Abstract
Background:
Breast cancer clinical outcome is affected by tumor molecular features, and the identification of subtype-specific prognostic biomarkers is relevant for breast cancer translational research. Gene expression signatures proved to be able to complement prognostic information provided by classical clinico-pathological features. Recently, microRNAs (miRNAs) have been causally linked to tumorigenesis and cancer progression and have been associated with patient outcome, also in breast cancer.
Methods:
MicroRNAs associated with the development of distant metastasis were identified in a cohort of 92 ESR1+/ERBB2− lymph node-negative breast cancers from patients not receiving adjuvant treatment. Results were confirmed and further investigated in a total of 1246 miRNA and gene expression profiles of the Molecular Taxonomy of Breast Cancer International Consortium data set. Moderated t-test, univariable and multivariable Cox regression models were used for statistical analyses.
Results:
miR-30e* was identified as independent protective prognostic factor in lymph node-negative untreated patients with ESR1+/ERBB2− tumours and retained a significant association with a good prognosis in treated patients with the same tumor subtype as well as in the ERBB2+ subtype, but not in ESR1−/ERBB2− tumours.
Conclusions:
We highlighted a relevant and subtype-specific role in breast cancer for miR-30e* and demonstrated that adding miRNA markers to gene signatures and clinico-pathological features can help for a better prognostication.
Keywords: miR-30e*, breast cancer, molecular subtypes, prognosis, gene expression profiles
Breast cancer is the most common cancer in women worldwide (Ferlay et al, 2010), with stage I-II tumours currently having on the average a relatively high 10-year survival, but showing a high inter-individual heterogeneity at clinical and molecular levels. An important area of current research aims at defining biologically personalised treatment strategies through the identification of new predictive and prognostic biomarkers that provide guidance in the choice of the therapeutic strategy. The development of microarray technology has further brought to light the molecular heterogeneity of breast cancer (Perou et al, 2000; Parker et al, 2009; Curtis et al, 2012), prompting several groups to identify prognostic and predictive gene expression signatures (Reis-Filho and Pusztai, 2011), some of which are currently under evaluation in phase III prospective trials in terms of clinical utility (Sparano, 2006; Cardoso et al, 2007). The prognostic information of such so-called ‘first generation' gene signatures is strictly associated to the expression of proliferation-related genes, and their prognostic relevance is generally limited to ER+HER2− tumours (Pusztai et al, 2006; Desmedt et al, 2008). Several studies have instead highlighted the prognostic role of immune-related signatures in all subtypes and, in particular, in highly proliferating tumours (Schmidt et al, 2008; Rody et al, 2009; Bianchini et al, 2010; Nagalla et al, 2013). We demonstrated a subtype-dependent prognostic role for a metagene comprising interferon (IFN)-stimulated genes whose overexpression was significantly associated with a worse outcome in the ER+HER2− subtype, independently of proliferation and immune metagenes (Callari et al, 2014). Importantly, the prognostic and predictive relevance of gene expression signatures has been shown to complement rather than replace traditional clinico-pathological parameters (Reis-Filho and Pusztai, 2011).
A new area of biomarker research opened up with the discovery of microRNAs (miRNAs). These small non-coding RNAs have a key role in post-transcriptional gene regulation and are being widely investigated in oncology (Jansson and Lund, 2012) using multiple experimental and bioinformatic approaches (De Cecco et al, 2013). MicroRNAs have been shown to be deregulated in many cancer types, including breast cancer (Iorio and Croce, 2009; Mulrane et al, 2013), and signatures associated with diagnosis, prognosis and response to treatment have been reported (Blenkiron et al, 2007; Janssen et al, 2010; Rothe et al, 2011; Jung et al, 2012). Recent studies have addressed the integrated analysis of miRNA and mRNA data, mainly to investigate their biological role (Buffa et al, 2011; Enerly et al, 2011), as in the comprehensive study recently reported by Dvinge et al (2013). Gene and miRNA expression patterns separately correlate with survival in breast cancer, which suggests that the development of models using miRNAs and gene markers together might improve their predictive performance. This would indicate a new concept of data integration not only aimed at obtaining information on the biological role of these small molecules, but also at predicting patients' prognosis.
In the present study, we performed a miRNA expression profile in a cohort of 92 lymph node-negative ESR1+/ERBB2− breast cancers from patients not receiving systemic treatment and either developing distant metastases within 5 years from surgery or remaining metastasis free for >5 years. Gene expression data from a previous study were also available for all the cases (Callari et al, 2014). MicroRNAs significantly associated with distant metastasis-free survival (DMFS) were identified, further investigated and confirmed on a total of 1246 breast cancer samples from the publicly available data set from the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC; Dvinge et al, 2013). A combined outcome prediction model using gene expression, miRNA expression and clinico-pathological features was implemented.
Materials and Methods
Case series
A case series of 92 lymph node-negative ESR1+/ERBB2− breast cancers obtained at the Fondazione IRCCS Istituto Nazionale dei Tumori (INT) was used to identify miRNAs associated with clinical outcome. The case series included 42 patients who developed distant metastasis within 5 years of surgery and 50 patients who were free of distant metastasis for at least 5 years, all were selected so they had a similar age and tumor size. The same case series has been investigated at the gene expression level, and clinico-pathological features have been already reported (Callari et al, 2014) complying with the Remark guidelines (McShane et al, 2005). A written informed consent signed by each patient authorised the use of material left over from the diagnosis for research purposes. The study was approved by the Independent Ethics Committee and INT Review Board.
As an independent set, the publicly available collection of both miRNA and mRNA expression profiles from 1246 clinically annotated primary fresh-frozen breast cancer specimens generated by the METABRIC (Curtis et al, 2012; Dvinge et al, 2013) were used. Breast cancer samples from patients without invasive carcinoma and without follow-up information were excluded. Clinico-pathological information for METABRIC data included in the present study is reported in Supplementary Table 1.
RNA extraction and miRNA microarrays hybridisation
In the case series obtained at the INT, representative frozen samples containing >60% of tumor cells were submitted to molecular analyses. Tissue was pulverised using a Mikrodismembrator (Braun Biotech International, Melsungen, Germany). Total RNA was extracted with the Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to manufacturer instructions and processed for microarray hybridisation by the INT Functional Genomics Core Facility. Briefly, 600 ng of RNA was amplified using the Illumina Human_v2 MicroRNA expression profiling kit based on the DASL (cDNA-mediated Annealing, Selection, Extension and Ligation) assay according to the manufacturer's recommendations (Illumina Inc., San Diego, CA, USA), fluorescence-labeled, and then hybridised to the Illumina miRNA BeadChip, which allows analysis of 1146 sequences (representing 97% of validated human miRNAs described in miRBase database version 12.0).
The Illumina BeadArray Reader (San Diego, CA, USA) was used for scanning the arrays, and the Illumina BeadScan (San Diego, CA, USA) software was used for image acquisition and recovery of primary signals.
MicroRNA profiling data analysis
Microarray raw data were obtained using Illumina BeadStudio 3.8 software and processed using the lumi package (Du et al, 2008) of Bioconductor (Gentleman et al, 2004). After quality control, Robust Spline Normalization was applied. Only probes targeting validated human miRNAs were selected. Experimental batch effects were eliminated by applying the Combat method to normalised expression data (Johnson et al, 2007). Raw and processed data were deposited in the Gene Expression Omnibus data repository (Barrett et al, 2011) with ID GSE59829.
Subtypes definition
Three main breast cancer subtypes were defined, as previously described (Callari et al, 2014). Briefly, the ILMN_15142 and ILMN_28003 probes in the case series of 92 breast cancers and ILMN_1678535 and ILMN_2352131 in the independent METABRIC collection were considered as reporters, respectively, for ESR1 and ERBB2 gene expression. The threshold values to define gene expression positivity were selected according to the strong bimodal distribution observed. All analyses were separately run for patients with ESR1−/ERBB2− (roughly corresponding to the basal-like subtype), with ESR1±/ERBB2+ (roughly corresponding to the HER2+ enriched subtype), and with ESR1+/ERBB2− (roughly corresponding to the luminal subtype) tumours.
Statistical analysis
All statistical analyses were performed using R, version 2.15.2 (http://www.R-project.org). The limma package (Smyth et al, 2004) was used for class comparison analysis in INT case series, and a two-tailed P-value <0.001 was considered statistically significant. Receiver operating characteristic (ROC) curve and area under the curve (AUC) were estimated using the pROC package (Robin et al, 2011). DMFS was the main clinical outcome considered in our case series.
Univariable and multivariable Cox proportional hazards regressions, as implemented in the survival and cmprsk packages (Gray, 2014), were used to correlate clinico-pathological and biological variables with outcome in the independent METABRIC data set. Hazard ratios (HR) and 95% confidence intervals were reported. The statistical significance of first order interactions between the variables taken into account and the main effects of such variables were investigated using the likelihood ratio test (LRT). Proportional hazard assumptions were evaluated using a goodness-of-fit testing procedure based on Schoenfeld residuals. P-value <0.05 was used to identify the statistically significant associations with clinical outcome and to test first order interactions and proportional hazard assumptions. Results were also plotted using the cumulative incidence curve, and survival differences were evaluated using the log-rank test. Disease-specific survival was the main end point in the METABRIC cohort. All observations were censored at 10 years of follow-up.
Experiments with cell lines
The human epithelial breast cell lines, MCF10A, were purchased from America Type Culture Collection and was cultured in Dulbecco's modified Eagle's medium (Lonza, Slough, UK) supplemented with 10% fetal bovine serum (Lonza), 0.01 mg ml−1 insulin, 0.02 ng μl−1 recombinant human epidermal growth factor (Peprotech, Rocky Hill, NJ, USA), 0.5 μg ml−1 hydrocortisone (Stem Cell Technologies, Vancouver, BC, Canada) and 1% penicillin/streptomycin (Lonza).
Cells were cultured at 37 °C in 95% humidified air in the presence of 5% CO2 and cell vitality was assessed by Trypan Blue exclusion assay (at least 95%) before starting experiments.
Authentication of cell lines by STR DNA profiling analysis was performed by the Functional Genomic Unit at Fondazione IRCCS Istituto Nazionale Tumori of Milano. For treatment with recombinant human TGF-β1 (Peprotech), MCF10A were plated in 24-well plate at a density of 0.07 × 106 in serum-free culture medium. Recombinant human TGF-β1 was added at a concentration of 10 ng ml−1 and cells were harvested after 3 days. Total RNA was isolated using Qiazol (Qiagen, Valencia, CA, USA) reagent. After a clean-up treatment with RNAeasy kit following the manufacturer's recommendations (Qiagen) and with RNase-free DNase to remove contaminating genomic DNA, RNA integrity and purity was assessed by Bioanalyzer (Agilent, Santa Clara, CA, USA). RNA concentration was spectrophotometrically defined with Nanodrop ND-2000C (Thermo Scientific, Waltham, MA, USA). Total RNA was reverse-transcribed using the MicroRNA Reverse Trascription Kit (Applied Biosystem, Foster City, CA, USA) for miR-30e* level and High-Capacity Reverse Trascription Kit (Applied Biosystem) for genes.
The expression level of miR-30e* was evaluated by qPCR using the TaqMan Fast Universal PCR Master Mix assay (Applied Biosystem) and employing RNU48B as housekeeping gene. Similarly expression levels for SNAI1, VIM, ZEB1, CDH1 and CDH2 were evaluated by qPCR with TaqMan Fast Universal PCR Master Mix assay (Applied Biosystem) and using GAPDH as housekeeping gene. Data were computed with the ΔΔCt method (Livak and Schmittgen, 2001).
Results
A workflow of the analyses performed in the study is reported in Supplementary Figure 1. Candidate outcome-related miRNAs were identified in our case series, confirmed and further investigated in the METABRIC cohort, which included other molecular subtypes and patients receiving adjuvant treatment.
Metastasis-associated miRNAs in lymph node-negative ESR1+/ERBB2− breast cancers
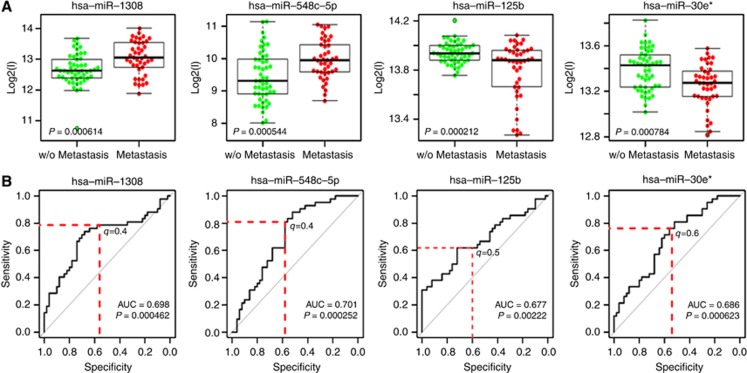
As it is well established that, in breast cancer, molecular features associated with outcome are subtype specific, we focused on 92 ESR+/ERBB2− tumours to identify outcome-related miRNAs in this subtype. The whole-genome miRNA expression profile was obtained, and 858 probes (corresponding to 858 validated human miRNAs) were retained after data normalisation and filtering. Four miRNAs were significantly expressed differentially when patients who developed metastasis within 5 years of surgery were compared with those free of any metastasis for more than 5 years. In particular, two miRNAs (miR-548c-5p and miR-1308) were upregulated in patients developing metastases and two (miR-125b and miR-30e*) were downregulated (Figure 1A).
Figure 1.
MicroRNAs associated with development of distant metastasis in the training set. (A) Boxplots of expression pattern of the four differentially expressed miRNAs in the training set for cases developing or not distant metastasis. (B) ROC curve analysis for the same four miRNAs; AUC and defined cutoffs (q-value) are reported.
In order to further investigate the discrimination power of these four miRNAs in predicting development of metastases, ROC curves were generated (Figure 1B). It can be noted that all AUC values were significantly >0.5. A specific cutoff was identified for each miRNA in order to attain sensitivity and specificity superior to 60% and 50%, respectively, as shown in Supplementary Table 2.
Confirmation in lymph node-negative patients with ESR1+/ERBB2− tumours
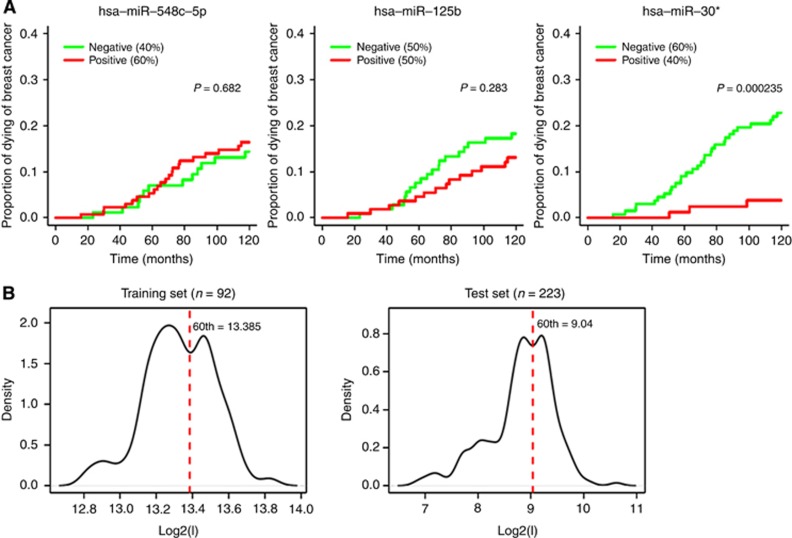
To confirm the role of the outcome-related miRNAs found in the first cohort, 223 node-negative women with ESR1+/ERBB2− tumours not receiving systemic treatment until relapse were selected in the independent METABRIC data set, and a univariable Cox proportional hazards model for disease-specific survival was fitted. The prognostic role of each miRNA was evaluated considering it as both a continuous and dichotomous variable. In the latter case, as a consequence of the different platforms used for miRNA profiling, the data categorisation in the METABRIC collection was done using the percentile threshold identified by ROC curves in the previous analysis.
Among the three miRNAs available in the METABRIC data set, miR-548c-5p did not have a significant effect on the rate of occurrence of death due to breast cancer, and only a trend of an association with survival was observed for miR-125b (Figure 2A and Supplementary Table 3). In contrast, miR-30e* resulted significantly associated with disease-specific survival either when it was considered as a dichotomous (β=−1.880, HR=0.153, P-value=0.0019) or as a continuous variable (β=−0.767, HR=0.464, P-value=0.0028). The estimated β-coefficients for miR-30e* suggested a significant protective effect for the high expression levels and a more marked association with outcome in the former case. In our cohort, this miRNA presented a peculiar bimodality with a local minimum roughly corresponding with the defined cutoff. In the METABRIC cohort, by selecting lymph node-negative untreated patients with ESR1+/ERBB2− tumours, it retained an intensity distribution characterised by two evident peaks and a local minimum that can be again well-approximated by the 60th percentile, therefore supporting the investigation of the prognostic role of miR-30e* by considering it as a dichotomous variable (Figure 2B).
Figure 2.
Association with outcome in the test set. (A) Three out of four miRNAs were present in the independent data set, and the association with disease-specific survival in lymph node-negative untreated cases with ESR1+/ERBB2− tumours (total number=223, unfavourable events=33) is represented by cumulative incidence curves. (B) Expression levels distribution for miR-30e* in the training set (left) and test set (right).
The independence between the effect of miR-30e* on disease-specific survival and the classical clinico-pathological risk factors was investigated, and the advantages deriving from the use of a combined gene- and miRNA-based outcome prediction were explored. For these aims, the prognostic contribution of miR-30e* was assessed by multivariable analysis in the presence of both conventional clinical variables (age at diagnosis, tumor size and histological grade) and gene expression signatures known to be prognostic in this subtype, including the Genomic Grade Index (GGI; Sotiriou et al, 2006), an IFN-induced metagene and an immune response-related metagene (Callari et al, 2014). The combined model performed using miR-30e* as a dichotomous variable is shown in Table 1. All first-order interactions between the miRNA and the other variables were removed as they did not add a significant contribution (Supplementary Table 4).
Table 1. Multivariable Cox regression analysis in lymph node-negative patients with ESR1+/ERBB2− tumours not receiving systemic treatment until relapse and results of likelihood ratio test for the main effects of considered variables (total number=207, unfavourable events=31 and missing values=16).
| Variables | β | HR (95% CI) | P-value | P-value of LRT |
|---|---|---|---|---|
| hsa-miR-30e* (+, −) | −2.115 | 0.121 (0.028–0.523) | 0.00471 | 0.000132 |
| Age (>50 years, ≤50 years) | −0.369 | 0.692 (0.310–1.541) | 0.367 | 0.394 |
| Size (>2 cm, ≤2 cm) | 0.772 | 2.163 (1.041–4.498) | 0.0388 | 0.0534 |
| Grade (3, 1–2) | 0.066 | 1.068 (0.496–2.303) | 0.866 | 0.772 |
| T-cell metagene (+, −) | −1.064 | 0.345 (0.148–0.804) | 0.0137 | 0.00662 |
| IFN metagene (+, −) | −0.202 | 0.817 (0.387–1.726) | 0.597 | 0.559 |
| GGI (>0, ≤0) | 1.372 | 3.943 (1.744–8.918) | 0.000982 | 0.000556 |
Abbreviations: CI=confidence interval; GGI=Genomic Grade Index; HR=hazard ratio; LRT=likelihood ratio test. P-values under the threshold for statistical significance are reported in bold.
MiR-30e* retained its statistically significant prognostic contribution regardless of both clinical variables and gene signatures, suggesting the advantage of combining miRNA and gene markers, other than clinico-pathological risk factors, for a better prognostication. Again, estimates suggested a statistically significant protective effect for miR-30e* expression (HR=0.121, P-value=0.00471).
To further emphasise the utility and efficacy of combined outcome prediction, the LRT was performed for main effects of variables included in multivariable analysis (Table 1). MiR-30e*, GG1, immune response-related metagene and tumor size, even if borderline, added a statistically significant contribution to the prediction performance of the model, confirming that a combined model could help to improve the prediction of a patients prognosis.
The multivariable Cox regression analysis was also performed considering miR-30e* as a continuous variable (Supplementary Table 5).
miR-30e* and outcome in treated patients with ESR1+/ERBB2− tumours
After confirmation of the miR-30e* prognostic role in the absence of treatment, we also investigated whether the same association was present in patients with ESR1+/ERBB2− tumours receiving adjuvant endocrine therapy and/or chemotherapy. For this purpose, 637 node-negative and node-positive women with ESR1+/ERBB2− tumours receiving adjuvant treatment were selected in the METABRIC data set. Overall, this group is likely to be clinically different from node-negative untreated patients. Consequently, the threshold applied in the previous analysis might not be suitable in such a group. For this reason, univariable Cox regression analysis for disease-specific survival was performed considering miR-30e* as continuous variable. Results indicated that miR-30e* had a statistically significant prognostic effect on disease-specific survival even in this subgroup (HR=0.680, P-value=0.00183).
The association of miR-30e* with disease-specific survival was further investigated in a multivariable Cox analysis including the same covariates considered before and the lymph node status. Results of multivariable Cox regression analysis are shown in Table 2. miR-30e* retained its significant and favourable prognostic role on disease-specific survival also in the presence of clinico-pathological variables and gene signatures. However, the significant interaction between miR-30e* and age at diagnosis (LRT=8.562 on 1 degree of freedom, P-value=0.00343; Supplementary Table 6) suggests that the effect of miR-30e* is different according to patient's age at diagnosis, with an attenuation of miRNA effect in older patients. Also in this context, the benefit of using a combined model with both miRNA and gene markers for prediction of patient prognosis was confirmed (Supplementary Table 6).
Table 2. Multivariable Cox regression analysis in patients with ESR1+/ERBB2− tumours receiving adjuvant treatment (total number=607, unfavourable events=115 and missing values=30).
| Variables | β | HR (95% CI) | P-value |
|---|---|---|---|
| hsa-miR-30e* (continuous) | −1.289 | 0.276 (0.130–0.584) | 0.000765 |
| Age (>50years, ≤50 years) | −9.522 | 7.324E-05 (0–0.041) | 0.00319 |
| Size (>2 cm, ≤2 cm) | 0.743 | 2.103 (1.363–3.246) | 0.000784 |
| Grade (3, 1–2) | 0.32 | 1.377 (0.910–2.084) | 0.13 |
| Lymph node (+, −) | 0.891 | 2.438 (1.572–3.780) | 6.83E−05 |
| T-cell metagene (+, −) | 0.12 | 1.127 (0.758–1.675) | 0.554 |
| IFN metagene (+, −) | 0.104 | 1.110 (0.751–1.640) | 0.602 |
| GGI (>0, ≤0) | 0.326 | 1.385 (0.918–2.090) | 0.121 |
| hsa-miR-30e* : age | 1.193 | 3.296 (1.498–7.251) | 0.00303 |
Abbreviations: CI=confidence interval; GGI=Genomic Grade Index; HR=hazard ratio; LRT=likelihood ratio test. P-values under the threshold for statistical significance are reported in bold.
miR-30e* and outcome in the other subtypes
After characterisation of the prognostic role of miR-30e* in patients with ESR1+/ERBB2− tumours, we carried out an exploratory analysis in the two remaining breast cancer subtypes to evaluate whether its prognostic role was subtype specific. In the analysis, treated and untreated patients were considered together due to the limited sample size, including, however, treatment as a covariate in the multivariable analysis. GGI was not included because it is known to be not prognostic in these breast cancer subtypes (Desmedt et al, 2008). Finally, as the definition of a threshold would be arbitrary, miR-30e* was investigated as a continuous variable.
By univariable Cox regression analysis in 206 women with ESR1−/ERBB2− tumours, we found no significant association between miR-30e* and disease-specific survival (HR=0.977, P-value=0.904). Also in the presence of conventional clinical variables and gene signatures, miR-30e* was not significantly associated with survival (data not shown).
Finally, univariable Cox regression analysis was performed in 167 patients with ERBB2+ tumours. In the subgroup, miR-30e* showed a HR similar to that observed in treated women with ESR1+/ERBB2− primaries, although the protective effect associated with higher miR-30e* expression was only marginally significant probably due to the reduced sample size (HR=0.710, P-value=0.127). In the multivariable analysis with clinical variables and gene signatures shown in Table 3, miR-30e* maintained a significant protective role. Nevertheless, the interaction between miR-30e* and the IFN metagene, retained in the model as it added a relevant contribution (LRT=5.641 on 1 degree of freedom, P-value=0.0176; Supplementary Table 7), suggests that the effect of miR-30e* is opposite in cases with high or low expression of the IFN metagene, as graphically reported in Supplementary Figure 2 The utility of combined outcome prediction was highlighted also in this subtype (Supplementary Table 7).
Table 3. Multivariable Cox regression analysis in untreated and treated patients with ERBB2+ tumours (total number=160, unfavourable events=56 and missing values=7).
| Variables | β | HR (95% CI) | P-value |
|---|---|---|---|
| hsa-miR-30e* (continuous) | −0.674 | 0.510 (0.288–0.903) | 0.0209 |
| Age (>50 years, ≤50 years) | −0.231 | 0.794 (0.445–1.415) | 0.434 |
| Size (>2 cm, ≤2 cm) | 0.551 | 1.730 (0.888–3.389) | 0.107 |
| Grade (3, 1–2) | 0.097 | 1.100 (0.567–2.140) | 0.775 |
| Lymph node (+, −) | 1.038 | 2.820 (1.264–6.302) | 0.0113 |
| Treatment (yes, no) | 0.103 | 1.110 (0.432–2.847) | 0.83 |
| T-cell metagene (+, −) | −0.37 | 0.691 (0.383–1.245) | 0.219 |
| IFN metagene (+, −) | −9.013 | 0.000122 (0–0.332) | 0.0255 |
| hsa-miR-30e* : IFN metagene | 1.154 | 3.170 (1.197–8.395) | 0.0202 |
Abbreviations: CI=confidence interval; GGI=Genomic Grade Index; HR=hazard ratio; IFN=interferon. P-values under the threshold for statistical significance are reported in bold.
Suggestions on the mechanism of action of miR-30e*
The possible mechanism of action of miR-30e* was investigated using two distinct approaches. In the first one, we took advantage from literature data obtained in glioma by Jiang et al (2012). Jiang et al (2012) suggest that miR-30e* acts by inhibiting IκB, interfering in the negative regulation of NFκB, and that the consequent uncontrolled activity of this transcription factor leads to increased expression of MMP9 and VEGFC, and supports its association with poor prognosis. To verify if a similar mechanism was true in breast cancer too, we analysed correlations between expression of miR-30e* and the following genes NFkBIA (r=0.058; r=−0.059), NFkB1 (r=0.11; r=0.258), MMP9 (r=−0.175; r=−0.273) and VEGFC (r=−0.118; r=−0.008) in our clinical data set and in the METABRIC data set, respectively (Supplementary Figure 3). Lack of correlation between miR30-e* and the investigated genes in the breast cancer clinical data sets definitely suggest that miR-30e* has a different mechanism of action in this context compared with glioma, providing an explanation for its opposite clinical role in these two neoplasias.
In the second approach, a set of six tools, Diana_microT-CDS (Paraskevopoulou et al, 2013), microrna.org (Betel et al, 2008), miRDB (Wang, 2008), PITA (Kertesz et al, 2007), RNA22 (Miranda et al, 2006) and Targetscan (Lewis et al, 2005), for predicting miRNA targets were used and a single list of 654 genes predicted as miR-30e* targets by at least four tools was produced. Two distinct list of genes that negatively correlated with miR-30e* (Rho<−0.2, Spearman) were produced using the Metabric data set (1177 genes) and our data set (722 genes). Each group of anti-correlated genes was compared with the putative target gene list to identify common genes, and only genes shared between the two lists of overlapping genes were considered. This approach yielded 11 statistically significant genes representing possible targets: DYRK2, MTDH, MYO5A, DNAJA2, NRAS, OAS2, YTHDF1, CEP152, SLC36A1, GBP1 and ARMC1. Interestingly, OAS2, an IFN-stimulated gene, which was already included in our IFN signature associated with bad prognosis in luminal breast cancer (Callari et al, 2014) was also among shared putative targets. This result pinpoints an interesting possibility that may be also supported by the loss of prognostic significance (Table 1 and Supplementary Table 6) observed for luminal tumours when the IFN metagene was included in the model together with miR-30e*. On the other hand, it may give an explanation for the interaction observed between miR-30e* and the IFN metagene in patients with ERBB2+ tumours (Supplementary Table 7). For some of the other genes, a role in breast cancer has already been reported in the literature as reported under ‘Discussion' section.
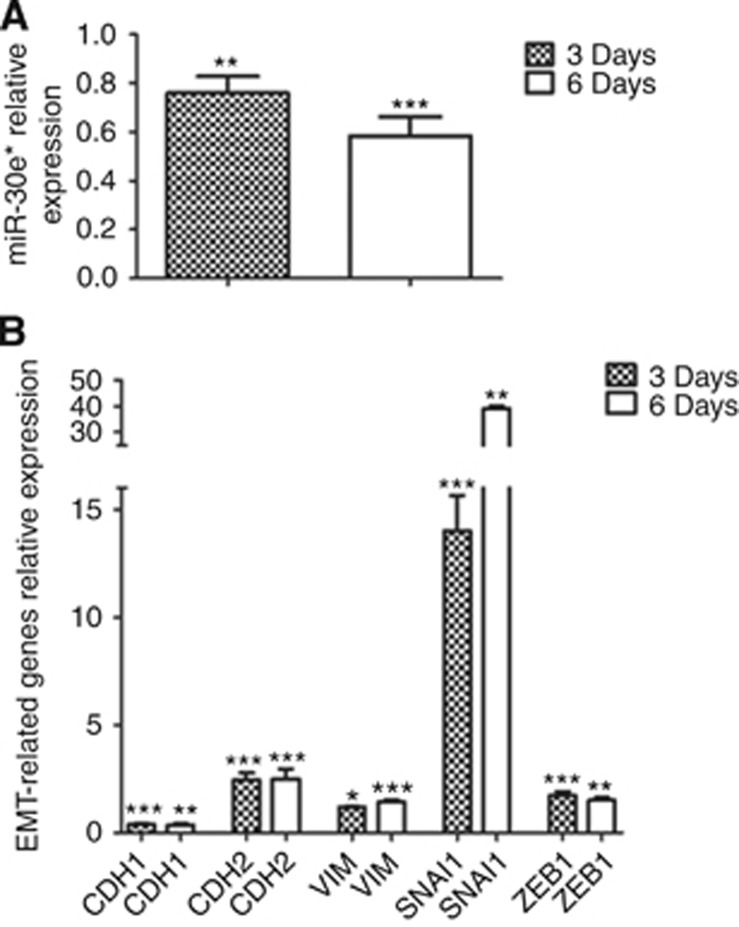
Finally, to gain insight into the mechanism of miR-30e* in breast cancer, the normal breast cell line MCF10A was treated with recombinant human TGF-β1 to induce epithelial–mesenchymal transition (EMT) and miR-30e* levels were measured. Recombinant human TGF-β1 induced an about 20% downregulation of miR-30e* expression with respect to controls (P<0.05), which reached about 40% at 6 days (P<0.0005) and was accompanied by a statistically significant upregulation of SNAI1, CDH2, VIM and ZEB1, and downregulation of CDH1 as expected for the EMT (Figure 3) This result suggests that at difference to what observed in glioma, lower levels of miR-30e* are associated with a more invasive phenotype even in normal breast cells, indirectly justifying the protective effect of miR-30e* identified in our clinical data set and validated on public data.
Figure 3.
In vitro experiments. (A) Relative expression of miR-30e* in the normal epithelial cell line MCF10A after a 3-day or 6-day treatment with 10 ng ml−1 of recombinant human TGF-β1 with respect to untreated controls. Bars represent the mean of three independent biological triplicates±s.d. Statistical significance of differences between miR-30e* in treated cells compared with controls was evaluated by Student's t-test. *P<0.05; **P<0.005 and ***P<0.0005. (B) Relative expression of CDH1, CDH2, VIM, SNAI1 and ZEB1 in the normal cell line MCF10A after a 3-day or 6-day treatment with 10 ng ml−1 of recombinant human TGF-β1 with respect to untreated controls. Bars represent the mean of three independent biological triplicates±s.d. Statistical significance of differences between gene expression in treated cells compared with controls was evaluated by Student's t-test. *P<0.05; **P<0.005 and ***P<0.0005.
Discussion
Prediction of risk of recurrence is an open issue in clinical management of early breast cancer. Much effort has been made to develop gene-based predictors, and some have been challenged for their clinical utility (Sparano, 2006; Cardoso et al, 2007). More recently, a new class of small RNAs, miRNAs, has been suggested to have a key role in breast cancer and to be able to give prognostic information (Foekens et al, 2008; Rothe et al, 2011; Falkenberg et al, 2013). Although several studies have integrated mRNA and miRNA data, mainly to better understand the biological role of the latter, few studies have investigated the possible advantages of an integrated outcome prediction. Falkenberg et al (2013) demonstrated the potential clinical impact of miR-221 regardless of clinical covariates, but not including any prognostic gene markers in the Cox regression model. Instead, Buffa et al (2011) identified the prognostic miRNAs in ER-positive and ER-negative breast cancer independently of clinical variables and key biological processes measured as gene expression signatures. However, they conducted the validation in independent cohorts of gene expression profiles by investigating cognate targets rather than identified miRNAs.
In the present study, using microarray technology, we aimed to identify pure prognostic miRNAs in breast cancer by analysing a homogenous case series of lymph node-negative untreated patients. As it is well-established that markers associated with patients' outcome can differ at least for the main breast cancer subgroups as defined by ER and HER2 status (Pusztai et al, 2006), the association of miRNAs with clinical outcome was investigated in the subgroup of ESR1+/ERBB2− tumours. Four miRNAs (including miR-1308) were identified as differentially expressed in the INT case series according to the development of distant metastasis, but miR-1308 was not among the measured miRNAs in the METABRIC cohort. Of the three remaining miRNAs, miR-30e* was confirmed in the independent data set, miR-548c-5p was not significantly associated with breast cancer-specific death, whereas miR-125b expression showed a weak association with good prognosis that, although not statistically significant, was still consistent with results obtained in our case series. The partial confirmation in the METABRIC data set of the results obtained in the first collection is not surprising and could be explained, at least in part, by the use of different platforms in the two cohorts. This is supported by results of platform comparison studies using clinical specimens (Callari et al, 2012).
Our findings suggest that, in general, high-expression levels of miR-30e* in primary tumours are significantly associated with a favourable prognosis. In node-negative untreated patients, this protective effect on disease-specific survival was demonstrated to be independent both of clinically relevant prognostic variables and of gene expression signatures, which suggests that miR-30* may identify a distinct dimension of tumor biology captured neither by clinical variables nor by the considered gene signatures. From a more general biological perspective, our results suggest that the combined analysis of miRNA and gene expression data can improve the prediction of patients' prognosis compared with the prediction achievable by considering only clinical variables and gene markers.
In women receiving adjuvant treatment with ESR1+/ERBB2− tumours and in those with ERBB2+ tumours, miR-30e* continued to be associated with good prognosis, although significant interactions were found with age at diagnosis and the IFN metagene, respectively. The interaction with age is not surprising in the context of breast cancer where menopausal status identifies two different types of diseases with distinct aetiology and outcome. Under such conditions, biomarkers might have slightly different roles. In contrast, miR-30e* expression did not affect the prognosis in patients with ESR1−/ERBB2− tumours. Hence, the prognostic role of this miRNA appears to be subtype specific.
A word of caution should be spent about the fact that the event considered in the INT case series was the occurrence of distant metastases, whereas only disease-specific death was available in the METABRIC data set. It is well known that the main cause of death for breast cancer is the development of metastasis at distant sites, rather than the primary tumor (Weigelt et al, 2005), which justifies our analyses in the second cohort. However, it should be noted that also patients untreated after surgery were likely to have received several treatments after relapse and before dying, and no information is available on this aspect.
Another consideration should be made about the IFN metagene, whose high expression in node-negative ESR1+/ERBB2− breast cancers from patients not receiving systemic treatment was reported as associated with increased metastasis risk, also in the presence of other prognostic factors (Callari et al, 2014). In the same subgroup in the METABRIC collection, the IFN metagene did not have a significant effect on disease-specific survival, although it showed a HR in univariable Cox analysis (HR=1.545, P-value=0.217) similar to that previously observed. Such a result could be a consequence of a different clinical outcome evaluated in the METABRIC cohort and the likely administration of systemic treatment after relapse and before dying to patients developing metastasis after surgery.
From a biological point of view, only a few studies have characterised the function of miR-30e* in cancer. Most studies focused miRNAs from the same family, and only one study in glioma considered the specific role of miR-30e* in a clinical context (Jiang et al, 2012). Opposite to our findings, in such studies miR-30e* expression predicted a poor survival. Studies on the mechanism of action revealed a possible role of miR-30e* in deregulation of the NFkB pathway, by targeting the inhibitory protein IFκBα. The loss of negative regulation was associated with upregulation of MMP9 and VEGFC linking this way the poor prognosis observed in glioma patients with a possible increase in invasion and neo-angiogenesis. Although data from our laboratory support an overall activation of the NFκB pathway in luminal breast cancer cells when stimulated by factors released by cancer-associated fibroblasts (CAFs), no modulation of miR-30e* was observed in the CAF-stimulated cells (manuscript in preparation) indirectly suggesting a different mechanism in breast tumours compared with gliomas. Indeed the putative target of miR-30e* suggested by Jiang et al (2012), NFκBIA, and the downstream genes NFκB1, NFκB2, MMP9 and VEGFC were also not modulated in our breast cancer model (manuscript in preparation). Conversely, some literature data in breast cancer support a protective role of this miRNA, as shown in our study, but none regards directly miR-30*, but rather other members of its family such as miR-30 whose ectopic expression reduced tumorigenesis (Yu et al, 2010) promoting apoptosis and interfering with self-renewal in breast cancer-initiating cell xenografts. The involvement of miR-30e in regulating non-attachment growth of breast cancer and its possible role in maintenance of self-renewal capacity was also reported by Ouzounova et al (2013). The role of miR-30e in breast cancer was explored (along with other miRNAs) also at the isomiR level (Wu et al, 2015) showing that in the miR-30e locus, at difference with others loci, there was only one specific dominantly expressed isomiR (Wu et al, 2015). Finally, in triple negative breast cancer miR-30e, together with miR-155, miR-493 and miR-27a, was found to be associated with response to taxanes (Gasparini et al, 2014). Additional studies suggested a role of the miR-30 family in EMT (Joglekar et al, 2009; Braun et al, 2010) and replicative senescence (Martinez et al, 2011), processes closely linked to stem cell biology and tumor suppression, respectively. This miRNA family was also found to be part of a metastatic signature in a series of breast, bladder, colon and lung cancers (Baffa et al, 2009).
It is, however, worth to mention that sometimes opposite clinical roles are observed also for much more studied miRNAs, as is the case for miR-21 in prostate cancer (Folini et al, 2010). In this sense our in vitro experiments with a normal breast cancer cell lines suggesting that EMT induction is associated with a downregulation of miR-30e*, although not fully explaining the mechanism of miR-30e* by identifying specific targets, do support its protective role in clinical tumours.
The final attempt to identify targets with a bioinformatics approach identified a list of genes worth of further studies. MTDH, coding for metadherin, was described in breast cancer as the target of a miRNA with oncosuppressive activity, miR-153 (Li et al, 2015). The authors showed that ectopic expression of miR-153 inhibited MTDH-induced EMT. Overexpression of the protein coded by AEG1 (alias for MTDH) was reported to be associated with poor survival in a clinical data set (Li et al, 2008). Similar data were obtained in triple negative breast cancer by Liu et al (2015), but this time MTDH, which was associated with lymph node metastasis and poor survival, was reported as the target of another oncosuppressive miRNA, miR-26a. Another study described MTDH as a gene coding for a protein able to mediate lung homing of breast cancer cells in experimental models (Brown and Ruoslahti, 2004). GBP1, may have a dual role in breast cancer, either favourable due to its participation in an NK signature associated with a longer disease-free survival (Ascierto et al, 2012), but also to its enclosure in a signature associated with tamoxifen resistance (Elias et al, 2015). MYO5A, an actin-dependent molecular motor under snail control, also has a role in cancer cell migration and metastasis (Lan et al, 2010) possibly justifying the favourable role played by miR-30e* upregulation in breast cancer.
In conclusion, our study identified expression of miR-30e* as a protective prognostic marker in breast cancer, mainly in the ESR1+/ERBB2− subtype, and demonstrated that a combined analysis of different molecular features can help to obtain a better prognostication in breast cancer.
Acknowledgments
We wish to thank all the patients who participated in the study and their families. We also thank Dr Roberto Agresti and the Surgical Department of the Fondazione IRCCS Istituto Nazionale dei Tumori for collaboration in biological sample collection, personnel and Committees of the INT Tissue Bank for providing breast cancer specimens, and the Institutional Functional Genomics Core Facility for microarray hybridisation. The study was supported by grants from the Associazione Italiana per la Ricerca sul Cancro (AIRC, the Italian Association for Cancer Research, grant number 10611 to MGD and Call for Proposals on Innovative tools for cancer risk assessment and early diagnosis), by the Italian Ministry of Health (Special Project on Female Cancers) and by Eurocan Platform. The study used data generated by the Molecular Taxonomy of Breast Cancer International Consortium. Funding for the project was provided by Cancer Research UK and by the British Columbia Cancer Agency Branch.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
Supplementary Material
References
- Ascierto ML, Kmieciak M, Idowu MO, Manjili R, Zhao Y, Grimes M, Dumur C, Wang E, Ramakrishnan V, Wang XY, Bear HD, Marincola FM, Manjili MH. A signature of immune function genes associated with recurrence-free survival in breast cancer patients. Breast Cancer Res Treat. 2012;131:871–880. doi: 10.1007/s10549-011-1470-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baffa R, Fassan M, Volinia S, O'Hara B, Liu CG, Palazzo JP, Gardiman M, Rugge M, Gomella LG, Croce CM, Rosenberg A. MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J Pathol. 2009;219:214–221. doi: 10.1002/path.2586. [DOI] [PubMed] [Google Scholar]
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Muertter RN, Holko M, Ayanbule O, Yefanov A, Soboleva A. NCBI GEO: archive for functional genomics data sets–10 years on. Nucleic Acids Res. 2011;39:D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchini G, Qi Y, Alvarez RH, Iwamoto T, Coutant C, Ibrahim NK, Valero V, Cristofanilli M, Green MC, Radvanyi L, Hatzis C, Hortobagyi GN, Andre F, Gianni L, Symmans WF, Pusztai L. Molecular anatomy of breast cancer stroma and its prognostic value in estrogen receptor-positive and -negative cancers. J Clin Oncol. 2010;28:4316–4323. doi: 10.1200/JCO.2009.27.2419. [DOI] [PubMed] [Google Scholar]
- Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, Barbosa-Morais NL, Teschendorff AE, Green AR, Ellis IO, Tavare S, Caldas C, Miska EA. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8:R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun J, Hoang-Vu C, Dralle H, Huttelmaier S. Downregulation of microRNAs directs the EMT and invasive potential of anaplastic thyroid carcinomas. Oncogene. 2010;29:4237–4244. doi: 10.1038/onc.2010.169. [DOI] [PubMed] [Google Scholar]
- Brown DM, Ruoslahti E. Metadherin, a cell surface protein in breast tumors that mediates lung metastasis. Cancer Cell. 2004;5:365–374. doi: 10.1016/s1535-6108(04)00079-0. [DOI] [PubMed] [Google Scholar]
- Buffa FM, Camps C, Winchester L, Snell CE, Gee HE, Sheldon H, Taylor M, Harris AL, Ragoussis J. MicroRNA-associated progression pathways and potential therapeutic targets identified by integrated mRNA and microRNA expression profiling in breast cancer. Cancer Res. 2011;71:5635–5645. doi: 10.1158/0008-5472.CAN-11-0489. [DOI] [PubMed] [Google Scholar]
- Callari M, Dugo M, Musella V, Marchesi E, Chiorino G, Grand MM, Pierotti MA, Daidone MG, Canevari S, De Cecco L. Comparison of microarray platforms for measuring differential microRNA expression in paired normal/cancer colon tissues. PLoS One. 2012;7:e45105. doi: 10.1371/journal.pone.0045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callari M, Musella V, Di Buduo E, Sensi M, Miodini P, Dugo M, Orlandi R, Agresti R, Paolini B, Carcangiu ML, Cappelletti V, Daidone MG. Subtype-dependent prognostic relevance of an interferon-induced pathway metagene in node-negative breast cancer. Mol Oncol. 2014;8 (7:1278–1289. doi: 10.1016/j.molonc.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso F, Piccart-Gebhart M, Van't VL, Rutgers E. The MINDACT trial: the first prospective clinical validation of a genomic tool. Mol Oncol. 2007;1:246–251. doi: 10.1016/j.molonc.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Graf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S, Langerod A, Green A, Provenzano E, Wishart G, Pinder S, Watson P, Markowetz F, Murphy L, Ellis I, Purushotham A, Borresen-Dale AL, Brenton JD, Tavare S, Caldas C, Aparicio S. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cecco L, Dugo M, Canevari S, Daidone MG, Callari M. Measuring microRNA expression levels in oncology: from samples to data analysis. Crit Rev Oncog. 2013;18:273–287. doi: 10.1615/critrevoncog.2013007207. [DOI] [PubMed] [Google Scholar]
- Desmedt C, Haibe-Kains B, Wirapati P, Buyse M, Larsimont D, Bontempi G, Delorenzi M, Piccart M, Sotiriou C. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin Cancer Res. 2008;14:5158–5165. doi: 10.1158/1078-0432.CCR-07-4756. [DOI] [PubMed] [Google Scholar]
- Du P, Kibbe WA, Lin SM. ‘lumi: a pipeline for processing Illumina microarray'. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- Dvinge H, Git A, Graf S, Salmon-Divon M, Curtis C, Sottoriva A, Zhao Y, Hirst M, Armisen J, Miska EA, Chin SF, Provenzano E, Turashvili G, Green A, Ellis I, Aparicio S, Caldas C. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature. 2013;497:378–382. doi: 10.1038/nature12108. [DOI] [PubMed] [Google Scholar]
- Elias D, Vever H, Laenkholm AV, Gjerstorff MF, Yde CW, Lykkesfeldt AE, Ditzel HJ. Gene expression profiling identifies FYN as an important molecule in tamoxifen resistance and a predictor of early recurrence in patients treated with endocrine therapy. Oncogene. 2015;34:1919–1927. doi: 10.1038/onc.2014.138. [DOI] [PubMed] [Google Scholar]
- Enerly E, Steinfeld I, Kleivi K, Leivonen SK, Aure MR, Russnes HG, Ronneberg JA, Johnsen H, Navon R, Rodland E, Makela R, Naume B, Perala M, Kallioniemi O, Kristensen VN, Yakhini Z, Borresen-Dale AL. MiRNA-mRNA integrated analysis reveals roles for miRNAs in primary breast tumors. PLoS One. 2011;6:e16915. doi: 10.1371/journal.pone.0016915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenberg N, Anastasov N, Rappl K, Braselmann H, Auer G, Walch A, Huber M, Hofig I, Schmitt M, Hofler H, Atkinson MJ, Aubele M. MiR-221/-222 differentiate prognostic groups in advanced breast cancers and influence cell invasion. Br J Cancer. 2013;109:2714–2723. doi: 10.1038/bjc.2013.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- Foekens JA, Sieuwerts AM, Smid M, Look MP, de Weerd V, Boersma AW, Klijn JG, Wiemer EA, Martens JW. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc Natl Acad Sci USA. 2008;105:13021–13026. doi: 10.1073/pnas.0803304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folini M, Gandellini P, Longoni N, Profumo V, Callari M, Pennati M, Colecchia M, Supino R, Veneroni S, Salvioni R, Valdagni R, Daidone MG, Zaffaroni N. miR-21: an oncomir on strike in prostate cancer. Mol Cancer. 2010;9:12. doi: 10.1186/1476-4598-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparini P, Cascione L, Fassan M, Lovat F, Guler G, Balci S, Irkkan C, Morrison C, Croce CM, Shapiro CL, Huebner K. MicroRNA expression profiling identifies a four microRNA signature as a novel diagnostic and prognostic biomarker in triple negative breast cancers. Oncotarget. 2014;5:1174–1184. doi: 10.18632/oncotarget.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray B.2014cmprsk: Subdistribution Analysis of Competing Risks. R package version 2.2-7 . http://CRAN.R-project.org/package=cmprsk .
- Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen EA, Slewa A, Gudlaugsson E, Jonsdottir K, Skaland I, Soiland H, Baak JP. Biologic profiling of lymph node negative breast cancers by means of microRNA expression. Mod Pathol. 2010;23:1567–1576. doi: 10.1038/modpathol.2010.177. [DOI] [PubMed] [Google Scholar]
- Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6:590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Lin C, Song L, Wu J, Chen B, Ying Z, Fang L, Yan X, He M, Li J, Li M. MicroRNA-30e* promotes human glioma cell invasiveness in an orthotopic xenotransplantation model by disrupting the NF-kappaB/IkappaBalpha negative feedback loop. J Clin Invest. 2012;122:33–47. doi: 10.1172/JCI58849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar MV, Patil D, Joglekar VM, Rao GV, Reddy DN, Mitnala S, Shouche Y, Hardikar AA. The miR-30 family microRNAs confer epithelial phenotype to human pancreatic cells. Islets. 2009;1:137–147. doi: 10.4161/isl.1.2.9578. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Jung EJ, Santarpia L, Kim J, Esteva FJ, Moretti E, Buzdar AU, Di LA, Le XF, Bast RC, Jr., Park ST, Pusztai L, Calin GA. Plasma microRNA 210 levels correlate with sensitivity to trastuzumab and tumor presence in breast cancer patients. Cancer. 2012;118:2603–2614. doi: 10.1002/cncr.26565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- Lan L, Han H, Zuo H, Chen Z, Du Y, Zhao W, Gu J, Zhang Z. Upregulation of myosin Va by Snail is involved in cancer cell migration and metastasis. Int J Cancer. 2010;126:53–64. doi: 10.1002/ijc.24641. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li J, Zhang N, Song LB, Liao WT, Jiang LL, Gong LY, Wu J, Yuan J, Zhang HZ, Zeng MS, Li M. Astrocyte elevated gene-1 is a novel prognostic marker for breast cancer progression and overall patient survival. Clin Cancer Res. 2008;14:3319–3326. doi: 10.1158/1078-0432.CCR-07-4054. [DOI] [PubMed] [Google Scholar]
- Li W, Zhai L, Zhao C, Lv S. miR-153 inhibits epithelial-mesenchymal transition by targeting metadherin in human breast cancer. Breast Cancer Res Treat. 2015;150:501–509. doi: 10.1007/s10549-015-3346-y. [DOI] [PubMed] [Google Scholar]
- Liu P, Tang H, Chen B, He Z, Deng M, Wu M, Liu X, Yang L, Ye F, Xie X. miR-26a suppresses tumour proliferation and metastasis by targeting metadherin in triple negative breast cancer. Cancer Lett. 2015;357:384–392. doi: 10.1016/j.canlet.2014.11.050. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Martinez I, Cazalla D, Almstead LL, Steitz JA, DiMaio D. miR-29 and miR-30 regulate B-Myb expression during cellular senescence. Proc Natl Acad Sci USA. 2011;108:522–527. doi: 10.1073/pnas.1017346108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. REporting recommendations for tumour MARKer prognostic studies (REMARK) Br J Cancer. 2005;93:387–391. doi: 10.1038/sj.bjc.6602678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Mulrane L, McGee SF, Gallagher WM, O'Connor DP. miRNA dysregulation in breast cancer. Cancer Res. 2013;73:6554–6562. doi: 10.1158/0008-5472.CAN-13-1841. [DOI] [PubMed] [Google Scholar]
- Nagalla S, Chou JW, Willingham MC, Ruiz J, Vaughn JP, Dubey P, Lash TL, Hamilton-Dutoit SJ, Bergh J, Sotiriou C, Black MA, Miller LD. Interactions between immunity, proliferation and molecular subtype in breast cancer prognosis. Genome Biol. 2013;14:R34. doi: 10.1186/gb-2013-14-4-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouzounova M, Vuong T, Ancey PB, Ferrand M, Durand G, Le-Calvez KF, Croce C, Matar C, Herceg Z, Hernandez-Vargas H. MicroRNA miR-30 family regulates non-attachment growth of breast cancer cells. BMC Genomics. 2013;14:139. doi: 10.1186/1471-2164-14-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, Filippidis C, Dalamagas T, Hatzigeorgiou AG. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41:W169–W173. doi: 10.1093/nar/gkt393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, Quackenbush JF, Stijleman IJ, Palazzo J, Marron JS, Nobel AB, Mardis E, Nielsen TO, Ellis MJ, Perou CM, Bernard PS. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de RM, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Pusztai L, Mazouni C, Anderson K, Wu Y, Symmans WF. Molecular classification of breast cancer: limitations and potential. Oncologist. 2006;11:868–877. doi: 10.1634/theoncologist.11-8-868. [DOI] [PubMed] [Google Scholar]
- Reis-Filho JS, Pusztai L. Gene expression profiling in breast cancer: classification, prognostication, and prediction. Lancet. 2011;378:1812–1823. doi: 10.1016/S0140-6736(11)61539-0. [DOI] [PubMed] [Google Scholar]
- Rody A, Holtrich U, Pusztai L, Liedtke C, Gaetje R, Ruckhaeberle E, Solbach C, Hanker L, Ahr A, Metzler D, Engels K, Karn T, Kaufmann M. T-cell metagene predicts a favorable prognosis in estrogen receptor-negative and HER2-positive breast cancers. Breast Cancer Res. 2009;11:R15. doi: 10.1186/bcr2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, Müller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe F, Ignatiadis M, Chaboteaux C, Haibe-Kains B, Kheddoumi N, Majjaj S, Badran B, Fayyad-Kazan H, Desmedt C, Harris AL, Piccart M, Sotiriou C. Global microRNA expression profiling identifies MiR-210 associated with tumor proliferation, invasion and poor clinical outcome in breast cancer. PLoS One. 2011;6:e20980. doi: 10.1371/journal.pone.0020980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Bohm D, von TC, Steiner E, Puhl A, Pilch H, Lehr HA, Hengstler JG, Kolbl H, Gehrmann M. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 2008;68:5405–5413. doi: 10.1158/0008-5472.CAN-07-5206. [DOI] [PubMed] [Google Scholar]
- Smyth GK.2004Linear models and empirical Bayes methods for assessing differential expression in microarray experiments Stat Appl Genet Mol Biol 3(1): article 3. [DOI] [PubMed] [Google Scholar]
- Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J, Nordgren H, Farmer P, Praz V, Haibe-Kains B, Desmedt C, Larsimont D, Cardoso F, Peterse H, Nuyten D, Buyse M, Van de Vijver MJ, Bergh J, Piccart M, Delorenzi M. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- Sparano JA. TAILORx: trial assigning individualized options for treatment (Rx) Clin Breast Cancer. 2006;7:347–350. doi: 10.3816/CBC.2006.n.051. [DOI] [PubMed] [Google Scholar]
- Wang X. miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA. 2008;14:1012–1017. doi: 10.1261/rna.965408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt B, Peterse JL, van 't Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer. 2005;5:591–602. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- Wu X, Zeng R, Wu S, Zhong J, Yang L, Xu J. Comprehensive expression analysis of miRNA in breast cancer at the miRNA and isomiR levels. Gene. 2015;557:195–200. doi: 10.1016/j.gene.2014.12.030. [DOI] [PubMed] [Google Scholar]
- Yu F, Deng H, Yao H, Liu Q, Su F, Song E. Mir-30 reduction maintains self-renewal and inhibits apoptosis in breast tumor-initiating cells. Oncogene. 2010;29:4194–4204. doi: 10.1038/onc.2010.167. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.