Summary
Genome-wide identification of the mechanism of action (MoA) of small-molecule compounds characterizing their targets, effectors, and activity modulators, represents a highly relevant yet elusive goal, with critical implications for assessment of compound efficacy and toxicity. Current approaches are labor-intensive and mostly limited to elucidating high-affinity binding target proteins. We introduce a regulatory network-based approach that elucidates genome-wide MoA proteins based on the assessment of the global dysregulation of their molecular interactions following compound perturbation. Analysis of cellular perturbation profiles identified established MoA proteins for 70% of the tested compounds and elucidated novel proteins that were experimentally validated. Finally, unknown-MoA compound analysis revealed altretamine, an anticancer drug, as an inhibitor of glutathione peroxidase 4 lipid repair activity, which was experimentally confirmed, thus revealing unexpected similarity to the activity of sulfasalazine. This suggests that regulatory network analysis can provide valuable mechanistic insight into the elucidation of small molecule MoA and compound similarity.
Introduction
The mechanism of action of a compound (MoA) is defined as the set of target and effector proteins necessary to produce its pharmacological effect in a specific cellular context. Its elucidation is critical in assessing both on-target compound activity as well as off-target effects associated with potential toxicity, thus providing critical insight into the two major challenges of drug development (Scannell et al., 2012). Since most compounds in clinical trials fail due to toxicity or lack of efficacy (Wehling, 2009), any improvements in systematic MoA characterization may increase the yield of pharmacological discovery pipelines.
MoA characterization remains a major challenge that is only partially addressed by experimental and computational strategies. Most experimental approaches rely on direct binding assays, such as affinity purification (Hirota et al., 2012; Ito et al., 2010) or affinity chromatography (Aebersold and Mann, 2003). These methods are labor-intensive and generally limited to the identification of high-affinity binding targets, rather than of all proteins responsible for compound activity. They may thus miss important indirect effectors, as well as lower-affinity targets responsible for both desirable and undesirable pharmacological properties. For instance, compounds can be effectively screened against all protein kinases, while missing equally relevant targets, as shown by the recent reclassification of the MET inhibitor tivantinib as a microtubule inhibitor (Basilico et al., 2013). In addition, these assays work in vitro and may miss effects from tissue specific interactions and signals.
Chemo-informatics methods have also been developed. Yet, these are mostly designed to assess compound MoA similarity or specific compound/target interactions (Keiser et al., 2009; Lomenick et al., 2009; Miller, 2002), by leveraging the integration of structural and genomic information (Yamanishi et al., 2008), text-mining algorithms (Li et al., 2009), or machine learning methods for data-mining (Hansen et al., 2009). As such, they rely on detailed three-dimensional structures of both compound and target proteins or on prior literature or database knowledge of related MoA compounds. More recently, assembly of large reference compendia by systematic gene expression profiles (GEP) analysis of cells following compound perturbations has spurred development of MoA analysis methods (Ganter et al., 2005; Lamb et al., 2006; Wolpaw et al., 2011). In general, however, these methods are mostly comparative in nature and thus poorly suited to de novo MoA elucidation or to recognize subtle MoA differences that may induce unexpected toxicity. Network-based methods have also been recently proposed (Bansal et al., 2006; di Bernardo et al., 2005; Gardner et al., 2003; Mani et al., 2008). Rather than focusing on individual genes, these methods perform integrative analyses over interacting gene subsets or pathways. Yet, these methods either rely on prior knowledge of the pathways that mediate compound activity, making them unsuitable for genome-wide analyses, or require very large samples sizes (n > 100), thus making them impractical even for small compound libraries. As a result, there is still a pressing need for experimentally validated methodologies for the de novo prediction of genome-wide compound targets and effectors or to mechanistically elucidate MoA proteins associated with differential activity or toxicity.
To address this challenge, we introduce DeMAND (Detecting Mechanism of Action by Network Dysregulation), a hybrid computational and experimental approach for MoA analysis. DeMAND elucidates compound MoA by interrogating tissue-specific regulatory networks using small-size GEP datasets (n ≥ 6 samples) representing in vitro or in vivo, compound perturbations (Figure 1). Using GEPs from human lymphoma cells perturbed with libraries of 14 and 92 compounds, respectively, we systematically assessed the algorithm’s ability to infer known compound targets (from public databases) and then experimentally validated novel compound activity effector and modulator predictions (hereafter MoA-proteins). DeMAND identified established MoA proteins for >70% of these compounds, as well as novel proteins that were experimentally validated, such as RPS3A, VHL, and CCNB1 for the mitotic spindle inhibitor vincristine and JAK2 for mitomycin C. We also tested the algorithm’s ability to assess compound MoA similarity. More than 50% of top predicted compound pairs were confirmed by literature and database analysis or by experimental validation. For instance DeMAND identified altretamine, an unknown MoA compound, as a novel GPX4 inhibitor based on predicted MoA similar to sulfasalazine, a system xc− cystine-glutamate antiporter mediated GPX4 inhibitor (Yang et al., 2014).
Figure 1. Schematics of the DeMAND algorithm.
(A) DeMAND requires both a regulatory network and a set of gene expression profiles from compound perturbed and control samples, as an input.
(B) DeMAND evaluates the dysregulation of each interaction in the regulatory network.
(C) To evaluate interaction dysregulation co-expression scatter plots for the two interacting genes are smoothed using a Gaussian Kernel method to generate an interaction probability density. The probability density difference before and after compound perturbation is evaluated using the KL-divergence. The top example illustrates no change in probability density (i.e., no dysregulation). The other three examples illustrate various examples of compound dysregulation, including correlation inversion, gain, and loss (top to bottom, respectively).
(D) The statistical significance of the KL-divergence is assessed by gene pair shuffling.
(E) The global dysregulation of each gene is determined by integrating the p-values of all its network interactions, while accounting for their dependencies (see also Figure S1 and Extended Experimental Procedures).
(F) DeMAND produces a list of all network genes and the statistical significance of their dysregulation.
DeMAND is freely available to the research community, both as a Bioconductor package (Gentleman et al., 2004) and as a web based geWorkbench module (Floratos et al., 2010).
Results
Overview of DeMAND algorithm
Consider the regulon of a gene G, i.e., all its interactions (G ↔ Gi) with other genes Gi, including transcriptional, signaling, and protein-complex interactions. If G belongs to a compound’s MoA, then it is reasonable to assume that its regulon gene interactions will be dysregulated by the compound. This can be optimally assessed by measuring changes in the joint gene expression probability density p(G, Gi), for each of its regulon genes. Such analysis can capture direct effects on gene expression and more importantly modulation of the interacting partner’s expression via either direct or indirect regulatory mechanisms (e.g., feedback loops). Consider for instance a transcription factor regulating a set of targets. A targeted inhibitor will significantly alter the joint expression probabilities p(G, Gi), as the expression of the targets will be dysregulated even though the expression of G is not generally affected (see Figure 1 and Experimental Procedures).
The Kullback-Leibler divergence (KLD) (Kullback and Leibler, 1951) provides an ideal metric to quantitatively assess probability density changes in one or more variables. From information theory, the KLD is easily interpreted as the loss of information resulting from using a probability density as a surrogate for another. For each regulon interaction (G ↔ Gi), we estimate the KLD of each probability density p(G, Gi), before and after compound perturbation. Their statistical significance is then integrated, thus producing a global statistical assessment of the compound-mediated dysregulation of G. To avoid overestimating such integrative significance, due to interaction dependencies, we use a modification of Brown’s method that compensates for the integration of correlated evidence (Brown, 1975). All genes are then ranked based on their global KLD statistics.
To identify the regulon of each gene-product of interest, we used a set of established network reverse engineering algorithms (see Experimental Procedures). However, DeMAND is agnostic to the specific approach and can use networks generated by any alternative means, both computational and experimental.
DeMAND predictions are enriched in established high-affinity binding targets
We first evaluated the accuracy of DeMAND-inferred MoA genes for 14 selected compounds, using the perturbation dataset (DP14) from the DREAM/NCI compound synergy challenge (Bansal et al., 2014). This includes 276 GEPs of diffuse large B-cell lymphoma cells (OCI-LY3), following perturbation with 14 distinct compounds, of which 11 have established primary targets (see Extended Experimental Procedures and Table S1), and DMSO as control media, at two concentrations and three time points, in triplicate. The network for these analyses was produced as described in (Lefebvre et al., 2010), using a published dataset of 226 U133p2 GEPs representing both normal and tumor related human B-cells (Basso et al., 2010) (see Extended Experimental Procedures). Although DeMAND is designed to predict both compound targets (i.e., high-affinity binding proteins) and effectors/modulators, its performance can only be systematically benchmarked against the former, because gold-standard datasets to systematically assess the latter are not yet available.
DeMAND identified the known primary targets of 7 of the 11 tested compounds as statistically significant, at a 10% False Discovery Rate (FDR) (see Experimental Procedures, Figure S2A, and Table S2). Since the GEPs used in this analysis were obtained at multiple time points (6h, 12h and 24h), we further assessed whether individual time points may be more informative. Intriguingly, several targets were best predicted at specific time points (Figure S2B), consistent with expectations that compound activity may be mediated over different time scales. Yet, integration over all time points performed as well or better than the optimal time point for all but 2 compounds (monastrol and doxorubicin). For these, the direct target was significant only when specific time point GEPs were used. In total, targets for 9 of the 11 compounds could be elucidated either from multi-point or single time point analysis. Replacing interaction dysregulation with the differential expression of neighbors reduces the performance (see Extended Experimental Procedures).
Differential expression analysis has been proposed to elucidate compound substrates (Ganter et al., 2005; Lamb et al., 2006; Wolpaw et al., 2011). We thus compared DeMAND’s performance with differential expression analysis, by t-test statistics. DeMAND systematically outperformed t-test analysis, except for blebbistatin for which neither method identified myosin II as statistically significant (Figure S2A). Indeed, DeMAND had an almost 5-fold better sensitivity in the top 100 predictions, compared to t-test analysis (15% vs. 3%), which was highly statistically significant (p = 5×10−4, and p = 0.06 by χ2 test, respectively) (see Extended Experimental Procedures and Figure 2A). Furthermore, any targets that were significant by t-test analysis were also significant by DeMAND analysis, but not the opposite. Considering the full area under the receiver operator characteristic (ROC) curve (AUC), DeMAND also consistently outperformed the t-test, AUC = 0.70 (p = 2×10−16 by Fisher integration of individual Mann-Whitney p-values for each compound) vs. AUC = 0.60 (p = 3.5×10−7), respectively, reflecting higher overall sensitivity and specificity (Figure S2C).
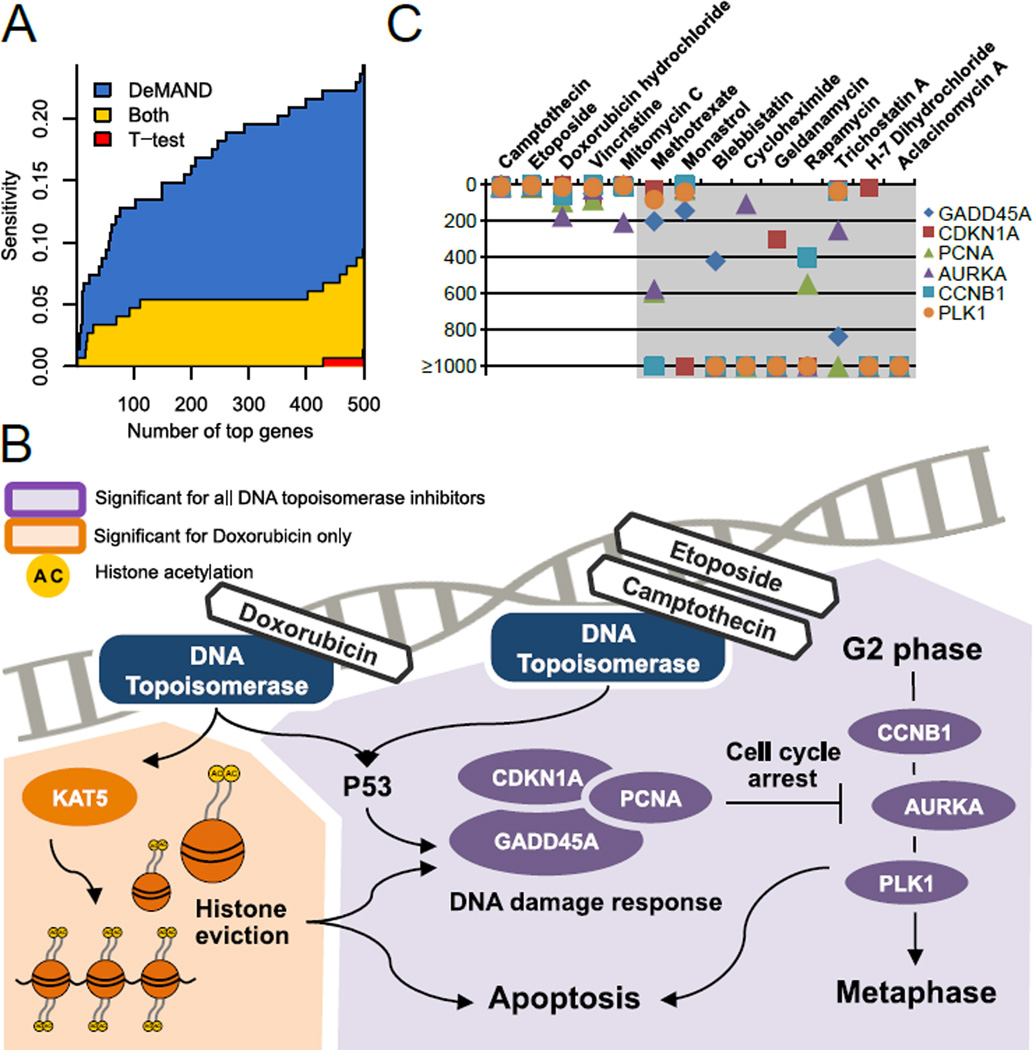
Figure 2. DP14 dataset analysis, see also Figure S2.
(A) The average sensitivity (true-positive rate) for identifying known direct targets in all DP14 compounds, as a function of the number of top selected predictions, using either DeMAND (blue+yellow areas) or t-test analysis (red+yellow areas). DeMAND consistently outperforms t-test. For instance, DeMAND achieves ~15% sensitivity across the top 100 predictions, compared to only 3% for t-test. Furthermore, virtually all targets that are significant by t-test analysis are also significant by DeMAND analysis (no red area for up to 400 genes). In contrast, DeMAND identifies many targets that are missed by t-test (large blue area).
(B) Comparative schematics of established MoA genes for camptothecin, doxorubicin, and etoposide. Doxorubicin specific DeMAND inferred MoA genes are shown with an orange background, while common inferred MoA genes for all compounds are shown with a purple background. The common genes include the core DNA-damage repair machinery (GADD45A, PCNA, and CDNK1A), and cell-cycle arrest genes (CCNB1, AURKA, PLK1). Doxorubicin’s specific MoA includes KAT5, a mediator of histone eviction.
(C) Rank of DNA damage response genes across all DP14 compounds. DeMAND predicts GADD45A, the canonical DNA-damage-inducible gene and its well-known partners CDKN1A, PCNA, CCNB1, AURKA, and PLK1 among the most significant genes only for the 5 DNA damaging agents (i.e., camptothecin, doxorubicin, etoposide, mitomycin C, and vincristine).
To assess DeMAND’s performance on MoA proteins other than high-affinity targets, we focused on two of the four compounds, whose direct targets were missed, including camptothecin (a TOP1 inhibitor) and doxorubicin (a TOP2A inhibitor), which severely disrupt DNA repair and mitosis. DeMAND identified GADD45A, CDKN1A, PCNA, AURKA, PLK1, and CCNB1 among the most statistically significant genes for both compounds (mostly in the top 20), which are known key downstream effectors of TOP1 and TOP2A inhibition (Figure 2B). DeMAND, therefore identifies key MoA proteins for both these compounds. More specifically, GADD45A (growth arrest and DNA damage-inducible gene 45A), an established DNA damage response effector (Goldwasser et al., 1996), acts by forming protein complexes with CDKN1A (Cyclin-Dependent Kinase Inhibitor 1A), and PCNA (proliferating cell nuclear antigen), a processivity factor of DNA polymerase delta required for high-fidelity DNA replication and excision repair (Smith et al., 1994). In turn, if DNA damage is detected, CDKN1A, PCNA, and GADD45A regulate the activity of CCNB1 (cyclin B1, a critical effector of the G2/M cell-cycle checkpoint) (Zhan et al., 1999), PLK1 (polo-like kinase 1), and AURKA (Aurora Kinase A, a mitosis regulator) either at the RNA or protein level (Shao et al., 2006). Of these six genes, only GADD45A and CDKN1A were differentially expressed, albeit at a much lower rank.
DeMAND identifies specific differences in compounds with similar MoA
Detailed assessment highlighted key differences and commonalities in DeMAND-inferred MoA of compounds with similar targets, which were undetectable by t-test analysis. For instance, camptothecin (TOP1), doxorubicin (TOP2A), and etoposide (TOP2A) are all topoisomerase (TOP) inhibitors, which induce single or double strand breaks following covalent trapping of the TOP-DNA cleavable complex (Gilbert and Hemann, 2010). Consistently, DeMAND identified a significant common footprint in their inferred MoA, as shown in the previous section. However, it also identified highly specific effectors, such as KAT5/TIP60 for doxorubicin (ranked 4th), suggesting potentially relevant MoA differences (Figure 2B). Indeed, contrary to etoposide and camptothecin, doxorubicin is also a strong DNA intercalator, inducing KAT5-dependent histone acetylation and release from open chromatin (histone eviction) (Choi et al., 2009; Ikura et al., 2000), leading to cell cycle arrest (Pang et al., 2013). Similarly, DeMAND identified SIK1 as a doxorubicin specific effector (ranked 36th), which is required for cardiac progenitor cell maintenance (CPCs) (Romito et al., 2010), thus pinpointing the compound’s key adverse event, i.e., cardiomyopathy followed by congestive heart failure (Zhang et al., 2012b). Both KAT5 and SIK1 were completely missed by t-test analysis.
Finally, DeMAND successfully stratified compounds based on MoA gene overlap, further emphasizing its specificity. For instance, for all DNA damaging agents, including camptothecin, doxorubicin, etoposide, mitomycin C, and vincristine, DeMAND predicted GADD45A, the canonical DNA-damage-inducible gene, and its well-known interactors (CDKN1, CCNB1 PCNA and AURKA) among the most significant genes (Figure 2C). Yet, these genes were not significant for other compounds (Figure 2C), confirming the algorithm’s specificity.
Validation of novel effectors and modulators of compound activity
To assess whether DeMAND can identify novel compound effectors and modulators, we validated novel predictions for vincristine and mitomycin C, an inhibitor of microtubule formation in mitotic spindle and an antineoplastic antibiotic, respectively. DeMAND successfully identified the known high-affinity target of vincristine (TUBB), as well as CCNB1, VHL, RPS3A and NFKBIA, in the top 5 predictions. While RPS3A and VHL, are known to affect mitotic spindle assembly (Jang et al., 2012; Thoma et al., 2009) and CCNB1 is a microtubule activity marker, their function in mediating/modulating vincristine’s activity is unknown.
Probing the microtubule network with an anti-tubulin antibody, following siRNA-mediated silencing of these genes, confirmed that loss of RPS3A (but not of VHL, CCNB1 or NFKBIA) disrupts microtubules in adherent U-2-OS cells (Figure 3A). To further validate the role of these genes in mediating vincristine’s activity, we performed dose-response curve assays in U-2-OS cells, following silencing of each gene (see Extended Experimental Procedures). These assays confirmed that all of these genes, except for NFKBIA, are key vincristine activity effectors and mediators. Specifically, VHL silencing increased vincristine sensitivity by more than two-fold (Figure 3B), while RPS3A and CCNB1 silencing had the opposite effect. Thus, 4 out of 5 of the top DeMAND-inferred genes were confirmed vincristine activity modulators, including its primary target (TUBB), suggesting that, for some compounds, false positive rates may be as low as 20%. None of these genes were significant by t-test analysis.
Figure 3. Validation of novel effectors of vincristine and mitomycin C.
(A) Immunohistochemistry-based imaging of microtubule networks in cells treated with DMSO, vincristine, non-target siRNA, and siRNA targeting RPS3A. Non-target siRNA is indistinguishable from DMSO controls. Both vincristine and siRPS3A significantly alter the microtubule network in U-2-OS cells (4nM of vincristine for 24h).
(B) Vincristine dose response curves in U-2-OS following transfection with non-target siRNA (blue) or siRNA targeting CCNB1 (orange), VHL (red), NFKBIA (black), and RPS3A (green). RPS3A and CCNB1 silencing reduces cell sensitivity to vincristine, while VHL silencing increases sensitivity by two-folds.
(C) Mitomycin C dose response curves in OCI-LY3 normalized to DMSO treatment (black) or following treatment with TG101348 (a JAK2 inhibitor), at 0.2uM (green), 0.4 uM (cyan), and 0.6uM (blue). JAK2 inhibition induces loss of sensitivity to mitomycin C.
DeMAND also inferred the JAK2 kinase as an exclusive mitomycin C MoA protein (i.e. JAK2 was not significant by DeMAND analysis for any other compound). This is of potential importance since constitutive activity of JAK2 causes chemo-resistance in lymphocytes (Gupta et al., 2012), while constitutive JAK2 activity may also affect DNA damage, repair and recombination outcome (Hoser et al., 2003). Confirming the prediction, dose-response curves for mitomycin C, following treatment with varying amounts of TG101348 (a JAK2 inhibitor), revealed highly significant, dose-dependent antagonism between JAK2 inhibition and mitomycin C activity (Figure 3C, see Experimental Procedures).
Finally, we analyzed DeMAND-inferred results for rapamycin. While DeMAND could not predict the highest-affinity targets, MTOR and FKBP1A, many genes downstream of MTOR pathways (Hsieh et al., 2012) were highly enriched in the top DeMAND-inferred genes (Figure S2E), including many ribosomal genes. The only other compound with significant ribosomal gene enrichment was cycloheximide, a known ribosomal activity inhibitor, thus further highlighting the algorithm’s specificity.
Algorithm robustness and requirements
We then benchmarked DeMAND’s performance as a function of network accuracy and size, as well as of the number of samples in the perturbation dataset. First, we compared the results obtained using an independent B-cell gene regulatory network, reconstructed from a distinct dataset of 254 Affymetrix U95av2 GEPs (see Experimental Procedures). We tested the enrichment of statistically significant DeMAND-inferred genes (FDR ≤ 0.1), using the U95av2 network, against those inferred using the U133p2 network, by Gene Set Enrichment Analysis (GSEA) (Subramanian et al., 2005). The analysis confirmed that DeMAND predictions were almost identical, independent of network model (p < 1×10−9 by GSEA, Figure S3A). Furthermore predictions were virtually unaffected when up to 60% of the network interactions were randomly removed (see Experimental Procedures, Figure S3B). Similarly, predictions were virtually identical, as long as 6 or more GEPs representative of compound perturbation were used (see Extended Experimental Procedures and Figure S3C). Taken together, these data suggest that DeMAND is highly robust to network noise and especially to false negative interactions, and that it can be applied to datasets with as few as 6 treatment and 6 untreated controls GEPs.
We then selected 13 datasets representing compound perturbations (GEO13) from the gene expression omnibus (GEO) database (Table 1, and Table S3). Only compounds with established targets with at least 6 treatment/control GEPs were selected, including 7 human breast cancer and 6 human B-cell lymphoma datasets. Confirming results on the DP14 dataset, DeMAND inferred known direct targets for 62% of these compound perturbations (FDR ≤ 0.1, Figure S4A), while still significantly outperforming t-test based methods (AUC = 0.82 vs. 0.74, respectively, p-value = 2.2×10−16 vs. p-value = 5.9×10−8, respectively, by Fisher integration of individual Mann-Whitney p-values for each compound) (Figure S4B). Among top predicted MoA proteins, DeMAND again achieved roughly 5-fold better performance than t-test (Figure S4C).
Table 1.
13 compound perturbation datasets from the GEO database, see also Figure S4.
| Compound | Cellular context | GEO ID |
|---|---|---|
| Zoledronate | Metastatic breast cancer cell lines (MDA-MB-231) | GSE33552 |
| Valproic Acid | Chronic lymphocytic leukemia (Patient derived B cells) | GSE14973 |
| Genistein | Breast cancer cell lines (MCF-7) | GSE9936 |
| S-Equol | Breast cancer cell lines (MCF-7) | GSE9936 |
| Estradiol | Breast cancer cell lines (MCF-7) | GSE9936 |
| Rituximab | B-cell non-Hodgkin’s lymphoma cell lines (K422) | GSE7292 |
| Thapsigargin | lytic-permissive lymphoblastoid cell lines | GSE31447 |
| Fluvastatin | Metastatic breast cancer cell lines (MDA-MB-231) | GSE33552 |
| MALT1 Inhibitor | Diffuse large B-cell lymphoma (Patient derived B cells) | GSE40003 |
| Docetaxel | Breast cancer cell lines (MCF-7) | GSE5149 |
| γ-Secretase Inhibitor | MCL cell lines | GSE34602 |
| Triptolide | Breast cancer cell lines (MCF-7) | GSE28662 |
| Actinomycin D | Breast cancer cell lines (MCF-7) | GSE28662 |
DeMAND-inferred MoA stratifies pharmacological effect
We then assessed whether DeMAND-inferred MoA overlap was predictive of pharmacological compound similarity. We first computed the significance of MoA overlap for each DP14 compound pair (FDR ≤ 0.1 by Fisher’s Exact Test, FET) (see Experimental Procedures, Figure 4A, and Table S4). Among all 91 possible compound pairs, the six most similar ones included only topoisomerase inhibitors and other DNA-damaging agents (etoposide, doxorubicin, camptothecin, and mitomycin C). Thus, DeMAND successfully assessed high compound MoA similarity between topoisomerase inhibitors and other DNA-damaging agents even though it could not identify TOP1 or TOP2A among the inferred MoA genes, suggesting that key effector proteins may be as informative as direct targets in terms of compound similarity.
Figure 4. Compound similarity inference, see also Figure S5.
(A) Compound similarity is assessed based on the statistical significance (by FET) of the overlap of their DeMAND-inferred MoA proteins.
(B) DeMAND-inferred compound similarity in the DP92 dataset is assessed by (a) the overlap of known direct targets between two compounds (orange), (b) compound sensitivity profile similarity based on CTD2 data (green), (c) overlap in compound classification, according to the Anatomical Therapeutic Chemical (ATC) Classification (blue), or (d) any of the above evidences (black).
To further evaluate this hypothesis, we applied the method to a much larger compound perturbation dataset (DP92), representing GEPs from three B-cell lymphoma cell lines (OCI-LY3, OCI-LY7 and U-2932), following perturbation with 92 unique FDA-approved, late-stage experimental, and tool compounds (see Extended Experimental Procedures, and Table S5). Since only three GEPs per compound and cell line are available in this dataset, we used it only for compound-pair similarity assessment (see Experimental Procedures).
DeMAND performance was objectively evaluated by comparison with three independent data sources: (a) compounds sharing established targets; (b) compounds sharing therapeutic and chemical characteristics, according to the Anatomical Therapeutic Chemical classification system (ATC) and (c) compounds with correlated drug-response profiles, as assessed by the Cancer Target Discovery and Development (CTD2) consortium (Basu et al., 2013) (see Extended Experimental Procedures). The latter dataset recapitulates dose-response curve vectors representing 338 unique compounds profiled against 257 distinct cancer lines. We evaluated the fraction of validated similar pairs (precision), based on each of the three evidence datasets, as a function of the number of significant pairs (precision curves, Figure 4B). DeMAND-inferred pairs were highly enriched in pairs from three evidence datasets, as assessed by each of the evidences individually (i.e., p-value = 2×10−8, 1.4×10−5, and 9×10−4, by GSEA, for pairs sharing the same ATC class, common established targets, and high dose-response vector correlation in the CTD2 dataset, respectively, Figure S5A), and also when taken together (GSEA p-value = 7.6×10−7). For instance, 8 of the top 10 and 43 of the top 100 DeMAND-inferred pairs were validated by at least one of the three datasets (p = 2.2×10−16 by FET).
DeMAND outperformed predictions using similarity obtained by overlapping statistically significant differential expressed genes (e.g., by t-test statistics) by consistently achieving higher sensitivity at any precision value (Figure S5B). DeMAND also outperformed another state of the art method, (MANTRA) (Iorio et al., 2010), which uses mutual gene set enrichment analysis (Subramanian et al., 2005) to compute similarity, again by achieving higher sensitivity at almost any desired precision value (Figure S5B).
Finally, we evaluated the correlation between compound-pair similarity as predicted by each method and their CTD2-based similarity. DeMAND prediction achieved significant Spearman correlation (ρ = 0.59, p-value=7.8×10−5, Figure S5C), while both the t-test and MANTRA methods did not achieve statistically significant correlation (Figures S5D, S5E). Thus, DeMAND could predict compounds with similar pharmacological effect and activity profile using only the GEP following their treatment in a single cell line.
DeMAND identifies GPX4 as a novel MoA effector for altretamine
We identified altretamine and sulfasalazine as the compound pair with the highest DeMAND-inferred MoA similarity (p-value = 9.91×10−81), among all pairs where the MoA of at least one compound was unknown. Altretamine is an FDA-approved antineoplastic drug with no established targets or effectors. Instead, sulfasalazine is an inhibitor of system xc−, the cystine-glutamate antiporter (Dixon et al., 2014), required for the biosynthesis of glutathione (GSH). Thus sulfasalazine inactivates enzymes that rely on reduced glutathione (GSH) as a cofactor, including glutathione peroxidase 4 (GPX4) (Dixon et al., 2012; Yang et al., 2014), leading to toxic accumulation of lipid reactive oxygen species (ROS).
We thus tested whether altretamine may also modulate the system xc−-GPX4 pathway. U-2932 cells were treated with altretamine and their GSH levels were assessed using Ellman’s reagent (Figure 5A and Extended Experimental Procedures). Sulfasalazine was used as a positive control for GSH depletion in U-2932 cells, confirming depletion of GSH levels following compound treatment. In contrast, altretamine did not deplete GSH levels, even after doubling its IC50 at 24h concentration, suggesting that the compound may target mechanisms downstream of GSH in this pathway. We thus treated U-2932 cells with altretamine, and prepared cell lysates for an LC-MS based GPX4 assay. Phosphatidylcholine hydroperoxide (PC-OOH), a specific substrate for GPX4 (Brigelius-Flohe and Maiorino, 2013), was added to cell lysates and PC-OOH to PC-OH reduction was assessed by the mass chromatogram of the [PC-OOH + H+] ion (m/z = 790.5). As shown in Figure 5B, lysates of untreated cells reduced PC-OOH levels completely, leaving no residual signal for the [PC-OOH + H+] ion (m/z = 790.5). In sharp contrast, lysates from altretamine treated cells displayed a significant [PC-OOH + H+] signal, indicating that abrogation of PC-OOH reduction was mediated by GPX4 inhibition (Experimental Procedures). Indeed, since GPX4 is the only enzyme capable of reducing lipid hydroperoxides (Yang et al., 2014), GPX4 inhibition is necessary to increases lipid-ROS levels (Thomas et al., 1990). As expected, both sulfasalazine and altretamine were confirmed to induce lipid-ROS accumulation in U-2932 cells, as assessed by BODIPY-C11 staining and flow cytometry (see Figure 5C and Experimental Procedures). Thus, DeMAND correctly predicted the unexpected mechanistic similarity between the MoA of two previously unrelated drugs (see Figure 5D), showing altretamine as a new GPX4 inhibitor and suggesting a potential mechanism for its antineoplastic activity.
Figure 5. DeMAND identifies the MoA of altretamine.
(A) GSH concentration following treatment of cells by negative control (DMSO, gray), sulfasalazine as a positive control (red), and altretamine (blue) show that sulfasalazine reduces active GSH levels compared to control, while altretamine results in active GSH levels indistinguishable from the control.
(B) The level of a GPX4-specific substrate (PC-OOH) is measured by mass spectrometry (a) without cell lysate (gray), (b) with untreated cell lysate (green), and (c) with cell lysate from altretamine treated cells (blue). PC-OOH levels in altretamine treated cells are similar to no-lysate, and markedly different from untreated lysate, indicating that altretamine inhibits GPX4 activity.
(C) Lipid reactive oxidative species (ROS) levels were measured by flow cytometry using DMSO treated cells (black curve, as control) and compound treated cells (red curve). Both altretamine and sulfasalazine significantly increases lipid-ROS levels, confirming the predicted similarity in their functional effect.
(D) Sulfasalazine is a known inhibitor of the System xc− cystine/glutamate antiporter. Its downstream effect on Glutathione (GSH) and GPX4 leads to accumulation of lipid ROS. DeMAND predicted significant similarity between sulfasalazine and altretamine and GPX4 but not GSH as altretamine specific MoA proteins, as experimentally confirmed panels (A–C).
Discussion
DeMAND elucidates compound MoA by assessing compound-mediated dysregulation of gene-gene interactions on a genome-wide basis, from gene expression profiles of compound perturbations. DeMAND reliably identifies compound targets, effectors and activity modulators, allowing effective assessment of compound MoA and MoA similarity. Indeed, DeMAND identified known and novel MoA genes for vincristine, mitomycin C, and altretamine that were experimentally validated. DeMAND also elucidated a novel MoA for altretamine, confirming its predicted similarity to sulfasalazine.
DeMAND was shown to be highly robust to network and sample variability. More importantly, unlike previous methods (di Bernardo et al., 2005; Mani et al., 2008), DeMAND can reliably predict compound MoA using as few as 6 control and 6 perturbation samples. This allows unprecedented applicability of the methods to elucidate MoA for novel developmental compounds within specific cellular contexts of interest, including in vivo.
DeMAND leverages integration of GEPs obtained at multiple time points and at multiple compound concentrations, thus simplifying experimental design when the precise concentration or time points at which the MoA may be revealed is unknown. Indeed, absent prior knowledge, compound MoA was optimally revealed by integrating multi-time-point compound perturbations for all but two of the tested compounds (Figure S2B).
DeMAND predictions are highly specific, allowing classification of compounds into groups of similar function and identification of pathways that are relevant to compound MoA. For instance, for DNA-damaging compounds (camptothecin, doxorubicin, etoposide, vincristine and mitomycin C), DeMAND correctly predicted several of the hallmark genes involved in DNA-damage-induced response. The specificity was evidenced by the fact that relevant MoA proteins were inferred only for DNA-damage inducing compounds and not for any other compound (including compounds exhibiting significant polypharmacology like H-7 dihydrochloride or cycloheximide). In other examples, high MoA specificity was shown for doxorubicin, where DeMAND identified KAT5, consistent with recent findings of KAT5-mediated histone eviction, as well as SIK1, a gene required for cardiac progenitor cells maintenance, providing a potential mechanistic link between doxorubicin and its known cardiac toxicity. Critically, SIK1 was also detected in the MoA of other DNA damaging agents, albeit at much lower rank/significance, suggesting that these compounds should also be monitored for cardiac toxicity. Taken together, these findings suggest that the algorithm is equally effective in predicting both direct targets and indirect compound effectors, thus helping elucidate both on-target pharmacology and off-target toxicity. Overall, DeMAND identified known MoA proteins for >70% of tested compound, while experimental validation suggests that false discovery rates (FDR) may be as low as 20%, although more extensive FDR estimate is impossible at this time because compound MoA in databases is largely incomplete, producing significant FDR overestimate. For instance, following experimental validation, FDR for vincristine went from 80%, as only TUBB was an established compound target/effector, to 20%.
DeMAND relies on the existence of high quality context-specific gene regulatory networks, which may represent a limitation for specific cellular contexts. However, given the abundance of data generated by large-scale projects such as the Cancer Genome Atlas (TCGA) and other related consortia, as well as the availability of increasingly accurate and comprehensive methods for context-specific network reverse engineering (Califano et al., 2012; Zhang et al., 2012a), this limitation is at best temporary. However, network availability does not guarantee identification of MoA proteins that are poorly represented. For instance, for blebbistatin (a myosin II inhibitor), using the U95av2 network, DeMAND identified PTK2B, GRB2 and FYN, all of which are both direct regulators of myosin II phosphorylation, and responders to myosin II perturbation (Sieg et al., 1998) (see Figure S2D). Yet, due to lack of GRB2 representation in the U133p2 network, this gene could not be inferred. It is also important to highlight that DeMAND analysis of the DP14 and DP92 datasets, using a high quality context-free network from the STRING database (Franceschini et al., 2013), was still able to identify ubiquitous targets and effectors (e.g., those involved in cell cycle and DNA damage repair mechanisms) with high precision and sensitivity, but exhibited lower performance both in compound similarity analysis and in the identification of genes with context specific function/expression. This suggests that non-context-specific networks can still be used for DeMAND analyses, albeit with an increase in false positive and negative predictions.
An important, albeit not critical, limitation of the current methodology is the lack of prediction of compound activity sign, i.e., whether a compound will induce increase or decrease in an inferred MoA protein activity. Conversely, the method cannot predict whether inhibiting an inferred MoA protein will likely either increase or decrease drug activity. Presently, the only way to resolve this question is by follow-up experimental assays. In addition, the need for at least six GEPs at multiple concentrations and time points is a potential limitation when assessing MoA for large compound libraries. Despite these limitations, however, DeMAND has proven highly effective in the de-novo identification of context-specific targets and effectors for arbitrary compounds of interest, providing important insight into the prioritization of novel compounds for development, or into the repositioning of previously approved compounds.
Experimental Procedures
Networks used in the analysis
We generated context specific gene-regulatory networks with both protein-DNA and protein-protein interactions (see Table S6). The analysis used both context specific GEPs and context independent information from multiple experimental and computational databases, which was integrated into a final interactome using Naïve Bayes Classifiers (see (Lefebvre et al., 2010) and Extended Experimental Procedures for detailed information). B Cell and breast cancer specific networks as well as the STRING database can be downloaded from http://wiki.c2b2.columbia.edu/califanolab/index.php/Software/DeMAND
Evaluating interaction dysregulation
For each pair of interacting genes in the network, we compute a two-dimensional probability density from their discrete rank-transformed expression in a given condition (treatment or control), by Gaussian kernel smoothing, using Silverman's approach (Silverman, 1986). The sum of the Gaussian probabilities densities from treatment samples, computed at each point of the discrete rank space, provides the perturbation probability distribution P, while that from control samples provides the control probability distribution Q. The distance between the two discrete probability distributions is evaluated using a symmetric form of the Kullback-Leibler divergence (KLD), obtained by averaging KLD(P|Q) and KLD(Q|P).
KLD statistical significance is determined using a null distribution generated by 105 KLD values generated from random gene pairs (regardless of whether they share a network edge), providing individual edge dysregulation p-values. These are integrated across all the interactions in a specific gene regulon, using the Fisher’s method, and corrected using a modification of Brown’s method for correction of p-value dependence (Brown, 1975), using the covariance between the residuals from a linear fit to the common gene, a (see Figure S1A). A more detailed description of this method is available in the Extended Experimental Procedures.
Determining known direct targets of compounds
Established targets for tested compounds were obtained from DrugBank (Wishart et al., 2008), MATADOR (Gunther et al., 2008), and literature searches. For MATADOR, only genes annotated as ‘direct’ or ‘direct-indirect’ were considered as compound targets, while genes labeled as ‘indirect’ were discarded. For a list of compound targets see Table S1.
Assessing drug similarity
To evaluate compound similarity, we first selected statistically significant MoA genes (FDR ≤ 0.1) for each compound. We then computed the significance of their overlap by FET analysis. Many genes were not significant for any compounds, thus biasing this analysis. To reduce this effect, we removed these genes from the analysis. Notably this correction did not affect compound pair ranking but only their absolute similarity p-values, by avoiding p-value underestimation.
To compute similarity p-values using the DP92 dataset we calculated p-values for each of the three cell lines independently and used Fisher’s method to combine them.
Robustness analysis
To evaluate the effect of network accuracy on DeMANDs’ performance we gradually removed interactions at random, in 10% increments, and compared the overlap of significant perturbed and unperturbed MoA protein predictions by FET analysis. To evaluate the effect of sample size we subsampled i samples (i=3..18) from the compound-treated and from the control samples, and compared these results with the result obtained using all samples. Both analyses were performed independently on each of the 14 compounds in the DP14 dataset. See Extended Experimental Procedure for additional information and Figure S3 for the results of the analysis.
Cell culture
Diffuse large B-cell lymphoma (DLBCL) OCI-LY3 and OCI-LY7 cells were obtained from University Health Network (Toronto, Canada); the U-2932 DLBCL cell line was purchased from the Leibniz-Institute DSMZ German Collection of Microorganisms and Cell Cultures; the U-2-OS osteosarcoma cell line was obtained from ATCC (Cat# ATCC HTB-96). OCI-LY3, OCI-LY7, U-2932 cells were cultured in Iscove’s Modified Dulbecco Medium (IMDM) supplemented with 10% fetal bovine serum at 37°C in a 5% CO2 atmosphere. U-2-OS cells were cultured in McCoy’s 5A medium, supplemented with 10% fetal bovine serum.
Dose response curves
The 92 compounds were selected based on primary activity screen of FDA-approved, late-stage experimental, and tool compounds. OCI-LY3, OCI-LY7 and U-2932 cells were seeded in white tissue culture treated 96-well plates, at a density of 5×104 cells per well in 100µL total volume using the Janus automated liquid handling system (Perkin Elmer Inc.). After 12h of incubation at 37°C plates were allowed to cool to room temperatur e, prior to compound addition via the Janus. Compounds were diluted in DMSO as a 7 point dilution curves in a stock plate, 1µL of these stock solutions where transferred into assay plates, in triplicate. These were subsequently placed on an orbital shaker for 5minutes and then back in the incubator. At 24h plates were removed from the incubator and equilibrated to room temperature before addition of 50µL of CellTiter-Glo Luminescent Cell Viability Assay (Promega Corp.) per well. Plates were shaken 5minutes on an orbital shaker before data acquisition in an Envision (PerkinElmer Inc.) (0.5 second read time, enhanced luminescence). IC20 values were assessed using a four parameter fit model (IDBS Activity Base).
Compound treatment for gene expression profiling
Cells were seeded in tissue culture-treated 96-well plates at a density of 5×104 cells per well using the Janus automated liquid handling system (Perkin Elmer, Inc.). They were then treated with the 24h IC20 of each compound (by DMSO dilution) for 6h, 12h, and 24h at 37°C, 5% CO2 under humidified conditions. For each compound/condition combination one single data point was analyzed and 0.2% DMSO vehicle treated samples were used as controls. Viability assay was run in parallel to monitor the compound effectiveness.
Generation of gene expression profiles
Total RNA was isolated with the RNAqueous-96 Automated Kit (Ambion) on the Janus automated liquid handling system (Perkin Elmer Inc.), quantified by NanoDrop 6000 spectrophotometer and quality checked by Agilent Bioanalyzer. 300ng of each of the samples with RIN value >7 were converted to biotinylated cRNA with the Illumina TotalPrep-96 RNA Amplification Kit (Ambion) using a standard T7-based amplification protocol and hybridized on the Human Genome U219 96-Array Plate (Affymetrix). Hybridization, washing, staining and scanning of the array plates were performed on the GeneTitan Instrument (Affymetrix) according to manufacturer’s protocols.
GPX4 enzymatic activity assay
GPX4 enzymatic activity assay was performed as described in (Yang et al., 2014). Briefly, 1×106 cells were re-suspended in the cell lysis buffer. Sonication was used to make cell lysates followed by centrifugation at 14,000 rpm for 10 minutes. Protein concentration of the cleared cell lysates was determined using a Bradford protein assay (Bio-Rad). Two hundred micrograms of cellular proteins was mixed with phosphatidyl choline hydroperoxide (PC-OOH), the GPX4 specific substrate, and reduced glutathione, a GPX4 cofactor. The mixture was incubated at 37°C for 30 minutes followed by lipid extraction using a chloroform:methanol (2:1) solution. The lipid extract was evaporated using a rotary evaporator, and re-dissolved in 100% ethanol before injecting into LC-MS instrument for PC-OOH quantitation.
Analysis of lipid reactive oxygen species (ROS) generation
U-2932 cells (2×105) were seeded in 6-well plates and incubated at 37°C for 16h. Cells were treated with test compounds for the indicated time, then harvested, pelleted and washed once with PBS. For lipid ROS detections, cells were re-suspended with Hanks Balanced Salt Solution (HBSS, Life Technologies) containing C11-BODIPY (581/591) (2µM) (Life Technologies) and incubated for 10 minutes at 37°C. Cells were then pelleted, re-suspended in 500µL HBSS, strained through 40µM cell strainer (BD Falcon), and analyzed using BD Accuri C6 flow cytometer (BD Biosciences). C11-BODIPY signal was measured using FL1 channel. Experiments were done in biological triplicates, and a representative result was shown.
Co-treatment with mitomycin C and a JAK2 inhibitor
The JAK2-selective inhibitor TG101348 (Wernig et al., 2008) and Mitomycin C were purchased from Selleckchem and Tocris Bioscience respectively and were dissolved in DMSO. OCI-LY3 cells were treated with the indicated compounds in 96-well plates and their growth was determined using the CellTiter-Glo Luminescent Cell Viability Assay (Promega Corp). Typically, 30,000 OCI-Ly3 cells per well in 200 µL of growth medium were grown for 48h in the presence or absence (DMSO alone) of the desired compounds, and then assayed with CellTiter Glo according to manufacturer’s instructions.
Supplementary Material
Acknowledgement
We thank Katia Basso for providing the U-2932 cell line for experimental validation, Wei Keat Lim for GEP normalization, Beatrice Salvatori for helpful feedback on the manuscript. This work is supported in part by the CTD2 (5U01CA168426), LINCS (1U01CA164184-02 and 3U01HL111566-02), and MAGNet (5U54CA121852-08) grants to AC. Raw CEL files and normalized data for DP92 data can be accessed from GEO (GSE60408). BRS is supported by the National Institute of Health (5R01CA097061, R01CA161061), and New York Stem Cell Science (C026715) and is an Early Career Scientist of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
MB and AC conceived the idea; JHW, YS, MB, AC developed the method; JHW, YS, WSY, PS, BRS, MB, AC wrote the manuscript; MM, RR, CK generated the data; WSY, PS, BRS validated the predictions; GL generated interactomes; MRM performed statistical analysis leading to method development; PN, AI, PS performed literature based analysis to established the connection of DeMAND’s predictions with the MoA of the compounds.
References
- 1.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 2.Bansal M, Della Gatta G, di Bernardo D. Inference of gene regulatory networks and compound mode of action from time course gene expression profiles. Bioinformatics. 2006;22:815–822. doi: 10.1093/bioinformatics/btl003. [DOI] [PubMed] [Google Scholar]
- 3.Bansal M, Yang J, Karan C, Menden MP, Costello JC, Tang H, Xiao G, Li Y, Allen J, Zhong R, et al. A community computational challenge to predict the activity of pairs of compounds. Nat Biotechnol. 2014;32:1213–1222. doi: 10.1038/nbt.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basilico C, Pennacchietti S, Vigna E, Chiriaco C, Arena S, Bardelli A, Valdembri D, Serini G, Michieli P. Tivantinib (ARQ197) Displays Cytotoxic Activity That Is Independent of Its Ability to Bind MET. Clinical Cancer Research. 2013;19:2381–2392. doi: 10.1158/1078-0432.CCR-12-3459. [DOI] [PubMed] [Google Scholar]
- 5.Basso K, Saito M, Sumazin P, Margolin AA, Wang K, Lim WK, Kitagawa Y, Schneider C, Alvarez MJ, Califano A, et al. Integrated biochemical and computational approach identifies BCL6 direct target genes controlling multiple pathways in normal germinal center B cells. Blood. 2010;115:975–984. doi: 10.1182/blood-2009-06-227017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basu A, Bodycombe NE, Cheah JH, Price EV, Liu K, Schaefer GI, Ebright RY, Stewart ML, Ito D, Wang S, et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell. 2013;154:1151–1161. doi: 10.1016/j.cell.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brigelius-Flohe R, Maiorino M. Glutathione peroxidases. Biochimica et biophysica acta. 2013;1830:3289–3303. doi: 10.1016/j.bbagen.2012.11.020. [DOI] [PubMed] [Google Scholar]
- 8.Brown MB. Method for Combining Non-Independent, One-Sided Tests of Significance. Biometrics. 1975;31:987–992. [Google Scholar]
- 9.Califano A, Butte AJ, Friend S, Ideker T, Schadt E. Leveraging models of cell regulation and GWAS data in integrative network-based association studies. Nature genetics. 2012;44:841–847. doi: 10.1038/ng.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi J, Heo K, An WJ. Cooperative action of TIP48 and TIP49 in H2A.Z exchange catalyzed by acetylation of nucleosomal H2A. Nucleic Acids Res. 2009;37:5993–6007. doi: 10.1093/nar/gkp660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.di Bernardo D, Thompson MJ, Gardner TS, Chobot SE, Eastwood EL, Wojtovich AP, Elliott SJ, Schaus SE, Collins JJ. Chemogenomic profiling on a genome-wide scale using reverse-engineered gene networks. Nat Biotechnol. 2005;23:377–383. doi: 10.1038/nbt1075. [DOI] [PubMed] [Google Scholar]
- 12.Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, et al. Ferroptosis: An Iron-Dependent Form of Nonapoptotic Cell Death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dixon SJ, Patel D, Welsch M, Skouta R, Lee E, Hayano M, Thomas AG, Gleason C, Tatonetti N, Slusher BS, et al. Pharmacological inhibition of cystine-glutamate exchange induces endoplasmic reticulum stress and ferroptosis. Elife. 2014;3 doi: 10.7554/eLife.02523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Floratos A, Smith K, Ji Z, Watkinson J, Califano A. geWorkbench: an open source platform for integrative genomics. Bioinformatics. 2010;26:1779–1780. doi: 10.1093/bioinformatics/btq282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ganter B, Tugendreich S, Pearson CI, Ayanoglu E, Baumhueter S, Bostian KA, Brady L, Browne LJ, Calvin JT, Day GJ, et al. Development of a large-scale chemogenomics database to improve drug candidate selection and to understand mechanisms of chemical toxicity and action. Journal of biotechnology. 2005;119:219–244. doi: 10.1016/j.jbiotec.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 17.Gardner TS, di Bernardo D, Lorenz D, Collins JJ. Inferring genetic networks and identifying compound mode of action via expression profiling. Science. 2003;301:102–105. doi: 10.1126/science.1081900. [DOI] [PubMed] [Google Scholar]
- 18.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert LA, Hemann MT. DNA damage-mediated induction of a chemoresistant niche. Cell. 2010;143:355–366. doi: 10.1016/j.cell.2010.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldwasser F, Bae I, Fornace AJ, Jr, Pommier Y. Differential GADD45, p21CIP1/WAF1, MCL-1 and topoisomerase II gene induction and secondary DNA fragmentation after camptothecin-induced DNA damage in two mutant p53 human colon cancer cell lines. Oncology research. 1996;8:317–323. [PubMed] [Google Scholar]
- 21.Gunther S, Kuhn M, Dunkel M, Campillos M, Senger C, Petsalaki E, Ahmed J, Urdiales EG, Gewiess A, Jensen LJ, et al. SuperTarget and Matador: resources for exploring drug-target relationships. Nucleic Acids Res. 2008;36:D919–D922. doi: 10.1093/nar/gkm862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta M, Han JJ, Stenson M, Maurer M, Wellik L, Hu G, Ziesmer S, Dogan A, Witzig TE. Elevated serum IL-10 levels in diffuse large B-cell lymphoma: a mechanism of aberrant JAK2 activation. Blood. 2012;119:2844–2853. doi: 10.1182/blood-2011-10-388538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen NT, Brunak S, Altman RB. Generating genome-scale candidate gene lists for pharmacogenomics. Clin Pharmacol Ther. 2009;86:183–189. doi: 10.1038/clpt.2009.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirota T, Lee JW, St John PC, Sawa M, Iwaisako K, Noguchi T, Pongsawakul PY, Sonntag T, Welsh DK, Brenner DA, et al. Identification of Small Molecule Activators of Cryptochrome. Science. 2012;337:1094–1097. doi: 10.1126/science.1223710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoser G, Majsterek I, Romana DL, Slupianek A, Blasiak J, Skorski T. Fusion oncogenic tyrosine kinases alter DNA damage and repair after genotoxic treatment: role in drug resistance? Leuk Res. 2003;27:267–273. doi: 10.1016/s0145-2126(02)00163-7. [DOI] [PubMed] [Google Scholar]
- 26.Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 28.Iorio F, Bosotti R, Scacheri E, Belcastro V, Mithbaokar P, Ferriero R, Murino L, Tagliaferri R, Brunetti-Pierri N, Isacchi A, et al. Discovery of drug mode of action and drug repositioning from transcriptional responses. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14621–14626. doi: 10.1073/pnas.1000138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito T, Ando H, Suzuki T, Ogura T, Hotta K, Imamura Y, Yamaguchi Y, Handa H. Identification of a Primary Target of Thalidomide Teratogenicity. Science. 2010;327:1345–1350. doi: 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- 30.Jang CY, Kim HD, Zhang X, Chang JS, Kim J. Ribosomal protein S3 localizes on the mitotic spindle and functions as a microtubule associated protein in mitosis. Biochem Biophys Res Commun. 2012;429:57–62. doi: 10.1016/j.bbrc.2012.10.093. [DOI] [PubMed] [Google Scholar]
- 31.Keiser MJ, Setola V, Irwin JJ, Laggner C, Abbas AI, Hufeisen SJ, Jensen NH, Kuijer MB, Matos RC, Tran TB, et al. Predicting new molecular targets for known drugs. Nature. 2009;462:175–181. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kullback S, Leibler RA. On Information and Sufficiency. Ann Math Stat. 1951;22:79–86. [Google Scholar]
- 33.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 34.Lefebvre C, Rajbhandari P, Alvarez MJ, Bandaru P, Lim WK, Sato M, Wang K, Sumazin P, Kustagi M, Bisikirska BC, et al. A human B-cell interactome identifies MYB and FOXM1 as master regulators of proliferation in germinal centers. Molecular systems biology. 2010;6:377. doi: 10.1038/msb.2010.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J, Zhu X, Chen JY. Building disease-specific drug-protein connectivity maps from molecular interaction networks and PubMed abstracts. PLoS computational biology. 2009;5:e1000450. doi: 10.1371/journal.pcbi.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lomenick B, Hao R, Jonai N, Chin RM, Aghajan M, Warburton S, Wang J, Wu RP, Gomez F, Loo JA, et al. Target identification using drug affinity responsive target stability (DARTS) Proceedings of the National Academy of Sciences of the United States of America. 2009;106:21984–21989. doi: 10.1073/pnas.0910040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mani KM, Lefebvre C, Wang K, Lim WK, Basso K, Dalla-Favera R, Califano A. A systems biology approach to prediction of oncogenes and molecular perturbation targets in B-cell lymphomas. Molecular systems biology. 2008;4:169. doi: 10.1038/msb.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller MA. Chemical database techniques in drug discovery. Nat Rev Drug Discov. 2002;1:220–227. doi: 10.1038/nrd745. [DOI] [PubMed] [Google Scholar]
- 39.Pang BX, Qiao XH, Janssen L, Velds A, Groothuis T, Kerkhoven R, Nieuwland M, Ovaa H, Rottenberg S, van Tellingen O, et al. Drug-induced histone eviction from open chromatin contributes to the chemotherapeutic effects of doxorubicin. Nat Commun. 2013;4 doi: 10.1038/ncomms2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Romito A, Lonardo E, Roma G, Minchiotti G, Ballabio A, Cobellis G. Lack of sik1 in mouse embryonic stem cells impairs cardiomyogenesis by down-regulating the cyclin-dependent kinase inhibitor p57kip2. PloS one. 2010;5:e9029. doi: 10.1371/journal.pone.0009029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scannell JW, Blanckley A, Boldon H, Warrington B. Diagnosing the decline in pharmaceutical R&D efficiency. Nat Rev Drug Discov. 2012;11:191–200. doi: 10.1038/nrd3681. [DOI] [PubMed] [Google Scholar]
- 42.Shao S, Wang Y, Jin S, Song Y, Wang X, Fan W, Zhao Z, Fu M, Tong T, Dong L, et al. Gadd45a interacts with aurora-A and inhibits its kinase activity. J Biol Chem. 2006;281:28943–28950. doi: 10.1074/jbc.M600235200. [DOI] [PubMed] [Google Scholar]
- 43.Sieg DJ, Ilic D, Jones KC, Damsky CH, Hunter T, Schlaepfer DD. Pyk2 and Src-family protein-tyrosine kinases compensate for the loss of FAK in fibronectin-stimulated signaling events but Pyk2 does not fully function to enhance FAK- cell migration. The EMBO journal. 1998;17:5933–5947. doi: 10.1093/emboj/17.20.5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silverman BW. Density estimation for statistics and data analysis. London ; New York: Chapman and Hall; 1986. [Google Scholar]
- 45.Smith ML, Chen IT, Zhan QM, Bae IS, Chen CY, Gilmer TM, Kastan MB, Oconnor PM, Fornace AJ. Interaction of the P53-Regulated Protein Gadd45 with Proliferating Cell Nuclear Antigen. Science. 1994;266:1376–1380. doi: 10.1126/science.7973727. [DOI] [PubMed] [Google Scholar]
- 46.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thoma CR, Toso A, Gutbrodt KL, Reggi SP, Frew IJ, Schraml P, Hergovich A, Moch H, Meraldi P, Krek W. VHL loss causes spindle misorientation and chromosome instability. Nat Cell Biol. 2009;11:994–1001. doi: 10.1038/ncb1912. [DOI] [PubMed] [Google Scholar]
- 48.Thomas JP, Geiger PG, Maiorino M, Ursini F, Girotti AW. Enzymatic reduction of phospholipid and cholesterol hydroperoxides in artificial bilayers and lipoproteins. Biochimica et biophysica acta. 1990;1045:252–260. doi: 10.1016/0005-2760(90)90128-k. [DOI] [PubMed] [Google Scholar]
- 49.Wehling M. Assessing the translatability of drug projects: what needs to be scored to predict success? Nat Rev Drug Discov. 2009;8:541–546. doi: 10.1038/nrd2898. [DOI] [PubMed] [Google Scholar]
- 50.Wernig G, Kharas MG, Okabe R, Moore SA, Leeman DS, Cullen DE, Gozo M, McDowell EP, Levine RL, Doukas J, et al. Efficacy of TG101348, a selective JAK2 inhibitor, in treatment of a murine model of JAK2V617F-induced polycythemia vera. Cancer Cell. 2008;13:311–320. doi: 10.1016/j.ccr.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 51.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolpaw AJ, Shimada K, Skouta R, Welsch ME, Akavia UD, Pe'er D, Shaik F, Bulinski JC, Stockwell BR. Modulatory profiling identifies mechanisms of small molecule-induced cell death. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E771–E780. doi: 10.1073/pnas.1106149108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamanishi Y, Araki M, Gutteridge A, Honda W, Kanehisa M. Prediction of drug-target interaction networks from the integration of chemical and genomic spaces. Bioinformatics. 2008;24:i232–i240. doi: 10.1093/bioinformatics/btn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, Cheah JH, Clemons PA, Shamji AF, Clish CB, et al. Regulation of Ferroptotic Cancer Cell Death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhan Q, Antinore MJ, Wang XW, Carrier F, Smith ML, Harris CC, Fornace AJ., Jr Association with Cdc2 and inhibition of Cdc2/Cyclin B1 kinase activity by the p53-regulated protein Gadd45. Oncogene. 1999;18:2892–2900. doi: 10.1038/sj.onc.1202667. [DOI] [PubMed] [Google Scholar]
- 56.Zhang QC, Petrey D, Deng L, Qiang L, Shi Y, Thu CA, Bisikirska B, Lefebvre C, Accili D, Hunter T, et al. Structure-based prediction of protein-protein interactions on a genome-wide scale. Nature. 2012a;490:556–560. doi: 10.1038/nature11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang S, Liu X, Bawa-Khalfe T, Lu LS, Lyu YL, Liu LF, Yeh ET. Identification of the molecular basis of doxorubicin-induced cardiotoxicity. Nat Med. 2012b;18:1639–1642. doi: 10.1038/nm.2919. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.