ABSTRACT
Influenza infection causes severe disease and death in humans. In traditional vaccine research and development, a single high-dose virus challenge of animals is used to evaluate vaccine efficacy. This type of challenge model may have limitations. In the present study, we developed a novel challenge model by infecting mice repeatedly in short intervals with low doses of influenza A virus. Our results show that compared to a single high-dose infection, mice that received repeated low-dose challenges showed earlier morbidity and mortality and more severe disease. They developed higher vial loads, more severe lung pathology, and greater inflammatory responses and generated only limited influenza A virus-specific B and T cell responses. A commercial trivalent influenza vaccine protected mice against a single high and lethal dose of influenza A virus but was ineffective against repeated low-dose virus challenges. Overall, our data show that the repeated low-dose influenza A virus infection mouse model is more stringent and may thus be more suitable to select for highly efficacious influenza vaccines.
IMPORTANCE Influenza epidemics and pandemics pose serious threats to public health. Animal models are crucial for evaluating the efficacy of influenza vaccines. Traditional models based on a single high-dose virus challenge may have limitations. Here, we describe a new mouse model based on repeated low-dose influenza A virus challenges given within a short period. Repeated low-dose challenges caused more severe disease in mice, associated with higher viral loads and increased lung inflammation and reduced influenza A virus-specific B and T cell responses. A commercial influenza vaccine that was shown to protect mice from high-dose challenge was ineffective against repeated low-dose challenges. Overall, our results show that the low-dose repeated-challenge model is more stringent and may therefore be better suited for preclinical vaccine efficacy studies.
INTRODUCTION
Influenza viruses, through annual outbreaks and occasional pandemics, pose a significant threat to public health. Each year, influenza causes the hospitalization of millions of people and is linked to ∼250,000 to 500,000 deaths worldwide (1). Influenza virus infection causes acute respiratory disease in humans and the sudden onset several symptoms, such as high fever, coryza, cough, headache, prostration, malaise, and inflammation of the upper respiratory tree and trachea, which can progress to pneumonia (2–4).
Vaccines can prevent influenza virus infections (5, 6). They are relatively ineffective at protecting highly vulnerable populations such as immunocompromised or aged individuals. They also perform poorly in years when the vaccine strains are mismatched to the circulating strains. A universal vaccine against all subtypes and strains of influenza virus would provide broader protection, but such constructs are not yet commercially available (7–10).
Novel vaccines, prior to their testing in humans, are evaluated in experimental animal models, which have limitations, as they incompletely mirror human infections and disease progression (11, 12). In humans, influenza virus replication reaches a peak at ∼48 h after infection in both the upper and lower respiratory tracts and then decreases slowly; virus shedding declines by nearly 1 week after infection (2). The virus is transmitted mainly through airborne droplets and direct contact of virus with mucosa surfaces. Intriguingly, there is evidence that aerosol transmission of influenza viruses in a low infectious dose may result in more severe disease (3, 4).
Traditionally, influenza vaccines have been evaluated preclinically in animals that upon vaccination are challenged with a single high dose of virus (8, 10, 13). This procedure uses more virus than is typically transmitted in natural infections. We therefore developed a model of repeated low-dose influenza virus challenge to more closely mimic viral doses transmitted during natural infections of humans. Such models of repeated low-dose infection are already being used for vaccines for other viruses, such as human immunodeficiency virus type 1 (HIV-1)/simian immunodeficiency virus (SIV) (14–16) and hepatitis B virus (HBV) (17). As our results show, this new animal challenge model provides a more stringent platform for influenza vaccine evaluation. Our results show that mice that received repeated low-dose challenges showed earlier morbidity and mortality and more severe disease than with a single high-dose infection. These mice developed higher vial loads and more serious lung pathology. In addition, they had greater inflammasome responses and developed only limited influenza A virus-specific B and T cell responses. A commercial trivalent influenza vaccine (TIV) protected mice against a single high dose of influenza A virus but was ineffective against repeated low-dose virus challenges.
MATERIALS AND METHODS
Ethics statement.
All animal procedures in this study were performed in strict accordance with the regulations in the guide for the care and use of laboratory animals by the Ministry of Science and Technology of the People's Republic of China (http://www.most.gov.cn/fggw/zfwj/zfwj2006/200609/t20060930_54389.htm). The protocol was approved by the Institutional Animal Care and Use Committee of the Institut Pasteur of Shanghai, Chinese Academy of Science (permit number A2012001).
Influenza A virus and commercial flu vaccine.
A/Puerto Rico/8/1934 H1N1 (A/PR/8) influenza virus was stored in our laboratory. Reassortant pandemic H1N1 (pdm H1N1) virus was generated with the surface glycoproteins hemagglutinin (HA) and neuraminidase (NA) from California/7/2009 H1N1 virus and the other six internal genes from A/PR/8 by using an eight-plasmid reverse-genetics system. The viruses were grown in the chorioallantoic fluid of 9-day-old specific-pathogen-free (SPF) embryonated chicken eggs (Merial Vital Laboratory Animal Technology Co., Ltd., Beijing, China) and titrated in 6- to 8-week-old mice to determine the median lethal dose (LD50) upon intranasal (i.n.) infection.
A trivalent influenza vaccine (Fluarix) was purchased from GlaxoSmithKline (GSK). This inactivated split influenza vaccine contains three different strains: A/Texas/50/2012, A/Christchurch/16/2010, and B/Massachusetts/02/2012.
Mice.
Six- to eight-week-old female C57BL/6 mice were purchased from Shanghai Laboratory Animal Center, China. All mice were housed at the Institut Pasteur of Shanghai Animal Facility.
Virus infection, vaccine immunization, and measurement of weight loss as well as survival.
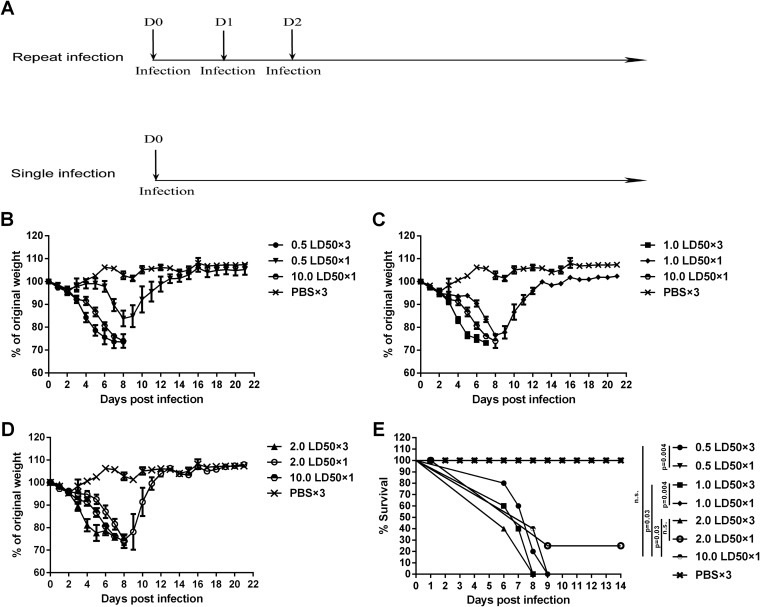
Groups of mice were anesthetized with 0.5% (wt/vol) pentobarbital sodium dissolved in phosphate-buffered saline (PBS) by intraperitoneal injection (18 μl/g body weight). Anesthetized mice were infected intranasally with 30 μl influenza A/PR/8 virus diluted in PBS at doses of 0.5 LD50, 1.0 LD50, and 2.0 LD50 once or three times on three consecutive days or at a dose of 10.0 LD50 once (Fig. 1A). Mice in the control group were anesthetized and inoculated with PBS on three consecutive days (Fig. 1A). Animals were weighed once daily for 3 weeks. Mice with a weight loss of >30% of their initial body weight were euthanized and recorded as dead.
FIG 1.
Repeated low-dose influenza virus infection causes earlier weight loss and mortality. (A) Virus infection experiment schedule. C57BL/6 mice were intranasally infected with 0.5, 1.0, and 2.0 LD50 of A/PR/8 virus once or three times or with 10.0 LD50 of virus once. Control mice were inoculated with PBS three times. (B to D) Weight loss of mice infected with different doses of A/PR/8 virus once or repeatedly. The weight of each mouse was monitored daily until day 21 postinfection. Data are shown as means ± standard errors of the means. (B) Weight loss of the 0.5 LD50 single- and repeated-infection groups, the 10.0 LD50 single-infection group, and the PBS group. (C) Weight loss of the 1.0 LD50 single- and repeated-infection groups, the 10.0 LD50 single-infection group, and the PBS group. (D) Weight loss of the 2.0 LD50 single- and repeated-infection groups, the 10.0 LD50 single-infection group, and the PBS group. (E) All infected mice were monitored daily for survival until day 14 postinfection. Each graph represents the combined results from 3 separate experiments, each with 6 mice per group per experiment.
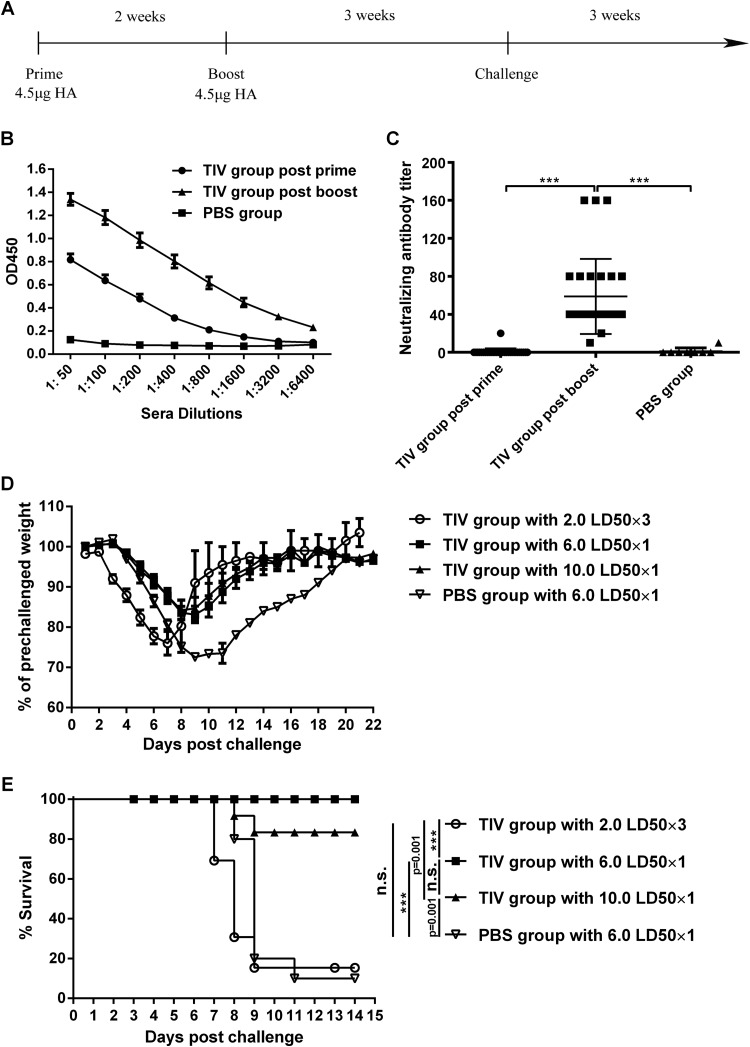
In the vaccination experiment, 6- to 8-week-old female C57BL/6J mice were immunized intramuscularly with the inactivated split influenza vaccine (Fluarix) with a dose of 1.5 μg HA per strain (total, 4.5 μg HA) twice within a 2-week interval, and control mice were injected with PBS (see Fig. 6A). Three weeks after the last immunization, vaccinated groups (TIV groups) of mice were anesthetized with pentobarbital sodium and challenged with pdm H1N1 virus by intranasal inoculation at doses of 2.0 LD50 three times at 24-h intervals, 6.0 LD50 once, and 10.0 LD50 once (see Fig. 6A). Mice in the control group were anesthetized and challenged with 6.0 LD50 pdm H1N1 virus once intranasally (see Fig. 6A). Survival and body weight were monitored daily after challenge for 21 days, and mice were euthanized when they lost in excess of 30% of their prechallenge body weight.
FIG 6.
TIV cannot protect mice from repeated low-dose infection with influenza virus. (A) Immunization and challenge schedule. Mice (n = 12 to 13) were immunized intramuscularly with an inactivated split influenza vaccine (Fluarix; GSK) twice at a 2-week interval. Three weeks after the boost, mice were challenged with repeated low-dose infection and single high-dose infection with pandemic H1N1 virus. PBS-injected mice (n = 10) were used as controls. (B) Antibody titers of postprime and postboost sera were measured by an ELISA with serial serum dilutions. Sera of the PBS group served as negative controls. Data are presented as means ± standard errors of the means. OD450, optical density at 450 nm. (C) Sera of all mice from the vaccine groups and the control group were collected 2 weeks after the prime and boost. Neutralizing antibody titers in MDCK cells were determined by a microneutralization assay. Data are presented as means ± standard errors of the means. (D) Weight loss of all mice was monitored every day for 21 days. Data are presented as means ± standard errors of the means. (E) Survival of all mice was monitored for 2 weeks after challenge. ***, P < 0.0001; n.s., no significance.
All infection work was performed in the biosafety level 2 facility of the Institut Pasteur of Shanghai and approved by the Biosafety Committee of the institute.
Viral load measurements.
The viral load measurement assay was adopted from a previously reported method (10). Lung tissue samples were harvested from mice 5 days after the first infection, and the weights of tissues were recorded. Lung tissue samples were mechanically homogenized by using a Precellys 24 instrument (Bertin Technologies, France), and RNA was isolated by using TRIzol reagent (Invitrogen, Carlsbad, CA). The RNA concentration of each sample was determined spectrophotometrically at an absorbance of 260 nm. cDNA was obtained from 1 μg of total RNA by using the Transcriptor First Strand cDNA synthesis kit (Roche). Reactions were run on a thermal cycler (Mastercycler; Eppendorf, Hamburg, Germany) with one cycle at 65°C for 10 min, 25°C for 10 min, 50°C for 60 min, and 85°C for 5 min. The M genes of the A/PR/8 viruses were cloned into the pMD18-T vector, which was used to create a standard curve by 10-fold serial dilution. Viral cDNA was quantified by using a SYBR green real-time PCR assay on a 7900HT real-time PCR system (Applied Biosystems, USA). A SYBR Premix Ex Taq kit (TaKaRa, Dalian, China) was used for real-time PCR, according to the manufacturer's instructions. The primers for viral cDNA quantification were specific for the influenza A virus matrix protein (MP) gene, which were MP sense primer 5′-AAG ACC AAT CCT GTC ACC TCT GA-3′ and MP antisense primer 5′-CAA AGC GTC TAC GCT GCA GTC C-3′. The cDNA samples from each individual were quantified in triplicate. Real-time PCR was performed with a 20-μl solution containing 10 μl SYBR Premix Ex Taq (Tli RNase H Plus) (2×), 0.4 μl PCR forward primer (10 μM), 0.4 μl PCR reverse primer (10 μM), 0.4 μl ROX reference dye II (50×), 2 μl cDNA, and 6.8 μl double-distilled water (ddH2O). The real-time PCR was performed according to the following protocol: an initial denaturation step at 95°C for 30 s followed by 40 cycles at 95°C for 5 s and 60°C for 30 s and a dissociation stage at 60°C for 1 min and 95°C for 15 s. All of the samples were analyzed in triplicate for each reaction. The data were analyzed by using the 7900HT SDS version 2.4 system (Applied Biosystems, Foster City, CA, USA). For analysis of the spectral curves, the cycle threshold was defined just above the emission baseline to stay within the exponential amplification phase of the PCR. Viral copy numbers were normalized to the original tissue sample masses and calculated based on the standard curve described above.
To determine the virus titer in lung tissues of infected mice by a 50% tissue culture infective dose (TCID50) assay, lungs from 4 mice per group were aseptically extracted at days 3 and 5 after the first infection and homogenized in virus growth medium (VGM) (10% wt/vol) in which the Dulbecco modified Eagle medium (DMEM) contains an antibiotic-antimycotic (HyClone; Thermo Scientific, USA) as well as 1% bovine serum albumin (BSA). Tenfold serial dilutions of the sample were added in quadruplicate to Madin-Darby canine kidney (MDCK) cells seeded in microwell plates 1 day earlier and allowed to absorb for 2 h at 37°C in an incubator. Fresh VGM was then added to the cells, and the cells were incubated at 37°C for another 48 h. The viral titer was determined by a hemagglutination inhibition assay. Briefly, the culture supernatants were mixed with the same volume of 1% (vol/vol) SPF chicken red blood cells (RBCs) (in PBS) and incubated for 15 to 20 min at room temperature. The virus titers were calculated by the Reed-Muench method (48) and expressed as the log10 TCID50 per milliliter of lung tissue.
Histology.
Mice were euthanized 5 days after the first infection. The lung tissues were perfused and washed with PBS containing 1% BSA. The lungs were then gently inflated with a buffered formaldehyde solution (pH 7.4), fixed for 24 h at 4°C in the buffered formaldehyde solution, dehydrated in a series of graded ethanol, and embedded in paraffin. Sections (5 μm) were cut and stained with hematoxylin and eosin, and two random sections of each lung sample were examined. Histopathological changes were scored by an investigator “blind” to sample identity as follows: a score of 1 indicates no pathology, 2 indicates perivascular infiltrates, 3 indicates perivascular and interstitial infiltrates affecting <20% of the lobe section, 4 indicates perivascular and interstitial infiltrates affecting 20 to 50% of the lobe section, and 5 indicates perivascular and interstitial infiltrates affecting >50% of the lobe section.
Microneutralization assay.
A microneutralization (MN) assay was performed as described previously (18), with some modifications. Briefly, MDCK cells were seed into microwell plates at 25,000 cells/well in complete DMEM (Invitrogen Life Technologies) supplemented with 10% fetal bovine serum (FBS; Invitrogen Life Technologies), 2 mM l-glutamine, 1 mM sodium pyruvate, penicillin (100 U/ml), and streptomycin (100 μg/ml) and cultured at 37°C overnight. On the second day, mouse sera were serially diluted in DMEM supplemented with 1% BSA and antibiotics (HyClone; Thermo Scientific, USA) and mixed with influenza virus A/PR/8 or pdm H1N1. After 1 h of incubation at room temperature, virus-serum mixtures were added to MDCK cells prewashed with PBS and incubated at 37°C for 2 days. The culture supernatants were then cultured with same volume of 1% (vol/vol) chicken RBCs (in PBS) and incubated for 15 to 20 min at room temperature. The neutralizing titer was the highest dilution of serum that inhibited virus-induced hemagglutination.
ELISA for detection of vaccine-specific antibody in sera.
Sera were collected 2 weeks after prime and boost. They were tested on plates coated with the trivalent inactivated influenza vaccine. Sera from the PBS-infected mouse group were used as a control. Briefly, wells of a flat-bottom enzyme-linked immunosorbent assay (ELISA) plate (Costar, NY, USA) were coated overnight at 4°C with the trivalent inactivated influenza vaccine diluted in 50 μl of coating buffer (0.05 M sodium bicarbonate buffer, pH 8.6). The plates were then washed 3 times with PBST (PBS with 0.1% Tween 20) and blocked with 100 μl 5% skim milk at 37°C. Two hours later, plates were washed with PBST 3 times, and 100 μl of serially diluted sera (1:50 to 1:6,400) was added to the plates and incubated at 37°C for 2 h. Plates were washed, a 1:10,000 dilution of horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Sigma-Aldrich, MO, USA) was then added to the plates, and the plates were incubated for 1 h at 37°C. After the final wash, 50 μl 3,3′,5,5′-tetramethylbenzidine (TMB) (eBioscience, CA, USA) was added to the plates for 5 min. The reaction was stopped by adding 50 μl 2 M H3PO4 to each well. The plates were read at 450 nm by using a Varioskan Flash multimode reader (Thermo Scientific, USA). All samples were tested in triplicate.
Tetramer staining and FACS analysis.
Virus-specific CD8+ T cell responses were evaluated by using nucleoprotein (NP)-specific tetramer staining and flow cytometry, as previously described (19). Lymphocytes were isolated from the blood of each mouse on day 7 after the first infection. Cells were stained with an allophycocyanin-labeled major histocompatibility complex class I (MHC-I) NP peptide tetramer (ASNENTETM) (Tetramer Core Facility, Emory University, GA, USA) and with an anti-CD8a-peridinin chlorophyll protein (PerCP)-Cy5.5 antibody (BD Biosciences, CA, USA) for 45 min at 4°C. Flow cytometry was performed on a BD LSRII instrument (BD Biosciences, CA, USA), and fluorescence-activated cell sorter (FACS) data were analyzed by using FlowJo software (TreeStar, Ashland, OR, USA).
Analysis of inflammasome responses.
Inflammasome responses were determined by detection of interleukin-1β (IL-1β) and IL-18 production in both bronchoalveolar lavage fluid (BALF) and lung homogenates using ELISA kits purchased from eBioscience (San Diego, CA, USA), according to the manufacturer's instructions. Lung homogenates and BALF were collected 5 days after the first infection, as previously described (20).
Statistical analysis.
Statistical significance between groups was analyzed by using one-way analysis of variance (ANOVA). A Kaplan-Meier curve was used for analysis of survival rates, and statistical significance was determined by chi-square tests. In all statistical analyses, P values of <0.05 were considered statistically significant. GraphPad Prism v6.0 software (GraphPad, San Diego, CA, USA) was used for statistical analysis.
RESULTS
Repeated low-dose infection with influenza virus causes earlier morbidity and mortality in mice.
Groups of 6- to 8-week-old female C57BL/6 mice were infected intranasally (i.n.) with A/Puerto Rico/8/1934 H1N1 (A/PR/8) influenza virus at low doses (0.5, 1.0, and 2.0 LD50) once or three times at 24-h intervals. Another group of mice was inoculated once i.n. with a high dose of A/PR/8 virus (10.0 LD50) (Fig. 1A). PBS-inoculated mice served as negative controls. The groups that received repeated low-dose challenge began to lose weight by day 3 after infection, and most died by days 7 to 8 (Fig. 1B to D). Mice that received a single low dose of virus lost weight later, and most of them survived (Fig. 1B to D). As expected, mice that received high doses of A/PR/8 virus (10.0 LD50) lost weight although with a delay compared to the group that received a repeated low-dose challenge; they then died within the same time frame. The weight loss of the group that received repeated infection with 0.5 LD50 (0.5 LD50 repeated-infection group) was significantly different from that of the matched-dose single-infection group from days 4 to 7 (P < 0.0001 at days 4 to 6 and P = 0.0006 at day 7) (Fig. 1B), and the same was true for the 1.0 LD50 repeated-infection group compared to the matched-dose, single-infection group (P = 0.0001 at day 4, P < 0.0001 at days 5 to 6, and P = 0.0014 at day 7) (Fig. 1C), while the weight loss of the 2.0 LD50 repeated-infection group was significantly different from that of the matched single-dose infection group only at day 4 (P = 0.0003) and day 5 (P = 0.0014) but not at day 6 or 7 (Fig. 1D). Compared with the 10.0 LD50 single-infection group, the weight loss of the 0.5 LD50 repeated-infection group was significantly different at day 4 (P = 0.0418) and day 5 (P = 0.0401); weight loss of the 1.0 LD50 repeated-infection group was significantly different at days 4 to 6 (P = 0.0013 at day 4, P = 0.0001 at day 5, and P = 0.0478 at day 6), and weight loss of the 2.0 LD50 repeated-infection group was significantly different only at day 4 (P = 0.0044) (Fig. 1B to D). Differences in survival between the 0.5 LD50 and 1.0 LD50 repeated-infection groups compared to those that received a matched single dose were significant, while survival rates were not different between 2.0 LD50 single- and repeated-infection groups (Fig. 1E). Compared to the high-dose (10.0 LD50) single-infection group, survival of the 1.0 LD50 and 2.0 LD50 but not the 0.5 LD50 repeated-infection groups was significantly different (Fig. 1E). Overall, these data show that repeated infection with low doses of influenza virus causes significant morbidity and mortality in mice.
Groups with repeated low-dose virus infection have higher viral loads and more severe pathological changes.
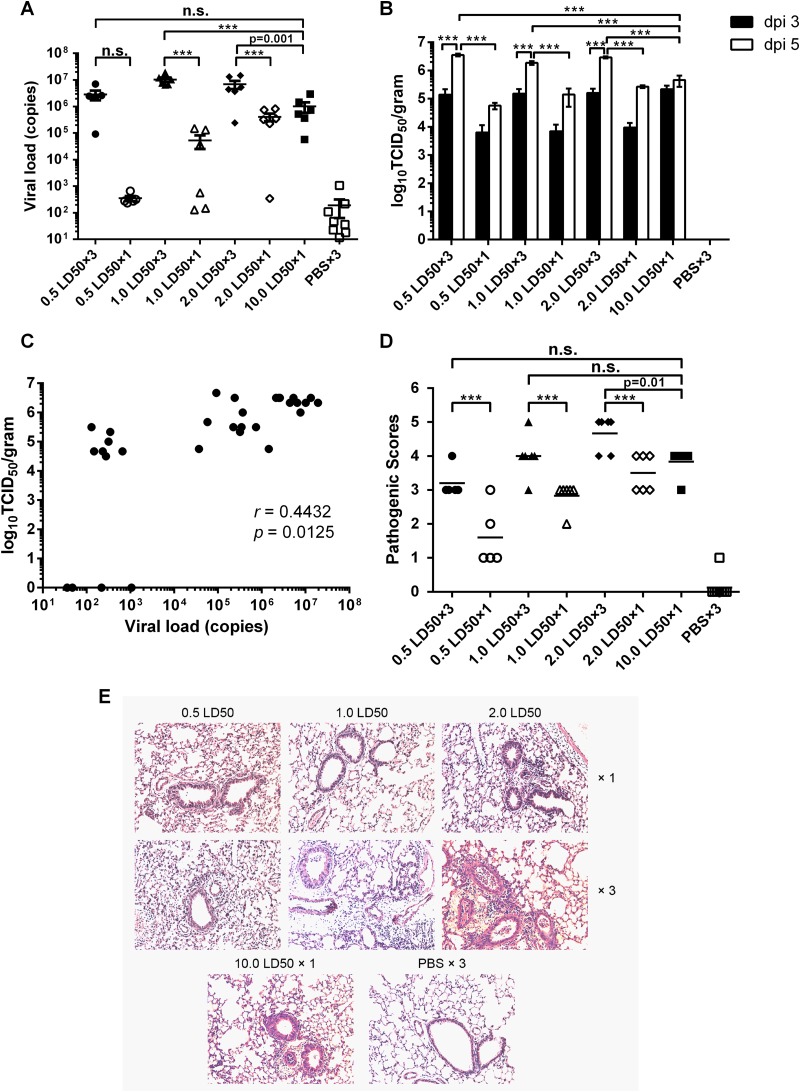
Viral loads were measured by real-time reverse transcription-PCR (RT-PCR) of lung homogenates from mice in the same groups described above 5 days after the first virus inoculation as well as by determination of the TCID50 in lung homogenates at days 3 and 5 after the first infection. As shown in Fig. 2A, lung virus titers in mice that had been repeatedly challenged with 1.0 or 2.0 LD50 of A/PR/8 virus were significantly higher than those in mice that had received the same doses once or the highest dose of 10.0 LD50 of A/PR/8 virus. The viral titers in the 0.5 LD50 repeated-infection group, however, were not significantly different from those in the matched-dose single-infection group or the high-dose (10.0 LD50) single-infection group (Fig. 2A). TCID50 assays showed that viral titers in the repeated-infection groups at day 5 were significantly higher than those at day 3 and also much higher than those of either the group infected with matched doses or the 10.0 LD50 single-infection group at day 5 (Fig. 2B). Vial titers of single-infection groups showed no significant difference between days 3 and 5, and viral titers at day 3 showed no significant difference among infection groups (Fig. 2B). Further analysis showed that the viral genome copy numbers and TCID50 titers at day 5 in lung tissues had a positive correlation (r = 0.4432; P = 0.0125) (Fig. 2C). Lung lobes harvested at day 5 after the first infection were stained with hematoxylin and eosin and scored for pathology (Fig. 2D and E). The low-dose repeated-infection groups had significantly higher pathological sores than did the groups that received the matching virus dose once (Fig. 2D). Mice injected repeatedly with 2.0 LD50 of virus also had more severe pathology (average score of 4.67) than did those that were inoculated once with 10.0 LD50 of virus, with a mean score of 3.83 (P = 0.01) (Fig. 2D). Histological analysis revealed that the low-dose repeated-infection groups with higher viral titers and higher pathology scores had more severe perivascular infiltrates and interstitial infiltrates in lung tissues than did the low-dose single-infection groups (Fig. 2E). The 10.0 LD50 single-infection group showed similar pathology changes, while the mice from the PBS group had no sign of inflammation (Fig. 2E). These results demonstrate that mice in the low-dose repeated-infection groups develop higher vial titers in lung tissues 5 days after the first infection, with more severe virus-associated pathology.
FIG 2.
Mice with repeated low-dose influenza virus infection have higher virus titers in lung tissues and more severe lung pathology. (A) Five days after infection, lung virus titers were determined by quantitative PCR. The graph shows titers of viral genomes (copies) per microgram of total RNA in lung tissue of individual mice (n = 6 to 8) in each group, with mean values ± standard errors of the means. (B) Viral titers lung tissues at days 3 and 5 after the first infection were determined by a TCID50 assay. Data are shown as mean values ± standard errors of the means. dpi, days postinfection. (C) Correlation between viral genome copy numbers and TCID50 titers in lung tissues on day 5 after the first infection. (D) Histological scoring for virus-infected mice (n = 6 to 8). Mean values for each group are shown. ***, P < 0.0001; n.s., no significance. (E) Representative hematoxylin- and eosin-stained sections were derived from mice (n = 6 to 8) from each group killed on day 5 after the first infection. ×1, mice were infected once; ×3, mice were infected three times. Original magnification, ×100.
Antibody responses.
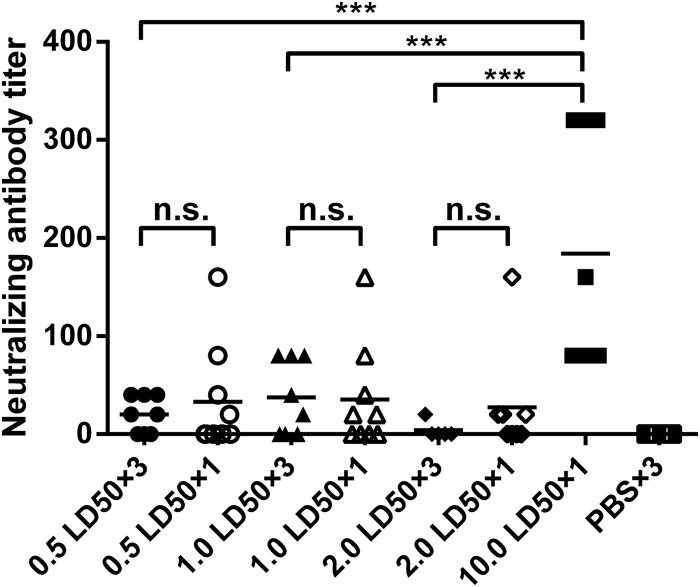
To evaluate humoral immune responses to influenza virus infection, mice of all groups were bled 1 week after infection. Sera were tested for influenza A virus-specific antibodies by microneutralization assays. As shown in Fig. 3, only the 10.0 LD50 single-infection group had significantly higher neutralizing titers (ranging from 1:80 to 1:320) than those of the low-dose repeated-infection groups (<1:80). In the low-dose single-infection groups, some mice had neutralizing titers of >1:80, indicating that a single low-dose virus infection could elicit neutralizing antibody responses to some extent. These data show that repeated low-dose infection in most mice failed to induce antibody responses within 7 days, while a single infection with low or higher virus doses elicited only very limited neutralizing antibody responses.
FIG 3.
Antibody responses in mice after virus infection. Mice (n = 8 to 10) from each group were bled at day 7 after the first infection, and sera were collected to determine antibody responses by a microneutralization assay. Graphs show titers of individual mice in each group, with mean values indicated by lines. Sera from PBS-inoculated mice served as negative controls. ***, P < 0.0001; n.s., no significance.
Virus-specific CD8+ T cell response.
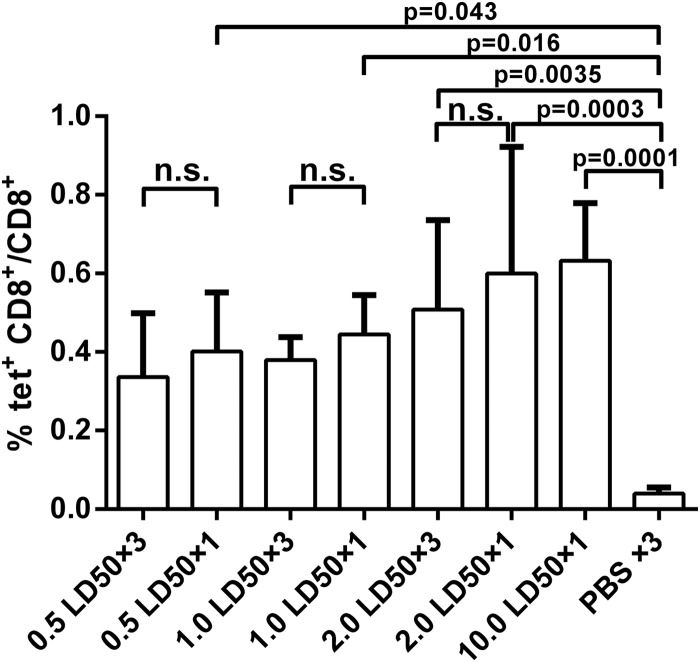
Mouse peripheral blood mononuclear cells (PBMCs) were collected 7 days after challenge, and virus-specific CD8+ T cell responses were analyzed by staining with an NP-specific MHC-I tetramer. As shown in Fig. 4, all mice developed virus-specific CD8+ T cell responses after virus infection, and there were no significant differences among the infection groups. Compared to the PBS-treated group, however, the frequencies of NP tetramer-specific CD8+ T cells in all single-infection groups as well as the 2.0 LD50 repeated -infection group were significantly higher (Fig. 4). These data demonstrate that virus-specific CD8+ T cell responses were induced after virus infection. The magnitude of T cell responses, however, did not correlate with virus dose or the number of infections.
FIG 4.
Virus-specific CD8+ T cell responses in mice after virus infection. Seven days after the first infection, PBMCs were isolated, and tetramer staining was performed to analyze the CD8+ T cell responses to the immunodominant epitope of NP in mice (n = 8 to 10). PBS-inoculated mice (n = 8) served as negative controls. tet+, tetramer positive.
Inflammasome responses in lungs.
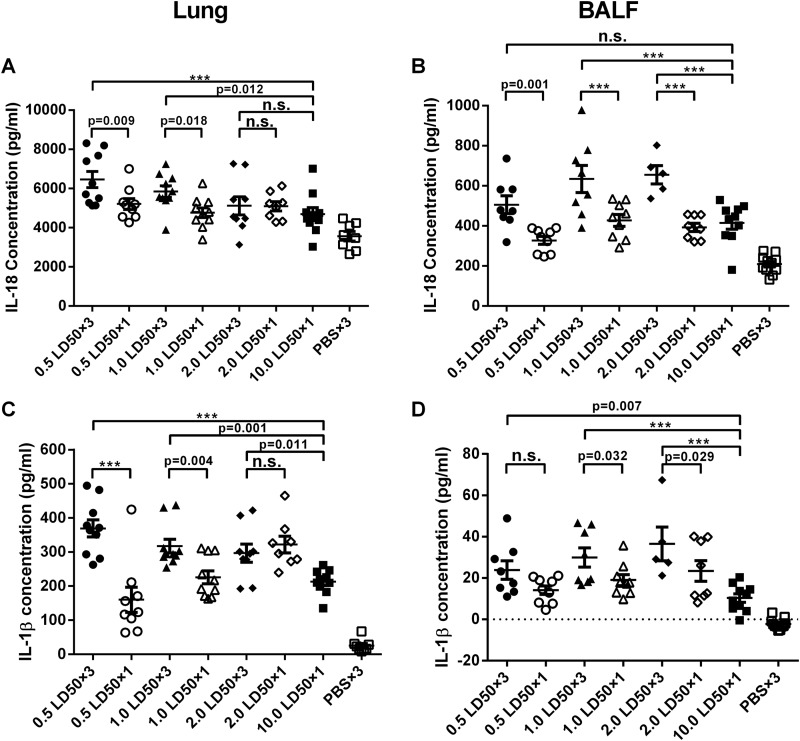
It has been reported that the NLRP3 inflammasome response is activated during influenza virus infection, causing increased secretion of the proinflammatory cytokines IL-1β and IL-18 (21). To define the relationship of the inflammasome response to the virus dose used for challenge, we measured levels of IL-1β and IL-18 expression in lung tissues 5 days after single or repeated challenges with different doses of A/PR/8 virus. Lung homogenates and bronchoalveolar lavage fluid (BALF) were collected after virus infection. As expected, the high-dose challenge with 10.0 LD50 of A/PR/8 virus given once increased IL-18 and IL-1β levels at both sites. Repeated low-dose challenges further increased the levels of IL-18 and IL-1β (Fig. 5). In lung homogenates, the levels of IL-18 in the 0.5 LD50 and 1.0 LD50 repeated-infection groups were significantly different from those in the matched single-dose infection groups, while there was no statistical difference between the 2.0 LD50 repeated- and single-infection groups (Fig. 5A). Similar results for IL-1β expression were obtained with lung homogenates (Fig. 5C). In lung homogenates, IL-18 concentrations in the 0.5 LD50 and 1.0 LD50 repeated-infection groups were significantly higher than those in the 10.0 LD50 single-infection groups, while IL-18 levels in the 2.0 LD50 repeated-infection group were comparable (Fig. 5A). In lung homogenates, IL-1β levels in the three repeated-infection groups showed significant differences from those in the 10.0 LD50 single-infection group (Fig. 5C). In BALF, IL-18 levels in the three repeated-infection groups showed significant differences from the corresponding matched-dose single-infection groups, and similar patterns were found for IL-1β levels in BALF, except for the 0.5 LD50 infection group (Fig. 5B and D). Levels of IL-1β in BALF in the repeated-infection groups were significantly higher than those in the 10.0 LD50 single-infection group (Fig. 5D); the same was observed for IL-18 levels in BALF, except for the 0.5 LD50 group (Fig. 5B). Overall, these data show that repeated low-dose challenges with influenza A virus cause more pronounced inflammasome responses than a single challenge with a high dose of virus.
FIG 5.
Repeated low-dose infection elicits greater inflammasome responses in mice. Mice (n = 8 to 10) were killed on day 5 after the first infection. Lung homogenates and BALF were collected for inflammasome response analysis using an ELISA. (A) IL-18 levels in lung homogenates. (B) IL-18 levels in BALF. (C) IL-1β expression in lung homogenates. (D) IL-1β concentrations in BALF. Data are presented as means ± standard errors of the means. ***, P < 0.0001; n.s., no significance.
Commercial influenza vaccine protects mice from a single high-dose virus infection but not repeated low-dose infection.
To assess whether repeated low-dose challenges of mice provide a more stringent model for preclinical efficacy testing of vaccines, we immunized mice (10 to 13 mice per group) with a commercial trivalent influenza vaccine (Fluarix; GSK) twice at a 2-week interval. As shown in Fig. 6A, mice were injected intramuscularly with a vaccine dose equal to 1.5 μg HA per strain (total, 4.5 μg HA). Mice inoculated with PBS served as controls. Three weeks after the boost, all mice in the vaccine groups were challenged with pandemic H1N1 virus given at 10.0 LD50 once, 6.0 LD50 once, and 2.0 LD50 three times. The PBS group was challenged with 6.0 LD50 of the same virus once.
HA-specific antibody responses were significantly greater in sera from immunized mice after the prime (P < 0.05) and the boost (P < 0.0001) than those in sera of the PBS controls (Fig. 6B). The neutralizing antibody titer was not detected postprime, but 2 weeks after the boost, the serum neutralization activities in vaccine-immunized mice were increased and significantly different from those in sera from either the control group or those after the prime (Fig. 6C). After challenge, the vaccine group (TIV group) infected repeatedly with a low dose of virus lost weight more rapidly than did mice in the high-dose-challenge vaccine groups (Fig. 6D). Most mice died, and survival curves were similar to those of PBS-injected control mice (Fig. 6E). In contrast, all of the TIV-immunized mice challenged with 6.0 LD50 of H1N1 virus survived; comparable results were obtained upon challenge with 10.0 LD50 of H1N1 virus (80% survived) (Fig. 6E).
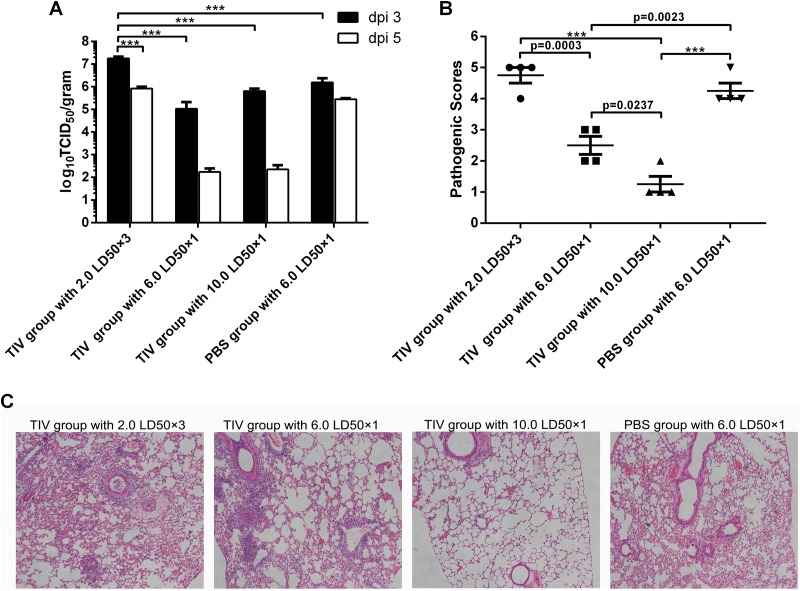
Three days after the first challenge, viral titers were significantly higher in lung tissues of mice with repeated infection than in those of the immunized groups with single infection and the control group (Fig. 7A). Although not reaching statistical significance, the viral titers of immunized mice with a single challenge dropped dramatically at day 5, with ∼102 TCID50 per g, approaching the lower limit of detection of the assay, while the viral titers were still high in the control group and even higher in immunized mice with repeated challenge (Fig. 7A). Pathogenic scores of immunized mice with repeated challenge were significantly higher than those of immunized groups with a single challenge but not those of the control group (Fig. 7B). Histology analysis revealed that the immunized mice with repeated challenge and the control group had more severe lung tissue damage (Fig. 7C). Although the immunized mice challenged with 6.0 LD50 had some pathological changes and higher pathogenic scores than did those challenged with 10.0 LD50 of virus, both groups showed protection after vaccination, with lower viral titers, lower pathogenic sores, and limited pathology changes in lungs at day 5 (Fig. 7). Taken together, these results lead us to conclude that the commercial vaccine can protect mice from a single high-dose virus infection but is ineffective against repeated low-dose challenges, confirming the higher stringency of this challenge protocol.
FIG 7.
TIV-immunized mice with repeated virus challenge have higher viral titers and more pathogenic damage in the lungs. (A) Viral titers were determined by a TCID50 assay of lung tissues from immunized mice and controls at days 3 and 5 after the first challenge. Data are presented as mean values ± standard errors of the means. dpi, days postinfection. (B) Mouse lung tissue damage was analyzed and scored at day 5 after the first challenge. Data are shown as mean values ± standard errors of the means. ***, P < 0.0001; n.s., no significance. (C) Representative hematoxylin- and eosin-stained sections from each group of mice scarified on day 5 after the first infection. Original magnification, ×100.
DISCUSSION
In the present study, we developed a novel preclinical influenza infection model by repeatedly inoculating mice intranasally with low doses of influenza A virus. Compared to a single high-dose infection, we found that repeated low-dose infections caused more severe symptoms, such as early morbidity and mortality as well as more rapid weight loss (Fig. 1). A more detailed analysis showed that mice repeatedly inoculated with low-dose influenza virus developed higher viral titers and more serious pathological changes in their lung tissues than did mice that received a single infection (Fig. 2). In humans, influenza A virus is shed from nasal secretions very rapidly within the first 24 h after infection; viral shedding peaks on day 2, and symptoms are commonly most severe by day 3 after infection (3, 4). In our study, we observed that mice in the low-dose repeated-infection groups began to lose weight within 24 h after infection, and from day 3 on, weight loss then accelerated, followed by early mortality compared to mice that received a single dose of virus.
The more rapid onset of severe symptoms in mice infected repeatedly with low doses of A/PR/8 virus could have been caused by higher viral loads resulting in more pronounced lung tissue damage. Several pathways could contribute to higher viral loads in mice that received influenza A virus repeatedly. First of all, receptors that allow for the uptake of virus in the mucosa of the respiratory tract may be limiting, thus restricting the uptake of the virus. This is rather unlikely, as morbidity and mortality rates increased in the single-dose groups with increasing doses of challenge virus. More likely, the inflammatory response elicited by the first dose of influenza A virus damaged the mucosal barrier, thus allowing the virus given as a second or third dose to penetrate more deeply into the lungs, as has been described for other antigens (22).
The viral loads of A/PR/8 infection at day 5 were higher than those at day 3 and even reached statistical significance for mice with repeated infection (Fig. 2B). In contrast, the PBS group challenged with pdm H1N1 appeared to have higher viral titers at day 3 than at day 5, although this was statistically insignificant (Fig. 7A). The reason for this observation might be that different virus strains have different kinetics of replication in lungs after infection. The inflammatory response was markedly greater in mice that received several doses of influenza A virus than in mice that received a single dose. Innate immune responses to influenza A viruses are triggered by 3 different types of pathogen recognition receptors, i.e., endosomal Toll-like receptor 3 (TLR-3) and TLR-7, the cytoplasmic RNA helicase RIG-1, and nucleotide-binding domain- and leucine-rich-repeat-containing proteins (NLRP3) (21, 23). NLRP3 senses influenza viruses through an M2 ion channel as well as M2-mediated perturbation of ionic concentrations or viral RNA (21, 24–28) and can be activated to recruit caspase-1 to the inflammasome, which in turn cleaves immature cytokines such as pro-IL-1β and pro-IL-18, resulting in their secretion into the extracellular space (21, 28–30). In our study, we found that inflammasome responses were elicited in mice after virus infection and that the low-dose repeated-infection groups had greater inflammasome responses, as indicated by increased IL-1β and IL-18 concentrations in lung tissues and BALF (Fig. 5). The intensity of the inflammasome responses correlated with viral loads, as shown previously (24). Others have shown that exaggerated innate immune responses can lead to enhanced pathology, including influenza-induced acute respiratory distress syndrome (ARDS) (23, 31). It has been reported that there are two types of innate immune cytokine responses to influenza infection: the proinflammatory response and the antiviral response. The proinflammatory cytokine response is likely to recruit immune effector cells that help clear the virus and cells that participate in adaptive immunity (21, 32). The antiviral response facilitates the intracellular control of viral replication through the induction of interferons, interferon-mediated antiviral signaling, and hundreds of interferon-stimulated genes (ISGs) (21, 33–35).
Virus infections, including those with influenza viruses, elicit adaptive immune responses to clear virus-infected cells and prevent subsequent infections (36–38). Our data show that prior to death, infected mice developed low titers of neutralizing antibodies. Seven days after infection, we could detect influenza virus-specific T cells; however, this response was detectable by 2 weeks after infection in surviving animals (data not shown). Similar results were found in HBV studies, where high-risk individuals were exposed to multiple small doses of HBV, as well as in the woodchuck model, in which animals were repeatedly exposed to small quantities of infectious HBV. In both cases, viral infection was molecularly evident and there were detectable virus-specific CD8+ T cell responses but undetectable virus-specific antibody responses. These data are in agreement with clinical findings in humans (17). Animals that survived the initial challenge(s) were protected against a subsequent challenge (data not shown), further confirming that the initial challenges had induced protective immunity.
The TIV Fluarix (GSK) is one of the vaccines approved for influenza virus infection with proven efficacy (39–41). In our present study, we found that vaccination with a TIV protected mice from a single high-dose virus infection but was inefficacious against repeated low-dose influenza virus infection (Fig. 6). Although the viral titers in lungs decreased from day 3 to day 5 in all the challenged groups, only immunized single-dose infection groups had viral titers that decreased below the limit of detection and manifested little lung pathology (Fig. 7). This result indicates that repeated low-dose challenges provide a more stringent mode to evaluate vaccine efficacy. Vaccines for other viruses, such as HIV-1/SIV, HBV, and others, rely on animal models of repeated low-dose virus infection, as they may more closely resemble natural infections in humans. These models have been used in assessments of HIV vaccine efficiency in experimental models (14, 17, 42–44). More studies are needed to test this influenza virus model, including the testing of other strains of influenza A virus and the use of other animal models such as ferrets, the animal model of choice for preclinical influenza vaccine studies (3, 4, 45–47).
In summary, here we describe a novel mouse model of repeated low-dose influenza virus infection. This type of challenge causes increased morbidity and mortality and may thus be more suitable for preclinical evaluation of vaccine efficacy.
ACKNOWLEDGMENTS
We gratefully acknowledge Youbing Zhang at The Third People's Hospital of Suzhou, Jiangsu, China, for pathology analysis and scoring. We also thank Guiqin Wang from the Institut Pasteur of Shanghai, Chinese Academy of Science, Shanghai, China, for help in the determination of viral titers in lungs.
This work was supported by grants from the Knowledge Innovation Program and the 100 Talent Program of the Chinese Academy of Sciences and the Shanghai Pasteur Foundation to D.Z. and by grants from the National Science Foundation of China (31300736) and the Science and Technology Innovation Plan of the Shanghai Municipal Science and Technology Commission for Animal Model Research (14140901002) to Y.S.
REFERENCES
- 1.Tumpey TM, Belser JA. 2009. Resurrected pandemic influenza viruses. Annu Rev Microbiol 63:79–98. doi: 10.1146/annurev.micro.091208.073359. [DOI] [PubMed] [Google Scholar]
- 2.Taubenberger JK, Morens DM. 2008. The pathology of influenza virus infections. Annu Rev Pathol 3:499–522. doi: 10.1146/annurev.pathmechdis.3.121806.154316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thangavel RR, Bouvier NM. 2014. Animal models for influenza virus pathogenesis, transmission, and immunology. J Immunol Methods 410:60–79. doi: 10.1016/j.jim.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouvier NM, Lowen AC. 2010. Animal models for influenza virus pathogenesis and transmission. Viruses 2:1530–1563. doi: 10.3390/v20801530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moscona A. 2008. Medical management of influenza infection. Annu Rev Med 59:397–413. doi: 10.1146/annurev.med.59.061506.213121. [DOI] [PubMed] [Google Scholar]
- 6.Pica N, Palese P. 2013. Toward a universal influenza virus vaccine: prospects and challenges. Annu Rev Med 64:189–202. doi: 10.1146/annurev-med-120611-145115. [DOI] [PubMed] [Google Scholar]
- 7.Poland GA. 2006. Vaccines against avian influenza—a race against time. N Engl J Med 354:1411–1413. doi: 10.1056/NEJMe068047. [DOI] [PubMed] [Google Scholar]
- 8.Wang G, Zhou F, Buchy P, Zuo T, Hu H, Liu J, Song Y, Ding H, Tsai C, Chen Z, Zhang L, Deubel V, Zhou P. 2014. DNA prime and virus-like particle boost from a single H5N1 strain elicits broadly neutralizing antibody responses against head region of H5 hemagglutinin. J Infect Dis 209:676–685. doi: 10.1093/infdis/jit414. [DOI] [PubMed] [Google Scholar]
- 9.Villari P, Manzoli L, Boccia A. 2004. Methodological quality of studies and patient age as major sources of variation in efficacy estimates of influenza vaccination in healthy adults: a meta-analysis. Vaccine 22:3475–3486. doi: 10.1016/j.vaccine.2004.01.068. [DOI] [PubMed] [Google Scholar]
- 10.Zhou D, Wu TL, Lasaro MO, Latimer BP, Parzych EM, Bian A, Li Y, Li H, Erikson J, Xiang Z, Ertl HC. 2010. A universal influenza A vaccine based on adenovirus expressing matrix-2 ectodomain and nucleoprotein protects mice from lethal challenge. Mol Ther 18:2182–2189. doi: 10.1038/mt.2010.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bodewes R, Rimmelzwaan GF, Osterhaus AD. 2010. Animal models for the preclinical evaluation of candidate influenza vaccines. Expert Rev Vaccines 9:59–72. doi: 10.1586/erv.09.148. [DOI] [PubMed] [Google Scholar]
- 12.Kim JI, Park S, Lee S, Lee I, Heo J, Hwang MW, Bae JY, Kim D, Jang SI, Park MS. 2013. DBA/2 mouse as an animal model for anti-influenza drug efficacy evaluation. J Microbiol 51:866–871. doi: 10.1007/s12275-013-3428-7. [DOI] [PubMed] [Google Scholar]
- 13.Zhou F, Wang G, Buchy P, Cai Z, Chen H, Chen Z, Cheng G, Wan XF, Deubel V, Zhou P. 2012. A triclade DNA vaccine designed on the basis of a comprehensive serologic study elicits neutralizing antibody responses against all clades and subclades of highly pathogenic avian influenza H5N1 viruses. J Virol 86:6970–6978. doi: 10.1128/JVI.06930-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Regoes RR, Longini IM, Feinberg MB, Staprans SI. 2005. Preclinical assessment of HIV vaccines and microbicides by repeated low-dose virus challenges. PLoS Med 2:e249. doi: 10.1371/journal.pmed.0020249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Regoes RR. 2012. The role of exposure history on HIV acquisition: insights from repeated low-dose challenge studies. PLoS Comput Biol 8:e1002767. doi: 10.1371/journal.pcbi.1002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banyard AC, Healy DM, Brookes SM, Voller K, Hicks DJ, Nunez A, Fooks AR. 2014. Lyssavirus infection: ‘low dose, multiple exposure’ in the mouse model. Virus Res 181:35–42. doi: 10.1016/j.virusres.2013.12.029. [DOI] [PubMed] [Google Scholar]
- 17.Gujar SA, Mulrooney-Cousins PM, Michalak TI. 2013. Repeated exposure to trace amounts of woodchuck hepadnavirus induces molecularly evident infection and virus-specific T cell response in the absence of serological infection markers and hepatitis. J Virol 87:1035–1048. doi: 10.1128/JVI.01363-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang R, Rong X, Pan W, Peng T. 2011. Determination of serum neutralization antibodies against seasonal influenza A strain H3N2 and the emerging strains 2009 H1N1 and avian H5N1. Scand J Infect Dis 43:216–220. doi: 10.3109/00365548.2010.539258. [DOI] [PubMed] [Google Scholar]
- 19.Lamoreaux L, Roederer M, Koup R. 2006. Intracellular cytokine optimization and standard operating procedure. Nat Protoc 1:1507–1516. doi: 10.1038/nprot.2006.268. [DOI] [PubMed] [Google Scholar]
- 20.Monticelli LA, Sonnenberg GF, Abt MC, Alenghat T, Ziegler CG, Doering TA, Angelosanto JM, Laidlaw BJ, Yang CY, Sathaliyawala T, Kubota M, Turner D, Diamond JM, Goldrath AW, Farber DL, Collman RG, Wherry EJ, Artis D. 2011. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat Immunol 12:1045–1054. doi: 10.1038/ni.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu S, Metcalf JP, Wu W. 2011. Innate immune response to influenza virus. Curr Opin Infect Dis 24:235–240. doi: 10.1097/QCO.0b013e328344c0e3. [DOI] [PubMed] [Google Scholar]
- 22.Sakamoto M, Ida S, Takishima T. 1984. Effect of influenza virus infection on allergic sensitization to aerosolized ovalbumin in mice. J Immunol 132:2614–2617. [PubMed] [Google Scholar]
- 23.Pulendran B, Maddur MS. 2015. Innate immune sensing and response to influenza. Curr Top Microbiol Immunol 386:23–71. doi: 10.1007/82_2014_405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ichinohe T, Pang IK, Iwasaki A. 2010. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat Immunol 11:404–410. doi: 10.1038/ni.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ichinohe T. 2010. Respective roles of TLR, RIG-I and NLRP3 in influenza virus infection and immunity: impact on vaccine design. Expert Rev Vaccines 9:1315–1324. doi: 10.1586/erv.10.118. [DOI] [PubMed] [Google Scholar]
- 26.Iwasaki A, Pillai PS. 2014. Innate immunity to influenza virus infection. Nat Rev Immunol 14:315–328. doi: 10.1038/nri3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stout-Delgado HW, Vaughan SE, Shirali AC, Jaramillo RJ, Harrod KS. 2012. Impaired NLRP3 inflammasome function in elderly mice during influenza infection is rescued by treatment with nigericin. J Immunol 188:2815–2824. doi: 10.4049/jimmunol.1103051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allen IC, Scull MA, Moore CB, Holl EK, McElvania-TeKippe E, Taxman DJ, Guthrie EH, Pickles RJ, Ting JP. 2009. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity 30:556–565. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas PG, Dash P, Aldridge JR Jr, Ellebedy AH, Reynolds C, Funk AJ, Martin WJ, Lamkanfi M, Webby RJ, Boyd KL, Doherty PC, Kanneganti TD. 2009. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity 30:566–575. doi: 10.1016/j.immuni.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schroder K, Tschopp J. 2010. The inflammasomes. Cell 140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 31.McAuley JL, Tate MD, MacKenzie-Kludas CJ, Pinar A, Zeng W, Stutz A, Latz E, Brown LE, Mansell A. 2013. Activation of the NLRP3 inflammasome by IAV virulence protein PB1-F2 contributes to severe pathophysiology and disease. PLoS Pathog 9:e1003392. doi: 10.1371/journal.ppat.1003392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kao W, Costanzo MR. 2000. Sudden death in heart failure patients: effects of optimized medical therapy. J Heart Lung Transplant 19:S32–S37. doi: 10.1016/S1053-2498(99)00108-4. [DOI] [PubMed] [Google Scholar]
- 33.Ronni T, Sareneva T, Pirhonen J, Julkunen I. 1995. Activation of IFN-alpha, IFN-gamma, MxA, and IFN regulatory factor 1 genes in influenza A virus-infected human peripheral blood mononuclear cells. J Immunol 154:2764–2774. [PubMed] [Google Scholar]
- 34.Ronni T, Matikainen S, Sareneva T, Melen K, Pirhonen J, Keskinen P, Julkunen I. 1997. Regulation of IFN-alpha/beta, MxA, 2′,5′-oligoadenylate synthetase, and HLA gene expression in influenza A-infected human lung epithelial cells. J Immunol 158:2363–2374. [PubMed] [Google Scholar]
- 35.Dienz O, Rud JG, Eaton SM, Lanthier PA, Burg E, Drew A, Bunn J, Suratt BT, Haynes L, Rincon M. 2012. Essential role of IL-6 in protection against H1N1 influenza virus by promoting neutrophil survival in the lung. Mucosal Immunol 5:258–266. doi: 10.1038/mi.2012.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quinones-Parra S, Loh L, Brown LE, Kedzierska K, Valkenburg SA. 2014. Universal immunity to influenza must outwit immune evasion. Front Microbiol 5:285. doi: 10.3389/fmicb.2014.00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quan FS, Compans RW, Nguyen HH, Kang SM. 2008. Induction of heterosubtypic immunity to influenza virus by intranasal immunization. J Virol 82:1350–1359. doi: 10.1128/JVI.01615-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soboll G, Horohov DW, Aldridge BM, Olsen CW, McGregor MW, Drape RJ, Macklin MD, Swain WF, Lunn DP. 2003. Regional antibody and cellular immune responses to equine influenza virus infection, and particle mediated DNA vaccination. Vet Immunol Immunopathol 94:47–62. doi: 10.1016/S0165-2427(03)00060-6. [DOI] [PubMed] [Google Scholar]
- 39.Manuela Q, Gianbattista L, Gianpiero L, Antonella DD. 2011. Evaluation of immune responses to seasonal influenza vaccination in healthy volunteers in South Apulia, Italy: a pilot study. J Clin Med Res 3:291–295. doi: 10.4021/jocmr647w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Couch RB, Bayas JM, Caso C, Mbawuike IN, Lopez CN, Claeys C, El Idrissi M, Herve C, Laupeze B, Oostvogels L, Moris P. 2014. Superior antigen-specific CD4+ T-cell response with AS03-adjuvantation of a trivalent influenza vaccine in a randomised trial of adults aged 65 and older. BMC Infect Dis 14:425. doi: 10.1186/1471-2334-14-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carmona Martinez A, Salamanca de la Cueva I, Boutet P, Vanden Abeele C, Smolenov I, Devaster JM. 2014. A phase 1, open-label safety and immunogenicity study of an AS03-adjuvanted trivalent inactivated influenza vaccine in children aged 6 to 35 months. Hum Vaccin Immunother 10:1959–1968. doi: 10.4161/hv.28743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Otten RA, Adams DR, Kim CN, Jackson E, Pullium JK, Lee K, Grohskopf LA, Monsour M, Butera S, Folks TM. 2005. Multiple vaginal exposures to low doses of R5 simian-human immunodeficiency virus: strategy to study HIV preclinical interventions in nonhuman primates. J Infect Dis 191:164–173. doi: 10.1086/426452. [DOI] [PubMed] [Google Scholar]
- 43.Vaccari M, Keele BF, Bosinger SE, Doster MN, Ma ZM, Pollara J, Hryniewicz A, Ferrari G, Guan Y, Forthal DN, Venzon D, Fenizia C, Morgan T, Montefiori D, Lifson JD, Miller CJ, Silvestri G, Rosati M, Felber BK, Pavlakis GN, Tartaglia J, Franchini G. 2013. Protection afforded by an HIV vaccine candidate in macaques depends on the dose of SIVmac251 at challenge exposure. J Virol 87:3538–3548. doi: 10.1128/JVI.02863-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kersh EN, Luo W, Adams DR, Srinivasan P, Smith JM, Promadej-Lanier N, Ellenberger D, Garcia-Lerma JG, Butera S, Otten R. 2009. Repeated rectal SHIVSF162P3 exposures do not consistently induce sustained T cell responses prior to systemic infection in the repeat-low dose preclinical macaque model. AIDS Res Hum Retroviruses 25:905–917. doi: 10.1089/aid.2008.0287. [DOI] [PubMed] [Google Scholar]
- 45.Belser JA, Szretter KJ, Katz JM, Tumpey TM. 2009. Use of animal models to understand the pandemic potential of highly pathogenic avian influenza viruses. Adv Virus Res 73:55–97. doi: 10.1016/S0065-3527(09)73002-7. [DOI] [PubMed] [Google Scholar]
- 46.Belser JA, Katz JM, Tumpey TM. 2011. The ferret as a model organism to study influenza A virus infection. Dis Model Mech 4:575–579. doi: 10.1242/dmm.007823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Belser JA, Gustin KM, Pearce MB, Maines TR, Zeng H, Pappas C, Sun X, Carney PJ, Villanueva JM, Stevens J, Katz JM, Tumpey TM. 2013. Pathogenesis and transmission of avian influenza A (H7N9) virus in ferrets and mice. Nature 501:556–559. doi: 10.1038/nature12391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27:493–497. [Google Scholar]