Abstract
The repair of cutaneous wounds in the adult body involves a complex series of spatially and temporally organized processes to prevent infection and restore homeostasis. Three characteristic phases of wound repair (inflammation, proliferation including re-epithelialization and remodelling) overlap in time and space. We have utilized a human skin wound-healing model to correlate changes in genotype and pheno-type with infrared (IR) and confocal Raman spectroscopic images during the re-epithelialization of excisional wounds. The experimental protocols validated as IR images clearly delineate the keratin-rich migrating epithelial tongue from the collagen-rich wound bed. Multivariate statistical analysis of IR datasets acquired 6 days post-wounding reveal subtle spectral differences that map to distinct spatial distributions, which are correlated with immunofluorescent staining patterns of different keratin types. Images computed within collagen-rich regions expose complementary spatial patterns and identify elastin in the wound bed. The temporal sequence of events is explored through a comparison of gene array analysis with confocal Raman microscopy. Our approach demonstrates the feasibility of acquiring detailed molecular structure information from the various proteins and their subclasses involved in the wound-healing process.
Keywords: wound healing, migrating epithelial tongue, confocal Raman microscopy, infrared imaging
Introduction
The response of the adult body to cutaneous wounding involves the organization of complex spatially and temporally coordinated processes to initiate rapid sealing of the site, prevention of infection and restoration of the injured tissue [1]. Re-epithelialization of skin wounds begins within hours following the injury [2]. Keratinocytes at the wound edge become activated, start to proliferate and migrate, aiming to cover the wound [3]. Within the first 2 days, a layer of epidermal cells known as the migrating epithelial tongue (MET) forms [2]. The process is essentially complete (∼7–9 days following the injury) when the denuded wound bed is covered by a layer of keratinocytes, followed by the re-establishment of the stratified epidermal layer. These major organizational changes have been substantially studied with genomic and proteomic approaches.
To date, methods that directly characterize the molecular structure and spatial distribution of the newly synthesized tissue constituents have been lacking. Such information would be of particular interest for processes such as the evaluation of therapeutic modalities and the study of interfaces between native skin and 3-D skin equivalents.
Vibrational microspectroscopic imaging is becoming widely applied for analysis of complex organized systems, including cells and tissues [4-6]. Both Raman and infrared (IR) spectroscopy provide direct molecular structure information, and possess the advantages of requiring no staining, no probe molecules and (for Raman) minimal sample preparation. In addition, for Raman microscopy spectral images may be acquired in a confocal manner. These approaches have allowed us to evaluate the permeation of drugs and enhancers into the skin and to monitor reversible solvent-induced structure changes in the keratin of single corneocytes [5, 6]. In the current study, we demonstrate the feasibility of tracking changes in the spatial distribution of the major skin proteins within the first several days after wounding in an organ culture wound-healing model and of correlating these changes with immunofluo-rescent staining patterns and data from microarray analysis.
Materials and methods
Human organ culture wound-healing model
Human skin specimens, obtained from reduction abdominoplasty in accordance with approved institutional protocol, were used to generate acute wounds as previously described [7]. A 3-mm biopsy punch was used to create an acute wound. Specimens were maintained at the air–liquid interface with Dulbecco's Modified Eagle's Medium (DMEM) (BioWhittaker Walkersville, MD, USA), antibiotic/antimycotic and foetal bovine serum (Gemini Bio – Products, West Sacramento, CA, USA) at 37°C, 5% CO2- and 95% relative humidity for 6 days.
Histology and immunohistochemistry
Paraffin-embedded normal skin and acute wound specimens were cut on a microtome (Carl Zeiss, Thornwood, NY, USA) and 5-μm thick sections were stained with haematoxylin and eosin (H&E). For immunofluorescent stainings, sections were de-waxed in xylene, re-hydrated and washed with 1x phosphate buffered saline (PBS). For antigen retrieval, paraffin sections were heated in a 95°C water bath in Target Retrieval Solution (DAKO Corporation, Carpinteria, CA, USA) and washed. Sections were blocked with 5% Bovine Serum Albumin (BSA) (Sigma, St. Louis, MO, USA) in 1x PBS for 30 min. Incubation with specific antibody against keratin 17 (gift from Dr. Coulombe) and keratin 14 (gift from Dr. Lane) diluted in 5% BSA was carried over night at 4°C. Slides were then rinsed in PBS and incubated with a secondary fluorescent – conjugated goat anti-mouse IgG Alexa Fluor 594 (Invitrogen, Eugene, OR, USA) or goat anti-rabbit Alexa Fluor 488 (Invitrogen) for 1 hr at room temperature. After a final wash in PBS, the sections were mounted using media containing 4'-6-diamidino-2-phenylindole, DAPI (Vector Labs, Burlingame, CA, USA), and examined under a Carl Zeiss microscope (Carl Zeiss, Thornwood, NY, USA). Digital images were collected using the Adobe Photoshop 4.0 TWAIN 32 program (Adobe Systems Incorporated, San Jose, CA, USA) and processed using Powerpoint (Microsoft, Corporation, Redmond, WA, USA).
Preparation and hybridization of probes
Briefly, unwounded and wounded skin specimens maintained at air-liquid interface for 48 and 96 hrs were homogenized and total RNA was isolated using an RNeasy Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer's protocol. Approximately 5 mg of total RNA was reverse transcribed, amplified and labelled as previously described [8]. Labelled cRNA was hybridized to HG-U95-set Gene Chip (Affymetrix, Santa Clara, CA, USA). The arrays were washed and stained with avidin-biotin streptavidin-phycoerythin labelled antibody using Affymetrix fluidics station and then scanned using the Agilent GeneArray Scanner system (Hewlett-Packard, Palo Alto, CA, USA) as described by Affymetrix.
Gene array data analysis
Microarray Suite 5.0 (Affymetrix) was used for data extraction and for further analysis. Data mining tool 3.0 (Affymetrix) and GeneSpring™ software 7.3.1 (Silicon Genetics, Redwood City, CA, USA) were used for normalization and degree of change and P-value calculations. Samples were normalized per chip to the 50th percentile and per gene to a median. Statistical comparisons of expression level between each condition were performed using anova test. Only genes with a P-value less than 0.05 were considered to be statistically significant. Differential expressions of transcripts were determined by calculating the degree of change. Genes were considered regulated if the expression levels differed more than two-fold relative to healthy, unwounded skin.
IR and Raman experiments
Unwounded and wounded skin samples maintained at air–liquid interface for specific periods of time (up to 7 days) were selected for IR imaging and confocal Raman microspectroscopy measurements.
For IR imaging, samples were flash frozen with liquid nitrogen and stabilized at −30°C on a chuck within a Bright/Hacker 5030 Microtome (Bright Instrument Company, Huntington, UK/Hacker Instruments, Fairfield, NJ, USA) oriented so that sections (5-μ(τι thick) were cut perpendicular to the stratum corneum (SC). The sections, placed on BaF2 IR windows, were mapped using a Perkin-Elmer Spotlight system equipped with an essentially linear array (16 × 1 detector elements) mercury-cad-mium-telluride (MCT) detector. An automated high-precision XY sample stage permitted the collection of IR images (pixel area of 6.25 μπι2) over ∼0.5 × 0.5 mm sample areas. Spectral resolution was 8 cm−1.
For confocal Raman imaging, samples were gently pressed, SC side up, into a milled brass cell and sealed with a microscope coverslip to prevent sample dehydration. Confocal Raman maps were acquired with a Kaiser Optical System Raman Microprobe described in detail previously [5]. Briefly, a solid-state diode laser (785 nm) is focused with a 100X oil immersion objective generating ∼7 mW of single mode power at the sample. The backscattered light is collected over a volume of ∼2 μm3 within the sample and illuminates a near-IR CCD (ANDOR Technology, Belfast, Ireland, UK, Model DU 401 -BR-DD). Spectral resolution is 4 cm−1 over the 100–3450 cnT−1 region. Spectra are acquired using a 60-sec. exposure time, two accumulations and cosmic ray correction.
IR and Raman spectra were analysed using ISys software version 3.1.1.17 (Malvern Instruments, LTD, UK). Image planes were generated after linear baselines were applied over spectral regions of interest and factor analysis was conducted. Details of the ISys factor analysis protocol (score segregation) are given elsewhere [5, 6].
Results and discussion
Validation of the experimental protocol: IR imaging, immunofluorescent staining and gene array analysis of a healing wound
The visible image (Fig. 1A) of a skin section acquired 4 days following wounding clearly reveals a MET (marked with the red arrow) progressing towards the exposed wound bed on the left side of the image. The mature SC is distinguishable at the top of the unwounded area to the right of the marked edge (partially torn from the underlying epidermis). The observation of the MET (Fig. 1A) demonstrates that healing processes evidently persist in the explant, even after 4 days. Factor analysis was conducted on an IR imaging dataset obtained from the same skin section. Score images (Fig. 1B and C) were generated based on factor loadings derived from the protein Amide I and II spectral region (1475–1720 cm−1), where the vibrational modes are sensitive to secondary structure. Factor loadings 1 and 2 (f1, f2) (Fig. 1D) represent collagen and keratin, respectively, as identified from spectra of model compounds and IR images from uninjured skin tissue [6]. In Fig. 1D, factor loadings compare well with average spectra acquired from regions where the corresponding loadings have high scores (e.g. spectra are averaged in the red areas of Fig. 1B and compared to loading f1). The Amide I mode (∼1650 cm−1) arises predominantly from peptide bond C = 0 stretch. Factor 1 reveals a major Amide I peak at 1660 cm−1 and a shoulder at -1644 cm−1, both characteristic of collagen [6]. Factor 2 depicts a typical Amide I contour for α-helical keratin at ∼1652 cm−1. Thus, the scores of f1 and f2, as depicted in Fig. 1B and C, depict the collagen-rich dermis and the keratin-rich epidermis, respectively.
Fig 1.
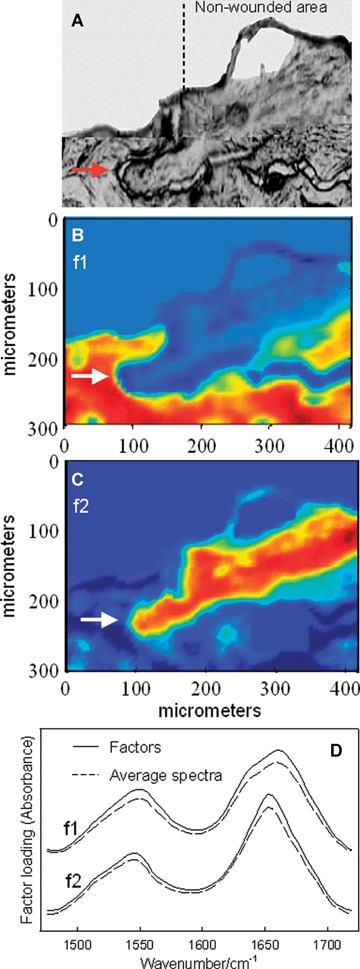
Detection and IR characterization (factor analysis conducted over the 1475–1720 cm−1 region) of a migrating epithelial tongue (MET) in a wound-healing model, 4 days post-wounding. (A) Visible image of an unstained skin section depicting the wounded and non-wounded areas, along with the MET (marked with arrow). IR images were acquired of this same skin section. (B) IR score image for factor loading 1 (f1) highlighting collagen-rich areas of the sample. (C) IR score image for factor loading 2 (f2) highlighting keratin-rich areas. (D) Factor loadings overlaid with spectra averaged from corresponding regions of high scores. Factor loading 1 displays the typical Amide I and II features of collagen and f2 is typical for keratin. Colour coding of all score images: red > yellow > blue.
Additional aspects of the molecular structure/composition changes that occur during wound healing are deduced from analysis of the 1185–1475 cm−1 region in a spatial area containing a wound edge and MET at 6 days post-wounding. The visible image of the skin section used for IR imaging is presented in Fig. 2A. The MET progresses from the unwounded area to the right side of the image where it appears as if collagen has been removed from the top of the MET during microtoming. Six factors (Fig. 2H) from the IR dataset and their score images are presented in Fig. 2B–G. The main spectral features in this wave number region arise from methyl and methylene group deformations (−1450 cm−1), carboxylate symmetric stretch (−1400 cm−1), a collagen ring mode (−1335 cm−1) and the Amide III mode (−1235 cm−1) comprised mainly of C-N stretching and N-H in plane bending. Factors 1–4 (f1–4) with similar spectral features (Fig. 4H) map to four spatially distinct keratin-rich areas; f5 and f6 are spatially restricted to collagen-rich regions. The score image for f1 (Fig. 2B) highlights the SC and provides a clear marker for the original wound edge as labelled in Fig. 2A. On the left side of the image for f1 (Fig. 2B), high scores continue throughout the SC and carry over into the viable epidermal region. Score images for f2 and f3 expose the suprabasal and basal epidermal regions (Fig. 2C and D), respectively, in the unwounded area with high scores extending over a large portion of the MET for f2. The score image for the basal layer also reveals a thin line of spectra with somewhat high scores extending into the lower region of the MET. The 4th score image with keratin-like spectral features appears to be spatially restricted to the outer and leading edges of the MET (Fig. 2E). Spectra located in the outer edges of the MET 4 days post-wounding (Fig. 1) share similar characteristics, i.e. a downward frequency shift in the carboxylate symmetric stretch, with loading f4 in the 6-day section. To begin to correlate the spectral variations in the keratin factors with changes in molecular composition taking place during re-epithelialization, we examined the expression of two different keratin types in our wound-healing model.
Fig 2.
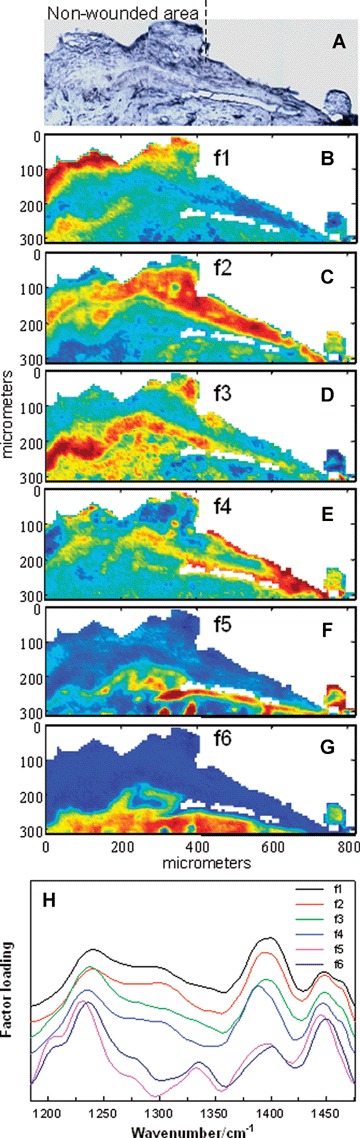
IR characterization (factor analysis conducted over the 1185–1475 cm−1 region) of wounded and non-wounded areas 6 days post-wounding. (A) Optical image of an unstained section with the edge of the wounded area marked. The MET is shown covering the wounded area on the right side of the micrograph. A tear is evident in the image of the thin section between the MET and the underlying provisional matrix. IR spectra were acquired of the same section and pixels corresponding to the tear were masked prior to statistical analysis. (B) The score image for f1 depicting the stratum corneum and part of the viable epidermis (red) of the non-wounded area in the skin section. (C) The score image for f2 highlights the spatial area of the suprabasal epidermis (red) proximal to the wound and in the MET. (D) The spatial distribution of high scores for f3 predominantly marks the basal epidermal layer in the non-wounded area. Scores are also somewhat higher (yellow) in the lower regions of the MET. (E) The score image for f4 illuminates the outer, leading edges of the MET (red). (F) The score image of f5 highlights a collagen-rich area directly under the MET. (G) The score image for f6 highlights a collagen-rich area that is continuous in both non-wounded and wounded (under the MET) regions. (H) Factor loadings used to generate the score images (B-G). Loadings f1-f4 are characteristic for keratin and map to keratin-rich spatial regions. High scores for f5 and f6 highlight collagen-rich areas.
Fig 4.
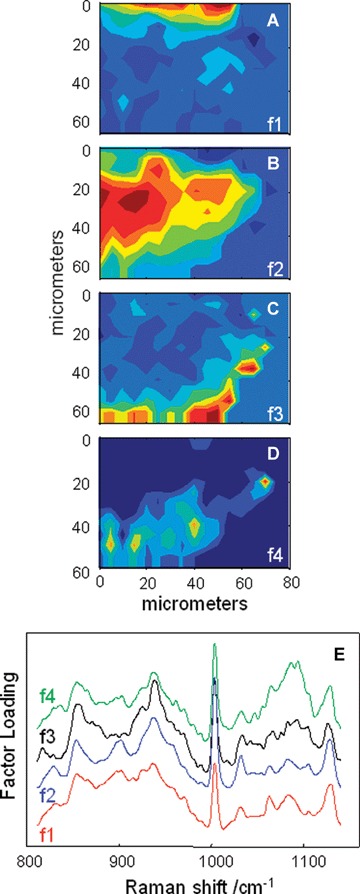
Factor analysis of a confocal Raman dataset delineates skin regions near a wound edge 0.5 days post-wounding. Factor analysis was conducted over the 800–1140 cm−1 region yielding four loadings that map to anatomically distinct regions in skin. (A) The spatial distribution of scores for f1 highlights the stratum corneum region of the skin, rich in keratin-filled corneocytes and lipids. (B) Factor loading 2 shows high scores in the underlying epidermal region while high scores for f3 (C) reside near the dermal-epidermal boundary region. (D) The size, location and spatial distribution of several smaller regions with high scores for f4 are identified as cell nuclei. (E) Factor loadings reveal several spectral features specific to the microanatomy of the epidermis in human skin.
In healthy, unwounded skin, keratinocytes in the basal layer express keratins 5 and 14, while suprabasal keratinocytes express keratins 1 and 10. Epithelialization of the wound begins when keratinocytes at the wound edge become activated, characterized by the expression of keratins 6,16 and 17 (K6, K16 and K17) [3]. To confirm the presence of activated keratinocytes at the wound edge in our model, we stained sections of acute wounds with K17-specific antibody. Sections of wounds and healthy skin were also stained with H&E, DAPI and K14-specific antibody (see Fig. 3). Keratin 17 was not present at the wound edge immediately after the wounding (Fig. 3B, 0 hr). However, 24 hrs after wounding we observed strong suprabasal K17 staining at the wound edge indicating activation of keratinocytes. Strong K17 staining persisted over 4 days after the wounding. Furthermore, keratinocytes composing the MET were positive for K17 (see 72- and 96-hr images), thus confirming migration of activated suprabasal keratinocytes over the wound bed. As anticipated, we did not observe K17 staining in healthy, unwounded skin. When we stained acute wounds with K14, we observed positive staining in the basal layer of the wound edge immediately after the wounding (Fig. 3C, 0 hr). A layer of keratinocytes covering the wound bed was also positive for K14 throughout the 4-day period indicating the presence of a basal keratin type (K14) in the MET. The immunofluorescent staining patterns shown in Fig. 3 are in agreement with recent studies [9, 10].
Fig 3.
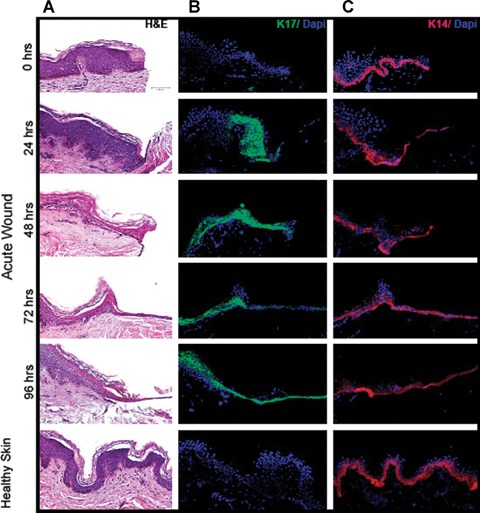
H&E, DAPI, and immunoflu-orescent keratin staining of healthy and acute wound sections as a function of healing time. (A) H&E stained sections. (B) Sections stained with K17-specific antibody and DAPI. (C) Sections stained with K14-specific antibody and DAPI.
A comparison of the IR imaging analysis with keratin immunofluorescent staining reported herein and previously [9, 10] suggests the following. Factor loading 1 and its corresponding score image (Fig. 2H and B, respectively) are characteristic of differentiating keratinocytes, rich in K1/10. The high-score areas of f2 (Fig. 2C) represent regions abundant in K17, whereas high scores for f3 (Fig. 2D) depict areas rich in K14. Furthermore, the high-score areas for f4 (Fig. 2E), encompassing the leading/outer edges of the MET, partially overlap high-score regions for both f2 (tip of MET) and f3 (bottom of MET extending back into non-wounded area). It follows that regions with high scores for f4 may represent areas with a higher content of both K14 and K17 consistent with the current staining and previous reports [9, 10]. Comparison of f4 with f2 and f3 also reveals the presence of an additional component or a change in a structural feature in f4 indicated by a shift to lower frequency in the ∼1400 cm−1 car-boxylate stretching mode. Utilizing a single skin section to elucidate the spatial distribution of multiple components is advantageous as section-to-section variability is avoided.
Gene microarray analysis did not detect the variation in keratin types possibly due to the fact that keratins are highly expressed in the skin and as a result, they exceed a linear range of hybridization thus leading to variable microarray readings [11, 12].
IR imaging also reveals two distinct collagen-like factor loadings (f5 and f6, Fig. 2H) with complementary spatial distributions (Fig. 2F and G). The highest scores for f5 predominantly reside directly under the MET, whereas high scores for f6 are distributed throughout the dermal area including a deeper region underneath the MET. Thus, f5 may contain features specific to an early provisional matrix enabling keratinocyte migration [13].
In healthy skin, type I collagen is the major component of the dermal extracellular matrix (80%), while type III collagen comprises only 10%. However 48–72 hrs post-wounding, collagen III becomes the dominant collagen in the dermis [13]. Data obtained from microarray analysis confirm the reported changes in collagen expression 48 and 96 hrs after wounding (Table 1). An increase in collagen type IV was also observed as previously reported [14]. In addition, we see an increase in other collagen subtypes (see Table 1). However, the significance of this is yet to be determined, as these subtypes are not known as major contributors. Post-wounding, collagen I decreases ∼1.5-fold compared to healthy skin and type III has a ∼2.2-fold increase after 96 hrs. Overall it appears as if the observed variation in factors (or spectra) obtained from IR imaging provides useful information about the spatial and sequential distribution of keratin and collagen types during the first 6 days of the wound-healing process in our model. In addition to elucidating fundamentals of the wound-healing process, it is easy to envision the technique's potential to evaluate therapeutic interventions.
Table 1.
Results of gene array analysis
| Fold change Wound/Control | Gene symbol | Unlgene comment | Function | |
|---|---|---|---|---|
| 48 hrs | 96 hrs | |||
| 1.05 | −1.47 | COL1A2 | collagen, type I, alpha 2 | ECM |
| 1.19 | 2.19 | COL3A1 | collagen, type III, alpha 1 | ECM |
| 2.4 | 2.86 | COL4A1 | collagen, type IV, alpha 1 | ECM |
| 3.02 | 2.38 | COL4A2 | collagen, type IV, alpha 2 | ECM |
| 1.41 | 3.35 | COL5A2 | collagen, type V, alpha 2 | ECM |
| 1.2 | −1.03 | COL6A1 | collagen, type VI, alpha 1 | ECM |
| 1.78 | 1.87 | COL6A2 | collagen, type VI, alpha 2 | ECM |
| 1.13 | 1.83 | COL6A3 | collagen, type VI, alpha 3 | ECM |
| −1 | 4.71 | COL9A3 | collagen, type IX, alpha 3 | ECM |
| 1.86 | −1.11 | COL13A1 | collagen, type XIII, alpha 1 | ECM |
| 2.26 | 1.24 | COL15A1 | collagen, type XV, alpha 1 | ECM |
| 1.68 | 2.59 | COLQ | collagen-like tail subunit | ECM |
| 1.1 | 1.23 | MFAP4 | microfibrillar-associated protein 4 | ECM |
| 1.69 | −6.09 | CTSS | cathepsin S | Proteolysis |
| 4.06 | 6.08 | MMP7 | Matrix metalloproteinase 7 | Proteolysis |
| 2.48 | −1.08 | LOX | lysyl oxidase | AA metabolism |
Confocal Raman microspectroscopy and gene array analysis of a healing wound
Whereas FTIR imaging samples relatively large areas of micro-tomed sections rapidly, Raman spectroscopy provides data in a confocal manner (‘Optical sectioning’) from full thickness skin. To amplify spectral differences in unwounded areas of skin approaching a wound edge, a confocal Raman image was acquired starting at ∼60–80 μm from the left edge of a wound in the non-wounded area. Spectra were acquired 12 hrs post-wounding and the results from factor analysis, conducted in the 800–1140 cm−1 spectral region, are shown in Fig. 4 (score images in Fig. 4A-D and loadings in Fig. 4E). Factors 1 and 2 are assigned to the SC and the viable epidermis, respectively, where keratin is the main protein. Factor loadings in these two layers are similar. By normalizing the spectral intensity to the phe ring breathing mode at 1004 cm−1, it is clear that f1 has stronger bands at 1060 cm−1,1083 cm−1 and 1125 −−1 compared to f2. These three bands arise from lipid C-C skeletal stretching modes [5]. The observation of slightly stronger lipid bands in f1 is consistent with the increased lipid concentration in the SC. Factor 3 is assigned to the epidermal basal layer-dermal boundary. Consistent with the fact that keratin synthesis commences in the spinosum directly above the basal layer, the factor loading from the basal layer displays a diminution of the characteristic keratin band at 900 cm−1 (compare f3 to f1 and f2 in Fig. 4E). This keratin element is replaced by traces of characteristic collagen features at −922 cm−1 and 877 cm−1 (f3, in Fig. 4E). Finally, f4 is suggested to arise from cell nuclei with strong bands at 1092 cnT1 (arising from the phosphodiester moiety of DNA) and a cytosine band at 785 cm−1 (not shown) [5]. The ability to identify cell nuclei along with their spatial distribution raises the possibility of monitoring the temporal sequence of cellular events during re-epithelialization.
The time-dependence of the spatial distribution of components during healing was also investigated by confocal Raman spec-troscopy. Spectra were collected at the centre of a wound that had been healing for 2, 4 and 6 days and compared to a non-wounded area of skin. The results of factor analysis, conducted over the 800–1140 cm−1 region, are shown in Fig. 5 (factor loadings in Fig. 5A and score images in Fig. 5B-D). In the loading plots (Fig. 5A), f1 differs from the others by the presence of a shoulder at 906 cnT1 and a more intense feature at 1105 cm−1. To explore the origin of this factor, the average spectrum of pixels having low scores in the area surrounding the higher scoring regions (in Fig. 5B) was subtracted from the average spectrum of pixels with high scores for ft The resulting difference spectrum (Fig. 5E) is essentially identical to that of bovine elastin powder [15]. It is likely that the elastin component was not identified as a single factor due to the small diameter of the elastin fibre bundles (∼0.25–1.0 μm) in the papillary dermis [16] compared to the spatial resolution (2–5 μm) in the current Raman experiment. Hence, f1 contains features of both elastin and collagen. One marked change observed in the temporal series is the variation in the elastin spatial distribution. Elastin levels in the wound bed or regenerating tissue have greatly decreased by the second day compared to the normal tissue and the levels remain essentially the same throughout the 6-day healing period. Although reports concerning elastic fibre degradation and regeneration during wound healing are available, there is considerable inconsistency in both the experimental models used and the findings [17, 18].
Fig 5.
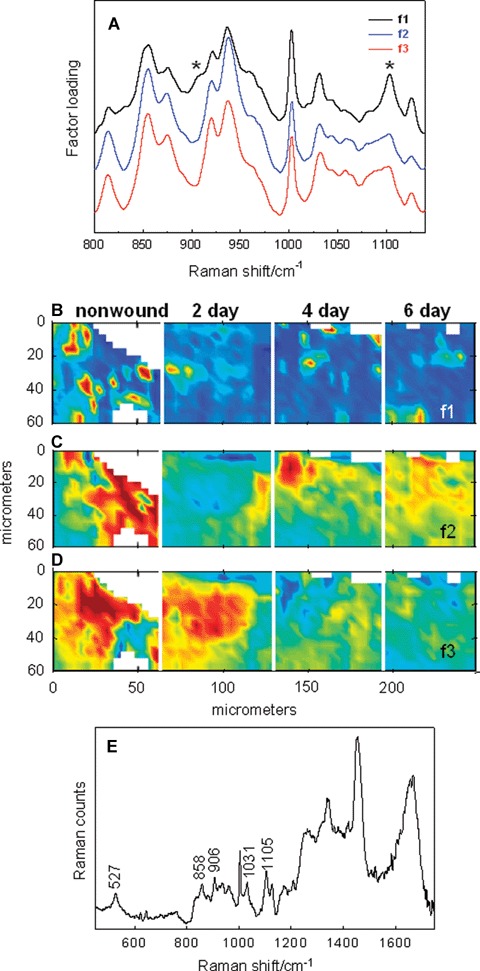
Time-dependent events during wound healing monitored by confocal Raman microscopy and factor analysis (800–1140 cm"1 region). Spectra acquired in a non-wounded area (30-50 μm beneath the epidermis) and 2, 4 6 days post-wounding (in the centre the wound bed) are collectively analysed. (A) Three factor loadings for the concatenated dataset are overlaid. (B) Score image for f1 depicts the time-dependence of the elastin distribution pre- and post-wounding. (C and D) Temporal and spatial distribution of score images for f2 and f3 collagen-rich factors. (E) A difference spectrum based on the score image of f1 identifies elastin in the wound bed (see text for details).
Proteolytic degradation of elastin has been associated with cathepsin S and matrix metalloproteinase 7 (MMP7) activities [19]. Microarray analysis shows increased expression of cathepsin S and MMP-7 48 hrs after wounding and MMP7 expression continues to significantly increase up to 96 hrs (Table 1). It has been shown that lysyl oxidases are essential for the elastic fibre homeostasis and maintenance at adult ages [20]. Four days after the wounding, we see suppression of lysyl oxide in our wounded skin explants. Diminished levels of elastin fibres in the dermis during wound healing in our model can be possibly explained by changes in cathepsin S, MMP-7 and lysyl oxidase.
Furthermore, the factor loadings f2 and f3 (Fig. 5A) depict the characteristic doublet of doublets pattern in the region 830–980 cm−1 specific to collagen. These bands arise from Pro and Hyp amino acid side chain modes. Although the band positions for these factors are largely the same, changes in the relative intensities of several bands are sufficient so that two factor loadings, possibly indicative of different collagen types and/or levels of hydration (unpublished work), are computed. The score images of f2 and f3 partially complement each other (compare Fig. 5C to D). In particular, the scores for f2 are shown to increase with healing time as the scores for f3 decrease.
In conclusion, we used a novel approach to track changes in the major skin proteins and their spatial distribution during wound healing. The spectroscopic methods complement the microarray array analysis and immunofluorescent staining by providing a molecular structure-based characterization of several factors, mostly protein, in a single skin section. Further development in the spectral analysis of additional wound-healing samples and purified materials under controlled environmental conditions will provide us with the opportunity to exploit the full realm of molecular information inherent in the data. This in turn may begin to provide a basis for understanding changes in the observed spatial distributions of particular proteins during therapeutic interventions for wound healing.
Notice of Retraction: “lntron-1 rs3761548 is Related to the Defective Transcription of Foxp3 in Psoriasis through Abrogating E47/c-Myb Binding”
This paper, by Zhu Shen, Ling Chen, Fei Hao, Gang Wang, Pingshen Fan and Yufeng Liu has been retracted at the request of authors Yufeng Liu and Gang Wang. The retraction has been agreed due to the use of the names of Professors Yufeng Liu and Gang Wang on the article without their knowledge or consent.
The Editor of Journal of Cellular and Molecular Medicine
Acknowledgments
We thank Prof. Joan Morrell, Center for Molecular and Behavioral Neuroscience, Rutgers University, for her expert guidance in microtoming protocols. We thank EPSRC, UK, for supporting KLAC (EP/D066859/1). This work was also supported by NIH grants, NR008029 and AG030673, to M. T-C.
References
- 1.Coulombe PA. Wound epithelialization: accelerating the pace of discovery. J Invest Derinatol. 2003;121:219–30. doi: 10.1046/j.1523-1747.2003.12387.x. [DOI] [PubMed] [Google Scholar]
- 2.Singer AJ, Clark RAF. Cutaneous wound healing. NEngl JMed. 1999;341:738–46. doi: 10.1056/NEJM199909023411006. [DOI] [PubMed] [Google Scholar]
- 3.Freedberg IM, Tomic-Canic M, Komine M, Blumenberg M. Keratins and the ker-atinocyte activation cycle. J Invest Derinatol. 2001;116:633–40. doi: 10.1046/j.1523-1747.2001.01327.x. [DOI] [PubMed] [Google Scholar]
- 4.van Manen H-J, Kraan YM, Roos D, Otto C. Single-cell Raman and fluorescence microscopy reveal the association of lipid bodies with phagosomes in leukocytes. Proc Natl Acad Sci USA. 2005;102:10159–64. doi: 10.1073/pnas.0502746102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang G, Moore DJ, Flach CR, Mendelsohn R. Vibrational microscopy and imaging of skin: from single cells to intact tissue. Anal Bloanal Chem. 2007;387:1591–9. doi: 10.1007/s00216-006-0852-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang G, Chan KLA. Flach CR, Mendelsohn R. Interplay of univariate and multivariate analysis in vibrational microscopic imaging of mineralized tissue and skin. In: Lasch P, Kneipp J, editors; Biomedical vibrational spectroscopy. John Wiley and Sons, Inc; 2008. pp. 357–78. In:, editors.. NJ:. [Google Scholar]
- 7.Tomic-Canic M, Mamber SW, Stojadlnovlc O, Lee B, Radoja N, McMichael J. Streptolysin 0 enhances keratinocyte migration and proliferation and promotes skin organ culture wound healing in vitro. Wound Repair Regen. 2007;15:71–9. doi: 10.1111/j.1524-475X.2006.00187.x. [DOI] [PubMed] [Google Scholar]
- 8.Brem H, Stojadlnovlc O, Dlegelmann RF, Entero H, Lee B, Pastar I, Golinko M, Rosenberg H, Tomic-Canic M. Molecular markers in patients with chronic wounds to guide surgical debridement. Mol Med. 2007;13:30–9. doi: 10.2119/2006-00054.Brem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel GK, Wilson CH. Harding KG, Flnlay AY, Bowden PE. Numerous keratinocyte subtypes involved in wound re-epithelial-ization. J Invest Dermatol. 2006;126:497–502. doi: 10.1038/sj.jid.5700101. [DOI] [PubMed] [Google Scholar]
- 10.Usul ML, Underwood RA, Mansbrldge JN, Muffley LA, Carter WG, Olerud JE. Morphological evidence for the role of suprabasal keratinocytes in wound reep-ithelialization. Wound Rep Reg. 2005;13:468–79. doi: 10.1111/j.1067-1927.2005.00067.x. [DOI] [PubMed] [Google Scholar]
- 11.Gazel A, Ramphal P, Rosdy M, De Wever B, Tornler C, Hoseln N, Lee B, Tomic-Canic M, Blumenberg M. Transcriptional profiling of epidermal keratinocytes: comparison of genes expressed in skin, cultured keratinocytes, and reconstituted epidermis, using large DNA microarrays. J Invest Dermatol. 2003;121:1459–68. doi: 10.1111/j.1523-1747.2003.12611.x. [DOI] [PubMed] [Google Scholar]
- 12.Blumenberg M, Tomic-Canic M. Gene profiling: implications in dermatology. Future Drugs. 2007;2:763–8. doi: 10.1586/17469872.2.6.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, Chen J, Kirsner R. Pathology of acute wound healing. Clln Dermatol. 2007;25:9–18. doi: 10.1016/j.clindermatol.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 14.Laplante AF, Germain L, Auger FA, Moulin V. Mechanisms of wound reepithe-lialization: hints from a tissue-engineered reconstructed skin to long-standing questions. FASEBJ. 2001;15:2377–89. doi: 10.1096/fj.01-0250com. [DOI] [PubMed] [Google Scholar]
- 15.Debelle L, Alix AJP, Jacob M-P, Huvenne J-P, Berjot M, Sombret B, Legrand P. Bovine elastin and κ-elastin secondary structure determination by optical spectroscopies. J Biol Chem. 1995;270:26099–103. doi: 10.1074/jbc.270.44.26099. [DOI] [PubMed] [Google Scholar]
- 16.Odland GF. Structure of the skin. In: Goldsmith LA, editor. Physiology, biochemistry, and molecular biology of the skin. Oxford University Press; 1991. pp. 1–62. In:, editor.. New York:. [Google Scholar]
- 17.Ashcroft GS. Kielty CM, Horan MA, Ferguson MWJ. Age-related changes in the temporal and spatial distribution so fib-rillin and elastin mRNAs and proteins in acute cutaneous wounds of healthy humans. J Pathol. 1997;183:80–9. doi: 10.1002/(SICI)1096-9896(199709)183:1<80::AID-PATH1104>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 18.Amadeu TP, Braune AS, Porto LC, DesmouliereA A, Costa AM. Fibrillin-1 and elastin are differentially expressed in hypertrophic scars and keloids. Wound Repair Regen. 2004;12:169–74. doi: 10.1111/j.1067-1927.2004.012209.x. [DOI] [PubMed] [Google Scholar]
- 19.Novinec M, Grass RN, Stark WJ, Turk V, Baici A, Lenarcic B. Interactions between human cathepsins K, L, and S and elastins. Mechanism of elastinolysis and inhibition by macromolecular inhibitors. J Biol Chem. 2007;282:7893–902. doi: 10.1074/jbc.M610107200. [DOI] [PubMed] [Google Scholar]
- 20.Maki JM, Sormunen R, Lippo S, Kaarteenaho-Wiik R, Soininen R, Myllyharju J. Lysyl oxidase is essential for normal development and function of the respiratory system and for the integrity of elastic and collagen fibers in various tissues. Am J Pathol. 2005;167:921–2. doi: 10.1016/S0002-9440(10)61183-2. [DOI] [PMC free article] [PubMed] [Google Scholar]


