Abstract
Cancer cell proliferation and progression require sufficient supplies of nutrients including carbon sources, nitrogen sources, and molecular oxygen. Particularly, carbon sources and molecular oxygen are critical for the generation of ATP and building blocks, and for the maintenance of intracellular redox status. However, solid tumors frequently outgrow the blood supply, resulting in nutrient insufficiency. Accordingly, cancer cell metabolism shows aberrant biochemical features that are consequences of oncogenic signaling and adaptation. Those adaptive metabolism features, including the Warburg effect and addiction to glutamine, may form the biochemical basis for resistance to chemotherapy and radiation. A better understanding of the regulatory mechanisms that link the signaling pathways to adaptive metabolic reprogramming may identify novel biomarkers for drug development. In this review we focus on the regulation of carbon source utilization at a cellular level, emphasizing its relevance to proliferative biosynthesis in cancer cells. We summarize the essential needs of proliferating cells and the metabolic features of glucose, lipids, and glutamine, and we review the roles of transcription regulators (i.e.,HIF-l, c-Myc, and p53) and two major oncogenic signaling pathways (i.e., PI3K-Akt and MAPK) in regulating the utilization of carbon sources. Finally, the effects of glucose on cell proliferation and perspective from both biochemical and cellular angles are discussed.
Keywords: c-Myc, glutaminolysis, HIF-1, MAPK, metabolism, PI3K, Warburg effect
I. INTRODUCTION
Normal cell physiology requires a sufficient supply of reduced carbon sources for the generation of ATP, building blocks and reducing power. The rapid proliferation of tumor cells increases these fundamental needs, which in turn demands accelerated utilization of carbon sources. However, solid tumors frequently outgrow their blood supply, which leads to insufficient carbon sources, nitrogen sources and molecular oxygen. One well-known adaptive strategy is angiogenesis, and the heterogeneous nature of tumor-associated angiogenesis leads to uneven distribution of blood flow, resulting in ischemic lesions in solid tumors. Therefore, cellular level metabolic adaptation to nutrient insufficiency is critical for tumor progression and invasion. Two prominent characteristics of tumor cell metabolism are the Warburg effect and glutaminolysis, which respectively illustrate the dependence of tumor cells on glucose and glutamine.
Distinguishable from the Pasteur effect, which reflects anaerobic fermentation of glucose, the Warburg effect is defined as the increased glucose consumption and lactate secretion in tumor cells even in the presence of sufficient oxygen.1 Based on experimental data, Warburg proposed that in cancer cells the glucose utilization was switched from oxidative phosphorylation to glycolysis.1 The Warburg effect successfully drew the attention of cancer biologists to the glucose metabolism in cancer cells. As increased glucose uptake was observed in various cancer cell lines with the application of the imaging technique 18 fluorodeoxyglucose (FdG) positron-emission tomography (PET), the Warburg effect has been recognized as a universal metabolic feature of a variety of cancers.2, 3
Glutamine has been considered as an important nutrient for cultured cells since the 1950s. Enhanced glutaminolysis has been observed in various types of tumors. It is generally believed that through glutaminolysis, glutamine provides another type of reduced carbon source, which facilitates the biosynthesis of macromolecules, energy production, and redox maintenance.4–8
Here, we review carbon source metabolism in cancer cells, focusing on recent literature that has advanced our understanding of how different types of carbon sources satisfy the needs of cancer cell survival, growth, and proliferation. We summarize how oncogenic pathways regulate carbon source utilization by reprogramming the gene transcription or by altering the activity of metabolic enzymes, and we discuss how cancer cells sense and respond to changes of glucose availability.
II. TYPES OF CARBON SOURCES AND PHYSIOLOGICAL ROLES
Three types of organic molecules are utilized by human cells as carbon sources: carbohydrates, amino acids, and lipids. Cell proliferation dictates the generation of ATP and organic molecules of cell components as well as the reducing power in the form of NADPH from the carbon sources. Although the utilization of carbon sources by cells in vivo is a rather complicated issue, the establishment of cell culture models has simplified the analysis of nutrient needs to support cell survival and proliferation. Fatty acids can be used directly for the biosynthesis of phospholipids or for the production of acetyl-CoA, which can be used for ATP production or biosynthesis. It is generally accepted that all cells can synthesize fatty acids to meet the needs of biomembrane biosynthesis during cell proliferation, and acetyl-coA can be obtained from carbohydrates and amino acids. Therefore, it is not surprising to note that fatty acids are not needed in cell culture media. In optimized cell culture media, in addition to essential nutrients, abundant amounts of glucose and glutamine are added. Notably, while animal sera are added generally to meet the needs of growth hormones, they also contain glucose, glutamine, fatty acids and other nutrients. When dialyzed sera are used in studies, it has been shown that both glucose and glutamine are absolutely required for most cell types,9 suggesting that each has an indispensible role in supporting cell proliferation. Therefore, glucose and glutamine, two non-essential nutrients at the organismal level, are essential for most cells cultured ex vivo.
A quick review of the biochemical and metabolic features of different types of carbon sources may help our understanding of their importance and their unique roles in cancer cell metabolism (Figure 1). To facilitate the discussion, we have listed the major enzymes (including transporters) implicated in the utilization of glucose and glutamine as carbon sources in Table 1. Upon entering cells, the majority of glucose is phosphorylated to form glucose-6-phosphate (G6P), which is the substrate for multiple metabolic pathways, including the glycolytic and pentose phosphate pathways. Through sequential glycolysis and pyruvate dehydrogenation, glucose provides acetyl-CoA, which can be used either for biosynthesis or for ATP production through the tricarboxylic acid (TCA) cycle and oxidative phosphorylation. Even under hypoxic conditions, glucose can support ATP production through fermentation. Through the pentose pathway, glucose satisfies the needs of NADPH. ATP, acetyl-CoA, NADPH and other intermediate metabolites of glycolysis, the pentose phosphate pathway, and the TCA cycle satisfy the needs for biosynthesis of a variety of biomolecules, including non-essential amino acids, fatty acids, and nucleotides. On the other hand, the use of fatty acids is limited to being oxidized to acetyl-CoA, which can be used to generate ATP or can participate in biosynthesis. Therefore, metabolically, glucose is a versatile reduced carbon source that should be sufficient to meet all cellular needs for carbon. In addition to carbohydrates and lipids, amino acids represent another type of carbon source. Particularly, glutamine provides carbon through glutaminolysis and subsequent conversion of glutamate to α-ketoglutarate. Given the roles of glutamine and glutamate in nitrogen anabolism, it is not difficult to appreciate their essentiality in cell culture systems.9 Then why cannot glutamate, which is eventually catabolized to α-ketoglutarate, be directly used in cell culture to replace glutamine? One obvious explanation could be that cells do not have effective transporters to uptake sufficient amounts of glutamate. In fact, cells need to maintain high levels of glutamate intracellularly. In addition to serving as a central hub to balance the amino acid pool through various transamination reactions, glutamate is required for multiple biosynthetic pathways that need nitrogen. Biosynthesis of glutathione and polyamines are two rather important examples. Upon entering the TCA cycle, the glutamine-derived α-ketoglutarate may end up in numerous organic metabolites; some may serve as a substrate for the production of NADPH.10 However, in the presence of glucose and pentose pathway, this function does not justify its essentiality for cell proliferation. Theoretically, α-ketoglutarate from glutaminolysis may be used in gluconeogenesis, or it may be converted to other organic molecules through anaplerotic pathways. In most cases, however, glutamine cannot support cell proliferation in glucose-free media, suggesting that the carbons from glutamine catabolism are not capable of completely replacing glucose. Moreover, replenishing intracellular α-ketoglutarate or oxaloacetate levels cannot rescue cell growth in glutamine-free media, either.9 If alternative pathways to generate glutamate in cells are provided, cells become capable of growing in glutamine-free media, indicating that the carbons from glutamine catabolism are not absolutely necessary for cell proliferation. Therefore, although glutaminolysis provides an alternative source of carbon, its essential role in supporting cell proliferation may be as a precursor of glutamate and, as such, participates in various nitrogen anabolic pathways.
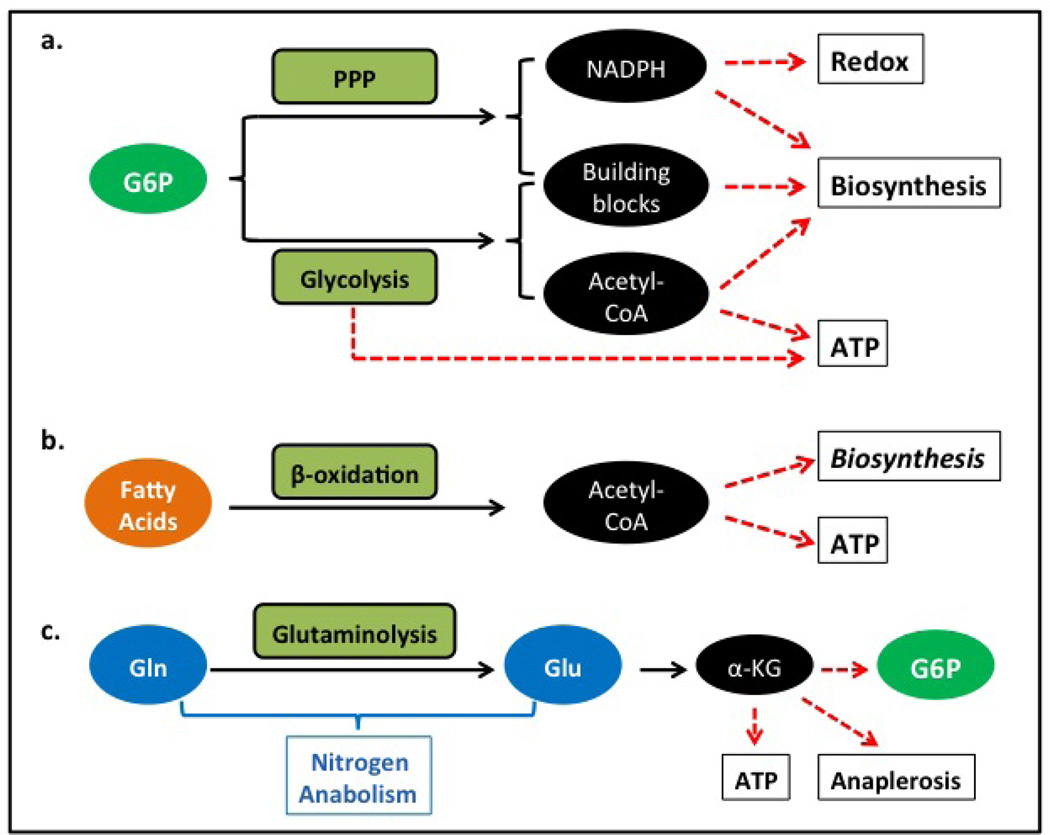
FIGURE 1.
Summary of the major types of carbon sources for cancer cells. A. Glucose is a universal carbon source that may fulfill all cell needs for carbon. Glucose-6 phosphate (G6P), derived from glucose, is the common substrate for multiple metabolic pathways. Specifically, the pentose phosphate pathway (PPP) regulated by G6PD activity is responsible for the production of NADPH, riboses, and other metabolites for biosynthesis. Glycolysis, regulated by PFK1 activity, generates pyruvate, which may be either oxidized to acetyl-CoA, or reduced to lactate. Pyruvate, acetyl CoA, and other metabolites of the glycolytic pathway can be used for biosynthesis. B. The function of fatty acids as a carbon source is limited to the generation of acetyl CoA, which can be used either for oxygen-dependent, electron transfer chain-dependent generation of ATP, or for some biosynthetic pathways. Note that fatty acids can be directly used for the biosynthesis of phospholipids. C. Glutamine (Gln) serves as a precursor of glutamate; both glutamate and glutamine have important roles in nitrogen-required anabolic pathways, including the synthesis of nucleotides, proteins, glutathione, heme, polyamines, and non-essential amino acids. Importantly, through transamination or oxidative deamination, glutamate can be converted to α-ketoglutarate (α-KG), which links amino acid metabolism to carbon source metabolism. By entering the Krebs cycle, α-KG can be either directly oxidized as an energy source or used in anaplerotic reactions and converted to other metabolites such as malate and isocitrate, which can be used to generate NADPH. In some types of cells, α-KG can be converted to G6P through gluconeo-genesis. If the conversion of Gln to glucose is sufficiently efficient, theoretically Gin should be able to replace glucose as a universal carbon source.
TABLE 1.
Currently available TB diagnostics
|
Glucose and glutamine update and trapping | |
| Glucose transporters | Updake glucose |
| Hexokinases | Phoshorylate glucose to G6P |
| Glutamine transporters | Uptake of glutamine |
| Glutaminases | Convert glutamine to glutamate |
| Glutamate dehydrogenase | Convert glutamate to α-ketoglutarate |
| Glutamate transaminases | Convert glutamate to α-ketoglutarate |
|
Glycolytic ATP Production | |
| Phosphofructose kinase 1 (PFK1) | Glycolytic enzyme, rate limiting |
| Pyruvate kinase | Glycolytic enzyme, phosphoenolpyruvate (PEP) to pyruvate |
| Phosphofructose kinase 2 (PFK2) | Glycolysis regulator, generate F2, 6BP |
| Lactate dehydrogenase | Interconversion between pyruvate and lactate |
|
Oxidative phosphorylation | |
| Pyruvate dehydrogenase kinase (PDK) | Repressor of pyruvate dehydrogenase complex |
| Pyruvate dehydrogenase complex | Pyruvate to acetyl CoA |
| TCA cycle enzymes | Oxidizing carbon sources, to generate NADH & FADH2 |
| Inter-conversion among carbon metabolites for biosynthesis | |
| Electron transfer complexes | Pass electrons from NADH/FADH2 to molecular O2 |
| ATP synthase | Regenerate ATP from ADP |
|
Cytosolic NADPH production | |
| Glucose-6-phospate dehydrogenase (G6PD) | Pentose phosphate pathway |
| Malic Enzyme | NADPH: malate to pyruvate |
| Isocitrate dehydrogenase 1, 2 | NADPH: isocitrate to α-ketoglutarate |
|
Biosynthesis of membrane | |
| ATP-dependent citrate lyase | Biosynthesis: acetyl CoA generation in cytosol |
| Acetyl CoA carboxylase (ACC) | Fatty acid synthesis: acetyl CoA to malonyl CoA |
| Fatty acid synthase (FASN) | Fatty acid synthesis: multiple activities, synthesize palmitate |
| HMG-CoA reductase | Cholesterol biosynthesis: HMG-CoA to mevalonate |
| Glycerol kinase | Phospholipid biosynthesis: glycerol to glycerol-3 |
| Glycerol-3-phosphate dehydrogenase (GPDH) | Phospholipid biosynthesis: DHAP to glycerol-3P |
| Phosphoenopyruvate carboxylkinases (PEPCK) | Convert oxalocaetate to PEP to DHAP to glycerol-3P |
| Acyl CoA transferases | Phospholipid biosynthesis: link fatty acid to glycerol-3P |
| Diacylglycerol (DAG) kinase | Phospholipid biosynthesis: DAG to phosphatidic acid |
| Serine C-palmitoyltransferases | Sphingosine biosynthesis |
|
Biosynthesis of other important metabolites for biosynthesis | |
| Pyruvate carboxylase | Convert pyruvate to oxaloacetate |
| Phosphoglycerate dehydrogenase (PHGDH) | Serine synthesis: 3-P-glycerate to 3-P-hydroxypyruvate |
| Serine hydroxymethyltransferase (SHMT) | Produce glycine and methylenetetrahydrofolate |
| Dihydrofolate reductase (DHFR) | Reduce dihydrofolate to tetrahydrofolate |
III. HOMEOSTATIC REGULATION OF CARBON SOURCE UTILIZATION
It is generally accepted that tumor cells require an increased, undisrupted supply of energy to support their survival, growth, and proliferation. Like normal cells, tumor cells continuously monitor the intracellular ATP levels, and accordingly they regulate the oxidation of the carbon source. The bioenergetic processes are homeostatically regulated by changes of ATP and ADP levels, which have been detailed in most biochemistry textbooks. In eukaryotic cells, the concentrations of ATP and ADP are much higher than that of AMP; a slight decrease of ATP level leads to a remarkable change of AMP levels. Therefore, change of AMP concentration is a more sensitive indicator of energy status. Accordingly, the AMP-activated protein kinase (AMPK) pathway has a very crucial role in the process. When the cellular ATP level is low, AMP concentration increases, which allosterically activates AMPK. Increased AMP levels also protect phosphorylated AMPK from dephosphorylation.11–15 Activated AMPK enhances ATP levels by coordinately reprogramming several cellular processes. First, AMPK inhibits the biosynthesis of fatty acids, cholesterol, glycogen, and proteins, conserving energy and diverting more carbon sources to ATP production.16–19 Second, AMPK activation stimulates catabolic pathways to increase ATP production; AMPK activates many catabolic enzymes participating in the glycolysis and fatty acid oxidation.20, 21 Third, AMPK activation can also cause a Gl phase cell cycle arrest and prevent the entry into S phase where a large amount of ATP is required.22 In normal myocytes and adipocytes, AMPK has been reported to collaborate with the insulin signaling pathway to promote the translocation of glucose transporter 4 (GLUT 4)23 and in the long term the expression of GLUT4.24 By increasing the transcriptional coactivator PGC-lα, AMPK can up-regulate mitochondrial biogenesis as well,25 facilitating ATP production and cellular adaptation. By enhancing ATP production and inhibiting ATP consumption, the AMPK pathway delicately maintains the energy homeostasis in tumor cells.
For all types of cells, a balance between reducing power and oxidizing power is crucial for normal cell function. In most cases, molecular oxygen serves as the final electron acceptor, being the major oxidizing power in cells. Two important molecules working as the reducing power are NADPH and glutathione (GSH). By donating electrons to oxidized glutathione (glutathione disulfide, GSSG), NADPH maintains the homeostatic ratio of [GSH]/[GSSG]. NADPH and GSH collaboratively protect enzymes and cellular structures from damage by free radicals or non-radical reactive oxygen species (ROSs). Although ionization and UV are considered the exogenous sources of free radicals or ROSs, incomplete reduction of oxygen during oxidative phophorylation in mitochondria serves as the major endogenous source of ROSs. In addition, oxidations catalyzed by other oxidoreductases such as NAD(P)H oxidases and xanthine oxidase may generate ROSs as well.26 Whereas adequate levels of ROS have various biological functions, ranging from signal transduction to regulation of gene expression,27 high levels of ROSs can cause damage to macromolecules, including DNA, proteins, and lipids.28 Recently it has been reported that ROSs also activate protein kinase C delta (PKC delta) to stimulate senescence29 and trigger the release of cytochrome c by permeating the mitochondria,30, 31 which eventually leads to apoptosis. Finally, the role of NADPH in reductive biosyn-thetic reactions as electron donor has been well established.
The ratio of [NADPH]/[NADP+] is dynamically maintained by oxidizing reduced carbon sources. Metabolically, NADP+ can be converted to NADPH in several ways: (1) the pentose phosphate pathway, which oxidizes phosphorylated glucose, G6P; (2) the reaction catalyzed by malic enzyme 1 (ME1) with malic acid as the substrate;10 and (3) the oxidation of glutamate catalyzed by glutamate dehydrogenase; and (4) the reaction converting isocitrate to α-ketoglutarate catalyzed by isocitrate dehydrogenase 1 and 2 (IDH 1 and 2) in cytosol, which has been recently discovered to use NADP+ as a cofactor, generating NADPH.32 It is generally accepted that the pentose pathway, which directly uses G6P as substrate, is a universal and the most important mechanism. Accordingly, this pathway is homeostatically regulated by the cytosolic NADPH levels. The rate-limiting enzyme of the pentose phosphate pathway (PPP) is glucose-6-phosphate dehydrogenase (G6PD), which catalyzes the first of the two oxidation steps of the PPP. An increase of NADPH allosterically represses G6PD activity, serving as a negative feedback mechanism.
Another major need of carbon sources is the biosynthesis and maintenance of biomembranes, which require the synthesis of phospholipids and sterols. The biosynthesis of phospholipids requires the synthesis of fatty acids, cholesterol, and glycerate-3-phosphate. The regulation of fatty acid and cholesterol biosynthesis has been extensively studied, mostly in hepatocytes and adipocytes in the context of adipogenesis. The de novo synthesis of sterols is controlled by sterol regulatory element (SRE) and SRE-binding proteins (SREBPs). SREBPs are a family of membrane-bound transcription factors. Normally SREBPs are inserted in the membranes of endoplasmic reticulum (ER), bound to the SREBP cleavage-activating protein (SCAP), which is both the escort of SREBP and the sterol sensor. When the intracellular sterol level is low, SREBP migrates to the Golgi apparatus, where SREBP is cleaved by site-1 and site-2 proteases (S1P and S2P) which are activated by SCAP. The cleaved SREBP then moves into the nucleus and acts as a transcription factor to up-regulate the transcription of more than 30 genes; most of which are involved in the synthesis or uptake of cholesterol, fatty acids, phospholipids, and triacylglycerols.33 The nuclear hormone receptors liver X receptors (LXRα and LXRβ interact with retinoid X factor (RXR) and function as heterodimeric transcription factors.34 In response to an increase of cholesterol and particularly oxysterols, LXR/RXR dimers bind to LXR-responsive elements (LXRE, AGGTCAnnnnAGGTCA) and up-regulate the expression of SREBP-lc, ChREBP, and genes involved in adipogenesis.35 Finally, the peroxisomal proliferator-activated receptors (PPARα , -β, -γ) are also known for regulating the utilization of glucose and lipids.36 However, it remains unclear whether these regulatory mechanisms for lipogenesis are fully or partly adopted for the production of biomembranes during cell division. How tumor cells sense the levels of fatty acids and glycerate-3-phosphate during cell proliferation also remains an interesting question.
Finally, several non-essential amino acids, including glutamine, glutamate, aspartate, serine, and glycine, are needed in large quantities in actively proliferating cells. These amino acids are either abundant in proteins or are required for the biosynthesis of nucleotides, heme, and other nitrogenous molecules. Glutamine fulfills cancer cells’ needs for glutamine and glutamate. Biosynthesis of alanine, aspartate, serine, and glycine make demands on the supply of carbon skeletons in the form of intermediate metabolites such as pyruvate, oxaloacetate, and 3-P-glycerate. Major enzymes required for the synthesis of these important metabolites are provided in Table 1 as well.
IV. TRANSCRIPTIONAL REGULATORS OF CARBON SOURCE METABOLISM IN TUMOR CELLS
Because cancer cell proliferation demands the synthesis of DNA, RNA, proteins, and biomembranes, oncogenic transformation should be able to promote the utilization of reduced carbon sources. Moreover, it is reasonable to argue that the oncogenic signaling pathways should coordinate the production of ATP, NADPH, acetyl-CoA and other organic metabolites that are needed in the biosynthesis of non-essential amino acids, nucleotides and phospholipids. Therefore, oncogenic transformation can be predicted to be associated with metabolic reprogramming, an essential process at least partly achieved by transcriptional reprogramming of genes involved in metabolism. Extensive studies have implicated many transcription factors in reprogramming gene expression in tumor cells.
HIF-1 is the major transcription factor regulating the expression of many genes in response to low oxygen conditions. HIF-1 is a heterodimer composed of the constitutively expressed HIF-1 β (also known as ARNT) and the function-determinant HIF-1 α. In the presence of oxygen, HIF-1 α is hydroxylated by oxygen-activated prolyl hydroxylase enzymes, then is recognized by tumor suppressor von Hippel-Lindau (VHL), an E3 ubiquitin ligase, and finally is degraded by the proteasome.37 HIF-1 can up-regulate the transcription of many genes to promote glucose utilization (Table 2), such as glucose transporter 1 (GLTU1) and glycolytic enzymes including lactate dehydrogenase A (LDH A), which diverges pyruvate to lactate.38–40 To promote cellular adaptation to the acidosis caused by increased fermentation, HIF-1 enhances the expression of the carbonic anhydrase CAIX and the lactate/H+ symporter MCT4.41, 42 In addition to promoting the glycolytic pathway, HIF-1 also activates the pyruvate dehydrogenase kinase (PDK1), which deactivate the mitochondria pyruvate dehydrogenase complex.43 As such, HIF-1 activation represses the generation of acetyl-CoA from pyruvate, slows down the Krebs cycle, and indirectly inhibits oxidative phosphorylation.
TABLE 2.
Major Metabolic Enzymes Up-regulated by HIF-1
The transcription factor c-Myc is responsible for many human cancers.44 Genes up-regulated by c-Myc also contribute to promoting the glycolytic phenotype in tumor cells (Table 3). It has been established that the expression of many glycolytic enzymes such as GLUT1, hexokinase 2 (HK2), phosphofructokinase (PFKM), and enolase 1 are stimulated by c-Myc.45–47 As HIF-1 also up-regulates these glycolytic enzymes, one can infer that c-Myc and HIF-1 are functionally related and interplay with each other to regulate glucose metabolism under various conditions. In addition to these enzymes, which are directly involved in glycolysis, c-Myc and HIF 1 also enhance the expression of lactate dehydrogenase (LDH), which facilitates the lactate formation and NAD+ regeneration. On the other hand, they inhibit mitochondrial oxidative phosphorylation by up-regulating pyruvate dehydrogenase kinase 1 (PDK1), which phosphorylates and inhibits pyruvate dehydrogenase (PDH), reducing the entry of pyruvate into the TCA cycle.46
TABLE 3.
Major Metabolic Enzymes Up-regulated by c-Myc
As a tumor suppressor and a transcription regulator, p53 is well known for its role in growth arrest, DNA damage response and apoptosis.48 Recent studies reveal that it is also an important regulator of cell metabolism.49, 50 Active p53 contributes to promoting oxidative phosphorylation and to slowing down glycolysis. Wild-type p53 inhibits glucose uptake by reducing the expression of GLUT1 and GLUT4. It also has been reported that p53 increases the expression of cytochrome c oxidase 2 (SC02), an enzyme required for assembly of the mitochondrial electron transport chain.51 As a target gene of p53, TP53-induced glycolysis and apoptosis regulator (TIGAR) decrease the level of fructose-2,6-biphosphate, which is an allosteric stimulator of phosphofructose kinase 1 (PFK1), the key regulatory enzyme of the glycolytic pathway.52, 53 Considering the inhibitory effect of p53 on glucose utilization, and the high incidence of p53 mutation in many tumors, p53 mutation likely not only promotes the malignant transformation and tumorigenesis but also facilitates the altered carbon utilization in tumor cells.
Both p53 and c-Myc regulate the utilization of amino acids. As an oncogene, c-Myc has been shown to promote the catabolism of glutamine in tumor cells. At the beginning of glutamine-utilizing processes, glutamine transporters are required for glutamine uptake by tumor cells. Upon entering the cells, glutamine is first hydrolyzed by a procedure termed glutaminolysis that is catalyzed by glutaminases. It has been shown that knockdown of c-Myc significantly decreased the expression of high-affinity glutamine transporters ASCT 2 and SN2, suggesting that c-Myc has an essential role in glutamine uptake.54 In addition, c-Myc has been also reported to activate glutaminolysis by repressing the expression of miR-23a/b, a microRNA that directly targets glutaminase 1 (GLS).55 On the other hand, as a tumor suppressor, p53 up-regulates glutaminase 2 (GLS2).56, 57 Whereas the biological significance of the differential regulation of glutaminase 1 and 2 remains elusive, up-regulation of glutaminase 1 is more closely related to tumor cell metabolism.
V. ONCOGENIC PATHWAYS PROMOTE THE UTILIZATION OF CARBON SOURCES IN TUMOR CELLS
Cell proliferation demands the coordinated production of energy, reductive equivalents, and building blocks.58 This coordination is mainly controlled by transcriptional reprogramming that results from the activation or inactivation of specific transcription factors. Therefore, it is not surprising to realize that oncogenic signaling pathways can regulate transcription factors and can eventually facilitate the carbon source utilization, rendering distinct metabolic features of tumor cells, such as the Warburg effect and active glutaminolysis.9 Among a variety of oncogenic signaling pathways, PI3K-AKT and MAPK represent the two most common ones activated in human tumors.
The PI3K-AKT signaling pathway is a critical cascade with various physiological functions, such as regulating cell survival, proliferation, motility, and promoting metabolism and angiogenesis.59, 60 It has been well-established that under physiological conditions, insulin activates the PI3K-AKT pathway. In tumors, other stimuli may activate this pathway in an insulin-independent manner; constitutive activation of the PI3K-AKT pathway induces tumorigenesis.61 Furthermore, maintaining the activation of PI3K-AKT pathway contributes to the coordination of tumor cell metabolism with rapid cell growth. The PI3K-AKT pathway is activated by the binding of growth factors or insulin to receptor tyrosine kinases (RTKs).59, 61 In addition to ligand binding, mutation of the tumor suppressor phosphatase and tensin homolog (PTEN) also leads to the activation of the PI3K-AKT pathway.62
AKT, also known as protein kinase B (PKB), has a central role in tumor cell metabolism. Activated AKT phosphorylates and inactivates TSC2, resulting in the activation of Rheb GTPase, which directly activates the mammalian target of the rapamycin complex 1 (mTORCl), an important factor in reprogramming the transcription and the metabolism of tumor cells.63 Particularly, activated mTORCl leads to the accumulation of HIF-lα,64 resulting in upregulation of HIF-responsive genes, such as GLUT1, HK2, and LDH.65 Increased glucose uptake and HK2 activity facilitates tumor cells to trap glucose as glucose-6-phosphate, which is the substrate for multiple metabolic pathways, including glycolysis and the pentose phosphate pathway.64 In addition, AKT enhances the transcription of phosphofructokinase-2 (PFK2)66 and directly phosphorylates and activates PFK2.67 A PFK2 product, fructose-2,6-bisphosphate, leads to allosteric activation of PFK1 that increases the glycolytic flow. The activation of the PI3K-AKT-mTORCl signaling pathway induces HIF-lα expression, which in turn enhances pyruvate kinase isoenzyme type M2 (PKM2) expression through collaboration with c-Myc-hnRNPs splicing regulators.68 Increased PKM2, along with other glycolytic enzymes, enhances aerobic glycolysis.68 Finally, activated AKT enhances the glucose uptake through upregulating the expression of GLUT1, GLUT2, and/or translocation of GLUT4 to the plasma membrane.69, 70 AKT also coordinates the carbon source metabolism to facilitate lipid synthesis. The accumulation of HIF-lα by mTORCl leads to the up-regulation of ATP-citrate lyase (ACL), which cleaves citrate from the TCA cycle for acetyl-CoA generation in cytosol, which can be used for the synthesis of fatty acids and cholesterol.11 Moreover, AKT may activate ACL by directly phosphorylating it.71 PI3K-AKT-mTORCl has been shown to activate SREBP through enhancing its trafficking from the endoplasmic reticulum to the Golgi apparatus and to subsequently facilitate proteolytic processing to produce its active form.72, 73 Additionally, PI3K-AKT has been reported to increase the expression of fatty acid synthase (FASN) to facilitate fatty acid synthesis in prostate cancer cell lines and primary tumors. 74, 75 The MAPK signaling pathway is critical in the regulation of cellular metabolism. It is activated by the sequential phosphorylation/activation of a cascade of kinases including MAPKKK, MAPKK, and MAPK.76 Eventually, activated MAPK phosphorylates the targeted proteins to perform biological functions. Four mammalian MAPK signaling pathways have been reported: extracellular signal-regulated kinase 1 and 2 (ERK1/2), c-Jun N-terminal kinase (JNK), p38, and ERK5.77 MAPK has been reported to regulate a series of cellular processes, such as cell survival, differentiation, proliferation, motility, and stress responses.78–80 In human MCF-7 breast cancer cells, the MAPK inhibitor PD98059 reduces insulin-induced glucose uptake and inhibits tumor cell proliferation.81 In simulated T lymphocytes, ERK1/2 has been shown to induce glucose uptake and glycolysis by increasing the expression and activation of HK.82 p44/p42 MAPK also regulates HIF transactivation through enhancing the formation of the HIF-p300/CBP complex.83, 84 Moreover, ERK1/2 has been reported to phosphorylate TSC2, leading to the dissociation of the TSC1-TSC2 complex and the activation of the mTOR signaling pathway.85 Activated mTOR phosphorylates S6K and 4E-BP1 that control translation.59, 72 Due to mTOR’s critical role in metabolism, it has been suggested that MAPK may control tumor cell metabolism through activating mTOR. ERK1/2 also regulates the uptake and metabolism of glutamine in activated T lymphocytes.86 ERK1/2 activation by CD28 induces the expression of the SNAT (SLC38) family of transporters that increase glutamine uptake.86 Moreover, ERK1/2 coordinates glutamine uptake with glutamine metabolism through increasing the activity of glutaminase, glutamine dehydrogenase, glutamic-oxaloacetic transaminase, and glutamic-pyruvic transaminase.86 Glutamine addiction and active glutaminolysis are metabolic characteristics of highly proliferating tumor cells.9 The constitutive activation of MAPK in human tumor indicates that it may participate in glutamine metabolism. In addition, p38 MAPK, activated by MKK6/3, has been shown to promote glucose transport through enhancing the expression of GLUT, 1 regardless of the stimulus.87 In addition to these factors, PI3K-AKT and ERK1/2 have been reported to up-regulate c-Myc expression in tumor cells,88, 89 which controls the expression and activity of enzymes involved in glucose or glutamine metabolism. Accordingly, c-Myc also has been proposed to lead to Warburg effect or active glutaminolysis.90–92
VI. ADAPTIVE METABOLISM OF TUMOR CELLS TO GLUCOSE AVAILABILITY
Whereas oncogenic transformation may promote utilization of glucose by a tumor cell, glucose availability may alter the metabolism of tumor cells. Normal cells respond to glucose concentrations via modulating the glucose transporter activity, hexokinase activity, and the activities of other enzymes of carbon metabolic pathways, depending on the cell types. These responses are generally triggered either directly by extracellular glucose concentrations or indirectly by glucose homeostatic hormones including insulin, glucagon, and epinephrine. The tissue-specific expression of receptors of the hormones, glucose transporters, and isoenzymes forms the biological bases for tissue-specific responses of normal cells to glucose fluctuation. Hormone-mediated systematic responses have been studied extensively; an emerging concept is that physiological responses to glucose availability also occur at the cellular level. While the precise mechanism underlying the glucose sensing at a cellular level remains elusive, two transcription complexes have been found to regulate gene expression in response to high concentrations of glucose: MondoA:Mlx and MondoB:Mlx. The basic helix-loop-helix-leucine zipper (bHL-HZip) dimeric transcription factor MondoA:Mlx complex shuttles between the outer mitochondrial membrane (OMM) and the nucleus, depending on the intracellular glucose concentration.93, 94 When the intracellular level of glucose increases, the MondoA:Mlx complex migrates into the nucleus and up-regulates its target genes, which are summarized in Table 4. MondoB is also called a carbohydrate response element binding protein (ChREBP); it was originally identified to physically interact with the carbohydrate response element (ChoRE). ChoRE was first found in a number of rodent genes involved in glycolysis and lipid metabolism, such as L-type pyruvate kinase (LPK) and acetyl-CoA carboxylase (ACC).95–97 Based on these DNA motifs, ChREBP was identified.98, 99 Elegant work from Ma et al. demonstrated that in response to high glucose, MondoB:Mlx plays a major role in stimulating glucose sensitive genes in liver.100 Whereas MondoA and MondoB up-regulate an overlapping repertoire of genes in response to high glucose levels (Table 5), their tissue-type-specific expressions are different. MondoA is highly expressed in skeletal muscle, whereas MondoB is highly expressed in the liver.101, 102 Consistently, knockout of Mondo B impairs the lipogenesis in mouse liver.103 Analysis of the functions of up-regulated genes revealed that the MondoA:Mlx and MondoB:Mlx-mediated transcriptional reprogramming leads to enhanced catabolism of glucose, enhanced utilization of glucose for carbon anabolism, but decreased glucose uptake. These responses clearly represent adaptive metabolic alterations.Transcriptional reprogramming to low glucose concentrations has not been extensively studied. As discussed, the AMP-activated protein kinase (AMPK) pathway serves as a prominent energy sensor. Any decrease in the ATP:AMP ratio upon glucose deprivation may trigger AMPK, which in turn inhibits biosynthesis activities. When glucose deprivation is continuous, AMPK can further work as an intrinsic regulator of the cell cycle that coordinates cellular proliferation with carbon source availability. In response to glucose deprivation, activated AMPK collaborates with cAMP-responsive element-binding protein (CREB) to transcriptionally regulate the expression of p53, a well-known inhibitory factor of cell proliferation.22, 104, 105 AMPK can also inhibit the mTOR signaling pathway by phosphorylating the tuberous sclerosis complex protein 2 (TSC 2), the negative regulator of mTOR. The inactivation of the mTOR pathway can then contribute to protecting tumor cells from apoptosis under glucose-deprived conditions.19 AMPK also protects tumor cells partially via induction of dual-specificity phosphatases (DUSPs), which suppress pro-apoptotic extracellular signal-regulated kinase (ERK).106 In prostate cancer cells, glucose deprivation activates c-Jun N-terminal kinase (JNK), which can promote cell survival in the early phase and induces apoptosis in the late phase, and the AMPK signaling pathway works as the key regulator of the dual function of JNK.107 In colorectal cancer cells, glucose deprivation induces the increased expression of receptors for adiponectin, which promotes cell survival through enhancing autophagy by activating AMPK and PPARα, but inhibiting PI3K-AKT pathway.108 In addition to activating the AMPK signaling pathway, glucose deprivation also induces AMPK-independent responses. Several genes have been reported to be triggered by low glucose via AMPK-independent mechanisms. In MCF-7 cells, glucose deprivation is reported to reduce synthesis of collagen, the main component of extracellular matrix.109 Follistatin, which is traditionally recognized as a secretory protein that inactivates extracellular activin, myostatin, and bone morphogenic proteins, has been reported to promote cancer cell survival under glucose-deprived conditions through inhibiting cellular rRNA synthesis.110–113 Glucose deprivation also stimulates O-GlcNaA modification of proteins through up-regulation of O-Linked N-acetylglucosaminyltransferase, which may play a role in nutrition sensing.114–116 At the protein level, glucose deprivation leads to Chkl degradation through the ubiquitin-proteasome pathway, which is a key regulator in the DNA replication checkpoint.117
TABLE 4.
Target Genes Up-regulated by MondoA:Mlx Complex
TABLE 5.
Target Genes Regulated by MondoB:Mlx Complex
| Genes | Swiss Prot | References | |
|---|---|---|---|
|
Carbon metabolism | |||
| Slc2a4 | Glucose transporter 4 | P19357 | 100 |
| Gckr | Glucokinase regulatory protein | Q07071 | 100 |
| Pklr | Liver-type pyruvate kinase | P12928 | 98 |
| Gpd1 | Glycerol-3-phosphate dehydrogenase 1 | O35077 | 100 |
| Acaca | Acetyl-CoA carboxylase 1 | P11497 | 123 |
| Fasn | Fatty acid synthase | P12785 | 124 |
| Gpam | Glycerol-3-phosphate acyltransferase 1, mitochondrial | 125 | |
|
Metabolic regulators | |||
| G0S2 | G0/G1 switch gene 2 | Q5M840 | 100 |
| Thrsp | Thyroid hormone-inducible hepatic protein | P04143 | 124 |
| Fgf21 | Fibroblast growth factor 21 | Q8VI80 | 100, 126 |
| Ppara* | Peroxisome proliferator-activated receptor á | P37230 | 127 |
| Arnt* | Aryl hydrocarbon receptor nuclear translocator | P41739 | 128 |
|
Redox-related protein | |||
| TXNIP | Thioredoxin-interacting protein | Q9H3M7 | 129 |
Ppara and Arnt are down-regulated by the MondoB:Mlx complex. All other genes are up-regulated.
VII. PERSPECTIVES AND CONCLUSION
Carbon source utilization is fundamental for cell proliferation and tumor growth. The Warburg effect is the most obvious biochemical feature of most tumor cells. Although it has been found for more than 80 years, its biological role in tumor progression remains elusive. Using glycolysis as the major way to produce ATP seems to be a disadvantage for tumor cells in competing with surrounding normal cells for carbon sources. One proposed explanation is that metabolites produced in glycolysis facilitate rapid carbon anabolism, such as biosynthesis of non-essential amino acids and lipids, to satisfy the increased demand for growth and proliferation of tumor cells. Under anaerobic conditions, fermentation is required to regenerate oxidized NAD+ so fermentation can continue to provide ATP. Stoichiometrical analysis shows that all glyceraldehyde-3-phosphate molecules entering into the oxidative phase need to be eventually reduced to lactate to maintain the balance between NAD+ and NADH. Therefore, if the Warburg effect provides metabolites for biosynthesis, the metabolites are limited to those available prior to entering the oxidative stage of glycolysis. Another explanation suggests that increased generation of lactate results in acidosis in the tumor microenvironment, which is advantageous for tumor metastasis and invasion, and facilitates the removal of surrounding normal cells.118 However, it is equally valid to consider the tumor metastasis and invasion as an adaptive consequence for tumor cells fighting the disadvantageous microenvironment of acidosis. Finally, lactate formation may be simply a consequence secondary to the inhibition of oxidative phosphorylation in tumor cells, representing an intriguing preference of tumor cells to use pyruvate instead of molecular oxygen as the oxidizing power. Further studies and systematic analyses of the carbon metabolic networks in tumor cells are required to understand the pathobiological significance of the Warburg effect in tumor cells.
Considering the complexity of carbon metabolic networks, both glucose and glutamine can be the ultimate substrates for NADPH production (Figure 1). However, when the glutamine carbon skeleton enters into the PPP, it requires an anaplerotic process followed by gluconeogenesis; its efficiency may be limited by cell-type-specific metabolic features. Generation of NADPH by ME1- or IDHl,2-catalyzed reactions requires the conversion of glutamine to malate or isocitrate via the synthetic function of the Krebs cycle; the efficiency of these reactions in cancer cells may vary. Similarly, the synthetic function of the Krebs cycle and glyceroneogenesis via oxaloacetate-PEP pathway are required for glutamine to form glycerol-3-phosphate for the biosynthesis of membrane phospholipids. The inefficiency of cells to use α-ketoglutarate as a substrate to generate NADPH and glycerol-3-phosphate could be the major reason that glutamine cannot fully compensate the lack of glucose as a carbon source in most cell types.9 These hypotheses remain to be tested.
Theoretically, glucose may satisfy cell needs for carbon sources, but the removal of glutamine stops the growth of most types of cells in cell culture models.9 Analysis of the key enzymes of carbon metabolic pathways revealed that glutamine starvation repressed their expression, suggesting that lack of glutamine represses the utilization of glucose.9 Because α-ketoglutarate cannot always rescue the repressed glucose utilization caused by glutamine starvation, it is likely that the limited availability of a nitrogen source inhibits the carbon source utilization (Figure 1). How cells sense the availability of a nitrogen source and coordinately regulate carbon source utilization remains to be investigated. Particularly, the coordinated utilization of nitrogen and carbon sources in tumor cells may become a potential target for cancer management in combination with chemotherapy and radiation.
Media with high levels of glucose (20 mM), high levels of glutamine (4 mM), and high levels of oxygen (21%) represent the conventional conditions for a cell culture system, which have been used for a long time and have contributed a great deal to our current understanding of tumor biology. However, it is noteworthy that in vivo tumor cells may receive much lower concentrations of glucose, glutamine, and oxygen; the physiologic ranges of glucose and oxygen in tissues are approximately 3–5 mM and 4–7%, respectively. Moreover, solid tumors usually have defective vasculature formed by angiogenesis, which further reduces the concentrations of glucose and oxygen in tumors and may also limit the availability of glutamine. Although oxygen sensing, energy sensing, adaptive mechanisms to hypoxia, and low energy status are well studied, how tumors sense low glucose status and respond to it at the cellular level remains unclear. In addition, how cancer cells interact with adjacent stromal cells in vivo to secure a nutrition supply remains to be fully investigated. A better understanding of the adaptive strategy of tumor cells to low glucose supply will pave a way toward better management of tumors as well as ischemic disorders where lack of glucose and energy are detrimental factors.
In conclusion, various oncogenic signaling pathways may lead to transcriptional reprogramming and metabolic reprogramming in cancer cells (Figure 2). As a result, cancer cells coordinate the utilization of carbon sources with the availability of nitrogen sources and molecular oxygen to balance the proliferative demands for ATP, redox powers, and building blocks. Furthermore, maintaining a balanced among pools of key building blocks includes mononucleotides, amino acids, heme, glycerol-3-phosphate, sphingosine, cholesterol, fatty acids, ubiquinone and polyamine is fundamental for cancer cell proliferation, which may turn out to be a novel target for future cancer therapy. The altered metabolic consequences may cause changes of the microenvironment, which further promote adaptation of cancer cells, and may eventually contribute to cancer cells’ resistance to chemotherapy and radiation.
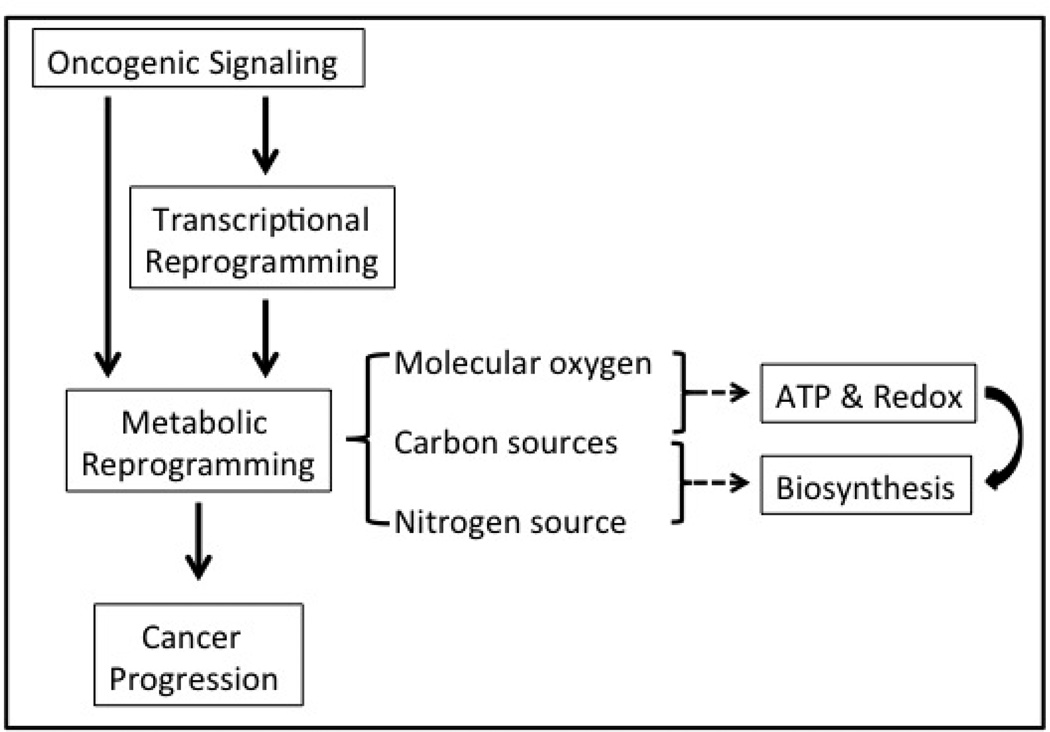
FIGURE 2.
Oncogenic signaling promotes cancer cell progression through transcriptional reprogramming and metabolic reprogramming. Based on our current understanding, oncogenic signaling pathways either directly alter the cancer cells’ metabolism or indirectly modulate the enzyme expression levels through transcriptional reprogramming. Eventually, carbon sources and molecular oxygen are utilized to generate ATP, reducing power and a variety of carbon metabolites, which together support the active biosynthesis of biomass. Note that a nitrogen source, in the form of amino acids, also is required for biosynthesis of nitrogenous molecules, and its availability may limit the general cell growth and utilization of carbon sources.
ACKNOWLEDGMENTS
Research in our lab is supported in part by Grant No. R01-CA129494 (to N.S.) from the National Cancer Institute (NCI), the National Institutes of Health (NIH), and a start-up fund from Drexel University.
REFERENCES
- 1.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 2.Gambhir SS. Molecular imaging of cancer with positron emission tomography. Nat Rev Cancer. 2002;2:683–693. doi: 10.1038/nrc882. [DOI] [PubMed] [Google Scholar]
- 3.Czernin J, Phelps ME. Positron emission tomography scanning: current and future applications. Annu Rev Med. 2002;53:8–112. doi: 10.1146/annurev.med.53.082901.104028. [DOI] [PubMed] [Google Scholar]
- 4.Eagle H. Nutrition needs of mammalian cells in tissue culture. Science. 1955;122:501–514. doi: 10.1126/science.122.3168.501. [DOI] [PubMed] [Google Scholar]
- 5.Coles NW, Johnstone RM. Glutamine metabolism in Ehrlich ascites-carcinoma cells. Biochem J. 1962;83:284–291. doi: 10.1042/bj0830284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reitzer LJ, Wice BM, Kennell D. Evidence that glutamine, not sugar, is the major energy source for cultured HeLa cells. J Biol Chem. 1979;254:2669–2676. [PubMed] [Google Scholar]
- 7.Zielke HR, Zielke CL, Ozand PT. Glutamine: a major energy source for cultured mammalian cells. Fed Proc. 1984;43:121–125. [PubMed] [Google Scholar]
- 8.Medina MA, Nunez de Castro I. Glutaminolysis and glycolysis interactions in proliferant cells. Int J Biochem. 1990;22:681–683. doi: 10.1016/0020-711x(90)90001-j. [DOI] [PubMed] [Google Scholar]
- 9.Meng M, Chen S, Lao T, Liang D, Sang N. Nitrogen anabolism underlies the importance of glutaminolysis in proliferating cells. Cell Cycle. 2010;9:3921–3932. doi: 10.4161/cc.9.19.13139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S. Thomson CB Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L, Jiao ZH, Zheng LS, Zhang YY, Xie ST, Wang ZX, Wu JW. Structural insight into the autoinhibition mechanism of AMP-activated protein kinase. Nature. 2009;459:1146–1149. doi: 10.1038/nature08075. [DOI] [PubMed] [Google Scholar]
- 12.Xiao B, Heath R, Saiu P, Leiper FC, Leone P, Jing C, Walker PA, Haire L, Eccleston JF, Davis CT, Martin SR, Carling D, Gamblin SJ. Structural basis for AMP binding to mammalian AMP-activated protein kinase. Nature. 2007;449:496–500. doi: 10.1038/nature06161. [DOI] [PubMed] [Google Scholar]
- 13.Hawley SA, Davison M, Woods A, Davies SP, Beri RK, Carling D, Hardie DG. Characterization of the AMP-activated protein kinase kinase from rat liver and identification of threonine 172 as the major site at which it phos-phorylates AMP-activated protein kinase. J Biol Chem. 1996;271:27879–27887. doi: 10.1074/jbc.271.44.27879. [DOI] [PubMed] [Google Scholar]
- 14.Davies P, Helps NR, Cohen PT, Hardie DG. 5’-AMPin-hibits dephosphorylation, as well as promoting phosphorylation, of the AMP-activated protein kinase. Studies using bacterially expressed human protein phosphatase-2C alpha and native bovine protein phosphatase-2AC. FEBS Lett. 1995;377:421–425. doi: 10.1016/0014-5793(95)01368-7. [DOI] [PubMed] [Google Scholar]
- 15.Sanders MJ, Grondin PO, Hegarty BD, Snowden MA, Carling D. Investigating the mechanism for AMP activation of the AMP-activated protein kinase cascade. Biochem J. 2007;403:139–148. doi: 10.1042/BJ20061520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corton JM, Gillespie JG, Hawley SA, Hardie DG. 5-ami-noimidazole-4-carboxamide ribonucleoside A specific method for activating AMP-activated protein kinase in intact cells? Eur J Biochem. 1995;229:558–565. doi: 10.1111/j.1432-1033.1995.tb20498.x. [DOI] [PubMed] [Google Scholar]
- 17.Henin N, Vincent MF, Gruber HE, Van den Berghe G. Inhibition of fatty acid and cholesterol synthesis by stimulation of AMP-activated protein kinase. FASEB J. 1995;9:541–546. doi: 10.1096/fasebj.9.7.7737463. [DOI] [PubMed] [Google Scholar]
- 18.Jorgensen SB, Nielsen JN, Birk JB, Olsen GS, Viollet B, Andreelli F, Schjerling P, Vaulont S, Hardie DG, Hansen BF, Richter EA, Wojtaszewski JF. The alpha2-5’AMP-activated protein kinase is a site 2 glycogen synthase kinase in skeletal muscle and is responsive to glucose loading. Diabetes. 2004;53:3074–3081. doi: 10.2337/diabetes.53.12.3074. [DOI] [PubMed] [Google Scholar]
- 19.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 20.Marsin AS, Bouzin C, Bertrand L, Hue L. The stimulation of glycolysis by hypoxia in activated monocytes is mediated by AMP-activated protein kinase and inducible 6-phosphofructo-2-kinase. J Biol Chem. 2002;277:30778–30783. doi: 10.1074/jbc.M205213200. [DOI] [PubMed] [Google Scholar]
- 21.Merrill GF, Kurth EJ, Hardie DG, Winder WW. AICA riboside increases AMP-activated protein kinase, fatty acid oxidation, and glucose uptake in rat muscle. Am J Physiol. 1997;273:E1107–E1112. doi: 10.1152/ajpendo.1997.273.6.E1107. [DOI] [PubMed] [Google Scholar]
- 22.Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, Birnbaum MJ, Thompson CB. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 23.Kurth-Kraczek EJ, Hirshman MF, Goodyear LJ, Winder WW. 5’ AMP-activated protein kinase activation causes GLUT4 translocation in skeletal muscle. Diabetes. 1999;48:1667–16671. doi: 10.2337/diabetes.48.8.1667. [DOI] [PubMed] [Google Scholar]
- 24.Holmes BF, Kurth-Kraczek EJ, Winder WW. Chronic activation of 5’-AMP-activated protein kinase increases GLUT-4, hexokinase, and glycogen in muscle. J Appl Physiol. 1999;87:1990–1995. doi: 10.1152/jappl.1999.87.5.1990. [DOI] [PubMed] [Google Scholar]
- 25.Zong H, Ren JM, Young LH, Pypaert M, Mu J, Birnbaum MJ, Shulman GI. AMP kinase is required for mitochondrial biogenesis in skeletal muscle in response to chronic energy deprivation. Proc Natl Acad Sci U S A. 2002;99:15983–15987. doi: 10.1073/pnas.252625599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maraldi T, Prata C, Caliceti C, Vieceli Dalla Sega F, Zambonin L, Fiorentini D, Hakim G. VEGF-induced ROS generation from NAD(P)H oxidases protects human leu-kemic cells from apoptosis. Int J Oncol. 2010;36:1581–1589. doi: 10.3892/ijo_00000645. [DOI] [PubMed] [Google Scholar]
- 27.Bell EL, Klimova TA, Eisenbart J, Schumacker PT, Chandel NS. Mitochondrial reactive oxygen species trigger hypoxia-inducible factor-dependent extension of the replicative life span during hypoxia. Mol Cell Biol. 2007;27:5737–5745. doi: 10.1128/MCB.02265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Devasagayam TP, Tilak JC, Boloor KK, Sane KS, Ghas-kadbi SS, Lele RD. Free radicals and antioxidants in human health: current status and future prospects. J Assoc Physicians India. 2004;52:794–804. [PubMed] [Google Scholar]
- 29.Takahashi A, Ohtani N, Yamakoshi K, Iida S, Tahara H, Nakayama K, Nakayama KI, Ide T, Saya H, Hara E. Mi-togenic signalling and the pl6INK4a-Rb pathway cooperate to enforce irreversible cellular senescence. Nat Cell Biol. 2006;8:1291–1297. doi: 10.1038/ncb1491. [DOI] [PubMed] [Google Scholar]
- 30.Phaneuf S, Leeuwenburgh C. Cytochrome c release from mitochondria in the aging heart: a possible mechanism for apoptosis with age. Am J Physiol Regul Integr Comp Physiol. 2002;282:R423–R430. doi: 10.1152/ajpregu.00296.2001. [DOI] [PubMed] [Google Scholar]
- 31.Han D, Antunes F, Canali R, Rettori D, Cadenas E. Voltage-dependent anion channels control the release of the superoxide anion from mitochondria to cytosol. J Biol Chem. 2003;278:5557–5563. doi: 10.1074/jbc.M210269200. [DOI] [PubMed] [Google Scholar]
- 32.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baranowski M. Biological role of liver X receptors. J Physiol Pharmacol. 2008;59(Suppl 7):31–55. [PubMed] [Google Scholar]
- 35.Zelcer N, Tontonoz P. Liver X receptors as integrators of metabolic and inflammatory signaling. J Clin Invest. 2006;116:607–614. doi: 10.1172/JCI27883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nunn AV, Bell J, Barter P. The integration of lipid-sensing and anti-inflammatory effects: how the PPARs play a role in metabolic balance. Nucl Recept. 2007;5:1. doi: 10.1186/1478-1336-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 38.Semenza GL. HIF-1: upstream and downstream of cancer metabolism. Curr Opin Genet Dev. 2010;20:51–56. doi: 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 40.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 41.Brahimi-Horn MC, Bellot G, Pouyssegur J. Hypoxia and energetic tumour metabolism. Curr Opin Genet Dev. 2011;21:67–72. doi: 10.1016/j.gde.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 42.Chiche J, Ilc K, Laferriere J, Trottier E, Dayan F, Mazure NM, Brahimi-Horn MC, Pouyssegur J. Hypoxia-inducible carbonic anhydrase IX and XII promote tumor cell growth by counteracting acidosis through the regulation of the intracellular pH. Cancer Res. 2009;69:358–368. doi: 10.1158/0008-5472.CAN-08-2470. [DOI] [PubMed] [Google Scholar]
- 43.Lu CW, Lin SC, Chen KF, Lai YY, Tsai SJ. Induction of pyruvate dehydrogenase kinase-3 by hypoxia-inducible factor-1 promotes metabolic switch and drug resistance. J Biol Chem. 2008;283:28106–28114. doi: 10.1074/jbc.M803508200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wokolorczyk D, Gliniewicz B, Sikorski A, Zlowocka E, Masojc B, Debniak T, Matyjasik J, Mierzejewski M, Medrek K, Oszutowska D, Suchy J, Gronwald J, Teodorczyk U, Huzarski T, Byrski T, Jakubowska A, Gorski B, van de Wetering T, Walczak S, Narod SA, Lubinski J, Cybulski C. A range of cancers is associated with the rs6983267 marker on chromosome 8. Cancer Res. 2008;68:9982–9986. doi: 10.1158/0008-5472.CAN-08-1838. [DOI] [PubMed] [Google Scholar]
- 45.Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, Dalla-Favera R, Dang CV. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci U S A. 1997;94:6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim JW, Gao P, Liu YC, Semenza GL, Dang CV. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol Cell Biol. 2007;27:7381–7393. doi: 10.1128/MCB.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Osthus RC, Shim H, Kim S, Li Q, Reddy R, Mukherjee M, Xu Y, Wonsey D, Lee LA, Dang CV. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem. 2000;275:21797–21800. doi: 10.1074/jbc.C000023200. [DOI] [PubMed] [Google Scholar]
- 48.Lakin ND, Jackson SP. Regulation of p53 in response to DNA damage. Oncogene. 1999;18:7644–7655. doi: 10.1038/sj.onc.1203015. [DOI] [PubMed] [Google Scholar]
- 49.Puzio-Kuter AM. The Role of p53 in Metabolic Regulation. Genes Cancer. 2011;2:385–391. doi: 10.1177/1947601911409738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- 51.Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 52.Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 2004;64:2627–2633. doi: 10.1158/0008-5472.can-03-0846. [DOI] [PubMed] [Google Scholar]
- 53.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 54.Wise DRDR, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, Thompson CB. Myc regulates a transcriptional pro-gram that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, Dang CV. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci U S A. 2010;107:7455–7460. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suzuki S, Tanaka T, Poyurovsky MV, Nagano H, Mayama T, Ohkubo S, Lokshin M, Hosokawa H, Nakayama T, Suzuki Y, Sugano S, Sato E, Nagao T, Yokote K, Tatsuno I, Prives C. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci U S A. 2010;107:7461–7466. doi: 10.1073/pnas.1002459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dang CV. Glutaminolysis: supplying carbon or nitrogen or both for cancer cells? Cell Cycle. 2010;9:3884–3886. doi: 10.4161/cc.9.19.13302. [DOI] [PubMed] [Google Scholar]
- 59.Markman B, Dienstmann R, Tabernero J. Targeting the PI3K/Akt/mTOR pathway—beyond rapalogs. Oncotaget. 2010;1:530–543. doi: 10.18632/oncotarget.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moritz A, Li Y, Guo A, Villen J, Wang Y, MacNeill J, Kornhauser J, Sprott K, Zhou J, Possemato A, Ren JM, Hornbeck P, Cantley LC, Gygi SP, Rush J, Comb MJ. Akt-RSK-S6 kinase signaling networks activated by oncogenic receptor tyrosine kinases. Sci Signal. 2010;3:ra64. doi: 10.1126/scisignal.2000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Darido C, Georgy SR, Wilanowski T, Dworkin S, Auden A, Zhao Q, Rank G, Srivastava S, Finlay MJ, Papenfuss AT, Pandolfi PP, Pearson RB, Jane SM. Targeting of the tumor suppressor GRHL3 by a miR-21-dependent proto-oncogenic network results in PTEN loss and tumorigenesis. Cancer cell. 2011;20:635–648. doi: 10.1016/j.ccr.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 62.Jiang BH, Liu LZ. PI3K/PTEN signaling in angiogenesis and tumorigenesis. Adv Cancer Res. 2009;102:19–65. doi: 10.1016/S0065-230X(09)02002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yecies JL, Manning BD. mTOR links oncogenic signaling to tumor cell metabolism. J Mol Med (Berl) 2011;89:221–228. doi: 10.1007/s00109-011-0726-6. [DOI] [PubMed] [Google Scholar]
- 64.Brugarolas JB, Vazquez F, Reddy A, Sellers WR, Kaelin WG., Jr. TSC2 regulates VEGF through mTOR-dependent and -independent pathways. Cancer Cell. 2003;4:147–158. doi: 10.1016/s1535-6108(03)00187-9. [DOI] [PubMed] [Google Scholar]
- 65.Robey RB, Hay N. Is Akt the “Warburg kinase”?-Akt-energy metabolism interactions and oncogenesis. Semin Cancer Biol. 2009;19:25–31. doi: 10.1016/j.semcancer.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fernandez de Mattos S, de los Pinos EE, Joaquin M, Tauler A. Activation of phosphatidylinositol 3-kinase is required for transcriptional activity of F-type 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase: assessment of the role of protein kinase B and p70 S6 kinase. Biochem J. 2000;349:59–65. doi: 10.1042/0264-6021:3490059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Deprez J, Vertommen D, Alessi DR, Hue L, Rider MH. Phosphorylation and activation of heart 6-phospho-fructo-2-kinase by protein kinase B and other protein kinases of the insulin signaling cascades. J Biol Chem. 1997;272:17269–17275. doi: 10.1074/jbc.272.28.17269. [DOI] [PubMed] [Google Scholar]
- 68.Sun Q, Chen X, Ma J, Peng H, Wang F, Zha X, Wang Y, Jing Y, Yang H, Chen R, Chang L, Zhang Y, Goto J, Onda H, Chen T, Wang MR, Lu Y, You H, Kwiatkowski D, Zhang H. Mammalian target of rapamycin up-regula-tion of pyruvate kinase isoenzyme type M2 is critical for aerobic glycolysis and tumor growth. Proc Natl Acad Sci U S A. 2011;108:4129–4134. doi: 10.1073/pnas.1014769108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Welsh GI, Hers I, Berwick DC, Dell G, Wherlock M, Birkin R, Leney S, Tavare JM. Role of protein kinase B in insulin-regulated glucose uptake. Biochem Soc Trans. 2005;33:346–349. doi: 10.1042/BST0330346. [DOI] [PubMed] [Google Scholar]
- 70.Huang YC, Chang WL, Huang SF, Lin CY, Lin HC, Chang TC. Pachymic acid stimulates glucose uptake through enhanced GLUT4 expression and translocation. Eur J Pharmacol. 2010;648:39–49. doi: 10.1016/j.ejphar.2010.08.021. [DOI] [PubMed] [Google Scholar]
- 71.Berwick DC, Hers I, Heesom KJ, Moule SK, Tavare JM. The identification of ATP-citrate lyase as a protein kinase B (Akt) substrate in primary adipocytes. J Biol Chem. 2002;277:33895–33900. doi: 10.1074/jbc.M204681200. [DOI] [PubMed] [Google Scholar]
- 72.Duvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, Vander Heiden MG, MacKeigan JP, Finan PM, Clish CB, Murphy LO, Manning BD. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, Griffiths JR, Chung YL, Schulze A. SREBP activity is regulated by mTORCl and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Van de Sande T, De Schrijver E, Heyns W, Verhoeven G, Swinnen JV. Role of the phosphatidylinositol 3’-kinase/ PTEN/Akt kinase pathway in the overexpression of fatty acid synthase in LNCaP prostate cancer cells. Cancer Res. 2002;62:642–646. [PubMed] [Google Scholar]
- 75.Van de Sande T, Roskams T, Lerut E, Joniau S, Van Pop-pel H, Verhoeven G, Swinnen JV. High-level expression of fatty acid synthase in human prostate cancer tissues is linked to activation and nuclear localization of Akt/PKB. J Pathol. 2005;206:214–219. doi: 10.1002/path.1760. [DOI] [PubMed] [Google Scholar]
- 76.Plotnikov A, Zehorai E, Procaccia S, Seger R. The MAPK cascades: signaling components, nuclear roles and mechanisms of nuclear translocation. Biochim Biophys Acta. 2011;1813:1619–1633. doi: 10.1016/j.bbamcr.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 77.Huang P, Han J, Hui L. MAPK signaling in inflammation-associated cancer development. Protein Cell. 2010;1:218–226. doi: 10.1007/s13238-010-0019-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shaul YD, Seger R. The MEK/ERK cascade: from signaling specificity to diverse functions. Biochim Biophys Acta. 2007;1773:1213–1226. doi: 10.1016/j.bbamcr.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 79.Raman M, Chen W, Cobb MH. Differential regulation and properties of MAPKs. Oncogene. 2007;26:3100–3112. doi: 10.1038/sj.onc.1210392. [DOI] [PubMed] [Google Scholar]
- 80.Pimienta G, Pascual J. Canonical and alternative MAPK signaling. Cell Cycle. 2007;6:2628–2632. doi: 10.4161/cc.6.21.4930. [DOI] [PubMed] [Google Scholar]
- 81.Harmon AW, Patel YM. Naringenin inhibits glucose uptake in MCF-7 breast cancer cells: a mechanism for impaired cellular proliferation. Breast Cancer Res Treat. 2004;85:103–110. doi: 10.1023/B:BREA.0000025397.56192.e2. [DOI] [PubMed] [Google Scholar]
- 82.Marko AJ, Miller RA, Kelman A, Frauwirth KA. Induction of glucose metabolism in stimulated T lymphocytes is regulated by mitogen-activated protein kinase signaling. PloS One. 2010;5:el5425. doi: 10.1371/journal.pone.0015425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Richard DE, Berra E, Gothie E, Roux D, Pouyssegur J. p42/p44 mitogen-activated protein kinases phosphory-late hypoxia-inducible factor 1 alpha (HTF-lalpha) and enhance the transcriptional activity of HTF-1. J Biol Chem. 1999;274:32631–32637. doi: 10.1074/jbc.274.46.32631. [DOI] [PubMed] [Google Scholar]
- 84.Sang N, Stiehl DP, Bohensky J, Leshchinsky I, Srinivas V, Caro J. MAPK signaling up-regulates the activity of hypoxia-inducible factors by its effects on p300. J Biol Chem. 2003;278:14013–14019. doi: 10.1074/jbc.M209702200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121:179–193. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
- 86.Carr EL, Kelman A, Wu GS, Gopaul R, Senkevitch E, Aghvanyan A, Turay AM, Frauwirth KA. Glutamine uptake and metabolism are coordinately regulated by ERK/ MAPK during T lymphocyte activation. J Immunol. 2010;185:1037–1044. doi: 10.4049/jimmunol.0903586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fujishiro M, Gotoh Y, Katagiri H, Sakoda H, Ogihara T, Anai M, Onishi Y, Ono H, Funaki M, Inukai K, Fukushima Y, Kikuchi M, Oka Y, Asano T. MKK6/3 and p38 MAPK pathway activation is not necessary for insulin-induced glucose uptake but regulates glucose transporter expression. J Biol Chem. 2001;276:19800–19806. doi: 10.1074/jbc.M101087200. [DOI] [PubMed] [Google Scholar]
- 88.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 89.Baselga J, Swain SM. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer. 2009;9:463–475. doi: 10.1038/nrc2656. [DOI] [PubMed] [Google Scholar]
- 90.Shanware NP, Mullen AR, DeBerardinis RJ, Abraham RT. Glutamine: pleiotropic roles in tumor growth and stress resistance. J Mol Med (Berl) 2011;89:229–236. doi: 10.1007/s00109-011-0731-9. [DOI] [PubMed] [Google Scholar]
- 91.Dang CV. Rethinking the Warburg effect with Myc micromanaging glutamine metabolism. Cancer Res. 2010;70:859–862. doi: 10.1158/0008-5472.CAN-09-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, Thompson CB. Myc regulates a transcriptional program that stimulates mitochondrial glutami-nolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stoltzman CA, Peterson CW, Breen KT, Muoio DM, Billin AN, Ayer DE. Glucose sensing by MondoA:Mlx complexes: a role for hexokinases and direct regulation of thioredoxin-interacting protein expression. Proc Natl Acad Sci U S A. 2008;105:6912–6917. doi: 10.1073/pnas.0712199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Peterson CW, Stoltzman CA, Sighinolfi MP, Han KS, Ayer DE. Glucose controls nuclear accumulation, promoter binding, and transcriptional activity of the Mon-doA-Mlx heterodimer. Mol Cell Biol. 2010;30:2887–2895. doi: 10.1128/MCB.01613-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Harmon JS, Mariash CN. Identification of a carbohydrate response element in rat S14 gene. Mol Cell Endocrinol. 1996;123:37–44. doi: 10.1016/0303-7207(96)03896-8. [DOI] [PubMed] [Google Scholar]
- 96.Towle HC, Kaytor EN, Shih HM. Regulation of the expression of lipogenic enzyme genes by carbohydrate. Annu Rev Nutr. 1997;17:405–433. doi: 10.1146/annurev.nutr.17.1.405. [DOI] [PubMed] [Google Scholar]
- 97.Vaulont S, Vasseur-Cognet M, Kahn A. Glucose regulation of gene transcription. J Biol Chem. 2000;275:31555–31558. doi: 10.1074/jbc.R000016200. [DOI] [PubMed] [Google Scholar]
- 98.Yamashita H, Takenoshita M, Sakurai M, Bruick RK, Henzel WJ, Shillinglaw W, Arnot D, Uyeda K. A glucose-responsive transcription factor that regulates carbohydrate metabolism in the liver. Proc Natl Acad Sci U S A. 2001;98:9116–9121. doi: 10.1073/pnas.161284298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Uyeda K, Repa JJ. Carbohydrate response element binding protein, ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metab. 2006;4:107–110. doi: 10.1016/j.cmet.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 100.Ma L, Robinson LN, Towle HC. ChREBP*Mlx is the principal mediator of glucose-induced gene expression in the liver. J Biol Chem. 2006;281:28721–28730. doi: 10.1074/jbc.M601576200. [DOI] [PubMed] [Google Scholar]
- 101.Billin AN, Eilers AL, Coulter KL, Logan JS, Ayer DE. MondoA, a novel basic helix-loop-helix-leucine zipper transcriptional activator that constitutes a positive branch of a max-like network. Mol Cell Biol. 2000;20:8845–8854. doi: 10.1128/mcb.20.23.8845-8854.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Postic C, Dentin R, Denechaud PD, Girard J. ChREBP, a transcriptional regulator of glucose and lipid metabolism. Annu Rev Nutr. 2007;27:179–192. doi: 10.1146/annurev.nutr.27.061406.093618. [DOI] [PubMed] [Google Scholar]
- 103.Iizuka K, Bruick RK, Liang G, Horton JD, Uyeda K. Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc Natl Acad Sci U S A. 2004;101:7281–7286. doi: 10.1073/pnas.0401516101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Okoshi R, Ando K, Suenaga Y, Sang M, Kubo N, Kizaki H, Nakagawara A, Ozaki T. Transcriptional regulation of tumor suppressor p53 by cAMP-responsive element-binding protein/AMP-activated protein kinase complex in response to glucose deprivation. Genes Cells. 2009;14:1429–1440. doi: 10.1111/j.1365-2443.2009.01359.x. [DOI] [PubMed] [Google Scholar]
- 105.Ide T, Brown-Endres L, Chu K, Ongusaha PP, Ohtsuka T, El-Deiry WS, Aaronson SA, Lee SW. GAMT, a p53-in-ducible modulator of apoptosis, is critical for the adaptive response to nutrient stress. Mol Cell. 2009;36:379–392. doi: 10.1016/j.molcel.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 106.Kim MJ, Park IJ, Yun H, Kang I, Choe W, Kim SS, Ha J. AMP-activated protein kinase antagonizes pro-apoptotic extracellular signal-regulated kinase activation by inducing dual-specificity protein phosphatases in response to glucose deprivation in HCT116 carcinoma. J Biol Chem. 2010;285:14617–14627. doi: 10.1074/jbc.M109.085456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yun H, Kim HS, Lee S, Kang I, Kim SS, Choe W, Ha J. AMP kinase signaling determines whether c-Jun N-terminal kinase promotes survival or apoptosis during glucose deprivation. Carcinogenesis. 2009;30:529–537. doi: 10.1093/carcin/bgn259. [DOI] [PubMed] [Google Scholar]
- 108.Habeeb BS, Kitayama J, Nagawa H. Adiponectin supports cell survival in glucose deprivation through enhancement of autophagic response in colorectal cancer cells. Cancer Sci. 2011;102:999–1006. doi: 10.1111/j.1349-7006.2011.01902.x. [DOI] [PubMed] [Google Scholar]
- 109.Cechowska-Pasko M, Kretowski R, Bankowski E. Glucose deficiency reduces collagen synthesis in breast cancer MCF7 cells. Cell Biol Int. 2011;35:141–145. doi: 10.1042/CBI20090383. [DOI] [PubMed] [Google Scholar]
- 110.Amthor H, Nicholas G, McKinnell I, Kemp CF, Sharma M, Kambadur R, Patel K. Follistatin complexes Myo-statin and antagonises Myostatin-mediated inhibition of myogenesis. Dev Biol. 2004;270:19–30. doi: 10.1016/j.ydbio.2004.01.046. [DOI] [PubMed] [Google Scholar]
- 111.Glister C, Kemp CF, Knight PG. Bone morphogenetic protein (BMP) ligands and receptors in bovine ovarian follicle cells: actions of BMP-4, -6 and -7 on granulosa cells and differential modulation of Smad-1 phosphorylation by follistatin. Reproduction. 2004;127:239–254. doi: 10.1530/rep.1.00090. [DOI] [PubMed] [Google Scholar]
- 112.Phillips DJ, de Kretser DM. Follistatin: a multifunctional regulatory protein. Front Neuroendocrinol. 1998;19:287–322. doi: 10.1006/frne.1998.0169. [DOI] [PubMed] [Google Scholar]
- 113.Gao X, Wei S, Lai K, Sheng J, Su J, Zhu J, Dong H, Hu H, Xu Z. Nucleolar follistatin promotes cancer cell survival under glucose-deprived conditions through inhibiting cellular rRNA synthesis. J Biol Chem. 2010;285:36857–36864. doi: 10.1074/jbc.M110.144477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Taylor RP, Parker GJ, Hazel MW, Soesanto Y, Fuller W, Yazzie MJ, McClain DA. Glucose deprivation stimulates O-GlcNAc modification of proteins through up-regulation of O-linked N-acetylglucosaminyltransferase. J Biol Chem. 2008;283:6050–6057. doi: 10.1074/jbc.M707328200. [DOI] [PubMed] [Google Scholar]
- 115.Kang JG, Park SY, Ji S, Jang I, Park S, Kim HS, Kim SM, Yook JI, Park YI, Roth J, Cho JW. O-GlcNAc protein modification in cancer cells increases in response to glucose deprivation through glycogen degradation. J Biol Chem. 2009;284:34777–34784. doi: 10.1074/jbc.M109.026351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Taylor RP, Geisler TS, Chambers JH, McClain DA. Up-regulation of O-GlcNAc transferase with glucose deprivation in HepG2 cells is mediated by decreased hexos-amine pathway flux. J Biol Chem. 2009;284:3425–3432. doi: 10.1074/jbc.M803198200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kim AJ, Kim HJ, Jee HJ, Song N, Kim M, Bae YS, Chung JH, Yun J. Glucose deprivation is associated with Chkl degradation through the ubiquitin-proteasome pathway and effective checkpoint response to replication blocks. Biochim Biophys Acta. 2011;1813:1230–1238. doi: 10.1016/j.bbamcr.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 119.Chen C, Pore N, Behrooz A, Ismail-Beigi F, Maity A. Regulation of glutl mRNA by hypoxia-inducible factor-1 Interaction between H-ras and hypoxia. J Biol Chem. 2001;276:9519–9525. doi: 10.1074/jbc.M010144200. [DOI] [PubMed] [Google Scholar]
- 120.Riddle SR, Ahmad A, Ahmad S, Deeb SS, Malkki M, Schneider BK, Allen CB, White CW. Hypoxia induces hexokinase II gene expression in human lung cell line A549. Am J Physiol Lung Cell Mol Physiol. 2000;278:L407–L416. doi: 10.1152/ajplung.2000.278.2.L407. [DOI] [PubMed] [Google Scholar]
- 121.Iyer NV, Kotch LE, Agani F, Leung SW, Laughner E, Wenger RH, Gassmann M, Gearhart JD, Lawler AM, Yu AY, Semenza GL. Cellular and developmental control of 02 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sans CL, Satterwhite DJ, Stoltzman CA, Breen KT, Ayer DE. MondoA-Mx heterodimers are candidate sensors of cellular energy status: mitochondrial localization and direct regulation of glycolysis. Mol Cell Biol. 2006;26:4863–4871. doi: 10.1128/MCB.00657-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.O’Callaghan BL, Koo SH, Wu Y, Freake HC, Towle HC. Glucose regulation of the acetyl-CoA carboxylase promoter PI in rat hepatocytes. J Biol Chem. 2001;276:16033–16039. doi: 10.1074/jbc.M101557200. [DOI] [PubMed] [Google Scholar]
- 124.Rufo C, Teran-Garcia M, Nakamura MT, Koo SH, Towle HC, Clarke SD. Involvement of a unique carbohydrate-responsive factor in the glucose regulation of rat liver fatty-acid synthase gene transcription. J Biol Chem. 2001;276:21969–21975. doi: 10.1074/jbc.M100461200. [DOI] [PubMed] [Google Scholar]
- 125.Guha P, Aneja KK, Shilpi RY, Haldar D. Transcriptional regulation of mitochondrial glycerophosphate acyltrans-ferase is mediated by distal promoter via ChREBP and SREBP-1. Arch Biochem Biophys. 2009;490:85–95. doi: 10.1016/j.abb.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Iizuka K, Takeda J, Horikawa Y. Glucose induces FGF21 mRNA expression through ChREBP activation in rat hepatocytes. FEBS Lett. 2009;583:2882–2886. doi: 10.1016/j.febslet.2009.07.053. [DOI] [PubMed] [Google Scholar]
- 127.Boergesen M, Poulsen LC, Schmidt SF, Frigerio F, Maechler P, Mandrup S. ChREBP mediates glucose repression of peroxisome proliferator-activated receptor alpha expression in pancreatic beta-cells. J Biol Chem. 2011;286:13214–13225. doi: 10.1074/jbc.M110.215467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Noordeen NA, Khera TK, Sun G, Longbottom ER, Pullen TJ, da Silva Xavier G, Rutter GA, Leclerc I. Carbohydrate-responsive element-binding protein (ChREBP) is a negative regulator of ARNT/HIF-lbeta gene expression in pancreatic islet beta-cells. Diabetes. 2010;59:153–160. doi: 10.2337/db08-0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Minn AH, Hafele C, Shalev A. Thioredoxin-interacting protein is stimulated by glucose through a carbohydrate response element and induces beta-cell apoptosis. Endocrinology. 2005;146:2397–2405. doi: 10.1210/en.2004-1378. [DOI] [PubMed] [Google Scholar]