Abstract
Endopeptidase classification based on catalytic mechanism and evolutionary history has proven to be invaluable to the study of proteolytic enzymes. Such general mechanistic- and evolutionary- based groupings have launched experimental investigations, because knowledge gained for one family member tends to apply to the other closely related enzymes. The serine endopeptidases represent one of the most abundant and diverse groups, with their apparently successful proteolytic mechanism having arisen independently many times throughout evolution, giving rise to the well-studied soluble chemotrypsins and subtilisins, among many others. A large and diverse family of polytopic transmembrane proteins known as rhomboids has also evolved the serine protease mechanism. While the spatial structure, mechanism, and biochemical function of this family as intramembrane proteases has been established, the cellular roles of these enzymes as well as their natural substrates remain largely undetermined. While the evolutionary history of rhomboid proteases has been debated, sorting out the relationships among current day representatives should provide a solid basis for narrowing the knowledge gap between their biochemical and cellular functions. Indeed, some functional characteristics of rhomboid proteases can be gleaned from their evolutionary relationships. Finally, a specific case where phylogenetic profile analysis has identified proteins that contain a C-terminal processing motif (GlyGly-Cterm) as co-occurring with a set of bacterial rhomboid proteases provides an example of potential target identification through bioinformatics.
Keywords: Rhomboid protease, intramembrane proteolysis, classification, evolution, structure, bioinformatics
Graphical Abstract
1 Introduction
Proteolytic enzymes were initially classified over fifty years ago into four types based on their chemical mechanism of catalysis: serine, aspartic, metallo-, and cysteine [1]. With increasing availability of sequence and structure information in the early 1990’s, evolutionary grouping of peptidases became possible [2] giving rise to the protease classification and nomenclature scheme implemented in the MEROPS database [3]. The hierarchical classification within MEROPS includes both the mechanistic type indicated by a letter (i.e. S, D, M or C for the four classical mechanistic types) followed by a letter denoting a clan of evolutionarily related families, which are numbered. Although the current database includes several additional mechanistic types (i.e. Glutamic, Asparagine, Threonine, and mixed), the four classic types encompass the majority of known peptidases. Considering the multiple defined clans within each type, peptidases appear to have arisen multiple independent times converging on similar mechanistic activities. Thus, the four classical chemical types can successfully accomplish proteolysis of various different peptide substrates within numerous and diverse protein architectures.
A new paradigm of proteolysis followed the discovery of regulated intramembrane proteolysis (RIP) by the transmembrane metalloprotease S2P[4]. S2P represented the first polytopic membrane protein that could catalyze cleavage of a transmembrane substrate within the lipid bilayer, releasing a soluble cleavage product into the cytoplasm. Given the hydrolytic nature of peptide bond cleavage, such an environment was a surprising addition to the repertoire of successful proteolytic mechanisms achieved in nature. Nevertheless, identification of additional intramembrane protease activities of the aspartic type (signal peptide peptidase [5] and presenelin [6]) and the serine type (rhomboid [7]) closely followed the discovery of S2P-mediated RIP.
Despite their differing mechanistic types, the intramembrane proteases belonging to the rhomboid (serine), S2P (metallo), and signal peptide peptidase/presenelin (aspartic) share some common features. Each of the intramembrane protease families possesses representatives throughout the three kingdoms of life [8–10], potentially representing ancient enzymes that arose with the creation of the membrane bilayer. Each family 1) cleaves transmembrane substrates within the lipid membrane bilayer and 2) retains conserved catalytic machinery within a core polytopic membrane protein architecture. This machinery is marked by sequence motifs that extend to the soluble proteases within the same mechanistic type. The short motifs could have arisen independently within the transmembrane folds, representing convergent evolution of proteolytic activity. Alternatively, the intramembrane proteases could have acquired hydrophobic components surrounding ancestral motifs. Such a scenario seems less likely, as is would require a fold with considerable plasticity that could switch easily between transmembrane and soluble states, with few substitutions. This review will focus on combining rhomboid protease evolution with its mechanism and structure and highlighting the resulting implications for bioinformatic analysis of function.
2 Rhombiod Classification and Evolution
Rhomboid proteases are present almost ubiquitously in all forms of life and are suggested to represent the most widely distributed membrane proteins in nature [11]. However, defining and classifying members of the rhomboid protease family has been challenging due to the existence of numerous paralogous groups that display relatively low sequence identity, the widespread occurrence of inactive enzymes that lack key active site signatures, and the presence of additional TMHs and soluble domains that decorate the universally present 6TMH rhomboid protease catalytic core [10–14]. Accordingly, the evolutionary history of rhomboid proteases has been difficult to assess, and their origin has been debated [11, 14]. While the near-universal presence of rhomboid proteases in all three kingdoms of life suggests that the family existed in the last universal common ancestor (LUCA), rigorous phylogenetic analysis has challenged this expectation. Instead, an alternate scenario of rhomboid protease origination in bacteria and acquisition by archaea and eukaryotes through multiple ancient horizontal gene transfers (HGT) was proposed [11]. Subsequent to this work, evolutionary analyses of rhomboid proteases has concentrated on eukaryotic members [14], with some independent considerations of gene expansions in apicomplexan parasites [12] and in plants [15]. While only one study has focused on bacteria, limiting analysis to mycobacterial species [13].
2.1 Phylogenetic Analysis Leads to Conflicting Views of Evolutionary History
Phylogenetic analysis of the conserved six-TMH rhomboid protease core from sequence representatives among the three branches of life revealed a complex tree topology with two major eukaryotic subfamilies (RHO and PARL) positioned among different prokaryotic branches. This unexpected topology lead to an interpretation that the rhomboid proteases had not been inherited from the last universal common ancestor (LUCA), as would be expected from such widespread existence in nature. A proposal that eukaryotes acquired rhomboid proteases through multiple ancient horizontal gene transfers from bacteria emerged [11]. Alternatively, phylogenetic analysis using a more complete eukaryotic subset of rhomboid protease sequences limited to the TMHs containing signature motifs (Loop1, TMH2, TMH4, and TMH6) produced a slightly different grouping, where the yeast secretory pathway rhomboid protease (Rbd2) partitioned with the secretase-type rhomboid proteases (previously termed RHO) as opposed to the mitochondrial PARL sequences. The eukaryotic tree included a distinct clade of previously missed inactive iRhoms and split the RHO secretases into more groups, including one speculated to represent an ancient rhomboid protease form (denoted as secretase-B class containing human RHBDL4 and yeast Rbd2) [14], challenging the multiple-HGT interpretation of rhomboid protease evolution.
Rhomboid proteases have also expanded in apicomplexan parasites and in land plants, and their phylogenetics have been considered independently by several groups [12, 14–16]. Apicomplexan parasites contain multiple diverse copies of rhomboid proteases, including a single widely distributed PARL-type protease (ROM6) that localizes to the mitochondria and has duplicated in Plasmodium (ROM9) [12, 14]. The remaining rhomboid protease members segregate with ROM4/ROM5 forming a close group that separates from the less widely distributed ROM7 and ROM1-3. These non-PARL rhomboid proteases were described as being unique to apicomplexans, possibly functioning in parasite specific processes [12]. However, the question remains as to whether these families are indeed distinct from other eukaryotic groups, originating independently in parasites, or they have diverged so much from the eukaryotic RHOs that the family groupings are not evident. The fact that the universal parasite ROM4/5 group localizes to the plasma membrane [12] and displays a 6+1 TMD topology similar to the other eukaryotic secretory RHOs suggests the latter. The ROM4 protease from T. gondi (TgROM4) has been experimentally characterized as cleaving several plasma membrane component adhesins (TgMIC2, TgAMA1, and possibly TgMIC8) that facilitate host cell invasion [17].
Phylogenetic analysis of plant rhomboid proteases was limited to those sequences that retained all catalytic residues and were presumed to be active. The active plant rhomboid proteases grouped into two classes: the secretory RHOs (AtRBL1-AtRBL7) and a more divergent class (AtRBL10-AtRBL15) that included mitochondrial PARL (AtRBL12) [15]. Consistent with this grouping, the plant RHO-like AtRBL1 and AtRBL2 that are most similar to Drosophila rhomboid-1 (Rho-1) were localized to the Golgi [18]. The divergent plant sequences were further subdivided in another phylogenetic study [14] that distinguished the PARL-like sequences from the rest. Like the other eukaryotic PARL rhomboid proteases, plant AtRBL12 localizes to mitochondria [15]. However, AtRBL12 lacks a predicted N-terminal TMH that is present in all the other eukaryotic PARL-like sequence 1+6 TMD topologies and does not appear to cleave the yeast PARL substrates. Plant AtRBL10 and AtRBL11 both localized to the chloroplast, although their physiological substrates remain unknown [15, 19].
Most sequenced bacterial genomes contain at least one rhomboid protease, with many species possessing multiple copies. A study of rhomboid protease sequences from sequenced mycobacterial genomes revealed two distinct groups represented by RV0110 and RV1337 from Mycobacteria tuberculosis H37Rv. Phylogenetic analysis confirmed the two paralogous groups, but could not distinguish their progenitor[13]. The RV1337 orthologs displayed a universal presence among mycobacterial species and retained similar genomic neighborhood organizations, while the RV0110 orthologs appeared to be less evolutionarily stable and were lost in some species. The RV0110 genome neighborhoods were not preserved, but retained conservations that mirrored the mycobacterial species tree. These orthologs clustered with eukaryotic rhomboid proteases, and displayed 6+1 TMD topology similar to that of the eukaryotic secretase-type RHO subfamily, perhaps providing an example of a bacterial progenitor to the eukaryotic rhomboid proteases as suggested in [11]. Alternatively, this bacterial group may represent an ancient family of rhomboid proteases present in the LUCA. The presence of intact rhomboid protease catalytic signatures in the mycobacterial members suggests the two orthologous groups cleave transmembrane proteins. The RV0110 orthologs reside in close proximity to the experimentally characterized P. stuartii AarA sequence as well as the GlpG structure representatives, and all retain the eukaryotic-like 6+1 TMD topology (although structures are limited to the 6TMH core). AarA has been shown to cleave the first seven residues of TatA, activating the P. stuartii twin-arginine translocase (Tat) protein secretion pathway. However, these residues are unique to the P. stuartii TatA substrate, suggesting that despite its similarity, RV0110 cleaves another transmembrane protein or proteins.
2.2 Network-Based Clustering Suggests a Possible Alternate Rhomboid Protease History
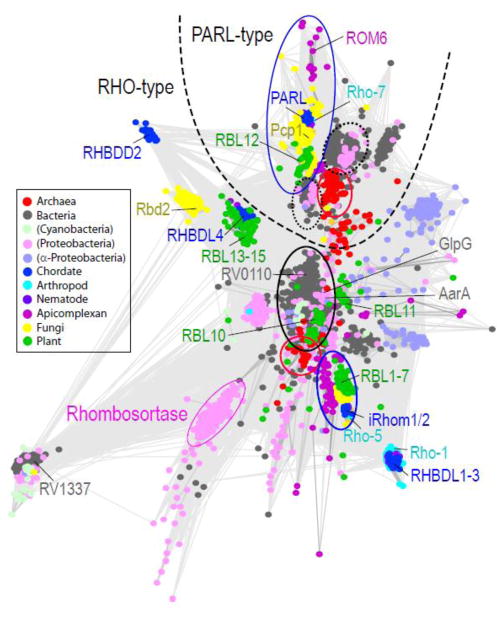
Interpretations of rhomboid protease phylogenetic trees have yielded differing views on their evolutionary history. While network-based grouping does not implicitly consider evolutionary models, the method allows analysis of the highly divergent rhomboid protease sequences that pose a challenge for multiple sequence alignment and phylogenetic tree reconstructon. A more complete network-based grouping of currently known rhomboid sequences from all kingdoms of life reveals a similar complex topology as previous studies (Figure 1). As noted with the mycobacterial orthologs, rhomboid proteases have expanded in some bacteria and tend to form various distinct groups. Notably, rhomboid proteases from proteobacteria form several groups, with some having expanded in a class-specific manner. For example, the alpha-proteobacteria (slate circles) rhomboid proteases belong to 3 groups comprised of diverse species, including both groups defined by the mycobacterial orthologs and a group that clusters near eukaryotic PARL. Representatives from alpha-proteobacteria also form class specific groups that probably arose from more recent duplication events. One of the groups containing diverse bacterial species forms a central cluster within the various eukaryotic RHOs (circled in black, sequences listed in Supplemental Table), any may represent an ancient form of the protease. Both the eukaryotic RHOs and the closely grouping bacterial sequences retain the rhomboid protease 6+1 TMD topology. While mycobacterial species do not possess a PARL-like rhomboid protease, two smaller bacterial groups with diverse species representatives cluster centrally near the eukaryotic PARLs (circled in black dots, sequences listed in Supplemental Table), with a third, more divergent group formed by bacterial sequences lacking key active site residues. Unlike the eukaryotic PARL sequences, which have acquired an additional N-terminal TMH, the PARL-like bacterial sequences are limited to the 6TMH rhomboid protease core.
Figure 1. Network-based rhomboid protease clusters.
Network-based CLANS [47] clustering of rhomboids defined by PFAM [48]. Divergent sequences such as derlins were excluded and bacterial representatives were filtered at 80% identity. The resulting 3345 nodes represent individual rhomboid sequences. Nodes are connected by lines that stand for pairwise BLAST E-values (cutoff 1e-7) and are colored according to taxonomy: chordate (blue), arthropod (cyan), nematode (purple), apicomplexan (magenta), fungi (yellow), plant (green), archaea (red), cyanobacteria (light green), proteobacteria (pink), alpha-proteobacteria (slate), and all other bacteria (gray); key rhomboid protease representatives are labeled to the side of their clusters and colored as above. A dashed line separates two major rhomboid protease groupings into PARL-type and RHO-type. Each broad group includes central clusters that segregate according to taxonomy: bacteria circled in black or dotted black, archaea circled in red, and eukaryota circled in blue. Sequences that fall within these centralized groups are listed in the Supplemental table.
Two smaller groupings of archaeal sequences mirror this class-based distribution (circled in red, sequences listed in Supplemental Table), perhaps supporting an alternative scenario to the multiple HGT hypothesis of rhomboid protease evolution [11]. Both the RHO-like and the PARL-like archaeal groups include sequences that distribute according to two phyla: Euryarchaeota and Crenarchaeota. An additional PARL-like group is formed by Halobacteria class specific duplications. The archaeal sequences all display a 6TMH rhomboid protease core topology, and distribute into either the PARL-like or the RHO-like groups, but not both. The RHO-like group includes most of the Crenarchaeota, as well as two of euryarchaeotal genus: Thermococcus and Pyrococcus; while the PARL-like group includes two Crenarchaeota from the genus Vulcanisaeta and many of the Euryarchaeota.
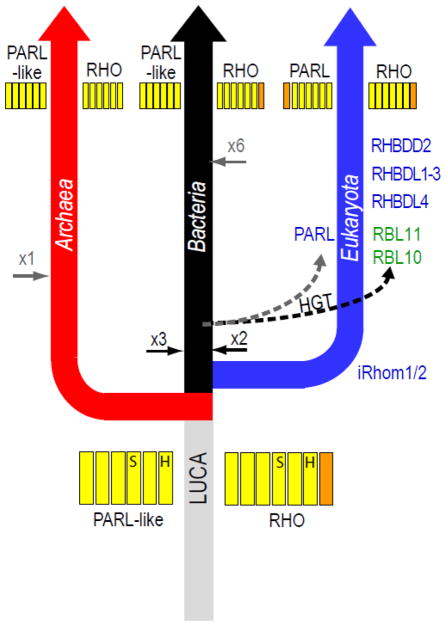
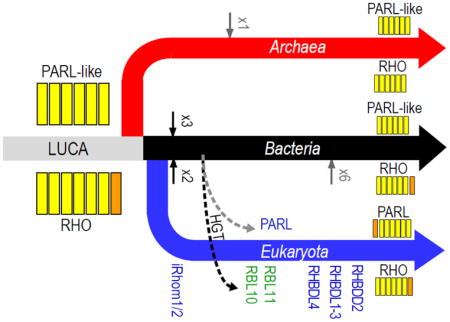
An alternative scenario for rhomboid protease evolution is suggested by clustering (Figure 2), where the LUCA may have contained an ancient rhomboid protease duplication. The ancient PARL-like rhomboid protease was probably limited to the 6TMD core that is reflected in present day bacterial and archaeal sequences, with the eukaryotic PARL sequences acquiring an N-terminal TMD. The ancient RHO-like rhomboid protease could have possessed either the 6+1 TMD topology reflected in present day bacterial and eukaryotic secretase A sequences (with loss of the C-terminal TMH in archaea) or the 6TMD core topology reflected in present day archaeal rhomboid proteases (with a less parsimonious independent acquisition of C-terminal TMH in eukaryotes and bacteria). The expansion of rhomboid proteases into diverse groupings probably arose from a combination of duplication events occurring early after branching, those occurring later among various classes of bacteria, or those occurring all along the eukaryotic clade. Consistent with the theory that chloroplasts originated in plants from bacterial endosymbionts, plant rhomboid proteases functioning in these organelles cluster within (RBL10) or near (RBL11) the central bacterial RHO-like sequences. While the PARL-like sequences from archaea and bacteria, and the PARL sequences from eukaryotes form distinct groups, the eukaryotic and bacterial sequences tend to display greater similarity.
Figure 2. Schematic cladogram illustrates possible scenario for rhomboid protease evolution.
Colors correspond to kingdoms outlined in A. The LUCA may have contained a rhomboid protease duplication: including ancient RHO-type and PARL-type forms. Both ancient forms duplicated early in the bacterial branch (black arrows), while the RHO-type expanded again later in several bacterial classes (gray arrow). The PARL-type duplicated early in the archaeal branch (black arrow), and the RHO-type expanded in eukaryotes (Names appear in order of expansion) and was acquired by plant (RBL10 and RBL11) through bacterial chloroplast-forming endosymbionts. Stronger similarity between the bacterial and eukaryotic PARL sequences might support its origination from bacterial mitochondria-forming endosymbionts.
These clusters suggest that the ancient RHO class is represented by chordate iRhoms, which have lost their ability to catalyze cleavage. Although this group has not previously been considered as ancient, it includes members from all eukaryotic subkingdoms that segregate according to the species tree, and the representative fungi and plant sequences have mostly retained their catalytic residues. Potentially, the chordate-specific iRhom inactivation could have occurred upon duplication and subsequent divergence into the next closest chordate group containing human RHBDL1-3. The relatively close proximity of RHBDL1 and RHBDF1 (iRhom1) on the human telomeric region of chromosome 16 (16p13.3) supports this notion. Regardless of the questions that remain concerning this rhomboid protease evolutionary scenario, the relative diversity of species with rhomboid protease representatives present in the RHO and PARL groups supports the presence of one (RHO-type) or both (RHO-type and PARL-type) in the LUCA.
3 Rhomboid Protease Structure and Mechanism
Dozens of structures of the bacterial rhomboid protease GlpG have been solved, including several mutants. These studies have revealed the transmembrane protein architecture, the nature of the active site, and the mechanism of substrate binding typified for the 6TMH catalytic core of the rhomboid protease serine proteases [20–27]. The GlpG structure reveals the transmembrane core to adopt a general up and down topology running perpendicular to the membrane, establishing the position of both termini facing the cytosol. The loop (L1) connecting TMH1 and TMH2 forms an unusual α-helical hairpin running perpendicular to the TMH core that is partially submerged within the outer membrane leaflet (Figure 3A, gray cartoon). The characteristic active site serine resides at the N-terminus of a shortened TMH4 that starts deep within the membrane. The S forms a catalytic dyad with a neighboring H from TMH6 (Figure 3A, black sticks). Another conserved motif in TMH6 (GxxxG) allows tight packing between the two TMHs that harbor the catalytic S-H dyad. Notably, the active site is accessible to water required for nucleophilic cleavage through a cleft facing the extracellular region that is capped by loop L5 [20, 21, 25–27]. Several atoms have been proposed to contribute to an oxyanion hole that stabilizes the developing negative charge on the carbonyl oxygen of the scissile bond during catalysis, including the main chain amide of the catalytic S and the side chain amides of two conserved TMH2 sidechains from the motif HxxxN (Figure 3A, shown in gray stick). Mutagenesis data combined with the inhibitor-bound structures tend to support the notion that the side chain amide of the catalytic serine provides the major oxyanion role, with some variable redundancy in the contribution of the surrounding conserved H and N residues.
Figure 3. Rhomboid protease structure and mechanism suggests convergence of serine protease activity.
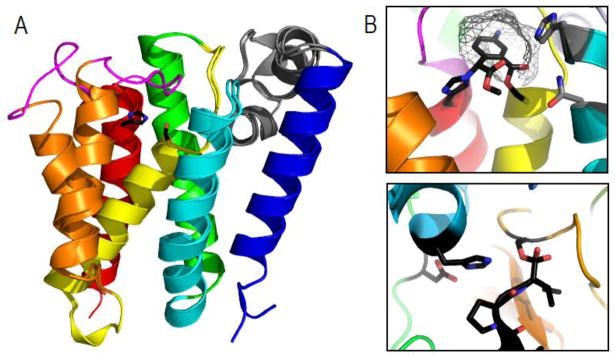
A) GlpG 6TMH transmembrane rhomboid protease core (TMHs colored in rainbow) forms open (PDB ID: 2nrf_A) and closed (PDB ID: 2xov) states dictated by movement of TMH5 (orange) and the loop5 cap (magenta) that covers the active site containing a serine-histidine catalytic dyad (black sticks). An unusual helical loop L1 (gray) is partially submerged in the membrane bilayer. B) Upper panel illustrates a zoom of the GlpG active site (PDB ID: 2oxw) covalently modified by an inhibitor (black) that highlights proximity of catalytic dyad (black stick) to residues that may assist oxyanion formation during catalysis (gray sticks), the presumed S1 pocket (gray wireframe), and the relative orientation of the serine nucleophile with respect to the inhibitor suggesting a si-face attack. C) A zoom of a chymotrypsin active site (PDB ID: 1haz) covalently modified by an inhibitor (black) highlights the catalytic triad (black sticks) with its nucleophilic serine on the opposite face of the inhibitor.
GlpG structures from E. coli as well as H. influenzae suggest a gating mechanism whereby TMH5 moves to allow lateral access to the active site by the transmembrane-spanning substrate [20, 21, 24–27]. Figure 3A illustrates GlpG structures adopting open and closed conformations. Movement of Loop5 (magenta) and TMH5 (orange) are thought to allow substrate access to the active site [21]. Although no structures exist with substrate bound, inhibitor complexes have given insight to the substrate binding mode and catalysis [23, 27]. A presumed S1 subsite cavity formed in the presence of inhibitor [23] is consistent with the protease preference for relatively small residues (A, C, S, or G) in the substrate P1 subsite (the residue N-terminal to the scissile bond) [28], while inhibitor binding to the S’ side opens the TMH5 helix and loop5 gating cap [27]. The strongest preference for rhomboid protease substrate specificity arises from the P1 subsite, which allows binding of several different small residues. Thus instead of a traditional sequence recognition motif, rhomboid protease specificity is driven by the propensity of the transmembrane helix substrate to destabilize [29]. This relative lack of substrate specificity may help explain the observation of rhomboid protease loss upon duplication that has occurred during evolution (for example in Mycobacterial orthologs [13]).
3.1 Relationship to soluble serine proteases
The rhomboid proteases form a separate clan in the MEROPS database[3], which represents the broadest level of homology in the classification scheme. When compared to other serine protease clan representatives, the rhomboid proteases differ in both structure and catalytic mechanism (Table 1). Not only does the rhomboid protease GlpG structure represent the only membrane protein among serine proteases, but none of the other serine protease clans exhibit an all-α fold. The closest serine protease is classified as a multidomain protein, with the active site SxxK located within an N-terminal helical bundle domain that forms a completely different topology than the GlpG helical arrangement. Similarly, none of the MEROPS-defined serine protease clans catalyze cleavage with a similar S-H dyad as found in the rhomboid protease. Most of the soluble serine proteases that catalyze cleavage with a dyad use S-K (i.e. clan SF, SJ, and SO use S-K, while clan SR uses K-S). Two unusual cases of catalytic S/H dyads do include the cytomegalovirus protease serine protease, whose catalytic S-H-H triad of can lose its bridging H to form a catalytic S-H dyad with relatively little effect on catalysis, and the autoprocessing protease activity of nucleoporin Nup98, whose catalytic dyad originates from the motif HxS. Finally, the assumed substrate binding mode of rhomboid proteases establishes cleavage of the scissile bond on the si-face [21], as opposed to the alternate approach on the re-face that is typically observed in serine proteases with known structure (except for bacterial signal peptidase). These structural and mechanistic properties adopted by rhomboid proteases differ significantly from their soluble serine protease counterparts, suggesting independent convergent evolution of rhomboid protease peptide bond cleavage.
Table 1.
MEROPS Serine Protease Clans
| MEROPS Clan | Family Number | Structure Example | SCOP Class | SCOP Fold | Catalytic Motif |
|---|---|---|---|---|---|
| SB | 2 | 1scn | α/β | Subtilisin-like | D-H-S |
| SC | 6 | 1qtr | α/β | α/β hydrolase | S-H-D |
| SE | 3 | 1es4 | multi | β-lactamase-like | SxxK |
| SF | 2 | 1umu | all-β | LexA/Signal peptidase | S-K |
| SH | 5 | 1lay | all-β | Herpes virus serine proteinase, assemblin | H-S-H |
| SJ | 3 | 1rr9 | α+β | Ribosomal protein S5 domain 2-like | S-K |
| SK | 3 | 1tyf | α/β | ClpP/crotonase | variable |
| SO | 1 | 3gw6 | na | na | S-K |
| SP | 1 | 1ko6 | all-β | C-terminal autoproteolytic domain of nucleoporin nup98 | HxS |
| SR | 1 | 1lct | α/β | Periplasmic binding protein-like II | K-S |
| SS | 1 | 1zrs | α/β | Flavodoxin-like; “swivelling” β/β/α domain | S-E-H |
| ST | 1 | 2ic8 | membrane | rhomboid-like | S-H |
| PA | 14 | 1bra | all-β | Trypsin-like | H-D-S |
| PB | 2 | 1pnk | α+β | Ntn hydrolase-like | S |
| PC | 1 | 1fye | α/β | Flavodoxin-like | S-H-E |
4 Functional Implications of Rhomboid Protease Evolution
Despite differing opinions about the ancient history of rhomboid proteases, phylogenetic studies tend to support a recurring theme: eukaryotic rhomboid proteases segregate broadly into PARL and RHO-type proteases according to their membrane localization. For the RHO-type rhomboid proteases that have duplicated multiple times throughout evolution, the functional implications of distinctions between various families remain difficult to assess given the relative lack of existing experimental information about their various biological substrates and cellular pathways. Despite this general lack of functional detail, some generalizations can be drawn from various rhomboid protease family relationships. In fact, phylogenetic profiling-based bioinformatics method has already led to a testable functional hypothesis about one distinct family of rhomboid proteases called rhombosortases.
4.1 PARL-type Rhomboid Proteases
The mammalian mitochondrial PARL sequences group with Drosophila mitochondrial Rho-7, yeast mitochondrial Pcp1 [14], and plant mitochondrial AtRBL12 [15], with Toxoplasma mitochondrial TgRom6 in a nearby group [12]. Branching of the mitochondrial PARL-type sequences reflects the phylogenetic species tree [14], and all members of the family tend to retain the same TMH domain topology (with the above-mentioned exception of plant AtRBL12), possessing an N-terminal TMH prior to the 6 TMH rhomboid protease core (1+6 TMD topology) [14]. Segregation of these mitochondrial rhomboid proteases likely reflects the conserved 1+6 TMD topology, but may also manifest from the bacterial-like lipid composition of the mitochondrial inner membrane surroundings. For the yeast PARL-type rhomboid protease Pcp1, proteolysis releases its substrate dynamin-like GTPase Mgm1 from the membrane of healthy mithochondria, excluding unhealthy mitochondria from membrane fusion [30]. A similar function has been described in Drosophila: Rho-7 cleaves the metazoan Mgm1 ortholog Opa1 and is required for normal mitochondrial dynamics [31]. Metazoan PARL-type rhomboid proteases (Rho-7 and PARL) also participate in the Parkin/PINK1 pathway [32–34], whereby unhealthy mitochondrial membranes tagged with uncleaved PINK1 are cleared from cells. Similar to Mgm1/Opa1 substrate cleavage, PARL cleavage of the PINK1 substrate appears to provide a checkpoint for maintaining healthy mitochondria. Similar to previous phylogenetic analysis [11], network-based clustering groups this eukaryotic PARL subfamily that cleaves dynamin-like GTPases near mixed prokaryotic clusters that are comprised of sequences that have yet unknown function. Dynamin-like GTPases exist in these prokaryotic species, with bacterial mitofusins retaining TMH anchors that could potentially be cleaved. Relatively little is known about the function of such proteins in bacterial membrane dynamics, although parallels with eukaryotic systems have led to speculation of a role in membrane tethering [35].
4.2 RHO-type Rhomboid Proteases
The founding member of the rhomboid proteases, Drosophila rhomboid (Rho-1), was initially implicated as an upstream activator of the epidermal growth factor (EGF) spitz (reviewed in [10]). By combining numerous experimental observations with sequence analysis and mutagenesis, a model of rhomboid protease functioning as an intramembrane serine protease that cleaves the spitz TMH and releases the growth factor from the membrane emerged. Similar functions have been shown for C. elegans ROM-1, which regulates EGF signaling in vulval development through cleavage of LIN-3L[36]. Humans encode three genes that cluster with the rhomboid proteases (RHBDL1-3). RHBDL2 has been shown to cleave thrombomodulin [37] and EphrinB1,2,3 [38]in addition to EGF [39], although the physiological roles of cleavage remain unclear. Although few target substrates have been identified, several tools have been developed to study rhomboid protease TMH substrate cleavage and a sequence preference for relatively small residues (A, C, S, or G) in the substrate P1 subsite has been defined among a few other preferences for hydrophobic residues consistent with the composition of the transmembrane substrate [28]. Bioinformatics methods could combine this substrate-binding preference with additional information such as localization or expression profiling to narrow the field of potential new rhomboid protease substrates.
As discussed previously, the RHBDL1-3 rhomboid protease family appears to have duplicated and diverged from the iRhoms (RHBDF1/2), which are inactive as proteases in human. Instead, iRhoms appear to regulate release of cytokines and growth factors through protein interactions in the endoplasmic reticulum and Golgi. The inactive RHBDF2/iRhom2 promotes trafficking of the TNF-alpha converting enzyme (TACE) to the Golgi where it can be activated by processing [40]. Alternatively, the inactive RHBDF1/iRhom1 binds to immaturely glysosylated forms of EGF-like ectodomains from TGF-alpha ligands [41], targeting them for endoplasmic reticulum-associated cleavage (ERAD) [42]. During ERAD a membrane associated multiprotein complex that includes yet another distantly related inactive rhomboid family member (Der1) passes the cleavage targets to the soluble ubiquitin proteosome system for degradation.
4.3 Bacterial RHO-like Rhombosortases Identified Using Phylogenetic Profiling
For bacterial genomes where operon structures are frequently maintained, functional links between genes can often be identified using information gleaned from genomic neighborhoods, co-expression data, or taxonomic co-occurrence [43, 44]. One such approach has identified a distinct set of bacterial RHO-type of rhomboid proteases as co-occurring with a set of proteins that possess a newly identified C-terminal homology domain (GlyGly-CTERM), which consists of a Gly-Gly motif followed by a transmembrane helix and a cluster of basic residues at the protein C-terminus [45]. This identified domain architecture was described as resembling protein sorting recognition signals such as LPXTG-CTERM and PEP-CTERM, suggesting a sorting signal function for the GlyGly-CTERM motif. Partial phylogenetic profiling using GlyGly-CTERM genes as queries identified genes encoding members of the rhomboid proteases as top hits. The identified rhomboid proteases belong to a distinct clade of sequences from proteobacteria and were named rhombosortases following the sorting signal nomenclature. In addition to the taxonomic co-occurrance between rhombosortases and their potential GlyGly-CTERM targets, the genes tend to be adjacent in the genome and are identified as associating with significant scores in the STRING database [44]. Identified functional associations between rhombosortases and GlyGly-CTERM domains may suggest the TMH component of the sorting signal as a rhombosortase substrate. Given this example of functional association, perhaps additional rhomboid protease substrates could be identified using similar bioinformatics methods together with careful definition of rhomboid protease clades, which are currently rather broadly defined in classifications such as the PFAM database [46].
Supplementary Material
Highlights.
Rhombiod protease classification and phylogenetics studies have lead to conflicting views on evolutionary origins of the family
Network-based clustering of present day rhomboid protease sequences suggests the possibility of an alternate history
Phylogenetic profiling has led to identification of potential substrates for a subset of bacterial rhomboid proteases
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hartley BS. Proteolytic enzymes. Annu Rev Biochem. 1960;29:45–72. doi: 10.1146/annurev.bi.29.070160.000401. [DOI] [PubMed] [Google Scholar]
- 2.Barrett AJ, Rawlings ND. Types and families of endopeptidases. Biochem Soc Trans. 1991;19:707–715. doi: 10.1042/bst0190707. [DOI] [PubMed] [Google Scholar]
- 3.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012;40:D343–350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rawson RB, Zelenski NG, Nijhawan D, Ye J, Sakai J, Hasan MT, Chang TY, Brown MS, Goldstein JL. Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol Cell. 1997;1:47–57. doi: 10.1016/s1097-2765(00)80006-4. [DOI] [PubMed] [Google Scholar]
- 5.Weihofen A, Binns K, Lemberg MK, Ashman K, Martoglio B. Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science. 2002;296:2215–2218. doi: 10.1126/science.1070925. [DOI] [PubMed] [Google Scholar]
- 6.Wolfe MS, Xia W, Ostaszewski BL, Diehl TS, Kimberly WT, Selkoe DJ. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and gamma-secretase activity. Nature. 1999;398:513–517. doi: 10.1038/19077. [DOI] [PubMed] [Google Scholar]
- 7.Urban S, Lee JR, Freeman M. Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell. 2001;107:173–182. doi: 10.1016/s0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- 8.Kinch LN, Ginalski K, Grishin NV. Site-2 protease regulated intramembrane proteolysis: sequence homologs suggest an ancient signaling cascade. Protein Sci. 2006;15:84–93. doi: 10.1110/ps.051766506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Golde TE, Wolfe MS, Greenbaum DC. Signal peptide peptidases: a family of intramembrane-cleaving proteases that cleave type 2 transmembrane proteins. Semin Cell Dev Biol. 2009;20:225–230. doi: 10.1016/j.semcdb.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Urban S, Dickey SW. The rhomboid protease family: a decade of progress on function and mechanism. Genome Biol. 2011;12:231. doi: 10.1186/gb-2011-12-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koonin EV, Makarova KS, Rogozin IB, Davidovic L, Letellier MC, Pellegrini L. The rhomboids: a nearly ubiquitous family of intramembrane serine proteases that probably evolved by multiple ancient horizontal gene transfers. Genome Biol. 2003;4:R19. doi: 10.1186/gb-2003-4-3-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graindorge MSJA, Soldati-Favre D. New insights into parasite rhomboid proteases. Mol Biochem Parasitol. 2012;182:27–36. doi: 10.1016/j.molbiopara.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 13.Kateete DP, Okee M, Katabazi FA, Okeng A, Asiimwe J, Boom HW, Eisenach KD, Joloba ML. Rhomboid homologs in mycobacteria: insights from phylogeny and genomic analysis. BMC Microbiol. 2010;10:272. doi: 10.1186/1471-2180-10-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemberg MK, Freeman M. Functional and evolutionary implications of enhanced genomic analysis of rhomboid intramembrane proteases. Genome Res. 2007;17:1634–1646. doi: 10.1101/gr.6425307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kmiec-Wisniewska B, Krumpe K, Urantowka A, Sakamoto W, Pratje E, Janska H. Plant mitochondrial rhomboid, AtRBL12, has different substrate specificity from its yeast counterpart. Plant Mol Biol. 2008;68:159–171. doi: 10.1007/s11103-008-9359-8. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Lorenzo M, Sjodin A, Jansson S, Funk C. Protease gene families in Populus and Arabidopsis. BMC Plant Biol. 2006;6:30. doi: 10.1186/1471-2229-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buguliskis JS, Brossier F, Shuman J, Sibley LD. Rhomboid 4 (ROM4) affects the processing of surface adhesins and facilitates host cell invasion by Toxoplasma gondii. PLoS Pathog. 2010;6:e1000858. doi: 10.1371/journal.ppat.1000858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanaoka MM, Urban S, Freeman M, Okada K. An Arabidopsis Rhomboid homolog is an intramembrane protease in plants. FEBS Lett. 2005;579:5723–5728. doi: 10.1016/j.febslet.2005.09.049. [DOI] [PubMed] [Google Scholar]
- 19.Thompson EP, Smith SG, Glover BJ. An Arabidopsis rhomboid protease has roles in the chloroplast and in flower development. J Exp Bot. 2012;63:3559–3570. doi: 10.1093/jxb/ers012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ben-Shem A, Fass D, Bibi E. Structural basis for intramembrane proteolysis by rhomboid serine proteases. Proc Natl Acad Sci U S A. 2007;104:462–466. doi: 10.1073/pnas.0609773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brooks CL, Lazareno-Saez C, Lamoureux JS, Mak MW, Lemieux MJ. Insights into substrate gating in H. influenzae rhomboid. J Mol Biol. 2011;407:687–697. doi: 10.1016/j.jmb.2011.01.046. [DOI] [PubMed] [Google Scholar]
- 22.Lemieux MJ, Fischer SJ, Cherney MM, Bateman KS, James MN. The crystal structure of the rhomboid peptidase from Haemophilus influenzae provides insight into intramembrane proteolysis. Proc Natl Acad Sci U S A. 2007;104:750–754. doi: 10.1073/pnas.0609981104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vinothkumar KR, Strisovsky K, Andreeva A, Christova Y, Verhelst S, Freeman M. The structural basis for catalysis and substrate specificity of a rhomboid protease. EMBO J. 2010;29:3797–3809. doi: 10.1038/emboj.2010.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Ha Y. Open-cap conformation of intramembrane protease GlpG. Proc Natl Acad Sci U S A. 2007;104:2098–2102. doi: 10.1073/pnas.0611080104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Zhang Y, Ha Y. Crystal structure of a rhomboid family intramembrane protease. Nature. 2006;444:179–180. doi: 10.1038/nature05255. [DOI] [PubMed] [Google Scholar]
- 26.Wu Z, Yan N, Feng L, Oberstein A, Yan H, Baker RP, Gu L, Jeffrey PD, Urban S, Shi Y. Structural analysis of a rhomboid family intramembrane protease reveals a gating mechanism for substrate entry. Nat Struct Mol Biol. 2006;13:1084–1091. doi: 10.1038/nsmb1179. [DOI] [PubMed] [Google Scholar]
- 27.Xue Y, Chowdhury S, Liu X, Akiyama Y, Ellman J, Ha Y. Conformational change in rhomboid protease GlpG induced by inhibitor binding to its S’ subsites. Biochemistry. 2012;51:3723–3731. doi: 10.1021/bi300368b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strisovsky K, Sharpe HJ, Freeman M. Sequence-specific intramembrane proteolysis: identification of a recognition motif in rhomboid substrates. Mol Cell. 2009;36:1048–1059. doi: 10.1016/j.molcel.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moin SM, Urban S. Membrane immersion allows rhomboid proteases to achieve specificity by reading transmembrane segment dynamics. Elife. 2012;1:e00173. doi: 10.7554/eLife.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herlan M, Vogel F, Bornhovd C, Neupert W, Reichert AS. Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA. J Biol Chem. 2003;278:27781–27788. doi: 10.1074/jbc.M211311200. [DOI] [PubMed] [Google Scholar]
- 31.Rahman M, Kylsten P. Rhomboid-7 over-expression results in Opa1-like processing and malfunctioning mitochondria. Biochem Biophys Res Commun. 2011;414:315–320. doi: 10.1016/j.bbrc.2011.09.047. [DOI] [PubMed] [Google Scholar]
- 32.Whitworth AJ, Lee JR, Ho VM, Flick R, Chowdhury R, McQuibban GA. Rhomboid-7 and HtrA2/Omi act in a common pathway with the Parkinson’s disease factors Pink1 and Parkin. Dis Model Mech. 2008;1:168–174. doi: 10.1242/dmm.000109. discussion 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deas E, Plun-Favreau H, Gandhi S, Desmond H, Kjaer S, Loh SH, Renton AE, Harvey RJ, Whitworth AJ, Martins LM, Abramov AY, Wood NW. PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum Mol Genet. 2011;20:867–879. doi: 10.1093/hmg/ddq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meissner C, Lorenz H, Weihofen A, Selkoe DJ, Lemberg MK. The mitochondrial intramembrane protease PARL cleaves human Pink1 to regulate Pink1 trafficking. J Neurochem. 2011;117:856–867. doi: 10.1111/j.1471-4159.2011.07253.x. [DOI] [PubMed] [Google Scholar]
- 35.Bramkamp M. Structure and function of bacterial dynamin-like proteins. Biol Chem. 2012;393:1203–1214. doi: 10.1515/hsz-2012-0185. [DOI] [PubMed] [Google Scholar]
- 36.Dutt A, Canevascini S, Froehli-Hoier E, Hajnal A. EGF signal propagation during C. elegans vulval development mediated by ROM-1 rhomboid. PLoS Biol. 2004;2:e334. doi: 10.1371/journal.pbio.0020334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng TL, Wu YT, Lin HY, Hsu FC, Liu SK, Chang BI, Chen WS, Lai CH, Shi GY, Wu HL. Functions of rhomboid family protease RHBDL2 and thrombomodulin in wound healing. J Invest Dermatol. 2011;131:2486–2494. doi: 10.1038/jid.2011.230. [DOI] [PubMed] [Google Scholar]
- 38.Pascall JC, Brown KD. Intramembrane cleavage of ephrinB3 by the human rhomboid family protease, RHBDL2. Biochem Biophys Res Commun. 2004;317:244–252. doi: 10.1016/j.bbrc.2004.03.039. [DOI] [PubMed] [Google Scholar]
- 39.Adrain C, Strisovsky K, Zettl M, Hu L, Lemberg MK, Freeman M. Mammalian EGF receptor activation by the rhomboid protease RHBDL2. EMBO Rep. 2011;12:421–427. doi: 10.1038/embor.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adrain C, Zettl M, Christova Y, Taylor N, Freeman M. Tumor necrosis factor signaling requires iRhom2 to promote trafficking and activation of TACE. Science. 2012;335:225–228. doi: 10.1126/science.1214400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakagawa T, Guichard A, Castro CP, Xiao Y, Rizen M, Zhang HZ, Hu D, Bang A, Helms J, Bier E, Derynck R. Characterization of a human rhomboid homolog, p100hRho/RHBDF1, which interacts with TGF-alpha family ligands. Dev Dyn. 2005;233:1315–1331. doi: 10.1002/dvdy.20450. [DOI] [PubMed] [Google Scholar]
- 42.Zettl M, Adrain C, Strisovsky K, Lastun V, Freeman M. Rhomboid family pseudoproteases use the ER quality control machinery to regulate intercellular signaling. Cell. 2011;145:79–91. doi: 10.1016/j.cell.2011.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Basu MK, Selengut JD, Haft DH. ProPhylo: partial phylogenetic profiling to guide protein family construction and assignment of biological process. BMC Bioinformatics. 2011;12:434. doi: 10.1186/1471-2105-12-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, Jensen LJ, von Mering C. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haft DH, Varghese N. GlyGly-CTERM and rhombosortase: a C-terminal protein processing signal in a many-to-one pairing with a rhomboid family intramembrane serine protease. PLoS One. 2011;6:e28886. doi: 10.1371/journal.pone.0028886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frickey T, Lupas A. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics. 2004;20:3702–3704. doi: 10.1093/bioinformatics/bth444. [DOI] [PubMed] [Google Scholar]
- 48.Coggill P, Finn RD, Bateman A. Identifying protein domains with the Pfam database. Curr Protoc Bioinformatics. 2008;Chapter 2(Unit 2):5. doi: 10.1002/0471250953.bi0205s23. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.