ABSTRACT
Kaposi's sarcoma-associated herpesvirus (KSHV) is etiologically associated with Kaposi's sarcoma (KS), primary effusion lymphoma (PEL), and multicentric Castleman's disease. We have previously shown that KSHV utilizes the host transcription factor Nrf2 to aid in infection of endothelial cells and oncogenesis. Here, we investigate the role of Nrf2 in PEL and PEL-derived cell lines and show that KSHV latency induces Nrf2 protein levels and transcriptional activity through the COX-2/PGE2/EP4/PKCζ axis. Next-generation sequencing of KSHV transcripts in the PEL-derived BCBL-1 cell line revealed that knockdown of this activated Nrf2 results in global elevation of lytic genes. Nrf2 inhibition by the chemical brusatol also induces lytic gene expression. Both Nrf2 knockdown and brusatol-mediated inhibition induced KSHV lytic reactivation in BCBL-1 cells. In a series of follow-up experiments, we characterized the mechanism of Nrf2-mediated regulation of KSHV lytic repression during latency. Biochemical assays showed that Nrf2 interacted with KSHV latency-associated nuclear antigen 1 (LANA-1) and the host transcriptional repressor KAP1, which together have been shown to repress lytic gene expression. Promoter studies showed that although Nrf2 alone induces the open reading frame 50 (ORF50) promoter, its association with LANA-1 and KAP1 abrogates this effect. Interestingly, LANA-1 is crucial for efficient KAP1/Nrf2 association, while Nrf2 is essential for LANA-1 and KAP1 recruitment to the ORF50 promoter and its repression. Overall, these results suggest that activated Nrf2, LANA-1, and KAP1 assemble on the ORF50 promoter in a temporal fashion. Initially, Nrf2 binds to and activates the ORF50 promoter during early de novo infection, an effect that is exploited during latency by LANA-1-mediated recruitment of the host transcriptional repressor KAP1 on Nrf2. Cell death assays further showed that Nrf2 and KAP1 knockdown induce significant cell death in PEL cell lines. Our studies suggest that Nrf2 modulation through available oral agents is a promising therapeutic approach in the treatment of KSHV-associated malignancies.
IMPORTANCE KS and PEL are aggressive KSHV-associated malignancies with moderately effective, highly toxic chemotherapies. Other than ganciclovir and alpha interferon (IFN-α) prophylaxis, no KSHV-associated chemotherapy targets the underlying infection, a major oncogenic force. Hence, drugs that selectively target KSHV infection are necessary to eradicate the malignancy while sparing healthy cells. We recently showed that KSHV infection of endothelial cells activates the transcription factor Nrf2 to promote an environment conducive to infection and oncogenesis. Nrf2 is modulated through several well-tolerated oral agents and may be an important target in KSHV biology. Here, we investigate the role of Nrf2 in PEL and demonstrate that Nrf2 plays an important role in KSHV gene expression, lytic reactivation, and cell survival by interacting with the host transcriptional repressor KAP1 and the viral latency-associated protein LANA-1 to mediate global lytic gene repression and thus cell survival. Hence, targeting Nrf2 with available therapies is a viable approach in the treatment of KSHV malignancies.
INTRODUCTION
Kaposi's sarcoma-associated herpesvirus (KSHV) is a lymphotropic gammaherpesvirus and is the etiological agent of Kaposi's sarcoma (KS), primary effusion lymphoma (PEL), and the plasmablastic variant of multicentric Castleman's disease (MCD) (1–3). In immunocompetent individuals, KSHV is latent in B lymphocytes, whereas in immunocompromised patients it undergoes reactivation and dissemination throughout the body, often infecting several cell types, including endothelial cells. This uncontrolled KSHV dissemination results in the development of the highly vascular, endothelium-derived KS (4). Often, PEL arises in a monoclonal fashion from an infected, hyperproliferative, KSHV-infected B cell (1, 5). Despite aggressive treatments, PEL remains resistant to multidrug chemotherapies and is considered universally lethal (6).
In vitro, KSHV is isolated from PEL cell lines latently infected with the virus by inducing lytic reactivation with 12-O-tetradecanoyl-phorbol-13-acetate (TPA) or sodium butyrate (NaB) (5, 7, 8). Upon de novo infection of permissive cell types, such as human dermal microvascular endothelial cells (HMVEC-d), an initial burst of lytic gene expression with immunomodulatory and antiapoptotic functions is followed by establishment of latency (9). The mechanism through which KSHV induces these lytic genes during early infection and subsequently suppresses them in latency is poorly understood. Chromatin immunoprecipitation techniques coupled with KSHV genome-sequencing methods (ChIP-seq) have proved to be a remarkable tool in analyzing the chromatin landscape of the KSHV genome that is present during KSHV infection. Specifically, it has been shown that during latency establishment, immediate-early (IE) and early (E) lytic KSHV genes, including the lytic cycle regulator open reading frame 50 (ORF50/RTA), are heterochromatinized with the repressive histone marker H3K27me3 (10, 11). Concomitantly, these histones are also tagged with the activating marker H3K4me3 (10, 11). In a bivalent state, the repressive marker takes priority but can be quickly removed by histone demethylases, giving way to the activating markers (10). This dynamic bivalent state is observed during TPA and NaB reactivation, which reduces the repressive marker H3K27me3 on the IE and E genes (12). While these studies have shed light on the heterochromatic changes that lead to repression and derepression of lytic genes and constitutive expression of latent genes, the transcription factors that are involved in these modifications remain unclear, though some of them are analyzed in a recent summary (12).
Latency-associated nuclear antigen 1 (LANA-1), expressed by KSHV ORF73, is a major latency protein with pleiotropic functions. LANA-1 plays an important role in repressing lytic genes by binding to and repressing the ORF50/RTA promoter (13, 14). It was recently found that LANA-1 recruits the host transcriptional repressor KAP1 (Krüppel-associated box [KRAB]-associated protein 1; also known as TRIM28 or TIF1β), which repressed the ORF50 promoter (15, 16). However, the location, mechanism, and dynamics of LANA-1/KAP1 binding to the ORF50 promoter have yet to be investigated.
Nuclear factor E2-related factor 2 (Nrf2) is a member of the Kap'n'Collar basic leucine zipper (bZIP) family of transcription factors and plays a central role in the cellular response to oxidative stress (17). During cellular stress or proliferative signaling, Nrf2 dissociates from its inhibitor, kelch-like ECH-associated protein 1 (Keap1), allowing Nrf2 stabilization and accumulation (18). Additional signaling involving several reported kinases induces serine-40 phosphorylation of Nrf2, enhancing its nuclear translocation and transcriptional activity (19, 20). De facto Nrf2 target genes include the genes encoding NQO1 and HO1, two antioxidants that are key in maintaining redox homeostasis (21, 22). Novel Nrf2 target genes also include the genes encoding antiapoptotic Bcl-2/Bcl-xL (23); the proangiogenic HIF-1α/VEGF axis (24, 25); prometastatic MMP9 (26); the proliferative pentose phosphate pathway enzymes G6PD, TKT, and TALDO (27); the drug resistance proteins Mrp1 and Mrp2 (28, 29); and the proinflammatory cyclooxygenase 2 (COX-2), making constitutive Nrf2 activation perilous to the cell (30). Multiple cancer types have gain-of-function Nrf2 mutations or loss-of-function Keap1 mutations, confirming its oncogenic potential (31).
In a set of recent studies, we demonstrated that de novo KSHV infection of endothelial cells induced Nrf2 through multiple mechanisms to create a microenvironment conducive to infection (30, 32). In the current study, we further investigated the role of Nrf2 in KSHV gene expression, focusing mainly on latently infected, PEL-derived cell lines. We demonstrate that latent KSHV infection induces Nrf2, which plays an important role in the dynamic changes observed in ORF50 expression. In the absence of LANA-1, Nrf2 acts as a transcription activator, but it functions as a repressor in the presence of LANA-1. We determined that this switch in Nrf2's role on the ORF50 promoter is mediated by LANA-1-mediated recruitment of the transcriptional repressor KAP1, ultimately leading to ORF50 repression and establishment of latency. Nrf2 inhibition further resulted in increased KSHV lytic cycle gene expression, viral-DNA (virion) production, and PEL cell death. Collectively, this study demonstrates that KSHV induces Nrf2 to facilitate lytic gene expression during de novo infection and to later repress this induction by using LANA-1-mediated KAP1 recruitment to the Nrf2 binding site.
MATERIALS AND METHODS
Cells and tissues.
KSHV-positive BCBL-1 and BC-3 and KSHV-negative Ramos, Akata, and BJAB cells were cultured in RPMI 1640 GlutaMax (Gibco Life Technologies, Grand Island, NY). BJAB cells harboring KSHV in an episomal form (BJAB-KSHV), obtained from Blossom Damania (University of North Carolina, Chapel Hill) (33), were cultured in RPMI 1640 GlutaMax supplied with the eukaryotic selection factor hygromycin (200 μg/ml). All B cell media were supplied with 10% fetal bovine serum (FBS) and penicillin-streptomycin. HEK293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM), while TIVE and TIVE/LTC cells, obtained from Rolf Renne (University of Florida) (34), and HMVEC-d were cultured in EBM-2 medium (Lonza, Walkersville, MD) (30, 32). All endothelial cell media were supplied with endothelial growth factors (EGM2). Formalin-fixed, paraffin-embedded stomach tissue samples from healthy subjects and patients with PEL were obtained from the AIDS and Cancer Specimen Resource, San Francisco, CA (ACSR).
Plasmid transfections, lentivirus production, and transduction of B cells.
For transfection, subconfluent HEK293T cells were transfected for 24 to 48 h with 1 μg/ml of plasmid DNA using calcium phosphate precipitation prior to experimentation. Lentiviral vectors containing short hairpin RNA against Nrf2 (shNrf2) (TRCN0000007558) and KAP1 (shKAP1) (TRCN0000018002) were purchased from ThermoFisher Scientific (Waltham, MA), lentiviral hemagglutinin (HA)-tagged KAP1 (HA-KAP1) (plasmid 45569) was obtained from Addgene (Cambridge, MA), and lentiviral ORF73 was obtained from Chris Boshoff (35). Lentiviral particles containing the above-mentioned expression vectors were prepared using a four-vector system in HEK293T cells, as previously described (32). The supernatants containing the respective lentiviral particles were used to transduce B cells in the presence of Polybrene (5 μg/ml). Seventy-two to 144 h postransduction, the cells were observed for vector uptake efficiency by using the transduction reporter (green fluorescent protein [GFP]) present in the shRL (Renilla luciferase) construct, and only experiments where shRL expression was present in >80% of the cells were investigated further.
Antibodies and reagents.
The antibodies against total Nrf2 protein (tNrf2), NAD(P)H quinone oxidoreductase 1 (NQO1), protein kinase C zeta (PKCζ), and phosphorylated PKCζ (pPKCζ) (Thr410) were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA); the antibody against caspase-3 was from Cell Signaling Technologies (Danvers, MA); the Ser-40-phosphorylated Nrf2 (pNrf2), TATA-binding protein (TBP), and mouse KAP1 antibodies were from Abcam (Boston, MA); the antibodies against β-actin and tubulin were from Sigma-Aldrich (St. Louis, MO); the antibodies against COX-1 and COX-2 were from Cayman Chemicals (Ann Arbor, MI); the ORF50 antibody was from ABBIOTEC (San Diego, CA); and the goat antibody against KAP1 was from Bethyl Laboratories Inc. (Montgomery, TX). Horseradish peroxidase (HRP)-linked anti-mouse and anti-rabbit antibodies were from KPL Inc. (Gaithersburg, MD). DAPI (4′,6-diamidino-2-phenylindole) and anti-rabbit and anti-mouse Alexa-Fluor 594 or 488 secondary antibodies were from Molecular Probes (Carlsbad, CA). The chemical inhibitors myristoylated PKCζ (Myr-PKCζ), the prostaglandin E receptor (EP) antagonists (AH8809, GW627368X, and SC-51322), and synthetic prostaglandin E2 (PGE2) were from Cayman Chemicals. Nuclear extract and TransAM Nrf2 kits were from Active Motif (Carlsbad, CA). Celecoxib was from Tocris Biosciences (Ellisville, MO). The anti-LANA-1 rabbit polyclonal antibody (UK 183) is a glutathione S-transferase (GST)-fused recombinant antibody and was prepared in our laboratory as previously described (36). The anti-LANA-1 mouse monoclonal antibody (1D10C3) was also prepared in our laboratory and targets the specific LANA-1 peptide sequence 490 to 506 (CEPQQREPQQREPQQ).
Western blotting.
Cells were suspended in RIPA lysis buffer (25 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1% NP-40, 0.1% SDS, supplied with protease/phosphatase inhibitor cocktails), and Western blotting was performed as previously described (32).
Isolation of nuclear and cytoplasmic fractions for Western blot analysis.
B cells at a density of ∼1 million cells/ml were washed two times with phosphate-buffered saline (PBS), and the nuclear and cytoplasmic proteins were fractionated according to the manufacturer's protocol (Nuclear Isolation kit; Active Motif) and as previously described (32).
Coimmunoprecipitation.
Cells were lysed with either denaturing RIPA buffer or nondenaturing buffer (NETN) (100 mM NaCl, 20 mM Tris-HCl, pH 8.0, 0.5 mM EDTA, and 0.5% NP-40), and 200 μg of protein was incubated with 1 to 2 μg of primary antibody, along with protein G-Sepharose beads. After overnight incubation at 4°C, the beads were pelleted by centrifugation and washed three times with the lysis buffer, and the protein-bead complexes were disrupted by resuspension in SDS loading buffer solution containing β-mercaptoethanol (β-ME) and heating to 95°C for 5 to 10 min prior to SDS-PAGE loading and Western blot analysis.
Measurement of host and KSHV gene expression by real-time RT-PCR.
To detect host and KSHV gene expression, total RNA from infected and uninfected cells was isolated using an RNeasy minikit (Qiagen). Using a high-capacity cDNA reverse transcription (RT) kit (Life Technologies), we created a cDNA library of all the transcribed genes. Using gene-specific primers and Power SYBR green PCR Master Mix real-time RT-PCR, we determined the cycle threshold (CT) values for each gene. ΔCT values relative to tubulin were assessed for each condition. Gene expression under the control condition(s) was arbitrarily set to 1, and the fold change was based on the ΔCT differential relative to this condition (ΔΔCT).
RNA sequencing and analysis of the KSHV transcriptome.
BCBL-1 cells were transduced with either shRL or shNrf2 for 72 h prior to RNA isolation using Qiagen's RNeasy minikit. Prior to analysis, shRL cells were observed for GFP expression to ensure efficient lentiviral transduction, which was ∼80% of the cells. Two micrograms of DNase-treated RNA from each sample was sent for high-throughput RNA sequencing at the University of Nevada, Reno, NV, as previously described (37). Briefly, a cDNA library was created using a TrueSeq RNA sample preparation kit v2 (Illumina, Inc.) according to the manufacturer's instructions, and the libraries were sequenced using a HiSeq next-generation sequencer (Illumina, Inc.). The data were analyzed using the KSHV genome as a reference (accession number NC_009333.1) and the CLC Genomik Workbench 7 software. The ratio of the numbers of tubulin-normalized reads per kilobase per million mapped reads (RPKM) for shNrf2 versus that for shRL was calculated to determine the fold change between these conditions. Values whose ratio was below 1.0 are represented in graphs as the negative of the reciprocal value, indicating the fold decrease.
Immunofluorescence microscopy.
B cells spotted on 10-chamber glass slides were fixed and permeabilized with acetone, whereas TIVE/LTC cells grown in 8-chamber Permanox plastic slides were fixed with paraformaldehyde and permeabilized with 0.2% Triton X-100. The permeabilized cells were then blocked with Image-iT FX signal enhancer (Life Technologies) and incubated with specific primary antibodies and fluorescent Alexa Fluor-conjugated secondary antibodies prior to mounting with DAPI for nuclear staining. Regular imaging was performed with Nikon imaging systems, and figure analysis and deconvolution of the images were performed using Nikon NIS-Elements software. Confocal imaging was performed using an Olympus FV10i microscope, and image analysis was performed using Fluoview1000 (Olympus) software.
Nrf2 enzyme-linked immunosorbent assay (ELISA).
To assess the DNA-binding activity of nuclear Nrf2, we isolated the nuclear protein fraction using a nuclear extraction kit (Active Motif), and the DNA-binding activity of Nrf2 was assessed using a TransAM Nrf2 kit (Active Motif) according to the manufacturer's instructions and as previously described (32). Briefly, equal amounts of nuclear protein (25 μg/condition), were loaded in each chamber containing the Nrf2-binding oligonucleotide probes (sequence TGANNNNGC) for 1 h at room temperature, washed, incubated with anti-Nrf2 antibody for 1 h, and then probed with an HRP-conjugated anti-Nrf2 antibody for an additional hour prior to chemiluminescence detection. Twenty-five micrograms of control nuclear extract was used as a positive control for the assay. Wild type (WT) and mutated (Mut) oligonucleotide sequences provided by the manufacturer were used to assess the specificity of the assay.
PLA.
A proximity ligation assay (PLA) was performed using a DuoLink PLA kit (Sigma-Aldrich) to detect protein-protein interactions using fluorescence microscopy according to the manufacturer's protocol and as previously described (32). Briefly, B cells in 10-chamber suspension cell slides and TIVE/LTC cells in 8-chamber permanox slides were blocked with DuoLink blocking buffer; incubated with primary antibodies against LANA-1 (mouse monoclonal), pNrf2 (rabbit monoclonal), or KAP1 (goat polyclonal or rabbit monoclonal) diluted in DuoLink antibody diluents for 2 h; washed; and then further incubated for 1 h at 37°C with species-specific PLA probes, each conjugated to complementary oligonucleotides that hybridize at close proximity (<40 nm). The cells were then incubated with a ligation solution for 30 min at 37°C prior to the amplification step for 100 min at 37°C. Detection solution consisting of fluorescently labeled oligonucleotides was added, and the labeled oligonucleotides were hybridized to the amplified products. The signal was detected as distinct fluorescent dots in either the fluorescein isothiocyanate (FITC) or tetramethyl rhodamine isocyanate (TRITC) channel of fluorescence or confocal microscopy. Negative controls consisted of cell lines deficient in LANA-1 expression.
For the simultaneous PLA of pNrf2/KAP1 and LANA-1/KAP1, after the completion of one round of PLA using anti-pNrf2 (rabbit) and anti-KAP1 (goat) antibodies; the respective PLA probes; and ligation, amplification, and detection steps (FITC), the cells were reblocked and stored overnight at 4°C in DuoLink blocking solution. The following day, another round of PLA was performed using anti-KAP1 (mouse) and anti-LANA-1 (rabbit) antibodies and proceeded like the first PLA, using the TRITC filter. PLA dots were visualized using either light or confocal microscopy.
Luciferase assay.
HEK293T cells were transiently transfected by the calcium phosphate method with plasmids expressing the full-length ORF50 promoter and its truncated construct (pcDNA2500 and pcDNA950), obtained from George Miller (Yale University School of Medicine), preceding the firefly luciferase gene (38). In addition, a vector expressing the Renilla luciferase gene under the control of the simian virus 40 (SV40) promoter was concomitantly transfected into the cells to account for transfection variability and potential cytotoxicity. Twenty-four to 48 h posttransfection, the cells were harvested and lysed in passive lysis buffer (Promega, Madison, WI). Soluble extracts were assayed for firefly and Renilla luciferase expression by using the Dual-Glo system according to the manufacturer's instructions, and the results were expressed as firefly/Renilla luciferase ratios and normalized to their respective control conditions.
Cell death analysis by annexin V-PI staining.
Approximately 500,000 B cells were suspended and incubated in 100 μl of annexin V binding buffer containing 5 μl of FITC-tagged annexin V and 3 μl of propidium iodide (PI) for 15 min at room temperature. An additional 400 μl of the binding buffer was then added to the cells, and the solution was analyzed using an LSRII flow cytometer (Becton Dickinson) and FlowJo software at the Rosalind Franklin University of Medicine and Science (RFUMS) Flow Cytometry Core Facility.
Chromatin immunoprecipitation assay.
ChIP assays were performed using the SimpleChIP Enzymatic Chromatin IP kit (magnetic beads) from Cell Signaling Technology following the manufacturer's instructions. Briefly, BCBL-1 cells (4 × 107) were fixed with 1% (vol/vol) formaldehyde for 10 min at room temperature, followed by glycine quenching of the cross-linking reaction for 5 min at room temperature. The cells were then washed two times with ice-cold PBS, followed by isolation of the nucleus. Fragmentation of the chromatin (150 to 750 bp) was achieved by treating the isolated nucleus with micrococcal nuclease (MNase) for 20 min at 37°C, which was stopped by EDTA addition. The nuclei were centrifuged and resuspended in ChIP buffer, followed by sonication on ice to lyse the nuclear membrane. Cellular debris was removed by centrifugation at 10,000 rpm at 4°C. For ChIP, chromatin containing 10 μg of DNA was diluted in 500 μl of ChIP buffer and immunoprecipitated with the respective antibodies (2 μg) overnight at 4°C. Ten microliters of the diluted chromatin was kept aside as a 2% input sample before addition of antibodies. ChIP grade protein G magnetic beads were used to pull down the chromatin-antibody complex for 2 h at 4°C. The immunoprecipitated complex was then washed three times with low-salt and once with high-salt wash buffers. To elute the chromatin from the antibody-protein G bead complex, the beads were resuspended in ChIP elution buffer, followed by incubation at 65°C for 30 min on a thermomixer (1,200 rpm). Next, removal of all protein and reversal of cross-linking was achieved by incubating the eluted chromatin with NaCl and proteinase K for 2 h at 65°C. The 2% input samples were treated similarly. Subsequently, DNA was purified using spin columns. Both input and ChIP DNAs were measured by quantitative PCR (qPCR) using Power SYBR green PCR Master Mix (Applied Biosystems). Enrichment of proteins on specific genomic regions was calculated as a percentage of the immunoprecipitated DNA compared to the input DNA.
The primers used for ORF50 promoter ChIP studies and their nucleotide positions were as follows: ORF50 set 1 (68892 to 69015), forward primer, CAGGTGTTTCTGTGCGTTTATG, and reverse primer, CTGGGATTGGTTCACGAGTT; ORF50 set 2 (69448 to 69579), forward primer, CCTGACTGATGGATAGGGT, and reverse primer, GTTAGCGGAAGTCAGACTCG; ORF50 set 3 (69988 to 70113), forward primer, GACGGCAAATAGCGCAAAG, and reverse primer, CATGCGTGAACCCTCACTAT; ORF50 set 4 (70765 to 70860), forward primer, GACAGTCCGCCATACTCTTC, and reverse primer, CTGGCTCTACCACATCTTCATAG; ORF50 set 5 (71161 to 71279), forward primer, CAGTCATCCCAGATCAAAGTCA, and reverse primer, AACACCAGACTGAATCTACTTCC; ORF50 set 6 (71532 to 71677), forward primer, GCTACAGCTTATCCTCCACTAAAT, and reverse primer, CAGTATTCTCACAACAGACTACCC; ORF50 set 7 (71920 to 72250), forward primer, GAGTTAGGGACGTGCTGATTAT, and reverse primer, CGAGGACTTTCAGGATACAGATT.
Gel filtration chromatography.
Cell lysates obtained from BC-3 cells were separated by gel filtration chromatography on a Superdex 200 HR column with a fast protein liquid chromatography (FPLC) system (Pharmacia Biotech, Uppsala, Sweden) as described previously (39).
Intracellular KSHV DNA quantification.
BCBL-1 cells were transduced with lentivirus expressing shNrf2 or shRL, or treated with brusatol (100 nM), or TPA (20 ng/ml) for 96 h prior to DNA isolation using Qiagen's DNeasy minikit. Equal amounts of DNA (100 ng) were run per condition by DNA qPCR using SYBR green probe and ORF73- and tubulin gene-specific primers. The ORF73 CT values were normalized to tubulin for each condition.
De novo infection of HMVEC-d.
HMVEC-d were serum starved for 2 h prior to addition of KSHV (40 DNA copies/cell) at the indicated time points. The cells were then lysed with nondenaturing buffer (NETN), and protein was isolated and quantified using the bicinchoninic acid (BCA) quantification method, as previously described (30, 32).
Statistical analysis.
Data are expressed as means and standard deviations (SD) for at least three independent replicates. In all tests, a P value of <0.05 was considered statistically significant. Experiments in which the P value is <0.05 are marked with asterisks.
RESULTS
Latent KSHV infection induces Nrf2 activity in PEL tissue and PEL cell lines.
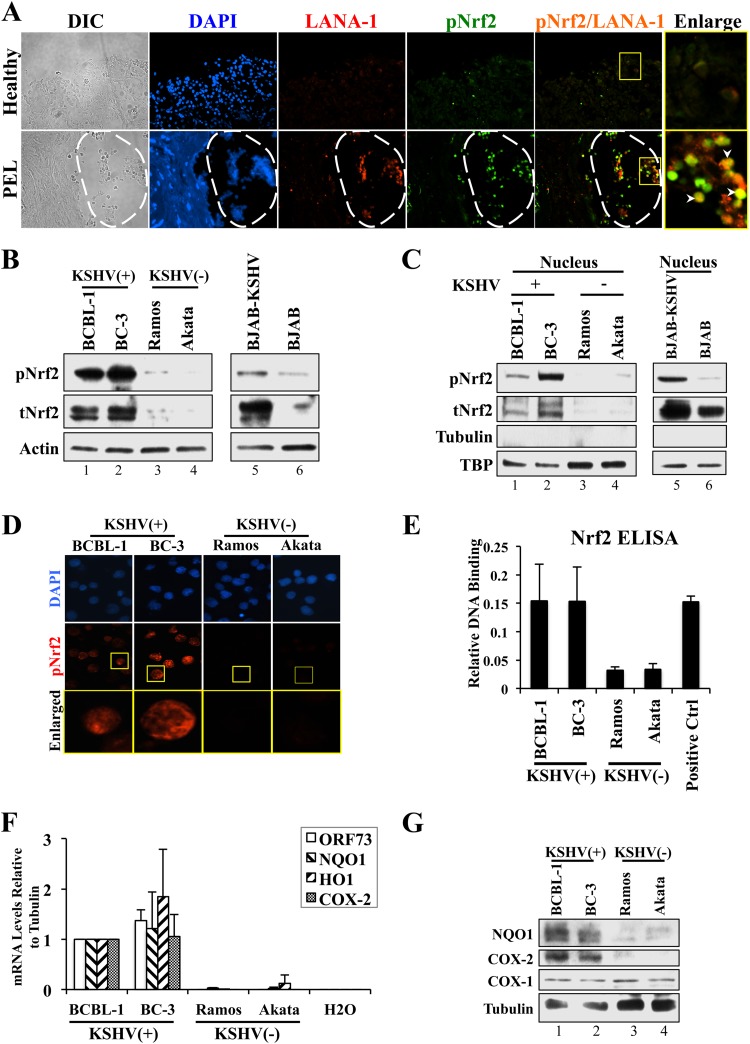
We recently observed that de novo and latent KSHV infections of endothelial cells induce Nrf2 activity and that KS tissue of the skin exhibited elevated nuclear Nrf2 levels (30, 32). Here, we investigated the role of prolonged latent KSHV infection of B cells in inducing total Nrf2 protein (tNrf2) levels and its Ser-40 phosphorylation (pNrf2). Immunofluorescence assays (IFA) showed that pNrf2 is undetectable in the lining of the normal gut mucosa (Fig. 1A, top row). In contrast, IFA of PEL of the stomach revealed significant Nrf2 phosphorylation, particularly in the area of the malignancy, with latently infected B cells as measured by the expression of the KSHV latency marker LANA-1 (Fig. 1A, bottom row, areas within the dashed lines).
FIG 1.
Assessment of Nrf2 activity in PEL and PEL-derived cell lines. (A) Stomach tissue from healthy individuals and from patients with primary effusion lymphoma of the stomach were stained for LANA-1 and pNrf2. The boxed areas were enlarged to determine colocalization. The arrowheads indicate sites of LANA-1/pNrf2 colocalization. The areas within the dashed lines show PEL effusion. DIC, differential interference contrast. (B) Whole-cell lysates from PEL- and non-PEL-derived KSHV-positive cell lines (BCBL-1, BC-3, and BJAB-KSHV) were compared to those from non-PEL-derived KSHV-negative cell lines (Ramos, Akata, and BJAB) by Western blotting for tNrf2 and pNrf2. (C) Nuclear proteins isolated from KSHV-positive and -negative cell lines were compared by Western blotting for tNrf2 and pNrf2. Tubulin and TBP were used as the cytoplasmic and nuclear controls. (D) KSHV-positive and -negative cell lines were stained for pNrf2 prior to fluorescence microscopy. The boxed areas are enlarged below. (E) DNA binding of the transcriptionally active Nrf2 in KSHV-positive and -negative cell lines was assessed by using the TransAM Nrf2 kit. The positive control (Ctrl) consisted of nuclear protein lysate enriched with transcriptionally active Nrf2. (F) RNA from KSHV-positive and -negative cell lines was analyzed by real-time RT-PCR using gene-specific primers. The fold inductions are relative to BCBL-1, which was arbitrarily set to 1. H2O was used as a control for primer dimer formation. (G) Western blot of whole-cell lysates from KSHV-positive and -negative cell lines for the Nrf2 target NQO1 and COX-2 genes. Constitutively expressed COX-1 was used as a negative control. The data are expressed as means and SD for at least three independent replicates.
We next investigated the levels of tNrf2 and transactivated pNrf2 in the PEL-derived, KSHV-positive cell lines BCBL-1 and BC-3 and compared them to those in the EBV- and KSHV-negative Burkitt's lymphoma cell lines Ramos and Akata. KSHV-positive PEL cell lines exhibited significantly elevated levels of tNrf2 and pNrf2 in the whole-cell protein lysate (Fig. 1B, lanes 1 to 4), as well as in the nucleus (Fig. 1C, lanes 1 to 4), compared to their KSHV-negative counterparts. Similarly, BJAB-KSHV cells exhibited enhanced tNrf2 and pNrf2 levels in the whole-cell lysate, as well as in the nucleus, compared to uninfected BJAB cells (Fig. 1B and C, lanes 5 and 6), suggesting that the observed Nrf2 upregulation is due to latent KSHV infection and not due to inherent genetic differences between the cell lines. We confirmed the increased nuclear presence of pNrf2 in KSHV-positive cell lines by IFA (Fig. 1D).
An ELISA designed to detect transcriptionally active Nrf2 in nuclear lysates using the antioxidant response element (ARE) (Nrf2-binding site) as the antigen further confirmed that the increase in nuclear Nrf2 in KSHV-positive cell lines resulted in increased DNA binding (Fig. 1E). Furthermore, mRNA expression of the de facto Nrf2 target NQO1 and heme oxygenase (HO1) genes showed that their expression was significantly upregulated in KSHV-positive cell lines (Fig. 1F). COX-2, whose optimal expression depends on Nrf2 induction during KSHV infection of endothelial cells (30), was highly expressed in KSHV-positive cell lines, as well (Fig. 1F). Western blotting confirmed the elevated protein expression of NQO1 and COX-2 in these cell lines, while no such correlation was observed between the constitutively active COX-1 and KSHV infection (Fig. 1G).
Collectively, these results demonstrated that the increased Nrf2 protein levels and phosphorylation in PEL and PEL-derived cell lines resulted in its direct nuclear translocation, DNA binding, and transactivation of de facto target genes.
The COX-2/PGE2 axis induces Nrf2 through prostaglandin E receptor 4 (EP4) signaling in PEL cell lines latently infected with KSHV.
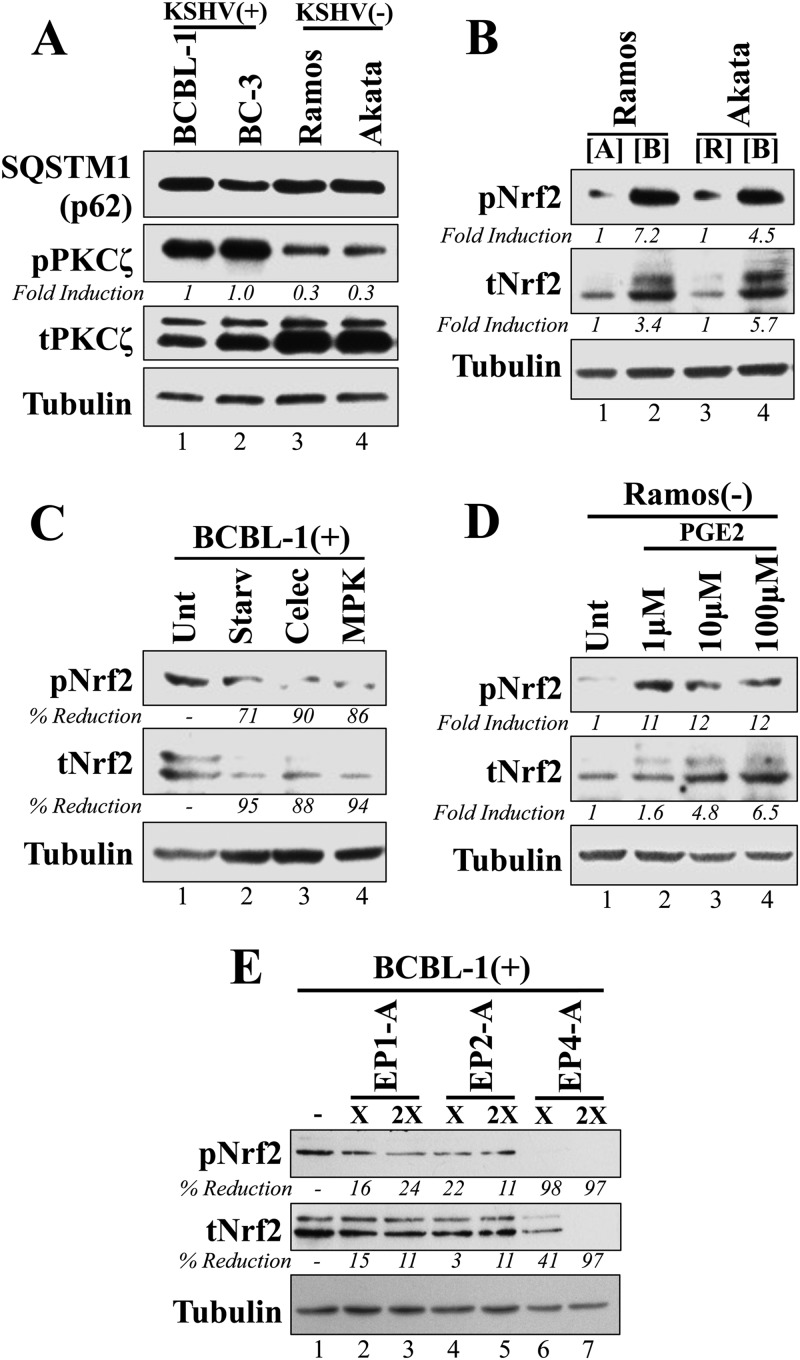
During the early stages of de novo endothelial cell infection, KSHV utilizes reactive oxygen species (ROS) to mediate Nrf2 stability and signaling kinases to mediate its phosphorylation (32). Interestingly, ROS are dispensable for Nrf2 induction during latency, where we have identified COX-2 as a major factor in sustained Nrf2 activity (30, 32). In this pathway, Nrf2 is important for optimal COX-2 expression, which in turn enzymatically activates PGE2 production and secretion. PGE2 autocrine/paracrine effects subsequently utilize PKCζ to induce Nrf2 activity, establishing a feed-forward loop between Nrf2 and COX-2. We also found that during prolonged endothelial latency, KSHV induces activation of the noncanonical Nrf2-activating pathway dependent on the autophagic protein p62 (sequestosome-1 [SQSTM1]) (30). In this pathway, KSHV-mediated inhibition of autophagy results in accumulation of p62, which competitively displaces Nrf2 from its inhibitor, Keap1.
Here, we investigated these mechanisms of Nrf2 induction during latent infection of B cells. We observed that p62 protein levels were similar regardless of KSHV infection, arguing against a role for the noncanonical pathway in Nrf2 activation in these cell lines (Fig. 2A). Investigation of the COX-2/PGE2/PKCζ axis showed that although total PKCζ levels were slightly reduced in KSHV-positive cells, those of its active, Thr-410-phosphorylated isoform, pPKCζ, were significantly elevated (Fig. 2A). Previously, we showed that pPKCζ is induced by the paracrine effect of PGE2 to mediate Nrf2 activation (30, 32). We designed a medium interchange experiment to confirm whether KSHV-mediated autocrine/paracrine signaling was also involved in Nrf2 activation in PEL-derived cell lines. We obtained supernatants of Ramos, Akata, and BCBL-1 cell cultures and used these supernatants to culture KSHV-negative cells, as shown in Fig. 2B. BCBL-1 media induced Nrf2 phosphorylation in Ramos and Akata cells significantly more than media from the uninfected Ramos and Akata cells (Fig. 2B), indicating that signaling agents in the BCBL-1 supernatant participate in paracrine signaling that is key to inducing Nrf2 activity, possibly through PKCζ activation.
FIG 2.
Analysis of KSHV-induced autocrine/paracrine signaling involved in Nrf2 activation in PEL cell lines. (A) KSHV-positive and -negative cell lines were Western blotted for the autophagic protein SQSTM1 (or p62), PKCζ, and its active, Thr-410-phosphorylated form. (B) Supernatants from BCBL-1 ([B]), Akata ([A]), and Ramos ([R]) cells cultured at a density of 250,000 cells/ml for 72 h were cleared of cell debris by centrifugation at 13,000 rpm for 30 min and used to culture Ramos or Akata cells at the same density for a total of 24 h prior to Western blotting for tNrf2 and pNrf2. (C) BCBL-1 cells were cultured in full medium (Unt), growth factor-free medium (Starv), or full medium containing either the COX-2-specific inhibitor celecoxib (Celec) (10 μM), or the PKCζ inhibitor MPK (10 μM). All treatments were performed for 12 h prior to Western blotting for tNrf2 and pNrf2 levels. (D) The KSHV-negative Ramos cells were treated for 12 h with increasing doses of PGE2, and whole-cell lysates were Western blotted for tNrf2 and pNrf2 levels. (E) BCBL-1 cells were mock treated with DMSO (−) or treated with EP antagonists for 12 h prior to Western blotting for tNrf2 and pNrf2. The EP1 antagonist (EP1-A) was SC-51322 (25 and 50 μM), the EP2-A was AH6809 (25 and 50 μM), and the EP4-A was GW627638X (2.5 and 5 μM). × and 2× indicate the low and high concentrations, respectively, for each drug. (A to E) All calculations were normalized to the loading control tubulin and are relative to the untreated/control condition.
To assess if paracrine signaling-mediated PKCζ activation is involved in Nrf2 activation similarly to its involvement in endothelial cells, we treated BCBL-1 cells with the myristoylated PKCζ pseudosubstrate inhibitor (MPK), the COX-2 inhibitor celecoxib, and starvation and observed a robust reduction in Nrf2 levels and phosphorylation under all conditions (Fig. 2C), suggesting that COX-2-mediated signaling is important for PKCζ-mediated Nrf2 activation. We further confirmed the involvement of PGE2 signaling in Nrf2 induction by treating Ramos cells with exogenous PGE2 and observed a dose-dependent induction of tNrf2 and pNrf2 levels (Fig. 2D). PGE2 mediates most of its paracrine effects by binding to and inducing the activities of EP1 to -4. Of the three overexpressed EPs in PEL, EP1, -2, and -4 (40), only treatment with the EP4 antagonist GW 627368 significantly reduced Nrf2 activity in a dose-dependent manner (Fig. 2E).
Collectively, these results demonstrated that KSHV-induced COX-2 mediates PGE2 production and secretion, which through autocrine/paracrine signaling amplifies EP4/PKCζ signaling to induce Nrf2 activation in KSHV-positive PEL cell lines.
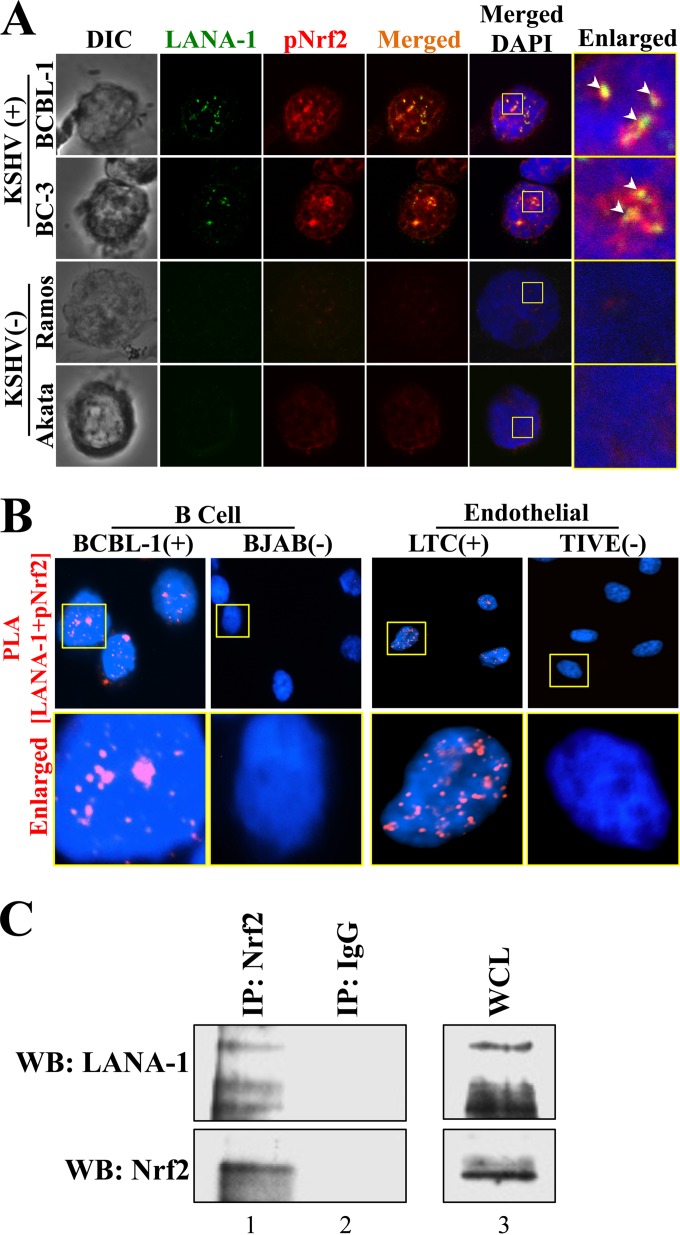
Nrf2 and the KSHV latency-associated protein LANA-1 interact with each other in PEL cell lines.
During de novo infection of endothelial cells, we observed that LANA-1 and pNrf2 colocalized with each other at latent time points (32). Since Nrf2 is activated in PEL, we next investigated the possibility of interaction between LANA-1 and Nrf2 and its implications in KSHV biology. By confocal microscopy, we observed that LANA-1 and pNrf2 also colocalize in multiple areas within the PEL cell nuclei (Fig. 3A). While pNrf2 staining revealed a characteristic nuclear distribution, there was a striking enrichment in areas surrounding LANA-1 puncta (Fig. 3A, arrowheads). Furthermore, we performed a sensitive PLA between pNrf2 and LANA-1, as this method detects two molecules in direct proximity at ∼40 nm or less (see Materials and Methods for details). PLA of LANA-1 and pNrf2 in BCBL-1 and TIVE/LTC cells showed significant LANA-1 and pNrf2 interaction spots in the nucleus, which were absent in the KSHV-negative cell lines (Fig. 3B). Lastly, coimmunoprecipitation (co-IP) of the BCBL-1 lysate with anti-Nrf2 antibody also pulled down LANA-1, confirming that LANA-1 and Nrf2 interact (Fig. 3C).
FIG 3.
Demonstration of Nrf2 and LANA-1 interaction in PEL cell lines. (A) KSHV-negative and -positive cell lines were stained for pNrf2 and LANA-1 prior to visualization by confocal microscopy. The arrowheads indicate sites of LANA-1/pNrf2 colocalization. The boxes indicate the enlarged area. (B) PLA determines pNrf2/LANA-1 colocalization in the KSHV-positive BCBL-1 and LTC cell lines. The KSHV-negative cell lines BJAB and TIVE were used as negative controls, as they do not have LANA-1 expression. Red staining indicates positive colocalization between pNrf2 and LANA-1. The boxes indicate the enlarged areas. (C) BCBL-1 whole-cell protein lysate (WCL) was immunoprecipitated with anti-Nrf2 (lane 1) or IgG control (lane 2) antibodies and Western blotted (WB) for tNrf2 and LANA-1. BCBL-1 input (WCL) (lane 3) is shown for comparison to the other lanes.
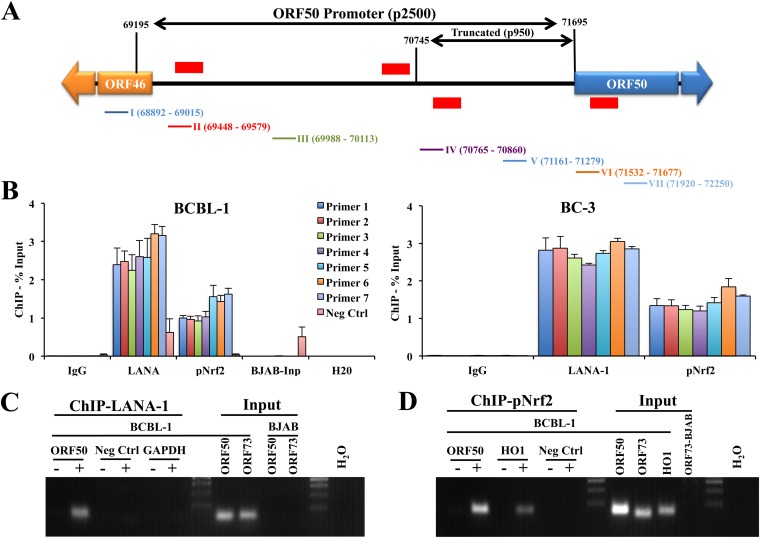
Nrf2 and LANA-1 bind to similar regions of the KSHV lytic switch ORF50 promoter.
Previous PLA experiments revealed that pNrf2 and LANA-1 colocalized with the KSHV genome during de novo latency in endothelial cells and that Nrf2 knockdown abrogated the lytic gene burst during de novo infection (32), indicating that Nrf2 might play an important role in the interplay between lytic gene expression and latency. Therefore, we hypothesized that pNrf2 may bind to the ORF50 promoter to regulate its transcriptional activity. To test this, we analyzed the 2,500 bp upstream of the ORF50 gene, widely accepted as the ORF50 promoter region, and identified four putative Nrf2-binding sites (AREs) on the sense and antisense strands (Fig. 4A, red lines). To assess whether Nrf2 binds to the ORF50 promoter, we performed a set of ChIP experiments using anti-LANA-1 (mouse) and anti-pNrf2 (rabbit) antibodies on chromatin isolated from BCBL-1 and BC-3 cells. Using a set of seven primers spanning the ORF50 promoter, as illustrated in Fig. 4A, we observed that LANA-1 bound throughout the ORF50 promoter (Fig. 4B), consistent with previous reports (16). Interestingly, pNrf2 also bound throughout the ORF50 promoter, with an observable enrichment around primer sets 5 to 7 (Fig. 4B). The promoter binding of pNrf2 and LANA-1 were remarkably similar in pattern in BCBL-1 cells, both peaking at a region covered by primer sets 5 to 7 (Fig. 4B, left). Overall, these results demonstrated that pNrf2 binds to the ORF50 promoter in a pattern that resembles that of LANA-1 binding, indicating the possibility that pNrf2 and LANA-1 bind to similar regions of the ORF50 promoter as a complex.
FIG 4.
Determination of pNrf2 and LANA-1 binding to the ORF50 promoter. (A) ORF50 promoter schematic spanning 2,500 bp upstream of the transcriptional start site (69195 to 71695). The red horizontal lines indicate putative Nrf2 binding sites (AREs; TGANNNNGC). The ARE sites indicated above the schematic are on the antisense DNA strand, while those below the schematic are on the sense DNA strand. The exact locations of the ARE sites are, from left to right, 69184 to -92, 70670 to -78, 70721 to -29, and 71571 to -79. The colored lines and numbers below the schematic correspond to the primer sets used in panels B to D and their locations on the promoter. p2500 and p950 indicate the luciferase promoter constructs used in Fig. 9. (B) Chromatin immunoprecipitation of KSHV-positive cells (BCBL-1 and BC-3) using anti-LANA-1, anti-pNrf2, and IgG control antibodies. The primers shown in panel A were used to PCR amplify the immunoprecipitated and input DNA. The negative-control (Neg Ctrl) primers amplify a region of the human DNA that is highly heterochromatinized and devoid of many DNA-binding factors and that was used to determine the background, nonspecific chromatin binding of the anti-LANA-1 and anti-pNrf2 antibodies. The results are expressed as percentages of DNA pulled down compared to the respective input chromatin. BJAB input genomic DNA was used to demonstrate the specificity of the designed ORF50 primers, while H2O was used to observe potential primer dimer formation. (C and D) DNA PCR for LANA-1 and pNrf2 pulldowns was carried out with ORF50 primer set V in the ChIP samples from panel B. −, IgG control was used for the pulldown; +, anti-LANA-1 (C) or anti-pNrf2 (D) antibody was used for the pulldown. Neg Ctrl and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) primers were used as negative controls. The HO1 gene, a de facto Nrf2 target gene, was used as a positive control for pNrf2 pulldown. The data are expressed as means and SD for at least three independent replicates.
Several controls were performed to ensure that the ChIP results were reliable. To ensure that the PCR results obtained by the LANA-1 and pNrf2 pulldown were specific and not a consequence of background DNA binding, we utilized a set of DNA primers that amplify a highly heterochromatinized region of human DNA devoid of Nrf2-binding sites, which showed negligible enrichment by pNrf2 pulldown compared to the ORF50 primer sets (Fig. 4B, left, Neg Ctrl [negative control]). Moreover, while LANA-1 pulldown did provide Neg Ctrl region amplification to a higher degree than either IgG or pNrf2, the enrichment of the ORF50 region was severalfold higher (Fig. 4B, left). DNA PCR on BJAB cell chromatin input revealed no amplification with the seven ORF50 primer sets but did show amplification of the Neg Ctrl primers, confirming the designed primers' specificity. The primers did not form any observable primer dimers in the H2O control (Fig. 4B, left). Lastly, agarose gel electrophoresis further confirmed the real-time PCR experiments, demonstrating single-product formation, specificity, and positive controls for these findings in both LANA-1 and pNrf2 pulldowns (Fig. 4C and D).
Nrf2 knockdown and chemical inhibition result in a global increase in lytic KSHV gene expression.
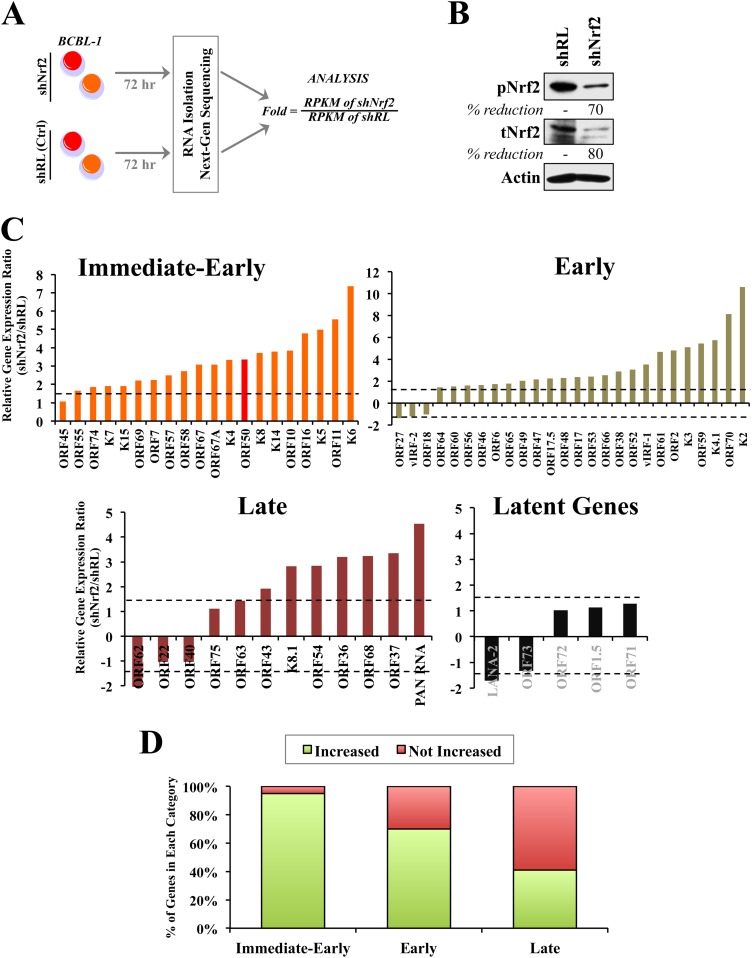
Next, we assessed the role of Nrf2 in KSHV gene expression in PEL cell lines. We transduced BCBL-1 cells with lentivirus containing shNrf2 to reduce endogenous Nrf2 activity in target cells (Fig. 5A and B); shRL was used as a negative control. We confirmed by Western blotting that the Nrf2 knockdown was efficient (Fig. 5B). Seventy-two hours postransduction, DNase-treated mRNA from both conditions was reverse transcribed and sequenced by RNA-seq for assessment of global viral-gene expression (Fig. 5A). The results are reported as the shNrf2/shRL RPKM ratio, and any induction/reduction beyond 1.5-fold was considered significant (Fig. 5A). The grouping of genes in IE, E, and late (L) lytic genes was performed as in a previous review (41). Interestingly, we observed a global increase in lytic KSHV gene expression when Nrf2 was knocked down (Fig. 5C). This increase in expression encompassed all classes of KSHV lytic genes, IE, E, and L (Fig. 5C). In contrast, latent KSHV genes were not significantly affected by Nrf2 knockdown (Fig. 5C). When the viral-gene expression was plotted in a 100% stacked column, we observed an interesting association between the proportion of KSHV genes that were induced and the lytic gene category that these genes belonged to. Specifically, 95%, 70%, and 41% of the IE, E, and L genes, respectively, were upregulated by Nrf2 knockdown, suggesting a temporal gene reprogramming that initiates with IE genes, followed by induction of E and then L genes (Fig. 5D).
FIG 5.
Next-generation sequencing of KSHV gene expression during Nrf2 modulation. (A) Schematic of the experiment. KSHV-positive BCBL-1 cells transduced with lentiviral shRL (control) and shNrf2 for 72 h were subjected to RNA isolation and sequenced for the whole KSHV transcriptome. A cDNA library was created using a TrueSeq RNA-seq library kit. The sequencing was performed using HiSeq and the KSHV genome as a reference to determine the relative abundances of the viral transcripts, which were expressed as RPKM. The results are expressed as the ratio of RPKM of shNrf2 to those of shRL. (B) Western blot of tNrf2 and pNrf2 levels in the lysates used in the sequencing experiment showing efficient Nrf2 knockdown. (C) Relative induction levels of KSHV immediate-early, early, late-lytic, and latent genes. The dashed horizontal lines indicate the 1.5-fold mark, set as the threshold. (D) Quantification of the percentages of genes that were increased or not increased (unchanged, undetected, or decreased) in each of the lytic gene classes. (C and D) Certain genes whose RPKM values were undetected in shRL but detected in shNrf2 could not be included in panel C, as division by 0 is not mathematically possible. These genes were included in panel D as part of the “increased” gene pool.
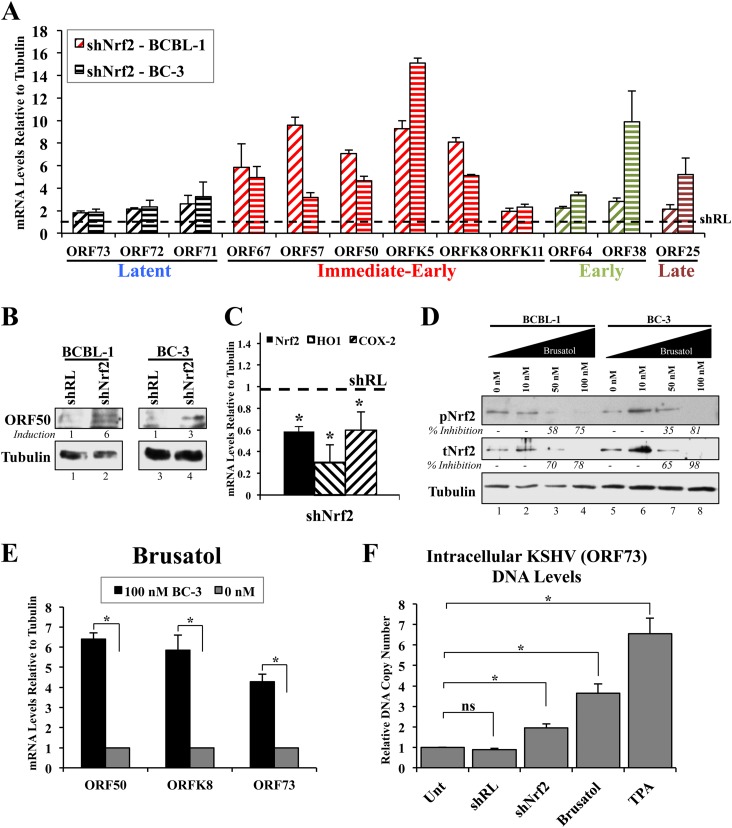
To confirm the sequencing findings, we performed real-time RT-PCR analysis on selected KSHV genes from BCBL-1 and BC-3 cells transduced with either shRL or shNrf2 for 72 h. We observed a robust induction of IE, E, and L genes during Nrf2 knockdown in both cell lines, demonstrating that this was not a cell line-specific effect (Fig. 6A). Interestingly, latent gene expression also increased moderately during Nrf2 knockdown, although not to the same degree as most lytic genes (Fig. 6A). Western blotting of ORF50/RTA showed that the increase in mRNA is concomitantly associated with an increase in protein expression (Fig. 6B). Real-time RT-PCR analysis confirmed that Nrf2 mRNA and mRNAs for Nrf2 target genes were significantly downregulated by shNrf2 treatment (Fig. 6C).
FIG 6.
Confirmation of the effect of Nrf2 modulation on lytic gene induction. (A) Real-time RT-PCR using gene-specific primers from the four major gene groups on RNA extracted from BCBL-1 and BC-3 cells lentivirally transduced with either shRL or shNrf2 for 72 h. The dashed horizontal line indicates the baseline level of the genes under the shRL condition for each cell line (arbitrarily set to 1 for each gene). (B) Protein lysates from cells in panel A were Western blotted for the KSHV lytic gene ORF50. (C) Real-time RT-PCR of endogenous BCBL-1 genes confirming Nrf2 mRNA decrease by shNrf2, as well as decreased mRNA levels of the Nrf2 target HO1 and COX-2 genes. The dashed horizontal line indicates the baseline level of each gene under the shRL condition (arbitrarily set to 1). *, P < 0.05. (D) BCBL-1 and BC-3 cells were treated with increasing doses of the Nrf2 inhibitor brusatol for 12 h prior to Western blot analysis of tNrf2 and pNrf2 levels. Percent inhibition is indicated. Calculations were normalized to the loading control tubulin and are relative to the 0 nM condition for each cell line. (E) RNA from BC-3 lysates in panel D was analyzed by real-time RT-PCR using KSHV gene-specific primers. (F) DNA qPCR using ORF73- and tubulin-specific primers on DNA extracts from BCBL-1 cells treated with shRL, shNrf2, brusatol (100 nM), or TPA (20 ng/ml) for 96 h. *, P < 0.05; ns, not significant. The data are expressed as means and SD for at least three independent replicates.
Brusatol is a novel chemical inhibitor of Nrf2 that mediates its proteasomal degradation at nanomolar concentrations (42). When we treated BCBL-1 and BC-3 cells with increasing doses of brusatol, we observed a significant reduction in Nrf2, especially at 100 nM (Fig. 6D). We then determined ORF50, ORFK8, and ORF73 gene expression in brusatol-treated BC-3 cells and observed a robust increase in their expression (Fig. 6E), consistent with the shNrf2 results. Nrf2 inhibition through either lentiviral knockdown or brusatol induced KSHV DNA replication in BCBL-1 cells (Fig. 6F).
Overall, these experiments demonstrated that Nrf2 inhibition results in a global increase in KSHV lytic gene expression, which proceeds in a temporal fashion and ultimately results in increased KSHV lytic replication.
KAP1 is expressed in PEL cell lines, and its inhibition induces lytic gene expression.
The sequencing findings were surprising to us, as they indicated that Nrf2 plays a repressive role in ORF50 and other lytic gene expression in latently infected PEL cells. This was in contrast to the findings observed during de novo infection, where we identified Nrf2 as a positive regulator of ORF50 and the lytic burst (30). We therefore hypothesized that the cellular microenvironment is modulated by latent KSHV to exploit the positive transcriptional activity of Nrf2 on ORF50 during early infection and to use it to repress ORF50 during latency. Specifically, we focused on KAP1, a transcriptional corepressor in the human genome. Recent reports have suggested that LANA-1 recruits KAP1 on the ORF50 promoter to mediate its repression in endothelial cells (16). In addition to its role in ORF50 repression through LANA-1 binding, KAP1 was also shown to bind Nrf2 in HeLa cells (43). We therefore investigated the possibility that KAP1 modulates the role of Nrf2 on the ORF50 promoter activity during KSHV infection.
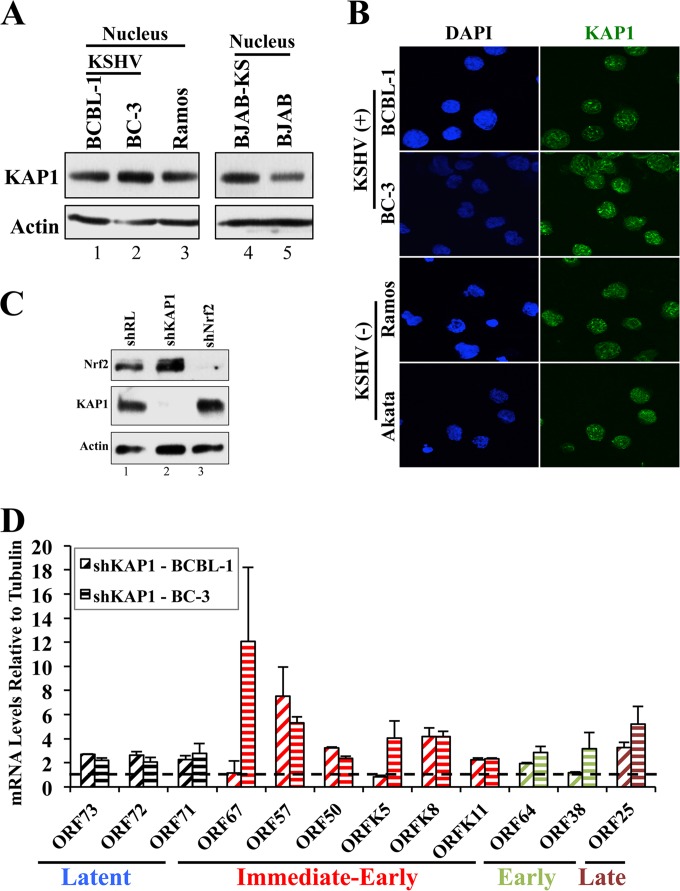
We initially investigated the pattern of KAP1 expression in PEL cell lines, which showed similar KAP1 protein levels and cellular distributions in KSHV-positive and -negative cell lines, residing mainly in the nucleus (Fig. 7A and B). IFA showed that the nuclear distributions of KAP1 were similar in the two cell lines (Fig. 7B). However, when we effectively and specifically knocked down KAP1 in the KSHV-positive BCBL-1 and BC-3 cells (Fig. 7C), we observed a robust induction of KSHV IE, E, and L lytic genes (Fig. 7D) and a moderate increase in latent KSHV genes (Fig. 7D). ChIP analysis confirmed previous reports that KAP1 binds to the ORF50 promoter (data not shown). Overall, these results suggested that while latent KSHV infection does not influence overall KAP1 protein levels in PEL cells as it does with Nrf2, KAP1 activity is important in repressing lytic gene expression, similar to Nrf2.
FIG 7.
Assessment of KAP1 levels in PEL cell lines. (A) Nuclear protein extracts from KSHV-positive and -negative cell lines were Western blotted for KAP1 levels. (B) Immunofluorescence analysis of KSHV-positive and -negative cell lines for KAP1 localization. (C) Determination of KAP1 knockdown during lentiviral transduction with shKAP1 (72 h), as well as the specificity of shKAP1 and shNrf2 for the respective proteins. (D) Real-time RT-PCR for selected genes in each of the four major KSHV gene groups performed on RNA extracted from BCBL-1 and BC-3 cells knocked down for KAP1 by lentiviral transduction for 72 h. The dashed horizontal line indicates the baseline level of the genes under the shRL condition for each cell line (arbitrarily set to 1). The data are expressed as means and SD for at least three independent replicates.
KAP1 and Nrf2 interact in two distinct molecular mass complexes in PEL cells.
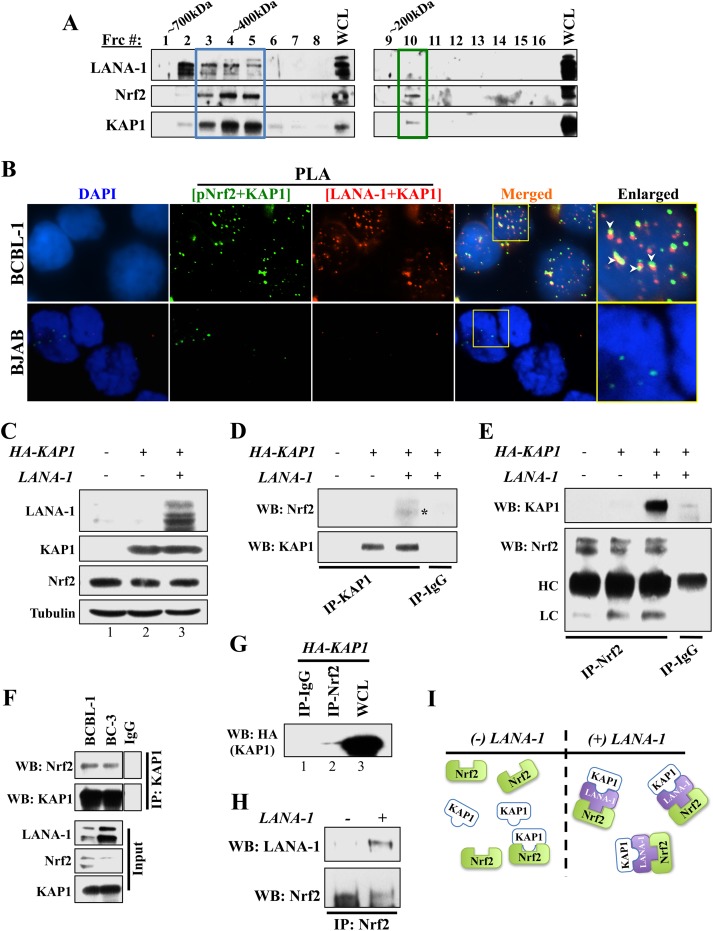
Because LANA-1 binds to Nrf2 and KAP1 and because KAP1 was reported to bind to Nrf2 in KSHV-negative independent systems (43), we hypothesized that KAP1, LANA-1, and pNrf2 may form a complex transcriptional machinery that binds to and affects ORF50 promoter activity differently during the spatiotemporal stages of KSHV infection. After determining that KAP1 binds to both pNrf2 and LANA-1 in PEL-derived cell lines (data not shown), we investigated the possibility of the three proteins interacting in one complex. To test this, initially, we fractionated whole-cell BC-3 protein lysate by high-resolution column gel filtration chromatography, which elutes molecular complexes in fractions by size (39, 44). Western blot analysis of these fractions showed that KAP1 and Nrf2 cofractionated in two separate molecular fractions (Frc). At high molecular masses, KAP1 and Nrf2 cofractionated in abundance, together with LANA-1 (Fig. 8A, Frc 3 to 5, blue box). As LANA-1 levels drop abruptly at fraction 6, so do Nrf2 and KAP1. Interestingly, cofractionation of Nrf2 and KAP1 was observed again in lower-molecular-mass fractions without LANA-1 (Fig. 8A, Frc 10, green box), suggesting a possible LANA-1-independent interaction between the two proteins.
FIG 8.
Investigation of the Nrf2/LANA-1/KAP1 complex. (A) Equal volumes of protein fractions from the KSHV-positive cell line BC-3 were isolated by size using gel filtration chromatography and were subsequently Western blotted for LANA-1, KAP1, and Nrf2. Blue box, the large molecular complex that includes LANA-1; green box, the small molecular complex that excludes LANA-1. (B) Double PLA determining triple colocalization of LANA-1, Nrf2, and KAP1. Initially, PLA was performed using anti-pNrf2 (rabbit) and anti-KAP1 (goat) antibodies and detected using a green-fluorescent PLA probe. After blocking, a second round of PLA was performed using anti-LANA-1 (rabbit) and anti-KAP1 (mouse) antibodies and detected using a red-fluorescent PLA probe. The arrowheads point to areas of significant red and green signal overlap, suggesting triple colocalization of pNrf2, LANA-1, and KAP1. The boxed areas are enlarged on the right. (C to E) HEK293T cells in 10-cm petri dishes were transfected with HA-KAP1 only or with LANA-1. A total of 5 μg of each plasmid was used, and an empty pcDNA vector was used to equalize the total amount of DNA. (C) Whole-cell lysates showing total protein levels of endogenous Nrf2 and tubulin and overexpressed LANA-1 and HA-tagged KAP1. (D and E) KAP1 (D) and Nrf2 (E) pulldowns were performed using 100 μg of protein lysate and 1 μg of the respective antibody. Normal rabbit IgG was used as a negative control. HC, heavy chain; LC, light chain. (F) KAP1 pulldown from BCBL-1 and BC-3 protein lysates demonstrating efficient co-IP between KAP1 and Nrf2 in these cells. (G) HEK293T cells transfected with 10 μg of HA-KAP1 plasmid for 24 h were pulled down with anti-Nrf2 antibody and Western blotted for HA-KAP1 adjacent to the whole-cell lysate to demonstrate the poor interaction levels between Nrf2 and KAP1 in the absence of LANA-1. (H) HEK293T cells transfected with LANA-1 (5 μg) for 24 h were pulled down for Nrf2, showing that Nrf2 and LANA-1 can readily interact in the absence of KAP1. (I) Schematic showing the interaction model between Nrf2, KAP1, and LANA-1.
Furthermore, when we performed a double PLA staining, we found several KAP1/pNrf2 (green) spots colocalized with LANA-1/KAP1 (red) spots in BCBL-1 cells, suggesting triple colocalization of the three proteins (Fig. 8B, arrowheads). We also observed a small number of KAP1/pNrf2 spots that did not colocalize with LANA-1, suggesting that pNrf2 and KAP1 can also interact in the absence of LANA-1 (Fig. 8B). Consistent with this interpretation, we also observed a reduced number of KAP1/pNrf2 (green) spots in the KSHV-negative BJAB cells, which contain no LANA-1 (Fig. 8B).
Overall, the results of these experiments suggested that Nrf2 and KAP1 interact in two separate molecular complexes, a high-molecular-mass complex with LANA-1 and a low-molecular-mass complex without LANA-1.
LANA-1 enhances Nrf2 and KAP1 binding.
To further test the hypothesis that LANA-1 modulates KAP1 and Nrf2 interaction, we used human embryonic kidney 293T (HEK293T) cells. We transfected these cells with HA-KAP1 alone or together with LANA-1 and performed a series of co-IP experiments. A Western blot demonstrating efficient expression of each construct is shown in Fig. 8C. It is important to note that LANA-1 expression did not affect HA-KAP1 plasmid expression or basal endogenous Nrf2 (Fig. 8C) and that HEK293T cells have undetectable endogenous KAP1 expression (Fig. 8C, lane 1). KAP1 pulldown efficiently coimmunoprecipitated Nrf2 only in the presence of LANA-1, while no Nrf2 was coimmunoprecipitated from lysates lacking LANA-1 expression (Fig. 8D, asterisk). Similarly, in the reciprocal assay, Nrf2 pulldown efficiently coimmunoprecipitated KAP1 only in the presence of LANA-1, while no co-IP was observed in its absence (Fig. 8E). Therefore, Nrf2 was easily detectable in KAP1 pulldowns from BC-3 and BCBL-1 lysates, which contain abundant levels of LANA-1 (Fig. 8F). In contrast, when we performed a Western blot for HA-KAP1 in HEK293T lysates pulled down for Nrf2 adjacent to a lane of whole-cell lysate from these cells, we observed that although Nrf2 and KAP1 did interact with one another in the absence of LANA-1, such interaction was difficult to detect and minimal compared to the whole-cell protein levels of KAP1 (Fig. 8G, compare lanes 2 and 3). Separately, Nrf2 and LANA-1 readily coimmunoprecipitated even in the absence of KAP1 (Fig. 8H).
Collectively, these results indicated that in the absence of LANA-1, Nrf2 and KAP1 have a weakly detectable affinity for one another, explaining the low-molecular-mass association of these proteins and the few pNrf2/KAP1 PLA spots [Fig. 8I, (−) LANA-1]. In the presence of LANA-1, however, the KAP1/Nrf2 interaction increases significantly, with LANA-1 acting as a facilitator of the interaction, explaining the high-molecular-mass association [Fig. 8I, (+) LANA-1].
LANA-1 and KAP1 modulate the effect of Nrf2 on the ORF50 promoter.
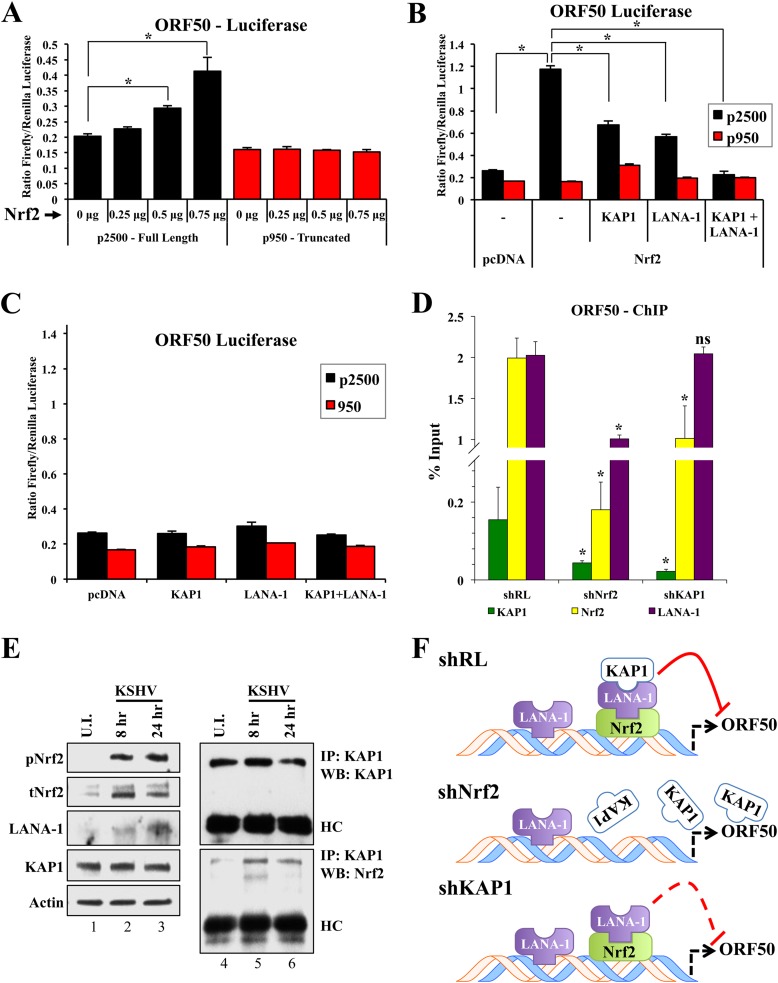
Through a series of luciferase assays, we investigated the effect of the complex interaction between Nrf2, LANA-1, and KAP1 on the ORF50 promoter. Using two ORF50 promoter constructs, the full-length promoter (p2500) and the truncated proximal 950 bp lacking the two distal AREs (p950) (Fig. 4A), we performed a luciferase assay. Overexpression of Nrf2 along with these constructs resulted in a dose-dependent induction of p2500 but no induction of p950 (Fig. 9A), suggesting that the two distal, but not the proximal, AREs are essential for the Nrf2-mediated effects on ORF50 expression.
FIG 9.
Luciferase and ChIP analyses of the ORF50 promoter. (A) Firefly luciferase constructs containing either the full-length ORF50 promoter (p2500) or its truncated counterpart lacking the distal ARE (p950) were transfected with increasing doses of an Nrf2-expressing vector for 24 h prior to assaying for luciferase expression. (B and C) p2500 and p950 ORF50 luciferase constructs were transfected along with various combinations of vectors expressing Nrf2, KAP1, and LANA-1 (0.25 μg each) and assayed for luciferase expression 48 h posttransfection. (A to C) Transfections were performed in HEK293T cells in a 24-well plate at ∼70% confluence. A cumulative amount of 1 μg plasmid DNA was added to each well. To account for variable transfection efficiency, a dual-glow luciferase system was utilized, using a constitutively expressed Renilla luciferase reporter plasmid. The bars indicate the firefly/Renilla luciferase expression ratios under each condition. *, P < 0.05. (D) ChIP analysis using anti-Nrf2, KAP1, and LANA-1 antibodies was performed on chromatin obtained from BCBL-1 cells that had been transduced with shRL, shNrf2, or shKAP1 for 72 h. The results are expressed as percentages of DNA pulled down compared to the respective input chromatin under each condition. *, P < 0.05 in relation to the respective shRL condition. (E) De novo-infected HMVEC-d (40 DNA copies per cell) were coimmunoprecipitated using an anti-KAP1 antibody to assess the interaction levels between Nrf2 and KAP1 at various times of infection. Lanes 1 to 3, input/WCL; lanes 4 to 6, co-IP conditions. U.I., uninfected. (F) Schematic showing the orientation of the Nrf2, LANA-1, and KAP1 complex on the ORF50 promoter and the effect of such binding on its activity. The solid red line indicates full inhibitory activity, while the dashed red line indicates attenuated inhibitory activity. The data are expressed as means and SD for at least three independent replicates.
Although Nrf2 played a negative regulatory role in ORF50 gene expression in PEL cells (Fig. 5 and 6), it appeared to play a positive role on the ORF50 promoter in the luciferase assay, which is consistent with the positive effect of Nrf2 in the lytic burst observed during de novo infection of endothelial cells (30). We therefore hypothesized that different agents present in PEL cells but absent in HEK293T and endothelial cells early during de novo infection, LANA-1 being one of them, could manipulate the activating effects of Nrf2 on the ORF50 promoter into a repressive one. Indeed, when we overexpressed LANA-1 along with Nrf2, the p2500 luciferase activity was moderately reduced (Fig. 9B). Similarly, KAP1 overexpression also moderately reduced Nrf2-mediated p2500 induction (Fig. 9B). More importantly, when KAP1 and LANA-1 were simultaneously coexpressed with Nrf2, p2500 was suppressed to baseline levels (Fig. 9B). On the other hand, when KAP1 and LANA-1 were coexpressed in the absence of Nrf2, they exerted no effect on the p2500 promoter (Fig. 9C).
Collectively, these luciferase assays suggested that in the absence of LANA-1 and KAP1 interaction, Nrf2 activates the ORF50 promoter, an effect that is actively repressed by LANA-1 and KAP1.
Nrf2 recruits LANA-1 and KAP1 to the ORF50 promoter.
Since we showed that these three proteins can form a complex and can affect ORF50 promoter activity, we postulated that Nrf2, KAP1, and LANA-1 bind to the ORF50 promoter as a complex. To assess the orientation of this complex, we knocked down Nrf2 and KAP1 in BCBL-1 cells and performed ChIP using anti-Nrf2, -KAP1, and -LANA-1 antibodies. LANA-1 knockdown was not feasible because it affects KSHV genome integrity. As expected, Nrf2 knockdown reduced Nrf2 binding to the ORF50 promoter by ∼80% (Fig. 9D). More importantly, both KAP1 and LANA-1 levels of binding to the ORF50 promoter were significantly reduced by Nrf2 knockdown (∼75% and 50%, respectively), suggesting that Nrf2 is imperative for this complex to efficiently bind to the promoter (Fig. 9D). KAP1 knockdown did not affect LANA-1 binding to the ORF50 promoter while it moderately affected Nrf2 binding (Fig. 9D).
Indeed, when HMVEC-d were infected de novo with KSHV, we observed a significant increase in KAP1 and Nrf2 interaction in infected cells compared to uninfected cells (Fig. 9E, lanes 4 to 6). This increased interaction is likely due to the increased levels of LANA-1 expression during the course of infection (Fig. 9E, lanes 1 to 3) as KSHV establishes latency. These results, coupled with the knowledge that Nrf2, but not KAP1, has direct DNA-binding ability, strongly suggest a model where this complex utilizes Nrf2 to bind to the ORF50 promoter (Fig. 9F, shRL). In the absence of Nrf2, this complex is disassembled and cannot bind to the ORF50 promoter (Fig. 9F, shNrf2), although additional LANA-1 binding sites through other transcription factors are likely. In the absence of KAP1, the Nrf2/LANA-1 complex still retains its ability to bind to the ORF50 promoter, although at a decreased efficiency for Nrf2, and reduced the repressive activity of the ORF50 promoter (Fig. 9F, shKAP1).
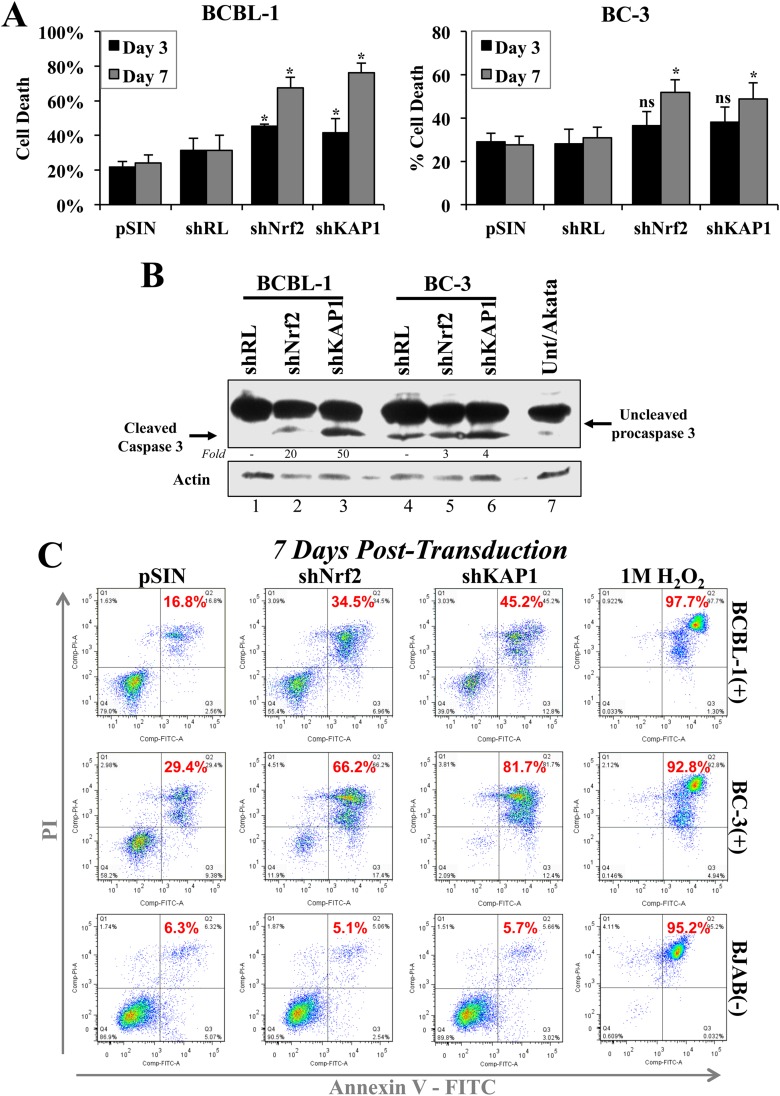
KAP1 and Nrf2 knockdown triggers PEL cell death.
Nrf2 and KAP1 inhibition has been associated with decreased proliferation and stress induction (45, 46). We undertook a series of assays to determine if inhibition of these two proteins in the context of PEL resulted in growth inhibition. Using trypan blue stain, we counted the cells of PEL cell lines BCBL-1 and BC-3 at 3 and 7 days postransduction with shNrf2 and shKAP1, and we observed a significant increase in the proportion of cell death (Fig. 10A). Western blot analysis revealed increased caspase-3 cleavage under both shKAP1 and shNrf2 conditions, although the increase was more prominent in BCBL-1 cell lines (Fig. 10B). Interestingly, BC-3 cells, with wild-type p53 status, exhibited elevated basal caspase-3 cleavage compared to BCBL-1 cells, which have an acquired loss-of-function p53 mutation (Fig. 10B, compare lanes 1 and 4) (47).
FIG 10.
Analysis of Nrf2 and KAP1 knockdown effects on PEL cell death. (A) BCBL-1 and BC-3 cells lentivirally transduced with a self-inactivating empty vector (pSIN), shRL, shNrf2, and shKAP1 for 3 and 7 days were stained with trypan blue to identify dead cells and counted in a hemocytometer. Two negative controls (pSIN and shRL) were used to ensure the baseline cell death observed was specific. The bars indicate the percentages of dead cells in the whole population of counted cells. *, P < 0.05 compared to the respective pSIN condition. (B) Western blot for caspase-3 and procaspase-3 staining from whole-cell lysates of BCBL-1 and BC-3 cells 3 days after lentiviral transduction with shRL, shNrf2, and shKAP1. Akata cells were included to observe baseline apoptosis in KSHV-negative lymphomas. (C) Annexin V and propidium iodide staining was performed on BCBL-1, BC-3, and BJAB cells to determine the levels of apoptosis (Q2, red numbers) in each cell line 7 days postransduction with pSIN, shNrf2, and shKAP1. shRL was avoided in this assay, as the green fluorescence of GFP expressed by shRL interfered with proper annexin V and PI gating. H2O2 (1 M) treatment for 30 min was used as a positive control for apoptosis in each cell line. The data are expressed as means and SD for at least three independent replicates.
Flow cytometric analysis of PEL cells further confirmed that the increased cell death in shKAP1 and shNrf2 was due to increased apoptosis, as we observed an increase in annexin V/PI-positive cells (Fig. 10C, quadrants 2 [Q2], red numbers). Interestingly, BC-3 cells were more prone to cell death by apoptosis than BCBL-1 cells, consistent with prior reports (47). Importantly, the KSHV-negative BJAB cell line did not respond to KAP1 and Nrf2 knockdown with increased cell death (Fig. 10C), suggesting that the apoptotic effects observed in PEL might be also due to lytic KSHV reactivation (Fig. 6F), which has been previously shown to induce PEL cell apoptosis (48).
DISCUSSION
Discovering targetable host factors involved in the KSHV life cycle is key to our understanding and treatment of KSHV-associated malignancies. Here, we extensively investigated the role of the transcription factor Nrf2 in KSHV latency, by focusing on PEL and PEL-derived cell lines, and its importance in global KSHV gene regulation. Specifically, we showed that (i) Nrf2 protein levels, phosphorylation, and transcriptional activity are induced in latently infected PEL cells; (ii) the COX-2/PGE2 paracrine signaling axis induces Nrf2 through the receptor EP4 (Fig. 11B); (iii) activated Nrf2 has an inducing effect on the ORF50 promoter in the absence of LANA-1; (iv) LANA-1 exploits Nrf2 binding on the ORF50 promoter to recruit KAP1 and mediate subsequent ORF50 repression (Fig. 11A); and (v) the Nrf2 inhibitor brusatol is a potential therapeutic agent to combat KSHV infection and associated malignancies. Below is a thorough analysis of our findings.
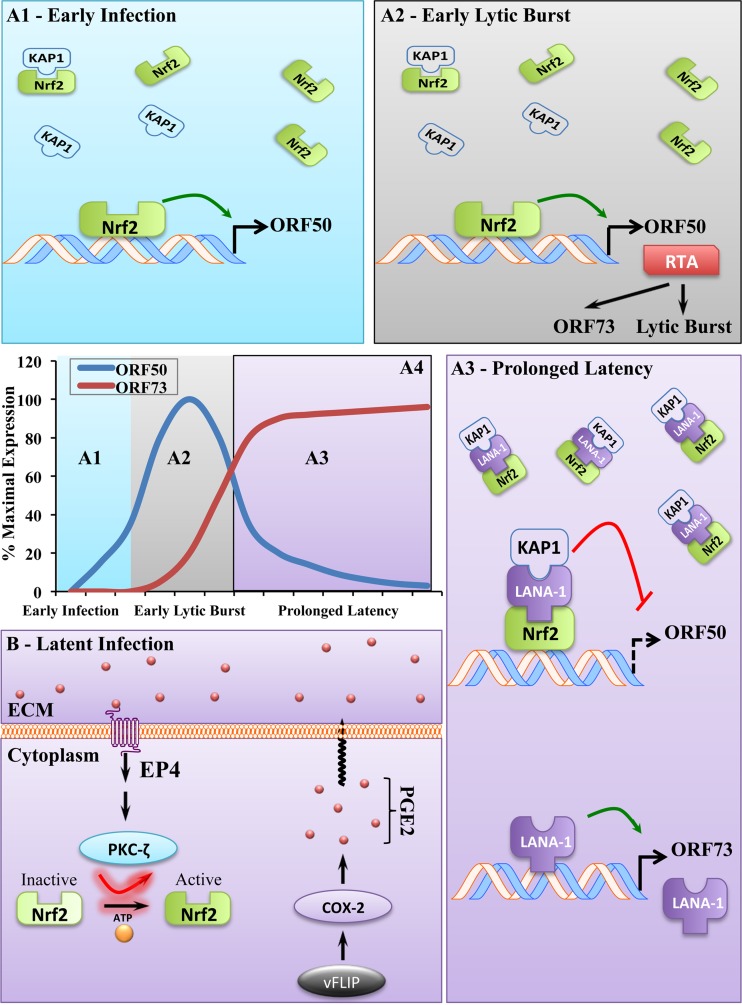
FIG 11.
Schematic of the role of Nrf2 in KSHV biology. (A) Effect of Nrf2 on lytic gene expression. (A1) During early de novo infection, when Nrf2 and KAP1 interaction is minimal due to LANA-1 absence, activated Nrf2 binds to and induces the ORF50 promoter. (A2) Therefore, when Nrf2 is inhibited prior to infection, the KSHV lytic burst is weakened, possibly exposing the virus to host responses in an in vivo system. During the lytic burst early in de novo infection, Nrf2-induced RTA binds to and transactivates LANA-1 through its inducible promoter, allowing its accumulation and initiation of latency. (A3) As latency is established and LANA-1 expression increases, LANA-1 binds to Nrf2 located on the ORF50 promoter and recruits KAP1 along with it, which inhibits the inducing transcriptional effects of Nrf2 on the region. (A4) Temporal representation of the expression of ORF50 and ORF73 during de novo KSHV infection and establishment of prolonged latency. (B) Mechanism of Nrf2 induction during KSHV latency. KSHV latent protein vFLIP-mediated constitutive COX-2 induction induces formation of PGE2, which, through autocrine and paracrine mechanisms, binds to EP4. Upon induction, EP4 augments PKCζ signaling and induces Nrf2 activation through Ser-40 phosphorylation, nuclear translocation, and DNA binding and results in target gene induction, lytic gene repression by LANA-1/KAP1 recruitment to the ORF50 promoter, and latency maintenance (A3).
Effect of Nrf2 on KSHV lytic gene expression.
De novo KSHV infection results in a burst of lytic gene expression with antiapoptotic and immune-evasive functions (9). After the lytic burst, ORF50 inhibition is required for latency establishment and maintenance, and these are achieved by the addition of repressive epigenetic markers (12). Inhibition of ORF50 is quickly reversible, however, and it is facilitated by the simultaneous addition of activating epigenetic markers (12). Such a dynamic change likely requires a preexisting protein complex(es) that can quickly switch the heterochromatic landscape of the ORF50 promoter observed during latency into the euchromatic state observed during lytic reactivation. We propose that the intricate Nrf2/LANA-1/KAP1 interaction model identified in this study is one of the important machineries that control the dynamic switch in ORF50 chromatin status. Activated Nrf2 likely binds to the ORF50 promoter throughout the course of infection and latency, acting as a constitutive “on” switch by recruiting several cofactors that mediate activating epigenetic modifications. During establishment of latency, Nrf2 binding to the ORF50 promoter is then exploited by LANA-1, which recruits KAP1 and possibly additional repressive factors to turn the ORF50 promoter “off.”
Potential transcriptional cofactors recruited by Nrf2, LANA-1, and KAP1.
Nrf2 transactivation of target genes requires assembly of high-order transcriptional machineries that contain the CBP/p300 acetyltransferase as a component, and such complexes may be utilized by Nrf2 to mediate ORF50 acetylation and promoter activation (49–51). Interestingly, LANA-1 inhibits CBP/p300 activity (52), and this may be the first step in ORF50 repression observed following the lytic burst. This interpretation is consistent with the luciferase assay findings in Fig. 9B, where LANA-1 moderately repressed Nrf2-mediated ORF50 activation even in the absence of KAP1.
KAP1 is a host transcriptional repressor that can inhibit KSHV lytic gene expression. The lytic viral protein kinase (vpk) inhibits the repressive function of KAP1 through Ser-824 phosphorylation (53), which competitively inhibits KAP1 SUMOylation, a modification required for its interaction with LANA-1 and ORF50 repression (15, 16, 54). Interestingly, a recent report by Sun et al. (16) demonstrated that LANA-1-mediated KAP1 recruitment to the ORF50 promoter resulted in repressive epigenetic modifications by decreasing activating histone acetylation (AcH3), possibly through recruitment of the histone deacetylase complexes HDAC1, HDAC2, and RBBP. This interpretation would explain the robust repressive effect that KAP1 recruitment to the ORF50 promoter had on the Nrf2-mediated ORF50 activation shown in Fig. 9B. Indeed, a bivalent system using an Nrf2-associated coactivating acetyltransferase (CBP/p300) and a KAP1-associated corepressing deacetylase (HDAC1/2) provides an appealing and plausible dynamic model, which should be investigated further. However, Nrf2 and KAP1 recruitment of additional chromatin-modifying agents, such as H3K4me3 and H3K9me3 methyltransferases/demethylases, is likely and would explain the quick enrichment in such markers on the ORF50 promoter following de novo infection.
Temporal analysis of the recruitment of Nrf2, LANA-1, and KAP1 to the ORF50 promoter.
Based on these studies, we propose a model in a temporal fashion (Fig. 11A4). During early de novo KSHV infection, activated Nrf2 binds to the naive ORF50 promoter and induces its expression, facilitating the lytic burst with antiapoptotic and immune-evasive functions (Fig. 11A1 and A2). At this early stage of infection, as LANA-1 is absent, KAP1 binds weakly to Nrf2 and is not capable of repressing the activating effects that pNrf2 exerts on the ORF50 promoter. As RTA accumulates, it binds to and activates the inducible ORF73 promoter (Lpi), resulting in a steady increase in LANA-1 transcription (Fig. 11A2). As LANA-1 accumulates, it recruits KAP1 to pNrf2 on the ORF50 promoter and represses its activating effects (Fig. 11A3).
Nrf2 induction through the COX-2/PGE2/EP4/PKCζ axis in KSHV-infected cells.
The induction of COX-2 during KSHV infection and its signaling through PGE2 production have been thoroughly investigated (40, 55–60). In our recent reports, we determined that Nrf2 induction during de novo KSHV infection of endothelial cells (HMVEC-d) was important for transcriptional upregulation of COX-2 expression (30, 61). More importantly, we determined that COX-2-mediated PGE2 signaling through PKCζ was required for prolonged Nrf2 activation by KSHV during de novo infection of endothelial cells. In the current study, we determined that Nrf2 activation in latently infected PEL cell lines relied mainly on COX-2/PGE2/PKCζ signaling (Fig. 11B). Furthermore, we discovered that EP4 was required for Nrf2 activation (Fig. 11B). Such findings are consistent with a report by Paul et al. (62), which determined that EP4, but not EP1 or EP2, activation mediated PKCζ activation, which we have shown to be crucial for Nrf2 activation by PGE2.
Implications for KSHV-infected PEL cell death and brusatol treatment.
We were surprised to observe the significant cell death by Nrf2 knockdown in the PEL cell lines. While the stress-inducing, antiproliferative effects of Nrf2 knockdown have been previously reported, these effects do not become apparent until Nrf2 inhibition is coadministered with cytotoxic chemotherapeutic drugs. Similarly, brusatol's anticancerous effects on A549, a lung adenocarcinoma cell line with elevated Nrf2, were not significant until coadministration with cisplatin, whereas its toxic effects on PEL cells at the same concentration were readily observable (data not shown) (42, 63–65). These data suggest that PEL cell lines are highly dependent on Nrf2 for proliferation and survival, explaining the high Nrf2 levels in these cell lines. It is tempting to speculate that increased lytic gene expression and virus production may contribute to the observed cell death, because such effects were not present during Nrf2 knockdown of the KSHV-negative BJAB cell line.
In conclusion, this study expands our recent findings on the role of Nrf2 in KSHV biology and establishes it as a molecular signaling hub during KSHV infection that initially induces the early lytic gene burst and subsequently facilitates the establishment of latency. The existence of orally available Nrf2 modulators, such as sulforaphane, tert-butylhydroquinone (tBHQ), and brusatol, make Nrf2 an appealing therapeutic agent in the fight against KSHV-associated malignancies, especially if used in combination with COX-2 and the lytic cycle inhibitors celecoxib and ganciclovir, respectively.
ACKNOWLEDGMENTS
This study was supported in part by Public Health Service grants CA 075911 and CA 168472 and by the Rosalind Franklin University of Medicine and Science H. M. Bligh Cancer Research Fund (to B.C.).
We thank Keith Philibert for critically reading the manuscript. We thank Subhash Verma for allowing us to use the facilities at the University of Nevada, Reno, NV, for the viral-RNA-sequencing experiments. We gratefully acknowledge Robert Dickinson (Flow Cytometry Core Facility, RFUMS) for assisting with flow cytometry.
REFERENCES
- 1.Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med 332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 2.Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 3.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay MF, Clauvel JP, Raphael M, Degos L, Sigaux F. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276–1280. [PubMed] [Google Scholar]
- 4.Costa J, Rabson AS. 1983. Generalised Kaposi's sarcoma is not a neoplasm. Lancet i:58. [DOI] [PubMed] [Google Scholar]
- 5.Moore PS, Gao SJ, Dominguez G, Cesarman E, Lungu O, Knowles DM, Garber R, Pellett PE, McGeoch DJ, Chang Y. 1996. Primary characterization of a herpesvirus agent associated with Kaposi's sarcomae. J Virol 70:549–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okada S, Goto H, Yotsumoto M. 2014. Current status of treatment for primary effusion lymphoma. Intractable Rare Dis Res 3:65–74. doi: 10.5582/irdr.2014.01010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med 2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 8.Miller G, Heston L, Grogan E, Gradoville L, Rigsby M, Sun R, Shedd D, Kushnaryov VM, Grossberg S, Chang Y. 1997. Selective switch between latency and lytic replication of Kaposi's sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J Virol 71:314–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krishnan HH, Naranatt PP, Smith MS, Zeng L, Bloomer C, Chandran B. 2004. Concurrent expression of latent and a limited number of lytic genes with immune modulation and antiapoptotic function by Kaposi's sarcoma-associated herpesvirus early during infection of primary endothelial and fibroblast cells and subsequent decline of lytic gene expression. J Virol 78:3601–3620. doi: 10.1128/JVI.78.7.3601-3620.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toth Z, Maglinte DT, Lee SH, Lee HR, Wong LY, Brulois KF, Lee S, Buckley JD, Laird PW, Marquez VE, Jung JU. 2010. Epigenetic analysis of KSHV latent and lytic genomes. PLoS Pathog 6:e1001013. doi: 10.1371/journal.ppat.1001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gunther T, Grundhoff A. 2010. The epigenetic landscape of latent Kaposi sarcoma-associated herpesvirus genomes. PLoS Pathog 6:e1000935. doi: 10.1371/journal.ppat.1000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toth Z, Brulois K, Jung JU. 2013. The chromatin landscape of Kaposi's sarcoma-associated herpesvirus. Viruses 5:1346–1373. doi: 10.3390/v5051346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeWire SM, Damania B. 2005. The latency-associated nuclear antigen of rhesus monkey rhadinovirus inhibits viral replication through repression of Orf50/Rta transcriptional activation. J Virol 79:3127–3138. doi: 10.1128/JVI.79.5.3127-3138.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu F, Day L, Gao SJ, Lieberman PM. 2006. Acetylation of the latency-associated nuclear antigen regulates repression of Kaposi's sarcoma-associated herpesvirus lytic transcription. J Virol 80:5273–5282. doi: 10.1128/JVI.02541-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai Q, Cai S, Zhu C, Verma SC, Choi JY, Robertson ES. 2013. A unique SUMO-2-interacting motif within LANA is essential for KSHV latency. PLoS Pathog 9:e1003750. doi: 10.1371/journal.ppat.1003750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun R, Liang D, Gao Y, Lan K. 2014. Kaposi's sarcoma-associated herpesvirus-encoded LANA interacts with host KAP1 to facilitate establishment of viral latency. J Virol 88:7331–7344. doi: 10.1128/JVI.00596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen T, Nioi P, Pickett CB. 2009. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J Biol Chem 284:13291–13295. doi: 10.1074/jbc.R900010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. 2004. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol 24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Apopa PL, He X, Ma Q. 2008. Phosphorylation of Nrf2 in the transcription activation domain by casein kinase 2 (CK2) is critical for the nuclear translocation and transcription activation function of Nrf2 in IMR-32 neuroblastoma cells. J Biochem Mol Toxicol 22:63–76. doi: 10.1002/jbt.20212. [DOI] [PubMed] [Google Scholar]
- 20.Bloom DA, Jaiswal AK. 2003. Phosphorylation of Nrf2 at Ser40 by protein kinase C in response to antioxidants leads to the release of Nrf2 from INrf2, but is not required for Nrf2 stabilization/accumulation in the nucleus and transcriptional activation of antioxidant response element-mediated NAD(P)H:quinone oxidoreductase-1 gene expression. J Biol Chem 278:44675–44682. doi: 10.1074/jbc.M307633200. [DOI] [PubMed] [Google Scholar]
- 21.Nioi P, McMahon M, Itoh K, Yamamoto M, Hayes JD. 2003. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. Biochem J 374:337–348. doi: 10.1042/BJ20030754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu R, Chen C, Mo YY, Hebbar V, Owuor ED, Tan TH, Kong AN. 2000. Activation of mitogen-activated protein kinase pathways induces antioxidant response element-mediated gene expression via a Nrf2-dependent mechanism. J Biol Chem 275:39907–39913. doi: 10.1074/jbc.M004037200. [DOI] [PubMed] [Google Scholar]
- 23.Niture SK, Jaiswal AK. 2012. Nrf2 protein up-regulates antiapoptotic protein Bcl-2 and prevents cellular apoptosis. J Biol Chem 287:9873–9886. doi: 10.1074/jbc.M111.312694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim TH, Hur EG, Kang SJ, Kim JA, Thapa D, Lee YM, Ku SK, Jung Y, Kwak MK. 2011. NRF2 blockade suppresses colon tumor angiogenesis by inhibiting hypoxia-induced activation of HIF-1alpha. Cancer Res 71:2260–2275. doi: 10.1158/0008-5472.CAN-10-3007. [DOI] [PubMed] [Google Scholar]
- 25.Zhou S, Ye W, Zhang M, Liang J. 2012. The effects of nrf2 on tumor angiogenesis: a review of the possible mechanisms of action. Crit Rev Eukaryot Gene Expr 22:149–160. doi: 10.1615/CritRevEukarGeneExpr.v22.i2.60. [DOI] [PubMed] [Google Scholar]
- 26.Pan H, Wang H, Zhu L, Mao L, Qiao L, Su X. 2013. The role of nrf2 in migration and invasion of human glioma cell u251. World Neurosurg 80:363–370. doi: 10.1016/j.wneu.2011.06.063. [DOI] [PubMed] [Google Scholar]
- 27.Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, Aburatani H, Yamamoto M, Motohashi H. 2012. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell 22:66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 28.Hayashi A, Suzuki H, Itoh K, Yamamoto M, Sugiyama Y. 2003. Transcription factor Nrf2 is required for the constitutive and inducible expression of multidrug resistance-associated protein 1 in mouse embryo fibroblasts. Biochem Biophys Res Commun 310:824–829. doi: 10.1016/j.bbrc.2003.09.086. [DOI] [PubMed] [Google Scholar]
- 29.Maher JM, Dieter MZ, Aleksunes LM, Slitt AL, Guo G, Tanaka Y, Scheffer GL, Chan JY, Manautou JE, Chen Y, Dalton TP, Yamamoto M, Klaassen CD. 2007. Oxidative and electrophilic stress induces multidrug resistance-associated protein transporters via the nuclear factor-E2-related factor-2 transcriptional pathway. Hepatology 46:1597–1610. doi: 10.1002/hep.21831. [DOI] [PubMed] [Google Scholar]
- 30.Gjyshi O, Bottero V, Veettil MV, Dutta S, Singh VV, Chikoti L, Chandran B. 2014. Kaposi's sarcoma-associated herpesvirus induces Nrf2 during de novo infection of endothelial cells to create a microenvironment conducive to infection. PLoS Pathog 10:e1004460. doi: 10.1371/journal.ppat.1004460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.No JH, Kim YB, Song YS. 2014. Targeting nrf2 signaling to combat chemoresistance. J Cancer Prev 19:111–117. doi: 10.15430/JCP.2014.19.2.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gjyshi O, Flaherty S, Veettil MV, Johnson KE, Chandran B, Bottero V. 2015. Kaposi's sarcoma-associated herpesvirus induces Nrf2 activation in latently infected endothelial cells through SQSTM1 phosphorylation and interaction with polyubiquitinated Keap1. J Virol 89:2268–2286. doi: 10.1128/JVI.02742-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nun TK, Kroll DJ, Oberlies NH, Soejarto DD, Case RJ, Piskaut P, Matainaho T, Hilscher C, Wang L, Dittmer DP, Gao SJ, Damania B. 2007. Development of a fluorescence-based assay to screen antiviral drugs against Kaposi's sarcoma associated herpesvirus. Mol Cancer Ther 6:2360–2370. doi: 10.1158/1535-7163.MCT-07-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.An FQ, Folarin HM, Compitello N, Roth J, Gerson SL, McCrae KR, Fakhari FD, Dittmer DP, Renne R. 2006. Long-term-infected telomerase-immortalized endothelial cells: a model for Kaposi's sarcoma-associated herpesvirus latency in vitro and in vivo. J Virol 80:4833–4846. doi: 10.1128/JVI.80.10.4833-4846.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vart RJ, Nikitenko LL, Lagos D, Trotter MW, Cannon M, Bourboulia D, Gratrix F, Takeuchi Y, Boshoff C. 2007. Kaposi's sarcoma-associated herpesvirus-encoded interleukin-6 and G-protein-coupled receptor regulate angiopoietin-2 expression in lymphatic endothelial cells. Cancer Res 67:4042–4051. doi: 10.1158/0008-5472.CAN-06-3321. [DOI] [PubMed] [Google Scholar]
- 36.Zhu L, Wang R, Sweat A, Goldstein E, Horvat R, Chandran B. 1999. Comparison of human sera reactivities in immunoblots with recombinant human herpesvirus (HHV)-8 proteins associated with the latent (ORF73) and lytic (ORFs 65, K81A, and K81B) replicative cycles and in immunofluorescence assays with HHV-8-infected BCBL-1 cells. Virology 256:381–392. doi: 10.1006/viro.1999.9674. [DOI] [PubMed] [Google Scholar]
- 37.Purushothaman P, Thakker S, Verma SC. 2015. Transcriptome analysis of Kaposi's sarcoma-associated herpesvirus during de novo primary infection of human B and endothelial cells. J Virol 89:3093–3111. doi: 10.1128/JVI.02507-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye J, Shedd D, Miller G. 2005. An Sp1 response element in the Kaposi's sarcoma-associated herpesvirus open reading frame 50 promoter mediates lytic cycle induction by butyrate. J Virol 79:1397–1408. doi: 10.1128/JVI.79.3.1397-1408.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarek G, Jarviluoma A, Ojala PM. 2006. KSHV viral cyclin inactivates p27KIP1 through Ser10 and Thr187 phosphorylation in proliferating primary effusion lymphomas. Blood 107:725–732. doi: 10.1182/blood-2005-06-2534. [DOI] [PubMed] [Google Scholar]
- 40.Paul AG, Chandran B, Sharma-Walia N. 2013. Cyclooxygenase-2-prostaglandin E2-eicosanoid receptor inflammatory axis: a key player in Kaposi's sarcoma-associated herpes virus associated malignancies. Transl Res 162:77–92. doi: 10.1016/j.trsl.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coscoy L. 2007. Immune evasion by Kaposi's sarcoma-associated herpesvirus. Nat Rev Immunol 7:391–401. doi: 10.1038/nri2076. [DOI] [PubMed] [Google Scholar]
- 42.Ren D, Villeneuve NF, Jiang T, Wu T, Lau A, Toppin HA, Zhang DD. 2011. Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Proc Natl Acad Sci U S A 108:1433–1438. doi: 10.1073/pnas.1014275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maruyama A, Nishikawa K, Kawatani Y, Mimura J, Hosoya T, Harada N, Yamamato M, Itoh K. 2011. The novel Nrf2-interacting factor KAP1 regulates susceptibility to oxidative stress by promoting the Nrf2-mediated cytoprotective response. Biochem J 436:387–397. doi: 10.1042/BJ20101748. [DOI] [PubMed] [Google Scholar]
- 44.Paudel N, Sadagopan S, Chakraborty S, Sarek G, Ojala PM, Chandran B. 2012. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen interacts with multifunctional angiogenin to utilize its antiapoptotic functions. J Virol 86:5974–5991. doi: 10.1128/JVI.00070-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng CT, Kuo CY, Ann DK. 2014. KAPtain in charge of multiple missions: emerging roles of KAP1. World J Biol Chem 5:308–320. doi: 10.4331/wjbc.v5.i3.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geismann C, Arlt A, Sebens S, Schafer H. 2014. Cytoprotection “gone astray”: Nrf2 and its role in cancer. Onco Targets Ther 7:1497–1518. doi: 10.2147/OTT.S36624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petre CE, Sin SH, Dittmer DP. 2007. Functional p53 signaling in Kaposi's sarcoma-associated herpesvirus lymphomas: implications for therapy. J Virol 81:1912–1922. doi: 10.1128/JVI.01757-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X, Feng J, Sun R. 2011. Oxidative stress induces reactivation of Kaposi's sarcoma-associated herpesvirus and death of primary effusion lymphoma cells. J Virol 85:715–724. doi: 10.1128/JVI.01742-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Katoh Y, Itoh K, Yoshida E, Miyagishi M, Fukamizu A, Yamamoto M. 2001. Two domains of Nrf2 cooperatively bind CBP, a CREB binding protein, and synergistically activate transcription. Genes Cells 6:857–868. doi: 10.1046/j.1365-2443.2001.00469.x. [DOI] [PubMed] [Google Scholar]
- 50.Zhu M, Fahl WE. 2001. Functional characterization of transcription regulators that interact with the electrophile response element. Biochem Biophys Res Commun 289:212–219. doi: 10.1006/bbrc.2001.5944. [DOI] [PubMed] [Google Scholar]
- 51.Ohta K, Ohigashi M, Naganawa A, Ikeda H, Sakai M, Nishikawa J, Imagawa M, Osada S, Nishihara T. 2007. Histone acetyltransferase MOZ acts as a co-activator of Nrf2-MafK and induces tumour marker gene expression during hepatocarcinogenesis. Biochem J 402:559–566. doi: 10.1042/BJ20061194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lim C, Gwack Y, Hwang S, Kim S, Choe J. 2001. The transcriptional activity of cAMP response element-binding protein-binding protein is modulated by the latency associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J Biol Chem 276:31016–31022. doi: 10.1074/jbc.M102431200. [DOI] [PubMed] [Google Scholar]
- 53.Chang PC, Fitzgerald LD, Van Geelen A, Izumiya Y, Ellison TJ, Wang DH, Ann DK, Luciw PA, Kung HJ. 2009. Kruppel-associated box domain-associated protein-1 as a latency regulator for Kaposi's sarcoma-associated herpesvirus and its modulation by the viral protein kinase. Cancer Res 69:5681–5689. doi: 10.1158/0008-5472.CAN-08-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang L, Zhu C, Guo Y, Wei F, Lu J, Qin J, Banerjee S, Wang J, Shang H, Verma SC, Yuan Z, Robertson ES, Cai Q. 2014. Inhibition of KAP1 enhances hypoxia-induced Kaposi's sarcoma-associated herpesvirus reactivation through RBP-Jkappa. J Virol 88:6873–6884. doi: 10.1128/JVI.00283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sharma-Walia N, Patel K, Chandran K, Marginean A, Bottero V, Kerur N, Paul AG. 2012. COX-2/PGE2: molecular ambassadors of Kaposi's sarcoma-associated herpes virus oncoprotein-v-FLIP. Oncogenesis 1:e5. doi: 10.1038/oncsis.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharma-Walia N, Paul AG, Bottero V, Sadagopan S, Veettil MV, Kerur N, Chandran B. 2010. Kaposi's sarcoma associated herpes virus (KSHV) induced COX-2: a key factor in latency, inflammation, angiogenesis, cell survival and invasion. PLoS Pathog 6:e1000777. doi: 10.1371/journal.ppat.1000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharma-Walia N, Raghu H, Sadagopan S, Sivakumar R, Veettil MV, Naranatt PP, Smith MM, Chandran B. 2006. Cyclooxygenase 2 induced by Kaposi's sarcoma-associated herpesvirus early during in vitro infection of target cells plays a role in the maintenance of latent viral gene expression. J Virol 80:6534–6552. doi: 10.1128/JVI.00231-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shelby BD, LaMarca HL, McFerrin HE, Nelson AB, Lasky JA, Sun G, Myatt L, Offermann MK, Morris CA, Sullivan DE. 2007. Kaposi's sarcoma associated herpesvirus G-protein coupled receptor activation of cyclooxygenase-2 in vascular endothelial cells. Virol J 4:87. doi: 10.1186/1743-422X-4-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paul AG, Chandran B, Sharma-Walia N. 2013. Concurrent targeting of eicosanoid receptor 1/eicosanoid receptor 4 receptors and COX-2 induces synergistic apoptosis in Kaposi's sarcoma-associated herpesvirus and Epstein-Barr virus associated non-Hodgkin lymphoma cell lines. Transl Res 161:447–468. doi: 10.1016/j.trsl.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paul AG, Sharma-Walia N, Chandran B. 2011. Targeting KSHV/HHV-8 latency with COX-2 selective inhibitor nimesulide: a potential chemotherapeutic modality for primary effusion lymphoma. PLoS One 6:e24379. doi: 10.1371/journal.pone.0024379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gjyshi O, Chandran B. 2015. Manipulating host signaling for pathogenic purposes: hijacking of the transcriptional activity of the host protein Nrf2 by Kaposi's sarcoma-associated herpesvirus. Sci Proc 2:e599. doi: 10.14800/sp.599. [DOI] [Google Scholar]
- 62.Paul AG, Sharma-Walia N, Kerur N, White C, Chandran B. 2010. Piracy of prostaglandin E2/EP receptor-mediated signaling by Kaposi's sarcoma-associated herpes virus (HHV-8) for latency gene expression: strategy of a successful pathogen. Cancer Res 70:3697–3708. doi: 10.1158/0008-5472.CAN-09-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu T, Harder BG, Wong PK, Lang JE, Zhang DD. 23 August 2014. Oxidative stress, mammospheres and Nrf2—new implication for breast cancer therapy? Mol Carcinog doi: 10.1002/mc.22202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tao S, Wang S, Moghaddam SJ, Ooi A, Chapman E, Wong PK, Zhang DD. 2014. Oncogenic KRAS confers chemoresistance by upregulating NRF2. Cancer Res 74:7430–7441. doi: 10.1158/0008-5472.CAN-14-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Olayanju A, Copple IM, Bryan HK, Edge GT, Sison RL, Wong MW, Lai ZQ, Lin ZX, Dunn K, Sanderson CM, Alghanem AF, Cross MJ, Ellis EC, Ingelman-Sundberg M, Malik HZ, Kitteringham NR, Goldring CE, Park BK. 2015. Brusatol provokes a rapid and transient inhibition of Nrf2 signaling and sensitizes mammalian cells to chemical toxicity—implications for therapeutic targeting of Nrf2. Free Radic Biol Med 78:202–212. doi: 10.1016/j.freeradbiomed.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]