ABSTRACT
CD4+ T cells play a pivotal role in the control of chronic viral infections. Recently, nontraditional CD4+ T cell functions beyond helper effects have been described, and a role for cytolytic CD4+ T cells in the control of HIV infection has been suggested. We define here the transcriptional, phenotypic, and functional profiles of HIV-specific cytolytic CD4+ T cells. Fluidigm BioMark and multiparameter flow cytometric analysis of HIV-specific cytolytic CD4+ T cells revealed a distinct transcriptional signature compared to Th1 CD4+ cells but shared similar features with HIV-specific cytolytic CD8+ T cells. Furthermore, HIV-specific cytolytic CD4+ T cells showed comparable killing activity relative to HIV-specific CD8+ T cells and worked cooperatively in the elimination of virally infected cells. Interestingly, we found that cytolytic CD4+ T cells emerge early during acute HIV infection and tightly follow acute viral load trajectory. This emergence was associated to the early viral set point, suggesting an involvement in early control, in spite of CD4 T cell susceptibility to HIV infection. Our data suggest cytolytic CD4+ T cells as an independent subset distinct from Th1 cells that show combined activity with CD8+ T cells in the long-term control of HIV infection.
IMPORTANCE The ability of the immune system to control chronic HIV infection is of critical interest to both vaccine design and therapeutic approaches. Much research has focused on the effect of the ability of CD8+ T cells to control the virus, while CD4+ T cells have been overlooked as effectors in HIV control due to the fact that they are preferentially infected. We show here that a subset of HIV-specific CD4+ T cells cooperate in the cytolytic control of HIV replication. Moreover, these cells represent a distinct subset of CD4+ T cells showing significant transcriptional and phenotypic differences compared to HIV-specific Th1 cells but with similarities to CD8+ T cells. These findings are important for our understanding of HIV immunopathology.
INTRODUCTION
The pivotal role of CD4+ T cells in the control of chronic viral infections is well established. In particular, robust and functional CD4+ T cell responses are critical to maintain the efficacy of virus-specific CD8+ T cell responses and to facilitate memory formation. However, the simplified view of CD4+ T cells as “helpers” and CD8+ T cells as “killers” has allowed other important CD4+ T cell functions to be overlooked. Since the 1980s, observations consistently reoccur that CD4+ T cells are not merely helpers but can also directly contribute to the control of viral infection through the killing of infected cells (1). An important role for these cytotoxic CD4+ T cells has been described for both acute influenza virus infection, as well as conferring improved clinical responses following expansion and readmission of an expanded autologous cytolytic CD4+ T cell clone in cancer (2, 3). Moreover, it has also been shown that cytolytic CD4+ T cells may play a prominent role in chronic viral infection, as evidenced by their influence in the containment of viral replication in Epstein-Barr virus and cytomegalovirus (CMV) infection (4). The ability of CD4+ T cells to directly assist in control of acute and chronic viral infections, as well as cancers, therefore represents a novel and intriguing possibility for immune interventions.
The importance of cytolytic CD4+ T cells in controlling infections suggests that they may play a role in the pathogenesis and progression of HIV infection. We were recently able to demonstrate that a distinct HIV-specific CD4+ T cell population, expressing the degranulatory marker CD107a, emerges early during acute HIV infection in individuals able to spontaneously control HIV replication for a prolonged period of time (5). These HIV-specific CD4+ T cell responses exhibited a human lymphocyte antigen (HLA) class II-dependent cytolytic phenotype, characterized by the expression of high levels of granzymes A and B, as well as perforin. Interestingly, the presence of these HIV-specific CD4+ T cell responses in acute HIV infection was highly predictive for disease outcome (5). Although the results of these studies are remarkable, little is known about the nature, phenotype, function, and lineage commitment of cytolytic CD4+ T cells in contrast to other CD4+ T cell subsets and CD8+ T cells. Furthermore, it is not known whether HIV-specific CD8+ T cells and HIV-specific cytolytic CD4+ T cells can act in concert in the control of HIV viremia. Here, we describe—phenotypically, transcriptionally, and functionally—a population of HIV-specific cytolytic CD4+ T cell responses that are distinct from HIV-specific Th1 CD4+ T cells but which show striking cytolytic similarities to HIV-specific CD8+ T cells. We demonstrate that HIV-specific cytolytic CD4+ and CD8+ T cells exhibit a strong cooperative antiviral effect, suggesting an important in vivo role for these cells in the control of HIV infection. These results further our understanding of HIV disease progression, give insight into the ability of certain patients to control viral replication, and have implications for the targeted elicitation of cytolytic CD4+ cells through vaccination.
MATERIALS AND METHODS
Study subjects.
Frozen, leukapheresed peripheral blood mononuclear cells (PBMCs) of 11 subjects with chronic HIV infection enrolled in study RV149 were used as positive HIV samples. Frozen PBMCs of 24 subjects used in the acute studies were enrolled in study RV217. Viral load data were clinically established and provided for the acutely infected individuals in RV217. The early viral set point was calculated by taking the average viral load between days 80 and 365 prior to the initiation of antiretroviral therapy and requiring at least two consecutive time points. In addition, cryopreserved specimens of 14 elite controllers, 4 viremic controllers, and 15 chronic progressors from the Massachusetts General Hospital (MGH) were used in the present study. All study subjects gave informed consent, and institutional review board (IRB) approval was obtained.
T cell activation for phenotyping experiments.
PBMCs from HIV-infected patients, either acutely or chronically infected, were stimulated with HIV-Gag peptide pools at 2 μg/ml (AIDS reagents) for 6 h at 37°C in the presence of CD28-49d costimulatory antibodies and anti-human CD107a PE-CF594 (clone H4A3; BD Biosciences). After 1 h of stimulation, a cocktail of Golgi Stop (monensin; BD) and brefeldin A (Sigma) was added to the PBMCs for the rest of the incubation. As a positive control, PBMCs also stimulated with staphylococcal enterotoxin B (List Biologicals) at 5 μg/ml. The cells were washed with phosphate-buffered saline (PBS) and stained with amine-reactive viability dye (Live/Dead Aqua; Life Technologies) for 20 min at room temperature. The cells were washed with staining buffer (PBS with 2% fetal calf serum [FCS]) and then stained for 20 min at 4°C with the surface markers CD8 allophycocyanin-H7 (APC-H7; clone SK1; BD) and CD127 brilliant violet 785 (BV785; clone A019D5, BioLegend) and any of the following combinations: CD57-fluorescein isothiocyanate (FITC; clone HNK-1; BioLegend) and KLRG1-phycoerythrin (PE; clone 2F5; BioLegend); CD45RO-eFluor650 (clone UCHL1; eBioscience), CD161-FITC (clone HP-3G10; BioLegend), and CD223-PE (polyclonal; R&D Systems); CD94-FITC (clone DX22; BioLegend) and CD314 (clone 1D11; BioLegend); and NKp80-PE (clone 5D12; BioLegend) and CCR5-PE (clone 2D7; BD). PBMCs were subsequently washed with staining buffer and then fixed and permeabilized with Foxp3/transcription factor staining buffer (eBioscience) for half an hour at room temperature. PBMCs were washed with 1× permeabilization buffer (eBioscience) and then stained intracellularly with CD3 Qdot605 (clone UCHT1; Life Technologies), CD4 PerCP-Cy5.5 (clone RPA-T4; BioLegend), granzyme B-Alexa Fluor 700 (AF700; clone GB11; BD), gamma interferon (IFN-γ)-PE-Cy7 (clone B27; BioLegend), Eomesodermin-eFluor 660 (clone WD1928; eBioscience), perforin-BV421 (clone B-D48; BioLegend), granulysin AF488 (clone RB1; BD), GATA3 PE (clone L50-823; BD), and T-bet-AF488 (clone O4-46; BD) at room temperature for 30 min. A four-laser LSR II flow cytometer running FACSDiVa software (BD) was used to collect data from T-cell activation experiments. The data were analyzed with FlowJo (v9.410; Tree Star, Inc.).
Gene expression analysis.
Similar to the T cell activation, the PBMCs of HIV-infected patients were stimulated with HIV-Gag peptide pools in the presence of CD28-49d costimulatory antibodies and anti-human CD107a PE-CF594 and Golgi Stop (monensin; BD) for 5 h. After being washed in cold medium, the cells were incubated with IFN-γ+ catch reagent (IFN-γ secretion assay; Miltenyi Biotec) for 5 min on ice, and then warm medium was added to the tube for a 45-min incubation at 37°C. PBMCs were subsequently washed with PBS and stained with amine-reactive viability dye (Live/Dead Aqua) for 20 min at room temperature. Cells were washed with staining buffer and stained for 20 min at 4°C with IFN-γ+ APC (IFN-γ secretion assay), CD8 FITC (clone RPA-T8; BD), CD4 brilliant violet 421 (RPA-T4; BioLegend), and CD3 Qdot 605 (clone UCHT1; Life Technologies). After being washed twice with staining buffer, the PBMCs were resuspended in sorting buffer (PBS, 1% FCS, 10 mM HEPES). An index sort was performed on a FACSAria (BD) running on FACSDiVa software (BD). Cells were sorted using a stringent gating strategy, excluding doublets and CD14+, CD19+, CD3−, and nonviable cells (see Fig. S1 in the supplemental material). Both CD4+ and CD8+ T cells were sorted into wells of 100 cells of 96-well plates containing 10 μl of reverse transcription preamplification reaction mix, composed of Superscript III platinum Taq (Life Technologies), SUPERase-In (Ambion), 0.2× assays (mixture of 96 primers and FAM-conjugated probes; Life Technologies). The reverse transcription reaction (15 min at 50°C and then 2 min at 95°C) was followed by a preamplification step of 16 cycles of 95°C for 15 s and 60°C for 4 min. The cDNA produced was diluted 1:5 in DNA suspension buffer (Teknova). Experimental assays (20× assay mixes; TaqMan primers and probe) were diluted 1:1 with 2× assay loading buffer (Fluidigm) and samples (diluted cDNA mixed with TaqMan Universal PCR Master mix [Life Technologies] and 20× GE sample loading reagent [Fluidigm]) were loaded onto a primed 96.96 Dynamic Array chip (Fluidigm). The chip was then loaded into the IFC Controller (Fluidigm) to fill the matrix with both samples and assays. The chip was transferred into a Biomark (Fluidigm) for thermocycling and fluorescence acquisition using the GE 96×96 standard v1 program. For quality control, quantitative PCR (qPCR) amplification curves were first validated using gene expression Fluidigm BioMark real-time PCR analysis software on a Biomark (Fluidigm). The analysis was done with the linear derivative mode with a cycle threshold (CT) check, which allows the threshold to be specific to each individual assay. The amplification curves were validated based on the intensity of the signal. Statistical analysis was performed on JMP10 software (SAS). The results are plotted as the expression threshold (ET), where ET = Cmax – CT, with Cmax being the maximum number of cycles, which in these experiments was 40 (6). A table of genes used in the Fluidigm analysis is displayed in Table S1 in the supplemental material.
qPCR measurement of viral loads.
qPCR was performed using the following primers and a probe for the HIV long terminal repeat: forward primer, GCCTCAATAAAGCTTGCCTTGA; reverse primer, GGGCGCCACTGCTAGAGA; and probe, 6-FAM-CCAGAGTCACACAACAGACGGGCACA (BHQ1a-6FAM). An ABI Prism was used to perform the qPCR, using a one-step qPCR Superscript kit (Invitrogen), according to the manufacturer's instructions. Reverse transcription was performed at 50°C for 15 min, with a hot start of 95°C for 2 min, followed by 45 cycles of PCR at 95°C for 15 s and 60°C for 30 s. The standards used were derived from plasmid NL4-3, with the dilution curve displaying an R2 of 0.9921 and a slope of −3.6. This curve was confirmed against known virus titers of stock NL4-3 virus.
Cell subset enrichment.
PBMCs were thawed and sorted using a Dynabeads CD8 positive selection kit or untouched CD4 selection kit (Life Technologies). To separate CD57, CD161, or HLA-DR positive and negative populations, CD4+ cells were stained with the respective PE-conjugated antibodies (CD57, CD161, and HLA-DR) and sorted using anti-PE MACS beads and MACS columns (Miltenyi Biotec), according to the manufacturer's instructions.
Viral inhibition assay.
A panel of four R5-tropic viruses was created by isolating viruses from HIV-infected individuals, cloning the sequences into expression vectors, and transfecting cells with the plasmid to grow virus stocks. One clone was found to be nevirapine resistant by comparison of the viral sequence with the Stanford HIV Drug Resistance Database, possessing the variant DQB106. Nevirapine resistance was confirmed by in vitro outgrowth in the presence of nevirapine.
293T cells were transfected with 1 μg of viral plasmid using calcium phosphate in Dulbecco's modified Eagle medium (DMEM) plus 10% FCS with 1 mM HEPES. The cells were incubated at 37°C with 5% CO2 for 1 day, after which the medium was removed and replaced with DMEM containing 10% FCS. Two days later, the virus-containing supernatant was harvested and filtered. A total of 50,000 PBMCs were then spinoculated at 800 × g for 30 min with the indicated virus at a multiplicity of infection (MOI) of 0.1. The cells were cultured in RPMI–10% FCS and incubated at 37°C with 5% CO2. After 2 h, virus was washed off the cells, and the cells were incubated at 37°C in 5% CO2 in the required assay.
Target cells (HLA-DR+) were infected with the nevirapine-resistant virus at an MOI of 0.01 for 2 h. Excess virus was washed off, and 100,000 cells were added to a 96-well plate in RPMI–10% FCS. Effector cells (CD4+ or CD8+ populations) were added to target cells at a ratio of 1:1. For coculture experiments, two populations of effectors were added in equal ratios, standardized for the total amount of effector cells. Nevirapine (AIDS reagents) was added to cultures on day 0 at a concentration of 10 μg/ml from a stock solution of 0.1 mg/ml.
Statistical analysis.
Mann-Whitney U tests were used to compare the gene expression levels in CD4+ and CD8+ T cell subsets, and a P value of <0.05 was considered significant in all scenarios unless otherwise indicated. GraphPad Prism v6 was used for statistical analysis, and JMP version 11 software was used for multivariate and principal component analysis. A D'Agostino-Pearson omnibus test was used to verify the normal distribution for Pearson correlation. Clustering analysis was performed in Gene-E as an unsupervised hierarchical clustering using Pearson correlation matrices. Both genes and flow markers, as well as sample identifications, were clustered for ease of presentation. Z scores in Fig. 6 were derived from the formula: Z = (raw scores – the mean)/the standard deviation.
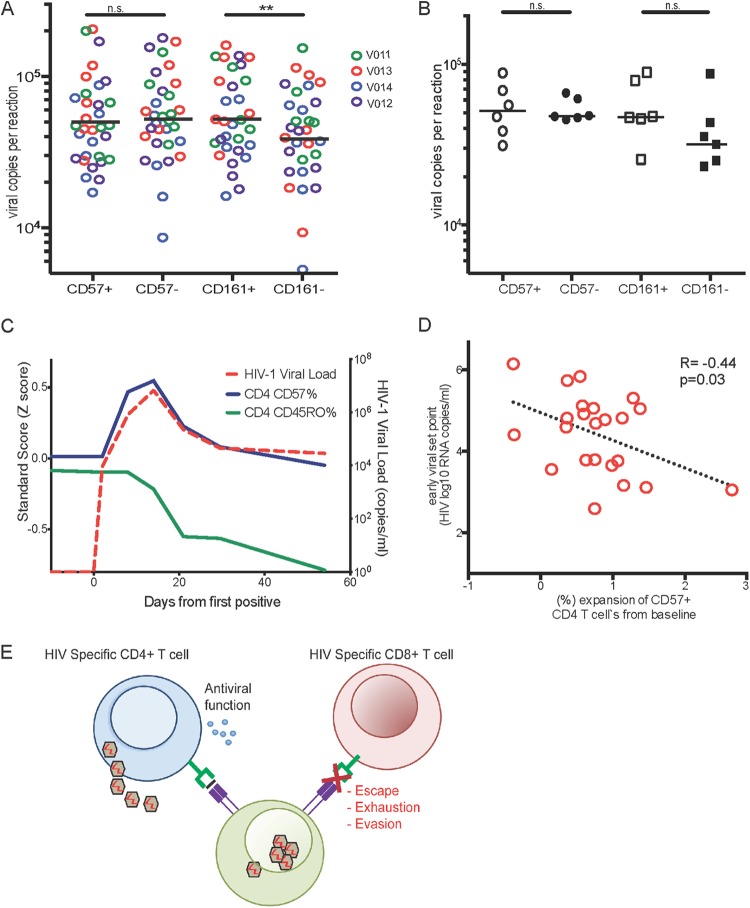
FIG 6.
Acute CD57+ CD4+ T cell kinetics suggest involvement of cytolytic CD4+ T cells in HIV control. (A and B) CD57+, CD57−, CD161+, and CD161− CD4+ T cells were purified via negative selection from PBMCs of HIV− donors (n = 10) using MACS columns and anti-PE beads. A total of 50,000 cells were infected with an MOI of 0.01 of four R5-tropic viruses, numbered V011 to V014 (A), or an X4-tropic virus strain (B). Supernatants were removed on day 3 postinfection and assessed for viral load directly via quantitative real-time PCR. CD57+ CD4+ T cells showed slightly lower susceptibility to HIV infection than CD57− CD4+ T cells. CD161+ CD4+ T cells were more susceptible to HIV infection than CD161− CD4+ T cells (P < 0.05; Student paired t tests). Different colors represent different R5 viruses used. (B) No difference in the susceptibility to HIV infection of CD57+ CD57− or CD161+ CD161− CD4+ T cells from HIV− donors (n = 6) to CXCR4 viruses. (C) Longitudinal analysis of the emergence of CD57+ CD4+ T cells during acute HIV infection in acutely infected individuals (n = 24) from preinfection to early viral set point. CD57+ CD4+ T cells are indicated in blue, and the general CD45RO+ CD4+ T cell population is indicated in green. The average viral load of all individuals is indicated by red dotted lines. (D) Association of the early viral set point with the expression level of CD57+ CD4+ T cells shows an inverse correlation of viral set point and CD57+ CD4+ T cells (R = −0.44, P = 0.03). (E) Schematic drawing of proposed model of cooperativity between HIV-specific cytolytic CD4+ and CD8+ T cell responses.
Ethics statement.
Specimens from the Stanford blood bank provided HIV-negative blood samples. Cryopreserved specimens of individuals from the Walter Reed Army Institute of Research (WRAIR) and the MGH were used. All study subjects gave written informed consent prior to participating in the study, and approval was obtained from the respective IRBs (WRAIR and MGH).
RESULTS
Cytolytic CD4+ T cells are a distinct subset from Th1 helper CD4+ T cells.
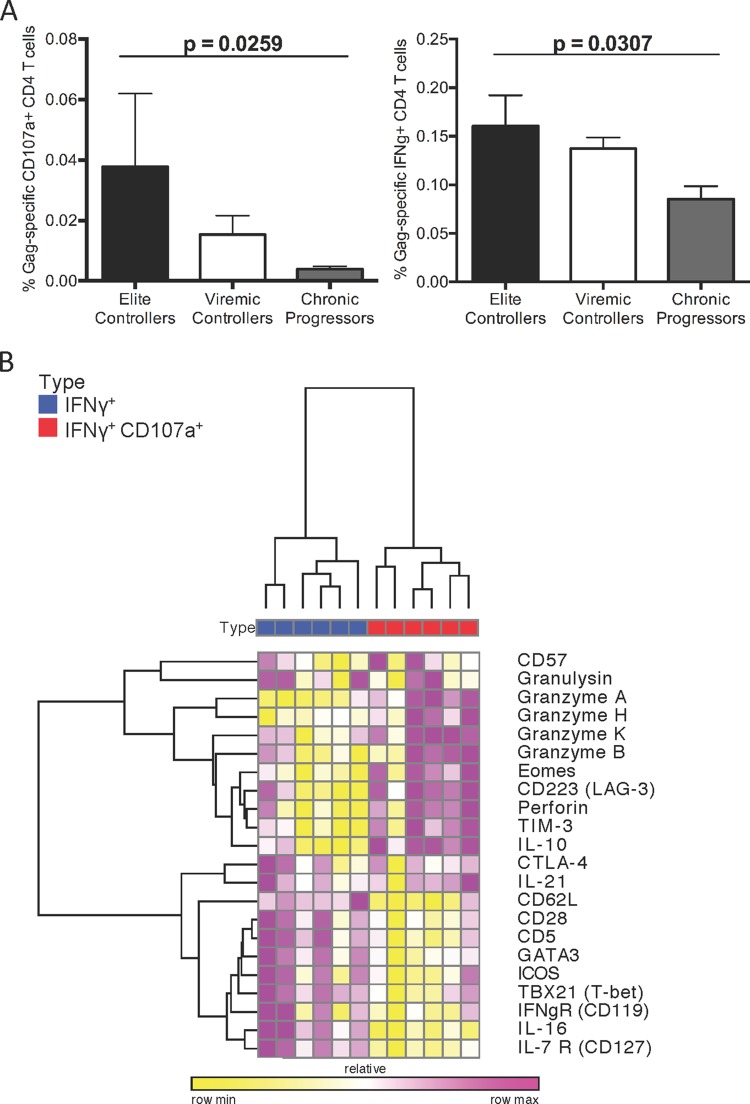
We have previously demonstrated that the emergence of HIV-specific cytolytic CD4+ T cell responses during acute HIV infection is associated with a low viral set point and a significant delay in disease progression, suggesting a potential role in HIV control (5). Indeed, examination of individuals able to spontaneously control HIV replication below the limit of detection (<50 HIV RNA copies/ml [termed elite controller]) in comparison to individuals with controlled viremia but with viral loads ranging up to 2,000 HIV RNA copies/ml (termed viremic controller) or individuals with high viremia (termed chronic progressor) revealed significant enrichment of HIV-specific cytolytic or IFN-γ+ CD4+ T cell responses in elite controllers compared to chronic progressors (Fig. 1A). Interestingly, elite controllers displayed 9.5-fold-higher levels of cytolytic CD4+ T cells relative to chronic progressors, while they only displayed 1.6-fold-higher levels of IFN-γ+ cells, highlighting the potential importance of cytolytic CD4+ T cells in viral control. Given the potential importance of these HIV-specific cytolytic CD4+ T cell responses in the long-term control of HIV viremia, we sought to determine factors involved in driving a cytotoxic program in HIV-specific CD4+ T cells. We first utilized a surface capture technique to isolate HIV-specific CD4+ T cells based on their ability to respond by either secreting IFN-γ alone (termed Th1) or in combination with degranulatory function (CD107a+ IFN-γ+) (termed cytolytic) after stimulation with HIV Gag peptides. These cells were then probed by Fluidigm BioMark HD analyses to dissect the gene profiles either associated with either cytotoxicity or classical Th1 function.
FIG 1.
Cytolytic CD4+ T cells are a distinct subset from Th1 helper CD4+ T cells. (A) CD4+ cells from HIV-infected patients were stimulated with Gag peptide pools and stained for CD107a (left) or IFN-γ (right). EC, elite controllers (n = 16); VC, viremic controllers (n = 4); CP, chronic progressors (n = 15). Mann-Whitney U tests were used for comparisons of subsets (*, P < 0.05). (B) CD4+ cells from HIV-infected patients (n = 6, numbered 1 to 6) were stimulated with Gag peptide pools and sorted into CD107a+ IFN-γ+ (red) and IFN-γ+ CD107a− (blue) populations. These cells, 100 per well, were then subjected to gene expression analysis via Fluidigm BioMark. Unsupervised hierarchical clustering of select gene expression from these cell populations was then performed using Pearson correlation matrices. Color indicates the relative minimum expression (yellow) to relative maximum expression (violet).
We found striking differences in the transcriptional program of Gag-specific CD107a+ IFN-γ+ CD4+ T cells compared to IFN-γ+ CD107a− CD4+ T cells (Fig. 1B). Performing unsupervised hierarchical clustering analyses on resultant transcriptional profiles revealed that CD107a+ IFN-γ+ displayed a transcriptional profile similar to the cytolytic effector functions that have been described for CD8+ T cells and natural killer (NK) cells, including granzymes A, B, and K and perforin. Besides these effector molecules, the surface natural killer 1 receptor (HNK-1; also known as CD57), which has also been previously linked to cytolytic activity of CD8+ T cells and NK cells, was prominently displayed, further suggesting cytolytic function in this subset (7–9). In contrast, Gag-specific Th1 CD4+ T cells showed higher expression of several surface markers associated with helper CD4+ T cell functions, including ICOS, interleukin-7 (IL-7) receptor, and CD161 (Fig. 1B). Together, these data demonstrate significant differences in the gene expression profile of HIV-specific cytolytic CD4+ T cells and HIV-specific Th1 cells and implicate CD107a+ IFN-γ+ cells as a unique cellular subset to classically defined Th1 cells.
CD57 phenotypically defines HIV-specific cytolytic CD4+ T cells.
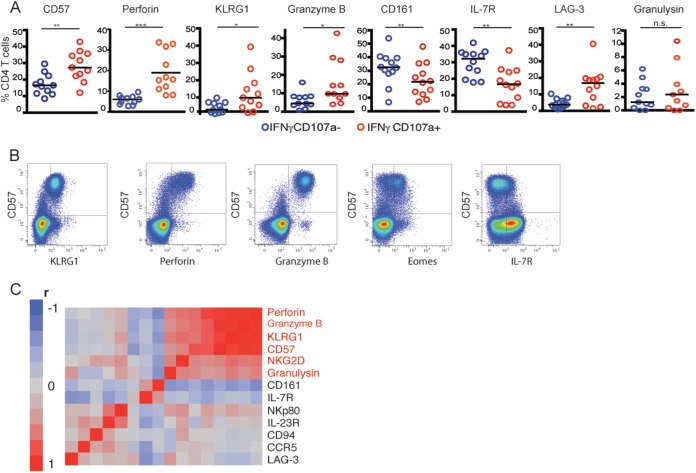
To further explore cellular phenotype associated with cytotoxic CD4+ T cells, we next analyzed phenotypic markers on the protein level of HIV-specific cells utilizing multiparameter flow cytometry. A panel of 16 putative biomarkers, which were significantly expressed in cytolytic CD4+ T cells versus Th1 cells by our previous Fluidigm BioMark analyses were assessed for protein expression via flow cytometry. Similar to prior transcriptional results, we found that perforin, granzyme B, and CD57 were expressed at significantly higher levels in HIV-specific cytolytic CD4+ T cells relative to Th1 cells, whereas IL-7 receptor and CD161 showed significantly lower expression (Fig. 2A). Moreover, in contrast to the mRNA results, we also found significantly higher expression of the killer-cell lectin-like receptor G1 (KLRG1) on HIV-specific cytolytic CD4+ T cells compared to HIV-specific Th1 CD4+ T cells. KLRG-1 has been previously associated with cytolytic effector function of CD8 T cells, further defining the phenotypic profile of cytolytic CD4+ T cells (10, 11). Interestingly, we found that CD57 surface expression could serve as a good surrogate to phenotypically describe HIV-specific cytolytic CD4+ T cells, since it showed close coexpression with other phenotypic and functional markers associated with cytolytic CD4+ T cell function, such as perforin and granzyme B expression (Fig. 2B). Moreover, we found that a set of six phenotypic markers tightly correlated with the cytolytic CD4+ T cell program (Fig. 2C), while seven phenotypic markers were instead associated with Th1 CD4+ T cell function. Thus, our data suggest that HIV-specific cytolytic CD4+ T cells have a phenotypically distinct profile compared to HIV-specific Th1 CD4+ T cells.
FIG 2.
CD57 is a marker of HIV-specific cytolytic CD4+ T cells. CD4+ cells from HIV-infected subjects (n = 12) were stimulated with Gag peptide pools and sorted into CD107a+ IFN-γ+ and IFN-γ+ CD107a− populations. These cells were then subjected to multiparameter flow cytometry. (A) Expression levels, presented as a percentage of CD4+ T cells, of CD57 and additional cytolytic markers. (B) Exemplary flow plots of CD57 coexpression with KLRG-1, perforin, granzyme B, Eomes, and IL-7R. (C) Coexpression of flow cytometric parameters from cytolytic (CD107a+ IFN-γ+) CD4+ T cell populations in Fig. 2A were determined by clustering by Pearson correlation matrices. Genes identified in red correspond to the identified cytolytic phenotype.
The cytolytic CD4+ T cell program is associated with the coexpression of two T-box transcription factors.
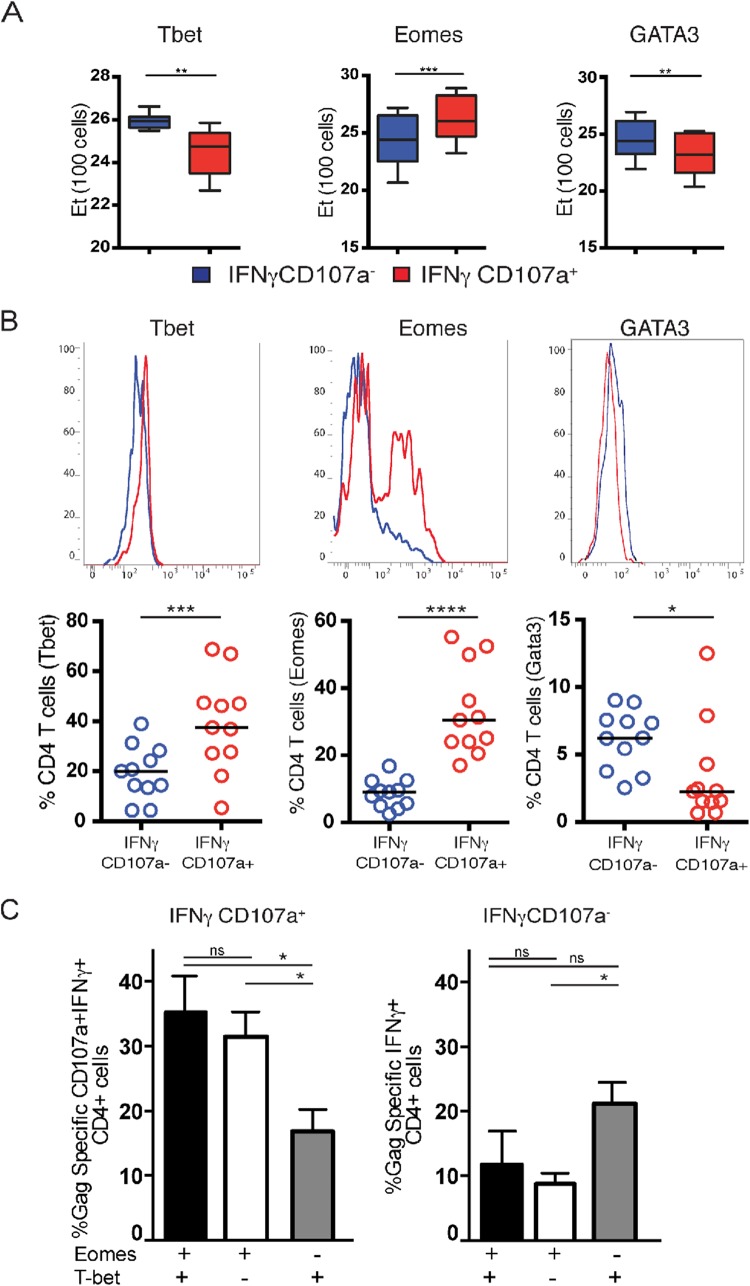
A CD4+ T cell lineage commitment is generally established through the expression of different characteristic transcription factors. The lineage commitment of Th1 cells has been previously linked to the expression of the transcription factor tbx21 (TBET) (12, 13). We therefore were interested in whether HIV-specific Th1 cells and HIV-specific cytolytic CD4+ T cells differ in their expression levels of T-box transcription factors, potentially further demonstrating differences in their lineage commitment. Using Fluidigm BioMark analysis as described above, we explored the expression of a panel of 12 characteristic CD4+ T cell lineage transcription factors in HIV-specific cytolytic CD4+ T cells compared to HIV-specific Th1 CD4+ T cells (Fig. 3A). Interestingly, we found significantly lower expression of TBET mRNA expression in HIV-specific cytolytic CD4+ T cells compared to Th1 cells (P = 0.003) (Fig. 3A). In contrast, the T-box transcription factor Eomesodermin (EOMES), which has been previously linked to cytolytic functions of CD8+ T cells (14), was significantly higher expressed in cytolytic CD4+ T cells compared to Th1 CD4+ T cells (P < 0.001). Other transcription factors that have been previously linked to cytolytic activity, including runx3 or THPOK displayed no differences between Th1 and cytolytic CD4+ T cells (data not shown). These differences in transcription factor mRNA expression were confirmed at the protein level by flow cytometry (Fig. 3B). Cytolytic CD4+ T cells were found to express higher levels of EOMES relative to Th1 cells, in agreement with our mRNA data (P < 0.0001). Unexpectedly, however, we observed higher levels of T-bet in cytolytic relative to Th1 CD4+ T cells as well (P < 0.05), while the Th2 transcription factor GATA3 was expressed at slightly higher levels in Th1 CD4+ T cells (P < 0.05), although these differences were relatively lower than that observed for Eomes. Moreover, we found that the coexpression of Eomes and T-bet, and to a lesser extent singular expression of Eomes, most closely associated with the cytolytic CD4+ T cell program, as previously suggested for mice (15–17) (Fig. 3C). As is the case for most transcription factors, however, a strict delineation of cytolytic versus Th1 CD4+ T cells was not achieved by comparing Eomes and T-bet. This may also indicate a role for additional transcription factors in maintenance of the cytolytic phenotype that have not been tested in the present study. Together, these data indicate that cytolytic and Th1 CD4+ T cells differ in their transcription factor expression, highlighting differences in their effector function.
FIG 3.
The cytolytic CD4+ T cell program is associated with T-box transcription factors. CD4+ T cells from HIV+ individuals (n = 12) were stimulated with Gag peptide pools and sorted into cytolytic (CD107a+ IFN-γ+) and Th1 (IFN-γ+ CD107a−) CD4+ T cell populations. These cells were then subjected to gene expression analysis via Fluidigm BioMark or flow cytometry. (A) Expression levels of transcription factors mRNA sorted HIV-specific cytolytic CD4+ T cells versus HIV-specific Th1 CD4+ T cells (blue, Th1 CD4+ T cells; red, cytolytic CD4+ T cells). (B) Protein expression of T-bet and Eomes in CD4+ cytolytic T cells and Th1 CD4+ T cells measured by flow cytometry. (C) Coexpression of T-bet and Eomes in cytolytic CD4+ versus Th1 CD4+ T cells measured by flow cytometry as in panel B.
Cytolytic CD4+ and CD8+ T cells share common cytolytic features.
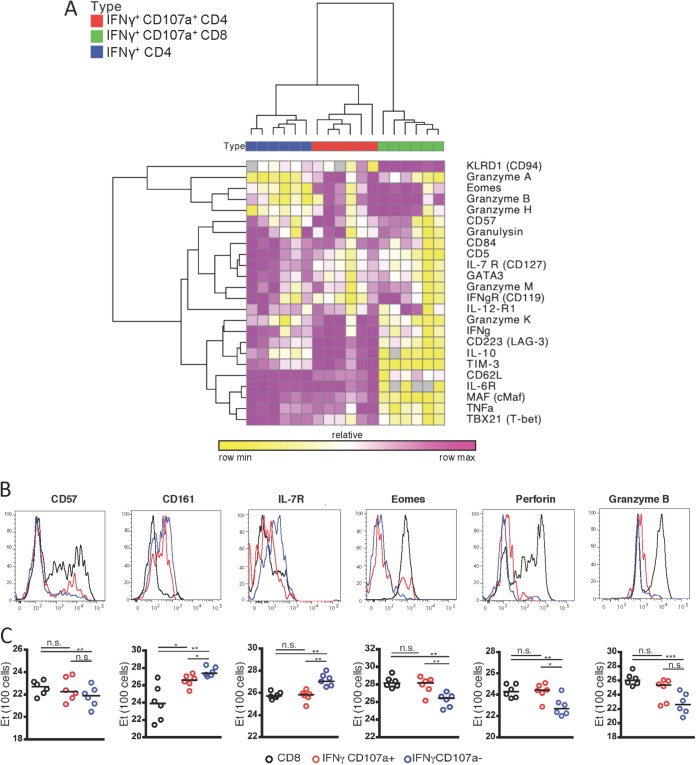
Given the expression of transcriptional factors associated with cytolytic function in cytolytic CD4+ T cells, we next sought to compare their transcriptional profile with classically defined cytolytic CD8+ T cells. We began by selecting 94 genes involved in T cell effector functions (see Table S1 in the supplemental material) and analyzed expression of these markers by Fluidigm BioMark analyses as described above for HIV-specific Th1 CD4+, cytolytic CD4+, and cytolytic CD8+ T cells. Neither CD4+ mRNA in CD8+ T cells nor CD8+ mRNA in CD4+ T cells, respectively, were detectable, confirming CD4+ and CD8+ T cell lineage commitment and sort purity (data not shown). Genes showing the broadest differences from the data set were then used in an unsupervised hierarchical clustering analysis (Fig. 4A). As expected, all three cell populations formed distinct clusters, and both Th1 and cytolytic CD4+ T cells clustered with one another, separate from cytolytic CD8+ T cells. However, despite the overall similarities in gene expression between the CD4+ T cell subsets, cytolytic CD4+ T cells uniquely shared a module of genes, including CD57, granzyme B, and granzyme H with cytolytic CD8+ T cells. Therefore, although there are fundamental differences between CD4+ and CD8+ T cell lineages, cytolytic CD4+ T cells express a module of genes classically associated with cytolytic function in CD8+ T cells.
FIG 4.
Cytolytic CD4+ cells and CD8+ T cells share common cytolytic features. PBMCs from HIV-infected patients (n = 6) were stimulated with Gag peptide pools and sorted into CD8+ T cells, cytolytic CD4+ T cells, or Th1 CD4+ T cell populations as defined above. These cells were then subjected to gene expression analysis via Fluidigm BioMark or flow cytometry. (A) Unsupervised hierarchical clustering analysis using Pearson coefficient of gene expression levels of selected genes associated with effector functions from HIV-specific CD8+ T cells (green), HIV-specific cytolytic CD4+ T cells (red), and HIV-specific Th1 CD4+ T cells (blue) for all six individuals. (B and C) Comparison of protein expression histograms (B) and gene expression of 100 cells via Fluidigm BioMark (C) of CD8+ (black), cytolytic CD4+ T cells (red), and Th1 CD4+ T cells (blue) for six cytolytic markers.
We next compared phenotypic differences between HIV-specific cytolytic CD4+ and CD8+ T cells. Using multicolor flow cytometry we found that CD8+ T cells shared on many levels common features with cytolytic CD4+ T cells but significantly differed from Th1 cells. Specifically, we found that cytolytic CD4+ and CD8+ T cells had a similar expression profile to that of the cytotoxic markers that we found describing cytolytic CD4+ T cells, including CD57, perforin, granzyme B, KLRG1, and the transcription factor Eomes (Fig. 4B). In contrast, these markers were significantly less expressed in Th1 CD4+ T cells. Notably, only the surface receptor CD161 was differently expressed in all three populations, with cytolytic CD4+ T cells exhibiting an intermediate expression level between CD8+ and Th1 CD4+ T cells. Taken together, our data demonstrate a striking similarity in the gene and surface expression profile of a cytotoxic program in cytolytic CD4+ T cells and CD8+ T cells.
Temporal cooperativity between HIV-specific cytolytic CD4+ T cells and HIV-specific CD8+ T cells in the clearance of virus-infected cells.
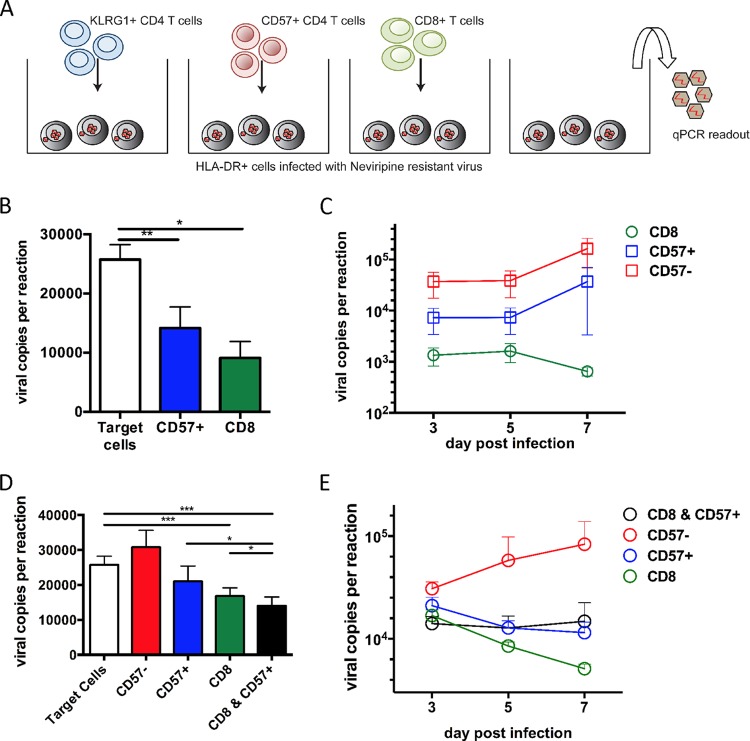
Previous studies have demonstrated the predominant role of HIV-specific CD8+ T cells in the control of HIV infection (18). However, given the transcriptional and phenotypic similarities between HIV-specific cytolytic CD4+ T cells and HIV-specific CD8+ T cells, we next sought to assess the ability of HIV-specific cytolytic CD4+ T cells and cytolytic CD8+ T cells to contribute to control of viral replication. To do so, we purified cytolytic CD4+ T cells based on the phenotypic marker CD57 and CD8+ T cells for use as effector cells in a modified viral inhibition assay (19). Activated HLA-DR+ CD4+ T cells were used as target cells and infected with nevirapine-resistant virus in culture, and effector CD4+ or CD8+ T cells were added at a ratio of 1:1 (Fig. 5A). As expected, after 3 days of coculture the CD8+ T cells showed robust viral inhibition of >2 logs (Fig. 5B). Cytolytic CD4+ T cells did not display significant differences in their viral inhibitory activity compared to CD8+ T cells, with both being able to reduce resultant virus titers by 32% (P = 0.0193) versus 43% (P = 0.0047), respectively. The ability to reduce viral replication between cytolytic CD4+ T cells and CD8+ T cells was not significantly different (P = 0.243) at day 3 after coculture. Importantly, noncytolytic CD4+ T cells (CD57−) had no effect on viral inhibition (Fig. 5B and C). These data suggest that while CD8+ T cells provide greater control of viral replication in coculture, cytolytic CD4+ T cells are also able to effect significant viral inhibition.
FIG 5.
Temporal cooperativity between HIV-specific cytolytic CD4+ T cells and HIV-specific CD8+ T cells in clearance of virus-infected cells. (A) Schematic drawing of the experimental setup. PBMCs from HIV+ donors were separated into multiple populations using MACS columns and Dynabead selection kits. For a target cell population, HLA-DR+ CD4+ T cells were infected at an MOI of 0.01 with a nevirapine-resistant R5-tropic virus. Effector cells and target cells were combined at a ratio of 1:1 with the respective population: CD8+, CD57+ cytolytic CD4+ T cells and CD57− noncytolytic CD4+ T cells. (B) Comparable levels of viral inhibition by CD8+ T cells and cytolytic CD57+ CD4+ T cells at 3 days postculture (n = 10). (C) Viral inhibition with increasing viral loads in cytolytic CD4+ T cells but decreasing viral loads in CD8+ T cells at 7 days postculture (n = 5). (D) Robust cooperativity between CD8+ T cells and cytolytic CD4+ T cells in the viral inhibition of HIV-infected cells. The addition of cytolytic CD4+ T cells to CD8+ T cells increased viral inhibition by 18% (P = 0.0089) at day 3 postcoculture (n = 5; Mann-Whitney U tests). (E) Cooperative short-term viral inhibition by CD8+ T cells and cytolytic CD4+ T cells (n = 5).
We next probed the possibility of whether cytolytic CD4+ T cells and cytolytic CD8+ T cells could display cooperativity in the control of HIV replication. To test this possibility, we repeated our coculture viral inhibition assay adding cytolytic CD4+ T cells and CD8+ T cells in equal ratio and controlled for the overall cell number. Strikingly, the addition of cytolytic CD4+ T cells further increased viral inhibition relative to cytolytic CD8+ T cells alone by on average of an additional 18% on day 3 (P = 0.009) (Fig. 5D and E). This effect was most potent at early time points in infection and waned over time. Only in the cultures of cytolytic CD4+ T cells in which CD8+ T cells were present was the inhibitory effect still observed. However, at day 7 of coculture, the addition of cytolytic CD4+ T cells to CD8+ T cells did not have an additional effect of suppressive activity. We speculate that this may occur due to cytolytic CD4+ T cells themselves becoming infected in the coculture assay and subsequently losing their protective activity. Thus, our data suggest a concerted action of cytolytic CD4+ and CD8+ T cells, which may be diminished if early viral infection is not controlled.
Acute CD57+ CD4+ T cell kinetics suggest involvement of cytolytic CD4+ T cells in HIV control.
During acute HIV infection, a massive depletion of activated CD4+ T cells occurs (20), this observation is mirrored by an increased infection of cytolytic CD4+ T cells by day 7 of our coculture experiments. We therefore investigated whether cytolytic CD4+ T cells are preferentially infected and depleted in the course of HIV infection. We first assessed the susceptibility of cytolytic CD4+ T cells to HIV infection in vitro. Cytolytic CD4+ T and Th1 CD4+ cells were enriched based on CD57 or CD161 expression as surrogate phenotypic markers for cytolytic CD4+ T cells and Th1 CD4+ T cells, respectively, from PBMCs of 10 HIV-uninfected individuals. We next infected each population with a panel of R5-tropic primary isolates, as well as NL4-3 virus (21). Although we observed that for some R5 viruses the CD57+ cytolytic CD4+ T cells to be less susceptible to infection than CD57− CD4+ T cells, this difference was not statistically significant for all four viruses tested (P = 0.58) (Fig. 6A). Similarly, we found that CD161+ Th1 CD4+ T cells showed slightly higher levels of infectibility by R5 viruses compared to CD161− CD4+ T cells (P = 0.021). There were no differences in the susceptibility to the X4 virus NL4-3 between cytolytic and Th1 CD4+ T cells (Fig. 6B). These data are in line with previous data that suggested that CD57+ CD4 T cells are slightly less infectible (22).
Due to the correlation between the emergence of cytolytic CD8+ T cells and the acute control of HIV viremia, we finally explored whether cytolytic CD4+ T cells similarly emerge and assist in viral control in acute HIV infection (18). We began by measuring early kinetics of CD57+ CD4+ T cells in a cohort of 24 acutely HIV infected at days 0, 5, 9, 17, 32, and 52 of infection. Strikingly, the emergence of cytolytic CD57+ CD4+ T cells was tightly associated with viral replication (Fig. 6C). In contrast, the memory CD4+ T cell population as CD45RO+ CD4+ T cells markedly dropped during acute HIV infection. These data are in agreement with our infectibility assays showing decreased infection of cytolytic CD57+ CD4+ T cells and furthermore suggest a role for these cells in acute HIV infection. Finally, in order to assess whether cytolytic CD4+ T cells may have a role in control of viremia and subsequent viral set point, we performed a correlation analysis on the degree of CD57+ CD4+ T cell expansion and subsequent early viral set point. Early emergence of CD57+ CD4+ T cells displayed a moderate inverse correlation with the early viral set point, again suggesting an active role of cytolytic CD4+ T cells in primary HIV infection (R = −0.44, P = 0.03) (Fig. 6D). We found, based on these results, that despite the susceptibility of cytolytic CD4+ T cells to HIV infection, the emergence of these cells during acute HIV infection is associated with early control of viremia. Thus, we describe here a model (Fig. 6E) wherein the HIV-specific cytolytic CD4+ T cells and CD8+ T cells cooperate in the control of early viral replication. Since cytolytic CD4+ T cells are still prone to infection, CD8+ T cells are required to keep overall viral replication in check, and yet cytolytic CD4+ T cells can contribute additively to the overall direct antiviral response.
DISCUSSION
Although CD4+ T cells exhibit a host of different helper functions that contribute to the HIV-specific immune response, a growing body of evidence supports the idea that direct cytolytic effector activity by CD4+ T cells is also important for HIV control and clearance (1, 23, 24). Indeed, besides HIV-specific CD8+ T cells, the emergence of HIV-specific cytolytic CD4+ T cells during acute HIV infection has been linked to control of viral replication (5, 25). Furthermore, a publication studying the effect of CD4+ T cell depletion prior to SIV infection had the unexpected result of the loss of post-peak SIV control despite the presence of HIV-specific CD8+ T cells (24). Subsequent analysis showed that this lack of control may be due to the depletion of cytolytic CD4+ T cells. In contrast, depletion of CD8+ T cells and control of SIV viremia was associated with the emergence of humoral and cytolytic CD4+ T cell responses (23). Beyond natural HIV infection, the ability to degranulate upon antigenic recognition and release effector molecules such as perforin and granzyme B has been shown to be a key feature of antigen-specific CD4+ T cells in other viral infections such as CMV and hepatitis C (26), as well as in cases of cancer (27) and after vaccination (2, 28).
We therefore sought to describe more fully the biology of HIV-specific cytolytic CD4+ T cells and understand their underlying biology and potential cooperation with HIV-specific CD8+ T cells in the control of HIV viremia. We found that cytolytic CD4+ T cells represent a distinct subset of CD4+ T cells that differ significantly in their transcriptional and phenotypic profile compared to Th1 cells, characterized by the expression of a module of genes associated with cytolytic function, as well as the T-box transcription factors T-bet and Eomes. Interestingly, the combinational expression of both have been previously shown to be linked to cytolytic function of cytolytic CD4+ T cells in lymphochoriomeningitis virus-infected mice (14), which has also been shown to be linked to the cytolytic activity of CD8+ T cells (29, 30). We observed that the transcriptional and phenotypic profile of CD8+ T cells and cytolytic CD4+ T cells were very similar in many respects. We found that the surface molecule human natural killer-1 receptor (HNK-1/CD57), which has been previously linked to cytolytic activity of CD8+ T cells, also best characterizes cytolytic CD4+ T cells (7, 31, 32). Interestingly, CD57 has been previously noted to denote a nonproliferating CD4+ T cell population that is present in higher numbers in HIV+ untreated individuals and predominantly secrete IFN-γ (33). We also found that levels of effector molecules of granzyme B and perforin were not significantly different between cytolytic CD4+ and cytolytic CD8+ T cells, further stressing their role and cooperativity in the control of HIV replication between both cell types. Taken together, these data indicate that cytolytic CD4+ T cells are a genuinely separate lineage of CD4+ T cells, which display phenotypic and transcriptional similarities to classically defined cytolytic functions of CD8+ T cells. The specific signals and pathways that lead to the induction of such cells and whether they arise from a particular helper progenitor subset is therefore of great interest and warrants further investigation.
We observed that CD57+ cytolytic CD4+ T cells dramatically expand during acute HIV infection, tightly following the viral load trajectory. Interestingly, the set point viral load was inversely correlated with the presence of CD57+ cytolytic CD4+ T cells during acute infection. This observation is particularly interesting in light of a recent study that also found that the frequency of CD57+ expressing CD8+ T cells is inversely correlated with HIV progression, suggesting a cooperative role for these cytotoxic cell subsets to control HIV loads in chronic infection (34). To probe this possibility, we tested the ability of CD57+ CD4+ T cells and cytotoxic CD8+ T cells to cooperatively inhibit viral replication. We showed that cytolytic CD4+ T cells are functional and able to inhibit viral replication to a similar extent as CD8+ T cells and that they displayed cooperativity in viral control. However, this effect was seen only early during the viral inhibition assay when the effect of CD8+ T cell inhibition was less pronounced than after 5 or 7 days. The effect of CD4+ T cell killing was later masked by the infection of CD4+ T cells themselves. The cooperativity observed between CD8+ and CD4+ T cells in viral inhibition is even more interesting when one considers that the efficacy of HIV-specific CD8+ T cells in controlling viral replication is more susceptible to HIV evasion strategies, including escape from targeted epitopes. The peptide-binding modalities of major histocompatibility complex class II (MHC-II) allow for much greater sequence diversity, reducing the ability of the virus to escape from HIV-specific CD4+ T cell responses (35, 36). Although MHC-II epitope escape has been previously reported (37, 38), it is less often observed. The fact that CD4+ T cells and CD8+ T cells commonly use different antigen recognition pathways may potentially allow for dual targeting of the same infected cell and thereby reduce the ability of HIV escape by epitope mutation. A key question is therefore whether individuals better able to control HIV replication display evidence of MHC-II-mediated selection pressure and whether such responses can be augmented by therapeutic interventions.
Taken together, our data show that HIV-specific cytolytic CD4+ T cells are distinct from Th1 CD4+ cells. These cytolytic CD4+ T cells share common features with cytolytic CD8+ T cells also and act in concert with CD8+ T cells to control viral replication. These data suggest a role for the combined activity of both cell types in the long-term control of HIV infection and provide crucial insight into the processes underlying HIV control, as well as putative protection from infection by vaccination.
Supplementary Material
ACKNOWLEDGMENTS
We thank Matt Creegan for technical support.
This study was funded by the U.S. National Institutes of Health (NIH; R01 AI091450-01 and R01 AI094602-01) and a cooperative agreement (W81XWH-11-2-0174) between the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., and the U.S. Department of Defense. The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: nevirapine; human rIL-2 from Maurice Gately, Hoffmann-La Roche, Inc.; and pNL4-3 from Malcolm Martin.
The views expressed are those of the authors and should not be construed to represent the positions of the U.S. Army or the Department of Defense.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00438-15.
REFERENCES
- 1.Soghoian DZ, Streeck H. 2010. Cytolytic CD4+ T cells in viral immunity. Expert Rev Vaccines 9:1453–1463. doi: 10.1586/erv.10.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilkinson TM, Li CK, Chui CS, Huang AK, Perkins M, Liebner JC, Lambkin-Williams R, Gilbert A, Oxford J, Nicholas B, Staples KJ, Dong T, Douek DC, McMichael AJ, Xu XN. 2012. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med 18:274–280. doi: 10.1038/nm.2612. [DOI] [PubMed] [Google Scholar]
- 3.Hunder NN, Wallen H, Cao J, Hendricks DW, Reilly JZ, Rodmyre R, Jungbluth A, Gnjatic S, Thompson JA, Yee C. 2008. Treatment of metastatic melanoma with autologous CD4+ T cells against NY-ESO-1. N Engl J Med 358:2698–2703. doi: 10.1056/NEJMoa0800251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casazza JP, Betts MR, Price DA, Precopio ML, Ruff LE, Brenchley JM, Hill BJ, Roederer M, Douek DC, Koup RA. 2006. Acquisition of direct antiviral effector functions by CMV-specific CD4+ T lymphocytes with cellular maturation. J Exp Med 203:2865–2877. doi: 10.1084/jem.20052246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soghoian DZ, Jessen H, Flanders M, Sierra-Davidson K, Cutler S, Pertel T, Ranasinghe S, Lindqvist M, Davis I, Lane K, Rychert J, Rosenberg ES, Piechocka-Trocha A, Brass AL, Brenchley JM, Walker BD, Streeck H. 2012. HIV-specific cytolytic CD4 T cell responses during acute HIV infection predict disease outcome. Sci Transl Med 4:123ra125. doi: 10.1126/scitranslmed.3003165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McDavid A, Finak G, Chattopadyay PK, Dominguez M, Lamoreaux L, Ma SS, Roederer M, Gottardo R. 2013. Data exploration, quality control and testing in single-cell qPCR-based gene expression experiments. Bioinformatics 29:461–467. doi: 10.1093/bioinformatics/bts714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee SA, Sinclair E, Hatano H, Hsue PY, Epling L, Hecht FM, Bangsberg DR, Martin JN, McCune JM, Deeks SG, Hunt PW. 2014. Impact of HIV on CD8+ T cell CD57 expression is distinct from that of CMV and aging. PLoS One 9:e89444. doi: 10.1371/journal.pone.0089444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petrovas C, Chaon B, Ambrozak DR, Price DA, Melenhorst JJ, Hill BJ, Geldmacher C, Casazza JP, Chattopadhyay PK, Roederer M, Douek DC, Mueller YM, Jacobson JM, Kulkarni V, Felber BK, Pavlakis GN, Katsikis PD, Koup RA. 2009. Differential association of programmed death-1 and CD57 with ex vivo survival of CD8+ T cells in HIV infection. J Immunol 183:1120–1132. doi: 10.4049/jimmunol.0900182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White MJ, Nielsen CM, McGregor RH, Riley EH, Goodier MR. 2014. Differential activation of CD57-defined natural killer cell subsets during recall responses to vaccine antigens. Immunology 142:140–150. doi: 10.1111/imm.12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang W, Kachapati K, Adams D, Wu Y, Leung PS, Yang GX, Zhang W, Ansari AA, Flavell RA, Gershwin ME, Ridgway WM. 2014. Murine autoimmune cholangitis requires two hits: cytotoxic KLRG1+ CD8 effector cells and defective T regulatory cells. J Autoimmun 50:123–134. doi: 10.1016/j.jaut.2014.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beyersdorf NB, Ding X, Karp K, Hanke T. 2001. Expression of inhibitory “killer cell lectin-like receptor G1” identifies unique subpopulations of effector and memory CD8 T cells. Eur J Immunol 31:3443–3452. doi:. [DOI] [PubMed] [Google Scholar]
- 12.Miller SA, Weinmann AS. 2010. Molecular mechanisms by which T-bet regulates T-helper cell commitment. Immunol Rev 238:233–246. doi: 10.1111/j.1600-065X.2010.00952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Placek K, Coffre M, Maiella S, Bianchi E, Rogge L. 2009. Genetic and epigenetic networks controlling T helper 1 cell differentiation. Immunology 127:155–162. doi: 10.1111/j.1365-2567.2009.03059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eshima K, Chiba S, Suzuki H, Kokubo K, Kobayashi H, Iizuka M, Iwabuchi K, Shinohara N. 2012. Ectopic expression of a T-box transcription factor, Eomesodermin, renders CD4+ Th cells cytotoxic by activating both perforin- and FasL-pathways. Immunol Lett 144:7–15. doi: 10.1016/j.imlet.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 15.Workman AM, Jacobs AK, Vogel AJ, Condon S, Brown DM. 2014. Inflammation enhances IL-2 driven differentiation of cytolytic CD4 T cells. PLoS One 9:e89010. doi: 10.1371/journal.pone.0089010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qui HZ, Hagymasi AT, Bandyopadhyay S, St Rose MC, Ramanarasimhaiah R, Menoret A, Mittler RS, Gordon SM, Reiner SL, Vella AT, Adler AJ. 2011. CD134 plus CD137 dual costimulation induces Eomesodermin in CD4 T cells to program cytotoxic Th1 differentiation. J Immunol 187:3555–3564. doi: 10.4049/jimmunol.1101244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Curran MA, Geiger TL, Montalvo W, Kim M, Reiner SL, Al-Shamkhani A, Sun JC, Allison JP. 2013. Systemic 4-1BB activation induces a novel T cell phenotype driven by high expression of Eomesodermin. J Exp Med 210:743–755. doi: 10.1084/jem.20121190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borrow P, Lewicki H, Hahn BH, Shaw GM, Oldstone MB. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J Virol 68:6103–6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saez-Cirion A, Shin SY, Versmisse P, Barre-Sinoussi F, Pancino G. 2010. Ex vivo T cell-based HIV suppression assay to evaluate HIV-specific CD8+ T-cell responses. Nat Protoc 5:1033–1041. doi: 10.1038/nprot.2010.73. [DOI] [PubMed] [Google Scholar]
- 20.Douek DC, Brenchley JM, Betts MR, Ambrozak DR, Hill BJ, Okamoto Y, Casazza JP, Kuruppu J, Kunstman K, Wolinsky S, Grossman Z, Dybul M, Oxenius A, Price DA, Connors M, Koup RA. 2002. HIV preferentially infects HIV-specific CD4+ T cells. Nature 417:95–98. doi: 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- 21.Spentzou A, Bergin P, Gill D, Cheeseman H, Ashraf A, Kaltsidis H, Cashin-Cox M, Anjarwalla I, Steel A, Higgs C, Pozniak A, Piechocka-Trocha A, Wong J, Anzala O, Karita E, Dally L, Gotch F, Walker B, Gilmour J, Hayes P. 2010. Viral inhibition assay: a CD8 T cell neutralization assay for use in clinical trials of HIV-1 vaccine candidates. J Infect Dis 201:720–729. doi: 10.1086/650492. [DOI] [PubMed] [Google Scholar]
- 22.Brenchley JM, Ruff LE, Casazza JP, Koup RA, Price DA, Douek DC. 2006. Preferential infection shortens the life span of human immunodeficiency virus-specific CD4+ T cells in vivo. J Virol 80:6801–6809. doi: 10.1128/JVI.00070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von Gegerfelt A, Valentin A, Alicea C, Van Rompay KK, Marthas ML, Montefiori DC, Pavlakis GN, Felber BK. 2010. Emergence of simian immunodeficiency virus-specific cytotoxic CD4+ T cells and increased humoral responses correlate with control of rebounding viremia in CD8-depleted macaques infected with Rev-independent live-attenuated simian immunodeficiency virus. J Immunol 185:3348–3358. doi: 10.4049/jimmunol.1000572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ortiz AM, Klatt NR, Li B, Yi Y, Tabb B, Hao XP, Sternberg L, Lawson B, Carnathan PM, Cramer EM, Engram JC, Little DM, Ryzhova E, Gonzalez-Scarano F, Paiardini M, Ansari AA, Ratcliffe S, Else JG, Brenchley JM, Collman RG, Estes JD, Derdeyn CA, Silvestri G. 2011. Depletion of CD4+ T cells abrogates post-peak decline of viremia in SIV-infected rhesus macaques. J Clin Invest 121:4433–4445. doi: 10.1172/JCI46023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenberg ES, Billingsley JM, Caliendo AM, Boswell SL, Sax PE, Kalams SA, Walker BD. 1997. Vigorous HIV-1-specific CD4+ T cell responses associated with control of viremia. Science 278:1447–1450. doi: 10.1126/science.278.5342.1447. [DOI] [PubMed] [Google Scholar]
- 26.Aslan N, Yurdaydin C, Wiegand J, Greten T, Ciner A, Meyer MF, Heiken H, Kuhlmann B, Kaiser T, Bozkaya H, Tillmann HL, Bozdayi AM, Manns MP, Wedemeyer H. 2006. Cytotoxic CD4 T cells in viral hepatitis. J Viral Hepat 13:505–514. doi: 10.1111/j.1365-2893.2006.00723.x. [DOI] [PubMed] [Google Scholar]
- 27.Akhmetzyanova I, Zelinskyy G, Schimmer S, Brandau S, Altenhoff P, Sparwasser T, Dittmer U. 2013. Tumor-specific CD4+ T cells develop cytotoxic activity and eliminate virus-induced tumor cells in the absence of regulatory T cells. Cancer Immunol Immunother 62:257–271. doi: 10.1007/s00262-012-1329-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Souza MS, Ratto-Kim S, Chuenarom W, Schuetz A, Chantakulkij S, Nuntapinit B, Valencia-Micolta A, Thelian D, Nitayaphan S, Pitisuttithum P, Paris RM, Kaewkungwal J, Michael NL, Rerks-Ngarm S, Mathieson B, Marovich M, Currier JR, Kim JH. 2012. The Thai phase III trial (RV144) vaccine regimen induces T cell responses that preferentially target epitopes within the V2 region of HIV-1 envelope. J Immunol 188:5166–5176. doi: 10.4049/jimmunol.1102756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, Banica M, DiCioccio CB, Gross DA, Mao CA, Shen H, Cereb N, Yang SY, Lindsten T, Rossant J, Hunter CA, Reiner SL. 2003. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science 302:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 30.Hersperger AR, Martin JN, Shin LY, Sheth PM, Kovacs CM, Cosma GL, Makedonas G, Pereyra F, Walker BD, Kaul R, Deeks SG, Betts MR. 2011. Increased HIV-specific CD8+ T-cell cytotoxic potential in HIV elite controllers is associated with T-bet expression. Blood 117:3799–3808. doi: 10.1182/blood-2010-12-322727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chattopadhyay PK, Betts MR, Price DA, Gostick E, Horton H, Roederer M, De Rosa SC. 2009. The cytolytic enzymes granyzme A, granzyme B, and perforin: expression patterns, cell distribution, and their relationship to cell maturity and bright CD57 expression. J Leukoc Biol 85:88–97. doi: 10.1189/jlb.0208107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopez-Verges S, Milush JM, Pandey S, York VA, Arakawa-Hoyt J, Pircher H, Norris PJ, Nixon DF, Lanier LL. 2010. CD57 defines a functionally distinct population of mature NK cells in the human CD56dimCD16+ NK-cell subset. Blood 116:3865–3874. doi: 10.1182/blood-2010-04-282301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palmer BE, Blyveis N, Fontenot AP, Wilson CC. 2005. Functional and phenotypic characterization of CD57+ CD4+ T cells and their association with HIV-1-induced T cell dysfunction. J Immunol 175:8415–8423. doi: 10.4049/jimmunol.175.12.8415. [DOI] [PubMed] [Google Scholar]
- 34.Lee SA, Sinclair E, Jain V, Huang Y, Epling L, Van Natta M, Meinert CL, Martin JN, McCune JM, Deeks SG, Lederman MM, Hecht FM, Hunt PW. 2014. Low proportions of CD28− CD8+ T cells expressing CD57 can be reversed by early ART initiation and predict mortality in treated HIV infection. J Infect Dis 210:374–382. doi: 10.1093/infdis/jiu109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muraille E. 2014. Generation of individual diversity: a too neglected fundamental property of adaptive immune system. Front Immunol 5:208. doi: 10.3389/fimmu.2014.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vossen MT, Westerhout EM, Soderberg-Naucler C, Wiertz EJ. 2002. Viral immune evasion: a masterpiece of evolution. Immunogenetics 54:527–542. doi: 10.1007/s00251-002-0493-1. [DOI] [PubMed] [Google Scholar]
- 37.Jones RB, Yue FY, Gu XX, Hunter DV, Mujib S, Gyenes G, Mason RD, Mohamed R, MacDonald KS, Kovacs C, Ostrowski MA. 2009. Human immunodeficiency virus type 1 escapes from interleukin-2-producing CD4+ T-cell responses without high-frequency fixation of mutations. J Virol 83:8722–8732. doi: 10.1128/JVI.00433-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burwitz BJ, Giraldo-Vela JP, Reed J, Newman LP, Bean AT, Nimityongskul FA, Castrovinci PA, Maness NJ, Leon EJ, Rudersdorf R, Sacha JB. 2012. CD8+ and CD4+ cytotoxic T cell escape mutations precede breakthrough SIVmac239 viremia in an elite controller. Retrovirology 9:91. doi: 10.1186/1742-4690-9-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.