SUMMARY
Mitochondrial diseases include a group of maternally inherited genetic disorders caused by mutations in mitochondrial DNA (mtDNA). In most of these patients, mutated mtDNA coexists with wild type mtDNA, a situation known as mtDNA heteroplasmy. Here we report on a strategy towards preventing germline transmission of mitochondrial diseases by inducing mtDNA heteroplasmy shift through the selective elimination of mutated mtDNA. As a proof of concept we took advantage of NZB/BALB heteroplasmic mice, which contain two mtDNA haplotypes, BALB and NZB, and selectively prevented their germline transmission using either mitochondria targeted restriction endonucleases or TALENs. In addition, we successfully reduced human mutated mtDNA levels responsible for Leber’s hereditary optic neuropathy (LHOND), and neurogenic muscle weakness, ataxia and retinitis pigmentosa (NARP), in mammalian oocytes using mito-TALENs. Altogether, our approaches represent a potential therapeutic avenue for preventing the transgenerational transmission of human mitochondrial diseases caused by mutations in mtDNA.
INTRODUCTION
Mitochondria are double-membrane cellular organelles of bacterial origin that play fundamental roles in multiple cellular processes including energy production, calcium homeostasis, cellular signaling, and apoptosis (Dyall et al., 2004). Mitochondria contain their own mitochondrial DNA (mtDNA) encoding 13 polypeptides of the mitochondrial respiratory chain as well as tRNAs and rRNAs necessary for their synthesis (Anderson et al., 1981). Mitochondrial DNA is present in multiple copies per cell, ranging from approximately 1000 copies in somatic cells to several 100,000 copies in oocytes, with an average 1-10 copies per organelle (Shoubridge and Wai, 2007). In contrast to nuclear DNA, mtDNA is exclusively transmitted through maternal inheritance. Diseases resulting from mitochondrial dysfunction caused by mtDNA mutations affect 1 in 5,000 children (Haas et al., 2007), and it is estimated that 1 in 200 women could be a mitochondrial disease carrier. Due to the fundamental role of mitochondria in energy production, mitochondrial diseases correlate with degeneration of tissues and organs with high energy demands. This leads to myopathies, cardiomyopathies, and encephalopathies, among other phenotypes (Taylor and Turnbull, 2005). Currently, there is no cure for mitochondrial diseases. Genetic counseling and pre-implantation genetic diagnosis (PGD) represent the only therapeutic options for preventing transmission of mitochondrial diseases caused by mtDNA mutations. However, due to the non-Mendelian segregation of mtDNA, PGD can only partially reduce the risk of transmitting the disease (Brown et al., 2006). Moreover, analysis of multiple blastomeres may compromise embryo viability. Recently, mitochondrial replacement techniques by spindle, pronuclear or polar body genome transfer into healthy enucleated donor oocytes or embryos have been reported (Craven et al., 2010; Paull et al., 2013; Tachibana et al., 2012; Wang et al., 2014). Application of these techniques implies combining genetic material from three different individuals, which has raised ethical, safety and medical concerns (Hayden, 2013; Vogel, 2014). Therefore, alternative and complementary approaches that alleviate or eliminate these concerns should be investigated when devising feasible clinical paths towards preventing the transmission of mitochondrial diseases caused by mtDNA mutations.
Due to the thousands of copies of mtDNA contained within a cell, the levels of mutated mtDNA can vary. The term homoplasmy refers to the presence of a single mtDNA haplotype in the cell, whereas heteroplasmy refers to the coexistence of more than one mtDNA haplotype. When the percentage of mutated mtDNA molecules exceeds a threshold that compromises mitochondrial function, a disease state may ensue (Taylor and Turnbull, 2005; Wallace and Chalkia, 2013). Threshold levels for biochemical and clinical defects are generally in the range of 60-95% mutated mtDNA depending on the severity of the mutation (Russell and Turnbull, 2014). Changes in the relative levels of heteroplasmic mtDNA can be referred to as mtDNA heteroplasmy shifts. Despite the fact that mitochondria posses all the necessary machinery for homologous recombination and non-homologous end joining, they do not seem to represent the major pathway for mtDNA repair in mammalian mitochondria (Alexeyev et al., 2013). Previous studies have demonstrated that the relative levels of mutated and wild type mtDNA can be altered in patient somatic cells containing the m.8993T>G mtDNA mutation responsible for the NARP and MILS syndromes, where elimination of mutated mtDNA led to the restoration of normal mitochondrial function (Alexeyev et al., 2008). Similarly, using the heteroplasmic NZB/BALB mouse model that carries two different mtDNA haplotypes (NZB and BALB), BALB mtDNA, which contains a unique ApaLI site, has been specifically reduced in vivo using a mitochondria targeted ApaLI (Bacman et al., 2012; 2010). Recently, transcription activator-like effector nucleases (TALENs) and zinc finger nucleases (ZFNs) targeted to mitochondria have being utilized for the specific elimination of mitochondrial genomes carrying mutations responsible for mitochondrial diseases (Bacman et al., 2013; Gammage et al., 2014; Minczuk et al., 2006; 2008). These novel approaches allow for the targeting of a wider spectrum of mutations against which restriction endonucleases could not be used. However, these approaches do not provide mechanisms for preventing the transmission of mutated mtDNA nor do they allow for a complete systemic clearance of mtDNA mutations in subsequent generations.
Here we report on the specific reduction of mitochondrial genomes in the germline for preventing transmission of mitochondrial diseases. As a proof of concept, and by using the heteroplasmic NZB/BALB mouse model, we specifically reduced BALB or NZB mitochondrial genomes in the germline using mitochondria targeted restriction endonucleases and TALENs and prevented their transmission to the next generation. Moreover, we successfully reduced mutated mitochondrial genomes responsible for human mitochondrial diseases in mouse oocytes using mitochondria targeted nucleases. The approaches presented here may be applied and developed to prevent the transgenerational transmission of human mitochondrial diseases.
RESULTS
Specific reduction of mitochondrial genomes in oocytes and embryos using restriction endonucleases
With the goal of establishing an alternative therapeutic approach for preventing the germline transmission of mitochondrial diseases caused by mtDNA mutations, we tested the specific elimination of BALB mtDNA in NZB/BALB oocytes and one-cell embryos. For this purpose, we generated a mammalian codon optimized ApaLI targeted to mitochondria by the ATP5b mitochondria targeting sequence and the ATP5b 5′ and 3′ UTRs to promote co-translation import from mitochondrial associated ribosomes (Marc et al., 2002). An enhanced green fluorescence protein (EGFP) reporter was also included in the construct to monitor expression (Figure 1A). First, we tested the mitochondrial localization of the ApaLI protein generated from the construct by immunostaining in NZB/BALB tail tip fibroblasts (TTFs) and observed robust co-localization of mitochondria targeted ApaLI (mito-ApaLI) with the mitochondrial dye Mitotracker (Figure S1A). In contrast, we failed to observe mitochondrial localization of non-mitochondria targeted ApaLI (Figure S1A). Analysis of mtDNA by “last-cycle hot” PCR and restriction fragment length polymorphism (RFLP) demonstrated induction of heteroplasmy shift by specific reduction of BALB mtDNA in cells transfected with mito-ApaLI compared to control cells transfected with mito-GFP after 72 h (Figure S1B). In addition, we found normal mtDNA copy number in mito-ApaLI transfected cells, which resulted from the replication of the remaining NZB mtDNA that compensated for the reduction of BALB mtDNA (Figure S1C).
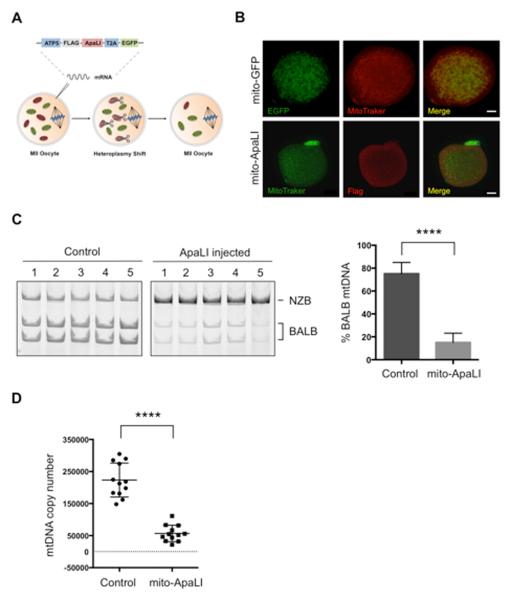
Figure 1. Heteroplasmy shift in NZB/BALB MII oocytes using mito-ApaLI.
(A) Injection of mito-ApaLI mRNA in oocytes for induction of heteroplasmy shift.
(B) Mitochondrial co-localization of mito-GFP and mito-ApaLI with Mitotracker in injected oocytes by immunofluorescence. Scale bars, 10μm.
(C) RFLP analysis and quantification of mtDNA heteroplasmy in control and mito-ApaLI injected MII oocytes after 48 h (Control n=16; mito-ApaLI n=12). Representative gel.
(D) Quantification of mtDNA copy number by qPCR in control and mito-ApaLI injected oocytes MII after 48 h (Control n=12; mito-ApaLI n=12).
Error bars represent ± SEM. ****p<0.0001. See also Figure S1.
We next decided to test whether a similar approach could be used in oocytes to specifically eliminate BALB mtDNA (Figure 1A). First, we confirmed the mitochondrial localization of mito-ApaLI in NZB/BALB metaphase II (MII) oocytes injected with mRNA encoding mito-ApaLI by immunostaining (Figure 1B). As expected, mito-ApaLI co-localized with Mitotracker in MII oocytes (Figure 1B). RFLP analysis 48 h after mito-ApaLI mRNA injection demonstrated the specific reduction of BALB mtDNA and a consequential increase in the relative NZB mtDNA levels (Figure 1C). In agreement with the lack of mtDNA replication in mature oocytes and pre-implantation embryos (Wai et al., 2010), analysis of mtDNA copy number by qPCR revealed a decrease in mtDNA copy number following mito-ApaLI injection proportional to the initial levels of BALB mtDNA (Figure 1D). To verify the reduction of BALB mtDNA we performed RFLP and qPCR analyses by amplification of an independent region of the mtDNA containing a unique HindIII site, exclusively present in BALB mtDNA. These analyses confirmed the specific reduction of BALB mtDNA upon injection of mito-ApaLI in NZB/BALB MII oocytes (Figure S1D and S1E). Injection of mito-ApaLI in BALB or NZB single haplotype oocytes resulted in complete depletion of mtDNA in BALB oocytes and did not affect mtDNA levels in NZB oocytes reinforcing the specificity of mito-ApaLI (Figure S1F). Collectively, these results suggest the potential of this approach for the specific reduction of mtDNA in the germline.
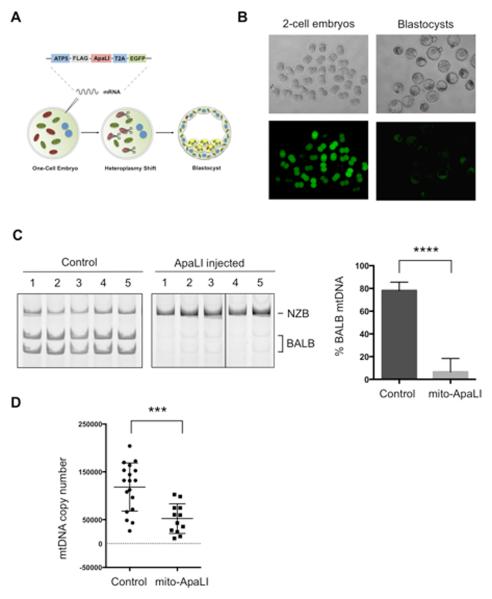
In addition to oocytes, we tested whether mtDNA heteroplasmy shift could be applied to one-cell embryos without affecting their normal development until the blastocyst stage (Figure 2A). For this purpose, NZB/BALB one-cell embryos were injected with mito-ApaLI mRNA. Time-lapse fluorescent microscopy images revealed the expression of mito-ApaLI indicated by EGFP expression, and more importantly, normal development of mito-ApaLI injected embryos through the different developmental stages analyzed (Figure 2B). Similarly to the results observed in oocytes, RFLP analysis of mito-ApaLI blastocysts demonstrated specific reduction of BALB mtDNA, and an increase in the relative levels of NZB mtDNA (Figure 2C). Moreover, due to the lack of mtDNA replication until the blastocyst stage (Wai et al., 2010), analysis of mtDNA copy number by qPCR showed a decrease in mtDNA levels proportional to the BALB mtDNA levels (Figure 2D). RFLP and qPCR analyses at the HindIII region confirmed the specific reduction of BALB mtDNA upon injection of mito-ApaLI in NZB/BALB embryos (Figure S2A and S2B).
Figure 2. Heteroplasmy shift in NZB/BALB embryos using mito-ApaLI.
(A) Injection of mito-ApaLI mRNA in one-cell embryos for induction of heteroplasmy shift.
(B) In vitro development of mito-ApaLI injected embryos to blastocyst stage. Time-lapse images of EGFP reporter expression at different developmental stages.
(C) RFLP analysis and quantification of mtDNA heteroplasmy in control and mito-ApaLI injected embryos (Control n=10; mito-ApaLI n=8). Representative gel.
(D) Quantification of mtDNA copy number by qPCR in control and mito-ApaLI injected embryos (Control n=18; mito-ApaLI n=12).
Error bars represent ± SEM. ***p<0.001. ****p<0.0001. See also Figure S2.
Preventing the transmission of mitochondrial genomes using mitochondria targeted restriction endonucleases
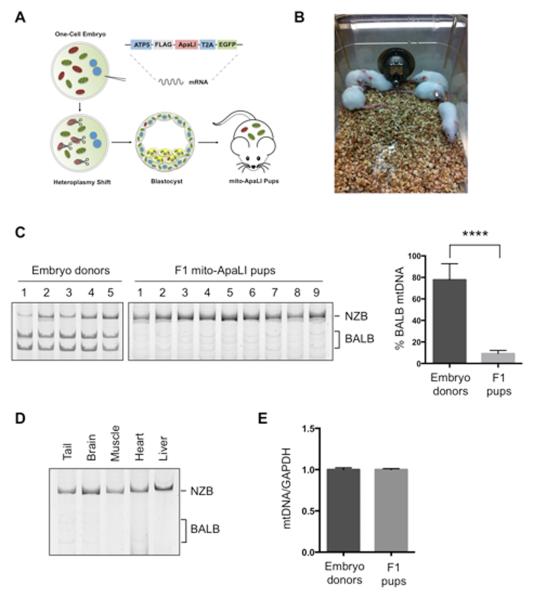
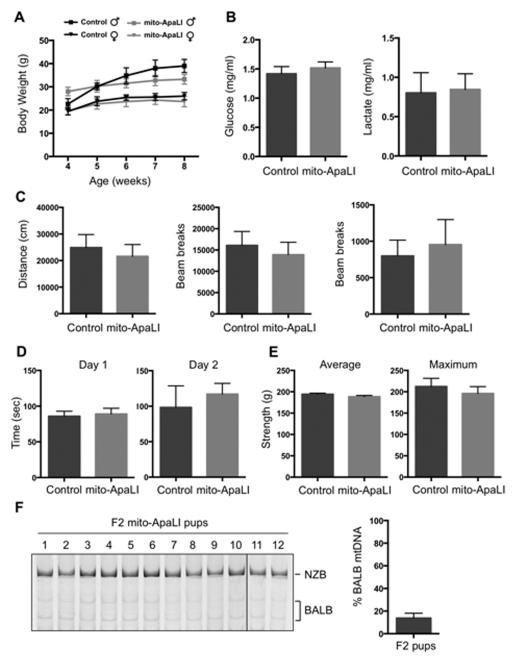
Next, we investigated whether induction of mtDNA heteroplasmy shift could be utilized for preventing the transmission of mitochondrial diseases to the next generation. NZB/BALB one-cell embryos injected with mito-ApaLI mRNA were cultured in vitro until the blastocyst stage and transferred to pseudopregnant mice (Figure 3A). After a standard gestation period, pseudopregnant mice gave birth to live pups through natural delivery (Figure 3B). Most importantly, RFLP analysis of total DNA from F1 mito-ApaLI animals revealed a significant reduction of BALB mtDNA (Figure 3C). Further analysis demonstrated reduction of BALB mtDNA in the brain, muscle, heart and liver. These data indicate the systemic clearance of a specific mtDNA in the offspring of heteroplasmic mothers (Figure 3D). Similarly, analysis at the HindIII region confirmed the specific reduction of BALB mtDNA in F1 mito-ApaLI animals (Figure S3A and S3B). Furthermore, analysis of mtDNA copy number showed normal mtDNA levels resulting from NZB mtDNA replication upon embryo implantation (Figure 3E). Comprehensive characterization of mito-ApaLI animals, both males and females, showed normal development, weight gain (Figure 4A), complete blood count (Table S1) as well as normal blood levels of glucose and lactate, all potential indicators of mitochondrial dysfunction (Haas et al., 2007) (Figure 4B). Moreover, typical behavioral studies indicative of central nervous system defects (Ross et al., 2013), including open field, rotor-rod, grip strength and sensory neuron screening, showed normal performance of mito-ApaLI animals (Figure 4C-4E).
Figure 3. Generation of live animals after induction of heteroplasmy shift in NZB/BALB embryos using mito-ApaLI.
(A) Outline for the generation of live animals after injection of mito-ApaLI mRNA in one-cell embryos.
(B) Representative photograph of F1 mito-ApaLI mice.
(C) RFLP analysis and quantification of mtDNA heteroplasmy in tail tip biopsies of embryo donors and generated F1 mito-ApaLI pups. (Donor n=10; mito-ApaLI n=9).
(D) RFLP analysis and quantification of mtDNA heteroplasmy in tail, brain, muscle, heart and liver of F1 mito-ApaLI mice.
(E) Quantification of mtDNA copy number by qPCR in F1 mito-ApaLI pups (Donor n=10; F1 mito-ApaLI n=9).
Error bars represent ± SEM. ****p<0.0001. See also Figure S3.
Figure 4. Characterization of F1 mito-ApaLI mice.
(A) Body weight of mito-ApaLI males (Control n=5 and mito-ApaLI n=3) and mito-ApaLI females (Control n=5 and mito-ApaLI n=6) at different time points. ns.
(B) Biochemical analysis of glucose and lactate in blood of control (n=10) and mito-ApaLI (n=9) mice. ns.
(C) Open field test measuring baseline levels of locomotor activity in freely moving mice quantifying distance traveled, ambulatory counts and vertical counts.
(D) Rotarod test evaluating locomotor coordination based on the latency at which a fall occurs on a gradually accelerating spinning rod.
(E) Grip strength test measuring average and maximum grip force in the forelimbs.
(F) RFLP analysis and quantification of mtDNA heteroplasmy in tail tip biopsies of F2 mito-ApaLI pups. (F2 mito-ApaLI n=12).
Error bars represent ± SEM. See also Figure S4 and Table S1.
To assess potential off-target effects on the nuclear genome, we performed comparative hybridization genomic (CHG) array and exome sequencing. CGH array indicated normal genomic integrity of mito-ApaLI animals (Figure S3C). Confirming this result, exome sequencing demonstrated variant rates in ApaLI containing exomic regions comparable to non-ApaLI exomic regions, excluding the possibility of off-target effects of mito-ApaLI (0.0014 vs. 0.0047 variants per hundred base pairs, respectively). Furthermore, mito-ApaLI animals were fertile, and RFLP analyses showed barely detectable levels of BALB mtDNA in the F2 generation (Figure 4F and S4). These results confirm the feasibility of mtDNA heteroplasmy shift to prevent the transgenerational transmission of mitochondrial diseases.
Preventing the transmission of mitochondrial genomes using mito-TALENs
Despite the broad range of over 200 mtDNA mutations associated with mitochondrial diseases, only the human mutation m8993T>G responsible for two mitochondrial diseases: neurogenic muscle weakness, ataxia and retinitis pigmentosa (NARP) and maternally inherited Leigh syndrome (MILS) generates a unique restriction site that can be targeted using the naturally occurring restriction endonuclease XmaI. For these reasons, alternative approaches to induce heteroplasmy shift based on the use of mitochondria targeted transcription activator-like effector nucleases (TALENs) and zinc finger nucleases (ZFNs), which could be designed against virtually any mutation, have been recently developed by us and other groups (Bacman et al., 2013; Gammage et al., 2014; Minczuk et al., 2006; 2008). In order to evaluate the use of mito-TALENs to prevent the transmission of mitochondrial diseases, we tested the specific elimination of NZB mtDNA in NZB/BALB oocytes. For this purpose, we first generated a collection of TALENs against NZB mtDNA and screened for a TALEN with the highest specificity against NZB mtDNA (Figure S5A-S5C). Under our design, the left monomer of the TALEN will bind to the common sequence of NZB and BALB mtDNA while the right monomer will preferentially recognize and bind to NZB mtDNA, dictating the specific cleavage of NZB mtDNA upon dimerization of the FokI nuclease (Figure S5A). NZB TALEN monomers were targeted to mitochondria by the human ATP5b and SOD2 mitochondria targeting sequence and the ATP5b and SOD2 5′ and 3′ UTRs to promote co-translation import from mitochondrial associated ribosomes (Marc et al., 2002). In addition, an EGFP or mCherry reporter was also included in the constructs encoding each TALEN monomer (Figure 5A). Once again, we tested the mitochondrial localization of the NZB TALEN by immunostaining in NZB/BALB tail tip fibroblasts (TTFs) and observed robust co-localization of mitochondria targeted NZB TALEN monomers (hereafter NZB mito-TALEN) with the mitochondrial dye Mitotracker (Figure S5D). Analysis of mtDNA by RFLP demonstrated induction of heteroplasmy shift in NZB/BALB cells by a specific reduction of NZB mtDNA after 72 h in cells transfected with NZB mito-TALENs compared to control cells transfected with mito-GFP (Figure S5E). In addition, similar to mito-ApaLI, we found normal mtDNA copy number in NZB mito-TALEN transfected cells resulting from the replication of the remaining BALB mtDNA that compensated for the reduction of NZB mtDNA (Figure S5F).
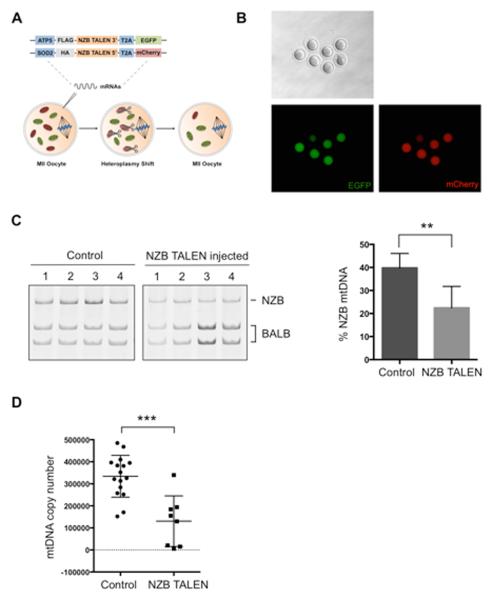
Figure 5. Heteroplasmy shift in NZB/BALB MII oocytes using NZB mito-TALEN.
(A) Injection of NZB mito-TALEN mRNA in oocytes for induction of heteroplasmy shift.
(B) Expression of fluorescent reporters of NZB TALEN monomer in MII oocytes.
(C) RFLP analysis and quantification of mtDNA heteroplasmy in control and NZB TALEN injected oocytes after 48 h (Control n=9; NZB TALEN n=7). Representative gel.
(D) Quantification of mtDNA copy number by qPCR in control and NZB TALEN injected oocytes after 48 h (Control n=16; NZB TALEN n=8).
Error bars represent ± SEM. **p<0.01. ***p<0.001. See also Figure S5.
We next decided to test whether mito-TALENs could be used in oocytes to specifically eliminate NZB mtDNA (Figure 5A). Fluorescent microscopy images revealed the expression of both NZB mito-TALEN monomers as indicated by EGFP and mCherry expression in oocytes (Figure 5B). RFLP analysis 48 h after NZB mito-TALEN mRNA injection demonstrated the specific decrease of NZB mtDNA and a consequential increase in the relative BALB mtDNA levels (Figure 5C). RFLP analysis at the HindIII region confirmed the specific reduction of NZB mtDNA upon injection of NZB mito-TALEN in NZB/BALB MII oocytes (Figure S5G). Analysis of mtDNA copy number by qPCR revealed a decrease in mtDNA copy number following NZB mito-TALEN injection in oocytes in agreement with the lack of mtDNA replication in oocytes (Figure 5D). These results demonstrate the potential of custom designed mito-TALENs for the specific elimination of mitochondrial genomes in the germline aimed at preventing the transmission of mitochondrial diseases.
Specific reduction of human mutated mitochondrial genomes responsible for mitochondrial diseases in mammalian oocytes
In order to evaluate the potential of our approach to prevent the transmission of human mitochondrial diseases we decided to test the use of mitochondria targeted nucleases against mutated mitochondrial genomes responsible for two mitochondrial diseases: Leber’s hereditary optic neuropathy and dystonia (LHOND) and NARP (Jun et al., 1994; Taylor and Turnbull, 2005). Due to the limited number of available patients and the difficulty in obtaining oocytes from these patients, we generated artificial mammalian oocytes carrying mutated genomes by cellular fusion of patient cells and mouse oocytes using Sendai virus (Figure 6A). Although this model has limitations compared to patient oocytes, it helped us to test the potential of our methodology for the specific elimination of pathogenic human mtDNAs in mammalian oocytes. For this purpose, we first tested the fusion of 143B osteosarcoma cybrid cells harboring the LHOND m.14459G>A mutation to mouse MII oocytes (Figure 6B). After 3 h, complete fusion was observed and no individual cells were detected under the zona pellucida of oocytes (Figure 6B). LHOND fused oocytes were incubated for 48 h and collected for analysis. PCR analysis using primers specific against the human mtDNA region containing the LHOND m.14459G>A mutation allowed for the detection of LHOND mtDNA in fused oocytes (Figure S6A). Next, we tested whether the LHOND mito-TALEN that we have recently reported could be used for the specific elimination of LHOND mtDNA in oocytes (Bacman et al., 2013). For this purpose, MII oocytes harboring LHOND mtDNA were injected with mRNA encoding the LHOND mito-TALEN 3 h after cell fusion. Fluorescent microscopy images revealed the expression of both LHOND mito-TALEN monomers as indicated by EGFP and mCherry expression (Figure S6B). RFLP analysis 48 h after mRNA injection demonstrated the specific reduction of LHOND mtDNA in fused oocytes (Figure 6C). Analysis of mtDNA copy number by qPCR confirmed a significant reduction of human mutated LHOND mtDNA upon injection of LHOND mito-TALENs in fused oocytes (Figure 6D). Finally, to demonstrate the potential of this approach against other mitochondrial diseases we decided to used a similar strategy to test the elimination of human mitochondrial genomes carrying the mutation NARP m.9176T>C. For this purpose, we first generated a collection of TALENs against NARP mtDNA and screened for a TALEN with the highest specificity against the mutation NARP m.9176T>C (Figure S6C-S6E). NARP mito-TALEN monomers were targeted to mitochondria by the ATP5b and SOD2 mitochondria targeting sequence and the ATP5b and SOD2 5′ and 3′ UTRs (Figure 6A). Immunostaining in NARP patient cells revealed a robust co-localization of mitochondria targeted NARP mito-TALEN monomers with the mitochondrial dye Mitotracker (Figure S6F). Subsequently, we tested the induction of heteroplasmy shift by NARP mito-TALEN using immortalized NARP patient cells. Analysis of mtDNA by RFLP demonstrated induction of heteroplasmy shift in NARP cells with a reduction in NARP mtDNA after 72 h in cells transfected with the NARP mito-TALEN compared to cells transfected with mito-GFP (Figure S6G). In addition, we found normal mtDNA copy numbers in NARP mito-TALEN transfected cells resulting from the replication of the remaining mtDNA (Figure S6H). Next, similar to LHOND, we tested the specific elimination of NARP mitochondrial genomes in oocytes. As before, patient cells harboring the NARP m.9176T>C mutation were fused to MII oocytes using Sendai virus and injected with NARP mito-TALEN 3 h after fusion. Fluorescent reporters for both NARP mito-TALEN monomers were observed in oocytes as indicated by EGFP and mCherry expression (Figure S6I). RFLP analysis 48 h after mRNA injection demonstrated the specific reduction of NARP mtDNA in fused oocytes (Figure 6E). Analysis of mtDNA copy number by qPCR confirmed a significant reduction of human mutated NARP mtDNA upon injection of NARP mito-TALENs in fused oocytes (Figure 6F). We speculate that the low levels of wild-type mtDNA carried by the NARP patient cells, together with the lack of mtDNA replication in oocytes, might be the reason why we fail to detect a significant increase in wild-type human mtDNA upon NARP mito-TALEN injection. Collectively, these results confirm the potential of custom-designed mito-TALENs for the specific elimination of clinically relevant mutated mitochondrial genomes responsible for human mitochondrial diseases in the germline.
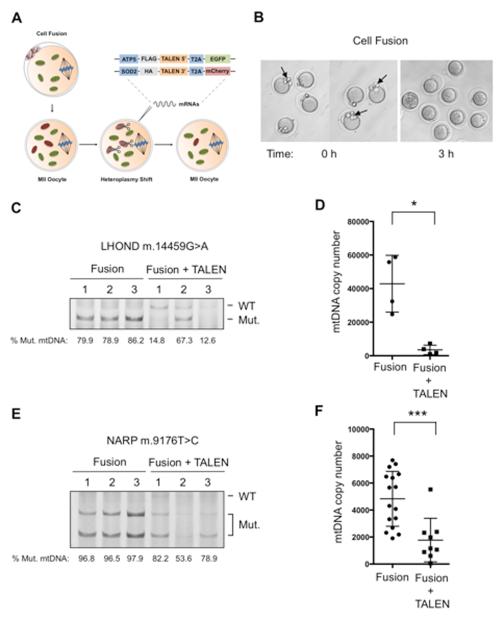
Figure 6. Specific elimination of human LHOND m.14459G>A and NARP m.9176T>C mutations in mammalian oocytes using mito-TALENs.
(A) Fusion of human cells harboring LHOND m.14459G>A and NARP m.9176T>C mutations with mouse MII oocytes followed by the injection of mito-TALENs for induction of heteroplasmy shift.
(B) Representative images of MII oocytes before and after cell fusion.
(C) RFLP analysis and quantification of LHOND heteroplasmy in individual MII oocytes with and without LHOND TALEN injection after 48 h (Fusion n=3; Fusion + TALEN n=3).
(D) Quantification of human mtDNA copy number by qPCR in individual MII oocytes with and without LHOND TALEN injection after 48 h (Fusion n=4; Fusion + TALEN n=4).
(E) RFLP analysis and quantification of NARP heteroplasmy in individual MII oocytes with and without NARP TALEN injection after 48 h (Fusion n=7; Fusion + TALEN n=3).
(F) Quantification of human mtDNA copy number by qPCR in individual MII oocytes with and without NARP TALEN injection after 48 h (Fusion n=17; Fusion + TALEN n=9).
Error bars represent ± SEM. *p<0.05. ***p<0.001. See also Figure S6.
DISCUSSION
In summary, we report here on novel strategies for preventing germline transmission of mitochondrial diseases through the induction of mtDNA heteroplasmy shift in oocytes and embryos. As a proof of concept, we used a heteroplasmic mouse model carrying two different mtDNA haplotypes: NZB and BALB. First, we demonstrated that injection of mRNA encoding mitochondria targeted ApaLI restriction enzyme into oocytes, as well as into one-cell embryos, led to the generation of live animals with significantly reduced levels of the BALB mtDNA haplotype. These animals displayed normal behavior, development, gross genomic integrity and fertility. Moreover, their progeny (F2 generation) maintained significantly reduced levels of BALB mtDNA. These results demonstrate the potential of germline heteroplasmy shift to prevent the transgenerational transmission of mitochondrial genomes. In addition, injection of mRNA encoding mitochondria targeted NZB mito-TALEN into oocytes led to a significant reduction of NZB mtDNA levels. Finally, fusion of human patient cells carrying mtDNA mutations to mouse oocytes followed by injection of mito-TALENs against human mtDNA mutations demonstrated a specific reduction in the levels of mutated mtDNA.
The use of restriction nucleases for the induction of heteroplasmy shift has been previously demonstrated in the NZB/BALB mouse as well as in patient somatic cells by us and other groups (Alexeyev et al., 2008; Bacman et al., 2010; 2012). However, the application of restriction enzymes to target clinically relevant mutations is limited to only m8993T>G, which is responsible for some cases of NARP and MILS, a mutation that generates a unique restriction site that can be targeted using the restriction endonuclease XmaI (Alexeyev et al., 2008). The use of other approaches using different types of nucleases including TALENs might allow for the custom-designed targeting of a wider range of human mitochondrial mutations responsible for mitochondrial diseases. Along this line, several reports have recently demonstrated the use of mitochondria targeted TALENs and zinc finger nucleases (ZFNs) for the specific elimination of mutated mitochondrial genomes in somatic cells (Bacman et al., 2013; Gammage et al., 2014; Minczuk et al., 2006; 2008). Although, when compared to mitochondria targeted restriction endonucleases, the use of mito-TALENs for preventing transmission of mitochondrial diseases in the germline may be less robust. However, we speculate that their therapeutic use will achieve specific reduction of mutated mitochondrial genomes below he threshold levels (60-95%) required for biochemical and clinical defects to manifest (Russell and Turnbull, 2014). In addition, we anticipate that the future development and application of more specific and efficient gene editing technologies will allow for a higher significant reduction of mutated mtDNA levels in the germline.
Transmission of mitochondrial diseases by female carriers directly correlates with the levels of mutated mtDNA present in oocytes. In many cases, asymptomatic female carriers with intermediate levels of mutant load may produce oocytes with different ranges of mutated mtDNA (Chinnery et al., 2000; Cree et al., 2009). Due to the lack of mtDNA replication in oocytes and pre-implantation embryos, targeting of mutated mtDNA in oocytes with high mutant loads using the approach presented here may lead to a dramatic reduction in mtDNA copy number. In mice, embryos with mitochondria levels below a specific threshold develop normally during the pre-implantation stages but subsequently fail to implant in the uterus or undergo development arrest (Wai et al., 2010). Consequently, oocytes containing high levels of mutated mtDNA that are subjected to heteroplasmy shift may result in embryos with low mtDNA copy number that may fail to implant in the uterine wall. In this case, though heteroplasmy shift may not result in a viable embryo, it would attain the goal of hampering the development and implantation of embryos with high mutant loads, thereby preventing the transmission of mitochondrial diseases to the next generation. In this scenario, PGD could be used as a complementary approach to select embryos with mtDNA copy numbers sufficient for implantation.
Due to the non-Mendelian segregation of mtDNA, current therapeutic approaches, including genetic counseling and pre-implantation genetic diagnosis (PGD), can only partially reduce, but not eliminate, the risk of transmission of mitochondrial diseases (Brown et al., 2006). The recent development of mitochondrial replacement techniques based on spindle, pronuclear or polar body transfer into healthy enucleated donor oocytes or embryos, currently under review by US and UK regulatory agencies, represent a valid and powerful alternative to current approaches (Craven et al., 2010; Paull et al., 2013; Tachibana et al., 2012; Wang et al., 2014). Mitochondrial replacement techniques involve a series of complex technical manipulations of nuclear genome between patient and donor oocytes that will result in the generation of embryos carrying genetic material from three different origins. For these reasons, mitochondrial replacement techniques have raised some biological, medical and ethical concerns (Hayden, 2013; Reinhardt et al., 2013). Despite their great potential, more studies are still required to show that these techniques are safe in human oocytes. The approach presented here relies on a single injection of mRNA into patient oocytes, which is technically simpler and less traumatic to the oocyte compared to mitochondrial replacement techniques (Craven et al., 2010; Paull et al., 2013; Tachibana et al., 2012; Wang et al., 2014). Importantly, it does not require healthy donor oocytes, thus avoiding ethical issues related to the presence of donor mtDNA.
Induction of mtDNA heteroplasmy shift using restriction endonucleases or TALENs has the potential to eliminate mutated mitochondrial genomes in the germline, and consequently, prevent the transgenerational transmission of mitochondrial diseases. In addition, since mtDNA mutations in the germline have been recently linked to aging (Ross et al., 2013), this strategy could also be applied to prevent the transmission of mtDNA variants with potential roles in aggravating aspects of human aging and age-associated diseases.
EXPERIMENTAL PROCEDURES
Plasmids
A synthetic gene coding for the ApaLI restriction endonuclease with a C-terminal HA (Hemagglutinin antigen) tag was purchased from Integrated DNA Technologies (Coralville) with codon usage optimized for mammalian translation. For the generation of the mito-ApaLI construct, ApaLI was subcloned into the pVAX plasmid containing the mitochondria localization signal derived from ATP5b, a unique Flag immunotag in the N-terminus, 5’ and 3’ untranslated regions from ATP5b to localize the mRNA to ribosomes associated with mitochondria, an independent fluorescent marker to select for expression (enhance green fluorescent protein (EGFP)) and a recoded picornaviral 2A-like sequence (T2A′) between the mito-ApaLI and the fluorescent marker. Subsequently, the fragment described was subcloned into the pcDNA3 plasmid containing a T7 promoter for in vitro transcription. For the generation of the mito-GFP construct, EGFP was subcloned into the previously described pVAX construct lacking the independent fluorescent marker and the recoded picornaviral 2A-like sequence (T2A′) but containing a T7 promoter. For the generation of ApaLI construct, ApaLI RE was subcloned into the previously described pVAX plasmid lacking the N-terminus mitochondria localization signal derived from ATP5b and the 5’ and 3’ untranslated regions from ATP5b with a T7 promoter. Cloning was done using the using the In-Fusion HD cloning kit (Clontech Laboratories).
Construction of mitochondria targeted TALEN (mito-TALENs)
TALEN target sites for NZB and NARP m.9176T>C were identified using the TAL effector-Nucleotide Targeter (TALE-NT) software (Christian et al., 2010). To increase TALEN specificity, TALEN with targeting sequences of various lengths ranging from 7.5 to 13.5 base pairs were designed. TALENs were constructed into the TALEN cloning vector of the TALE Toolbox kit from Addgene (cat#1000000019) (Sanjana et al., 2012), and the TALENs recognizing the target sites were constructed using the Golden Gate Assembly method. Mitochondria targeted TALEN (mito-TALEN), were constructed by addition of mitochondria localization signals derived from ATP5b or SOD2 mitochondria localization signal, inclusion of a unique immuno-tag in the N terminus of the mature protein (hemagglutinin (HA) or Flag), inclusion of a 3′ untranslated region from a mitochondrial gene known to localize mRNA to ribosomes on mitochondria, inclusion of an independent fluorescent marker to select for expression (EGFP in one monomer and mCherry in the other) and inclusion of a recoded picornaviral 2A-like sequence (T2A′) between the mitoTALEN and the fluorescent marker
Animals
All animal procedures were performed according to NIH guidelines and approved by the Committee on Animal Care at Salk Institute. NZB/BALB heteroplasmic founder females were originally generated (Jenuth et al., 1996). NZB/BALB colony was maintained by breeding the females with BALB/cByJ males. Tail tip genotyping was routinely performed in order to exclude females carrying low levels of one of the two mtDNA haplotypes. BALB/c, BALB/cByJ and NZB mice were obtained from Jackson laboratory.
Cells, transfection and sorting
Simian virus 40 (SV40) immortalized NZB/BALB fibroblasts containing NZB and BALB mitochondrial DNA were derived from tail tip of NZB/BALB mice. Human patient cells harboring the NARP m.9176T>C mutation were obtained by skin biopsy after signed informed consent of the donor and with the approval of the Institutional Review Board of the Hospital Clinic, Spain. Cells were immortalized using SV40 and cultured at 37 °C in DMEM (Invitrogen) containing GlutaMAX, non-essential amino acids and 10% fetal bovine serum (FBS). 143B osteosarcoma cybrid cells harboring the LHOND m.14459G>A mutation were obtained and cultured as previously described (Bacman et al., 2013). Cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. After 72 h, cells were sorted using a BD Influx (Becton, Dickinson and Company) by gating on single-cell fluorescence using a 488-nm laser with a 505LP, 530/40 filter set for EGFP and a 561-nm laser with a 600LP, 610/20 filter set for mCherry. Total DNA was extracted from sorted cells using the DNeasy Blood and Tissue Kit (Qiagen) following the protocol suggested by the manufacturer.
Single strand annealing (SSA) reporter assay
Please refer to Extended Experimental Information.
Production of mRNA
In vitro transcription of mRNA was performed using mMESSAGE mMACHINE T7 ULTRA kit (Life Technologies) according to the manufacturer’s instructions using linearized and gel purified (Qiagen) plasmid template. The mRNA was purified using MEGAclear kit (Life Technologies) and quantified using NanoDrop 8000 (Thermo Scientific).
Oocyte collection and mRNA Injection
Female mice were superovulated with pregnant mares serum gonadotropin (PMSG) and human chorionic gonadotropin (hCG). MII oocytes were collected 14 h after hCG injection in M2 medium (Millipore) and freed of cumulus cells using hyaluronidase. For collection of 1-cell embryos, superovulated female mice were mated to BALB/c males and fertilized embryos were collected 18-20 h after hCG injection from oviduct. mRNA (50 – 250ng/μl) was injected into the cytoplasm of MII oocytes and fertilized embryos in M2 medium using Eppendorf micromanipulator. The injected MII oocytes were in vitro cultured in KSOM (Millipore) for 48 h before analysis. The injected embryos were cultured in KSOM at 37°C under 5% CO2 in air until blastocyst stage. Subsequently, blastocysts were collected for analysis or transferred to BALB/c pseudopregnant females. Live pups were obtained by natural delivery.
Cell fusion
Cell fusion was achieved by using inactivated Sendai virus (GenomeOne, Cosmo Bio). Sendai virus stock solutions were prepared according to the manufacturer instructions and further diluted 1:20 in cell fusion buffer. The 143B osteosarcoma cybrid cells harboring LHON m.14459G>A mutation and patient cells harboring NARP m9176T>C mutation were used for fusion with mature MII oocytes. Cells were cultured for 48 h in DMEM no glucose medium supplemented with galactose before using for cell fusion to increase mtDNA content. On the day of fusion, cells were trypsinized and resuspended in M2 medium. For each MII oocyte, 5 cells briefly placed in Sendai virus were injected under the zona pellucida. After 3 h successfully fused oocytes were selected for mito-TALEN mRNA injection. Lastly, surviving oocytes were cultured in KSOM for 48 h before analysis.
Immunofluorescence
Cells were seeded on coverslips before transfection. 48 h after of transfection cells were incubated in the presence of 350nM Mitotracker (Invitrogen) for 30 min. Subsequently, cells were fixed and permeabilized with 4% PFA and 0.1% Triton X-100, respectively. After fixation, cells were blocked for 1 h at room temperature with 1% BSA/PBS. Next, cells were incubated with an anti-Flag M2 primary antibody (Sigma) or anti-HA antibody (Millipore) overnight at 4 °C. The next day, cells were washed three times with PBS and incubated for 1 h at room temperature with Alexa Fluor 488-conjugated donkey antibodies to goat IgG (Molecular Probes) or Alexa Fluor 647-conjugated donkey antibodies to mouse IgG and 10 min with Hoechst 33342 (0.5 μg ml−1 in PBS) (Invitrogen). Finally, cells were washed three times with PBS and mounted using Fluoromount-G (Southernbiotech). Confocal image acquisition was performed using a Zeiss LSM 780 laser-scanning microscope (Carl Zeiss Jena).
‘Last-cycle hot’ PCR and Restriction Fragment Length Polymorphism (RFLP)
Total DNA from cells, tail biopsies and oocytes/embryos were used to determine mtDNA heteroplasmy by ‘Last-cycle hot’ PCR using the mtDNA 5’ Fluorescein amidite (FAM) labeled primers as listed in Table S2. NZB/BALB PCR products were digested with ApaLI or HindIII, which digests BALB mtDNA at positions 5461 (ApaLI targeting site) and 9136 respectively. NARP PCR products were digested with BsrI which digest mutated NARP mtDNA at position 9176. The levels of LHON m.14459G>A were determined as previously reported (Bacman et al., 2013). Digested PCR products were subjected to electrophoresis in an 12% polyacrylamide gel. The fluorescein signal was quantified using a Typhoon 8600 system (Molecular Dynamics) and gels were quantified using ImageQuant 5.2 (Molecular Dynamics).
Quantification of mtDNA copy number
Absolute mtDNA copy numbers were quantified by real-time PCR using iQSyber Green on Bio-Rad iCycler (Bio-Rad). Individual oocytes and embryos were transferred into lysis buffer (200mM KOH) and incubated for 10 min at 65 °C. The reaction was neutralized by addition of 200mM HCl. Absolute mtDNA copy number per 1μl of lysate was calculated using a standard curve derived from the Q-PCR amplification of a fragment of mtDNA genome. Briefly, primers listed in Table S2 were used to quantify the absolute level of mtDNA in samples. First, a standard curve was generated by a 10-fold serial dilution of a PCR product obtained using Standard curve primers for the different regions of mtDNA analyzed. Subsequently, to quantify the absolute level of mtDNA, quantitative real-time PCR was performed using qPCR primers listed in Table S2.
Blood and plasma parameters
Blood collection was performed by sub-mandibular bleeding. Whole EDTA blood samples were analyzed in duplicates for Complete Blood Count (CBC) on a Hemavet 950FS Multi Species Hematology System (Drew Scientific). For glucose and lactate analysis was determined using the Glucose (GO) Assay Kit (Sigma) according to the manufacturer’s instructions. Plasma lactate concentration was determined using the Lactate Assay Kit (Sigma) according to the manufacturer’s instructions. Please refer to Extended Experimental Information.
Behavioral analysis
Behavioral testing was carried out at the Salk Institute for Biological Studies Behavioral Testing Core. Basic sensorimotor function was assessed in the Open Field Test, Rotarod, Grip Strength and Neurological Screen. Please refer to Extended Experimental Information.
Array comparative genomic hybridization (aCGH)
aCGH was performed following Agilent Oligonucleotide Array-Based CGH for Genomic DNA Analysis (Agilent Technologies, Santa Clara, CA). Please refer to Extended Experimental Information
Exome capture and high-throughput sequencing
Exome capture was using the SeqCap EZ Mouse Exome Design probe pool (54 Mb, NimbleGen) according to the manufacturer's protocol. Please refer to Extended Experimental Information
Statistical evaluation
Statistical analyses were performed by using standard unpaired Student t test with Welch’s correction using Prism 6 software (GraphPad). All data are presented as mean ± SEM and represent a minimum of two independent experiments. Statistical significance is displayed as *p<0.05, **p<0.01, ***p<0.001 and ****p<0.0001.
Supplementary Material
ACKNOWLEDGEMENTS
We thank M. Schwarz for administrative support. We thank S. Heinz and M.M. Ku from H. A. and Mary K. Chapman Charitable Foundations Genomic Sequencing Core at the Salk Institute for help with sequencing analysis. We thank M. Chang from the Integrative Genomics and Bioinformatics Core at the Salk Institute for sequencing data analysis. We thank C. B. Farrokhi from the Behavioral Testing Core at the Salk Institute for help with behavioral studies. We thank Eric Shoubridge from McGill University, Montreal, Canada for sharing with us the NZB/BALB/c heteroplasmic mice. We thank Julio Montoya Villarroya from the University of Zaragoza, Zaragoza, Spain and Ma Ángeles Ruiz Gómez from the Hospital de Son Espases, Palma de Mallorca, Spain. Financial support. M.M. Ku and S. Heinz are supported by the Leona M. and Harry B. Helmsley Charitable Trust. A. Ocampo was partially supported by a US National Institutes of Health Ruth L. Kirschstein National Research Service Award Individual Postdoctoral Fellowship. G.-H. Liu was supported by National Basic Research Program of China (973 Program, 2015CB964800; 2014CB964600), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA01020312), National Natural Science Foundation of China (NSFC: 81271266; 31222039; 31201111; 81371342). C.T. Moraes was supported by US National Institutes of Health grants 5R01EY010804, 1R01AG036871, The JDM Fund, The Muscular Dystrophy Association and The United Mitochondrial Disease Foundation. S.L. Williams is supported by Florida Department of Health Grant 3KN09. Work in the laboratory of J.C. Izpisua Belmonte was supported by the G. Harold and Leila Y. Mathers Charitable Foundation and The Leona M. and Harry B. Helmsley Charitable Trust (2012-PG-MED002).
Footnotes
ACCESSION NUMBERS
aCGH data sets are available in the Gene Expression Omnibus database under the accession number (pending) . Exome sequencing data sets are available in the Gene Expression Omnibus database under the accession number (SRP056327).
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, six figures, and two tables and can be found with this article online at
AUTHORS CONTRIBUTIONS
P.R., A.O, C.T.M. and J.C.I.B. designed all experiments. P.R, A.O, I.S-M. and J.C.I.B. prepared the figures and wrote the manuscript. P.R., A.O., K.S., S.R.B., Y.T., J.W., D.L., X.X., N.M., and C.R., performed and analyzed all in vitro experiments. P.R., A.O., J.L., A.S. and D.O. performed and analyzed all in vivo experiments. S.R.B., S.L.W., G.H.L, D.M., S.C., M.M.C., H.Z. and C.T.M. provided reagents. F. C., J. C. and J.M.C. contributed to the design of the project.
REFERENCES
- Alexeyev MF, Venediktova N, Pastukh V, Shokolenko I, Bonilla G, Wilson GL. Selective elimination of mutant mitochondrial genomes as therapeutic strategy for the treatment of NARP and MILS syndromes. Gene Ther. 2008;15:516–523. doi: 10.1038/gt.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexeyev M, Shokolenko I, Wilson G, LeDoux S. The Maintenance of Mitochondrial DNA Integrity--Critical Analysis and Update. Cold Spring Harb Perspect Biol. 2013;5:a012641–a012641. doi: 10.1101/cshperspect.a012641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S, Bankier AT, Barrell BG, Debruijn M, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and Organization of the Human Mitochondrial Genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Bacman SR, Williams SL, Duan D, Moraes CT. Manipulation of mtDNA heteroplasmy in all striated muscles of newborn mice by AAV9-mediated delivery of a mitochondria-targeted restriction endonuclease. Gene Ther. 2012;19:1101–1106. doi: 10.1038/gt.2011.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacman SR, Williams SL, Garcia S, Moraes CT. Organ-specific shifts in mtDNA heteroplasmy following systemic delivery of a mitochondria-targeted restriction endonuclease. Gene Ther. 2010;17:713–720. doi: 10.1038/gt.2010.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacman SR, Williams SL, Pinto M, Peralta S, Moraes CT. Specific elimination of mutant mitochondrial genomes in patient-derived cells by mitoTALENs. Nat. Med. 2013;19:1111–1113. doi: 10.1038/nm.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DT, Herbert M, Lamb VK, Chinnery PF, Taylor RW, Lightowlers RN, Craven L, Cree L, Gardner JL, Turnbull DM. Transmission of mitochondrial DNA disorders: possibilities for the future. Lancet. 2006;368:87–89. doi: 10.1016/S0140-6736(06)68972-1. [DOI] [PubMed] [Google Scholar]
- Chinnery PF, Thorburn DR, Samuels DC, White SL, Dahl HM, Turnbull DM, Lightowlers RN, Howell N. The inheritance of mitochondrial DNA heteroplasmy: random drift, selection or both? Trends Genet. 2000;16:500–505. doi: 10.1016/s0168-9525(00)02120-x. [DOI] [PubMed] [Google Scholar]
- Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven L, Tuppen HA, Greggains GD, Harbottle SJ, Murphy JL, Cree LM, Murdoch AP, Chinnery PF, Taylor RW, Lightowlers RN, et al. Pronuclear transfer in human embryos to prevent transmission of mitochondrial DNA disease. Nature. 2010;465:82–85. doi: 10.1038/nature08958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cree LM, Samuels DC, Chinnery PF. The inheritance of pathogenic mitochondrial DNA mutations. Biochim. Biophys. Acta. 2009;1792:1097–1102. doi: 10.1016/j.bbadis.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall SD, Brown MT, Johnson PJ. Ancient invasions: from endosymbionts to organelles. Science. 2004;304:253–257. doi: 10.1126/science.1094884. [DOI] [PubMed] [Google Scholar]
- Gammage PA, Rorbach J, Vincent AI, Rebar EJ, Minczuk M. Mitochondrially targeted ZFNs for selective degradation of pathogenic mitochondrial genomes bearing large-scale deletions or point mutations. EMBO Mol Med. 2014;6:458–466. doi: 10.1002/emmm.201303672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas RH, Parikh S, Falk MJ, Saneto RP, Wolf NI, Darin N, Cohen BH. Mitochondrial disease: a practical approach for primary care physicians. Pediatrics. 2007;120:1326–1333. doi: 10.1542/peds.2007-0391. [DOI] [PubMed] [Google Scholar]
- Hayden EC. Regulators weigh benefits of “three-parent” fertilization. Nature. 2013;502:284–285. doi: 10.1038/502284a. [DOI] [PubMed] [Google Scholar]
- Jenuth JP, Peterson AC, Fu K, Shoubridge EA. Random genetic drift in the female germline explains the rapid segregation of mammalian mitochondrial DNA. Nature Genetics. 1996;14:146–151. doi: 10.1038/ng1096-146. [DOI] [PubMed] [Google Scholar]
- Jun AS, Brown MD, Wallace DC. A mitochondrial DNA mutation at nucleotide pair 14459 of the NADH dehydrogenase subunit 6 gene associated with maternally inherited Leber hereditary optic neuropathy and dystonia. Proc. Natl. Acad. Sci. U.S.a. 1994;91:6206–6210. doi: 10.1073/pnas.91.13.6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc P, Margeot A, Devaux F, Blugeon C, Corral-Debrinski M, Jacq C. Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep. 2002;3:159–164. doi: 10.1093/embo-reports/kvf025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minczuk M, Papworth MA, Kolasinska P, Murphy MP, Klug A. Sequence-specific modification of mitochondrial DNA using a chimeric zinc finger methylase. Proc. Natl. Acad. Sci. U.S.a. 2006;103:19689–19694. doi: 10.1073/pnas.0609502103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minczuk M, Papworth MA, Miller JC, Murphy MP, Klug A. Development of a single-chain, quasi-dimeric zinc-finger nuclease for the selective degradation of mutated human mitochondrial DNA. Nucleic Acids Res. 2008;36:3926–3938. doi: 10.1093/nar/gkn313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull D, Emmanuele V, Weiss KA, Treff N, Stewart L, Hua H, Zimmer M, Kahler DJ, Goland RS, Noggle SA, et al. Nuclear genome transfer in human oocytes eliminates mitochondrial DNA variants. Nature. 2013;493:632–637. doi: 10.1038/nature11800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt K, Dowling DK, Morrow EH. Medicine. Mitochondrial replacement, evolution, and the clinic. Science. 2013;341:1345–1346. doi: 10.1126/science.1237146. [DOI] [PubMed] [Google Scholar]
- Rogers DC, Fisher EM, Brown SD, Peters J, Hunter AJ, Martin JE. Behavioral and functional analysis of mouse phenotype: SHIRPA, a proposed protocol for comprehensive phenotype assessment. Mamm. Genome. 1997;8:711–713. doi: 10.1007/s003359900551. [DOI] [PubMed] [Google Scholar]
- Ross JM, Stewart JB, Hagström E, Brené S, Mourier A, Coppotelli G, Freyer C, Lagouge M, Hoffer BJ, Olson L, et al. Germline mitochondrial DNA mutations aggravate ageing and can impair brain development. Nature. 2013;501:412–415. doi: 10.1038/nature12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell O, Turnbull D. Mitochondrial DNA disease-molecular insights and potential routes to a cure. Exp. Cell Res. 2014;325:38–43. doi: 10.1016/j.yexcr.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, Zhang F. A transcription activator-like effector toolbox for genome engineering. Nat Protoc. 2012;7:171–192. doi: 10.1038/nprot.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoubridge EA, Wai T. Mitochondrial DNA and the mammalian oocyte. Curr. Top. Dev. Biol. 2007;77:87–111. doi: 10.1016/S0070-2153(06)77004-1. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Amato P, Sparman M, Woodward J, Sanchis DM, Ma H, Gutierrez NM, Tippner-Hedges R, Kang E, Lee H-S, et al. Towards germline gene therapy of inherited mitochondrial diseases. Nature. 2012;493:627–631. doi: 10.1038/nature11647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel G. Assisted reproduction. FDA considers trials of 'three-parent embryos'. Science. 2014;343:827–828. doi: 10.1126/science.343.6173.827. [DOI] [PubMed] [Google Scholar]
- Wai T, Ao A, Zhang X, Cyr D, Dufort D, Shoubridge EA. The role of mitochondrial DNA copy number in mammalian fertility. Biology of Reproduction. 2010;83:52–62. doi: 10.1095/biolreprod.109.080887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC, Chalkia D. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb Perspect Biol. 2013;5:a021220. doi: 10.1101/cshperspect.a021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Sha H, Ji D, Zhang HL, Chen D, Cao Y, Zhu J. Polar body genome transfer for preventing the transmission of inherited mitochondrial diseases. Cell. 2014;157:1591–1604. doi: 10.1016/j.cell.2014.04.042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.