Summary
The tumor suppressor BRCA2 is thought to facilitate the handoff of ssDNA from replication protein A (RPA) to the RAD51 recombinase during DNA break and replication fork repair by homologous recombination. However, we find that RPA-RAD51 exchange requires the BRCA2 partner DSS1. Biochemical, structural, and in vivo analyses reveal that DSS1 allows the BRCA2-DSS1 complex to physically and functionally interact with RPA. Mechanistically, DSS1 acts as a DNA mimic to attenuate the affinity of RPA for ssDNA. A mutation in the solvent-exposed acidic domain of DSS1 compromises the efficacy of RPA-RAD51 exchange. Thus, by targeting RPA and mimicking DNA, DSS1 functions with BRCA2 in a two-component homologous recombination mediator complex in genome maintenance and tumor suppression. Our findings may provide a paradigm for understanding the roles of DSS1 in other biological processes.
Graphical Abstract
Introduction
Homologous recombination (HR) is a conserved, generally error-free mechanism for the elimination of DNA double-strand breaks (DSBs). Moreover, HR is essential for successful navigation through S phase, wherein it restores replication forks that have stalled or encountered a lesion. As such, HR is essential for maintaining genome integrity and its dysfunction leads to disease, cancer in particular (Hoeijmakers, 2009; Negrini et al., 2010).
DNA joint formation during HR is mediated by the RAD51 recombinase. RAD51 forms a right-handed helical filament (presynaptic filament) on ssDNA generated by nucleolytic resection of a primary lesion, e.g. a DSB (San Filippo et al., 2008; Symington and Gautier, 2011). In vitro and in cells, presynaptic filament assembly is a rate-limiting step. This stems from the slow nucleation of RAD51 onto ssDNA, thus rendering presynaptic filament assembly prone to interference by the abundant ssDNA binding protein RPA. Inhibition posed by RPA is relieved by recombination mediator proteins, most notably, yeast Rad52 and human BRCA2 (San Filippo et al., 2008). While the function of yeast Rad52 has been defined (San Filippo et al., 2008), much remains to be learned regarding the mechanism of BRCA2.
BRCA2 deficient cells are hypersensitive to genotoxic agents and replication stress (Holloman et al., 2008; San Filippo et al., 2008). Recently, BRCA2 has been implicated in protecting perturbed DNA replication forks against nucleolytic attrition (Hashimoto et al., 2010; Schlacher et al., 2011). Individuals with BRCA2 mutations exhibit genomic instability and are predisposed to breast, ovarian, and other cancers (Gayther et al., 1997; Wooster et al., 1995). BRCA2 interacts with RAD51 through eight BRC (Breast Cancer) repeats and a region located at its C-terminus (Esashi et al., 2005; San Filippo et al., 2008; Wong et al., 1997). BRCA2 also possesses a DNA binding domain (DBD), consisting of three OB (oligonucleotide binding) folds, named OB1, OB2, and OB3 (Yang et al., 2002). BRCA2 or a polypeptide that harbors two BRC repeats and the DBD is able to enhance RAD51 presynaptic assembly on RPA-coated ssDNA, leading to the suggestion that BRCA2 promotes RPA-RAD51 exchange (Jensen et al., 2010; Liu et al., 2010; San Filippo et al., 2006). However, unlike yeast Rad52 (Seong et al, 2008 and references therein), BRCA2 does not bind RPA (Jensen et al., 2010), so it remains possible that a BRCA2 partner targets RPA. Herein, we show that the BRCA2-associated DSS1 protein mediates RPA interaction and functions as a DNA mimic to promote RPA-RAD51 exchange on ssDNA.
DSS1, a candidate gene for split hand/split foot syndrome (Crackower et al., 1996; Ignatius et al., 1996), is a biomarker for different cancers (Ma et al., 2013; Rezano et al., 2013; Wei et al., 2003). It is small (harboring 70 residues) and highly acidic. Like BRCA2, DSS1 is crucial for DSB and replication fork repair (Gudmundsdottir et al., 2004; Jeyasekharan et al, 2013; Kojic et al., 2003; Kristensen et al., 2010; Li et al., 2006; Pispa et al., 2008). Interestingly, DSS1 and its orthologs play an important role in other biological processes, including proteasome assembly and various aspects of RNA metabolism (Garncarz et al., 2013; Pick et al., 2009). S. cerevisiae Dss1 acts as a chaperone in proteasome assembly (Tomko and Hochstrasser, 2014) and S. pombe Dss1 functions as a ubiquitin-binding subunit of the proteasome (Paraskevopoulos et al, 2014). However, the mechanisms by which DSS1 mediates HR other biological processes are unknown.
DSS1 associates with OB1 and an adjoining helical region in BRCA2 (Marston et al., 1999; Yang et al., 2002). As revealed by X-ray crystallography, many acidic residues are located in a solvent-exposed loop in DSS1. Importantly, by a combination of biochemical, structural, and cell-based analyses, we show that DSS1 targets RPA via this solvent-exposed acidic loop domain and acts as a DNA mimic to promote the assembly of the RAD51 presynaptic filament. Thus, DSS1 is an integral, indispensable component of a two-subunit HR mediator complex with BRCA2.
Results
Expression and purification of BRCA2 proteins
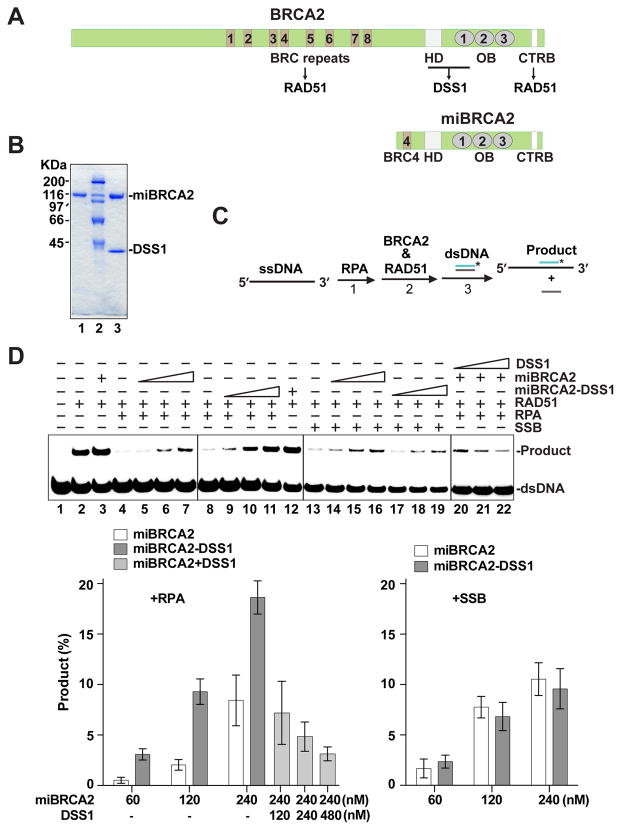
The expression and purification of BRCA2 protein had remained an insurmountable challenge until recently, when mammalian cell- and yeast cell-based systems were developed to achieve this goal (Jensen et al., 2010; Liu et al., 2010; Thorslund et al., 2010). Working independently, we have constructed baculoviruses for the expression of full-length BRCA2 and also complexes of BRCA2 with wild type or mutant DSS1 in insect cells, and devised a simple method for their purification to near homogeneity (see below). Importantly, the initial characterization of DSS1 was performed with a functional BRCA2 multi-domain construct, termed mini-BRCA2 (miBRCA2), which harbors the BRC4 repeat, the DBD, and the C-terminal domain, and which retains the ability to form a stable complex with either wild type or mutant DSS1 (Figure 1A & B). The major findings were then verified using full-length BRCA2 alone or in complex with wild type or mutant DSS1 (see below).
Figure 1. Enhancement of miBRCA2 activity by DSS1.
See also Figure S1.
(A) Schematic of BRCA2 and miBRCA2 with various functional domains.
(B) SDS-PAGE of purified miBRCA2 (lane 1) and miBRCA2-DSS1 complex (lane 3). Size markers were run in lane 2.
(C) Schematic of the homologous DNA pairing reaction.
(D) The miBRCA2-DSS1 complex or miBRCA2 without and with DSS1 tested in homologous pairing reactions with ssDNA coated with RPA or SSB. The mean values (± s.d.) of data from 3 independent experiments were plotted.
Enhancement of presynaptic filament assembly by DSS1
We used a well-established homologous DNA pairing assay (Figure 1C) to test miBRCA2 and the miBRCA2-DSS1 complex (San Filippo et al., 2006). As expected, homologous pairing by RAD51 was suppressed upon preincubation of the ssDNA substrate with RPA (Figure 1D; lanes 2 and 4). This inhibitory effect of RPA was alleviated upon the addition of an increasing amount of miBRCA2 (Figure 1D; lane 5–7). Importantly, the miBRCA2-DSS1 complex proved to be much more effective than miBRCA2 in the restoration of homologous pairing efficiency (Figure 1D; lanes 9–11).
We next applied a magnetic bead-based pulldown assay to monitor the loading of RAD51 onto ssDNA (see Figure S1A for schematic). RAD51 loading became strongly inhibited when ssDNA was pre-incubated with RPA (Figure S1B). Consistent with results from the homologous DNA pairing assay, even though both miBRCA2 and miBRCA2-DSS1 were able to restore RAD51-ssDNA association, the latter was much more adept in this regard (Figure S1B). Finally, we used electron microscopy (EM) to provide physical evidence that miBRCA2 and miBRCA2-DSS1 act by re-establishing RAD51 presynaptic filament assembly (Figure S1C). The EM results also revealed a greater efficacy of miBRCA2-DSS1 compared to miBRCA2 in restoring RAD51-ssDNA association (Figure S1C).
DSS1 alone is devoid of recombination mediator activity (data not shown). Moreover, we observed that (i) purified DSS1 fails to associate with miBRCA2 (data not shown) and (ii) co-addition of separately purified DSS1 with miBRCA2 does not enhance the activity of the latter (Figure 1D).
Overall, the above results suggested that DSS1 plays a critical role in presynaptic filament assembly in conjunction with stably associated BRCA2, a premise that was independently verified with the use of full-length BRCA2 in complex with DSS1 (see below). Consistent with what we observed with miBRCA2 and DSS1, we note that in order to assemble a stable BRCA2-DSS1 complex, the two proteins need to be co-expressed in insect cells (see below).
Specificity of the recombination mediator activity of BRCA2-DSS1
Preincubation of the ssDNA template with the E. coli single-strand DNA binding protein SSB also led to a strong suppression of RAD51-mediated homologous pairing (Figure 1D). As reported (San Filippo et al., 2006), miBRCA2 is adept at overcoming the inhibitory effect of SSB (Figure 1D). However, we found that the miBRCA2-DSS1 complex is no more effective than miBRCA2 in the restoration of homologous pairing efficiency (Figure 1D). Thus, DSS1 appears to facilitate the RAD51-mediated homologous pairing reaction by targeting human RPA but not E. coli SSB.
DSS1-RPA interaction and attenuation by DSS1 of RPA ssDNA binding
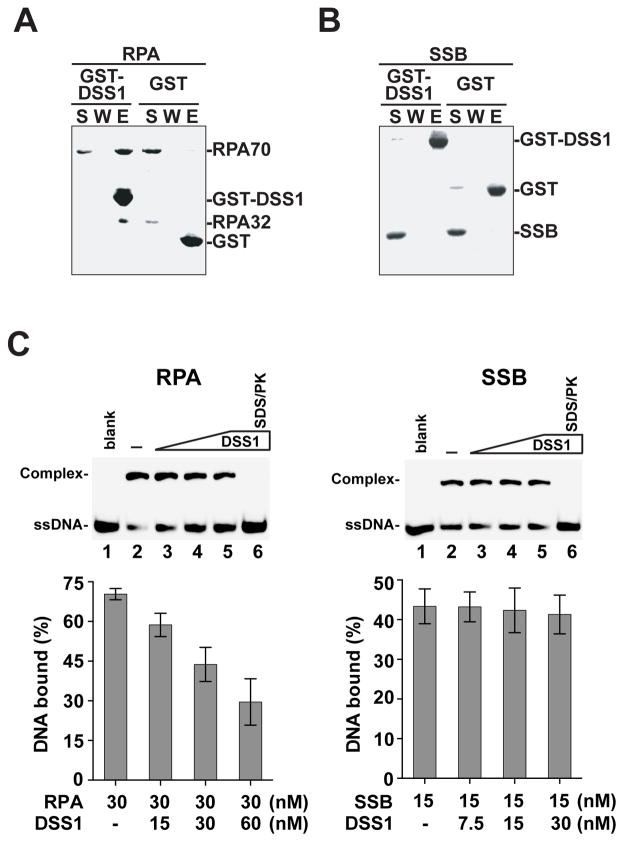
DSS1 does not bind ssDNA or dsDNA, nor does it interact with RAD51 (data not shown). We have also verified that DSS1 does not interfere with miBRCA2-RAD51 interaction (Figure S1D). Instead, the results above provide evidence that DSS1, in a specific fashion, targets the BRCA2-RAD51 complex to RPA-coated ssDNA. Indeed, by affinity pulldown, we found that (i) DSS1 physically interacts with RPA, but not with E. coli SSB (Figure 2A & B), (ii) only when miBRCA2 is complexed with DSS1 is it able to interact with RPA (Figure S2A), and (iii) DSS1 can also associate with the RPA-ssDNA complex (Figure S2B).
Figure 2. DSS1-mediated attenuation of ssDNA binding by RPA.
See also Figure S2 and Figure S4.
(A) GST-pulldown assay to test for the interaction of RPA with DSS1. The supernatant (S), the wash (W), and the eluate (E) fractions were analyzed.
(B) Pulldown assay as in (A) to test for interaction of DSS1 with E. coli SSB.
(C) The effect of DSS1 on DNA binding by RPA or SSB was examined. The mean values (± s.d.) of data from 3 independent experiments were plotted. Note treatment with SDS and proteinase K (SDS/PK) released DNA from RPA or SSB.
Since DSS1 directly interacts with RPA and the RPA-ssDNA complex, we inquired whether it impacts RPA binding of ssDNA. To this end, we used a DNA mobility shift assay with E. coli SSB as control. The experiments revealed that DSS1 weakens the ability of RPA to bind 30-mer oligo dT (dT30) either when RPA is free or pre-bound to the DNA (Figure 2C and Figure S4C & D), but does not affect the activity of SSB (Figure 2C). Taken together, these results provide evidence for a dual role of DSS1 in the HR reaction, namely, to deliver BRCA2-RAD51 to RPA-bound ssDNA and also to attenuate the ssDNA binding affinity of RPA.
Targeting of RPA70 by DSS1
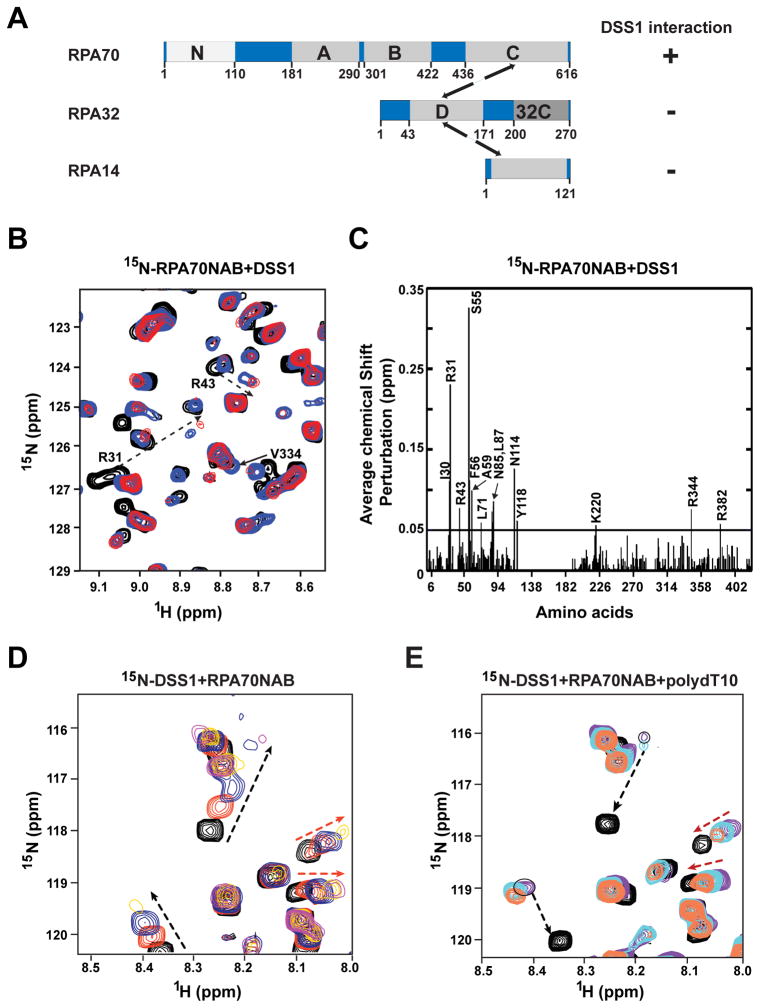
Next, we sought to understand the nature of the DSS1-RPA complex and the mechanism by which DSS1 attenuates ssDNA binding by RPA. RPA consists of three subunits: RPA70, RPA32, and RPA14 (Figure 3A). RPA70 harbors a protein interaction module at its N-terminus (RPA70N), and three of the four DNA binding OB fold domains (designated RPA70A, B, and C) of RPA. The fourth DNA binding OB fold domain resides in RPA32 (RPA32D), and RPA32 also possesses a protein interaction module at its C-terminus (RPA32C).
Figure 3. Evidence for RPA70 interaction activity and DNA mimicry in DSS1.
See also Figure S2 and Figure S3.
(A) Schematic of the RPA subunits RPA70, RPA32, and RPA14 with functional domains.
(B) Overlay of 15N-1HSQC spectra of 15N-enriched RPA70NAB as the solution was titrated with DSS1. The circles identify peaks belonging to RPA70N. The solid and dotted arrows indicate significant chemical shift perturbations upon binding of DSS1 for residues within RPA70AB and 70N, respectively.
(C) Chemical shift changes plotted against the sequence of RPA70NAB. The mean value of chemical shift changes is shown by the solid horizontal line.
(D) Overlay of 15N-1HSQC spectra of 15N-enriched DSS1 in complex with unlabeled RPA70NAB at molar ratios of 1:0 (black), 1:0.2 (red), 1:0.4 (blue), and 1:1 (yellow). The black and red dotted arrows identify peaks with chemical shift preturbations arsing from interaction with 70N and with 70AB, respectively.
(E) Overlay of 15N-1HSQC spectra of 15N-enriched DSS1 in complex with unlabeled RPA70NAB and ssDNA. Spectra were acquired at DSS1:RPA70NAB:ssDNA ratios of 1:0:0 (black), 1:1:0 (purple), 1:1:0.5 (cyan) and 1:1:1 (red) molar ratios. Red arrows identify two peaks that revert to the position of free DSS1 upon addition of ssDNA due to release of DSS1 by ssDNA binding to RPA70AB. Black arrows identify two peaks that are not affected by addition of ssDNA because they interact with RPA70N.
Using an affinity pulldown assay, we examined possible interactions of GST-tagged DSS1 with a range of RPA domain constructs (Figure S2C). The results revealed an affinity of DSS1 for RPA70N, RPA70A, RPA70B, RPA70AB, RPA70NAB and RPA70C-RPA32D-RPA14, but not for RPA32C or RPA32D-RPA14. Thus, DSS1 associates with RPA primarily if not exclusively through the RPA70 subunit via multiple interactions with its N, A, B, and C domains (Figure 3A). Using isothermal titration calorimetry (ITC), we verified that DSS1 binds RPA70N with a 1:1 stoichiometry and determined a Kd of ~23 μM (Figure S2D). Consistent with results from the affinity pulldown, ITC measurements revealed that RPA70NAB has a higher affinity for DSS1 than RPA70N alone (Kd ~ 3 μM) (Figure S2D).
DNA mimicry as revealed by NMR analysis
To obtain insights into the DSS1-RPA70 interaction, we performed NMR chemical shift perturbation experiments. Changes in NMR chemical shifts of RPA70N induced by the binding of DSS1 were determined from 15N-1H HSQC spectra of 15N-enriched RPA70N recorded as the solution was titrated with unlabeled DSS1. Significant chemical shift perturbations were observed over the course of the titration to a 1:1 molar ratio (Figure S3A). The observation of only a limited number of perturbations in the spectra indicates there is a specific interaction interface between RPA70N and DSS1. The largest effects were observed for RPA70N residues: I30, R31, R43, L45, S55, F56, L58, A59, L87, K88, R92, V93, N114, Y118, and E120 (Figure S3B).
A similar NMR analysis was performed for both RPA70AB and RPA70NAB. As for RPA70N, titration with DSS1 induced a select number of perturbations in RPA70AB (Figure S3C). However, the interaction between DSS1 and RPA70AB was noticeably weaker (Figure S3C & D). The titration of DSS1 into 15N-labeled 70NAB gave rise to a substantial number of peak perturbations, as expected based on DSS1 binding to both RPA70N and RPA70AB (Figure 3B). In addition to chemical shift changes, many of the perturbed peaks began to lose their intensity over the course of titration to a 1:1 molar ratio (Figure 3B). The reduction in peak intensity is attributed to line broadening arising from a shift from independent tumbling of RPA70N and RPA70AB to coordinated tumbling of all three domains through their mutual binding to DSS1. Coordinated binding of all three domains is also reflected in the magnitude of the chemical shift perturbations, which are larger for DSS1 binding to RPA70NAB than for binding to the individual RPA70N and RPA70AB domains. The differences are most evident for the residues with the largest perturbations (I30, R31, R43, S55, F56, A59, L71, N85, L87, N114, Y118, K220, R344, and R382) (Figure 3C). The observation of stronger binding of DSS1 by the multi-valent RPA70NAB is anticipated on the basis of the linkage (chelation) effect (Stauffer and Chazin, 2004).
Mapping the perturbed residues onto the crystal structure of 70N revealed that DSS1 binds in the basic cleft of 70N (Figure S3E, left panel). Similarly, the residues of 70AB that were significantly perturbed by DSS1 map to both basic clefts, which in these domains are also the site of ssDNA binding (Figure S3E, right panel). To confirm that DSS1 interacts in the DNA binding site, we examined whether ssDNA competes for DSS1 binding to 70AB using the NMR chemical shift perturbation approach. For this experiment, we monitored the chemical shift changes in the 15N-1H HSQC spectrum of DSS1 upon titration first with unlabeled RPA70NAB (Figure 3D), and then with ssDNA (Figure 3E). As expected, the binding of RPA70NAB induces chemical shift perturbations in the spectrum of DSS1 in the first phase of the titration. In the second phase of the titration, the peaks that were perturbed due to 70AB binding reverted to their original position (Figure 3E). These data indicate that DSS1 associates with RPA70AB in a manner that mimics its binding of ssDNA. We also observed that the changes in DSS1 peaks associated with 70N binding were largely unchanged in the second phase of the titration upon the addition of ssDNA (Figure 3E), consistent with the negligible ssDNA binding affinity of RPA70N. Overall, the above results provide evidence that DSS1 utilizes a DNA mimicry mechanism to drive its interactions with the ssDNA binding OB folds of RPA70.
DSS1 mutants defective in RPA-binding
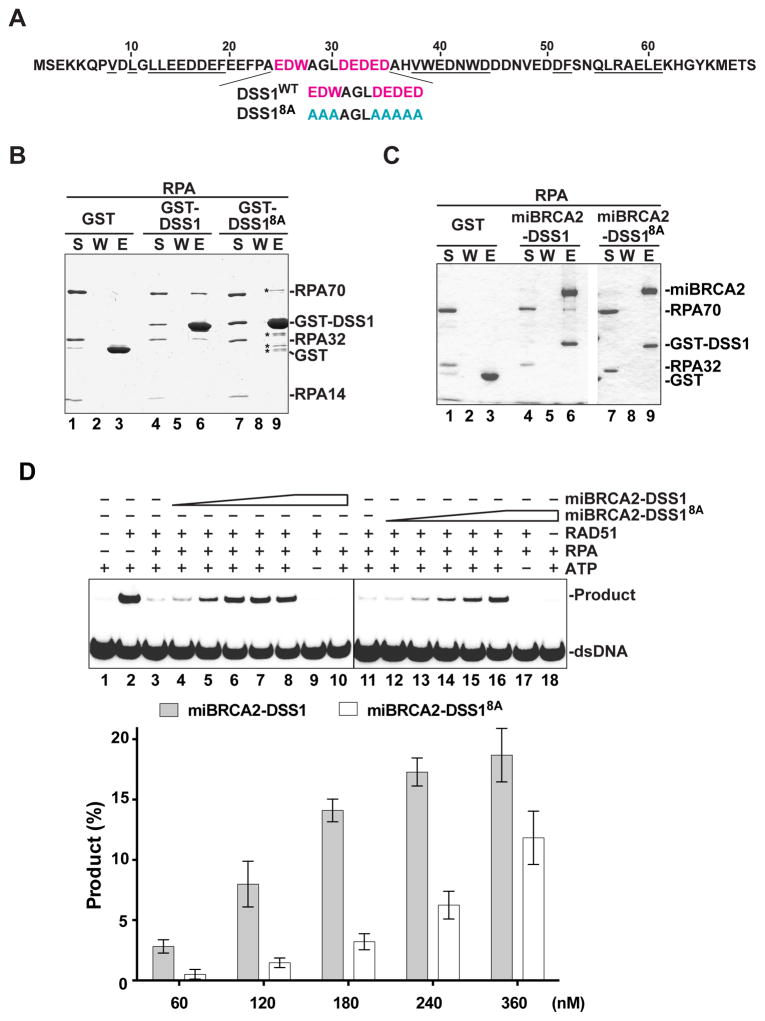
In order to address the significance of the DSS1-RPA complex in HR and to further the hypothesis of DNA mimicry (see above sections), we sought to isolate a RPA-interaction defective DSS1 mutant. The crystal structure of the mouse Dss1-Brca2 DBD complex highlights interaction interfaces (Yang et al., 2002; Figure 4A). This structure also reveals a solvent-exposed, disordered segment within the middle portion of Dss1 (Figure 4A). This DSS1 segment contains a high concentration of acidic residues (seven in total) that we hypothesized to be involved in RPA interaction and relevant for the DNA mimetic function of DSS1. To test this idea, these acidic residues (E25, D26, D31, E32, D33, E34, and D35) and an amphipathic residue (W27) located therein were changed to alanine, to yield a compound point mutant that we refer to as the DSS18A mutant (Figure 4A). We note that DSS18A can be readily expressed in insect cells alone or in a stable complex with miBRCA2, and that the miBRCA2-DSS18A complex can be purified to near homogeneity using the procedure developed for the wild type complex (Figure S1D and Figure 4C). We found that DSS18A is strongly impaired in the interaction with RPA (Figure 4B) or the various RPA domain constructs (Figure S4A). In addition, DSS18A is incapable of associating with the RPA-ssDNA complex (Figure S2B) and exhibits a much lower affinity for RPA70N (75 μM) and RPA70NAB (36 μM) in ITC analysis (Figure S4B). As expected, miBRCA2-DSS18A is defective in RPA interaction (Figure 4C) but proficient in RAD51 association (Figure S1D). Moreover, unlike DSS1, the DSS18A mutant exerts no negative effect on ssDNA binding by RPA (Figure S4C & D). We also tested two additional DSS1 mutants with fewer changes of the residues in the solvent-exposed acidic patch, namely, the DSS13A mutant (harboring the change of E25, D26, and W27 to alanine) and the DSS15A mutant (harboring the change of D31, E32, D33, E34, and D35 to alanine) (Figure S4E). These mutants are less impaired for RPA association (Figure S4F) and the ability to affect ssDNA binding by the latter (Figure S4C & D). These findings indicate that the solvent-exposed, acidic region of DSS1 is indispensable for its interaction with RPA and for the attenuation of ssDNA binding by RPA.
Figure 4. Importance of DSS1-RPA interaction in efficacy of miBRCA-DSS1.
See also Figure S1, Figure S4 and Figure S5.
(A) DSS1 residues (underlined) involved in BRCA2 interaction and solvent-exposed residues (red) in the Brca2-Dss1 crystal structure (Yang et al., 2002) targeted for mutagenesis.
(B) GST-pulldown assay to test for the interaction of RPA with DSS1 or the DSS18A mutant. The supernatant (S), the wash (W), and the eluate (E) fractions were analyzed. The asterisks highlight minor proteolytic products and contaminants in the GST-DSS18A preparation.
(C) GST-pulldown assay as in (B) to test for the interaction of RPA with miBRCA2-DSS1 and miBRCA2-DSS18A.
(D) miBRCA2-DSS1 and miBRCA2-DSS18A were tested for their recombination mediator activity. The mean values (± s.d.) of data from 3 independent experiments were plotted.
DSS1-RPA interaction is indispensable for recombination mediator function
Next, we tested the miBRCA2-DSS18A complex in the homologous DNA pairing reaction. Even though the addition of an increasing amount of miBRCA2-DSS18A to RPA-coated ssDNA led to restoration of homologous pairing, its efficacy was much reduced when compared to miBRCA2-DSS1 (Figure 4D) and no greater than that of miBRCA2 (see Figure 1D for data on miBRCA2 alone). In contrast, both protein complexes enhanced homologous DNA pairing to the same degree when RPA was absent (Figure S5A) and exhibited comparable efficacy in homologous pairing restoration when SSB-coated ssDNA was used (Figure S5B). By EM and the magnetic bead-based pulldown assay, we verified that miBRCA2-DSS18A is no more efficacious than miBRCA2 in the restoration of RAD51 presynaptic filament assembly (Figure S1B & C). Thus, the recombination mediator activity of the miBRCA2-DSS1 complex is strongly dependent on the ability of DSS1 to associate with RPA.
Examination of full-length BRCA2 alone and in complex with DSS1
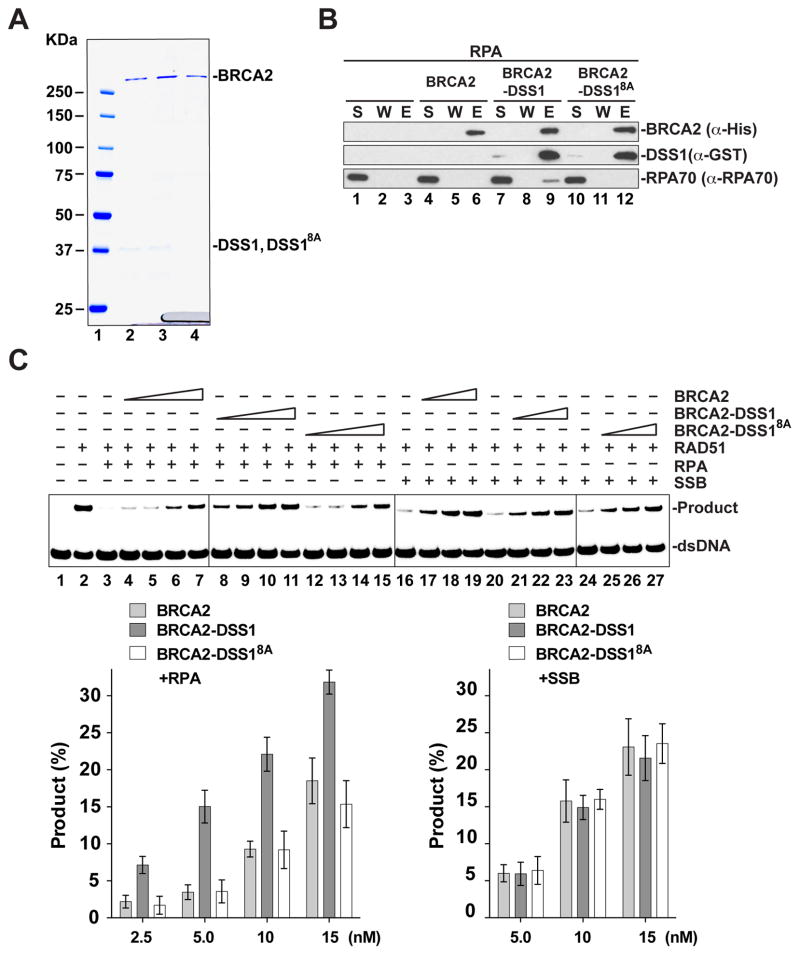
To verify the findings for the miBRCA2 model, we performed key experiments with full-length BRCA2. Specifically, full-length BRCA2 was tested alone or in complex with either DSS1 or DSS18A for RPA interaction and in the homologous DNA pairing reaction (Figure 5A). As reported previously (Jensen et al., 2010) and confirmed herein, BRCA2 by itself does not interact with RPA (Figure 5B). Importantly, we observed that the purified BRCA2-DSS1 complex associates with RPA, but the BRCA2-DSS18A mutant complex fails to do so (Figure 5B). Moreover, BRCA2-DSS18A is much less able to bind the RPA-ssDNA complex than does BRCA2-DSS1 (data not shown). These results thus reveal the solvent-exposed acidic loop domain in DSS1 as being essential for RPA association.
Figure 5. Relevance of DSS1-RPA interaction for the recombination mediator activity of BRCA2-DSS1.
See also Figure S5.
(A) SDS-PAGE of purified BRCA2-DSS1 (lane 2), BRCA2-DSS18A (lane 3) and BRCA2 (lane 4). Size markers were run in lane 1.
(B) MPB-pulldown assay to test for the interaction between RPA and BRCA2, BRCA2-DSS1 or BRCA2-DSS18A.
(C) BRCA2, BRCA2-DSS1 and BRCA2-DSS18A were tested for their recombination mediator activity with RPA-coated ssDNA or SSB-coated ssDNA. The mean values (± s.d.) of data from 3 independent experiments were plotted.
The BRCA2 and BRCA2-DSS1 species were examined for their ability to restore homologous DNA pairing efficiency using RPA-coated ssDNA as the starting substrate. As expected (Jensen et al., 2010; San Filippo et al., 2006), RAD51 would utilize the RPA-coated DNA template for homologous pairing in a BRCA2 concentration-dependent manner. Importantly, the results showed that BRCA2-DSS1 is much more adept than BRCA2 in the promotion of homologous DNA pairing (Figure 5C). In sharp contrast, we found that BRCA2-DSS18A is no more efficacious than BRCA2 (Figure 5C). Moreover, neither BRCA2-DSS1 nor BRCA2-DSS18A is more effective than BRCA2 when tested on a SSB-coated ssDNA template (Figure 5C). Together, these results provide strong evidence that DSS1 enhances the recombination mediator activity of the BRCA2-DSS1 complex through its ability to interact with RPA and DNA mimicry.
We found that, as with miBRCA2, mixing purified full-length BRCA2 and DSS1 fails to reconstitute the BRCA2-DSS1 complex (Figure S5C). Accordingly, addition of DSS1 does not enhance the recombination mediator activity of full-length BRCA2 (Figure S5D), in contrast to the BRCA2-DSS1 complex (Figure 5C). In fact, an excess of free DSS1, reduces the level of homologous DNA pairing (Figure S5D, data not shown). These observations indicate that stable association with BRCA2 is a prerequisite for DSS1 to exert its beneficial effect in RAD51 presynaptic filament assembly, and they further suggest that (i) the folding of BRCA2 in the absence of DSS1 results in its DSS1 binding site being occluded, and (ii) DSS1 interferes with BRCA2-RAD51 action possibly because of its high acidic content.
DSS1-RPA interaction is required for HR efficiency in vivo
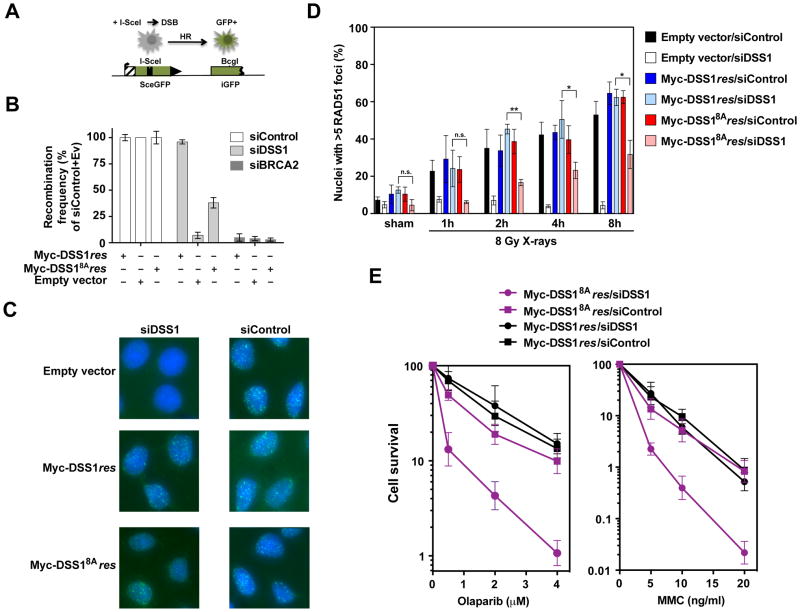
We utilized U2OS cells harboring the DR-GFP HR reporter (Nakanishi et al., 2005; Xia et al., 2006) to query whether the interaction of DSS1 and RPA is critical for HR efficiency (Figure 6A). In congruence with published work (Adamson et al., 2012; Kristensen et al., 2010), knockdown of endogenous DSS1 by one of three different siRNA species impaired HR repair (Figure S6A; Table S1). Importantly, the degree of HR impairment correlated well with the relative reduction of DSS1 protein level (Figure S6A). We saw a diminished BRCA2 protein level upon DSS1 knockdown. However, even though transient expression of BRCA2 could restore the BRCA2 protein level (Figure S6B), it failed to correct the HR deficiency caused by DSS1 knockdown, while transient expression of Myc-tagged DSS1 that is resistant to siDSS1#3 (Myc-DSS1res) complemented the deficiency well (Figure S6B). Next, cell lines that stably express Myc-DSS1res or Myc-DSS18Ares were generated, and clones with a comparable level of ectopically expressed protein were selected for testing (Figure S6C). Upon knockdown of the endogenous DSS1 (Figure S6D), we observed that cells expressing Myc-DSS1res remain proficient in HR, whereas cells harboring Myc-DSS18Ares are impaired in this regard (Figure 6B). We note that BRCA2 protein remains in the nucleus of cells harboring only Myc-DSS18Ares (Figure S6E), and that these cells exhibit the same cell cycle profile as their Myc-DSS1res counterpart (Figure S6F). Together, the above results show that the DSS1-RPA interaction is important for efficient DSB repair by HR in cells but is dispensable for the nuclear localization of BRCA2.
Figure 6. Biological relevance of the DSS1-RPA complex.
See also Figure S6.
(A) Schematic of the HR assay with the DR-GFP reporter in DR-U2OS cells (adapted from Schlacher et al, 2011)
(B) Quantification of results from the HR assays in DR-U2OS cells with stable expression of Myc-DSS1res or Myc-DSS18Ares and after DSS1 or BRCA2 knockdown and I-SceI expression. GFP-positive cells indicate the fraction of successfully completed HR events. The mean values (± s.d.) from 3 independent experiments were plotted.
(C) Representative micrographs of RAD51 foci (green) in HeLa cell nuclei at 8 h after exposure to 8 Gy X-rays. Blue: DAPI.
(D) Quantification of RAD51 foci at various time points after exposure to 8 Gy X-rays or sham irradiation. The mean values ± s.e.m. of at least 3 independent experiments are shown. n.s., not significant; *, P<0.05 and **, P<0.01.
(E) Survival curves of HeLa cells with stable expression of Myc-DSS1res or Myc-DSS18Ares after the treatment with increasing concentrations of Olaparib or MMC. The mean values (± s.d.) from 3 independent experiments were plotted.
DSS1 is required for optimal DNA damage-induced assembly of nuclear RAD51 foci (Gudmundsdottir et al., 2004). It was of interest to investigate whether this requirement extends to interaction between DSS1 and RPA. We established HeLa cell lines stably expressing Myc-DSS1res or mutant Myc-DSS18Ares, or that harbored the empty vector (Myc-vector). After exposure to 8 Gy of X-rays, the effects of treatment on RAD51 focus formation were examined in cells treated with either siDSS1#3 to deplete endogenous DSS1 or control siRNA (Figure S6G). In cells with control siRNA, no effect was observed on the frequency or intensity of RAD51 foci in all three cell lines (Figure 6C & D; Table S2). Treatment with DSS1 siRNA impaired RAD51 focus formation in cells with the Myc-vector. These cells were also deficient in spontaneous RAD51 focus formation (Figure 6D). In cells expressing Myc-DSS1res, DSS1 knockdown did not affect the levels of RAD51 foci, either spontaneously or after X-rays (Figure 6C & D). In cells expressing mutant Myc-DSS18Ares, RAD51 focus formation was suppressed up to 8 h after X-rays when DSS1 was knocked down (Figure 6C & D). Specifically, at 8 h post X-ray exposure, 31.7±7.4% of the nuclei contained > 5 RAD51 foci with DSS1 knockdown compared to 62.4±3.4% of such nuclei in cells treated with control siRNA (P=0.02; unpaired Student’s t-test). The remaining RAD51 foci in Myc-DSS18Ares cells with DSS1 knockdown are reduced in intensity (Table S2). We sought further evidence that the impairment in RAD51 focus assembly in response to ionizing radiation is related to diminished DNA repair by monitoring the formation of pRPA(S4/S8) foci which are reflective of persistent DNA damage (Marechal and Zou, 2015). As shown in Figure S6I & J, both pRPA foci and the pRPA protein levels were increased in Myc-DSS18Ares cells with DSS1 knockdown compared to Myc-DSS1res cells with DSS1 knockdown at 1–8 h post radiation exposure, and for the same time points at which RAD51 foci formation was delayed (see Figure 6D). These results provide support for the premise that the DSS1-RPA interaction is required for the optimal assembly of RAD51 foci. Notably, our observation that RAD51 focus formation is not fully dependent on the DSS1-RPA interaction is consistent with biochemical data presented above and elsewhere (Jensen et al., 2010; Liu et al., 2010; San Filippo et al., 2006) that show BRCA2 can by itself mediate RAD51 presynaptic filament assembly, albeit at a much reduced efficiency than when DSS1 is present.
Next, the cytotoxic effects of MMC and of Olaparib were assessed in clonogenic survival assays using HeLa Myc-DSS1res and HeLa Myc-DSS18Ares cells treated with DSS1 or control siRNA (Figure S6H). In contrast to Myc-DSS1res, which upon knockdown of endogenous DSS1 affords wild type levels of protection against these two genotoxic agents, Myc-DSS1res8A is largely defective in this regard (Figure 6E). Similarly, Myc-DSS1res8A was much less adept than Myc-DSS1res in maintaining cell viability after 8 Gy X-irradiation (Figure S6K). Thus, the DSS1-RPA interaction is important for efficient DNA damage repair by HR in human cells.
Discussion
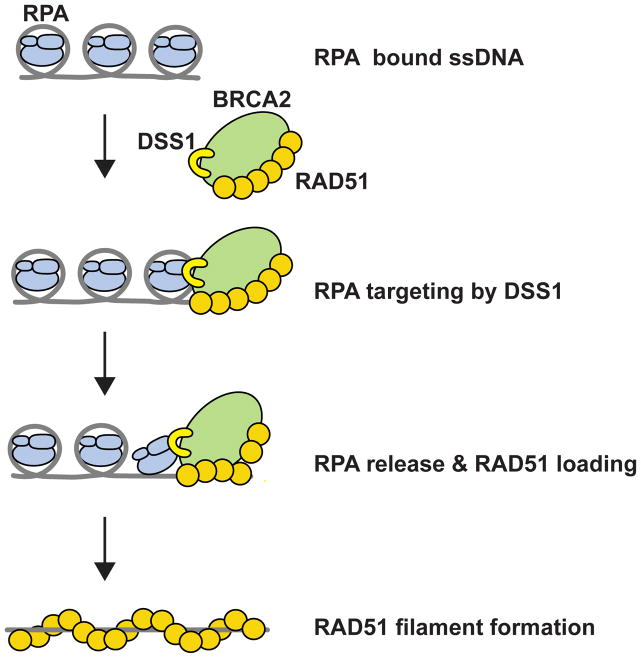
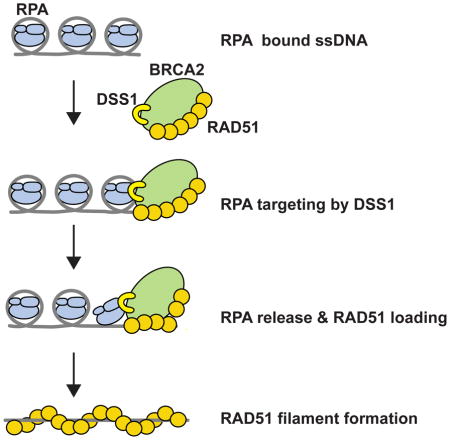
We have provided insights into how DSS1 affects RAD51 presynaptic filament assembly. Our results, as summarized in Figure 7, reveal roles of DSS1 as the RPA targeting entity and also as a DNA mimic that attenuates the affinity of RPA for ssDNA.
Figure 7. Model for DSS1 functions in BRCA2-RAD51 mediated HR.
See also Figure S7.
DSS1 promotes the loading of BRCA2-RAD51 onto RPA-coated ssDNA to facilitate RAD51-ssDNA filament assembly through its ability to (i) physically interact with RPA and (ii) attenuate the affinity of RPA for ssDNA.
Distinct means for the promotion of presynaptic filament assembly
Much of our understanding of eukaryotic recombination mediators has originated from studies done in model organisms such as the budding yeast S. cerevisiae and the corn smut fungus U. maydis (Holloman et al., 2008; San Filippo et al., 2008; Symington, 2002; Yang et al., 2005). S. cerevisiae Rad52, through its abilities to bind DNA and physically interact with Rad51 and RPA, facilitates the nucleation of Rad51 onto RPA-coated ssDNA (San Filippo et al., 2008). Accordingly, the DNA binding and protein interaction activities of Rad52 are indispensable for its recombination mediator function (Krejci et al., 2002; Plate et al., 2008; Seong et al., 2008). Moreover, the physical interaction of Rad52 with ssDNA-bound RPA allows it to mediate (i) the annealing of RPA-coated complementary ssDNA in the single-strand annealing (SSA) pathway of HR, and (ii) the capture of the second, RPA-coated ssDNA end during DSB repair (Lao et al., 2008; Mortensen et al., 1996; Sugiyama et al., 2006).
In contrast, BRCA2 has no RPA interaction attribute (Jensen et al., 2010; this work) and does not appear to function in SSA (Stark et al., 2004; Tutt et al., 2001). Thus, even though BRCA2 and BRCA2-polypeptides can enhance RAD51-mediated homologous DNA pairing (Jensen et al., 2010; San Filippo et al., 2006) with RPA-coated ssDNA, this does not appear to follow the same mechanism as yeast Rad52. Rather, the BRCA2 recombination mediator activity may stem from the stabilization of the nascent presynaptic filament. That several BRC repeats limit the association of RAD51 with dsDNA may also contribute to its recombination mediator activity (Carreira et al., 2009; Carreira and Kowalczykowski, 2011; Shivji et al., 2009). Consistent with this premise, we have found no specificity in BRCA2 action in RAD51-mediated homologous DNA pairing regardless of whether RPA-coated or SSB-coated ssDNA is employed (Jensen et al., 2010; San Filippo et al., 2006; this work). Thus, our work showing DSS1 as the RPA targeting entity and acting via DNA mimicry provide crucial insights into the function of BRCA2-DSS1 complex during HR.
We note that studies by us and others have uncovered important differences between the human BRCA2-DSS1 and the presumptive U. maydis ortholog Brh2-Dss1. First, even though Brh2-Dss1 is dispensable for cell viability (Kojic et al., 2002; Kojic et al., 2003), Brca2 null mutation results in early embryonic lethality in mice and depletion of either BRCA2 or DSS1 in human cells inhibits cell proliferation (Feng et al., 2011; Ma et al., 2013; Sharan et al., 1997; data not shown). Brh2 has a much simpler architecture, possessing only a single BRC-like repeat and a DNA binding domain within the C-terminal region that harbors only two OB folds (Holloman et al., 2008). Notably, Brh2 also possesses a distinct DNA binding domain at its N-terminus. It has been suggested that this N-terminal DNA binding domain provides the DNA binding activity needed for the biological efficacy of Brh2, whereas the C-terminal DNA binding domain serves only a regulatory role (Zhou et al., 2009a). Consistent with this premise, U. maydis Dss1, which only loosely associates with OB1 in the C-terminal DNA binding domain, attenuates the DNA binding potential of Brh2. Interestingly, it has been suggested that both the amino-terminal domain of Brh2 and DNA act together to evict Dss1 from its C-terminal interaction surface to reveal the full DNA binding potential of Brh2 (Zhou et al., 2012; Zhou et al., 2009b). In contrast, the DBD of BRCA2 is indispensable for function both in vitro and in cells (Al Abo et al., 2014; San Filippo et al., 2006; Siaud et al., 2011). The results described herein enhance our understanding of these differences, and also furnish strong evidence that DSS1 must be stably associated with BRCA2 to be functional (Figure 1 & 5; Figure S5), and that it targets the RPA-ssDNA complex via physical interaction with RPA and acts as a DNA mimic to promote RPA-RAD51 exchange on ssDNA (Figure 2, 3, 7 & S7).
Implications for DSS1 functions in meiotic HR and other biological processes and for cancer therapy
BRCA2 physically interacts with the conserved, meiosis-specific recombinase DMC1 via a motif with the sequence FVPP and also the C-terminal domain (Thorslund et al., 2007). Given that BRCA2 functions in DMC1-dependent meiotic HR (Seeliger et al., 2012; Siaud et al., 2004), it seems reasonable to suggest that the BRCA2-DSS1 complex helps mediate DMC1 presynaptic filament assembly as well (Figure S7A). In this regard, the experimental systems that we have established in this and other studies (Sehorn et al., 2004) should be of value in testing the roles of BRCA2 and DSS1 in DMC1-mediated homologous DNA pairing, in particular with respect to the formation of the crossover recombinants that are indispensable for chromosome segregation in the first meiotic division (Neale and Keeney, 2006).
Genetic and other studies have unveiled a role of DSS1 in the biogenesis and function of the 26S proteasome and also in RNA metabolic pathways, including RNA export, editing, and splicing (Garncarz et al., 2013; Pick et al., 2009). Recent work on Sem1 (Tomko and Hochstrasser, 2014), the yeast DSS1 ortholog, has provided evidence for a chaperone-like function of this protein in ensuring the orderly incorporation of Rpn3 and Rpn7 into the proteasome lid. In this role, Sem1 tethers Rpn3 and Rpn7 (Figure S7D) through conserved domains that in human DSS1 are involved in complex formation with BRCA2. However, the precise functions of DSS1 in RNA metabolic processes have not yet been elucidated. The ability of DSS1 to act as a ssDNA mimic suggests a more general nucleic acid mimicry property that may also be relevant for protein handoff in this context (Wilmes et al., 2008). For instance, DSS1 could promote the exchange of the budding yeast Sub2 protein (UAP56 in mammals) for the Mex67 protein (NXF1 in mammals) on the newly synthesized mRNA to facilitate its nuclear export ((Wilmes et al., 2008); Figure S7B). Alternatively, the solvent-exposed acidic DSS1 domain could help shepherd the reshuffling of protein components in macromolecular complexes by weakening specific protein interfaces in favor of the establishment of a new set of interactions (Figure S7C). In this regard, DSS18A may also affect one of more of the other functions of DSS1 mentioned here, and it remains possible that the biological effects engendered by this mutant may not stem entirely from compromising the DNA mimicry role of DSS1 in HR.
DSS1 is elevated in tumor samples and, conversely, depletion of DSS1 can suppress cellular proliferation (Ma et al., 2013; Rezano et al., 2013; data not shown). Moreover, amplification of DSS1 correlates with increased therapy resistance (Ma et al., 2013; Rezano et al., 2013), and breast cancer cohorts with higher DSS1 expression exhibit poor prognosis and relapse-free survival rates (Rezano et al., 2013). Given the amplification of DSS1 in a variety of tumor types, a potentially beneficial therapeutic avenue would be to inhibit DSS1 expression in these tumors. Blockade of the DSS1-RPA interaction would interfere with BRCA2/RAD51-dependent HR to alleviate resistance to therapy. Small molecule inhibitors or peptides that block DSS1 function may act as radio-sensitizers or be utilized in combination with standard chemotherapy to enhance tumor cell killing.
Experimental Procedures
Recombination proteins
BRCA2, BRCA2-DSS1/DSS18A, miBRCA2, and miBRCA2-DSS1/DSS18A were expressed in Hi5 insect cells infected by one or more of the recombinant baculoviruses. Proteins were purified making use of their affinity tags. DSS1 and mutants were expressed in E. coli and purified via their affinity tags. Other proteins were expressed and purified following our published procedures. The details are provided in the Extended Experimental Procedures.
Biochemical assays
The various biochemical assays are described in the Extended Experimental Procedures.
Cell-based experiments
U2OS and HeLa cells were treated with the indicated siRNA to transiently deplete DSS1 or BRCA2. For some experiments, HeLa and U2OS cell lines stably expressing a siRNA-resistant form of DSS1 or DSS18A were used. The measurement of HR frequency using the DR-GFP reporter, immunofluorescence microscopy and image analyses, the clonogenic survival assay, preparation of cytoplasmic and nuclear extracts, and cell cycle analysis are provided in the Extended Experimental Procedures. The siRNAs used are listed in Table S1.
Supplementary Material
Highlights.
DSS1 works in conjunction with BRCA2 to facilitate RPA-RAD51 exchange on ssDNA.
DSS1 interacts with RPA via its solvent-exposed acidic loop.
DSS1 binds the RPA70 subunit of RPA and attenuates ssDNA-binding by it.
DSS1 could also function as a nucleic acid mimic in other biological processes.
Acknowledgments
We thank Stephen Kowalczykowski for the phCMV1-BRCA2 vector and Alan Ashworth for the pEGFP-DSS1 vector. This work was supported by US National Institutes of Health grants ES015252, ES007061, CA168635, CA92584, ES021454 and GM65484. Access to facilities was supported by P30 ES00267 and P30 CA068485. NMR instrumentation was supported by grants from the NSF (0922862), NIH (S10 RR025677) and Vanderbilt University matching funds.
Footnotes
Supplemental Information includes Supplemental Data (seven Supplemental Figures and two Supplemental Tables), Extended Experimental Procedures and Supplemental References.
Author Contributions
P.S., W.Z., C.W. and W.J.C. conceived the study. W.Z., S.V., D.G.M., G.V.F., W.J.C., C.W., and P.S. designed the experiments and analysed the data. W.Z., S.V., C.W., J.S.F., D.G.M., J.J.-S., G.V.F., Y.K., S.G.L., L.L. and R.B.J. executed the experiments. W.Z., W.J.C., C.W., and P.S. wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adamson B, Smogorzewska A, Sigoillot FD, King RW, Elledge SJ. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat Cell Biol. 2012;14:318–328. doi: 10.1038/ncb2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Abo M, Dejsuphong D, Hirota K, Yonetani Y, Yamazoe M, Kurumizaka H, Takeda S. Compensatory Functions and Interdependency of the DNA-Binding Domain of BRCA2 with the BRCA1-PALB2-BRCA2 Complex. Cancer Res. 2014;74:797–807. doi: 10.1158/0008-5472.CAN-13-1443. [DOI] [PubMed] [Google Scholar]
- Carreira A, Hilario J, Amitani I, Baskin RJ, Shivji MK, Venkitaraman AR, Kowalczykowski SC. The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51. Cell. 2009;136:1032–1043. doi: 10.1016/j.cell.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira A, Kowalczykowski SC. Two classes of BRC repeats in BRCA2 promote RAD51 nucleoprotein filament function by distinct mechanisms. Proc Natl Acad Sci U S A. 2011;108:10448–10453. doi: 10.1073/pnas.1106971108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crackower MA, Scherer SW, Rommens JM, Hui CC, Poorkaj P, Soder S, Cobben JM, Hudgins L, Evans JP, Tsui LC. Characterization of the split hand/split foot malformation locus SHFM1 at 7q21.3-q22.1 and analysis of a candidate gene for its expression during limb development. Hum Mol Genet. 1996;5:571–579. doi: 10.1093/hmg/5.5.571. [DOI] [PubMed] [Google Scholar]
- Esashi F, Christ N, Gannon J, Liu Y, Hunt T, Jasin M, West SC. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- Feng Z, Scott SP, Bussen W, Sharma GG, Guo G, Pandita TK, Powell SN. Rad52 inactivation is synthetically lethal with BRCA2 deficiency. Proc Natl Acad Sci U S A. 2011;108:686–691. doi: 10.1073/pnas.1010959107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garncarz W, Tariq A, Handl C, Pusch O, Jantsch MF. A high-throughput screen to identify enhancers of ADAR-mediated RNA-editing. RNA Biol. 2013;10:192–204. doi: 10.4161/rna.23208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayther SA, Mangion J, Russell P, Seal S, Barfoot R, Ponder BA, Stratton MR, Easton D. Variation of risks of breast and ovarian cancer associated with different germline mutations of the BRCA2 gene. Nat Genet. 1997;15:103–105. doi: 10.1038/ng0197-103. [DOI] [PubMed] [Google Scholar]
- Gudmundsdottir K, Lord CJ, Witt E, Tutt AN, Ashworth A. DSS1 is required for RAD51 focus formation and genomic stability in mammalian cells. EMBO Rep. 2004;5:989–993. doi: 10.1038/sj.embor.7400255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Ray Chaudhuri A, Lopes M, Costanzo V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat Struct Mol Biol. 2010;17:1305–1311. doi: 10.1038/nsmb.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- Holloman WK, Schirawski J, Holliday R. The homologous recombination system of Ustilago maydis. Fungal Genet Biol. 2008;45(Suppl 1):S31–39. doi: 10.1016/j.fgb.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatius J, Knuutila S, Scherer SW, Trask B, Kere J. Split hand/split foot malformation, deafness, and mental retardation with a complex cytogenetic rearrangement involving 7q21.3. J Med Genet. 1996;33:507–510. doi: 10.1136/jmg.33.6.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyasekharan AD, Liu Y, Hattori H, Pisupati V, Jonsdottir AB, Rajendra E, Lee M, Sundaramoorthy E, Schlachter S, Kaminski CF, et al. A cancer-associated BRCA2 mutation reveals masked nuclear export signals controlling localization. Nat Struct Mol Biol. 2013;20:1191–1198. doi: 10.1038/nsmb.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojic M, Kostrub CF, Buchman AR, Holloman WK. BRCA2 homolog required for proficiency in DNA repair, recombination, and genome stability in Ustilago maydis. Mol Cell. 2002;10:683–691. doi: 10.1016/s1097-2765(02)00632-9. [DOI] [PubMed] [Google Scholar]
- Kojic M, Yang H, Kostrub CF, Pavletich NP, Holloman WK. The BRCA2-interacting protein DSS1 is vital for DNA repair, recombination, and genome stability in Ustilago maydis. Mol Cell. 2003;12:1043–1049. doi: 10.1016/s1097-2765(03)00367-8. [DOI] [PubMed] [Google Scholar]
- Krejci L, Song B, Bussen W, Rothstein R, Mortensen UH, Sung P. Interaction with Rad51 is indispensable for recombination mediator function of Rad52. J Biol Chem. 2002;277:40132–40141. doi: 10.1074/jbc.M206511200. [DOI] [PubMed] [Google Scholar]
- Kristensen CN, Bystol KM, Li B, Serrano L, Brenneman MA. Depletion of DSS1 protein disables homologous recombinational repair in human cells. Mutat Res. 2010;694:60–64. doi: 10.1016/j.mrfmmm.2010.08.007. [DOI] [PubMed] [Google Scholar]
- Lao JP, Oh SD, Shinohara M, Shinohara A, Hunter N. Rad52 promotes postinvasion steps of meiotic double-strand-break repair. Mol Cell. 2008;29:517–524. doi: 10.1016/j.molcel.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zou C, Bai Y, Wazer DE, Band V, Gao Q. DSS1 is required for the stability of BRCA2. Oncogene. 2006;25:1186–1194. doi: 10.1038/sj.onc.1209153. [DOI] [PubMed] [Google Scholar]
- Liu J, Doty T, Gibson B, Heyer WD. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol. 2010;17:1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma YY, Lin H, Chang FM, Chang TC, Trieu T, Pridgen HI, Zhang Y, Huang J, Patino-Guzman K, Diab N, et al. Identification of the deleted in split hand/split foot 1 protein as a novel biomarker for human cervical cancer. Carcinogenesis. 2013;34:68–78. doi: 10.1093/carcin/bgs279. [DOI] [PubMed] [Google Scholar]
- Marechal A, Zou L. RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 2015;25:9–23. doi: 10.1038/cr.2014.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston NJ, Richards WJ, Hughes D, Bertwistle D, Marshall CJ, Ashworth A. Interaction between the product of the breast cancer susceptibility gene BRCA2 and DSS1, a protein functionally conserved from yeast to mammals. Mol Cell Biol. 1999;19:4633–4642. doi: 10.1128/mcb.19.7.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen UH, Bendixen C, Sunjevaric I, Rothstein R. DNA strand annealing is promoted by the yeast Rad52 protein. Proc Natl Acad Sci U S A. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K, Yang YG, Pierce AJ, Taniguchi T, Digweed M, D’Andrea AD, Wang ZQ, Jasin M. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc Natl Acad Sci U S A. 2005;102:1110–1115. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MJ, Keeney S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature. 2006;442:153–158. doi: 10.1038/nature04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- Paraskevopoulos K, Kriegenburg F, Tatham MH, Rosner HI, Medina B, Larsen IB, Brandstrup R, Hardwick KG, Hay RT, Kragelund BB, et al. Dss1 Is a 26S Proteasome Ubiquitin Receptor. Mol Cell. 2014;56:453–461. doi: 10.1016/j.molcel.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick E, Hofmann K, Glickman MH. PCI complexes: Beyond the proteasome, CSN, and eIF3 Troika. Mol Cell. 2009;35:260–264. doi: 10.1016/j.molcel.2009.07.009. [DOI] [PubMed] [Google Scholar]
- Pispa J, Palmen S, Holmberg CI, Jantti J. C. elegans dss-1 is functionally conserved and required for oogenesis and larval growth. BMC Dev Biol. 2008;8:51. doi: 10.1186/1471-213X-8-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plate I, Hallwyl SC, Shi I, Krejci L, Muller C, Albertsen L, Sung P, Mortensen UH. Interaction with RPA is necessary for Rad52 repair center formation and for its mediator activity. J Biol Chem. 2008;283:29077–29085. doi: 10.1074/jbc.M804881200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezano A, Kuwahara K, Yamamoto-Ibusuki M, Kitabatake M, Moolthiya P, Phimsen S, Suda T, Tone S, Yamamoto Y, Iwase H, et al. Breast cancers with high DSS1 expression that potentially maintains BRCA2 stability have poor prognosis in the relapse-free survival. BMC Cancer. 2013;13:562. doi: 10.1186/1471-2407-13-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J, Chi P, Sehorn MG, Etchin J, Krejci L, Sung P. Recombination mediator and Rad51 targeting activities of a human BRCA2 polypeptide. J Biol Chem. 2006;281:11649–11657. doi: 10.1074/jbc.M601249200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeliger K, Dukowic-Schulze S, Wurz-Wildersinn R, Pacher M, Puchta H. BRCA2 is a mediator of RAD51- and DMC1-facilitated homologous recombination in Arabidopsis thaliana. New Phytol. 2012;193:364–375. doi: 10.1111/j.1469-8137.2011.03947.x. [DOI] [PubMed] [Google Scholar]
- Sehorn MG, Sigurdsson S, Bussen W, Unger VM, Sung P. Human meiotic recombinase Dmc1 promotes ATP-dependent homologous DNA strand exchange. Nature. 2004;429:433–437. doi: 10.1038/nature02563. [DOI] [PubMed] [Google Scholar]
- Seong C, Sehorn MG, Plate I, Shi I, Song B, Chi P, Mortensen U, Sung P, Krejci L. Molecular anatomy of the recombination mediator function of Saccharomyces cerevisiae Rad52. J Biol Chem. 2008;283:12166–12174. doi: 10.1074/jbc.M800763200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan SK, Morimatsu M, Albrecht U, Lim DS, Regel E, Dinh C, Sands A, Eichele G, Hasty P, Bradley A. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature. 1997;386:804–810. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- Shivji MK, Mukund SR, Rajendra E, Chen S, Short JM, Savill J, Klenerman D, Venkitaraman AR. The BRC repeats of human BRCA2 differentially regulate RAD51 binding on single- versus double-stranded DNA to stimulate strand exchange. Proc Natl Acad Sci U S A. 2009;106:13254–13259. doi: 10.1073/pnas.0906208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siaud N, Barbera MA, Egashira A, Lam I, Christ N, Schlacher K, Xia B, Jasin M. Plasticity of BRCA2 function in homologous recombination: genetic interactions of the PALB2 and DNA binding domains. PLoS Genet. 2011;7:e1002409. doi: 10.1371/journal.pgen.1002409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siaud N, Dray E, Gy I, Gerard E, Takvorian N, Doutriaux MP. Brca2 is involved in meiosis in Arabidopsis thaliana as suggested by its interaction with Dmc1. Embo J. 2004;23:1392–1401. doi: 10.1038/sj.emboj.7600146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol. 2004;24:9305–9316. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauffer ME, Chazin WJ. Structural mechanisms of DNA replication, repair, and recombination. J Biol Chem. 2004;279:30915–30918. doi: 10.1074/jbc.R400015200. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Kantake N, Wu Y, Kowalczykowski SC. Rad52-mediated DNA annealing after Rad51-mediated DNA strand exchange promotes second ssDNA capture. Embo J. 2006;25:5539–5548. doi: 10.1038/sj.emboj.7601412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- Thorslund T, Esashi F, West SC. Interactions between human BRCA2 protein and the meiosis-specific recombinase DMC1. Embo J. 2007;26:2915–2922. doi: 10.1038/sj.emboj.7601739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorslund T, McIlwraith MJ, Compton SA, Lekomtsev S, Petronczki M, Griffith JD, West SC. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat Struct Mol Biol. 2010;17:1263–1265. doi: 10.1038/nsmb.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomko RJ, Jr, Hochstrasser M. The Intrinsically Disordered Sem1 Protein Functions as a Molecular Tether during Proteasome Lid Biogenesis. Mol Cell. 2014;53:433–443. doi: 10.1016/j.molcel.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutt A, Bertwistle D, Valentine J, Gabriel A, Swift S, Ross G, Griffin C, Thacker J, Ashworth A. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. Embo J. 2001;20:4704–4716. doi: 10.1093/emboj/20.17.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei SJ, Trempus CS, Cannon RE, Bortner CD, Tennant RW. Identification of Dss1 as a 12-O-tetradecanoylphorbol-13-acetate-responsive gene expressed in keratinocyte progenitor cells, with possible involvement in early skin tumorigenesis. J Biol Chem. 2003;278:1758–1768. doi: 10.1074/jbc.M206328200. [DOI] [PubMed] [Google Scholar]
- Wilmes GM, Bergkessel M, Bandyopadhyay S, Shales M, Braberg H, Cagney G, Collins SR, Whitworth GB, Kress TL, Weissman JS, et al. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol Cell. 2008;32:735–746. doi: 10.1016/j.molcel.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AK, Pero R, Ormonde PA, Tavtigian SV, Bartel PL. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J Biol Chem. 1997;272:31941–31944. doi: 10.1074/jbc.272.51.31941. [DOI] [PubMed] [Google Scholar]
- Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH, Zheng N, Chen PL, Lee WH, Pavletich NP. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433:653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Kojic M, Holloman WK. DNA-binding Domain within the Brh2 N Terminus Is the Primary Interaction Site for Association with DNA. J Biol Chem. 2009a;284:8265–8273. doi: 10.1074/jbc.M809226200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Kojic M, Holloman WK. Dss1 release activates DNA binding potential in Brh2. Biochemistry. 2012;51:9137–9146. doi: 10.1021/bi3011187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Mazloum N, Mao N, Kojic M, Holloman WK. Dss1 regulates interaction of Brh2 with DNA. Biochemistry. 2009b;48:11929–11938. doi: 10.1021/bi901775j. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.