Abstract
Purpose
Osteosarcoma (OS) is the most common cancer of bone occurring mostly in teenagers. Despite rapid advances in our knowledge of the genetics and cell biology of OS, significant improvements in patient survival have not been observed. The identification of effective therapeutics has been largely empirically based. The identification of new therapies and therapeutic targets are urgently needed to enable improved outcomes for OS patients.
Experimental Design
We have used genetically engineered murine models of human OS in a systematic, genome wide screen to identify new candidate therapeutic targets. We performed a genome wide siRNA screen, with or without doxorubicin. In parallel a screen of therapeutically relevant small molecules was conducted on primary murine and primary human OS derived cell cultures. All results were validated across independent cell cultures and across human and mouse OS.
Results
The results from the genetic and chemical screens significantly overlapped, with a profound enrichment of pathways regulated by PI3K and mTOR pathways. Drugs that concurrently target both PI3K and mTOR were effective at inducing apoptosis in primary OS cell cultures in vitro in both human and mouse OS, while specific PI3K or mTOR inhibitors were not effective. The results were confirmed with siRNA and small molecule approaches. Rationale combinations of specific PI3K and mTOR inhibitors could recapitulate the effect on OS cell cultures.
Conclusions
The approaches described here have identified dual inhibition of the PI3K/mTOR pathway as a sensitive, druggable target in OS and provide rationale for translational studies with these agents.
Keywords: Osteosarcoma, PI3K, mTOR, siRNA screen, chemical screen
Introduction
Osteosarcoma (OS) is the most common primary tumor of bone. It occurs mostly in the second decade of life and is the fifth most prevalent cancer in children. Patients with localized disease have 5 year survival rates of ~70%. However, at presentation ~20% of patients have metastases and almost all patients with recurrent OS have metastatic disease (1, 2). The 5-year survival for patients with metastatic disease is 20% (3, 4). Treatment relies on the use of chemotherapy and surgery, which cause considerable morbidity (5). While substantial advances in our understanding of OS biology and genetics have occurred, there have been no significant advances in therapy for the past 30 years (6, 7). The first whole genome sequence of mutations in human OS was recently generated, revealing TP53 pathway mutations in all samples assessed and recurrent somatic changes in RB1, ATRX and DLG2 in 29–53% of the tumors (8). This analysis highlighted the complexity of the OS genome and provides invaluable information for improving preclinical modeling of OS, however it does not immediately reveal actionable strategies for improving therapy for patients.
To date, research has led to only a limited number of clinically relevant biologic insights (9, 10). Empirical evaluation of novel agents in human xenografts has not to date yielded any major translational advances (10). The only agent to show promise from these studies has been mTOR inhibitors such as rapamycin (11). Improvements in the delivery and application of existing treatments, rather than the introduction of new therapies, have seen some improvement in the management of OS. Novel approaches to drug target identification are needed alongside robust pre-clinical testing platforms. The development of genetically engineered mouse models (GEMM), reflective of the human OS, represent a critical component to improving patient outcomes and preclinical target validation (12). We previously developed a GEMM of the fibroblastic OS subtype, through deletion of p53 and pRb from the osteoblast-lineage that has been independently validated (13–15). We recently described the first murine model of osteoblastic OS, the most common subtype of human OS (16).
Advances in technology and the capacity for high-throughput phenotypic and chemical screens offer considerable promise for identifying new therapeutic agents. Screening approaches afford an opportunity for an unbiased, saturation coverage of the genome. Genome-wide siRNA screens offer an unmatched probing of the genetics of OS, but the immediate clinical utility of many identified candidates, such as transcription factors, may be limited as they are not modifiable using current therapeutic approaches. Chemical screens with drugs that are in clinical or pre-clinical use interrogate a limited spectrum of targets but, if validated, offer the prospect of a more rapid clinical application if validated (17). Given the limited advances in translating basic knowledge of OS biology to patient benefit an approach utilizing systematic screening of drugs or focusing on defining genetic susceptibilities of OS could offer a new means to identify new potential candidates for either preclinical testing or further development.
Here we report results from parallel screens using primary cell cultures derived from murine OS models which faithfully replicate the human disease (13, 16). Firstly, a whole genome siRNA screen for enhancers of cell death was performed. The screen was conducted with either siRNA alone or as siRNA with doxorubicin, a standard of care chemotherapy for OS (5). Secondly, a curated drug/chemical library predominantly targeting kinases or known targets was screened against three independent primary cell cultures derived from paired primary and metastatic OS. Validation across mouse and human biopsy derived primary OS cell cultures established the robustness of our analyses. These complementary chemical and genetic strategies have converged to provide independent evidence that concurrent targeting of protein translation and growth control pathways, in particular the PI3KCA and mTOR pathways, represent tractable targets in OS.
Materials and Methods
OS cell cultures and mouse models
Primary mouse OS cell cultures were derived from tumors generated in either Osx-Cre p53fl/flpRbfl/fl mice (fibroblastic OS model) (13) or Osx-Cre TRE-p53.1224 pRbfl/fl model (osteoblastic OS model) (16) (all obtained directly from mouse models of OS). Experiments using mouse OS models were approved by the St Vincent’s Hospital AEC. Primary human OS (OS17) was derived from a primary human OS xenograft generously provided by Dr Peter Houghton (Nationwide Children’s Hospital, Ohio). Excess, de-identified tumor material was collected from patients with osteosarcoma at St Jude Children’s Research Hospital (SJCRH) in agreement with local institutional ethical regulations and IRB approval. Primary human OS (SJOS001112_X1, SJOS010929_X1, SJOS001107_X1, SJOS001107_X2) were derived from primary human xenografts collected from patients with osteosarcoma at SJCRH. Human OS cell lines MG-63, Saos-2, U2OS, G-292, 143B and SJSA-1 were purchased from ATCC (MG63 from ECACC cell line remainder from ATCC; no authentication performed by the authors). Normal human osteoblasts were derived from bone marrow aspirates from the posterior iliac crest of healthy human adult donors (17–35 years of age), with informed consent (IMVS/SA Pathology normal bone-marrow donor program RAH#940911a). The cells were outgrown from the bony spicules that were collected following filtration of the BM through a 70μm filter. Cell cultures were grown in αMEM with 10% FCS (non-heat inactivated), 1% Penicillin/Streptomycin and 1% Glutamax supplement.
High-throughput siRNA whole genome screen
OS cell line 494H (Cre:lox model derived cell line representative of fibroblastic OS passage 8 from derivation from primary tumor) were grown and transfected with SMARTpool siRNAs using DharmaFECT lipid 3 transfection reagents (Dharmacon GE). The mouse siRNA library (Dharmacon GE) consists of 16,874 individual protein coding genes consisting of the following gene families all designed to RefSeq52. Cells were reverse transfected (18) at 400 cells per well with 40nM of the siGENOME SMARTpool library complexed for 20 minutes with DharmaFECT 3 using the Sciclone ALH3000 (Caliper Life Sciences) and BioTek 406 (BioTek) liquid handling robotics. Cell lines were transfected in quadruplicate (day 0) and one set of duplicates treated as control (siRNA only) and a second set of duplicates treated after 48hrs (day 2) with 100nM doxorubicin. At 120hrs post transfection (day 5) the cell viability for each well was quantified on the basis of the direct measurement of intracellular ATP using the CellTiter-Glo luminescent assay (Promega) at a 1:2 dilution. Full analysis methods are provided in the supplemental information. RNAi screen data is available from Pubchem with PubChem accession ID #1053208 (primary screen) and PubChem AID# 1053209 (validation screen) and in Supplemental Dataset 1. RNA-seq data from mouse OS models is deposited in GEO (GSE58916).
Small molecule screen against primary mouse OS paired samples
Three independent sets of paired primary and metastatic OS cell lines (from fibroblastic OS model, less than or equal to passage 8 from derivation (13)) were plated at 400 cell/well in 384-well plates. After 24hrs the cells were treated with the 131 different compounds in an 11-point dose response ranging from 0.01–10mM. All assays were performed in duplicate. At 48hr post treatment the cell viability for each well was quantified on the basis of the direct measurement of intracellular ATP using the CellTiter-Glo luminescent assay (Promega) at a 1:2 dilution. All luminescent measurements were taken on the EnSpire plate reader (PerkinElmer, Waltham, MA, USA). Data was plotted and the IC50 calculated using Prism 6 software.
Small molecule screen against primary human xenograft-derived OS
Excess, de-identified tumor material was collected from patients with osteosarcoma at SJCRH in agreement with local institutional ethical regulations and institutional review board approval. Small molecule/drug screening was performed as previously described (19, 20).
Cell death assays
Cells were treated with the indicated drug or vehicle control for 48hrs prior to harvesting. Cells were washed with PBS and 100,000 cells were stained in 1xAnnexin Binding buffer (BD Biosciences) diluted 1:20 with Annexin V-APC (1mg/ml) (BD Pharmingen) and 7-Aminoactinomycin D (7AAD) (100μg/ml) (Life Technologies) for 15 min protected from light. Following the addition of 4 volumes of 1xAnnexin Binding buffer, apoptotic cells were quantified using FACS (LSRFortessa, BD Biosciences). Live cells (Annexin V negative, 7AAD low) and cells in early and late stages of apoptosis (Annexin V positive, 7AAD low/high) were analyzed using FlowJo software (v8.8.7) (Ashland, OR, USA). Each cell line was assessed in duplicate in three separate biological experiments.
Statistical analysis (excluding siRNA screen which is described separately)
Each experiment is represented as the mean±SEM calculated from biological replicates unless otherwise stated. Unpaired parametric Student’s t-test was used to compare results unless otherwise stated. Where indicated in legends, unpaired nonparametric Mann Whitney ranks test or 2-way analysis of variance (2-way ANOVA) were used to compare groups. Data was log transformed to normalize variances before 2-way ANOVA. In all analyses, statistical significance of p<0.05, p<0.01, p<0.001, and p<0.0001 are represented as *, **, ***, **** respectively. Prism 6.0e software was used for statistical analyses.
Results
Whole OS genome siRNA screen to identify OS susceptibilites
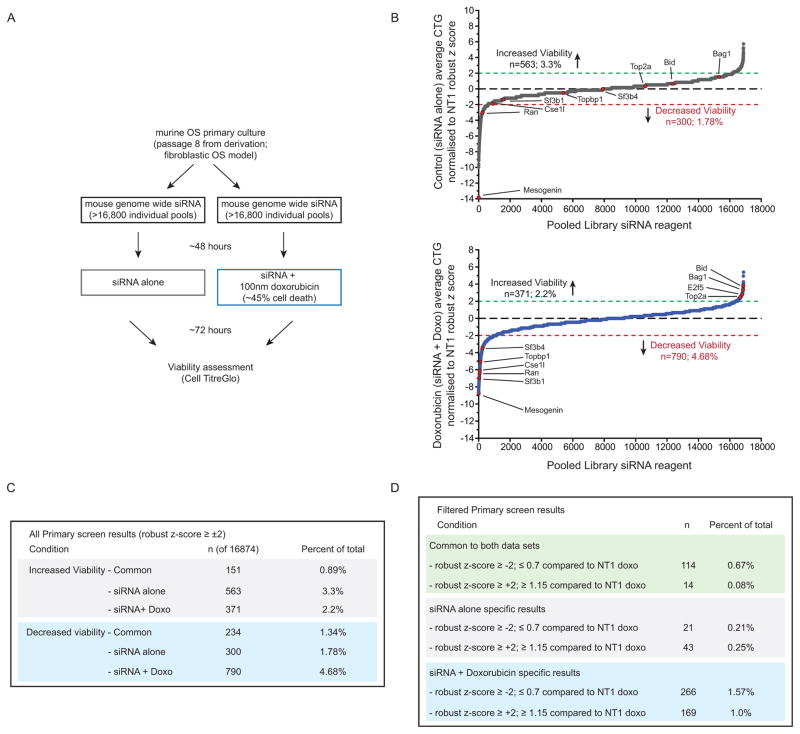
We sought to identify, in a genome-wide fashion, inducers of OS cell death and in parallel sensitizers to doxorubicin-induced cell death. Doxorubicin is a standard of care agent in the management of OS (5). The screen was performed in early passage cell cultures established from the murine Osx-Cre p53fl/flpRbfl/fl OS model (fibroblastic OS) (13). A genome wide screen was performed using a library containing >16,000 siRNAs targeting murine protein-coding genes (each siRNA pool has 4 individual siRNA against the same target). Mouse OS cells were treated with either siRNA alone or, on a duplicate plate, siRNA in combination with 100nM doxorubicin (siRNA + doxorubicin; Figure 1A). The dose of doxorubicin was titrated to achieve ~45% cell death as a single agent. All plates were screened in duplicate under both conditions. A robust z-score sample based normalization strategy was performed across the screen. To be classified as a hit a candidate had to have a robust z-score ≤−2 and a viability of equal to or less than 70% (fold change of ≤0.7) when compared to the respective non-targeting siRNA control for each treatment arm (NT1, Figure 1B–C). A total of ~270 hits were identified from the primary screen that passed statistical analysis and robust z-score stratification. A difference of at least 25% between siRNA alone and siRNA+doxorubicin was categorized as an interaction after filtering based on robust z-scores (Figure 1D). We further triaged candidates based on their expression in RNA-seq/microarray datasets from GEMM OS models. The top 299 candidates that enhanced cell death were then validated by deconvolution of the SMARTpool into its constituent 4 individual siRNAs and re-screening using the same assay. Of the primary screen hits, 113 validated with 3 or more of the individual siRNA recapitulating the primary screen result (37.7%), considered high confidence targets. A further 38 candidates were validated with 2 of the individual siRNA, and were classified as medium confidence hits. In total approximately 50% of the primary screen hits validated (Figure 2A–C).
Figure 1. Whole genome siRNA screen against murine OS.
(A) Schematic outline of the siRNA screen. (B) Primary screen results from either siRNA alone (top panel, grey line) or siRNA + 100nM doxorubicin (lower panel, blue line). Each assay was performed in duplicate and is plotted as robust z-score normalized to non-targeting (NT1) control treated cells. Total numbers and selected candidates that increased viability (robust z-score of ≥+2) or decreased viability (robust z-score of ≤−2). (C) Primary screen results based on robust z-score for the indicated groups; (D) stratification of siRNA candidates based on normalized z-score and a difference of at least 30% between control and doxorubicin treated plates.
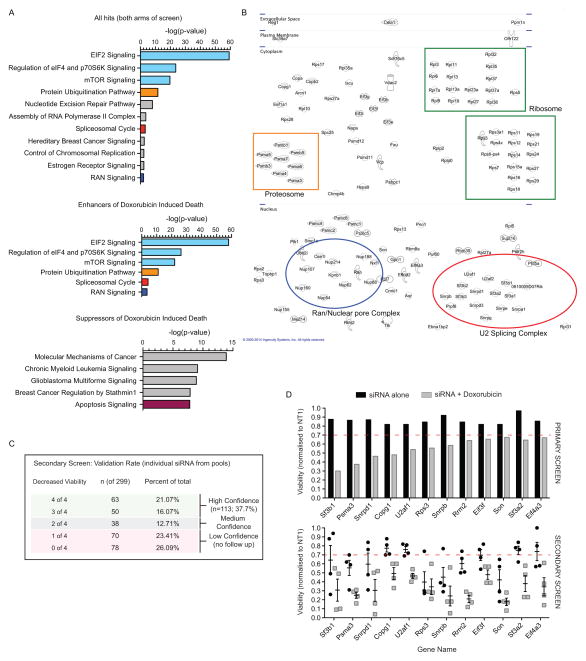
Figure 2. Protein translation and RNA splicing enhance OS cell death.
(A) Analysis of pathways enriched in the primary screen from both arms (+/− doxorubicin; upper panel), those enriched within hits that enhanced doxorubicin induced cell death (middle panel) and those that increased cell survival in the presence of doxorubicin (lower panel). Data plotted as log fold enrichment (p-value) based on Ingenuity Pathway Analysis. (B) Visualization of the individual candidate hits based on subcellular localization (from Ingenuity Pathway Analysis software) with specific pathways that are enriched indicated within each colored box based on n the validated primary screen results described in part A. (C) Secondary screen deconvolution analysis of the candidates identified in the primary screen. Primary screen candidates were secondarily screened as individual siRNAs and those with 3 or 4 of 4 validating the primary screen were considered high confidence candidates; 2 of 4 medium confidence and 1 or less not considered further. (D) Examples of individual target effects in the primary and secondary screen. Upper panel: mean viability of duplicate samples for each treatment arm from the primary siRNA screen; lower panel: viability following transfection of the individual siRNA validation for each target in the secondary screen. In the lower panel each point represents an individual siRNA from within the pooled complex used for the primary screen with mean ± SEM for each gene.
A range of candidates that reduced cell viability independently of whether doxorubicin was present in addition to those that specifically enhanced the toxicity of doxorubicin were identified (Figure 2A–D). While not strongly powered to detect loss of function alleles involved in resistance to doxorubicin, several candidates were identified (Figure 2A) including siRNA against topoisomerase 2a (Top2a), the cellular target of doxorubicin (21, 22), and a number of pro-apoptotic genes such as Bid (Figure 1B, Supp Figure 1A–B). A significantly higher false positive rate on the candidates that prevented cell death was apparent, with only Top2a and Bid robustly validating of the 60 candidates tested. From the enhancers of doxorubicin induced death we identified 266 siRNA candidate genes (1.576%) that fulfilled the classification criteria (subset visualized in Figure 2B). Analysis of the hits interacting with doxorubicin revealed a marked enrichment of candidates on chromosome 7 (p=4.77×10−11, FDR=3.68×10−8; statistics derived from DAVID analysis), relevant as the loss of one to two copies of chromosome 7 from a tetraploid karyotype was recurrently noted in the murine OS models (16). Using Ingenuity Pathway Analysis software, we tested if the identified hits functioned in overlapping pathways/gene expression signatures (Figure 2A–B). Pathways enriched included those associated with RNA transport (Ran) and RNA splicing, in particular the U2 splicing complex (Figure 2A–B, Supp Figure 1C). Eleven of the 13 identified U2 snRNP complex members in mammals were identified in the screen. In contrast, we did not observe enrichment for any other snRNP complexes, suggesting a specific requirement for the U2 snRNP complex in OS pathogenesis (Figure 2B). The most significantly enriched pathways were associated with protein translation and mTOR signaling (Figure 2A–D).
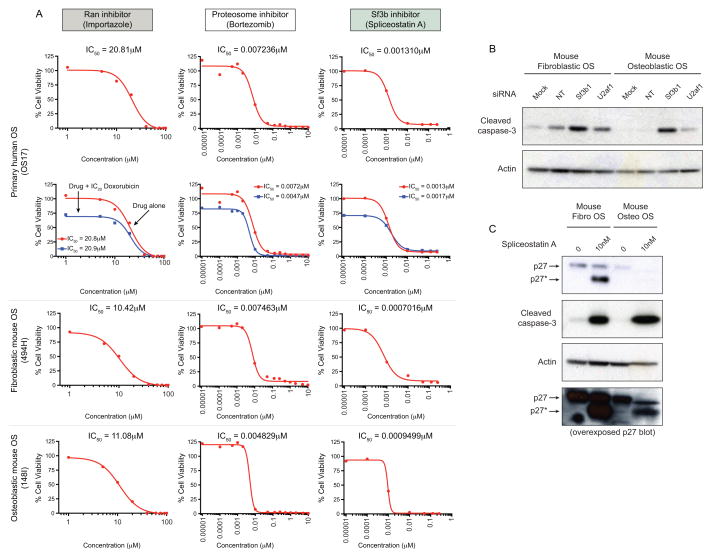
Next we tested if small molecules targeting the identified pathways demonstrated toxicity against OS cells. The agents were tested against a primary human OS xenograft-derived cell culture (OS17) and murine primary tumor derived cultures from both fibroblastic (494H) and osteoblastic (148I) OS models. We tested a Ran inhibitor, an inhibitor of the U2 splicing complex member Sf3b spliceostatin A (SSA, (23)), and the proteosome inhibitor bortezomib (Figure 3A–C). All agents showed activity against the OS cells, although the specificity of the bortezomib response was indeterminate given its reported in vitro activity against a large panel of solid tumors. Given the enrichment of the U2 snRNP specifically in the siRNA screen and the potent activity of SSA, we sought to further understand the effects of impaired U2 snRNP activity in OS. We treated fibroblastic and osteoblastic mouse OS cells with non-targeting or siRNA against S3fb1 and U2af1, both components of the U2 snRNP (Figure 3B). In both cases, cell death as measured by cleaved capsase-3 levels was observed. Treatment of human or mouse OS cells with SSA resulted in impaired splicing of p27 and detection of an aberrant protein isoform (p27*), a known consequence of Sf3b1 inhibition (Figure 3C, Supp Figure 2). Consistent with the siRNA results, SSA treatment resulted in massive induction of apoptosis in fibroblastic and osteoblastic murine OS, consistent with the low nM IC50 in these cells. These studies identify a range of susceptibilities in OS cells that had not been previously observed. Furthermore, these targets were validated by independent means using chemical or drugs that target these processes.
Figure 3. Small molecule validation of siRNA candidates.
(A) Dose response viability curves of human (OS17) and mouse fibroblastic (494H) or osteoblastic (148I) OS cell cultures treated with the Ran inhibitor Importazole either alone (red line) or in combination with doxorubicin (blue line). Data is presented as IC50 values (n=2–4 independent replicates); sensitivity of human (+/− doxorubicin), murine fibroblastic and osteoblastic OS to the proteosome inhibitor bortezomib; sensitivity of human (+/− doxorubicin), murine fibroblastic and osteoblastic OS to the Sf3b1 inhibitor spliceostatin A (SAA). (B) Treatment of murine fibroblastic and osteoblastic OS for 72 hours with mock, non-targeting (NT), Sf3b1 or U2af1 siRNA. Western blots were probed for the presence of cleaved caspase-3 and actin as a control. (C). Murine fibroblastic and osteoblastic OS were treated for 48 hours with SSA. Western blot analysis demonstrates the presence of an abnormally spliced p27 isoform (indicated as p27*) accompanied by cleaved caspase-3 in SSA treated cells.
Kinase Inhibitor screen: dual PI3K/mTOR inhibitors show strong activity against OS
The activity of a chemical library of 131 primarily kinase inhibitors, either currently in clinical trial and those that either failed trial or were not developed further, as single agents was also assessed. The screen was conducted in duplicate on 3 sets of paired primary and metastatic early passage OS cell cultures derived from the murine fibroblastic OS model (Figure 4A). OS culture (494H) was also used for the siRNA screen. Cell viability was determined after 48hrs exposure to the agents and a dose response curve established.
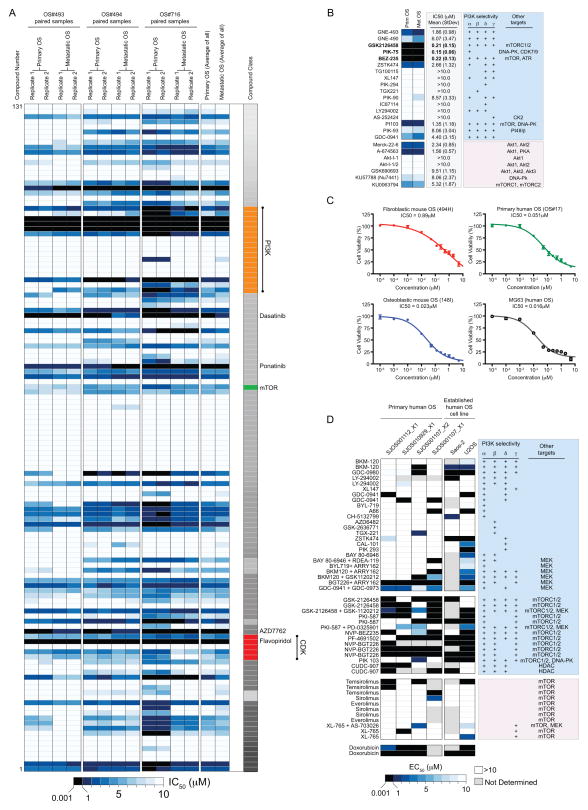
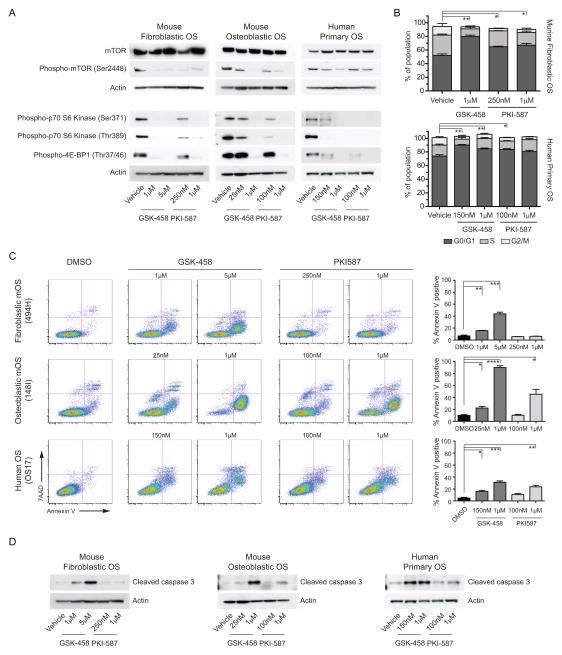
Figure 4. Specific sensitivity to PI3K/mTOR inhibition in both mouse and human primary OS cells.
(A) Three independent murine fibroblastic primary and metastatic paired OS cell cultures were screened in an 11-point dose response against 131 compounds (full list of compounds and screen results in Supplemental Dataset 2). Data are presented as a heat map of sensitivity for replicate assays or pooled by primary or metastatic location. The greatest sensitivity is in black (0.001–1μM), graded to least/no sensitivity in white (>10μM). Individual agents with sub-1μM activity are listed on the left hand side of the data. (B) All agents that have activity against PI3K are listed and the IC50 (mean± StDev) and target specificity against PI3K subunits and other targets listed. (C). Testing of GSK2126458 against murine fibroblastic (494H, red), murine osteoblastic (148I, blue), primary human (OS17, green) and the established human OS cell line (MG63, grey) with separate drug stocks to those used in the initial screen. Data is presented as mean IC50 values ± SEM (n=3 independent assays for 494H, 148I and OS17; n=2 for MG63). (D) Four primary human biopsy derived cultures and the established human OS cell lines Saos-2 and U2OS were treated with the indicated agents. Data are presented as a heat map of sensitivity with replicates as indicated by multiple listings of the same agent. The greatest sensitivity is in black (0.001–1μM), graded to least/no sensitivity in white (>10μM) with grey not tested. Target specificity against PI3K subunits and other targets is listed.
The chemical screen identified 8 compounds with activity as single agents in the sub-1μM range (6.1%, Figure 4A). Of the active agents there were 2 multi-kinase inhibitors (Ponatinib and Dasatinib), a CHK inhibitor (AZD7762), flavopiridol (CDK inhibitor) and a PLK-1 inhibitor (BI-2536(R-)). Plk-1 siRNA was used as a positive control for our siRNA screen so it was known that inhibition of PLK was toxic to OS cells. Of particular interest were 3 compounds with an overlapping spectrum of targets. PIK-75, GSK2126458 and BEZ-235. All target phosphatidylinositide 3-kinases (PI3K) together with mTOR and/or DNA-PK (Supp Figure 3). The effect was specific to these dual inhibitors as we did not observe low IC50 values with an additional 14 agents with PI3K restricted activity. Additionally, the specific mTORC1/2 inhibitor (KU0063794), the DNA-PK inhibitor (NU7441) and several specific Akt inhibitors (Akt-I-1; Akt-I-1/2) did not exhibit potent activity (sub-1μM, Figure 4B). These findings suggested that concurrent inhibition of both the PI3K and another pathway (mTOR or DNA-PK) was most likely responsible for the observed activity. We did not observe any differential sensitivity between the primary and metastatic paired samples. The combined overlap of the results from chemical and genetic screening revealed a profound enrichment for cell growth/protein translation regulation pathways as a vulnerability in OS. We further tested PIK-75, GSK2126458 and BEZ-235 across a panel of OS cultures encompassing murine osteoblastic OS, murine fibroblastic OS, an established human OS line (MG63) and a primary human OS xenograft-derived cell culture (OS17). The activity was confirmed with independent stocks of drug and facilities and on all OS cell cultures tested (Figure 4C).
Primary human OS also display sensitivity to dual PI3K/mTOR inhibitors
Our data indicated that dual targeting of the PI3K/mTOR pathway represents an effective strategy to induce OS cell death. To extend our analysis and expand the genetic diversity of the primary human OS that were assessed, we tested a targeted range of small molecule inhibitors against a panel of acute short-term cultures of primary OS material derived from orthotopic xenografts (8, 20). The established human OS cell lines Saos-2 and U2OS were also included. Strikingly the most active agents against primary human OS cultures were dual PI3K and mTOR inhibitors (GSK2126458, PKI-587, BEZ-235 and BGT-226; Figure 4D). Therefore the sensitivity to concurrent inhibition of PI3K and mTOR pathways is conserved across species and subtypes. These data provide a strong rationale, obtained from systematic screening, for the testing of dual PI3K/mTOR pathway inhibitors in OS.
Induction of OS cell apoptosis by dual inhibition PI3K and mTOR pathways
Of the small molecule inhibitors identified, PIK-75 cannot be directly clinically applied and BEZ-235 has ceased clinical development. Therefore, we tested compounds that could be clinically translated including GSK2126458 and included a second PI3K/mTOR active agent PKI-587 (24, 25) (Supp Figure 3A–C). GSK2126458 and PKI-587 could be combined with doxorubicin in vitro with no reduced effectiveness of either agent (Supp Figure 3D–G). Both agents inhibited phosphorylation of known targets downstream of PI3K/mTOR in a dose dependent manner in mouse OS subtypes and human primary OS cell cultures (Figure 5A). Next we assessed if GSK2126458 and PKI-587 were inducing changes in cell cycle distribution or cell death. There was a significant increase in cells in the G0/G1 phase of the cell cycle in both mouse and human OS cells treated with GSK2126458 and to a lesser magnitude with PKI-587 (Figure 5B). In all cells tested there was a dose dependent increase in the induction of apoptosis as determined by Annexin-V/7AAD staining or the presence of cleaved caspase-3 (Figure 5C–D). In all three OS cell models tested, GSK2126458 was more potent at inducing apoptosis than PKI-587, despite similar effectiveness at inhibiting downstream signaling events (Figure 5C–D).
Figure 5. Dual PI3K/mTOR inhibitors induce apoptosis in OS cells.
(A) Analysis of target inhibition downstream of PI3K/mTOR activity was assessed by western blot of the indicated proteins against murine fibroblastic, murine osteoblastic and primary human OS. Each cell type was treated with the calculated IC50 and Emax respectively for each drug. (B) Cell cycle distribution of murine fibroblastic OS or primary human OS cells treated with the indicated concentrations of GSK2126458 and PKI587 for 24hrs, statistical comparison performed comparing the proportions of G0/G1 phase cells between vehicle and each treatment. (C) GSK2126458 and PKI587 induce apoptosis as assessed by annexin V/7AAD staining (D) or cleaved caspase-3 western blot in a dose dependent manner. Where indicated data presented as mean ± SEM, *P<0.05, **P<0.01, ***P<0.001 (n=3 technical replicates per cell line, 3 independent cell lines tested as indicated).
Combining PIK3CA and mTOR inhibitors as a therapeutic approach
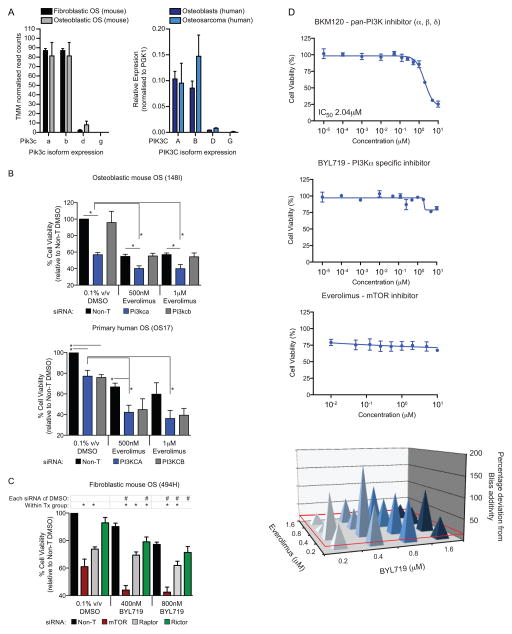
The activity of the dual PI3K/mTOR inhibitors corresponded most closely with activity against PIK3CA (Figure 4B). We determined the expression of the different catalytic subunits of PI3K in murine OS subtypes and in human OS and normal human osteoblasts. The murine OS tumors had the highest expression of Pik3ca and Pik3cb with low to undetectable levels of Pik3cd and Pik3cg (Figure 6A). This pattern of expression was mirrored in normal human osteoblasts and OS samples (Figure 6A). Given the relative isoform specificity of PIK-75, GSK2126458 and BEZ-235, this indicated that the activity was most likely primarily mediated by inhibition of PIK3CA together with mTOR. To directly test the requirement for inhibition of PIK3CA, we used siRNA to knockdown the PIK3CA and PIK3CB. Knockdown of Pik3ca in mouse OS and PIK3CA in human OS cells resulted in significantly reduced viability (Figure 6B). When the siRNA was combined with the specific mTOR inhibitor everolimus, an increased cell death compared to that achieved with either agent alone was observed. The increased death from concurrent inhibition was more pronounced with PIK3CA compared to PIK3CB knockdown, although the reduced viability with PIK3CB siRNA would suggest some contribution from this isoform to the overall effect (Figure 6B, Supp Figure 4). We tested if knockdown of mTOR, Raptor or Rictor could cooperate with inhibition of Pik3ca. The knockdown of mTor was the most effective and when combined with the PIK3CA specific inhibitor BYL719 resulted in significantly reduced cell viability compared to the siRNA alone (Figure 6C, Supp Figure 4). These data demonstrate that specific targeting of either the PI3K or mTOR pathways in isolation is not as effective at inducing OS cell death as dual inhibition.
Figure 6. PIK3CA and mTOR inhibition mediate the sensitivity of OS cells to agents targeting these pathways.
(A) Transcript levels of Pik3c isoforms were quantified from RNA-seq of murine fibroblastic and osteoblastic OS or by real-time PCR in human OS cells compared to normal human osteoblast cells. Data is presented as TMM (trimmed mean of M values) normalized read counts (n=3 per subtype) for RNA-seq or mean relative expression to PGK1 ± SEM (n=4–7) for QPCR. (B) Human and mouse OS cells were transfected with 25nm SMARTpool siRNAs targeting Pik3ca/b or PIK3CA/B respectively or a pool of non-targeting siRNAs (NT2) for 24 hours then treated with the indicated concentrations of the mTOR inhibitor everolimus for 48 hours. Cell viability was assessed relative to Non-T DMSO treated cells. Data is presented as mean ± SEM (n=3). (C) Mouse fibroblastic OS cells (494H) were transfected with siRNAs targeting mTor, Raptor or Rictor or a pool of non-targeting siRNAs (Non-T) for 24 hours then treated with the indicated concentrations of the PIK3CA inhibitor BYL719 for 48 hours. Cell viability was assessed relative to Non-T DMSO treated cells. Data is presented as mean ± SEM (n=3).). *P<0.05, #P<0.05 calculated using students t-tests. (D) Treatment of murine OS cells (494H) with the pan-PI3K inhibitor BKM120, PI3KCA specific inhibitor BYL719 or the mTOR inhibitor Everolimus as single agents. Data is presented as mean IC50 values ± SEM (n=3 per treatment); Needle graphs show synergy effects in mouse fibroblastic OS cells treated with combinations of Everolimus and BYL719 for 72 hours. Synergy is represented as percent deviation from a predicted additive response (Bliss additivity). Deviations greater than 15% were considered synergistic (red baseline). Data is presented as mean deviation (n=3).
Finally, we sought to determine if combinations of specific inhibitors of the PI3K and mTOR pathway could be used to recapitulate the activity of the dual inhibitors. This may be an approach amenable to translation as several pathway specific inhibitors have progressed in clinical trial or, in the case of the everolimus, been approved (26). We tested combinations of either the pan-PI3K inhibitor BKM120 or the PIK3CA specific inhibitor BYL719 (27) with everolimus. When tested individually all agents failed to demonstrate significant activity (Figure 6D). When combined, we observed a synergistic interaction between the BLY719 and everolimus based on BLISS synergy scores (Figure 6D, Supp Figure 5). Both agents were tolerated alone or in combination by normal osteoblasts in vitro, consistent with in vivo reports with these and similar agents (Supp Figure 6; (28, 29)). Therefore agents currently in clinical trial with activity against PIK3CA and mTOR could be combined and demonstrate efficacy against OS in vitro. These data demonstrate the feasibility of dual targeting of the PI3K/mTOR pathway in OS and propose a new therapeutic approach in this cancer.
Discussion
To identify new therapeutic targets for OS we have made use of murine OS models combined with primary human OS xenograft derived cell cultures (13, 16). These independent approaches converged to identify dual inhibition of the PI3K/mTOR pathways as a species conserved sensitivity in OS. The use of the primary tumor derived material from multiple species strengthens the confidence in our data and provides a new approach to integrating mouse models into preclinical target identification and validation. We observed in vivo activity against OS of the dual PI3K/mTOR inhibitor GSK2126458 (Supp Figure 7). Supporting our results are previous data testing a range of compounds that target single arms of these pathways with evidence for some, albeit limited, in vivo activity against human OS xenografts (11, 30–33). Dual PI3K/mTOR inhibitors were far more potent that PI3K selective agents on established OS cell lines, further validating our observations with primary OS derived cells (34). Most recently, exome capture of human OS and shRNA screening independently confirmed that the PI3K/mTOR pathway is an attractive candidate in OS, supported by the data from the screening and validation described here (35).
Of the pathways identified in our siRNA screen, the regulation of RNA processing and splicing in OS is an intriguing finding. We recovered 11 of 13 members of the U2 snRNP complex without significant enrichment for other complexes of the RNA splicing machinery in cells treated with siRNA and doxorubicin (36). The OS cells were highly sensitive to inhibition of Sf3b1 with SSA (23). Changes in RNA splicing are recognized as major contributors to cancer, particularly hematological malignancy (37). Recently, other genetics screens have identified RNA splicing machinery as preferentially required in cancer cells, with PRPF6 important in colon cancer (38) and PHF5a in brain cancer (39). These data suggest that aberrant RNA splicing in OS are important in maintaining the tumor state and warrant further investigation. The identity of the aberrantly spliced RNAs causing OS cell death OS cells is currently unknown, however one candidate group of transcripts encode pro-death proteins (40). We identified pro-apoptotic effectors as suppressors of doxorubicin-induced cell death in our siRNA screen, providing a plausible link to the sensitivity of OS cells to U2 snRNP inhibition. Another intriguing candidate was Mesogenin (Msgn), a bHLH transcription factor that was the most potent inducer of OS cell death irrespective of the presence of doxorubicin. While there is limited information relating to Msgn, the Msgn−/− mouse has defects in skeletal development and Msgn act downstream of the Wnt and β-catenin pathways, both implicated in OS (41, 42). It would be interesting to determine if Msgn is required for normal osteoblast survival once the lineage is established, or analogous to the requirement for another bHLH transcription factor SCL/tal-1 in hematopoiesis, it is only required developmentally for lineage specification (43). If this were the case then developing strategies to target Msgn may be of value.
Given the overlap between the siRNA and chemical/drug screen results we focused our attention on understanding the contribution of the PI3K/mTOR pathway in OS. Recently several groups have reported that agents working within this pathway have some activity in OS (28, 29). Our pre-clinical screening has indicated the requirement for concurrent inhibition of PI3K, in particular PIK3CA, and mTOR pathways. The screens reported herein provide a rationale for combining PI3KCA and mTOR inhibitors, rather than the use of either inhibitor alone, in OS. Recent OS studies from other groups have drawn attention to this pathway (28, 29, 44–46), yet our study is distinct from these as we have identified this pathway as a result of independent chemical and genetic screens and can demonstrate that dual pathway inhibition is the most potent. We further define that PI3KCA activity is the driver of this PI3K activity in both human OS and mouse models of OS. The sensitivity to PI3K/mTOR inhibitors is conserved across OS subtypes and across species, greatly strengthening the case that this pathway should be tested clinically.
Our results using GSK2126458 and PKI-587 contrast with those described by Gobin et al, who reported the effects of BEZ-235 (28). We identified BEZ-235 as active in our chemical screen. We observe robust induction of apoptosis with both GSK2126458 and PKI-587 in murine and in primary human OS derived cells at significantly lower doses than that reported on established murine or human OS cell lines (28, 29). Gobin et al reported a cell cycle arrest in the absence of apoptosis when treating with up to 25μM BEZ-235 or the PI3KCA specific agent BYL719. The discrepancy in effects is most likely a reflection of the cells used for the respective studies. Our study has utilized early passage primary OS cell cultures derived from either GEMM (at least 3 independent cultures of each subtype) or multiple independent primary human OS cultures derived from xenografts. In contrast, Gobin et al utilized a range of long established OS derived cell lines. Therefore, the age and nature of the cells used may led to divergent effects on cell survival. The anti-OS activity of the PI3K/mTOR inhibitors on primary OS cells is mediated via induction of apoptosis and cell cycle arrest.
A large Phase II trial with single agent mTOR inhibitor ridaforolimus in sarcomas noted some, albeit limited, activity in OS. Two partial responses in OS patients were seen, which is interesting, but modest when a total of 54 patients with bone sarcomas were treated (47). BEZ-235 has in vivo activity in preclinical OS models, supporting our preliminary data (28). However BEZ-235 has been discontinued from further development, emphasizing our focus on clinically utilizable agents. We demonstrate in vitro activity of the dual inhibitors GSK2126458 and PKI-587 and the combination of the PIK3CA specific inhibitor BLY719 and the mTORC1 inhibitor everolimus. BYL719 and everolimus are currently being tested together clinically a Phase I/II trial in patients with in breast cancers, based on the observation that dual inhibition of PIK3CA and mTORC reversed resistance of PIK3CA mutant breast cancer to BYL719 monotherapy (48). This will act as a good indicator of how readily combinable these two agents are in terms of clinical potency, manageable off-target effects, and possible pharmacological interactions.
Through combining siRNA screening with targeted chemical library screens we have identified a number of candidate targets in OS. The most directly translatable pathway is PI3K/mTOR dual inhibition, with a number of agents already in clinical trial or approved. The screens described herein provide a rational basis for detailed in vivo orthotopic pharmacokinetic and pharmacodynamic studies and the assessment of these agents in preclinical Phase I, II and III studies that will be required. Furthermore, using independent approaches and validation across species, OS subtypes and facilities, we provide evidence that PI3K/mTOR dual inhibition is an attractive therapeutic candidate for OS.
Supplementary Material
Statement of Translational Relevance.
Current therapy for osteosarcoma relies on surgery and conventional chemotherapy. In recent decades, survival rates have plateaued. Improvements in patient outcome have largely been derived from improved delivery of existing agents, rather than the introduction of novel agents. To probe for new susceptibilities, we have undertaken independent genome wide siRNA and chemical screens in murine models of human OS, identifying PI3K and mTOR pathways as therapeutic vulnerabilities. Our studies demonstrate that dual inhibition of PI3K and mTOR pathways, but not either pathway independently, led to apoptosis of OS cells. These findings were recapitulated in genetically diverse primary human OS samples in separate facilities. These data demonstrate that dual PI3K/mTOR inhibitors warrant prioritization for development as a strategy to treat osteosarcoma.
Acknowledgments
We thank the SVH BioResources Centre; L Purton, J Martin and J Heierhorst for comments and discussion; D Thomas and Y Handoko (Victorian Centre for Functional Genomics, Peter MacCallum Cancer Centre) for technical assistance and screening support; P Houghton (Nationwide Children’s Hospital, Ohio) for providing human xenograft material; M Yoshida (RIKEN) for generously providing Spliceostatin A; G Philip and C Sloggett at the Victorian Life Sciences Computation Initiative for help with RNA-seq analysis and pipelines.
Footnotes
Author contribution statement:
C.W., A.M., K.S. and D.H. conceived study; A.G., E.B., C.G., A.C., S.T., S.W, K.L, J.D., P.M., A.Z. C.B., D.H., K.S. and C.W. performed experiments, analyzed and interpreted data; drug screening against primary human OS xenograft derived cells was performed at St. Jude’s Children’s Hospital by A.L., E.S., A.S., A.K. and M.D.; J.D., C.B., M.D., D.H., K.S. and C.W. provided intellectual input and conceptual advice; C.W. wrote the manuscript; All authors reviewed the manuscript.
Conflict of Interest Statement: Dr Jayesh Desai is a consultant for Novartis, Pfizer, GSK, Bayer, Sanofi, Circadian, and Bionomics and receives research support from Novartis, GSK, and Roche.
The remaining author discloses no conflicts of interest.
Financial Information: This work was supported by grants from the AACR-Aflac Inc, Career Development Award for Pediatric Cancer Research (C.W.; 12-20-10-WALK); Colin North (CW); Zig Inge Foundation (C.W.); 5point foundation (to E.B); Cancer Council of Victoria (to C.W. and E.B.; Grant-in-Aid APP1047593); NHMRC Career Development Award (C.W.; APP559016); NHMRC Research Fellowship (D.H.; APP1043149); Cure Cancer Australia Foundation Fellowship (E.B); NHMRC Program grant (D.H.; APP1016701); LLS Specialized Center of Research (C.B., D.H.; 7001-13); NHMRC IRIIS grant (to SVI and WEHI); in part by the Victorian State Government Operational Infrastructure Support Program (to SVI and WEHI); The Victorian Centre for Functional Genomics is funded by the Australian Cancer Research Foundation (ACRF), the Victorian Department of Industry, Innovation and Regional Development (DIIRD), the Australian Phenomics Network (APN) and supported by funding from the Australian Government’s Education Investment Fund through the Super Science Initiative, the Australasian Genomics Technologies Association (AMATA), the Brockhoff Foundation and the Peter MacCallum Cancer Centre Foundation; M.A.D is an Investigator of the Howard Hughes Medical Institute; C.W. is the Philip Desbrow Senior Research Fellow of the Leukaemia Foundation.
References
- 1.Sandberg AA, Bridge JA. Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors: osteosarcoma and related tumors. Cancer Genet Cytogenet. 2003;145:1–30. [PubMed] [Google Scholar]
- 2.Meyers PA, Schwartz CL, Krailo M, Kleinerman ES, Betcher D, Bernstein ML, et al. Osteosarcoma: a randomized, prospective trial of the addition of ifosfamide and/or muramyl tripeptide to cisplatin, doxorubicin, and high-dose methotrexate. J Clin Oncol. 2005;23:2004–11. doi: 10.1200/JCO.2005.06.031. [DOI] [PubMed] [Google Scholar]
- 3.Janeway KA, Barkauskas DA, Krailo MD, Meyers PA, Schwartz CL, Ebb DH, et al. Outcome for adolescent and young adult patients with osteosarcoma: a report from the Children’s Oncology Group. Cancer. 2012;118:4597–605. doi: 10.1002/cncr.27414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer. 2009;115:1531–43. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janeway KA, Grier HE. Sequelae of osteosarcoma medical therapy: a review of rare acute toxicities and late effects. Lancet Oncol. 2010;11:670–8. doi: 10.1016/S1470-2045(10)70062-0. [DOI] [PubMed] [Google Scholar]
- 6.Allison DC, Carney SC, Ahlmann ER, Hendifar A, Chawla S, Fedenko A, et al. A meta-analysis of osteosarcoma outcomes in the modern medical era. Sarcoma. 2012;2012:704872. doi: 10.1155/2012/704872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khanna C, Fan TM, Gorlick R, Helman LJ, Kleinerman ES, Adamson PC, et al. Toward a drug development path that targets metastatic progression in osteosarcoma. Clin Cancer Res. 2014;20:4200–9. doi: 10.1158/1078-0432.CCR-13-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X, Bahrami A, Pappo A, Easton J, Dalton J, Hedlund E, et al. Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep. 2014;7:104–12. doi: 10.1016/j.celrep.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorlick R, Anderson P, Andrulis I, Arndt C, Beardsley GP, Bernstein M, et al. Biology of childhood osteogenic sarcoma and potential targets for therapeutic development: meeting summary. Clin Cancer Res. 2003;9:5442–53. [PubMed] [Google Scholar]
- 10.Sampson VB, Gorlick R, Kamara D, Anders Kolb E. A review of targeted therapies evaluated by the pediatric preclinical testing program for osteosarcoma. Front Oncol. 2013;3:132. doi: 10.3389/fonc.2013.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Houghton PJ, Morton CL, Kolb EA, Gorlick R, Lock R, Carol H, et al. Initial testing (stage 1) of the mTOR inhibitor rapamycin by the pediatric preclinical testing program. Pediatr Blood Cancer. 2008;50:799–805. doi: 10.1002/pbc.21296. [DOI] [PubMed] [Google Scholar]
- 12.Sellers WR. A blueprint for advancing genetics-based cancer therapy. Cell. 2011;147:26–31. doi: 10.1016/j.cell.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 13.Walkley CR, Qudsi R, Sankaran VG, Perry JA, Gostissa M, Roth SI, et al. Conditional mouse osteosarcoma, dependent on p53 loss and potentiated by loss of Rb, mimics the human disease. Genes Dev. 2008;22:1662–76. doi: 10.1101/gad.1656808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin PP, Pandey MK, Jin F, Raymond AK, Akiyama H, Lozano G. Targeted mutation of p53 and Rb in mesenchymal cells of the limb bud produces sarcomas in mice. Carcinogenesis. 2009;30:1789–95. doi: 10.1093/carcin/bgp180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berman SD, Calo E, Landman AS, Danielian PS, Miller ES, West JC, et al. Metastatic osteosarcoma induced by inactivation of Rb and p53 in the osteoblast lineage. Proc Natl Acad Sci U S A. 2008;105:11851–6. doi: 10.1073/pnas.0805462105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mutsaers AJ, Ng AJ, Baker EK, Russell MR, Chalk AM, Wall M, et al. Modeling distinct osteosarcoma subtypes in vivo using Cre:lox and lineage-restricted transgenic shRNA. Bone. 2013;55:166–78. doi: 10.1016/j.bone.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 17.North TE, Goessling W, Walkley CR, Lengerke C, Kopani KR, Lord AM, et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447:1007–11. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ziauddin J, Sabatini DM. Microarrays of cells expressing defined cDNAs. Nature. 2001;411:107–10. doi: 10.1038/35075114. [DOI] [PubMed] [Google Scholar]
- 19.Stewart E, Goshorn R, Bradley C, Griffiths Lyra M, Benavente C, Twarog Nathaniel R, et al. Targeting the DNA Repair Pathway in Ewing Sarcoma. Cell Reports. 2014;9:829–40. doi: 10.1016/j.celrep.2014.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X, Stewart E, Shelat AA, Qu C, Bahrami A, Hatley M, et al. Targeting oxidative stress in embryonal rhabdomyosarcoma. Cancer Cell. 2013;24:710–24. doi: 10.1016/j.ccr.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Srikantan S, Abdelmohsen K, Lee EK, Tominaga K, Subaran SS, Kuwano Y, et al. Translational control of TOP2A influences doxorubicin efficacy. Mol Cell Biol. 2011;31:3790–801. doi: 10.1128/MCB.05639-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burgess DJ, Doles J, Zender L, Xue W, Ma B, McCombie WR, et al. Topoisomerase levels determine chemotherapy response in vitro and in vivo. Proc Natl Acad Sci U S A. 2008;105:9053–8. doi: 10.1073/pnas.0803513105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol. 2007;3:576–83. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]
- 24.Knight SD, Adams ND, Burgess JL, Chaudhari AM, Darcy MG, Donatelli CA, et al. Discovery of GSK2126458, a Highly Potent Inhibitor of PI3K and the Mammalian Target of Rapamycin. ACS Medicinal Chemistry Letters. 2010;1:39–43. doi: 10.1021/ml900028r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mallon R, Feldberg LR, Lucas J, Chaudhary I, Dehnhardt C, Santos ED, et al. Antitumor efficacy of PKI-587, a highly potent dual PI3K/mTOR kinase inhibitor. Clin Cancer Res. 2011;17:3193–203. doi: 10.1158/1078-0432.CCR-10-1694. [DOI] [PubMed] [Google Scholar]
- 26.Rodon J, Dienstmann R, Serra V, Tabernero J. Development of PI3K inhibitors: lessons learned from early clinical trials. Nat Rev Clin Oncol. 2013;10:143–53. doi: 10.1038/nrclinonc.2013.10. [DOI] [PubMed] [Google Scholar]
- 27.Furet P, Guagnano V, Fairhurst RA, Imbach-Weese P, Bruce I, Knapp M, et al. Discovery of NVP-BYL719 a potent and selective phosphatidylinositol-3 kinase alpha inhibitor selected for clinical evaluation. Bioorg Med Chem Lett. 2013;23:3741–8. doi: 10.1016/j.bmcl.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 28.Gobin B, Battaglia S, Lanel R, Chesneau J, Amiaud J, Redini F, et al. NVP-BEZ235, a dual PI3K/mTOR inhibitor, inhibits osteosarcoma cell proliferation and tumor development in vivo with an improved survival rate. Cancer Lett. 2014;344:291–8. doi: 10.1016/j.canlet.2013.11.017. [DOI] [PubMed] [Google Scholar]
- 29.Gobin B, Huin MB, Lamoureux F, Ory B, Charrier C, Lanel R, et al. BYL719, a new alpha-specific PI3K inhibitor: single administration and in combination with conventional chemotherapy for the treatment of osteosarcoma. Int J Cancer. 2015;136:784–96. doi: 10.1002/ijc.29040. [DOI] [PubMed] [Google Scholar]
- 30.Gorlick R, Maris JM, Houghton PJ, Lock R, Carol H, Kurmasheva RT, et al. Testing of the Akt/PKB inhibitor MK-2206 by the Pediatric Preclinical Testing Program. Pediatr Blood Cancer. 2012;59:518–24. doi: 10.1002/pbc.23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carol H, Morton CL, Gorlick R, Kolb EA, Keir ST, Reynolds CP, et al. Initial testing (stage 1) of the Akt inhibitor GSK690693 by the pediatric preclinical testing program. Pediatr Blood Cancer. 2010;55:1329–37. doi: 10.1002/pbc.22710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorlick R, Kolb EA, Houghton PJ, Morton CL, Neale G, Keir ST, et al. Initial testing (stage 1) of the cyclin dependent kinase inhibitor SCH 727965 (dinaciclib) by the pediatric preclinical testing program. Pediatr Blood Cancer. 2012;59:1266–74. doi: 10.1002/pbc.24073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Houghton PJ, Morton CL, Kolb EA, Lock R, Carol H, Reynolds CP, et al. Initial testing (stage 1) of the proteasome inhibitor bortezomib by the pediatric preclinical testing program. Pediatr Blood Cancer. 2008;50:37–45. doi: 10.1002/pbc.21214. [DOI] [PubMed] [Google Scholar]
- 34.Klijn C, Durinck S, Stawiski EW, Haverty PM, Jiang Z, Liu H, et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotechnol. 2015;33:306–12. doi: 10.1038/nbt.3080. [DOI] [PubMed] [Google Scholar]
- 35.Perry JA, Kiezun A, Tonzi P, Van Allen EM, Carter SL, Baca SC, et al. Complementary genomic approaches highlight the PI3K/mTOR pathway as a common vulnerability in osteosarcoma. Proc Natl Acad Sci U S A. 2014;111:E5564–73. doi: 10.1073/pnas.1419260111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–18. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 37.Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–9. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 38.Adler AS, McCleland ML, Yee S, Yaylaoglu M, Hussain S, Cosino E, et al. An integrative analysis of colon cancer identifies an essential function for PRPF6 in tumor growth. Genes Dev. 2014;28:1068–84. doi: 10.1101/gad.237206.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hubert CG, Bradley RK, Ding Y, Toledo CM, Herman J, Skutt-Kakaria K, et al. Genome-wide RNAi screens in human brain tumor isolates reveal a novel viability requirement for PHF5A. Genes Dev. 2013;27:1032–45. doi: 10.1101/gad.212548.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore MJ, Wang Q, Kennedy CJ, Silver PA. An alternative splicing network links cell-cycle control to apoptosis. Cell. 2010;142:625–36. doi: 10.1016/j.cell.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoon JK, Wold B. The bHLH regulator pMesogenin1 is required for maturation and segmentation of paraxial mesoderm. Genes Dev. 2000;14:3204–14. doi: 10.1101/gad.850000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chalamalasetty RB, Dunty WC, Jr, Biris KK, Ajima R, Iacovino M, Beisaw A, et al. The Wnt3a/beta-catenin target gene Mesogenin1 controls the segmentation clock by activating a Notch signalling program. Nat Commun. 2011;2:390. doi: 10.1038/ncomms1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mikkola HK, Klintman J, Yang H, Hock H, Schlaeger TM, Fujiwara Y, et al. Haematopoietic stem cells retain long-term repopulating activity and multipotency in the absence of stem-cell leukaemia SCL/tal-1 gene. Nature. 2003;421:547–51. doi: 10.1038/nature01345. [DOI] [PubMed] [Google Scholar]
- 44.Kuijjer ML, van den Akker BE, Hilhorst R, Mommersteeg M, Buddingh EP, Serra M, et al. Kinome and mRNA expression profiling of high-grade osteosarcoma cell lines implies Akt signaling as possible target for therapy. BMC Med Genomics. 2014;7:4. doi: 10.1186/1755-8794-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moriceau G, Ory B, Mitrofan L, Riganti C, Blanchard F, Brion R, et al. Zoledronic acid potentiates mTOR inhibition and abolishes the resistance of osteosarcoma cells to RAD001 (Everolimus): pivotal role of the prenylation process. Cancer Res. 2010;70:10329–39. doi: 10.1158/0008-5472.CAN-10-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Allen EM, Wagle N, Stojanov P, Perrin DL, Cibulskis K, Marlow S, et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat Med. 2014;20:682–8. doi: 10.1038/nm.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chawla SP, Staddon AP, Baker LH, Schuetze SM, Tolcher AW, D’Amato GZ, et al. Phase II study of the mammalian target of rapamycin inhibitor ridaforolimus in patients with advanced bone and soft tissue sarcomas. J Clin Oncol. 2012;30:78–84. doi: 10.1200/JCO.2011.35.6329. [DOI] [PubMed] [Google Scholar]
- 48.Elkabets M, Vora S, Juric D, Morse N, Mino-Kenudson M, Muranen T, et al. mTORC1 inhibition is required for sensitivity to PI3K p110alpha inhibitors in PIK3CA-mutant breast cancer. Sci Transl Med. 2013;5:196ra99. doi: 10.1126/scitranslmed.3005747. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.