Abstract
Clinical progression of B cell chronic lymphocytic leukemia (B-CLL) reflects the clone’s Ag receptor (BCR) and involves stroma-dependent B-CLL growth within lymphoid tissue. Uniformly elevated expression of TLR-9, occasional MYD88 mutations, and BCR specificity for DNA or Ags physically linked to DNA together suggest that TLR-9 signaling is important in driving B-CLL growth in patients. Nevertheless, reports of apoptosis after B-CLL exposure to CpG oligodeoxynucleotide (ODN) raised questions about a central role for TLR-9. Because normal memory B cells proliferate vigorously to ODN+IL-15, a cytokine found in stromal cells of bone marrow, lymph nodes, and spleen, we examined whether this was true for B-CLL cells. Through a CFSE-based assay for quantitatively monitoring in vitro clonal proliferation/survival, we show that IL-15 precludes TLR-9–induced apoptosis and permits significant B-CLL clonal expansion regardless of the clone’s BCR mutation status. A robust response to ODN+IL-15 was positively linked to presence of chromosomal anomalies (trisomy-12 or ataxia telangiectasia mutated anomaly + del13q14) and negatively linked to a very high proportion of CD38+ cells within the blood-derived B-CLL population. Furthermore, a clone’s intrinsic potential for in vitro growth correlated directly with doubling time in blood, in the case of B-CLL with Ig H chain V region–unmutated BCR and <30% CD38+ cells in blood. Finally, in vitro high-proliferator status was statistically linked to diminished patient survival. These findings, together with immunohistochemical evidence of apoptotic cells and IL-15–producing cells proximal to B-CLL pseudofollicles in patient spleens, suggest that collaborative ODN and IL-15 signaling may promote in vivo B-CLL growth.
Introduction
B cell chronic lymphocytic leukemia (B-CLL) is the most prevalent adult leukemia in the United States, Europe, and Australia, and it targets mainly elderly adults (1). Its incidence will undoubtedly increase as the population aged >60 y grows in future decades. Although recent therapeutic advances have notably improved the outcome for many patients (2, 3), B-CLL remains incurable for the following reasons: 1) diverse sites for B-CLL compartmentalization in the body, 2) important ancillary effects of the stromal environment, 3) mutagenic mechanisms for generating variants able to escape therapy, and 4) a possible leukemic stem cell compartment that remains unaffected upon depletion of mature leukemic cells. Thus, continued insights are needed regarding how to control this disorder.
A major advance in understanding B-CLL biology was the definition of a B-CLL proliferative component (4, 5), despite blood manifestation as small, relatively quiescent cells. Proliferative foci, often termed pseudofollicles or proliferation centers, are typically found within secondary lymphoid tissue and the bone marrow (6, 7). Growth not only expands leukemic cell numbers, but additionally introduces genetic instability through diverse routes, including division-related upregulation of activation-induced cytosine deaminase (8, 9).
Pseudofollicle growth depends on features of the leukemic clone, as well as stimuli within the leukemic milieu (10, 11). Ag receptors (BCR) expressed by the leukemic clone appear to play a critical role as suggested by the strong linkage between leukemia in vivo growth and Ig H chain V region (IGHV) mutation status (4, 5), and by the recent effectiveness of Btk inhibitors in treating B-CLL (2). Although the mechanism(s) whereby BCR fosters B-CLL growth remain poorly understood (12–16), the generally modest in vitro B-CLL proliferation elicited by most BCR cross-linking agents (17) suggests that the BCR of many B-CLL are anergic (18) and that costimuli from other receptors are important.
However, the concept that BCR might function more in mediating the internalization of immunostimulatory CpG DNA, the ligand for intracellular TLR9, has several lines of support. The BCRs of many B-CLL, particularly the IGHV unmutated B-CLL (U-CLL) subgroup, bind to intracellular proteins expressed on apoptotic cell membranes (19–21) that coexpress DNA (19, 22). Furthermore, numerous B-CLL BCRs show specificity for DNA (23–25), and at least two stereotyped BCRs of the IGHV mutated B-CLL (M-CLL) subgroup, one specific for IgG (rheumatoid factor specificity) (26, 27) and the other specific for β-(1, 6)-glucan of yeasts and filamentous fungi (28), engage Ags physically linked to CpG DNA (29, 30). All of these could elicit intracellular TLR-9 signaling upon BCR internalization of Ag (31, 32). Providing additional support for a role of BCR and TLR-9 signaling in driving B-CLL pseudofollicle growth is the evidence that malignant cells from lymph nodes express more BCR- and TLR-pathway genes than those taken from blood (11). Furthermore, a number of B-CLL bear function-enhancing mutations in MYD88, a critical protein in the TLR-9 signaling pathway (33). B-CLL progression in TCL-transgenic mice is also accelerated when SIGIRR (TIR8), a known negative regulator of TLR-9 signaling, is deleted (34).
Interest in a possible B-CLL dependency on TLR-9 signals led several laboratories to investigate whether in vitro exposure to CpG oligodeoxynucleotide (ODN) triggers blood-derived B-CLL to proliferate (35–37). Unexpectedly, although ODN did trigger increased [3H]thymidine uptake in a subset of B-CLL (those characterized by stronger activation of Akt, ERK, JNK, p38 MAPK, and NF-κB) (35, 36), another subset succumbed to ODN-induced apoptosis (37, 38). This was not linked to TLR-9 expression because all B-CLL express relatively high TLR-9 (35, 39). Rather, as recently reviewed (40), B-CLL cell diversity in TLR-9 responsiveness correlates with IGHV mutation status, with M-CLL clones showing significant ODN-induced apoptosis.
The stromal environment plays key roles in promoting B-CLL growth (10, 11), and it is warranted to consider which costimuli might make TLR-9 signals uniformly stimulatory for all B-CLL. Signals from nearby activated CD4+ T cells might be important given earlier evidence that the B-CLL response to ODN is boosted with CD40L and IL-2 (36, 39, 41). Also important may be in vivo signals from lymphoid tissue stromal cells, follicular dendritic cells (FDCs), and endothelial cells, each of which has been reported to affect B-CLL survival/growth under other conditions (reviewed in Ref. 42).
IL-15, an inflammatory cytokine produced by each of the earlier mentioned nonlymphoid cells (43–46), is a plausible candidate for promoting TLR-9–triggered growth of B-CLL. Although the cytokine is best known for its major effects on the development/growth/survival of NK cells, CD8 T cells, and intraepithelial γ/δ T cells (47, 48), human memory B cells exhibit vigorous in vitro proliferation upon exposure to both IL-15 and CpG DNA (49). Evidence that B-CLL are more homologous to memory B cells than naive B cells in gene expression arrays (50) suggests that B-CLL might exhibit a similar response. This possibility is heightened by past findings that B-CLL cells express all three chains of the trimeric IL-15R: high-affinity IL-15–specific IL-15Rα, lower-affinity IL-2/15Rβ (CD122), and common γ-chain, γc (CD132) (51, 52); furthermore, IL-15 increases B-CLL survival and proliferation in response to either CD40L (52) or Staphylococcus aureus Cowan strain 1 cells (53). One particularly compelling reason for considering that IL-15 might foster B-CLL growth within patients is the recent finding that IL-15 is constitutively produced by stromal cells within bone marrow, spleen, and lymph nodes (43–45), which are sites for B-CLL growth in patients. Furthermore, like the incidence of B-CLL, the levels of stromal cell–expressed IL-15 increase with age in both mouse spleens (44) and human bone marrow (54).
Taken together, these findings pose the question of whether IL-15 might boost the response of B-CLL to TLR-9 signals and drive B-CLL clonal expansion. In this study, we investigated this question by monitoring the growth and survival of CFSE-labeled B-CLL cells with approaches that have yielded important insights concerning the clonal growth and activation-induced death of normal human B lymphocytes (55–57). We found that B-CLL populations almost without exception show heightened in vitro viability and clonal expansion upon exposure to both ODN and IL-15. Nonetheless, because diversity between individual B-CLL populations in the degree of survival and/or cycling was apparent, we explored whether these differences associate with IGHV U versus M status, with the expression of chromosomal anomalies commonly linked to B-CLL, and with patient clinical outcome. Finally, we undertook an immunohistological study to discern whether IL-15–producing cells are present within and proximal to B-CLL pseudofollicles in patient spleens.
Materials and Methods
Ethics statement
These studies were approved by the Institutional Review Board of the North Shore–Long Island Jewish Health System (08-202A). Written, informed consent was obtained from CLL-bearing patients and normal subjects in accordance with the Declaration of Helsinki before blood collection.
CLL patient samples and characterization
B-CLL specimens for in vitro study were obtained, in nearly all cases (39/40), from patient peripheral blood (PB); in one case, B-CLL cells were obtained from lymphoid tissue after splenectomy in the course of treatment. Most samples were obtained from B-CLL patients prior to commencing chemotherapy or treatment with biologicals (33/40 cases). For the 7 B-CLL cases that received prior treatment, the interval between therapy and sample isolation is indicated in Table I. Clinical information, laboratory data including results from fluorescence in situ hybridization (FISH) analyses for common B-CLL chromosomal anomalies, and IGHV DNA sequence mutation status were available for these patients (Table I). The cases were either not complex, being solely del13q14 or del11q22 or trisomy-12 (TRI-12), or when they were complex involved primarily del11q22 + del13q14; none involved del17p (p53 deletion). In this study, the term B-CLL clone is used to connote a CD19+/CD5+ B-CLL population expressing a uniform IGHV sequence. This does not exclude the presence of subclones with other differing characteristics. Indeed, heterogeneity in the proportion of leukemic PB cells displaying FISH genetic anomalies (Table I) shows that subclones were present at the time of sampling.
Table I. Clinical and biological characteristics of tumor-bearing patients and B-CLL clones.
| CLL Clone | Age (y) | Sex | RAI Stagea | IGHV mut Status | IGHV Gene | del13q14b | TRI-12b | del11q22b | del17pb | ATM Mutation | % CD38+ in Blood B-CLL | % CD19/CD5+ B-CLL in Blood lcc | WBC Count ×103/μl (% lc) | TFT (mo) | OS (mo) | Functional Study: Time from Therapyd |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 186 | 71 | Female | 1 (1) | M | 3-34*04 | Neg | Neg | Neg | Neg | nd | 1.0 | 82 | 26.4 (80) | >193 | >215 | — |
| 275 | 75 | Female | 0 | M | 3-30 | + (16% ho; 6% het) | Neg | Neg | Neg | nd | nd | nd | 35.6 (76) | >203 | >203 | — |
| 321 | 60 | Male | 1 (3) | U | 4-34 | Neg | + (53%) | Neg | Neg | nd | 9.1 | 100 | 94.3 (nd) | 140 | >183 | — |
| 346 | 92 | Female | 1 (1) | M | 3-7*01 | Neg | Neg | Neg | Neg | WT | 6.0 | 71 | 14.8 (64) | 58 | >522 | 276 mo |
| 430 | 68 | Male | 2 (1) | U | 1-69*01 | + (97% het) | Neg | + (10% het) | Neg | c.8876_8879 del GACT (exon 64) | 4.5 | 99.5 | 288 (85) | 58 | 141 | 12 mo |
| 529 | 65 | Male | 0 (1) | U | 1-69*01 | Neg | Neg | Neg | Neg | nd | 80.8 | 79 | 50.4 (84) | >112 | >136 | — |
| 543 | 63 | Male | 1 (2) | U | 4-59 | Neg | Neg | Neg | Neg | nd | 2.0 | 99/99/ | 104.0 (73) | 79 | >128 | — |
| 600 | 72 | Female | 0 (0) | M | 4-34*01 | + (99% ho) | Neg | Neg | Neg | nd | 0.7 | 96/99/3 | 113 (75) | >77 | >119 | — |
| 624 | 63 | Male | 1 | M | 3-7*01 | Neg | + (10%) | Neg | Neg | WT | 77 | 82 | 16.4 (nd) | >108 | >116 | — |
| 625 | 62 | Male | 2 | U | 1-69 | Neg | Neg | + (80% het) | Neg | nd | 76 | 99 | 3.9 (7) | 22 | >116 | — |
| 631 | 61 | Male | 1 | U | 3-20 | Neg | Neg | + (90% het) | Neg | nd | 70 | 75/80/15 | 36.6 (83) | >68 | >115 | — |
| 675 | 65 | Female | 0 (1) | U | 3-23*01 | + (80% het) | Neg | + (19% het) | Neg | nd | 74 | 99.6 | 10.8 (61) | 86 | 174 | 39 mo |
| 693 | 65 | Female | 0 | M | 3-7 | Neg | Neg | Neg | Neg | nd | 2.0 | 99.8 | 224.1 (87) | 128 | >186 | — |
| 770 | 66 | Female | 1 | U | 3-15*01 | + (89% het) | Neg | Neg | Neg | 5890 A > G (exon 41) + c6997-6998insA mut (exon 50) | 2 | 95/89/3 | 112.9 (79) | 69 | >115 | — |
| 849 | 81 | Female | 3 (3) | M | 3-72 | + (68% het) | + (96%) | Neg | Neg | nd | 81 | 99/47/1 | 217 (65) | 12 | >168 | 72 mo |
| 887 | 74 | Male | 1 (1) | U | 3-30-3*01 | + (30% het) | Neg | + (27% het) | Neg | nd | 2.5 | 98.4 | 91.9 (76) | >62 | >96 | — |
| 922 | 52 | Male | 1 (1) | M | 4-34*07 | + (45% het) | Neg | Neg | Neg | WT | 1.1 | 99.6 | 38.1 (79) | >62 | >99 | — |
| 945 | 55 | Female | 0 | M | 3-7 | + (96% het) | Neg | Neg | Neg | nd | nd | 71/72/11 | 49.2 (85) | >77 | >96 | — |
| 949 | 73 | Male | 1 | M | 3-72 | Neg | Neg | + (8.5%) | Neg | nd | 4.6 | 97/97/2 | 216 (76) | 0 | 485 | 418 mo |
| 950 | 77 | Male | 0 (0) | U | 2-5*10 | + (89% het) | Neg | Neg | Neg | nd | 4.0 | 88/95/7 | 19.4 (87) | 29 | >92 | 8 mo |
| 967 | 55 | Male | 2 (0) | M | 3-7*01 | Neg | Neg | Neg | Neg | nd | 79 | 98.6 | 20.8 (62) | 25 | >90 | — |
| 996 | 84 | Male | 0 | U | 1-69 | Neg | Neg | Neg | Neg | nd | 45.8 | 95.1 | 178.4 (87) | > 36 | 45 | — |
| 1013 | 85 | Male | 0 (2) | U | 3-33 | Neg | + (52%) | Neg | Neg | nd | 44.8 | 83.8 | 49.9 (77) | 52 | 75 | — |
| 1031 | 73 | Female | 1 | M | 4-39 | Neg | Neg | Neg | Neg | nd | 2.7 | 99.9 | 225 (91) | >326 | >356 | — |
| 1081 | 72 | Female | 1 (2) | U | 3-11*03 | Neg | Neg | + (96% het) | Neg | nd | 47.4 | 100 | 77.1 (71) | 8 | >192 | 31 mo/chemo 8 mo/Ritux |
| 1086 | 68 | Male | 1 | U | 4-34*01 | + (94% het) | Neg | Neg | Neg | WT | 1.5 | 99.8 | 32.8 (82) | 78 | >157 | — |
| 1158 | 69 | Female | 1 | U | 3-15*01 | + (97% het) | Neg | + (48%) | Neg | nd | 15.1 | 91 | 246.9 (88) | 210 | 252 | — |
| 1238 | 54 | Male | 0 | U | 3-30-3*01 | + (55% het) | Neg | + (69% het) | Neg | nd | 86.9 | 98.2 | 26.0 (73) | 54 | >70 | — |
| 1239 | 67 | Female | 0 | U | 3-30*03 | Neg | Neg | Neg | Neg | nd | 7.0 | 99.6 | 159.2 (90) | >148 | >164 | — |
| 1252 | 69 | Male | 0 | M | 3-33*01 | Neg | Neg | Neg | Neg | nd | 0.6 | 93/97/5 | 22.6 (80) | >73 | >79 | — |
| 1258 | 66 | Male | 0 (0) | M | 3-23*01 | Neg | Neg | Neg | Neg | nd | nd | nd | 18.7 (67) | >72 | >80 | — |
| 1260 | 65 | Female | 0 | M | 4-4*02 | Neg | Neg | Neg | Neg | WT | 18.4 | 81.9 | not avail | >77 | >129 | — |
| 1300 | 64 | Male | 1 | M | 3-7*02 | Neg | Neg | Neg | Neg | WT | 89.5 | 82/98/15 | 35.6 (78) | >75 | >86 | — |
| 1328 | 82 | Male | 0 | M | 4-61*01 | + (86%) | Neg | Neg | Neg | WT | 3.2 | 81/92/15 | 25.5 (77) | >9 | >61 | — |
| 1331 | 72 | Female | 3 | U | 4-39*01 | Neg | Neg | + (76% het) | Neg | nd | 100 | 99.9 | 109.2 (70) | 1 | >61 | — |
| 1380 | 64 | Male | 1 | M | 3-7*01 | + (50% ho; 22% het) | Neg | Neg | Neg | nd | 4.5 | 100 | 111.4 (86) | >37 | >71 | — |
| 1413 | 61 | Female | 1 | M | 4-39*01 | Neg | + (20%) | Neg | Neg | nd | 1.3 | 80/84/14 | 13.9 (61) | >52 | >55 | — |
| 1529 | 51 | Male | 1 | M | 4-59*01 | + (30%ho; 60% het) | Neg | Neg | Neg | nd | 0.9 | 99/99/1 | 141.2 (75) | >70 | >111 | — |
| 1615 | 68 | Female | 0 | U | 1-69 | Neg | Neg | Neg | Neg | nd | nd | nd | 96.8 (89) | >46 | >48 | — |
| 1692 | 56 | Male | 1 (3) | U | 2-7 | Neg | + (85%) | Neg | Neg | nd | 30 | 98 | 82.7 (78) | >33 | >33 | — |
RAI stage at the time of test sample acquisition (RAI stage at the last time the patient was clinically monitored).
Designates the results of FISH-determined chromosomal analyses. If deletions were present, the % of cells showing either heterozygous (het) or homozygous (ho) deletions is indicated in parentheses. Chromosomal defects were considered present if the anomaly was present in ≥5% of the cells. Although in most cases, FISH/ATM mutation analyses were performed within 1 y of the blood sampling for later functional studies, in the case of the following B-CLL, FISH analyses were performed from 2 to 5 y before the sample for culture: CLL IDs: 321, 430, 600, 624, 887, and 950.
B-CLL blood had been screened for % of CD19+/CD5+ doubly stained cells in the lymphocyte (lc) population after diagnosis and generally before the time of blood sampling for B cell isolation. In some cases, single staining for CD19, CD5, and CD3 was performed; in the latter, % of the blood lymphocyte population positive for CD19/CD5/CD3, respectively, is shown.
Shown is the time separation (in months) between any earlier chemotherapy and the acquisition of PB for B-CLL purification and growth experiments. Note that 84% of the B-CLL clones tested were obtained from patients prior to the beginning of therapy. Dashes indicate that B-CLL specimens were obtained prior to any patient therapy.
chemo, chemotherapy; del, deletion; lc, lymphocytes; M, IGHV mutated; mut, mutation; nd, not determined; Neg, negative; not avail, not available; Ritux, Rituximab; U, IGHV unmutated; WT, wild-type ATM gene.
Ataxia telangiectasia mutated mutation status
In a selected group of B-CLL (9/40), the entire ataxia telangiectasia mutated (ATM) coding region, consisting of 62 exons, was screened using HPLC. Sequence changes were considered to be: 1) truncating mutations, if they were predicted to cause premature termination of the protein or small in-frame deletion; or 2) missense mutations, if they were either reported in ATM patients or predicted to cause an amino acid substitution in the residue located within the region encoding the functional domain of the ATM protein and/or conserved between human and mouse and not observed in 200 alleles of healthy controls. This method was described in an earlier publication (58).
B-CLL cell isolation
B-CLL cells from each patient’s PB, or in some cases from normal subjects, were isolated by negative selection using RosetteSep Human B Cell Enrichment Cocktail (Stemcell Technologies, Vancouver, BC). Whole PB was incubated with the mixture, then diluted with 2% FBS (Atlanta Biologicals, Flowery Branch, GA) in PBS and centrifuged over RosetteSep DM-L Density Medium (Stemcell Technologies). When 31 B-CLL PB specimens initially purified by this technique were tested for purity by the Center for CLL Research, it was determined that of the events collected, 81 ± 10% was in the lymphocyte scatter gate and 92.7 ± 9% of the latter was CD19+/CD5+. Based on scatter, some of the nonlymphocytes were contaminating RBCs; others may have been aggregates. These were not apparent when isolated B-CLL B cell preparations were frozen (−150°C), defrosted, and collected at the interface after Ficoll-Hypaque density centrifugation. Ten of the frozen B-CLL B cell preparations evaluated in this study were subjected to immunofluorescent Ab staining after defrosting, and the following profile was noted: CD19/CD5+ = 92 ± 5% (mean ± SD); CD3+ = <1%; CD14 = <1%; CD16 = <1%. Minor contamination with CD19+, CD5− normal B cells was on occasion evident but always <10%; only in one case (CLL1300) was CD3 (8%) and CD14 (2%) contamination seen. Isolation of a single spleen-derived B-CLL (CLL967) involved Ficoll-Hypaque density centrifugation of a single-cell suspension of splenocytes before B cell negative selection with magnetic beads and freezing; defrosted cells were subsequently centrifuged through a 40/50/55/75% Percoll density gradient to enrich for uniformly small lymphocytes at the 55/75% interface; when analyzed by flow cytometry, these were 98% CD19+CD5+, 1% CD3+, and 1% negative for B/T markers. Defrosted samples were routinely subjected to Ficoll-Hypaque centrifugation to eliminate any dying/dead cells and to ensure that all studies began with uniformly viable B-CLL. Viability assessments (trypan blue dye exclusion) of defrosted PB-derived B-CLL B cells made prior to Ficoll-Hypaque centrifugation in a random group of B-CLL (n = 10) showed viability values ranging from 74 to 99% (92 ± 3% = mean ± SEM). In the total samples studied (n = 40), the average post-Ficoll recovery of viable B-CLL cells from those initially frozen was 57 ± 5% (mean ± SEM). Frozen B-CLL specimens that routinely showed poor recovery/viability were excluded.
In vitro culture conditions for B-CLL replication
CFSE-labeled B-CLL cells were cultured in an enriched medium used for normal B cell replication in long-term cultures (55) with added insulin/transferrin/selenium supplement (catalog no. 17-8387; BioWhittaker). Notably, this medium contains the reducing agent, 2-ME (5 × 10−5 M). The latter replaces an important function of bone marrow stromal cells in converting cystine to cysteine, which is needed for B-CLL uptake and use in the glutathione synthesis needed for retained viability (59). To minimize interexperimental differences, we freshly prepared medium for each experiment using stock additives whose expiration date was carefully monitored and, in the case of FCS and the insulin/transferrin/selenium supplement, kept frozen in aliquots until use. Cultures were routinely established in 96-well plates (Falcon; Costar) at 105 cells per 200-μl volume with triplicates for each culture condition. Recombinant human IL-15 (R&D Systems) and CpG DNA TLR-9 ligand (ODN-2006; Invivogen) (frozen in aliquots until use) were added at final culture concentrations of 15 ng/ml and 0.2 μM (1.5 μg/ml), respectively. In designated cultures, the pan-caspase inhibitor, Z-VAD-FMK (40 μM final; Sigma-Aldrich, St. Louis, MO), or DMSO vehicle was added.
Assays for B-CLL replication and viability
Previously described methods for quantifying the growth/viability of CFSE-labeled lymphoblasts within in vitro–activated normal human B cell cultures were used (55–57) and are outlined in schematic form in Supplemental Fig. 1. When the absolute number of viable lymphoblasts present in culture was quantified, cultures were pulsed with a known number of CountBright absolute counting beads (Molecular Probes, Life Technologies) just before harvesting (56, 57). For cell harvest, culture plates were routinely placed on wet ice for 10 min to facilitate detachment of activated B-CLL from the culture wells; without this step, B-CLL recovery in ODN+IL-15 cultures was often compromised because of cell adhesion to wells. Cells (and accompanying beads) were harvested and washed through cold PBS before immediate fixation in 1.5% formaldehyde for subsequent analysis by flow cytometry (FACSCalibur or FACS-Verse with CellQuest or FlowJo 7.6.1 data analysis software). Selection of intact cells (viable + apoptotic) as opposed to debris was based on a consistent threshold set in each experiment based on the forward light scatter (FSC) intensity of the standardizing CellQuant beads (note that the side scatter of the beads is significantly greater than that of B-CLL cells, enabling clear segregation). Quantification of beads is performed on the basis of their high allophycocyanin fluorescence. Gates for delineating undivided B-CLL cells were established on the basis of CFSE intensity of B-CLL from cultures supplemented with IL-15 only, in which the majority of cells do not replicate at the used dose of IL-15; gates for other divisions were based both upon a 50% reduction in CFSE fluorescence upon each successive division and visual inspection of CFSE peaks from ODN+IL-15–stimulated cultures. Percent viability of activated B-CLL within each division subset was discerned by assessing CFSE-gated cells on the basis of light scatter (55–57) or exclusion of Fixable Viability Stain 450 (FVS450; BD Biosciences). As described earlier for studies on activated normal B lymphoblasts (55, 57, 60), percentage viability values of ODN-activated B-CLL obtained through light scatter and viability dye exclusion are virtually identical (see Supplemental Fig. 1). Nonetheless, viable dye exclusion assays are more reliable when small, relatively quiescent B-CLL cells cultured with medium or IL-15 alone are monitored.
Two-color flow-cytometric assays for assessing CD38 expression as a function of cell division
CFSE-labeled cultures were harvested in cold PBS, resuspended in PBS + 1% BSA + 0.1% sodium azide assay buffer, surface stained with allophycocyanin anti-human CD38 or allophycocyanin-IgG control (Becton Dickinson), fixed with 1% paraformaldehyde, and analyzed by flow cytometry using methods described earlier (55).
Immunohistochemical analyses
Formalin-fixed, paraffin-embedded sections from human B-CLL spleens, removed for therapeutic reasons, were serially sliced into 4- to 6-μM sections and placed onto glass slides. Similarly prepared sections from two normal human spleens were obtained. One spleen had been determined by pathologists to have only passive congestion after removal from a patient with pancreatic carcinoma; the other was a spleen from a deidentified 30-y-old normal man, sectioned and commercially distributed by US Biomax (HuFPT082; Rockville, MD). All spleen tissue sections were deparaffinized and subjected to Ag retrieval in a heated pressure cooker (125°C, 3 min) followed by blocking for endogenous peroxidases with 3% H2O2 in methanol. The tissues were preblocked with 25% normal serum matching the secondary detection Ab’s species and further subjected to avidin/biotin blocking (kit from Vector Labs) before incubation at 4°C overnight in a humidified chamber with primary Ab: goat anti-human IL-15 (AF 315; R&D Systems), mouse anti-human Pax-5 (IS650, clone DAK; Dako), or mouse anti-human Ki-67 (N1633, clone Mib 1; Dako) at the recommended dilutions along with appropriate isotype controls. Sections from CLL-967 spleen were additionally exposed to mouse IgM anti-human FDC mAb (clone CNA.42; 1:500; Affymetrix; eBioscience) or rabbit IgG mAb specific for active (cleaved) caspase-3 (Asp175; 1:200; Cell Signaling 9664), or negative controls. The following secondary biotin-conjugated, affinity-purified F(ab′)2 reagents were used for Ig detection (each commercially absorbed against human serum proteins): donkey anti-goat IgG (code 705-066-147; Jackson Immunoresearch), donkey anti-rabbit IgG (code 711-066-152; Jackson Immunoresearch), horse anti-mouse IgG (BA-2000; Vector Laboratories), or goat anti-mouse IgM (μ-chain–specific; code 115-066-075; Jackson Immunoresearch). Each secondary Ab was followed by treatment with Vectastain ABC kit (Elite, pk6100-standard; Vector Laboratories) and detection achieved with the ImmPACT DAB Peroxidase substrate kit (Sk 4105; Vector laboratories). The tissues were counterstained with hematoxylin, dehydrated, and mounted using Vectamount permanent mounting media (H5000; Vector Laboratories). Most images were photographed at final magnifications of 100×, 200×, or 600× (the latter with a 60×/1.25 Oil Iris ∞/0.17) using an Olympus BX40 phase-contrast microscope and Olympus DP20 camera.
Determinations of B-CLL in vivo growth rate
Values for B-CLL growth rate (GR; number of doublings per month) were determined from the calculated time for doubling of lymphocyte counts within PB using an online doubling time/GR calculator based on exponential regression (Roth V. 2006 http://www.doubling-time.com/compute.php). GR determinations were made for CLL cases in which sequential values for absolute lymphocyte count (ALC) in blood were available (36/40). ALCs were obtained during a period that included the time blood was acquired for these functional studies, in all but two cases, over a time range of 1 to 80 mo (mean ± SD = 27 ± 23 mo). The incidence of ALC measurements for doubling time/GR determinations was as follows: 7 CLLs based on 2 ALCs, 18 based on 3, 5 based on 4, 4 based on 5, 1 based on 6, and 1 based on 10.
Kaplan–Meier analysis was used to test whether clinical progression, defined as time-to-first treatment (TFT), was a function of several parameters in ODN+IL-15–stimulated cultures: 1) viability in the divided (DIV) blasts (≤75% versus >75% viable); 2) replication (≤75% versus >75% of the total viable cells as DIV; and 3) extended replication (≤50 versus >50% of the total viable DIV cells representing >2 DIV). In each of these cases, the cohort was subdivided so that approximately equal numbers of clones were represented in each category. In addition, TFT and overall survival (OS) were monitored as a function of extended replication (≤50 versus >50% of the total viable cells representing >2 DIV). Finally, TFT was monitored as a function of the IGHV mutation and del11q22 status of the B-CLL clones. Log-rank statistical evaluation was used to determine significance of any differences noted in each pair of plots.
Other statistics
Box plots were used to show summed statistics for various cohorts of B-CLL studied. In box plots, the upper part of the box represents the third quartile (75th percentile) and the lower part of the box the first quartile (25th percentile); upper and lower whiskers represent error bars for the 10th and 90th percentiles, respectively; outliers are shown as individual points. Furthermore, the mean and median values for the grouped data are shown by solid and dashed lines, respectively, within each box. In nontreatment-related comparisons, when the samples were from the same B-CLL, the nonparametric test, Wilcoxon signed rank test for matched pairs, was used to determine whether medians of the differences between measurements differed significantly. When samples were independent, the nonparametric test, Wilcoxon rank sum test, was used to compare pairwise group differences, and the Kruskal–Wallis test was used when groups were ≥3. These tests were used because they do not assume the differences in the outcome are normally distributed. For the correlation analyses, both the Pearson correlation coefficients and the Spearman rank coefficients were calculated. The Pearson correlation coefficients measure the strength and direction of the linear relationship between the two variables, whereas the Spearman correlation is used when one or both of the variables are not assumed to be normally distributed. For the analyses evaluating significance in all pairwise FISH-determined genetic anomaly groups, the nonparametric Wilcoxon rank sum test was used with two-sided Monte Carlo estimation of the exact p values. Results are considered statistically significant if p < 0.05 on the two-sided tests, with the exception of the pairwise comparisons between responses of the various FISH subgroups in which Bonferroni correction methods for adjusting the p value for multiple comparisons was used (statistical significance if p < 0.005). Analyses were carried out in SAS Version 9.3 (Cary, NC). SigmaPlot 12.0 was used to determine linear regression plots and respective significance values for comparisons of B-CLL in vivo and in vitro growth.
Results
Effect of IL-15 on B-CLL viability and growth in ODN-stimulated B-CLL cultures
We initiated these studies with the hypothesis that B-CLL growth responses to CpG DNA will be significantly augmented by the inflammation-linked cytokine, IL-15. Purified B cells from the PB of 12 B-CLL patients were CFSE labeled and cultured (6–7 d) in medium supplemented with either IL-15 or ODN, or both, to test this hypothesis. The number of division cycles and the viability during successive divisions were determined, as described in Supplemental Fig. 1 and detailed elsewhere (55, 57). Assessing the impact of division on viability was deemed important given earlier evidence that normal cycling B lymphoblasts are prone to p53-regulated death during responses to T cell–independent stimuli (55, 57).
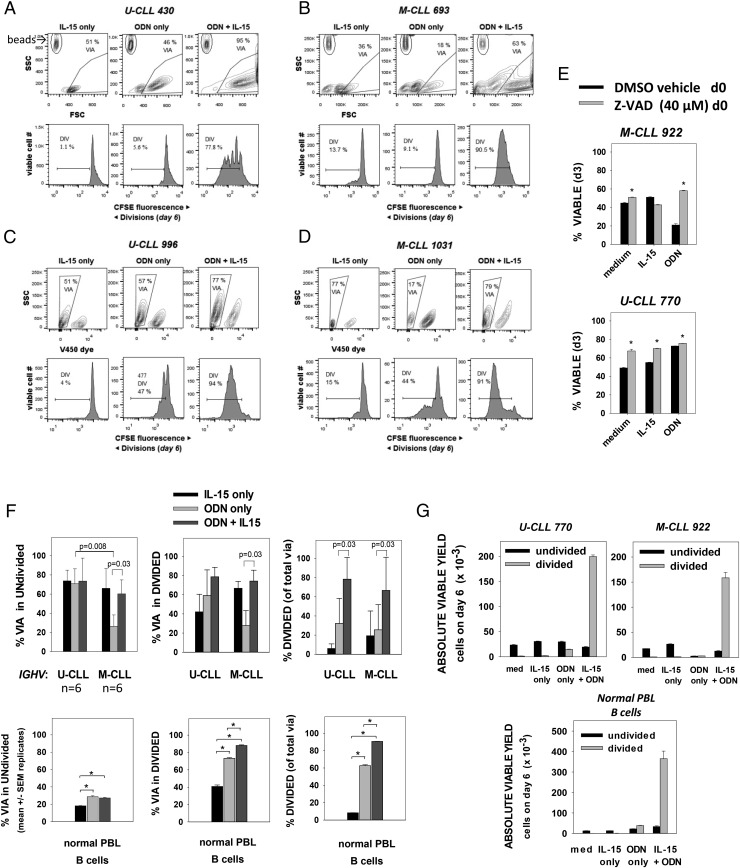
Fig. 1A–D shows representative flow-cytometric data from cultures of four leukemic B-CLL clones: two with unmutated and two with mutated IGHVs (U-CLL-430/U-CLL-996 and M-CLL-1380/M-CLL-1031, respectively). We found that B-CLL exposed to IL-15 alone (15 ng/ml) remained relatively small (low FSC; Fig. 1A, 1B) and generally did not proliferate, as illustrated by the single major peak in the CFSE histograms (Fig. 1A–D, lower left). At this dose of IL-15, there was only a slight improvement in viability above cells in medium alone (111 ± 12%, mean ± SD in n = 10 experiments using V450 dye exclusion to monitor viability; see also Fig. 1E). Stimulation by ODN alone augmented B-CLL size (Fig. 1A, 1B), but only slightly increased division over the span of 6–7 d (Fig. 1A–D, lower middle histograms). Consistent with earlier reports, the viability of U-CLL and M-CLL cultures after ODN stimulation was quite different (U-CLL = 46–57% versus M-CLL = 15–17%; Fig. 1A–D, upper middle dot plots). ODN-induced death in M-CLL cultures was blocked if the pan-caspase inhibitor, Z-VAD-FMK, was present (Fig. 1E), reflecting ODN-induced apoptotic death (38). Importantly, when ODN-exposed U-CLL or M-CLL concomitantly received IL-15 signals, culture viability increased (Fig. 1A–D, upper right dot plots). Furthermore, clonal expansion was clearly evident (Fig. 1A–D, lower right histograms). Other experiments show that IL-15 doses as low as 1 ng/ml can significantly augment ODN-triggered B-CLL division (data not shown).
FIGURE 1.
ODN+IL-15 signaling induces M-CLL and U-CLL clonal expansion. (A–D) Plots showing viability gating and CFSE division histograms (for viable cells) in IGHV U-CLL 430 and 996 (A and C) and IGHV M-CLL 693 and 1031 (B and D) clones at day 6 after culture with IL-15 alone, ODN alone, or both ODN+IL-15. Plots in (A) and (B) reveal the presence of calibration beads (upper left gate) added for quantification of absolute yield of viable cells (as for G) and as a means of discerning the threshold between debris and intact (via or dead) cells (see Supplemental Fig. 1). (E) Pan-caspase inhibitor (Z-VAD-FMK) inhibits ODN-induced death in M-CLL, consistent with an apoptotic mechanism. Immediately after B-CLL culture with the indicated stimuli, sets of parallel cultures were pulsed with Z-VAD-FMK (40 μM) or vehicle control (DMSO). Cells were harvested at day 3, stained with viable cell exclusion Dye 450, and fixed and analyzed by flow cytometry for viable/dead cells. Data are expressed as mean ± SEM values of triplicate cultures. Asterisks indicate that differences between DMSO versus Z-VAD–treated cultures were statistically significant (p < 0.01) on the basis of a two-sided, unpaired Student t test. (F, top panels) Pooled analysis (mean ± SD) of experiments with six IGHV U and six IGHV M B-CLL clones showing (top left panel) % viability in undivided gate of total viable+apoptotic CFSE-labeled cells, (top middle panel) % viability in DIV gate of total viable+apoptotic cells, and (top right panel) % DIV cells within all gated viable cells after 6–7 d of culture with stimulants shown. The p values for two-sided significance are indicated. Bottom panels, Similar analysis of the ODN ± IL-15 responses of purified normal human PB B cells (determined to be 98% CD19+ with 27% CD27+ memory B cells before culture). Values represent the mean ± SEM of quadruplicate cultures; asterisks indicate statistically significant differences. (G) Absolute yield of viable B-CLL cells (undivided or DIV) from representative experiments with IGHV U-CLL 770, IGHV M-CLL 922, or normal PB B cells. Each was cultured with the indicated stimuli for 6 d before addition of calibration beads and harvest. Bars represent mean ± SEM values from three to four replicate cultures.
A summary of IL-15 effects on the ODN-stimulated responses of six M-CLL and six U-CLL is shown in Fig. 1F (top row). Concomitant IL-15 signaling significantly augmented survival in both noncycling and cycling M-CLL (p = 0.03 for both). This prosurvival effect appeared to fully eliminate the striking difference between U-CLL and M-CLL viability seen in the nondivided fraction when IL-15 was absent (p = 0.008). Furthermore, IL-15 significantly boosted the proportion of viable cells evidencing division, both in M-CLL and in U-CLL cultures (p = 0.03 for both). Additional evidence of the division-potentiating effects of this cytokine is shown in Fig. 1G (top panels), where data are represented as the absolute yield of viable cells per culture well (determined by pulsing cultures with standardizing beads at the time of harvest). Exposure to IL-15 at the time of ODN stimulation increased the yield of DIV B-CLL lymphoblasts from 14-fold to 56-fold in the case of U-CLL 770 and M-CLL 922, respectively.
An example of the effect of ODN ± IL-15 signaling on normal human B cells (27% positive for memory marker, CD27) is presented in Fig. 1F (bottom row) and 1G (bottom panel). This experiment confirms that IL-15 can significantly augment ODN-induced B cell growth (49, 61), and additionally shows that IL-15 boosts the survival of DIV normal B cell blasts, as was noted earlier for U-CLL and M-CLL. Normal B cell responses are more representative of U-CLL than M-CLL: ODN signaling alone enhanced B cell survival above that seen in IL-15–only cultures (or medium-only cultures; data not shown). Contrasting with the generally high viability of undivided CLL cells (exception is ODN-treated M-CLL) was the low viability of undivided normal B cells (<30% viable after 6 d of culture; Fig. 1F). This difference likely reflects the elevated baseline expression of several prosurvival molecules in B-CLL (62).
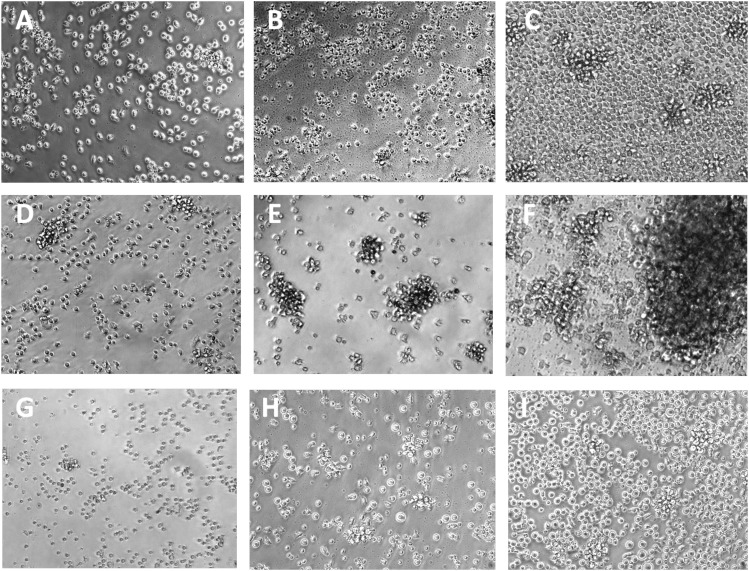
Phase microscopic evidence of the cellular changes induced by ODN ± IL-15 is presented in Fig. 2. M-CLL cultures receiving the TLR-9 stimulus alone show substantially more cell shrinkage and debris formation (Fig. 2B) than comparably stimulated cultures of U-CLL (Fig. 2E) or PB B cells (Fig. 2H). In contrast, when ODN and IL-15 are jointly present, both U-CLL and M-CLL cultures show significant cell enlargement and aggregate formation (Fig. 2C, 2F). The clustering was comparable with or greater than that seen in normal B cell cultures (Fig. 2G–I). Differences were noted between B-CLL clones in the degree of clustering, but the reason for this is unexplored.
FIGURE 2.
Photomicrographs of M-CLL 1031 (A–C), U-CLL 996 (D–F), and normal B cells from PB (G–I) after 6 d of culture in IL-15 alone (A, D, and G), ODN alone (B, E, and H), or both ODN+IL-15 (C, F, and I). Culture of M-CLL with ODN alone (B) typically induces prominent loss of cell volume and formation of debris, not seen with similarly cultured U-CLL (E) or normal B cells (F). Stimulation with ODN+IL-15 induces notable cell enlargement, larger clump formation, and significant cell proliferation in M-CLL (C), U-CLL (F), and normal B cell (I) cultures. Photographs were taken at original magnification ×200 using an Olympus BX40 phase-contrast microscope equipped with an Olympus DP20 camera.
Taken together, our study confirms earlier findings that IGHV-mutated B-CLL are prone to apoptosis upon receiving TLR-9 stimuli alone (37, 38, 40), and importantly shows for the first time, to our knowledge, that additional IL-15 signaling not only abrogates this outcome but can foster significant clonal expansion within both M-CLL and U-CLL, as earlier reported for normal memory B cells (49).
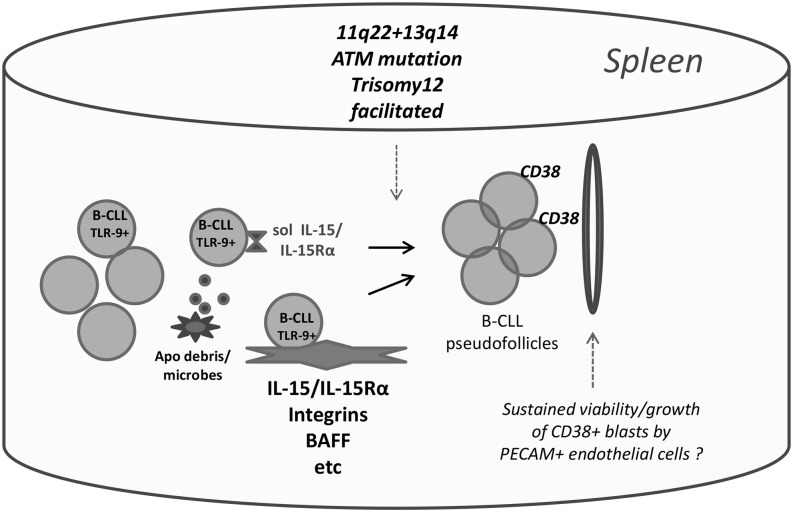
Immunohistochemical evidence for IL-15–expressing cells and apoptotic cells/debris within B-CLL–infiltrated human spleens with pseudofollicles
The recent evidence that the IL-15 gene is constitutively active within several nonlymphoid cell lineages (macrophages and stromal, endothelial, and dendritic cells) in normal mouse primary and secondary lymphoid tissues (44) suggests that IL-15 should be present in known sites of human B-CLL clonal expansion. This possibility is further supported by reports of IL-15 protein in isolated myofibroblasts from normal human spleen (63), FDCs within human tonsils (43), and stromal cells within human bone marrow (45, 64). Nonetheless, there is no direct evidence showing that IL-15–producing cells are present within B-CLL–infiltrated tissues harboring pseudofollicles, and it remains possible that B-CLL infiltration might suppress IL-15 production in these sites, perhaps as a means of preventing the expansion of cytotoxic T cells and NK cells that rely on this cytokine (47, 48).
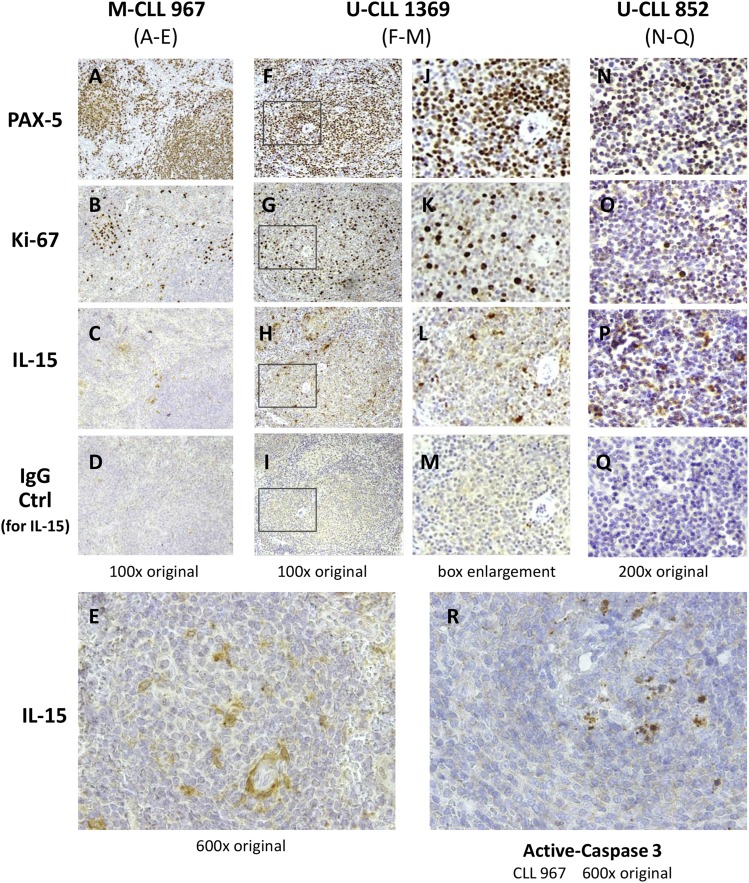
To discern the extent that IL-15–producing cells localize proximal to areas of B-CLL replication in patient lymphoid tissue, we undertook an immunohistological study of spleen sections from three B-CLL patients subjected to therapeutic splenectomy. Fig. 3 shows the results from staining sequential slices of each spleen for IL-15, the B cell–specific transcription factor, PAX-5, and the proliferation marker, Ki-67. The white pulp of spleens infiltrated with M-CLL 967 and U-CLL 1369 was hyperplastic and possessed PAX-5+ pseudofollicles (Fig. 3A, 3F) with numerous Ki-67+ proliferating cells (Fig. 3B, 3G). The sections of spleen with U-CLL 852 had less defined pseudofollicles, but Ki-67+ proliferating cells were scattered uniformly throughout (Fig. 3O). In each of these spleens, cells expressing cytoplasmic IL-15 were prevalent throughout the white pulp, as represented by Fig. 3C and 3E (M-CLL967), Fig. 3H and 3L (U-CLL 1369), and Fig. 3P (U-CLL 852). Although IL-15+ cells could be found within the Ki-67+ zones of many pseudofollicles (Fig. 3B, 3C, 3G, 3H, 3K, 3L), others were present at the margins of pseudofollicles. From the higher magnification of the CLL-967–infiltrated spleen (Fig. 3E), it can be discerned that some IL-15+ cells possess the elongated processes characteristic of FDCs or possibly reticular stromal cells. In addition, in some IL-15–stained cells, the cytokine appears to be localized in small vesicles or in a perinuclear pattern, both of which have been reported (65, 66).
FIGURE 3.
IL-15–producing cells are present, proximal to pseudofollicles, in B-CLL–infiltrated spleens harboring apoptotic cells. Serial sections of fixed and embedded splenic tissue from three B-CLL patients (M-CLL 967 with negative FISH and both U-CLL 1369 and 852, positive for del17p) were stained for the B cell–specific transcription factor, PAX-5 (A, F, J, and N), proliferation marker Ki-67 (B, G, K, and O); or IL-15 (C, E, H, L, and P). A separate section of the same tissue stained for IL-15 was stained with goat IgG control on the same slide (D, I, M, and Q). In addition, a section of the U-CLL 967–infiltrated spleen was stained for active (cleaved) caspase-3 (Asp175), a marker for apoptotic cells (R). (E and R) Represent higher magnification (600×) photographs of differing sections of CLL-967 from (A) to (D), whereas (J)–(M) represent enlargements of the boxed pseudofollicle section in (F)–(I), respectively (from U-CLL 1369–infiltrated spleen). The surgical pathology report (case number: FS08-8095; Genzyme specimen number: 08-50886842-MH) for the M-CLL 967 spleen noted: “A markedly enlarged spleen (>2 kg) in a patient with a history of CLL/SLL. Infiltrated lymphocytes are small to medium with round to oval to nuclear contours and inconspicuous nucleoli. Cells are located in both white pulp and red pulp. They are positive for CD20, PAX5, CD5, CD43, Bcl-2, with kappa l.c. restriction and negative for CD10, CD23, Bcl-1, Bcl-6, MUM-1 and T cell markers.” The pathology reports for U-CLL 1369/U-CLL 852 were not available, but it was evident that little normal splenic architecture remained in U-CLL 852. Photographs were taken from an Olympus BX40 microscope using 10×, 20× and 60× UPLanFl oil immersion objectives and an Olympus DP20 camera. The images involving thicker sections of U-CLL852 spleen were brightened to better distinguish the specific brown staining from the very dark hematoxylin staining of nuclei.
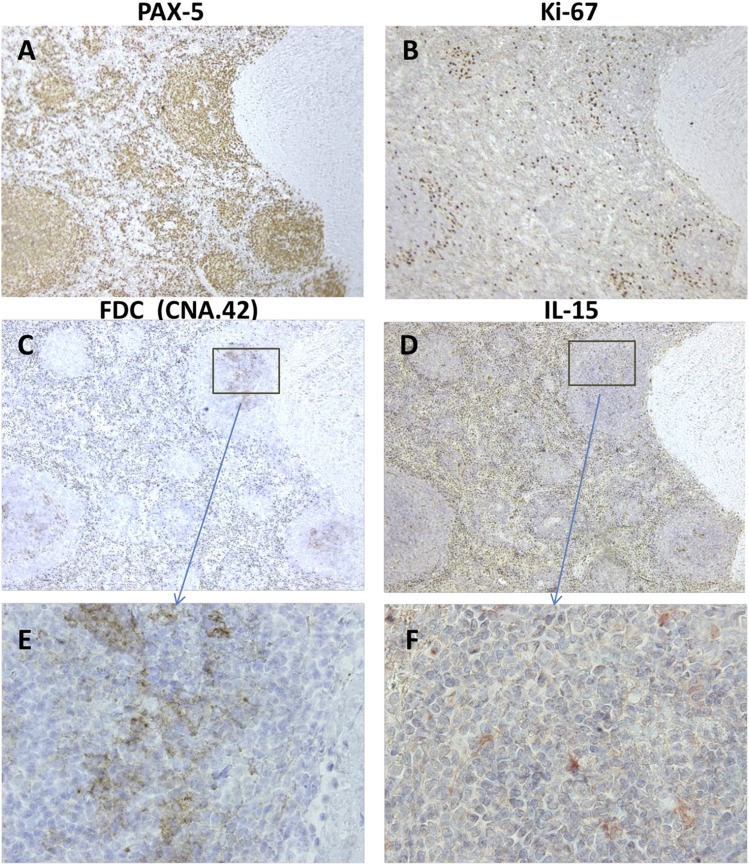
Because of reports that FDCs within tonsils produce IL-15 (43), we investigated whether IL-15+ cells colocalized with FDCs using sequential tissue slices and a staining mAb specific for human FDCs in fixed, paraffin-embedded tissues, CNA.42 (67) (Fig. 4). From this analysis, we concluded that both IL-15–producing cells and FDCs are present in the same PAX-5+, Ki-67+ pseudofollicles (Fig. 4A–D). Nonetheless, a definitive conclusion that splenic FDCs are IL-15 producers could not be made with this approach. (Two-color fluorescent staining was precluded because of a very high RBC fluorescence background in the available B-CLL sections.)
FIGURE 4.
FDCs colocalize with IL-15–producing cells in pseudofollicles of a B-CLL–infiltrated spleen. Serial sections of CLL-967–infiltrated spleen harboring numerous PAX-5+ (A) and Ki-67+ (B) pseudofollicles (stained as in Fig. 3) were stained for IL-15 (D and F) or for a carbohydrate Ag unique to human FDCs (CNA.42) (67) (C and E). A region of the spleen with clearly identifiable landmarks was selected for the visualization of these respective molecules. (A–D) Panels represent original magnifications ×100 of the region of interest. (E and F) Panels represent original magnifications ×600 of the boxed regions shown in (C) and (D) for FDC-specific CNA.42 and IL-15, respectively. (F) Image was hue-enhanced to convert yellow-brown stained cells to red, which is better visualized against a background of blue nuclei.
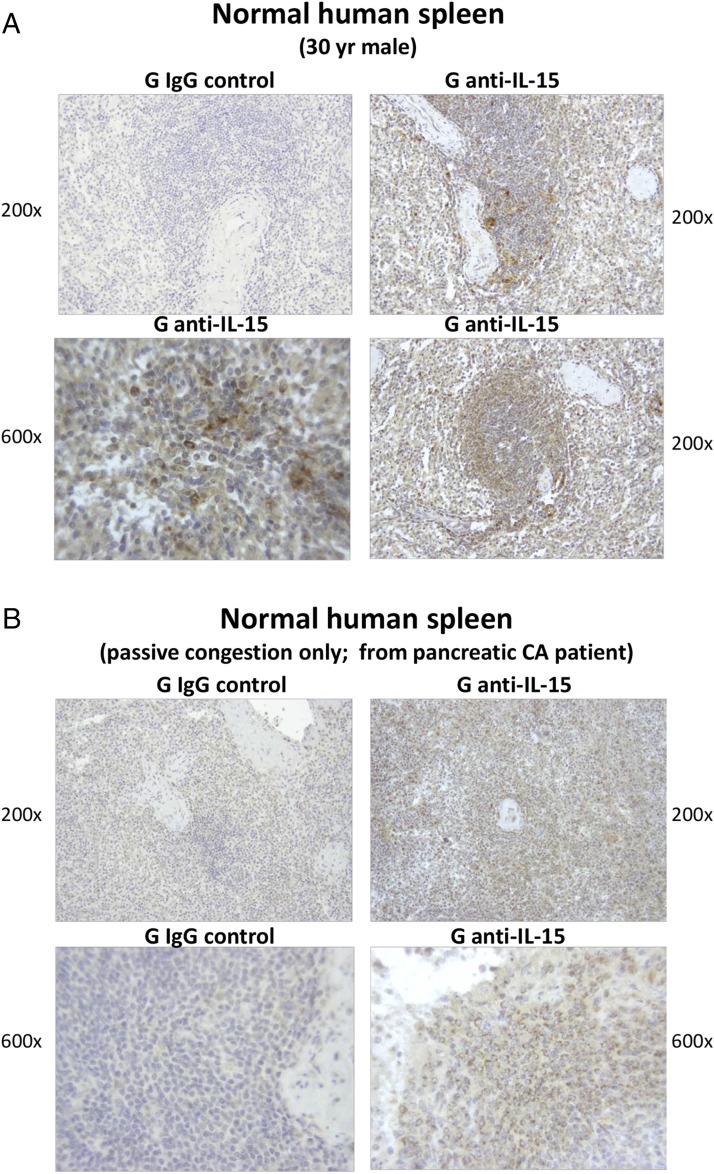
IL-15–producing cells are also found within normal human spleens, as evidenced by the images in Fig. 5. These represent 200× and 600× photographs of IL-15–stained spleen sections from a normal 30-y-old man (see Materials and Methods) (Fig. 5A) and from a patient with pancreatic cancer (Fig. 5B). The latter spleen was determined to be normal, with the exception of passive congestion, upon evaluation by a clinical pathology laboratory. Both of these spleens had IL-15–stained cells scattered throughout the examined white pulp, albeit the staining intensity was diminished in the spleen with passive congestion. The 600× image in Fig. 5A shows that IL-15 appeared localized throughout the cytoplasm in some cells and also expressed in a perinuclear pattern in others. Both the reports that IL-15 localization patterns can vary in differing cell populations (66, 68, 69) and the varying morphologies of IL-15+ cells in all the above spleens suggest that more than one splenic population is IL-15+, but this will require further investigation. Our finding of IL-15 expression within normal human spleen is consistent with images found online in the Protein Atlas (http://www.proteinatlas.org/ENSG00000164136-IL15/tissue/spleen).
FIGURE 5.
IL-15–producing cells can be detected within the white pulp of normal human spleens. Tissue sections were derived from two human spleens (A and B), determined to be normal upon pathological examination (see Materials and Methods). They were stained and examined for human IL-15, as described for Fig. 3. Cells positive for cytoplasmic IL-15 were found scattered throughout the white pulp of each spleen, in patterns similar to those shown in the present images. A rim of more dense IL-15+ cells often appeared to encircle follicles [as in the lower right panel in (A)], suggesting possible marginal zone localization. In addition, cells with more intense IL-15 expression were sometimes found proximal to the splenic artery [as in the upper right panel in (A)].
Taken together, the earlier immunohistochemical studies show that IL-15–producing cells are localized within the splenic white pulp of both B-CLL patients (3/3) and individuals without B-CLL (2/2). In the case of the three B-CLL–infiltrated spleens, IL-15+ cells were found proximal to areas of proliferating B cells, albeit they were not exclusive to this location. The latter findings support the possibility that IL-15 might be providing important signals to B-CLL cells within the patient. If findings in studies with NK cells and T cells apply to B-CLL, it is likely that IL-15 signaling in the spleen involves highly functional, soluble IL-15Rα/IL-15 complexes (70, 71) or IL-15Rα–bound IL-15 on the surfaces of IL-15–producing cells (72), both of which are more effective than soluble IL-15.
Membrane CD38 rises on B-CLL cells during ODN+IL-15–induced cycling
CD38, a type II transmembrane protein, belongs to a family of nucleotide-metabolizing enzymes and catalyzes the conversion of NAD+ to cyclic ADP ribose, with a resulting Ca2+ flux. It is notably upregulated in normal germinal center B cells, plasma cells, and several malignancies (reviewed in Ref. 73). In B-CLL, CD38 is linked to an activated phenotype (74–76), greater Ki-67 expression (6, 75), and typically, but not uniformly, worse patient prognosis (77–79). Very recent evidence suggests that CD38 enzymatic function is needed for in vivo binding to PECAM-expressing endothelial cells and CLL migration (79).
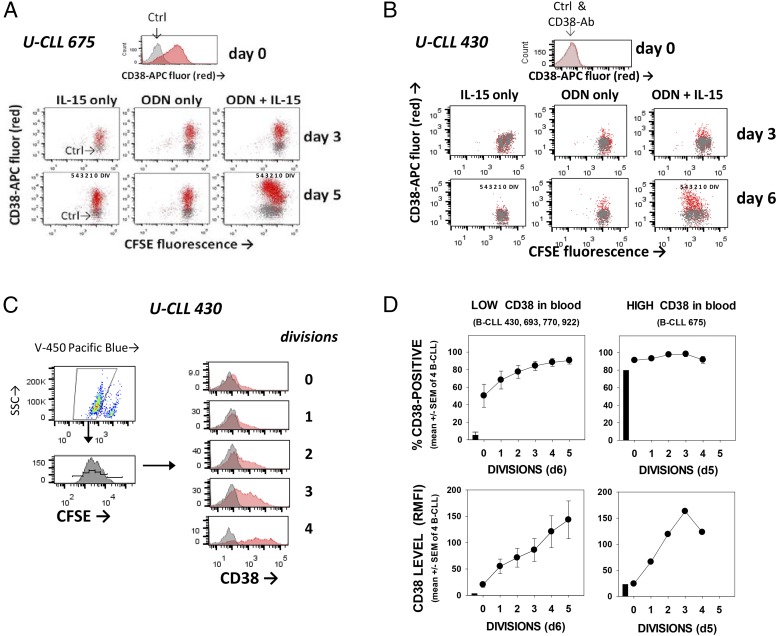
If collaborative ODN+IL-15 signaling is a driver of in vivo B-CLL growth, one would anticipate these stimuli to upregulate CD38. Through two-color flow cytometry of CFSE-labeled B-CLL stained for surface CD38 after activation, we found that both CD38 expression levels and the percentage of CD38+ cells progressively increase with increased division cycles (Fig. 6). On average, CD38 levels in cycling blasts were 1–2 logs greater than in cells exposed to either ODN or IL-15 (Fig. 6A, 6B). Furthermore, CD38 expression rose upon division, even when the blood-isolated quiescent B-CLL population was predominantly CD38+ (U-CLL 675 = 74% positive). Even so, not all proliferating B-CLL uniformly became CD38+, as can be seen in the case of U-CLL 430, a clone with few CD38+ cells in blood (Fig. 6C, 6D). Taken together, we conclude that exposure to ODN+IL-15 is sufficient for inducing division-linked CD38 expression in B-CLL leukemic cells. Nonetheless, expression of CD38 is not restricted to these stimuli because it is also inducible after B-CLL stimulation by IL-2 (80), and particularly CD40L + IL-4 (81) and autologous activated T cells (6). In addition, and of interest, although most extensively DIV B-CLL blasts express CD38, intraclonal heterogeneity can exist, leaving a fraction of cells to undergo at least three division cycles without upregulating the surface enzyme. This latter evidence that division can proceed without the upregulation of CD38 is consistent with results of a study using 2H (deuterium)-labeled water to monitor B-CLL divisions within patients (82). In that study, division was detected in both CD38+ and CD38− B-CLL, although CD38+ cells appeared to proliferate more rapidly (82).
FIGURE 6.
Membrane expression of CD38 rises in replicating blasts during ODN+IL–15-induced B-CLL clonal expansion. (A and B) Cell-surface CD38 levels were assessed on gated viable cells at time 0 and after 3–6 d of CFSE-labeled B-CLL culture with stimulant: IL-15 only, ODN only, or both ODN+IL-15. U-CLL 675 and U-CLL 430 represented B-CLL with relatively high and low proportions, respectively, of CD38+ cells in blood (anti-CD38 = darker shading; IgG control = light). Both populations appear to notably upregulate CD38 expression during divisions triggered by ODN+IL-15. (C) ODN+IL-15–stimulated U-CLL 430 cells were stained for CD38 and gated for viability and division status (on the basis of CFSE fluorescence). Histograms represent the fluorescence intensity of CD38-stained (or IgG control-stained) cells within each division subset and show definitively that CD38 expression rises with each incremental division. (D) Summary of experiments monitoring CD38 expression in ODN+IL-15–stimulated B-CLL cultures (n = total of 5 CLL; top panels represent % CD38+ cells in each division subset; bottom panels represent CD38 level, represented by the ratio of allophycocyanin anti-CD38 fluorescence intensity/allophycocyanin-IgG control fluorescence intensity).
Factors that regulate strength of the B-CLL proliferative response to ODN+IL-15
Initial experiments to test for ODN+IL-15 synergy in promoting B-CLL growth revealed considerable diversity between clones in the extent of cycling. Diversity could not be simply explained by whether the B-CLL specimen was isolated from fresh blood or after storage in liquid nitrogen: in several B-CLL that underwent replicate testing under both conditions, relatively minimal, if any, interexperimental differences in viability and/or division potential were noted (Supplemental Fig. 2). Therefore, other explanations were investigated for the differences between B-CLL clones.
No influence of IGHV mutated/unmutated status.
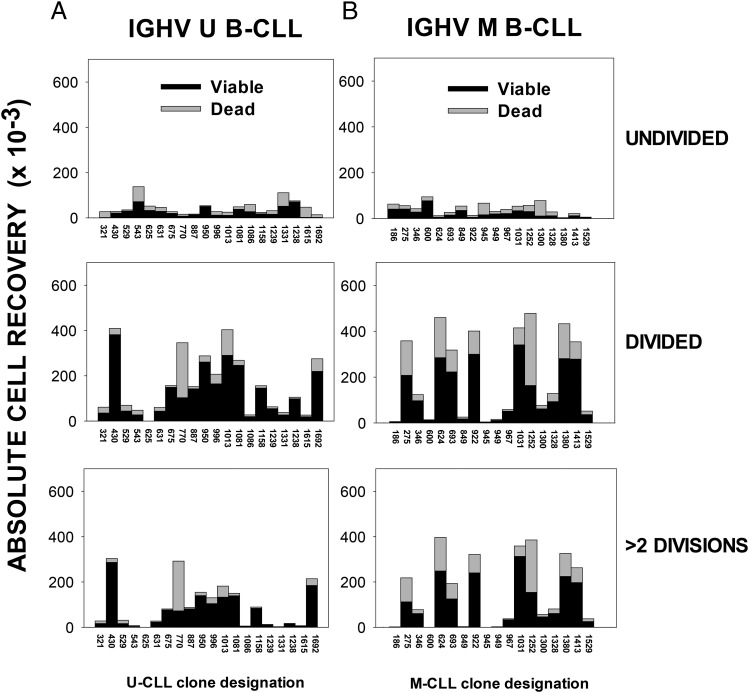
As indicated earlier, IGHV mutation status is a strong predictor of B-CLL in vivo growth (4, 5, 83): U-CLL > M-CLL. Therefore, despite early impressions that U-CLL and M-CLL could manifest relatively similar responses to combined ODN + IL-15 signaling (Fig. 1), it was critical to use a larger group of clones to adequately test the null hypothesis. Characteristics of the larger group of 40 CLL (20 U-CLL and 20 M-CLL) are shown in Table I, and a summary of our findings concerning the in vitro growth of these clones is presented in Fig. 7A and 7B, respectively. Results are shown in stacked bar format as absolute recovery of viable (black) and dead (gray) cells within subpopulations gated on the basis of division: undivided (Fig. 7, top row), total DIV (middle row), and more than two divisions (bottom row). Individual CFSE histogram profiles of each B-CLL population’s response can be found in Supplemental Fig. 3. Together, these studies showed that M-CLL and U-CLL both exhibit significant interclonal variation in responses. A statistical analysis of the viable or dead cell yield within each of the division-based groups, using the Mann–Whitney rank sum test, showed no statistically significant differences between the U-CLL and M-CLL subgroups. In addition, when U-CLL and M-CLL cohorts were compared on the basis of percentage viability within the undivided or DIV subsets or percentage DIV within total viable cells (criteria used in prior studies with normal B cells) (55–57), no statistically significant difference emerged (data not shown). Taken together, we find no evidence that the B-CLL clone’s BCR mutation status is independently linked to vigor of the ODN+IL-15–induced in vitro response. These findings suggest that if adequate amounts of CpG DNA and IL-15 are found within patient lymphoid tissues, leukemic cells from both U-CLL and M-CLL subsets should be driven to replicate, survive, and generate CD38+ progeny, unless otherwise suppressed or negatively regulated by other stimuli or cell interactions.
FIGURE 7.
IGHV unmutated and mutated B-CLL subsets both display notable interclonal diversity in ODN+IL-15–induced growth. CFSE-labeled (A) U-CLL cells (n = 20) and (B) M-CLL cells (n = 18), each at 105 cells/culture, were stimulated for 6–7 d with ODN+IL-15 before harvest with standardization beads, fixation, and FACS analysis for division status and viability (as in Supplemental Figs. 1, 3). A few cultures manifesting earlier peak division were harvested at day 5 to prevent death from nutrient deprivation. Experimental data shown represent the absolute recovery of gated viable or dead cells within the designated categories: undivided, total DIV, or DIV with more than two divisions. The data are shown in stacked bar format for each B-CLL clone (indicated by ID on the abscissa). (Note: M-CLL 1258 and M-CLL 1260 are not represented because experiments with the latter B-CLL did not include standardization beads). SEM of triplicate/quadruplicate cultures was nearly always <15% of the mean value, and most typically <5% (as in Fig. 1G). On rare occasions when the calculated absolute cell yield in one replicate varied >2-fold from the others, it was determined to be an outlier and excluded. Mann–Whitney rank sum tests were performed to evaluate whether the U-CLL and M-CLL subgroups varied in their responses within any one measurement category. No statistically significant differences were noted.
Effect of B-CLL chromosomal anomalies on the in vitro response to ODN + IL-15.
Several chromosomal anomalies are frequently found in B-CLL: del13q14, TRI-12, del11q22, and del17p. Possession of monoallelic del13q14 alone is generally, albeit not uniformly, linked to less aggressive disease (84, 85), whereas TRI-12, del11q22, and del17p more typically predict worse prognosis (86). We hypothesized that the relative potential of a B-CLL clone to mount a robust in vitro response to ODN+IL-15 might be linked to expression of pathogenic anomalies in at least a fraction of the total IGHV-determined clonal population. Because of the evidence that p53 activation negatively affects the survival of normal replicating human B lymphoblasts (57, 87), it warrants noting that TRI-12 increases the gene dose for Mdm2 (88), a major p53 inhibitor (89). Furthermore, del11q22 removes the gene for ATM (90, 91), a major p53 activator after DNA damage or oxidative stress. Deletion of 17p removes the p53 gene itself (86). Finally, del13q14 removes miR15/16, which negatively regulate Bcl-2, Mcl-1, and certain cell cycle proteins, as well as often removing DLeu7, an NF-κB inhibitor (reviewed in Ref. 92).
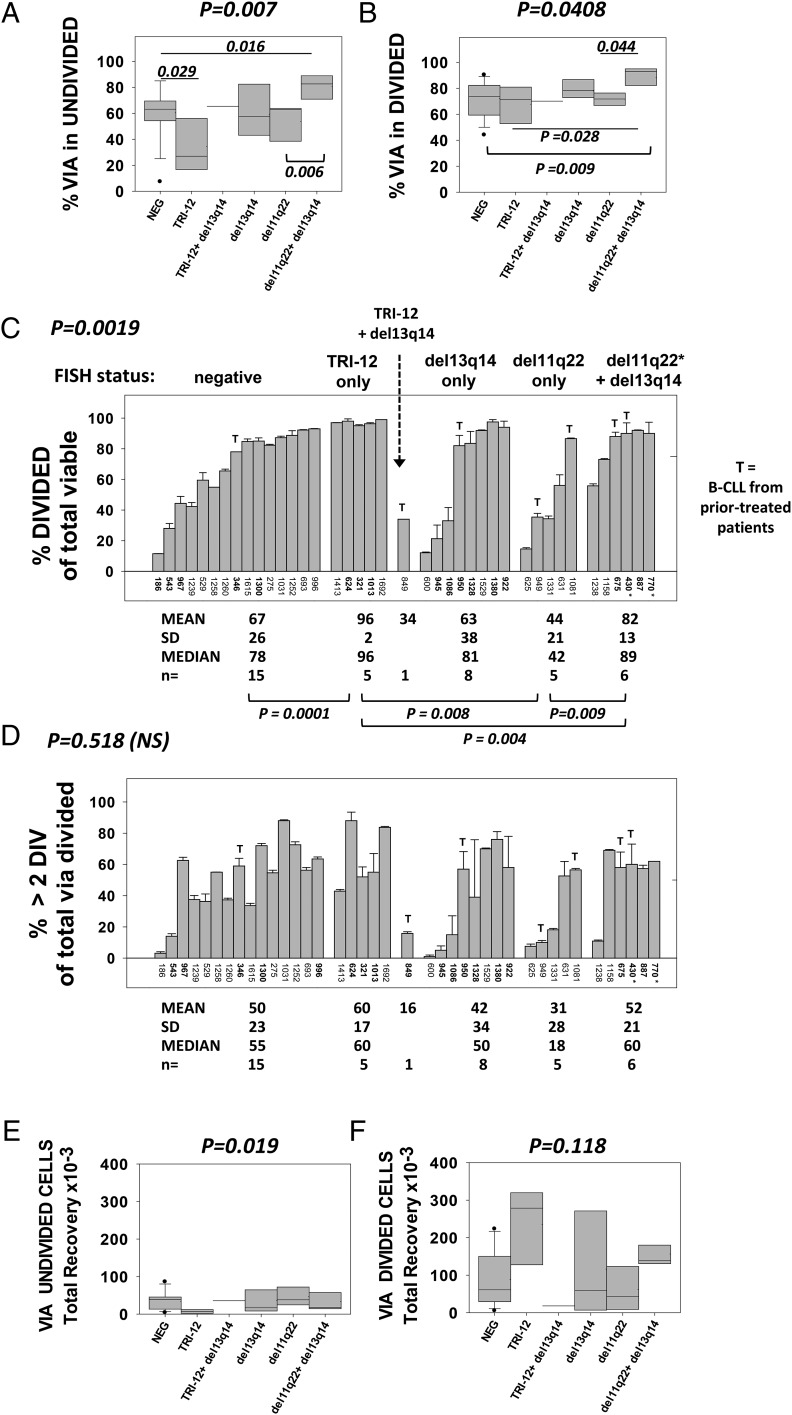
Insights into how genetic anomalies found in our cohort (Table I) affect the ODN+IL-15–induced response came from the data in Fig. 8. Populations were grouped into those with: 1) no FISH-determined anomalies (n = 15), 2) TRI-12 alone (n = 5), 3) TRI-12 and del13q14 (n = 1), 4) del13q14 alone (n = 8), 5) del11q22 alone (n = 5), and 6) both del11q22 (or ATM mutation) and del13q14 (n = 6). The viability of undivided and DIV B-CLL cells after 6–7 d of culture is shown in Fig. 8A and 8B, respectively, whereas Fig. 8C–F reveals the extent of B-CLL cycling, as represented in three diverse ways: 1) percentage DIV cells within the total viable gate (Fig. 8C); 2) percentage of total viable DIV cells with >2 divisions (Fig. 8D); and 3) absolute yield of viable DIV cells per culture (Fig. 8F). The absolute yield of viable undivided cells is also shown (Fig. 8E).
FIGURE 8.
Effect of common B-CLL genetic anomalies on survival/growth properties of ODN+IL-15–stimulated B-CLL clones. B-CLL clones in Table I were subdivided on the basis of FISH analyses for TRI-12, del13q14, and del11q22 (ATM). B-CLL849 was the only TRI-12+ population additionally positive for del13q14. It was considered independent of the group expressing TRI-12 alone (n = 5) and not included in any of the statistical evaluations. *CLL770 was placed in the group with both del13q14 and del11q22 on the basis of harboring two ATM mutations. *CLL430 possessed both del11q22 and del13q14 and one ATM mutation. B-CLL subgroups were compared for % viability in the (A) undivided and (B) DIV cell gates; (C) % of viable-gated cells showing evidence of division; (D) % of DIV viable cells with more than two divisions; (E) absolute recovery of viable, undivided cells per culture; and (F) absolute recovery of viable DIV blasts per culture. (C and D) The responses of B-CLL evaluated in a single experiment are shown by bars representing the intraexperimental mean ± SD of triplicate cultures; values for other B-CLL in bold (ID 346, 1300, 624, 321, 1013, 945, 1086, 940, 1328, 1380, 922, 675, 430, 887, and 770) represent the mean ± SD value from two to four separate experiments with the same B-CLL. The T atop certain bars indicates that this B-CLL population derived from a patient who received treatment at ≥8 mo prior to acquisition of leukemic B cells from blood (see Table I). The p values placed above each plot are derived from the Kruskal–Wallis test for differences within the multiple groups and represent Monte Carlo estimates for the exact test. The p values within (or below) the plots are derived from Wilcoxon rank sum test for differences between paired groups, as indicated in Materials and Methods.
Several statistical analyses were performed to discern whether differences between B-CLL clones in each of these functional parameters could be attributed to the FISH subgroups. An initial Kruskal–Wallis statistical test was performed to assess whether there was statistical variation between the five major groups (see Fig. 8 legend). This revealed significant differences among the subgroups in percent viability within undivided cells (Fig. 8A, p = 0.007), percent viability within DIV blasts (Fig. 8B, p = 0.0408), percentage DIV within total viable cells (Fig. 8C, p = 0.0019), and absolute yield of viable undivided cells (Fig. 8E, p = 0.019). Although variation was apparent in the absolute yield of viable DIV cells (Fig. 8F), this did not reach statistical significance (p = 0.118). Further statistical evaluations using a Wilcoxon rank sum test, which made pairwise comparisons of the subgroups, delineated which genetic anomalies were linked to notably greater viability and/or growth.
A trend toward heightened viability in undivided cells (Fig. 8A) and DIV blasts (Fig. 8B), above that in the FISH-negative group, was seen in B-CLL clones jointly expressing del11q22 (± ATM mutation) and del13q14. For both undivided and DIV cells, this was unlikely due to chance (p = 0.016 and p = 0.009, respectively), although with a Bonferroni correction for multiple comparisons set at 0.005, these were not statistically significant. B-CLL clones bearing both an ATM anomaly and del13q14 also trended toward greater viability in the undivided and DIV fractions than those expressing del11q22 alone (p = 0.006 and p = 0.044, respectively; Fig. 8A).
Interestingly, B-CLL expressing only TRI-12 showed significantly greater propensity for division in response to ODN+IL-15 (Fig. 8C; p = 0.0001 for TRI-12 versus FISH−, p = 0.004 for TRI-12 versus del11q22 (± ATM mut) + del13q14, and p = 0.008 for TRI-12 versus del11q22 alone). No other group differed significantly from the FISH− group in terms of percent division in the viable-gated fraction, although joint expression of an ATM anomaly and del13q14 appeared to favor greater division when compared with clones with del11q22 only (p = 0.009; Fig. 8C). Taken together, this analysis suggests that TRI-12 enhances the potential for in vitro ODN+IL-15–stimulated B-CLL division, whereas an ATM anomaly combined with del13q14 promotes greater viability and perhaps greater division.
The frequency of FISH-determined anomalies within cells of a clone is quite heterogenous (Table I). This diversity likely contributes to the intraclonal differences observed in percentage viability and/or extent of cycling (e.g., in Fig. 1A and Supplemental Fig. 3). It is additionally possible that intraclonal variations represent inherent differences in subclonal members that relate to differing surface membrane receptors or signaling status, that are either fixed or temporary, for example, based on time since last replication (93). It is unlikely that most differences in the extent of cycling within a given B-CLL population represent contaminating nonleukemic cells, because diversity was evidenced in cultures that were 99–100% CD19+/CD5+ before activation, as well as in day 5 activated cells stained positively for CD19/CD5 (data not shown).
Effect of prior treatment on ODN+IL-15–induced in vitro B-CLL clonal expansion.
Prior therapy has been linked to the emergence of more malignant B-CLL subclones. Because a minor proportion of our cohort (7/40) represented leukemic cells taken from patients with earlier treatment (8 to 418 mo prior to blood sampling; Table I), it was deemed relevant to examine whether a more robust ODN+IL-15 response was statistically linked to prior treatment. In Fig. 8, those B-CLL clones taken from patients receiving earlier treatment are represented by T above the bar delineating each clone’s response. From the dotted line thresholds for separating relatively high versus relatively low growth (Fig. 8C, 8D), it can be discerned that 71% (5/7) of the B-CLL from pretreated patients showed extensive ODN+IL-15–induced replication. Nonetheless, there was no statistically significant linkage between prior treatment and the assessed growth parameters using a Fisher exact test.
Because malignancies may develop successive mutations over time, even independent of therapy, it was additionally relevant to examine whether sampling time after initial diagnosis affected the propensity to proliferate in response to ODN+IL-15. From regression analyses involving either the total B-CLL cohort (n = 40) or B-CLL segregated on the basis of FISH anomalies, we found no definitive link between time after diagnosis and greater division in response to ODN+IL-15 (data not shown). Further statistical evaluations with the Fisher exact test showed that B-CLL cells (both U-CLL and M-CLL) obtained at ≤50 or >50 mo postdiagnosis exhibit no significant difference in growth potential, as discerned by percentage of total viable cells with more than two divisions (data not shown). Thus, within the presently studied B-CLL cohort, there was no statistical evidence that B-CLL populations taken from a late point in the disease process showed a significantly more robust response to ODN+IL-15. Nonetheless and importantly, this assessment may be compromised by limited sample number. Better insights on whether in vitro growth potential is altered with time after diagnosis will require a longitudinal study of individual B-CLL populations.
Does ODN+IL-15–stimulated in vitro growth of B-CLL clones correlate with in vivo GR in the patient?
B-CLL in vivo GR was estimated from B-CLL doubling time in blood (see Materials and Methods). To most accurately relate a population’s in vitro growth to its replication in vivo, we took measurements at or near the time blood was acquired for subsequent in vitro experiments; this could be done in all but two cases (latter = 1–2 y after sample). Nonetheless, it must be kept in mind that other factors influence blood doubling time, for example, rates of survival, release from lymphatic tissue, and retention in nontissue compartments.
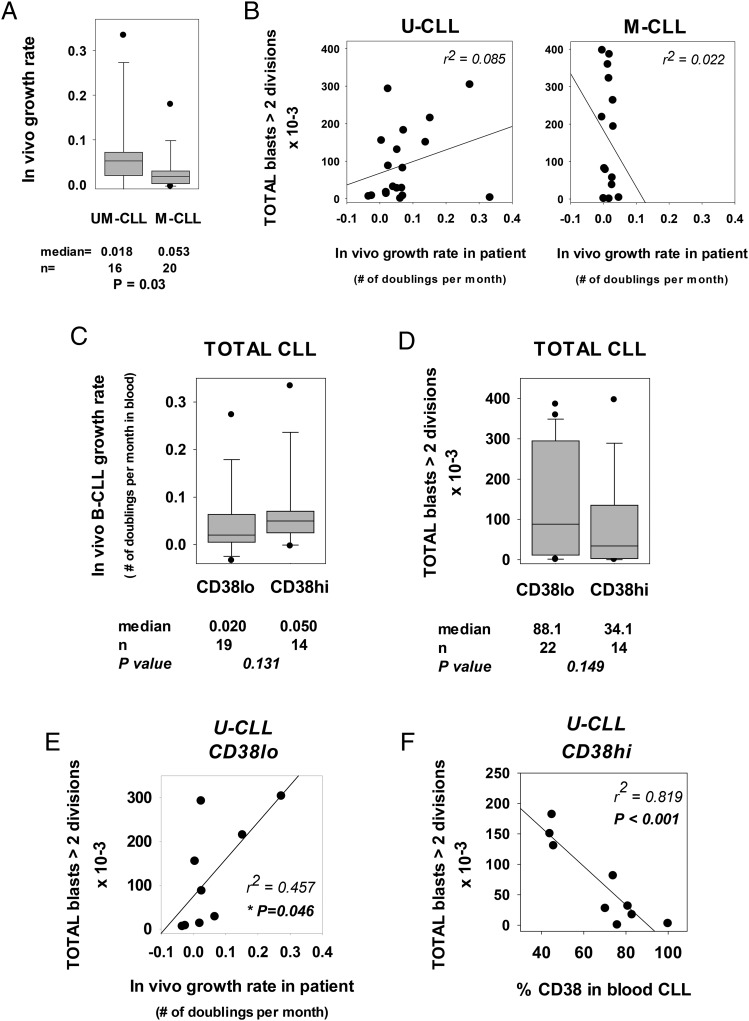
A regression analysis comparing in vivo GR with in vitro growth (absolute yields of CLL lymphoblasts with more than two divisions) is shown in Fig. 9. Evaluations were made for U-CLL and for M-CLL separately, because GRs of the latter two subsets differ significantly (4), as confirmed in our CLL cohort (Fig. 9A). Importantly, when in vivo GR of individual clones is compared with respective levels of in vitro growth, very different regression curves were obtained for the U-CLL and M-CLL subsets. Within the M-CLL subset, there was no correlation between in vivo GR and robustness of the ODN+IL-15–induced response (Fig. 9B, right panel). In fact, M-CLL clones with vigorous and with weak ODN+IL-15–induced responses had equally low GR. The latter indicates that the characteristic slow clinical progression of M-CLL must, at least in some cases, reflect factor(s) other than a clone’s intrinsic ability to respond to growth-promoting stimuli. In contrast, for U-CLL clones, there was a suggestion that in vivo and in vitro growth are directly linked, although statistical significance was not reached (Fig. 9A, left panel).
FIGURE 9.
B-CLL intrinsic potential for ODN+IL-15–induced growth versus in vivo GR and expression of CD38+ cells in blood. (A) The in vivo GR of U-CLL (n = 16) and M-CLL (n = 20) clones are compared in box-plot format; they were statistically different (p = 0.03). These clones represent those from which serial blood lymphocyte counts at or near the time of blood acquisition for functional study were available. (B) Potential for in vitro clonal expansion, as indicated by the absolute yield of total lymphoblasts (viable + dead) in ODN+IL-15–stimulated cultures (y-axis) was compared with the in vivo B-CLL GR (x-axis), calculated from the lymphocyte doubling time in blood (see Materials and Methods). No statistically significant relationship was observed, although the suggestion of a positive relationship was seen in some U-CLL clones. (C and D) B-CLL clones, whose blood leukemic cells had been assayed for CD38 expression at or near the time of blood acquisition for these studies, were subdivided into CD38high and CD38low subsets and compared for (C) in vivo GR and (D) ODN+IL-15–induced in vitro clonal expansion. These comparisons show a trend (albeit not of statistical significance) for CD38high B-CLL to show greater in vivo GR and lesser in vitro growth, as compared with CD38low B-CLL. We note that when the total CLL cohort was first subdivided into U-CLL and M-CLL subsets and then examined in a similar manner, similar trends were observed: 1) in vivo GR: for U-CLL, medians = 0.025 and 0.063 for CD38low and CD38high, respectively; for M-CLL, medians = 0.018 and 0.027, respectively; and 2) in vitro yield of highly DIV cells: for U-CLL, medians = 119,400 and 68,900 for CD38low and CD38high, respectively; for M-CLL, medians = 137,000 and 37,200, respectively. No statistical significance was reached with the limiting sample size. (E) Regression analysis showing the relationship between in vitro and in vivo growth within CD38low U-CLL. A statistically significant relationship was seen (p = 0.046). (F) Regression analysis showing the relationship between absolute frequency of CD38+ cells within CD38high U-CLL and ODN+IL-15–induced clonal expansion. A highly significant inverse relationship was found (p < 0.001).
Recent in vivo activation of B-CLL appears to compromise ODN+IL-15–induced in vitro clonal expansion
We wished to understand why a clone’s intrinsic potential for ODN+IL-15–stimulated growth was not in greater alignment with its apparent growth in the patient. One possibility was that the relatively recent emergence of blood B-CLL from a proliferative experience in tissue might be a factor influencing the robustness of their in vitro response. Recent activation might either accelerate (prime) responses to ODN+IL-15 or alternatively dampen them, for example, as a result of proliferative senescence (94).
As an indicator of relatively recent in vivo proliferation, we considered the proportion of blood leukemic cells expressing CD38. Although blood B-CLL are uniformly small, expression of this marker has been strongly linked to recent B-CLL growth (75, 76). B-CLL clones were subdivided into CD38high and CD38low groups, on the basis of a 30% threshold. The latter is often used to distinguish CLL with fast and slow clinical progression, CD38high generally > CD38low (77, 95, 96). CD38high and CD38low subgroups were compared in box-plot format for in vivo GR (Fig. 9C) and separately for in vitro expansion in response to ODN+IL-15 (Fig. 9D). As expected, CD38high B-CLL had a median in vivo GR that was 2.5-fold greater than CD38low B-CLL (Fig. 9C), although the difference did not reach statistical significance, likely because of limiting cohort size. Unexpectedly and to the contrary, extended in vitro growth in ODN+IL-15–stimulated CD38high clones was 39% of that seen with CD38low clones (Fig. 9D). This trend for CD38high clones to proliferate less vigorously than CD38low clones was seen in both U-CLL and M-CLL subsets, although as expected with diminished sample size, statistical significance was not reached (data not shown).
Based on the above insight, we re-examined whether a direct relationship exists between in vitro and in vivo growth using only CD38low U-CLL clones. The U-CLL clones were of particular interest because we envisioned that the BCR specificity of this group makes it less likely that limited access to stimuli will be a critical factor controlling in vivo growth. Unlike the more restricted BCR specificities of M-CLL clones, U-CLL exhibit polyreactive BCR that can bind several self-Ags, including many expressed on stressed and apoptotic cell membranes (19–21, 28). Interestingly, with this approach, a statistically significant link was found between GR in the patient and ODN+IL-15–triggered growth (p = 0.046; Fig. 9E). When the CD38high U-CLL subset was considered, no such relationship was apparent (data not shown). Rather, a clone (CLL 1331) with the most rapid in vivo GR (0.3337), and the correspondingly highest percentage CD38+ cells in blood (100%), showed negligible growth in response to ODN+IL-15 (yield of 2410 blasts with >2 divisions).
The latter two observations prompted us to examine whether having a very high proportion of CD38+ cells might compromise a U-CLL clone’s in vitro response to ODN+IL-15. Indeed, when clones with evidence of recent activation history were examined (those >30% CD38+), a highly significant inverse relationship was found between the percent of CD38+ cells and B-CLL in vitro growth (p < 0.001; Fig. 8F). Parenthetically, when M-CLL were similarly examined, four of five (80%) of the CD38high clones were very poor responders (data not shown).
Taken together, the earlier CD38-based assessments suggest that if the U-CLL blood population possesses relatively few CD38+ cells (due to either a substantial interval between the last proliferative burst or poor CD38 upregulation during recent cycling), then the leukemic population’s in vitro growth response to ODN+IL-15 is a good indicator of the U-CLL clone’s overall GR in the patient. In contrast, if most blood leukemic cells represent CD38+ progeny of a relatively recent proliferative burst, then they are compromised in responding to ODN+IL-15 with a new burst of growth. The reasons for these differences can presently only be speculated (see Discussion) and will require further study.
Impact of genetic anomalies on in vitro growth of CD38high and CD38low U-CLL
Although our cohort size is too limited to undertake a multivariate statistical analysis, we believe some discussion of the distribution of chromosomal anomalies in the CD38low and CD38high subsets is warranted. The earlier analysis in Fig. 8 showed that a robust B-CLL response to ODN+IL-15 is linked to TRI-12 and to the combination of del11q22 + del13q14. Therefore, it was possible that these anomalies mitigate the negative influences of elevated CD38 expression on ODN+IL-15–induced cycling.
When the CD38low U-CLL subgroup in Fig. 9E was inspected for FISH anomalies, we found that 80% of those clones with strong in vitro growth possessed either TRI-12 or both del11q22 (or ATM mutations) and del13q14. The latter clones (CLL 1692, 430, 770, 950, and 887) are represented by the black circles on or above the regression line. Only 25% (1/4) of this subgroup’s members with poor in vitro growth possessed these anomalies. The earlier findings substantiate the link between vigor of the in vitro response and these genetic anomalies. Importantly, they also indicate that expression of these anomalies within a clone is not uniformly linked to high levels of CD38+ cells in blood.
Within the CD38high U-CLL subgroup (plot not shown), 75% (3/4) of the clones with the greatest in vitro growth (81–182 × 103 blasts with more than two divisions) expressed either TRI-12 or the combination of deletions in 11q11 and 13q14 (CLL 1013, 675, 1081). This contrasted with this subgroup’s clones manifesting the weakest in vitro growth (0.02–31 × 103 blasts >2 divisions) (CLL 631, 529, 1238, 625, and 1331). Of interest, 80% (4/5) of the latter expressed del11q22 alone, without accompanying del13q14, in 70–100% of the blood leukemic cells (Table I), as well as a very elevated frequency of blood CD38+ cells (mean ± SD = 82 ± 11% positive). The possibility that TRI-12 or combined deletions in 11q and 13q override the negative influences of CD38 will need to be examined through future FISH analyses of leukemic blasts sorted on the basis of CFSE expression. Nonetheless, there are strong indications that CD38+ B-CLL bearing del11q22 alone are compromised in their in vitro growth to ODN+IL-15.
Comparison of in vitro viability/growth characteristics of ODN+IL-15–stimulated B-CLL clones with clinical progression in patients
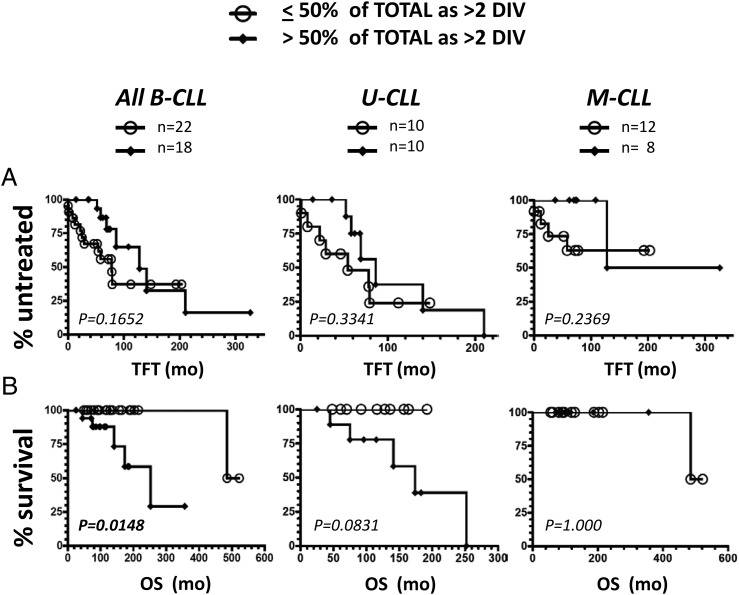
The best evidence to date that in vivo leukemic cell birth rate is linked to disease activity comes from experiments that measured leukemic cell uptake of deuterium in patients drinking heavy water (4, 83). Prompted by the strong suggestions that IL-15 and ODN signaling could contribute to pseudofollicle growth, we examined whether a clone’s intrinsic potential for in vitro ODN+IL-15–induced clonal expansion correlates with the patient’s disease progression. Kaplan–Meier plots, with accompanying log-rank statistical evaluations, were used to assess whether TFT or OS of the patient is linked to the B-CLL clone’s high-proliferator versus low-proliferator status in vitro. Of note, CD38high B-CLL with compromised in vitro growth were not excluded from these analyses because of the need to retain a relatively large sample size.
In Fig. 10, we compare the clinical outcome of the full CLL cohort, and segregated U-CLL and M-CLL subsets, with the frequency of highly DIV viable blasts in ODN+IL-15–stimulated cultures. Both TFT (Fig. 10A) and OS (Fig. 10B) were considered. The Kaplan–Meier plots reveal no statistically significant TFT difference within patients harboring high- versus low-proliferating clones for the full B-CLL cohort or its U-CLL or M-CLL subsets (Fig. 10A). Nonetheless, significantly worse OS was noted in patients with high-proliferator clones, when the full cohort of CLL was assessed (p = 0.015; Fig. 10B, left plot). Separation of the full cohort into its U-CLL and M-CLL components suggests that accelerated patient death associated with high-proliferating clones is a characteristic of U-CLL and not M-CLL, even though statistical significance was not reached in the case of U-CLL clones (p = 0.083). A lack of statistical significance for the U-CLL comparison likely reflects reduced sample size (n = 20 versus n = 40 for the full cohort). This conclusion is supported by a significant difference in OS values for high- versus low-proliferator U-CLL clones (p = 0.04), when a slightly different discrimination threshold was used (≤75% or >75% DIV within total viable cells). Furthermore, high and low proliferators in the M-CLL subset showed no significant difference with either threshold (p = 1.000).
FIGURE 10.
In vitro high-proliferator status of U-CLL, but not M-CLL, is statistically linked to overall patient survival. The 40 B-CLL clones tested for in vitro clonal expansion in response to ODN+IL-15 were analyzed for clinical progression using (A) TFT and (B) OS as indicators. The total cohort and U-CLL and M-CLL subsets were each subdivided into high-proliferator clones (>50% of total viable cells with more than two DIV) or low-proliferator clones (≤50% of total with more than two DIV). For OS, the differing groups were distributed in the following manner: total B-CLL: low = 21 censored, 1 death; high = 13 censored, 5 deaths; U-CLL: low = 10 censored, 0 deaths; high = 5 censored, 5 deaths; M-CLL: low = 11 censored, 1 death; high = 8 censored, 0 deaths. The U-CLL populations with diminished OS included CLL 1013 (TRI-12), CLL 430 (del11q22 + del13q14 + ATM mutation), CLL 675 (del11q22 + del13q14), CLL 1158 (del11q22 + del13q14), and CLL 996 (negative for tested FISH anomalies). Kaplan–Meier analysis was performed to discern whether the high-proliferator subgroup significantly differed from the low-proliferator subgroup regarding TFT or OS. No significant difference between subgroups was noted for TFT. Nonetheless, patients with high proliferator clones (total cohort and U-CLL cohort) appeared to have shortened OS as compared with those that did not (p = 0.015 for total clones was statistically significant; p = 0.083 for U-CLL clones approached significance). There was no significant difference in the OS of M-CLL clones with high- or low-proliferator status (p = 1.00).
Discussion
Despite their blood manifestation as relatively quiescent lymphocytes, a fraction of each B-CLL clone undergoes significant proliferation in the patient, associated with varying degrees of clonal contraction (4, 5). Bursts of proliferation within lymphoid tissue pseudofollicles are linked to clinical progression (11, 97), and this continues to elicit interest in defining the mechanisms for localized B-CLL growth. We report for the first time, to our knowledge, that two stimuli, which are likely present in all primary and secondary lymphoid tissues, display notable synergy in promoting in vitro B-CLL cycling. In lymphoid tissues, CpG DNA could be available from apoptotic cells/debris and/or microbes (98–102), and IL-15 is anticipated from stromal cells, dendritic cells, and macrophages (43–45). In support of the above, to our knowledge, this study provides the first immunohistochemical evidence that apoptotic cells and IL-15–producing cells are present in B-CLL–infiltrated spleens. Our in vitro functional experiments with CFSE-labeled B-CLL show that significant ODN+IL-15–induced growth can be elicited from both IGHV-unmutated and IGHV-mutated B-CLL subsets, the latter of which is characteristically vulnerable to apoptosis upon exposure to CpG DNA alone (35, 37, 40, and this report). The novel evidence for CpG DNA + IL-15 synergy in promoting B-CLL growth is in harmony with past findings that normal human memory B cells cycle extensively in response to CpG and IL-15 (49) and further evidence that B-CLL cells manifest gene expression profiles resembling memory B cells (50). In addition, our observations are consistent with evidence that both U-CLL and M-CLL manifest high levels of TLR-9 (35, 39) and that B-CLL express high- and low-affinity receptors for IL-15 (51, 52). IL-15 is best recognized as a growth-promoting cytokine for cells involved in immune surveillance: NK cells and CD8 cytotoxic cells (73, 103). In the case of patients with B-CLL, the malignant clone could usurp IL-15 for its own expansion (schematic in Fig. 11).
FIGURE 11.
Schematic illustrating hypothesized role for TLR and IL-15 signaling and genomic aberrations that mitigate p53 axis function for B-CLL growth within lymphoid tissue pseudofollicles. Synergy between CpG DNA from microbes and/or apoptotic cells and IL-15 bound to membranes of IL-15–producing cells (43, 103) and/or in the form of cleaved IL-15Rα/IL-15 complexes (71) may constitute an important external stimulus for promoting the growth of B-CLL leukemic cells within tissue pseudofollicles. B-CLL clonal expansion is likely strongly influenced by factors intrinsic to the leukemic cells themselves, for example, presence of genomic aberrations that counteract activation-linked, lymphocyte apoptosis and cell-cycle control. In addition, interactions with PECAM-expressing endothelial cells may protect recently proliferated CD38high cells from functional senescence and/or help sustain their survival (150).
The intracellular mechanisms responsible for the synergy between CpG ODN and IL-15 in eliciting B-CLL growth are presently under investigation. Nonetheless, important insights can be derived from past investigations. M-CLL clones, predisposed to apoptosis after early TLR-9 activation (104), show weaker ODN-induced activation of Akt and MAPKs (ERK, JNK, and p38) than do U-CLL clones characterized by more sustained viability (35). Other observations that treatment of B-CLL with IL-15 induces phosphorylation of ERK1/2 (52) and AKT (R. Gupta and P. Mongini, manuscript in preparation) suggest that IL-15 signals might compensate for this poor ODN-induced signaling. In addition, because a TLR-9→NF-κB→IL-10→Stat1 pathway can trigger ODN-induced B-CLL apoptosis (38), it is likely relevant that IL-15 promotes the upregulation of several survival molecules, Bcl-2, Bcl-x, and Mcl-1, in other cell lineages (47, 105, 106). Furthermore, in CD8 T cells, IL-15 promotes increases in c-Myc (107) and telomerase (108), two molecules linked to in vivo B-CLL pseudofollicle growth (11, 109, 110). An IL-15–related cytokine, IL-2, augments CLL responses to ODN (41, 104) through a mechanism involving the heightened expression of cyclin D2, D3, and cyclin-dependent kinase, cdk4 (41). Given that both IL-15 and IL-2 signaling involves the shared IL-2/IL-15R β− (CD122) and common γ (CD132)-chains (72), IL-15–promoted growth might involve similarly modulated cell-cycle proteins. Nonetheless, we find that IL-15 is superior to IL-2 in both functioning at lower doses and yielding more robust responses than IL-2 in ODN-triggered B-CLL cultures (R. Gupta and P. Mongini, manuscript in preparation).
An important discovery was that spleens of B-CLL patients possess IL-15–producing cells scattered throughout the B-CLL–infiltrated white pulp. Through examining serial tissue sections, we detected cells with cytoplasmic IL-15 located proximal to and often within B-CLL pseudofollicles bearing Ki-67+ cells. This suggests an in vivo role for IL-15 in promoting B-CLL growth. Our additional finding that two normal human spleens also possessed IL-15+ cells scattered throughout the white pulp is quite consistent with the recent findings of Cui et al. (44) in mouse spleen. Using novel IL-15 reporter mice, the latter investigators found the IL-15 promoter is active within nonlymphoid cells of normal spleens; this was particularly noted in VCAM-1+ stromal cells, where reporter expression increased as mice aged (44). In our studies, some of the splenic IL-15+ cells possessed an elongated appearance, consistent with past reports of IL-15+ myofibroblasts (111), dendritic cells (43), and endothelial cells (46, 112, 113) within lymphoid tissues. Other IL-15–producing cells might represent macrophages (114, 115), but further studies are needed for their identification.
Numerous studies have shown that nurse cells within the stroma of lymphoid tissue play critical roles in promoting B-CLL survival and growth (reviewed in Ref. 116). Stromal cell lines are often used to provide such prosurvival signals in vitro. In a set of recent experiments, not detailed in this article, we examined whether IL-15 was expressed and functional in HS-5 cells, a human bone marrow–derived stromal cell line with known prosurvival effects on B-CLL cells (117, 118). Using a commercial ELISA kit for human IL-15, we found that HS-5 culture supernatants contained low but significant levels of IL-15 (611 ± 163 pg/ml in three separate lots). Furthermore, an IL-15 neutralizing Ab significantly reduced B-CLL viability in cultures containing U-CLL (n = 2) and irradiated HS-5 cells (20:1 ratio) (R. Gupta and P. Mongini, unpublished observations). Thus, at least some stromal cell lines with nurse activity do produce functional IL-15.
Although in vivo B-CLL growth is primarily localized in primary and secondary lymphoid tissues, the lungs and skin of B-CLL patients often show atypical B-CLL infiltration. It may be important that the latter are locales where IL-15 levels rapidly increase postinfection. Within lungs, IL-15 is produced in alveolar macrophages and epithelial cells as a sequel to influenza and pneumococcal infections (119–121). High-affinity IL-15 binding to IL-15Rα on the membranes of all IL-15–producing cells (Kd = 10−11 M) (122) makes lungs particularly effective reservoirs for IL-15 (103). In the context our study’s findings, one might suspect that a combined surge in levels of both IL-15 and CpG DNA, for example, from microbes and apoptotic netting neutrophils, during lung infections could provoke ectopic B-CLL growth. Indeed, a statistical link between development of B-CLL and earlier pneumococcal lung infections has been reported (123). Possibly, synergy between TLR-9 and IL-15 signaling in pulmonary tissue explains why ∼30% of B-CLL patients show lung parenchymal infiltration with leukemic cells upon autopsy (124). Within skin, IL-15 levels have been elevated in Langerhans cells and keratinocytes after several routes of stimulation, including viral infection (125–127). Although speculative, synergy between TLR-9 (or related TLR) and this cytokine might contribute to the occasional emergence of skin-localized B-CLL follicles (124, 128, 129) at sites of mosquito bites and former herpes simplex or zoster lesions (130, 131).
Despite the uniform evidence of IL-15 and CpG DNA synergy, quite importantly, we also noted substantial diversity between individual B-CLL clones in the vigor of the ODN+IL-15–induced response. Through a systematic comparison of the leukemic populations for several parameters—IGHV mutation status, common B-CLL chromosomal anomalies, and recent in vivo proliferative history—we gained insights into the basis for this diversity and, correspondingly, factors important in regulating B-CLL clonal expansion.
First, we found that IGHV mutation status has no obvious effect on a leukemic population’s intrinsic responsiveness to CpG DNA and IL-15. Both U-CLL and M-CLL subsets contained individual clones that were either weak or strong responders. We believe it is important to emphasize that these findings are not in opposition to findings that U-CLL and M-CLL exhibit very different GRs in vivo (5, 77, 95). Rather, examples of robust in vitro proliferation in M-CLL clones with slow in vivo GRs support the notion that the availability of stimulatory signals within the in vivo milieu may be as relevant as a malignant cell’s intrinsic potential for cell-cycle turnover in regulating clinically relevant growth. In particular, the more highly specific BCRs of M-CLL (26–28) make it likely that the latter subset less frequently internalizes Ag-linked TLR ligand (e.g., CpG DNA) than the U-CLL subset, whose BCR are polyreactive and bind with low affinity to numerous Ags exposed on apoptotic/stressed cells (19–21). This interpretation is consistent with evidence that apoptotic cells/debris express surface CpG DNA (19, 22) and that the latter can foster the activation of normal B cells with low BCR affinity for self-Ags (32, 132). It is also consistent with gene array studies showing that the TLR-9 pathway is activated within B-CLL pseudofollicles (11).
A robust B-CLL response to ODN+IL-15 was statistically linked to the presence of chromosomal anomalies often found in B-CLL (133): TRI-12 alone or, alternatively, del11q22 (or ATM mutation) together with del13q14. These observations are consistent with past clinical evidence that TRI-12, del11q22, and ATM mutations are linked to bulky disease (significant lymphadenopathy; splenomegaly) and/or elevated leukemic blood counts, and worse patient prognosis (134–136). Nonetheless, they add an important dimension in providing more direct evidence that genetic anomalies influence the intrinsic potential of B-CLL cells to survive/expand. Although a link between the latter and OS might be expected, this is not necessarily the case and needs experimental support. Attributes other than the malignant cell’s intrinsic growth potential could influence patient survival. These include communications between B-CLL cells and their milieu (stroma, T cells, etc.) that affect leukemic cell survival, prevent immune rejection of the B-CLL, and block the development of normal hematopoietic cells.
We found that TRI-12 alone was linked to greater division potential, but not heightened viability of dividing blasts. Given the strong link between proliferation and mutagenesis, the greater replication seen in TRI-12+ B-CLL is consistent with the suggestion that TRI-12 is a driver mutation that facilitates the accrual of further downstream mutations (33). Heightened growth of TRI-12+ clones is also in agreement with recent findings that TRI-12+ B-CLL express elevated CD38 in blood (137). Interestingly, two recent studies found B-CLL with TRI-12 (or del11q22+) to express elevated levels of insulin growth factor receptor 1 (138, 139). The latter has pathobiological implications for B-CLL (140) and might additionally contribute to the heightened growth within our in vitro cultures with supplementary insulin (Materials and Methods).
In contrast with TRI-12, combined expression of an ATM anomaly (del11q22 ± ATM mutation) with del13q14 significantly improved the viability of cycling B-CLL. Although these combined anomalies also appeared to favor relatively robust division, this was below statistical significance. It was intriguing to find that B-CLL clones with del11q22 alone showed significantly lesser viability/growth than did clones with deletions in both 11q22 and 13q14. Given the numerous roles for ATM during cellular stress (91), it is possible that the consequences of a 13q14 deletion might be of particular advantage to leukemic cells with compromised ATM function. Providing some support to the latter is evidence from a past clinical study of B-CLL from untreated patients. Despite the fact that the cohort under study was composed of a greater number of clones with TRI-12 than with del11q, deletion of 13q was more frequently found together with del11q (141). In addition, the same study’s assessment of clonal evolution, by sequential FISH analyses of B-CLL populations, revealed one del13q+ clone that later acquired del11q+, and conversely, one del11q+ clone that later acquired del13q (141).
How might TRI-12 or ATM deletion/mutation directly influence B-CLL viability/growth during clonal expansion? The genes responsible for heightened in vivo pathogenesis of TRI-12+ B-CLL clones are poorly defined. Nonetheless, a past gene array analysis showed E2F1, BAX, P27, and CDK4 to be consistently overexpressed in TRI-12+ clones (86), with only two of these (CDK4 and P27) explained by a gene dosage effect (86). The study’s investigators suggested that the noted aberrations were most consistent with TRI-12 potentiating the proliferation of affected B-CLL (86). Our findings support this. Surprisingly, in the former study, cyclin D2 and Mdm2 (both encoded on Chr12) were not elevated when compared with other B-CLL (86). Nonetheless, because most B-CLL clones overexpress the latter two proteins (142, 143), it needs to be considered that distinct B-CLL use differing mechanisms for boosting the expression of these important genes, and that the gene dosage effect of TRI-12 on cyclin D2 and Mdm2 may have been missed.
Some insight into why TRI-12 and del11q22 aberrations are positively selected within leukemic clones can be gleaned from the evidence that an activated p53 axis plays an important role in curtailing the viability/growth of normal dividing B lymphoblasts, particularly in the absence of T cell help (57, 87). Levels of p53 protein are known to increase upon DNA damage and cell stress, resulting in cell-cycle blocks, impaired glycolytic metabolism, and/or apoptosis (57, 144, 145). TRI-12 and del11q22 likely compromise the p53 axis in diverse ways. Mdm2, whose gene dose is increased by TRI-12, is ubiquitinase with a major role in reducing p53 protein levels. Conversely, ATM, the major gene downregulated in del11q22+ B-CLL (86), is a kinase with a critical role in stabilizing p53 protein after DNA damage or oxidative stress (91).
During our investigations, we unexpectedly discovered that ODN+IL-15–induced in vitro clonal expansion was impaired if the blood B-CLL population showed signs suggestive of recent proliferative history (high proportion of CD38+ cells). One possible explanation centers on the hypothesis that these in vitro cultures are deficient in providing the critical signals needed for prolonged survival and/or a new burst of growth in this population. In vivo, there is strong evidence that CD38 promotes B-CLL migration and survival through binding its endothelial cell–expressed ligand, PECAM (CD31) (146) (Fig. 11 schematic). Furthermore, there is in vitro evidence that isolated CD38high B lineage cells are vulnerable to apoptosis. Upon BCR cross-linking, CD38high B-CLL isolated from blood show a significant increase in iCa2+ followed by rapid apoptosis, whereas CD38low B-CLL do not (147). In addition, isolated nonmalignant CD38high germinal center cells undergo rapid apoptosis in vitro (148), unless treated by an agonist anti-CD38 mAb that elevates Bcl-2 levels (149). An alternative hypothesis is that CD38high cells taken from blood are either permanently and/or temporarily compromised in their growth potential. Additional studies are needed to examine these possibilities. To date, a clear correlation between ODN+IL-15–stimulated culture viability and the CD38high versus CD38low status of B-CLL taken from blood has not yet emerged (P. Mongini, unpublished observations).
A final observation within this study that merits discussion is that a U-CLL clone’s high-proliferator status to ODN+IL-15 stimulation was statistically linked to clinical progression. This was evidenced by both a direct link between in vitro growth and worse OS and a direct link between in vitro growth and in vivo GR (as determined by doubling time in blood). The latter association, but not the former, required that CD38high U-CLL populations be excluded from analysis. Although this appears to be inconsistent, there is a possible explanation that integrates both the failure to see an association of in vitro growth and TFT and the need to exclude CD38high clones for evidencing a link between in vitro and in vivo growth. This explanation is based on the strong indications that CD38high status in blood reflects in vivo B-CLL clonal expansion within tissues at the time of the analysis (75, 82), as well as the fact that many CD38high clones in our cohort prompted patient therapy after our blood sampling. It is possible that some of the same properties of CD38high clones that contributed to their refractoriness to in vitro stimuli also fostered greater in vivo responsiveness to treatment. The latter would mean that CD38high clones with impaired in vitro growth would score as having prolonged patient survival. An alternative hypothesis is that the CD38high status of certain monitored clones was only temporary and did not represent sustained growth in the patient. Both of these speculations are consistent with findings that high levels of blood CD38+ cells are not uniformly a sign of poor patient outcome (150).
Our study’s discovery of synergy between TLR-9 and IL-15 signaling pathways in eliciting B-CLL clonal expansion provides a new perspective on what may trigger pseudofollicle growth in vivo. The hypothesized pathway, illustrated in Fig. 11, is supported by several lines of evidence. B-CLL express both relatively high levels of TLR-9 and high- and low-affinity receptors for IL-15. Furthermore, TLR-9 ligands, in the form of microbial DNA or DNA from apoptotic cells, are very likely present within tissues where B-CLL cells multiply, and internalization of these ligands will be facilitated by the characteristic BCR specificities of both U-CLL and M-CLL. In addition, IL-15–producing cells appear to be intrinsic to all normal tissues where B-CLL growth is centered and, as shown here, are detectable near or within B-CLL pseudofollicles in patient spleens. Finally and importantly, IL-15 can convert a suboptimal TLR-9–induced stimulus into a strong signal for B-CLL clonal expansion.
During numerous cycles of proliferation, new mutations can be generated through oxidation, enzymatic activity of activation-induced cytosine deaminase, and/or chemotherapy, even in normal human B cells (151). Thus, it is expected that a B-CLL population's greater intrinsic potential for growth, facilitated by certain common chromosomal anomalies, will promote the continued generation of further pathogenic variants. Certainly, finding ways to curb B-CLL growth cycles within lymphoid tissues is critical.
Inhibitors of IL-15 signaling are being developed as therapeutics for other disorders and these might be efficacious in B-CLL, particularly in association with inhibitors of TLR-9 and BCR signaling pathways. In the context of this study’s observations, it is perhaps surprising that infusion therapy with either ODN alone (38) or IL-15 alone (152) has been advocated as a treatment for B-CLL. Because of the likelihood that synergistic IL-15 and ODN signaling in vivo may promote B-CLL clonal expansion, we suggest that these approaches might not be appropriate.
Supplementary Material
Acknowledgments
We are grateful to Drs. Rajendra Damle and Xiao Jie Yan for guiding the characterization of B-CLL surface markers and IGHV, respectively, in this CLL cohort; Dr. Barbara Sherry for providing tissue slides from two of the B-CLL spleens; Joseph You for contributing to the histological staining; Dr. Xinmin Zhang for additional pathological consultation regarding the B-CLL spleen tissue sections; and Dr. Betty Diamond for supporting the continuation of this study and encouragement.
This work was supported by the National Cancer Institute, National Institutes of Health (Grant CA081554 to N.C.); National Institute of Arthritis and Musculoskeletal and Skin Diseases, National Institutes of Health (Grant AR061653 to P.K.A.M.); philanthropic contributions to the Center for Chronic Lymphocytic Leukemia Research from The Karches Foundation, the Marks Foundation, the Jerome Levy Foundation, the Leon Levy Foundation, and the Frank and Muriel Feinberg Foundation; and research support from Dr. Betty Diamond (the Center for Autoimmunity and Musculoskeletal Diseases, Manhasset, NY).
The online version of this article contains supplemental material.
- ATM
- ataxia telangiectasia mutated
- B-CLL
- B cell chronic lymphocytic leukemia
- DIV
- divided
- FDC
- follicular dendritic cell
- FISH
- fluorescence in situ hybridization
- FSC
- forward light scatter
- GR
- growth rate
- IGHV
- Ig H chain V region
- M-CLL
- IGHV mutated B-CLL
- ODN
- oligodeoxynucleotide
- OS
- overall survival
- PB
- peripheral blood
- TFT
- time-to-first treatment
- TRI-12
- trisomy-12
- U-CLL
- IGHV unmutated B-CLL.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Redaelli A., Laskin B. L., Stephens J. M., Botteman M. F., Pashos C. L. 2004. The clinical and epidemiological burden of chronic lymphocytic leukaemia. Eur. J. Cancer Care (Engl.) 13: 279–287. [DOI] [PubMed] [Google Scholar]
- 2.Byrd J. C., Furman R. R., Coutre S. E., Flinn I. W., Burger J. A., Blum K. A., Grant B., Sharman J. P., Coleman M., Wierda W. G., et al. 2013. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N. Engl. J. Med. 369: 32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Furman R. R., Sharman J. P., Coutre S. E., Cheson B. D., Pagel J. M., Hillmen P., Barrientos J. C., Zelenetz A. D., Kipps T. J., Flinn I., et al. 2014. Idelalisib and rituximab in relapsed chronic lymphocytic leukemia. N. Engl. J. Med. 370: 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Messmer B. T., Messmer D., Allen S. L., Kolitz J. E., Kudalkar P., Cesar D., Murphy E. J., Koduru P., Ferrarini M., Zupo S., et al. 2005. In vivo measurements document the dynamic cellular kinetics of chronic lymphocytic leukemia B cells. J. Clin. Invest. 115: 755–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Gent R., Kater A. P., Otto S. A., Jaspers A., Borghans J. A., Vrisekoop N., Ackermans M. A., Ruiter A. F., Wittebol S., Eldering E., et al. 2008. In vivo dynamics of stable chronic lymphocytic leukemia inversely correlate with somatic hypermutation levels and suggest no major leukemic turnover in bone marrow. Cancer Res. 68: 10137–10144. [DOI] [PubMed] [Google Scholar]
- 6.Patten P. E., Buggins A. G., Richards J., Wotherspoon A., Salisbury J., Mufti G. J., Hamblin T. J., Devereux S. 2008. CD38 expression in chronic lymphocytic leukemia is regulated by the tumor microenvironment. Blood 111: 5173–5181. [DOI] [PubMed] [Google Scholar]
- 7.Damle R. N., Calissano C., Chiorazzi N. 2010. Chronic lymphocytic leukaemia: a disease of activated monoclonal B cells. Best Pract. Res. Clin. Haematol. 23: 33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patten P. E., Chu C. C., Albesiano E., Damle R. N., Yan X. J., Kim D., Zhang L., Magli A. R., Barrientos J., Kolitz J. E., et al. 2012. IGHV-unmutated and IGHV-mutated chronic lymphocytic leukemia cells produce activation-induced deaminase protein with a full range of biologic functions. Blood 120: 4802–4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oppezzo P., Vuillier F., Vasconcelos Y., Dumas G., Magnac C., Payelle-Brogard B., Pritsch O., Dighiero G. 2003. Chronic lymphocytic leukemia B cells expressing AID display dissociation between class switch recombination and somatic hypermutation. Blood 101: 4029–4032. [DOI] [PubMed] [Google Scholar]
- 10.Burger J. A., Ghia P., Rosenwald A., Caligaris-Cappio F. 2009. The microenvironment in mature B-cell malignancies: a target for new treatment strategies. Blood 114: 3367–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herishanu Y., Pérez-Galán P., Liu D., Biancotto A., Pittaluga S., Vire B., Gibellini F., Njuguna N., Lee E., Stennett L., et al. 2011. The lymph node microenvironment promotes B-cell receptor signaling, NF-kappaB activation, and tumor proliferation in chronic lymphocytic leukemia. Blood 117: 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dühren-von Minden M., Übelhart R., Schneider D., Wossning T., Bach M. P., Buchner M., Hofmann D., Surova E., Follo M., Köhler F., et al. 2012. Chronic lymphocytic leukaemia is driven by antigen-independent cell-autonomous signalling. Nature 489: 309–312. [DOI] [PubMed] [Google Scholar]
- 13.Woyach J. A., Johnson A. J., Byrd J. C. 2012. The B-cell receptor signaling pathway as a therapeutic target in CLL. Blood 120: 1175–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burger J. A., Chiorazzi N. 2013. B cell receptor signaling in chronic lymphocytic leukemia. Trends Immunol. 34: 592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petlickovski A., Laurenti L., Li X., Marietti S., Chiusolo P., Sica S., Leone G., Efremov D. G. 2005. Sustained signaling through the B-cell receptor induces Mcl-1 and promotes survival of chronic lymphocytic leukemia B cells. Blood 105: 4820–4827. [DOI] [PubMed] [Google Scholar]
- 16.Chen S. S., Batliwalla F., Holodick N. E., Yan X. J., Yancopoulos S., Croce C. M., Rothstein T. L., Chiorazzi N. 2013. Autoantigen can promote progression to a more aggressive TCL1 leukemia by selecting variants with enhanced B-cell receptor signaling. Proc. Natl. Acad. Sci. USA 110: E1500–E1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Damle R. N., Temburni S., Banapour T., Paul S., Mongini P. K., Allen S. L., Kolitz J. E., Rai K. R., Chiorazzi N. 2012. T-cell independent, B-cell receptor-mediated induction of telomerase activity differs among IGHV mutation-based subgroups of chronic lymphocytic leukemia patients. Blood 120: 2438–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Packham G., Krysov S., Allen A., Savelyeva N., Steele A. J., Forconi F., Stevenson F. K. 2014. The outcome of B-cell receptor signaling in chronic lymphocytic leukemia: proliferation or anergy. Haematologica 99: 1138–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lanemo Myhrinder A., Hellqvist E., Sidorova E., Söderberg A., Baxendale H., Dahle C., Willander K., Tobin G., Bäckman E., Söderberg O., et al. 2008. A new perspective: molecular motifs on oxidized LDL, apoptotic cells, and bacteria are targets for chronic lymphocytic leukemia antibodies. Blood 111: 3838–3848. [DOI] [PubMed] [Google Scholar]
- 20.Catera R., Silverman G. J., Hatzi K., Seiler T., Didier S., Zhang L., Hervé M., Meffre E., Oscier D. G., Vlassara H., et al. 2008. Chronic lymphocytic leukemia cells recognize conserved epitopes associated with apoptosis and oxidation. Mol. Med. 14: 665–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chu C. C., Catera R., Zhang L., Didier S., Agagnina B. M., Damle R. N., Kaufman M. S., Kolitz J. E., Allen S. L., Rai K. R., Chiorazzi N. 2010. Many chronic lymphocytic leukemia antibodies recognize apoptotic cells with exposed nonmuscle myosin heavy chain IIA: implications for patient outcome and cell of origin. Blood 115: 3907–3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pisetsky D. S., Gauley J., Ullal A. J. 2011. Microparticles as a source of extracellular DNA. Immunol. Res. 49: 227–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bröker B. M., Klajman A., Youinou P., Jouquan J., Worman C. P., Murphy J., Mackenzie L., Quartey-Papafio R., Blaschek M., Collins P., et al. 1988. Chronic lymphocytic leukemic (CLL) cells secrete multispecific autoantibodies. J. Autoimmun. 1: 469–481. [DOI] [PubMed] [Google Scholar]
- 24.Sthoeger Z. M., Wakai M., Tse D. B., Vinciguerra V. P., Allen S. L., Budman D. R., Lichtman S. M., Schulman P., Weiselberg L. R., Chiorazzi N. 1989. Production of autoantibodies by CD5-expressing B lymphocytes from patients with chronic lymphocytic leukemia. J. Exp. Med. 169: 255–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hervé M., Xu K., Ng Y. S., Wardemann H., Albesiano E., Messmer B. T., Chiorazzi N., Meffre E. 2005. Unmutated and mutated chronic lymphocytic leukemias derive from self-reactive B cell precursors despite expressing different antibody reactivity. J. Clin. Invest. 115: 1636–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kostareli E., Gounari M., Janus A., Murray F., Brochet X., Giudicelli V., Pospisilova S., Oscier D., Foroni L., di Celle P. F., et al. 2012. Antigen receptor stereotypy across B-cell lymphoproliferations: the case of IGHV4-59/IGKV3-20 receptors with rheumatoid factor activity. Leukemia 26: 1127–1131. [DOI] [PubMed] [Google Scholar]
- 27.Hoogeboom R., Wormhoudt T. A., Schipperus M. R., Langerak A. W., Dunn-Walters D. K., Guikema J. E., Bende R. J., van Noesel C. J. 2013. A novel chronic lymphocytic leukemia subset expressing mutated IGHV3-7-encoded rheumatoid factor B-cell receptors that are functionally proficient. Leukemia 27: 738–740. [DOI] [PubMed] [Google Scholar]
- 28.Hoogeboom R., van Kessel K. P., Hochstenbach F., Wormhoudt T. A., Reinten R. J., Wagner K., Kater A. P., Guikema J. E., Bende R. J., van Noesel C. J. 2013. A mutated B cell chronic lymphocytic leukemia subset that recognizes and responds to fungi. J. Exp. Med. 210: 59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leadbetter E. A., Rifkin I. R., Hohlbaum A. M., Beaudette B. C., Shlomchik M. J., Marshak-Rothstein A. 2002. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature 416: 603–607. [DOI] [PubMed] [Google Scholar]
- 30.Kasperkovitz P. V., Khan N. S., Tam J. M., Mansour M. K., Davids P. J., Vyas J. M. 2011. Toll-like receptor 9 modulates macrophage antifungal effector function during innate recognition of Candida albicans and Saccharomyces cerevisiae. Infect. Immun. 79: 4858–4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marshak-Rothstein A., Rifkin I. R. 2007. Immunologically active autoantigens: the role of toll-like receptors in the development of chronic inflammatory disease. Annu. Rev. Immunol. 25: 419–441. [DOI] [PubMed] [Google Scholar]
- 32.Viglianti G. A., Lau C. M., Hanley T. M., Miko B. A., Shlomchik M. J., Marshak-Rothstein A. 2003. Activation of autoreactive B cells by CpG dsDNA. Immunity 19: 837–847. [DOI] [PubMed] [Google Scholar]
- 33.Landau D. A., Carter S. L., Stojanov P., McKenna A., Stevenson K., Lawrence M. S., Sougnez C., Stewart C., Sivachenko A., Wang L., et al. 2013. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell 152: 714–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bertilaccio M. T., Simonetti G., Dagklis A., Rocchi M., Rodriguez T. V., Apollonio B., Mantovani A., Ponzoni M., Ghia P., Garlanda C., et al. 2011. Lack of TIR8/SIGIRR triggers progression of chronic lymphocytic leukemia in mouse models. Blood 118: 660–669. [DOI] [PubMed] [Google Scholar]
- 35.Longo P. G., Laurenti L., Gobessi S., Petlickovski A., Pelosi M., Chiusolo P., Sica S., Leone G., Efremov D. G. 2007. The Akt signaling pathway determines the different proliferative capacity of chronic lymphocytic leukemia B-cells from patients with progressive and stable disease. Leukemia 21: 110–120. [DOI] [PubMed] [Google Scholar]
- 36.Decker T., Schneller F., Sparwasser T., Tretter T., Lipford G. B., Wagner H., Peschel C. 2000. Immunostimulatory CpG-oligonucleotides cause proliferation, cytokine production, and an immunogenic phenotype in chronic lymphocytic leukemia B cells. Blood 95: 999–1006. [PubMed] [Google Scholar]
- 37.Jahrsdörfer B., Wooldridge J. E., Blackwell S. E., Taylor C. M., Griffith T. S., Link B. K., Weiner G. J. 2005. Immunostimulatory oligodeoxynucleotides induce apoptosis of B cell chronic lymphocytic leukemia cells. J. Leukoc. Biol. 77: 378–387. [DOI] [PubMed] [Google Scholar]
- 38.Liang X., Moseman E. A., Farrar M. A., Bachanova V., Weisdorf D. J., Blazar B. R., Chen W. 2010. Toll-like receptor 9 signaling by CpG-B oligodeoxynucleotides induces an apoptotic pathway in human chronic lymphocytic leukemia B cells. Blood 115: 5041–5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tromp J. M., Tonino S. H., Elias J. A., Jaspers A., Luijks D. M., Kater A. P., van Lier R. A., van Oers M. H., Eldering E. 2010. Dichotomy in NF-kappaB signaling and chemoresistance in immunoglobulin variable heavy-chain-mutated versus unmutated CLL cells upon CD40/TLR9 triggering. Oncogene 29: 5071–5082. [DOI] [PubMed] [Google Scholar]
- 40.Efremov D. G., Bomben R., Gobessi S., Gattei V. 2013. TLR9 signaling defines distinct prognostic subsets in CLL. Front. Biosci. (Landmark Ed). 18: 371–386. [DOI] [PubMed] [Google Scholar]
- 41.Decker T., Schneller F., Hipp S., Miething C., Jahn T., Duyster J., Peschel C. 2002. Cell cycle progression of chronic lymphocytic leukemia cells is controlled by cyclin D2, cyclin D3, cyclin-dependent kinase (cdk) 4 and the cdk inhibitor p27. Leukemia 16: 327–334. [DOI] [PubMed] [Google Scholar]
- 42.Dal Bo M., Bomben R., Zucchetto A., Del Poeta G., Gaidano G., Deaglio S., Efremov D. G., Gattei V. 2012. Microenvironmental interactions in chronic lymphocytic leukemia: hints for pathogenesis and identification of targets for rational therapy. Curr. Pharm. Des. 18: 3323–3334. [DOI] [PubMed] [Google Scholar]
- 43.Park C. S., Yoon S. O., Armitage R. J., Choi Y. S. 2004. Follicular dendritic cells produce IL-15 that enhances germinal center B cell proliferation in membrane-bound form. J. Immunol. 173: 6676–6683. [DOI] [PubMed] [Google Scholar]
- 44.Cui G., Hara T., Simmons S., Wagatsuma K., Abe A., Miyachi H., Kitano S., Ishii M., Tani-ichi S., Ikuta K. 2014. Characterization of the IL-15 niche in primary and secondary lymphoid organs in vivo. Proc. Natl. Acad. Sci. USA 111: 1915–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mrózek E., Anderson P., Caligiuri M. A. 1996. Role of interleukin-15 in the development of human CD56+ natural killer cells from CD34+ hematopoietic progenitor cells. Blood 87: 2632–2640. [PubMed] [Google Scholar]
- 46.Oppenheimer-Marks N., Brezinschek R. I., Mohamadzadeh M., Vita R., Lipsky P. E. 1998. Interleukin 15 is produced by endothelial cells and increases the transendothelial migration of T cells In vitro and in the SCID mouse-human rheumatoid arthritis model In vivo. J. Clin. Invest. 101: 1261–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malamut G., El Machhour R., Montcuquet N., Martin-Lannerée S., Dusanter-Fourt I., Verkarre V., Mention J. J., Rahmi G., Kiyono H., Butz E. A., et al. 2010. IL-15 triggers an antiapoptotic pathway in human intraepithelial lymphocytes that is a potential new target in celiac disease-associated inflammation and lymphomagenesis. J. Clin. Invest. 120: 2131–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fehniger T. A., Caligiuri M. A. 2001. Interleukin 15: biology and relevance to human disease. Blood 97: 14–32. [DOI] [PubMed] [Google Scholar]
- 49.Bernasconi N. L., Traggiai E., Lanzavecchia A. 2002. Maintenance of serological memory by polyclonal activation of human memory B cells. Science 298: 2199–2202. [DOI] [PubMed] [Google Scholar]
- 50.Klein U., Tu Y., Stolovitzky G. A., Mattioli M., Cattoretti G., Husson H., Freedman A., Inghirami G., Cro L., Baldini L., et al. 2001. Gene expression profiling of B cell chronic lymphocytic leukemia reveals a homogeneous phenotype related to memory B cells. J. Exp. Med. 194: 1625–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trentin L., Cerutti A., Zambello R., Sancretta R., Tassinari C., Facco M., Adami F., Rodeghiero F., Agostini C., Semenzato G. 1996. Interleukin-15 promotes the growth of leukemic cells of patients with B-cell chronic lymphoproliferative disorders. Blood 87: 3327–3335. [PubMed] [Google Scholar]
- 52.de Totero D., Meazza R., Capaia M., Fabbi M., Azzarone B., Balleari E., Gobbi M., Cutrona G., Ferrarini M., Ferrini S. 2008. The opposite effects of IL-15 and IL-21 on CLL B cells correlate with differential activation of the JAK/STAT and ERK1/2 pathways. Blood 111: 517–524. [DOI] [PubMed] [Google Scholar]
- 53.Söderberg O., Christiansen I., Nilsson G., Carlsson M., Nilsson K. 1997. Interleukin-15 + thioredoxin induce DNA synthesis in B-chronic lymphocytic leukemia cells but not in normal B cells. Leukemia 11: 1298–1304. [DOI] [PubMed] [Google Scholar]
- 54.Herndler-Brandstetter D., Landgraf K., Tzankov A., Jenewein B., Brunauer R., Laschober G. T., Parson W., Kloss F., Gassner R., Lepperdinger G., Grubeck-Loebenstein B. 2012. The impact of aging on memory T cell phenotype and function in the human bone marrow. J. Leukoc. Biol. 91: 197–205. [DOI] [PubMed] [Google Scholar]
- 55.Mongini P. K., Inman J. K., Han H., Kalled S. L., Fattah R. J., McCormick S. 2005. Innate immunity and human B cell clonal expansion: effects on the recirculating B2 subpopulation. J. Immunol. 175: 6143–6154. [DOI] [PubMed] [Google Scholar]
- 56.Mongini P. K., Inman J. K., Han H., Fattah R. J., Abramson S. B., Attur M. 2006. APRIL and BAFF promote increased viability of replicating human B2 cells via mechanism involving cyclooxygenase 2. J. Immunol. 176: 6736–6751. [DOI] [PubMed] [Google Scholar]
- 57.Lee H., Haque S., Nieto J., Trott J., Inman J. K., McCormick S., Chiorazzi N., Mongini P. K. 2012. A p53 axis regulates B cell receptor-triggered, innate immune system-driven B cell clonal expansion. J. Immunol. 188: 6093–6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Austen B., Powell J. E., Alvi A., Edwards I., Hooper L., Starczynski J., Taylor A. M., Fegan C., Moss P., Stankovic T. 2005. Mutations in the ATM gene lead to impaired overall and treatment-free survival that is independent of IGVH mutation status in patients with B-CLL. Blood 106: 3175–3182. [DOI] [PubMed] [Google Scholar]
- 59.Zhang W., Trachootham D., Liu J., Chen G., Pelicano H., Garcia-Prieto C., Lu W., Burger J. A., Croce C. M., Plunkett W., et al. 2012. Stromal control of cystine metabolism promotes cancer cell survival in chronic lymphocytic leukaemia. Nat. Cell Biol. 14: 276–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mongini P. K., Vilensky M. A., Highet P. F., Inman J. K. 1998. Membrane IgM-stimulated human B lymphocytes succumb to activation-related apoptosis at a G1→S transition: influence of ligand affinity and valency. Cell. Immunol. 188: 137–150. [DOI] [PubMed] [Google Scholar]
- 61.Jiang W., Lederman M. M., Harding C. V., Rodriguez B., Mohner R. J., Sieg S. F. 2007. TLR9 stimulation drives naïve B cells to proliferate and to attain enhanced antigen presenting function. Eur. J. Immunol. 37: 2205–2213. [DOI] [PubMed] [Google Scholar]
- 62.Packham G., Stevenson F. K. 2005. Bodyguards and assassins: Bcl-2 family proteins and apoptosis control in chronic lymphocytic leukaemia. Immunology 114: 441–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Giron-Michel J., Giuliani M., Fogli M., Brouty-Boyé D., Ferrini S., Baychelier F., Eid P., Lebousse-Kerdilès C., Durali D., Biassoni R., et al. 2005. Membrane-bound and soluble IL-15/IL-15Ralpha complexes display differential signaling and functions on human hematopoietic progenitors. Blood 106: 2302–2310. [DOI] [PubMed] [Google Scholar]
- 64.Herndler-Brandstetter D., Landgraf K., Jenewein B., Tzankov A., Brunauer R., Brunner S., Parson W., Kloss F., Gassner R., Lepperdinger G., Grubeck-Loebenstein B. 2011. Human bone marrow hosts polyfunctional memory CD4+ and CD8+ T cells with close contact to IL-15-producing cells. J. Immunol. 186: 6965–6971. [DOI] [PubMed] [Google Scholar]
- 65.Ouyang S., Hsuchou H., Kastin A. J., Pan W. 2013. TNF stimulates nuclear export and secretion of IL-15 by acting on CRM1 and ARF6. PLoS ONE 8: e69356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sugiura T., Harigai M., Kawaguchi Y., Takagi K., Fukasawa C., Ohsako-Higami S., Ohta S., Tanaka M., Hara M., Kamatani N. 2002. Increased IL-15 production of muscle cells in polymyositis and dermatomyositis. Int. Immunol. 14: 917–924. [DOI] [PubMed] [Google Scholar]
- 67.Maeda K., Matsuda M., Suzuki H., Saitoh H. A. 2002. Immunohistochemical recognition of human follicular dendritic cells (FDCs) in routinely processed paraffin sections. J. Histochem. Cytochem. 50: 1475–1486. [DOI] [PubMed] [Google Scholar]
- 68.Liu Z., Geboes K., Colpaert S., D’Haens G. R., Rutgeerts P., Ceuppens J. L. 2000. IL-15 is highly expressed in inflammatory bowel disease and regulates local T cell-dependent cytokine production. J. Immunol. 164: 3608–3615. [DOI] [PubMed] [Google Scholar]
- 69.Saeed S., Revell P. A. 2001. Production and distribution of interleukin 15 and its receptors (IL-15Ralpha and IL-R2beta) in the implant interface tissues obtained during revision of failed total joint replacement. Int. J. Exp. Pathol. 82: 201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bergamaschi C., Rosati M., Jalah R., Valentin A., Kulkarni V., Alicea C., Zhang G. M., Patel V., Felber B. K., Pavlakis G. N. 2008. Intracellular interaction of interleukin-15 with its receptor alpha during production leads to mutual stabilization and increased bioactivity. J. Biol. Chem. 283: 4189–4199. [DOI] [PubMed] [Google Scholar]
- 71.Rubinstein M. P., Kovar M., Purton J. F., Cho J. H., Boyman O., Surh C. D., Sprent J. 2006. Converting IL-15 to a superagonist by binding to soluble IL-15Ralpha. Proc. Natl. Acad. Sci. USA 103: 9166–9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Waldmann T. A., Tagaya Y. 1999. The multifaceted regulation of interleukin-15 expression and the role of this cytokine in NK cell differentiation and host response to intracellular pathogens. Annu. Rev. Immunol. 17: 19–49. [DOI] [PubMed] [Google Scholar]
- 73.Malavasi F., Deaglio S., Funaro A., Ferrero E., Horenstein A. L., Ortolan E., Vaisitti T., Aydin S. 2008. Evolution and function of the ADP ribosyl cyclase/CD38 gene family in physiology and pathology. Physiol. Rev. 88: 841–886. [DOI] [PubMed] [Google Scholar]
- 74.Damle R. N., Ghiotto F., Valetto A., Albesiano E., Fais F., Yan X. J., Sison C. P., Allen S. L., Kolitz J., Schulman P., et al. 2002. B-cell chronic lymphocytic leukemia cells express a surface membrane phenotype of activated, antigen-experienced B lymphocytes. Blood 99: 4087–4093. [DOI] [PubMed] [Google Scholar]
- 75.Damle R. N., Temburni S., Calissano C., Yancopoulos S., Banapour T., Sison C., Allen S. L., Rai K. R., Chiorazzi N. 2007. CD38 expression labels an activated subset within chronic lymphocytic leukemia clones enriched in proliferating B cells. Blood 110: 3352–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pittner B. T., Shanafelt T. D., Kay N. E., Jelinek D. F. 2005. CD38 expression levels in chronic lymphocytic leukemia B cells are associated with activation marker expression and differential responses to interferon stimulation. Leukemia 19: 2264–2272. [DOI] [PubMed] [Google Scholar]
- 77.Damle R. N., Wasil T., Fais F., Ghiotto F., Valetto A., Allen S. L., Buchbinder A., Budman D., Dittmar K., Kolitz J., et al. 1999. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood 94: 1840–1847. [PubMed] [Google Scholar]
- 78.Del Principe M. I., Del Poeta G., Buccisano F., Maurillo L., Venditti A., Zucchetto A., Marini R., Niscola P., Consalvo M. A., Mazzone C., et al. 2006. Clinical significance of ZAP-70 protein expression in B-cell chronic lymphocytic leukemia. Blood 108: 853–861. [DOI] [PubMed] [Google Scholar]
- 79.Deaglio S., Aydin S., Grand M. M., Vaisitti T., Bergui L., D’Arena G., Chiorino G., Malavasi F. 2010. CD38/CD31 interactions activate genetic pathways leading to proliferation and migration in chronic lymphocytic leukemia cells. Mol. Med. 16: 87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Deaglio S., Capobianco A., Bergui L., Dürig J., Morabito F., Dührsen U., Malavasi F. 2003. CD38 is a signaling molecule in B-cell chronic lymphocytic leukemia cells. Blood 102: 2146–2155. [DOI] [PubMed] [Google Scholar]
- 81.Willimott S., Baou M., Huf S., Deaglio S., Wagner S. D. 2007. Regulation of CD38 in proliferating chronic lymphocytic leukemia cells stimulated with CD154 and interleukin-4. Haematologica 92: 1359–1366. [DOI] [PubMed] [Google Scholar]
- 82.Calissano C., Damle R. N., Hayes G., Murphy E. J., Hellerstein M. K., Moreno C., Sison C., Kaufman M. S., Kolitz J. E., Allen S. L., et al. 2009. In vivo intraclonal and interclonal kinetic heterogeneity in B-cell chronic lymphocytic leukemia. Blood 114: 4832–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Murphy, E. J., D. Neuberg, K. R. Rai, L. Rassenti, C. Emson, K. Li, N. E. Kay, W. G. Wierda, G. M. Hayes, J. R. Brown, et al. 2014. Kinetic measurement of leukemia-cell proliferation rate by deuterium labeling predicts time to initial treatment of patients with chronic lymphocytic leukemia. Blood. 21: 829. [Google Scholar]
- 84.Veronese A., Pepe F., Chiacchia J., Pagotto S., Lanuti P., Veschi S., Di Marco M., D’Argenio A., Innocenti I., Vannata B., et al. 2015. Allele-specific loss and transcription of the miR-15a/16-1 cluster in chronic lymphocytic leukemia. Leukemia 29: 86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ouillette P., Collins R., Shakhan S., Li J., Li C., Shedden K., Malek S. N. 2011. The prognostic significance of various 13q14 deletions in chronic lymphocytic leukemia. Clin. Cancer Res. 17: 6778–6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kienle D. L., Korz C., Hosch B., Benner A., Mertens D., Habermann A., Kröber A., Jäger U., Lichter P., Döhner H., Stilgenbauer S. 2005. Evidence for distinct pathomechanisms in genetic subgroups of chronic lymphocytic leukemia revealed by quantitative expression analysis of cell cycle, activation, and apoptosis-associated genes. J. Clin. Oncol. 23: 3780–3792. [DOI] [PubMed] [Google Scholar]
- 87.Martinez-Valdez H., Guret C., de Bouteiller O., Fugier I., Banchereau J., Liu Y. J. 1996. Human germinal center B cells express the apoptosis-inducing genes Fas, c-myc, P53, and Bax but not the survival gene bcl-2. J. Exp. Med. 183: 971–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Elnenaei M. O., Gruszka-Westwood A. M., A’Hernt R., Matutes E., Sirohi B., Powles R., Catovsky D. 2003. Gene abnormalities in multiple myeloma; the relevance of TP53, MDM2, and CDKN2A. Haematologica 88: 529–537. [PubMed] [Google Scholar]
- 89.Manfredi J. J. 2010. The Mdm2-p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes Dev. 24: 1580–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dickinson J. D., Gilmore J., Iqbal J., Sanger W., Lynch J. C., Chan J., Bierman P. J., Joshi S. S. 2006. 11q22.3 deletion in B-chronic lymphocytic leukemia is specifically associated with bulky lymphadenopathy and ZAP-70 expression but not reduced expression of adhesion/cell surface receptor molecules. Leuk. Lymphoma 47: 231–244. [DOI] [PubMed] [Google Scholar]
- 91.Shiloh Y., Ziv Y. 2013. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 14: 197–210. [PubMed] [Google Scholar]
- 92.Pekarsky Y., Croce C. M. 2015. Role of miR-15/16 in CLL. Cell Death Differ. 22: 6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Calissano C., Damle R. N., Marsilio S., Yan X. J., Yancopoulos S., Hayes G., Emson C., Murphy E. J., Hellerstein M. K., Sison C., et al. 2011. Intraclonal complexity in chronic lymphocytic leukemia: fractions enriched in recently born/divided and older/quiescent cells. Mol. Med. 17: 1374–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Collado M., Serrano M. 2010. Senescence in tumours: evidence from mice and humans. Nat. Rev. Cancer 10: 51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hamblin T. J., Orchard J. A., Ibbotson R. E., Davis Z., Thomas P. W., Stevenson F. K., Oscier D. G. 2002. CD38 expression and immunoglobulin variable region mutations are independent prognostic variables in chronic lymphocytic leukemia, but CD38 expression may vary during the course of the disease. Blood 99: 1023–1029. [DOI] [PubMed] [Google Scholar]
- 96.Schroers R., Griesinger F., Trümper L., Haase D., Kulle B., Klein-Hitpass L., Sellmann L., Dührsen U., Dürig J. 2005. Combined analysis of ZAP-70 and CD38 expression as a predictor of disease progression in B-cell chronic lymphocytic leukemia. Leukemia 19: 750–758. [DOI] [PubMed] [Google Scholar]
- 97.Pileri S. A., Ascani S., Sabattini E., Fraternali-Orcioni G., Poggi S., Piccioli M., Piccaluga P. P., Gamberi B., Zinzani P. L., Leoncini L., Falini B. 2000. The pathologist’s view point. Part I—indolent lymphomas. Haematologica 85: 1291–1307. [PubMed] [Google Scholar]
- 98.den Haan J. M., Kraal G. 2012. Innate immune functions of macrophage subpopulations in the spleen. J. Innate Immun. 4: 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Scott R. S., McMahon E. J., Pop S. M., Reap E. A., Caricchio R., Cohen P. L., Earp H. S., Matsushima G. K. 2001. Phagocytosis and clearance of apoptotic cells is mediated by MER. Nature 411: 207–211. [DOI] [PubMed] [Google Scholar]
- 100.Miyake Y., Asano K., Kaise H., Uemura M., Nakayama M., Tanaka M. 2007. Critical role of macrophages in the marginal zone in the suppression of immune responses to apoptotic cell-associated antigens. J. Clin. Invest. 117: 2268–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bell D. A., Morrison B., VandenBygaart P. 1990. Immunogenic DNA-related factors. Nucleosomes spontaneously released from normal murine lymphoid cells stimulate proliferation and immunoglobulin synthesis of normal mouse lymphocytes. J. Clin. Invest. 85: 1487–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Casciola-Rosen L. A., Anhalt G., Rosen A. 1994. Autoantigens targeted in systemic lupus erythematosus are clustered in two populations of surface structures on apoptotic keratinocytes. J. Exp. Med. 179: 1317–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sato N., Patel H. J., Waldmann T. A., Tagaya Y. 2007. The IL-15/IL-15Ralpha on cell surfaces enables sustained IL-15 activity and contributes to the long survival of CD8 memory T cells. Proc. Natl. Acad. Sci. USA 104: 588–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jahrsdorfer B., Mühlenhoff L., Blackwell S. E., Wagner M., Poeck H., Hartmann E., Jox R., Giese T., Emmerich B., Endres S., et al. 2005. B-cell lymphomas differ in their responsiveness to CpG oligodeoxynucleotides. Clin. Cancer Res. 11: 1490–1499. [DOI] [PubMed] [Google Scholar]
- 105.Oh S., Perera L. P., Terabe M., Ni L., Waldmann T. A., Berzofsky J. A. 2008. IL-15 as a mediator of CD4+ help for CD8+ T cell longevity and avoidance of TRAIL-mediated apoptosis. Proc. Natl. Acad. Sci. USA 105: 5201–5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shenoy A. R., Kirschnek S., Häcker G. 2014. IL-15 regulates Bcl-2 family members Bim and Mcl-1 through JAK/STAT and PI3K/AKT pathways in T cells. Eur. J. Immunol. 44: 2500–2507. [DOI] [PubMed] [Google Scholar]
- 107.Bianchi T., Gasser S., Trumpp A., MacDonald H. R. 2006. c-Myc acts downstream of IL-15 in the regulation of memory CD8 T-cell homeostasis. Blood 107: 3992–3999. [DOI] [PubMed] [Google Scholar]
- 108.Li Y., Zhi W., Wareski P., Weng N. P. 2005. IL-15 activates telomerase and minimizes telomere loss and may preserve the replicative life span of memory CD8+ T cells in vitro. J. Immunol. 174: 4019–4024. [DOI] [PubMed] [Google Scholar]
- 109.Krysov S., Dias S., Paterson A., Mockridge C. I., Potter K. N., Smith K. A., Ashton-Key M., Stevenson F. K., Packham G. 2012. Surface IgM stimulation induces MEK1/2-dependent MYC expression in chronic lymphocytic leukemia cells. Blood 119: 170–179. [DOI] [PubMed] [Google Scholar]
- 110.Damle R. N., Batliwalla F. M., Ghiotto F., Valetto A., Albesiano E., Sison C., Allen S. L., Kolitz J., Vinciguerra V. P., Kudalkar P., et al. 2004. Telomere length and telomerase activity delineate distinctive replicative features of the B-CLL subgroups defined by immunoglobulin V gene mutations. Blood 103: 375–382. [DOI] [PubMed] [Google Scholar]
- 111.Briard D., Brouty-Boyé D., Azzarone B., Jasmin C. 2002. Fibroblasts from human spleen regulate NK cell differentiation from blood CD34(+) progenitors via cell surface IL-15. J. Immunol. 168: 4326–4332. [DOI] [PubMed] [Google Scholar]
- 112.Estess P., Nandi A., Mohamadzadeh M., Siegelman M. H. 1999. Interleukin 15 induces endothelial hyaluronan expression in vitro and promotes activated T cell extravasation through a CD44-dependent pathway in vivo. J. Exp. Med. 190: 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Angiolillo A. L., Kanegane H., Sgadari C., Reaman G. H., Tosato G. 1997. Interleukin-15 promotes angiogenesis in vivo. Biochem. Biophys. Res. Commun. 233: 231–237. [DOI] [PubMed] [Google Scholar]
- 114.Musso T., Calosso L., Zucca M., Millesimo M., Ravarino D., Giovarelli M., Malavasi F., Ponzi A. N., Paus R., Bulfone-Paus S. 1999. Human monocytes constitutively express membrane-bound, biologically active, and interferon-gamma-upregulated interleukin-15. Blood 93: 3531–3539. [PubMed] [Google Scholar]
- 115.Neely G. G., Robbins S. M., Amankwah E. K., Epelman S., Wong H., Spurrell J. C., Jandu K. K., Zhu W., Fogg D. K., Brown C. B., Mody C. H. 2001. Lipopolysaccharide-stimulated or granulocyte-macrophage colony-stimulating factor-stimulated monocytes rapidly express biologically active IL-15 on their cell surface independent of new protein synthesis. J. Immunol. 167: 5011–5017. [DOI] [PubMed] [Google Scholar]
- 116.Burger J. A., Gribben J. G. 2014. The microenvironment in chronic lymphocytic leukemia (CLL) and other B cell malignancies: insight into disease biology and new targeted therapies. Semin. Cancer Biol. 24: 71–81. [DOI] [PubMed] [Google Scholar]
- 117.Plander M., Seegers S., Ugocsai P., Diermeier-Daucher S., Iványi J., Schmitz G., Hofstädter F., Schwarz S., Orsó E., Knüchel R., Brockhoff G. 2009. Different proliferative and survival capacity of CLL-cells in a newly established in vitro model for pseudofollicles. Leukemia 23: 2118–2128. [DOI] [PubMed] [Google Scholar]
- 118.Schulz A., Toedt G., Zenz T., Stilgenbauer S., Lichter P., Seiffert M. 2011. Inflammatory cytokines and signaling pathways are associated with survival of primary chronic lymphocytic leukemia cells in vitro: a dominant role of CCL2. Haematologica 96: 408–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hocke A. C., Lampe M. P., Witzenrath M., Mollenkopf H., Zerrahn J., Schmeck B., Kessler U., Krüll M., Hammerschmidt S., Hippenstiel S., et al. 2007. Cell-specific interleukin-15 and interleukin-15 receptor subunit expression and regulation in pneumococcal pneumonia—comparison to chlamydial lung infection. Cytokine 38: 61–73. [DOI] [PubMed] [Google Scholar]
- 120.Hocke A. C., Hartmann I. K., Eitel J., Optiz B., Scharf S., Suttorp N., Hippenstiel S. 2008. Subcellular expression pattern and role of IL-15 in pneumococci induced lung epithelial apoptosis. Histochem. Cell Biol. 130: 165–176. [DOI] [PubMed] [Google Scholar]
- 121.Verbist K. C., Cole C. J., Field M. B., Klonowski K. D. 2011. A role for IL-15 in the migration of effector CD8 T cells to the lung airways following influenza infection. J. Immunol. 186: 174–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Giri J. G., Kumaki S., Ahdieh M., Friend D. J., Loomis A., Shanebeck K., DuBose R., Cosman D., Park L. S., Anderson D. M. 1995. Identification and cloning of a novel IL-15 binding protein that is structurally related to the alpha chain of the IL-2 receptor. EMBO J. 14: 3654–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Landgren O., Rapkin J. S., Caporaso N. E., Mellemkjaer L., Gridley G., Goldin L. R., Engels E. A. 2007. Respiratory tract infections and subsequent risk of chronic lymphocytic leukemia. Blood 109: 2198–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Barcos M., Lane W., Gomez G. A., Han T., Freeman A., Preisler H., Henderson E. 1987. An autopsy study of 1206 acute and chronic leukemias (1958 to 1982). Cancer 60: 827–837. [DOI] [PubMed] [Google Scholar]
- 125.Rückert R., Asadullah K., Seifert M., Budagian V. M., Arnold R., Trombotto C., Paus R., Bulfone-Paus S. 2000. Inhibition of keratinocyte apoptosis by IL-15: a new parameter in the pathogenesis of psoriasis? J. Immunol. 165: 2240–2250. [DOI] [PubMed] [Google Scholar]
- 126.Loser K., Mehling A., Apelt J., Ständer S., Andres P. G., Reinecker H. C., Eing B. R., Skryabin B. V., Varga G., Schwarz T., Beissert S. 2004. Enhanced contact hypersensitivity and antiviral immune responses in vivo by keratinocyte-targeted overexpression of IL-15. Eur. J. Immunol. 34: 2022–2031. [DOI] [PubMed] [Google Scholar]
- 127.Romano E., Cotari J. W., Barreira da Silva R., Betts B. C., Chung D. J., Avogadri F., Fink M. J., St Angelo E. T., Mehrara B., Heller G., et al. 2012. Human Langerhans cells use an IL-15R-α/IL-15/pSTAT5-dependent mechanism to break T-cell tolerance against the self-differentiation tumor antigen WT1. Blood 119: 5182–5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cerroni L., Zenahlik P., Höfler G., Kaddu S., Smolle J., Kerl H. 1996. Specific cutaneous infiltrates of B-cell chronic lymphocytic leukemia: a clinicopathologic and prognostic study of 42 patients. Am. J. Surg. Pathol. 20: 1000–1010. [DOI] [PubMed] [Google Scholar]
- 129.Robak E., Robak T. 2007. Skin lesions in chronic lymphocytic leukemia. Leuk. Lymphoma 48: 855–865. [DOI] [PubMed] [Google Scholar]
- 130.Cerroni L., Zenahlik P., Kerl H. 1995. Specific cutaneous infiltrates of B-cell chronic lymphocytic leukemia arising at the site of herpes zoster and herpes simplex scars. Cancer 76: 26–31. [DOI] [PubMed] [Google Scholar]
- 131.Bairey O., Goldschmidt N., Ruchlemer R., Tadmor T., Rahimi-Levene N., Yuklea M., Shvidel L., Berrebi A., Polliack A., Herishanu Y., Israeli Chronic Lymphocytic Leukemia Study Group (ICLLSG) 2012. Insect-bite-like reaction in patients with chronic lymphocytic leukemia: a study from the Israeli Chronic Lymphocytic Leukemia Study Group. Eur. J. Haematol. 89: 491–496. [DOI] [PubMed] [Google Scholar]
- 132.Avalos A. M., Busconi L., Marshak-Rothstein A. 2010. Regulation of autoreactive B cell responses to endogenous TLR ligands. Autoimmunity 43: 76–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Puiggros A., Blanco G., Espinet B. 2014. Genetic abnormalities in chronic lymphocytic leukemia: where we are and where we go. Biomed. Res. Int. 2014: 435983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Matutes E., Oscier D., Garcia-Marco J., Ellis J., Copplestone A., Gillingham R., Hamblin T., Lens D., Swansbury G. J., Catovsky D. 1996. Trisomy 12 defines a group of CLL with atypical morphology: correlation between cytogenetic, clinical and laboratory features in 544 patients. Br. J. Haematol. 92: 382–388. [DOI] [PubMed] [Google Scholar]
- 135.Döhner H., Stilgenbauer S., Benner A., Leupolt E., Kröber A., Bullinger L., Döhner K., Bentz M., Lichter P. 2000. Genomic aberrations and survival in chronic lymphocytic leukemia. N. Engl. J. Med. 343: 1910–1916. [DOI] [PubMed] [Google Scholar]
- 136.Van Bockstaele F., Verhasselt B., Philippé J. 2009. Prognostic markers in chronic lymphocytic leukemia: a comprehensive review. Blood Rev. 23: 25–47. [DOI] [PubMed] [Google Scholar]
- 137.Riches J. C., O’Donovan C. J., Kingdon S. J., McClanahan F., Clear A. J., Neuberg D. S., Werner L., Croce C. M., Ramsay A. G., Rassenti L. Z., et al. 2014. Trisomy 12 chronic lymphocytic leukemia cells exhibit upregulation of integrin signaling that is modulated by NOTCH1 mutations. Blood 123: 4101–4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Maura F., Mosca L., Fabris S., Cutrona G., Matis S., Lionetti M., Agnelli L., Barbieri M., D’Anca M., Manzoni M., et al. 2015. Insulin growth factor 1 receptor expression is associated with NOTCH1 mutation, trisomy 12 and aggressive clinical course in chronic lymphocytic leukaemia. PLoS ONE 10: e0118801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yaktapour N., Übelhart R., Schüler J., Aumann K., Dierks C., Burger M., Pfeifer D., Jumaa H., Veelken H., Brummer T., Zirlik K. 2013. Insulin-like growth factor-1 receptor (IGF1R) as a novel target in chronic lymphocytic leukemia. Blood 122: 1621–1633. [DOI] [PubMed] [Google Scholar]
- 140.Saiya-Cork K., Collins R., Parkin B., Ouillette P., Kuizon E., Kujawski L., Erba H., Campagnaro E., Shedden K., Kaminski M., Malek S. N. 2011. A pathobiological role of the insulin receptor in chronic lymphocytic leukemia. Clin. Cancer Res. 17: 2679–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shanafelt T. D., Witzig T. E., Fink S. R., Jenkins R. B., Paternoster S. F., Smoley S. A., Stockero K. J., Nast D. M., Flynn H. C., Tschumper R. C., et al. 2006. Prospective evaluation of clonal evolution during long-term follow-up of patients with untreated early-stage chronic lymphocytic leukemia. J. Clin. Oncol. 24: 4634–4641. [DOI] [PubMed] [Google Scholar]
- 142.Delmer A., Ajchenbaum-Cymbalista F., Tang R., Ramond S., Faussat A. M., Marie J. P., Zittoun R. 1995. Overexpression of cyclin D2 in chronic B-cell malignancies. Blood 85: 2870–2876. [PubMed] [Google Scholar]
- 143.Watanabe T., Hotta T., Ichikawa A., Kinoshita T., Nagai H., Uchida T., Murate T., Saito H. 1994. The MDM2 oncogene overexpression in chronic lymphocytic leukemia and low-grade lymphoma of B-cell origin. Blood 84: 3158–3165. [PubMed] [Google Scholar]
- 144.Efeyan A., Serrano M. 2007. p53: guardian of the genome and policeman of the oncogenes. Cell Cycle 6: 1006–1010. [DOI] [PubMed] [Google Scholar]
- 145.Maddocks O. D., Vousden K. H. 2011. Metabolic regulation by p53. J. Mol. Med. 89: 237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Vaisitti T., Audrito V., Serra S., Buonincontri R., Sociali G., Mannino E., Pagnani A., Zucchetto A., Tissino E., Vitale C., et al. 2015. The enzymatic activities of CD38 enhance CLL growth and trafficking: implications for therapeutic targeting. Leukemia 29: 356–368. [DOI] [PubMed] [Google Scholar]
- 147.Zupo S., Isnardi L., Megna M., Massara R., Malavasi F., Dono M., Cosulich E., Ferrarini M. 1996. CD38 expression distinguishes two groups of B-cell chronic lymphocytic leukemias with different responses to anti-IgM antibodies and propensity to apoptosis. Blood 88: 1365–1374. [PubMed] [Google Scholar]
- 148.Lebecque S., de Bouteiller O., Arpin C., Banchereau J., Liu Y. J. 1997. Germinal center founder cells display propensity for apoptosis before onset of somatic mutation. J. Exp. Med. 185: 563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zupo S., Rugari E., Dono M., Taborelli G., Malavasi F., Ferrarini M. 1994. CD38 signaling by agonistic monoclonal antibody prevents apoptosis of human germinal center B cells. Eur. J. Immunol. 24: 1218–1222. [DOI] [PubMed] [Google Scholar]
- 150.Malavasi F., Deaglio S., Damle R., Cutrona G., Ferrarini M., Chiorazzi N. 2011. CD38 and chronic lymphocytic leukemia: a decade later. Blood 118: 3470–3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Haque S., Yan X. J., Rosen L., McCormick S., Chiorazzi N., Mongini P. K. 2014. Effects of prostaglandin E2 on p53 mRNA transcription and p53 mutagenesis during T-cell-independent human B-cell clonal expansion. FASEB J. 28: 627–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Laprevotte E., Voisin G., Ysebaert L., Klein C., Daugrois C., Laurent G., Fournie J. J., Quillet-Mary A. 2013. Recombinant human IL-15 trans-presentation by B leukemic cells from chronic lymphocytic leukemia induces autologous NK cell proliferation leading to improved anti-CD20 immunotherapy. J. Immunol. 191: 3634–3640. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.