Abstract
Organisms are adapted to the relentless cycles of day and night, because they evolved timekeeping systems called circadian clocks, which regulate biological activities with ~24-h rhythms. The clock of cyanobacteria is driven by a three-protein oscillator comprised of KaiA, KaiB, and KaiC, which together generate a circadian rhythm of KaiC phosphorylation. We show that KaiB flips between two distinct three-dimensional folds, and its rare transition to an active state provides a time delay that is required to match the timing of the oscillator to that of earth’s rotation. Once KaiB switches folds, it binds phosphorylated KaiC and captures KaiA, initiating a phase transition of the circadian cycle, and regulates components of the clock-output pathway, providing the link that joins the timekeeping and signaling functions of the oscillator.
Endogenous circadian (~24 h) rhythms are found in diverse organisms, arising as an adaptation to the earth’s persistent cycles of night and day (1). To uncover the molecular mechanism of a circadian clock, we chose the cyanobacterial system because its oscillator can be reconstituted in vitro (2). The oscillator is composed of only three proteins KaiA, KaiB, and KaiC (3), which together generate a circadian rhythm of KaiC phosphorylation at residues S431 and T432 in the CII domain (4). KaiA promotes KaiC (auto)phosphorylation during the subjective day (4, 5), whereas KaiB provides negative feedback to inhibit KaiA (6, 7), promoting KaiC (auto)dephosphorylation during the subjective night. KaiB is also involved in regulating two antagonistic clock-output proteins – SasA (8) and CikA (9), which reciprocally control the master regulator of transcription, RpaA (10).
To determine the structure of KaiB in its KaiC-bound state, we used a monomeric variant of the KaiB-binding domain of KaiC, CI*, and a dimeric KaiB variant (11), KaiB*, with enhanced KaiC binding. Dimeric forms of free KaiB retain the same tertiary structure in crystals as tetrameric forms (12). Free KaiB has been shown by X-ray crystallography (13) to adopt a fold found in no other proteins (14), despite clear sequence similarity with the thioredoxin-like fold at the N-terminus of SasA, N-SasA (8). For structural studies we used proteins from Thermosynechococcus elongatus (denoted byte), because they are more stable than those from Synechococcus elongatus (15). For functional studies we used proteins from S. elongatus (denoted byse), the standard model for investigating in vivo circadian rhythms (16). Analytical ultracentrifugation experiments indicated that KaiBte* binds to CIte* as a monomer with a stoichiometric ratio of 1:1 (fig. S1A). Secondary chemical shifts of backbone resonances (17) of KaiBte* in a complex with CIte* (fig. S1) revealed a thioredoxin-like secondary structure (βαβαββα) (18), rather than the secondary structure of free KaiB (βαββααβ) found in protein crystals (Fig. 1A). Hereafter, we refer to the βαββααβ form of KaiB as the ground state (gsKaiB), and the βαβαββα state as fold switched (fsKaiB). Fewer than 10 proteins are known to switch reversibly between distinct folds under native conditions, and they are collectively known as metamorphic proteins (19). KaiB is the only metamorphic protein known to function in biological clocks.
Fig. 1. KaiB switches its fold to bind KaiC.
(A) Plots of chemical-shift based secondary structures of KaiBte*, KaiBte* + CIte*, and G89A, D91R-KaiBte* determined by TALOS+ (17). Unassigned proline and non-proline residues are indicated by small red and blue dots along the horizontal axis at y=0. The secondary structures of KaiBte and G89A, D91R-KaiBte*, are shown for comparison. KaiBte* residues Q52 – E56 in the KaiBte*–CIte* complex were not assignable, probably due to exchange broadening. Vertical dashed lines are visual guides separating the N terminal and C-terminal halves of KaiB. (B) Structural comparisons of KaiBte, G89A, D91R-KaiBte*, and N-SasAse. Residues K58, G89 and D91 are highlighted for their roles in fold switching.
Along a β strand, side chains typically alternate ↑↓↑↓⋯. In the β4 strand of gsKaiB, the side chain pattern is ↑↓–↑, where the dash is G89; in fsKaiB, G89 lies in the α3 helix. We reasoned that a G89A substitution would destabilize β4 in gsKaiB but not α3 in fsKaiB (Fig. 1B). A D91R substitution should also destabilize gsKaiB. NMR secondary chemical shift analysis revealed that, unlike KaiBte*, the two single-point mutants had populations of both gsKaiB and fsKaiB states, but the double mutant was ≥98% in the fsKaiB state (figs. S2–S6). A structural model of G89A, D91R-KaiBte* determined by CS-Rosetta (20) using chemical shifts and backbone amide 1HN–1HN nuclear Overhauser effects (NOEs) as restraints confirmed that G89A, D91R-KaiBte* adopted a thioredoxin-like fold (fig. S7), similar to that of N-SasA (21).
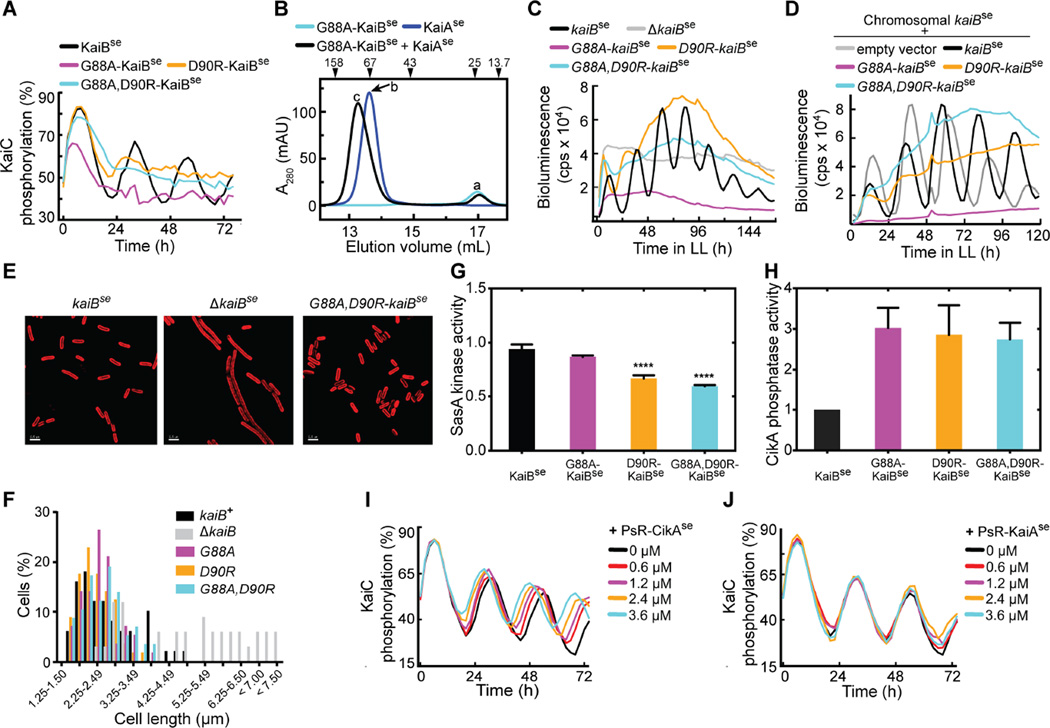
The corresponding KaiB variants in S. elongatus, G88A-KaiBse, D90R-KaiBse, and G88A, D90R-KaiBse also promoted the fsKaiB state relative to WT KaiBse (figs. S8 and S9), although to a lesser extent than in the corresponding T. elongatus variants. G88A, D90R-KaiBse formed a complex with CIse*, with near complete binding within 5 min (fig. S10). In contrast, WT KaiBse bound CIse* marginally, even after 24 h (fig. S11). In vitro oscillation assays showed that the KaiBse variants disrupted KaiCse phosphorylation rhythms (Fig. 2A, and fig. S12).
Figure 2. KaiB fold switching regulates oscillator function and clock output.
(A) In vitro KaiC phosphorylation assays using KaiCse, KaiAse, and KaiBse, G88A-KaiBse, D90R-KaiBse, or G88A, D90R-KaiBse. (B) Gel-filtration profiles of G88A-KaiBse, KaiAse, and G88A-KaiBse + KaiAse. Peaks a–c were analyzed by SDS-PAGE (fig. S13). (C) Bioluminescence from strains that carry a PkaiB luc reporter for circadian rhythmicity. Cells harbored kaiBse, G88AkaiBse, D90R-kaiBse, or G88A, D90R-kaiBse, or cells with kaiBse deletion. (D) Bioluminescence from strains that carry a PkaiB luc reporter expressing kaiBse, G88A-kaiBse, D90R-kaiBse, G88A, D90R-kaiBse, or empty vector, in addition to chromosomal kaiBse. (E) Representative micrographs of cells expressing kaiBse, lacking kaiBse, or harboring G88A, D90R-kaiBse. Cellular autofluorescence in red, scale bars=2.5 microns. (F) Histograms showing cell-length distributions of strains expressing kaiBse, ΔkaiBse, G88A-kaiBse, D90R-kaiBse, or G88A, D90R kaiBse as the only copy of kaiB. (G) SasAse kinase activities in the presence of S431E-KaiCse and KaiBse, G88A-KaiBse, D90R-KaiBse, or G88A, D90R-KaiBse. The mixtures were incubated for 2 hours before SasAse, RpaAse and [γ-32P]-labeled ATP were added. Relative kinase activities compare the mean steady-state amount of 32P-labeled RpaAse to that of a reaction of S431EKaiCse alone (n = 4, error bars denote SEM). One-way ANOVA gives p-value < 0.001, and **** denotes Bonferroni corrected p-values < 0.001 for pairwise comparisons against kinase activity with KaiBse (α = 0.05). (H) CikA phosphatase activity toward phosphorylated RpaA in the presence of KaiCse and KaiBse, G88A-KaiBse, D90R-KaiBse, or G88A, D90R-KaiBse (n = 4–5, error bars denote SEM). KaiCse alone or KaiBse alone did not activate CikA phosphatase activity (fig. S17). (I) In vitro KaiCse phosphorylation assays as a function of concentration of PsRCikAse. (J) Same as (I) except for using PsR-KaiAse instead of PsR-CikAse. The black curves in (I) and (J) are identical.
Amounts of KaiCse phosphorylation were higher in the presence of D90R-KaiBse or G88A, D90R-KaiBse than they were with G88A-KaiBse. G88A-KaiBse formed a complex with KaiAse (Fig. 2B), whereas the two KaiBse mutants containing D90R did not (fig. S13), indicating that although the D90R mutation promotes the fsKaiB state, it also disrupts binding. Higher amounts of unsequestered KaiA would be expected to lead to higher amounts of KaiC phosphorylation, accounting for the observed differences in the KaiC phosphorylation profiles when using the D90R mutants. In vivo bioluminescence rhythms from cyanobacterial luciferase reporter strains harboring kaiBse variants (Fig. 2C) were also disrupted, with phenotypes that closely matched the in vitro phosphorylation patterns (Fig. 2A). A chromosomal copy of kaiB+ in addition to the mutant kaiB variants did not restore bioluminescence rhythms in vivo, indicating a dominant-negative effect of the fold-switch mutations (Fig. 2D and fig. S14). KaiB fold switch-stabilizing mutants are less abundant in vivo than WT KaiB (fig. S15), so at equilibrium in vivo fsKaiB is probably rare.
Although KaiBse variants disrupted rhythms in vitro and in vivo, each of them restored the celllength phenotype in kaiB− strains (Fig. 2E and F) in which the SasA-RpaA output pathway was hyperstimulated and cell division was inhibited (22). These functional variants indicate that fsKaiB regulates the clock-output enzymes SasA (8) and CikA (9, 22). SasA is activated when it binds KaiC (23), and SasA and KaiB compete for the CI domain of KaiC (24). Preincubation with the D90R- and G88A, D90R-KaiBse variants inhibited the ability of S431E-KaiCse (a KaiC variant that mimics phosphorylation at the S431 residue) to trigger output signaling through SasAse (Fig. 2G, and fig. S16). Also, mixtures of KaiB and KaiC activate the phosphatase activity of CikA that dephosphorylates RpaA (25). Relative to WT KaiBse, fsKaiBse variants enhanced CikAse phosphatase activity by approximately three-fold in vitro (Fig. 2H and fig. S17), and suppressed RpaAse phosphorylation in vivo (fig. S18). We propose that fsKaiB forms a complex with KaiC that both activates signaling through CikA and inhibits signaling through SasA by outcompeting SasA for binding to KaiC. It is likely that fsKaiB interacts with the CikA pseudo-receiver domain (PsR-CikA), because adding PsR-CikA to the in vitro oscillator shortened the period and reduced the amplitude (Fig. 2I and fig. S19). Overexpression of just the PsR-CikA domain in cyanobacteria similarly shortened the period of bioluminescence rhythms (26). Interaction with PsR-CikA was detected for G88A, D90R-KaiBse, but not KaiBse (fig. S20). Addition of the PsR domain of KaiA did not affect phosphorylation rhythms (Fig. 2J), even though it has the same tertiary structure as PsR-CikA (5, 27).
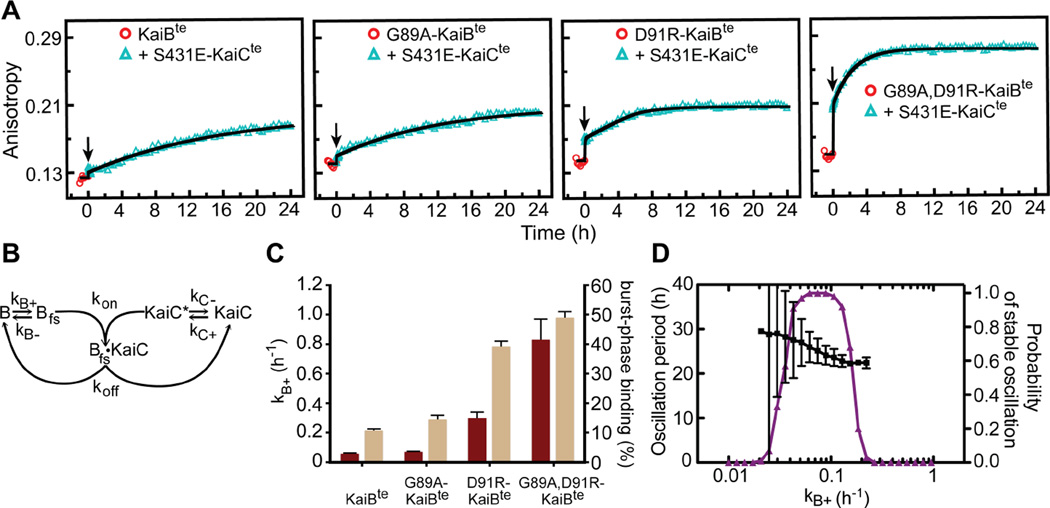
fsKaiB/gsKaiB equilibrium constants, K = kB+/kB−, were estimated by fitting the kinetics of binding of KaiBte variants to CIte* (fig. S21). Equilibrium constants were larger for G89A-KaiBte (K = 0.13 ± 0.02), D91R-KaiBte (1.2 ± 0.1), and G89A, D91R-KaiBte (6.7 ± 0.4), relative to KaiBte (0.08 ± 0.01). For KaiBte variants binding S431E-KaiCte, a distinct fast-binding phase was followed by slow binding (Fig. 3A). These multiphase binding kinetics were reproduced in a gsKaiB ⇔ fsKaiB fold-switching model (Fig. 3B), in which KaiB was initially at equilibrium between the two folds. The pool of fsKaiB bound rapidly upon adding S431E-KaiC (Fig. 3C), followed by a slow gsKaiB → fsKaiB population shift. As shown by our computational model (Fig. 3B and fig. S22), the slow formation of the KaiB-KaiC complex contributes to the delay that allows a population of KaiC proteins to become highly phosphorylated under continued stimulation by KaiA. Increasing the rate of KaiB-KaiC binding in our model causes the in silico phosphorylation rhythm to fail (Fig. 3D), similar to that observed in vitro. Comparing the kinetics of binding to CI (fig. S21) and full-length KaiC (Fig. 3A) by KaiB mutants shows that full-length KaiC contributes to the slow phase as well, and is likely due to the CI ATPase (28) exposing the KaiB-binding site upon its activation by CI-CII ring stacking (11).
Figure 3. KaiB fold switching regulates slow formation of the KaiB-KaiC complex.
(A) Fluorescence anisotropies of 6-IAF (6-iodoacetamidofluorescein)-labeled KaiBte, G89AKaiBte, D91R-KaiBte, and G89A, D91R-KaiBte in the presence of S431E-KaiCte. KaiB samples were incubated for 1-h (circles) before addition (arrow) of S431E-KaiCte. A54C mutation was introduced to all KaiB for fluorescence labeling. (B) Scheme for modeling. (C) Forward fold-switching rate constants, kB+ (maroon), and burst-phase binding to S431E-KaiCte (tan). Burst-phase binding, defined as the percentage of KaiBte-S431E-KaiCte complexes formed at t = 0.1 h in the model relative to steady state binding at t = 24 h, were derived from fitting data after adding S431E-KaiCte in (A), to the model shown in (B). Burst-phase error bars show the standard deviation from model calculations by bootstrap resampling the raw data (n = 20). kB+ values used in these fits were pre-determined from analysis of the kinetics of binding of KaiBte variants to the isolated CIte* domain (fig. S21), a condition where we assumed the rate-limiting step in complex formation is due only to KaiB fold switching. Error bars for kB+ were estimated by bootstrap resampling the original dataset 500 times. (D) Mathematical modeling of KaiC phosphorylation period (black) and probability of stable oscillation (purple) as a function of, kB+. The black bars indicate the standard deviation in the model period from 100 oscillator calculations at each value of kB+, with the other parameters randomly varied as described in Supplementary Materials.
N-SasA, which also adopts a thioredoxin-like fold (21), competes with KaiB for binding KaiC (24) (fig. S23). Intermolecular distances for the complexes CIte*-G89A, D91R-KaiBte* and CIte*- N-SasAte (figs. S24 and S25) were measured using the pulsed electron paramagnetic resonance method of double electron-electron resonance (DEER), in combination with mutagenesis studies (figs. S26–S29). The higher quality DEER data for the CIte*-N-SasAte complex allowed structural modeling (Fig. 4A, and figs. S30 and S31). In this model, the α2 helix of N-SasA binds to the B-loop region of the CI domain (residues A109 – D123), with additional interactions involving the CI α-helix that follows. Hydrogen/deuterium exchange mass spectrometry (HDXMS) data on both complexes (Fig. 4A and B, and figs. S32 to S35) also suggest that this region of KaiC is a common binding site for KaiB and SasA. This model is consistent with a report that truncation of the B-loop abolished binding (24). B-loop truncation also restored the cell-length phenotype of a strain that lacked kaiB (fig. S36). Furthermore, a F121A mutation in the B-loop of full-length KaiC (F121A-S431E-KaiCse) abolished KaiC binding to full-length SasAse, KaiBse, and G88A, D90R-KaiBse (figs. S37–S40). HDX-MS data (Fig. 4A and B) indicated that N-SasA and KaiB induce long-range perturbations in isolated CI domains that, in the context of fulllength KaiC, may affect intersubunit interactions. Another HDX-MS derived model (29) indicated that KaiB most likely binds to CII, based on a calculation minimizing the docking energy between KaiC and gsKaiB, not fsKaiB.
Figure 4. KaiB and SasA bind to similar sites on CI.
(A) EPR-restrained model of the CIte*-N-SasAte complex. The HADDOCK model of the complex with the best score is superimposed on the crystal structure of KaiCte (PDB ID: 4o0m). (B) Qualitative structural model of the interaction of CIte* and fsKaiB (G89A, D91R-KaiBte*), based on HDX-MS data and mutagenesis. Dark blue and cyan spheres represent CI residues whose mutations strongly or moderately weaken binding, respectively. Dark blue and cyan ribbons represent protection against H/D exchange upon complex formation that are >1.5 and 0.5 – 1.5 standard deviations above the average, respectively, as determined by HDX-MS (figs. S32–S35).
Naturally occurring variations at residue positions 58, 89, and 91 of KaiB (fig. S41A) suggest that the ancestral KaiB protein had the fsKaiB thioredoxin-like fold, and the gsKaiB fold evolved later with the circadian clock. In support of this notion, a homolog of KaiB from Legionella pneumophila, with no known circadian rhythms, has an alanyl residue at position 89, and crystallizes in the fsKaiB fold (30) (fig. S41B). Rare excursions of KaiB between two distinct folds are essential for a robust circadian period, and reciprocally regulate mutually antagonistic clock output-signaling pathways (Fig. 5).
Figure 5. Model of KaiB fold switching as linchpin for the cyanobacterial clock.
Excursion of KaiB to the rare fold-switch state causes fsKaiB to displace SasA for binding to KaiC. KaiC-stabilized fsKaiB captures KaiA, initiating the dephosphorylation phase of the cycle. These aspects control oscillator period. CikA and KaiA compete for binding to fsKaiB, further linking oscillator function related to KaiA and output activity via CikA-mediated dephosphorylation of RpaA. The competitive interactions of fsKaiB with SasA, and KaiA with CikA, implicate “output components” CikA and SasA as parts of an extended oscillator.
Supplementary Material
Acknowledgments
We thank R. Greenspan, J.-P. Changeux, M. Paddock, Y. Shen, R. Peterson, S. Chou, and N.-W. Kuo for discussions, and A. Chavan for figure preparation. Work was supported by AFOSR grant 13RSL012 and NIH grant GM107521 to A.L., a Burroughs-Wellcome Career Award at the Scientific interface to M.J.R., NIH grants GM100116 and GM062419 to S.S.G., AI081982 and AI101436 for support of the UCSD DXMS Laboratory, American Cancer Society Postdoctoral Fellowship PF-12-262-01-MPC to S.E.C., and NSF Graduate Research Fellowship to R.T. The data reported in this paper are tabulated in the Supporting Online Material.
References and Notes
- 1.Dunlap JC. Molecular basis for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 2.Nakajima M, et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 3.Ishiura M, et al. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 4.Iwasaki H, Nishiwaki T, Kitayama Y, Nakajima M, Kondo T. KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc. Natl. Acad. Sci. USA. 2002;99:15788–15793. doi: 10.1073/pnas.222467299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams SB, Vakonakis I, Golden SS, LiWang A. Structure and function from the circadian clock protein KaiA of Synechococcus elongatus: A potential clock input mechanism. Proc. Natl. Acad. Sci. USA. 2002;99:15357–15362. doi: 10.1073/pnas.232517099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kitayama Y, Iwasaki H, Nishiwaki T, Kondo T. KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J. 2003;22:2127–2134. doi: 10.1093/emboj/cdg212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Y, Mori T, Johnson CH. Cyanobacterial circadian clockwork: roles of KaiA, KaiB and the kaiBC promoter in regulating KaiC. EMBO J. 2003;22:2117–2126. doi: 10.1093/emboj/cdg168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iwasaki H, et al. A KaiC-interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell. 2000;101:223–233. doi: 10.1016/S0092-8674(00)80832-6. [DOI] [PubMed] [Google Scholar]
- 9.Schmitz O, Katayama M, Williams SB, Kondo T, Golden SS. CikA, a bacteriophytochrome that resets the cyanobacterial circadian clock. Science. 2000;289:765–768. doi: 10.1126/science.289.5480.765. [DOI] [PubMed] [Google Scholar]
- 10.Markson J, Piechura J, Puszynska A, O'Shea E. Circadian Control of Global Gene Expression by the Cyanobacterial Master Regulator RpaA. Cell. 2013;155:1396–1408. doi: 10.1016/j.cell.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang Y, Tseng R, Kuo N, LiWang A. Rhythmic ring-ring stacking drives the circadian oscillator clockwise. Proc. Natl. Acad. Sci. USA. 2012;109:16847–16851. doi: 10.1073/pnas.1211508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garces RG, Wu N, Gillon W, Pai EF. Anabaena circadian clock proteins KaiA and KaiB reveal a potential common binding site to their partner KaiC. EMBO J. 2004;23:1688–1698. doi: 10.1038/sj.emboj.7600190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Villarreal SA, et al. CryoEM and Molecular Dynamics of the Circadian KaiB–KaiC Complex Indicates That KaiB Monomers Interact with KaiC and Block ATP Binding Clefts. J. Mol. Biol. 2013;425:3311–3324. doi: 10.1016/j.jmb.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vakonakis I, et al. NMR structure of the KaiC-interacting C-terminal domain of KaiA, a circadian clock protein: implications for the KaiA-KaiC interaction. Proc. Natl. Acad. Sci. USA. 2004;101:1479–1484. doi: 10.1073/pnas.0305516101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong G, Kim Y, Golden SS. Simplicity and complexity in the cyanobacterial circadian clock mechanism. Curr. Opin. Genet. Dev. 2010;20:619–625. doi: 10.1016/j.gde.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin JL. Thioredoxin - a fold for all reasons. Structure. 1995;3:245–250. doi: 10.1016/s0969-2126(01)00154-x. [DOI] [PubMed] [Google Scholar]
- 19.Murzin AG. Metamorphic Proteins. Science. 2008;320:1725–1726. doi: 10.1126/science.1158868. [DOI] [PubMed] [Google Scholar]
- 20.Raman S, et al. NMR Structure Determination for Larger Proteins Using Backbone-Only Data. Science. 2010;327:1014–1018. doi: 10.1126/science.1183649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vakonakis I, Klewer DA, Williams SB, Golden SS, LiWang AC. Structure of the N-terminal Domain of the Circadian Clock-associated Histidine Kinase SasA. J. Mol. Biol. 2004;342:9–17. doi: 10.1016/j.jmb.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 22.Dong G, et al. Elevated ATPase Activity of KaiC Applies a Circadian Checkpoint on Cell Division in Synechococcus elongatus. Cell. 2010;140:529–539. doi: 10.1016/j.cell.2009.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith RM, Williams SB. Circadian rhythms in gene transcription imparted by chromosome compaction in the cyanobacterium Synechococcus elongatus. Proc. Natl. Acad. Sci. USA. 2006;103:8564–8569. doi: 10.1073/pnas.0508696103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tseng R, et al. Cooperative KaiA–KaiB–KaiC Interactions Affect KaiB/SasA Competition in the Circadian Clock of Cyanobacteria. J. Mol. Biol. 2014;426:389–402. doi: 10.1016/j.jmb.2013.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gutu A, O’Shea E. Two Antagonistic Clock-Regulated Histidine Kinases Time the Activation of Circadian Gene Expression. Mol. Cell. 2013;50:288–294. doi: 10.1016/j.molcel.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, Dong G, Golden SS. The pseudo-receiver domain of CikA regulates the cyanobacterial circadian input pathway. Mol. Microbiol. 2006;60:658–668. doi: 10.1111/j.1365-2958.2006.05138.x. [DOI] [PubMed] [Google Scholar]
- 27.Gao T, Zhang X, Ivleva NB, Golden SS, LiWang A. NMR structure of the pseudo-receiver domain of CikA. Protein Sci. 2007;16:465–475. doi: 10.1110/ps.062532007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phong C, Markson JS, Wilhoite CM, Rust MJ. Robust and tunable circadian rhythms from differentially sensitive catalytic domains. Proc. Natl. Acad. Sci. USA. 2013;110:1124–1129. doi: 10.1073/pnas.1212113110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Snijder J, et al. Insight into cyanobacterial circadian timing from structural details of the KaiB–KaiC interaction. Proc. Natl. Acad. Sci. USA. 2014;111:1379–1384. doi: 10.1073/pnas.1314326111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loza-Correa M, et al. The Legionella pneumophila kai operon is implicated in stress response and confers fitness in competitive environments. Environ. Microbiol. 2013;16:359–381. doi: 10.1111/1462-2920.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.