SUMMARY
The liver is thought to utilize facultative stem cells, also known as “oval cells” or “atypical ductal cells” (ADCs), for regeneration following various types of injury. However, this notion has been based largely on in vitro studies and transplantation models; where lineage tracing has been used, results have been conflicting and effect sizes have been small. Here, we used genetic and nucleoside analog-based tools to mark and track the origin and contribution of various cell populations to liver regeneration in vivo following several ADC-inducing insults. We report that, contrary to prevailing stem-cell-based models of regeneration, virtually all new hepatocytes come from preexisting hepatocytes.
INTRODUCTION
It is well established that organs can maintain homeostasis via either cellular replication or differentiation from stem cells. In addition, it has been proposed that select tissues contain a population of so-called “facultative stem cells,” which contribute to tissue homeostasis under special circumstances. By definition, facultative stem cells lack stem cell activity during normal tissue turnover but are recruited during specific types of injury to function as stem or progenitor cells (Yanger and Stanger, 2011). The mammalian liver has stood as the major paradigm for regeneration via a facultative stem-cell-mediated recovery. In response to various disease states and toxin-induced injuries, rodents and humans exhibit an accumulation of atypical ductal cells (ADCs)—commonly referred to as “oval cells”—within the liver parenchyma (Farber, 1956; Popper et al., 1957). ADCs have a ductal morphology, but their arrangement into an intricate anastomosing configuration that extends into the hepatic lobule gives them a histologic appearance that is distinct from that of normal biliary epithelial cells (BECs) (Desmet, 1985). ADCs are thought to arise from BECs within the Canals of Hering, structures that reside at the interface of the intrahepatic bile ducts and hepatocyte-lined canaliculi (Figure 1A) (Factor et al., 1994; Preisegger et al., 1999). This putative mode of recovery stands in contrast to liver regeneration following surgical resection, which is mediated largely through cell growth and division (Miyaoka et al., 2012).
Figure 1. BECs Lack Detectable Progenitor Cell Activity In Vivo.
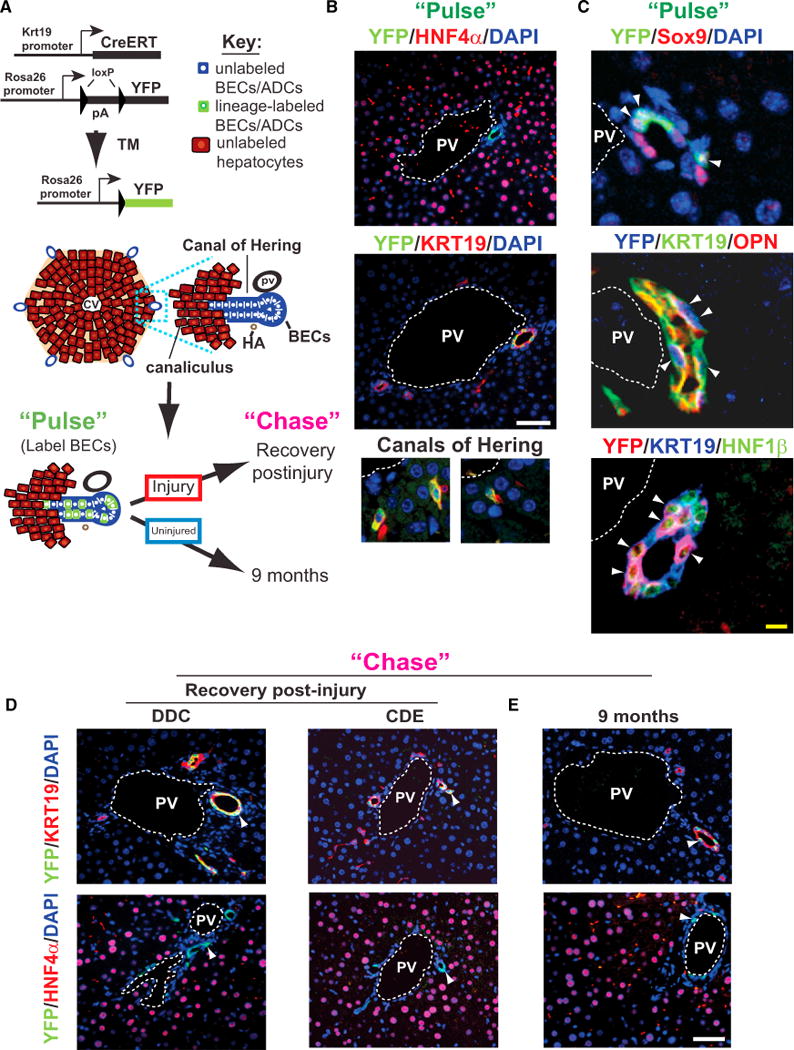
(A) Schematic view of BEC labeling using Krt19-CreER;R26YFP mice. An idealized hexagonally shaped lobule is shown. A blow-up of one portal tract illustrates the interface between BECs (blue) and hepatocytes (crimson). Cells are marked in a mosaic fashion on tamoxifen injection (“Pulse”), and the ability of labeled BECs to give rise to hepatocytes is assessed following injury and recovery or under long-term (9 month) homeostatic conditions (“Chase”).
(B) YFP-labeling at “pulse.” Labeling occurs exclusively in BECs that are negative for HNF4a (top panel) and positive for Krt19 in ducts (middle panel) or within the Canals of Hering (bottom panels).
(C) Costaining of the YFP label with the BEC markers Sox9, OPN, and HNF1β (arrowheads mark double positive/triple cells).
(D) Following injury-recovery, a similar pattern and degree of labeling is observed after the chase; no YFP label is observed in hepatocytes (“Chase”–DDC and CDE).
(E) Livers from Krt19-CreER; R26YFP mice examined 9 months after tamoxifen injection do not show any hepatocytes bearing the YFP label (“Chase”–9 months). Images shown are representative of multiple experiments (pulse: n = 7; DDC: n = 8; CDE: n = 3; 9 months chase group: n = 5).
TM, tamoxifen; CV, central vein; PV, portal vein; HA, hepatic artery. White scale bars, 50 mm, yellow scale bar, 5 mm. See also Figure S1.
Based on in vitro studies, ultrastructural analyses, and cell transplantation assays, ADCs have been proposed to function as bipotent facultative stem cells, giving rise to both hepatocytes and BECs, during toxin-mediated liver injury, although this issue is controversial (Español-Suñer et al., 2012; Fausto and Campbell, 2003; Friedman and Kaestner, 2011; Furuyama et al., 2011; Huch et al., 2013; Malato et al., 2011; Zaret and Grompe, 2008). Furthermore, adult hepatocytes exhibit significant plasticity in vivo, a phenomenon that may give the appearance of stem-cell-mediated differentiation (Michalopoulos et al., 2005; Tanimizu et al., 2014; Yanger et al., 2013). In order to obtain direct evidence for liver stem cell activity in vivo, we labeled three distinctive cell populations in the liver—BECs, hepatocytes, and rapidly dividing cells—using both direct genetic and unbiased nucleoside analog-based lineage labeling tools under multiple ADC-inducing injury conditions. Our studies demonstrate that hepatocytes, not ADCs, serve as the major, if not exclusive, source for hepatocyte renewal and regeneration in the adult liver, regardless of the type of injury.
RESULTS
BECs Do Not Exhibit Progenitor Cell Activity In Vivo
BECs residing within the Canals of Hering are thought to serve as precursors of liver progenitor cells (Factor et al., 1994; Wang et al., 2003). Such BECs, in both rodent and human studies, are characterized as single or small groups of cells positive for the BEC marker, cytokeratin-19 (KRT19), and located within the hepatic lobule separated from the larger bile ducts (Crawford et al., 1998; Roskams et al., 2004; Saxena and Theise, 2004; Theise et al., 1999). To directly address the ability of these cells to differentiate into hepatocytes in vivo, we crossed inducible KRT19 promoter (Krt19) CreER knockin mice (Means et al., 2008) to Rosa26YFP reporter mice (Srinivas et al., 2001) to label cells from the BEC lineage prior to injury (Figure 1A). Bigenic Krt19-CreER/R26YFP mice were given tamoxifen, resulting in pulse labeling completely restricted to BECs as previously reported, with no yellow-fluorescent-protein-positive (YFP+) cells staining for the hepatocyte marker hepatocyte nuclear factor 4 α (HNF4α) (Figure 1B, top panel; Scholten et al., 2010). The efficiency of labeling was 36.2% ± 8.7% of Krt19+ cells (Figure 1B, middle panel; n = 4) and included single/small groups of cells within the Canals of Hering (Figure 1B, bottom panels).
To ensure that labeling was not limited to a specific subset of BECs, we examined the expression of TROP2, a widely used oval cell/ADC marker (Okabe et al., 2009; Yamazaki et al., 2011), and found that Krt19+/YFP+ cells and Krt19+/YFP− cells stained positively for TROP2 with equal frequency (Figures S1A and S1B available online). We further assessed the spectrum of cellular labeling by costaining with YFP and a number of other BEC/ADC markers. This analysis showed that colabeling occurred with a similar efficiency to that observed with Krt19, as 39.5% of Sox9+ cells (n = 3), 31.6% of osteopontin+ (Opn+) cells (n = 3), and 40.8% of HNF1β+ cells (n = 3) were labeled (Figure 1C). Considering that Krt19 staining overlaps completely with staining for these BEC markers (353/353 Sox9+ cells, n = 3; 352/352 OPN+ cells, n = 3; and 349/349 HNF1β+ cells, n = 2), these results suggest that tamoxifen treatment of Krt19-CreER/R26YFP mice results in specific labeling of BECs, including the putative progenitor cells that reside within the Canals of Hering.
We then tested whether labeled BECs give rise to hepatocytes under injury or homeostatic conditions. Six- to eight-week-old bigenic mice were given tamoxifen and, after a washout period of two weeks, were subjected to an injury-recovery protocol with various ADC-inducing injury models including 3,5-diethoxycarbonyl-1,4-dihydrocollidine (DDC); a choline-deficient ethionine-supplemented (CDE) diet; CCl4 administration; and alpha-naphthyl-isothiocyanate (ANIT) diet. Under all injury-recovery circumstances, all YFP+ cells costained for the biliary marker Krt19 but not the hepatocyte marker HNF4α (Figure 1D) (DDC: 1,191 cells counted, n = 3; CDE: 1,157 counted, n = 3; for ANIT and CCl4, data not shown). Thus, any contribution of YFP+ BECs to hepatocytes during regeneration from these injuries was below the limit of detection.
To determine whether BECs might generate hepatocytes over longer periods of time, as has been previously reported for Sox9+ BECs (Furuyama et al., 2011), we examined Krt19-CreER/R26YFP mice 9 months after tamoxifen labeling. Following this long “chase” period, all YFP+ cells continued to stain for biliary markers but not hepatocyte markers (Figure 1E; 1,154 YFP+ cells examined for biliary markers, 4,773 YFP+ cells examined for hepatocyte markers). Likewise, no YFP+ cells exhibited morphological features of hepatocytes. Hence, KRT19-expressing BECs do not appear to give rise to hepatocytes following injury or during normal liver turnover.
ADCs Do Not Exhibit Progenitor Cell Activity In Vivo
It is possible that KRT19+ADCs arise from a unique cell population that is not labeled in the quiescent state. To this end, we administered tamoxifen to Krt19-CreER/R26YFP mice during the second half of treatment with ADC-inducing injuries including DDC and CDE, thus labeling newly formed ADCs (Figure 2A). Injury alone (in the absence of tamoxifen) did not induce recombination of the reporter allele (Figure 2B). Tamoxifen administration resulted in YFP labeling of KRT19+ cells during DDC and CDE injuries (pulse; n = 3; Figures 2C and 3D, top panels), including A6+ ADCs (Figure S2A). The labeling encompassed cells within large Krt19+ mature-appearing ductal structures, as well as isolated ductal cells that penetrated the lobule, including Sox9+ cells (pulse; Figures 2C and 2D, middle panels). Labeling during the injury was highly specific for ADCs, as all labeled cells (DDC: 1,010 counted, n = 6; CDE: 3,904 counted, n = 2) exhibited a biliary morphology and stained with ADC/biliary markers but lacked a hepatocyte morphology and were negative for HNF4α (pulse; Figures 2C and 2D, bottom panels).
Figure 2. ADCs Do Not Give Rise to Hepatocytes.
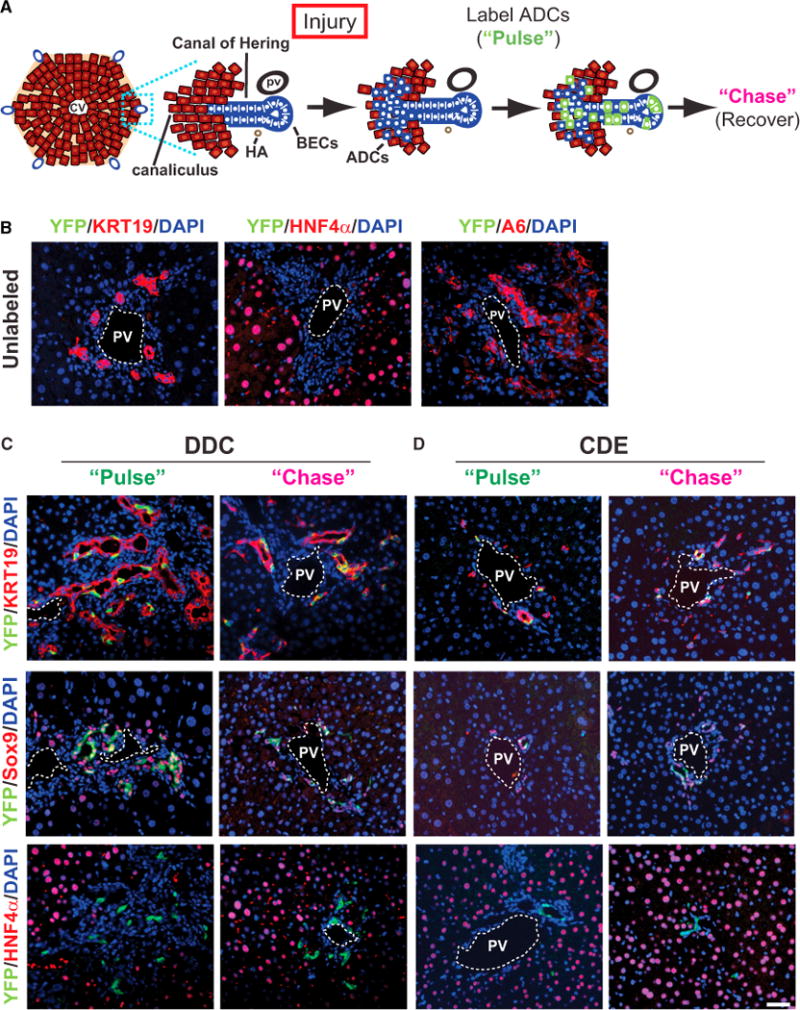
(A) Schematic view of ADC labeling using Krt19-CreER;R26YFP mice. Treatment of mice with toxin (Injury) leads to the emergence of ADCs in the lobule (blue cells mingled with hepatocytes). Lineage labeling (“Pulse”) results in the heritable marking of ADCs (green) but not hepatocytes; labeled progeny can be followed after recovery (“Chase”).
(B) DDC-treated Krt19-CreER;R26YFP mice do not exhibit YFP expression in the absence of tamoxifen.
(C) The Krt19-CreER transgene permits specific labeling of Krt19+ and Sox9+ ADCs after DDC treatment. No label-bearing hepatocytes were observed during the pulse (“Pulse”) or following recovery (“Chase”).
(D) The Krt19-CreER transgene permits specific labeling of KRT19+ and Sox9+ ADCs after CDE treatment. No label-bearing hepatocytes were observed during the pulse (“Pulse”) or following recovery (“Chase”). Scale bar, 50 μm.
See also Figure S2.
Figure 3. A Pulse-Chase System for Determining the Origin of Regenerating Hepatocytes.
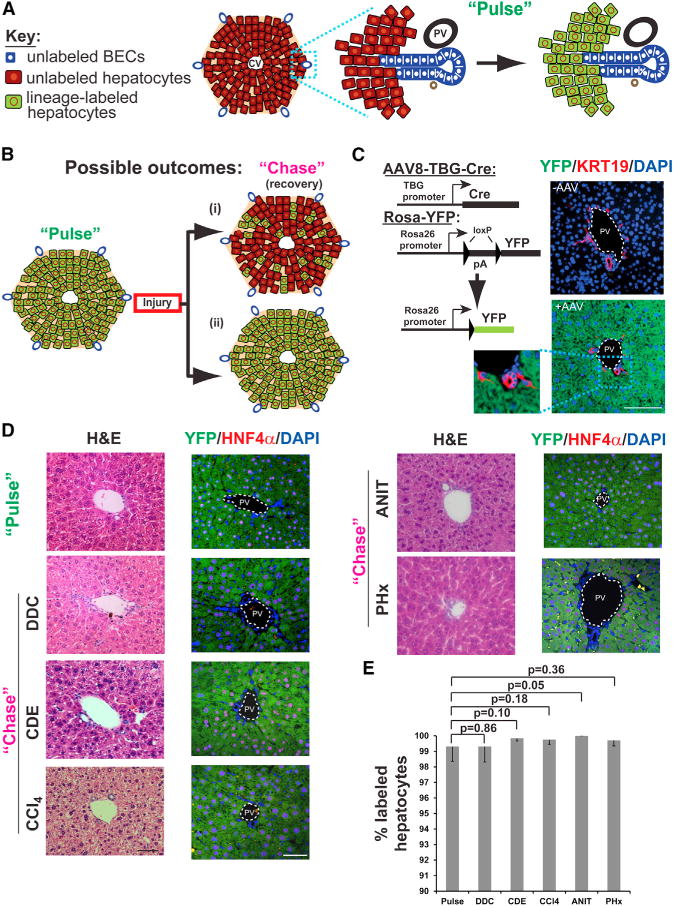
(A) Schematic view of hepatocyte labeling using AAV8-based lineage tracing. Labeling via viral infection (“Pulse”) results in the heritable marking of hepatocytes (green), but not BECs.
(B) Predictions from different models of liver regeneration. After injury and recovery (“Chase”), stem-cell-based repair would result in a decrease in the hepatocyte labeling index (i), while hepatocyte-mediated recovery would result in no change in labeling index (ii).
(C) Hepatocyte labeling was achieved by administering AAV8-TBG-Cre to R26YFP mice; immunofluorescent images show specific and efficient labeling of hepatocytes. Labeling of BECs was never observed following AAV (lower panel, n = 6).
(D) Liver histology returns to normal following injuries with DDC, CDE, CCl4, ANIT, and partial hepatectomy (PHx) (H&E, hematoxylin and eosin, left panels), with no appreciable change in the frequency of hepatocyte labeling. Scale bar, 50 μm.
(E) Quantification of hepatocyte labeling following AAV injection (“Pulse”) and recovery from DDC, CDE, CCl4, ANIT, and PHx injuries. Labeling index was quantified from six, six, four, four, two, and two mice for each of the conditions, respectively (means ± SD). Absolute numbers of cells counted for pulse and chase are provided in Results.
Following 2–3 weeks of recovery (Chase), YFP+ cells were readily detected in the livers of Krt19-CreER/R26YFP mice. YFP+ cells in the recovery group resembled normal BECs, exhibiting a biliary morphology and staining with biliary markers (chase; Figures 2C and 2D, top and middle panels). Consistent with our results from BEC labeling prior to injury, YFP expression was never observed in cells with a hepatocyte morphology or HNF4a expression (chase; Figures 2C and 2D, bottom panels; DDC: 2,474 counted, n = 5; CDE: 4,252 counted, n = 4). As some stem/progenitor cell populations undergo replication rates that differ from those of surrounding cells (Blanpain et al., 2007), we ensured there was equal YFP labeling of proliferating and nonproliferating ADCs using Ki-67 (Figure S2B). These results indicate that ADCs labeled under multiple-injury conditions do not give rise to hepatocytes. Taken together, the results reveal that neither ADCs nor BECs (their presumptive cells of origin) give rise to hepatocytes under homeostatic conditions or after toxin-mediated injury.
Hepatocytes Are Derived from Preexisting Hepatocytes In Vivo
These experiments do not rule out the possibility that hepatocytes arise from progenitor cells that were not marked by the Krt19-CreER labeling approach (including “marker-negative” cells). To test this possibility, we labeled differentiated hepatocytes and determined the contribution of “nonhepatocytes” to recovery following injury (Figure 3A). The rationale for the approach follows from the expectation that unlabeled stem cells contributing to liver regeneration would give rise to unlabeled progeny, resulting in decreased hepatocyte-labeling index (Figure 3Bi). Alternatively, if new hepatocytes are derived solely from existing hepatocytes, then the hepatocyte-labeling index would remain unchanged (Figure 3Bii). This method has been used as a general means of determining the degree to which putative stem/progenitor cells contribute to tissue regeneration when markers of such cells are lacking (Dor et al., 2004).
To label hepatocytes—defined here as postnatal cells expressing HNF4a but not expressing BEC markers—we utilized a replication-incompetent, recombinant adeno-associated virus serotype 2/8 expressing Cre recombinase driven by the hepatocyte-specific promoter (thyroid hormone-binding globulin, AAV8-TBG-Cre). One of the reasons for using this method is that the tamoxifen used to induce recombination in Cre-based inducible models has been shown to have a “toxin-like” effect, leading to ectopic activation of BEC genes in hepatocytes such as Sox9 (Carpentier et al., 2011). We thus wanted to circumvent this potential mislabeling by using the AAV8-TBG-Cre instead to label hepatocytes. This transduction was highly specific, as all YFP+ cells were HNF4α+ as previously shown (Wang et al., 2010; Yanger et al., 2013). Moreover, labeling was efficient, as more than 99% of hepatocytes were genetically marked when R26YFP mice were infected (Figures 3C–3E), with no labeling of nonhepatocytes (Yanger et al., 2013). We have previously shown that the AAV8 serotype provides a hepatocyte-specific tropism within the liver, as only hepatocytes are labeled by infection with viruses carrying Cre recombinase under the control of a ubiquitously expressed cytomegalovirus promoter (Yanger et al., 2013). Thus, AAV8-TBG-Cre permits labeling in the liver that is highly efficient and specific for hepatocytes.
We then subjected lineage-labeled AAV8-TBG-Cre; R26YFP mice (pulse) to the injury-recovery protocols described earlier (chase). Under these conditions, the percentage of labeled hepatocytes remained unchanged (Figures 3D and 3E). Specifically, the labeling index for the pulse group (99.36% ± 0.96%, 11,359 counted) did not decrease following recovery after DDC (99.31% ± 1.00%, 5,830 counted), CDE (99.83% ± 0.17%, 1,838 counted), CCl4 (99.75% ± 0.38%, 1,677 counted), or ANIT (100%, 1,944 counted). As a control, we performed 2/3 partial hepatectomy, which also showed no change in the YFP labeling index (99.71% ± 0.36%, 695 counted). Thus, by this sensitive labeling technique, we failed to find evidence that hepatocytes arise from nonhepatocytes after recovery from multiple types of ADC-inducing injuries.
Previous studies have shown that the AAV8 viral genome is cleared from the body within days of infection (Zincarelli et al., 2008). However, we sought to further confirm that new, unlabeled hepatocytes were not transduced and labeled by potential AAV8 persistence in the system. To this end, we performed a serum transfer experiment in which we assessed the infective activity of the virus by isolating serum from mice given AAV8 2 or 30 weeks previously and injecting it into Cre-naive R26YFP mice. Livers of these serum-recipient mice were assessed 1 or 6 weeks later, and no YFP signal was detected (Figure 4; data not shown). These results indicate that the durability of high (>99%) levels of hepatocyte labeling after the injury-recovery period is not due to labeling of progenitor-derived hepatocytes that are infected by latent AAV8 viral particles.
Figure 4. New Hepatocytes Are Not Labeled on AAV Serum Transfusion.
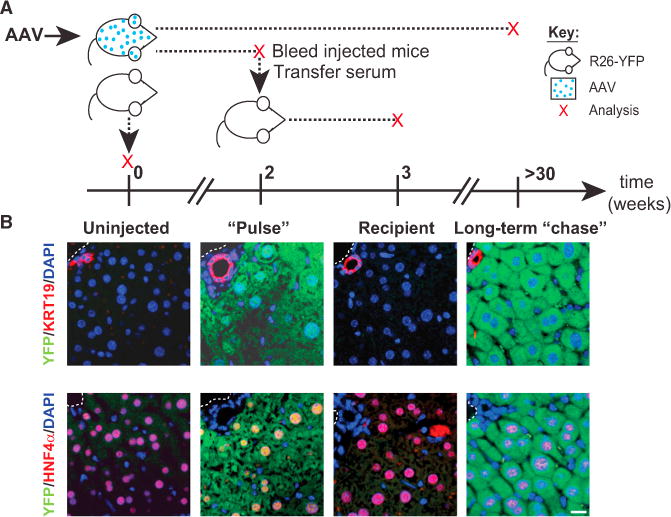
(A) Schematic showing the timing of infection, serum transfer, and analysis of injected R26YFP mice; each red “x” indicates a sampling point. AAV8-TBG-Cre-injected mice were analyzed either 2 weeks (“Pulse”) or >30 weeks after injection (Long-term “chase”). Prior to analysis at 2 weeks, mice were bled, and serum was injected into naive, uninfected R26YFP mice whose livers were assessed after 1 week (Recipient). Uninfected mice served as a control (Uninjected).
(B) Images showing immunoflurescence staining for YFP (green) and Krt19 or HNF4α (red) in livers from each of the four groups analyzed.
Rapidly Proliferating Cells in the Liver Do Not Give Rise to Hepatocytes
Finally, we complemented the two genetic lineage tracing methods with an unbiased labeling approach that marks rapidly cycling cells during injury and identifies their progeny. Stem cells contribute to tissue homeostasis and regeneration by generating rapidly dividing progeny—commonly referred to as transit-amplifying (TA) cells—which expand prior to final differentiation (Blanpain et al., 2007). ADCs with their high proliferative index are characterized as liver TA cells (Alison et al., 2004). Teta et al. have previously reported that TA cells can be labeled by incorporation of two thymidine analogs (i.e., dual labeling) when administered in succession (Teta et al., 2007).
We first showed that we could track double-labeled progenitor cells using the thymidine analogs iododeoxyuridine (IdU) and chlorodeoxyuridine (CldU) in the intestine (Figures S3A and S3B) and verified long-term stability of the label in the liver (Figure S3C). We next applied this labeling method to livers subjected to injury induced by CDE diet and DDC treatment, as these injuries have been reported to promote “liver progenitor cell” activity (Bird et al., 2008; Wang et al., 2003). Administration of IdU and CldU resulted in the labeling of both hepatocytes and ADCs in CDE-treated mice, with a significant number of ADCs exhibiting dual labeling (Figure 5A). We treated this dual labeling as a pulse condition for lineage tracing, reasoning that if new hepatocytes are derived from rapidly cycling, then the frequency of double-labeled hepatocytes should be higher in the chase samples than in the pulse samples (Figure 5Bi). By contrast, if new hepatocytes are derived by self-duplication, the frequency of double-labeled hepatocytes should not increase (Figure 5Bii).
Figure 5. TA Cells Do Not Give Rise to Hepatocytes.
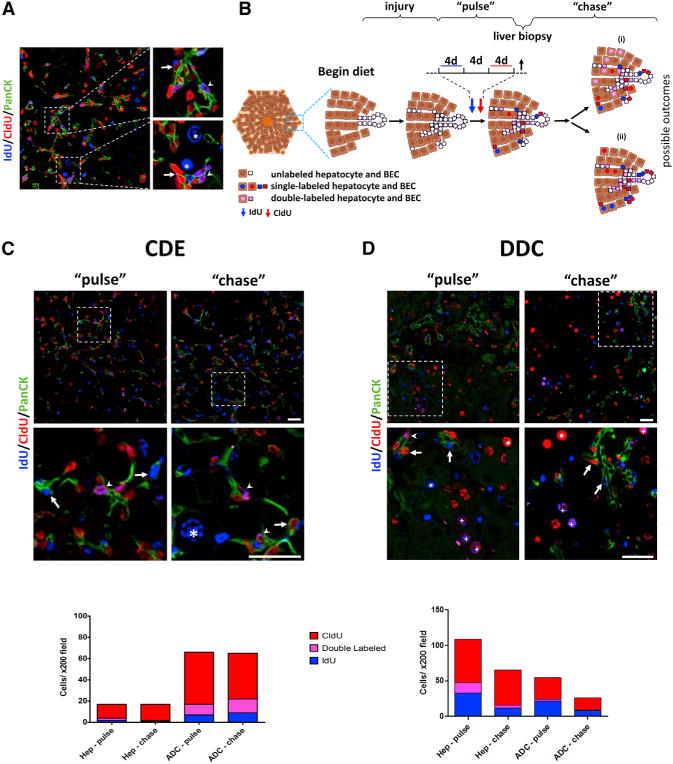
(A) Representative image of dual-labeled cells, obtained by liver biopsy following the “pulse.”
(B) Schematic view of the double-label pulse-chase experiment and the possible outcomes (see Results). d, days.
(C) Top panels: representative immunofluorescent images of labeled cells in livers from “pulse” (left) and “chase” (right) mice following treatment with a CDE diet. Middle panels: high magnification view of the areas indicated in the top panels. Bottom panel: quantification of single- and dual-labeled hepatocytes (Hep) and ADCs following pulse or chase (≥6 200× fields counted per mouse, n = 6).
(D) Top panels: representative immunofluorescent images of labeled cells in livers from “pulse” (left) and “chase” (right) mice following treatment with DDC. Middle panels: high-magnification view of the areas indicated in the top panels. Bottom panel: quantification of single- and double-labeled hepatocytes (Hep) and ADCs following pulse or chase (≥6 200× fields counted per mouse, n = 7). Arrows denote single-labeled ADCs; arrowheads denote double-labeled ADCs; the asterisk indicates a single-labeled hepatocyte; and the plus sign indicates a double labeled hepatocyte. PanCK, pancytokeratin; Scale bars, 50 μm. See also Figure S3.
For this protocol, mice received the CDE diet followed by IdU and CldU administration (Figure 5B), with adequate washout between the two thymidine analogs to avoid overlap (Figure S3D). Postlabeling, a liver biopsy (~3% of liver mass) was obtained in order to quantify labeling indices for each animal (pulse). This minimal hepatectomy did not result in any measurable compensatory proliferation and was representative of the rest of the liver (Figure S3E). After a 4-week chase period, mice were sacrificed, and the number of single- and double-labeled hepatocytes and BECs in the pulse and chase groups were compared. As the response to CDE is heterogeneous, we included in the analysis only mice that exhibited negligible dual labeling of hepatocytes in the pulse biopsy (<4% of hepatocytes, n = 6).
Examination of the pulse biopsy revealed that 7.0% ± 6.5% of cycling ADCs were double labeled during CDE treatment, compared to 1.3% ± 1.4% of cycling hepatocytes (Figure 5C). We also detected double-labeled cells that were neither hepatocytes nor BECs, indicating that rapidly cycling cells exist in other liver populations (e.g., inflammatory or endothelial cells). At the chase, we observed a similar percentage of double-labeled ADCs (11% ± 8%), with no increase in the percentage of double-labeled hepatocytes (0.8% ± 0.9%; Figure 5C). Further experiments, in which a 4-week injury-free recovery period was used, yielded the same conclusion (Figure S3F).
To ensure that our failure to observe a contribution of rapidly proliferating cells to hepatocytes was not specific to the CDE injury model, we performed a similar experiment in DDC-treated mice. Dual nucleoside analog administration in the setting of DDC injury resulted in double labeling of 4.2% ± 1.7% of cycling ADCs and 13.4% ± 1.1% (n = 7) of cycling hepatocytes (Figure 5D). After a chase period of 4 weeks postinjury, the frequency of double-labeled hepatocytes had decreased (6.0% ± 1.6%, p < 0.001, n = 7; Figure 5D), and there was no change in the frequency of double-labeled ADCs (3.3% ± 2.4%; Figure 5D). Thus, these analyses suggest that there is no significant contribution of rapidly proliferating cells—either ADCs or other types of rapidly dividing cells—to the hepatocyte pool following CDE- or DDC-mediated liver damage.
DISCUSSION
In this study, we have used three distinct lineage tracing approaches to test the hypothesis that regeneration from toxin-induced liver damage is mediated by facultative stem cells. First, we genetically labeled KRT19-expressing BECs and ADCs— the tissue compartment in which liver stem cells are believed to reside—and found no evidence that these cells give rise to hepatocytes in the four injury models tested. Second, we labeled hepatocytes with high efficiency and specificity and found that there was no decrease in the labeling index with the same types of injury, a result that is consistent with the notion that new hepatocytes are derived from preexisting hepatocytes. Finally, we used nucleoside analog labeling to trace the fate of highly proliferative liver cells, finding no evidence that rapidly dividing nonhepatocytes differentiated into hepatocytes. Taken together, these data suggest that ADCs and other nonhepatocyte populations do not contribute significantly to hepatocyte neogenesis during liver regeneration.
The long-standing notion that ADCs are facultative stem cells has been based largely on static observations, in vitro studies, and cellular transplantation assays (Huch et al., 2013; Shin et al., 2011; Wang et al., 2003). However, these assays test cell potential rather than cell fate. Previous work from our lab and others have demonstrated significant plasticity between hepatocytes and BECs, both in vitro and in vivo (Limaye et al., 2008b; Michalopoulos et al., 2005; Nishikawa et al., 2005; Tanimizu et al., 2014; Yanger et al., 2013). Thus, while cell transplantation and in vitro culture can provide insight into cell potential under these experimental conditions, lineage tracing permits a more accurate indication of cell fate.
Several groups have used lineage tracing to characterize the origin and fate of ADCs. Initial work using [3H]-thymidine incorporation gave rise to discordant conclusions, with experimental evidence both favoring (Evarts et al., 1987, 1989) and refuting (Grisham and Porta, 1964; Tatematsu et al., 1984) an ADC-to-hepatocyte differentiation lineage relationship. In addition, more recent lineage tracing studies have also led to disparate results (Friedman and Kaestner, 2011). Specifically, Cre-based labeling using Osteopontin- or Lgr5-inducible drivers resulted in the inheritance of label in hepatocytes, although in both cases, the contribution to the hepatocyte pool was very low (Español-Suñer et al., 2012; Huch et al., 2013). More dramatic evidence for liver progenitors was provided by Furuyama et al. (2011), who reported that a significant percentage (up to 90%) of the liver was eventually marked following pulse labeling with a Sox9-CreER strain, a result that, on face value, is strong evidence for physiologically active stem cells. By contrast, other studies utilizing a similar but separately constructed Sox9-CreER strain did not find evidence for such liver progenitors (Carpentier et al., 2011; Tarlow et al., 2014). Additionally, in using a similar hepatocyte labeling method with adeno-associated virus, no decrease in labeling was observed following most forms of injury, and a scant 1.3% reduction in labeling was observed following CCl4 injury (an effect whose statistical significance from baseline was not reported), indicating that stem cells play a minor role, if any, in liver homeostasis and regeneration (Malato et al., 2011).
The ability to reach strong conclusions about lineage is deeply dependent on the specificity of the tracing tools used. For example, the OPN-, Lgr5-, and Sox9-based lineage tracing studies all rely on the assumption that Cre-mediated recombination never occurs in hepatocytes. If such a lack of specificity were to exist, even to a small extent, one would be left with the impression that hepatocytes are derived from BECs or ADCs, when in fact, no such progenitor-progeny relationship existed. We speculate that this technical point accounts for the discrepancy between our results and those of others who have reported liver progenitor cell activity in vivo. Indeed, when we tested the specificity of the same Sox9-CreER strain used by Furuyama and colleagues (Soeda et al., 2010) with a R26YFP reporter, we found that this strain confers substantial hepatocyte labeling (Figure S4). This result is consistent with the observation that Sox9 is induced in hepatocytes solely by tamoxifen administration (Carpentier et al., 2011; Tarlow et al., 2014) and under pathological conditions (Yanger et al., 2013). Hence, “ectopic” Sox9 expression in hepatocytes, resulting in their labeling, may have led to the erroneous conclusion that they were derived from biliary cells. A similar phenomenon may have contributed to the low level of hepatocytes marked with OPN-CreER (Español-Suñer et al., 2012) and Lgr5-CreER (Huch et al., 2013), as hepatocytes have a propensity for expressing certain biliary markers, including osteopontin, on stress (Coombes and Syn, 2013; Limaye et al., 2008a; Yanger et al., 2013). As a “terminal” biliary marker, Krt19 appears to be an exception to this specificity problem; in contrast to Sox9 and OPN, Krt19 is not expressed by hepatocytes on injury, and hepatocytes become Krt19+ only after prolonged toxin exposure as part of a hepatocyte-to-BEC reprogramming process (Yanger et al., 2013). Indeed, we have observed that OPN is normally expressed in a large subset of hepatocytes in injured conditions (data not shown). Hence, Krt19-CreER mice are likely to represent a more specific—and, hence, more reliable—tool for assessing the contribution of BECs and ADCs to liver repopulation.
Our results do not eliminate the possibility that, under more demanding circumstances, ADCs could exhibit bipotency. For instance, in lower vertebrates such as zebrafish, >95% hepatocyte ablation in young fish results in a BEC contribution to hepatocyte differentiation, a phenomenon that also occurs to a lesser extent in adult fish (Choi et al., 2014; He et al., 2014). However, in FAH−/−-deficient mice that exhibit gross hepatocyte death, hepatocyte transplantation rescues and repopulates the liver while the ADC-containing population does not (Tarlow et al., 2014). Similarly, we cannot rule out the possibility that the Krt19-CreER strain, which labeled 30%–40% of biliary cells, failed to mark a subset of ADCs with progenitor activity. However, a comparison of the biliary cells labeled by this strain to those cells that remained unmarked failed to reveal any differences between the two populations (with respect to markers or proliferation). In addition, the two independent lineage tracing approaches we used—hepatocyte labeling and dual-nucleoside labeling— further bolsters the interpretation that biliary cells do not contribute to hepatocyte neogenesis, at least in the injury models we examined.
Likewise, it remains possible that hepatocytes differ with respect to replicative ability or that a specialized subset of hepatocytes remains multipotent. Hepatocyte heterogeneity with respect to cell division has been recently reported (Miyaoka et al., 2012) although whether this reflects an intrinsic property of a subset of hepatocytes or simply stochastic differences has yet to be determined. In either case, our data support the view that new adult hepatocytes come from preexisting hepatocytes, not only following partial hepatectomy but also in the setting of toxin injuries as well. As hepatocytes are able to divide more than 100 times without losing function (Overturf et al., 1997) and can also differentiate into biliary cells on injury (Yanger et al., 2013; Tanimizu et al., 2014; Sekiya and Suzuki, 2014), hepatocytes rather than biliary cells would appear to constitute the facultative stem cell compartment of the liver.
EXPERIMENTAL PROCEDURES
Mice and Injury
Mice were maintained in a pathogen-free environment. Krt19-CreER and RosaYFP strains have been described elsewhere (Means et al., 2008; Srinivas et al., 2001). To generate an ADC response, 6- to 8-week-old mice were given 0.1% wt/wt DDC (Sigma-Aldrich) in PMI Mouse Diet #5015 (Harlan Teklad) for 2–3 weeks, at which point the diet was changed to regular chow for 3–5 weeks to allow mice to recover. A choline-deficient diet (MP Biomedicals) supplemented with 0.15% ethionine drinking water (Sigma-Aldrich, E5139) was administered for 2 weeks as described elsewhere (Carpentier et al., 2011), followed by 2 weeks of recovery. Mice were fed 0.25 g/kg of chow supplemented with ANIT (Dyets) as described elsewhere (Faa et al., 1998) for 2 weeks, followed by the same length of recovery. CCl4 administration and recovery was conducted as described elsewhere using the chronic injury protocol of Malato et al. (2011). Partial hepatectomy was performed as described elsewhere (Greenbaum et al., 1995), followed by a 2-week recovery period. As expected, we observed the emergence of numerous ADCs within 2 weeks of DDC, ANIT, and CCl4 treatment and within 4 days of CDE treatment.
Viral Infections and Tamoxifen Administration
Pulse labeling with AAV8-TBG-Cre virus was performed as described elsewhere in 6- to 8-week-old mice, followed by a 2-week washout period (Yanger et al., 2013). KRT19-CreER transgenes were achieved by giving mice 40 mg of tamoxifen over five doses. For labeling ADCs, KRT19-CreER; R26YFP mice were given three to five doses of tamoxifen (8 mg per dose) during the second half of DDC and CDE treatments. All studies were conducted in accordance with the policies of the National Institutes of Health and the University of Pennsylvania Institutional Animal Care and Use Committee guidelines.
Nucleoside Analog-Based Pulse-Chase Double Labeling
Six-week-old male mice (C57BL/6J, Harlan) were fed a CDE diet for two weeks or a DDC diet for three weeks. Following the aforementioned period, 1 mg/ml IdU (Sigma-Aldrich) was added to drinking water for 4 days. Following a washout period of 4 days, 1 mg/ml CldU (MP Biomedicals) was added to drinking water for 4 days. After another 2 days, mice were anesthetized, and one omental lobe was removed via laparotomy (Greene and Puder, 2003) for further analysis. For examining pulse-chase labeling after a “recovery period” (Figure S3F), mice were fed normal chow for another 4 weeks before sacrifice. For labeling experiments involving DDC treatment, DDC administration was performed as described earlier. Liver and small intestine tissue sections were triple stained with anti-bromodeoxyuridine antibodies as described elsewhere with minor modifications (Teta et al., 2007) and with anti-Pancytokeratin (DAKO). Every staining included mice that received only one of the thymidine analogs. Images were obtained with an inverted confocal microscope (Olympus). Cells were counted manually in a blinded fashion using ImageJ software (≥6 × 200 fields).
Immunostaining and Quantification
Antibody staining was performed as described elsewhere (Zong et al., 2009). Primary antibody sources and concentrations are listed in Table S1. The percentage of marker-positive cells was determined by taking representative images and directly counting cell number. For all quantitations, cell enumerations for each experiment are listed in the text or figure legends. Student’s t tests were used to calculate p values. Error bars show SD.
Supplementary Material
Acknowledgments
We thank V. Factor and D. Melton for providing A6 and Krt19 antibodies, respectively. We are grateful to A. Panikkar and C. Yang for technical assistance and the Abramson Family Center Research Institute histology core for sample processing. We are indebted to F. Camargo, K. Kaestner, K. Zaret, J. Rajagopal, and L. Greenbaum for helpful discussions and especially grateful to A. Penzo-Mendez for insightful comments on the manuscript. This work was supported in part by the National Institute of Diabetes and Digestive and Kidney Diseases Center for Molecular Studies in Digestive and Liver Diseases (P30DK050306) and its core facilities (Molecular Pathology and Imaging Core, Molecular Biology/Gene Expression Core, Transgenic and Chimeric Mouse Core, and Cell Culture Core), as well as by Public Health Service grant DK083355 (to B.Z.S.), a grant from the European Research Council within the FP-7 (281738 LIVERMICROENV to E.P.), and an award from the Pew Charitable Trusts (to B.Z.S.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes four figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.stem.2014.06.003.
AUTHOR CONTRIBUTIONS
K.Y., D.K., E.P., and B.S. designed the study, analyzed the data, and wrote the manuscript. K.Y. and D.K. conducted the majority of the experiments. Y.Z. and L.M. conducted additional experiments, and L.M. collected data. G.G. and H.A. provided reagents. E.P. and B.S. supervised the entire study.
References
- Alison MR, Vig P, Russo F, Bigger BW, Amofah E, Themis M, Forbes S. Hepatic stem cells: from inside and outside the liver? Cell Prolif. 2004;37:1–21. doi: 10.1111/j.1365-2184.2004.00297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird TG, Lorenzini S, Forbes SJ. Activation of stem cells in hepatic diseases. Cell Tissue Res. 2008;331:283–300. doi: 10.1007/s00441-007-0542-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C, Horsley V, Fuchs E. Epithelial stem cells: turning over new leaves. Cell. 2007;128:445–458. doi: 10.1016/j.cell.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpentier R, Suñer RE, van Hul N, Kopp JL, Beaudry JB, Cordi S, Antoniou A, Raynaud P, Lepreux S, Jacquemin P, et al. Embryonic ductal plate cells give rise to cholangiocytes, periportal hepatocytes, and adult liver progenitor cells. Gastroenterology. 2011;141:1432–1438. doi: 10.1053/j.gastro.2011.06.049. e1–e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi TY, Ninov N, Stainier DYR, Shin D. Extensive conversion of hepatic biliary epithelial cells to hepatocytes after near total loss of hepatocytes in zebrafish. Gastroenterology. 2014;146:776–788. doi: 10.1053/j.gastro.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes J, Syn WK. Utility of osteopontin in lineage tracing experiments. Gastroenterology. 2013;145:254–255. doi: 10.1053/j.gastro.2013.02.051. [DOI] [PubMed] [Google Scholar]
- Crawford AR, Lin XZ, Crawford JM. The normal adult human liver biopsy: a quantitative reference standard. Hepatology. 1998;28:323–331. doi: 10.1002/hep.510280206. [DOI] [PubMed] [Google Scholar]
- Desmet VJ. Intrahepatic bile ducts under the lens. J Hepatol. 1985;1:545–559. doi: 10.1016/s0168-8278(85)80752-2. [DOI] [PubMed] [Google Scholar]
- Dor Y, Brown J, Martinez OI, Melton DA. Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature. 2004;429:41–46. doi: 10.1038/nature02520. [DOI] [PubMed] [Google Scholar]
- Español-Suñer R, Carpentier R, Van Hul N, Legry V, Achouri Y, Cordi S, Jacquemin P, Lemaigre F, Leclercq IA. Liver progenitor cells yield functional hepatocytes in response to chronic liver injury in mice. Gastroenterology. 2012;143:1564–1575. doi: 10.1053/j.gastro.2012.08.024. e7. [DOI] [PubMed] [Google Scholar]
- Evarts RP, Nagy P, Marsden E, Thorgeirsson SS. A precursor-product relationship exists between oval cells and hepatocytes in rat liver. Carcinogenesis. 1987;8:1737–1740. doi: 10.1093/carcin/8.11.1737. [DOI] [PubMed] [Google Scholar]
- Evarts RP, Nagy P, Nakatsukasa H, Marsden E, Thorgeirsson SS. In vivo differentiation of rat liver oval cells into hepatocytes. Cancer Res. 1989;49:1541–1547. [PubMed] [Google Scholar]
- Faa G, Van Eyken P, Roskams T, Miyazaki H, Serreli S, Ambu R, Desmet VJ. Expression of cytokeratin 20 in developing rat liver and in experimental models of ductular and oval cell proliferation. J Hepatol. 1998;29:628–633. doi: 10.1016/s0168-8278(98)80158-x. [DOI] [PubMed] [Google Scholar]
- Factor VM, Radaeva SA, Thorgeirsson SS. Origin and fate of oval cells in dipin-induced hepatocarcinogenesis in the mouse. Am J Pathol. 1994;145:409–422. [PMC free article] [PubMed] [Google Scholar]
- Farber E. Similarities in the sequence of early histological changes induced in the liver of the rat by ethionine, 2-acetylamino-fluorene, and 3′-methyl-4-dimethylaminoazobenzene. Cancer Res. 1956;16:142–148. [PubMed] [Google Scholar]
- Fausto N, Campbell JS. The role of hepatocytes and oval cells in liver regeneration and repopulation. Mech Dev. 2003;120:117–130. doi: 10.1016/s0925-4773(02)00338-6. [DOI] [PubMed] [Google Scholar]
- Friedman JR, Kaestner KH. On the origin of the liver. J Clin Invest. 2011;121:4630–4633. doi: 10.1172/JCI59652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama K, Kawaguchi Y, Akiyama H, Horiguchi M, Kodama S, Kuhara T, Hosokawa S, Elbahrawy A, Soeda T, Koizumi M, et al. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat Genet. 2011;43:34–41. doi: 10.1038/ng.722. [DOI] [PubMed] [Google Scholar]
- Greenbaum LE, Cressman DE, Haber BA, Taub R. Coexistence of C/EBP alpha, beta, growth-induced proteins and DNA synthesis in hepatocytes during liver regeneration. Implications for maintenance of the differentiated state during liver growth. J Clin Invest. 1995;96:1351–1365. doi: 10.1172/JCI118170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene AK, Puder M. Partial hepatectomy in the mouse: technique and perioperative management. J Invest Surg. 2003;16:99–102. [PubMed] [Google Scholar]
- Grisham JW, Porta EA. Origin and Fate of Proliferated Hepatic Ductal Cells in the Rat: Electron Microscopic and Autoradiographic Studies. Exp Mol Pathol. 1964;86:242–261. doi: 10.1016/0014-4800(64)90057-7. [DOI] [PubMed] [Google Scholar]
- He J, Lu H, Zou Q, Luo L. Regeneration of liver after extreme hepatocyte loss occurs mainly via biliary transdifferentiation in zebrafish. Gastroenterology. 2014;146:789, e8. doi: 10.1053/j.gastro.2013.11.045. [DOI] [PubMed] [Google Scholar]
- Huch M, Dorrell C, Boj SF, van Es JH, Li VSW, van de Wetering M, Sato T, Hamer K, Sasaki N, Finegold MJ, et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494:247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limaye PB, Alarcón G, Walls AL, Nalesnik MA, Michalopoulos GK, Demetris AJ, Ochoa ER. Expression of specific hepatocyte and cholangiocyte transcription factors in human liver disease and embryonic development. Lab Invest. 2008a;88:865–872. doi: 10.1038/labinvest.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limaye PB, Bowen WC, Orr AV, Luo J, Tseng GC, Michalopoulos GK. Mechanisms of hepatocyte growth factor-mediated and epidermal growth factor-mediated signaling in transdifferentiation of rat hepatocytes to biliary epithelium. Hepatology. 2008b;47:1702–1713. doi: 10.1002/hep.22221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malato Y, Naqvi S, Schürmann N, Ng R, Wang B, Zape J, Kay MA, Grimm D, Willenbring H. Fate tracing of mature hepatocytes in mouse liver homeostasis and regeneration. J Clin Invest. 2011;121:4850–4860. doi: 10.1172/JCI59261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Means AL, Xu Y, Zhao A, Ray KC, Gu G. A CK19(CreERT) knockin mouse line allows for conditional DNA recombination in epithelial cells in multiple endodermal organs. Genesis. 2008;46:318–323. doi: 10.1002/dvg.20397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalopoulos GK, Barua L, Bowen WC. Transdifferentiation of rat hepatocytes into biliary cells after bile duct ligation and toxic biliary injury. Hepatology. 2005;41:535–544. doi: 10.1002/hep.20600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaoka Y, Ebato K, Kato H, Arakawa S, Shimizu S, Miyajima A. Hypertrophy and unconventional cell division of hepatocytes underlie liver regeneration. Curr Biol. 2012;22:1166–1175. doi: 10.1016/j.cub.2012.05.016. [DOI] [PubMed] [Google Scholar]
- Nishikawa Y, Doi Y, Watanabe H, Tokairin T, Omori Y, Su M, Yoshioka T, Enomoto K. Transdifferentiation of mature rat hepatocytes into bile duct-like cells in vitro. Am J Pathol. 2005;166:1077–1088. doi: 10.1016/S0002-9440(10)62328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe M, Tsukahara Y, Tanaka M, Suzuki K, Saito S, Kamiya Y, Tsujimura T, Nakamura K, Miyajima A. Potential hepatic stem cells reside in EpCAM+ cells of normal and injured mouse liver. Development. 2009;136:1951–1960. doi: 10.1242/dev.031369. [DOI] [PubMed] [Google Scholar]
- Overturf K, al-Dhalimy M, Ou CN, Finegold M, Grompe M. Serial transplantation reveals the stem-cell-like regenerative potential of adult mouse hepatocytes. Am J Pathol. 1997;151:1273–1280. [PMC free article] [PubMed] [Google Scholar]
- Popper H, Kent G, Stein R. Ductular cell reaction in the liver in hepatic injury. J Mt Sinai Hosp N Y. 1957;24:551–556. [PubMed] [Google Scholar]
- Preisegger KH, Factor VM, Fuchsbichler A, Stumptner C, Denk H, Thorgeirsson SS. Atypical ductular proliferation and its inhibition by transforming growth factor beta1 in the 3,5-diethoxycarbonyl-1,4-dihydrocollidine mouse model for chronic alcoholic liver disease. Lab Invest. 1999;79:103–109. [PubMed] [Google Scholar]
- Roskams TA, Theise ND, Balabaud C, Bhagat G, Bhathal PS, Bioulac-Sage P, Brunt EM, Crawford JM, Crosby HA, Desmet V, et al. Nomenclature of the finer branches of the biliary tree: canals, ductules, and ductular reactions in human livers. Hepatology. 2004;39:1739–1745. doi: 10.1002/hep.20130. [DOI] [PubMed] [Google Scholar]
- Saxena R, Theise N. Canals of Hering: recent insights and current knowledge. Semin Liver Dis. 2004;24:43–48. doi: 10.1055/s-2004-823100. [DOI] [PubMed] [Google Scholar]
- Scholten D, Osterreicher CH, Scholten A, Iwaisako K, Gu G, Brenner DA, Kisseleva T. Genetic labeling does not detect epithelial-to-mesenchymal transition of cholangiocytes in liver fibrosis in mice. Gastroenterology. 2010;139:987–998. doi: 10.1053/j.gastro.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya S, Suzuki A. Hepatocytes, rather than cholangiocytes, can be the major source of primitive ductules in the chronically injured mouse liver. Am J Pathol. 2014;184:1468–1478. doi: 10.1016/j.ajpath.2014.01.005. [DOI] [PubMed] [Google Scholar]
- Shin S, Walton G, Aoki R, Brondell K, Schug J, Fox A, Smirnova O, Dorrell C, Erker L, Chu AS, et al. Foxl1-Cre-marked adult hepatic progenitors have clonogenic and bilineage differentiation potential. Genes Dev. 2011;25:1185–1192. doi: 10.1101/gad.2027811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeda T, Deng JM, de Crombrugghe B, Behringer RR, Nakamura T, Akiyama H. Sox9-expressing precursors are the cellular origin of the cruciate ligament of the knee joint and the limb tendons. Genesis. 2010;48:635–644. doi: 10.1002/dvg.20667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimizu N, Nishikawa Y, Ichinohe N, Akiyama H, Mitaka T. Sry HMG box protein 9-positive (Sox9+) epithelial cell adhesion molecule-negative (EpCAM-) biphenotypic cells derived from hepatocytes are involved in mouse liver regeneration. J Biol Chem. 2014;289:7589–7598. doi: 10.1074/jbc.M113.517243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarlow BD, Finegold MJ, Grompe M. Hepatology. 2014. Clonal tracing of Sox9(+) liver progenitors in mouse oval cell injury. Published online February 22, 2014. http://dx.doi.org/10.1002/hep.27084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatematsu M, Ho RH, Kaku T, Ekem JK, Farber E. Studies on the proliferation and fate of oval cells in the liver of rats treated with 2-acetylaminofluorene and partial hepatectomy. Am J Pathol. 1984;114:418–430. [PMC free article] [PubMed] [Google Scholar]
- Teta M, Rankin MM, Long SY, Stein GM, Kushner JA. Growth and regeneration of adult beta cells does not involve specialized progenitors. Dev Cell. 2007;12:817–826. doi: 10.1016/j.devcel.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Theise ND, Saxena R, Portmann BC, Thung SN, Yee H, Chiriboga L, Kumar A, Crawford JM. The canals of Hering and hepatic stem cells in humans. Hepatology. 1999;30:1425–1433. doi: 10.1002/hep.510300614. [DOI] [PubMed] [Google Scholar]
- Wang X, Foster M, Al-Dhalimy M, Lagasse E, Finegold M, Grompe M. The origin and liver repopulating capacity of murine oval cells. Proc Natl Acad Sci USA. 2003;100(Suppl 1):11881–11888. doi: 10.1073/pnas.1734199100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang H, Bell P, McCarter RJ, He J, Calcedo R, Vandenberghe LH, Morizono H, Batshaw ML, Wilson JM. Systematic evaluation of AAV vectors for liver directed gene transfer in murine models. Mol Ther. 2010;18:118–125. doi: 10.1038/mt.2009.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki Y, Moore R, Negishi M. Nuclear receptor CAR (NR1I3) is essential for DDC-induced liver injury and oval cell proliferation in mouse liver. Lab Invest. 2011;91:1624–1633. doi: 10.1038/labinvest.2011.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanger K, Stanger BZ. Facultative stem cells in liver and pancreas: fact and fancy. Dev Dyn. 2011;240:521–529. doi: 10.1002/dvdy.22561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanger K, Zong Y, Maggs LR, Shapira SN, Maddipati R, Aiello NM, Thung SN, Wells RG, Greenbaum LE, Stanger BZ. Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes Dev. 2013;27:719–724. doi: 10.1101/gad.207803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Grompe M. Generation and regeneration of cells of the liver and pancreas. Science. 2008;322:1490–1494. doi: 10.1126/science.1161431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zincarelli C, Soltys S, Rengo G, Rabinowitz JE. Analysis of AAV serotypes 1–9 mediated gene expression and tropism in mice after systemic injection. Mol Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- Zong Y, Panikkar A, Xu J, Antoniou A, Raynaud P, Lemaigre F, Stanger BZ. Notch signaling controls liver development by regulating biliary differentiation. Development. 2009;136:1727–1739. doi: 10.1242/dev.029140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


