Abstract
Decitabine, a cancer therapeutic that inhibits DNA methylation, produces variable antitumor response rates in patients with solid tumors that might be leveraged clinically with identification of a predictive biomarker. In this study, we profiled the response of human ovarian, melanoma and breast cancer cells treated with decitabine, finding that RAS/MEK/ERK pathway activation and DNMT1 expression correlated with cytotoxic activity. Further, we showed that KRAS genomic status predicted decitabine sensitivity in low and high-grade serous ovarian cancer cells. Pre-treatment with decitabine decreased the cytotoxic activity of MEK inhibitors in KRAS-mutant ovarian cancer cells, with reciprocal downregulation of DNMT1 and MEK/ERK phosphorylation. In parallel with these responses, decitabine also upregulated the pro-apoptotic BCL-2 family member BNIP3, which is known to be regulated by MEK and ERK, and heightened the activity of pro-apoptotic small molecule navitoclax, a BCL-2 family inhibitor. In a xenograft model of KRAS-mutant ovarian cancer, combining decitabine and navitoclax heighted antitumor activity beyond administration of either compound alone. Our results define the RAS/MEK/DNMT1 pathway as a determinant of sensitivity to DNA methyltransferase inhibition, specifically implicating KRAS status as a biomarker of drug response in ovarian cancer.
Introduction
DNA methylation plays an active role in chromatin structure and gene expression and thus can significantly impact tumorigenicity (1–3). Decitabine is a cancer therapeutic that disrupts DNA methylation through inhibition of DNA methyltranferases, DNMT1, DNMT3a and DNMT3b (3). Decitabine is approved to treat hematological malignancies and in this context provides significant therapeutic benefit. Indeed, low-dose decitabine induced an objective response in 73% of patients with myelodysplasic (n=77) and chronic myelomonocytic leukemia (n=18) (4). Although the best clinical response occurred in patients who showed rapid hypomethylation in peripheral blood and bone marrow cells, the degree of hypomethylation failed to correlate with response (4).
In contrast to hematopoietic cancers, decitabine shows moderate to low response rates in patients with solid tumors. Treatment with low-dose decitabine in patients with female reproductive (n=35), melanoma (n=23) and breast (n=4) cancers demonstrated a combined response and stable disease count of 6%, 26% and 50%, respectively (2). Identifying stratification markers as well as optimal combination strategies for decitabine treatment may enhance this compound’s clinical benefit in patients with solid tumors.
Small-molecule sensitivity profiling of deeply characterized cancer cell lines is one approach to identify features that correlate with compound activity (5–8). For example, profiling experiments clearly identify the enhanced sensitivity of BRAF-mutant cells to BRAF inhibitors, and this association predicts response in patient populations (6–8). To date, sensitivity-profiling experiments have relied on three-day time points to measure viability. However, small molecules that target chromatin-modifying proteins are reported to decrease cellular viability at extended time points (9, 10). As such, longer time points may be more informative in studying dependencies targeted by chromatin-modifying agents, such as decitabine.
Here, we used a nine-day viability assay to demonstrate that a subset of solid tumor cell lines is sensitive to low-dose decitabine at clinically achievable concentrations. We showed that RAS/RAF/MEK pathway activation as well as DNMT1 expression correlates with sensitivity to decitabine. We demonstrated that mutation or amplification of KRAS predicts sensitivity to decitabine in ovarian cancer cell lines. We further observed changes in activity of navitoclax and MEK inhibitors following decitabine pre-treatment and showed that BCL-2 and MEK signaling may regulate decitabine’s activity in RAS-activated cancer cell lines. Finally, we showed that the combination of decitabine and navitoclax significantly decreased tumor volume to a greater extent than either agent alone in a cell line-derived xenograft model.
Methods
Reagents and cell lines
All cell lines were obtained from the Broad Institute Biological Samples Platform (BSP) or ATCC. All cell lines were purchased in 2012 and cultured as previously described (6). Cell line profiling was performed within six months of receiving the cell lines. The cell lines were authenticated by BSP or ATCC via SNP array and short tandem repeat profiling, respectively. Authentication of the cell lines after purchasing was not performed. Mutation and gene expression data for each cell line was obtained from the Cancer Cell Line Encyclopedia (8). Antibodies were purchased from Cell Signaling. DNMT3B antibody was purchased from Abcam. All compounds were dissolved in DMSO and stored at −20°C. For all six- and nine-day treatments, media and compound were replenished every 3 days.
Cell viability
Cell density was optimized in 384-well plates for three- or nine-day treatment independently using CellTiter Glo (Promega) per the manufacturer’s instructions (30 µL). Cell density was optimized such that cell growth for the duration of the treatment fell within the linear range. For co-treatment combination studies, cells were plated at a density of 250 cells per well in 96-well plates (100 µL). Cells were plated and allowed to adhere for 24 hours prior to administering the compound. Cell viability was measured using CellTiter Glo (1:2 with PBS) per the manufacturer’s instructions. Luminescence was measured using the Envision (Perkin Elmer). Background luminescence was subtracted, and the measured ATP level was normalized to the vehicle. For all six- and nine-day treatments, media and compound were replenished every 3 days.
Capase 3/7 activation
Cells were plated at a density of 250 cells per well in 96-well plates and allowed to adhere for 24 hours prior to treatment. Cells were treated for six days with decitabine. CellTiter Blue (Promega) and Caspase Glo (Promega) was used in combination per the manufacture’s instructions. Fluorescence (560Ex/590Em nm) and luminescence was measured using the SpectraMax M5 (Molecular Devices). Background luminescence was subtracted, and the measured caspase activation was normalized to cell number. For all six-day treatments, media and compound were replenished every 3 days.
Western blot analysis
Cells were lysed in RIPA buffer (Thermo) supplemented with PhosphoStop (Roche) and complete protease inhibitor (Roche). Lysate (50 µg) was loaded onto a NuPage Bis-Tris Gel (4–12%, Invitrogen) and transferred using the iBlot (Invitrogen). The membrane was blocked (5% milk/PBS/0.1% Tween) and incubated with primary antibody (3% BSA/PBS/0.1%Tween) overnight at 4°C or at room temperature for two hours. The membrane was washed three times (0.1% Tween/PBS) and incubated with the appropriate secondary antibody (3% BSA/PBS/0.1%Tween). Luminescence was captured after addition of FemtoSubstrate (Thermo) using the Image Station 4000 MM (Kodak) and Carestream MI software at various exposure times. Quantification of the western blots was performed using Image J.
Pre-treatment combination studies
Cells (0.1–0.25×106) were plated in 100 cm2 dishes. After 24 hours, cells were treated with decitabine (0.5 µM) or DMSO for three, six or nine days. Cell density was adjusted every three days to prevent over growth, and the media and compound treatment was replaced. Viable cells were harvested by trypsin and plated (30 µL) in 384 well plates in the presence of decitabine or DMSO at an optimized density and allowed to adhere for 24 hours. Combination compounds (100 nL) were administered for 72 hours, and cell viability was measured as described (11). Viability was normalized to the cell state tested, and the area under the curve was determined for treatment with decitabine (AUCdectabine) or DMSO (AUCDMSO) as previously described (6). Cell viability of decitabine or dmso pre-treated cells was confirmed by calcein AM staining.
Correlation analysis using differential mutual information
We used an estimator of the differential mutual information,
| (1) |
where x is the activity to decitabine profile and y is each of the genomic alterations or pathway profiles being compared against. The computation is based on kernel density estimation following a methodology similar to that used in Wood et al. but normalizing the mutual information (12, 13) as to produce an information coefficient,
| (2) |
To assess the significance of every Ic score we perform a permutation test randomly permuting the activity to decitabine profile and based on those scores compute nominal p-values and False Discover Rates (FDR) as those shown in Supplemental Tables S1 and S2.
Database of gene sets
The database of gene sets used in this study (7,762 gene sets) consists of the Molecular Signatures Database (MSigDB, release 4.0, www.broadinstitute.org/msigdb) sub-collections c2, c3, c5 and c6 (14), plus a preliminary unpublished collection of oncogenic signatures that will be incorporated in sub-collection c6 in a forthcoming release of MSigDB.
Xenograft assay
Four to five-week-old female athymic Nude-Foxn1nu mice were purchased from Harlan Laboratories and maintained in sterile housing throughout the experimental period. For this subcutaneous (SC) xenograft model, 5 × 106 OVCAR8 cells in 200 µL of a PBS/Matrigel (BD Biosciences) mixture (1:1 v/v) were injected SC into the right flank of the mice. After the tumors reached approximately 200 mm3 in volume, mice were randomized and treated with decitabine (0.2 mg/kg SC thrice weekly), navitoclax (100 mg/kg IP five times per week), decitabine and navitoclax as described or vehicle control (PBS) for 4 weeks before euthanasia and necropsy. Week four treatment of the combination study omitted one injection of decitabine and two injections of navitoclax in all cohorts. Groups of 7 mice were used. Tumor volume was calculated weekly from caliper measurements of the smallest (SD) and largest tumor diameter (LD) using the formula: volume = [LD × SD2] × π/6. Experiments performed received prior approval from the Vanderbilt University Institutional Animal Use and Care Committee, and all animals were maintained in accordance to guidelines of the American Association of Laboratory Animal Care.
Combined bisulfite restriction analysis (COBRA)
Cells were treated for six days with decitabine (0.5 µM) or DMSO. Cell lysis and bisulfite treatment was accomplished using the EpiTect Plus LyseAll Lysis Kit and the EpiTect Plus Bisulfite Kit by Qiagen per the manufacturer’s instructions. Combine bisulfite restriction analysis was performed as previously described (15). Briefly, PCR was carried out using HotStarTaq DNA polymerase (Qiagen) per the manufacturer’s instructions using bisulfite treated DNA (300 ng). Touchdown PCR was performed using BNIP3 primers surrounding the transcriptional start site (F: 5’-TTYGGTYGGAGGAATTTATAGGGTAG, R: 5’-CCCTCRCCCACCRCCAAAAC (where Y = C or T and R = A or G), temperature-cycle: 58-3, 56-4, 54-5, 52-26). The 156 bp PCR product (10 µL) was digested with the restriction enzyme, AfaI (Invitrogen), for one hour at 37°C, and the DNA was subjected to 2% agarose gel electrophoresis and imaged.
Results
Decitabine decreases cell viability in a time-dependent manner in a subset of solid tumor cell lines
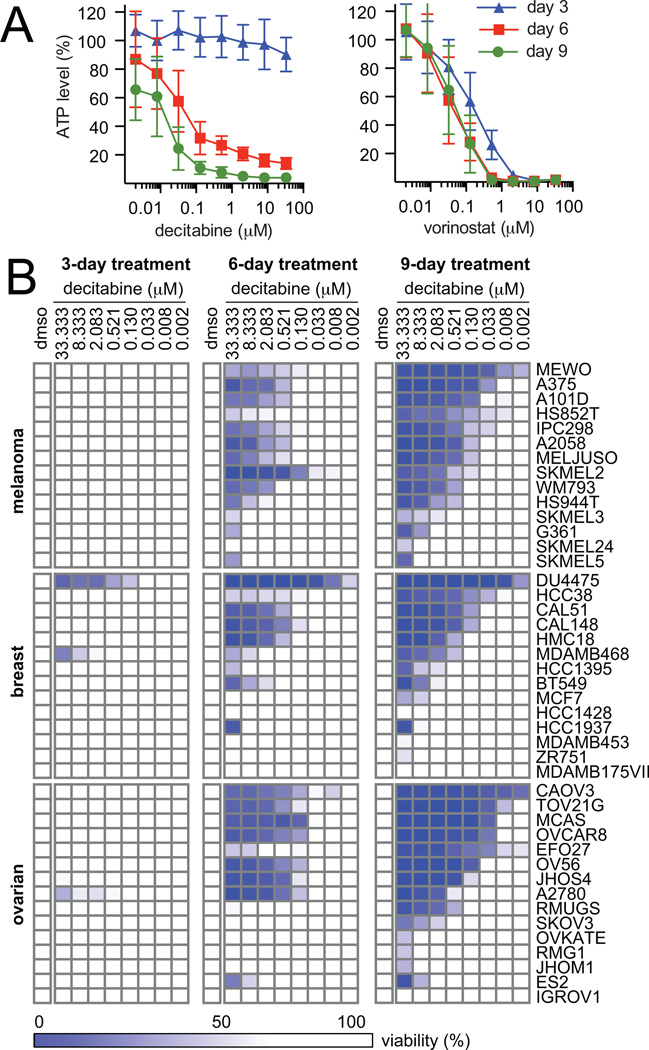
To study the effects of compounds that lead to changes in chromatin modifications, we used a nine-day viability assay. We targeted two distinct chromatin modifications, DNA methylation and histone deacetylation, using the FDA-approved small-molecule therapeutics, decitabine and vorinostat, in an ovarian cancer cell line (OVCAR8). Media and compound were replaced every three days to minimize nutrient deprivation and compound degradation. Although both compounds showed a dose-dependent decrease in viability, decitabine showed a marked change in activity over time. Specifically, activity improved ~1000-fold from three days (IC50day 3, > 33.3 µM) to nine days of treatment (IC50day 9, 22.2 nM). In contrast, treatment with vorinostat caused a stable decrease in cell viability at all three time points (Fig. 1A).
Figure 1.
Decitabine decreases cell viability in a time-dependent manner in a subset of cancer cell lines. A, cell viability was measured after treatment with decitabine (left) or vorinostat (right) in OVCAR8 cells. Data are representative of two independent experiments (14 replicates, mean ± SD). B, cell viability was measured for a panel of 45 solid tumor lines after treatment with decitabine. Data are representative of two independent experiments each measured in 14 replicates (mean).
To understand better decitabine’s activity in solid tumors, we profiled the response of 45 cancer cell lines in ovarian, melanoma and breast lineages over nine days of treatment with decitabine. In general, decitabine showed no activity at three days of treatment, but significantly decreased cell viability at six and nine days of treatment (Fig. 1B). A subset of cell lines in all three lineages showed substantial sensitivity to low doses of decitabine at nine days of treatment (IC50 < 150 nM), and decitabine elicited a dynamic range in sensitivity greater than 1000-fold (10 nM > IC50 > 10 µM) in all lineages after nine days of treatment (Supplemental Fig. S1).
KRAS mutation status predicts decitabine sensitivity in ovarian cancer cell lines
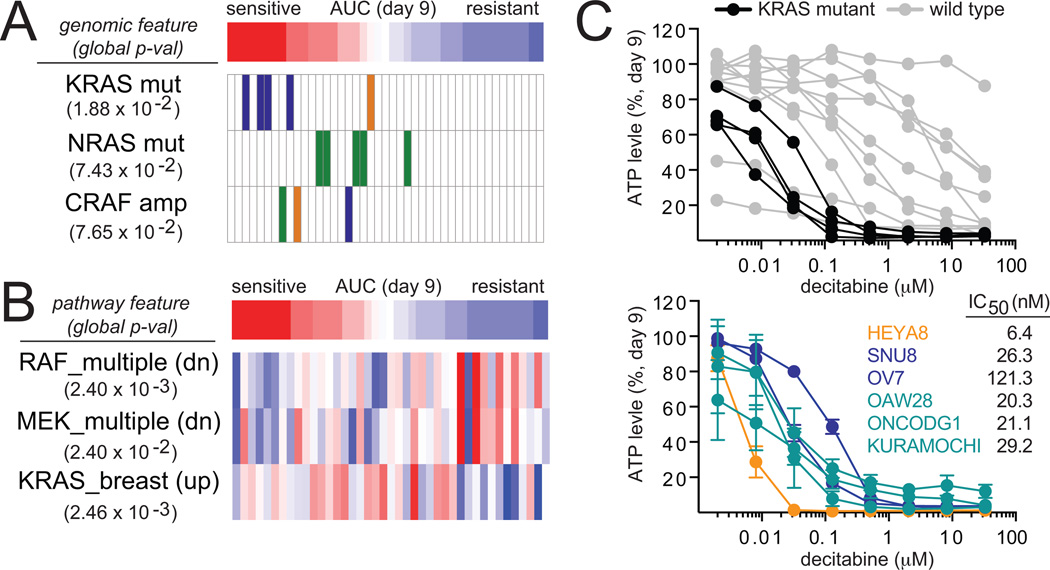
We correlated genomic alterations and pathway features to decitabine sensitivity and resistance in an unbiased manner using a differential mutual information estimator (see methods). We found that activating mutations in KRAS (p-val 1.88 × 10−2) and NRAS (p-val 7.43×10−2), but not BRAF strongly correlate with sensitivity to decitabine. Amplifications in CRAF (p-val 7.65×10−2), a downstream effector of RAS proteins, also correlated with sensitivity. Seven of the ten most sensitive cell lines contained genomic alterations in KRAS or CRAF, and the most sensitive cell lines were KRAS-mutant ovarian cancer cell lines (n=4) (Fig. 2A, Supplemental Table S1).
Figure 2.
KRAS genomic status predicts decitabine sensitivity in ovarian cancer cell lines. A, correlation analysis based on differential mutual information was used to correlate the cellular response of decitabine (nine-day treatment) with genomic features of ovarian (blue), melanoma (green) and breast (orange) cancer cell lines. B, correlation analysis based on differential mutual information was used to correlate the cellular response of decitabine (nine-day treatment) with pathway features as defined by gene sets with high gene expression (red) or low gene expression (blue). C, cell viability after nine days of treatment with decitabine are shown for the profiled KRAS-mutant (black) and wild-type (gray) ovarian cancer cell lines. Cell viability was measured in one additional KRAS-mutant low-grade serous ovarian cancer cell line (orange), two additional KRAS-mutant high-grade serous ovarian cancer cell lines (navy) and three additional KRAS-amplified high-grade serous ovarian cancer cell lines (teal). Data are representative of two independent experiments (14 replicates, mean ± SD).
We also found that genomic alterations in genes associated with the activation of p38 signaling correlated with resistance to decitabine. Specifically, amplification of MAP2K6 (p-val 1.9×10−3), which activates p38, and deletion of genes that inactivate p38, such as USP47 (p-val 1.4×10−3) and DUSP26 (p-val 1.0×10−3), correlated with resistance to decitabine (Supplemental Table S1). Activation of p38 is reported to oppose RAS-induced cancer cell proliferation (16, 17) and may represent a distinguishing feature of cancer cell lines that are resistant to decitabine. To mitigate confounding variables, we analyzed doubling time for each cell line. Importantly, sensitivity to decitabine failed to correlate with doubling time (Supplemental Fig. S2A).
Global gene-expression profiles available from the Cancer Cell Line Encyclopedia (8) were analyzed to score the activity of gene sets (see methods) that represent defined pathways for each cell line using single-sample gene set enrichment analysis (GSEA) analysis (18). These enrichment scores were correlated to decitabine sensitivity using the differential mutual information estimator to identify pathways implicated in the response to decitabine. Consistent with their association to genomic alterations in RAS pathway genes, sensitivity to decitabine also correlated with gene-set signatures derived from overexpression of KRAS (KRAS.Breast_HMLE.24_UP, p-val 2.46×10−3), CRAF (RAF_UP.V1_DN, p-val 2.40×10−3) and MEK (MEK_UP.V1_DN, p-val 2.40×10−2) (Fig. 2B). High expression of genes in gene-set signatures corresponding to cell cycle regulation (Mitotic_Cell_Cycle_Checkpoint, p-val 5.19×10−4, Cell_Cycle_Go_0007049, p-val 7.78×10−4) also strongly correlated with sensitivity to decitabine (Supplemental Table S2).
Given the impact of RAS/RAF/MEK signaling on decitabine activity, we hypothesized that KRAS genomic status may predict sensitivity to decitabine in ovarian cancer. Clinically, KRAS mutations occur in ~30% of low-grade serous ovarian cancers, and KRAS amplifications occur in ~11% of high-grade serous ovarian cancers (19). We profiled an additional low-grade KRAS-mutant (p53-wild type), two high-grade KRAS-mutant (p53-mutant) and three high-grade KRAS-amplifiied (p53-mutant) serous ovarian cancer cell lines. Low-grade and high-grade ovarian cancer cell lines that harbored mutations or genomic amplifications in KRAS were highly sensitive to decitabine (IC50 < 150 nM) compared to KRAS wild-type ovarian cancer cell lines (Fig. 2C). This sensitivity is consistent with decitabine’s activity in the previously profiled KRAS-mutant ovarian cancer cell lines. To extend these finding to other lineages, we tested the activity of decitabine against a limited panel of pancreatic and colon cancer cell lines. All four pancreatic cell lines tested harbor a G12 mutation in KRAS and were sensitive to decitabine after nine days of treatment. The IC50 values for the pancreatic cell lines, PANC1, KP2, L33 and SNU213, were 267.2 nM, 75.2 nM, 61.29 nM and 15 nM, respectively (Supplemental Fig. S2B). In contrast, KRAS mutation status failed to predict sensitivity to decitabine in colon cancer; although two KRAS-mutant cell lines (HCT116, RCM) were sensitive to decitabine, two (HCT15, LS513) displayed moderate sensitivity and one (SNUC2A) was resistant (Supplemental Fig. S2C).
DNMT1 plays a role in mediating decitabine’s activity in cancer cell lines
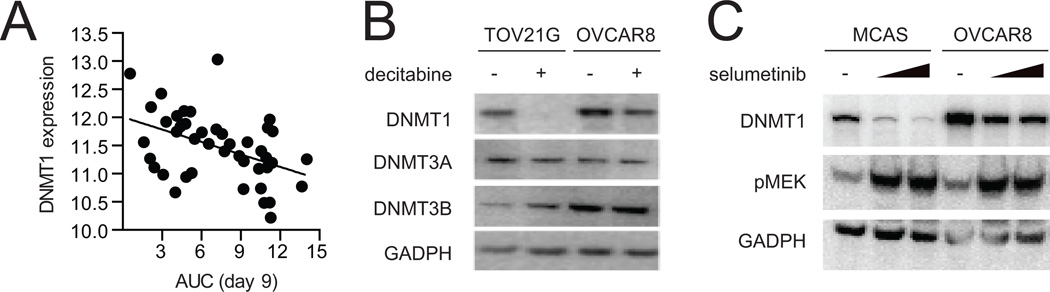
Decitabine targets DNA methyltransferases DNMT1, DNMT3a and DNMT3b. Given the role of DNMT1 as a downstream effector of RAS proteins (20, 21), we analyzed basal gene-expression levels available from the Cancer Cell Line Encyclopedia (8) and changes in protein levels of DNA methyltranserases after treatment with decitabine. Decitabine sensitivity correlated more strongly with high expression of DNMT1 (ρ, −0.443) than DNMT3A (ρ, −0.176) or DNMT3B (ρ, 0.187) as assessed by Pearson regression analysis (Fig. 3A, Supplemental Fig. S3A). In addition, decitabine clearly decreased DNMT1 levels in three KRAS-mutant ovarian cancer cell lines, OVCAR8, TOV21G and OV56 (Figure 3B, Supplemental Figure S3B). DNMT3A protein levels remained constant in all three cell lines, and DNMT3B protein level decreased only in OV56. KRAS-wild type ovarian cancer cell lines displayed a varied response to decitabine (Supplemental Fig. S3B).
Figure 3.
DNMT1 plays a role in mediating decitabine’s activity. A, cell viability after treatment with decitabine (AUC, x-axis) was plotted against DNMT1 mRNA expression (log2, y-axis). B, western blot analysis was performed after treatment with decitabine (0.5 µM) or DMSO for nine days in two KRAS-mutant ovarian cancer cell lines. C, DNMT1 protein level was probed by western blot after treatment with selumetinib for six days in two KRAS-mutant ovarian cancer cell lines. Data are representative of two independent experiments.
Given our observed connection between RAS/RAF/MEK pathway activation and DNMT1, we investigated the impact of MEK inhibition on DNMT1 protein levels. As such, we observed that treatment with selumetinib, a small-molecule inhibitor of MEK, decreased DNMT1 protein levels in two KRAS-mutant ovarian cancer cell lines (Fig. 3C, Supplemental Figure S3C). This observation is in accordance with the reported regulation of DNMT1 protein levels by the RAS/RAF/MEK signaling cascade (22).
Pre-treatment with decitabine alters the activity of a subset of small-molecule probes
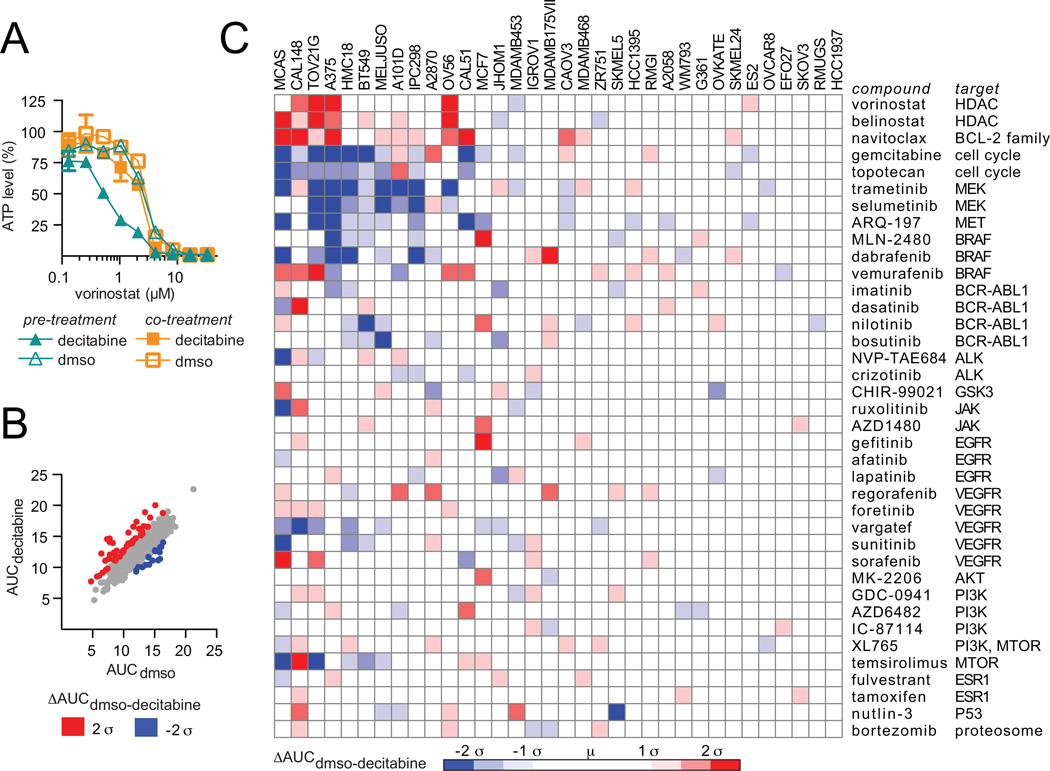
We used combination profiling to interrogate further the mechanism of action of decitabine in a panel of 34 decitabine-sensitive and -resistant cell lines. To determine the appropriate combination schedule, we measured the change in activity of the histone deacetylase inhibitor vorinostat, a compound known to synergize with decitabine (23). The human plasma maximum concentration (Cmax) value ranges from 0.3–1.6 µM for low-dose decitabine treatment (24), and thus, a clinically meaningful concentration of decitabine (0.5 µM) was chosen for all combination experiments. For co-treatment experiments, A375 cancer cells were treated for three days with vorinostat and low-dose decitabine (0.5 µM), and cell viability was measured. For pre-treatment experiments, A375 cancer cells were pre-treated with low-dose decitabine (0.5 µM) for nine days and live cells, as verified by staining with calcein AM, were treated with decitabine and vorinostat for three days. Pre-treatment with decitabine, but not co-treatment, increased activity of vorinostat (Fig. 4A).
Figure 4.
Pre-treatment with decitabine alters the activity of a subset of small-molecule probes. A, cell viability was measured after co-treatment with vorinostat and decitabine for three days or after pre-treatment with decitabine or DMSO for nine days followed by co-treatment with vorinostat and decitabine for three days (two biological replicates, mean ± SD). B, Thirty-four cancer cell lines were pre-treated with decitabine (0.5 µM) or DMSO for nine days. Viable cells were harvested and co-treated with decitabine (0.5 µM) or DMSO and the combination ‘informer set’. Cell viability was measured after three days and normalized to the cell state tested. The area under the curve was determined for treatment with decitabine (AUCdecitabine, y-axis) or DMSO (AUCDMSO, x-axis). Data are the mean of two biological replicates. C, the difference between AUCdecitabine and AUCDMSO was plotted for each cell line (x-axis) and each ‘informer set’ compound (y-axis).
Next, we measured the change in activity of a combination ‘informer set’ consisting of 38 small molecules with varying mechanisms of action after pre-treatment with low-dose decitabine (0.5 µM) for nine days followed by co-administration of decitabine and the combination ‘informer set’. Although the majority of compounds responded in a similar manner, a number of significant changes in compound activity were observed upon pre-treatment with decitabine (Fig. 4B). Specifically, our data confirmed that decitabine pre-treatment increased activity of HDAC inhibitors in a number of solid tumor cell lines. In addition, pre-treatment with decitabine increased the activity of the BCL-2 family inhibitor, navitoclax, and decreased activity of MEK inhibitors, such as trametinib and selumetinib, and DNA targeting agents (Fig. 4C). The decrease in activity of DNA-targeting agents, which require cell division to induce cell death, is consistent with the reported observation that decitabine can induce a quiescent cell population in certain contexts (25). Importantly, the majority of compounds, including GDC-0941 (pan-PI3K inhibitor), bortezomib (proteasome inhibitor) and NVP-TAE684 (ALK inhibitor), showed no change in activity after pre-treatment with decitabine despite showing activity as single agents (Fig. 4C, Supplemental Fig. S4A). Together, our data suggests that decitabine induces an altered cell state characterized by decreased sensitivity to MEK inhibitors and increased sensitivity to HDAC and BCL-2 family inhibitors. We hypothesize that MEK/ERK and BCL-2 family signaling may play a direct role in mediating decitabine sensitivity.
Decitabine modulates sensitivity to MEK inhibitors in KRAS-mutant ovarian cancer cell lines
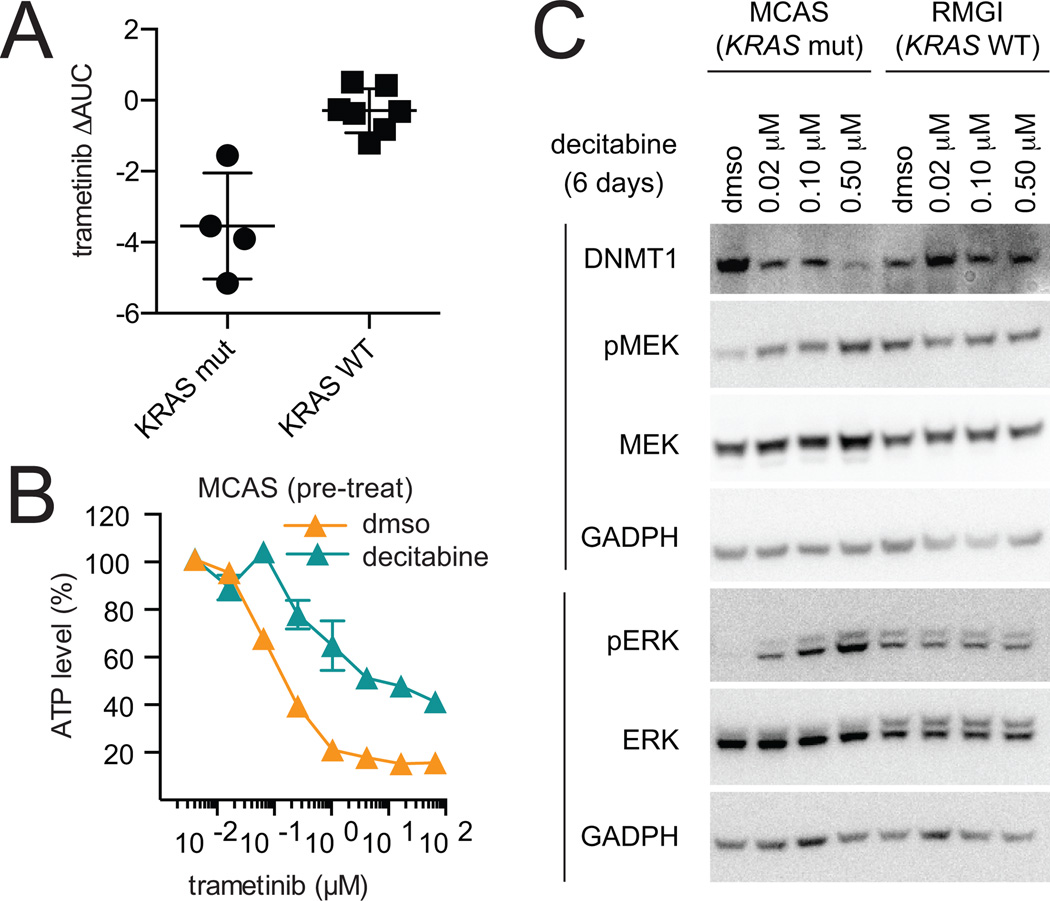
We investigated the relationship between KRAS status and changes in sensitivity to the MEK inhibitor, trametinib, after pre-treatment with decitabine. We observed that ovarian cancer cell lines harboring a mutation in KRAS had a decreased response to trametinib after pre-treatment with decitabine (Fig. 5A). We assessed the time point at which pre-treatment with decitabine altered compound activity. We found that pre-treatment with decitabine for three days, followed by co-treatment with decitabine and trametinib for three days, induced changes in compound sensitivity comparable to decitabine pre-treatment for nine days in a KRAS-mutant ovarian cancer cell line (MCAS) and a CRAF-amplified melanoma cell line (A375) (Fig. 5B, Supplemental Fig. S4B).
Figure 5.
Decitabine modulates MEK/ERK pathway activation. A, change in activity of the MEK inhibitor, trametinib, after pre-treatment for nine days with decitabine in KRAS-mutant and KRAS-wild type ovarian cancer cell lines. B, KRAS-mutant ovarian cancer cell line, MCAS, was pre-treated with decitabine for three days, followed by co-treatment with decitabine and trametinib for three days (two biological replicates, mean ± SD). Data were normalized to the ATP levels (CTG) measured for the corresponding DMSO- or decitabine-treated controls (6 days). C, phosphorylation of MEK and ERK was probed by western blot after treatment with decitabine for six days in a KRAS-mutant (MCAS) and KRAS-wild type (RMGI) ovarian cancer cell line. Data is representative of two independent experiments.
Re-activation of the MEK/ERK pathway as demonstrated by phosphorylation of MEK and ERK has been shown to reduce activity of MEK inhibitors (26). As such, we examined the effect of decitabine treatment on phosphorylation of MEK and ERK. Decitabine treatment for six days increased phosphorylation of both MEK and ERK in a KRAS-mutant ovarian cancer cell line (MCAS), but not a lineage-matched KRAS-wild type cell lines (RMGI) (Fig. 5C, Supplemental Fig. S5A). Decitabine also increased phosphorylation of MEK in OVCAR8, a high-grade serous ovarian cancer cell line that harbors a mutation in KRAS (Supplemental Fig. S5B).
Decitabine modulates sensitivity to the BCL-2 family inhibitor, navitoclax, in KRAS-mutant ovarian cancer cell lines
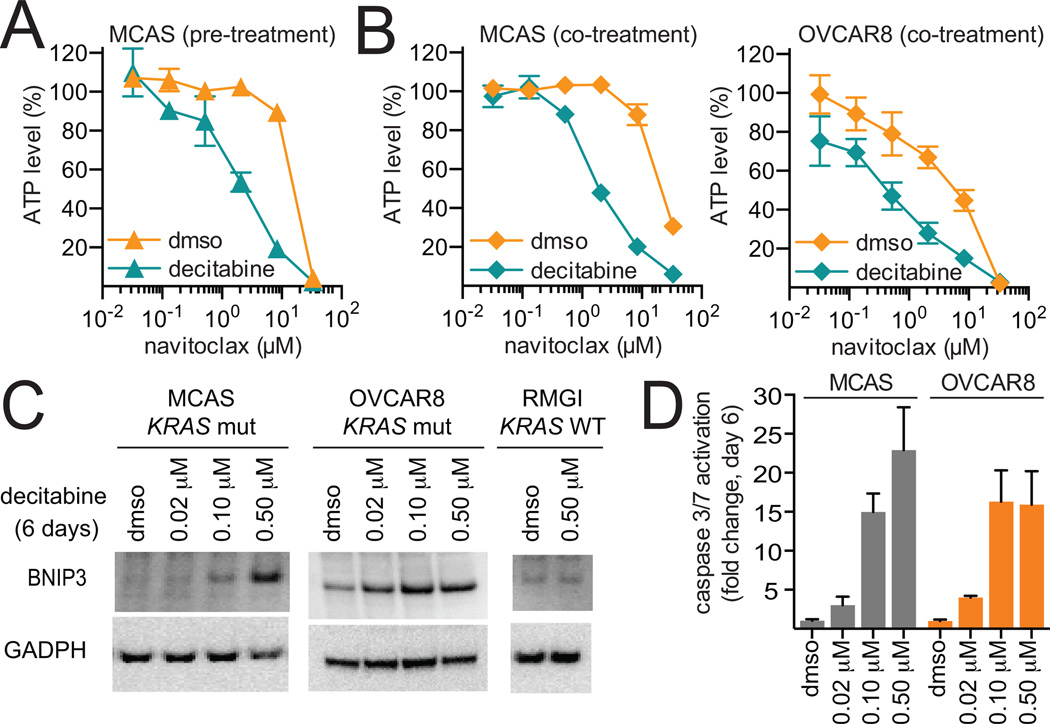
We confirmed that pre-treatment with decitabine increases sensitivity to the BCL-2 family inhibitor, navitoclax. Specifically, we observed that pre-treatment with decitabine for three days followed by co-treatment with decitabine and navitoclax for three days increased activity of navitoclax in a KRAS-mutant ovarian cancer cell line (MCAS) (Fig. 6A). In an effort to simplify dosing and scheduling, we assessed the effect of co-treatment with decitabine and navitoclax. Co-treatment with decitabine increased activity of navitoclax in MCAS and OVCAR8 cell lines (Fig. 6B, Supplemental Fig. S5C). Interestingly, decitabine co-treatment (Fig. 6B), but not pre-treatment (Fig. 4C) increased sensitivity to navitoclax in OVCAR8, a high-grade serous ovarian cancer.
Figure 6.
Decitabine modulates activity of the BCL-2 family inhibitor, navitoclax. A, KRAS-mutant ovarian cancer cell line, MCAS, was pre-treated with decitabine for three days, followed by co-treatment with decitabine and navitoclax for three days (two biological replicates, mean ± SD). Data were normalized to the ATP levels (CTG) measured for the corresponding DMSO- or decitabine-treated controls (6 days). Data is representative of two independent experiments. B, KRAS-mutant ovarian cancer cell lines, MCAS and OVCAR8, were co-treated with decitabine and navitoclax for six days (two biological replicates, mean ± SD). Data were normalized to the ATP levels (CTG) measured for the corresponding DMSO- or decitabine-treated controls (6 days). Data is representative of two independent experiments. C, BNIP3 was probed by western blot after treatment with decitabine for six days in KRAS-mutant (MCAS and OVCAR8) and KRAS-wild type (RMGI) ovarian cancer cell lines. D, caspase-3 and -7 activation was measured after three, six and nine days of treatment in KRAS-mutant ovarian cancer cell lines (six replicates, mean ± SD). Data is representative of two independent experiments.
Activation of the MEK/ERK pathway has previously been shown to increase expression of the BCL-2 family pro-apoptotic protein, BNIP3 (27–29). Consistent with this report, we found that decitabine treatment for six days increased expression of BNIP3 in MCAS and OVCAR8, KRAS-mutant ovarian cancer cell lines, but not in RMGI, a KRAS-wild type ovarian cancer cell line after treatment for six days (Fig. 6C). In addition, decitabine treatment for six days decreased methylation at the transcriptional start site of BNIP3 in MCAS (Supplemental Fig. S5D). Given the role of BNIP3 as a pro-apoptotic BCL-2 family member, we measured activation of caspase-3 and -7 as a marker for apoptosis after decitabine treatment for six days Decitabine induced a dose-dependent increase in caspase activation at time points corresponding to decreased cell viability in both MCAS and OVCAR8 (Fig. 6D). Together, these data suggest that MEK/ERK activation and upregulation of BNIP3 may play a role in mediating decitabine sensitivity in KRAS-activated ovarian cancer cell lines, and provide strong rationale for combining decitabine and navitoclax in this cancer context.
The combination of decitabine and navitoclax shows activity in a xenograft model derived from a KRAS-mutant ovarian cancer cell line
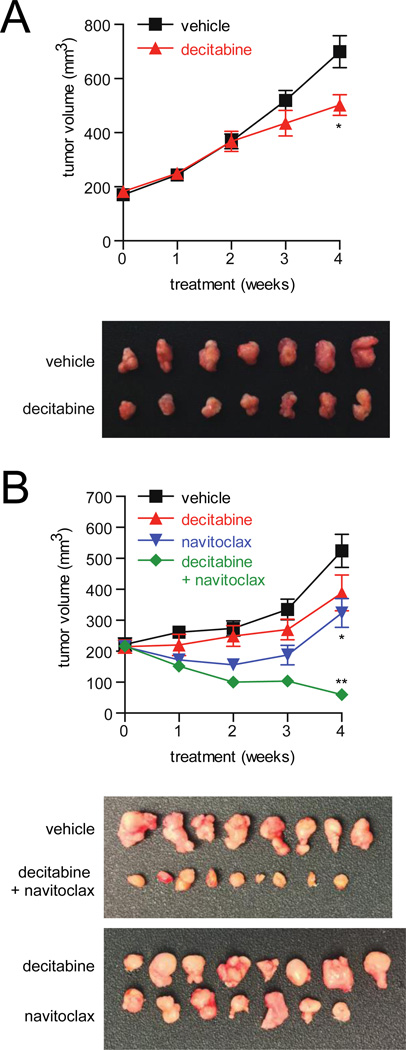
To extend these findings in vivo, we first tested the response of a KRAS-mutant ovarian cell line (OVCAR8) to decitabine as a single agent in a murine xenograft model. We used a subcutaneous OVCAR8 xenograft model, which displayed slower growth than an intraperitoneal xenograft and allowed for extended treatment with decitabine. Decitabine was administered at low doses as previously reported for four weeks following initial tumor formation (0.2 mg/kg SC, 3x per week) (30). Low-dose decitabine displayed a moderate decrease in tumor volume (Fig. 7A) and tumor mass (Supplemental Fig. S6A). No overt toxicities were observed.
Figure 7.
The combination of decitabine and navitoclax shows activity in a xenograft mouse model derived from a KRAS-mutant ovarian cancer cell line. A, tumor volume as measured by caliper of low dose decitabine (0.2 mg/kg, SC, 3x per week) or vehicle administered for four weeks in a subcutaneous xenograft model using a KRAS-mutant cell line (OVCAR8) (*, p = 0.02; Mann-Whitney). Tumors were excised after four weeks of treatment. B, tumor volume as measured by caliper of low-dose decitabine (0.2 mg/kg in 7A and 7B SC, 3x per week), navitoclax (100 mg/kg IP, 5x per week) or vehicle administered alone or in combination for four weeks in a subcutaneous xenograft model using a KRAS-mutant cell line (OVCAR8) (*, p < 0.03; t-test, **, p < 0.001; t-test). Week four treatment omitted one injection of decitabine and two injections of navitoclax. Tumors were excised after four weeks of treatment.
We next tested the effect of co-administration of decitabine and navitoclax in a murine xenograft model using OVCAR8. Decitabine (0.2 mg/kg SC, 3x per week) and navitoclax (100 mg/kg IP, 5x per week) were administered alone or in combination for four weeks following initial tumor formation. The combination of decitabine and navitoclax significantly reduced tumor volume (Fig. 7B) and tumor mass (Supplemental Fig. S6B) compared to either agent alone. No overt toxicities were observed.
Discussion
We profiled a panel of 45 solid tumor cell lines in ovarian, melanoma and breast cancer lineages for their response to decitabine over a nine-day period. We demonstrated that decitabine shows pronounced time-dependent activity in a subset of solid tumor cancer cell lines. For example, activity of decitabine in OVCAR8, an ovarian cancer cell line, improved ~1000-fold from three days of treatment (IC50day 3, > 33.3 µM) to nine days of treatment (IC50day 9, 22.2 nM). Overall, decitabine demonstrated activity at low doses (IC50 < 150 nM) in a subset of solid tumor cancer cell lines and elicited a large dynamic range in sensitivity (10 nM > IC50 > 10 µM) in all lineages. In contrast, a panel of hematopoietic cell lines showed a similar range of sensitivities to decitabine after three days of treatment (data not published). The maximum concentration (Cmax) of decitabine achieved in human plasma in response to low-dose decitabine treatment is 0.3–1.6 µM (24). Given the sensitivity of a subset of solid tumors to decitabine (IC50 < 150 nM ), these data suggest that a prolonged, low-dose treatment regimen may benefit patients with some solid tumors.
To understand if decitabine’s activity was stronger in certain cancer contexts, we correlated compound sensitivity to cell-line features. The absence of confounding correlations to lineage or doubling time enabled us to conduct an unbiased search for genomic features and activated pathways that are associated with sensitivity to decitabine. Genomic alterations in KRAS, NRAS and CRAF as well as activation of the RAS/RAF/MEK pathway significantly correlated with sensitivity to decitabine. In addition, cell cycle regulation correlated with sensitivity to decitabine. This pathway was previously shown to associate strongly with KRAS activation (30, 31).
Given the correlation of RAS/RAF/MEK pathway activation and decitabine activity, we tested the ability of KRAS genomic status to predict sensitivity to decitabine in vitro. KRAS mutations occur in 30% of low-grade serous ovarian cancers, and genomic amplifications of KRAS occur in 11% of high-grade serous ovarian cancer tumors (32). Six additional KRAS-mutant and -amplified ovarian cancer cell lines representing low- and high-grade serous ovarian cancer lines showed sensitivity to decitabine at low dose (IC50 < 150 nM). These data suggest that KRAS genomic status may serve as a potential biomarker for sensitivity to decitabine in ovarian cancer.
RAS/RAF/MEK signaling is reported to regulate DNMT1 and DNMT1-dependent DNA methylation (20–22, 33). Specifically, overexpression of RAS has been shown to increase DNMT1 protein levels (28), and inhibition of MEK by siRNA or small-molecule inhibitors decreased expression of DNMT1 (22). In accordance with our observation that decitabine sensitivity associated with RAS pathway activation, we observed that high gene expression of DNMT1, but not other DNA methyltransferases, correlated with sensitivity to decitabine. We further showed that inhibition of RAS/RAF/MEK signaling with the MEK inhibitor, selumetinib, decreased DNMT1 levels in KRAS-mutant ovarian cancer cell lines. The observed data align with the reported RAS-DNMT1 signaling pathway and imply that targeting DNA methyltransferases in RAS-activated cancers may be a useful therapeutic strategy.
The activity of compounds in the presence or absence of decitabine may provide mechanistic insight into the impact of KRAS signaling on decitabine activity. For example, the combination of decitabine and vorinostat are reported to induce synergistic activity attributed to the combined disruption of chromatin marks (23). This combination has provided a platform for clinical translation (23, 34, 35). We performed compound profiling in 34 solid tumor-derived cell lines with varying sensitivities to decitabine. Given our observation that pre-treatment with decitabine altered the activity of vorinostat, we profiled the response of 38 small molecules after pre-treatment with decitabine for nine days. We observed that decitabine altered the activity of select compounds targeting a subset of biological pathways, including RAS/RAF/MEK signaling and BCL-2 family proteins.
Interestingly, decitabine substantially decreased activity of MEK inhibitors in KRAS-mutant ovarian cancers. Re-activation of the MEK/ERK pathway has been implicated in resistance to BRAF and MEK inhibitors (26) and may account for the altered activity of MEK inhibitors. Indeed, decitabine activated the MEK/ERK pathway as indicated by increased phosphorylation of MEK and ERK. Previous studies show that activation of the MEK/ERK pathway increases pro-apoptotic BCL-2 family members, such as BNIP3 (27–29). The increased expression of this pro-apoptotic BCL-2 protein may contribute to the increased activity of the BCL-2 family inhibitor, navitoclax, after pre-treatment with decitabine. Consistent with these findings, we found that treatment with decitabine increased BNIP3 protein levels in two KRAS-mutant ovarian cancer cell lines and activated caspase-3 and -7 at time points corresponding to a reduction in cell viability. Together, these data suggest that upregulation of the MEK/ERK pathway and pro-apoptotic BCL-2 family proteins by decitabine contributes to reduced viability in RAS-activated cell lines and provides a rationale for the combination of decitabine and navitoclax in KRAS-mutant ovarian cancers.
We extended the observed cellular activity of decitabine and navitoclax to an in vivo xenograft model using a KRAS-mutant ovarian cancer cell line. We demonstrated that co-treatment with decitabine and navitoclax dramatically decreased tumor volume compared to administration of either agent alone. Further studies using multiple KRAS-mutant and KRAS-wild-type ovarian cancer cell lines in diverse in vivo settings are necessary to fully account for the effect of tumor heterogeneity and genotype heterogeneity on the efficacy of decitabine.
Precision medicine aspires to match cancer therapeutics to patients most likely to benefit from the treatment. We have shown that RAS/RAF/MEK pathway activation correlates with sensitivity to decitabine, a small molecule traditionally used in hematological cancers that may also have benefit in solid tumors. Cell line and compound profiling revealed a direct role of RAS/MEK/DNMT1 signaling and the BCL-2 family of proteins in mediating sensitivity to decitabine. We showed that KRAS mutations or amplifications predicted sensitivity to decitabine in ovarian cancer cell lines and suggest a potential biomarker for patient stratification. Further studies are necessary to test the therapeutic benefit of these strategies in preclinical models and ultimately in patients.
Supplementary Material
Acknowledgements
This work was supported by the NCI’s Cancer Target Discovery and Development grant (U01CA176152, awarded to S.L.S.) and an NCI Career Development Award (K08CA148887, awarded to D.K.). We gratefully acknowledge the following colleagues for providing a wonderfully collaborative environment and valuable comments: Drs. J. S. Boehm, W. C. Hahn, and J. P. Mesirov. We would like to thank the following colleagues for their thoughtful discussions and critique: Drs. J. Kotz, D. Adams, A. Basu, N. E. Bodycombe, S. Chattopadhyay, J. H. Cheah, P. A. Clemons, C. Hon, C. Johannessen, E.H. Leshchiner, M. Rees, G. I. Schaefer, B. Seashore-Ludlow, A. M. Stern and V. Sridhar. We thank the Biological Samples Platform for providing the cancer cell lines. The project was enabled by the Cancer Target Discovery and Development Centers at the Broad Institute and Dana-Farber Cancer Institute and the Broad Institute Center for the Science of Therapeutics. The authors are grateful for the leadership of the CTD2 Network by Daniela Gerhard (Director, Office of Cancer Genomics, NCI). S.L.S. is an Investigator at the Howard Hughes Medical Institute.
Footnotes
M.L.S., A.F.S., and S.L.S. designed research; M.L.S, T.P., A.J.W., S.W. and Y.M.C. performed research; T.P. and J.W.K. contributed new reagents/analytic tools; M.L.S, T.P, A.J.W., S.W., D.K., A.F.S, and S.L.S analyzed data; and M.L.S., T.P., A.J.W., D.K., A.F.S., and S.L.S. wrote the manuscript.
Contributor Information
ML Stewart, The Broad Institute of Harvard and MIT (Cambridge, MA).
P Tamayo, The Broad Institute of Harvard and MIT (Cambridge, MA).
AJ Wilson, Department of Obstetrics and Gynecology, Vanderbilt University, Nashville, Tennessee.
S Wang, The Broad Institute of Harvard and MIT (Cambridge, MA).
YM Chang, The Broad Institute of Harvard and MIT (Cambridge, MA).
JW Kim, The Broad Institute of Harvard and MIT (Cambridge, MA).
D Khabele, Department of Obstetrics and Gynecology, Vanderbilt University, Nashville, Tennessee.
AF Shamji, The Broad Institute of Harvard and MIT (Cambridge, MA).
SL Schreiber, The Broad Institute of Harvard and MIT (Cambridge, MA).
References
- 1.Barton CA, Hacker NF, Clark SJ, O'Brien PM. DNA methylation changes in ovarian cancer: implications for early diagnosis, prognosis and treatment. Gynecol Oncol. 2008;109(1):129–139. doi: 10.1016/j.ygyno.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 2.Cowan LA, Talwar S, Yang AS. Will DNA methylation inhibitors work in solid tumors? A review of the clinical experience with azacitidine and decitabine in solid tumors. Epigenomics. 2010;2(1):71–86. doi: 10.2217/epi.09.44. [DOI] [PubMed] [Google Scholar]
- 3.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358(11):1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 4.Kantarjian H1, Oki Y, Garcia-Manero G, Huang X, O'Brien S, Cortes J, Faderl S, et al. Results of a randomized study of 3 schedules of low-dose decitabine in higher-risk myelodysplastic syndrome and chronic myelomonocytic leukemia. Blood. 2007;109(1):52–57. doi: 10.1182/blood-2006-05-021162. [DOI] [PubMed] [Google Scholar]
- 5.Sharma SV, Haber DA, Settleman J. Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat Rev Cancer. 2010;10(4):241–253. doi: 10.1038/nrc2820. [DOI] [PubMed] [Google Scholar]
- 6.Basu A, Bodycombe NE, Cheah JH, Price EV, Liu K, Schaefer GI, et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell. 2013;154(5):1151–1161. doi: 10.1016/j.cell.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483(7391):570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim W, Bird GH, Neff T, Guo G, Kerenyi MA, Walensky LD, Orkin SH. Targeted disruption of the EZH2-EED complex inhibits EZH2-dependent cancer. Nat Chem Biol. 2013;9(10):643–650. doi: 10.1038/nchembio.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vedadi M, Barsyte-Lovejoy D, Liu F, Rival-Gervier S, Allali-Hassani A, Labrie V, Wigle TJ, et al. A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nat Chem Biol. 2011;7(8):566–574. doi: 10.1038/nchembio.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adams DJ, Dai M, Pellegrino G, Wagner BK, Stern AM, Shamji AF, Schreiber SL. Synthesis, cellular evaluation, and mechanism of action of piperlongumine analogs. Proc Natl Acad Sci U S A. 2012;109(38):15115–15120. doi: 10.1073/pnas.1212802109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joe H. Relative Entropy Measures of Multivariate Dependence. J Am Stat Assoc. 1989;84:157–164. [Google Scholar]
- 13.Wood KC, Konieczkowski DJ, Johannessen CM, Boehm JS, Tamayo P, Botvinnik OB, et al. MicroSCALE screening reveals genetic modifiers of therapeutic response in melanoma. Sci Signal. 2012;5(224):rs4.20. doi: 10.1126/scisignal.2002612. [DOI] [PMC free article] [PubMed] [Google Scholar]; Linfoot EH. An informational measure of correlation. Information and Control. 1985;1:85–89. [Google Scholar]
- 14.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abe T, Toyota M, Suzuki H, Murai M, Akino K, Ueno M, Nojima M, Yawata A, Miyakawa H, Suga T, Ito H, Endo T, Tokino T, Hinoda Y, Imai K. Upregulation of Bnip3 by 5-aza-2’-deoxycytidine sensitizes pancreatic cancer cells to hypoxia-mediated cell death. J Gastroenterol. 2005;40:504–510. doi: 10.1007/s00535-005-1576-1. [DOI] [PubMed] [Google Scholar]
- 16.Ellinger-Ziegelbauer H, Kelly K, Sienbenlist U. Cell cycle arrest and reversion of Ras-induced transformation by conditionally activated form of mitogen-activated protein kinase kinase kinase 3. Mol Cell Biol. 1999;19(5):3857–3868. doi: 10.1128/mcb.19.5.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen G, Hitomi M, Han J, Stacey DW. The p38 pathway provides negative feedback for Ras proliferative signaling. J Biol Chem. 2000;275(50):38973–38980. doi: 10.1074/jbc.M002856200. [DOI] [PubMed] [Google Scholar]
- 18.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462(7269):108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bell D, Berchuck A, Birrer M, Chien J, Cramer D, Dao F, et al. Integrated genomic analyses of ovarian carcinoma. Nature. 2012;490(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gazin C, Wajapeyee N, Gobeil S, Virbasius CM, Green MR. An elaborate pathway required for Ras-mediated epigenetic silencing. Nature. 2007;449(7165):1073–1077. doi: 10.1038/nature06251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wajapeyee N, Malonia SK, Palakurthy RK, Green MR. Oncogenic RAS directs silencing of tumor suppressor genes through ordered recruitment of transcriptional repressors. Genes Dev. 2013;27(20):2221–2226. doi: 10.1101/gad.227413.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu R, Wang X, Chen ZF, Sun DF, Tian ZQ, Fang JY. Inhibition of the extracellular signal-regulated kinase/mitogen-activated protein kinase pathway decreases DNA methylation in colon cancer cells. J Biol Chem. 2007;282(16):12249–12259. doi: 10.1074/jbc.M608525200. [DOI] [PubMed] [Google Scholar]
- 23.Stathis A, Hotte SJ, Chen EX, Hirte HW, Oza AM, Moretto P, et al. Phase I study of decitabine in combination with vorinostat in patients with advanced solid tumors and non-Hodgkin's lymphomas. Clin Cancer Res. 2011;17(6):1582–1590. doi: 10.1158/1078-0432.CCR-10-1893. [DOI] [PubMed] [Google Scholar]
- 24.Cashen AF, Shah AK, Todt L, Fisher N, DiPersio J. Pharmacokinetics of decitabine administered as a 3-h infusion to patients with acute myeloid leukemia (AML) or myelodysplastic syndrome (MDS) Cancer Chemother Pharmacol. 2008;61(5):759–766. doi: 10.1007/s00280-007-0531-7. [DOI] [PubMed] [Google Scholar]
- 25.Alcazar O, Achberger S, Aldrich W, Hu A, Negrotto S, Saunthararajah Y, Triozzi P. Epigenetic regulation by decitabine of melanoma differentiation in vitro and in vivo. Int J Cancer. 2012;131(1):18–29. doi: 10.1002/ijc.26320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Little AS, Smith PD, Cook SJ. Mechanisms of acquired resistance to ERK1/2 pathway inhibitors. Oncogene. 2013;32(10):1207–1215. doi: 10.1038/onc.2012.160. [DOI] [PubMed] [Google Scholar]
- 27.An JH, Lee H, Paik SG. Silencing of BNIP3 results from promoter methylation by DNA methyltransferase 1 induced by the mitogen-activated protein kinase pathway. Mol Cells. 2011;31(6):579–583. doi: 10.1007/s10059-011-0065-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.An JH, Maeng O, Kang KH, Lee JO, Kim YS, Paik SG, Lee H. Activation of Ras up-regulateds pro-apoptoic BNIP3 in nitric oxide-induced cell death. J Biol Chem. 2006;281(45):33939–33948. doi: 10.1074/jbc.M605819200. [DOI] [PubMed] [Google Scholar]
- 29.Kalas W, Swiderek E, Rapak A, Kopij M, Rak J, Strzadala L. H-ras up-regualates expression of BNIP3. Anticancer Res. 2011;m31(9):2869–2875. [PubMed] [Google Scholar]
- 30.Ebrahem Q, Negrotto S, Mahfouz RZ, Link KA, Hu Z, et al. Ng KP1. p53 independent epigenetic-differentiation treatment in xenotransplant models of acute myeloid leukemia. Leukemia. 2011;25(11):1739–1750. doi: 10.1038/leu.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, Wong K, Elledge SJ. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137(5):835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Domcke S, Sinha R, Levine DA, Sander C, Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nature Communications. 2013;4:2126. doi: 10.1038/ncomms3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pakneshan P, Szyf M, Rabbani SA. Methylation and inhibition of expression of uPA by the RAS oncogene: divergence of growth control and invasion in breast cancer cells. Carcinogenesis. 2005;26(3):557–664. doi: 10.1093/carcin/bgi009. [DOI] [PubMed] [Google Scholar]
- 34.Matei DE, Nephew KP. Epigenetic therapies for chemoresensitization of epithelial ovarian cancer. Gynecol Oncol. 2010;116(2):195–201. doi: 10.1016/j.ygyno.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matei D, Fang F, Shen C, Schilder J, Arnold A, Zeng Y, et al. Epigenetic resensitization to platinum in ovarian cancer. Cancer Res. 2012;72(9):2197–2205. doi: 10.1158/0008-5472.CAN-11-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.