Abstract
Purpose
Morphometry techniques were applied to quantify the normal tissue therapy response in patients receiving whole-brain radiation for intracranial malignancies.
Methods
Pre- and Post-irradiation magnetic resonance imaging (MRI) data sets were retrospectively analyzed in N = 15 patients. Volume changes with respect to pre-irradiation were quantitatively measured in the cerebrum and ventricles. Measurements were correlated with the time interval from irradiation. Criteria for inclusion included craniospinal irradiation, pre-irradiation MRI, at least one follow-up MRI, and no disease progression. The brain on each image was segmented to remove the skull and registered to the initial pre-treatment scan. Average volume changes were measured using morphometry analysis of the deformation Jacobian and direct template registration-based segmentation of brain structures.
Results
An average cerebral volume atrophy of −0.2 and −3 % was measured for the deformation morphometry and direct segmentation methods, respectively. An average ventricle volume dilation of 21 and 20 % was measured for the deformation morphometry and direct segmentation methods, respectively.
Conclusion
The presented study has developed an image processing pipeline for morphometric monitoring of brain tissue volume changes as a response to radiation therapy. Results indicate that quantitative morphometric monitoring is feasible and may provide additional information in assessing response.
Keywords: Image registration, Radiation therapy response, Morphometry, Diffeomorphism, Computational anatomy
Introduction
Approximately 211,000 patients present each year in the USA with brain tumors. Of these, 38,000 are benign primary tumors, 23,000 primary malignant tumors, and 150,000 metastatic cancer, originating largely from lung, breast, and melanoma [20,27]. The average life expectancy for patients with primary and metastatic malignancies in the brain, from time of diagnosis until death, is approximately 12–16 months. Five-year survival for primary brain malignancies is among the lowest of all cancers. Current treatment options are aggressive and typically include maximal surgical resection, stereotactic radiosurgery (SRS), and/or chemotherapy.
Increasingly effective therapies for treating malignancies in the brain is creating a growing population of survivors with cognitive impairment as a side effect of treatment. Since cognitive impairment can significantly impair quality of life and independence, therapeutic strategies to minimize radiation-induced cognitive impairment are becoming more important. The possibility of incurring toxicity that may impair the patients quality of life is increasingly influencing the choice of therapeutic modality. Although state-of-the-art radiation therapy techniques have nearly eliminated acute brain injury, cognitive impairment occurs in 50–90 % of patients who survive >6 months post-irradiation. This has resulted in over 100,000 patients yearly with intracranial malignancies who live long enough to develop irreversible radiation-induced brain injury [17]. In a period greater than 6 months, excluding identifiable tumor recurrence, complications of radiotoxicity include cognitive impairment, dizziness, fatigue, headache, mood changes, progressively severe dementia, gait disturbance, and incontinence.
Quantification of the radiation therapy response of normal tissue as well as malignant tissue has received significant attention [6,13,17,29]. This study develops an image processing pipeline to quantify the structural response associated with whole-brain radiation therapy. The success of image registration-based morphometry techniques in identifying key structural patterns associated with Alzheimer’s disease [22], schizophrenia [9], and abnormal brain development [34] motivates the developed approach.
Previous efforts have reported radiation-induced structural response in longitudinal CT and MR imaging studies [2,15,23,25,28,31,38]. Nieder et al. [28] measured ventricular dilation in millimeters from the laterolateral diameter; sulcal enlargement was qualitatively reported (N = 49 adult patients, age ≥18, median follow-up imaging = 10 months). However, the measured CT changes did not correlate with clinical symptoms of late radiation toxicity. Asai et al. [2] reported ventricle enlargement, widening of the cortical sulci, and attenuated CT numbers in white matter for N = 6 patients. Similarly, DeAngelis et al. [15] reported cortical atrophy and hypodense white matter in N = 12 patients. Zhang et al. [38] reported reduced density in gray and white matter using voxel-based morphometry techniques in N = 13 patients. Higher-level cognitive skills are known to be affected, suggesting that the cerebral cortex is affected by the therapy [23]. A decrease in white matter has also been correlated with radiation dose [25,31]. Multiple authors [1,24,35] have suggested avoidance of critical neuroanatomical targets such as the hippocampus and temporal lobes may preserve specific cognitive functions such as memory or protect special cell populations such as neural stem cells.
Within the context of this study, deformation-based morphometry is applied to MR images for quantitative analysis of volumetric anatomical changes in response to radiation therapy. The developed image processing pipeline provides quantitative information for further evaluation of correlated toxicities. Nonlinear deformable image registration techniques align all images within a longitudinal MRI study. The applied morphometry techniques provided quantitative measurements of the ventricular dilation as well as cerebral atrophy. The statistical significance of the anatomical volume change measurements is evaluated in N = 15 patients.
Methods
Image processing pipeline
A reference atlas or template is widely used within the field of neuroimaging to provide a standardized neuroanatomical space and probability priors for multiple segmentation methods [5]. Template-based segmentation is used within this study to facilitate morphometry calculations within the ventricles and cerebrum. As seen in Fig. 1, a deformation-based morphometry analysis pipeline was created in which post-treatment imaging was registered to the initial pre-treatment scan. The ICBM (International Consortium for Brain Mapping) high-resolution image and its label map were downloaded from the Laboratory of Neuro Imaging (LONI). A separate label map for the cerebrum and ventricles was created for this study by merging the corresponding labels of the ICBM label map. Segmentations of the initial pre-treatment image for each patient were generated from the image registration displacement field mapping the template to the respective pre-treatment image. The integral of the Jacobian determinant values of the displacement field was calculated across each segmented region of the label map to determine quantitative percent volume change, see Eq. (2). Representative images of the major steps of the pipeline are shown in Fig. 2.
Fig. 1.
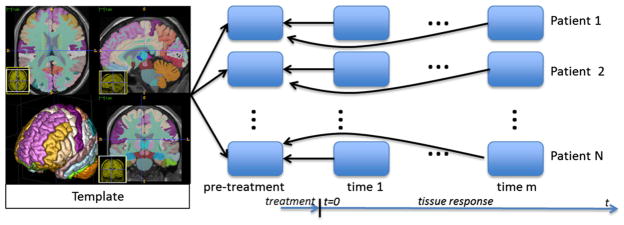
Overview of image processing pipeline. The time instance of the initial MR scan (prior to radiation therapy) for each patient is denoted ‘pre-treatment.’ A spatially uniform whole-brain radiation dose is delivered in multiple fractions following the pre-treatment scan. Post-treatment MR scans are taken at non-uniform time intervals, t > 0, with respect to the completion of the dose delivery. Details of the radiation dose, delivery time, and the time instance of the follow-up MR images are provided in Fig. 3. Post-treatment MR scans for each patient are registered to the initial pre-treatment scan. Relative volume change information was measured at each voxel of the pre-treatment image using the determinant of the displacement field Jacobian. Registration-based segmentation of the template image shown is used to compute the average value of the volume change observed in ventricles and the combined gray and white matter within the cerebrum
Fig. 2.
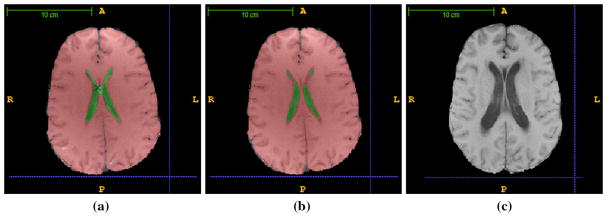
Representative images. Representative images for the processing pipeline from Fig. 1 are shown. Each image is skull stripped and bias corrected. The image template and corresponding cerebrum and ventricle labels are registered to the pre-treatment image for each patient. a An example of the image registration-based segmentation of the cerebrum and ventricles is shown. Statistics for the Jacobian maps as well as direct volume measurements is obtained using the labeled ventricles and cerebrum shown. An error noted in the registration-based segmentation is marked ‘×.’ b Errors observed in the labels of the cerebrum and ventricles were manually corrected on the baseline pre-treatment image by a medical expert for evaluation of the segmentation accuracy. c An example of the follow-up image, corresponding to (b), is shown; time = 347 days post-irradiation. The ventricle size has noticeably increased. Follow-up images are registered to both the pre-treatment image and the template. Registration to the pre-treatment image provides deformation Jacobian information that may be used to quantify volume change (2). Similarly, registration to the image template provides a direct estimate of volume change information
The accuracy of the registration-based segmentation was quantitatively evaluated against manual segmentations of the pre-treatment image for each patient. Errors observed in the labels of the cerebrum and ventricles were manually corrected on the baseline pre-treatment image by a medical expert. The Dice similarity coefficient [39], , was used calculate the overlap of the template-based segmentation, A ⊂ ℝ3, and the manual segmentation, B ⊂ ℝ3. The DSC was calculated in both the ventricles and cerebrum.
A direct registration-based segmentation of brain structures was also implemented in which the template was registered to each time point within the longitudinal study. Volume changes within the anatomical structures of interest for each patient may be directly determined from the volume information contained within the deformed label map. In addition to the Jacobian-based method, the analysis provides a second measure of the percent volume change observed, Eq. (2).
Brain extraction for each image was performed using the Brain Extraction Tool [19] to create a binary label mask which was then corrected manually using the GUI interface provided by itk-SNAP [37]. Each extracted brain was treated with N4 bias field correction [4] prior to registration to remove shading artifacts. Application of the bias field correction algorithm results in a bimodal distribution of intensity values within the T1W images due to the amplification of the individual distributions of the white and gray matter. Increase uniformity of the intensity values across the white and gray matter is visually confirmed.
Image registration
The presented deformation morphometry pipeline applies modern diffeomorphic image registration methods to measure the volume change. These techniques have been significantly influenced by the mathematical formalism of large deformation diffeomorphisms [7,14,16]. The diffeomorphic formalism provides a rigorous framework for finding a deformation field that satisfies the physics of an elastic or fluid deformation and guarantees sufficient smoothness expected of biological deformations. Diffeomorphic registration techniques rely on defining a differentiable, bijective mapping between the fixed and moving image and guarantee that the inverse displacement fields and Jacobian-based volume measurements will be well defined. The regularity on the solution imposed within the diffeomorphic framework preserves the topology of the brain and prevents the deformation in an uncontrolled manner which would otherwise make results difficult to interpret.
The well-known large deformation diffeomorphic metric mapping (LDDMM) algorithm [7] yields a geodesic solution that conforms to the space of diffeomorphic transformations. However, floating point operation and memory costs increase with multiple partial differential equation solves and image gradient calculations needed at each integration time point required by the diffeomorphic transformation. The symmetric normalization (SyN) approach [3,4] is a popular variant of the diffeomorphic transformation and is used in this study. A Gaussian convolution within the SyN approach provides a computationally efficient approximation for the physics of the deformation field [8,33]. SyN calculates image gradients only at the midpoint in time of the full diffeomorphic transformation and provides an explicit symmetry of the large deformation diffeomorphic metric mapping (LDDMM) formulation [7]. The distance between the current image, I, and reference image, J, mapped into the transformation midpoint provides an estimate of the image similarity and performance measure of the registration. Under a linear intensity transformation, i.e., I ≈ α J + β, the correlation coefficient (CC) is known to be optimal [18] and a localized version (1) was used in this study for the bias field corrected images.
| (1) |
The neighborhood radius, Nr, in computing cross-correlation was 5 voxels.
Specifically, the greedy SyN, ‘GR,’1, available from the ANTs package [3,4] was used. Algorithmic details may be found in [4,36]. The registration consisted of (1) an affine registration step and (2) a diffeomorphic step. A multi-resolution approach was applied in both steps. Intuitively, the lowest resolutions are computationally efficient and capture bulk movement. Each resolution increasingly recovers more fine-scale features. A five-level and three-level image pyramids were applied to the affine registration and diffeomorphic registration steps, respectively. The default gradient descent was used in the optimization of the affine and diffeomorphic registration. A Gaussian regularization kernel with an isotropic width of 3 voxels was used as an estimate of the Green’s kernel for the deformation operator and is further discussed in the Appendix.
The ANTs software is developed under significant open source infrastructure to facilitate reproducible research and includes significant documentation, working examples, and scripts used in previous successful studies [21,26]. The portable CMake build system, software version control, and unit testing environment provide quality assurance in the numerical results of the algorithms. Verbatim command line parameters used in previous studies, as well as this study, are available and provide confidence that the intended numerical algorithms, in which significant resources have been invested, are being properly applied in this study.
Assuming a constant volume, such as within the skull, the volume of an anatomical region of interest at a particular time instance may be related back to the reference image at t = 0 using the classical change in variables for integration. On a pixelated image, the volume [mm3] of a labeled region is the product of the number of voxels in the segmentation, NS, and the volume per voxel, ΔV. The new volume [mm3] at time t = τ may be calculated by integrating the Jacobian, J, of the deformation, φ, over the segmented region. The average Jacobian may be related to the percent volume change, μ.
| (2) |
Statistical analysis
Statistical analysis was computed using the R package [30]. A one-sided t-test was used to evaluate percent volumetric changes (2) from the baseline pre-treatment scans. One-sample calculations were computed using zero percent change, H0 : μ = 0, as the null hypothesis. As seen in Eq. (2), an integral Jacobian value greater than one implies expansion or dilation; a value less than one implies tissue atrophy or contraction. Correlation analysis was performed to assess relationships between the measured volume changes, radiation dose, and time lapse since radiation exposure. Correlation coefficients, r, were computed using a Spearman method. A one-sample t-test for the correlation coefficient was computed to estimate statistically significant correlation changes from zero.
Image data
Patients that were diagnosed with intracranial medulloblastoma and received a uniform whole-brain radiation to the cerebrum were identified under an institutionally approved IRB protocol for retrospective studies. Similar to CT imaging studies [2,15,28], the volume changes reported by the deformation-based morphometry are considered as the volume response of the combined gray and white matter within the cerebrum. Volume response in the ventricles is also considered. Criteria for inclusion in our study included craniospinal irradiation, pre-irradiation magnetic resonance imaging (MRI), at least one follow-up MRI, and no disease progression during follow-up. Summary statistics for the selected patient cohort is presented in Fig. 3.
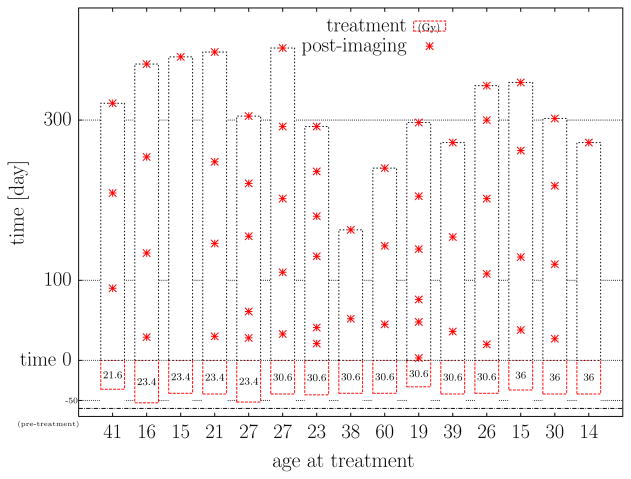
Fig. 3.
Patient cohort summary statistics (N = 15). A summary of the patient imaging times considered in this study is presented. The imaging times (day) of the 61 post-treatment images are denoted ‘*’ and shown relative to the completion of dose delivery, t = 0. The total time (day) and total dose (Gy) delivered for the fractionated therapy are also presented. Min/Median/Max summary statistics are: age (year) 14/26/60, dose (Gy) 21.6/30.6/36, treatment time (day) 33/42/53, and post-imaging (day) 3/154/390
At each time instance, axial T1-weighted images (median TR/TE = 550 ms/9 ms, 1mm×1mm pixel size, slice thickness 5 mm) were selected from the routine imaging protocols to assess the response of radiation therapy for treating patients with medulloblastoma.
Results
Representative study time instances and deformation-based segmentations for morphometry analysis are shown in Fig. 2. Registration of the template to a specific patient image at the initial pre-treatment scan of the study is shown in Fig. 2a. Figure 2b shows the manual label map of the cerebrum and ventricles for the pre-treatment image. The DSC values used to quantitatively evaluate the accuracy of the template registration-based segmentation against the manual segmentation are presented for the pre-treatment image of each patient in Fig. 4b. Figure 2c illustrates the same patient observed during a follow-up MR scan. For this particular case, the follow-up MR scan was 347 days post-irradiation and the ventricles have visibly dilated compared with the pre-treatment scan, Fig. 2b. Similar to the deformable registration-based cerebrum and ventricle segmentations shown in Fig. 2a, template-based segmentation of the follow-up image shown Fig. 2c provides volume change. Registration of Fig. 2b–c provides a second measure of the volume change observed using the Jacobian of the deformation field. A summary of the volume change characteristics for the patient cohort is presented in Figs. 4a and 5.
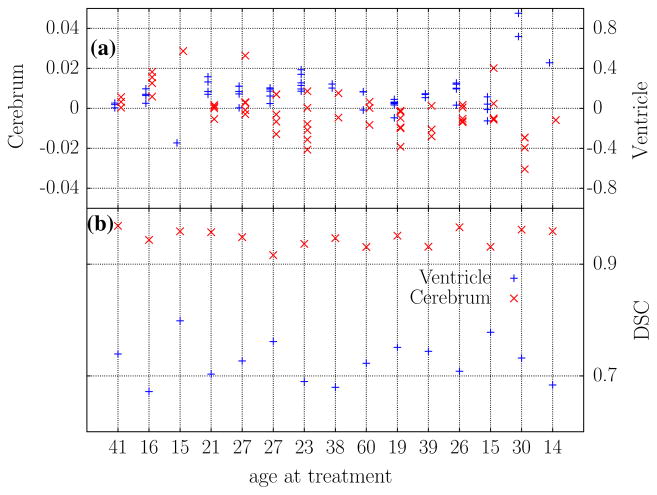
Fig. 4.
Analysis summary. The average Jacobian measurements from each post-treatment image included in the deformation morphometry analysis are shown in (a). Measurements and DSC calculations are aligned horizontally with the respective patient and correspond to Fig. 3. The left and right axes plot the measurements for the cerebrum ‘×’ and ventricles ‘+,’ respectively. DSC calculations comparing manual segmentations of the pre-treatment image of each patient to the registration-based segmentation approach are presented in (b). Min/Median/Max summary statistics in the ventricles and cerebrum are: DSC (ventricle) 0.67/0.72/0.79 and DSC (cerebrum) 0.91/0.94/0.96
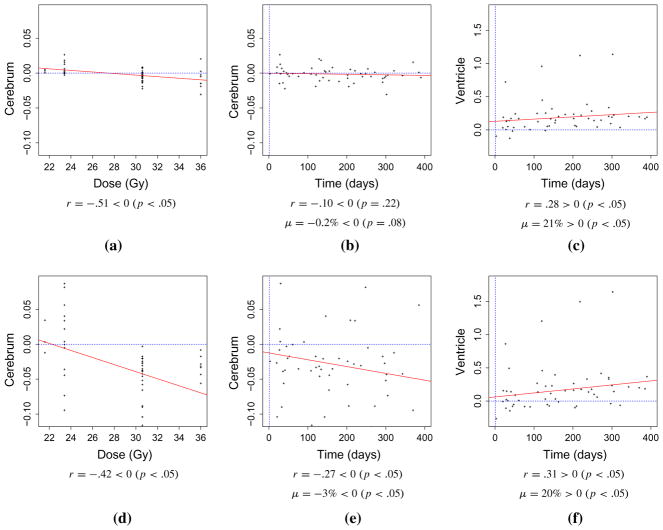
Fig. 5.
Volume change summary. A summary of the volume changes, μ, see Eq. (2), observed in the cerebrum and ventricles as well as correlation, r, with radiation dose and time instance of the follow-up MR imaging is presented. a–c Represent results from the deformation morphometry-based Jacobian analysis in which each patient image was registered to the respective pre-treatment image and template registration-based segmentation was applied to the pre-treatment image. d–f Represent results from the direct analysis in which the template was registered to each image within the analysis. The computed correlations coefficients, r, as well as p values estimating statistically significant correlation changes from zero are provided below each graph. t test results estimating statistically significant atrophy and dilation are also provided
Statistics observed in the patient cohort of the analysis is shown in the scatter plots in Fig. 5. For reference, a dotted line for zero volume change is plotted. Figure 5a–c represents results from the deformation morphometry-based Jacobian analysis in which each patient image was registered to the respective pre-treatment image and template registration-based segmentation was applied to the pre-treatment image. Figure 5d–f represents results from the direct analysis in which the template was registered to each image within the analysis and volume changes were interpreted directly from the volumes of the individual labels. The time instance of the follow-up MRI is taken with respect to the completion of the dose delivery for each patient. The hypothesis testing framework for the direction of the one-sided statistical inference is provided with each graph.
Figure 5a, d plots the correlations between the measured cerebral atrophy and radiation dose. Figure 5b, e plots the correlations between the measured cerebral atrophy and the time instance of the follow-up MR image. An average cerebral volume decrease of −0.2 and −3 % was measured for the deformation morphometry and direct methods, respectively. Statistically significant p values were observed in the t tests considered for the volume change in the direct method, p < 0.05. Cerebral volume loss was inversely correlated with amounts of irradiation, r = −0.51/−0.42, and inversely correlated with time since the initial scan, r = −0.10/−0.27.
Similarly, scatter plots Fig. 5c, f plot the correlations between the measured ventricle dilation and the time instance of the follow-up MR image. An average ventricle volume increase of 21 and 20 % was measured for the deformation morphometry and direct methods, respectively. Cerebral volume dilation was correlated with time since the initial scan, r = 0.28/0.31. Statistically significant p-values were observed for the t-test and correlation test considered, p < 0.05.
Discussion
Deformation morphometry-based image analysis techniques were applied to quantitatively measure anatomical volume change in brain as a therapy response in patients receiving whole-brain radiation for treating medulloblastoma. Sixty-one post-irradiation magnetic resonance imaging (MRI) data sets were retrospectively analyzed in N = 15 patients. Volume changes measured from (1) template-based segmentation at each time point in the longitudinal study and (2) intra-patient Jacobian determinant analysis at multiple time points post-irradiation both show promise as a quantitative imaging biomarker of the therapy. Quantitative morphometry measurements of ventricle dilation and cerebral atrophy agree with observations of previous longitudinal imaging studies [2,23,28,38].
The measured atrophy in the cerebrum and dilation in the ventricles within the patient cohort chosen, age [min, max] = [14, 60], is unlikely attributed to natural aging within the time period of this study, max time 390 days. Classical growth curves of the developing brain show the brain reaches 90 % of its maximum volume during the first five years of development [32]. Further, the volume of the skull is expected to be fixed after 6 year of age in which the cranial sutures are closed; an increase in ventricle volume likely corresponds to cerebrum atrophy. The brain is expected to have asymptotically reached the fully developed volume within the patient cohort studied; age-related bias and registration inaccuracies that may be attributed to anatomical differences between a developing brain and the image template used in this study are unlikely. However, significant further work is needed in establishing and processing an age-matched control cohort of normal patients as a quantitative reference for the radiation-induced volume change.
An isotropic 1-mm image template was repeatedly registered to the MRI of each patient at multiple time points. This provided a registration-based segmentation of the cerebrum and ventricles for each image including the initial time point for Jacobian determinant analysis. The template-based analysis implicitly assumes that the anatomical structures of the brain are faithfully preserved under the diffeomorphic registration with the routine protocol imaging data used in this study. Although current results demonstrate the potential of morphometry techniques to quantitatively measure the brain tissue response to radiation therapy, registration inaccuracies are expected to limit the applicability of this approach and are likely responsible for the noisy measurements observed. For example, under ideal conditions, both deformation-based morphometry and direct segmentation techniques are expected to provide the same quantitative information. The relatively larger variance in the statistics of the direct segmentation approach, Fig. 5d–f, indicates larger potential registration inaccuracies between the template and each individual time point as compared to intra-patient registrations. The segmentation error, as measured by the DSC Fig. 4b for the initial time point, is likely to have accumulated due to the segmentation at each time point and results in the greater variance for the direct segmentation approach. A larger DSC accuracy is observed in the cerebrum compared with the ventricle and indicates a relatively larger volume fraction error in the ventricles. An example of the segmentation error is qualitatively observed in Fig. 2a. Similar to large-scale Alzheimer’s studies [34], future efforts should strive to prospectively implement a strict imaging protocol on isotropic imaging data sets to enhance the accuracy of the statistical measurements.
The dilation observed in the ventricles, as measured by the deformation-based morphometry seen in Fig. 5c, provided the strongest signal in measuring the response to radiation therapy. Although less than zero, the volume change in the cerebrum was a relatively weaker signal. Visual inspection indicates that resolving registration inaccuracies in the gyri in future studies may further improve the signal. However, further efforts are needed to quantitatively determine sources of registration inaccuracy. Currently, the spatial recognition patterns of the human brain are considered the gold standard for registering a given set of images using landmark feature pairs [12] or segmenting identifiable structures [21]. Expert determined landmark feature pair correspondences have been shown to provide an accurate and repeatable spatial registration [11,12]. Generation of these data sets requires >40 man hours from a labor-intensive, meticulous and combined effort of expert and secondary observers to achieve a high-quality mapping of selected feature images and is outside the scope of this study. Future validations efforts involving these high-quality data sets promise to provide rigorous evaluation of the accuracy of the computer-generated solution; quantification of potential errors induced from the algorithmic approximations will provide quantitative confidence levels of the measured morphometry statistics. The effect of multiple image templates on the fidelity of the morphometry measurements should also be carefully studied. This will extend the analysis to a statistical setting in which probability maps are provided for the segmentations. Parallelized GPU-based algorithms [10] will also be useful in performing future morphometry studies on larger patient cohorts with multiple templates in an acceptable time frame on commodity resources.
Conclusion
A deformation morphometry image processing pipeline has been developed for monitoring of brain tissue volume changes as a response to radiation therapy. Results indicate that quantitative morphometric monitoring is feasible. Further efforts investigating correlations of quantitative volume change information with radiation dose, age, and cognitive studies may provide insight into monitoring the efficacy of new therapeutic approaches and analyzing toxicity.
Acknowledgments
This work is supported in part by the O’Donnell Foundation and NIH DP2OD007044, NIH DP2OD007044-01S1, and CPRIT RP101502 funding mechanisms. The authors would also like to thank the open source communities ITK, ANTs [4], itk-SNAP [37], and FSL [19] for providing enabling software for image processing and visualization.
Appendix: Gaussian approximation of the Green’s Kernel
Following a diffeomorphic image registration framework [4, 7,14], consider two images, I0 and I1, defined on an Eulerian reference domain, Ω ⊂ ℝ3. The goal of the image registration is to determine the motion, φ(x, t) : Ω × [0, 1] → Ω, that maps the reference image, I0, to the current image, I1.
A symmetric diffeomorphic registration is optimal with respect to a given image similarity metric, d : Ω × Ω → ℝ, and penalized by the velocity of the transformation.
| (3) |
Here, the deformations, φi, i = 1, 2, are defined with respect to the midpoint, t = .5, of the transformation. Time is parameterized in opposite directions between φ1 and φ2. The operator norm ||·||L is induced by a differential operator of the type, L = αΔ + I d, α ∈ ℝ [7]. The symmetric diffeomorphic formulation mappings are constructed sufficiently smooth such that the inverse of the motion is well defined, φ−1(x, t) : Ω × [0, 1] → Ω, and gives a consistent solution for the forward and inverse mapping, φ ∘ φ−1 = I d.
The Euler–Lagrange equations provide necessary conditions for which a solution of the optimization formulation (3) must satisfy.
| (4) |
Here, the gradient of the objective function (3) is with respect to the velocity of the transformation. The operator L represents a physics constraint on the deformation solution. Assuming the deformation behaves as a viscous fluid provides an intuitive solution field that may be understood to adhere to first principle conservation laws.
| (5) |
Further, compared with linear elastic displacement models that constrain the accuracy of large deformations because of internal elastic strain, accurate large deformations may be achieved within this viscous fluid model because internal forces disappear over time and the desired deformation can be fully achieved [8]. However, this approach leads to computationally expensive numerical solution schemes that couple the individual components of the deformation. Alternatively, assuming each deformation component is decoupled and diffuses along the respective gradient of the deformation field yields the algorithmically and numerically tractable Gaussian convolution kernel used in SyN [4]. The greedy update at the midpoint in time simplifies to a fixed point iteration on the velocity field [36], and the transformation field is iteratively updated through a finite difference approximation of the velocity
| (6) |
Here, K ★ represents the Gaussian convolution operation and is obtained from a Fourier transform solution of the heat equation. Solutions of the Euler–Lagrange equations may be interpreted as a decoupled component-wise solution to a isotropic heat transfer equation with initial conditions given by the gradient of the similarity metric.
Footnotes
ANTS -s CC[${fixed}${moving},1,5] -t SyN[0.25] -r Gauss[3,0] -i 30x90x20 –use-Histogram-Matching–number-of-affine-iterations10000x10000x10000x10000x10000 –MI-option 32x16000.
Conflict of interest The authors have no conflicts of interest to report.
Contributor Information
D. Fuentes, Email: dtfuentes@mdanderson.org, Department of Imaging Physics, The University of Texas M.D. Anderson Cancer Center, Houston, TX 77030, USA
J. Contreras, Department of Radiation Oncology, The University of Texas, M.D. Anderson Cancer Center, Houston, TX 77030, USA
J. Yu, Department of Radiation Oncology, The University of Texas, M.D. Anderson Cancer Center, Houston, TX 77030, USA
R. He, Department of Imaging Physics, The University of Texas M.D. Anderson Cancer Center, Houston, TX 77030, USA
E. Castillo, Department of Radiation Physics, The University of Texas M.D. Anderson Cancer Center, Houston, TX 77030, USA
R. Castillo, Department of Radiation Physics, The University of Texas M.D. Anderson Cancer Center, Houston, TX 77030, USA
T. Guerrero, Department of Radiation Oncology, The University of Texas, M.D. Anderson Cancer Center, Houston, TX 77030, USA
References
- 1.Armstrong C, Ruffer J, Corn B, DeVries K, Mollman J. Biphasic patterns of memory deficits following moderate-dose partial-brain irradiation: neuropsychologic outcome and proposed mechanisms. J Clin Oncol. 1995;13(9):2263–2271. doi: 10.1200/JCO.1995.13.9.2263. [DOI] [PubMed] [Google Scholar]
- 2.Asai A, Matsutani M, Kohno T, Nakamura O, Tanaka H, Fujimaki T, Funada N, Matsuda T, Nagata K, Takakura K. Subacute brain atrophy after radiation therapy for malignant brain tumor. Cancer. 1989;63(10):1962–1974. doi: 10.1002/1097-0142(19890515)63:10<1962::aid-cncr2820631016>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 3.Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal. 2008;12(1):26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avants BB, Tustison NJ, Song G. A reproducible evaluation of ANTs similarity metric performance in brain image registration. NeuroImage. 2011;54(3):44–2033. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avants BB, Yushkevich P, Pluta J, Minkoff D, Korczykowski M, Detre J, Gee JC. The optimal template effect in hippocampus studies of diseased populations. Neuroimage. 2010;49(3):2457–2466. doi: 10.1016/j.neuroimage.2009.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bayouth JE, Casavant TL, Graham MM, Sonka M, Muruganandham M, Buatti JM. Image-based biomarkers in clinical practice. Semin Radiat Oncol. 2011;21(2):157–166. doi: 10.1016/j.semradonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beg MF, Miller MI, Trouvé A, Younes L. Computing large deformation metric mappings via geodesic flows of diffeomorphisms. Int J Comput Vis. 2005;61(2):139–157. [Google Scholar]
- 8.Bro-Nielsen M, Gramkow C. Fast fluid registration of medical images. Proc Vis Biomed Comput. 1996;4:267. [Google Scholar]
- 9.Brown GG, Lee JS, Strigo IA, Caligiuri MP, Meloy M, Lohr J. Voxel-based morphometry of patients with schizophrenia or bipolar i disorder: a matched control study. Psychiatry Res Neuroimaging. 2011;194(2):149–156. doi: 10.1016/j.pscychresns.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castillo E, Castillo R, Fuentes D, Guerrero T. Computing global minimizers to a constrained B-spline image registration problem from optimal l1 perturbations to block match data. Med Phys. 2014;41(4):041, 904. doi: 10.1118/1.4866891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castillo R, Castillo E, Fuentes D, Ahmad M, Wood AM, Ludwig MS, Guerrero T. A reference dataset for deformable image registration spatial accuracy evaluation using the COPDgene study archive. Phys Med Biol. 2013;58(9):2861–2877. doi: 10.1088/0031-9155/58/9/2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castillo R, Castillo E, Guerra R, Johnson VE, McPhail T, Garg AK, Guerrero T. A framework for evaluation of deformable image registration spatial accuracy using large landmark point sets. Phys Med Biol. 2009;54(7):1849. doi: 10.1088/0031-9155/54/7/001. [DOI] [PubMed] [Google Scholar]
- 13.Cherrier MM, Anderson K, David D, Higano CS, Gray H, Church A, Willis SL. A randomized trial of cognitive rehabilitation in cancer survivors. Life Sci. 2013;93(17):617–622. doi: 10.1016/j.lfs.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christensen GE, Rabbitt RD, Miller MI. Deformable templates using large deformation kinematics. IEEE Trans Image Process. 1996;5(10):1435–1447. doi: 10.1109/83.536892. [DOI] [PubMed] [Google Scholar]
- 15.DeAngelis LM, Delattre JY, Posner JB. Radiation-induced dementia in patients cured of brain metastases. Neurology. 1989;39(6):789–789. doi: 10.1212/wnl.39.6.789. [DOI] [PubMed] [Google Scholar]
- 16.Dupuis P, Grenander U, Miller MI. Variational problems on flows of diffeomorphisms for image matching. Q Appl Math. 1998;56(3):587. [Google Scholar]
- 17.Greene-Schloesser D, Robbins ME, Peiffer AM, Shaw EG, Wheeler KT, Chan MD. Radiation-induced brain injury: a review. Frontiers Oncol. 2012;2(July):73. doi: 10.3389/fonc.2012.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holden M, Hill DLG, Denton ERE, Jarosz JM, Cox TCS, Rohlfing T, Goodey J, Hawkes DJ. Voxel similarity measures for 3-D serial MR brain image registration. IEEE Trans Med Imaging. 2000;19(2):94–102. doi: 10.1109/42.836369. [DOI] [PubMed] [Google Scholar]
- 19.Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. Fsl. NeuroImage. 2012;62(2):782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 20.Kalkanis SN, Linskey ME. Evidence-based clinical practice parameter guidelines for the treatment of patients with metastatic brain tumors: introduction. J Neuro-Oncol. 2010;96(1):7–10. doi: 10.1007/s11060-009-0065-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klein AEA. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. NeuroImage. 2009;46(3):786–802. doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehmann M, Crutch SJ, Ridgway GR, Ridha BH, Barnes J, Warrington EK, Rossor MN, Fox NC. Cortical thickness and voxel-based morphometry in posterior cortical atrophy and typical alzheimer’s disease. Neurobiol Aging. 2011;32(8):1466–1476. doi: 10.1016/j.neurobiolaging.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 23.Liu AK, Marcus KJ, Fischl B, Grant PE, Young Poussaint T, Rivkin MJ, Davis P, Tarbell NJ, Yock TI. Changes in cerebral cortex of children treated for medulloblastoma. Int J Radiat Oncol Biol Phys. 2007;68(4):992–998. doi: 10.1016/j.ijrobp.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 24.Merchant TE, Hua CH, Shukla H, Ying X, Nill S, Oelfke U. Proton versus photon radiotherapy for common pediatric brain tumors: comparison of models of dose characteristics and their relationship to cognitive function. Pediatr Blood Cancer. 2008;51(1):110–117. doi: 10.1002/pbc.21530. [DOI] [PubMed] [Google Scholar]
- 25.Mulhern RK, Palmer SL, Reddick WE, Glass JO, Kun LE, Taylor J, Langston J, Gajjar A. Risks of young age for selected neurocognitive deficits in medulloblastoma are associated with white matter loss. J Clin Oncol. 2001;19(2):472–479. doi: 10.1200/JCO.2001.19.2.472. [DOI] [PubMed] [Google Scholar]
- 26.Murphy K, Van Ginneken B, Reinhardt JM, Kabus S, Ding K, Deng X, Cao K, Du K, Christensen GE, Garcia V, et al. Evaluation of registration methods on thoracic CT: the EMPIRE10 challenge. IEEE Trans Med Imaging. 2011;30(11):1901–1920. doi: 10.1109/TMI.2011.2158349. [DOI] [PubMed] [Google Scholar]
- 27.National Cancer Institute. A snapshot of: Brain and central nervous system cancers. 2013. [Google Scholar]
- 28.Nieder C, Leicht A, Motaref B, Nestle U, Niewald M, Schnabel K. Late radiation toxicity after whole brain radiotherapy: the influence of antiepileptic drugs. Am J Clin Oncol. 1999;22(6):573–579. doi: 10.1097/00000421-199912000-00007. [DOI] [PubMed] [Google Scholar]
- 29.Patchell RA, Tibbs PA, Regine WF, Dempsey RJ, Mohiuddin M, Kryscio RJ, Markesbery WR, Foon KA, Young B. Postoperative radiotherapy in the treatment of single metastases to the brain. J Am Med Assoc. 1998;280(17):1485–1489. doi: 10.1001/jama.280.17.1485. [DOI] [PubMed] [Google Scholar]
- 30.R Core Team: R. A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna: 2013. [Google Scholar]
- 31.Reddick WE, Russell JM, Glass JO, Xiong X, Mulhern RK, Langston JW, Merchant TE, Kun LE, Gajjar A. Magn Reson Imaging. 2000;18(7):787–793. doi: 10.1016/s0730-725x(00)00182-x. [DOI] [PubMed] [Google Scholar]
- 32.Tanner JM. Growth at adolescence. Thomas; Springfield, IL: 1962. [Google Scholar]
- 33.Thirion JP. Tech rep, Research Report RR-2547, Epidure Project, INRIA Sophia. 1995. Fast non-rigid matching of 3D medical image. [Google Scholar]
- 34.Thompson PM, Hayashi KM, Sowell ER, Gogtay N, Giedd JN, Rapoport JL, de Zubicaray GI, Janke AL, Rose SE, Semple J, et al. Mapping cortical change in alzheimer’s disease, brain development, and schizophrenia. Neuroimage. 2004;23:S2–S18. doi: 10.1016/j.neuroimage.2004.07.071. [DOI] [PubMed] [Google Scholar]
- 35.Torres I, Mundt A, Sweeney P, Llanes-Macy S, Dunaway L, Castillo M, Macdonald R. A longitudinal neuropsychological study of partial brain radiation in adults with brain tumors. Neurology. 2003;60(7):1113–1118. doi: 10.1212/01.wnl.0000055862.20003.4a. [DOI] [PubMed] [Google Scholar]
- 36.Tustison NJ, Avants BB. Explicit B-spline regularization in diffeomorphic image registration. Front Neuroinformatics. 2013;7:39. doi: 10.3389/fninf.2013.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. User-guided 3d active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31(3):1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y, Zou P, Mulhern RK, Butler RW, Laningham FH, Ogg RJ. Brain structural abnormalities in survivors of pediatric posterior fossa brain tumors: a voxel-based morphometry study using free-form deformation. NeuroImage. 2008;42(1):218–29. doi: 10.1016/j.neuroimage.2008.04.181. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2591023&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zou K, Wells W, III, Kikinis R, Warfield S. Three validation metrics for automated probabilistic image segmentation of brain tumours. Stat Med. 2004;23(8):1259–1282. doi: 10.1002/sim.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]