was described previously (33). The cycloheximide (CHX) (Sigma-Aldrich, St. Louis, MO) stock concentration (in ethanol) was 15 mg/ml, and the working concentration was 75 μg/ml. The MG132 (Sigma) stock concentration (in dimethyl sulfoxide [DMSO]) was 10 mM, and the working concentration was 5 μM. Cells were pretreated with the drugs 2 h before infection or transfection, and the drug remained in the medium as the infection or transfection took place unless otherwise indicated. Poly(I · C) (Sigma) was resuspended in PBS and transfected into cells by using TransIT LT1 transfection reagent (Mirus, Madison, WI).
Purification of RRV virions.
RFs were infected with the indicated virus at a multiplicity of infection (MOI) of 0.01 PFU per cell and allowed to progress to complete cytopathic effect (CPE). The supernatant and cells were collected and separated by centrifugation at 1,000 × g for 15 min at 4°C. The supernatant, containing extracellular virus, and the pellet, containing intracellular and cell-associated virus, were processed separately. The extracellular-virus-containing supernatant was further centrifuged at 12,000 × g for 1 h at 4°C, after which samples were resuspended in DMEM and sonicated twice for 30 s each time. The intracellular-virus-containing pellet was freeze-thawed, sonicated 2 times for 30 s each time, and centrifuged at 1,000 × g for 15 min at 4°C, and the supernatant was saved for further processing. The supernatants containing either extracellular virus or intracellular virus were then pelleted through a 30% sorbitol cushion in a Beckman SW41 rotor at 18,000 rpm for 1 h at 4°C. The pellets were resuspended in 1 ml 1× PBS, layered on a 20 to 60% sorbitol step gradient, and spun in a Beckman SW41 rotor at 18,000 rpm for 2 h at 4°C. The virus band was collected at the 50%-60% interface, which coincides with infectious virus as defined by plaque assay. The gradient-purified preparation was diluted in 15 ml cold 1 mM Tris-HCl and then pelleted by centrifugation in the SW41 rotor at 18,000 rpm for 50 min at 4°C. The virus pellet was then resuspended in Hanks' balanced salt solution (HBSS) plus 2% FBS and stored at −80°C.
Plasmid transfections and virus infections of cell cultures.
tRFs and tRF-ISREs were transfected with the indicated expression plasmids using the TransIT-LT1 transfection reagent (Mirus Bio). Transfections proceeded for 40 h before transfection of poly(I · C). The transfection efficiency for the tRFs was approximately 10 to 40%, as defined using the pQ100 plasmid encoding enhanced green fluorescent protein (eGFP) under the transcriptional control of the EF1-alpha promoter. RFs, tRFs, and tRF-ISREs were infected at an MOI of 2.5 PFU per cell. Infected cells or coverslips were collected at different time points postinfection (p.i.) for further analysis.
Immunoprecipitation (IP), SDS-PAGE analysis, and Western blotting.
Cell lysates were immunoprecipitated with rabbit anti-CBP polyclonal antibody (PAb) (A-22; Santa Cruz Biotechnology, Santa Cruz, CA) in native lysis buffer (50 mM Tris-Cl [pH 8.0], 1% NP-40, and 150 mM NaCl supplemented with phosphatase inhibitors [100× cocktail; Sigma] and protease inhibitors [100× cocktail; Sigma]), followed by incubation with Protein A/G Plus-agarose (Santa Cruz) and eluted in radioimmunoprecipitation assay (RIPA) buffer (native lysis buffer with 0.1% SDS and 0.5% sodium deoxycholate). Whole-cell extracts were collected in RIPA buffer, and nuclear and cytoplasmic lysates were collected according to kit protocols (NE-PER; Thermo Scientific). Samples were analyzed with Novex 4% to 12% Tris-Bis Mini Gels (Life Technologies Inc., Carlsbad, CA), and proteins were then transferred onto a polyvinylidene difluoride (PVDF) membrane (Bio-Rad Laboratories Inc., Hercules, CA) via semidry transfer (30 min at 15 V at room temperature).
The membranes were blocked for 1 h in TBS-T (Tris-buffered saline with 0.1% Tween 20) containing 5% bovine serum albumin (BSA) and subsequently probed with the following antibodies. The primary antibodies used in this study were anti-HA PAb (Y-11; Santa Cruz), anti-HA monoclonal antibody (MAb) (HA-7; Sigma), anti-CBP PAb (A-22; Santa Cruz), anti-IRF3 (SL-12; Santa Cruz), anti-human phosphor-IRF3 (Ser396) (4D4G; Cell Signaling Technology, Beverly, MA), anti-TBK (M-375; Santa Cruz), anti-human poly(ADP-ribose) polymerase 1/2 (PARP1/2) PAb (H-250; Santa Cruz), anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) MAb (SC-51906; Santa Cruz), and anti-RRV major capsid protein (MCP) MAb (Monoclonal Antibody Core, Vaccine and Gene Therapy Institute, Beaverton, OR). Secondary antibodies consisted of horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Santa Cruz) and goat anti-rabbit IgG-HRP (Cell Signaling). The membranes were treated with SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific), and bands were detected by exposing the membranes to X-ray film (Kodak; BioMax XAR film). The films were developed and band intensities were determined using densitometry in ImageJ.
Immunofluorescence.
Cells were grown on glass coverslips in 12-well plates and fixed with 4% paraformaldehyde in PBS (20 min at room temperature). The cells were then permeabilized and blocked in 5% normal goat serum (NGS)-0.1% Triton X in PBS (PBST) (1 h at room temperature) prior to staining, and all subsequent steps were performed with 1% NGS-PBST. Cells on coverslips were stained with anti-human IRF3 MAb (clone SL012.1; BD Pharmingen, San Diego, CA) or anti-CBP PAb (A-22; Santa Cruz) overnight at 4°C and subsequently stained with anti-mouse IgG-Texas Red (Vector Laboratories, Burlingame, CA) or anti-rabbit IgG-Texas Red (Vector Laboratories), respectively. The cells were then stained with anti-HA-fluorescein isothiocyanate (FITC) (HA-7; Sigma) (2 h at room temperature), and nuclei and/or DNA was detected by using Hoechst 33258 dye. Cells on coverslips were mounted onto slides using Vectashield (Vector Laboratories) and examined on a Zeiss Axio Imager.M1 microscope (Zeiss Imaging Solutions, Thornwood, NY). Images were acquired by using a Zeiss Axiocam camera (MRm) with Axiovision software (version 4.6) and subsequently processed by using Adobe Photoshop (Adobe Systems, San Jose, CA).
RNA isolation and RT-PCR.
RNA was isolated from tRFs by using TRI reagent (Sigma), and DNA endonuclease (RQ1) was used to remove DNA from RNA preparations (Promega, Fitchburg, WI) according to commercial-kit protocols. Reverse transcription (RT)-PCR was performed by using Superscript III one-step RT-PCR with Platinum Taq (Life Technologies Inc., Carlsbad, CA). Transcripts were detected with the following primers: IFN-β 5′ primer (5′-GAC GCC GCA TTG ACC ATC TA-3′), IFN-β 3′ primer (5′-CCT TAG GAT TTC CAC TCT GAC T-3′), GAPDH 5′ primer (5′-GTG GAT ATT GTT GCC ATC AAT-3′), and GAPDH 3′ primer (5′-ATA CTT CTC ATG GTT CAC ACC-3′). All data were normalized to the level of GAPDH in each sample.
Construction of R6HA RRV.
The construction of R6HA RRV was achieved by using the WT RRV BAC (35) in conjunction with the galK recombination system in Escherichia coli SW105 cells (37). Initially, a BAC clone lacking R6 was generated by recombination with a galK cassette flanked by 50-bp arms homologous to the region outside the R6 ORF (nucleotides [nt] 76216 to 80463), and a clone lacking R6 was identified. Next, a DNA cassette possessing C-terminally HA-tagged R6 and 50-bp flanking homology arms was cloned into pcDNA3.1(−) and sequenced. The HA-tagged R6 cassette, along with the homology arms, was excised from the expression vector and used to replace the galK cassette. GalK-negative recombinants were selected for resistance to 2-deoxygalactose in minimal medium with glycerol as the sole carbon source. Individual R6HA RRV clones were analyzed via restriction digestion in conjunction with Southern blot analysis and were subsequently analyzed via comparative genome hybridization (CGH) (NimbleGen Systems, Inc., Madison, WI), as described previously (35). Single-step growth curve analysis (MOI = 2.5) revealed no difference between R6HA RRV and WTBAC RRV. RT-PCR analysis of total cellular RNA isolated from R6HA- or WTBACRRV-infected cells to detect R6, R7, ORF57, and cellular GAPDH transcripts was performed using ORF-specific primers as described previously (33).
Generation of a doxycycline-inducible stable cell line.
The pLVX lentivirus vector system was utilized for constructing a stable doxycycline (Dox)-inducible cell line. The cell lines and vectors used in the construction of this cell line were obtained from Victor DeFilippis (Vaccine and Gene Therapy Institute, Beaverton, OR). The pLVX-R6HA plasmid was constructed by subcloning full-length HA-tagged R6 from the pcDNA3.1(−) expression vector, described in reference (33), into the pLVX-Tight-Puro retroviral vector. Replication-defective recombinant retrovirus was produced by transfecting the retroviral vector into HEK 293T/17 cells, along with a 2nd-generation packaging system (packaging plasmid psPAX2 and envelope plasmid pMD2.g). The supernatant was harvested 48 h later, purified by centrifugation, and filtered through a 0.45-μm filter to remove cell debris. Target cells (tRFs containing a Dox-responsive transactivator [tRF-rtTAs]) were exposed to the purified retrovirus for 3 h in the presence of Polybrene to facilitate infection, and this process was repeated once more 48 h later. Once the cells reached confluence, they were grown in DMEM plus 10% Tet-free FBS containing 1.5 μg/ml puromycin (Sigma) and 150 μg/ml hygromycin B (Fisher Scientific, Pittsburgh, PA). The cells were continually passaged in the presence of increasing concentrations of puromycin and hygromycin B (3 μg/ml and 300 μg/ml, respectively) until the cells were fully resistant. In order to determine the optimal concentration of Dox and duration of Dox treatment, cells were experimentally treated with Dox at various concentrations and for various lengths of time. We found that 1 μg/ml of Dox for 24 h yielded the most protein expression.
EMSA.
Nuclear extracts were prepared after transfection or infection according to kit protocols (NE-PER; Thermo Scientific). Equivalent amounts of nuclear extract (20 μg) were assayed for IRF3 and CBP binding in gel shift analysis using a 3′-biotin-labeled double-stranded oligonucleotide corresponding to the PRDI-PRDIII region of the IFN-β promoter: 5′-GAAAACTGAAAGGAGAACTGAAAGTG-3′. Biotin labeling was performed using a DNA 3′-end biotinylation kit (Thermo). Complexes were formed by incubating the probe (final concentration, 20 fmol) with 20 μg of nuclear lysate in the presence or absence of the indicated antibodies. The binding reaction mixture (20 μl) contained 2.5% glycerol, 5 mM MgCl2, 0.05% NP-40, and 50 ng/μl poly(dI-dC) to reduce nonspecific binding. To demonstrate the specificity of protein-DNA complex formation, 500-fold molar excess of unlabeled wild-type oligonucleotide corresponding to the PRDI-PRDIII region of the IFN-β promoter was added before adding labeled probe or was preincubated with anti-CBP (H100; Santa Cruz) or anti-IRF3 (SL-12; Santa Cruz). After 20 min of incubation with probe, binding reaction mixtures were loaded on a Novex 6% DNA retardation gel (Life Technologies) and run for 1 h at 100 V. Samples were then transferred to a positively charged nylon membrane (Immobilon-Ny+; Millipore, Billerica, MA) via semidry transfer (30 min at 15 V at room temperature) and detected using streptavidin-HRP chemiluminescence for biotin-labeled probes (LightShift Chemiluminescent electrophoretic mobility shift assay [EMSA] kit; Thermo Scientific). The membranes were exposed to X-ray film, and band intensities were analyzed by densitometry in ImageJ.
Luciferase assay.
IRF3-mediated transcription was measured by using tRFs carrying the firefly luciferase gene driven by the ISRE in the promoter (tRF-ISREs), generously provided by Victor DeFilippis. The firefly luciferase readings were normalized to the constitutive expression of Renilla luciferase (pRL-SV40; Promega). tRF-ISREs were transfected for 40 h with 50 ng pcDNA3.1-R6HA or empty pcDNA3.1(−), along with 10 ng pRL-SV40. The cells were then transfected with 10 μg poly(I · C) for 8 h or the indicated time and analyzed with the Dual-Glo luciferase assay according to the manufacturer's protocol (Promega). For infection studies, tRF-ISREs were transfected with the pRL-SV40 plasmid to constitutively express Renilla, infected with individual RRV isolates, and analyzed with the Dual-Glo luciferase assay at defined times postinfection.
Immunogold EM.
Gradient-purified virus preparations of extracellular R6HA RRV were fixed in 2.5% gluteraldehyde, pelleted, and embedded in resin. Thin sections were made, incubated with anti-HA antibody (Y-11; Santa Cruz), and subsequently incubated with 10-nm gold-conjugated secondary goat anti-mouse IgG (Ted Pella Inc., Redding, CA). The percentage of virion-localized gold particles was determined by manually counting the gold particles associated with virions, as well as total gold particles, within three separate fields of view. This was then used to calculate the percentage. Samples were imaged at 120 kV on a FEI Tecnai Spirit transmission electron microscopy (TEM) system. Images were acquired as 2,048- by 2,048-pixel, 16-bit gray scale files using FEI's TEM imaging and analysis (TIA) interface on an Eagle 2K charge-coupled-device (CCD) multiscan camera.
Statistical analysis.
Data were analyzed using GraphPad Instat (GraphPad Software, La Jolla, CA), and significant differences were determined using a paired t test, with P values of ≤0.05 considered significant and P values from 0.05 to 0.1 considered to indicate a significant trend.
RESULTS
R6 vIRF is sufficient for the inhibition of IFN-β transcription.
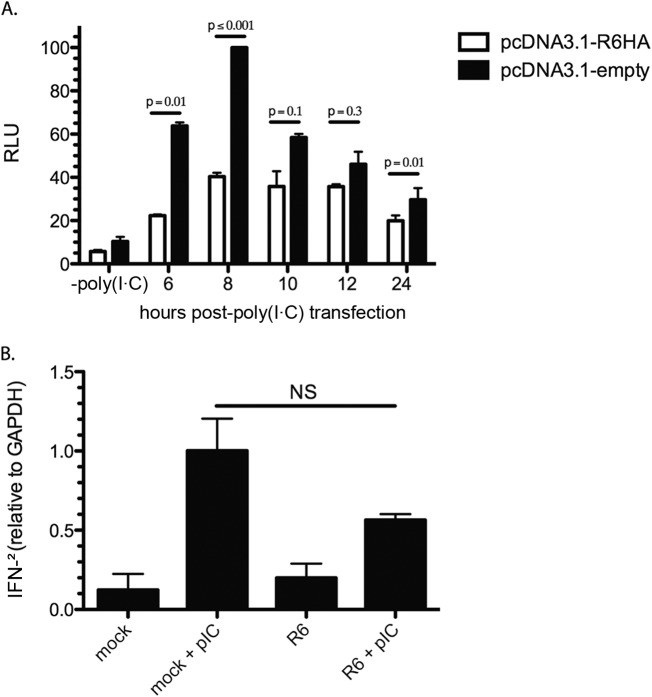
Since nuclear accumulation of pIRF3 was inhibited at time points earlier than 8 h p.i. with WT RRV and not vIRF-ko, it was important to characterize the early kinetics of R6 on IFN production that was previously observed (33) in order to determine if R6 was indeed responsible for pIRF3 modulation. To achieve this, tRF-ISREs were transfected with an R6 expression clone with a C-terminal HA tag or empty vector as a control, and the cells were then transfected 40 h later with poly(I · C) to induce the activation of IRF3. Luciferase levels were analyzed at 6, 8, 10, 12, and 24 h post-poly(I · C) transfection as a means of tracking the early effects on transcription. Relative luciferase units (RLU) were determined by defining 100% RLU as the output of poly(I · C)-transfected cells transfected with empty vector at 8 h. A significant decrease in luciferase was observed in the presence of RRV vIRF R6 at 6 and 8 h post-poly(I · C) transfection. Similar to previous results, R6 expression led to a reduction in luciferase levels at all time points post-poly(I · C) transfection compared to empty vector. (Fig. 1A). A decrease in luciferase was also evident pre-poly(I · C) transfection in R6HA-transfected cells compared to empty vector. We believe this is most likely due to the presence of R6HA.
FIG 1.
R6 inhibits the IFN response. (A) tRF-ISREs were transfected for 40 h with pRL-SV40 and either pcDNA3.1-R6HA or empty pcDNA3.1. The cells were then transfected with poly(I · C) and assayed at the indicated time points (bottom). Firefly luciferase levels were normalized to constitutively expressed Renilla luciferase levels in each well, and all samples were normalized to the positive control [empty vector plus poly(I · C) at 8 h post-poly(I · C) transfection]. The data are averages (and standard errors of the mean [SEM]) from 3 independent experiments and are represented as RLU. (B) Telomerized RFs were transfected with pcDNA3.1-R6HA or mock transfected for 40 h and subsequently transfected with poly(I · C) for 8 h. RNA was extracted and analyzed by RT-PCR. The values are relative to GAPDH levels. The data were analyzed using a paired t test. P values of less than 0.05 were considered significant, and values greater than 0.05 were not significant (NS).
Cellular IRF3, upon activation and nuclear translocation, induces IFN-β transcription by binding to the IRF element (IRF-E) present in the PRD of the IFN-β promoter (38). IFN-β then acts in an autocrine and paracrine fashion to stimulate the transcription of ISGs. To determine if the effect on IRF3-mediated transcription by R6 vIRF directly impacts IFN transcription, IFN-β transcripts were analyzed with or without ectopic expression of R6. We found that induction of IFN-β in R6-transfected tRFs was diminished by nearly 50% at 8 h post-poly(I · C) transfection, in contrast to cells transfected with empty vector (Fig. 1B). These data demonstrate that in cells transfected with poly(I · C) to induce IRF3 activation, R6 vIRF is sufficient for early inhibition of IFN-β transcription.
R6 affects IRF3 translocation to the nucleus and interacts with CBP.
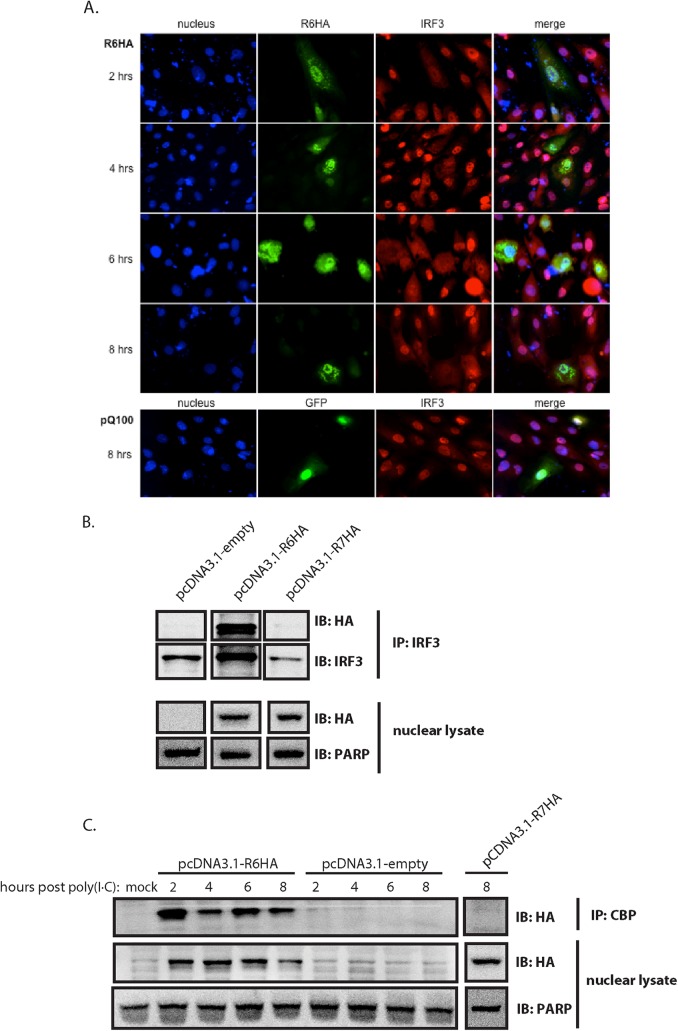
In order to delineate the mode of action of R6 in the inhibition of IFN-β transcription, the cellular localization of R6 was examined by Western blotting, as well as immunofluorescence. In transfected cells, R6 vIRF was localized to both the nucleus and the cytoplasm, with a majority of the protein expressed in the nucleus (Fig. 2A and B). Similar to previous results (33), colocalization was observed between IRF3 and R6 in poly(I · C)-transfected cells and was not restricted to either the cytoplasm or the nucleus. This was also observed by coimmunoprecipitation of R6 with IRF3 (33). We also evaluated the intracellular localization of IRF3 in the presence or absence of R6. Localization was addressed via immunofluorescence assay (IFA) over time (Fig. 2A). Poly(I · C)-transfected cells previously transfected with a green fluorescent protein (GFP)-expressing vector (pQ100) as a control for our transfection, showed accumulation of IRF3 within the nucleus. In the presence of R6 vIRF, however, IRF3 localization was diffuse throughout the cell and did not accumulate in the nucleus at any time point examined. These data indicate that R6 affects the localization of IRF3.
FIG 2.
R6 localizes to the nucleus and affects nuclear accumulation of pIRF3. (A, B, and C) Telomerized RFs were transfected with pcDNA3.1-R6HA, pcDNA3.1-R7HA, or empty pcDNA3.1 for 40 h and subsequently transfected with poly(I · C) for the indicated times. (A) Transfected cells were fixed and analyzed by immunofluorescence for the detection of R6 (anti-HA) (green) and cellular IRF3 (red) and stained with Hoechst (blue) for the detection of nuclei. (B) Nuclear lysates prepared from cells 8 h post-poly(I · C) transfection were immunoprecipitated with anti-IRF3 antibody and then subjected to SDS-PAGE and probed with anti-HA antibody. Nuclear lysates were probed for HA expression, with PARP as a loading control and as a control for purity of nuclear fractionation. IB, immunoblotting. (C) Nuclear lysates were immunoprecipitated with CBP antibody and then subjected to SDS-PAGE and probed with anti-HA antibody. The nuclear lysates were probed for HA expression, with PARP as a loading control and as a control for purity of nuclear fractionation.
We next wanted to investigate whether R6 interacts with other cellular proteins that associate with pIRF3 in the nucleus. Upon its phosphorylation and dimerization, pIRF3 translocates to the nucleus, where it associates with other components of the enhanceosome (i.e., NF-κB and ATF-2/c-Jun) and further recruits RNA polymerase, chromatin-remodeling complexes, and histone-modifying complexes, such as CBP/p300 (39). This complex of proteins is referred to as the transcriptional preinitiation complex and is responsible for the transcription of IFN-β. Interestingly, IRF3 lacks intrinsic transcriptional activity, and therefore, the specific interaction between IRF3 and CBP/p300 is critical for the activation of transcription (40–42). This complex has been shown to be targeted by both KSHV vIRF1 (18) and HSV-1 ICP0 (43) via direct interaction as a means of inhibiting IFN-β transcription. To examine the possibility of R6 and CBP interaction, coimmunoprecipitation experiments were performed on whole-cell lysates of tRFs transfected with vector expressing HA-tagged R6 vIRF and subsequently transfected with poly(I · C). Lysates were immunoprecipitated with anti-CBP PAb, followed by Western blotting using an anti-HA antibody. The results showed that R6 is able to interact with endogenous cellular CBP at all time points. Interestingly, we detected evidence of a second band with the anti-HA antibody that migrates slightly faster than the predominant band, which could suggest that R6 undergoes a posttranslational modification that leads to the slower-migrating band. The specificity of this interaction is supported by the lack of an interaction observed between HA-tagged R7 vIRF and CBP (Fig. 2B).
R6 prevents formation of a functional IRF3/CBP complex.
Since R6 vIRF was shown to inhibit IRF3-mediated transcription (33) (Fig. 1A) and to associate with CBP (Fig. 2B), it was of particular interest to determine if R6 could interact with CBP and impede CBP/IRF3 complex formation. To evaluate this, we transfected increasing amounts of HA-tagged R6 into tRFs and, 40 h later, transfected them with poly(I · C) to trigger IRF3 translocation. Cells were harvested at 6 h post-poly(I · C) transfection, and nuclear extracts were then isolated and immunoprecipitated with anti-CBP antibody, followed by Western blotting with anti-pIRF3 and anti-HA antibodies. The coimmunoprecipitation data revealed an interaction between CBP and pIRF3, as well as between CBP and R6, suggesting two distinct populations of CBP in the cell or a complex that is comprised of CBP/pIRF3/R6. Interestingly, we observed a slight decrease in binding of pIRF3 when cells were transfected with large amounts (20 μg) of R6, suggesting that, at increased concentrations, R6 can diminish pIRF3/CBP complex formation (Fig. 3A).
FIG 3.
R6 competes with IRF3 for binding to CBP. (A) Telomerized RFs were transfected with pcDNA3.1-R6HA (5, 10, or 20 μg DNA) or empty pcDNA3.1 for 40 h and subsequently transfected with poly(I · C) for 6 h. Nuclear lysates were immunoprecipitated with anti-CBP antibody and then subjected to SDS-PAGE and probed with anti-HA antibody or anti-pIRF3 antibody. The nuclear lysates were probed for HA expression, with PARP as a loading control and a control for purity of nuclear fractionation. (B) EMSA was performed on whole-cell extracts (20 μg) derived from telomerized RFs transfected with pcDNA3.1-R6HA or empty pcDNA3.1. The biotin-labeled probe corresponds to the PRDI-PRDIII motif (5′-GAAAACTGAAAGGAGAACTGAAAGTG-3′) of the IFN-β promoter. Anti-CBP antibody and anti-IRF3 antibody were added as indicated to demonstrate the presence of CBP and IRF3 in the DNA-protein complexes. For oligonucleotide competition, a 500-fold molar excess of unlabeled PRDI-PRDIII probe was added as indicated. (C) Telomerized RFs were pretreated with MG132 or left untreated. The cells were then transfected with pcDNA3.1-R6HA or empty pcDNA3.1 for 40 h and then transfected with poly(I · C) for 6 h. Whole-cell lysates were immunoprecipitated with anti-IRF3 or anti-TBK and then subjected to SDS-PAGE and probed with anti-IRF3 or anti-TBK to gauge total levels of IRF3 and TBK within the cells. The nuclear lysates were subjected to SDS-PAGE and probed with anti-pIRF3 antibody or PARP, which served as a loading control and a control for purity of nuclear fractionation.
In order to establish the functionality of pIRF3/CBP complexes in the presence of R6, we examined the DNA binding capability of the complex using the PRDI-PRDIII domain of the IFN-β promoter in EMSAs (Fig. 3B). Similar to previous experiments, tRFs were transfected with HA-tagged R6 or empty vector for 40 h and subsequently transfected with poly(I · C) for 6 h. Nuclear lysates were harvested and then tested in a biotin-labeled PRDI-PRDIII oligonucleotide binding assay and subjected to EMSA. In the absence of R6, cells expressing endogenous and basal levels of pIRF3 and CBP displayed a protein-DNA complex indicated by a probe shift (Fig. 3B, lane 1). To demonstrate the presence of IRF3 and CBP in the complex, nuclear extracts were preincubated with either anti-IRF3 or anti-CBP antibodies for 5, 10, or 20 min and then allowed to complex with the PRDI-PRDIII oligonucleotide (Fig. 3B, lanes 3 to 8). It is important to note that these antibodies have been previously shown to ablate the ability of IRF3 and CBP to bind to the PRDI-PRDIII domain of the IFN-β promoter (18, 40). A decrease in protein complex binding was observed after a 10-min preincubation with either antibody, with an even greater decrease after 20 min preincubation, indicating that both IRF3 and CBP are involved in binding the PRDI-PRDIII domain of the IFN-β promoter region. The specificity of the complex binding to the probe was determined by the addition of unlabeled PRDI-PRDIII oligonucleotide, which resulted in loss of binding to the biotin-labeled probe (Fig. 3B, lane 2). Interestingly, ectopic expression of R6 resulted in a drastic reduction in probe binding by IRF3/CBP (Fig. 3B, lane 9), suggesting that the presence of R6 disrupts the IRF3/CBP complex binding of the probe. This, along with the coimmunoprecipitation data, shows that in the presence of R6, IRF3/CBP complexes are less able to effectively bind the PRDI-PRDIII oligonucleotide. Increasing amounts of R6 did not fully abolish the interaction between IRF3 and CBP. This could be an effect of less than optimal transfection efficiency; however, it did result in decreased IRF3 bound to CBP. This suggests that R6 displaces IRF3 from CBP enough to impede its DNA binding activity, thus preventing the transcription of IFN-β.
To determine if the decrease in pIRF3 by R6 is mediated by the proteasome, we transfected cells with R6-HA that were grown in the presence or absence of the proteasome inhibitor MG132. Through this analysis, we found that pIRF3 levels sharply decrease in the presence of R6 and that this effect can be reversed with the addition of MG132 (Fig. 3C). This effect was specific to pIRF3, as total levels of IRF3 remained constant regardless of the addition of MG132.
We also evaluated changes in Tank-binding kinase (TBK) levels, as TBK is the main cellular kinase that is responsible for the phosphorylation and subsequent activation of pIRF3 upon Toll-like receptor (TLR) ligation. TBK levels were assessed by Western blotting and found to be stable regardless of R6 expression. These experiments demonstrate that R6 is capable of inhibiting IRF3/CBP complex binding to the IFN-β promoter by displacing pIRF3 from CBP and that the presence of R6 results in proteasome-mediated degradation of pIRF3.
R6 is associated with purified RRV virions.
As we observed early inhibition of IFN stimulation, we postulated that R6 must act early to impede IRF3 activity. Currently, the lack of antibodies specific to the RRV vIRFs makes it particularly difficult to characterize the roles of individual vIRFs during infection. To circumvent this shortcoming, we utilized BAC technology to generate a recombinant virus in which R6 possesses a C-terminal HA epitope tag, allowing its detection. Importantly, all 7 other vIRFs are fully intact in this virus and left untagged. Insertion of the HA tag sequence into the R6 sequence was accomplished using the RRV BAC coupled with a galK-based positive/negative selection system in E. coli (37), so that the R6 ORF was initially replaced with a galK cassette and then subsequently replaced with a recombinant R6-HA cassette containing HA sequence at the 3′ end of R6. After isolation of an R6-HA-containing RRV BAC clone, DNA was isolated and used to produce infectious virus, as previously described (33). The genome of the resultant virus was analyzed via CGH in order to compare the genomic sequence of R6HA RRV to that of WTBAC RRV17757 (Fig. 4A), confirming the correct insertion of the HA epitope sequence and demonstrating that the remainder of the genomic sequence outside the modified R6 ORF was identical to that of the parental WT BAC virus. Finally, the R6 HA virus was analyzed to confirm that modification of the R6 ORF did not have a negative effect on virus growth kinetics or transcription of neighboring ORFs compared to the WTBAC RRV17577. Our analysis revealed that the R6HA virus displayed growth kinetics similar to those of the WTBACRRV (Fig. 4B) and that transcription of ORF57 and R7 was not adversely impacted (Fig. 4C).
FIG 4.
Molecular and in vitro characterization of R6HA RRV. (A) Comparative genome hybridization was used to directly compare viral DNA from the R6HA RRV recombinant to that from WTBACRRV. Alterations within the R6HA RRV genome resulted in incomplete hybridization to the array, depicted by the ratio of R6HA to WTBACRRV, and signaled a potential nucleotide mismatch between the two viral genomes. This comparison identified the HA tag located at the C-terminal end of R6. A second mismatch, indicated with an asterisk, was incorrectly identified, and the identified sequence was confirmed to be similar to the WT via PCR and DNA sequence analysis. (B) RFs were infected with either WTBACRRV or R6HA RRV at an MOI of 2.5 for single-step growth curve analysis. Infected RFs were harvested at the specified time points and subjected to a serial-dilution plaque assay on RFs to determine viral titers. The data from 4 separate experiments were averaged (±SEM). (C) RFs were infected with WTBACRRV or R6HA RRV or left uninfected, and at 48 h postinfection, total cellular RNA was harvested. Equivalent amounts of total RNA were analyzed by RT-PCR for ORF57, R7, and R6 transcripts, with cellular GAPDH as a loading control. Samples were run simultaneously with Taq polymerase only (−RT) to control for input and purity. Numbers at top indicate nucleotides.
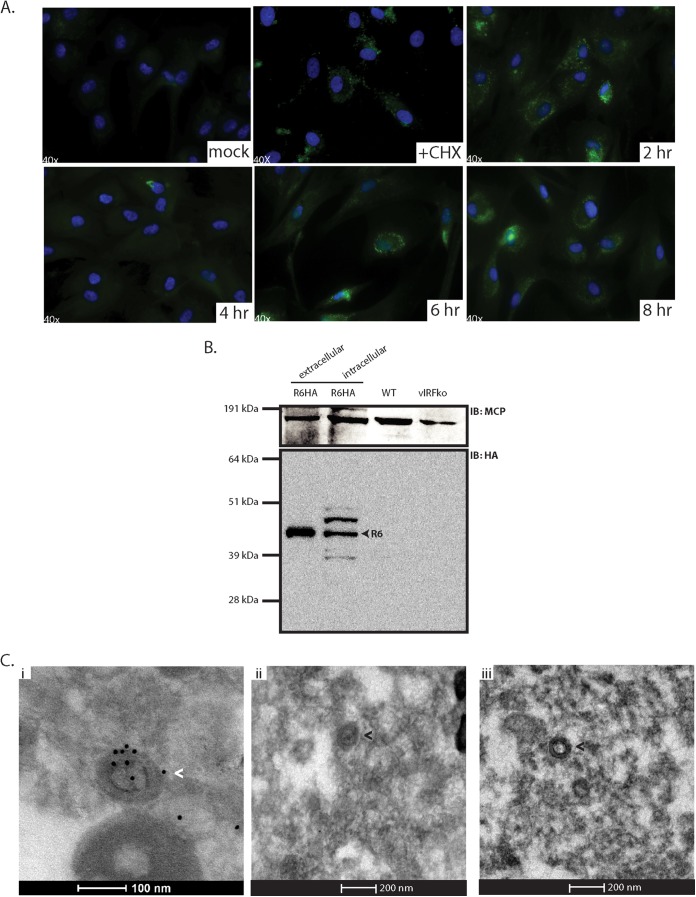
To evaluate the role of R6 during early RRV infection, it was first necessary to determine the expression kinetics of R6 in the first 8 h of infection. To that end, a CHX reversal assay was performed (Fig. 5A). Cells were first treated with CHX, a protein translation inhibitor, and subsequently infected with R6HA RRV at an MOI of 2.5 for 6 h in the presence of CHX. CHX was subsequently removed, and cells were immediately prepared and analyzed for R6HA expression via IFA or kept in culture for analysis at later time points for kinetic analysis of expression. It was previously shown that R6 transcripts are detected in RRV-infected cells at 6 h postinfection, but not at 3 h p.i. (33), and therefore, it was expected that the R6 protein would likely be present only at 6 h postinfection or later. Remarkably, however, the R6 protein was detected in infected cells even in the presence of CHX, suggesting that R6 came into the cells or that the conditions did not prevent de novo protein synthesis (Fig. 5A). After removal of the CHX block, we observed R6 at 2 h and again at 6 and 8 h. R6 signal was noticeably diminished at 4 h after removal of the CHX block, which could suggest that the initial R6 signal was degraded and new R6 expression was due to de novo transcription and subsequent translation of the R6 ORF, whereas the first wave could potentially represent R6 protein that entered the cell during infection.
FIG 5.
R6 is associated with RRV virions. (A) Primary RFs were pretreated with CHX and subsequently infected with R6HA-RRV at an MOI of 2.5. CHX was removed, and the cells were fixed at the indicated time points and analyzed by immunofluorescence for the detection of R6-HA (anti-HA) (green) and stained with Hoechst (blue) for the detection of nuclei. (B) 1 × 105 PFU of gradient-purified virus samples (extracellular R6HA-RRV, intracellular R6HA-RRV, WTBACRRV, and vIRFko-RRV) was subjected to SDS-PAGE and probed with anti-HA antibody and anti-MCP antibody as a control. (C) (i) Gradient-purified R6HA-RRV was fixed, pelleted, and sectioned. The sections were immunogold stained with anti-HA antibody and 10-nm gold-conjugated secondary antibody. (ii) Gradient-purified WTBACRRV was sectioned and immunogold stained with anti-HA antibody and 10-nm gold-conjugated secondary antibody as a control. (iii) R6HA-RRV sections were stained with 10-nm gold-conjugated secondary antibody alone as a control. Virus particles with gold particles are indicated by the white arrowhead, and virus particles with no gold particles are indicated by black arrowheads.
To determine if R6 entered the cell during the infection process, we analyzed equivalent numbers (1 × 105) of PFU of gradient-purified virus preparations of extracellular R6HA RRV, intracellular R6HA RRV, WTBACRRV, and vIRF-ko RRV by SDS-PAGE, followed by immunoblot analysis with an anti-HA antibody, as well as a control antibody against RRV MCP (Fig. 5B). As expected, MCP was found in all virus preparations. However, R6 was detected only in the extracellular and intracellular virus preparations of R6HA RRV, but not in WTBACRRV or vIRF-ko RRV preparations. Interestingly, we also observed additional bands within the intracellular virus fraction when immunoblotting for HA. They could be additional forms of R6, potentially due to posttranslational modifications or processing, that are found only intracellularly. To further evaluate virion-associated R6, we performed immunogold electron microscopy on extracellular R6HA RRV and WTBACRRV (Fig. 5C). Gradient-purified R6HA RRV preparations were fixed, embedded, sectioned, and stained with anti-HA antibody and subsequently with a secondary antibody conjugated with 10-nm-diameter gold particles. Gold particles were seen on and within R6HA RRV particles (Fig. 5C, i), but not in WTBACRRV (Fig. 5C, ii) or in R6HA RRV incubated with secondary gold-conjugated antibody alone (Fig. 5C, iii) to assess the level of background. In three fields of view, the gold particles that colocalized with virions were manually counted and calculated as a percentage of the total number of gold particles viewed. Approximately 50% of the gold particles were localized to RRV virion particles (Fig. 5C, i, arrowhead), with the majority of gold particles localizing to the virus tegument. No background was observed in R6HA RRV with secondary antibody alone. Some virions that were detected did not colocalize with gold particles. This could mean that not all virions contain R6 protein or that the section obtained did not contain R6, even though R6 may have been detected in other sections of the same virion. These data demonstrate that R6 is a virion-associated vIRF.
Virion-associated R6 is functional.
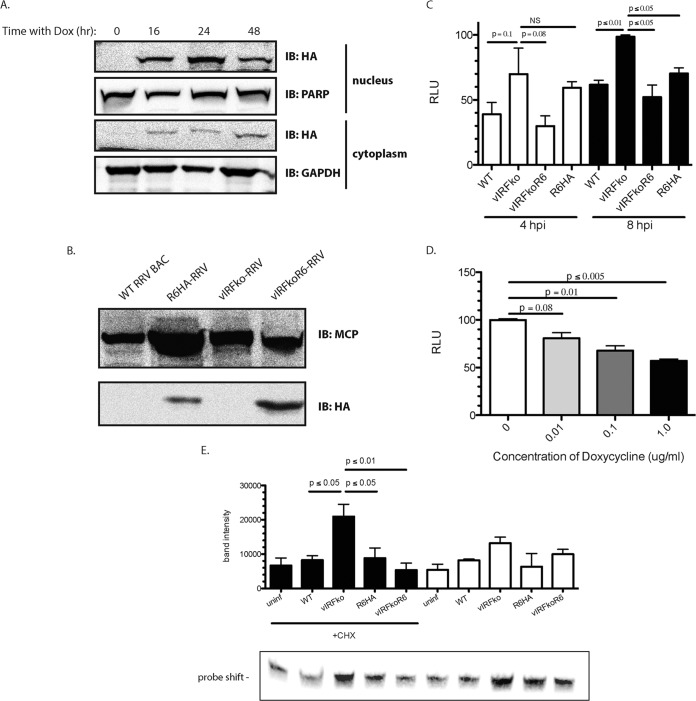
To determine if virion-associated R6 is functional, we sought to test whether an R6-expressing cell line could complement the lack of R6 in the vIRF-ko virus. To accomplish this, we generated an R6HA-expressing cell line that controls for R6HA expression with a tetracycline-inducible promoter. This Dox-inducible cell line expressing R6 (tRF-rtTA:R6HA) was constructed to evaluate if R6HA could be packaged in RRV virions to complement a vIRF-ko virus (Fig. 6A). We choose to treat cells with 1.0 μg/ml Dox for 24 h to allow the expression of HA-tagged R6 and then infect them with vIRF-ko RRV, as we found this concentration of Dox and time postinduction yielded higher R6HA production (data not shown). A total of 5 μg of gradient-purified virus preparation was used to examine the presence of R6HA by Western blotting. Analysis of extracellular virus isolated from the Dox-induced tRF-rtTA:R6HA cell line showed that R6 was effectively packaged into vIRF-ko RRV virions compared to vIRF-ko grown in normal RFs. As controls, we utilized WTBACRRV and R6HA RRV grown in normal RFs (Fig. 6B).
FIG 6.
Virion-associated R6 is functional. (A) Telomerized RF-rtTAs stably transduced with R6-HA were treated with Dox for the indicated times. The nuclear and cytoplasmic lysates were subjected to SDS-PAGE and probed with anti-HA antibody. GAPDH and PARP served as loading controls and as controls for purity of fractionation for the cytoplasmic and nuclear extracts, respectively. (B) Telomerized RF-rtTAs were treated with Dox and infected with vIRFko-RRV at an MOI of 0.01 per cell. The resultant virus (vIRFkoR6-RRV) was gradient purified from cell supernatants. Five micrograms of gradient-purified virus (WTBACRRV, vIRFko-RRV, R6HA-RRV, and vIRFkoR6-RRV) was subjected to SDS-PAGE and probed with anti-HA antibody and anti-MCP antibody as a control. (C) tRF-ISREs were infected for 4 or 8 h with the indicated virus at an MOI of 2.5 PFU per cell. The cells were then assayed for firefly luciferase activity. Firefly luciferase levels were normalized to constitutively expressed Renilla luciferase levels in each well. The data are averages (and SEM) from the results of 3 independent experiments. (D) tRF-rtTA:R6HAs were infected with vIRFko-RRV at an MOI of 0.01 PFU per cell after pretreatment with the indicated amounts of Dox. Virus was then harvested and gradient purified. The resultant virus was used to infect tRF-ISREs at an MOI of 2.5 PFU per cell for 8 h. The cells were assayed for firefly luciferase activity. Firefly luciferase levels were normalized to constitutively expressed Renilla luciferase levels in each well. The data are averages (and SEM) from the results of 3 independent experiments. The total levels of MCP and R6HA in virion preparations were assessed by Western blotting with anti-MCP and anti-HA antibodies. (E) Telomerized RFs were infected with the indicated virus at an MOI of 2.5 PFU per cell for 8 h in the presence or absence of CHX. EMSA was performed on nuclear extracts (20 μg). The biotin-labeled probe corresponded to the PRDI-PRDIII motif (5′-GAAAACTGAAAGGAGAACTGAAAGTG-3′) of the IFN-β promoter. The data were analyzed using a paired t test. P values of ≤0.05 were considered significant, and values greater than 0.05 were not significant.
To determine if the R6 protein associated with this complemented virus (vIRF-koR6 RRV) was capable of inhibiting the type I IFN response in infected cells, we infected the tRF-ISREs and measured the luciferase units (Fig. 6C). Infection with either WTBACRRV or R6HA RRV resulted in a significant decrease in luciferase expression compared to vIRF-ko RRV at 8 h p.i. Likewise, vIRF-koR6 RRV inhibited luciferase expression, similar to levels that were seen in WT infection. To determine if this effect was dose dependent, R6HA expression was induced in tRFs with increasing amounts of Dox (0, 0.01, 0.1, and 1.0 μg/ml), and each induced cell sample was subsequently infected with vIRF-ko RRV, thus potentially creating a panel of vIRF-koR6 RRVs containing increasing amounts of virion-associated R6. The resultant viruses were then tested in the luciferase assay by infecting the reporter cells at an MOI of 2.5. As anticipated, increased Dox-induced R6HA within the virion results in more of an inhibitory effect (Fig. 6D). These data suggest that virion-associated R6 is not only functional, but is also capable of hindering the type I IFN response.
To further support our result, we analyzed the ability of virion-associated R6 to inhibit binding of transcription factors to the IFN-β promoter (Fig. 6E). Cells were either pretreated with CHX or left untreated prior to and during virus infection. Treatment with CHX ensured that no new proteins would be translated, thus allowing direct examination of the effects of virion components. Infection with R6HA RRV in the presence of CHX led to decreased binding to the IFN-β promoter, indicating that intact virus carries within it an inhibitor that can ablate IRF3/CBP binding to PRDI-PRDIII. Conversely, vIRF-ko RRV infection of cells treated with CHX caused a 3-fold increase in the amount of probe that was shifted. These data, so far, show not only that RRV has a virion-associated protein that interferes with the IFN-β promoter, but also that this virion-associated protein is associated with the region of the vIRF ORFs. Finally, when cells were infected with vIRF-koR6 RRV, the level of protein complex binding to the IFN-β promoter decreased to nearly the levels observed in R6HA RRV infection. Overall, these results indicate R6 is an important virion-associated mediator of the type I IFN response and that at least one of its functions is to prevent the transcription of IFN-β.
DISCUSSION
Approximately 25% of the KSHV genome is dedicated to immunomodulation (5). Of the 22 immunomodulatory genes carried by KSHV, those encoding the viral interferon regulatory factors are of particular interest, as they are unique to both gamma-2 herpesviruses, KSHV and the closely related RRV (8, 9, 12). The eight vIRFs encoded by RRV display sequence homology to cellular IRFs, as well as KSHV vIRF1. In fact, R6, R7, R8, R10, and R11 all have similarities to KSHV vIRF1, ranging from 26 to 33% (12). These similarities have led to the hypothesis that the RRV vIRFs may be modulators of cellular IRFs, thus interfering with the type I IFN response. This hypothesis was validated when in vivo and in vitro comparisons of WTBACRRV and vIRF-ko RRV revealed significant differences in the antiviral state early during infection (33, 34). The RRV vIRFs were found to be responsible for a decrease in gene expression and IFN production, as well as decreased nuclear accumulation of pIRF3 during de novo infection with RRV. The findings on ectopic expression of R6 vIRF and its significant impact on the inhibition of IRF3-mediated transcription warranted further dissection of its potential immunomodulatory function.
The induction of IFN-β initiated by ligation of TLR3 and cytoplasmic sensors, such as RIG-I, MDA-5, and c-GAS, is largely orchestrated by IRF3, which is constitutively expressed in most cell types. IRF3 is rapidly activated after sensing these pathogen-associated molecular patterns (PAMPs), where they accumulate in the nucleus and commence transcription of type I IFNs. A reporter assay expressing firefly luciferase under the control of an ISRE was used to examine the kinetics of IFN induction upon independent expression of R6. These data showed a significant decrease in IFN production by R6 soon after poly(I · C) transfection, and the decrease was sustained even 8 h posttransfection. Additionally, analysis of IFN-β transcription revealed a nearly 50% decline in transcripts in the presence of R6, which further supports the immunomodulatory function of R6 vIRF.
The KSHV vIRFs target a variety of components in the pathways leading up to type I IFN induction. The vIRF1 protein suppresses the transcriptional activity of IRF3 (14, 18, 26, 27) and IRF1 (14, 24) by interfering with their binding to the transcriptional coactivator p300. vIRF2 disrupts NF-κB- and IRF1-dependent transcription (15), both of which are required for effective IFN-β transcription. Lastly, the DNA binding function of IRF7 has been shown to be inhibited by KSHV vIRF3 (22). Knowing that the KSHV vIRFs are multifunctional and have diverse targets, it was important to first define the cellular localization of the R6 vIRF in order to pinpoint its primary mode of action. By Western blotting and immunofluorescence assay, R6 was found to be concentrated in, but not limited to, the nucleus. R6 was previously shown to coimmunoprecipitate and colocalize with IRF3 in both the nucleus and cytoplasm. Additionally, the vIRFs in RRV were found to prevent nuclear accumulation of IRF3 without having an effect on IRF3 phosphorylation or dimerization (33). Therefore, the effect of R6 on phosphorylated IRF3 was explored. What was particularly interesting was the lack of pIRF3 concentration within the nuclei of cells expressing R6, especially at 6 and 8 h post-poly(I · C) transfection. Phosphorylated IRF3 was likely not being sequestered in the cytoplasm, as there was no observed accumulation of cytoplasmic pIRF3. The fact that pIRF3 was still able to enter the nucleus at early time points, and that R6 is primarily localized to the nucleus, led to the conclusion that R6 likely acts on the function of IRF3 within the nucleus. A vital step in the activation of IFN-β transcription is the binding of pIRF3 to the transcriptional coactivator and histone acetyltransferase CBP. Similar to KSHV vIRF1, RRV R6 binds to CBP and IRF3 and interferes with the DNA binding capacities of these IRF3/CBP complexes (Fig. 7). Interestingly, R6 does not inhibit IRF3 binding to CBP as potently as KSHV vIRF1 does. However, this may be a result of transfection efficiency.
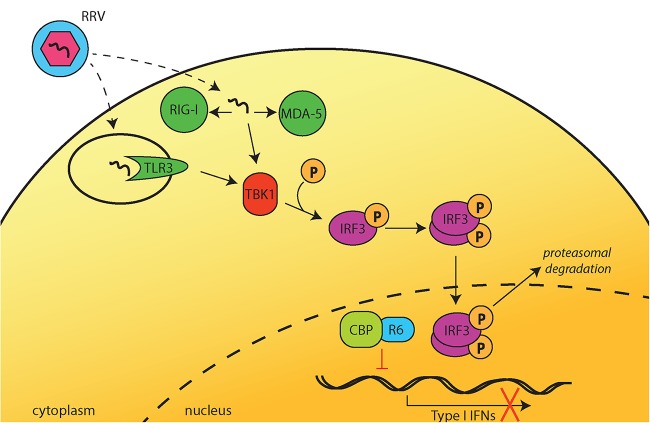
FIG 7.
Potential model of IFN inhibition by R6. Upon detection of RRV infection by TLR3, RIG-I, or MDA-5, TBK1 is activated and subsequently phosphorylates IRF3. pIRF3 then dimerizes and translocates to the nucleus. Within the nucleus, R6 binds to the transcriptional coactivator CBP. This interaction prevents pIRF3 from binding to CBP, and pIRF3 is exported from the nucleus and degraded by the proteasome. Nuclear R6 interferes with complex formation between pIRF3 and CBP and, as a result, decreases pIRF3/CBP complex binding to the IFN-β promoter and inhibits IFN-β production.
It is not unprecedented for a virus to prepackage antiviral mediators within the virion in order to quickly dampen the immune response. A prime example is KSHV ORF45, which is not only virion associated, but also inhibits type I IFN induction by specifically blocking the phosphorylation and nuclear accumulation of IRF7 (44–46). RRV does in fact encode a homologue of KSHV ORF45, and the RRV and KSHV ORF45s share approximately 24% amino acid identity. RRV ORF45 has also been found within the tegument of virion particles, but its immunomodulatory functions have yet to be explored (47). Given that ectopic expression of RRV vIRF R6 hampered IFN-β transcription very soon after poly(I · C) transfection, this led to our investigation of whether R6 might be a virion-associated protein. In this study, we show that R6 is indeed virion associated, as seen by immunofluorescence and electron microscopy. Furthermore, virion-associated R6 can inhibit IRF3/CBP DNA binding and therefore inhibit IFN-β transcription. The prompt inhibition of the type I IFN response may be especially crucial in endothelial and epithelial cells, as they are some of the first cells to be infected and are also key producers of IFN-β. The potent antiviral response mediated by type I IFNs is brought about by further activation of ISGs, whose products inhibit various stages of viral replication. It is therefore of the utmost importance for RRV to quickly downmodulate IFNs. Not only do type I IFNs play a significant role in the innate immune response to infection, but also, they are vital in the downstream development of the adaptive immune response to the infection at hand. For example, type I IFNs enhance the expression of major histocompatibility complex class I (MHC-I), thereby promoting antigen presentation and development of an effective CD8+ T cell response (48). In addition, type I IFNs have dramatic effects on natural killer (NK) cells (49, 50) and CD4+ and CD8+ T cells (51, 52), as well as dendritic cells (DCs) (53–56). In fact, it was previously shown that complete deletion of all 8 RRV vIRFs initiates an earlier T cell response (34), and therefore, it could be postulated that the IFN-β inhibition mediated by virion-associated R6 contributes to the delayed T cell response observed in WT RRV infection. It would be particularly interesting to create an R6 deletion mutant of RRV and to determine the phenotype of this virus and the ensuing immune response. These data show that RRV has a prepackaged vIRF for immediate delivery into target cells upon de novo infection, thus providing the virus with a direct mechanism to inhibit or slow the innate immune response. We postulate that virion-associated R6 functions to enable RRV to successfully establish infection and further progress in its viral life cycle, including transcription of the remaining vIRFs, without a robust type I IFN response from the host.
ACKNOWLEDGMENTS
Electron microscopy was performed at the Multi-Scale Microscopy Core (MMC) with technical support from the Oregon Health and Science University (OHSU)-FEI Living Laboratory and the OHSU Center for Spatial Systems Biomedicine (OCSSB).
This study was supported by National Institutes of Health grants 8P51OD011092-54 and CA075922 (S.W.W.). G.M. and B.A.R. were supported by training grants T32 AI74494 and T32 AI07472, respectively.
REFERENCES
- 1.Fitzgerald-Bocarsly P, Dai J, Singh S. 2008. Plasmacytoid dendritic cells and type I IFN: 50 years of convergent history. Cytokine Growth Factor Rev 19:3–19. doi: 10.1016/j.cytogfr.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ivashkiv LB, Donlin LT. 2014. Regulation of type I interferon responses. Nat Rev Immunol 14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sen GC. 2001. Viruses and interferons. Annu Rev Microbiol 55:255–281. doi: 10.1146/annurev.micro.55.1.255. [DOI] [PubMed] [Google Scholar]
- 4.Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. 2001. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol 19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 5.Mamane Y, Heylbroeck C, Genin P, Algarte M, Servant MJ, LePage C, DeLuca C, Kwon H, Lin R, Hiscott J. 1999. Interferon regulatory factors: the next generation. Gene 237:1–14. doi: 10.1016/S0378-1119(99)00262-0. [DOI] [PubMed] [Google Scholar]
- 6.Areste C, Blackbourn DJ. 2009. Modulation of the immune system by Kaposi's sarcoma-associated herpesvirus. Trends Microbiol 17:119–129. doi: 10.1016/j.tim.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Bowie AG, Unterholzner L. 2008. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol 8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rezaee SA, Cunningham C, Davison AJ, Blackbourn DJ. 2006. Kaposi's sarcoma-associated herpesvirus immune modulation: an overview. J Gen Virol 87:1781–1804. doi: 10.1099/vir.0.81919-0. [DOI] [PubMed] [Google Scholar]
- 9.Alexander L, Denekamp L, Knapp A, Auerbach MR, Damania B, Desrosiers RC. 2000. The primary sequence of rhesus monkey rhadinovirus isolate 26-95: sequence similarities to Kaposi's sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J Virol 74:3388–3398. doi: 10.1128/JVI.74.7.3388-3398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore PS, Gao SJ, Dominguez G, Cesarman E, Lungu O, Knowles DM, Garber R, Pellett PE, McGeoch DJ, Chang Y. 1996. Primary characterization of a herpesvirus agent associated with Kaposi's sarcoma. J Virol 70:549–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Russo JJ, Bohenzky RA, Chien MC, Chen J, Yan M, Maddalena D, Parry JP, Peruzzi D, Edelman IS, Chang Y, Moore PS. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc Natl Acad Sci U S A 93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Searles RP, Bergquam EP, Axthelm MK, Wong SW. 1999. Sequence and genomic analysis of a rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J Virol 73:3040–3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burysek L, Pitha PM. 2001. Latently expressed human herpesvirus 8-encoded interferon regulatory factor 2 inhibits double-stranded RNA-activated protein kinase. J Virol 75:2345–2352. doi: 10.1128/JVI.75.5.2345-2352.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burysek L, Yeow WS, Lubyova B, Kellum M, Schafer SL, Huang YQ, Pitha PM. 1999. Functional analysis of human herpesvirus 8-encoded viral interferon regulatory factor 1 and its association with cellular interferon regulatory factors and p300. J Virol 73:7334–7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burysek L, Yeow WS, Pitha PM. 1999. Unique properties of a second human herpesvirus 8-encoded interferon regulatory factor (vIRF-2). J Hum Virol 2:19–32. [PubMed] [Google Scholar]
- 16.Lubyova B, Pitha PM. 2000. Characterization of a novel human herpesvirus 8-encoded protein, vIRF-3, that shows homology to viral and cellular interferon regulatory factors. J Virol 74:8194–8201. doi: 10.1128/JVI.74.17.8194-8201.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lubyova B, Kellum MJ, Frisancho AJ, Pitha PM. 2004. Kaposi's sarcoma-associated herpesvirus-encoded vIRF-3 stimulates the transcriptional activity of cellular IRF-3 and IRF-7. J Biol Chem 279:7643–7654. doi: 10.1074/jbc.M309485200. [DOI] [PubMed] [Google Scholar]
- 18.Lin R, Genin P, Mamane Y, Sgarbanti M, Battistini A, Harrington WJ Jr, Barber GN, Hiscott J. 2001. HHV-8 encoded vIRF-1 represses the interferon antiviral response by blocking IRF-3 recruitment of the CBP/p300 coactivators. Oncogene 20:800–811. doi: 10.1038/sj.onc.1204163. [DOI] [PubMed] [Google Scholar]
- 19.Fuld S, Cunningham C, Klucher K, Davison AJ, Blackbourn DJ. 2006. Inhibition of interferon signaling by the Kaposi's sarcoma-associated herpesvirus full-length viral interferon regulatory factor 2 protein. J Virol 80:3092–3097. doi: 10.1128/JVI.80.6.3092-3097.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flowers C, Flowers S, Nabel G. 1998. Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor confers resistance to the antiproliferative effect of interferon-alpha. Mol Med 4:402–412. [PMC free article] [PubMed] [Google Scholar]
- 21.Gao SJ, Boshoff C, Jayachandra S, Weiss RA, Chang Y, Moore PS. 1997. KSHV ORF K9 (vIRF) is an oncogene which inhibits the interferon signaling pathway. Oncogene 15:1979–1985. doi: 10.1038/sj.onc.1201571. [DOI] [PubMed] [Google Scholar]
- 22.Joo CH, Shin YC, Gack M, Wu L, Levy D, Jung JU. 2007. Inhibition of interferon regulatory factor 7 (IRF7)-mediated interferon signal transduction by the Kaposi's sarcoma-associated herpesvirus viral IRF homolog vIRF3. J Virol 81:8282–8292. doi: 10.1128/JVI.00235-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wies E, Hahn AS, Schmidt K, Viebahn C, Rohland N, Lux A, Schellhorn T, Holzer A, Jung JU, Neipel F. 2009. The Kaposi's sarcoma-associated herpesvirus-encoded vIRF-3 inhibits cellular IRF-5. J Biol Chem 284:8525–8538. doi: 10.1074/jbc.M809252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zimring JC, Goodbourn S, Offermann MK. 1998. Human herpesvirus 8 encodes an interferon regulatory factor (IRF) homolog that represses IRF-1-mediated transcription. J Virol 72:701–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mutocheluh M, Hindle L, Areste C, Chanas SA, Butler LM, Lowry K, Shah K, Evans DJ, Blackbourn DJ. 2011. Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor-2 inhibits type 1 interferon signalling by targeting interferon-stimulated gene factor-3. J Gen Virol 92:2394–2398. doi: 10.1099/vir.0.034322-0. [DOI] [PubMed] [Google Scholar]
- 26.Li M, Damania B, Alvarez X, Ogryzko V, Ozato K, Jung JU. 2000. Inhibition of p300 histone acetyltransferase by viral interferon regulatory factor. Mol Cell Biol 20:8254–8263. doi: 10.1128/MCB.20.21.8254-8263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seo T, Lee D, Lee B, Chung JH, Choe J. 2000. Viral interferon regulatory factor 1 of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) binds to, and inhibits transactivation of, CREB-binding protein. Biochem Biophys Res Commun 270:23–27. doi: 10.1006/bbrc.2000.2393. [DOI] [PubMed] [Google Scholar]
- 28.Shin YC, Nakamura H, Liang X, Feng P, Chang H, Kowalik TF, Jung JU. 2006. Inhibition of the ATM/p53 signal transduction pathway by Kaposi's sarcoma-associated herpesvirus interferon regulatory factor 1. J Virol 80:2257–2266. doi: 10.1128/JVI.80.5.2257-2266.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rivas C, Thlick AE, Parravicini C, Moore PS, Chang Y. 2001. Kaposi's sarcoma-associated herpesvirus LANA2 is a B-cell-specific latent viral protein that inhibits p53. J Virol 75:429–438. doi: 10.1128/JVI.75.1.429-438.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamura H, Li M, Zarycki J, Jung JU. 2001. Inhibition of p53 tumor suppressor by viral interferon regulatory factor. J Virol 75:7572–7582. doi: 10.1128/JVI.75.16.7572-7582.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee HR, Toth Z, Shin YC, Lee JS, Chang H, Gu W, Oh TK, Kim MH, Jung JU. 2009. Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor 4 targets MDM2 to deregulate the p53 tumor suppressor pathway. J Virol 83:6739–6747. doi: 10.1128/JVI.02353-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirchhoff S, Sebens T, Baumann S, Krueger A, Zawatzky R, Li-Weber M, Meinl E, Neipel F, Fleckenstein B, Krammer PH. 2002. Viral IFN-regulatory factors inhibit activation-induced cell death via two positive regulatory IFN-regulatory factor 1-dependent domains in the CD95 ligand promoter. J Immunol 168:1226–1234. doi: 10.4049/jimmunol.168.3.1226. [DOI] [PubMed] [Google Scholar]
- 33.Robinson BA, Estep RD, Messaoudi I, Rogers KS, Wong SW. 2012. Viral interferon regulatory factors decrease the induction of type I and type II interferon during rhesus macaque rhadinovirus infection. J Virol 86:2197–2211. doi: 10.1128/JVI.05047-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson BA, O'Connor MA, Li H, Engelmann F, Poland B, Grant R, DeFilippis V, Estep RD, Axthelm MK, Messaoudi I, Wong SW. 2012. Viral interferon regulatory factors are critical for delay of the host immune response against rhesus macaque rhadinovirus infection. J Virol 86:2769–2779. doi: 10.1128/JVI.05657-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Estep RD, Powers MF, Yen BK, Li H, Wong SW. 2007. Construction of an infectious rhesus rhadinovirus bacterial artificial chromosome for the analysis of Kaposi's sarcoma-associated herpesvirus-related disease development. J Virol 81:2957–2969. doi: 10.1128/JVI.01997-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeFilippis VR, Sali T, Alvarado D, White L, Bresnahan W, Fruh KJ. 2010. Activation of the interferon response by human cytomegalovirus occurs via cytoplasmic double-stranded DNA but not glycoprotein B. J Virol 84:8913–8925. doi: 10.1128/JVI.00169-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res 33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawai T, Akira S. 2008. Toll-like receptor and RIG-I-like receptor signaling. Ann N Y Acad Sci 1143:1–20. doi: 10.1196/annals.1443.020. [DOI] [PubMed] [Google Scholar]
- 39.Genin P, Vaccaro A, Civas A. 2009. The role of differential expression of human interferon-a genes in antiviral immunity. Cytokine Growth Factor Rev 20:283–295. doi: 10.1016/j.cytogfr.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 40.Wathelet MG, Lin CH, Parekh BS, Ronco LV, Howley PM, Maniatis T. 1998. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol Cell 1:507–518. doi: 10.1016/S1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 41.Yang H, Lin CH, Ma G, Orr M, Baffi MO, Wathelet MG. 2002. Transcriptional activity of interferon regulatory factor (IRF)-3 depends on multiple protein-protein interactions. Eur J Biochem 269:6142–6151. doi: 10.1046/j.1432-1033.2002.03330.x. [DOI] [PubMed] [Google Scholar]
- 42.Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J 17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melroe GT, Silva L, Schaffer PA, Knipe DM. 2007. Recruitment of activated IRF-3 and CBP/p300 to herpes simplex virus ICP0 nuclear foci: potential role in blocking IFN-beta induction. Virology 360:305–321. doi: 10.1016/j.virol.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu FX, King SM, Smith EJ, Levy DE, Yuan Y. 2002. A Kaposi's sarcoma-associated herpesviral protein inhibits virus-mediated induction of type I interferon by blocking IRF-7 phosphorylation and nuclear accumulation. Proc Natl Acad Sci U S A 99:5573–5578. doi: 10.1073/pnas.082420599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu FX, Sathish N, Yuan Y. 2010. Antagonism of host antiviral responses by Kaposi's sarcoma-associated herpesvirus tegument protein ORF45. PLoS One 5:e10573. doi: 10.1371/journal.pone.0010573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu FX, Yuan Y. 2003. The ORF45 protein of Kaposi's sarcoma-associated herpesvirus is associated with purified virions. J Virol 77:4221–4230. doi: 10.1128/JVI.77.7.4221-4230.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu FX, Chong JM, Wu L, Yuan Y. 2005. Virion proteins of Kaposi's sarcoma-associated herpesvirus. J Virol 79:800–811. doi: 10.1128/JVI.79.2.800-811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Connor CM, Kedes DH. 2006. Mass spectrometric analyses of purified rhesus monkey rhadinovirus reveal 33 virion-associated proteins. J Virol 80:1574–1583. doi: 10.1128/JVI.80.3.1574-1583.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. 1998. How cells respond to interferons. Annu Rev Biochem 67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 50.Biron CA, Nguyen KB, Pien GC, Cousens LP, Salazar-Mather TP. 1999. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu Rev Immunol 17:189–220. doi: 10.1146/annurev.immunol.17.1.189. [DOI] [PubMed] [Google Scholar]
- 51.Nguyen KB, Salazar-Mather TP, Dalod MY, Van Deusen JB, Wei XQ, Liew FY, Caligiuri MA, Durbin JE, Biron CA. 2002. Coordinated and distinct roles for IFN-alpha beta, IL-12, and IL-15 regulation of NK cell responses to viral infection. J Immunol 169:4279–4287. doi: 10.4049/jimmunol.169.8.4279. [DOI] [PubMed] [Google Scholar]
- 52.Huber JP, Farrar JD. 2011. Regulation of effector and memory T-cell functions by type I interferon. Immunology 132:466–474. doi: 10.1111/j.1365-2567.2011.03412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marrack P, Kappler J, Mitchell T. 1999. Type I interferons keep activated T cells alive. J Exp Med 189:521–530. doi: 10.1084/jem.189.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ito T, Amakawa R, Inaba M, Ikehara S, Inaba K, Fukuhara S. 2001. Differential regulation of human blood dendritic cell subsets by IFNs. J Immunol 166:2961–2969. doi: 10.4049/jimmunol.166.5.2961. [DOI] [PubMed] [Google Scholar]
- 55.Luft T, Pang KC, Thomas E, Hertzog P, Hart DN, Trapani J, Cebon J. 1998. Type I IFNs enhance the terminal differentiation of dendritic cells. J Immunol 161:1947–1953. [PubMed] [Google Scholar]
- 56.Montoya M, Schiavoni G, Mattei F, Gresser I, Belardelli F, Borrow P, Tough DF. 2002. Type I interferons produced by dendritic cells promote their phenotypic and functional activation. Blood 99:3263–3271. doi: 10.1182/blood.V99.9.3263. [DOI] [PubMed] [Google Scholar]