Abstract
Simian hemorrhagic fever (SHF) is lethal for macaques. Based on clinical presentation and serological diagnosis, all reported SHF outbreaks were thought to be caused by different strains of the same virus, simian hemorrhagic fever virus (SHFV; Arteriviridae). Here we show that the SHF outbreaks in Sukhumi in 1964 and in Alamogordo in 1989 were caused not by SHFV but by two novel divergent arteriviruses. Our results indicate that multiple divergent simian arteriviruses can cause SHF.
TEXT
In late August 1964, a viral hemorrhagic fever epizootic occurred among captive macaques at the Institute of Experimental Pathology and Therapy in Sukhumi, Georgia, Soviet Union. All 64 infected animals succumbed. The disease was named simian hemorrhagic fever (SHF), as the etiological virus was not related to any other virus known at the time (1–3). In November 1964, an epizootic with a highly similar clinical presentation and case fatality rate occurred among macaques at the National Institutes of Health (NIH) Primate Quarantine Unit in Bethesda, MD, killing a total of 223 animals. Outbreak investigations revealed that the macaques affected at both facilities had been imported from the same primate supplier in India, suggesting that the two outbreaks were related (4).
A list of additional SHF outbreaks, the majority of which had connections to Indian suppliers of nonhuman primates, is provided in Table 1. The diagnosis of SHF during these additional outbreaks was based on the typical clinical and pathological presentation of sick macaques (4–6) and on serological assays such as indirect fluorescence assay (IFA) or complement fixation (CF) (7–10). Several observations suggested that different strains of SHFV may have caused the various outbreaks. For example, central nervous system (CNS) manifestations were much more commonly observed among macaques during the 1964 Sukhumi outbreak than during the 1964 Bethesda and 1967 Sukhumi outbreaks (11). The presence or absence of CNS involvement could be reproduced in macaques experimentally infected with clinical material from the different outbreaks (11). CF, IFA, and enzyme-linked immunosorbent assay studies performed with virus antigen or nonhuman primate sera from various outbreaks often yielded contradictory results, either confirming or refuting direct relationships between the etiological viruses (7–10, 12, 13). Unfortunately, virus isolation in cell culture was reported for only a few outbreaks, and with the exception of 1964 Bethesda isolate NIH LVR42-0/M6941, all of the isolates and most of the associated materials have been accidentally or deliberately destroyed (Table 1).
TABLE 1.
History of SHF outbreaks
| Yr of outbreak | Location of outbreak | Nonhuman primates affected (species) | Basis of SHF diagnosis | Isolate | Reference(s) |
|---|---|---|---|---|---|
| 1964 | Institute of Experimental Pathology and Therapy, Sukhumi, Georgia, Soviet Union | Assam macaques,a rhesus monkeys,b southern pig-tailed macaques,c stump-tailed macaquesd | Clinical presentation, serology, virus isolation, exptl reproduction | Sukhumi-64 (destroyed?) | 1–3, 30, 31 |
| 1964 | NIH Primate Quarantine Unit, Bethesda, MD | Rhesus monkeys | Clinical presentation, serology, virus isolation, exptl reproduction | NIH LVR42-0/M6941,g NIH LVR543/654 (destroyed), LVR M8930 (destroyed) | 4, 5, 18, 32 |
| 1965 | National Center for Primate Biology, Davis, CA | ? | Retrospective diagnosis based on clinical presentation | None | 8 |
| 1966-1967 | Shamrock Farm Ltd., Small Dole, West Sussex, United Kingdom | Crab-eating macaques,e rhesus monkeys | Clinical presentation | None | 10, 33 |
| 1967 | Institute of Experimental Pathology and Therapy, Sukhumi, Georgia, Soviet Union | Rhesus monkeys | Clinical presentation | None | 15, 34 |
| 1967 | National Center for Primate Biology, Davis, CA | Rhesus monkeys | Serology, exptl reproduction | None | 8, 10 |
| 1968 | Institute of Experimental Pathology and Therapy, Sukhumi, Georgia, Soviet Union | Rhesus monkeys | Clinical presentation | None | 15, 34 |
| 1968 | Shamrock Farm Ltd. Small Dole, West Sussex, United Kingdom | Rhesus monkeys | Clinical presentation, exptl reproduction | None | 9, 10, 35 |
| 1968 | Austria | Rhesus monkeys | Clinical presentation | None | 36 |
| 1969 | Shamrock Farm Ltd., Small Dole, West Sussex, United Kingdom | Crab-eating macaques, rhesus monkeys | Clinical presentation, serology, virus isolation, exptl reproduction | Unnamed isolate (destroyed) | 9, 10, 33, 37 |
| 1970 | Institute of Experimental Pathology and Therapy, Sukhumi, Georgia, Soviet Union | Rhesus monkeys | Clinical presentation | None | 15, 34 |
| 1972 | NIH Corbel Facility, Rockville, MD | Rhesus monkeys | Clinical presentation, serology, virus isolation | P-180 (destroyed), P-248 (destroyed), P-741 (destroyed) | 12, 14, 38, 39 |
| 1974 | Institute of Experimental Pathology and Therapy, Sukhumi, Georgia, Soviet Union | Patas monkeys,f southern pig-tailed macaques | Clinical Presentation | None | 15, 34, 40 |
| 1989 | Primate Research Institute of New Mexico State University, Alamogordo, NM | Crab-eating macaques | Clinical presentation, serology, electron microscopy, virus isolation | I-621 (destroyed), Ph0080 (destroyed) | 41, 42 |
| 1989 | Undisclosed facility, Miami, FL | ? | ? | ? | 43 |
| 1989 | Hazelton Research Products, Reston, VA | Crab-eating macaques | Clinical presentation, serology, virus isolation | 4 unnamed isolates (destroyed?) | 44 |
| 1996 | Hazelton Research Products, Alice, TX | Crab-eating macaques | Clinical presentation, virus isolation | 1 unnamed isolate (destroyed?) | 45 |
The origin of SHFV is unclear, although various African nonhuman primates, such as patas monkeys (Erythrocebus patas) (14, 15), Guinea baboons (Papio papio) (14), and olive baboons (Papio anubis) (16), have been implicated as possible carriers. This hypothesis is strengthened by the recent discovery in apparently healthy African nonhuman primates of several novel and highly divergent simian arteriviruses, all of which are more closely related to SHFV and each other than to the other known arteriviruses, i.e., equine arteritis virus, lactate dehydrogenase-elevating virus, porcine reproductive and respiratory syndrome virus, wobbly possum disease virus, and a yet-to-be-described arterivirus from a forest giant pouched rat. These simian arteriviruses include Kibale red colobus virus 1 (KRCV-1) and KRCV-2 found in the Ugandan red colobus (Procolobus [Piliocolobus] rufomitratus tephrosceles) (17, 18) and Kibale red-tailed guenon virus 1 (KRTGV-1) and KRTGV-2 found in red-tailed monkeys (Cercopithecus ascanius) (19) in Uganda's Kibale National Park, Mikumi yellow baboon virus 1 (MYBV-1) found in yellow baboons (Papio cynocephalus) in Tanzania's Mikumi National Park (20), and a yet-to-be-described virus from DeBrazza's monkeys (Cercopithecus neglectus) in the Democratic Republic of Congo. Another novel simian arterivirus, southwest baboon virus 1 (SWBV-1), was found in healthy captive olive baboons at the Southwest National Primate Research Center in San Antonio, TX (20).
We attempted to recover archived materials such as frozen or fixed tissues from past SHF outbreaks to identify and characterize the viruses responsible by next-generation sequencing (NGS). Thus far, we have been successful in two instances. We obtained a vial containing lyophilized brain tissue (10% homogenate) of a rhesus monkey (Macaca mulatta) that succumbed during the 1964 Sukhumi epizootic from the Russian State Collection of Viruses at the D. I. Ivanovsky Institute of Virology in Moscow, Russia. This vial was one of several provided to the collection by two of us (Z.V.S. and B.A.L.) in 1969. In the second instance, we obtained three vials containing serum of three rhesus monkeys (samples F1167, I621, and F628) experimentally infected with material from crab-eating macaques (Macaca fascicularis) that had succumbed during the 1989 Alamogordo epizootic from the United States Army Medical Research Institute of Infectious Diseases at Fort Detrick, Frederick, MD.
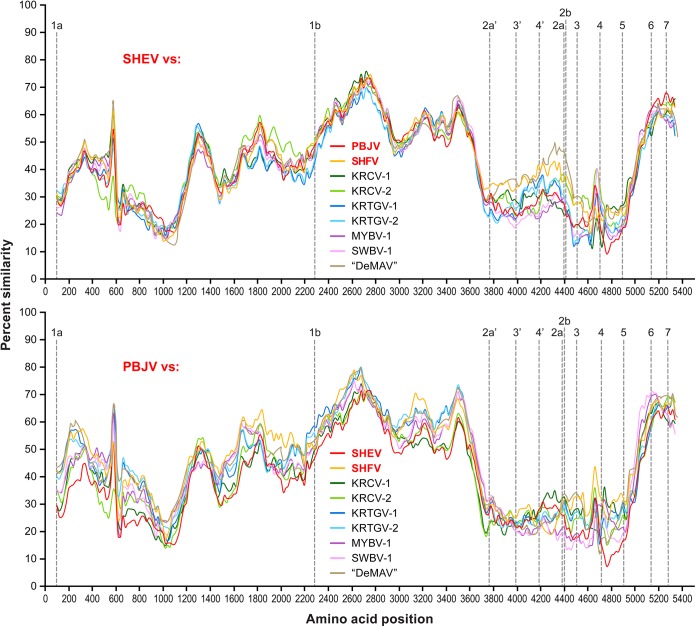
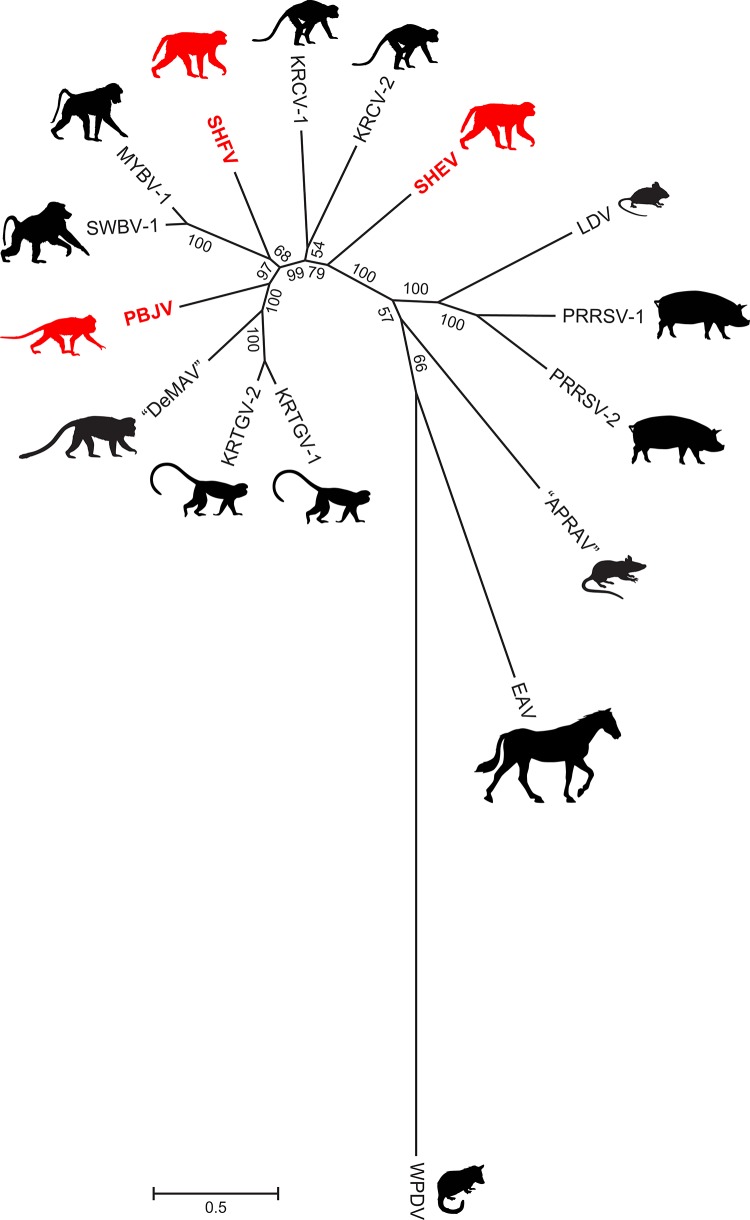
To determine the sequences of the Sukhumi-64, F1167, I621, and F628 viruses, we purified their total RNAs with TRIzol (Invitrogen) and the QIAamp MinElute virus spin kit (Qiagen) and removed host rRNA and tRNA with the GeneRead rRNA depletion kit (Qiagen). For the Sukhumi-64 virus, a DNA library for NGS was prepared from 10 ng of depleted RNA with the NEBnext Ultra RNA Library Prep kit (New England BioLabs). For the F1167, I621, and F628 viruses, a randomly primed double-stranded cDNA was synthesized as previously described (21) and NGS libraries were prepared with the Nextera XT kit (Illumina). Sequencing was performed on an Illumina MiSeq instrument with MiSeq Reagent kits V2 (300PE; Illumina Inc.). Low-quality (<Q30) and short (<100 bp) reads were removed, and the remaining reads were assembled de novo with CLC Genomics Workbench 7 (CLC bio). Sequence gaps in the genome were filled by Sanger sequencing on an Applied Biosystems 3130 genetic analyzer. The genomic 3′ and 5′ termini of the Sukhumi-64 virus were determined with the Mint RACE kit (Evrogen). Viral genomes were annotated in CLC Genomics Workbench 7.1. A query against the NCBI GenBank database (22) revealed two novel simian arteriviruses, here named simian hemorrhagic encephalitis virus (SHEV) and Pebjah virus (PBJV), from the samples obtained in Sukhumi, Russia, and Alamogordo, NM, respectively. The genomic organization of SHEV and PBJV is highly similar to that of all known simian arteriviruses. However, the SHEV and PBJV genomes differ from each other by >52.9% in pairwise analyses. KRCV-1 and KRCV-2 are the closest relatives of SHEV (45.4 and 45.6% identity) and PBJV (46.5 and 46.6% identity). In a pairwise sequence comparison with a modified basic local alignment search tool (BLAST) algorithm (23), the SHEV and PBJV genomes differed from each other by 54% and from KRCV-2 and SHFV, the most closely related simian arteriviruses, by ∼54 and ∼48%, respectively. The resulting histograms, visualizing the distribution of the number of arterivirus pairs at each identity percentage, can be found at http://www.ncbi.nlm.nih.gov/sutils/pasc/viridty.cgi?cmdresult=main&id=377. These stark differences are confirmed by amino acid similarity plots (Fig. 1). PBJV genomes from the three macaques that succumbed during the Alamogordo epizootic were highly similar to each other (99.89 to 99.96% identity on the genome level), suggesting that the outbreak was caused by a single etiological agent. Finally, phylogenetic analysis (Fig. 2) indicates that SHEV and PBJV indeed cluster with the simian arterivirus clade, although both viruses are distinct and divergent from their congenerics.
FIG 1.
Genomic and amino acid sequence comparisons of SHEV, PBJV, and other arteriviruses. Sliding-window similarity plots of percent amino acid identity of SHEV and PBJV compared to the coding genomes of other simian arteriviruses, i.e., SHFV, KRCV-1, KRCV-2, KRTGV-1, KRTGV-2, MYBV-1, SWBV-1, and “DeBrazza's monkey arterivirus” (“DeMAV”). Simian arteriviruses associated with SHF outbreaks are shown in red. Open reading frame 1a (ORF1a), ORF1b, ORF2a′, ORF3′, ORF4′, ORF2a, ORF2b, and ORF3 to ORF7 were individually aligned with other simian arteriviruses by a codon-based version of the multiple alignment with fast Fourier transform (MAFFT) algorithm (24) implemented in Translator X (25). Individual ORF alignments were then concatenated, and sliding-window plots of inferred amino acid sequence similarity were created with SimPlot v3.5.1 (window size, 200 amino acids; step size, 25 amino acids) (26). Dashed vertical lines represent the start positions of the ORFs. RefSeq/GenBank accession numbers of the sequences used for the analysis: SHEV, KM677927; PBJV, KR139839; SHFV, NC_003092; KRCV-1, HQ845737; KRCV-2, KC787631.1; KRTGV-1, JX473847; KRTGV-2, JX473849; MYBV-1, NC_025112.1; SWBV-1, NC_025113.1; “DeMAV,” NC_026509.
FIG 2.
Arterivirus phylogeny based on ORF1b nucleotide sequences. Sequences were aligned by a codon-based version of the MAFFT algorithm with regions of poor alignment removed by the Gblocks method implemented in the computer program Translator X (25). The tree was constructed by Phylogenetic Analysis Using Parsimony (PAUP*) version 4.0b10 (27) from a final alignment of 3,840 positions by the maximum-likelihood method with a best-fit substitution model of the form GTR+I+Γ (general time-reversible model with invariable sites parameter and gamma distribution accounting for rate variation among sites) and parameters identified through jModeltest (28, 29). Numbers beside branches represent bootstrap values (based on 1,000 bootstrap replicates of the data), and branch lengths are proportional to the numbers of nucleotide substitutions per site (scale bar). Silhouettes indicate hosts (see the text), with nonhuman primates associated with SHF outbreaks shown in red, as the natural hosts of those three viruses remain unknown. The virus name abbreviations and GenBank accession numbers shown are identical to those in Fig. 1 plus equine arteritis virus (EAV; accession no. NC_002532), lactate dehydrogenase-elevating virus (LDV; accession no. NC_001639), porcine reproductive and respiratory syndrome viruses 1 (PRRSV-1; accession no. M96262.2), PRRSV-2 (accession no. NC_001961), wobbly possum disease virus (WPDV; accession no. JN116253), and “African pouched rat arterivirus” (“APRAV”; accession no. NC_026439.1).
Our results indicate that on at least two occasions, SHF in captive macaques has, in all likelihood, been caused by viruses distinct from SHFV and distinct from each other (SHEV and PBJV). We conclude that SHF is caused not by a single etiological agent but rather by diverse members of the simian clade of the family Arteriviridae. Our findings raise the possibility that other recently discovered simian arteriviruses (17–20) may also be capable of infecting macaques and causing SHF.
Nucleotide sequence accession numbers.
The genomic sequences obtained in this study were submitted to GenBank and assigned accession no. KM677927 (Sukhumi-64), KR139840 (F1167), KR139839 (I621), and KR139838 (F628).
ACKNOWLEDGMENTS
We appreciate and are very grateful to T. F. F. Ng et al., who deposited sequences of “African pouched rat arterivirus” and “DeBrazza's monkey arterivirus” in GenBank before the publication of detailed descriptions of these viruses. We are also grateful to Laura Bollinger of the Integrated Research Facility at Fort Detrick, Frederick, MD, for critically editing the manuscript.
The content of this publication does not necessarily reflect the views or policies of the U.S. Department of the Army, the U.S. Department of Defense, the U.S. Department of Health and Human Services, or the institutions and companies with which we are affiliated. This work was funded in part through the Battelle Memorial Institute's prime contract with the U.S. National Institute of Allergy and Infectious Diseases under contract HHSN272200700016I. The subcontractors to Battelle Memorial Institute who performed this work are E.P. and J.H.K, employees of Tunnell Government Services, Inc.; M.G.L., an employee of Lovelace Respiratory Research Institute; and S.M., an employee of MRIGlobal. This work was also funded in part by NIH grant TW009237 as part of the joint NIH-National Science Foundation Ecology of Infectious Disease program, grant R01 AI077376, and by Office of Research Infrastructure Programs grant P51OD011106. This research was also supported in part by the Intramural Research Program of the NIH, National Library of Medicine (Y.B.).
REFERENCES
- 1.Lapin BA, Pekerman SM, Yakovleva LA, Dzhikidze EK, Shevtsova ZV, Kuksova MI, Dan'ko LV, Krylova RI, Akbroit E, Agrba VZ. 1967. Hemorrhagic fever in monkeys. Vopr Virusol 12:168–173. (In Russian.) [PubMed] [Google Scholar]
- 2.Shevtsova ZV. 1967. Study of the etiology of hemorrhagic fever in monkeys. Vopr Virusol 12:47–51. (In Russian.) [PubMed] [Google Scholar]
- 3.Shevtsova ZV, Kuksova MI, Dzhikidze EK, Krylova RI, Dan'ko LV. 1966. Experimental studies of hemorrhagic fever of monkeys, p 146–150. In Lapin BA. (ed), Biology and pathology of monkeys, studies of human diseases in experiments on monkeys. Materials of Symposium in Sukhumi, October 17 to 22. Academy of Medical Sciences of the USSR, Institute of Experimental Pathology and Therapy, Tbilisi, Georgia, Soviet Union: (In Russian.) [Google Scholar]
- 4.Palmer AE, Allen AM, Tauraso NM, Shelokov A. 1968. Simian hemorrhagic fever. I. Clinical and epizootiologic aspects of an outbreak among quarantined monkeys. Am J Trop Med Hyg 17:404–412. [PubMed] [Google Scholar]
- 5.Allen AM, Palmer AE, Tauraso NM, Shelokov A. 1968. Simian hemorrhagic fever. II. Studies in pathology. Am J Trop Med Hyg 17:413–421. [DOI] [PubMed] [Google Scholar]
- 6.Krylova RI. 1968. Pathologic anatomy of spontaneous hemorrhagic fever in monkeys, p 311–316. In Lagutina NI. (ed), Problems in physiology and experimental pathology. Proceedings of the Institute of Experimental Pathology and Therapy of the Academy of Medical Sciences of the USSR. Institute of Experimental Pathology and Therapy of the Academy of Medical Sciences of the USSR, Sukhumi, Georgia, Soviet Union (In Russian.) [Google Scholar]
- 7.Tauraso NM, Shelokov A, Allen AM, Palmer AE, Aulisio CG. 1968. Epizootic of simian haemorrhagic fever. Nature 218:876–877. doi: 10.1038/218876a0. [DOI] [Google Scholar]
- 8.España C. 1971. Review of some outbreaks of viral disease in captive nonhuman primates. Lab Anim Sci 21:1023–1031. [PubMed] [Google Scholar]
- 9.Tauraso NM, Aulisio CG, España CD, Wood OL, Liebhaber H. 1971. Simian hemorrhagic fever virus, p 208–215. In Martini GA, Siegert R (ed), Marburg virus disease. Springer-Verlag, Berlin, West Germany. [Google Scholar]
- 10.Tauraso NM, Myers MG, McCarty K, Tribe GW. 1970. Simian hemorrhagic fever, p 101–109. In Balner H, Beveridge WIB (ed), Infections and immunosuppression in subhuman primates. Munksgaard, Copenhagen, Denmark. [Google Scholar]
- 11.Shevtsova ZV, Krylova RI. 1971. A comparative study of 2 strains of simian hemorrhagic fever virus. Vopr Virusol 16:686–688. (In Russian.) [PubMed] [Google Scholar]
- 12.Gravell M, London WT, Rodriguez M, Palmer AE, Hamilton RS. 1980. Simian haemorrhagic fever (SHF): new virus isolate from a chronically infected patas monkey. J Gen Virol 51:99–106. doi: 10.1099/0022-1317-51-1-99. [DOI] [PubMed] [Google Scholar]
- 13.Lapin BA, Shevtsova ZV. 1971. On the identity of two simian hemorrhagic fever virus strains (Sukhumi and NIH). Z Versuchstierkd 13:21–23. [PubMed] [Google Scholar]
- 14.London WT. 1977. Epizootiology, transmission and approach to prevention of fatal simian haemorrhagic fever in rhesus monkeys. Nature 268:344–345. doi: 10.1038/268344a0. [DOI] [PubMed] [Google Scholar]
- 15.Shevtsova ZV, Pekerman SM, Krylova RI. 1977. Simian hemorrhagic fever: data from the Suhumi nursery, p 114–117. In Lapin BA. (ed), Modeling human pathological conditions, vol 2 Experimental studies of human diseases in monkeys. USSR Academy of Medical Sciences—Institute of Experimental Pathology and Therapy, Moscow, Soviet Union: (In Russian.) [Google Scholar]
- 16.Vatter HA, Donaldson EF, Huynh J, Rawlings S, Manoharan M, Legasse A, Planer S, Dickerson MF, Lewis AD, Colgin LM, Axthelm MK, Pecotte JK, Baric RS, Wong SW, Brinton MA. 2015. A simian hemorrhagic fever virus isolate from persistently infected baboons efficiently induces hemorrhagic fever disease in Japanese macaques. Virology 474:186–198. doi: 10.1016/j.virol.2014.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bailey AL, Lauck M, Weiler A, Sibley SD, Dinis JM, Bergman Z, Nelson CW, Correll M, Gleicher M, Hyeroba D, Tumukunde A, Weny G, Chapman C, Kuhn JH, Hughes AL, Friedrich TC, Goldberg TL, O'Connor DH. 2014. High genetic diversity and adaptive potential of two simian hemorrhagic fever viruses in a wild primate population. PLoS One 9:e90714. doi: 10.1371/journal.pone.0090714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lauck M, Hyeroba D, Tumukunde A, Weny G, Lank SM, Chapman CA, O'Connor DH, Friedrich TC, Goldberg TL. 2011. Novel, divergent simian hemorrhagic fever viruses in a wild Ugandan red colobus monkey discovered using direct pyrosequencing. PLoS One 6:e19056. doi: 10.1371/journal.pone.0019056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lauck M, Sibley SD, Hyeroba D, Tumukunde A, Weny G, Chapman CA, Ting N, Switzer WM, Kuhn JH, Friedrich TC, O'Connor DH, Goldberg TL. 2013. Exceptional simian hemorrhagic fever virus diversity in a wild African primate community. J Virol 87:688–691. doi: 10.1128/JVI.02433-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bailey AL, Lauck M, Sibley SD, Pecotte J, Rice K, Weny G, Tumukunde A, Hyeroba D, Greene J, Correll M, Gleicher M, Friedrich TC, Jahrling PB, Kuhn JH, Goldberg TL, Rogers J, O'Connor DH. 2014. Two novel simian arteriviruses in captive and wild baboons (Papio spp.). J Virol 88:13231–13239. doi: 10.1128/JVI.02203-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lauck M, Palacios G, Wiley MR, Lǐ Y, Fāng Y, Lackemeyer MG, Caì Y, Bailey AL, Postnikova E, Radoshitzky SR, Johnson RF, Alkhovsky SV, Deriabin PG, Friedrich TC, Goldberg TL, Jahrling PB, O'Connor DH, Kuhn JH. 2014. Genome sequences of simian hemorrhagic fever virus variant NIH LVR42-0/M6941 isolates (Arteriviridae: Arterivirus). Genome Announc 2:e00978-14. doi: 10.1128/genomeA.00978-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 23.Bao Y, Chetvernin V, Tatusova T. 2014. Improvements to pairwise sequence comparison (PASC): a genome-based web tool for virus classification. Arch Virol 159:3293–3304. doi: 10.1007/s00705-014-2197-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abascal F, Zardoya R, Telford MJ. 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res 38:W7–W13. doi: 10.1093/nar/gkq291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 73:152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swofford DL. 2003. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4.0b10. Sinauer Associates, Sunderland, MA. [Google Scholar]
- 28.Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 30.Lapin BA, Shevtsova ZV, Krylova RI. 1969. Experimental hemorrhagic fever of monkeys, p 196–203. In Hofer HO. (ed), Proceedings of the 2nd International Congress on Primatology, Atlanta, GA, 1968 Neurology, Physiology and Infectious Diseases, vol 3 S. Karger, Basel, Switzerland. [Google Scholar]
- 31.Shevtsova ZV. 1969. A further study of simian hemorrhagic fever virus. Vopr Virusol 14:604–607. (In Russian.) [PubMed] [Google Scholar]
- 32.Vatter HA, Di H, Donaldson EF, Baric RS, Brinton MA. 2014. Each of the eight simian hemorrhagic fever virus minor structural proteins is functionally important. Virology 462-463:351–362. doi: 10.1016/j.virol.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simpson DIH. 1972. Other virus diseases, p 702–714. In Fiennes RN. (ed), Pathology of simian primates, part II: infectious and parasitic diseases. S. Karger, Basel, Switzerland. [Google Scholar]
- 34.Lapin BA, Shevtsova ZV. 1992. Simian viral infections in the Sukumi monkey colony, p 417–424. In Matano S, Tuttle RH, Ishida H (ed), Topics in primatology, vol 3 University of Tokyo Press, Tokyo, Japan. [Google Scholar]
- 35.Shelokov A, Tauraso NM, Allen AM, España C. 1971. Epizootic, clinical, and pathological aspects of simian hemorrhagic fever, p 203–207. In Martini GA, Siegert R (ed), Marburg virus disease. Springer-Verlag, Berlin, West Germany. [Google Scholar]
- 36.Köhler H, Grünberg W. 1970. Febrile hemorrhagic diathesis in rhesus monkeys. Dtsch Tierarztl Wochenschr 77:83–88. (In German.) [PubMed] [Google Scholar]
- 37.Myers MG, Vincent MM, Hensen SA, Tauraso NM. 1972. Problems in the laboratory isolation of simian hemorrhagic fever viruses and isolation of the agent responsible for the Sussex-69 epizootic. Appl Microbiol 24:62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gravell M, London WT, Leon ME, Palmer AE, Hamilton RS. 1986. Differences among isolates of simian hemorrhagic fever (SHF) virus. Proc Soc Exp Biol Med 181:112–119. doi: 10.3181/00379727-181-42231. [DOI] [PubMed] [Google Scholar]
- 39.Madden DL, Fuccillo DA, Dorosz JA, London WT, Palmer AE, Castellano GA. 1978. Antigenic relationship of two strains of simian hemorrhagic fever virus. Lab Anim Sci 28:422–427. [PubMed] [Google Scholar]
- 40.Lapin BA, Krylova RI. 1980. Virus infections of monkeys in the Sukhumi colony, p 65–68. In Ippen R, Schröder H (ed), Erkrankungen der Zootiere—Verhandlungsbericht des Internationalen Symposiums über die Erkrankungen der Zootiere. Akademie Verlag, East Berlin, German Democratic Republic: (In Russian.) [Google Scholar]
- 41.Zack PM. 1993. Simian hemorrhagic fever, p 118–131. In Jones ITC, Mohr U, Hunt RD (ed), Nonhuman primates. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 42.Renquist D. 1990. Outbreak of simian hemorrhagic fever. J Med Primatol 19:77–79. [PubMed] [Google Scholar]
- 43.Plagemann PGW, Moennig V. 1992. Lactate dehydrogenase-elevating virus, equine arteritis virus, and simian hemorrhagic fever virus: a new group of positive-strand RNA viruses. Adv Virus Res 41:99–192. doi: 10.1016/S0065-3527(08)60036-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jahrling PB, Geisbert TW, Dalgard DW, Johnson ED, Ksiazek TG, Hall WC, Peters CJ. 1990. Preliminary report: isolation of Ebola virus from monkeys imported to U S A Lancet 335:502–505. [DOI] [PubMed] [Google Scholar]
- 45.Rollin PE, Williams RJ, Bressler DS, Pearson S, Cottingham M, Pucak G, Sanchez A, Trappier SG, Peters RL, Greer PW, Zaki S, Demarcus T, Hendricks K, Kelley M, Simpson D, Geisbert TW, Jahrling PB, Peters CJ, Ksiazek TG. 1999. Ebola (subtype Reston) virus among quarantined nonhuman primates recently imported from the Philippines to the United States. J Infect Dis 179:S108–S114. doi: 10.1086/514303. [DOI] [PubMed] [Google Scholar]