Abstract
Defining fiber orientation at each voxel within a 3D biomedical image stack is potentially useful for a variety of applications, including cancer, wound healing and tissue regeneration. Current methods are typically computationally intensive or inaccurate. Herein, we present a 3D weighted orientation vector summation algorithm, which is a generalization of a previously reported 2D vector summation technique aimed at quantifying collagen fiber orientations simultaneously at each voxel of an image stack. As a result, voxel-wise fiber orientation information with 4° to 5° accuracy can be determined, and the computational time required to analyze a typical stack with the size of 512x512x100 voxels is less than 5 min. Thus, this technique enables the practical extraction of voxel-specific orientation data for characterizing structural anisotropy in 3D specimens. As examples, we use this approach to characterize the fiber organization in an excised mouse mammary gland and a 3D breast tissue model.
OCIS codes: (100.2960) Image analysis, (100.6950) Tomographic image processing, (180.4315) Nonlinear microscopy
1. Introduction
Collagen, in the form of elongated fibrils, is the main protein of the various connective tissues in animals [1,2]. Tough bundles of collagen fibers are a major component of the extracellular matrix (ECM) that provides support for cells in most tissues [3,4]. The triple helical structure of collagen enables adhesiveness of cells and is important for the proper assembly of the ECM [5]. The levels of hydroxylation, glycosylation, cross-linking and the mechanical properties of collagen are dependent on a number of important tissue properties, including tissue type, age, and hormonal status [2]. In addition, collagen fiber orientation provides a potential biomarker for diagnosing diseased tissues, assessing response to treatments, localizing injuries, or monitoring engineered tissue development [6–10]. A number of techniques, such as ones based on the Fourier transform [11–13], Hough transform [10,14], and principal component analysis [15–17] are widely employed to quantify fiber orientation within two dimensional (2D) images. The applications of these 2D techniques have shown potential in providing orientation distribution information to assess collagenous tissues and understand interactions between cells and the ECM. Nevertheless, we expect that 3D analysis represents more accurately the architecture of collagen fibers than 2D analysis, and may be more sensitive to subtle changes in the micro-environment [18]. For example, recent research on collagenous fibrosis has shown that 3D analysis depicts pathological changes due to fibrosis progression more precisely than 2D analysis [19–21].
Although there are a number of 2D approaches developed for collagen fiber orientation analysis, only a small number of 3D techniques exist to quantify the fiber direction in an image stack. Sophisticated techniques such as fiber tracking approaches have been developed [22–25], in which fiber objects are defined and tracked through energy minimization (e.g., active contours) or line propagation algorithms. However, the performance of these advanced techniques strongly depends on the complexity of the image [26], and they are less feasible when applied to, for example, dense fibrillar objects. They utilize iterative processes, which are relatively time consuming and typically require some level of user interaction.
Fourier transform (FT) based approaches are commonly applied for 2D orientation analysis, and they have been extended to 3D fiber characterization of second harmonic generation (SHG) images [27]. For such methods, the image stack is first divided to create a set of regions of interest (ROIs), and the overall orientation of collagen fibers within each ROI is determined by the filter bank method [27]. The filter bank consists of various 3D orientation filters constructed in the Fourier space. By comparing the Fourier transform of each ROI with filter banks, the filter orientation that corresponds to the maximum likelihood is designated as the fiber orientation. However, the computational cost of this technique relies strongly on the fiber density of the image, and the size of ROI is a tradeoff between the determination accuracy and the computational time. Yet, the ability to automatically generate fiber orientation information at each voxel within an image would be advantageous for many applications. Further approaches include the chord length transform [28], Gaussian orientation space [29,30] and inertia moments [31]; however there are some restrictions on the characteristics of fibers (e.g., shape, cross section or thickness) when using these techniques.
In our previous work, we developed a weighted orientation vector summation algorithm which is capable of detecting fiber orientation simultaneously at each pixel within a 2D image [32]. This technique relies on changes in the intensity variations across a fiber in different directions. For example, intensity variations will be subtle along the direction of a fiber, and rather significant and abrupt while traversing the fiber diameter. As a result, pixel-specific fiber orientation information can be provided faster than pixel-wise Fourier analysis with comparable accuracy by an order of magnitude. This technique has been successfully applied for studying interactions between the matrix and human breast cells [33], and mapping scar formation in histological tissue sections [34,35]. Compared to edge detection algorithms that have been developed for rapidly detecting fiber orientation in micrographs, such as algorithms based on Sobel or Canny operators [36–38], this technique has improved accuracy.
In this study, we develop a 3D weighted orientation vector summation technique in MATLAB for the rapid and accurate quantification of fiber orientation at each voxel within an image stack. We describe our methodology, including an overview of the analysis algorithm and evaluation of the technique performance. We assess the accuracy of this detection algorithm by simulating images with known orientations, monitor computational time with varying image and window sizes, and make a comparison between 3D and 2D techniques. Finally, we present applications of this technique to analyze second harmonic generation (SHG) images acquired from an excised mouse mammary gland and a 3D breast tissue model.
2. Methods
2.1 Overview of the vector summation technique for 3D orientation detection
The algorithm developed in this study detects 3D fiber orientation by identifying the variability of intensities along all the directions surrounding each voxel within an image stack. In order to depict an orientation in 3D space, normally an angle in the transverse plane and an inclination angle are defined (Fig. 1(a)). To cover all the 3D orientations, the range of both and can vary between 0° and 180°. The red solid line in Fig. 1(a) represents a fiber of interest, with defined as the angle between the projection of the fiber in the plane and the axis, and defined as the angle between the fiber and the axis. In our algorithm, we define two additional angles, and : is the angle between the projection of the fiber in the plane and the axis, and is the angle between the projection in the plane and the axis. We assume the red dashed line to be the calculated fiber orientation, and then is defined as the angle between the defined and the calculated orientation (Fig. 1(a)).
Fig. 1.
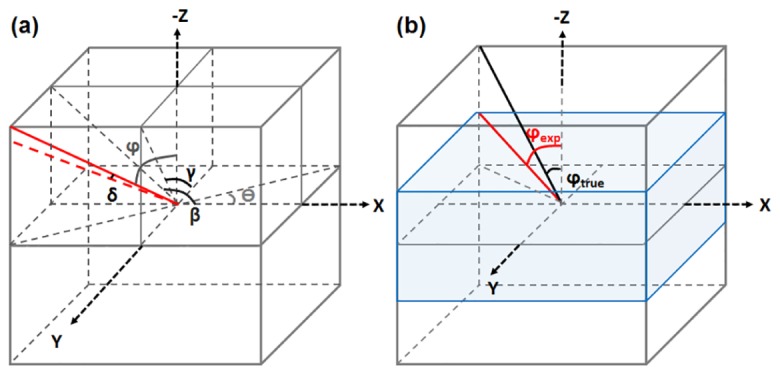
Schematic of the angle definition in this study. (a) Basic angles defined for orientation calculation and error assessment. (b) Angles defined to reveal the case of varied axial-to-lateral sampling ratio.
Vector summation obeying basic rules can provide orientation information of circular data (for which an orientation of 0° and 360° are equivalent). However, fiber orientations are axial data (for which 0° and 180° are equivalent), and the vector summation rules should be modified accordingly. In the 2D case, a single angle is sufficient to depict the fiber orientation. To account for the degeneracy of the axial data in 2D space, all vector angles are first transformed to circular data by multiplying the angular values by 2 prior to the summation of and components, and the resultant angle is then divided by two in order to determine the mean fiber orientation [39]. In the 3D case, the situation is much more complicated requiring that both and should be determined to characterize the fiber orientation. The determination of is achieved by projecting fibers to a fixed 2D plane, i.e., the plane (Fig. 1(a)), which means that the calculation of still obeys the 2D vector summation rules. However, according to the definition of which is the angle between the fiber of interest and the axis, there is no fixed 2D plane that can be projected to for the determination of all the possible orientations. Therefore, the 2D vector summation rules for axial data are no longer applicable to the determination of . In order to solve this problem, we transform our data into 3 sets of 2D data by projecting our 3D fiber images onto 3 orthogonal planes. , and are the 3 angles defined by the projections on the 3 orthogonal planes, and therefore we are able to describe their statistical properties using the vector summation rules for axial data mentioned above. According to Fig. 1(a), the inclination angle can be expressed using a combination of and as follows:
| (1) |
As a generalization of the 2D algorithm, the 3D orientation algorithm starts by generating a cube window so as to evaluate each voxel within the cube. We calculate , and for every voxel in the 3D window. We first define all the possible vectors passing through the center voxel, then weigh these vectors by intensity variations among voxels, and finally sum up these weighted vectors to obtain the orientation. Specifically, after weighing these vectors by the voxel intensity, we project them to the , and plane, and sum up projections of these vectors to determine angles , and , respectively. To save computational time, instead of assessing all the orientation vectors within a cube window to determine the orientation of the center voxel of the cube, we first average the cube along one of the three orthogonal planes to reduce the cube window to a square window, and then calculate the orientation of the center pixel of the square, following an approach identical to the 2D weighted orientation vector approach described previously [32]. In this manner, the algorithm dealing with a 3D cube is converted to an algorithm dealing with a set of 2D squares, and therefore the computational time is greatly reduced by approximately -fold (assuming that the cube window has a size).
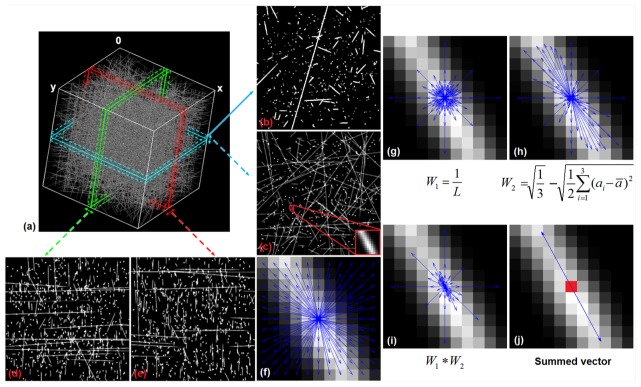
The overall approach is depicted in Fig. 2. Figure 2(a) is a 3D reconstruction of a test image stack, with one frame on the plane shown in Fig. 2(b). Figure 2(c), 2(d) and 2(e) are averaged projections of 11 frames in the , and planes respectively, for the determination of angles (c), (d) and (e), with the number of averaged frames corresponding to the width of the analysis window size. The inset in Fig. 2(c) zooms in a small region with a pixel window, which is used for the ensuing analysis steps. The determination of , and obeys the same rules, and therefore only the determination of is demonstrated in Fig. 2(f)-2(j). First, all the vector directions passing through the center pixel (as marked in red in Fig. 2(j)) within the window are defined (Fig. 2(f)). The perimeter of a circle with a diameter is , and therefore the number of lines with length passing through the center pixel of the window also scales in number by . Because the longer vectors corresponding to intensity fluctuations farther from the center pixel are more numerous, these vectors would have a greater collective weight when calculating the mean orientation of the center pixel using vector summation. To account for this effect, each vector is weighted by the inverse of its length (), which is the first weight factor (Fig. 2(g)). By doing this, pixels close to the center pixel of interest would have equal collective weight as pixels far away from the center (Fig. 2(g)). The second weight factor is based on the grayscale intensity fluctuations among the pixels in a given direction. As a vector is defined by two additional pixel locations symmetric about the center pixel, the second weight factor is determined by weighting the vectors according to the lack of variance among the intensities of the three pixel locations used to define the vector. The expression of the second weight factor is the difference between the maximum possible standard deviation among these three pixel locations and the measured standard deviation among them (Fig. 2(h)), where is the intensity of each pixel, and is the mean intensity of these three pixels. If the intensity of the image is converted to a range from 0 to 1, then the maximum standard deviation among three pixels is the square root of 1/3. After being weighted by these two factors (Fig. 2(i)), all the vectors are summed to determine an average orientation of , and surrounding the center pixel (Fig. 2(j)), and is then calculated according to Eq. (1).
Fig. 2.
Summary of 3D weighted orientation vector summation technique for voxel-wise fiber orientation detection. (a) 3D reconstruction of the test image stack. (b) One single frame in the plane. (c) The same frame as (b) but with neighboring frames averaged. (d) One averaged frame in the plane. (e) One averaged frame in the plane. (f) An analysis window with all the orientation vectors. (g) Vectors modified by weight factor 1 (). (h) Vectors modified by weight factor 2 (). (i) Vectors modified by combining weight factors 1 and 2. (j) The orientation of the center pixel calculated by summation of all the weighted orientation vectors.
2.2 Evaluation of the vector summation technique performance
To evaluate the accuracy of this vector summation technique to detect fiber orientation, images with known orientations were generated and assessed. First, to determine the relationship between measurement error and fiber thickness, 7 image stacks with a size of voxels with 28 cylinders were produced with different diameters of 3, 5, 7, 9, 11, 13 and 15 pixels. To generate these cylinders, the center of each cylinder could be a random voxel within the stack, and then the cylinder was formed by stretching across the space based on randomly defined and orientations (using the rand MATLAB function), which covered the full range of (0°, 180°) and known during the image generation. The only difference among these 7 stacks was the cylinder diameter. Cylinders were not permitted to overlap because a true orientation value at such a voxel would be unknown, but cylinders of different orientations were permitted to be located very close to each other, with only a 1-voxel gap in localized regions. To study the effects of fiber density, a similar generation method was employed to create three voxel cylinder stacks with the same diameter of 7 pixels, but different cylinder numbers of 27, 44 and 59, respectively.
The average absolute difference in the defined and calculated orientations of angles and , and the average absolute value of the angle between the defined and the calculated orientation () were calculated to assess the accuracy of the 3D vector summation technique. To define , let us assume that the unit vector for the defined orientation is : ( ), and the unit vector for the calculated orientation is : ( ). Based on the definition: , is expressed as follows:
| (2) |
where is the defined angle value, the calculated angle value, and is the difference between the defined and the calculated angle.
The orientation errors as a function of window size, image pixel resolution, and fiber orientation, were investigated using an additional voxel image created with 99 5-pixel diameter cylinders of varying orientations and lengths. We set the fiber diameter to be 5 pixels because it is close to the fiber size of the breast tissue sample that we analyze in this study. To assess the effects of window size, the orientation error was calculated for each voxel with the window size varying from voxels to voxels. In typical microscopy applications, the lateral resolution is always better than the axial resolution, and the sampling frequency in the direction is typically higher than that in the direction. If the axial sampling is -times as coarse as the lateral sampling, then the following equation can be used to convert between our orientation defined in voxel space, and the true orientation in physical space (Fig. 1(b)):
| (3) |
where is the measured value from the experimental image stack assuming cubic voxels, and is the true angle regardless of the axial and lateral resolutions, as shown in Fig. 1(b), where the axial-to-lateral sampling ratio is 2. To assess the effect from the inequality between the axial and lateral sampling frequency, we varied the axial-to-lateral sampling ratio from 1 to 6, and computed the measurement error accordingly. In the test of error versus fiber orientation, we computed the error as a function of cylinder orientation with a 10° interval. In addition to the error assessment, the computational time corresponding to a range of window and stack sizes was determined and compared to the computational time of a reported Fourier transform based technique.
These simplified collections of 3D stacks provided an ideal platform for assessing the accuracy of the algorithm over a range of parameters. Once we demonstrated the validity of the approach using these models, we generated more complex simulations to explore further the effects of various additional factors that we may encounter in real biological samples. Specifically, we created cylinder stacks with a size of voxels that included 334 fibers with randomly defined and orientations. The diameter of these fibers was either 3 or 5 pixels, and the length varied from approximately 5 to 400 pixels. For one stack, the intensity of these fibers was uniform. We created another stack with the same organization of fibers, but the intensity of individual fibers was randomly assigned values of 1, 1.5 and 3 arbitrary unit (a.u.). Finally, to evaluate the performance of the algorithm for the analysis of curvy collagen fibers, we created a voxel stack with spirals. The thickness of the spirals was 5 pixels in diameter.
Then we made a comparison between 3D and 2D techniques. We created 8 fiber stacks with a size of voxels. The diameter was 5 pixels, and each stack had approximately 35 fibers. To mimic the breast tissue model where parallel sheets of collagen fibers dominated the alignment, the orientation of approximately 72% of fibers in each stack was between 80° and 100°, and that of the rest of fibers was randomly generated within the range of (0°, 80°) and (100°,180°). For all the fibers, the orientation was randomly defined (the rand MATLAB function). To acquire the 3D analysis results, we calculated the distribution of each voxel within these voxel stacks using the 3D vector summation technique. Regarding the 2D analysis, we calculated the orientation using the 2D vector summation technique in a frame-by-frame manner, which was previously reported [33]. Specifically, we applied the 2D algorithm to 150 frames of each stack sequentially, and acquired the orientation for every voxel of these stacks too. Then we calculated the mean angle of each stack based on the defined, 3D acquired and 2D acquired orientations. For each stack, we obtained the absolute difference between the defined mean and the 3D acquired mean , as well as that between the defined mean and the 2D acquired mean . For statistical analysis, a t-test was used to determine the difference between 2D and 3D analyses based on the 8 sets of data, and results were considered significant at p < 0.05.
To illustrate the applicability of this approach, fiber alignment distributions were calculated from multi-photon microscopy images of a mammary gland and a hormone-sensitive 3D culture model of the breast epithelium. The mammary tissue was collected from the fourth mammary gland of non-parous 12 week old FVB/N female mice. The fresh tissue was embedded in optimal cutting temperature (OCT) compound and snap frozen in a methanol bath. It was immersed in phosphate buffered saline to remove the OCT compound before use. Images were also acquired from an engineered 3D breast tissue model described previously [40]. Briefly, human T47D cells were seeded in 3D culture using a 1 mg/ml collagen type 1 matrix and treated with mammogenic hormones (17β estradiol in combination with promegestone, an analogue of progesterone) for two weeks. The gels were whole-mounted and stained with Carmine Alum to visualize the epithelial structures [40]. Images of both samples were acquired using two-photon excitation fluorescence (TPEF) (ex: 800, em: 52525), together with SHG which resolved the collagen fiber structure (ex: 800, em: 40010). SHG images were analyzed using the 3D weighted orientation vector summation approach. Voxel-wise distribution histograms were calculated to reveal the and orientations of the collagen fibers.
3. Results and discussion
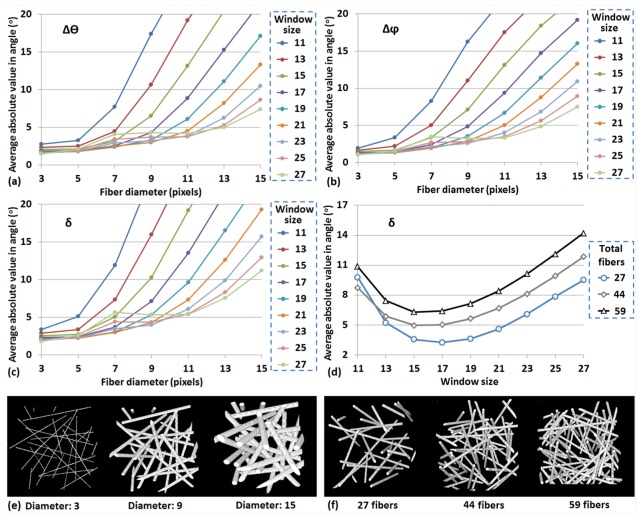
Fiber thickness and density are important factors that can affect the accuracy of the average fiber orientation measurement. For most window sizes, error in the fiber orientation measurements increases exponentially as the fiber diameter approaches, and eclipses the width of the window size (Fig. 3(a), 3(b) and 3(c)). However, if we focus on a specific fiber thickness, it is interesting that error values do not decrease monotonically as the window size increases. For fibers with a diameter of 7 pixels, a window size ranging from voxels to voxels offers the most accurate fiber orientation assessments. When the window increases further, the error increases. This happens because the proximity of fibers to each other is also a factor that affects the orientation measurement, and more than one of fibers were present in these larger windows. The error of as a function of fiber density and window size is shown in Fig. 3(d). These fibers have the same diameter of 7 pixels. With the increase of the total fiber number, there is an increase in the overall error. Nevertheless, they follow similar trends in terms of their accuracy dependence and the most accurate assessments are achieved at either or voxel window sizes. These results demonstrate that the fiber thickness is the major determinant of accuracy related to the choice of window size, which should be between 2 and 3 fold greater than the fiber diameter to achieve optimal results.
Fig. 3.
Errors of (a) , (b) and (c) in fiber orientation measurements with respect to different fiber thickness and window size. (d) Error of with respect to different fiber density and window size. (e) 3D reconstruction of stacks with different fiber thickness. (f) 3D reconstruction of stacks with different fiber density.
Analysis of an image stack of 99 cylinders with a 5-pixel diameter of varied lengths and randomly defined orientations results in significant improvements in the average voxel orientation accuracy between to voxel window sizes (Fig. 4(a)). Using a voxel window size, the average absolute values in , and are , and , respectively. For window sizes greater than voxels, not only the error increases, but also the standard deviation increases from a minimum of 8.5° to a maximum of 13.1° for a voxel window (for ). Although the average absolute values of , and are different, they follow the same trend as a function of window size. The 3D reconstructions of intensity (Fig. 4(b)), (Fig. 4(c)) and (Fig. 4(d)) stacks reveal the voxel-wise accuracy of this 3D orientation measurement technique. To show the distribution of the error within the whole stack, false color maps of with a window of voxel (Fig. 4(e)), voxel (Fig. 4(f)) and voxel (Fig. 4(g)) are shown, with insets zooming in a region with closely located fibers (Fig. 4(f) and 4(g)). With the increase of window size, the calculation error for the majority of the voxels approaches zero (as depicted in blue for these three graphs), suggesting an improvement in accuracy. However, it can be seen from the arrow-pointed regions in the inset (Fig. 4(g)) that an increase in error is produced in certain locations if the window size is large enough to include multiple fibers in close proximity to each other. This increased spatial variability in accuracy is the reason for the increase in the standard deviation.
Fig. 4.
Error in orientation detection using the 3D vector summation technique for a simulated image stack containing regions with varying orientations. (a) Errors of and , and the overall angle error in fiber orientation measurements with respect to different window size. 3D reconstructions of (b) intensity, (c) , and (d) map stacks. 3D reconstructions of with a window size of (e) voxels, (f) voxels and (g) voxels. The insets in (f) and (g) zoom in the same region in the image stack.
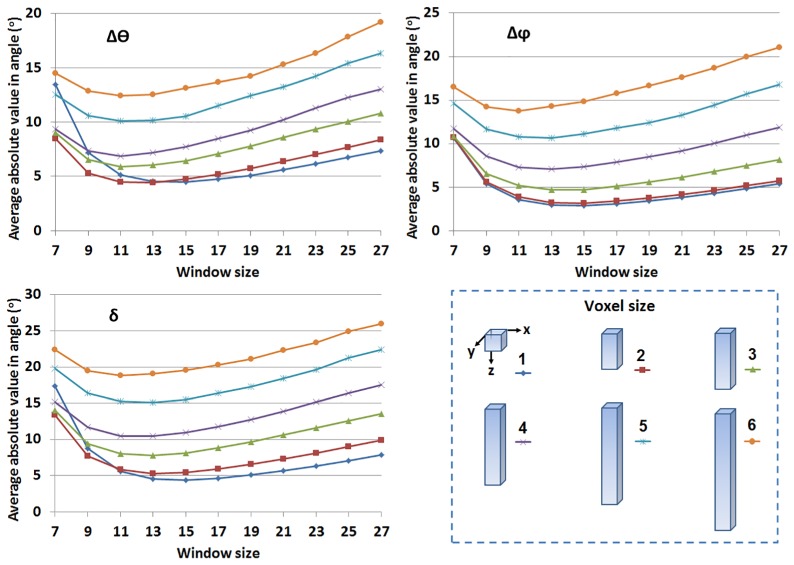
The effect of differences in the lateral and axial sampling frequencies typically found in biomedical microscopy images on the accuracy of this method was simulated and assessed. We assumed that the sampling for and directions (lateral resolutions) was equal, while the direction (axial) was sampled at a different (lower) frequency. We considered six independent situations with the axial-to-lateral sampling ratio ranging from 1 to 6. The error values of , and as a function of window size corresponding to different relative sampling ratios followed similar trends (Fig. 5). The accuracy decreased with an increase in the relative sampling ratio, because of the loss of details in the image stack. It is interesting to note that the error values change very slightly when the axial-to-lateral sampling ratio increases from 1 to 2, suggesting that image stacks in which the direction is sampled at a frequency that is twice as coarse as that of the dimension doesn’t lead to any significant effects in the accuracy with which we can extract the orientation of the collagen fibers. This of course, assumes that resolution of the images is sufficient to resolve individual fibers. Knowing that the axial direction can be sampled at a lower frequency is useful, because this decreases data acquisition time.
Fig. 5.
Errors of , and in fiber orientation measurements with respect to different axial-to-lateral sampling ratio and window size. Insets show voxel size during calculation.
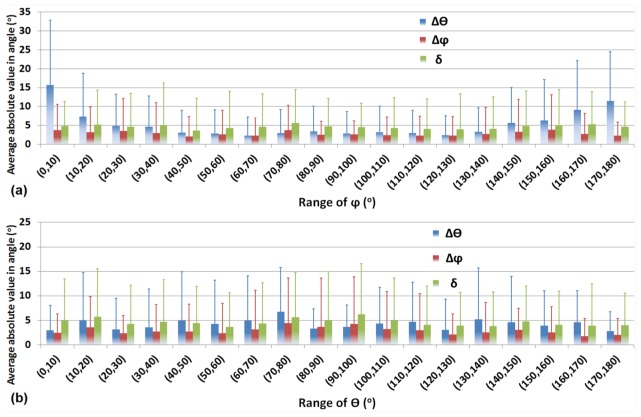
To have a better sense of error distribution as a function of fiber orientation, errors of and , and the overall error were determined with respect to both and covering the full range of (0°, 180°) in a 10° interval (Fig. 6). We found that the error of was dependent on the angle (Fig. 6(a)), and the average absolute value for could be more than 10° when approached 0° or 180°. This is because the orientation of the projection can be determined more accurately if the fiber is more parallel to the plane rather than perpendicular to it. In fact, is undefined when is equal to 0° or 180°. However, the average absolute value of is not dependent on the orientation (Fig. 6(a)). According to Eq. (2), if is accurately determined, will approach 1 (which means will approach 0) even though there are large fluctuations in near values of 0° and 180°. The average values of are close to or less than 5° for almost the full range of . As shown in Fig. 6(b), we do not observe a strong dependence of the error on orientation. The error values of and , as well as the value of are equal to or less than 5° for the full range of . Based on this error test, it is revealed that the actual error, , is independent of the fiber orientation. Therefore, there are no restrictions on the orientation of fibers to which the 3D vector summation technique can be applied.
Fig. 6.
Errors of , and in fiber orientation measurements with respect to different (a) and (b) range, corresponding to a window of voxels.
The computational time of this 3D vector summation technique is shown as a function of stack size and window size in Fig. 7. The time required to detect alignment at each voxel scales exponentially with stack size, and as expected, a larger window size requires more computational time. The inset zooms in the region with great stack sizes for a better reading of the exact computational cost. When compared to a Fourier-based algorithm [27], which provides an average orientation of each ROI and not each voxel, the 3D vector summation technique has improved computational time. Specifically, the computational time required by the Fourier-based algorithm to analyze a voxel stack is approximately 1.5 on a desktop computer with a 3.4 GHz processor and 8 GB of RAM [27], while the time required by the vector summation technique with exactly the same configuration of a computer is 59 , using a window of voxels. For a typical stack size of voxels, the analysis could be done within 5 min. As stated in the Methods section, instead of a direct generalization from the 2D to a 3D technique with the cube based algorithm, we first reduced the cube window to a square window by averaging neighboring frames in three orthogonal planes, so as to reduce the computational cost. To analyze the voxel stack shown in Fig. 4, the computational time was 2655 (44.3 min) when using the direct cube based technique with a voxel window, while only 268 (4.5 ) was needed using the 3D weighted orientation vector summation technique with the same window size. Using this window size, the overall error from the direct cube based technique is , compared to for the 3D weighted orientation vector summation technique. Considering the similar accuracy afforded by both approaches, the latter is better suited for biomedical image analysis due to its significantly improved speed.
Fig. 7.
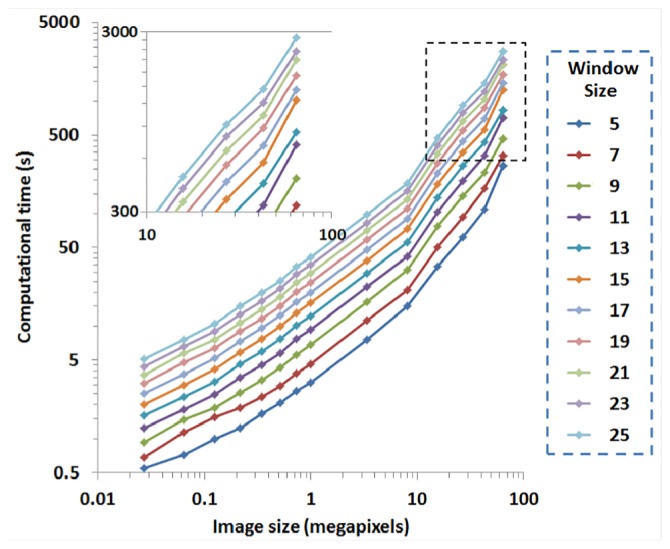
Computational time of this 3D technique with respect to different image and window sizes. Inset zooms in the region with large image sizes.
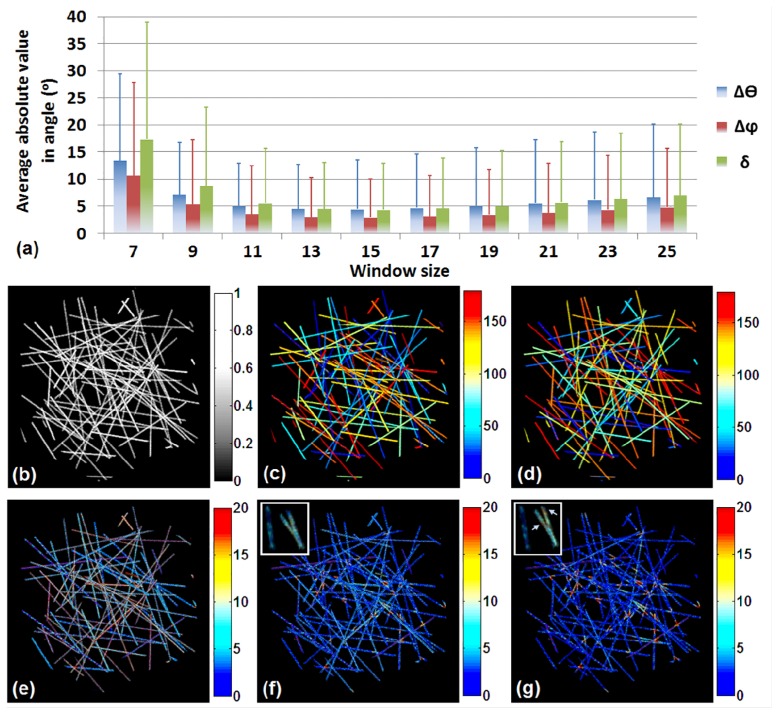
To assess the performance of the algorithm for the analysis of more challenging image stacks, we simulated a much denser stack of 334 fibers with different fiber thickness and length that have either uniform signal intensity (Fig. 8(a)) or intensity that varies from 1 to 3 a.u. (Fig. 8(b)). The overall error maps for these stacks are shown in Fig. 8(c) and 8(d), respectively, for a window size of voxels. The error as a function of window size is shown in Fig. 8(e), demonstrating that a minimum error of is achieved for the window of voxels with errors remaining less than 8.3° for window sizes as large as voxels. The optimal window size is a little smaller than the one for the less dense stacks, which is reasonable, since as the window size increases, we can have contributions from multiple fibers in dense matrices. Nevertheless, the error remains fairly small even for these more tissue-like simulations. The intensity variations do not appear to have a significant impact on the overall error.
Fig. 8.
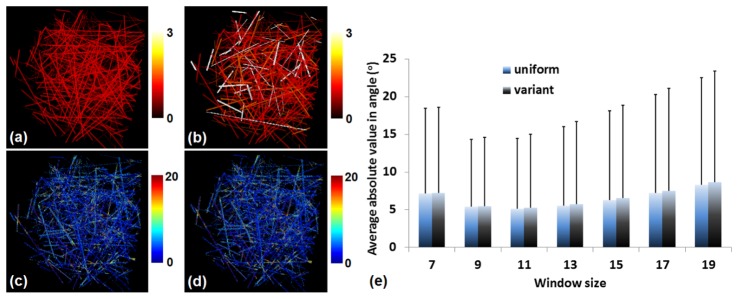
Error in orientation detection for simulated dense stacks. (a) A dense stack with 334 fibers with different fiber diameter and length, but uniform signal intensity. (b) The same stack as in (a), but variant signal intensity. (c) and (d) The error maps corresponding to (a) and (b), respectively, achieved by a window of voxels. (e) Errors of , corresponding to the uniform and variant intensity stacks, with respect to different window size.
In addition, we sought to assess the performance of the algorithm in the characterization of curvy structures by utilizing a 3D stack consisting of two sets of spirals. Figure 9(a) and 9(b) show 3D reconstructions of the and maps as determined by the 3D algorithm, with top-view images shown in insets. As can be seen from Fig. 9(c), the overall error is approaching 5° with the window of voxels and is only slightly higher than 7° for an voxel window, revealing that the 3D vector summation technique works well for the characterization of curvy features for window sizes that are similar to those that are optimal for the analysis of the straight features.
Fig. 9.
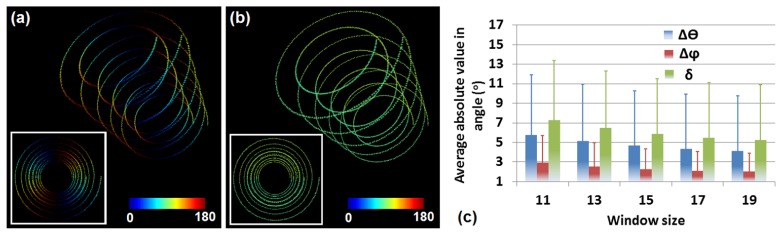
Error in orientation detection for simulated spiral stacks. (a) and (b) 3D reconstructions of and maps of the spiral fiber stack, respectively. Insets show top-view images. (c) Errors of and , and the overall angle error in spiral fiber orientation measurements with respect to different window size.
In Fig. 10, we show the comparison between the 2D and 3D analysis of a set of simulated images. Specifically, we created 8 stacks to compare the accuracy of determination using 2D and 3D techniques. Figure 10(a) shows the 3D reconstruction of these stacks. We implemented the 2D analysis in a frame-by-frame manner, as shown in Fig. 10(b), where the top 10 frames were zoomed in for illustration. After the orientation analysis, we calculated the mean angle using the defined, 2D acquired and 3D acquired information, and were able to extract the absolute difference between the defined mean and the calculated mean , either by the 2D or 3D technique. As shown in Fig. 10(c), the error by the 2D approach was less than 5°, and the accuracy was improved significantly by the 3D approach (p = 0.015). For fibers with great inclination to the transverse plane, they appear as individual spots on the cross section image, and therefore 2D analysis is difficult to compute the orientation accurately for these regions. In contrast, since 3D analysis considers the entire volume window, spots on the cross section images will stack up and form cylinders in 3D space. Therefore, the 3D technique is able to resolve such uncertainty that occurs in 2D analysis, and improve the accuracy.
Fig. 10.
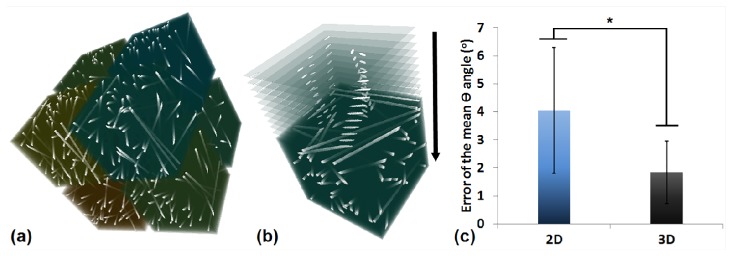
A comparison between 2D and 3D techniques. (a) 3D reconstruction of the 8 test stacks. (b) Schematic of the frame-by-frame manner of 2D analysis. Only the top 10 frames are zoomed in here for illustration. (c) Histograms showing the error of the mean determination. *, significant difference.
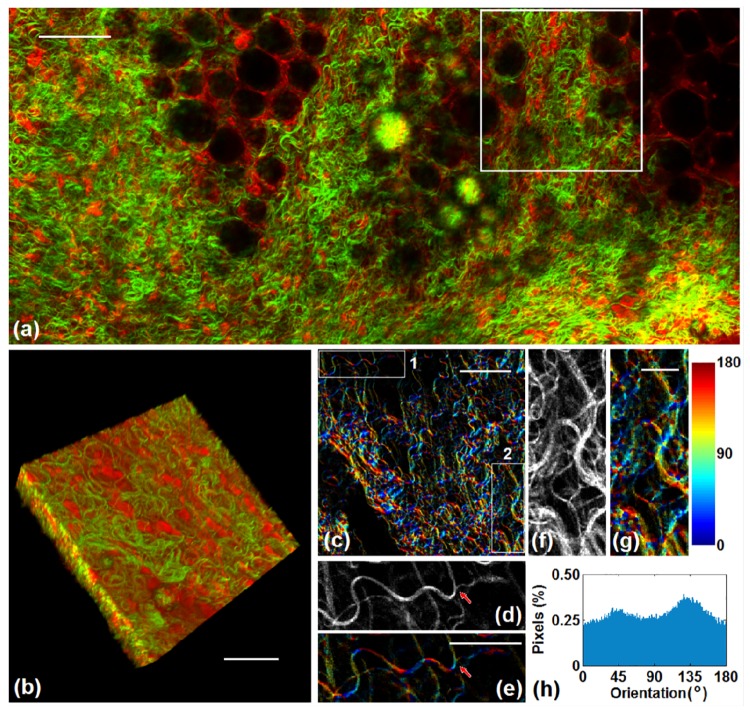
The applicability of this analysis approach is illustrated by utilizing it to analyze a 3D SHG image stack acquired from a mammary gland. As can be seen from the combined fluorescence (red) and SHG (green) image (Fig. 11(a)), the mammary gland is rich in curvy collagen fibers. The 3D reconstruction of the marked region in Fig. 11(a) is shown in Fig. 11(b), and the 3D vector summation technique was applied to the SHG signals of the entire 3D stack. Figure 11(c) shows the orientation map of collagen fibers for a certain frame in this stack, with two local regions zoomed in and shown in Fig. 11(d)-11(g), where Fig. 11(d) and 11(f) are the SHG intensity images of corresponding regions for reference. The curvy structures of collagen fibers are characterized well by the orientation map (Fig. 11(e) and 11(g)). The 3D algorithm also can provide orientation information of branched structures (indicated by arrows in Fig. 11(d) and 11(e)). Figure 11(h) shows the distribution histogram of for this frame which helps to characterize the varying orientation of these curvy structures.
Fig. 11.
Orientation analysis of fibers from the mammary gland sample. (a) Combined fluorescence (red) and SHG (green) image. Scale bar: 100 μm. (b) The 3D stack of the marked region in (a). Scale bar: 50 μm. (c) One frame of the orientation map from the 3D stack, calculated by the 3D algorithm. Scale bar: 50 μm. The two regions shown in (c) are zoomed in to show the details of fiber orientation. (d) Original SHG intensity of region 1. (e) orientation map of region 1. Scale bar: 30 μm. The arrows shown in (d) and (e) point out the branched structure. (f) Original SHG intensity of region 2. (g) orientation map of region 2. Scale bar: 15 μm. (h) The distribution histogram of corresponding to this frame.
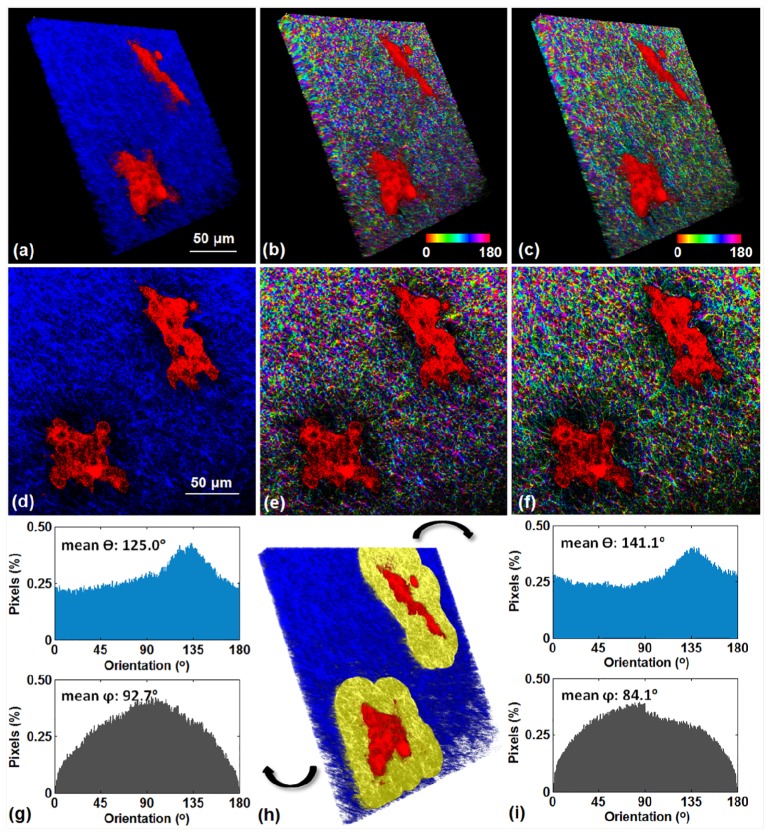
The applicability of this approach to analyze biomedical images was illustrated also by using it to determine the fiber alignment distributions within a 3D breast tissue culture model (Fig. 12). The image stack was acquired with a lateral resolution of 0.46 and an axial resolution of 1 . TPEF and SHG images were acquired simultaneously to gain a good understanding of the alignment of collagen fibers (SHG) relative to cells (TPEF) (Fig. 12(a) and 12(d)). Figure 12(a) is the 3D reconstruction of the combined images with false color coding (ImageJ), and Fig. 12(d) shows a certain frame of the stack. The 3D weighted orientation vector summation technique was employed to calculate the fiber orientation of each voxel based on the SHG signal. The 3D reconstructed stacks of distribution (Fig. 12(b)) and distribution (Fig. 12(c)), as well as corresponding frames (Fig. 12(e) and 12(f)) are shown. As can be seen from the and orientation maps, orientation information of each voxel was acquired and false colored.
Fig. 12.
Applicability of the 3D vector summation technique to the analysis of an image stack from a 3D breast tissue model. 3D reconstructions of (a) a combination of SHG and TPEF signals, (b) and (c) orientation maps. (d), (e), and (f) show one frame of these stacks. (h) The two regions surrounding the epithelial structures marked in yellow are investigated, and the distribution histograms of both and within these two regions are shown in (g) and (i), respectively, as pointed by the arrows.
We calculated the fiber orientation distribution histograms of two regions within 35 from the cell surface, shown in yellow in Fig. 12(h). The two cell clusters exhibited either a rounded (Fig. 12(g)) or a more elongated (Fig. 12(i)) morphology. The fiber alignments within these two regions were shown from and distribution histograms (Fig. 12(g) and 12(i)), where the mean and values corresponding to the entire region volume were provided. In both cases, the histogram of shows a peak centered at approximately 90°, indicating that parallel sheets of collagen fibers dominated the alignment. This example illustrates that the 3D vector summation technique developed in this study is able to identify the organization of ECM, which contributes enormously to the properties and function of each organ and tissue [33,41].
4. Conclusions
Herein, we present a 3D weighted orientation vector summation technique for determination of the 3D orientation of collagen fibers. It provides voxel-wise orientation information with an accuracy of 4° to 5°, and is able to analyze a typical image stack with a size of voxels within 5. We present a thorough analysis of the orientation measurement error dependence on fiber thickness and density, analysis window size, axial-to-lateral sampling ratio and fiber orientation. These results suggest that to achieve optimal results, the applied window size should be between 2 and 3 fold greater than the fiber diameter, while the sampling of the z direction can be twice as coarse as the sampling without affecting significantly the accuracy of the algorithm. The actual error, , is independent of either the or the orientation, revealing that there are no restrictions on the fiber orientation while using this technique. A comparison between 2D and 3D techniques in analyzing fiber stacks reveals that the 3D technique is able to acquire the orientation with higher accuracy than the 2D technique. Compared with traditional analyses that produce directional statistics based on the total orientation distribution of the image, the capability of this technique to provide voxel-wise information enables the extraction of a wider variety of potentially important diagnostic biomarkers. The local directional variance of orientation values within specific regions of an image, such as the ECM surrounding an epithelial structure, is an example of such a biomarker. We have previously found the collagen fiber directional variance defined within 2D images to be a useful metric in understanding the matrix’s role in breast epithelial morphogenesis [33] and in localizing burn wounds [34]. By acquiring 3D orientation information, such assessments can be easily extended to the analysis of 3D image stacks, which provide a more thorough and accurate representation of cell-matrix interactions than 2D images. While we illustrate the potential of our approach in analyzing 3D collagen fiber images acquired using SHG, the technique is suitable for analysis of images with fiber-like features from a variety of modalities, such as confocal microscopy, structure illumination microscopy (SIM), and two-photon excitation fluorescence (TPEF) microscopy, etc. This rapid 3D fiber orientation analysis technique capable of voxel-wise definition with competitive accuracy is expected to enable the practical characterization of unique directional biomarkers for a number of applications seeking to understand the role of collagen organization during normal and/or diseased tissue development.
Acknowledgments
This research was supported by NIBIB/NIH Grant R01EB007542 to IG, American Cancer Society Research Scholar Grant RSG-09-174-01-CCE to IG, NIBIB/NIH Grant K99EB017723 to KPQ, Avon Grant 02-2009-093 and 02-2011-095 to AMS, NIEHS/NIH Grant R01ES08314 to AMS, and Tufts Collaborates Seed Grant Program.
References and links
- 1.Di Lullo G. A., Sweeney S. M., Körkkö J., Ala-Kokko L., San Antonio J. D., “Mapping the Ligand-binding Sites and Disease-associated Mutations on the Most Abundant Protein in the Human, Type I Collagen,” J. Biol. Chem. 277(6), 4223–4231 (2002). 10.1074/jbc.M110709200 [DOI] [PubMed] [Google Scholar]
- 2.Eyre D. R., “Collagen: Molecular Diversity in the Body’s Protein Scaffold,” Science 207(4437), 1315–1322 (1980). 10.1126/science.7355290 [DOI] [PubMed] [Google Scholar]
- 3.Fratzl P., Collagen: Structure and Mechanics (Springer, 2008). [Google Scholar]
- 4.Buehler M. J., “Nature designs tough collagen: Explaining the nanostructure of collagen fibrils,” Proc. Natl. Acad. Sci. U.S.A. 103(33), 12285–12290 (2006). 10.1073/pnas.0603216103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cunniffe G. M., OBrien F. J., “Collagen scaffolds for orthopedic regenerative medicine,” J. Miner. Met. Mater. Soc. 63(4), 66–73 (2011). 10.1007/s11837-011-0061-y [DOI] [Google Scholar]
- 6.Thomopoulos S., Marquez J. P., Weinberger B., Birman V., Genin G. M., “Collagen fiber orientation at the tendon to bone insertion and its influence on stress concentrations,” J. Biomech. 39(10), 1842–1851 (2006). 10.1016/j.jbiomech.2005.05.021 [DOI] [PubMed] [Google Scholar]
- 7.Provenzano P. P., Eliceiri K. W., Campbell J. M., Inman D. R., White J. G., Keely P. J., “Collagen reorganization at the tumor-stromal interface facilitates local invasion,” BMC Med. 4(1), 38 (2006). 10.1186/1741-7015-4-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hadian M., Corcoran B. M., Han R. I., Grossmann J. G., Bradshaw J. P., “Collagen organization in canine myxomatous mitral valve disease: an x-ray diffraction study,” Biophys. J. 93(7), 2472–2476 (2007). 10.1529/biophysj.107.107847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robinson P. S., Tranquillo R. T., “Planar biaxial behavior of fibrin-based tissue-engineered heart valve leaflets,” Tissue Eng. Part A 15(10), 2763–2772 (2009). 10.1089/ten.tea.2008.0426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bayan C., Levitt J. M., Miller E., Kaplan D., Georgakoudi I., “Fully automated, quantitative, noninvasive assessment of collagen fiber content and organization in thick collagen gels,” J. Appl. Phys. 105(10), 102042 (2009). 10.1063/1.3116626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ayres C. E., Jha B. S., Meredith H., Bowman J. R., Bowlin G. L., Henderson S. C., Simpson D. G., “Measuring fiber alignment in electrospun scaffolds: a user’s guide to the 2D fast Fourier transform approach,” J. Biomater. Sci. Polym. Ed. 19(5), 603–621 (2008). 10.1163/156856208784089643 [DOI] [PubMed] [Google Scholar]
- 12.Sander E. A., Barocas V. H., “Comparison of 2D fiber network orientation measurement methods,” J. Biomed. Mater. Res. A 88(2), 322–331 (2009). 10.1002/jbm.a.31847 [DOI] [PubMed] [Google Scholar]
- 13.Sivaguru M., Durgam S., Ambekar R., Luedtke D., Fried G., Stewart A., Toussaint K. C., Jr., “Quantitative analysis of collagen fiber organization in injured tendons using Fourier transform-second harmonic generation imaging,” Opt. Express 18(24), 24983–24993 (2010). 10.1364/OE.18.024983 [DOI] [PubMed] [Google Scholar]
- 14.Zhou Y., Zheng Y. P., “Estimation of muscle fiber orientation in ultrasound images using revoting hough transform (RVHT),” Ultrasound Med. Biol. 34(9), 1474–1481 (2008). 10.1016/j.ultrasmedbio.2008.02.009 [DOI] [PubMed] [Google Scholar]
- 15.Josso B., Burton D. R., Lalor M. J., “Texture orientation and anisotropy calculation by Fourier transform and principal component analysis,” Mech. Syst. Signal Process. 19(5), 1152–1161 (2005). 10.1016/j.ymssp.2004.07.005 [DOI] [Google Scholar]
- 16.Feng X. G., Milanfar P., “Multiscale principal components analysis for image local orientation estimation,” in Proceedings of 36th Asilomar conference on signals, systems and computers, (2002), vol. 1, pp. 478–482. [Google Scholar]
- 17.Yi W., Marshall S., “Principal component analysis in application to object orientation,” Geo-Spat. Inf. Sci. 3(3), 76–78 (2000). [Google Scholar]
- 18.Morishige N., Takagi Y., Chikama T., Takahara A., Nishida T., “Three-dimensional analysis of collagen lamellae in the anterior stroma of the human cornea visualized by second harmonic generation imaging microscopy,” Invest. Ophthalmol. Vis. Sci. 52(2), 911–915 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Pena A. M., Fabre A., Débarre D., Marchal-Somme J., Crestani B., Martin J. L., Beaurepaire E., Schanne-Klein M. C., “Three-dimensional investigation and scoring of extracellular matrix remodeling during lung fibrosis using multiphoton microscopy,” Microsc. Res. Tech. 70(2), 162–170 (2007). 10.1002/jemt.20400 [DOI] [PubMed] [Google Scholar]
- 20.Sun T. L., Liu Y., Sung M. C., Chen H. C., Yang C. H., Hovhannisyan V., Lin W. C., Jeng Y. M., Chen W. L., Chiou L. L., Huang G. T., Kim K. H., So P. T. C., Chen Y. F., Lee H. S., Dong C. Y., “Ex vivo imaging and quantification of liver fibrosis using second-harmonic generation microscopy,” J. Biomed. Opt. 15(3), 036002 (2010). 10.1117/1.3427146 [DOI] [PubMed] [Google Scholar]
- 21.Morishige N., Yamada N., Teranishi S., Chikama T., Nishida T., Takahara A., “Detection of subepithelial fibrosis associated with corneal stromal edema by second harmonic generation imaging microscopy,” Invest. Ophthalmol. Vis. Sci. 50(7), 3145–3150 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Mori S., van Zijl P. C. M., “Fiber tracking: principles and strategies - a technical review,” NMR Biomed. 15(7-8), 468–480 (2002). 10.1002/nbm.781 [DOI] [PubMed] [Google Scholar]
- 23.Rodriguez A., Ehlenberger D. B., Hof P. R., Wearne S. L., “Three-dimensional neuron tracing by voxel scooping,” J. Neurosci. Methods 184(1), 169–175 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu J., Rajwa B., Filmer D. L., Hoffmann C. M., Yuan B., Chiang C. S., Sturgis J., Robinson J. P., “Analysis of orientations of collagen fibers by novel fiber-tracking software,” Microsc. Microanal. 9(6), 574–580 (2003). 10.1017/S1431927603030277 [DOI] [PubMed] [Google Scholar]
- 25.Park H. J., Kubicki M., Westin C. F., Talos I. F., Brun A., Peiper S., Kikinis R., Jolesz F. A., McCarley R. W., Shenton M. E., “Method for combining information from white matter fiber tracking and gray matter parcellation,” AJNR Am. J. Neuroradiol. 25(8), 1318–1324 (2004). [PMC free article] [PubMed] [Google Scholar]
- 26.Friman O., Farnebäck G., Westin C. F., “A Bayesian approach for stochastic white matter tractography,” IEEE Trans. Med. Imaging 25(8), 965–978 (2006). 10.1109/TMI.2006.877093 [DOI] [PubMed] [Google Scholar]
- 27.Lau T. Y., Ambekar R., Toussaint K. C., “Quantification of collagen fiber organization using three-dimensional Fourier transform-second-harmonic generation imaging,” Opt. Express 20(19), 21821–21832 (2012). 10.1364/OE.20.021821 [DOI] [PubMed] [Google Scholar]
- 28.Sandau K., Ohser J., “The chord length transform and the segmentation of crossing fibres,” J. Microsc. 226(1), 43–53 (2007). 10.1111/j.1365-2966.2007.01748.x [DOI] [PubMed] [Google Scholar]
- 29.Robb K., Wirjadi O., Schladitz K., “Fiber orientation estimation from 3D image data: practical algorithms, visualization, and interpretation,” in Proceedings of IEEE 7th International Conference on Hybrid Intelligent Systems, (IEEE, 2007), pp. 320–325. 10.1109/HIS.2007.26 [DOI] [Google Scholar]
- 30.Wirjadi O., Schladitz K., Rack A., Breuel T., “Applications of anisotropic image filters for computing 2D and 3D-fiber orientations,” in Stereology and Image Analysis – Proceedings of the 10th European Congress of ISS, Capasso V., et al., ed. (2009), vol. 4, pp. 107–112. [Google Scholar]
- 31.Altendorf H., Decencière E., Jeulin D., De sa Peixoto P., Deniset-Besseau A., Angelini E., Mosser G., Schanne-Klein M.-C., “Imaging and 3D morphological analysis of collagen fibrils,” J. Microsc. 247(2), 161–175 (2012). 10.1111/j.1365-2818.2012.03629.x [DOI] [PubMed] [Google Scholar]
- 32.Quinn K. P., Georgakoudi I., “Rapid quantification of pixel-wise fiber orientation data in micrographs,” J. Biomed. Opt. 18(4), 046003 (2013). 10.1117/1.JBO.18.4.046003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barnes C., Speroni L., Quinn K. P., Montevil M., Saetzler K., Bode-Animashaun G., McKerr G., Georgakoudi I., Downes C. S., Sonnenschein C., Howard C. V., Soto A. M., “From single cells to tissues: interactions between the matrix and human breast cells in real time,” PLoS ONE 9(4), e93325 (2014). 10.1371/journal.pone.0093325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quinn K. P., Golberg A., Broelsch G. F., Khan S., Villiger M., Bouma B., Austen W. G., Jr, Sheridan R. L., Mihm M. C., Jr, Yarmush M. L., Georgakoudi I., “An automated image processing method to quantify collagen fibre organization within cutaneous scar tissue,” Exp. Dermatol. 24(1), 78–80 (2015). 10.1111/exd.12553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Golberg A., Khan S., Belov V., Quinn K. P., Albadawi H., Broelsch G. F., Watkins M. T., Georgakoudi I., Papisov M., Mihm M. C., Austen W. G., Yarmush M. L., “Skin rejuvenation with non-invasive pulsed elecdtric fields,” Sci. Rep. (to be published). [DOI] [PMC free article] [PubMed]
- 36.Rao A. R., Schunck B. G., “Computing oriented texture fields,” CVGIP Graph. Model. Im. 53(2), 157–185 (1991). 10.1016/1049-9652(91)90059-S [DOI] [Google Scholar]
- 37.Hong L., Wan Y. F., Jain A., “Fingerprint image enhancement: algorithm and performance evaluation,” IEEE T. Pattern Anal. 20(8), 777–789 (1998). [Google Scholar]
- 38.Dabbah M. A., Graham J., Malik R. A., Efron N., “Detecting and analyzing linear structures in biomedical images: a case study using corneal nerve fibers,” in: Dougherty G. ed. Medical Image Processing: Techniques and Applications (Springer, New York, 2011), Chap. 7. [Google Scholar]
- 39.Mardia K. V., Jupp P. E., Directional Statistics (John Wiley and Sons Ltd., ed. 2, 2000). [Google Scholar]
- 40.Speroni L., Whitt G. S., Xylas J., Quinn K. P., Jondeau-Cabaton A., Barnes C., Georgakoudi I., Sonnenschein C., Soto A. M., “Hormonal regulation of epithelial organization in a 3D breast tissue culture model,” Tissue Eng. Part C Methods 20(1), 42–51 (2014). 10.1089/ten.tec.2013.0054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosso F., Giordano A., Barbarisi M., Barbarisi A., “From cell-ECM interactions to tissue engineering,” J. Cell. Physiol. 199(2), 174–180 (2004). 10.1002/jcp.10471 [DOI] [PubMed] [Google Scholar]