ABSTRACT
Hepatitis C virus (HCV) is a leading cause of chronic hepatitis C (CHC), liver cirrhosis, and hepatocellular carcinoma (HCC). Immunohistochemistry of archived HCC tumors showed abundant FBP1 expression in HCC tumors with the CHC background. Oncomine data analysis of normal versus HCC tumors with the CHC background indicated a 4-fold increase in FBP1 expression with a concomitant 2.5-fold decrease in the expression of p53. We found that FBP1 promotes HCV replication by inhibiting p53 and regulating BCCIP and TCTP, which are positive and negative regulators of p53, respectively. The severe inhibition of HCV replication in FBP1-knockdown Huh7.5 cells was restored to a normal level by downregulation of either p53 or BCCIP. Although p53 in Huh7.5 cells is transcriptionally inactive as a result of Y220C mutation, we found that the activation and DNA binding ability of Y220C p53 were strongly suppressed by FBP1 but significantly activated upon knockdown of FBP1. Transient expression of FBP1 in FBP1 knockdown cells fully restored the control phenotype in which the DNA binding ability of p53 was strongly suppressed. Using electrophoretic mobility shift assay (EMSA) and isothermal titration calorimetry (ITC), we found no significant difference in in vitro target DNA binding affinity of recombinant wild-type p53 and its Y220C mutant p53. However, in the presence of recombinant FBP1, the DNA binding ability of p53 is strongly inhibited. We confirmed that FBP1 downregulates BCCIP, p21, and p53 and upregulates TCTP under radiation-induced stress. Since FBP1 is overexpressed in most HCC tumors with an HCV background, it may have a role in promoting persistent virus infection and tumorigenesis.
IMPORTANCE It is our novel finding that FUSE binding protein 1 (FBP1) strongly inhibits the function of tumor suppressor p53 and is an essential host cell factor required for HCV replication. Oncomine data analysis of a large number of samples has revealed that overexpression of FBP1 in most HCC tumors with chronic hepatitis C is significantly linked with the decreased expression level of p53. The most significant finding is that FBP1 not only physically interacts with p53 and interferes with its binding to the target DNA but also functions as a negative regulator of p53 under cellular stress. FBP1 is barely detectable in normal differentiated cells; its overexpression in HCC tumors with the CHC background suggests that FBP1 has an important role in promoting HCV infection and HCC tumors by suppressing p53.
INTRODUCTION
Hepatitis C virus (HCV) infection is a leading cause of chronic liver diseases. More than a decade after the identification of HCV as the major causative agent of non-A, non-B hepatitis (1), molecular strategies for complete eradication of HCV infection are actively pursued. HCV is the major cause of chronic liver disease. According to new findings from the U.S. Centers for Disease Control and Prevention (CDC), the number of individuals in the U.S. living with chronic hepatitis C virus infection is about 2.7 million (2). Globally, the number of people with HCV is greater than 185 million (3). During the past 3 years, the U.S. Food and Drug Administration has approved four new medications (boceprevir, telaprevir, sofosbuvir, and simeprevir) for treatment of HCV infection, and many new drugs are under development. There has been a renewed effort by the CDC to prevent HCV-associated complications by improving treatment. However, the cost of HCV treatment is highly prohibitive; it costs $80,000 for a three-month treatment course with the recently approved sofosbuvir (Gilead Sciences, CA).
Although the majority of HCV-infected persons are unaware of their infection (4), 15 to 25% of them clear the virus without treatment, while the majority of infections persist, leading to chronic hepatitis C (CHC), which is closely linked with the risk of liver cirrhosis (LC) (5) and hepatocellular carcinoma (HCC). The molecular mechanisms that establish persistent HCV infection and its progression to LC and HCC are poorly understood. The HCV genome is a positive-strand RNA containing highly structured 5′ and 3′ nontranslated regions (NTRs) with multiple regulatory elements essential for viral replication and translation. We have identified many host cell factors associated with the viral RNA genome (6, 7); many of them were shown to be essential for HCV replication. One of the host factors essential for HCV replication was FBP1 (6), which is known to interact with the far-upstream element (FUSE) of the c-myc proto-oncogene and activates its transcription (8, 9). Earlier, we showed that FBP1 specifically interacts with HCV NS5A and the FUSE-like poly(UC)-rich region in the HCV 3′NTR and promotes HCV replication (10). Downregulation of FBP1 drastically inhibited HCV replication in hepatic cells, whereas its overexpression promotes robust viral replication (10). NS5A, which is coimmunoprecipitated with FBP1 (10), also interacts with tumor suppressor p53 (11), which significantly contributes to cellular antiviral defense against HCV (12).
Recently, we showed that FBP1 coimmunoprecipitates p53 and antagonizes p53 activity in Huh7 cells in which FBP1 is abundantly expressed (13). In approximately 80% of HCC tumors, FBP1 is overexpressed, and its expression in tumor cells is linked to poor patient survival (14, 15). p53 in a Huh7-derived cell line carries a mutation at codon 220 (Y220C). This mutation has been attributed to inactivation of p53 due to the loss of DNA binding activity (16–18). However, mutant p53Y220C has been shown to display wild-type p53 activity in the yeast system (19), which could be due to the absence of FBP1 homolog in the yeast system. Another study with truncated p53 containing the core domain has shown that mutant p53Y220C displays 17% and 45% of the wild-type DNA binding activity at physiological and subphysiological temperatures, respectively (20).
In the crystal structure of the p53-DNA binary complex, Tyr220 is 38 Å away from the DNA bound to the DNA binding motif. Therefore, it is highly intriguing that Tyr→Cys mutation at this position negatively influenced the DNA binding function of mutant p53. In this study, we have determined the DNA binding affinity of the mutant and wild-type p53, and we explored the mechanisms by which the activity of mutant p53Y220C in Huh7-derived cells remained suppressed by FBP1 and how this suppression of p53 promotes cell survival and persistent HCV replication/infection of hepatic cells.
MATERIALS AND METHODS
Immunohistochemistry of HCC tumors.
We carried out immunohistochemistry on archived HCC tumors with a polyclonal FBP1 antibody and isotype IgG (Santa Cruz Biotechnology) at a 1:100 dilution. The archived HCC tissues embedded in paraffin were obtained from the university hospital. All of the archived tumor tissues were from patients who had undergone a liver transplant. The slides were cut in triplicate; one slide was processed for immunohistochemistry with FBP1 antibody, the second slide was processed with isotype IgG as a control (sc-2028; Santa Cruz Biotechnology), and the third slide was stained with hematoxylin-eosin and examined by a pathologist, Kenneth M. Klein, for marking the tumor area. All of the slides were examined using a Nikon Eclipse E800 microscope. An Institution Review Board (IRB) approval (IRB protocol 0120080317) was obtained prior to obtaining archived HCC tumors for immunohistochemistry.
Cell culture, preparation of cell lysates, preparation of nuclear and cytoplasmic extract, immunoprecipitation, Western blotting, and construction of stably transduced cells knocked down for targeted proteins.
Cured MH14 and MH14 cells were gifts from Makoto Hijikata (Kyoto University, Japan). MH14 cells are a derivative of the Huh7 cell line, which carries stable HCV subgenomic replicons (21). Cured MH14 cells were prepared by treating MH14 cells with 5,000 IU/ml of alpha interferon (IFN-α) for 2 weeks. Huh7.5 cells and cured MH14 cells were maintained in Dulbecco's modified Eagle medium (DMEM; Sigma) supplemented with 10% fetal bovine serum (Sigma), 100 U/ml of nonessential amino acids (Gibco), and 100 μg/ml of penicillin and streptomycin sulfate (Gibco) (21, 22). MH14 cells were cultured in medium supplemented with 300 μg/ml of G418 (Calbiochem). Cells were grown at 37°C with 5% CO2. The HepG2 cells were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum, 4 mM l-glutamine, 100 U of penicillin, and 100 μg of streptomycin per ml at 37°C in 5% CO2-containing humidified air. The transfection of cells with short interfering RNA (siRNA), the preparation of cell lysates and nuclear and cytoplasmic extracts, coimmunoprecipitation, and Western blotting were done as described earlier (10). Huh7.5, MH14, and HepG2 cells stably knocked down for either FBP1, p53, or BCCIP were constructed by transducing with a lentivirus vector encoding short hairpin RNA (shRNA) against the mRNA of the targeted protein (Santa Cruz Biotechnology) by following the manufacturer's protocol.
Preparation of cell-free replication lysate.
We prepared the replicative cytoplasmic fractions from MH14 cells by following the protocol described previously, with minor modifications (10, 23, 24). In brief, MH14 cells were grown in 10-cm petri dishes and washed with ice-cold buffer containing 150 mM sucrose, 30 mM HEPES (pH 7.4), 33 mM ammonium chloride, 7 mM KCl, and 4.5 mM magnesium acetate. The washed cells first were treated with lysolecithin solution (250 μg/ml) in the washing buffer for 1 min, followed by washing with 3 ml of washing buffer. The cells were scraped from the plate after addition of 200 μl of replication buffer containing 100 mM HEPES (pH 7.4); 50 mM ammonium chloride; 7 mM KCl; 1 mM spermidine; 0.5 mM (each) ATP, GTP, UTP, and CTP; 1 mM dithiothreitol (DTT); and 10% glycerol. The cells were lysed gently by pipetting up and down several times, and then we centrifuged the lysed cells at 1,600 rpm for 5 min at 4°C. The supernatant fraction (replicative lysate) was stored at −80°C until use.
Endogenous HCV replication assay in cell-free replication lysate.
For endogenous HCV replication assay, an aliquot of normalized cell-free replication lysate (equivalent to 100 μg protein) containing 0.5 mM (each) four ribonucleoside triphosphates (rNTPs) and 10 μCi of [α-32P]CTP (specific activity, 400 mCi/mmol) was incubated for 1 h at 30°C. The reaction was terminated by adding 0.5% SDS in STE buffer (10 mM Tris-HCl, pH 7.5; 1 mM EDTA; 150 mM NaCl), and we extracted total RNA twice with phenol-chloroform-isoamyl alcohol (25:24:1) and twice with water-saturated ether. We precipitated the RNA with ethanol and dissolved it in diethyl pyrocarbonate (DEPC)-treated water. The total RNA from the reaction was purified and subjected to denatured agarose gel electrophoresis. The radioactive RNA products were visualized by autoradiography.
Plasmids and oligonucleotides.
Plasmids carrying the HCV subgenomic replicon (pMH14) (25) were obtained from Makoto Hijikata (Kyoto University, Japan). Huh7.5 and pFL-J6/JFH were a gift from Charles Rice (22). FBP1 shRNA lentiviral particles and P53 siRNA sense (GCA UGA ACC GGA GGC CCA UTT) and antisense (5′-AUG GGC CUC CGG UUC AUG CTT-3′) primers were purchased from Santa Cruz Biotechnology. Lentiviral particles expressing shRNAs targeting FBP1, p53, or BCCIP were purchased from Santa Cruz Biotechnology (CA). Plasmids expressing green fluorescent protein (GFP) fused to the N terminus of human CD81 (pTRIP-GFP-hCD81) or expressing miR-122 (pTRIP-Puro-miR122) and their negative vector controls were a generous gift from Matthew J. Evans (26).
The primers for reverse transcription-PCR (RT-PCR) and real-time RT-PCR of the HCV 5′NTR (up, 5′-CGG GAG AGC CAT AGT GG-3′), HCV 5′NTR (down, 5′-AGT ACC ACA AGG CCT TTC G-3′), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA (up, 5′-CTCTGCTCCTCCTGTTCGAC-3′; down, 5′-ATG GGT GGA ATC ATA TTG GAAC-3′), actin mRNA (up, 5′-CAGGCACCAGGGCGTGATGG-3′; down, 5′-AGG CGT ACA GGG ATA GCA CA-3′), BCCIP mRNA (up, 5′-ATG GCG TCC AGG TCT AAG-3′; down, 5′-TTA GAA AAA GCT GCT GC-3′), p21 mRNA (up, 5′-TAC CCT TGT GCC TCG CTC AG-3′; down, 5′-CGG CGT TTG GAG TGG TAG-3′), TCTP mRNA (up, 5′-GAT CGC GGA CGG GTT GT-3′; down, 5′-TTC AGC GGA GGC ATT TCC-3′), and p53 mRNA (up, TCA ACA AGA TGT T TT GCC AAC-3′; down, 5′-ATG TGC TGT GAC TGC TTG TAG ATG-3′) were obtained from Sigma. Double-stranded 30-bp p53 target wild-type p53-activated fragment (WAF)-side DNA (5′-CGA GGA ACA TGT CCC AAC ATG TTG CTC GAG-3′ and 5′-CTC GAG CAA CAT GTT GGG ACA TGT TCC TCG-3′) and p53 nontarget and nonspecific 30-bp double-stranded HIV-1 U5 PBS DNA (5′-CAG GGA CAA GCC CGC GGT GAC GAT CTC TAA-3′ and 5′-TTA GAG ATC GTC ACC GCG GGC TTG TCC CTG-3′) also were obtained from Sigma (27).
Preparation of infectious HCV virions.
Huh7.5 cells (2 × 106 cells) were grown overnight in 10-cm culture plates and then transfected with JFH1 HCV RNA transcribed from pJFH1 as described previously (22). Replication of HCV in cells was detected by RT-PCR for HCV RNA; release of infectious HCV virions in the culture supernatant was detected by Western blotting for HCV core protein. The culture supernatant was filtered through 0.45-μm pores (Millipore, USA); the filtrate was concentrated 10-fold by an Amicon Ultra-15 device, aliquoted, and stored at −80°C.
Infection of Huh7.5 cells with infectious HCV virions.
Huh7.5 cells grown overnight in 6-well plates were infected with 0.2 ml of concentrated culture supernatant in DMEM containing 5-μg/ml Polybrene. After 3 h, cells were washed two times with phosphate-buffered saline (PBS) and supplemented with fresh medium and then grown for the indicated periods of time.
Transient expression of FBP1 in FBP1-knockdown cells.
The FBP1 shRNA lentiviral vector contains shRNA-targeting FBP1 codons 248 to 254 and 560 to 567. We constructed shRNA-resistant FBP1 expression clones (pCIA-CMV-FBP1SHR) by point mutations in the degenerate codons without altering the amino acid sequence. The resistance to shRNA was confirmed by transient expression of FBP1 in FBP1-kd cells.
Expression and purification of recombinant FBP1.
A recombinant clone of His-FBP1 (pET28a-FBP1) was expressed in Escherichia coli Rosetta (DE3) and purified by affinity chromatography using nickel-nitrilotriacetic acid (Ni-NTA) and Hi-Trap heparin columns (Pharmacia). In brief, transformed E. coli Rosetta (DE3) cells were grown at 37°C in Luria broth (LB) medium containing 30 μg/ml of kanamycin until an optical density at 595 nm (OD595) of 0.4 was achieved. The medium was cooled to 18°C, supplemented with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and further incubated at 18°C for 16 h with vigorous shaking. The cells were harvested, washed, and resuspended in a lysis buffer containing 20 mM Tris-HCl, pH 7.4, 200 mM NaCl, 1 mM β-mercaptoethanol, 10% glycerol, 1% Triton-X 100, 5 mM imidazole, and 1× ProteoBlock protease inhibitor cocktail (Fermentas) containing 2 mg/ml lysozyme. The suspension was sonicated and centrifuged. The clear supernatant was applied to a Ni-NTA column preequilibrated with binding buffer (20 mM Tris-HCl, pH 7.4, 200 mM NaCl, 10% glycerol, 5 mM imidazole). The column was washed with the binding buffer containing 50 mM imidazole. His-FBP1 then was eluted with 200 mM imidazole in the same buffer. Fractions containing FBP1 were pooled and diluted 2-fold by adding an equal volume of a buffer containing 20 mM Tris-HCl (pH 7.5), 5% glycerol, 0.5% NP-40, and 1 mM β-mercaptoethanol. The diluted fraction then was applied to the Hi-Trap heparin column (Pharmacia). The column was washed extensively, and FBP1 was eluted with a linear gradient (0% to 80%) of 1 M KCl in the same buffer for 20 min (1 ml/min). Eluted fractions showing more than 95% purity on SDS-PAGE (8%) were pooled and dialyzed against buffer containing 50 mM Tris-HCl (pH 7.5), 2 mM DTT, 100 mM NaCl, and 50% glycerol.
Expression and purification of recombinant p53.
The recombinant clone of GST-p53 (pET11GTK-p53) was expressed in E. coli BL21 and purified by affinity chromatography using glutathione-Sepharose beads (Pharmacia) and Hi-Trap heparin columns (Pharmacia). In brief, E. coli cells transformed with pET11GTK-p53 plasmid were grown at 37°C in LB medium with 100 μg/ml ampicillin until the OD595 was 0.5. The culture medium was cooled to room temperature, supplemented with 0.4 mM IPTG, and further incubated at 25°C for 3 h with vigorous shaking. Cells were harvested, washed with 50 mM Tris-HCl containing 0.15 M NaCl, and resuspended in the lysis buffer (25 mM HEPES buffer, pH 7.6, 0.1 M KCl, 2 mM EDTA, 20% glycerol, 2 mM DTT, 0.1% NP-40, and 1× cocktail of protease inhibitors) containing 100 μg/ml of lysozyme. Cells were subjected to three cycles of freezing at −80°C and thawing and then sonicated at an amplitude of 40 with three 15-s pulses. The lysed cells were centrifuged. The supernatant was mixed with glutathione-Sepharose beads (0.5 mg beads/ml), incubated at 4°C for 1 h, and placed in a column, which was extensively washed with a wash buffer containing 50 mM Tris-HCl (pH 8.0) and 5% glycerol. GST-p53 was eluted from the column with 10 mM reduced glutathione (Sigma-Aldrich) and diluted to 15-fold with a dilution buffer containing 20 mM HEPES (pH 7.5), 50 mM NaCl, 0.1 mM EDTA, 10 mM β-mercaptoethanol, 10% glycerol, and protease cocktail inhibitor (Roche) and then applied to the fast protein liquid chromatography (FPLC) Hi-Trap heparin column. The column was washed extensively with the dilution buffer. p53 was eluted from the column at 20 min of application of a linear gradient (0% to 80%) of 1.5 M NaCl in the same buffer (1 ml/min). Eluted fractions showing greater than 95% purity on SDS-PAGE (8%) were dialyzed against isothermal titration calorimetry (ITC) buffer containing 50 mM sodium phosphate (pH 7.8) and 150 mM NaCl.
ITC of purified FBP1 with p53.
Protein-protein interaction between purified FBP1 and p53 were measured by ITC using an ITC200 microcalorimeter (Microcal). Both of the purified proteins were dialyzed overnight in ITC buffer containing 20 mM sodium phosphate (pH 7.8) and 150 mM NaCl and then concentrated by a Centricon-30 (Millipore). The sample cell was filled with 8 μM FBP1 and then injected 20 times with 2 μl of 50 μM p53 at 25°C. The ITC data then were applied to determine the binding isotherm of interaction between FBP1 and p53 using Origin7.
Streptavidin magnetic bead p53-DNA binding assay.
We used the biotin-labeled top strand of 30-bp duplex WAF-side DNA as the target DNA sequence for p53 binding. For the DNA binding assay, we first incubated 20 pmol of biotin-labeled double-stranded WAF-side DNA and 6 μg of nonspecific poly-d(AT) with the nuclear extract (equivalent to 200 μg of protein) in a binding buffer containing 20 mM HEPES buffer (pH 7.5), 1 mM EDTA, 1 mM DTT, 10 mM ammonium sulfate, 10 mM KCl, and 0.2% Tween 20 in a final volume of 100 μl. The mixture was incubated at 37°C for 30 min with shaking. DNA-protein complexes were captured by adding 70 μg of streptavidin paramagnetic beads and then further incubated for 30 min at 37°C. The beads were magnetically separated, extensively washed with the binding buffer, and boiled in Laemmli buffer for 5 min. The beads were separated, after which the supernatant was resolved by SDS-PAGE and Western blotted for p53.
Electrophoretic mobility shift assay (EMSA) for p53-DNA binding.
We used 32P-labeled top-strand WAF side DNA annealed with its complementary strand as the target DNA for p53 binding. An aliquot of nuclear extract (20 μg protein) or purified recombinant p53 protein (0.12 to 10 nM) was incubated with the 32P-labeled target WAF-side DNA (0.02 pmol; 40,000 cpm) at 37°C. We also used 32P-labeled 30-bp nontarget double-stranded HIV-1 U5-PBS DNA as the negative control. After incubation for 30 min at 37°C, the DNA-p53 complex was separated on 4% native polyacrylamide gel and visualized with a PhosphorImager. To measure p53-bound DNA at different concentrations of p53, each gel-retarded band was excised and counted for radioactivity. The dissociation constant was determined by GraphPad using a nonlinear regression curve fit for one-site binding of DNA with tetrameric p53.
Isothermal titration calorimetry of interaction between recombinant p53 and its target sequence (WAF-side DNA).
Interaction between purified p53 (wild-type and mutant p53) and WAF-side DNA was measured by isothermal titration calorimetry. The purified recombinant proteins and WAF-side DNA were in ITC buffer containing 50 mM sodium phosphate (pH 7.8) and 150 mM NaCl. The sample cell, filled with 0.5 μM wild-type or mutant p53, was injected 20 times with 2 μl of 5 μM WAF-side DNA at 37°C. The ITC data then were applied to determine the binding isotherm of interaction between WAF-side DNA and wild-type or mutant Y220C p53 using Origin7.
Luciferase reporter gene assay.
The reporter plasmids, p53-Luc premixed with constitutively expressing Renilla luciferase (Qiagen), were transfected into Huh7.5 cells by following the manufacturer's protocol. Transfected cells were grown for 48 h, γ irradiated (3 Gy), and, 6 h later, measured for luciferase activity.
Real-time quantitative RT-PCR.
We isolated total RNA from cells using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. One microgram of total RNA was used to synthesize cDNA corresponding to the mRNA of FBP1, p21, BCCIP, TCTP, p53, HCV 5′NTR, and GADPH by reverse transcription, as described earlier. We used 50 ng of cDNA in Fast SYBR green master mix (Applied Biosystems) with primers directed to specific mRNA to do real-time quantitative PCR using the Fast Real PCR system (Applied Biosystems) as described previously (27, 28). Data were analyzed using 7500 software (Applied Biosystems), and the relative fold change in specific mRNA copies was calculated by normalizing the amount of GAPDH mRNA in each sample. All experiments were done in triplicate for each data point.
Assessment of cell migration by wound-healing assay.
Control and FBP1-kd Huh7.5 cells were grown to 100% confluence in 6-well plates and then starved overnight in DMEM containing 0.1% fetal bovine serum (FBS). Cells then were washed with 1× PBS. Using a 200-μl sterilized tip, a scratch was made across each well. Cells then were washed two times with PBS supplemented with 2 ml of medium with 10% FBS. Images were taken at 0, 16, and 32 h of incubation. A landmark was made on the scratches to maintain the same fields for all time points of imaging. Areas covered in the scratches were quantified by ImageJ software (29) and statistically tested by two-way analysis of variance (ANOVA) and Bonferroni's post hoc tests.
Real-time migration assay using the RTCA DP Xcelligence system.
Real-time migration was done to test the migration dynamics of control and FBP1-kd Huh7.5 cells by a method described previously (30). Briefly, the cells were seeded in complete medium and grown to 70 to 80% confluence and then serum starved overnight in medium containing 0.5% FBS. After trypsinization, the cells were counted and resuspended in serum-free medium. In the lower chamber of the CIM plate, 1× DMEM containing 10% FBS was added as a chemoattractant. In the top chamber, 50 μl of 1× DMEM was added to all wells. Both chambers were assembled, loaded in the Xcelligence system, and run for 30 min for equilibration or background readings. One hundred microliters of each cell line suspended in 1× DMEM (40,000 cells/100 μl) then was added in triplicates in upper chamber wells. Readings for changes in the cell index (CI) were taken every 15 min for 36 h. Finally, migration was shown as a change in cell index versus time.
Bioinformatics meta-analysis for expression of FBP1 in RNA sequencing (RNA-seq) data from human cancers.
To delineate the gene regulation pattern of FBP1 in different cancer types, we meta-analyzed next-generation sequencing data from various published studies. The data analyzed were obtained from the Cancer Genome Project, which was made available through cBioPortal for Cancer Genomics and Oncomine databases. Based on the data available from different cancer centers, the genomic and gene expression data were analyzed for copy number variations and changes in mRNA expression levels in the FBP1 gene. The data sets that showed P values of less than 1E−4 were considered significant and were used for analysis. To eliminate bias arising from the experimental conditions, such as sample preparation changes used in each data set, we did these analyses in multiple different data sets from different studies. The data for the final result of the analysis are represented in box-whisker plots and copy numbers, and mRNA expression values are presented on a log2 scale.
Immunofluorescence and colocalization.
Cells (1 × 105 cells/well) were grown on BD Falcon 8-chamber tissue culture slides at 37°C in a 5% CO2 atmosphere for 24 h. Cultures were washed, fixed in 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and stained with primary monoclonal antibody as described before (10). Cells were washed with PBS containing 0.3% Triton X-100 and then incubated with anti-mouse IgG antibody conjugated with Alexa Fluor 568 (Invitrogen) at a 1:500 dilution. After 1 h of incubation at room temperature, cells were washed, treated with rabbit monoclonal antibody against another target protein (Santa Cruz Biotechnology), and incubated with anti-rabbit IgG antibody conjugated with Alexa Fluor 488 (Invitrogen) as described above in the same chambers for 1 h at room temperature. Cells then were washed and stained with 4′,6-diamidino-2-phenylindole (DAPI; Sigma Chemical) for 10 min, washed four times with PBS, and air dried. The slides were mounted with mounting medium (ProLong antifade kit; Molecular Probes, Eugene, OR) and visualized under a multiphoton confocal microscope system (Nikon A1R). Images were processed using NIS software.
RESULTS
FBP1 is expressed in HCC tumors with a history of chronic hepatitis C.
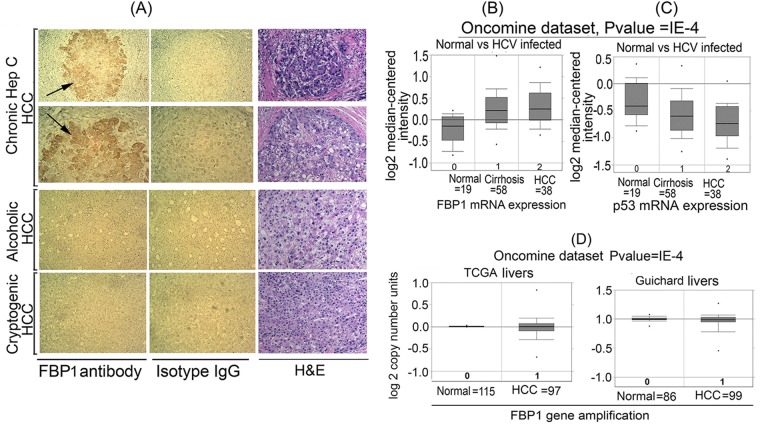
Earlier, we showed that FBP1 is an essential cell factor for HCV replication (10). In an immunochemical analysis of 109 HCC tumors, 83% were positive for FBP1 (15). However, it was not known how many of these HCC tumors had a background of chronic hepatitis C (CHC). We immunohistochemically stained archived HCC tumors with anti-FBP1 antibody, finding that 7 of these tumors had a history of CHC while 3 had an alcoholic background, and 2 were cryptogenic. FBP1 was specifically expressed in all HCC tumors having a history of CHC but not in non-HCV HCC tumors (Fig. 1A). The tumor area was confirmed by hematoxylin-eosin staining of the corresponding slide. An Oncomine expression data analysis of 19 normal human livers versus HCV-infected tumors, 58 of which were cirrhotic, and 38 HCC tumors (Fig. 1B) demonstrated an average 4-fold increase in FBP1 expression in LC and HCC samples compared to that of normal liver samples (31). In contrast to the 4-fold increase in FBP1 expression, there was a 2- to 2.5-fold decrease in the p53 mRNA level in the same set of the HCV-infected cirrhotic liver and HCC tumors (Fig. 1C). To determine whether increased FBP1 expression is linked with an amplified FBP1 gene, we analyzed two different data sets from Oncomine, one from TCGA and the other from Guichard (32) (Fig. 1D). We found no significant difference in amplification of the FBP1 gene in HCC tumors compared to that in normal livers. These observations suggest that a high level of expression of FBP1 in HCC tumors has implications concerning HCV-associated tumorigenesis.
FIG 1.
FBP1 is specifically overexpressed in human HCC tumors with CHC background. (A) Immunohistochemistry was done on archived HCC tumors; seven had a history of chronic hepatitis (Hep) C, three had the alcoholic background, and two were cryptogenic. Based on the Edmondson and Steiner nuclear grading scheme (79), 4 of the HCC-HCV tumors were well differentiated (grade1), 2 were moderately differentiated (grade 2), and 1 was moderately to poorly differentiated (grades 2 to 3). All of the alcoholic and cryptogenic HCC were moderately differentiated grade 2 tumors. Representative pictures are shown of HCC tumors treated with a polyclonal antibody against FBP1 (left) or isotype IgG (middle). The hematoxylin-eosin (H&E) staining of the corresponding slides is shown on the right. The alcoholic and cryptogenic HCC slides show only the cancer area. (B and C) Oncomine data analysis for FBP1 and p53 expression in 19 normal human livers versus 58 cirrhotic livers and 38 HCC tumors with a CHC background (31). (D) Oncomine data analysis of FBP1 gene amplification in normal livers versus HCC liver tumors from two data sets, one from TCGA livers (TCGA 2012) and the other from Guichard livers (32).
Endogenous HCV replication in cell-free replication lysates is reduced in FBP1-kd cells but significantly enhanced on FBP1 overexpression.
Earlier, we demonstrated that HCV replication is positively influenced by FBP1 expression level in MH14 cells (10). Downregulation of FBP1 by FBP1-siRNA severely impaired HCV replication, whereas overexpression of FBP significantly enhanced HCV replication but had a negative influence on HCV translation (10). In order to examine whether in vitro endogenous HCV replication in cell-free replication lysate also is affected by increased or decreased levels of FBP1 in cells, we used replicative cell-free lysates prepared from MH14 cells in which FBP1 was either stably knocked down or overexpressed. The reaction products were analyzed on a denaturing agarose gel and visualized by autoradiography. We also Western blotted cell-free replication lysates for FBP1, HCV NS5A, and actin after normalizing their protein concentrations. We found that in vitro endogenous HCV replication in replicative lysates from FBP1-kd cells was only 15% (Fig. 2, lane 3) compared to that in control MH14 cells (lane 2). In contrast, a 3-fold increase in endogenous HCV replication activity occurred in cell lysates in which FBP1 was overexpressed (lane 4). These findings suggest that FBP1 facilitates HCV replication by interacting with components of the HCV replication complex or by abrogating the inhibitory effect of some unknown cellular factor(s) on HCV replication. We also Western blotted normalized cell-free replication lysate for the expression levels of FBP1 and HCV NS5A, as well as that of actin as the loading control. We found that the level of NS5A in the replication lysate from control cells was greater than that from cells in which FBP1 was overexpressed. This is not surprising, since we showed earlier that the upregulation of FBP1 enhanced HCV replication but inhibited HCV translation. Since HCV is a positive-stranded RNA virus, its genomic RNA is the template for both translation and replication complexes that move in opposite directions on the RNA template. Thus, these two processes must be mutually exclusive. In poliovirus, a switch from translation to replication and vice versa has been demonstrated (33). Therefore, it is possible that FBP is a key factor in regulating this switch from translation to HCV replication.
FIG 2.
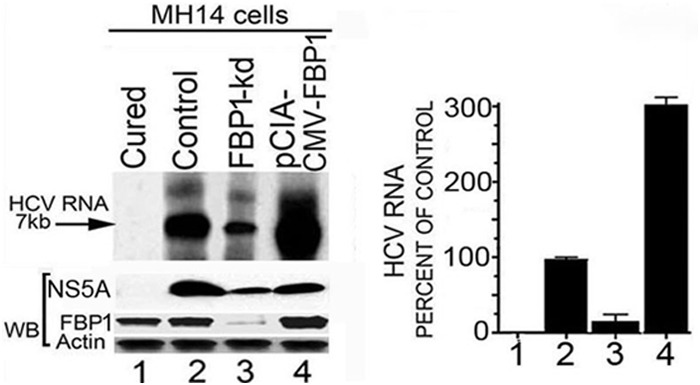
Endogenous HCV replication in cell-free replication lysates is severely reduced in FBP1-kd cells but increased in FBP1-overexpressing MH14 cells. An aliquot of normalized replication lysates from MH14 cells in which FBP1 was either knocked down or overexpressed was examined for endogenous HCV replication activity; the radiolabeled RNA products were analyzed by denaturing agarose gel electrophoresis and visualized by autoradiography. Normalized cell-free replication lysates also were Western blotted (WB) for the expression level of FBP1, HCV NS5A, and actin. (Left) Lane 1, cured MH14 cells without HCV replicons; lane 2, control MH14 cells with HCV replicons; lane 3, FBP1-kd MH14 cells; lane 4, MH14 cells in which FBP1 was overexpressed. (Right) The percentage of endogenous replication products relative to those in controls.
FBP1 interacts with p53 and viral proteins NS5A and NS5B.
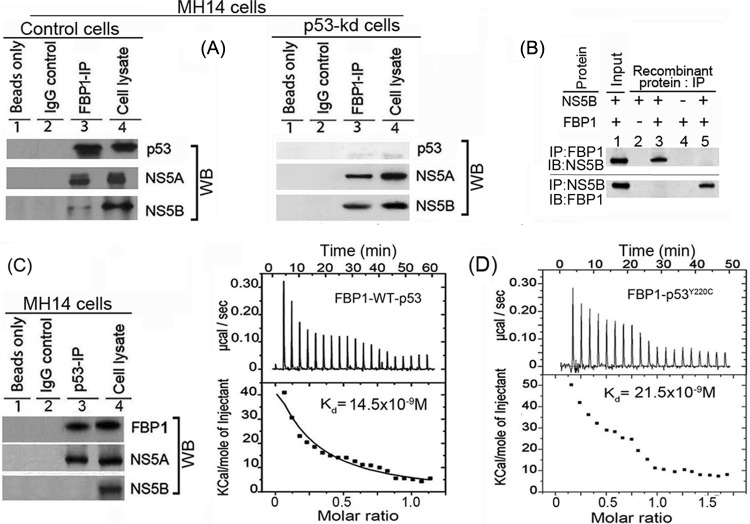
FBP1 interacts with NS5A and stimulates HCV replication (10), whereas NS5A interacts with p53 (11, 34, 35) and inhibits p53-mediated apoptosis (11). We showed earlier that FBP1 interacts with p53 and inhibits its transactivation activity in Huh 7 cells (13). In this study, we found FBP1 immunoprecipitation (IP) coimmunoprecipitates not only p53 and NS5A but also NS5B from the cell lysates of control MH14 cells (Fig. 3A, lane 3, left). We further confirmed that the interaction of FBP1 with viral proteins NS5A and NS5B is independent of p53. FBP IP in lysate from p53-kd cells also coprecipitated these viral proteins (Fig. 3A, lane 3, right), suggesting that it is an important component of the HCV replication complex. Since HCV NS5A has been shown to be in a complex with NS5B, it is possible that FBP1 IP pulls down NS5B via its interaction with NS5A. To rule out this possibility, we carried out immunoprecipitation on a mixture of recombinant purified FBP1 and NS5B and found that reciprocal IP coprecipitate each other (Fig. 3B). FBP1 also contains ATP-dependent helicase activity, which can unwind both DNA/DNA and RNA/RNA duplexes (36). This could be one of the mechanisms by which FBP1 displays a stimulatory effect on HCV replication. The p53 IP also coprecipitated FBP1 and NS5A but not NS5B (Fig. 3C, lane 3). We confirmed earlier the direct interaction of FBP1 with p53 by reciprocal IP on a mixture of purified recombinant proteins of FBP1 and p53 (13).
FIG 3.
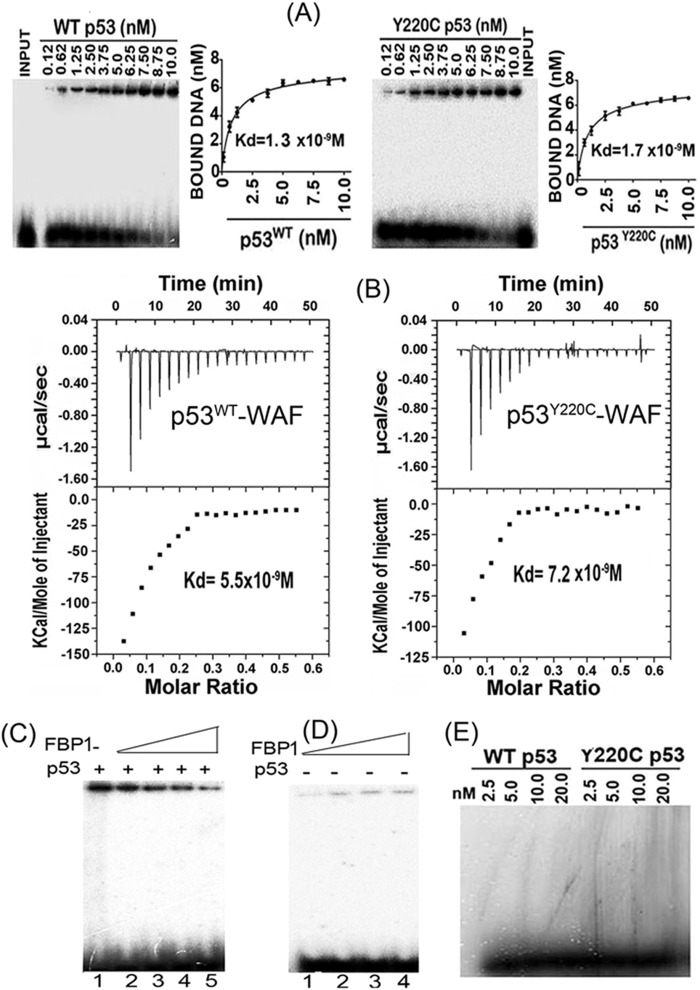
FBP1 physically interacts with p53 and HCV proteins NS5B and NS5A. (A) FBP1 IP was done on RNase-treated lysates from control MH14 cells (left) or p53-kd MH14 cells (right) and Western blotted for p53, NS5A, and NS5B. Lane 1, beads only; lane 2, isotype IgG control; lane 3, IP sample; lane 4, cell lysate. (B) FBP1 IP and NS5B IP on a mixture of purified proteins pull down each other. We used 2 μg each of purified recombinant FBP1 and NS5B and carried out reciprocal IP and Western blotting for either FBP1 or NS5A. (Upper) Lanes 2 to 5, FBP1 IP immunoblotted (IB) for NS5B. (Bottom) Lanes 2 to 5, NS5B IP immunoblotted for FBP1. Lane 1, input protein controls. (C) p53 IP on the lysates from control MH14 cells. Lane 1, beads only; lane 2, isotype IgG control; lane 3, IP sample; lane 4, cell lysate. (D) Isothermal titration calorimetry of FBP1 interaction with wild-type p53 (left) and Y220C mutant p53 (right). A syringe of ITC containing purified p53 was titrated into a cell containing purified FBP1 in ITC buffer containing 20 mM sodium phosphate buffer, pH 7.8, and 150 mM NaCl at 25°C. (Top) The calorimetric data on titration of FBP1 with p53 as a function of time. (Bottom) The integrated heat per injection versus the molar ratio of p53 to FBP1. The graph corresponds to the best fit of the experimental data to a one-site model, providing a dissociation constant (37) of 14.5 nM for wild-type p53 and 21.5 nM for the Y220C mutant.
ITC of FBP1 interaction with wild-type p53 and mutant p53Y220C.
MH14 cells are derived from the Huh7 cell line, in which p53 carries a Tyr→Cys mutation at position 220. We examined whether FBP1 binding to p53 is specific to mutant p53Y220C or also can interact with wild-type p53. We used ITC to characterize the interaction of recombinant FBP1 thermodynamically with Y220C mutant and wild-type p53. A syringe of ITC containing purified p53 was titrated into a cell containing purified FBP1. As the two proteins interacted, the heat was absorbed in direct proportion to the amount of binding that occurred. When FBP1 in the cell became saturated with added p53, the heat signal diminished. The absorbed heat was measured to determine the binding constant (K) or dissociation constant (37) of interaction between FBP1 and p53 (Fig. 3D). The top panel shows the calorimetric data; each injection produced upward spikes indicating endothermic interaction between p53 and FBP1. The top panel displays a plot of the total heat generated per injection as a function of the molar ratio of p53 to FBP1. The amount of heat absorbed per mole of p53 is shown as a function of the molar ratio of p53 to FBP1. The bottom panel shows the best fit to the experimental data for the one-binding-site model. We found that FBP1 interacted with both wild-type and mutant p53 with nearly similar affinity, displaying association constants of 1.72 × 107 M−1 and 1.15 × 107 M−1, respectively. With the standard state being equal to 1.0 M, the free energy change for the binding of FBP1 to both wild-type and mutant p53 was calculated (ΔG° = −RT ln Ka [where R is gas constant, T is temperature in Kelvin, and Ka is association constant]) to be −10.2 and −10.0 kcal mol−1, respectively.
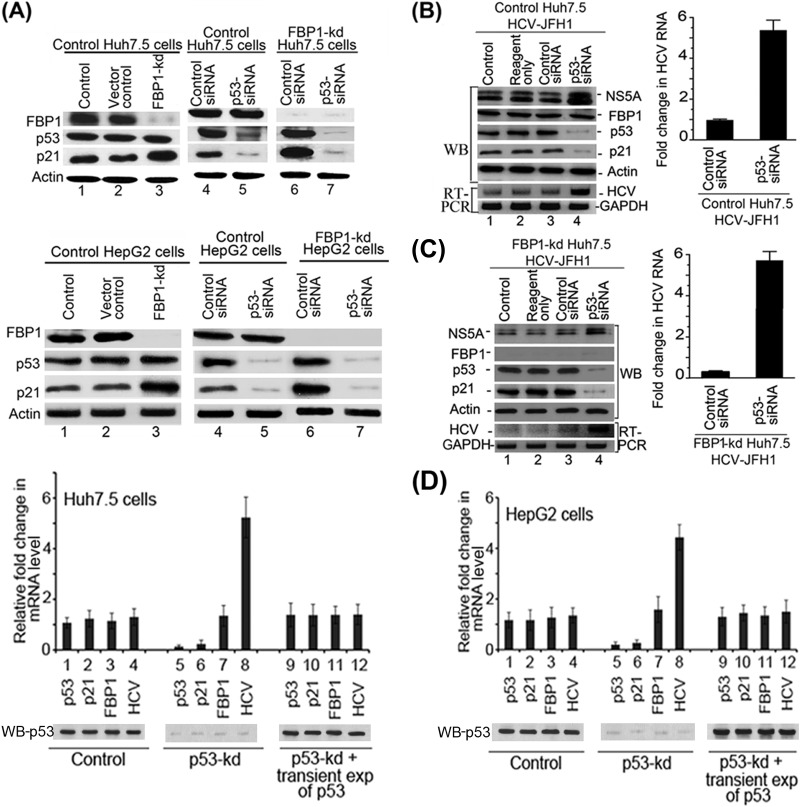
Downregulation of p53 abolishes the enhanced expression of p21 in FBP1-kd cells of both Huh7.5 and HepG2.
We showed earlier that FBP1-kd in Huh7.5 cells significantly enhanced p21 expression (13). Since the p21 expression is positively regulated by p53, we postulated that the enhanced expression of p21 in FBP1-kd was a result of the activation of p53 in the absence of FBP1 (13). To examine this postulation, we used both Huh7.5 cells expressing mutant p53Y220C (Fig. 4A, top) as well as HepG2 cells expressing wild-type p53 (Fig. 4A, bottom). We downregulated p53 expression in control and FBP1-kd cells of both cell lines and then analyzed the cell lysates for p21 expression. We found that p21 expression was significantly enhanced in FBP-kd cells of both cell lines (Fig. 4A, lane 3, top and bottom). We further observed that enhanced expression of p21 in FBP1-kd cells was nearly abolished upon downregulation of p53 in both cell lines (Fig. 4A, lane 7, top and bottom). These results clearly indicate that expression of p21 is under the positive control of p53 in both Huh7-derived cells as well as in HepG2 cells expressing wild-type p53. These findings also suggest that p53Y220C in Huh7.5 cells is functional and active but strongly suppressed by FBP1.
FIG 4.
Downregulation of p53 and FBP1 have opposite effects on p21 expression and HCV replication. (A) Upregulation of p21 in FBP1-kd Huh7.5 or HepG2 cells is abolished upon downregulation of p53. Control and FBP1-kd Huh7.5 cells (upper) or HepG2 cells (lower) were transfected with p53 siRNA and grown for 48 h. The normalized cell lysates were analyzed for the expression of FBP1, p53, p21, and actin by Western blotting. Lane 1, control cells; lane 2, vector control; lane 3, FBP1-kd cells; lanes 4 and 6, cells transfected with control siRNA; lanes 5 and 7, cells transfected with p53 siRNA. (B and C) Downregulation of p53 enhanced HCV replication in control and FBP1-kd Huh7.5 cells. Control Huh7.5 cells (B) and stable FBP1-kd Huh7.5 cells (C) were transfected with p53 siRNA and, 10 h later, infected with infectious HCV-JFH1 virions and then grown further for 72 h. Cell lysates were prepared, normalized for protein concentration, and Western blotted for the expression of NS5A, FBP1, p53, p21, and actin. Another set of cells was used for isolation of total RNA for quantitative real-time RT-PCR of JFH1 HCV RNA and GAPDH mRNA. (Left) Lane 1, control; lane 2, reagent control; lane 3, siRNA control; lane 4, p53 siRNA. (Right) Fold change in JFH1 HCV RNA concentration in control and FBP1-kd Huh7.5 cells quantified by real-time RT-PCR. (D) Transient (exp) expression of p53 suppresses HCV replication in p53-kd Huh7.5 and HepG2 cells. Stable p53-kd Huh7.5 cells (left) and HepG2 cells (right) were transfected with an shRNA-resistant expression clone of p53, and 10 h later, Huh7.5 cells were infected with HCV-JFH1 virions while HepG2 cells were transfected with MH14 HCV replicon RNA. Cells were grown for 72 h; total RNA was isolated. The quantitative real-time RT-PCR for HCV RNA and mRNA levels of p53, p21, and FBP1 were carried out with GAPDH mRNA as the internal control. The Western blot of p53 expression also is shown. Lanes 1 to 4, control cells; lane 5 to 8, p53-kd cells; lanes 9 to 12, p53-kd cells in which p53 was transiently expressed.
Inhibition of HCV replication in FBP1-kd Huh7.5 cells is reversed by downregulation of p53.
We showed earlier that HCV replication is severely impaired by downregulation of FBP1 (10). Since p53 inhibits HCV replication (12), we postulated that activation of p53 causes the inhibition of HCV replication in FBP1-kd cells. To test this hypothesis, we transfected control and FBP1-kd Huh7.5 cells with p53-siRNA and then infected the cells with JFH1 HCV virions. After 72 h of infection, cell lysates were Western blotted for NS5A, FBP1, p53, and p21; quantitative real-time RT-PCR was done on HCV RNA. We found that downregulation of p53 enhanced HCV replication by 5-fold in control Huh7.5 cells compared to the level of the siRNA control (Fig. 4B, lane 4, and right). Interestingly, the severe reduction in HCV replication observed in FBP1-kd cells was completely reversed by downregulation of p53 (Fig. 4C, lane 4). The quantitative RT-PCR data indicated that HCV replication in FBP1-kd cells in which p53 was downregulated is enhanced 6-fold compared to that of the FBP1-kd control (Fig. 4C, right). These results strongly suggest that mutant p53Y220C in Huh7.5 cells displays an inhibitory effect on HCV replication that is strongly suppressed in the presence of FBP1.
Promotion of HCV replication in p53-kd cells is significantly suppressed by transient expression of p53.
Since p53-kd cells are highly permissive to HCV replication, we examined whether restoring the p53 expression in p53-kd cells could suppress the enhanced viral replication. Therefore, we constructed an shRNA-resistant clone of p53 by point mutation in exon seven at codons 244 (GGC→GGT) and 245 (GGC→GGT) without altering the codon usage and transfected this clone into p53-kd Huh7.5 and HepG2 cells. The transfected cells were grown for 72 h, and the levels of mRNA of p53, p21, and FBP1 as well as HCV RNA were determined by quantitative RT-PCR. The results shown in Fig. 4D (left and right) indicate that enhanced HCV replication in p53-kd cells in both cell lines (lane 8) was significantly suppressed and restored to the control level upon transient expression of p53 (lane 12).
DNA binding and transactivation activities of mutant p53Y220C in Huh7.5 cells are activated by knockdown of FBP1.
Mutant p53Y220C in Huh7 cells has been shown to be defective in its ability to bind target DNA at physiological temperatures (16–18). However, in yeast systems, mutant p53Y220C has been shown to be transcriptionally active and displays wild-type target DNA binding activity at subphysiological temperatures (19). In contrast, another study with the truncated p53Y220C containing only the core domain showed that the DNA binding ability of mutant p53Y220C is impaired at both physiological and subphysiological temperatures (20).
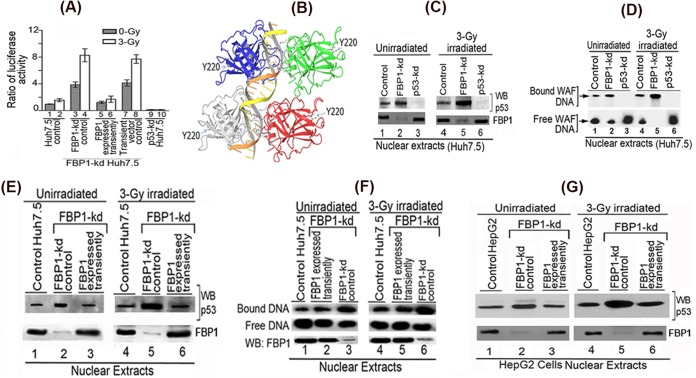
Our results indicated that p21 expression in Huh7.5 cells is under the positive regulation of p53 that is strongly suppressed in the presence of FBP1 (Fig. 4). This premise was further confirmed using reporter p21-Luc activity in control and FBP1-kd Huh7.5 cells under radiation-induced stress. We also included p53-kd Huh7.5 cells as negative controls. We found no activation of reporter activity in γ-irradiated control Huh7.5 cells compared to that in unirradiated cells (Fig. 5A, lanes 1 and 2). In contrast, in FBP1-kd Huh7.5 cells, reporter activity was significantly activated in both unirradiated and irradiated cells (lanes 3 and 4). Reporter activity was completely absent from p53-kd Huh7.5 cells (lanes 9 and 10).
FIG 5.
Transactivation activity and DNA binding ability of mutant p53Y220C in Huh7.5 cells is activated by knockdown of FBP1 expression. (A) Transcription activity of mutant p53Y220C in control and FBP1-kd Huh7.5 cells under radiation-induced stress. The p21-luc reporter and pRL-SV40 plasmids were cotransfected into control, FBP1-kd, and FBP1-kd cells in which FBP1 was transiently expressed via transfection of an shRNA-resistant FBP1-expressing clone (pCIA-cmv-FBP1SHR). Forty-eight hours later, cells were irradiated with a 3-Gy dose of gamma irradiation. Luciferase activity was measured after 6 h postirradiation. Experiments were done in triplicate; results are expressed as the ratio of firefly luciferase to Renilla luciferase activities in cell lysate. Lanes 1 and 2, control cells; lanes 3 and 4, FBP1-kd cells; lanes 5 and 6, FBP1 transiently expressed in FBP1-kd cells; lanes 7 and 8, FBP1-kd cells transfected with vector alone; lanes 9 and 10, p53 kd cells. (B) The three-dimensional structure of homotetrameric p53-DNA binary complex and position of Y220. Using Maestro molecular modeling software, version 9.3.5 (Schrodinger, Inc.), we downloaded the backbone structure of the p53-DNA binary complex from PDB entry 4HJE (80). The three-dimensional crystal structure of DNA-bound p53 is displayed without any modification. The backbone of duplex DNA bound to the tetrameric p53 and the position of Y220 located far away from the bound DNA are shown. (C) Streptavidin magnetic bead DNA binding assay indicates enhanced DNA binding activity of mutant p53Y220C in the nuclear extract from FBP1-kd Huh7.5 cells. Normalized nuclear extracts from unirradiated or 3-Gy-γ-irradiated control and FBP1-kd cells were incubated with 30 bp biotinylated WAF-side DNA at 37°C. The nuclear extract from p53-kd cells was included as a negative control. DNA-protein complexes were captured on streptavidin paramagnetic beads, resolved on SDS-PAGE, and Western blotted for p53. The normalized nuclear extract also was Western blotted for FBP1. Lanes 1 to 3, unirradiated; lanes 4 to 6, γ-irradiated. (D) EMSA showing enhanced binding of 30 bp WAF-side DNA by p53Y220C in the nuclear extract from FBP1-kd Huh7.5 cells. Normalized nuclear extracts from unirradiated and 3-Gy-γ-irradiated control and FBP1-kd cells were incubated with 32P-labeled WAF-side DNA and then subjected to EMSA on 4% native polyacrylamide gel. p53-kd Huh7.5 cells were included as a negative control. Lanes 1 to 3, unirradiated; lanes 4 to 6, γ-irradiated. (E and F) Transient expression of FBP1 in FBP1-kd cells strongly inhibits p53Y220C binding to its target DNA. DNA binding activity of mutant p53 in normalized nuclear extract from unirradiated and irradiated control, FBP1-kd, and transiently FBP1 expressing FBP1-kd Huh7.5 cells was examined by streptavidin magnetic bead DNA binding assay (E) and EMSA (F). The normalized nuclear extract also was Western blotted for FBP1. Lanes 1 to 3, unirradiated; lanes 4 to 6, irradiated. (G) Transient expression of FBP1 in FBP1-kd HepG2 cells inhibits binding of wild-type p53 to its target DNA. DNA binding activity of wild-type p53 in a normalized nuclear extract from unirradiated and irradiated control, FBP1-kd, and transiently FBP1-expressing FBP1-kd HepG2 cells was examined by streptavidin magnetic bead DNA binding assay. The p53 bound to the target DNA was captured on streptavidin magnetic beads and Western blotted for p53. The normalized nuclear extract also was Western blotted for FBP1. Lanes 1 to 3 represent unirradiated control, FBP-kd, and FBP-kd cells, respectively, transiently expressing FBP1; lanes 4 to 6 represent irradiated control, FBP-kd, and FBP-kd cells, respectively, transiently expressing FBP1.
These findings demonstrated that mutant p53Y220C in Huh 7.5 cells is transcriptionally active but strongly suppressed or inhibited in the presence of FBP1. To confirm whether observed activation of mutant p53Y220C indeed was due to the absence of FBP1, we transiently expressed FBP1 in FBP1-kd cells by transfecting the shRNA-resistant FBP1 expression clone and examined the activation of p53 by measuring p21-Luc reporter activity. We found that the activation of p53 in FBP1-kd cells was strongly suppressed when FBP1 was transiently expressed (Fig. 5A, lanes 5 and 6) but remained unaffected when transfected with empty vector alone (lanes 7 and 8).
The residue Y220 in p53 is 38 Å away from the DNA binding motif (Fig. 5B); it is intriguing that Tyr→Cys mutation at this position should affect the DNA binding function of p53. The observed activation of mutant p53Y220C in irradiated FBP1-kd Huh7.5 cells implies that the mutant p53 must be actively binding to its target DNA in the absence of FBP1. To confirm this possibility, we used a streptavidin-paramagnetic bead assay system to examine the DNA binding ability of mutant p53Y220C in the nuclear extracts of γ-irradiated and unirradiated controls, FBP1-kd cells, and p53-kd Huh7.5 cells. We incubated the normalized nuclear extracts with biotinylated 30-bp WAF-side DNA at 37°C, captured the p53-DNA complex on streptavidin magnetic beads, and analyzed the bound p53 by Western blotting. As shown in Fig. 5C, mutant p53 in the nuclear extract of both unirradiated and irradiated control Huh7.5 cells displays only 20% to 25% of the DNA binding ability (lanes 1 and 4) of the corresponding FBP1-Kd cells (Fig. 5C, lanes 2 and 5). We did not detect any DNA binding activity with the nuclear extract from both unirradiated and irradiated p53-kd Huh7.5 cells (lanes 3 and 6).
We also did an electrophoretic mobility shift assay (EMSA) to determine the DNA binding activity of mutant p53 in control and FBP1-kd Huh7.5 cells. We incubated the normalized nuclear extracts with 32P-labeled 30-bp WAF-side DNA at physiological temperature. The p53-bound DNA complexes were resolved by EMSA. The results (Fig. 5D) indicated that mutant p53Y220C in both unirradiated and irradiated Huh7.5 cells (lanes 1 and 4) poorly binds WAF-side DNA compared to the enhanced DNA binding observed in corresponding FBP1-kd cells (lanes 2 and 5). The nuclear extract from p53-kd Huh7.5 cells did not show binding to the target WAF-side DNA (lanes 3 and 6). These results confirm that the target DNA binding activity of mutant p53Y220C in Huh7.5 cells is strongly suppressed by FBP1.
Transient expression of FBP1 in FBP1-kd Huh7.5 cells strongly suppresses DNA binding activity of mutant and wild-type p53.
Our results strongly imply that the mutant p53Y220C in Huh7.5 cells is significantly activated to bind target DNA in the absence of FBP1. To ascertain this, we transiently expressed FBP1 in FBP1-kd Huh7.5 cells and examined DNA binding activity of p53 in the nuclear extracts of both unirradiated and irradiated cells. Results obtained with the streptavidin binding assay indicated that enhanced DNA binding of p53Y220C observed in the nuclear extract from FBP1-kd cells (Fig. 5E, lanes 2 and 5) is strongly inhibited by transient expression of FBP1 (Fig. 5E, lanes 3 and 6), and this inhibition was similar to that observed in control Huh7.5 cells (lanes 1 and 4). We obtained similar results by EMSA in the nuclear extracts from control, FBP1-kd, and FBP1-kd Huh7.5 cells in which FBP1 was transiently expressed (Fig. 5F). The enhanced DNA binding of mutant p53Y220C from the nuclear extract of FBP1-kd cells (lanes 3 and 6) is strongly suppressed when FBP1 is transiently expressed in the cells (lanes 2 and 5).
We also examined DNA binding activity of wild-type p53 in the nuclear extract of irradiated and unirradiated control and FBP1-kd HepG2 cells (Fig. 5G). We found significantly enhanced DNA binding activity of wild-type p53 in the nuclear extract of irradiated FBP-kd HepG2 cells (lane 5) compared to that of the irradiated control cells (lane 4). Similar to Huh7.5 cells, the enhanced DNA binding activity of p53 in the nuclear extract from FBP1-kd HepG2 cells (lanes 2 and 5) also was strongly inhibited by transient expression of FBP1 (lanes 3 and 6). These findings confirm that DNA binding activity of both mutant and wild-type p53 is negatively influenced by the presence of FBP1.
DNA binding ability of the recombinant wild-type p53 and its Y220C mutant derivative.
We also carried out EMSA with purified recombinant wild-type p53 and mutant p53Y220C to examine their binding affinities to 32P-labeled 30-bp WAF-side DNA (Fig. 6A) at physiological temperatures. The dissociation constants determined from the EMSA data were 1.3 nM and 1.7 nM, respectively, for the wild-type and mutant p53, indicating no significant difference in their binding affinities for the target WAF-side DNA. We also did ITC to determine the binding affinity of wild-type and mutant p53 to the WAF-side DNA (Fig. 6B). As p53 interacts with its target DNA, heat is absorbed in direct proportion to the amount of binding that occurs. The top panel of Fig. 6B shows the calorimetric data. Each injection produced downward spikes indicating exothermic interaction between p53 and its target DNA. The absorbed heat was measured to determine the binding constant (Ka) or dissociation constant (37) of interaction between p53 and the target DNA (Fig. 6B, lower). We found that the wild-type and mutant p53 displayed similar binding affinities for the target DNA, with respective dissociation constants of 5.5 nM and 7.2 nM. The free energy change for the binding of p53 to target DNA was calculated to be −14.5 kcal mol−1 for the wild-type p53 and −14.0 kcal mol−1 for the Y220C mutant p53.
FIG 6.
Target DNA binding affinity of recombinant wild-type p53 and mutant p53Y220C. (A) EMSA with purified wild-type p53 and mutant p53Y220C shows similar DNA binding affinity for the target DNA. A fixed concentration of 32P-labeled 30-bp WAF-side DNA was incubated with increasing concentrations of purified recombinant wild-type p53 (left) or mutant p53Y220C (right) at 37°C. The mixture was subjected to EMSA. The bound WAF-side DNA level versus the tetrameric p53 concentration was plotted using GraphPad, and the dissociation constant was determined using a nonlinear regression curve fit for one-site binding of DNA with tetrameric p53. (B) ITC of 30-bp WAF-side DNA binding to the wild-type p53 (left) and mutant p53Y220C (right) shows similar binding affinity. A syringe of ITC containing WAF-side DNA was titrated into a cell containing purified wild-type or mutant p53 in ITC buffer containing 20 mM sodium phosphate buffer, pH 7.8, and 150 mM NaCl at 37°C. (Top) The calorimetric data of titration of p53 with DNA as a function of time. (Bottom) The integrated heat per injection versus the molar ratio of tetrameric p53 to DNA. The graph corresponds to the best fit of the experimental data to the one-site model, providing a dissociation constant (37) of 5.5 nM for the wild-type p53 and 7.2 nM for the mutant p53Y220C. (C) Inhibition of DNA binding activity of wild-type p53 by recombinant FBP1. The purified recombinant p53 (7 nM) was incubated with increasing concentrations of FBP1 (25 to 100 nM) in a final volume of 20 μl. After 30 min of incubation at room temperature, a fixed concentration of 32P-labeled 30-bp WAF-side DNA (40,000 cpm) was added; the mixture was incubated for 30 min at 37°C. The p53-DNA complexes were resolved by EMSA. Lane 1, p53 binding to DNA in the absence of FBP1; lanes 2 through 5, p53 binding to DNA in the presence of 25, 50, 75, and 100 nM recombinant FBP1. (D) Binding of FBP1 alone to 32P-labeled WAF-side DNA is not significant. Lanes 1 through 4, increasing concentrations of FBP1 (25 to 100 nM) were incubated at room temperature with 32P-labeled WAF-side DNA (40,000 cpm) in a final volume of 20 μl for 30 min. The bound and unbound DNA were resolved by EMSA. (E) EMSA with a nontarget 32P-labeled 30-bp HIV-1 U5 PBS DNA was used to determine the target specificity of wild-type p53 and mutant p53Y220C.
FBP1 blocks the binding of recombinant wild-type p53 to the target WAF-side DNA.
Since recombinant proteins of both p53Y220C and wild-type p53 bind WAF-side p21 DNA with similar affinity, it is intriguing that basal levels of p21 expression in both Huh7.5 and HepG2 cells were significantly enhanced upon knockdown of FBP1. Since FBP1 physically interacts with p53, it is possible that, in the presence of FBP1, the binding of p53 to its target WAF-side DNA is compromised. Thus, we examined the binding of the wild-type p53 to the 30-bp WAF-side DNA in the absence and presence of FBP1. We preincubated a fixed concentration of tetrameric p53 (7 nM) with increasing concentrations of FBP1 (25 to 100 nM) and then incubated it with 32P-labeled WAF-side DNA. In a parallel experiment, we incubated labeled WAF-side DNA with increasing concentrations of FBP1 alone (25 to 100 nM). The binding of p53 to the target DNA was significantly inhibited in the presence of FBP1 (Fig. 6C). In the absence of p53, the extent of FBP1 binding to the 30-bp WAF-side DNA was insignificant (Fig. 6D). These results suggest a mechanism whereby FBP1 blocks the binding of p53 to its target DNA sequence and inhibits its transcription activity. We further confirmed that binding of both wild-type and mutant p53Y220C to the target WAF-side DNA is highly specific, since they did not bind to a 30-bp nontarget double-stranded HIV-1 U5PBS DNA (Fig. 6E).
FBP1 promotes HCV replication by regulating p53 regulatory proteins.
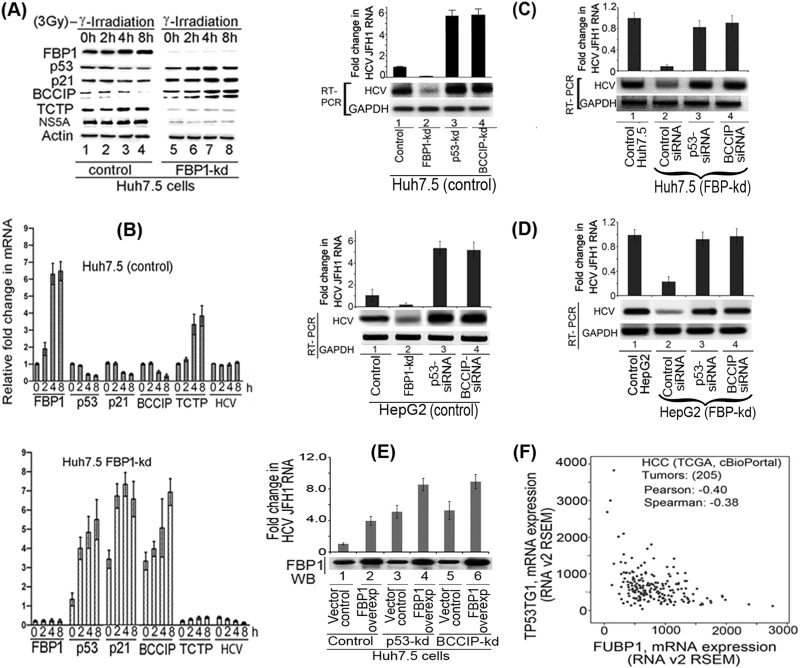
Although mutant p53Y220C binds DNA as efficiently as wild-type p53, it remained transcriptionally inactive in Huh7-derived cells. Our results suggest that FBP1, which is overexpressed in Huh7.5 cells, is involved in suppressing the function of p53 either directly or by regulating p53-regulatory proteins. The translationally controlled tumor protein (TCTP; also known as fortilin) is a negative regulator of p53 that functions as an antiapoptotic factor by interacting with and destabilizing p53 (38, 39). BCCIP, an important cofactor for BRCA2 in tumor suppression, is a positive regulator of p53 and modulates CDK2 kinase activity by interacting with p21 (40–42). To examine whether FBP1 has any influence on the expression of p53 and its regulatory proteins, we first transfected control and FBP1-kd Huh7.5 cells with JFH1-HCV virus; 72 h later, we irradiated cells with 3 Gy gamma irradiation and examined the expression of p21, p53, BCCIP, TCTP, and HCV NS5A at both protein and mRNA levels. An aliquot of the normalized cell lysates containing equivalent protein was subjected to SDS-PAGE and Western blotted for the expression of FBP1, p53, p21, BCCIP, and TCTP, as well as for HCV protein NS5A. Real-time RT-PCR determined their mRNA levels.
We found that in control Huh7.5 cells, FBP1 expression was only slightly increased by 4 h postirradiation. There was a corresponding marginal decrease in the expression of p53 and p21 without any significant change in expression of the HCV protein NS5A (Fig. 7A, left, lanes 3 and 4). In contrast, in FBP1-kd cells, expression of both p53 and p21 was significantly enhanced at 2 to 8 h postirradiation (Fig. 7A, right, lanes 6 to 8), while the NS5A level was drastically reduced at all time points. We further found that TCTP, a negative regulator of p53, was upregulated in control Huh7.5 cells at 4 to 8 h postirradiation (Fig. 7A, left, lanes 3 and 4), while its expression level in FBP1-kd cells was drastically reduced at all time points (Fig. 7A, right, lanes 5 to 8), suggesting that TCTP is positively regulated by FBP1. In contrast, the expression level of BCCIP, a positive regulator of p53, was strongly suppressed in control Huh7.5 cells after irradiation but significantly boosted in FBP1-kd cells (Fig. 7A, lanes 5 and 8). The upregulation of BCCIP in FBP1-kd cells suggests that FBP1 is a negative regulator of BCCIP. These results were confirmed by determining fold changes in the mRNA levels of p21, p53, BCCIP, and TCTP, as well as HCV RNA levels in irradiated control and FBP1-kd cells (Fig. 7B). These results also confirmed that FBP1 suppresses the p53-mediated response to cellular stress by regulating the expression of p53 and its regulatory factors, BCCIP and TCTP.
FIG 7.
FBP1 promotes HCV replication by regulating p53 and its regulatory proteins, TCTP and BCCIP. (A) FBP1 upregulates TCTP while it downregulates the expression of p53, p21, and BCCIP under cellular stress. The control and FBP1-kd Huh7.5 cells were transfected with JFH1-HCV virus; 72 h later, cells were irradiated with 3 Gy gamma irradiation and grown for the indicated times, and their cell lysates were examined for the expression of FBP1, p53, p21, BCCIP, TCTP, and NS5A by Western blotting. Lanes 1 to 4 (control Huh7.5 cells) and lanes 5 to 8 (FBP1-kd Huh7.5 cells) show results after growth for 0, 2, 4, and 8 h postirradiation. (B) Fold change in HCV RNA and mRNA level of FBP1, p53, p21, BCCIP, and TCTP in control (top) and FBP1-kd (bottom) Huh7.5 cells. Quantitative RT-PCR on total RNA isolated from another set of cells in the same experiment was done to determine relative fold changes in mRNA levels of FBP1, p53, p21, BCCIP, TCTP, and HCV RNA at 0, 2, 4, and 8 h postirradiation. (C, left) Control Huh7.5 cells knocked down for either p53Y220C or BCCIP are highly permissive to HCV replication. Lane 1, control cells; lane 2, FBP1-kd cells; lane 3, p53-kd cells; lane 4, BCCIP-kd cells. (Right) HCV replication in FBP1-kd Huh7.5 cells is restored by downregulation of either p53 or BCCIP. Lane 1, untransfected control Huh7.5 cells; lanes 2 to 4, FBP1-kd cells were transfected with control siRNA, p53-siRNA, and BCCIP-siRNA, respectively. (D, left) Downregulation of p53 or BCCIP enhances HCV replication in control HepG2/CD81/miR122 cells. HepG2 cells expressing CD81 and miR122 were transfected with either p53 siRNA or BCCIP siRNA, and 12 h later they were infected with JFH1 HCV virions. Cells were grown for 72 h and analyzed for HCV RNA level by quantitative real-time PCR. Lane 1, control cells; lane 2, FBP1-kd cells; lanes 3 and 4, cells transfected with p53-siRNA and BCCIP-siRNA, respectively. (Right) Downregulation of p53 or BCCIP restored HCV replication in FBP1-kd HepG2/CD81/miR122 cells. FBP1-kd HepG2 cells expressing CD81 and miR122 were transfected with p53-siRNA or BCCIP-siRNA. After 12 h posttransfection, cells were infected with JFH1 HCV and grown for 72 h. Total RNA was isolated, and quantitative real-time PCR determined the level of HCV RNA. Lane 1, untransfected control HepG2 cells; lanes 2 to 4, FBP1-kd cells were transfected with control siRNA, p53-siRNA, and BCCIP-siRNA, respectively. (E) The level of HCV replication in highly permissive p53-kd or BCCIP-kd cells is further increased by overexpression (overexp) of FBP1. The FBP1 overexpression plasmid (pCIA-CMV-FBP) was transfected into control, p53-kd, and BCCIP-kd Huh7.5 cells; 10 h later cells were infected with HCV-JFH1 virions. The cells transfected with vector alone were used as controls. The cells were grown for 72 h, and levels of HCV RNA were determined by quantitative RT-PCR. The FBP1 expression in cells was examined by Western blotting of the normalized cell lysates. Lanes 2, 4, and 6 are control, p53-kd, and BCCIP-kd cells, respectively, in which FBP1 was overexpressed. Lanes 1, 3, and 5 are the respective vector controls. (F) The cBioPortal coexpression analysis of FBP1 and p53-inducible gene TP53TG1. The cBioPortal coexpression analysis was done on data from 205 HCC tumors (TCGA, Provisional), indicating an inverse correlation between the expression of FBP1 and the p53-inducible gene, TP53TG1.
We then examined the level of HCV replication in control and FBP1-kd cells of both Huh7.5 and HepG2 cells in which either p53 or BCCIP had been knocked down or downregulated (Fig. 7C and D). We found that control Huh7.5 cells are highly permissive to HCV replication when knocked down for either p53 or BCCIP, resulting in a relative 5-fold increase in the virus replication with respect to the control (Fig. 7C, lanes 3 and 4, left). Interestingly, the drastic reduction of HCV replication observed in FBP1-kd Huh7.5 cells is completely restored to the control level by downregulation of either p53 or BCCIP (Fig. 7C, lanes 3 and 4, right). We found similar results with HepG2 cells in which downregulation of either p53 or BCCIP enhanced HCV replication in control cells (Fig. 7D, lanes 3 and 4, left) and restored the loss of HCV replication in FBP1-kd HepG2 cells (Fig. 7D, lanes 3 and 4, right). We further examined the effect of overexpression of FBP1 on HCV replication in Huh7.5 cells stably knocked down for the expression of p53 or BCCIP. We transfected control, p53-kd, and BCCIP-kd cells with FBP1 overexpression plasmid or empty vector alone; 10 h later, the cells were infected with HCV JFH1 virions and grown further for 72 h, and total RNA was isolated. The relative fold change in the level of HCV RNA was determined by quantitative RT-PCR. As shown in Fig. 7E, a 5-fold increase in HCV replication occurred in p53-kd cells (lane 3) with respect to the control (lane 1), which was further increased to 8-fold upon overexpression of FBP1 (lane 4). Similar results also were obtained with BCCIP-kd cells in which FBP1 was overexpressed (lane 6). These results clearly suggest that FBP1 promotes HCV replication by suppressing the function of p53 and its positive regulator, BCCIP. This contention also is supported by Oncomine expression data analysis for the expression of FBP1 and p53 in a large number of normal human liver versus HCV-infected cirrhotic livers and HCC tumors (31). In contrast to a 4-fold increase in FBP1 expression (Fig. 1B), there was a 2.5-fold decrease in the p53 mRNA level in the same set of cirrhotic livers and HCC tumors (Fig. 1C). That FBP1 suppresses p53 function also is supported by the cBioPortal coexpression analysis (43, 44) of data from 205 HCC tumors (TCGA, provisional), showing a significant decrease in the p53-inducible gene, TP53TG1, with an increase in the FBP1 expression level (Fig. 7F). This observation also establishes an inverse correlation of FBP1 expression with the expression of p53-targeted genes.
FBP1 enhances cell migration.
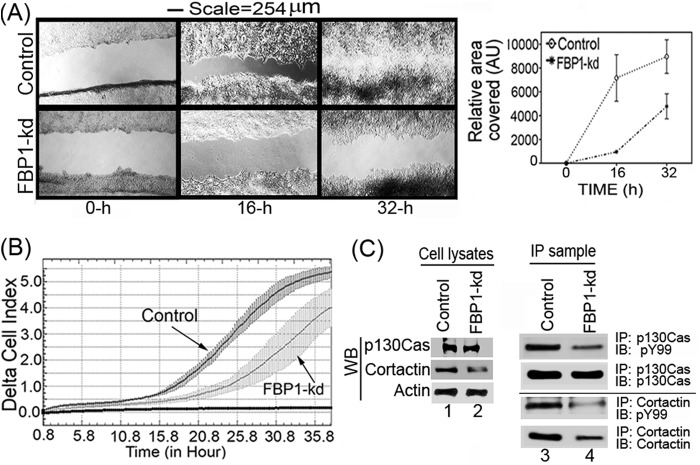
FBP1 has been shown to be involved in tumor metastasis through the regulation of the microtubule-destabilizing proteins Stathmin 1 and Stathmin 3 (45). These two proteins have been suggested to be involved in cell motility and metastasis, since they are abundantly expressed at the invasion front in non-small-cell lung carcinoma (45, 46). In a wound-healing assay, we found that the migration of the FBP1-kd Huh7.5 cells was drastically reduced compared to the level for the control cells (Fig. 8A, left and right). In order to further confirm the results obtained with the wound-healing assay, we also carried out real-time migration dynamics of control and FBP1-kd Huh7.5 cells using a real-time cell analyzer dual-plate RTCA DP xCELLigence system (30). The real-time migration assay was done in triplicate for up to 36 h, and changes in cell index were recorded at 15-min intervals. We found a severe reduction in the migration of FBP1-kd cells compared to that of control cells, as judged by the reduced cell index (Fig. 8B). We also analyzed the expression and phosphorylation levels of cortactin and p130Cas, which are markers for cell migration and invasion (47–49). Cortactin, involved in actin polymerization and cell motility, is an important biomarker for many invasive cancers, including HCC (49, 50). P130Cas, also known as BCAR1, promotes actin remodeling, actomyosin contraction, and cell migration (51). Both cortactin and p130Cas are Src substrates and, in the phosphorylated form, activate signaling events that are associated with cell migration (49, 52). We determined the expression level of these markers by Western blotting of cell lysates, and their phosphorylated form was determined by immunoblotting of their IP samples using an antibody against phosphotyrosine (pY99). The Western blot analysis of p130Cas in cell lysates indicated no significant change in its expression level in control and FBP1-kd cells (Fig. 8C, lanes 1 and 2), but compared to the control, the level of phosphorylated p130Cas was significantly decreased in FBP1-kd cells (Fig. 8C, lanes 3 and 4). In contrast, both cortactin expression and phosphorylation levels were significantly decreased in FBP-kd cells (Fig. 8C, lanes 2 and 4). This observation indicates that FBP1 not only promotes viral replication but also facilitates migration and metastasis of HCC tumors.
FIG 8.
Migration of FBP1-kd Huh7.5 cells is drastically reduced. (A) Wound-healing assay with control and FBP-kd cells. Cells grown to 100% confluence were starved by restricting FBS in the medium to 0.1% overnight. A wound-like simulation was done by making a scratch across each well. Cells then were supplemented with medium with 10% FBS. Images were taken at the indicated times (left), and the areas covered by the scratches were quantified by ImageJ software (right). AU, arbitrary units. (B) Real-time migration dynamics also indicated a drastic reduction in the migration of FBP1-kd cells. We used the RTCA DP Xcelligence system to measure the real-time migration of control and FBP1-kd Huh7.5 cells (30). Migration is shown as a change in delta cell index versus time. (C) Expression of cell migration markers in control and FBP1-kd cells. Control and FBP1-kd cells were grown to 50% confluence, starved overnight in 0.1% FBS, and then supplemented with 10% FBS. Cell were grown further for 32 h and lysed, and cell lysates were Western blotted for cortactin, p130Cas, and actin (lanes 1 and 2). To detect the level of the phosphorylated form of cortactin and p130Cas, we carried out IP using antibody against cortactin and p130Cas and immunoblotted them for total cortactin or p130Cas, as well as their phosphorylated forms, using antibody against phosphotyrosine (lanes 3 and 4).
Colocalization of FBP1 with p53 and BCCIP in Huh7.5 cells.
FBP1, being a transcription factor, is localized mainly in the nucleus. The tumor suppressor protein p53 is a nucleocytoplasmic shuttling protein localized mainly in the nucleus. Its functional regulation depends on its proper subcellular localization (53). Under cell stress, p53 expression is increased and concentrated within the nucleus; under normal unstressed conditions, it is present in low concentrations in a transcriptionally inactive form and distributed throughout the cell (54–57). BCCIP positively regulates p53-dependent p21 expression and is localized mainly in the nucleus (58). Since p53 and BCCIP independently interact with FBP1 (13), we examined whether these proteins also are colocalized with FBP1. We did immunofluorescence on irradiated Huh7.5 cells, finding that BCCIP and p53 are colocalized with FBP1 in the nucleus (Fig. 9A and B).
FIG 9.
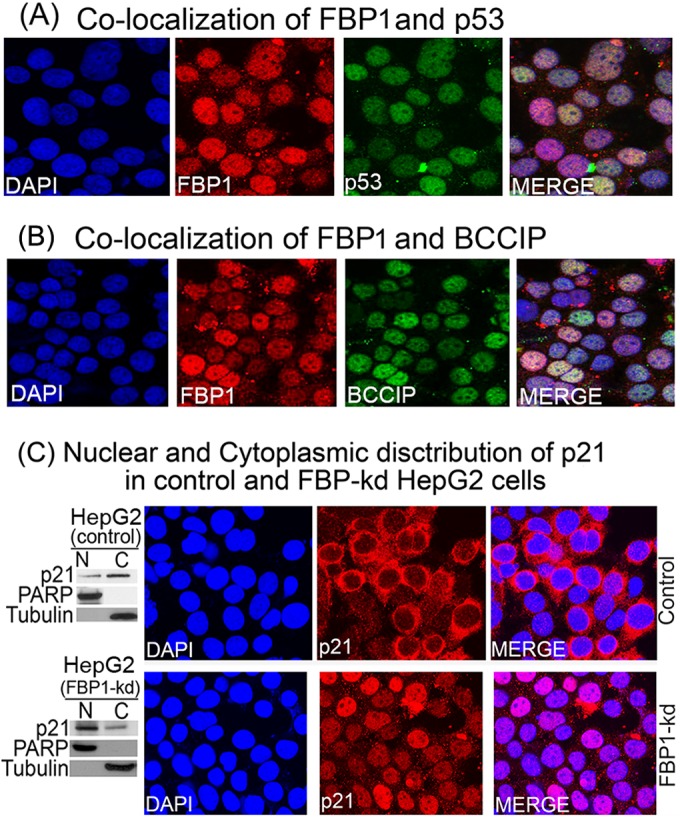
Colocalization of FBP1-p53 and FBP1-BCCIP and cellular distribution of p21 in control and FBP1-kd cells. Huh7.5 cells were grown on chamber slides and treated with goat anti-FBP1 antibody and Alexa 568-labeled secondary antibody (red), treated with mouse anti-p53 antibody (A) or mouse anti-BCCIP antibody (B), and then treated with Alexa 488-labeled secondary antibody (green). DAPI was used to stain the nuclei (blue). Cells were observed individually for FBP1, BCCIP, and p53 localization by a Nikon A1R confocal microscope. (C) The cellular distribution of p21 in control and FBP1-kd HepG2 cells. (Left) Distribution of p21 in the nuclear (N) and cytoplasmic (C) fractions of control and FBP1-kd HepG2 cells. Nuclear and cytoplasmic fractions from control and FBP1-kd cells were prepared and Western blotted for p21. PARP and tubulin also were Western blotted as specific markers for the nucleus and cytoplasm, respectively. (Right) Control and FBP-kd HepG2 cells were grown on a chamber slide for 24 h, fixed, and treated with anti-p21 antibody and Alexa 568-labeled secondary antibody (red). DAPI was used to stain the nuclei. The cellular distribution of p21 was observed under a confocal microscope.
p21 is localized mainly in the nucleus in FBP1-kd HepG2 cells.
BCCIP is known to influence the cellular distribution of p21 (59). Downregulation of BCCIP reduces nuclear p21 and enhances its distribution in the cytoplasm. Since FBP1 interacts with BCCIP and suppresses p21 expression (13), we hypothesize that FBP1-BCCIP interaction also influences the distribution of p21 in the cell. Therefore, we examined the distribution of p21 in control and FBP1-kd HepG2 cells expressing wild-type p53. We first prepared the nuclear and cytoplasmic fractions from control and FBP1-kd HepG2 cells and analyzed them for p21 expression by Western blotting along with tubulin and PARP as the markers for cytoplasmic and nuclear fractions, respectively. As shown in Fig. 9C (left), the majority of p21 in control HepG2 cells is localized in the cytoplasm, while in FBP1-kd cells, it is localized mainly in the nucleus. We then confirmed this observation by immunofluorescence of p21 in control and FBP1-kd cells (Fig. 9C, right). We found that in control cells, the distribution of p21 in the cytoplasm is significantly enhanced, whereas in FBP1-kd cells, p21 is localized mainly in the nucleus. The p21 in nuclei promotes cell cycle arrest by inhibiting cyclin-CDK2 or cyclin-CDK4 complexes; in the cytosol, it promotes cell survival by facilitating the assembly of cyclin D with CDK4 (60).
DISCUSSION
We have shown earlier that FBP1, which interacts with HCV NS5A and poly(UC)-rich regions in the 3′NTR of the viral RNA genome, is an essential cell factor for HCV replication (10). Downregulation and upregulation of FBP1 display strongly negative and positive effects, respectively, on HCV replication. How does FBP1 promote HCV replication? Does HCV use FBP1 to antagonize intrinsic cellular defense against its replication? Recently, Dharel et al. have demonstrated that p53 is involved in innate host defense against HCV replication (12). They found that Huh7 cells downregulated for p53 expression are highly permissive for HCV replication. Recently, we demonstrated that FBP1 interacts with p53 and antagonizes its transcription activity (13). In the present study, we demonstrated that the drastic reduction in HCV replication observed in FBP1-kd Huh7.5 cells is completely restored to control levels by downregulation of p53. These observations clearly suggest that inhibition of HCV replication in FBP1-kd cells is caused by the activation of p53 that may have been suppressed by FBP1 in control Huh7.5 cells.
FBP1 is considered a new proto-oncogene that promotes cell proliferation (46). In many HCC tumors, FBP1 is overexpressed (15), while in normal cells it is negligible or expressed at a very low background level (61). Immunohistochemistry of archived HCC tumors revealed that FBP1 is overexpressed in HCC tumors with a history of CHC (Fig. 1A). Oncomine expression data analysis of normal human livers versus HCV-infected LC and HCC tumors revealed a 4-fold increase in the expression of FBP1 in cirrhotic livers and HCC tumors (Fig. 1B) with a concomitant 2- to 2.5-fold decrease in the expression of p53 (Fig. 1C) (31). Interestingly, FBP1 also suppresses a cell cycle regulator, p21, which is tightly controlled by p53. The enhanced expression of p21 in FBP1-kd Huh7.5 cells (Fig. 4A, lane 6, upper) suggests that p53 is activated in the absence of FBP1. This postulation was supported by the fact that downregulation of p53 in FBP1-kd cells nearly decimated the expression of p21 (Fig. 4A, lane 7, top). These results provide convincing evidence to suggest that p53 activity is negatively regulated by FBP1 in Huh7-derived cell lines. The activation of p53 in FBP1-kd cells was further confirmed by enhanced reporter activity under radiation-induced stress.
In Huh7-derived cells, p53 contains a Tyr→Cys mutation at position 220, which inactivates p53 due to the loss of DNA binding activity (62). Although Y220C mutation is 38 Å away from the conserved DNA binding domain (63), it is intriguing how this mutation could affect the DNA binding of p53. Another study with the truncated p53 containing the core domain has shown that mutant p53Y220C is temperature sensitive; its binding to target DNA is nearly half that of wild-type p53 at subphysiological temperatures (20). Some reports have shown that mutant p53Y220C is transcriptionally inactive (62, 64), while others have found it transcriptionally active (12, 65–67). A recent study using a yeast-based assay system found mutant p53Y220C was as active as wild-type p53 (64).
Our results also confirm that mutant p53Y220C in the nuclear extract from Huh7.5 cells is inactive due to the poor binding to its target DNA sequence. Interestingly, this seemingly inactive p53Y220C is significantly activated in FBP1-kd Huh7.5 cells, as judged by the enhanced reporter activity (Fig. 5A). The activation of p53Y220C in FBP1-kd cells suggests that p53Y220C must be able to bind its target DNA in the absence of FBP1. Using various experimental approaches, we confirmed that the DNA binding ability of the mutant p53Y220C is strongly inhibited in the control Huh7.5 cells but not in FBP1-kd cells (Fig. 5C, D, and E, lanes 5). The transient expression of FBP1 in FBP1-kd cells restored the control phenotype in which the DNA binding ability of the p53Y220C is strongly inhibited (Fig. 5E, lane 6). When we compared the target DNA binding affinity of the purified recombinant wild-type p53 to that of mutant p53Y220C, we found no difference in their DNA binding affinity at physiological temperatures (Fig. 6A and B). However, in the presence of increasing concentrations of purified FBP1, the DNA binding ability of the recombinant wild-type p53 is strongly inhibited (Fig. 6C), while FBP1 alone displayed no affinity for the target DNA sequence (Fig. 6D).
Recent studies have shown that many of the p53 mutations that cause inactivation of p53 also lower the melting temperature of its DNA binding domain (68, 69). It has been proposed that the loss of DNA binding function of mutant p53Y220C is due to the loss of hydrophobic interaction, creating a solvent-accessible cleft at the site of mutation that decreases its thermodynamic stability (69, 70). Liu et al. have shown that the binding of a small molecule, PK7088, to mutant p53Y220C in Huh7 cells restored its transcription activity to a level similar to that of wild-type p53 (5). It will be interesting to examine whether PK7088 inhibits the interaction between p53 and FBP1. It is possible that the transactivation activity of mutant p53Y220C in Huh7 cells is not entirely inactivated by Y220C mutation. Some cell factors, such as FBP1, also may be responsible for suppressing p53 function. That the activation of p21 reporter activity observed in FBP1-kd cells was strongly inhibited by transient expression of FBP1 also supports this contention (Fig. 5A).
We found that FBP1 physically interacts with p53 (Fig. 3A, C, and D) and may be responsible for suppressing its transcription activity. The DNA binding ability of p53 is significantly inhibited in the presence of FBP1 (Fig. 6C), which may well explain why mutant p53Y220C is transcriptionally inactive in Huh7 cells in which FBP1 is abundantly expressed. FBP1 also regulates the transcription of p53, p21, BCCIP, and TCTP under cellular stress. In response to radiation-induced stress, the mRNA levels and expression of p53 and p21 were significantly upregulated in FBP1-kd Huh 7.5 cells, while their levels remained suppressed in control Huh7 cells (Fig. 7A and B). The transcription of TCTP and BCCIP, which function as negative and positive regulators of p53, respectively, also is regulated by FBP1 (Fig. 7A and B). The suppression of TCTP and upregulation of BCCIP in FBP1-kd cells indicated that they were under the positive and negative regulation of FBP1, respectively. Recently, TCTP was shown to promote ubiquitination of p53 via MDM2 and also to repress each other reciprocally (71). In the absence of FBP1, p53 may repress TCTP expression and prevent its degradation from TCTP-mediated ubiquitination via MDM2. Since BCCIP positively regulates p53 (58), its downregulation by FBP1 may be another mechanism of suppression of p53 activity in Huh7 cells. Since Huh7.5 cells knocked down for either p53 or BCCIP are highly permissive for HCV replication, FBP1 may suppress p53 via negative regulation of BCCIP (Fig. 7C). The fact that downregulation of either p53 or BCCIP in FBP1-kd cells of both Huh7.5 and HepG2 cell lines fully restored HCV replication to the control phenotype also supports the role of FBP1 in suppression of p53 via negative regulation of BCCIP. BCCIP also enhances the nuclear localization of p21, where p21 has an antiproliferative effect (59). In HepG2 cells, we found p21 is distributed more in cytosol, whereas in FBP1-kd cells it is distributed mainly in the nucleus (Fig. 9C). We found similar results in FBP1-kd Huh7.5 cells (data not shown). The interaction of FBP1 with BCCIP may prevent nuclear distribution of p21 and facilitate cell survival by abrogating p21-mediated inhibition of CDK2.
There are other pathways that may influence FBP1-mediated suppression of p53. Ubiquitin-specific protease 22 (USP22), which regulates cell proliferation, also suppresses p21 expression by deubiquitinating FBP1 (72). AMP2/p38, a cell factor that downregulates FBP1 (73), also enhances the stability of p53 and p21 (74). Another cell factor, nucleophosmin (NMP), which interacts with p21 and contributes to the stability of p21 and p53 (75, 76), is suppressed by FBP1 (63). FBP1 also is known as a positive regulator of Stathmin, a microtubule destabilizing protein that influences proliferation and migration of cancer cells (14, 45). Stathmin is a potential target of p53 transcriptional repression (77); its overexpression is necessary for the survival of cancer cells (78). Thus, FBP1 has multiple modes of suppressing p53 activity. It physically interacts with p53 and inhibits its binding to target DNA and suppresses p53 activity by regulating various cell factors; thus, it facilitates cell survival and promotes persistent HCV infection and associated pathogenesis. Inhibiting or dissociating the p53 and FBP1 interaction may be a promising target for therapy against HCC tumors. Since FBP1 is overexpressed in most HCC tumors but barely detectable in normal liver tissues, it could be a target for drug development.
ACKNOWLEDGMENTS
This work was supported by NIDDK, National Institutes of Health grant DK083560 (to V.N.P.).
We thank Dinesh Manvar for assisting in experiments with ITC.
REFERENCES
- 1.Choo QL, Weiner AJ, Overby LR, Kuo G, Houghton M, Bradley DW. 1990. Hepatitis C virus: the major causative agent of viral non-A, non-B hepatitis. Br Med Bull 46:423–441. [DOI] [PubMed] [Google Scholar]
- 2.Denniston MM, Jiles RB, Drobeniuc J, Klevens RM, Ward JW, McQuillan GM, Holmberg SD. 2014. Chronic hepatitis C virus infection in the United States, National Health and Nutrition Examination Survey 2003 to 2010. Ann Int Med 160:293–300. doi: 10.7326/M13-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. 2013. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology 57:1333–1342. doi: 10.1002/hep.26141. [DOI] [PubMed] [Google Scholar]
- 4.Denniston MM, Klevens RM, McQuillan GM, Jiles RB. 2012. Awareness of infection, knowledge of hepatitis C, and medical follow-up among individuals testing positive for hepatitis C: National Health and Nutrition Examination Survey 2001-2008. Hepatology 55:1652–1661. doi: 10.1002/hep.25556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu X, Wilcken R, Joerger AC, Chuckowree IS, Amin J, Spencer J, Fersht AR. 2013. Small molecule induced reactivation of mutant p53 in cancer cells. Nucleic Acids Res 41:6034–6044. doi: 10.1093/nar/gkt305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris D, Zhang Z, Chaubey B, Pandey VN. 2006. Identification of cellular factors associated with the 3′-nontranslated region of the hepatitis C virus genome. Mol Cell Proteomics 5:1006–1018. doi: 10.1074/mcp.M500429-MCP200. [DOI] [PubMed] [Google Scholar]
- 7.Upadhyay A, Dixit U, Manvar D, Chaturvedi N, Pandey VN. 2013. Affinity capture and identification of host cell factors associated with hepatitis C virus (+) strand subgenomic RNA. Mol Cell Proteomics 12:1539–1552. doi: 10.1074/mcp.M112.017020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avigan MI, Strober B, Levens D. 1990. A far upstream element stimulates c-myc expression in undifferentiated leukemia cells. J Biol Chem 265:18538–18545. [PubMed] [Google Scholar]
- 9.He L, Weber A, Levens D. 2000. Nuclear targeting determinants of the far upstream element binding protein, a c-myc transcription factor. Nucleic Acids Res 28:4558–4565. doi: 10.1093/nar/28.22.4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Z, Harris D, Pandey VN. 2008. The FUSE binding protein is a cellular factor required for efficient replication of hepatitis C virus. J Virol 82:5761–5773. doi: 10.1128/JVI.00064-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lan KH, Sheu ML, Hwang SJ, Yen SH, Chen SY, Wu JC, Wang YJ, Kato N, Omata M, Chang FY, Lee SD. 2002. HCV NS5A interacts with p53 and inhibits p53-mediated apoptosis. Oncogene 21:4801–4811. doi: 10.1038/sj.onc.1205589. [DOI] [PubMed] [Google Scholar]
- 12.Dharel N, Kato N, Muroyama R, Taniguchi H, Otsuka M, Wang Y, Jazag A, Shao RX, Chang JH, Adler MK, Kawabe T, Omata M. 2008. Potential contribution of tumor suppressor p53 in the host defense against hepatitis C virus. Hepatology 47:1136–1149. doi: 10.1002/hep.22176. [DOI] [PubMed] [Google Scholar]
- 13.Dixit U, Liu Z, Pandey AK, Kothari R, Pandey VN. 2014. Fuse binding protein antagonizes the transcription activity of tumor suppressor protein p53. BMC Cancer 14:925. doi: 10.1186/1471-2407-14-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malz M, Weber A, Singer S, Riehmer V, Bissinger M, Riener MO, Longerich T, Soll C, Vogel A, Angel P, Schirmacher P, Breuhahn K. 2009. Overexpression of far upstream element binding proteins: a mechanism regulating proliferation and migration in liver cancer cells. Hepatology 50:1130–1139. doi: 10.1002/hep.23051. [DOI] [PubMed] [Google Scholar]
- 15.Rabenhorst U, Beinoraviciute-Kellner R, Brezniceanu ML, Joos S, Devens F, Lichter P, Rieker RJ, Trojan J, Chung HJ, Levens DL, Zornig M. 2009. Overexpression of the far upstream element binding protein 1 in hepatocellular carcinoma is required for tumor growth. Hepatology 50:1121–1129. doi: 10.1002/hep.23098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bressac B, Galvin KM, Liang TJ, Isselbacher KJ, Wands JR, Ozturk M. 1990. Abnormal structure and expression of p53 gene in human hepatocellular carcinoma. Proc Natl Acad Sci U S A 87:1973–1977. doi: 10.1073/pnas.87.5.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu IC, Tokiwa T, Bennett W, Metcalf RA, Welsh JA, Sun T, Harris CC. 1993. p53 gene mutation and integrated hepatitis B viral DNA sequences in human liver cancer cell lines. Carcinogenesis 14:987–992. doi: 10.1093/carcin/14.5.987. [DOI] [PubMed] [Google Scholar]
- 18.Puisieux A, Ji J, Guillot C, Legros Y, Soussi T, Isselbacher K, Ozturk M. 1995. p53-mediated cellular response to DNA damage in cells with replicative hepatitis B virus. Proc Natl Acad Sci U S A 92:1342–1346. doi: 10.1073/pnas.92.5.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Como CJ, Prives C. 1998. Human tumor-derived p53 proteins exhibit binding site selectivity and temperature sensitivity for transactivation in a yeast-based assay. Oncogene 16:2527–2539. doi: 10.1038/sj.onc.1202041. [DOI] [PubMed] [Google Scholar]
- 20.Bullock AN, Henckel J, Fersht AR. 2000. Quantitative analysis of residual folding and DNA binding in mutant p53 core domain: definition of mutant states for rescue in cancer therapy. Oncogene 19:1245–1256. doi: 10.1038/sj.onc.1203434. [DOI] [PubMed] [Google Scholar]
- 21.Murata T, Ohshima T, Yamaji M, Hosaka M, Miyanari Y, Hijikata M, Shimotohno K. 2005. Suppression of hepatitis C virus replicon by TGF-beta. Virology 331:407–417. doi: 10.1016/j.virol.2004.10.036. [DOI] [PubMed] [Google Scholar]
- 22.Lindenbach BD, Meuleman P, Ploss A, Vanwolleghem T, Syder AJ, McKeating JA, Lanford RE, Feinstone SM, Major ME, Leroux-Roels G, Rice CM. 2006. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc Natl Acad Sci U S A 103:3805–3809. doi: 10.1073/pnas.0511218103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waris G, Sarker S, Siddiqui A. 2004. Two-step affinity purification of the hepatitis C virus ribonucleoprotein complex. RNA 10:321–329. doi: 10.1261/rna.5124404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ali N, Tardif KD, Siddiqui A. 2002. Cell-free replication of the hepatitis C virus subgenomic replicon. J Virol 76:12001–12007. doi: 10.1128/JVI.76.23.12001-12007.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyanari Y, Hijikata M, Yamaji M, Hosaka M, Takahashi H, Shimotohno K. 2003. Hepatitis C virus non-structural proteins in the probable membranous compartment function in viral genome replication. J Biol Chem 278:50301–50308. doi: 10.1074/jbc.M305684200. [DOI] [PubMed] [Google Scholar]
- 26.Narbus CM, Israelow B, Sourisseau M, Michta ML, Hopcraft SE, Zeiner GM, Evans MJ. 2011. HepG2 cells expressing microRNA miR-122 support the entire hepatitis C virus life cycle. J Virol 85:12087–12092. doi: 10.1128/JVI.05843-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yokota T, Sakamoto N, Enomoto N, Tanabe Y, Miyagishi M, Maekawa S, Yi L, Kurosaki M, Taira K, Watanabe M, Mizusawa H. 2003. Inhibition of intracellular hepatitis C virus replication by synthetic and vector-derived small interfering RNAs. EMBO Rep 4:602–608. doi: 10.1038/sj.embor.embor840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krieger N, Lohmann V, Bartenschlager R. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J Virol 75:4614–4624. doi: 10.1128/JVI.75.10.4614-4624.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsui T, Tanihara K, Date T. 2001. Expression of unphosphorylated form of human double-stranded RNA-activated protein kinase in Escherichia coli. Biochem Biophys Res Commun 284:798–807. doi: 10.1006/bbrc.2001.5039. [DOI] [PubMed] [Google Scholar]
- 30.Limame R, Wouters A, Pauwels B, Fransen E, Peeters M, Lardon F, De Wever O, Pauwels P. 2012. Comparative analysis of dynamic cell viability, migration and invasion assessments by novel real-time technology and classic endpoint assays. PLoS One 7:e46536. doi: 10.1371/journal.pone.0046536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mas VR, Maluf DG, Archer KJ, Yanek K, Kong X, Kulik L, Freise CE, Olthoff KM, Ghobrial RM, McIver P, Fisher R. 2009. Genes involved in viral carcinogenesis and tumor initiation in hepatitis C virus-induced hepatocellular carcinoma. Mol Med 15:85–94. doi: 10.2119/molmed.2008.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, Calderaro J, Bioulac-Sage P, Letexier M, Degos F, Clement B, Balabaud C, Chevet E, Laurent A, Couchy G, Letouze E, Calvo F, Zucman-Rossi J. 2012. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet 44:694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gamarnik AV, Andino R. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev 12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Majumder M, Ghosh AK, Steele R, Ray R, Ray RB. 2001. Hepatitis C virus NS5A physically associates with p53 and regulates p21/waf1 gene expression in a p53-dependent manner. J Virol 75:1401–1407. doi: 10.1128/JVI.75.3.1401-1407.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gong GZ, Jiang YF, He Y, Lai LY, Zhu YH, Su XS. 2004. HCV NS5A abrogates p53 protein function by interfering with p53-DNA binding. World J Gastroenterol 10:2223–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vindigni A, Ochem A, Triolo G, Falaschi A. 2001. Identification of human DNA helicase V with the far upstream element-binding protein. Nucleic Acids Res 29:1061–1067. doi: 10.1093/nar/29.5.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akinmade D, Talukder AH, Zhang Y, Luo WM, Kumar R, Hamburger AW. 2008. Phosphorylation of the ErbB3 binding protein Ebp1 by p21-activated kinase 1 in breast cancer cells. Br J Cancer 98:1132–1140. doi: 10.1038/sj.bjc.6604261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rho SB, Lee JH, Park MS, Byun HJ, Kang S, Seo SS, Kim JY, Park SY. 2011. Anti-apoptotic protein TCTP controls the stability of the tumor suppressor p53. FEBS Lett 585:29–35. doi: 10.1016/j.febslet.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 39.Chen Y, Fujita T, Zhang D, Doan H, Pinkaew D, Liu Z, Wu J, Koide Y, Chiu A, Lin CC, Chang JY, Ruan KH, Fujise K. 2011. Physical and functional antagonism between tumor suppressor protein p53 and fortilin, an anti-apoptotic protein. J Biol Chem 286:32575–32585. doi: 10.1074/jbc.M110.217836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J, Yuan Y, Huan J, Shen Z. 2001. Inhibition of breast and brain cancer cell growth by BCCIPalpha, an evolutionarily conserved nuclear protein that interacts with BRCA2. Oncogene 20:336–345. doi: 10.1038/sj.onc.1204098. [DOI] [PubMed] [Google Scholar]
- 41.Ono T, Kitaura H, Ugai H, Murata T, Yokoyama KK, Iguchi-Ariga SM, Ariga H. 2000. TOK-1, a novel p21Cip1-binding protein that cooperatively enhances p21-dependent inhibitory activity toward CDK2 kinase. J Biol Chem 275:31145–31154. doi: 10.1074/jbc.M003031200. [DOI] [PubMed] [Google Scholar]
- 42.Meng X, Yue J, Liu Z, Shen Z. 2007. Abrogation of the transactivation activity of p53 by BCCIP down-regulation. J Biol Chem 282:1570–1576. doi: 10.1074/jbc.M607520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao JJ, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun YC, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singer S, Malz M, Herpel E, Warth A, Bissinger M, Keith M, Muley T, Meister M, Hoffmann H, Penzel R, Gdynia G, Ehemann V, Schnabel PA, Kuner R, Huber P, Schirmacher P, Breuhahn K. 2009. Coordinated expression of stathmin family members by far upstream sequence element-binding protein-1 increases motility in non-small cell lung cancer. Cancer Res 69:2234–2243. doi: 10.1158/0008-5472.CAN-08-3338. [DOI] [PubMed] [Google Scholar]
- 46.Zhang J, Chen QM. 2013. Far upstream element binding protein 1: a commander of transcription, translation and beyond. Oncogene 32:2907–2916. doi: 10.1038/onc.2012.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cunningham-Edmondson AC, Hanks SK. 2009. p130Cas substrate domain signaling promotes migration, invasion, and survival of estrogen receptor-negative breast cancer cells. Breast Cancer 1:39–52. doi: 10.2147/BCTT.S6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamada S, Yanamoto S, Kawasaki G, Mizuno A, Nemoto TK. 2010. Overexpression of cortactin increases invasion potential in oral squamous cell carcinoma. Pathol Oncol Res 16:523–531. doi: 10.1007/s12253-009-9245-y. [DOI] [PubMed] [Google Scholar]
- 49.Chuma M, Sakamoto M, Yasuda J, Fujii G, Nakanishi K, Tsuchiya A, Ohta T, Asaka M, Hirohashi S. 2004. Overexpression of cortactin is involved in motility and metastasis of hepatocellular carcinoma. J Hepatol 41:629–636. doi: 10.1016/j.jhep.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 50.MacGrath SM, Koleske AJ. 2012. Cortactin in cell migration and cancer at a glance. J Cell Sci 125:1621–1626. doi: 10.1242/jcs.093781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meenderink LM, Ryzhova LM, Donato DM, Gochberg DF, Kaverina I, Hanks SK. 2010. P130Cas Src-binding and substrate domains have distinct roles in sustaining focal adhesion disassembly and promoting cell migration. PLoS One 5:e13412. doi: 10.1371/journal.pone.0013412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sharma A, Mayer BJ. 2008. Phosphorylation of p130Cas initiates Rac activation and membrane ruffling. BMC Cell Biol 9:50. doi: 10.1186/1471-2121-9-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karni-Schmidt O, Friedler A, Zupnick A, McKinney K, Mattia M, Beckerman R, Bouvet P, Sheetz M, Fersht A, Prives C. 2007. Energy-dependent nucleolar localization of p53 in vitro requires two discrete regions within the p53 carboxyl terminus. Oncogene 26:3878–3891. doi: 10.1038/sj.onc.1210162. [DOI] [PubMed] [Google Scholar]
- 54.Giaccia AJ, Kastan MB. 1998. The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev 12:2973–2983. doi: 10.1101/gad.12.19.2973. [DOI] [PubMed] [Google Scholar]
- 55.Appella E, Anderson CW. 2001. Post-translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem 268:2764–2772. doi: 10.1046/j.1432-1327.2001.02225.x. [DOI] [PubMed] [Google Scholar]
- 56.Liang SH, Clarke MF. 2001. Regulation of p53 localization. Eur J Biochem 268:2779–2783. doi: 10.1046/j.1432-1327.2001.02227.x. [DOI] [PubMed] [Google Scholar]
- 57.Prives C, Manley JL. 2001. Why is p53 acetylated? Cell 107:815–818. doi: 10.1016/S0092-8674(01)00619-5. [DOI] [PubMed] [Google Scholar]
- 58.Meng X, Lu H, Shen Z. 2004. BCCIP functions through p53 to regulate the expression of p21Waf1/Cip1. Cell Cycle 3:1457–1462. doi: 10.4161/cc.3.11.1213. [DOI] [PubMed] [Google Scholar]
- 59.Fan J, Wray J, Meng X, Shen Z. 2009. BCCIP is required for the nuclear localization of the p21 protein. Cell Cycle 8:3019–3024. doi: 10.4161/cc.8.18.9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.LaBaer J, Garrett MD, Stevenson LF, Slingerland JM, Sandhu C, Chou HS, Fattaey A, Harlow E. 1997. New functional activities for the p21 family of CDK inhibitors. Genes Dev 11:847–862. doi: 10.1101/gad.11.7.847. [DOI] [PubMed] [Google Scholar]
- 61.Duncan R, Bazar L, Michelotti G, Tomonaga T, Krutzsch H, Avigan M, Levens D. 1994. A sequence-specific, single-strand binding protein activates the far upstream element of c-myc and defines a new DNA-binding motif. Genes Dev 8:465–480. doi: 10.1101/gad.8.4.465. [DOI] [PubMed] [Google Scholar]
- 62.Kubicka S, Trautwein C, Niehof M, Manns M. 1997. Target gene modulation in hepatocellular carcinomas by decreased DNA-binding of p53 mutations. Hepatology 25:867–873. doi: 10.1002/hep.510250414. [DOI] [PubMed] [Google Scholar]
- 63.Cho Y, Gorina S, Jeffrey PD, Pavletich NP. 1994. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science 265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 64.Dearth LR, Qian H, Wang T, Baroni TE, Zeng J, Chen SW, Yi SY, Brachmann RK. 2007. Inactive full-length p53 mutants lacking dominant wild-type p53 inhibition highlight loss of heterozygosity as an important aspect of p53 status in human cancers. Carcinogenesis 28:289–298. doi: 10.1093/carcin/bgl132. [DOI] [PubMed] [Google Scholar]
- 65.Hsieh JL, Wu CL, Lee CH, Shiau AL. 2003. Hepatitis B virus X protein sensitizes hepatocellular carcinoma cells to cytolysis induced by E1B-deleted adenovirus through the disruption of p53 function. Clin Cancer Res 9:338–345. [PubMed] [Google Scholar]
- 66.Chiba T, Yokosuka O, Arai M, Tada M, Fukai K, Imazeki F, Kato M, Seki N, Saisho H. 2004. Identification of genes up-regulated by histone deacetylase inhibition with cDNA microarray and exploration of epigenetic alterations on hepatoma cells. J Hepatol 41:436–445. doi: 10.1016/j.jhep.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 67.Terui T, Murakami K, Takimoto R, Takahashi M, Takada K, Murakami T, Minami S, Matsunaga T, Takayama T, Kato J, Niitsu Y. 2003. Induction of PIG3 and NOXA through acetylation of p53 at 320 and 373 lysine residues as a mechanism for apoptotic cell death by histone deacetylase inhibitors. Cancer Res 63:8948–8954. [PubMed] [Google Scholar]
- 68.Boeckler FM, Joerger AC, Jaggi G, Rutherford TJ, Veprintsev DB, Fersht AR. 2008. Targeted rescue of a destabilized mutant of p53 by an in silico screened drug. Proc Natl Acad Sci U S A 105:10360–10365. doi: 10.1073/pnas.0805326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Joerger AC, Ang HC, Fersht AR. 2006. Structural basis for understanding oncogenic p53 mutations and designing rescue drugs. Proc Natl Acad Sci U S A 103:15056–15061. doi: 10.1073/pnas.0607286103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rauf SM, Endou A, Takaba H, Miyamoto A. 2013. Effect of Y220C mutation on p53 and its rescue mechanism: a computer chemistry approach. Protein J 32:68–74. doi: 10.1007/s10930-012-9458-x. [DOI] [PubMed] [Google Scholar]
- 71.Amson R, Pece S, Lespagnol A, Vyas R, Mazzarol G, Tosoni D, Colaluca I, Viale G, Rodrigues-Ferreira S, Wynendaele J, Chaloin O, Hoebeke J, Marine JC, Di Fiore PP, Telerman A. 2012. Reciprocal repression between P53 and TCTP. Nat Med 18:91–99. doi: 10.1038/nm.2546. [DOI] [PubMed] [Google Scholar]
- 72.Atanassov BS, Dent SY. 2011. USP22 regulates cell proliferation by deubiquitinating the transcriptional regulator FBP1. EMBO Rep 12:924–930. doi: 10.1038/embor.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim MJ, Park BJ, Kang YS, Kim HJ, Park JH, Kang JW, Lee SW, Han JM, Lee HW, Kim S. 2003. Downregulation of FUSE-binding protein and c-myc by tRNA synthetase cofactor p38 is required for lung cell differentiation. Nat Genet 34:330–336. doi: 10.1038/ng1182. [DOI] [PubMed] [Google Scholar]
- 74.Han JM, Park BJ, Park SG, Oh YS, Choi SJ, Lee SW, Hwang SK, Chang SH, Cho MH, Kim S. 2008. AIMP2/p38, the scaffold for the multi-tRNA synthetase complex, responds to genotoxic stresses via p53. Proc Natl Acad Sci U S A 105:11206–11211. doi: 10.1073/pnas.0800297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xiao J, Zhang Z, Chen GG, Zhang M, Ding Y, Fu J, Li M, Yun JP. 2009. Nucleophosmin/B23 interacts with p21WAF1/CIP1 and contributes to its stability. Cell Cycle 8:889–895. doi: 10.4161/cc.8.6.7898. [DOI] [PubMed] [Google Scholar]
- 76.Colombo E, Marine JC, Danovi D, Falini B, Pelicci PG. 2002. Nucleophosmin regulates the stability and transcriptional activity of p53. Nat Cell Biol 4:529–533. doi: 10.1038/ncb814. [DOI] [PubMed] [Google Scholar]
- 77.Ahn J, Murphy M, Kratowicz S, Wang A, Levine AJ, George DL. 1999. Down-regulation of the stathmin/Op18 and FKBP25 genes following p53 induction. Oncogene 18:5954–5958. doi: 10.1038/sj.onc.1202986. [DOI] [PubMed] [Google Scholar]
- 78.Sonego M, Schiappacassi M, Lovisa S, Dall'Acqua A, Bagnoli M, Lovat F, Libra M, D'Andrea S, Canzonieri V, Militello L, Napoli M, Giorda G, Pivetta B, Mezzanzanica D, Barbareschi M, Valeri B, Canevari S, Colombatti A, Belletti B, Del Sal G, Baldassarre G. 2013. Stathmin regulates mutant p53 stability and transcriptional activity in ovarian cancer. EMBO Mol Med 5:707–722. doi: 10.1002/emmm.201201504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Edmondson HA, Steiner PE. 1954. Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer 7:462–503. doi:. [DOI] [PubMed] [Google Scholar]
- 80.Chen Y, Zhang X, Dantas Machado AC, Ding Y, Chen Z, Qin PZ, Rohs R, Chen L. 2013. Structure of p53 binding to the BAX response element reveals DNA unwinding and compression to accommodate base-pair insertion. Nucleic Acids Res 41:8368–8376. doi: 10.1093/nar/gkt584. [DOI] [PMC free article] [PubMed] [Google Scholar]