Abstract
In this issue of Developmental Cell, Drake and colleagues (2014) report that Ras signaling results in Dicer phosphorylation, which induces its nuclear localization and modulates its function. This regulatory strategy, conserved in mammals, allows dynamic control of miRNA function required for C. elegans germline development and oogenesis.
While the initially proposed schematics of miRNA biogenesis implied an inexorable and linear pathway, we now understand there to exist (1) multiple alternative ribonucleolytic strategies that generate functional miRNAs (Yang and Lai, 2011), and (2) diverse intervention points to regulate small RNA biogenesis factors and RNA substrates (Ha and Kim, 2014). However, most of our understanding comes from cultured cells and in vitro systems, and there is still relatively little information on how such regulation is implemented in developmental or physiological contexts. In this issue of Developmental Cell, Drake et al., (2014) links Ras signaling to inhibition of miRNA biogenesis, via ERK-mediated phosphorylation of Dicer and induction of its nuclear translocation. This regulatory mechanism is deployed dynamically during the C. elegans oogenesis-to-embryogenesis transition.
Extracellular stimuli activate the Ras pathway via a receptor tyrosine kinase (RTK), which in turn activates the small GTPase Ras and initiates a kinase cascade that includes MAP Kinase (MPK-1 in C. elegans, ERK in mammals). The RTK/Ras/MAPK pathway is not only a conserved developmental cell-signaling system, but is also deregulated in a substantial proportion of human cancers. Previous work showed that Ras/ERK signaling and Dicer regulate C. elegans oogenesis, and identified Dicer as a putative ERK substrate (Arur et al., 2009). Building upon this, the authors first showed that murine ERK2 can phosphorylate C. elegans DCR-1 in vitro, via two well-conserved Ser residues, S1705 (in the RNase IIIb domain) and S1833 (in the dsRNA-binding domain: dsRBD). Using antibodies specific for phospho-DCR-1, they visualized phosphorylated Dicer in C. elegans and showed its dependency on MPK-1. Interestingly, phosphorylated Dicer localizes to the nucleus and is present during most of oogenesis, but is rapidly lost from the terminal oocyte shortly before fertilization. Using GFP-fused WT-DCR-1 and DCR-1 phosphomimetic (S>E) and non-phosphorylatable (S>A) variants, the authors showed that phosphorylation of DCR-1 is necessary and sufficient for nuclear localization (Figure 1).
Figure 1. Stimulus-induced translocation of RNase III enzymes.
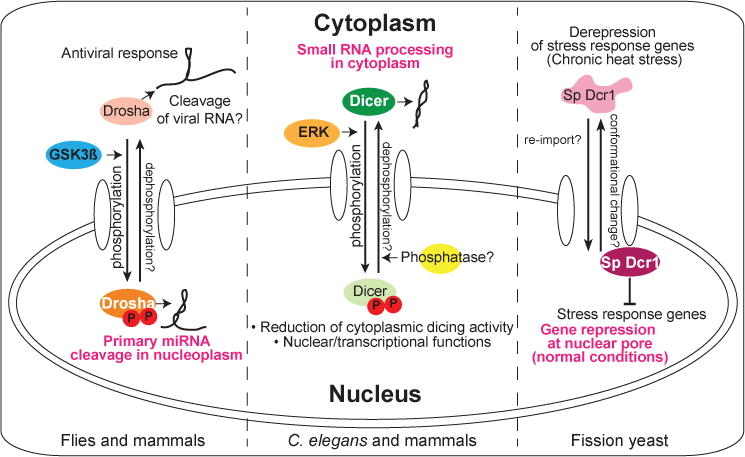
The primary locations of RNase III enzymes under normal conditions or in most cell types are designated by white lettering. (Left) Drosha is normally phosphorylated (by GSK3ß) for nuclear localization and cleaves primary miRNA. Viral infection induces its cytoplasmic accumulation, where it has antiviral activity. (Middle) Dicer is generally cytoplasmically localized to process small RNA precursors. During C. elegans oogenesis or in FGF-stimulated mammalian cells, activated MAPK/ERK phosphorylates Dicer for nuclear translocation, leading to reduced dicing activity in the cytoplasm. Nuclear functions of Dicer have also been proposed. (Right) In fission yeast, stress response genes are suppressed by Dicer (Sp Dcr1) via cotranscriptional silencing at nuclear pores. Chronic heat shock triggers Dcr1 translocation to the cytoplasm, resulting in gene derepression. Current studies of these examples have focused on “one-way” trafficking, but in principle, all of these translocations might be reversible (as designated by question marks).
Genetic mosaic analyses demonstrated that the oogenesis and ovulation defects of dcr-1(0) mutants were attributable to somatic, rather than germline, function of DCR-1. Phosphomimetic DCR-1 variants (single and double mutations) complemented oogenesis defects in dcr-1(0) mutants, but resulting oocytes died during embryogenesis, similar to germline dcr-1(0) mutants. This suggests that DCR-1 needs to be dephosphorylated for embryogenesis. On the other hand, expression of DCR-1S1833A largely complemented the oogenesis and embryogenesis phenotypes in dcr-1(0). However, levels of activated MPK-1 were elevated in the loop region of the germline, indicating that phosphorylation of DCR-1 leads to inhibition of MPK-1 activity. The other variant DCR-1S1705A failed to complement developmental phenotypes in dcr-1(0), and even showed semi-dominant effects in wild-type, suggesting that phosphorylation of S1705 is necessary for Dicer activity inhibition and normal oogenesis progression.
Since Dicer is primarily known as a cytoplasmic miRNA processing enzyme, one may hypothesize that nuclear translocation should impact its activity. However, the constraints of C. elegans as a biochemical system limit the conclusions reached presently. Small RNA sequencing analysis showed no dramatic changes in general miRNA abundance in dcr-1(0) mutant, due to maternal contributions of DCR-1 and/or miRNAs. Still, an informative observation was that dcr-1(0) libraries contained “pseudo-miRNAs”, which carry 2–5 nt extensions to either ends of reference mature species. The authors speculate these represent products of an alternative pathway when canonical dicing was unavailable. “Pseudo-miRNAs” accumulated in dcr-1(0) mutants rescued with either phosphomimetic Dicer or with non-phosphorylatable S1705A Dicer, suggesting that these mutations alter dicing activity. Nevertheless, the meaning of the pseudo-miRNA readout is not concretely established, and it is not fully evident why some phosphomimetic and non-phosphorylatable variants should be similarly deficient, instead of acting oppositely. Therefore, the precise effects of phosphorylation and/or nuclear localization on Dicer activity require further insights from in vitro dicing assays, perhaps using purified proteins or from appropriate subcellular fractions.
The study of Drake and colleagues (2014) raises many interesting questions for understanding molecular machineries involved in C. elegans development. For example, a negative feedback loop between DCR-1 and MPK-1 is inferred and remains to be understood at the mechanistic level. Another notable direction regards the reversibility of Dicer phosphorylation during embryogenesis. If this is true, there may exist a phosphatase whose function is relevant to both the Ras and miRNA pathways.
Going beyond worms, the authors present evidence that the MAPK-mediated switch in Dicer localization is conserved in vertebrates. Murine ERK2 can phosphorylate human Dicer in vitro at the conserved serine residues. Conveniently, the phospho-specific worm Dicer antibody also recognizes modified mammalian Dicer. The authors used this reagent to demonstrate nuclear phospho-Dicer signals in human cells as well as in mouse tissues. Therefore, the phospho-specific Dicer antibody may provide both immunohistochemical and biochemical access to a specific population of modified Dicer.
Stimulus-induced relocalization of RNase III enzymes may represent an emerging theme (Figure 1). In fission yeast, Dcr1 is normally nuclearly localized, where it represses stress response genes via cotranscriptional gene silencing. Chronic heat shock induces cytoplasmic translocation of Dcr1, accompanied by derepression of stress response genes (Woolcock et al., 2012). In animals, the RNase III enzyme Drosha mediates nuclear primary miRNA cleavage, and phosphorylation of Drosha by Glycogen Synthase Kinase 3ß (GSK3ß) at two serine residues is important for its nuclear localization (Tang et al., 2011). This mechanism may permit dynamic control of Drosha localization. For example, cytoplasmic accumulation of Drosha was observed following viral infection, and phosphomimetic mutations at the serine residues abolished infection-induced translocation. As cytoplasmic Drosha exhibits antiviral activity potentially via direct cleavage of viral RNAs and/or regulation of cellular RNAs, translocation of Drosha may switch between miRNA processing and antiviral defense systems (Shapiro et al., 2014).
The evolutionarily conserved, signaling-induced translocation of Dicer raises another question: does nuclear Dicer function apart from its major role in the cytoplasm? More analyses are needed to address this, especially as nuclear roles of RNAi machineries in higher animals remain controversial. However, various studies report detection of core “cytoplasmic” RNAi factors in mammalian nuclei, at least under certain conditions (Schraivogel and Meister, 2014). For example, animal Dicers are proposed to play various chromatin-related roles during transcription, termination and/or clearance of nuclear dsRNA (Schraivogel and Meister, 2014; Skourti-Stathaki et al., 2014; White et al., 2014). The study by Drake et al. (2014) reporting dynamic relocalization of Dicer protein in vivo encourages careful examination of potential nuclear functions of RNAi factors in perhaps unsuspected settings.
References
- Arur S, Ohmachi M, Nayak S, Hayes M, Miranda A, Hay A, Golden A, Schedl T. PNAS. 2009;106:4776–4781. doi: 10.1073/pnas.0812285106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake M, Furuta T, Man KS, Gonzalez G, Liu B, Kalia A, Ladbury J, Fire AZ, Skeath JB, Arur S. Developmental Cell. 2014 doi: 10.1016/j.devcel.2014.11.004. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Kim VN. Nature Reviews Molecular Cell Biology. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- Schraivogel D, Meister G. Trends in Biochemical Sciences. 2014;39:420–431. doi: 10.1016/j.tibs.2014.07.004. [DOI] [PubMed] [Google Scholar]
- Shapiro JS, Schmid S, Aguado LC, Sabin LR, Yasunaga A, Shim JV, Sachs D, Cherry S, tenOever BR. PNAS. 2014;111:7108–7113. doi: 10.1073/pnas.1319635111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Kamieniarz-Gdula K, Proudfoot NJ. Nature. 2014 doi: 10.1038/nature13787. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Li M, Tucker L, Ramratnam B. PloS One. 2011;6:e20391. doi: 10.1371/journal.pone.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E, Schlackow M, Kamieniarz-Gdula K, Proudfoot NJ, Gullerova M. Nature Structural & Molecular Biology. 2014;21:552–559. doi: 10.1038/nsmb.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolcock KJ, Stunnenberg R, Gaidatzis D, Hotz HR, Emmerth S, Barraud P, Buhler M. Genes & Development. 2012;26:683–692. doi: 10.1101/gad.186866.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Lai EC. Molecular Cell. 2011;43:892–903. doi: 10.1016/j.molcel.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]


