Abstract
Histopathological diagnosis in most of the world's hospitals is based upon formalin-fixed and paraffin-embedded (FFPE) tissues. Although this standard fixation and embedding procedure keeps the tissue in excellent form for morphological and immunohistological analysis, FFPE is inappropriate for nucleic acids and protein studies. We investigated the potential value of RCL2, a new non-toxic fixative, for sparing proteins preserved in paraffin-embedded tissues. Normal colonic mucosa tissue was fixed in RCL2 prior to paraffin embedding (RCL2P), and then processed for quality and quantity of protein conservation, as compared to frozen and FFPE tissues using complementary proteomic analysis approaches. Using 4 different protein extraction protocols, RCL2P tissue consistently showed the highest protein yield. Similar protein patterns were observed with RCL2P and frozen tissues using mono and bi-dimensional electrophoresis. Moreover, membrane, cytoplasmic and nuclear proteins, as well as phosphorylated proteins, were successfully detected using western-blot. Furthermore, protein patterns observed by mass spectrometry analysis after laser-captured microdissection were found to be identical for frozen and RCL2-fixed tissues. At last, immunohistochemistry using various antibodies showed comparable results between both tissue storage methods. We concluded that RCL2 has great potential for performing both morphological and molecular analyses on the same archival paraffin-embedded tissue sample, and can be a new method for investigating protein biomarkers.
Keywords: paraffin-embedded tissue, fixative, proteomics
Introduction
Human tissues are an important biological material for the discovery of biomarkers and identification of novel therapeutic targets. Formalin fixed and paraffin-embedded (FFPE) tissue represents the most abundant supply of archival material for clinical and molecular analyses. FFPE is the standard-processing methodology practiced in histopathology laboratories worldwide, resulting in a highly stable and easily stored form of tissue. Although FFPE preserves the cellular and architectural morphologic details in tissue sections, it alters and fragments nucleic acids. It also impairs the extraction efficiency and quality of DNA, and more strikingly, RNA, preventing molecular analyses [1]. In addition, as a highly reactive dipolar compound, formalin facilitates the formation of protein–protein cross-links in vitro, and it renders FFPE tissues refractory to many protein studies.
Recently, Palmer-Toy et al. and Crockett et al. reported suitable alternatives for protein studies from formalin fixation, and Becker et al. described the commercialized Qproteome FFPE tissue kit as an optimal solution for proteomic analyses of formalin-fixed tissues [2–5]. These approaches represent new improvements for FFPE tissue exploitation. However, they are only appropriate for a restrictive number of proteomic methodologies, precluding routine western blot or bi-dimensional electrophoresis analyses of protein expression. Moreover, protein modifications induced by formalin would render many proteins unidentifiable or lead to misidentifications, thus hindering reproducible quantification of proteomic analysis [5]. Although immunohistochemistry lacks sensitivity and quantification, it possesses the more useful capability of providing proteomic information from these samples. Unfixed fresh or snap-frozen tissues could represent an ideal alternative, allowing for complete molecular analyses, but does not provide accurate morphological details and may impair histological diagnosis.
Alternative tissue fixation procedures are critical to preserving the morphologic details, DNA, RNA and proteins of tissue. Methacarn, a solution of methanol, chloroform, and acetic acid, is a non-cross-linking organic solvent that was used to maintain tissue morphology and preserve nucleic acid and protein integrities [6, 7]. Interestingly, methacarn-fixed tissues have been successfully used for quantitative expression analysis of mRNAs after microdissection [8]. Morales et al. and Vincek et al. described UMFIX, a mixture of methanol and polyethylene glycol, with properties similar to Methacarn. Also, FineFIX has been proposed for standard molecular analysis [9–11]. Although these fixatives are appropriate for DNA or RNA analysis, the related procedures have been poorly investigated in the proteomic fields. In a previous study, we have shown that a new fixative, RCL2, protects tissue morphology, DNA, and RNA [12]. The aim of this study was to assess the feasibility of proteomic investigations on RCL2-tissue embedded using a comprehensive panel of proteomic methods.
Materials and methods
Tissue Samples
Two normal colonic mucosa samples were obtained following colostomy by the Department of Pathology (Montpellier). One part of the tissue was fixed in NBF (Neutral buffered formaldehyde, 4%) for 24 hr at room temperature (RT), dehydrated and paraffin embedded using a TissueTek VIP automated processor (Bayer HealthCare Diagnosis Division) according to the standard protocol used for diagnosis. The remaining tissue was divided in two samples. One sample was immediately snap-frozen in liquid nitrogen and stored at (80°C. The other sample was fixed overnight at 4°C in RCL2 (Alphelys, plaisir, France) before paraffin embedding. This protocol consisted of dehydration in ethanol (4 baths, 37°C, 4.25 hr total time), followed by xylene (3 baths, 37°C, 3.5 hr total time) and paraffin-immersion (4 baths, 58–59°C, 4.25 hr total). FFPE tissue was conserved at RT, and RCL2 paraffin (RCL2P) blocks were maintained at–20°C.
Protein extraction
Three 10-μm-thick sections from frozen, FFPE and RCL2P tissues were cut directly from the blocks into a 1.5 ml tube. Paraffin-embedded tissue sections were de-waxed by three 5-min extractions in 100% xylene at RT prior two washing with 100% ethanol. Proteins were extracted 20 min at +4°C in 150 μl of:
lysis buffer from protocol A: 50 mM Tris-HCI buffer, pH 7.5, containing 7 M urea, 2 M thiourea, 2% CHAPS, 1% Mega, 0.5% Triton X 100 1%OGP and 50 mMDTT,
lysis buffer from protocol B: 7 M urea, 2 M thiourea, 4% CHAPS andlM DTT,
lysis buffer from protocol C: 50 mM Tris-HCI buffer pH 7,5 containing 150 mM NaCI, 0,5% Triton X-100 and 0,5% deoxycholate.
All buffers contained a mixture of protease inhibitor cocktail (Complete, Roche Diagnostics).
Protein extracts were clarified at 15,000 g for 15 min at 4°C, and super-natants were recovered for subsequent proteomic analysis.
For protocol D, proteins were extracted using Qproteome FFPE Tissue Kit – Qiagen according to the manufacturer's protocol.
Protein assay
Protein concentration was measured using EZQ Protein quantitation kit (Molecular Probes) according to the manufacturer's protocol. We have used the tissue size as a normalization factor as all of the tissue sections showed approximately identical dimension (0.5 cm2 with 10 μm thickness). Protein concentration was expressed as μg/cm2 of tissue. Three replicates were conducted, and the performance of protein extraction protocols was evaluated.
One- and two-dimensional electrophoresis (1 and 2-DE) analysis
For 1-DE, 10 μg of protein were precipitated and re-suspended in loading buffer. Electrophoresis was conducted on a 12%-SDS polyacrylamide gel, and silver staining was performed according to Shevchenko's procedure [13].
2-DE reagents and materials were purchased from GE Healthcare. Protein extractions from both FFPE and RCL2P samples were performed using protocols A and D, whereas for frozen samples, proteins were extracted only with protocol A. Nucleic acids, lipids and salts were removed with the 2-DE Clean-Up Kit. Proteins (150 μg) were solubilized in 350 μl of isoelectrofocusing medium, as previously described [14]. For the second dimension, the strips were loaded onto vertical 10–17% SDS polyacrylamide gradient gels prior to silver staining. Spot detection and gel alignment were performed with the Image Master 2D Platinum software. For protein identification, spots were excised from 2D gels, digested with trypsin (Gold Promega) as previously described [15]. Digest products were completely dehydrated in a vacuum centrifuge and re-suspended in 10 μl formic acid (2%), desalted using ZipTips C18 (Millipore), eluted with 10 μl acetonitrile-trifluoroacetic acid (ACN-TFA) (50–0.1%) and concentrated to 2 pi. Aliquots (0.5 pi) were mixed with the same volume of a-cyano-4-hydroxy-frans-cin-namic acid (Laserbiolabs, 10 mg/ml in ACN-TFA, 50–0.1%) before applying samples to target plates and analysed by the matrix-assisted laser desorption ionization-tandem time-of-flight (MALDI-TOF/TOF) method using the 4800 Plus MALDI T0F/T0F(TM) Analyser (Applied biosystems). Identification of proteins was performed using the MASCOT software (version 2.1, Matrixscience) against the Swiss-Prot database. MASCOT score greater than 63 was considered significant (P<0.01) for Swiss-Prot database interrogation.
Western-blot
30 μg of protein extracts were loaded onto a 12% polyacrylamide separating gel. After protein transfer, PVDF membranes were blocked and incubated overnight at 4°C with several antibodies: mouse monoclonal anti-E-cadherin (1:2500, BD transduction Laboratories), anti-α-tubulin (1:10000, Sigma-Aldrich), rabbit polyclonal anti-Sp1 (1:500, Santa Cruz Biotechnology, Inc) anti-phospho-MEK1/2 antibodies (1:1000, Cell Signaling Technology), and anti-phospho-p44/42 MAP kinase antibodies (Thr202/Tyr204) (1:1000,Cell Signaling Technology). The peroxidase-conjugated secondary antimouse IgG (Jackson ImmunoResearch, Laboratories) or antirabbit IgG (Santa Cruz) antibodies were diluted at 1:5000. The blots were developed using SuperSignal West Pico Chemiluminescent Substrate Kit (Pierce) and Hyperfilm ECL (Amersham).
Laser capture microdissection (LCM)
LCM of frozen and RCL2P tissues were performed using the PixCell II LCM system (Acturus Engineering). Briefly, 7-μm-thick sections were stained according to the manufacturer's protocol. The staining do not modified tissue architecture. Paraffin-embedded tissue sections were de-paraffinized before staining in 100% ethanol and stored in a desiccated container for at least 15 min. before LCM. For each analysis, 500–1500 colon cells were laser-captured from the same tissue. Microdissection efficiency was assessed by examining cells present on the LCM cap. Microdissected cells were immediately lysed (protocol A) prior to SELDI analysis.
Histology and immunohistochemistry
4-μm-thick sections of FFPE and RLC2P tissues were de-paraffinized with xylene and re-hydrated with several graded ethanols before Haematoxylin-Eosin-Saffron staining or immunohistostaining. Immunohistochemical analyses were performed by using the Dako autostainer (Dako). According to the tested antibody, and whenever needed, tissue sections were treated for 45 min at 95°C with citrate or ethylenediaminetetraacetic acid for antigen retrieval. Slides were treated with a peroxidase inhibitor (Dako) for 10 min. to quench the endogenous peroxidase activity. The detection of the antibody binding was visualized, either with the streptavidin–biotin peroxidase complex (LSAB™ 2, Dako) or with a peroxidase-conjugated polymer backbone (Envision™+ dual link, Dako) using diaminobenzidine as a chromogen. The sections were then counterstained with haematoxylin. Antibodies and staining conditions are listed in Table 1. Same E-Cadherin and phosphorylated-MAPK/extracellular signal-regulated kinase (p-MEK) antibodies were used for both western blot and immunohistostaining.
Table 1.
Sources and methods for the antibodies used.
| Antibody (clone) | Source | Immuno-localization | Fixative | Pre-treatment | Dilution (incubation time) | Detection |
|---|---|---|---|---|---|---|
| hMLH1 (G168-15) | BD Bio sciences | nuclear | NBF | EDTA pH9 | 1/30 (90 min.) | Envision |
| RCL2 R | EDTA pH9 | 1/100 (30 min.) | Envision | |||
| Cytokeratin AE1 AE3 (AE1/AE3) | Dako | Cytoplasmic | NBF | Citrate pH6 | 1/100 (45 min.) | LSAB2 |
| RCL2 R | Citrate pH6 | 1/100 (45 min.) | LSAB2 | |||
| E-Cadherin (36) | BD Bio sciences | Cell membrane | NBF | Citrate pH6 | 1/500 (45 min.) | LSAB2 |
| RCL2 R | Citrate pH6 | 1/500 (45 min.) | LSAB2 | |||
| p-MEK (rabbit polyclonal) | Cell signaling | Cytoplasmic and | NBF | Citrate pH6 | 1/500 (60 min.) | Envision |
| nuclear | RCL2 R | Citrate pH6 | 1/50 (90 min.) | Envision |
Mass spectrometry analysis
For ProteinChip array binding, CM10 surfaces were equilibrated with binding buffer (100 mM Na Acetate, pH 4). Six micrograms of total protein extracts (protocol A), or 500 and 1500 cells from LCM tissues, were loaded on each spot. After 30 minutes, the arrays were washed three times with binding buffer for 5 min, and then twice with water. Sinapinic acid solution was applied twice to each spot, as an energy-absorbing matrix. Arrays were read on a PBS II ProteinChip reader, and peak detection was performed using the ProteinChip and Biomarker Wizard software (Ciphergen Biosystem). Automatic peak detection was performed in the range of 2.5–50 kD, with a signal-to-noise ratio cut-off at 4 for the first pass and 2 for the second pass, and a cluster mass window at 0.5% of mass.
Results
Proteins can be efficiently extracted from RCL2P tissue
To reliably extract high amounts of non-degraded, full length and immunoreactive proteins, including membrane proteins and low-abundance proteins, four extraction methods were initially used (Table 2). Quantitative comparisons of extracted proteins from RLC2P, FFPE and frozen tissues revealed that protocol A, which contains a combination of non-ionic detergents, gave significantly greater protein yields than either protocol B or the more commonly used protocol C in both colon tissue samples. Surprisingly, protocol D, which corresponds to the commercialized kit, does not substantially increase the quantity of protein extract, even with FFPE tissues, as previously described [2]. Overall, protein yield obtained from frozen tissue was found to be lower than from RCL2P, regardless of which extraction protocol was used. No protein yield could be measured in FFPE tissue, except with protocol D.
Table 2.
Protein yield from RCL2P, FFPE and frozen tissues using four protein extraction protocols. Values are the means of three independent experiments with standard deviations in parentheses.
| Protocol | Protein yield (μg/cm2)/tissue section | |||||
|---|---|---|---|---|---|---|
| Tissue 1 | Tissue 2 | |||||
| RCL2P | FFPE | FROZEN | RCL2P | FFPE | FROZEN | |
| A | 9.16 (1.49) | ND | 1.28 (0.44) | 7.11 (3.61) | ND | 0.59 (0.12) |
| B | 3.75 (2.33) | ND | 0.87 (0.14) | 4.81 (0.97) | ND | 0.91 (0.15) |
| C | 0.07 (0.06) | ND | 0.57 (0.10) | 0.16 (0.07) | ND | 0.48 (0.06) |
| D | 2.9 (0.83) | 0.36 (0.05) | 1.04 (0.27) | 2.26 (0.54) | 1.26 (0.54) | 0.64 (0.2) |
RCL2P and frozen tissues revealed similar protein profiles on 1-DE and 2-DE gels
The overall protein profile extraction was evaluated, for each extraction protocol and tissue storage, by 1-DE analysis and sensitive silver staining. Representative pictures from replicated experiments are shown in Figure 1. No major difference in the protein pattern was observed between protocols A, B or C in the range of 25–250 kD. Distinct bands were observed with both frozen and RCL2P samples, whereas no protein pattern could be detected using FFPE material. However, protocol D, which gave low-protein yield, showed typical aggregated profiles, revealing the poor quality of protein extraction. Using breast and prostate tissue samples, we also observed a similar protein pattern between RCL2P and frozen tissues (data not shown).
Fig 1.
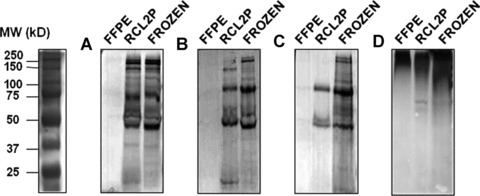
SDS-PAGE gels of extracted proteins from formalin and RCL2-fixed paraffin-embedded tissues compared to frozen colonic tissues. Proteins were extracted using protocols A (A), B (B), C (C) and Qproteome FFPE kit (D).
Protein profiles were then extensively analysed using 2-DE gels and silver staining. Protocol A showed the best protein yield and was thus chosen for protein extraction of FFPE, RCL2P and frozen tissues. Proteins, from both FFPE and RCL2P materials, were also extracted using the Qproteome FFPE kit (protocol D). The FFPE protein extraction with protocol A showed a degraded pattern without any distinct spots, confirming the unsuitability of formalin-fixed tissue for 2-DE analysis (Fig. 2A). Proteins extracted using protocol D did not increase the protein yield (Fig. 2B), and gave similar results to RCL2P tissue processing (Fig. 2D). Frozen tissue showed excellent protein quality and quantity, as expected, with a mean number of 600 detected spots (Fig. 2E). Interestingly, RCL2P tissue revealed the same pattern, with a mean number of 500 proteins spots (Fig. 2C). In addition, when compared to proteins extracted from frozen tissues, protein mass and p/from RCL2P tissues were not affected by the tissue-processing method, though some spots did appear slightly fuzzy. Finally, we were able to identify several protein spots extracted from RCL2 tissue separated onto 2-DE gel by mass spectrometry. Figure 3 showed the mass spectra obtained from one of these spots using MALDI-TOF/TOF method. We identified the heat shock protein β-1 by mass spectrometry analysis and confirmed this result by direct sequencing of tryptic peptides in both RCL2P and frozen samples. Too more proteins (Fig. 2C and D, circle 2 and 3) were also identified as proteasome activator complex subunit 2 and the lactate dehydrogenase B chain.
Fig 2.
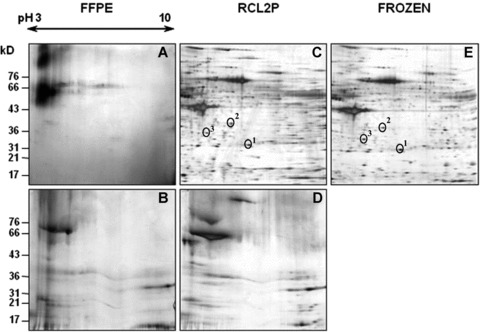
Representative silver stained two-dimensional electrophoresis maps obtained from FFPE tissues (panels A and B), RCL2P tissues (panels C and D), and frozen colonic mucosa tissues (panel E). Proteins were extracted using either protocol A (panels A, C, E) or protocol D (panels B and D). On panels C and E, the selected protein spots cut from the gels and analysed by MALDI TOF/TOF method were indicated.
Fig 3.
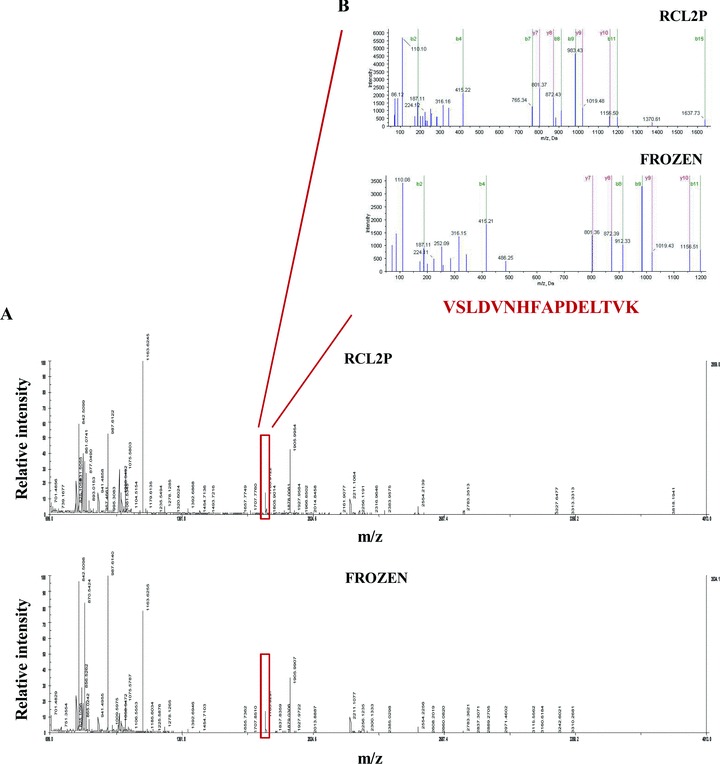
MS analysis of spot 1 excised from 2-DE gels obtained from RCL2P and frozen tissues respectively were presented on panel A, and the two subsequent MS/MS analysis of the 1783.92 kD protein peak are shown on panel B. Via Swiss-Prot database searching the Heat Shock protein β-1 (accession number P04792, theoretical pl value 5.98 and theoretical Mr 22826 Da) was identified. Mascott scores obtained were 74 and 71 for protein extracted from RCL2P and Frozen maps, respectively and the sequence coverage were 54 and 76%. The two MS/MS spectra of m/z 1783.92 generated a partial y ion series which led to the identification via Swiss-Prot database search of VSLDVNHFAPDELTVK from Heat Shock protein beta-1.
RCL2 allowed detection of different cellular localization and post-translationally modified proteins by western-blot
To evaluate RCL2P tissue for protein expression, proteins with different cellular localization and after translational modifications were analysed by western-blot (Fig. 4). Membrane E-cadherin, cytoplasmic α-tubulin, and nuclear Sp1 protein expressions were similar between RCL2P and frozen tissues (Fig. 4A). Phosphorylated MAPK/extracellular signal-related kinase (MEK) and p44/42 mitogen activated protein kinase (MAPK) were also detected by immunoblot in both tissues (Fig. 4B). In agreement with our previous experiments, no signal could be detected in FFPE tissue using protocol D, which was then excluded from all further proteomic investigtions.
Fig 4.
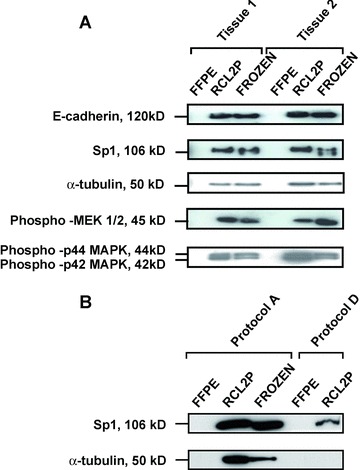
Western blot performed using antibodies against E-cadherin, Sp1, α-tubulin, phospho MEK 1/2, phospho-p42 and phospho-p44 MAPK using protocol A extraction on two colonic mucosa tissue samples (A). Western blot performed using Sp1 and α-tubulin antibodies on proteins extracted using protocol D, compared to protocol A, of one colonic mucosa tissue sample (B).
RCL2P tissue was suitable for morphological and immunohistochemical analyses
We next analysed tissue morphology and immunohistochemical reactivity of colon tissue after fixation and paraffin-embedding using either formalin or RCL2 fixatives. RCL2 clearly preserved tissue integrity compared to the reference fixative of formalin (Fig. 5A and B).
Fig 5.
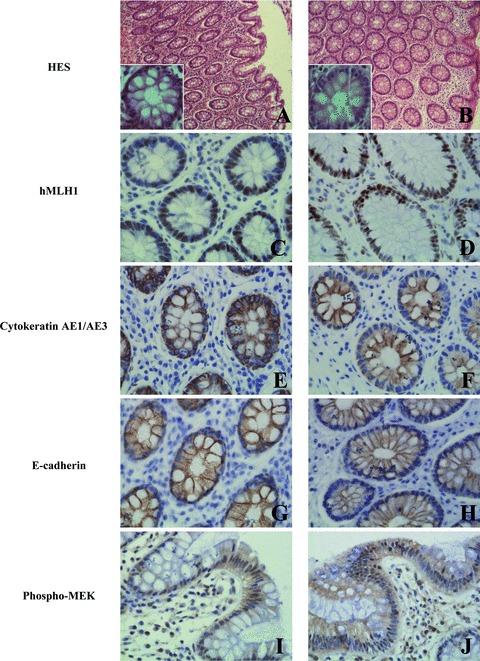
Morphology and immunohistochemical staining of hMLH1, cytokeratin AE1/AE3 and E-cadherin of normal colonic mucosa fixed with formalin and RCL2 on the left and right panels, respectively. HES stained sections (A, B); nuclear hMLH1 (C, D); cytoplasmic cytokeratin AE1/AE3 (E, F); membrane E-cadherin (G, H) and cytoplasmic and nuclear phospho-MEK (I, J) immunoreactivities are presented. Original magnification x 400. For panels A and B, original magnification x100, insets x400.
We then performed immunohistochemistry to compare antigen integrity and accessibility in both formalin and RCL2-fixed tissues. RCL2 fixative required optimization of the immunostaining procedures (i.e. antibody concentration dilutions), likely due to better antigenic preservation and accessibility. Immunoreactivities for various antibodies, including hMLH1 (Fig. 5C and D), cytoker-atin AE1/AE3 (Fig. 5E and F), E-cadherin (Fig. 5G and H) and phospho-MEK (Fig. 5I and J), were similar in RCL2P samples, as compared to FFPE tissue.
Whole and laser-microdissected RCL2P tissues allowed mass spectrometry profiling
To assess the effect of RCL2 on protein integrity and its compatibility with mass spectrometry analysis, whole frozen and RCL2-fixed tissues were analysed by surface-enhanced laser desorption/ionization (SELDI). Total protein extracts were loaded onto CM10 arrays, and SELDI profiles were obtained in duplicate for each sample. Figure 6A shows the spectrum of proteins retained on the CM10 arrays for both conditions. In the 2–50 kD range, 85 protein peaks were detected in frozen tissues, as already described [16]. As expected, the number of protein peaks in RCL2P tissue was similar to that detected in frozen tissue, with a mean of 81 peaks detected in the same mass range (Fig. 6A). Of note, the observed peaks were at the same position in the frozen and RCL2P samples, suggesting good protein preservation. Similar results were obtained with different surface arrays, such as H50, Q10 or immobilized metal affinity chromatography (IMAC) (data not shown).
Fig 6.
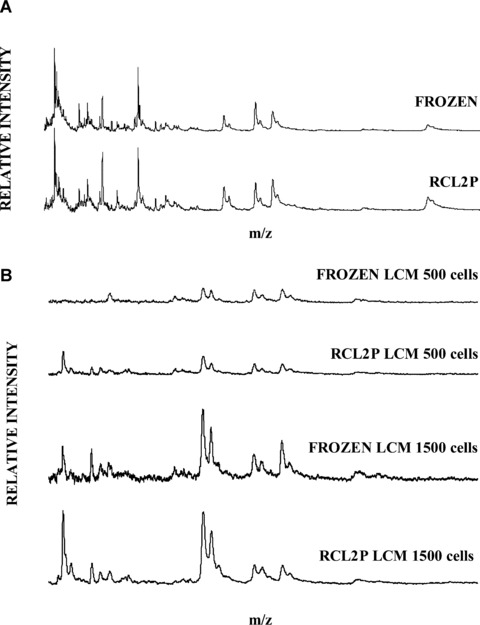
Mass spectrometry analysis of proteins extracted from whole RCL2P compared with frozen normal colonic tissue (A). LCM and mass spectrometry analysis of proteins extracted from 500 and 1500 cells from RCL2-fixed and frozen tissues (B).
LCM with ProteinChips analysis was then performed. Areas corresponding to 500 or 1500 cells were excised and lysed using protocol A. Five hundred cells from frozen and RCL2P tissues were sufficient to detect more than 59 and 56 peaks, respectively. However, with 1500 cells, up to 80 protein peaks were detected with higher intensities (Fig. 6B). By limiting cell numbers and increasing the reproducibility of protein profiles, RCL2P usefully displayed a sufficient number of peaks for differential expression analysis.
Discussion
FFPE tissue has been used for decades for tumour diagnosis and staging via light microscopic evaluation. Most tissues stored in hospitals and clinical laboratories across the world are then fixed in formalin, enabling retrospective biomarker investigation [17]. However, major drawbacks exist concerning FFPE tissue storage and fixation. First, formalin does not allow reproducible nucleic acid analysis, since it alters and fragments nucleic acids, and also impairs the extraction efficiency and quality of both DNA and RNA [6, 18–20]. Second, formalin leads to chemical reactions in tissues, such as the formation of methylenic bridges between protein side chains [21], rendering formalin fixation processes inappropriate to high-quality protein extraction, and more generally to proteomic investigations [22]. Thus, immunohistochemistry represents the only approach for elucidating protein expression and cellular localization in specific cellular populations [23], though with very poor absolute quantitative information [24, 25]. Third, formalin is an extremely toxic fixative; re-exposure by nasal, oral or dermal routes is a human health risk, with outcomes ranging from minor irritation of the eyes, nose and throat, to more severe complications including dysphagia, bronchitis, pneumonia and squamous cell carcinoma of the nose and pharynx [26–28]. RCL2, a promising new fixative, has great potential for concomitantly performing morphological and molecular analyses on the same tissue sample. We previously demonstrated that the analyses of RCL2P human breast tumour samples were highly suitable for tissue morphology, DNA preservation and RNA preservation, even after several months of tissue storage [12].
The aim of our study was to demonstrate that RCL2 was also suitable for proteomic analysis, providing RCL2P tissue as an alternative source of archival clinical material for biomarker investigations. We showed that the highest protein yield was obtained from RCL2P tissue, with different protein yields depending on the extraction protocols. The difference observed in protein yields with RLC2P versus the gold standard of frozen tissue can be largely explained by tissue retraction, which is usually observed with an ethanol-fixative reagent [29]. However, this effect did not impair the global tissue architecture, nor the cellular details of the tissue analysed. For RCL2P and frozen tissues, comprehensive protein preservation was confirmed from gel electrophoresis results with highly similar patterns. Western-blotting clearly revealed that RCL2 fixation associated with extraction protocol A enabled the recovery and detection of membrane, cytoplasmic and nuclear proteins. To our knowledge, this is the first demonstration of a fixative that enables the detection of after translational modifications, such as phosphorylation, suggesting that RLC2P induces limited or no alterations in protein structure. Moreover, since immunohistochemistry was performed using diluted antibody concentrations, the protein epitopes were rendered more accessible in RCL2P tissue.
Using the Qproteome FFPE kit, Becker et al. have shown that they were able to detect membrane, cytoplasmic and nuclear proteins by reverse phase protein microarray [2]. Surprisingly, using the same protocol, we observed a completely degraded protein profile on 1-DE and 2-DE gels (Figs 1 and 2). Nuclear and cytoplasmic proteins, such as Sp1 and α-tubulin, could not be detected by western-blot (Fig. 4B). Other recent studies developed conditions for formalin-tissue proteomic analysis based on a ‘bottom-up’ strategy for protein identification in FFPE samples, using trypsin digestion and a subsequent ‘shotgun’ liquid-chromatography-mass spectrometry (LC-MS) for protein identification [5, 22, 25, 30]. Although these approaches are relevant, one may be concerned by the possibility of introducing biases related to the effects of formalin on protein extraction and MS analysis. Particularly, some protein modifications can render them unidentifiable or misidentifiable using the LC-MS approach [5]. Interestingly, when we performed protein array mass spectrometry analysis on RCL2P, no differences in the protein peak positions were observed between frozen and RCL2P samples, suggesting appropriate preservation of these proteins without any mass modification. In conclusion, although FFPE-tissue archives enable retrospective proteomic analysis, as well as the discovery of new diseases or therapeutic markers, formalin does not represent the ideal fixative. It is necessary to modernize pathology and propose new alternatives for tissue archival. New fixatives should be designed to preserve histomorphologic features, similar to those seen in FFPE material, while protecting DNA, RNA, and protein in a manner comparable to fresh-frozen tissue. In our study, we performed a careful evaluation of RCL2 fixative using paraffin-embedded tissue samples and found that it possesses high performance regarding histomorphology and protein preservation, including protein extracted from microdis-sected tissue samples. In addition, analysis of 8-year old tissue sections showed that no morphological detail and protein degradation were seen by microscopic observation and immunohistochemistry, respectively, which strongly supports its use in long-term tissue storage (data not shown). According to previous results, we believe that the non-toxic and non-volatile reagent, RCL2, represents an easy-to-use alternative to FFPE, since it simultaneously protects the histomorphology and the integrity of macromolecules.
References
- 1.Lehmann U, Kreipe H. Real-time PCR analysis of DNA and RNA extracted from formalin-fixed and paraffin-embedded biopsies. Method. 2001;25:409–18. doi: 10.1006/meth.2001.1263. [DOI] [PubMed] [Google Scholar]
- 2.Becker KF, Schott C, Hipp S, Metzger V, Porschewski P, Beck R, Nahrig J, Becker I, Hofler H. Quantitative protein analysis from formalin-fixed tissues: implications for translational clinical research and nanoscale molecular diagnosis. J Pathol. 2007;211:370–8. doi: 10.1002/path.2107. [DOI] [PubMed] [Google Scholar]
- 3.Crockett DK. Lin Z. Vaughn CP. Lim MS, Elenitoba-Johnson KS. Identification of proteins from formalin-fixed paraffin-embedded cells by LC-MS/MS. Lab Invest. 2005;85:1405–15. doi: 10.1038/labinvest.3700343. [DOI] [PubMed] [Google Scholar]
- 4.Hwang SI, Thumar J, Lundgren DH, Rezaul K, Mayya V, Wu L, Eng J, Wright ME, Han DK. Direct cancer tissue pro-teomics: a method to identify candidate cancer biomarkers from formalin-fixed paraffin-embedded archival tissues. Oncogene. 2007;26:65–76. doi: 10.1038/sj.onc.1209755. [DOI] [PubMed] [Google Scholar]
- 5.Palmer-Toy DE, Krastins B, Sarracino DA, Nadol JB, Jr, Merchant SN. Efficient method for the proteomic analysis of fixed and embedded tissues. J Proteome Res. 2005;4:2404–11. doi: 10.1021/pr050208p. [DOI] [PubMed] [Google Scholar]
- 6.Lewis F, Maughan NJ, Smith V, Hillan K, Quirke P. Unlocking the archive–gene expression in paraffin-embedded tissue. J Pathol. 2001;195:66–71. doi: 10.1002/1096-9896(200109)195:1<66::AID-PATH921>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 7.Shibutani M, Uneyama C, Miyazaki K, Toyoda K, Hirose M. Methacarn fixation: a novel tool for analysis of gene expressions in paraffin-embedded tissue specimens. Lab Invest. 2000;80:199–208. doi: 10.1038/labinvest.3780023. [DOI] [PubMed] [Google Scholar]
- 8.Takagi H, Shibutani M, Kato N, Fujita H, Lee KY, Takigami S, Mitsumori K, Hirose M. Microdissected region-specific gene expression analysis with methacarn-fixed paraffin-embedded tissues by real-time RT-PCR. JHistochem Cytochem. 2004;52:903–13. doi: 10.1369/jhc.3A6215.2004. [DOI] [PubMed] [Google Scholar]
- 9.Morales AR, Essenfeld H, Essenfeld E, Duboue MC, Vincek V, Nadji M. Continuous-specimen-flow, high-throughput, 1-hour tissue processing. A system for rapid diagnostic tissue preparation. Arch Pathol Lab Med. 2002;126:583–90. doi: 10.5858/2002-126-0583-CSFHTH. [DOI] [PubMed] [Google Scholar]
- 10.Stanta G, Mucelli SP, Petrera F, Bonin S, Bussolati G. A novel fixative improves opportunities of nucleic acids and proteomic analysis in human archive's tissues. Diagn Mol Pathol. 2006;15:115–23. doi: 10.1097/00019606-200606000-00009. [DOI] [PubMed] [Google Scholar]
- 11.Vincek V, Nassiri M, Nadji M, Morales AR. A tissue fixative that protects macro-molecules (DNA, RNA, and protein) and histomorphology in clinical samples. Lab Invest. 2003;83:1427–35. doi: 10.1097/01.lab.0000090154.55436.d1. [DOI] [PubMed] [Google Scholar]
- 12.Delfour C, Roger P, Bret C, Berthe ML, Rochaix P, Kalfa N, Raynaud P, Bibeau F, Maudelonde T, Boulle N. RCL2, a new fixative, preserves morphology and nucleic acid integrity in paraffin-embedded breast carcinoma and microdissected breast tumor cells. J Mol Diagn. 2006;8:157–69. doi: 10.2353/jmoldx.2006.050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. AnalChein. 1996;68:850–8. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 14.Solassol J, Marin P, Demettre E, Rouanet P, Bockaert J, Maudelonde T, Mange A. Proteomic detection of prostate-specific antigen using a serum fractionation procedure: potential implication for new low-abundance cancer biomarkers detection. Anal Biochem. 2005;338:26–31. doi: 10.1016/j.ab.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 15.Becamel C, Galeotti N, Poncet J, Jouin P, Dumuis A, Bockaert J, Marin P. A proteomic approach based on peptide affinity chromatography, 2-dimensional elec-trophoresis and mass spectrometry to identify multiprotein complexes interacting with membrane-bound receptors. Blol Proced Online. 2002;4:94–104. doi: 10.1251/bpo39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melle C, Bogumil R, Ernst G, Schimmel B, Bleul A, von Eggeling F. Detection and identification of heat shock protein 10 as a biomarker in colorectal cancer by protein profiling. Proteomics. 2006;6:2600–8. doi: 10.1002/pmic.200500427. [DOI] [PubMed] [Google Scholar]
- 17.Hood BL, Conrads TP, Veenstra TD. Unravelling the proteome of formalin-fixed paraffin-embedded tissue. Brief Fund Genomic Proteomic. 2006;5:169–75. doi: 10.1093/bfgp/ell017. [DOI] [PubMed] [Google Scholar]
- 18.Frank TS. Svoboda-Newman SM. Hsi ED. Comparison of methods for extracting DNA from formalin-fixed paraffin sections for nonisotopic PCR. Diagn Mol Pathoi. 1996;5:220–4. doi: 10.1097/00019606-199609000-00012. [DOI] [PubMed] [Google Scholar]
- 19.Masuda N, Ohnishi T, Kawamoto S, Monden M, Okubo K. Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res. 1999;27:4436–43. doi: 10.1093/nar/27.22.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shibata D. Extraction of DNA from paraffin-embedded tissue for analysis by poly-merase chain reaction: new tricks from an old friend. Hum Pathoi. 1994;25:561–3. doi: 10.1016/0046-8177(94)90219-4. [DOI] [PubMed] [Google Scholar]
- 21.Werner M, Chott A, Fabiano A, Battifora H. Effect of formalin tissue fixation and processing on immunohisto-chemistry. Am J Surg Pathoi. 2000;24:1016–9. doi: 10.1097/00000478-200007000-00014. [DOI] [PubMed] [Google Scholar]
- 22.Hood BL, Conrads TP. Veenstra TD. Mass spectrometric analysis of formalin-fixed paraffin-embedded tissue: unlocking the proteome within. Proteomics. 2006;6:4106–14. doi: 10.1002/pmic.200600016. [DOI] [PubMed] [Google Scholar]
- 23.Cheuk W, Chan JK. Subcellular localization of immunohistochemical signals: knowledge of the ultrastructural or biologic features of the antigens helps predict the signal localization and proper interpretation of immunostains. IntJ Surg Pathoi. 2004;12:185–206. doi: 10.1177/106689690401200301. [DOI] [PubMed] [Google Scholar]
- 24.Grube D. Constants and variables in immunohistochemistry. Arch Histol Cytoi. 2004;67:115–34. doi: 10.1679/aohc.67.115. [DOI] [PubMed] [Google Scholar]
- 25.Shi SR, Liu C, Balgley BM, Lee C, Taylor CR. Protein extraction from formalin-fixed paraffin-embedded tissue sections: quality evaluation by mass spectrometry. J Histochem Cytochem. 2006;54:739–43. doi: 10.1369/jhc.5B6851.2006. [DOI] [PubMed] [Google Scholar]
- 26.Kilburn KH, Warshaw R, Boylen CT, Johnson SJ, Seidman B, Sinclair R, Takaro T., Jr Pulmonary and neurobehavioral effects of formaldehyde exposure. Arch Environ Health. 1985;40:254–60. doi: 10.1080/00039896.1985.10545928. [DOI] [PubMed] [Google Scholar]
- 27.Kilburn KH, Warshaw R, Thornton JC. Formaldehyde impairs memory equilibrium, and dexterity in histology technicians: effects which persist for days after exposure. Arch Environ Health. 1987;42:117–20. doi: 10.1080/00039896.1987.9935806. [DOI] [PubMed] [Google Scholar]
- 28.Morgan KT. A brief review of formaldehyde carcinogenesis in relation to rat nasal pathology and human health risk assessment. Toxicol Pathoi. 1997;25:291–307. doi: 10.1177/019262339702500307. [DOI] [PubMed] [Google Scholar]
- 29.Ahram M, Flaig MJ, Gillespie JW, Duray PH, Linehan WM, Ornstein DK, Niu S, Zhao Y, Petricoin EF, 3rd, Emmert-Buck MR. Evaluation of ethanol-fixed, paraffin-embedded tissues for proteomic applications. Proteomics. 2003;3:413–21. doi: 10.1002/pmic.200390056. [DOI] [PubMed] [Google Scholar]
- 30.Bagnato C, Thumar J, Mayya V, Hwang SI, Zebroski H, Claffey KP, Haudenschild C, Eng JK, Lundgren DH, Han DK. Proteomics analysis of human coronary atherosclerotic plaque: a feasibility study of direct tissue proteomics by liquid chromatography and tandem mass spectrometry. Mol Cell Proteomics. 2007;6:1088–102. doi: 10.1074/mcp.M600259-MCP200. [DOI] [PubMed] [Google Scholar]


