Abstract
Certain cells have the ability to block retroviral infection at specific stages of the viral cycle by the activities of well-characterized factors and transcriptional silencing machinery. Infection of murine stem cells (MSCs) by the murine leukemia viruses (MLVs) is profoundly blocked postintegration by transcriptional silencing. Here, we show that a dominant point of restriction of HIV-1 in human CD34+ cells is prior to integration of viral DNA and that HIV-1 restriction by human CD34+ cells is fundamentally different from MLV restriction by mouse cells.
TEXT
There is currently significant controversy regarding whether human hematopoietic stem cells (HSCs) can become infected with HIV-1; some but not all investigators have detected HIV-1 in human HSCs of infected patients (1, 2). Lentiviruses do successfully infect nondividing cells, but HIV-1 infection of human HSCs in culture is inefficient compared to that in activated T cells (3–5). A significant body of work has identified postintegration silencing as a major mechanism employed by murine stem cells (MSCs) to restrict retroviruses, but human HSCs may be different. The point in the retroviral replication cycle at which retroviruses are blocked in human HSCs remains unknown, and whether this involves a preintegration block or is due to transcriptional silencing of integrated proviruses has not been studied in detail (6–10).
This investigation evaluated infection of primary cord-derived human CD34+ cells, a mixed population composed of true human HSCs (Lin− 34+ 38− 90+ 45RAdim) as well as several hematopoietic progenitor cell (HPC) types. We identified a block occurring prior to viral DNA integration as a major factor in the nonpermissiveness of cord-derived primary CD34+ cells for HIV-1 infection.
Transduction of primary cord-derived CD34+ cells by lentiviral vectors is inefficient at low MOIs.
Primary human cord-derived CD34+ cells were used to study the efficiency of lentiviral transduction under various conditions. Each experimental replicate was performed independently with primary human CD34+ cells from different donors. CD34+ cells were isolated with magnetic beads from cord blood samples obtained from the Long Island Blood Service or obtained preisolated from AllCells (11, 12). CD34+ cells were put into culture in serum-free VIVO-X 20 medium. These cells were either cultured in the absence of cytokines or prestimulated for 24 h with various cytokines, singly or in combination, prior to exposure to preparations of pseudotyped lentiviral vectors. Vesicular stomatitis virus G (VSV-G)-pseudotyped virus with a modified pNL4.3 HIV-1-based core was prepared by cotransfecting 293T cells with a pNL4.3.mCherry.R−.E− HIV-1-based plasmid lacking env and vpr genes and having an mCherry open reading frame (ORF) instead of the nef gene, together with a VSV-G expression plasmid (13). We used a VSV-G envelope to bypass cell entry limitations and to look specifically at postentry events. Multiplicities of infection (MOIs) were calculated based on viral titers of mCherry transduction units, determined by infection of the permissive 293T cell line with serial dilutions of the virus preparation. The cells were preincubated for 24 h with no cytokines, Flt3 ligand (FLT3L) at 100 ng/ml, stem cell factor (SCF) at 100 ng/ml, thrombopoietin (TPO) at 100 ng/ml, or all three cytokines at 100 ng/ml. These cells were then exposed to pseudotyped virus by spinoculation at 37°C and 2,000 rpm for 60 min. Cells were then left in culture for 72 h at 37°C and evaluated using flow cytometry and PCR. All experiments were performed in triplicate, and controls were all done using nevirapine at 50 μg/ml when specified.
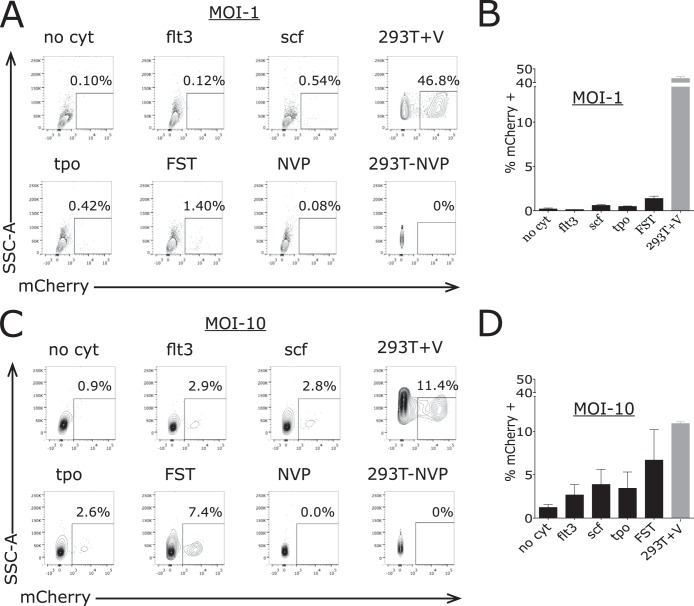
When CD34+ cells were incubated in the absence of cytokines and exposed to pseudotyped virus at an MOI of 1, only 0.2% ± 0.1% (mean ± standard error of the mean [SEM]) cells expressed mCherry (Fig. 1). When the cells were incubated with FLT3L, SCF, or TPO individually, less than 1% of the cells scored positive for mCherry; when cells were incubated with FLT3L, SCF, and TPO together, still only 1.4% ± 0.3% cells expressed mCherry. In contrast, the permissive 293T cells were efficiently transduced under these conditions, with more than 40% expressing mCherry (Fig. 1). These findings indicate a major block to transduction in primary cord-derived stem cells with or without cytokine stimulation.
FIG 1.
Variable percentages of CD34+ cells express mCherry after exposure to pseudotyped HIV-1 vector at a relative MOI of 1 in the absence of and with incubation with cytokines. Primary human CD34+ cells were incubated with no cytokines (no cyt), Flt3 ligand (flt3), stem cell factor (scf), thrombopoietin (tpo), all three cytokines (FST) or virus with nevirapine (NVP). 293T cells were similarly incubated with virus (293T+V) or virus plus NVP. (A) Cells were evaluated by flow cytometry for expression of mCherry after 72 h, and a representative example for each condition is shown for a viral MOI of 1. (B) Percent cells expressing mCherry. Values are means and error bars indicate SEM (n = 3) for samples incubated with a viral MOI of 1. (C) A representative example for CD34+ cells incubated under each condition is shown for a viral MOI of 10. (D) Percent cells expressing mCherry. Values are means and error bars indicate SEM (n = 3) for samples incubated with a viral MOI of 10.
Transduction of primary cord-derived CD34+ cells by lentiviral vectors is modestly increased by infection at high MOI.
Primary CD34+ cells were then exposed to pseudotyped virus at a calculated MOI of 10. At these very high concentrations of virus, we actually observed a decrease in the transduction efficiency of 293T cells relative to the experiments using an MOI of 1, with 11.1% ± 0.2% of cells expressing mCherry (Fig. 1). We consistently saw this decrease with 293T as well as HeLa cells. The basis of the reduction in percentage of 293T cells expressing mCherry is not clear, but it may reflect toxicity of the high concentration of the VSV-G glycoprotein being applied to the cells or a competition by defective virus particles with infectious virus at any limiting step of infection. It should be noted that a nominal MOI of 10, based on titer of virus in permissive cells, likely corresponds to the application of hundreds or thousands of physical virion particles per cell, due to the high particle-to-PFU ratio typical of retroviruses. Inspection of the cells at the time of scoring suggested some visible changes in morphology. We note that in spite of this small drop in efficiency, a very high portion of the 293T cells were successfully transduced in this setting.
Under these conditions of high multiplicity, we observed a significant increase in the transduction efficiency of CD34+ cells. Without cytokines, 1.2% ± 0.35% (mean ± SEM) cells expressed mCherry after 72 h. Addition of various cytokines led to significant increases in transduction above the MOI of 1 case (5- to 10-fold). Incubation with FLT3L led to 2.7% ± 1.2% cells being mCherry positive, incubation with SCF led to 3.9% ± 1.7% cells being mCherry positive, and incubation with TPO led to 3.4% ± 1.9% cells being mCherry positive (Fig. 1). Thus, exposure of the cells to high concentrations of virus could result in modest efficiencies of transduction when augmented by cytokine stimulation, particularly in the presence of a cytokine cocktail, as is typically done in gene therapy protocols.
Intracellular lentiviral DNA forms are profoundly reduced in primary cord-derived CD34+ cells exposed to pseudotyped virus at MOI of 1.
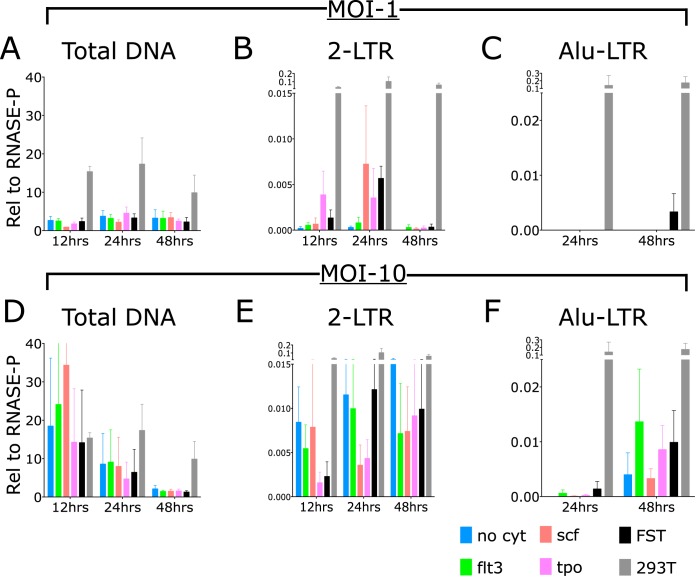
We evaluated the efficiency of various steps in the course of infection of cord-derived CD34+ cells relative to the efficiency in permissive 293T cells using established primer sequences and using the chromosomal RNase P gene as our reference (14). We looked specifically at the levels of total viral DNA, 2-long-terminal-repeat (LTR) circular DNA, and integrated DNA present at 12 h, 24 h, and 48 h (Fig. 2). The DNA copy numbers were calculated relative to the copy number of the RNase P gene. Without or with cytokines, infection of the CD34+ cells at an MOI of 1 produced 1 to 3 copies of total viral DNA relative to the single-copy gene. Infection of the permissive 293T cells under these conditions yielded 10 to 20 copies of total viral DNA per single-copy gene (Fig. 2A). Thus, while there was some reduction in efficiency of total viral DNA synthesis in CD34+ cells, the decrease was only to a level 3- to 10-fold below that in permissive cells. Tests of infections carried out in the presence of the reverse transcriptase (RT) inhibitor nevirapine (NVP) revealed only background levels of viral DNA, confirming that the DNA detected was due to true infection and was not attributable to contaminating plasmid DNA present in the virus preparations.
FIG 2.
Kinetics of production of total viral DNA, 2-LTR circles, and integrated DNAs in CD34+ cells after exposure to pseudotyped HIV-1 vector at MOIs of 1 and 10. Primary human CD34+ cells were incubated with no cytokines (no cyt), Flt3 ligand (flt3), stem cell factor (scf), thrombopoietin (tpo), or all three cytokines (FST). Cells were lysed at 12 h, 24 h, or 48 h, and DNA was extracted and analyzed by TaqMan PCR analysis for products relative to RNase P (n = 3 per condition and per time point). (A to C) DNA levels relative to RNase P. Values are means, and error bars indicate SEM. (D to F) Cells were analyzed at 12 h, 24 h, and 48 h after infection at an MOI of 10. Results are shown relative to RNase P; bars represent means, and error bars indicate SEM.
Analysis of circular viral DNAs containing the 2-LTR junction sequence revealed only very low levels of DNA in the CD34+ cells (Fig. 2). Without cytokines, or with Flt3 alone, the levels were in the range of 2 × 10−4 to 3 × 10−4 DNA copies compared to the single-copy standard. Incubation with stem cell factor, TPO, or the cocktail of three cytokines increased the copy numbers of 2-LTR circles to 3 × 10−3 to 6 × 10−3 copies—a significant increase, but still far below the levels in permissive cells. The levels in the permissive 293T cells were dramatically higher, in the range of 0.1 to 0.2 copy per single-copy standard. Thus, across the various conditions, there was a reduction of 2-LTR circular DNAs in the range of 30- to 100-fold compared to the permissive cells. This indicates a strong block in infection of CD34+ cells, likely at the stage of nuclear entry.
Analysis of the copy numbers of the integrated proviral DNA in the CD34+ cells revealed an even more profound reduction. We assessed integration using a TaqMan-based assay, scoring for linkage between host Alu repeats and the vector LTR sequences (referred to as Alu-LTR levels). We could not detect any amplified products in the CD34+ cells after 50 cycles of PCR. The permissive cells yielded approximately 0.1 to 0.2 DNA copies relative to the single-copy gene standard at 24 h and 48 h postinfection. Thus, although some viral DNA was synthesized in the CD34+ cells, very little entered the nucleus to form proviral DNAs.
Intracellular lentiviral DNA forms are also profoundly reduced in primary cord-derived CD34+ cells exposed to pseudotyped virus at an MOI of 10.
CD34+ cells and control permissive 293T cells were exposed to pseudotyped virus at an MOI of 10, with and without cytokines, and DNA was extracted at various times postinfection and analyzed by PCR as before. Infection at these high MOIs resulted in substantial increases in the level of total viral DNA over that seen at lower MOIs. The DNA copy number of total viral DNA was in the range of 10 to 30 copies relative to the chromosomal single-copy standard and was comparable or even higher than that in the permissive 293T cells (Fig. 2D). These high levels of viral DNA were seen without cytokines and were not significantly or consistently increased by the addition of Flt3, SCF, TPO, or all three together. The levels were highest at 12 h postinfection and fell gradually at 24 h and 48 h, more rapidly than was seen in the 293T cells, suggesting that the viral DNA was less stable in CD34+ cells. At an MOI of 1, these levels peaked at 24 h, suggesting differences in stability and production at different MOIs (Fig. 2D).
Assays of the infected CD34+ cells for 2-LTR circular DNA revealed a major decrease relative to that seen in the permissive 293T cells (Fig. 2E). While the copy number for total DNA relative to the single-copy standard was approximately 20 for both CD34+ cells and permissive 293T cells, the copy number for 2-LTR circular DNA was only 0.01 copy relative to the single-copy standard in CD34+ cells, as contrasted with the copy number near 0.1 in the permissive cells. Thus, CD34+ cells' circular DNA levels (2-LTR) were 2,000 times lower than linear total viral DNA levels, while in permissive 293T cells, circular DNA levels (2-LTR) were 200 times lower than linear total viral DNA levels. Therefore, there was a 10-fold decrease in formation of circular DNA seen in the CD34+ cells compared with permissive cells. The levels were not significantly increased by the addition of cytokines. The levels did not seem to fall over time, perhaps reflecting continuing synthesis and movement of DNA into the circular pool over the time course of the experiment. Given the high levels of total DNA and very low levels of circles formed in these infections, the data suggest a major block at the step of nuclear entry.
PCR assays for integrated proviral DNA confirmed and extended these findings. Here, only very low, barely detectable levels of Alu-LTR DNA products were detected in the CD34+ cells: roughly 0.001 copy relative to the single-copy standard at 24 h and 0.03 to 0.015 copy at 48 h postinfection. The levels of integrated DNA did seem to consistently increase with time, suggesting that integration may occur more slowly than in permissive cells. There was no consistent increase with cytokines. These numbers are in contrast to the levels in permissive cells: roughly 0.1 to 0.2 copy relative to the standard. Thus, the levels of integrated DNAs in CD34+ cells are about 10-fold lower than in permissive cells. This decrease is comparable to that seen in the circular DNA pool, suggesting that there is no specific block to integration once the DNAs have entered the nucleus; the major block is probably at nuclear entry, and the small amount that is able to enter the nucleus can continue to integrate.
The rare successfully transduced cord-derived primary CD34+ cells do contain integrated proviral DNAs.
To test the notion that there is truly a correlation between integration and expression, primary cord-derived CD34+ cells were incubated with Flt3 ligand (FLT3L), stem cell factor (SCF), and thrombopoietin (TPO) for 24 h prior to and during exposure to mCherry pseudotyped virus at an MOI of 10, as described above. On day 5, cells were sorted into those expressing and those not expressing mCherry by fluorescence-activated cell sorting (FACS). DNA was extracted from each population and then evaluated for integrated viral DNA copy number using Alu-LTR TaqMan assays, with results calculated relative to RNase P as before. CD34+ cells that expressed mCherry had an Alu-LTR DNA copy number of 0.4 ± 0.07 relative to the single-copy standard, while the CD34+ pool that was negative for expression of mCherry had 20-fold less Alu-LTR DNA, at 0.02 ± 0.01 copies (P < 0.009), showing a correlation between integration and expression.
In conclusion, these findings demonstrate that primary cord-derived human CD34+ cells, when exposed to lentivirus vector pseudotypes with or without cytokine stimulation, demonstrate significant restriction of retroviral infection. When we increased the multiplicity of infection, we still saw that only a minority of cells, in the range of 3 to 6%, expressed the mCherry reporter. The analysis of viral DNAs suggests that human CD34+ cell restriction occurs prior to integration.
Infection of primary human CD34+ cells at an MOI of 1 resulted in lower levels of total viral DNA products than that seen in 293T cells, but the decrease was only in the range of 3- to 10-fold. There was a much larger reduction in the levels of circular and integrated DNAs. Increasing the viral MOI to 10 resulted in a major increase in the levels of total viral DNA products, up to levels equal to or even above those seen in 293T cells (Fig. 2). However, the production of 2-LTR circles and integrated DNA was consistently found to be profoundly reduced in CD34+ cells, to levels below 1% of those seen in permissive 293T cells. Thus, even at high MOIs, formation of circles and proviral DNAs was very low in CD34+ cells. The differences between the modest 3- to 10-fold change in viral DNA products relative to 293T cells and the much larger reduction in production of 2-LTR circles and integrated DNA suggest that HIV-1 is restricted prior to integration in primary cord CD34+ cells.
A close examination of our data supports the idea that there may be multiple points of preintegration restriction. There is a significant block in viral DNA synthesis at low MOIs, and this may be due to low levels of deoxynucleoside triphosphate s (dNTPs) in the CD34+ cells. Restriction factors such as Trex1, which degrades RT products, SAMHD1, which depletes NTPs and may degrade viral RNA, and APOBEC3G, which deaminates cytidines and may block reverse transcription, could all contribute to this restriction. This block seems to be substantially overcome at high MOIs. This is a surprising finding, and it is not clear how this is achieved; one possibility is that the large amount of incoming virus saturates out a restriction factor, much as a high MOI can overcome the block to infection of mouse cells imposed by the Fv1 gene (15, 16). A more significant point of restriction occurs after viral DNA synthesis at the time of nuclear entry of the DNA, as evidenced by the low levels of 2-LTR circles relative to permissive 293T cells. There may be an additional minor restriction at the time of integration, though the proviral DNA levels are not always significantly reduced below that of the 2-LTR circles.
Ultimately, knowledge regarding how cells are able to restrict HIV-1 is fundamental to a better understanding of the role of human HSCs as a reservoir of HIV-1 and essential in progress toward a cure. Understanding the mechanism behind HSCs' ability to block HIV-1 may potentially be exploited in protecting human HSCs, HPCs, and other cells from HIV-1 infection.
ACKNOWLEDGMENTS
We thank Yosef Sabo for providing the mCherry expressing lentiviral vector.
This work was supported by NIH grant R01 AI 106629 from the National Institute of Allergy and Infectious Diseases. S.P.G. is an Investigator of the Howard Hughes Medical Institute. D.O.G. was supported by the Donald and Barbara Zucker Family Foundation.
We have no conflicting financial interests to declare.
REFERENCES
- 1.Carter CC, Onafuwa-Nuga A, McNamara LA, Riddell J IV, Bixby D, Savona MR, Collins KL. 2010. HIV-1 infects multipotent progenitor cells causing cell death and establishing latent cellular reservoirs. Nat Med 16:446–451. doi: 10.1038/nm.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Josefsson L, Eriksson S, Sinclair E, Ho T, Killian M, Epling L, Shao W, Lewis B, Bacchetti P, Loeb L, Custer J, Poole L, Hecht FM, Palmer S. 2012. Hematopoietic precursor cells isolated from patients on long-term suppressive HIV therapy did not contain HIV-1 DNA. J Infect Dis 206:28–34. doi: 10.1093/infdis/jis301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang J, Scadden DT, Crumpacker CS. 2007. Primitive hematopoietic cells resist HIV-1 infection via p21. J Clin Invest 117:473–481. doi: 10.1172/JCI28971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Logan AC, Lutzko C, Kohn DB. 2002. Advances in lentiviral vector design for gene-modification of hematopoietic stem cells. Curr Opin Biotechnol 13:429–436. doi: 10.1016/S0958-1669(02)00346-4. [DOI] [PubMed] [Google Scholar]
- 5.Scherr M, Eder M. 2002. Gene transfer into hematopoietic stem cells using lentiviral vectors. Curr Gene Ther 2:45–55. doi: 10.2174/1566523023348237. [DOI] [PubMed] [Google Scholar]
- 6.Wolf D, Goff SP. 2009. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458:1201–1204. doi: 10.1038/nature07844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolf D, Cammas F, Losson R, Goff SP. 2008. Primer binding site-dependent restriction of murine leukemia virus requires HP1 binding by TRIM28. J Virol 82:4675–4679. doi: 10.1128/JVI.02445-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolf D, Goff SP. 2008. Host restriction factors blocking retroviral replication. Annu Rev Genet 42:143–163. doi: 10.1146/annurev.genet.42.110807.091704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolf D, Hug K, Goff SP. 2008. TRIM28 mediates primer binding site-targeted silencing of Lys1,2 tRNA-utilizing retroviruses in embryonic cells. Proc Natl Acad Sci U S A 105:12521–12526. doi: 10.1073/pnas.0805540105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schlesinger S, Goff SP. 2013. Silencing of proviruses in embryonic cells: efficiency, stability and chromatin modifications. EMBO Rep 14:73–79. doi: 10.1038/embor.2012.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffin DO, Rothstein TL. 2012. Human B1 cell frequency: isolation and analysis of human B1 cells. Front Immunol 3:122. doi: 10.3389/fimmu.2012.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffin DO, Holodick NE, Rothstein TL. 2011. Human B1 cells in umbilical cord and adult peripheral blood express the novel phenotype CD20+ CD27+ CD43+ CD70. J Exp Med 208:67–80. doi: 10.1084/jem.20101499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sabo Y, Walsh D, Barry DS, Tinaztepe S, de Los Santos K, Goff SP, Gundersen GG, Naghavi MH. 2013. HIV-1 induces the formation of stable microtubules to enhance early infection. Cell Host Microbe 14:535–546. doi: 10.1016/j.chom.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butler SL, Hansen MS, Bushman FD. 2001. A quantitative assay for HIV DNA integration in vivo. Nat Med 7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- 15.Pincus T, Hartley JW, Rowe WP. 1971. A major genetic locus affecting resistance to infection with murine leukemia viruses. I. Tissue culture studies of naturally occurring viruses. J Exp Med 133:1219–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jolicoeur P, Baltimore D. 1975. Effect of the Fv-1 locus on the titration of murine leukemia viruses. J Virol 16:1593–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]