ABSTRACT
Respiratory syncytial virus (RSV) is the leading cause of pediatric respiratory disease. RSV has an RNA-dependent RNA polymerase that transcribes and replicates the viral negative-sense RNA genome. The large polymerase subunit (L) has multiple enzymatic activities, having the capability to synthesize RNA and add and methylate a cap on each of the viral mRNAs. Previous studies (H. Xiong et al., Bioorg Med Chem Lett, 23:6789–6793, 2013, http://dx.doi.org/10.1016/j.bmcl.2013.10.018; C. L. Tiong-Yip et al., Antimicrob Agents Chemother, 58:3867–3873, 2014, http://dx.doi.org/10.1128/AAC.02540-14) had identified a small-molecule inhibitor, AZ-27, that targets the L protein. In this study, we examined the effect of AZ-27 on different aspects of RSV polymerase activity. AZ-27 was found to inhibit equally both mRNA transcription and genome replication in cell-based minigenome assays, indicating that it inhibits a step common to both of these RNA synthesis processes. Analysis in an in vitro transcription run-on assay, containing RSV nucleocapsids, showed that AZ-27 inhibits synthesis of transcripts from the 3′ end of the genome to a greater extent than those from the 5′ end, indicating that it inhibits transcription initiation. Consistent with this finding, experiments that assayed polymerase activity on the promoter showed that AZ-27 inhibited transcription and replication initiation. The RSV polymerase also can utilize the promoter sequence to perform a back-priming reaction. Interestingly, addition of AZ-27 had no effect on the addition of up to three nucleotides by back-priming but inhibited further extension of the back-primed RNA. These data provide new information regarding the mechanism of inhibition by AZ-27. They also suggest that the RSV polymerase adopts different conformations to perform its different activities at the promoter.
IMPORTANCE Currently, there are no effective antiviral drugs to treat RSV infection. The RSV polymerase is an attractive target for drug development, but this large enzymatic complex is poorly characterized, hampering drug development efforts. AZ-27 is a small-molecule inhibitor previously shown to target the RSV large polymerase subunit (C. L. Tiong-Yip et al., Antimicrob Agents Chemother, 58:3867–3873, 2014, http://dx.doi.org/10.1128/AAC.02540-14), but its inhibitory mechanism was unknown. Understanding this would be valuable both for characterizing the polymerase and for further development of inhibitors. Here, we show that AZ-27 inhibits an early stage in mRNA transcription, as well as genome replication, by inhibiting initiation of RNA synthesis from the promoter. However, the compound does not inhibit back priming, another RNA synthesis activity of the RSV polymerase. These findings provide insight into the different activities of the RSV polymerase and will aid further development of antiviral agents against RSV.
INTRODUCTION
Worldwide, respiratory syncytial virus (RSV) is the major cause of respiratory disease in infants under the age of one, and it is the leading cause of infant hospitalization in the United States (1, 2). RSV also is recognized as a significant cause of morbidity and mortality in the elderly (3). Significant efforts to develop a safe and effective vaccine against RSV are ongoing, but this has proven difficult, and currently none is licensed (4, 5). The only effective antiviral drug is palizivumab, a humanized monoclonal antibody against the viral fusion protein, but this drug is costly and effective only if administered prophylactically (6). Currently there are no licensed, effective antiviral treatments. However, studies in human subjects showed that there is a correlation between virus load and disease severity, suggesting that administration of effective RSV inhibitors early in the disease course would reduce morbidity (7–9), and a recent human trial of a candidate RSV drug confirmed that a small-molecule inhibitor of viral replication ameliorated RSV-induced disease (10). Thus, there is a window during infection in which it is possible to treat RSV with antiviral drugs. This highlights the need to develop a detailed understanding of viral molecular mechanisms that drive viral replication to allow the development of small-molecule inhibitors.
The RSV polymerase is increasingly recognized as an attractive target for antiviral drug development. RSV has a nonsegmented, negative-sense (NNS) RNA genome. The viral RNA-dependent RNA polymerase (RdRp) is responsible for transcribing the viral genes to produce capped and polyadenylated mRNAs and for replicating the RNA genome via a positive-sense RNA replicative intermediate. Several RSV proteins are involved in these processes, including the large RdRp subunit (L) and its cofactor, phosphoprotein (P), M2-1 protein, a transcription elongation factor necessary for production of full-length mRNAs, and nucleoprotein (N), which is required to encapsidate newly synthesized replicative RNAs (11). The enzymatic activities required for RNA synthesis and mRNA capping all are contained within the 2,165-amino-acid L protein (12). There is no three-dimensional structure available for the RSV L protein, but there is information regarding the locations of the different enzymatic domains based on amino acid sequence alignments and comparison with other viruses, mutation analysis of the RSV L protein, and testing in functional assays, and a study with a capping inhibitor (13–18). These studies have shown that the L protein contains six regions that are highly conserved among all NNS RNA viruses. Regions II and III contain the RNA polymerization domain (14, 16, 18). Region V contains a domain required for addition of a cap to the mRNA termini, and this likely occurs by an unusual polyribonucleotidyl transferase activity (15, 19, 20). Region VI contains a methyltransferase domain for methylating the mRNA cap (13). Polyadenylation of mRNAs is not thought to involve a specific enzymatic domain but to occur by RdRp stuttering on a uridine tract during mRNA synthesis (21, 22).
Although the structure of the RdRp complex remains poorly characterized, the steps involved in RSV transcription and RNA replication are relatively well defined (23, 24). The RSV genome contains a single promoter located within the first 12 nucleotides of the leader (le) region at the 3′ end of the RNA, which signals initiation of both mRNA transcription and RNA replication (25, 26). Although the 10 viral genes do not have their own independently functioning promoters, they each are flanked by conserved cis-acting elements: a gene start (gs) signal at the beginning of each gene and a gene end (ge) signal at the end (27, 28). Transcription likely begins when the RdRp binds the promoter and initiates RNA synthesis opposite position 3 of the le region (29). When the RdRp initiates at this site, it synthesizes and releases a short RNA. It is then thought to scan the template and reinitiate RNA synthesis at the gs signal of the first gene (29). The RdRp caps the RNA cotranscriptionally, and this enables the RdRp to become processive and to elongate the mRNA to the ge signal (15). The ge signal causes the RdRp to stutter and polyadenylate and then release the mRNA. Most of the RdRp can remain attached to the template and reinitiate RNA synthesis at the next gs signal; it then continues in this fashion along the genome (28). By stopping and restarting RNA synthesis at the gene junctions, the RdRp is able to sequentially synthesize the 10 individual mRNAs from the single le promoter. To perform RNA replication, the RdRp binds to the same promoter sequence, but in this case it initiates RNA synthesis opposite position 1 (29, 30). Replicative RNA becomes encapsidated with N protein as it is synthesized, and this is thought to cause the RdRp to become superprocessive, enabling it to override the ge signals and elongate the RNA to the end of the genome to generate a complementary antigenome RNA (31, 32). It currently is unclear in what ways the RdRps that engage in mRNA and antigenome synthesis differ from each other or when the distinction arises.
The 3′ end of the antigenome contains a trailer (tr) promoter region that allows the RdRp to initiate genome synthesis (33, 34). The first 12 nucleotides (nt) of the tr promoter are almost identical to those of the le promoter and signal initiations from positions +1 and +3. Similar to events at the le promoter, the RNA initiated at +1 is extended into a full-length replication product, whereas the RdRp that initiates at +3 produces a small RNA (16, 33). There is no gs sequence adjacent to the tr promoter on the antigenome, so unlike the le promoter, the tr promoter does not signal production of capped and polyadenylated mRNAs. Nonetheless, the tr promoter also can be considered to signal initiation of replication (from +1) and transcription (from +3), but in this case, the transcription pathway leads to synthesis of a short RNA only (which might play a role in inhibiting the cellular stress granule response) (35). In addition to initiating de novo RNA synthesis from the +1 and +3 initiation sites, when the RdRp engages with the tr promoter, it also can modify the tr RNA by folding the RNA into a secondary structure, and adding 1 to 3 nucleotides (in the order GUC) to the RNA 3′ end by using the folded RNA as a template (16, 36). This activity also occurs in RSV-infected cells (16), but its significance during infection is not understood.
Inhibitors not only are useful as potential leads for developing antiviral drugs but also can be used to dissect different RdRp functions, providing insight into RdRp molecular biology that might otherwise be elusive and furthering our understanding of molecular mechanisms that drive infection. Several compounds that inhibit RSV RdRp activity have been described (15, 37–41, 54). In this study, we investigated AZ-27 (Fig. 1), a benzothienoazepine compound derived from YM-53403 (41). Resistance to YM-53403 was conferred by replacement of Y1631 of the L protein, suggesting that the compound directly targets the L protein (40). AZ-27 has approximately 75-fold increased potency compared to that of YM-53403, with a 50% effective concentration (EC50) of 10 nM against RSV strain A2 under multicycle growth conditions (42). Consistent with being an RdRp inhibitor, AZ-27 was able to inhibit gene expression of an intracellular, stably maintained RSV replicon, and resistance to AZ-27 also was conferred by mutations at Y1631 in both infectious virus and the replicon system (42). Surprisingly, YM-53403 and AZ-27 are significantly less potent against subgroup B strains of RSV than subgroup A strains, despite the fact that the site where resistance mutations arise is conserved in both subgroups (42). While there is compelling evidence that AZ-27 targets the RSV RdRp, the site of resistance is not in a putative enzymatic domain but lies between conserved regions V and VI of L, the regions involved in capping mRNA; thus, the specific step(s) that it inhibits remains unclear.
FIG 1.
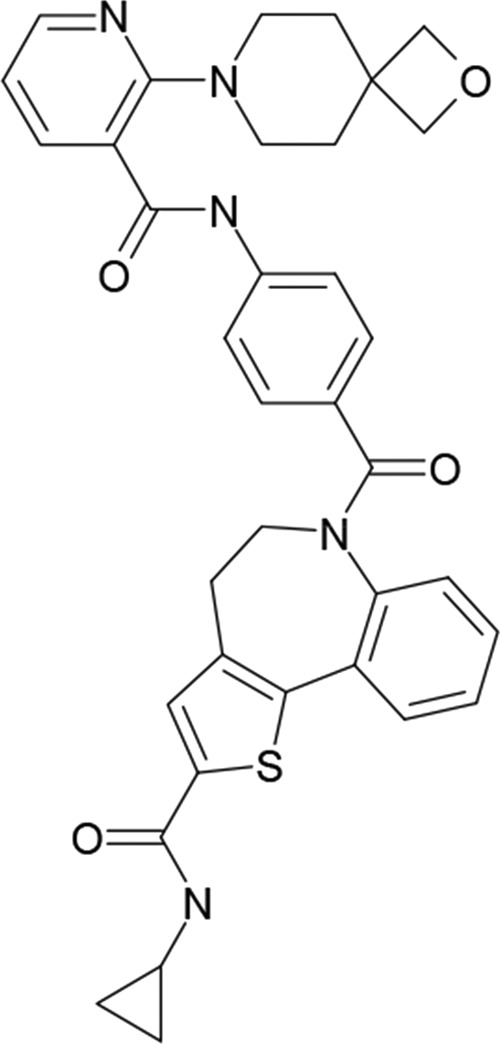
Chemical structure of AZ-27.
In this study, we examined AZ-27 activity against RSV with two goals in mind: first, to gain more information regarding the mechanism of action of this compound to aid development of small-molecule inhibitors against RSV, and second, to use AZ-27 as a chemical probe to help us understand different RdRp activities and how transcription and RNA replication become differentiated. As described above, the RSV RdRp is involved in numerous activities, so there were multiple points at which the inhibitor could have an effect. The data obtained show that AZ-27 inhibits the initiation of de novo RNA synthesis at the promoter, and that this inhibits both transcription and RNA replication. However, AZ-27 does not inhibit back-priming, suggesting that although de novo initiation and back-priming both involve RdRp interacting with the promoter, they might be performed by different structural forms of the RdRp.
MATERIALS AND METHODS
Cells and virus.
HEp-2 cells (ATCC) were grown in Opti-MEM reduced serum medium (Invitrogen) supplemented with 2% fetal bovine serum (FBS). RSV strains A2 and B9320 were used as examples of subgroups A and B, respectively. Recombinant RSV containing AZ-27 resistance mutations in the L gene (Y1631H and Y1631C) were generated by reverse genetics using a previously described RSV A2 backbone (43), which contains a position 4G-to-C substitution relative to the 3′ terminus of the le. This le substitution has been shown to have no effect on the efficacy of AZ-27 (37). The resistant virus stocks that were used in the experiments that were described here were sequenced to confirm that the Y1631 mutations were present and that there were no second-site mutations.
Minigenome assays.
Minigenome-expressing plasmids were derived from plasmid MP28, which has been described previously (26). Each minigenome cassette was flanked with a T7 promoter at the 5′ end and a hepatitis delta virus ribozyme to generate the RNA template 3′ end. The le-containing minigenome, used to measure transcription activity, contained a chloramphenicol acetyltransferase (CAT) gene divided into two segments of 580 and 190 nt by the N-P gene junction. The 3′ end of the minigenome contained the 44-nt le, NS1 gs signal, and 37-nt NS1 nontranslated region, and the 5′ end contained the last 12 nt of the L nontranslated region, L ge signal, and 155-nt 5′ trailer region. The trailer region had a C-to-G substitution at position 2 relative to the 5′ end to inhibit minigenome synthesis from the replicative intermediate antigenome. The tr-containing minigenome, used to measure replication activity, had a similar minigenome structure, except that the 3′ end of the minigenome contained nucleotides 1 to 36 of the tr promoter region directly abutting the nonspecific CAT sequence. In addition, the terminal 22 nt from the 5′ trailer region was deleted to limit RNA replication to a single step, and a hammerhead ribozyme was inserted between the remaining trailer region and the T7 promoter. RNA synthesis was reconstituted in HEp-2 cells as described previously (44). Cells were transfected with 0.2 μg of minigenome DNA, 0.4 μg of pTM1 N, 0.2 μg of pTM1 P, 0.1 μg of pTM1 M2-1, and 0.1 μg of pTM1 L plasmids using Lipofectin (Invitrogen) according to the manufacturer's instructions. Cells were infected simultaneously with MVA-T7 to express T7 RNA polymerase and drive plasmid expression (45). Immediately following addition of the transfection mixture to the cells, AZ-27 serially diluted in dimethyl sulfoxide (DMSO) (such that the concentration of DMSO was equivalent in each well) was added at the concentration indicated; an equivalent volume of DMSO without compound was added as a control. After 18 h, the transfection mixture was replaced with Opti-MEM containing 2% FBS and either fresh DMSO or AZ-27 at the appropriate concentration. Approximately 40 h posttransfection, RNA samples were harvested.
RNA analysis.
Total intracellular RNA was isolated using an RNeasy kit (Qiagen) according to the manufacturer's instructions. Northern blot transfer, probe preparation, and probe hybridization were performed as described previously (26). For minigenome analyses, negative- or positive-sense 32P-labeled, CAT-specific riboprobes were synthesized with T7 RNA polymerase as described previously (32). RSV N mRNA was detected with a [32P]ATP-incorporated DNA probe, specific to positive-sense N RNA, prepared as described previously (46). Primer extension analysis was performed using a 32P-end-labeled primer that hybridized at positions nt 12 to 31 of the CAT 1 gene to detect CAT 1 mRNA. Primer extension reactions were performed as described previously (33). The position of the gs initiation site was determined by comigration of an end-labeled DNA oligonucleotide, corresponding in sequence to the expected primer extension product. Products were visualized by autoradiography and quantified by phosphorimage analysis using a Bio-Rad personal molecular imager system and Quantity One quantification software.
Immunoprecipitation of L-P complexes and effect of AZ-27 on complex integrity.
HEp-2 cells were transfected with 0.5 μg of pTM1 P and/or 0.25 μg pTM1 V5-tagged L plasmids using Lipofectin (Invitrogen) according to the manufacturer's instructions. The cells were infected simultaneously with MVA-T7 to express T7 RNA polymerase and drive plasmid expression. After ∼18 h, the transfection mixture was replaced with Opti-MEM containing 2% FBS and cells were harvested at 48 h posttransfection. Cell pellets were washed with phosphate-buffered saline and lysed in 20 mM Tris, pH 7.4, 150 mM NaCl, 0.5% NP-40, 1 mM EDTA, incubated on ice for 15 min, and then centrifuged for 10 min at 16,000 × g at 4°C to clear the lysate. The supernatant was collected and combined with 1 μl mouse anti-V5 antibody (Invitrogen) and 50 μl of μMACs protein G beads (Miltenyi Biotec) and incubated at 4°C with rotation for 2 h to precipitate L-P complexes. Magnetic μ columns were placed in the magnetic field of a μMACs separator and washed once with 200 μl of lysis buffer. Samples were applied to the column and washed four times with 200 μl of lysis buffer to purify L/P complexes. The flowthrough was discarded. To test the impact of AZ-27 on V5-L interactions with P, columns were washed three to five times (depending on the experiment) with 30 μl of in vitro transcription buffer (50 mM Tris, pH 7.4, 8 mM MgCl2, 10% glycerol, no dithiothreitol [DTT]) containing either AZ-27 (dissolved in DMSO) at a final concentration of 10 μM or an equivalent volume of DMSO. The flowthrough from each wash was collected in separate tubes and combined with 30 μl of SDS-PAGE loading buffer. To harvest L-P protein complexes remaining on the column, 20 μl of 1× SDS-PAGE loading buffer, preheated to 95°C, was added to each column and incubated for 5 min. A further 50 μl of preheated SDS-PAGE loading buffer was applied to each column, and the flowthrough containing eluted protein was collected. Proteins were separated by SDS-PAGE and transferred to nitrocellulose. Proteins were detected using a goat anti-RSV antibody (ab20745; 1:500; Abcam) to detect P and mouse anti-V5 antibody (1:1,000; Invitrogen) to detect V5-L.
In vitro transcription run-on assay.
RSV transcription in crude cell extracts was analyzed as described previously (47). Briefly, HEp-2 cells were infected or mock infected with RSV at a multiplicity of infection (MOI) of 5 for 17 to 22 h (as specified in the figure legend), following which the medium was replaced with medium containing 2 μg/ml actinomycin D. After 1 h, cells were gently lysed and scraped into transcription buffer (50 mM Tris-acetate [pH 8], 8 mM Mg-acetate, 300 or 100 mM K-acetate [as specified in the figure legends], 2 mM DTT, 1 mM spermidine, 10 mM creatine phosphate, 1 μg/ml aprotinin, 16 U creatine phosphokinase, 1 mM ATP, 1 mM CTP, 1 mM GTP, 50 μM UTP, 2 μg/ml actinomycin D), and cell debris was removed by centrifugation. Aliquots of soluble extract were combined with additional transcription buffer containing 10 μCi [α-32P]UTP, RNase inhibitor, and either DMSO or compound AZ-27 serially diluted in DMSO (such that the concentration of DMSO was equivalent in each reaction) in a total reaction volume of 50 μl. Reaction mixtures were incubated for 1 to 4 h at 30°C (as specified in the figure legends), following which total RNA was extracted using a Qiagen RNeasy kit according to the manufacturer's instructions. Purified total RNA samples were further processed by RNase H digestion to remove poly(A) tails from mRNA transcripts, allowing resolution of individual mRNA bands. RNase H-treated RNA samples were subjected to denaturing gel electrophoresis on a 4% acrylamide gel containing 7 M urea. RNA products were analyzed by autoradiography and quantified by phosphorimage analysis.
In vitro assay of RdRp activities at the tr promoter.
The RSV L/P complex was purified from insect cells as described previously (16). RNA synthesis reactions were performed using two different conditions. For the first condition, reaction mixtures consisted of 2 μM RNA oligonucleotide representing nt 1 to 25 of tr promoter sequence (Dharmacon); 50 mM Tris-HCl at pH 7.4; 8 mM MgCl2; 5 mM DTT; 10% glycerol; ATP, CTP, and GTP each at 1 mM and UTP at 50 μM with 10 μCi of [α-32P]UTP. DMSO or various concentrations of AZ-27, serially diluted in DMSO (such that the concentration of DMSO was equivalent in each reaction mix) as indicated, were included in the reaction mixtures. Reaction mixtures were incubated at 30°C for 10 min, and then RSV L-P (containing ∼200 ng of L protein) was added to the transcription mix last, such that the final reaction volume was 50 μl. Reaction mixtures were incubated at 30°C for 3 h and then heated to 90°C for 3 min to inactivate the RdRp and cooled briefly on ice. Reaction mixtures were diluted in an equal volume of stop buffer (deionized formamide containing 20 mM EDTA, bromophenol blue, and xylene cyanol) and analyzed by electrophoresis on a 20% polyacrylamide gel containing 7 M urea in Tris-borate-EDTA buffer. Under the other condition, reaction mixes contained 2 μM RNA oligonucleotide representing nt 1 to 16 of tr promoter sequence (Dharmacon); 50 mM Tris, pH 7.4; 8 mM MgCl2; 5 mM DTT; 10% glycerol; 500 μM (each) GTP and UTP and 10 μM ATP with 2 μl [α-32P]ATP. DMSO or AZ-27 was included in each reaction mixture, as described above. Reaction mixtures were preincubated prior to addition of L-P protein as described above. Reaction mixtures then were incubated at 30°C for 2 h, heated to 90°C for 3 min to inactivate the RdRp, and cooled briefly on ice, and then an equal volume of stop buffer was added. Reactions were analyzed by electrophoresis on a 25% polyacrylamide gel containing 6 M urea in Tris-taurine-EDTA buffer. The nucleotide lengths of the RNA products were determined by comparison to a molecular weight ladder generated by alkali hydrolysis of a 32P end-labeled RNA oligonucleotide representing the anticipated RNA products from the +1 and/or +3 sites. RNA products were analyzed by autoradiography and quantified by phosphorimager analysis.
Infection and treatment of cells for analysis of RNA and protein levels under single-cycle growth conditions.
Cells in 6-well plates were infected with RSV at an MOI of 4 PFU/cell with serum-free Opti-MEM containing either DMSO or various concentrations of AZ-27 (serially diluted in DMSO, such that the concentration of DMSO was equivalent in each well) as indicated. The virus was allowed to adsorb for ∼1 h, after which the inoculum in each well was removed and replaced with 1 ml of Opti-MEM containing 2% FBS and fresh DMSO or various concentrations of AZ-27 as appropriate. At 18 h postinfection, cells were scraped into the medium and collected by centrifugation. The cell pellets were processed for RNA analysis by Northern blotting as described above.
RESULTS
AZ-27 inhibits RSV mRNA transcription and genome replication.
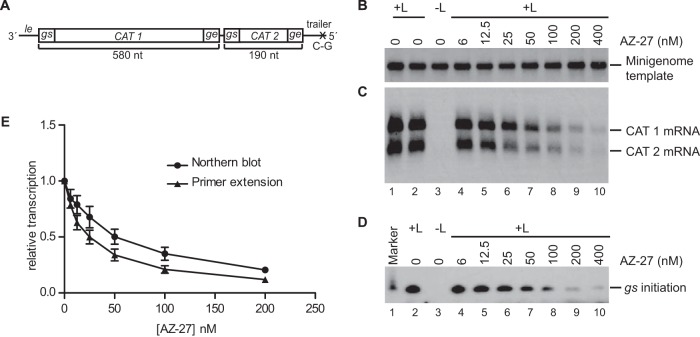
The RSV RdRp is a multifunctional complex, and there were many potential steps at which AZ-27 could exert its inhibitory effect, including promoter recognition and transcription or replication initiation, RNA synthesis elongation, mRNA cap addition and/or methylation, or encapsidation of replicative RNA. It also was possible that AZ-27 interfered with expression or stability of the RdRp complex. AZ-27 previously had been shown to inhibit RSV gene expression in the RSV replicon system (42). This system consists of cells stably transfected with a subgenomic RSV replicon that contains the RSV genes required for transcription and genome replication, as well as an antibiotic resistance marker. Cells containing an actively transcribing and replicating replicon are selected and maintained by virtue of the antibiotic resistance. Thus, while this assay provided evidence to support the hypothesis that AZ-27 is an RdRp inhibitor, it could not distinguish between any of the possible inhibition mechanisms described above. Therefore, our first step was to determine if AZ-27 could specifically inhibit either transcription or replication, as this would provide insight as to which RdRp function AZ-27 was affecting. For example, if it were a capping inhibitor, it would be expected to inhibit transcription but not replication. To analyze transcription and replication as independent events, we performed experiments in a transient-transfection assay that employs a nonreplicating minigenome. In this system, mRNA transcription and RNA replication are uncoupled from each other, as described previously (26, 48). To analyze mRNA transcription, cells were transiently transfected with a plasmid expressing a minigenome containing a 3′ le promoter and gs signal (Fig. 2A) and plasmids expressing the RSV N, P, L, and M2-1 proteins. AZ-27 was added to the cells at a range of concentrations immediately following transfection and maintained in the medium until the cells were harvested. RNA was isolated and analyzed by Northern blotting using probes specific to either the input minigenome template or the positive-sense RNA products it would generate (Fig. 2B and C). Analysis of the input template indicated that its levels were not affected by increasing concentrations of AZ-27 (Fig. 2B), confirming that AZ-27 did not impact either vaccinia virus replication or T7 polymerase activity, both of which are necessary to drive the minigenome system. In contrast, AZ-27 inhibited expression of minigenome-specific mRNAs in a concentration-dependent manner (Fig. 2C and E). Similar results were obtained by primer extension analysis (Fig. 2D and E). These findings demonstrate that AZ-27 inhibits mRNA transcription independently of any effect it has on RNA replication.
FIG 2.
Effect of AZ-27 on mRNA transcription by the RSV RdRp in the minigenome system. (A) Schematic diagram of the RSV minigenome used to measure transcription. The minigenome contains two reporter genes, CAT 1 and CAT 2 (each gene contains a different portion of the CAT open reading frame sequence), each flanked with RSV gs and ge sequences, and the 3′ and 5′ ends of the minigenome contain the 44-nt le and 155-nt tr sequences, respectively. The trailer region contained a C-to-G substitution at position 2 relative to the 5′ end to inactivate the tr promoter that typically would be present at the 3′ end of the replication product. (B and C) Effect of various concentrations of AZ-27 on minigenome template and RSV transcription products in transfected cells, as determined by Northern blotting. Panel B shows input minigenome template, and panel C shows CAT 1 and CAT 2 mRNAs. (D) Primer extension analysis of the same RNA samples as those used for panels B and C, performed using a primer that hybridized to nucleotides 12 to 31 relative to the 5′ end of the CAT 1 mRNA, to detect initiation from the first gs signal. (E) Quantification of the CAT 1 mRNA product, as determined by Northern blotting and primer extension analysis, with the numbers expressed relative to the mean levels of CAT 1 mRNA generated from the minigenome in the absence of compound. Each point represents the means from three independent experiments, with standard errors shown.
In the experiment described above, the levels of mini-antigenome produced from the minigenome template were too low to be readily detected. Therefore, to examine the effect of AZ-27 on RNA replication, a modified version of the minigenome template was used in which the le promoter and first gs signal were deleted and replaced with the tr promoter (Fig. 3A). The tr promoter is a stronger replication promoter than the le promoter, and because transcription-specific signals near the promoter were removed, no mRNAs were synthesized and the replication product could be more readily detected and quantified. Minigenome RNA replication was reconstituted in the presence of increasing concentrations of AZ-27, as described above. Northern blot analysis of the replication product showed that its levels of accumulation were reduced with increasing concentrations of AZ-27 (Fig. 3C), with a concentration inhibition curve similar to that for transcription (compare Fig. 2E and 3D). Thus, AZ-27 inhibits an RdRp activity that is required for both transcription and RNA replication.
FIG 3.
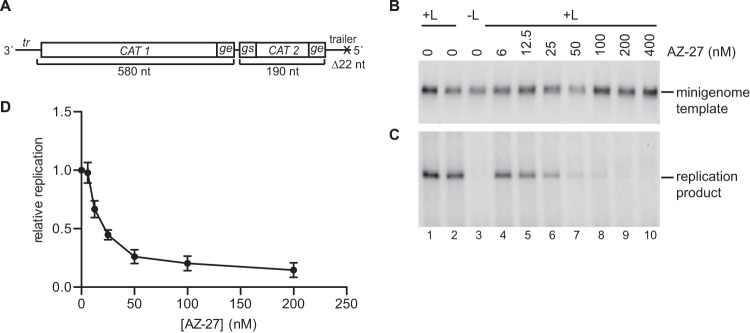
Effect of AZ-27 on RNA replication by the RSV RdRp in the minigenome system. (A) Schematic diagram of the minigenome used to measure RNA replication. In this minigenome, all transcription signals from the 3′ end, including the le and first gs signal, were removed and replaced with nucleotides 1 to 36 of the tr promoter. The trailer region at the 5′ end of the minigenome contained a deletion of the 5′ terminal 22 nucleotides to avoid terminal complementarity and to inactivate the tr promoter that typically would be present at the 3′ end of the replication product. (B and C) Effect of various concentrations of AZ-27 on minigenome template and RSV replication product in transfected cells, as determined by Northern blotting. Panel B shows the input minigenome template, and panel C shows the replication product generated by the RSV RdRp. (D) Quantification of the replication product, as determined by Northern blotting, with the numbers expressed relative to the mean levels of replicative RNA generated from the minigenome in the absence of compound. Each point represents the means from three independent experiments, with standard errors shown.
AZ-27 does not disrupt L-P interaction.
One possible explanation for why AZ-27 inhibits transcription and replication is that it disrupts the integrity of RdRp complexes. As a step toward determining if this was the case, we determined if AZ-27 disrupts the interaction between the two RdRp subunits, L and P, which comprise the minimal RdRp. V5-tagged L protein and untagged P protein were expressed in HEp-2 cells in a transient transfection. Cell lysates were passed through a column charged with anti-V5 antibody to isolate V5-L-P complexes, and the columns were washed multiple times with lysis buffer. The columns then were washed with a buffer that supports RSV transcription in vitro (see below), containing AZ-27 at a final concentration of 10 μM (a significantly higher concentration than that required to detect an effect on RdRp activity in cells) or an equivalent volume of DMSO, and the fractions were collected. Following the wash steps, the L (and L-P) complexes remaining on the column were eluted with SDS-PAGE lysis buffer. The column eluate containing the proteins that had remained attached to the beads following the washes and the AZ-27 and DMSO washes were analyzed by Western blotting using antibodies to detect P or V5-L (Fig. 4). This analysis showed that essentially all of the P that was present in the L-P complexes remained attached to L on the column throughout the AZ-27 washes and was eluted only by SDS-PAGE lysis buffer (Fig. 4, upper, compare lane 9 with lanes 10 to 14), similar to what was observed with the DMSO-only washes (Fig. 4, upper, lanes 3 to 8). This result shows that AZ-27 does not disrupt L-P interaction.
FIG 4.
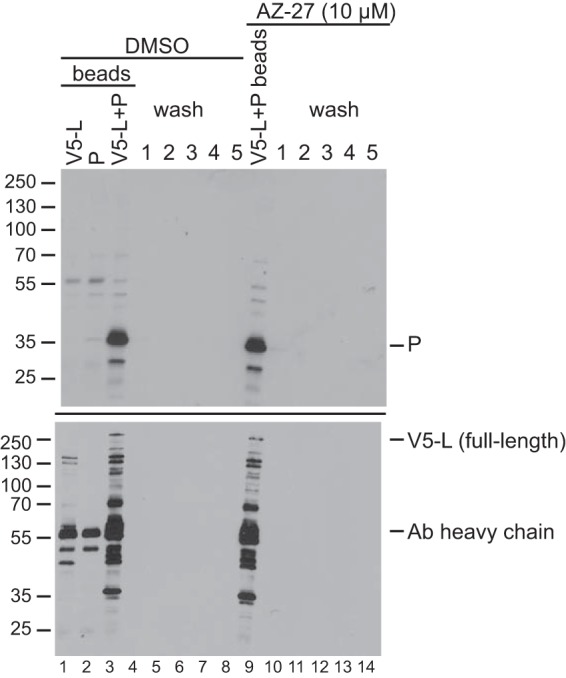
Effect of AZ-27 on L-P interaction. RSV L and P proteins were expressed in HEp-2 cells, and L-P complexes were purified by virtue of a V5 tag on the L protein. V5-L-P complexes were subjected to five washes with transcription buffer containing either DMSO (lanes 3 to 8) or 10 μM AZ-27 (lanes 9 to 14) such that the concentration of DMSO was equivalent in both conditions. L-P complexes remaining attached to the columns were eluted with heated SDS-PAGE lysis buffer (lanes 3 and 9); the wash fractions are shown in lanes 4 to 8 and 10 to 14. Aliquots of each sample were analyzed by Western blotting using an RSV-specific polyclonal antibody (Ab) to detect P (top; note that this antibody does not detect L) or a V5-specific antibody to detect V5-L (bottom). Lanes 1 and 2 show control samples in which only V5-L or untagged P were expressed in cells (lanes 1 and 2, respectively). The band at 55 kDa is the heavy chain of the V5 antibody. The bands smaller than that of full-length V5-L detected by the V5 antibody are truncated L proteins (note that the L gene used for this experiment was not codon optimized). This result is representative of three independent experiments.
AZ-27 has a more potent inhibitory effect on transcription of 3′- than 5′-proximal RSV genes.
The minigenome experiments described above indicated that AZ-27 can inhibit an early step in RSV transcription and genome replication. However, because the compound was added immediately following transfection and because the accumulation of RSV minigenome-encoded RNAs was measured, these experiments did not provide information to indicate if RdRp already associated with the nucleocapsid template was susceptible to inhibition with AZ-27. To address this, we examined the effect of AZ-27 in an in vitro transcription run-on assay, which utilizes crude cell extracts from RSV-infected cells. In this experiment, transcription/replication complexes are already preformed and active on the template prior to addition of the compound. Thus, this assay monitors the ability of RdRps already associated with the template to continue RNA synthesis, as well as for free and released RdRp to initiate and reinitiate transcription. Cells were mock infected or infected with RSV. At approximately 18 h postinfection, when nucleocapsids have accumulated to a high level in the cytoplasm, the cells were harvested and clarified lysates were incubated in a buffer containing actinomycin D, to inhibit cellular (but not RSV) transcription, and nucleoside triphosphates (NTP), including [α-32P]UTP. AZ-27 was included at various concentrations in the transcription mixes. Following incubation and treatment to remove their poly(A) tails (to allow resolution of the different gene transcripts), RSV mRNA transcripts were separated by denaturing gel electrophoresis and detected by autoradiography (Fig. 5A). No labeled transcripts were detected in reaction mixtures containing lysate from mock-infected cells, whereas a series of RNA transcripts were detected in reaction mixtures containing lysate from RSV-infected cells, confirming that the transcripts that were detected were products of the RSV RdRp (Fig. 5A, compare lanes 1 and 2). The identities of the RSV transcripts were assigned based on their known nucleotide length and comparison to a 900-nt size marker (data not shown). The strong doublet at the top of the gel presumably includes L mRNA and genome/antigenome RNAs, as these RNAs would be expected to have a strong signal due to their length (which allows a greater level of incorporation of the radiolabeled nucleotide). However, this doublet was not clearly observed in all gels, possibly due to this large RNA becoming trapped in the wells in some cases. Inclusion of AZ-27 in the transcription reaction mixes caused a concentration-dependent decrease in the accumulation of all RSV transcripts, consistent with inhibition of RdRp activity (Fig. 5A). However, not all transcripts were affected similarly. For example, examination of the bands near the top of the gel showed that with increasing AZ-27 concentration, the levels of N transcript were reduced to a greater extent than those of the M and M2 transcripts. Likewise, near the bottom of the gel, the levels of NS1 and NS2 transcripts were affected more than that of the SH transcript. Thus, AZ-27 affected the synthesis of different RSV gene transcripts to different extents. This differential effect clearly was not dependent on transcript length. Instead, it appeared that AZ-27 had a more potent effect on the accumulation of transcripts synthesized from the 3′-proximal than 5′-proximal genes (the RSV gene order from 3′ to 5′ is NS1, NS2, N, P, M, SH, G, F, M2, and L) (Fig. 5B). This finding suggests that AZ-27 inhibited RdRp initiating at the 3′ end of the template to a greater extent than RdRp that already was associated with the nucleocapsid.
FIG 5.
Effect of AZ-27 on RSV subgroup A RdRp activity in a run-on transcription assay. (A) RNA synthesis in the presence of AZ-27. Mock- or RSV A2-infected cells were harvested at 18 h postinfection. Reactions were performed using cell lysates in a buffer containing 300 mM potassium acetate, with radiolabeled UTP included in the reaction mixtures for incorporation into newly synthesized RNA. AZ-27 was included in the RNA synthesis reaction mixtures at the indicated concentrations, and the mixtures were incubated for 1 h. Labeled RNAs were analyzed by urea acrylamide gel electrophoresis. The identities of the RSV transcripts were deduced by comparison to a 900-nt RNA size marker and the gene lengths (which are indicated in parentheses). (B) Quantification of the NS1, NS2, N, and F mRNA transcripts with various concentrations of AZ-27. The data are plotted relative to the level of transcripts produced in DMSO-only control reactions. The results show the means and standard errors for three independent experiments.
AZ-27 has a differential effect on RSV RdRp activities at the promoter.
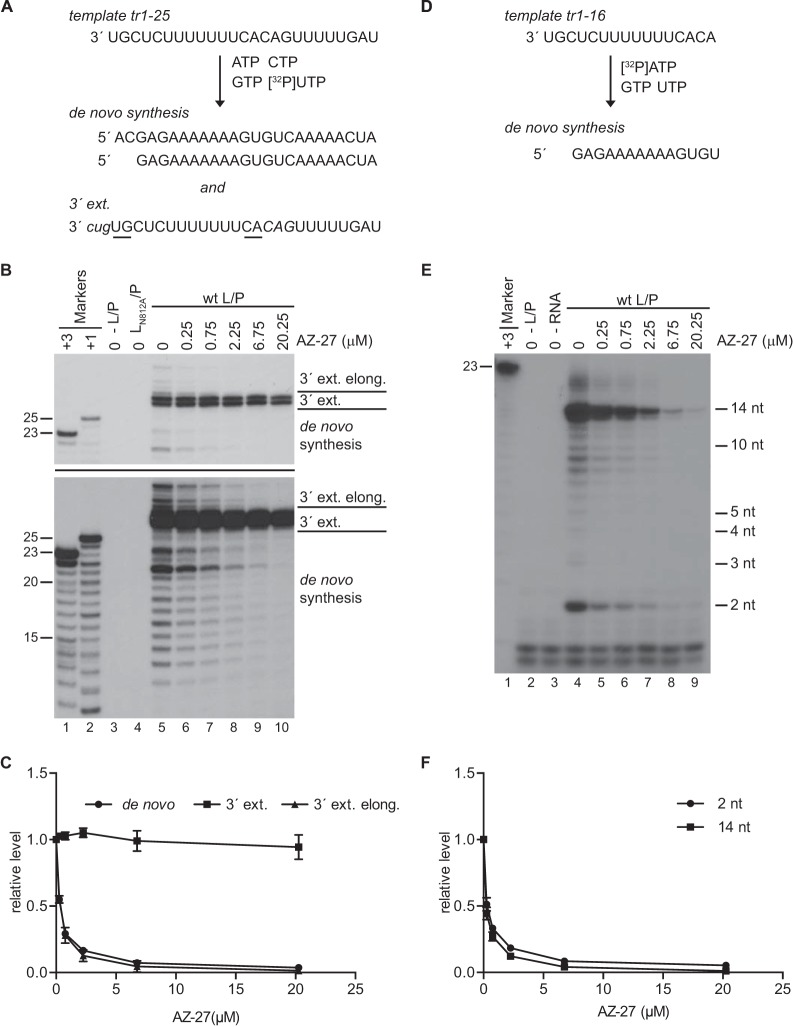
The results described above suggest that AZ-27 inhibits an early stage in RSV mRNA transcription. They also suggest that AZ-27 inhibits a polymerase activity that is common to both mRNA transcription and RNA replication. Together, these findings suggest that AZ-27 inhibits one or more of the steps that occur as the RdRp engages with the promoter and begins RNA synthesis. To determine if this is the case, we examined the effect of AZ-27 in an in vitro assay, involving NTPs, purified recombinant RdRp (L-P complexes), and an RNA oligonucleotide template consisting of the RSV tr promoter.
As described in Introduction, the RSV promoters contain two initiation sites at positions +1 and +3, which are likely to be the replication and transcription initiation sites, respectively. We have shown previously that the RdRp initiates at both of these sites in the in vitro assay to generate RNA (de novo RNA synthesis) (16, 36). The RdRp also can interact with the tr promoter sequence to perform a back-priming and 3′ extension reaction in which it folds the tr RNA into a secondary structure and extends the 3′ end. This activity yields a characteristic pattern of products: the dominant modification is the addition of one to three nucleotides, added in the order G then U then C (3′ extension). Occasionally, the RdRp can extend the 3′ extension products to the end of the template (3′ extension elongation). The de novo RNA synthesis and 3′ extension activities were assayed in parallel by performing reactions with a template consisting of 25 nt of tr sequence and a reaction mix containing [α32P]UTP (Fig. 6A). De novo RNA synthesis products ranged in size from 12 to 25 nt, reflecting the facts that the RdRp can initiate at position +3 and the first UTP incorporation site is at nucleotide 14 (Fig. 6B, lower, lane 5). With increasing concentrations of AZ-27, the accumulation of all RNA transcripts ≤25 nt in length was inhibited (Fig. 6B, lower, lanes 5 to 10), demonstrating that AZ-27 inhibited de novo RNA synthesis from both the +1 and +3 sites. 3′ Extension activity was monitored by examining products of 27 and 28 nt in length (Fig. 6B, upper and lower, lane 5). In contrast to its effect on de novo RNA synthesis, AZ-27 had little or no detectable effect on addition of 2 or 3 nt by back-priming and 3′ extension (Fig. 6B, upper, lanes 5 to 10). However, the compound did inhibit further extension of the back-primed RNA, inhibiting this process to the same extent as de novo RNA synthesis (Fig. 6B and C). These data show that AZ-27 did not inhibit the addition of 1 to 3 nucleotides by back-priming but did inhibit de novo RNA synthesis and further elongation of the back-primed RNA.
FIG 6.
Effect of AZ-27 on RSV RdRp activities at the promoter. (A) Depiction of the template representing nt 1 to 25 of tr promoter sequence and the products generated by the RSV RdRp that would be detected in the experimental conditions used for panels B and C. RNA is generated by de novo synthesis from positions 1U and 3C, and the template is modified by back-priming and 3′ extension (3′ ext.). Back-priming and 3′ extension occurs as a result of nucleotides 13C and 14A of the template base-pairing with nucleotides 2G and 1U, respectively (underlined). Templated extension opposite an internal sequence (in italics) typically results in the addition of 1 to 3 nucleotides to the 3′ end (the nucleotides that are added are in lowercase and italicized). The RdRp can also elongate (elong.) this RNA to the end of the folded template (not shown). (B) Effect of AZ-27 on de novo RNA synthesis and 3′ extension. Purified wt (lanes 5 to 10) or mutant (lane 4) RdRp was added to reaction mixtures containing either DMSO or various concentrations of AZ-27 (serially diluted in DMSO) as indicated. The labeled products were separated by denaturing gel electrophoresis and visualized by autoradiography. Bands of ≤25 nt in length represent products of de novo synthesis, bands of 27 and 28 nt in length represent the major products of 3′ extension, and bands of 29 to 36 nt in length represent products of 3′ extension and then further elongation. The upper panel is a shorter exposure of the lower panel to allow clearer visualization of the 27- and 28-nt products of the 3′ extension. Lanes 1 and 2 show the molecular size ladder, representing nt 3 to 25 and 1 to 25 of the anticipated products, respectively, and lane 3 is a negative control in which RdRp was not included in the reaction. (C) Quantification of the 21-nt band, representing de novo RNA synthesis, the 27- and 28-nt bands, representing 3′ extension, and the uppermost band on the gel, representing elongated 3′ extension products, from replicates of the data shown in panel B, with the numbers expressed relative to the mean levels of RNA generated in the absence of compound. (D) Depiction of the tr nt 1 to 16 template and the product generated by the RSV RdRp that would be detected under the experimental conditions used for panels E and F. In this case, only RNA generated by de novo synthesis from position +3C would be detected. (E) Effect of AZ-27 on initiation of de novo RNA synthesis by the RSV RdRp. Lane 1 shows the molecular size ladder, representing RNA products initiated from position +3. It should be noted that the small products synthesized by the RSV RdRp do not migrate identically to their counterparts in the ladder because the RNA transcripts in the ladder contained a 5′ monophosphate rather than triphosphate. Lanes 2 and 3 are negative controls in which RdRp or RNA template, respectively, were not included in the reaction. The bands of >14 nt in length are generated as a consequence of RdRp stuttering on the U tract in the template (36). (F) Quantification of the 2- and 14-nt bands from replicates of the data shown in panel E, with the numbers expressed relative to the mean levels of RNA generated in the absence of compound. In panels C and F, each point represents the means from three independent experiments, with standard errors shown.
Studies with T7 RNA polymerase have shown that shortly after beginning RNA synthesis, the polymerase undergoes a conformational change to allow it to break promoter contacts and extrude the nascent RNA (49), and it appears that this sequence of events also applies to RNA-dependent RNA polymerases (50). Back-priming and 3′ extension of the RSV tr promoter RNA might not involve the same sequence of events as initiation of RNA synthesis at a promoter; nonetheless, one plausible explanation for why the addition of 1 to 3 nucleotides by back-priming was refractory to AZ-27 activity, whereas further elongation was inhibited, is that AZ-27 inhibits the ability of the RdRp to undergo conformational changes necessary to transition into an elongation mode. If so, this also could be the case during de novo RNA synthesis. To determine if the initiation and elongation steps of de novo RNA synthesis were differentially affected by AZ-27, an experiment was performed under conditions that allowed detection of very short RNA products (Fig. 6D). Previous studies using the in vitro RSV RNA synthesis assay have shown that +3C is the dominant initiation site in the tr promoter. Therefore, reactions were performed with [α-32P]ATP as the radiolabeled NTP. As ATP is the fourth nucleotide in the tr template, this allowed detection of 2-nt products generated following initiation at position +3. To simplify interpretation of the data, CTP was omitted from the reactions to prevent initiation from the +1 site, and the assay was performed using a template consisting of tr nucleotides 1 to 16. Analysis of the products generated under these conditions in the absence of AZ-27 showed that two dominant products were generated: a 14-nt transcript, which is consistent with initiation of the RdRp at position +3 and extension to the end of the template, and a 2-nt product (Fig. 6E, lane 4). The abundance of the 2-nt product, compared to the level for longer RNAs, suggests that extension beyond 2 nt is a rate-limiting step in de novo RNA synthesis from the RSV promoter. Increasing concentrations of AZ-27 inhibited production of the 2-nt product to the same extent as inhibition of the 14-nt product (Fig. 6E, lanes 4 to 9, and F). These results show that AZ-27 inhibited de novo RNA synthesis by the RdRp at a step prior to or including formation of the first phosphodiester bond. This is in contrast to the finding that AZ-27 did not inhibit the ability of the RdRp to add 1 to 3 nucleotides to the tr RNA by a back-priming mechanism. Thus, AZ-27 had a differential effect on RSV RdRp activities at the promoter.
AZ-27 is less potent against the RdRp of RSV B than against that of RSV A.
Previous studies have shown that while AZ-27 is effective against subgroup B strains of RSV, it is less potent than against RSV subgroup A strains (41). This difference was observed by assaying RSV replication under multicycle growth conditions using an enzyme-linked immunosorbent assay (ELISA)-based assay and virus release assay. One possible explanation for the difference between subgroups A and B is that their RdRps could be sufficiently different in structure that AZ-27 affects their activities differently. However, RSV A and B exhibit slightly different patterns of cytopathic effect, and RSV B replicates more slowly in cell culture. Therefore, an alternative explanation is that AZ-27 inhibits RSV A and B RdRps to a similar extent, but there is a bottleneck in some aspect of RSV B replication such that viral RNA accumulation and virus release are not correlated in RSV B infection. If this were the case, the concentration of AZ-27 required to inhibit RSV B replication might be greater. To distinguish between these possibilities, we compared the effects of AZ-27 on RSV strains A2 and B9320 as representatives of subgroups A and B, respectively, under single-cycle growth conditions. HEp-2 cells were infected with RSV subgroup A or B at a target MOI of 4 PFU/cell and incubated in the presence of various concentrations of AZ-27. Cells were harvested at 18 h postinfection, which is before significant virus release occurs, and analyzed for RSV RNA expression by Northern blotting using an RSV N-specific probe. This analysis showed that N mRNA accumulation was inhibited by AZ-27 in both RSV A and B infections, but that an approximately 100-fold greater concentration of AZ-27 was required to inhibit mRNA production from RSV B than RSV A (Fig. 7A and B). Analysis of intracellular RSV protein expression by Western blotting using an anti-RSV polyclonal antibody yielded a similar result (data not shown). These results, combined with the previous findings regarding AZ-27 EC50s during multicycle growth, show that there is a correlation between RSV RNA accumulation and virus spread and confirm that AZ-27 has a differential effect on the RdRps of RSV A and B.
FIG 7.
Comparison of the effect of AZ-27 on RSV subgroup A and B RNA synthesis. (A) Effect of AZ-27 on RSV mRNA production under single-cycle growth conditions. HEp-2 cells were infected with RSV A2 or B9320 and incubated in the presence of various concentrations of AZ-27, as indicated. N mRNA levels were measured by Northern blotting with a specific probe. The lanes labeled mock are negative controls of RNA from mock-infected cells. (B) Quantification of replicates of the data shown in panel A with the AZ-27 concentration plotted on a log10 scale. The graphs show the means and standard errors from sets of three independent experiments. (C) Effect of AZ-27 on RSV B RdRp in a run-on transcription assay. RNA synthesis assays were performed using lysate from RSV B-infected cells, as described for Fig. 5, except that cells were harvested at 22 h postinfection and reaction mixtures were incubated for 4 h. The identities of the bands were deduced as described for Fig. 5.
To confirm that the difference in the susceptibility of RSV A and B was due to differences in the RNA synthesis activities of the RdRp, the effect of AZ-27 on RSV subgroup B was tested in the in vitro run-on transcription assay. Similar to what was observed in the analysis of RNA accumulation in cell culture, AZ-27 was required at a 100-fold higher concentration to cause a 50% reduction in transcription by RSV subgroup B RdRp than that of RSV subgroup A (compare Fig. 5A with 7C). This result confirms that the RdRp complexes of RSV subgroups A and B are differentially sensitive to inhibition by AZ-27. Unlike the situation observed with RSV A, there was no differential inhibition of 3′- versus 5′-proximal gene expression. It is possible that this apparent difference is because of the longer incubation time required to detect products in transcription run-on assays using RSV B, which would bias the assay toward measuring initiation from the promoter, rather than a combination of initiation and continued transcription by preassociated RdRp.
Mutations in L confer resistance to AZ-27 in the transcription run-on assay.
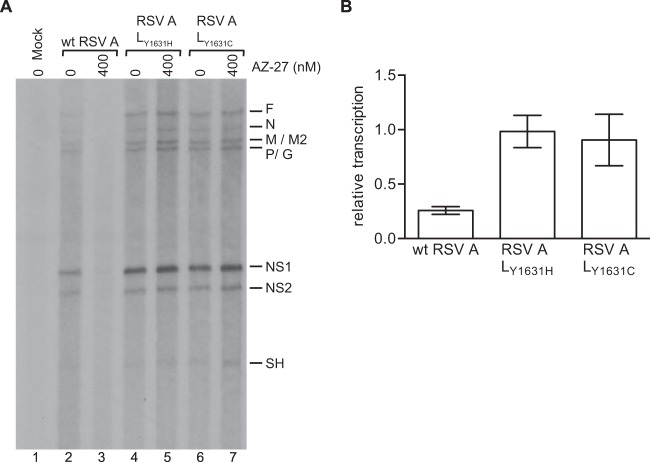
Lastly, we examined if resistance mutations in L could restore transcription by the subgroup A RdRp in the presence of AZ-27. Recombinant mutant viruses that contained substitutions of either Y1631H or Y1631C were tested. These two substitutions previously have been shown to confer resistance to AZ-27 in the context of virus infection and the replicon system, respectively (37, 42). These mutant viruses were compared to wild-type (wt) RSV subgroup A in the in vitro transcription run-on assay in the presence of DMSO carrier or 400 nM AZ-27. In this experiment, the assay incubation time was increased from 1 to 3 h to bias the assay toward detecting transcription initiation by RdRps in the cell extract, as opposed to transcription performed by preloaded RdRp, and thereby to amplify differences between the viruses in the presence of AZ-27. Under these conditions, AZ-27 almost completely inhibited transcription in extracts from cells infected with wt RSV A2 (Fig. 8A, compare lanes 2 and 3, and B). In contrast, AZ-27 had no detectable effect on transcription in extracts from cells infected with either of the two mutant viruses (Fig. 8A, lanes 4 to 7, and B). These results confirm that substitutions at position 1631 of L enable the RdRp to overcome the barrier to transcription initiation that is imposed by AZ-27.
FIG 8.
Effect of AZ-27 on RSV A resistance mutants in a run-on transcription assay. (A) RNA synthesis assays were performed using lysate from cells infected with either wt RSV (lanes 2 and 3) or RSV variants containing either a Y1631H (lanes 4 and 5) or Y1631C (lanes 6 and 7) substitution in the L gene. Reactions were performed as described for Fig. 5, except cells were harvested at 22 h postinfection and reaction mixtures contained 100 mM potassium acetate and were incubated for 3 h. The identities of the bands were deduced as described for Fig. 5. (B) Bar chart showing quantification of the NS1 bands from replicates of the data shown in panel A, with the numbers expressed relative to the mean levels of RNA generated for each virus in the absence of compound. Each bar represents the means from three independent experiments, with standard errors shown.
DISCUSSION
The goal of this study was to elucidate the mechanism of RSV RdRp inhibition by a small-molecule inhibitor, AZ-27, with the hope that this also would provide insight into the mechanism of action of the compound and the molecular biology of the RdRp complex. By analogy with other NNS RNA virus RdRps, the RSV RdRp is a multifunctional machine that is able to synthesize and modify RNA and presumably must transition between different structural forms to perform different aspects of RNA synthesis. The data presented in this study indicate that AZ-27 has a differential effect on RSV RdRp activities and inhibits mRNA transcription and genome replication by inhibiting initiation of RNA synthesis.
Evidence indicating that AZ-27 inhibits the initiation step of RNA synthesis comes from the in vitro analyses of RSV RdRp activity (Fig. 5 and 6). An in vitro transcription run-on assay showed that AZ-27 had a more potent inhibitory effect on synthesis of transcripts synthesized from genes at the 3′ end of the genome than on those synthesized from the 5′ end (Fig. 5). This finding indicates that AZ-27 inhibits RNA synthesis from the promoter. In RSV, the steps involved in initiating RNA synthesis at the promoter could be parsed as follows. First, the RdRp interacts with the 3′ end of the encapsidated template and displaces N protein. It then binds to the promoter and initiates RNA synthesis. Presumably, it undergoes conformational changes to enable it to move away from the promoter and begin elongation, similar to other polymerases. The assay used for the experiments shown in Fig. 6 allowed the analysis of RdRp activities at the promoter at a stage following displacement of the N protein from the RNA. The finding that AZ-27 inhibited de novo RNA synthesis in an assay involving a naked RNA template (with no N protein present in the reaction) indicates that AZ-27 can inhibit a step that occurs after relaxation of the N-RNA structure. AZ-27 inhibited synthesis of RNA initiated at either the +3 or +1 site of the promoter (Fig. 6B), consistent with the finding that the compound inhibited both mRNA transcription and genome replication (Fig. 2 and 3). Analysis of small RNA products indicated that it inhibited the formation of even the first phosphodiester bond in RNA synthesis (Fig. 6E and F). Therefore, AZ-27 suppresses RSV transcription and genome replication by inhibiting promoter binding and/or phosphodiester bond formation during the initiation step of these two RNA synthesis processes. So far, we have not been able to establish a promoter-binding assay for the RSV RdRp, so we cannot distinguish between these possibilities. It is intriguing that the RSV RdRp is able to initiate transcription and replication from two sites within a single promoter, and we are currently investigating the mechanism by which this occurs. The data presented here provide some insight by indicating that there is a common step or steps involved in both initiation events.
The most surprising finding in the study was that, despite inhibiting de novo RNA synthesis, AZ-27 had no effect on the ability of the RdRp to add one to three nucleotides to the 3′ end of the tr RNA by back-priming and 3′ extension, although it did inhibit further elongation of this RNA (Fig. 6B and C). The 3′ extension activity of the RSV RdRp is an enigmatic phenomenon, as the role of this activity in the viral replication cycle is not known. However, recent analysis suggests that it is not an artifact of coincidental secondary structure but rather is a relatively controlled process, depending on sequence beyond the first 20 nt of the tr promoter to occur accurately (36). Similar to de novo RNA synthesis, the back-priming and 3′ extension activity depends on the polymerization domain of the RdRp (Fig. 6B, lane 4). The fact that AZ-27 did not inhibit addition of 1 to 3 nucleotides by 3′ extension indicates that this compound does not impair RNA binding or phosphodiester bond formation per se. One possible explanation is that AZ-27 specifically inhibits incorporation of ATP during RNA synthesis; if so, this would inhibit initiation of de novo RNA synthesis, as well as elongation of the 3′ extension product (Fig. 6A). However, AZ-27 is not a nucleoside analog and bears no obvious similarity to ATP in its structure (Fig. 1) (41). Furthermore, if it were inhibiting ATP incorporation, in the transcription runoff assay, this would be expected to lead to equal inhibition of transcription of all RSV genes rather than preferential inhibition of 3′-proximal genes (Fig. 5). Therefore, we think that this explanation is unlikely. The other possibility is that there is something distinct between initiation of de novo RNA synthesis and the initial steps of 3′ extension that results in differential susceptibility to AZ-27. In this scenario, we suggest two possible explanations for these findings. One is that there are two conformations of RdRp, one responsible for de novo RNA synthesis and the elongation phase of 3′ extension and another responsible for addition of 1 to 3 nucleotides by 3′ extension, and that these two pools of RdRp differ in their susceptibility to AZ-27 binding. The other possibility is that initiation of de novo RNA synthesis and the transition to the elongation phase of 3′ extension both require conformational changes in the RdRp, and it is RdRp conformational change that is inhibited by AZ-27 binding.
Resistance to AZ-27 is not conferred by substitutions in a known enzymatic domain but instead by a change in amino acid Y1631, which lies between conserved regions V and VI, the putative polyribonucleotidyltransferase and methyltransferase domains, respectively (Fig. 8) (42). Although this region of the L protein is thought to be involved in capping, previous studies with Sendai virus L protein have shown that mutations in conserved regions V and VI can affect RNA synthesis (51, 52). Y1631 lies adjacent to a region that is highly variable (amino acids 1730 to 1750) and which has been shown to accept large insertions and to be a site where L can be split into two (14, 53). Thus, this resistance site is adjacent to a likely hinge region in the RdRp, which might be important for RdRp flexibility and dynamic transitions. This structural information regarding the resistance site is consistent with the hypothesis that AZ-27 impairs the ability of the RSV RdRp to adopt a particular structural form required for RNA synthesis and/or to transition between different structural states. It should be noted that AZ-27 also might affect the polyribonucleotidyltransferase and/or methyltransferase functions of the RdRp, but this has not yet been determined.
Previously, it had been shown that RSV subgroup B is less susceptible to inhibition by AZ-27 than subgroup A (42). Here, we confirmed that this difference is due to differential susceptibility of the two viral RdRps to the inhibitor, with at least a 100-fold higher concentration of AZ-27 being required for inhibition of RNA synthesis by RSV B than RSV A (compare Fig. 5A and 7C). This difference in RdRp susceptibility resulted in a differential reduction in viral mRNA accumulation in cells (Fig. 7A and B) and in an in vitro assay (Fig. 7C). The amino acid that can be changed to confer resistance to AZ-27 (Y1631) is identical between RSV A and B (42). If AZ-27 inhibits RSV RdRp activity by affecting structural transitions, it is possible that structural differences between the L proteins of RSV A and B, and in particular in the hinge region around amino acids 1730 to 1750, account for the differential susceptibility of these proteins. Structural analysis of the RSV A and B RdRps will be helpful in determining if this is the case. It should be noted that the data comparing susceptibility of RSV A and B to AZ-27 are reassuring, because there appears to be a direct correlation between inhibition of RdRp activity and virus multicycle growth (Fig. 7) (42). This finding provides confirmatory evidence that the RSV RdRp is a valuable target for intervention with antiviral drugs.
In summary, the results presented here show that AZ-27 inhibits the initiation step of transcription and RNA replication by RSV A2. Interestingly, they also suggest either that there are two pools of RSV RdRp that exists in at least two conformational states, and that these pools have different activities at the tr promoter, or that the initiation of RNA synthesis from the +1 and +3 sites of the promoter requires structural transitions in the RdRp that can be inhibited by AZ-27.
ACKNOWLEDGMENTS
This research was funded through a sponsored research agreement between Boston University Medical School and AstraZeneca.
We thank Peter Collins for plasmids used for the minigenome experiments and Bernard Moss for MVA-T7. We also thank Peter Doig, Manos Perros, and Adam Shapiro for helpful discussions regarding this work.
REFERENCES
- 1.Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O'Brien KL, Roca A, Wright PF, Bruce N, Chandran A, Theodoratou E, Sutanto A, Sedyaningsih ER, Ngama M, Munywoki PK, Kartasasmita C, Simoes EA, Rudan I, Weber MW, Campbell H. 2010. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 375:1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robinson RF. 2008. Impact of respiratory syncytial virus in the United States. Am J Health Syst Pharm 65:S3–S6. doi: 10.2146/ajhp080438. [DOI] [PubMed] [Google Scholar]
- 3.Falsey AR, Hennessey PA, Formica MA, Cox C, Walsh EE. 2005. Respiratory syncytial virus infection in elderly and high-risk adults. N Engl J Med 352:1749–1759. doi: 10.1056/NEJMoa043951. [DOI] [PubMed] [Google Scholar]
- 4.Collins PL, Melero JA. 2011. Progress in understanding and controlling respiratory syncytial virus: still crazy after all these years. Virus Res 162:80–99. doi: 10.1016/j.virusres.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krilov LR. 2011. Respiratory syncytial virus disease: update on treatment and prevention. Expert Rev Anti Infect Ther 9:27–32. doi: 10.1586/eri.10.140. [DOI] [PubMed] [Google Scholar]
- 6.Chu HY, Englund JA. 2013. Respiratory syncytial virus disease: prevention and treatment. Curr Top Microbiol Immunol 372:235–258. [DOI] [PubMed] [Google Scholar]
- 7.DeVincenzo JP, El Saleeby CM, Bush AJ. 2005. Respiratory syncytial virus load predicts disease severity in previously healthy infants. J Infect Dis 191:1861–1868. doi: 10.1086/430008. [DOI] [PubMed] [Google Scholar]
- 8.DeVincenzo JP, Wilkinson T, Vaishnaw A, Cehelsky J, Meyers R, Nochur S, Harrison L, Meeking P, Mann A, Moane E, Oxford J, Pareek R, Moore R, Walsh E, Studholme R, Dorsett P, Alvarez R, Lambkin-Williams R. 2010. Viral load drives disease in humans experimentally infected with respiratory syncytial virus. Am J Respir Crit Care Med 182:1305–1314. doi: 10.1164/rccm.201002-0221OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El Saleeby CM, Bush AJ, Harrison LM, Aitken JA, Devincenzo JP. 2011. Respiratory syncytial virus load, viral dynamics, and disease severity in previously healthy naturally infected children. J Infect Dis 204:996–1002. doi: 10.1093/infdis/jir494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeVincenzo JP, Whitley RJ, Mackman RL, Scaglioni-Weinlich C, Harrison L, Farrell E, McBride S, Lambkin-Williams R, Jordan R, Xin Y, Ramanathan S, O'Riordan T, Lewis SA, Li X, Toback SL, Lin SL, Chien JW. 2014. Oral GS-5806 activity in a respiratory syncytial virus challenge study. N Engl J Med 371:711–722. doi: 10.1056/NEJMoa1401184. [DOI] [PubMed] [Google Scholar]
- 11.Collins PL, Fearns R, Graham BS. 2013. Respiratory syncytial virus: virology, reverse genetics, and pathogenesis of disease. Curr Top Microbiol Immunol 372:3–38. doi: 10.1007/978-3-642-38919-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morin B, Kranzusch PJ, Rahmeh AA, Whelan SP. 2013. The polymerase of negative-stranded RNA viruses. Curr Opin Virol 3:103–110. doi: 10.1016/j.coviro.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bujnicki JM, Rychlewski L. 2002. In silico identification, structure prediction and phylogenetic analysis of the 2′-O-ribose (cap 1) methyltransferase domain in the large structural protein of ssRNA negative-strand viruses. Protein Eng 15:101–108. doi: 10.1093/protein/15.2.101. [DOI] [PubMed] [Google Scholar]
- 14.Fix J, Galloux M, Blondot ML, Eleouet JF. 2011. The insertion of fluorescent proteins in a variable region of respiratory syncytial virus L polymerase results in fluorescent and functional enzymes but with reduced activities. Open Virol J 5:103–108. doi: 10.2174/1874357901105010103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liuzzi M, Mason SW, Cartier M, Lawetz C, McCollum RS, Dansereau N, Bolger G, Lapeyre N, Gaudette Y, Lagace L, Massariol MJ, Do F, Whitehead P, Lamarre L, Scouten E, Bordeleau J, Landry S, Rancourt J, Fazal G, Simoneau B. 2005. Inhibitors of respiratory syncytial virus replication target cotranscriptional mRNA guanylylation by viral RNA-dependent RNA polymerase. J Virol 79:13105–13115. doi: 10.1128/JVI.79.20.13105-13115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noton SL, Deflube LR, Tremaglio CZ, Fearns R. 2012. The respiratory syncytial virus polymerase has multiple RNA synthesis activities at the promoter. PLoS Pathog 8:e1002980. doi: 10.1371/journal.ppat.1002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poch O, Blumberg BM, Bougueleret L, Tordo N. 1990. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J Gen Virol 71(Part 5):1153–1162. doi: 10.1099/0022-1317-71-5-1153. [DOI] [PubMed] [Google Scholar]
- 18.Poch O, Sauvaget I, Delarue M, Tordo N. 1989. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J 8:3867–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, Rahmeh A, Morelli M, Whelan SP. 2008. A conserved motif in region V of the large polymerase proteins of nonsegmented negative-sense RNA viruses that is essential for mRNA capping. J Virol 82:775–784. doi: 10.1128/JVI.02107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogino T, Banerjee AK. 2007. Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol Cell 25:85–97. doi: 10.1016/j.molcel.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 21.Barr JN, Wertz GW. 2001. Polymerase slippage at vesicular stomatitis virus gene junctions to generate poly(A) is regulated by the upstream 3′-AUAC-5′ tetranucleotide: implications for the mechanism of transcription termination. J Virol 75:6901–6913. doi: 10.1128/JVI.75.15.6901-6913.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barr JN, Whelan SP, Wertz GW. 1997. Cis-acting signals involved in termination of vesicular stomatitis virus mRNA synthesis include the conserved AUAC and the U7 signal for polyadenylation. J Virol 71:8718–8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cowton VM, McGivern DR, Fearns R. 2006. Unravelling the complexities of respiratory syncytial virus RNA synthesis. J Gen Virol 87:1805–1821. doi: 10.1099/vir.0.81786-0. [DOI] [PubMed] [Google Scholar]
- 24.Noton SL, Fearns R. 2015. Initiation and regulation of paramyxovirus transcription and replication. Virology 479-480C:545–554. doi: 10.1016/j.virol.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dickens LE, Collins PL, Wertz GW. 1984. Transcriptional mapping of human respiratory syncytial virus. J Virol 52:364–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fearns R, Peeples ME, Collins PL. 2002. Mapping the transcription and replication promoters of respiratory syncytial virus. J Virol 76:1663–1672. doi: 10.1128/JVI.76.4.1663-1672.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collins PL, Dickens LE, Buckler-White A, Olmsted RA, Spriggs MK, Camargo E, Coelingh KV. 1986. Nucleotide sequences for the gene junctions of human respiratory syncytial virus reveal distinctive features of intergenic structure and gene order. Proc Natl Acad Sci U S A 83:4594–4598. doi: 10.1073/pnas.83.13.4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuo L, Grosfeld H, Cristina J, Hill MG, Collins PL. 1996. Effects of mutations in the gene-start and gene-end sequence motifs on transcription of monocistronic and dicistronic minigenomes of respiratory syncytial virus. J Virol 70:6892–6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tremaglio CZ, Noton SL, Deflube LR, Fearns R. 2013. Respiratory syncytial virus polymerase can initiate transcription from position 3 of the leader promoter. J Virol 87:3196–3207. doi: 10.1128/JVI.02862-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noton SL, Fearns R. 2011. The first two nucleotides of the respiratory syncytial virus antigenome RNA replication product can be selected independently of the promoter terminus. RNA 17:1895–1906. doi: 10.1261/rna.2813411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gubbay O, Curran J, Kolakofsky D. 2001. Sendai virus genome synthesis and assembly are coupled: a possible mechanism to promote viral RNA polymerase processivity. J Gen Virol 82:2895–2903. [DOI] [PubMed] [Google Scholar]
- 32.McGivern DR, Collins PL, Fearns R. 2005. Identification of internal sequences in the 3′ leader region of human respiratory syncytial virus that enhance transcription and confer replication processivity. J Virol 79:2449–2460. doi: 10.1128/JVI.79.4.2449-2460.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noton SL, Cowton VM, Zack CR, McGivern DR, Fearns R. 2010. Evidence that the polymerase of respiratory syncytial virus initiates RNA replication in a nontemplated fashion. Proc Natl Acad Sci U S A 107:10226–10231. doi: 10.1073/pnas.0913065107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peeples ME, Collins PL. 2000. Mutations in the 5′ trailer region of a respiratory syncytial virus minigenome which limit RNA replication to one step. J Virol 74:146–155. doi: 10.1128/JVI.74.1.146-155.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanley LL, McGivern DR, Teng MN, Djang R, Collins PL, Fearns R. 2010. Roles of the respiratory syncytial virus trailer region: effects of mutations on genome production and stress granule formation. Virology 406:241–252. doi: 10.1016/j.virol.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noton SL, Aljabr W, Hiscox JA, Matthews DA, Fearns R. 2014. Factors affecting de novo RNA synthesis and back-priming by the respiratory syncytial virus polymerase. Virology 462-463:318–327. doi: 10.1016/j.virol.2014.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laganas VA, Dunn EF, McLaughlin RE, Tiong-Yip CL, Yuzhakov O, Isabella VM, Hill P, Yu Q. 2015. Characterization of novel respiratory syncytial virus inhibitors identified by high throughput screen. Antiviral Res 115:71–74. doi: 10.1016/j.antiviral.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 38.Mason SW, Lawetz C, Gaudette Y, Do F, Scouten E, Lagace L, Simoneau B, Liuzzi M. 2004. Polyadenylation-dependent screening assay for respiratory syncytial virus RNA transcriptase activity and identification of an inhibitor. Nucleic Acids Res 32:4758–4767. doi: 10.1093/nar/gkh809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matharu DS, Flaherty DP, Simpson DS, Schroeder CE, Chung D, Yan D, Noah JW, Jonsson CB, White EL, Aube J, Plemper RK, Severson WE, Golden JE. 2014. Optimization of potent and selective quinazolinediones: inhibitors of respiratory syncytial virus that block RNA-dependent RNA-polymerase complex activity. J Med Chem 57:10314–10328. doi: 10.1021/jm500902x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sudo K, Miyazaki Y, Kojima N, Kobayashi M, Suzuki H, Shintani M, Shimizu Y. 2005. YM-53403, a unique anti-respiratory syncytial virus agent with a novel mechanism of action. Antiviral Res 65:125–131. doi: 10.1016/j.antiviral.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 41.Xiong H, Foulk M, Aschenbrenner L, Fan J, Tiong-Yip CL, Johnson KD, Moustakas D, Fleming PR, Brown DG, Zhang M, Ferguson D, Wu D, Yu Q. 2013. Discovery of a potent respiratory syncytial virus RNA polymerase inhibitor. Bioorg Med Chem Lett 23:6789–6793. doi: 10.1016/j.bmcl.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 42.Tiong-Yip CL, Aschenbrenner L, Johnson KD, McLaughlin RE, Fan J, Challa S, Xiong H, Yu Q. 2014. Characterization of a respiratory syncytial virus L protein inhibitor. Antimicrob Agents Chemother 58:3867–3873. doi: 10.1128/AAC.02540-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jin H, Clarke D, Zhou HZ, Cheng X, Coelingh K, Bryant M, Li S. 1998. Recombinant human respiratory syncytial virus (RSV) from cDNA and construction of subgroup A and B chimeric RSV. Virology 251:206–214. doi: 10.1006/viro.1998.9414. [DOI] [PubMed] [Google Scholar]
- 44.Grosfeld H, Hill MG, Collins PL. 1995. RNA replication by respiratory syncytial virus (RSV) is directed by the N, P, and L proteins; transcription also occurs under these conditions but requires RSV superinfection for efficient synthesis of full-length mRNA. J Virol 69:5677–5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wyatt LS, Moss B, Rozenblatt S. 1995. Replication-deficient vaccinia virus encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology 210:202–205. doi: 10.1006/viro.1995.1332. [DOI] [PubMed] [Google Scholar]
- 46.Carlos TS, Fearns R, Randall RE. 2005. Interferon-induced alterations in the pattern of parainfluenza virus 5 transcription and protein synthesis and the induction of virus inclusion bodies. J Virol 79:14112–14121. doi: 10.1128/JVI.79.22.14112-14121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Munday DC, Wu W, Smith N, Fix J, Noton SL, Galloux M, Touzelet O, Armstrong SD, Dawson JM, Aljabr W, Easton AJ, Rameix-Welti MA, de Oliveira AP, Simabuco FM, Ventura AM, Hughes DJ, Barr JN, Fearns R, Digard P, Eleouet JF, Hiscox JA. 2015. Interactome analysis of the human respiratory syncytial virus RNA polymerase complex identifies protein chaperones as important cofactors that promote L-protein stability and RNA synthesis. J Virol 89:917–930. doi: 10.1128/JVI.01783-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fearns R, Peeples ME, Collins PL. 1997. Increased expression of the N protein of respiratory syncytial virus stimulates minigenome replication but does not alter the balance between the synthesis of mRNA and antigenome. Virology 236:188–201. doi: 10.1006/viro.1997.8734. [DOI] [PubMed] [Google Scholar]
- 49.Yin YW, Steitz TA. 2002. Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science 298:1387–1395. doi: 10.1126/science.1077464. [DOI] [PubMed] [Google Scholar]
- 50.van Dijk AA, Makeyev EV, Bamford DH. 2004. Initiation of viral RNA-dependent RNA polymerization. J Gen Virol 85:1077–1093. doi: 10.1099/vir.0.19731-0. [DOI] [PubMed] [Google Scholar]
- 51.Cortese CK, Feller JA, Moyer SA. 2000. Mutations in domain V of the Sendai virus L polymerase protein uncouple transcription and replication and differentially affect replication in vitro and in vivo. Virology 277:387–396. doi: 10.1006/viro.2000.0615. [DOI] [PubMed] [Google Scholar]
- 52.Feller JA, Smallwood S, Horikami SM, Moyer SA. 2000. Mutations in conserved domains IV and VI of the large (L) subunit of the Sendai virus RNA polymerase give a spectrum of defective RNA synthesis phenotypes. Virology 269:426–439. doi: 10.1006/viro.2000.0234. [DOI] [PubMed] [Google Scholar]
- 53.Dochow M, Krumm SA, Crowe JE Jr, Moore ML, Plemper RK. 2012. Independent structural domains in paramyxovirus polymerase protein. J Biol Chem 287:6878–6891. doi: 10.1074/jbc.M111.325258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang G, Deval J, Hong J, Dyatkina N, Prhavc M, Taylor J, Fung A, Jin Z, Stevens SK, Serebryany V, Liu J, Zhang Q, Tam Y, Chanda SM, Smith DB, Symons JA, Blatt LM, Beigelman L. 2015. Discovery of 4′-chloromethyl-2′-deoxy-3′, 5′-di-O-isobutyryl-2′-fluorocytidine (ALS-8176), a first-in-class RSV polymerase inhibitor for treatment of human respiratory syncytial virus infection. J Med Chem 58:1862–1878. doi: 10.1021/jm5017279. [DOI] [PubMed] [Google Scholar]