ABSTRACT
The human interferon-inducible IFI16 protein, an innate immune sensor of intracellular DNA, was recently demonstrated to act as a restriction factor for human cytomegalovirus (HCMV) and herpes simplex virus 1 (HSV-1) infection by inhibiting both viral-DNA replication and transcription. Through the use of two distinct cellular models, this study provides strong evidence in support of the notion that IFI16 can also restrict human papillomavirus 18 (HPV18) replication. In the first model, an immortalized keratinocyte cell line (NIKS) was used, in which the IFI16 protein was knocked down through the use of small interfering RNA (siRNA) technology and overexpressed following transduction with the adenovirus IFI16 (AdVIFI16) vector. The second model consisted of U2OS cells transfected by electroporation with HPV18 minicircles. In differentiated IFI16-silenced NIKS-HPV18 cells, viral-load values were significantly increased compared with differentiated control cells. Consistent with this, IFI16 overexpression severely impaired HPV18 replication in both NIKS and U2OS cells, thus confirming its antiviral restriction activity. In addition to the inhibition of viral replication, IFI16 was also able to reduce viral transcription, as demonstrated by viral-gene expression analysis in U2OS cells carrying episomal HPV18 minicircles and HeLa cells. We also provide evidence that IFI16 promotes the addition of heterochromatin marks and the reduction of euchromatin marks on viral chromatin at both early and late promoters, thus reducing both viral replication and transcription. Altogether, these results argue that IFI16 restricts chromatinized HPV DNA through epigenetic modifications and plays a broad surveillance role against viral DNA in the nucleus that is not restricted to herpesviruses.
IMPORTANCE Intrinsic immunity is mediated by cellular restriction factors that are constitutively expressed and active even before a pathogen enters the cell. The host nuclear factor IFI16 acts as a sensor of foreign DNA and an antiviral restriction factor, as recently demonstrated by our group for human cytomegalovirus (HCMV) and herpes simplex virus 1 (HSV-1). Here, we provide the first evidence that IFI16 inhibits HPV18 replication by repressing viral-gene expression and replication. This antiviral restriction activity was observed in immortalized keratinocytes transfected with the religated genomes and in U2OS cells transfected with HPV18 minicircles, suggesting that it is not cell type specific. We also show that IFI16 promotes the assembly of heterochromatin on HPV DNA. These changes in viral chromatin structure lead to the generation of a repressive state at both early and late HPV18 promoters, thus implicating the protein in the epigenetic regulation of HPV gene expression and replication.
INTRODUCTION
Many recent studies point to the importance of cell-type- and host-specific expression of antiviral factors in limiting viral infection (1–4). Some of the very early antiviral responses include the activation of intrinsic restriction factors at high enough levels to inhibit the first stages of viral replication. Such factors include proteins that localize to the nucleus and mediate the transcriptional repression of viruses that replicate within this subcellular compartment (5–9). Many restriction factor genes are also interferon (IFN)-stimulated genes, consistent with the fundamental role of this class of genes in antiviral responses (10–14).
Human hematopoietic interferon-inducible nuclear proteins with a 200-amino-acid repeat (HIN200) domain-containing proteins, AIM2, IFI16, myeloid cell nuclear differentiation antigen (MNDA), and IFIX, have long been known to be transcriptional regulators involved in apoptosis, autoimmunity, and cell cycle regulation and differentiation (reviewed in references 15 and 16). Recently, a role in microbial DNA sensing was also found for AIM2 and IFI16 (17–22). The latter is predominantly nuclear, although it has been shown to translocate to the cytoplasm following the recognition of certain stimuli, including viral infections and UVB irradiation, while AIM2 is usually cytoplasmic (23–25). Both AIM2 and IFI16 contain pyrin and HIN domains (PYHINs); they can associate with ASC and other proteins through their pyrin domains and with DNA in the cytoplasm (AIM2) or in the nucleus (IFII16) through their HIN200 domains (9, 17, 25–27). IFI16 cooperatively binds double-stranded DNA (dsDNA) in a length-dependent manner and clusters into distinct protein filaments through the pyrin domain, even in the presence of excess dsDNA (28). Moreover, IFI16 has been demonstrated to interact directly with STING in a DNA-dependent manner, leading to the recruitment of TBK1, IRF3 activation, and the stimulation of beta interferon (IFN-β) production (17, 29–31).
We recently demonstrated that the replication of some herpesviruses, in particular, human cytomegalovirus (HCMV), is significantly enhanced in the absence of functional IFI16 (32). We showed that IFI16 acts as a restriction factor for HCMV infection by inhibiting both viral-DNA replication and transcription. In order to exert its antiviral activity, IFI16 binds to and displaces the Sp1 transcription factor interacting with the responsive IR-1 element present in the HCMV UL54 promoter. Sp1 detachment from its DNA cognate element was found to lead to a decrease in HCMV DNA synthesis and, as a consequence, the inhibition of virus replication.
IFI16 can also restrict herpes simplex virus 1 (HSV-1) replication by repressing viral-gene expression independently of its roles in the immune response. Using a permanently IFI16-negative cell line that we generated, we demonstrated that IFI16 reduces the association of important transcription factors on HSV-1 promoters and promotes global histone modifications by increasing the markers for repressive chromatin and decreasing the markers for activating chromatin on viral and cellular genes (33).
The finding that the nuclear DNA sensor IFI16 controls virus growth was fundamental in advancing our understanding of the intrinsic mechanisms that drive viral infections sustained by DNA viruses that replicate in the nucleus, such as herpesviruses. Human papillomaviruses (HPV) are small nonenveloped dsDNA viruses that selectively infect keratinocytes in stratified epithelia of both skin and mucosa (34, 35). Like herpesviruses, they replicate in the nucleus, and their target cells—keratinocytes—constitutively express IFI16, endowing the cells with the ability to control HPV replication through IFI16 activity (23, 36).
In the present study, we examined the role of IFI16 in controlling human papillomavirus 18 (HPV18) replication in primary human keratinocytes (NIKS) and U2OS cells. The latter is a human osteosarcoma cell line that provides an invaluable cellular assay system for the study of HPV genome replication due to its unique ability to support the transient and stable replication of HPV (37). Using small interfering RNA (siRNA) technology, we show that IFI16 protein knockout in NIKS cells results in increased viral replication. Accordingly, IFI16 overexpression by recombinant adenovirus infection decreased both viral-DNA copy numbers and transcription in NIKS and U2OS cells. IFI16 restriction activity was also observed in U2OS cells with gene-edited IFI16 with CRISPR (clustered regularly interspaced short palindromic repeat) (IFI16 knocked out). Finally, we provide evidence indicating that IFI16 overexpression results in changes in viral chromatin structure that lead to the generation of a repressive state at both early and late HPV18 promoters, implicating the protein in epigenetic regulation of HPV gene expression and replication.
MATERIALS AND METHODS
Cell culture, plasmids, and transfection.
The NIKS cell line (Stratatech Corporation, Madison, WI, USA) was cultured in the presence of J2 3T3 fibroblast feeders that were maintained at low passage numbers in selected growth medium—Ham's F-12 medium–Dulbecco's modified Eagle's medium (DMEM) (3:1) supplemented with 2.5% fetal bovine serum (FBS), 0.4 μg hydrocortisone per ml, 8.4 μg cholera toxin per ml, 5 μg insulin per ml, 24 μg adenine per ml, 10 μg epidermal growth factor per ml, 100 units penicillin, and 100 μg streptomycin per ml (F medium)—in the presence of mitomycin C-treated feeders. U2OS, HeLa, and C33A cells were grown in DMEM (Life Technologies Italia, Monza, Italy) supplemented with 10% FBS (Sigma-Aldrich, Milan, Italy).
To prepare the religated genomes, the HPV18 genome was released from the pBluescript vector by EcoRI digestion. Linear genomes were gel purified, recircularized using a dilute ligation reaction, and introduced into NIKS cells by cotransfection with a blasticidin-resistant plasmid using Effectene transfection reagent (Qiagen Srl, Milan, Italy), as previously described (38). Following treatment with blasticidin (5 μg/ml for 1 week), resistant colonies were pooled, expanded, and named NIKS-HPV18. Differentiation was induced with 1.5% methylcellulose as previously described (39).
HPV18 minicircle viral genomes were produced as previously described (40).
U2OS-HPV18 cells were obtained by electroporation of U2OS cells with a Microporator MP-100 (Life Technologies Italia) with 0.7 μg of HPV18 minicircles according to the manufacturer's instructions (1,230 V; 10-ms pulse width; 4 impulses).
For luciferase assays, cells were transfected using Effectene transfection reagent (Qiagen) according to the manufacturer's instructions. After 24 h, the cells were infected with adenovirus IFI16 (AdVIFI16) or the control indicator plasmid AdVLacZ (multiplicity of infection [MOI], 30 PFU/ml), and 24 h later, luciferase activity was measured using the Dual Luciferase Reporter Assay System kit (Promega Italia, Milan, Italy) on a Lumino luminometer (Stratec Biomedical Systems, Birkenfeld, Germany), as previously described by Baggetta et al. (41).
pGL4-LCRHPV18 (Addgene plasmid 22859) has been previously described (42). The pGL4-LCRHPV18 Sp1 mutant was generated by site-directed mutagenesis using pGL4-LCRHPV18 as a template for amplification by PCR and the following primers: mutSp1fw, 5′-ATATAAAAAAACTAGTAACCGAAAAC-3′, and mutSp1rev, 5′-GTTTTCGGTTACTAGTTTTTTTATAT-3′. The underlined bases represent the introduced mutations to the Sp1 binding site, which abolish Sp1 binding as described by Hoppe-Seyler and Butz (43). The sequence of the plasmid construct was confirmed by automated sequencing to ensure the designated mutations were successfully introduced.
For the inhibition of IFI16 expression, NIKS cells were transiently transfected using a Microporator according to the manufacturer's instructions (1,350 V; 30-ms pulse width; single impulse) with a pool of IFI16 small interfering RNAs or control siRNA (siCtrl) as a negative control (final concentration, 300 nM; Qiagen). The IFI16 siRNA sequences are available upon request. IFI16 siRNA-induced blockade was checked by immunoblotting with rabbit anti-IFI16 antibodies and by real-time PCR.
IFI16-null U2OS clone 67 cells.
IFI16-negative U2OS cells were maintained in DMEM supplemented with 10% FBS and 5% penicillin-streptomycin. Their generation has been described by Johnson et al. (clone 67) (33). Briefly, a guided RNA plasmid was constructed with target sequence GAAAAGTTCCGAGGTGATGCTGG within a guided RNA scaffold (44) in pGEMT, using NheI sites. Using Lipofectamine LTX and Plus reagent (Life Technologies), U2OS cells were transfected with 3 plasmids carrying guide RNA, Cas9 (Addgene plasmid 41815), and green fluorescent protein (GFP) at a ratio of 4:1:1, respectively. At 48 h posttransfection, GFP-positive cells were sorted individually into 96-well plates containing complete growth medium. Lack of IFI16 expression in each clone was screened by dot blotting and confirmed by Western blotting.
Adenoviral vectors and infection.
The adenovirus transfer vector pAC-CMV IFI16 was constructed as described previously (45). For cell transduction, postconfluent NIKS-HPV18, HeLa, U2OS-mcHPV18, and C33A cells were washed once with phosphate-buffered saline (PBS) and incubated with AdVIFI16 or AdVLacZ at an MOI of 30 in F medium or DMEM. After 2 h at 37°C, the virus was washed off and fresh medium was applied.
Immunoblotting.
Whole-cell protein extracts were prepared and subjected to immunoblot analysis as described previously (45). Rabbit polyclonal anti-C-terminal IFI16 antibodies (diluted 1:1,000) were used, with monoclonal antibody (MAb) against α-tubulin (39527; Active Motif; 1:4,000) as a control for protein loading. Immunocomplexes were detected using sheep anti-mouse or donkey anti-rabbit immunoglobulin antibodies conjugated to horseradish peroxidase (GE Healthcare Europe GmbH, Milan, Italy) and visualized by enhanced chemiluminescence (Super Signal West Pico; Pierce-Thermo Fischer Scientific, Rockford, IL, USA). Images were acquired, and densitometry of the bands was performed using Quantity One software (version 4.6.9; Bio-Rad Laboratories). Densitometry values were normalized using the corresponding loading controls.
Quantitative nucleic acid analysis.
Real-time quantitative reverse transcription (qRT)-PCR analysis was performed on an Mx3000P apparatus (Agilent Technologies Italia, Milan, Italy). Total RNA was extracted using a NucleoSpin RNA kit (Macherey-Nagel GmbH, Düren, Germany), and 1 μg was retrotranscribed using a RevertAid H Minus First Strand cDNA synthesis kit (Fisher Scientific Italy, Milan, Italy). Reverse-transcribed cDNAs were amplified in duplicate using SsoAdvanced Universal SYBR green Supermix (Bio-Rad Laboratories Srl, Milan, Italy) for viral genes, as well as cellular genes. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) housekeeping gene was used to normalize for variations in cDNA levels. Total cellular DNA was isolated from cells with TRI Reagent (Sigma-Aldrich). A 600-ng DNA sample was digested with DpnI to remove the unreplicated input DNA. After digestion, 15 ng was analyzed by quantitative PCR (qPCR) using 10 nM primers and SsoAdvanced Universal SYBR green Supermix (Bio-Rad Laboratories Srl). The reaction conditions consisted of a 15-min 95°C activation cycle, 40 cycles of 10 s denaturation at 95°C and 30 s annealing at 60°C, and elongation at 72°C for 10 min. Copy number analysis was completed by comparing the unknown samples to standard curves of linearized HPV18 DNA. The GAPDH DNA copy number was used as an endogenous control. The primer sequences are available upon request. The specificity of the L2 primers was tested in nontransfected cells where no amplification occurred.
Nuclear extract isolation and EMSA.
Nuclear extracts were obtained using a Nuclear Extract kit (Active Motif) according to the manufacturer's instructions, and an electrophoretic mobility shift assay (EMSA) was carried out as previously described (46). Briefly, nuclear extracts (15 μg of protein) were incubated in binding buffer (10 mM Tris-HCl [pH 7.5], 50 mM NaCl, 0.5 mM EDTA, 0.5 mM dithiothreitol, 1 mM MgCl2, 5% glycerol) containing 2 μg of poly(dI-dC) and the 32P-labeled double-stranded Sp1 consensus oligonucleotide (Promega). Unlabeled 30-bp annealed oligonucleotide was added as competitor DNA in 100-fold molar excess above the level of the probe. The oligonucleotide probe was labeled with [γ-32P]ATP (PerkinElmer Italia) and T4 polynucleotide kinase according to the manufacturer's protocol and finally column purified on G-25 Sephadex (Bio-Rad Laboratories). Complexes were analyzed by nondenaturing 4% PAGE, dried, and detected by autoradiography.
ChIP assay.
The suitability of each antibody (Ab) for the chromatin immunoprecipitation (ChIP) assay was confirmed by immunoprecipitation-Western blotting (data not shown). ChIP assays were performed using the Shearing Optimization kit and the OneDay ChIP kit (Diagenode Europe, Seraing, Belgium). Extracts were sonicated using the BioruptorH Twin (Diagenode) for 10 cycles (30 s on and 30 s off) at the high power setting. Immunoprecipitation was performed with 2 μg of unmodified histone H3 (catalog number 06-755), acetyl-histone H3 (catalog number 06-599), dimethyl-histone H3 (Lys4; catalog number 07-030), and dimethyl-histone H3 (Lys9; catalog number 07-441), all purchased from Merck Millipore (Merck Millipore SpA, Milan, Italy). DNA solution (1 μl/reaction) was used for qPCR. Threshold cycle (CT) values for the samples were equated to input CT values to give percentages of input for comparison. The primers used to amplify the long control region (LCR) and the p811 promoters are available upon request. Ten microliters of each sample (corresponding to 10% of input) were loaded and immunoblotted with anti-H3 antibody to verify equal loading.
Southern blot analysis.
Total genomic DNA was prepared by suspending the cell pellet in lysis buffer (400 mM NaCl, 10 mM Tris-HCl [pH 7.4], 10 mM EDTA) and digesting with RNase A (50 μg/ml) for 1 h at 37°C, followed by incubation with proteinase K (50 μg/ml) and 0.2% sodium dodecyl sulfate (SDS) at 37°C overnight. Five micrograms of genomic DNA was digested with DpnI to remove any residual input DNA and with HindIII (with no restriction site in HPV18) or DpnI and EcoRI (with a single restriction site in HPV18 or two restriction sites in HPV18 minicircles) to linearize HPV DNA. The digested DNA was separated on a 0.8% agarose gel, agitated in 0.25 N HCl for 20 min, and then transferred to a Hybond-XL membrane, using the alkaline transfer method as instructed by the manufacturer (GE Healthcare). The HPV18 probe was synthesized using 25 ng of the HPV genome released from pBS-HPV18 by EcoRI digestion, followed by gel purification. The HPV18 fragment was subjected to random-prime labeling by using Ready-To-Go DNA Labeling Beads (without dCTP) (GE Healthcare) and 5 μl of [α-32P]dCTP (3,000 Ci/mmol; PerkinElmer Italia SpA, Monza, Italy). The membrane was prehybridized in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-5× Denhardt's solution-0.5% SDS-100 μg/ml of denatured salmon sperm DNA; 10 × 106 cpm of probe was added to fresh prehybridization solution after heat denaturation and hybridized overnight. The membrane was washed twice in 2× SSC-0.1% SDS at room temperature and once at 65°C for 15 min. If necessary, additional 15-min washes were performed at 65°C in 1× SSC-0.1% SDS and in 0.1× SSC-0.1% SDS. The results were quantitated using a Personal Molecular Imager (PM) System (Bio-Rad) equipped with Quantity One software.
IFN assay and neutralization of type I IFNs.
IFN assays were performed by microtitration assay of vesicular stomatitis virus (VSV) cytopathology on human fibroblasts as previously described (47). Briefly, 3-fold dilutions of samples in DMEM supplemented with 2% fetal calf serum (FCS) were added to cell monolayers. After 16 h of incubation, the medium was removed and the cell monolayers were infected with VSV (strain Indiana) in 1% FCS-DMEM. Titers are expressed in laboratory units. In our system, 1 unit corresponds to 1 international reference unit (NIH standard G-002-9045511; a mixture of leukocyte and fibroblast mouse IFNs). Samples were tested in duplicate. Each experiment was repeated at least three times.
To neutralize the activity of type I IFNs, specific blocking antibodies against the common receptor IFNAR (clone MMHAR-2; PBL Assay Science, Piscataway, NJ, USA) were added to culture media at a concentration of 5 μg/ml for 12 h prior to infection with AdV or treatment with poly(I·C) (25 μg/ml) and then left in the supernatant until the end of the respective experiment. Mouse IgG2a (clone MOPC-173; BD Biosciences Europe, Milan, Italy) was used as an isotype control.
FISH and immunofluorescence analysis.
To perform fluorescence in situ hybridization (FISH) coupled with immunofluorescence analysis, cells were grown on microscope slides, fixed with 4% paraformaldehyde, and permeabilized with 0.5% Triton X-100 in PBS for 20 min at 4°C. FISH probe was generated by using Biotin Nick Translation Mix (Roche Diagnostics SpA, Monza, Italy) according to the manufacturer's protocol with the HPV18 full-length genome as a template. The samples were then sequentially dehydrated in a series of ethanol washes (70%, 90%, 95%, and 100% ethanol) and air dried. The probe was added to the hybridization buffer (50% formamide in 5× SSC, 0.1% Tween 20, and 0.2 ng/μl yeast tRNA) at a concentration of 2 ng/μl and then incubated at 72°C for 5 min in order to denature the probe and the sample. Hybridization was performed overnight at 37°C in a humidified chamber. After stringent washing, the cells were blocked with 10% normal goat serum, incubated with monoclonal anti-IFI16 antibody (clone 1G7; Santa Cruz Biotechnology, Heidelberg, Germany; 1:100) for 1 h at room temperature in 5% normal goat serum, washed, and incubated with Alexa Fluor-conjugated secondary antibody for 1 h in PBS. HPV18 probe was simultaneously detected using a Tyramide Signal Amplification kit, according to the manufacturer's instructions (PerkinElmer Life and Analytical Sciences Inc.). The coverslips were mounted with Vectashield mounting medium (Vector Laboratories Ltd.), and the cells were visualized with a Leica TCS SP2 confocal microscope equipped with a UV laser (351 to 364 nm) and an argon-krypton laser (457 to 675 nm) (Leica Microsystems Srl), using a 63× oil immersion objective, numerical aperture (NA) 1.4.
Statistical analysis.
All statistical tests were performed using GraphPad Prism version 5.00 for Windows (GraphPad Software, San Diego, CA, USA). The data are presented as means ± standard deviations (SD). For comparisons consisting of two groups, means were compared using two-tailed Student's t tests; for comparisons consisting of three groups, means were compared using one-way or two-way analysis of variance (ANOVA) with Bonferroni's posttest. Differences were considered statistically significant at a P value of <0.05.
RESULTS
The IFI16 protein is a restriction factor of HPV18 replication in NIKS cells.
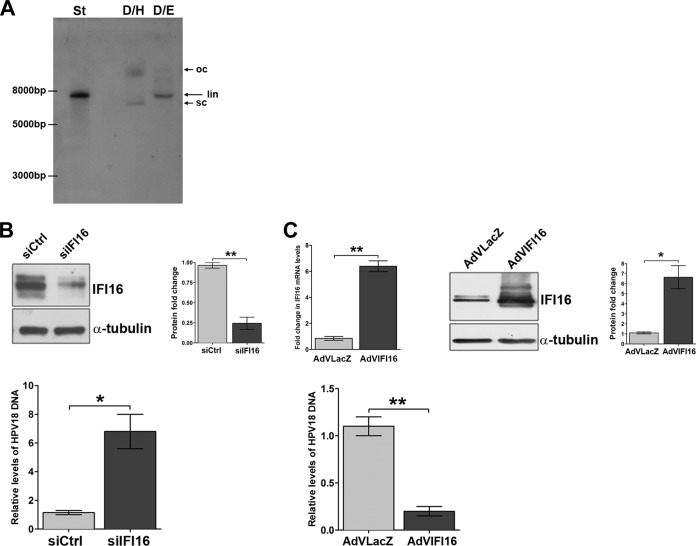
To study HPV replication and gene expression in the context of the viral life cycle, linear 8-kb HPV18 genomes were recircularized using a dilute ligation reaction mixture and introduced into the isogenic keratinocyte cell line (NIKS) by cotransfection with a blasticidin-resistant plasmid. Following selection, colonies were expanded, pooled, and used for the experiments approximately 1 month after transfection. To confirm the episomal status of HPV18, harvested DNA from pooled cells (passage 4) was digested with DpnI (to remove any residual input DNA) and HindIII (DpnI/HindIII [D/H]) (with no restriction site in HPV18) or DpnI and EcoRI (DpnI/EcoRI [D/E]), cutting the HPV genome once to linearize the viral DNA, and then subjected to Southern blot analysis with an HPV18 probe, as shown in the representative blot in Fig. 1A. With D/H-digested DNA, a pattern of migration was observed for HPV18 sequences that was consistent with open circular and supercoiled forms of episomal DNA (48). Digestion with EcoRI resulted in all hybridizing species migrating at the position of the ∼8-kb linearized nonintegrated HPV genome as the linearized pBluescript-HPV18 plasmid digested with EcoRI (Fig. 1A, lane St). The copy number of HPV18 present in pooled cells was estimated to be 10 copies per nucleus by comparison with standards. The viral-load number was also confirmed to be in the range of 10 genomes/cell by qPCR with primers designed to amplify a short fragment that spanned two DpnI sites in the L2 gene and therefore amplified DpnI-resistant viral DNA only. For IFI16 silencing, these cells were transfected with either pools of siRNAs targeting IFI16 or an siRNA control and 24 h later transferred into methylcellulose medium for 72 h to allow differentiation. Suspension of keratinocytes in semisolid medium recapitulates the differentiation process required for HPV life cycle completion and viral-DNA replication (39). To determine the efficiency of siRNA depletion, protein lysates were harvested and analyzed by Western blotting. As shown in Fig. 1B (top), the IFI16 protein was reduced ∼80% in IFI16 siRNA-treated NIKS-HPV18 cells compared with siCtrl-treated cells. Total cellular DNA was also isolated, DpnI digested, and analyzed for viral replication by qPCR. Compared to siCtrl-treated cells, viral-DNA loads were significantly (6-fold) higher in IFI16 siRNA-treated cells (Fig. 1B, bottom).
FIG 1.
The IFI16 protein is a negative regulator of HPV replication. (A) Genomic DNA was extracted from pooled NIKS-HPV18 cells and examined by Southern blotting. All samples contained 5 μg of genomic DNA and were digested with DpnI to remove any residual input DNA and with HindIII (D/H) (with no restriction site in HPV18) or DpnI and EcoRI (D/E) (with a single restriction site in HPV18) to linearize HPV DNA. The positions of open-circle (oc), linear (lin), and supercoiled (sc) forms of HPV DNA are indicated on the right of the blots. A standard (St) (pBluescript-HPV18 plasmid digested with EcoRI) corresponding to 50 copies of the HPV18 genome per cell was included in the left lane. (B) NIKS-HPV18 cells were electroporated with a mixture of four different small interfering RNAs (siIFI16) or scrambled control siRNA (siCtrl). After 24 h, the cells were transferred into methylcellulose medium for 72 h to allow differentiation. (Top left) The efficiency of siRNA depletion was analyzed by Western blotting with anti-IFI16 polyclonal antibody, and α-tubulin was included as a loading control. One representative blot of 3 independent experiments is shown. (Top right) Densitometric analysis predicting the fold change in expression (means ± SD) from three independent experiments is represented by histograms. (Bottom) To measure viral replication, total cellular DNA was analyzed by qPCR after DpnI digestion. The levels of glyceraldehyde 3-phosphate dehydrogenase were used to normalize the HPV18 levels. The data shown are the means of the results of three experiments ± SD (*, P < 0.05; **, P < 0.01; unpaired t test). (C) NIKS-HPV18 cells were infected for 24 h with AdVIFI16 or AdVLacZ and then transferred into methylcellulose medium for 72 h to allow differentiation. The efficiency of IFI16 overexpression was analyzed by both qRT-PCR and Western blotting using anti-IFI16 polyclonal antibody and α-tubulin (included as a loading control). (Top left) Total RNA was extracted and analyzed by qRT-PCR to measure the efficiency of IFI16 overexpression. The values were normalized to glyceraldehyde 3-phosphate dehydrogenase and are shown as fold changes relative to AdVLacZ-infected cells. The data shown are the averages of the results of three experiments ± SD (**, P < 0.01; unpaired t test). (Top middle) One representative Western blot is shown. (Top right) Densitometric analysis predicting the fold change in expression (means ± SD) from three independent experiments is represented by histograms (*, P < 0.05; unpaired t test). (Bottom) To measure viral replication, total cellular DNA was analyzed by qPCR after DpnI digestion. The levels of glyceraldehyde 3-phosphate dehydrogenase were used to normalize the HPV18 levels. The data shown are the means of the results of three experiments ± SD (**, P < 0.01; unpaired t test).
To provide further evidence supporting the physiological relevance of IFI16 in the control of HPV replication, the IFI16 protein was overexpressed by infection with the adenovirus recombinant vector AdVIFI16 or with AdVLacZ as a control at an MOI of 30 in NIKS-HPV18 cells. At 24 h after AdV infection, cells were grown in methylcellulose medium for 72 h and then checked for IFI16 expression at both the mRNA and protein levels. Real-time RT-PCR analysis demonstrated an elevation in IFI16 mRNA that was about 6-fold compared with AdVLacZ-infected cells (Fig. 1C, top left), and quantitation of the Western blot assays showed that 5-fold increases were achieved compared with AdVLacZ-infected cells (Fig. 1C, top right). As expected for a viral restriction factor, qPCR analysis revealed that the viral-load values were significantly decreased (5-fold) in AdVIFI16-infected cells compared with AdVLacZ-infected cells (Fig. 1C, bottom).
Altogether, these results demonstrate that reducing IFI16 activity with siRNA increases the rate of HPV replication, whereas overexpression of the full-length protein downregulates HPV replication, indicating IFI16 is a novel restriction factor in the replication cycles of these viruses.
IFI16 overexpression also impairs HPV18 transient replication and transcription in U2OS cells.
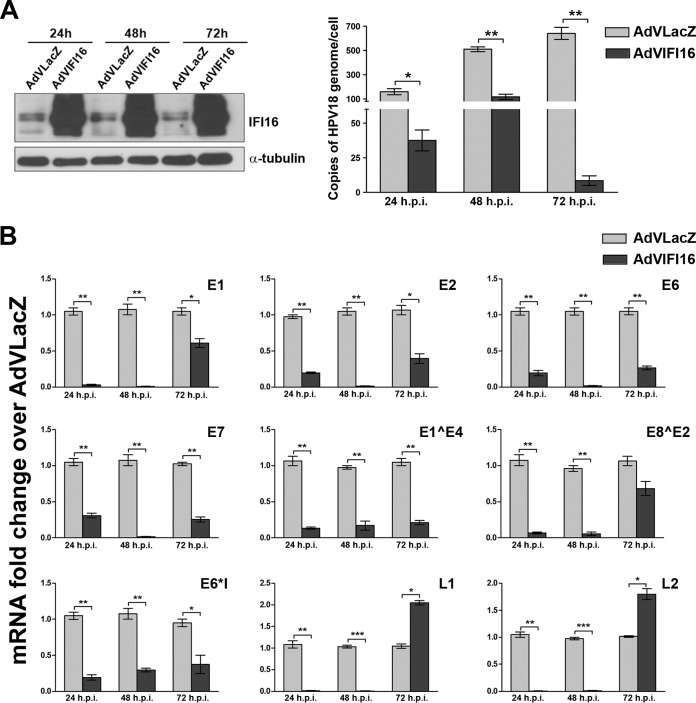
To increase the efficiency of our HPV replication model, religated HPV18 genomes were replaced with minicircles, which were then electroporated into U2OS cells as previously described by Reinson et al. (40). Cells were electroporated with HPV18 minicircle DNA and infected at passage 1 with either AdVIFI16 or AdVLacZ, as an internal control, at an MOI of 30, and the IFI16 protein content was monitored by Western blotting. As shown in Fig. 2A, left, IFI16 protein levels started to increase as early as 24 h after AdV infection, and they continued to increase for at least 72 h. Total cellular DNA was also isolated and analyzed for viral replication by qPCR after DpnI digestion. Consistent with the results obtained in NIKS cells, viral-DNA loads were reduced ∼75% at 24 h after AdVIFI16 infection compared with AdVLacZ-infected cells (Fig. 2A, right). This inhibitory effect was maintained at the 48- and 72-h time points, with reductions of about 78 and 97%, respectively, compared to AdVLacZ-infected cells.
FIG 2.
IFI16 overexpression impairs HPV18 replication by inhibiting both early and late viral-gene expression. (A) U2OS cells were electroporated with HPV18 minicircle DNA and, at passage 1, infected with AdVIFI16 or AdVLacZ. (Left) Total protein extracts were harvested at 24, 48, and 72 h after AdV infection (p.i.) and analyzed for IFI16 expression by Western blotting. α-Tubulin was included as a loading control. Experiments were repeated at least three times, and one representative result is shown. (Right) Total DNA was extracted at 24, 48, and 72 h after AdV infection to measure replicated (DpnI-resistant) viral DNA by qPCR. Levels of glyceraldehyde 3-phosphate dehydrogenase were used to normalize the HPV18 levels. The data shown are the averages of the results of three experiments ± SD (*, P < 0.05; **, P < 0.01; unpaired t test). (B) Total RNA was extracted at 24, 48, and 72 h after AdV infection and analyzed by real-time RT-PCR to measure the viral mRNAs. The values were normalized to glyceraldehyde 3-phosphate dehydrogenase and are shown as fold changes relative to AdVLacZ-infected cells for each time point. The data shown are the averages of the results of three experiments ± SD (*, P < 0.05; **, P < 0.01; ***, P < 0.001; unpaired t test).
To investigate whether IFI16 overexpression would also modulate viral transcription, we analyzed the levels of early and late viral mRNAs, including spliced messages, at different time points after IFI16 transduction by recombinant AdV infection. qRT-PCR analysis for viral transcripts showed they were always reduced in AdVIFI16-infected cells compared with AdVLacZ-infected cells (Fig. 2B). This inhibitory effect was most evident at 24 and 48 h after AdVIFI16 infection, and the level of viral transcription did not necessarily depend on the number of viral templates available. Despite the strong inhibition observed for the viral L1 and L2 mRNAs at the early time points, their levels increased over those detected in AdVLacZ-infected cells at the 72-h time point in AdVIFI16-infected cells. This change may be related to the reduced availability of the early regulatory genes, leading to altered regulation of the late genes.
These results show that (i) IFI16 overexpression inhibits HPV replication in the U2OS cell replication model, (ii) IFI16 inhibitory activity is not cell type dependent, and (iii) its repressive activity affects viral-mRNA transcription in a manner that is independent of the number of viral templates available for transcription.
IFI16 repressive activity is also functional on the integrated HPV18 genome and the LCR-driven reporter vector.
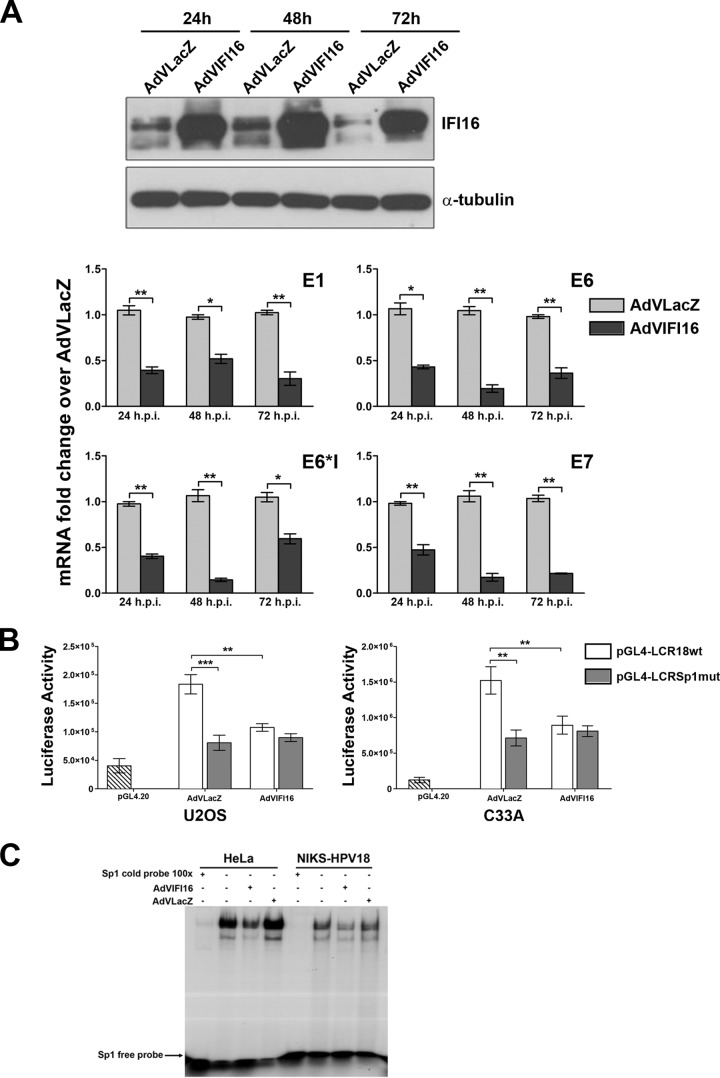
To corroborate the data obtained in the U2OS cell model and definitively exclude the possibility that differences in the levels of viral-gene expression are caused by variations in DNA template availability rather than in IFI16 itself, the inhibitory activity of IFI16 was also tested in HeLa cells carrying the integrated HPV18 genome (49, 50). Cells were infected with AdVIFI16 or AdVLacZ, as an internal control, at an MOI of 30, and IFI16 protein levels were determined by Western blotting at different time points after AdV infection. As shown in Fig. 3A, the trend in IFI16 protein expression observed in HeLa cells highly resembled that seen in U2OS cells. qRT-PCR analysis for early viral transcripts showed that their expression was significantly lower in AdVIFI16-infected cells than in AdVLacZ-infected cells, especially at 48 h.
FIG 3.
Effects of IFI16 overexpression on HPV18 promoter activity. (A) HeLa cells were infected with AdVIFI16 or AdVLacZ. (Top) Total protein extracts were harvested at 24, 48, and 72 h after AdV infection to demonstrate IFI16 overexpression by Western blotting with anti-IFI16 polyclonal antibody. α-Tubulin was included as a loading control. Experiments were repeated at least three times, and one representative result is shown. (Bottom) Total RNA was extracted at 24, 48, and 72 h after AdV infection and analyzed by real-time RT-PCR to measure the viral mRNAs. The values were normalized to glyceraldehyde 3-phosphate dehydrogenase and are shown as fold changes relative to AdVLacZ-infected cells for each time point. The data shown are the averages of the results of three experiments ± SD (*, P < 0.05; **, P < 0.01; unpaired t test). (B) C33A and U2OS cells were transfected with luciferase reporter plasmids containing either the wild-type HPV18 LCR promoter (pGL4-LCR18wt) or the promoter with a specific mutation in the Sp1 binding site (pGL4-LCR18Sp1mut). Twenty-four hours later, the cells were infected with AdVIFI16 or AdVLacZ at an MOI of 30 PFU/cell, and luciferase activity was assayed after 48 h. Luciferase activities are depicted relative to pGL4.20. The data represent the means of the relative activities from three independent experiments, each performed in duplicate, ± SD (**, P < 0.01; ***, P < 0.001; unpaired t test). (C) Nuclear protein extracts from HeLa and NIKS-HPV18 cells infected with AdVIFI16 or AdVLacZ at an MOI of 30 PFU/cell for 24 h or left uninfected were incubated with a radiolabeled oligonucleotide containing the consensus Sp1 binding site. Competition was done with 100-fold excess of cold specific oligonucleotide. Each experiment was repeated three times, and one representative result is shown.
The observed effect of IFI16 on the integrated HPV genome is consistent with its activity on viral and cellular promoters alongside its interaction with histone modification machinery. Indeed, IFI16 has been demonstrated to induce changes in markers for heterochromatin and euchromatin on viral and cellular promoters (21, 33, 51).
Next, the pGL4 reporter vector containing the HPV18 LCR region (nucleotides [nt] 6943 to 7857), namely, pGL4LCR18, was transfected into U2OS and C33A cells, as was a vector carrying a mutation in the HPV18 Sp1 binding site. After 24 h, the cells were once again infected with either AdVIFI16 or AdVLacZ, as an internal control, at an MOI of 30, and luciferase activity was assessed following an additional 48 h in order to assess the repressive effect of IFI16 activity on the LCR-driven luciferase reporter vectors. Measurements were normalized to the levels detected with empty pGL4.20 vector, which exhibited detectable luciferase activity above background levels. As shown in Fig. 3B, transfection with HPV18 LCR resulted in similar levels of luciferase activity in the two cell lines, which were 4.5- and 12-fold higher than that of the empty vector, respectively. This basal activity was reduced ∼2-fold by IFI16 overexpression compared to AdVLacZ-infected cells.
Since we have previously demonstrated that the IFI16 inhibitory activity in the HCMV model is mediated by the displacement of Sp1 binding to its cognate site in the viral DNA polymerase (UL54) promoter (32), transient reporter assays were also performed using an LCR carrying a mutated Sp1 binding site. Consistent with previous findings reporting Sp1 to be a key activator of RNA polymerase II-dependent promoters in HPV (43, 52), introduction of a mutation into its binding site caused a significant decrease in LCR promoter activity, which was no longer repressed by IFI16 overexpression. These results indicate that the Sp1 binding site could constitute a target of IFI16. Consistent with this hypothesis, the blocking of Sp1 binding to its cognate DNA by IFI16 overexpression was demonstrated by EMSA in AdVIFI16-infected cells (Fig. 3C), while inconsistent results were obtained when we tried to confirm Sp1 displacement on the HPV promoter by ChIP assay, which was at least partially explained by the low percentage of input obtained with the anti-Sp1 antibodies tested.
Although these findings confirm that the inhibitory action of IFI16 upon HPV18 LCR activity involves the Sp1 binding site, they do not exclude the possibility that other, more general mechanisms of transcription regulation may also exist.
IFI16 promotes heterochromatin association with HPV18 DNA at both early and late promoters.
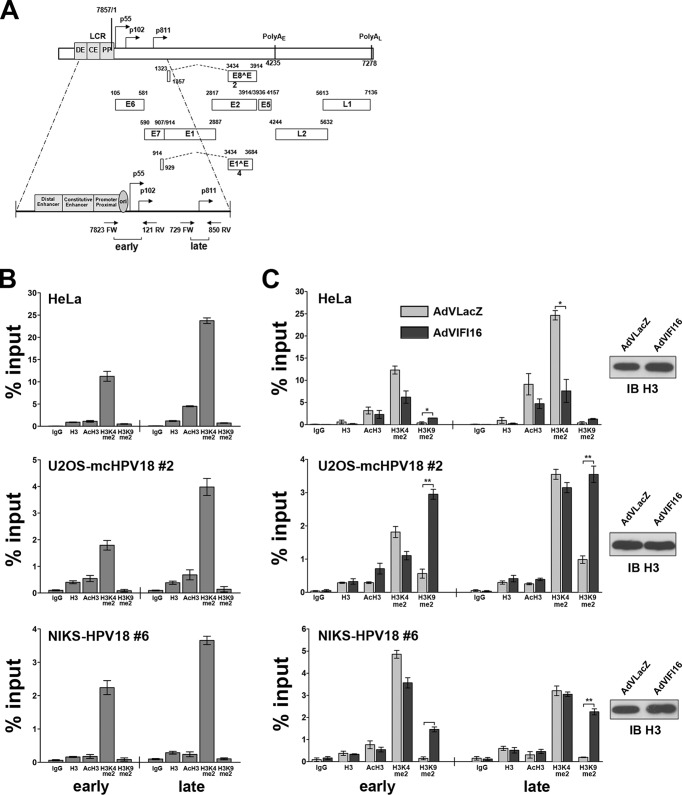
In addition to sequence-specific binding proteins, gene expression is also regulated by changes in chromatin structure through the specific modification of core histones (53–55). Modifications, such as the dimethylation of histone H3 lysine 4 (H3K4), are associated with actively transcribing genes, whereas the dimethylation of histone H3 lysine 9 (H3K9) is a mark of heterochromatin. To determine whether IFI16 could mediate the repressive activity described above through changes in HPV18 chromatin structure, we used a ChIP assay to first examine histone association with viral DNA in all three experimental settings described above. Lysates from formaldehyde-fixed cells were subjected to immunoprecipitation using antibodies specific for histone H3, acetylated H3 (AcH3), H3K4, or H3K9. The fraction of viral DNA immunoprecipitated was determined by qPCR. The first primer set used encompassed nucleotides 7823 to 121, which spans the p55 and p102 proximal promoter region (Fig. 4A) (56). The second set of primers was directed at nucleotides 729 to 850, which correspond to the late p811 promoter region within the E7 open reading frame (ORF) of the virus genome. These two regions are separated by approximately 1 kb; our analysis therefore reflects binding to two distinct regions. Both regions bound acetylated H3 and dimethylated H3K4, indicating that they were in an active chromatin state (Fig. 4B). Interestingly, the levels of dimethylated H3K4 found to bind to the late promoter region were almost 2-fold higher than those found associated with the sequences around the early promoter. The chromatin states around the early and late HPV18 promoters were very similar in all three cellular models (HeLa, U2OS-mcHPV18, and NIKS-HPV18). To examine whether IFI16 expression could modulate chromatin structure, cells were infected with AdVLacZ or AdVIFI16 for 24 h, formaldehyde fixed, and subjected to chromatin immunoprecipitation with the same set of antibodies (Fig. 4C). The levels of dimethylated H3K4 bound to the early promoter decreased by 2-fold in AdVIFI16-infected HeLa cells compared to AdVLacZ-infected HeLa cells. A similar trend was found at the late promoter, for which the reduction in AdVIFI16-infected cells was even higher (3.4-fold versus AdVLacZ). AdVIFI16-infected cells had higher levels (3.5-fold) of dimethylated H3K9 at both the early and late promoters than AdvLacZ-infected cells. U2OS-mcHPV18 cells displayed a similar pattern of chromatin structure with a slight reduction in the levels of bound dimethylated H3K4 (especially at the early promoter) and induction of dimethylated H3K9 in IFI16-expressing cells in comparison to control AdVLacZ-infected cells at both promoter sites (5- and 3.6-fold, respectively). The chromatin structure in NIKS-HPV18 cells mirrored that found in U2OS cells, with an ∼10-fold increase of bound dimethylated H3K9 at both promoters in IFI16-overexpressing cells in comparison with AdVLacZ-infected cells. These changes were not the result of increased amounts of viral DNA resulting from amplification, as our analysis was normalized with respect to the levels of HPV18 genomes in the sample.
FIG 4.
IFI16 promotes heterochromatin association with viral DNA. (A) Linear depiction of the HPV18 genome with the early (p55 and p102) and late (p811) regulatory regions expanded. (B) Extracts were prepared from HeLa, U2OS-mcHPV18, and NIKS-HPV18 cells, and ChIP was carried out using antibodies specific to unmodified histone H3 (H3), AcH3, dimethylated lysine 4 of H3 (H3K4me2), dimethylated lysine 9 of H3 (H3K9me2), or IgG as a control. Immunoprecipitated early and late promoter sequences were measured by qPCR, and CT values for the samples were equated to input CT values to give the percentages of input values for comparison. The data shown are the averages of the results of three experiments ± SD. (C) HeLa, U2OS-mcHPV18 at 2nd passage (#2), and NIKS-HPV18 cells at 6th passage (#6) were infected with AdVIFI16 or AdVLacZ. Cell extracts were prepared at 24 h after AdV infection, and ChIP was carried out as described for panel B. The data shown are the averages of the results of three experiments ± SD (*, P < 0.05; **, P < 0.01; unpaired t test). (Right) Lysate samples taken prior to immunoprecipitation (10 μl) were used for Western blot analysis (IB) with the antibodies for H3 to monitor equal loading.
Together, these results support the hypothesis that IFI16 restricts viral-gene expression by promoting heterochromatin association with viral promoters.
A high rate of HPV18 genome replication in active viral chromatin configuration occurs in IFI16-knocked-out cells, which is strongly inhibited by IFI16 restoration.
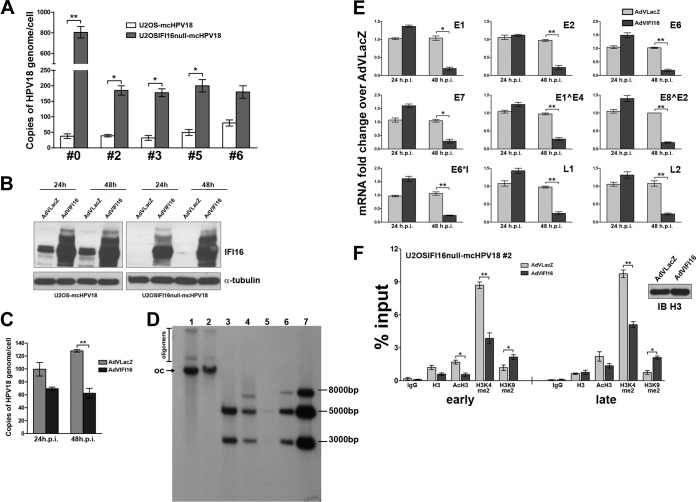
To prove conclusively that IFI16 exerts restriction activity against HPV18, a gain of function in viral-DNA replication in stable knocked-down cells that could subsequently be inhibited by the addition of IFI16 would have to be demonstrated. To this end, we used a permanently IFI16-negative U2OS cell line (IFI16-null U2OS clone 67 cells), which was generated by CRISPR Cas9-mediated genome editing (33). These U2OSIFI16null cells, as well as their wild-type (wt) parental cells, were electroporated with HPV18 minicircles, and viral replication was evaluated by qPCR over time. It is known that when using the U2OS cell-based model system, the initial transient-amplification phase develops into the stable maintenance of HPV genomes, which can progress to the secondary amplification phase, when high cell density is achieved. Consistent with this model, at 7 days postelectroporation, HPV18 viral loads underwent an initial amplification that was 20-fold higher in null cells than in the parental cell line and reached very high copy numbers (up to 103 copies/cell) (Fig. 5A). Cultures were then cultivated with regular passaging under subconfluent conditions, and the viral-load values analyzed at different passages revealed drops in their values in both cell lines, but the 4-fold difference between null cells and the parental cell line was maintained over time, at least up to the fifth passage. Altogether, these findings indicate that in the absence of IFI16 restriction activity, viral replication is significantly enhanced.
FIG 5.
Stable knockout of the IFI16 gene in U20S cells enhances HPV18 replication and inhibits viral-gene expression through histone modifications. (A) U2OS IFI16-knockout cells and the parental cell line were electroporated with HPV18 minicircle DNA and, at the indicated passage (#0, #2, #3, #5, or #6), total cellular DNA was extracted. Viral replication was measured by qPCR of the DpnI-digested DNA. The levels of glyceraldehyde 3-phosphate dehydrogenase were used to normalize the HPV18 levels. The data shown are the averages of the results of three experiments ± SD (*, P < 0.05; **, P < 0.01; unpaired t test). (B) U2OS IFI16-knockout cells at the 2nd passage after HPV18 minicircle DNA electroporation and the parental cell line were infected with AdVIFI16 or AdVLacZ, and at 24 and 48 h after AdV infection, total protein extracts were prepared for Western blot analysis with anti-IFI16 polyclonal antibody or α-tubulin as a loading control. Experiments were repeated at least three times, and one representative result is shown. (C) U2OS IFI16-knockout cells at the 2nd passage after HPV18 minicircle DNA electroporation were infected with AdVIFI16 or AdVLacZ, and at 24 and 48 h after AdV infection, total DNA was extracted and used to measure replicated (DpnI-resistant) viral DNA by qPCR. Levels of glyceraldehyde 3-phosphate dehydrogenase were used to normalize the HPV18 levels. The data shown are the averages of the results of three experiments ± SD (**, P < 0.01; unpaired t test). (D) U2OS IFI16-knockout cells at the 2nd passage after HPV18 minicircle DNA electroporation were infected with AdVLacZ (lanes 1 and 3) or AdVIFI16 (lanes 2 and 4), and 48 h later, total DNA extracts were prepared for Southern blot analysis. All samples contained 5 μg of genomic DNA and were digested with DpnI to remove any residual input DNA and with HindIII (with no restriction site in HPV18) (lanes 1 and 2) or DpnI and EcoRI to linearize HPV DNA (with one site in the HPV genome and one in the linker, which gave two bands of 5,113 bp and 2,835 bp) (lanes 3 and 4). The positions of HPV open-circle (oc) and oligomer forms are indicated on the left of the blots. Standards (HPV18 minicircles digested with EcoRI) corresponding to 10, 100, and 1,000 copies of the HPV18 genome per cell are included on the right (lanes 5, 6, and 7, respectively). The ∼8,000-bp band corresponds to partially digested minicircle DNA. The Southern blot was hybridized using the complete HPV18 genome as a probe. (E) Total RNA was extracted at 24 and 48 h after AdV infection in U2OS IFI16-knockout cells at the 2nd passage after HPV18 minicircle DNA electroporation and analyzed by real-time RT-PCR to measure the viral mRNAs. The values were normalized to glyceraldehyde 3-phosphate dehydrogenase and are shown as fold changes relative to AdVLacZ-infected cells for each time point. The data shown are the averages of the results of three experiments ± SD (*, P < 0.05; **, P < 0.01; unpaired t test). (F) ChIP assay performed with formalin-fixed extracts obtained from U2OS IFI16-knockout cells, at the 2nd passage after HPV18 minicircle electroporation, infected with AdVIFI16 or AdVLacZ for 24 h. ChIP was carried out using antibodies specific to unmodified histone H3 (H3), AcH3, H3K4me2, H3K9me2, or IgG as a control. Immunoprecipitated early and late promoter sequences were measured by qPCR; CT values for the samples were equated to input CT values to give the percentages of input values for comparison. The data shown are the averages of the results of three experiments ± SD (*, P < 0.05; **, P < 0.01; unpaired t test). (Right) Lysate samples taken prior to immunoprecipitation (10 μl) were used for Western blot analysis with the antibodies for H3 to monitor equal loading.
Next, to verify whether it was possible to restore IFI16 restriction activity, at the second passage after HPV18 electroporation, U2OSIFI16null cells were infected with AdVIFI16 or AdVLacZ and analyzed 24 and 48 h later. As shown in Fig. 5B, Western blot analysis demonstrated the complete restoration of IFI16 expression following AdVIFI16 infection. Viral-load values were also assessed (Fig. 5C), and the IFI16 repressive activity of the restored protein was very evident, especially at 48 h (about 45% reduction compared to AdVLacZ-infected cells). The reduction in viral replication efficiency was confirmed by Southern blotting in IFI16-transduced U2OS clone 67 cells (Fig. 5D). In addition, the patterns obtained were consistent with the episomal status of the HPV genomes, as digestion with EcoRI (one site in the HPV genome and one in the linker) gave the expected two bands of 5,113 bp and 2,835 bp (Fig. 5D, lanes 3 and 4). Finally, viral-gene expression was also analyzed at the same time points, and the results obtained mirrored those already found for the viral replication assay, showing a delayed but strong inhibitory effect of IFI16 upon viral mRNAs at 48 h after AdVIFI16 infection (both early and late genes) (Fig. 5E).
Having established that the IFI16 repressive activity is mediated by epigenetic modifications of the viral promoters, we wanted to investigate the viral chromatin configuration in the absence of the IFI16 protein and after its restoration. To this end, second-passage U2OSIFI16null-mcHPV18 cells were analyzed by ChIP assay (as described above), and the amounts of histones and modified histones were evaluated at both the early and late promoters. Consistent with the higher viral replication and transcription rates detected in null cells, considered to be associated with an active chromatin state, the amounts of acetylated and, in particular, dimethylated K4 were significantly (4-fold) higher in IFI16-null cells at both promoter regions than in parental cells (data not shown). In contrast, no significant changes were found in the heterochromatin mark K9. Definitive proof of the repressive IFI16 activity was obtained from ChIP assays after IFI16 restoration, which revealed a significant reduction in dimethylated-K4 levels in AdVIFI16-infected cells compared with AdVLacZ-infected cells (2-fold for both promoters). Dimethylated-K9 levels were also increased in AdVIFI16-infected cells compared with AdVLacZ-infected cells (1.8- and 2.5-fold for the early and late promoters, respectively) (Fig. 5F).
These data confirm the results obtained by transient transfection with siRNA molecules targeting the IFI16 mRNA in NIKS cells and show that IFI16 can repress both viral transcription and replication through epigenetic modifications of the viral promoters.
IFI16 does not colocalize with the HPV18 genome, and its viral restriction activity is independent of IFN response.
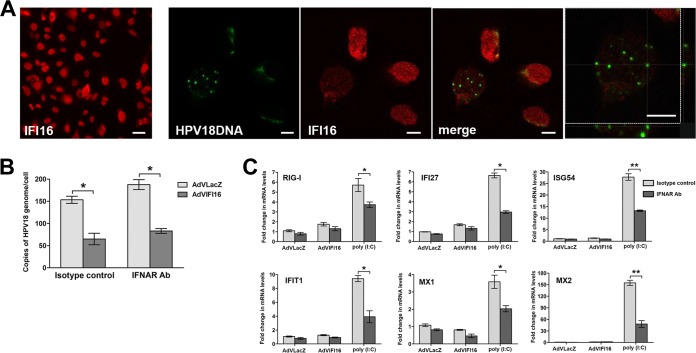
IFI16 has previously been shown to interact with HSV-1, as well as HCMV, DNA early during infection (24, 32). To determine HPV18 genome recognition by IFI16 and colocalization, U2OSIFI16null-mcHPV18 or U2OSIFI16null cells were infected with AdVIFI16 and 8 h later analyzed by combined immunofluorescence and FISH that was performed using anti-IFI16 antibodies and a probe for the HPV18 genome. As shown in Fig. 6A, HPV18-FISH-HPV-positive nuclei usually displayed lower expression levels of IFI16 than cells in which the HPV genomes were undetectable by FISH. However, even in the cells where IFI16 was still evident, we failed to detect any colocalization between FISH signal and anti-IFI16 immunofluorescence. The lack of colocalization of IFI16 with the viral genome was also determined by cross-sectional analysis of z-stack image series (Fig. 6A, far right). This difference in IFI16 expression levels was not observed in nonelectroporated U2OSIFI16null cells, where the efficiency of IFI16 transduction by AdV infection was very high in all the cells (Fig. 6A, far left). Since FISH-positive cells are usually regarded as cells with a high number of viral genomes resembling the late stage of infection, we cannot exclude the possibility that transient interactions might occur very early during infection that are not detected by this technique.
FIG 6.
IFI16 does not colocalize with the HPV18 genome, and its viral restriction activity is independent of IFN response. (A) U2OS IFI16-knockout cells at the 2nd passage after HPV18 minicircle DNA electroporation or nonelectroporated cells were grown at high density, infected with AdVIFI16 for 8 h, and then fixed in 4% paraformaldehyde and subjected to combined FISH with HPV18 probe (green dots) and immunofluorescence analysis with anti-IFI16 antibodies (red). The images were taken by confocal microscopy; the far-left image shows IFI16 expression in nonelectroporated cells (scale bar = 40 μm), while all the other images were taken in U2OSIFI16null-mcHPV18 cells; the far-right image shows cross-sectional analysis of a z-stack image series. Ten fields were digitally analyzed, and one representative image is shown. Scale bars = 10 μm. (B) U2OS IFI16-knockout cells at the 2nd passage after HPV18 minicircle DNA electroporation were infected with AdVLacZ or AdVIFI16 in the presence of anti-IFNAR antibodies or an isotype control, as detailed in Materials and Methods. At 48 h after AdV infection, total DNA was extracted and used to measure replicated (DpnI-resistant) viral DNA by qPCR. The levels of glyceraldehyde 3-phosphate dehydrogenase were used to normalize the HPV18 levels. The data shown are the averages of the results of three experiments ± SD (*, P < 0.05; unpaired t test). (C) Total RNA was also extracted from U2OS IFI16-knockout cells at the 2nd passage after HPV18 minicircle DNA electroporation infected with AdVLacZ or AdVIFI16 (24 h) or treated with poly(I·C) (8 h) in the presence of anti-IFNAR antibodies or isotype control, as detailed in Materials and Methods. Representative expression of a set of ISGs is shown. The values were normalized to glyceraldehyde 3-phosphate dehydrogenase and are shown as fold changes relative to AdVLacZ-infected cells for each time point. The data shown are the averages of the results of three experiments ± SD (*, P < 0.05; **, P < 0.01; unpaired t test).
Since emerging evidence indicates that the IFI16 protein is a mediator of type I IFN induction upon recognition of viral DNA, the supernatants of the cell cultures shown in Fig. 1, 2, 4, and 5 were analyzed for type I IFN production. As a positive control, supernatants of U2OS cells infected with HSV-1 for 6 h were included in the assay. None of the HPV18-transduced cells expressed type I IFN even after AdVLacZ or AdVIFI16 infection, while it was produced as a consequence of HSV-1 infection (27 U). To definitely exclude the possibility that the interferon response was involved in the observed phenotypes, U2OSIFI16null-mcHPV18 cells were treated for 12 h with anti-IFNAR Ab or an isotype control, infected with AdVLacZ or AdVIFI16, and analyzed for viral replication by qPCR following an additional 48 h of incubation with anti-IFNAR Ab or an isotype control (Fig. 6B). As expected, viral-DNA loads were reduced ∼55% at 48 h after AdVIFI16 infection compared with AdVLacZ-infected cells, and this inhibitory effect was not affected by the presence of anti-IFNAR Ab. Total RNA was also extracted from these cell cultures at 24 h after AdV infection and used to detect expression levels of interferon-stimulated genes (ISGs) by qRT-PCR. As a control, U2OSIFI16null-mcHPV18 cells were treated for 12 h with anti-IFNAR Ab or with an isotype control and stimulated with synthetic dsRNA, poly(I·C), for an additional 8 h in the presence or absence of anti-IFNAR Ab, and then total RNA was extracted. We determined the expression levels of RIG-I, IFI27, ISG54, IFIT1, MX1, and MX2 genes, all of which are known ISGs (Fig. 6C). Following treatment with poly(I·C), the mRNA levels of all the ISGs tested were increased and significantly inhibited by the presence of anti-IFNAR neutralizing antibody. Consistent with the lack of type I IFN production, we failed to detect any induction of the same ISGs in IFI16-overexpressing cells, and their expression levels were unaffected by the presence of anti-IFNAR neutralizing antibody.
Altogether, these studies suggested that the role of IFI16 in the inhibition of HPV18 replication was independent of its roles as a viral-DNA sensor and interferon inducer.
DISCUSSION
Through the use of two distinct cellular models, this study provides strong evidence in support of the notion that IFI16 acts as a restriction factor for HPV18 replication. In the first model, an immortalized keratinocyte cell line (NIKS) was used, in which the IFI16 protein was either knocked out through the use of siRNA technology or overexpressed following transduction with the AdVIFI16 vector. In these cells, HPV18 religated genomes were introduced by transfection, and replication was achieved by methylcellulose-induced differentiation. In these differentiated IFI16-silenced NIKS-HPV18 cells, viral-load values were significantly increased compared with differentiated control cells. The second model consisted of U2OS cells transfected by electroporation with HPV18 minicircles, which sustain high levels of viral replication without the need for any differentiation stimuli (40, 57), thus providing a suitable model for the assessment of IFI16 antiviral activity. In both cell lines, viral genomes were established as stably replicating extrachromosomal plasmids, as demonstrated by Southern blotting. In these cell cultures, IFI16 overexpression by recombinant adenovirus infection severely impaired HPV18 replication, thus confirming its antiviral restriction activity. This activity has also been observed in permanently IFI16-negative cells, where HPV replication was higher and maintained at higher levels during passages in culture than in the parental cell line. IFI16 restoration in these cells was as effective as in the transiently silenced cells with respect to inhibition of HPV replication and transcription. In addition to the inhibition of viral replication, IFI16 was also able to reduce viral transcription, as demonstrated by viral-gene expression analysis in U2OS cells carrying episomal HPV18 minicircles. IFI16 overexpression in these cells affected both early and late mRNAs, as shown by qPCR, independent of the number of viral templates available.
Definitive support for the inhibitory action of IFI16 on viral transcription came from the experiments performed using the integrated HPV18 genome in HeLa cells. Once again, we showed that the transcriptional activity of the HPV18 promoter became inhibited following IFI16 overexpression by recombinant AdV infection. Accordingly, transient transfection of an LCR-driven luciferase reporter vector showed reduced transcriptional activity upon IFI16 overexpression. A mutation in the Sp1 binding site reduced the promoter activity by about 50%, as well as the inhibitory activity of IFI16.
In a previous study, we demonstrated that IFI16 restricted HCMV replication through a mechanism involving the IFI16-mediated blockade of Sp1-dependent transcription of the gene encoding the viral DNA polymerase (32). However, emerging evidence indicates that IFI16 may exert its effects via various mechanisms that similarly result in genome regulation (58). Consistent with this scenario, two recent papers demonstrated that nuclear IFI16 promotes the addition of heterochromatin marks and yet reduces the number of euchromatin marks on the HSV-1 genome (33, 59). Accordingly, in the present study, we found that (i) both the early promoter (containing the Sp1 binding site) and the late promoter (lacking any canonical Sp1 binding site) were inhibited by IFI16 overexpression, indicating that its action was not restricted to Sp1 interference, and (ii) the chromatin states at both the early and late promoters were substantially modified by IFI16 overexpression, exhibiting a reduction in the levels of acetylated H3 and dimethylated K4, and both promoters were associated with increased levels of the repressive forms of histone modifications, such as dimethylated K9.
Our results are fully consistent with those of a previous study that analyzed the chromatin states at both the early and late promoters using cells that stably maintain HPV31 episomes (55). The study showed that in undifferentiated cells the chromatin surrounding both early and late viral promoter regions revealed an open, transcriptionally active state with dimethylated forms of histone H3K4, as well as acetylated histone H3. A similar pattern of chromatin structure was found here in our own experimental setting, indicating that the HPV genomes are promptly chromatinized once introduced into the cells by electroporation or other transfection techniques. The chromatin structure of HPV DNA is crucial, because viral DNA entering the nucleus might have different epigenetic or chromatin marks that could be detected by intrinsic viral defenses, whereas long-term-replicating genomes might have acquired different chromatin marks that make it indistinguishable from host DNA. Here, we show that as early as the first passage after electroporation in U2OS cells, the HPV genomes contain modified H3 histones consistent with those observed in HeLa cells containing integrated HPV18 genomes and previously reported for HPV31 and other genotypes (55, 60, 61); moreover, this configuration was maintained over time. In contrast to previous reports arguing that IFI16 can silence only naked DNA (59), our results clearly demonstrate that it can also inhibit viral DNA in the form of nucleosomal chromatin. One possible explanation for this discrepancy could lie in the different kinetics of the experiments and the cell lines used. In the study by Orzalli and colleagues (59), IFI16 activity was evaluated at 48 h following the transfection of simian virus 40 (SV40) DNA into human fibroblasts, while in our case, keratinocytes or U2OS osteosarcoma cells were used and later time points were studied following transfection. Here, we propose that IFI16 promotes the addition of heterochromatin marks and the reduction of euchromatin marks on viral chromatin, thus reducing both viral replication and transcription. It remains to be explained how IFI16, which is already present in the nuclei of keratinocytes and U2OS cells, can distinguish foreign from self-DNA and selectively engage foreign DNA. Consistent with what we observed for HSV-1 replication, we show that IFI16 inhibits HPV18 replication in multiple cell types, suggesting that this effect is general and not likely to be cell type dependent (33).
Another aspect that needs to be further investigated is whether IFI16 binds the HPV genome and whether this interaction might lead to any activation of innate immunity, such as inflammasome or IFN production. We failed to detect any type I IFN production, as well as ISG induction, in our in vitro setting, and addition of neutralizing anti-IFNAR antibodies did not affect the IFI16 antiviral activity. Combined immunofluorescence with anti-IFI16 antibodies and FISH analysis with the HPV18 genome did not reveal any colocalization between IFI16 and HPV18-positive FISH signal. Although further studies are required to exclude any transient interactions occurring at the earlier times of infection, it seems that, at least in our experimental model, the IFI16 inhibitory activity is independent of its roles as a DNA sensor and IFN inducer. These results are fully consistent with those previously reported by our groups for HCMV and HSV-1, demonstrating that IFI16 viral restriction is independent of the roles it plays in the inflammasome and interferon responses (32, 33).
IFI16 is composed of two domains, namely, one N-terminal pyrin domain (PYD) (IFI16PYD) followed by two HIN200 domains (IFI16HinA and IFI16HinB). The HIN200 domains nonspecifically bind various single-stranded DNA (ssDNA) and dsDNA fragments on the phosphate backbone via electrostatic interactions, with a footprint of 8 or 9 bases, thus confirming that the DNA sequence (e.g., the CpG island) is not a recognition element (58, 62). On the other hand, the PYD constitutes a homotypic protein-protein interaction domain whose function is thought to be limited to recruiting downstream effectors (63). IFI16 binds dsDNA in a nonlinear length-dependent manner and forms oligomers that are clearly different from entities resembling beads on a string (28). Although the viral genome is packaged into chromatin with host histones, it should be less dense and much more loosely packed than the nuclear self-DNA, thus allowing robust filament assembly of IFI16 and formation of signaling foci.
This is the first report showing IFI16 antiviral activity against a member of the papillomavirus family and the fourth to describe its activity as a viral restriction factor, while many studies have reported its role in innate immune responses against herpesviruses (18, 20, 21, 29). The first report, by Orzalli et al. (59), found that nuclear IFI16 acts as a restriction factor against ICP0-null HSV-1 in fibroblasts, limiting viral replication and immediate-early (IE) gene expression. The second paper was the result of work by our group and demonstrated that another member of the herpesvirus family, namely, HCMV, is restricted by this nuclear protein in fibroblasts (32). The third study showed that IFI16 prevents association of important transcriptional activators with wt HSV-1 promoters and suggested potential mechanisms of IFI16 restriction of wt HSV-1 replication and a direct or indirect role for IFI16 in histone modification (33).
Papillomaviruses and herpesviruses share a very important commonality, that is, nuclear replication in target cells that naturally express IFI16, thus providing IFI16 with the opportunity to interfere with their replication and transcription. Orzalli and coworkers propose a model in which nuclear IFI16 in fibroblasts binds to nonnucleosomal DNA when introduced into the nucleus by transfection or herpesvirus infection (59). Thereafter, IFI16 undergoes a conformational change upon DNA binding that allows the recruitment of histone modification enzymes that promote heterochromatic modifications on the exogenous DNA, compaction of the chromatin, and silencing of viral IE gene promoters. Our working hypothesis is very consistent with this, although in our case, the IFI16 repressive activity, through the reduction of euchromatin marks and addition of heterochromatin marks, is also active on nucleosomal HPV DNA. Considering that the papillomavirus genomes packed into virions are organized in the form of specifically positioned nucleosomes packaged into chromatin while during herpesvirus infection the viral genomic DNA is nuclesome free, one would expect that IFI16 could also silence chromatinized viral DNA in order to protect cells from the variety of viruses that replicate in the nuclear compartment. In accordance with this hypothesis, IFI16-induced modifications in the chromatin structure of the HPV18 genomes were observed in every cell line used in this work, including HeLa cells harboring integrated genomes. Reduced levels of dimethylated H3K4 in IFI16-overexpressing cells were consistently found in all experimental settings, while increased levels of the heterochromatin mark (dimethylated H3K9) were more prominent in U2OS cells.
The IFI16 restriction activity seems to be different and more long lasting than that reported for Sp100, which was shown to be confined to the early phases of HPV replication, since once viral genomes were established as stably replicating extrachromosomal plasmids, Sp100 downregulation had no effect on viral transcription and replication (64).
Overall, the results presented in this study congruently demonstrate that the actions of IFI16 contribute to a cell's intrinsic repression mechanism for HPV replication and gene expression. It remains to be determined, however, how the virus counteracts IFI16 activity and shifts the balance toward viral evasion and its consequent growth.
ACKNOWLEDGMENTS
We thank Mart Ustav, Eve Sankovsky, and Marit Orav, Estonian Biocenter-Tartu, for providing us with the minicircle system for HPV18 genome generation and Peter Howley, Harvard Medical School—Boston, for the pGL4-LCRHPV18 plasmid. We also acknowledge Salvatore Oliviero and Stefania Rapelli, Human Genetics Foundation (HuGeF)—Turin, and Gianluca Baldanzi, Novara Medical School, Biochemistry Unit, for their technical assistance.
This work was supported by the Italian Ministry for University and Research-MIUR (FIRB-Futuro in Ricerca 2008 to M.D.A.; PRIN 2012 to C.B. and S.L.), Compagnia di San Paolo (grant CSP2012 to M.G.), Associazione Italiana per la Ricerca sul Cancro-AIRC (grant IG 2012 to M.G.), and the European Society of Clinical Microbiology and Infectious Diseases-ESCMID (research grant 2013 to C.B.).
REFERENCES
- 1.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 2.Bieniasz PD. 2004. Intrinsic immunity: a front-line defense against viral attack. Nat Immunol 5:1109–1115. doi: 10.1038/ni1125. [DOI] [PubMed] [Google Scholar]
- 3.Duggal NK, Emerman M. 2012. Evolutionary conflicts between viruses and restriction factors shape immunity. Nat Rev Immunol 12:687–695. doi: 10.1038/nri3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan N, Chen ZJ. 2012. Intrinsic antiviral immunity. Nat Immunol 13:214–222. doi: 10.1038/ni.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takeuchi H, Matano T. 2008. Host factors involved in resistance to retroviral infection. Microbiol Immunol 52:318–325. doi: 10.1111/j.1348-0421.2008.00040.x. [DOI] [PubMed] [Google Scholar]
- 6.Tavalai N, Stamminger T. 2011. Intrinsic cellular defense mechanisms targeting human cytomegalovirus. Virus Res 157:128–133. doi: 10.1016/j.virusres.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Tavalai N, Stamminger T. 2008. New insights into the role of the subnuclear structure ND10 for viral infection. Biochim Biophys Acta 1783:2207–2221. doi: 10.1016/j.bbamcr.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 8.Adler M, Tavalai N, Müller R, Stamminger T. 2011. Human cytomegalovirus immediate-early gene expression is restricted by the nuclear domain 10 component Sp100. J Gen Virol 92:1532–1538. doi: 10.1099/vir.0.030981-0. [DOI] [PubMed] [Google Scholar]
- 9.Unterholzner L, Bowie AG. 2011. Innate DNA sensing moves to the nucleus. Cell Host Microbe 9:351–353. doi: 10.1016/j.chom.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Neil SJD, Zang T, Bieniasz PD. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 11.Tokarev A, Skasko M, Fitzpatrick K, Guatelli J. 2009. Antiviral activity of the interferon-induced cellular protein BST-2/tetherin. AIDS Res Hum Retroviruses 25:1197–1210. doi: 10.1089/aid.2009.0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sadler AJ, Williams BRG. 2008. Interferon-inducible antiviral effectors. Nat Rev Immunol 8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fensterl V, Sen GC. 2009. Interferons and viral infections. BioFactors 35:14–20. doi: 10.1002/biof.6. [DOI] [PubMed] [Google Scholar]
- 14.Brennan K, Bowie AG. 2010. Activation of host pattern recognition receptors by viruses. Curr Opin Microbiol 13:503–507. doi: 10.1016/j.mib.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 15.Gariglio M, Mondini M, De Andrea M, Landolfo S. 2011. The multifaceted interferon-inducible p200 family proteins: from cell biology to human pathology. J Interferon Cytokine Res 31:159–172. doi: 10.1089/jir.2010.0106. [DOI] [PubMed] [Google Scholar]
- 16.Ludlow LEA, Johnstone RW, Clarke CJP. 2005. The HIN-200 family: more than interferon-inducible genes? Exp Cell Res 308:1–17. doi: 10.1016/j.yexcr.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 17.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. 2010. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol 11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B. 2011. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi sarcoma-associated herpesvirus infection. Cell Host Microbe 9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veeranki S, Choubey D. 2012. Interferon-inducible p200-family protein IFI16, an innate immune sensor for cytosolic and nuclear double-stranded DNA: regulation of subcellular localization. Mol Immunol 49:567–571. doi: 10.1016/j.molimm.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li T, Chen J, Cristea IM. 2013. Human cytomegalovirus tegument protein pUL83 inhibits IFI16-mediated DNA sensing for immune evasion. Cell Host Microbe 14:591–599. doi: 10.1016/j.chom.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson MR, Sharma S, Atianand M, Jensen SB, Carpenter S, Knipe DM, Fitzgerald KA, Kurt-Jones EA. 2014. Interferon gamma inducible protein (IFI)16 transcriptionally regulates type I interferons and other interferon stimulated genes and controls the interferon response to both DNA and RNA viruses. J Biol Chem 289:23568–23581. doi: 10.1074/jbc.M114.554147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goubau D, Rehwinkel J, Reis e Sousa C. 2010. PYHIN proteins: center stage in DNA sensing. Nat Immunol 11:984–986. doi: 10.1038/ni1110-984. [DOI] [PubMed] [Google Scholar]
- 23.Costa S, Borgogna C, Mondini M, De Andrea M, Meroni PL, Berti E, Gariglio M, Landolfo S. 2011. Redistribution of the nuclear protein IFI16 into the cytoplasm of ultraviolet B-exposed keratinocytes as a mechanism of autoantigen processing. Br J Dermatol 164:282–290. doi: 10.1111/j.1365-2133.2010.10097.x. [DOI] [PubMed] [Google Scholar]
- 24.Dell'Oste V, Gatti D, Gugliesi F, De Andrea M, Bawadekar M, Lo Cigno I, Biolatti M, Vallino M, Marschall M, Gariglio M, Landolfo S. 2014. Innate nuclear sensor IFI16 translocates into the cytoplasm during the early stage of in vitro human cytomegalovirus infection and is entrapped in the egressing virions during the late stage. J Virol 88:6970–6982. doi: 10.1128/JVI.00384-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaw N, Liu Z-J. 2014. Role of the HIN domain in regulation of innate immune responses. Mol Cell Biol 34:2–15. doi: 10.1128/MCB.00857-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh VV, Kerur N, Bottero V, Dutta S, Chakraborty S, Ansari MA, Paudel N, Chikoti L, Chandran B. 2013. Kaposi's sarcoma-associated herpesvirus latency in endothelial and B cells activates gamma interferon-inducible protein 16-mediated inflammasomes. J Virol 87:4417–4431. doi: 10.1128/JVI.03282-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ansari MA, Singh VV, Dutta S, Veettil MV, Dutta D, Chikoti L, Lu J, Everly D, Chandran B. 2013. Constitutive interferon-inducible protein 16-inflammasome activation during Epstein-Barr virus latency I, II, and III in B and epithelial cells. J Virol 87:8606–8623. doi: 10.1128/JVI.00805-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morrone SR, Wang T, Constantoulakis LM, Hooy RM, Delannoy MJ, Sohn J. 2014. Cooperative assembly of IFI16 filaments on dsDNA provides insights into host defense strategy. Proc Natl Acad Sci U S A 111:E62–E71. doi: 10.1073/pnas.1313577111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orzalli MH, DeLuca NA, Knipe DM. 2012. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc Natl Acad Sci U S A 109:E3008–E3017. doi: 10.1073/pnas.1211302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jakobsen MR, Bak RO, Andersen A, Berg RK, Jensen SB, Tengchuan J, Laustsen A, Hansen K, Ostergaard L, Fitzgerald KA, Xiao TS, Mikkelsen JG, Mogensen TH, Paludan SR. 2013. IFI16 senses DNA forms of the lentiviral replication cycle and controls HIV-1 replication. Proc Natl Acad Sci U S A 110:E4571-E4580. doi: 10.1073/pnas.1311669110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Connolly DJ, Bowie AG. 2014. The emerging role of human PYHIN proteins in innate immunity: implications for health and disease. Biochem Pharmacol 92:405–414. doi: 10.1016/j.bcp.2014.08.031. [DOI] [PubMed] [Google Scholar]
- 32.Gariano GR, Dell'Oste V, Bronzini M, Gatti D, Luganini A, De Andrea M, Gribaudo G, Gariglio M, Landolfo S. 2012. The intracellular DNA sensor IFI16 gene acts as restriction factor for human cytomegalovirus replication. PLoS Pathog 8:e1002498. doi: 10.1371/journal.ppat.1002498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson KE, Bottero V, Flaherty S, Dutta S, Singh VV, Chandran B. 2014. IFI16 restricts HSV-1 replication by accumulating on the HSV-1 genome, repressing HSV-1 gene expression, and directly or indirectly modulating histone modifications. PLoS Pathog 10:e1004503. doi: 10.1371/journal.ppat.1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maglennon GA, Doorbar J. 2012. The biology of papillomavirus latency. Open Virol J 6:190–197. doi: 10.2174/1874357901206010190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quint KD, Genders RE, de Koning MNC, Borgogna C, Gariglio M, Bouwes Bavinck JN, Doorbar J, Feltkamp MC. 2015. Human beta-papillomavirus infection and keratinocyte carcinomas. J Pathol 235:342–354. doi: 10.1002/path.4425. [DOI] [PubMed] [Google Scholar]
- 36.Gariglio M, Azzimonti B, Pagano M, Palestro G, De Andrea M, Valente G, Voglino G, Navino L, Landolfo S. 2002. Immunohistochemical expression analysis of the human interferon-inducible gene IFI16, a member of the HIN200 family, not restricted to hematopoietic cells. J Interferon Cytokine Res 22:815–821. doi: 10.1089/107999002320271413. [DOI] [PubMed] [Google Scholar]
- 37.Geimanen J, Isok-Paas H, Pipitch R, Salk K, Laos T, Orav M, Reinson T, Ustav M, Ustav E. 2011. Development of a cellular assay system to study the genome replication of high- and low-risk mucosal and cutaneous human papillomaviruses. J Virol 85:3315–3329. doi: 10.1128/JVI.01985-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Isaacson Wechsler E, Wang Q, Roberts I, Pagliarulo E, Jackson D, Untersperger C, Coleman N, Griffin H, Doorbar J. 2012. Reconstruction of human papillomavirus type 16-mediated early-stage neoplasia implicates E6/E7 deregulation and the loss of contact inhibition in neoplastic progression. J Virol 86:6358–6364. doi: 10.1128/JVI.07069-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson R, Laimins LA. 2005. Differentiation of HPV-containing cells using organotypic “raft” culture or methylcellulose. Methods Mol Med 119:157–169. doi: 10.1385/1-59259-982-6157. [DOI] [PubMed] [Google Scholar]
- 40.Reinson T, Toots M, Kadaja M, Pipitch R, Allik M, Ustav E, Ustav M. 2013. Engagement of the ATR-dependent DNA damage response at the human papillomavirus 18 replication centers during the initial amplification. J Virol 87:951–964. doi: 10.1128/JVI.01943-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baggetta R, De Andrea M, Gariano GR, Mondini M, Rittà M, Caposio P, Giovarelli M, Gariglio M, Landolfo S. 2010. The interferon-inducible gene IFI16 secretome of endothelial cells drives the early steps of the inflammatory response. Eur J Immunol 40:2182–2189. doi: 10.1002/eji.200939995. [DOI] [PubMed] [Google Scholar]
- 42.Schweiger M-R, Ottinger M, You J, Howley PM. 2007. Brd4-independent transcriptional repression function of the papillomavirus e2 proteins. J Virol 81:9612–9622. doi: 10.1128/JVI.00447-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoppe-Seyler F, Butz K. 1992. Activation of human papillomavirus type 18 E6-E7 oncogene expression by transcription factor Sp1. Nucleic Acids Res 20:6701–6706. doi: 10.1093/nar/20.24.6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013. RNA-guided human genome engineering via Cas9. Science 339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gugliesi F, Mondini M, Ravera R, Robotti A, De Andrea M, Gribaudo G, Gariglio M, Landolfo S. 2005. Up-regulation of the interferon-inducible IFI16 gene by oxidative stress triggers p53 transcriptional activity in endothelial cells. J Leukoc Biol 77:820–829. doi: 10.1189/jlb.0904507. [DOI] [PubMed] [Google Scholar]
- 46.Caposio P, Gugliesi F, Zannetti C, Sponza S, Mondini M, Medico E, Hiscott J, Young HA, Gribaudo G, Gariglio M, Landolfo S. 2007. A novel role of the interferon-inducible protein IFI16 as inducer of proinflammatory molecules in endothelial cells. J Biol Chem 282:33515–33529. doi: 10.1074/jbc.M701846200. [DOI] [PubMed] [Google Scholar]
- 47.Stewart WE II, Lin LS, Wiranowska-Stewart M, Cantell K. 1977. Elimination of size and charge heterogeneities of human leukocyte interferons by chemical cleavage. Proc Natl Acad Sci U S A 74:4200–4204. doi: 10.1073/pnas.74.10.4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Orav M, Henno L, Isok-Paas H, Geimanen J, Ustav M, Ustav E. 2013. Recombination-dependent oligomerization of human papillomavirus genomes upon transient DNA replication. J Virol 87:12051–12068. doi: 10.1128/JVI.01798-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rösl F, Westphal EM, zur Hausen H. 1989. Chromatin structure and transcriptional regulation of human papillomavirus type 18 DNA in HeLa cells. Mol Carcinog 2:72–80. doi: 10.1002/mc.2940020205. [DOI] [PubMed] [Google Scholar]
- 50.Meissner JD. 1999. Nucleotide sequences and further characterization of human papillomavirus DNA present in the CaSki, SiHa and HeLa cervical carcinoma cell lines. J Gen Virol 80:1725–1733. [DOI] [PubMed] [Google Scholar]
- 51.Kang HJ, Lee MH, Kang HL, Kim SH, Ahn JR, Na H, Na TY, Kim YN, Seong JK, Lee MO. 2014. Differential regulation of estrogen receptor α expression in breast cancer cells by metastasis-associated protein 1. Cancer Res 74:1484–1494. doi: 10.1158/0008-5472.CAN-13-2020. [DOI] [PubMed] [Google Scholar]
- 52.Rose B, Steger G, Dong XP, Thompson C, Cossart Y, Tattersall M, Pfister H. 1998. Point mutations in SP1 motifs in the upstream regulatory region of human papillomavirus type 18 isolates from cervical cancers increase promoter activity. J Gen Virol 79:1659–1663. [DOI] [PubMed] [Google Scholar]
- 53.Cosma MP. 2002. Ordered recruitment: gene-specific mechanism of transcription activation. Mol Cell 10:227–236. doi: 10.1016/S1097-2765(02)00604-4. [DOI] [PubMed] [Google Scholar]
- 54.Narlikar GJ, Fan H-Y, Kingston RE. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475–487. doi: 10.1016/S0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 55.Wooldridge TR, Laimins LA. 2008. Regulation of human papillomavirus type 31 gene expression during the differentiation-dependent life cycle through histone modifications and transcription factor binding. Virology 374:371–380. doi: 10.1016/j.virol.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang X, Meyers C, Wang H-K, Chow LT, Zheng Z-M. 2011. Construction of a full transcription map of human papillomavirus type 18 during productive viral infection. J Virol 85:8080–8092. doi: 10.1128/JVI.00670-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sankovski E, Männik A, Geimanen J, Ustav E, Ustav M. 2014. Mapping of betapapillomavirus human papillomavirus 5 transcription and characterization of viral-genome replication function. J Virol 88:961–973. doi: 10.1128/JVI.01841-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jakobsen MR, Paludan SR. 2014. IFI16: at the interphase between innate DNA sensing and genome regulation. Cytokine Growth Factor Rev 25:649–655. doi: 10.1016/j.cytogfr.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 59.Orzalli MH, Conwell SE, Berrios C, DeCaprio JA, Knipe DM. 2013. Nuclear interferon-inducible protein 16 promotes silencing of herpesviral and transfected DNA. Proc Natl Acad Sci U S A 110:E4492–E4501. doi: 10.1073/pnas.1316194110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.You J. 2010. Papillomavirus interaction with cellular chromatin. Biochim Biophys Acta 1799:192–199. doi: 10.1016/j.bbagrm.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Johannsen E, Lambert PF. 2013. Epigenetics of human papillomaviruses. Virology 445:205–212. doi: 10.1016/j.virol.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brázda V, Coufal J, Liao JCC, Arrowsmith CH. 2012. Preferential binding of IFI16 protein to cruciform structure and superhelical DNA. Biochem Biophys Res Commun 422:716–720. doi: 10.1016/j.bbrc.2012.05.065. [DOI] [PubMed] [Google Scholar]
- 63.Liao JC, Lam R, Brazda V, Duan S, Ravichandran M, Ma J, Xiao T, Tempel W, Zuo X, Wang YX, Chirgadze NY, Arrowsmith CH. 2011. Interferon-inducible protein 16: insight into the interaction with tumor suppressor p53. Structure 19:418–429. doi: 10.1016/j.str.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stepp WH, Meyers JM, McBride AA. 2013. Sp100 provides intrinsic immunity against human papillomavirus infection. mBio 4:e00845–13. doi: 10.1128/mBio.00845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]