SUMMARY
Multiple sequence alignment (MSA) is an essential tool with many applications in bioinformatics and computational biology. Accurate MSA construction for divergent proteins remains a difficult computational task. The constantly increasing protein sequences and structures in public databases could be used to improve alignment quality. PROMALS3D is a tool for protein MSA construction enhanced with additional evolutionary and structural information from database searches. PROMALS3D automatically identifies homologs from sequence and structure databases for input proteins, derives structure-based constraints from alignments of 3-dimensional structures, and combines them with sequence-based constraints of profile-profile alignments in a consistency-based framework to construct high-quality multiple sequence alignments. PROMALS3D output is a consensus alignment enriched with sequence and structural information about input proteins and their homologs. PROMALS3D web server and package are available at http://prodata.swmed.edu/PROMALS3D.
Keywords: Multiple sequence alignment, database searches, 3-dimensional structural alignment, consistency-based scoring, probabilistic model of profile-profile alignment
1. INTRODUCTION
Multiple sequence alignment (MSA) is fundamentally important for a variety of tasks in bioinformatics and computational biology, including homology-based structure modeling, prediction of structural properties, sequence similarity searches, phylogenetic reconstruction, and identification of functionally important sites. For a set of protein sequences, MSA construction involves placement of gap characters in sequences so that each position (column) contains evolutionarily or structurally equivalent amino acid residues. Such a biologically meaningful representation of multiple sequences not only facilitates their visualization and inspection, but also helps extraction of valuable information such as sequence conservation and residue preferences on a positional basis.
Accurate and fast MSA construction has been under extensive research with significant progress made in the last decade [1–5]. Dynamic programming algorithms [6–7] are effective in aligning of a pair of sequences (pairwise alignment), while such techniques are too time and memory consuming to align a large number of sequences [8–9]. Many MSA methods resort to a heuristic, the progressive alignment technique [10–11], that reduces the task of aligning multiple sequences to a hierarchical series of pairwise alignments of sequence subsets. In early progressive methods, aligning two subsets of sequences only used information from these two subsets, and mistakes introduced in this process were fixed and propagated to later steps. One way to improve the alignment quality is through refinements after MSA assembly, often conducted by repeatedly dividing the MSA into sub-alignments and realigning the sub-alignments [12–13]. Another popular alignment technique uses consistency-based scoring functions [14–16] to improve alignment quality by exploring information from the entire set of sequences when aligning subsets of sequences.
While various MSA methods generally produce high-quality alignments when sequence similarity is high (e.g., sequence identity above 40%), it is still difficult to achieve accurate results for distantly related proteins. It is not uncommon for evolutionarily related proteins to have highly divergent sequences (e.g., sequence identity below 20%) while maintaining similar structures and related functions. Alignments constructed with information from divergent sequences themselves are often prone to mistakes. Additional evolutionary information from homologous sequences is useful to enhance alignment quality. First, a protein sequence can be augmented by information from its homologs by using sequence profile, a numerical representation of positional amino acid usage. Profile-to-profile alignment is generally more accurate than sequence-to-sequence alignment [17–18]. Secondly, positional structural properties such as secondary structures and solvent accessibilities can be predicted from sequence profile, and scoring functions incorporating predicted structural information can lead to better alignment quality [19–20]. As protein spatial structures are generally more conserved than sequences [21], comparison of available 3-dimensional (3D) structures can offer high-quality alignment constraints for MSA construction [22–24].
PROMALS3D [23,25] is a tool for MSA construction that integrates various sources of evolutionary and structural information, such as sequence profile derived from database homologs, predicted secondary structures, and available 3D structures. PROMALS3D combines profile-derived alignment constraints and structure-derived alignment constraints within a consistency-based framework to produce protein MSAs of improved quality.
2. METHODS
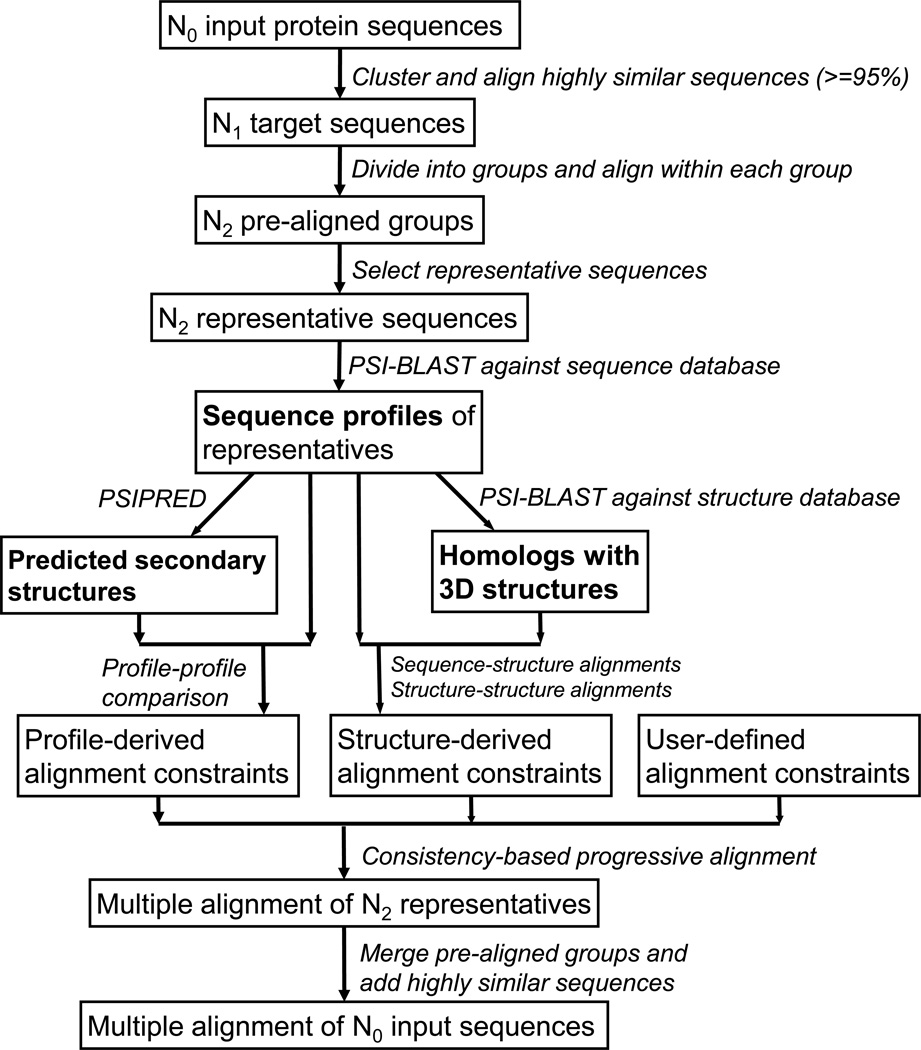
PROMALS3D is a progressive multiple protein sequence alignment tool. A key feature of PROMALS3D is that it uses different strategies to align subsets of sequences with different levels of difficulty to properly balance alignment accuracy and speed. Relatively similar sequences are aligned by a fast algorithm to form pre-aligned groups without retrieving additional information from databases. To align the relatively divergent pre-aligned groups, PROMALS3D resorts to more elaborate alignment techniques and uses additional information from homologous sequences and structures found by database searches. Such a method of using different strategies at different aligning stages allows PROMALS3D to align thousands of protein sequences in manageable time, since the time- and memory-consuming steps of database search and consistency computation are only applied to a subset consisting of representative sequences of the pre-aligned groups instead of the entire set of input sequences. The flowchart of PROMALS alignment procedure is shown in Fig. 1.
Fig. 1.
Flowchart of the PROMALS3D method.
2.1 Initial clustering and reducing sequence redundancy
For an input set with N0 sequences, PROMALS3D first rapidly clusters sequences using the program CD-HIT [26] with sequence identity cutoff of 95% (-c option) and alignment coverage for the longer sequence of 0.95 (-aL option). This initial step results in N1 clusters (N1 <= N0) of highly similar sequences. Clusters with more than one sequence are individually aligned in a fast way by MAFFT (with --auto option) [27]. This step could significantly reduce computation for datasets with a large number of near-identical sequences. One target sequence is selected from each cluster. The N1 target sequences after initial filtering of highly similar sequences are subject to further alignment steps described below.
2.2 Dividing target sequences to groups and obtaining pre-aligned groups
PROMALS3D divides the N1 non-redundant target sequences into a set of N2 groups (N2 <= N1) and aligns each group without information from sequence and structure databases. Two methods are used to obtain the groups. If N1 is no more than 200, PROMALS3D uses the UPGMA method to build a tree based on a crude measure of distances (k-mer counting) [12] among the sequences. Given a distance cutoff (-id_thr option, default: 0.6) the tree is divided into a set of subtrees, and the sequences in each subtree forming a group [28]. If the number of formed groups is larger than the maximum number of groups set by PROMALS3D (-max_group_number option, default: 60), PROMALS3D automatically adjusts the distance cutoff so that the number of formed groups is the same as the maximum number of groups allowed.
We observed that the UPGMA method for deducing groups can produce large groups (groups with too many sequences) when the input data set is large (e.g., thousands of sequences). These large groups may not be properly aligned without using additional information. For large input datasets (more than 200 sequences), we used a second method based on K-center clustering to divide the target sequences into a number of groups when the number of target sequences is more than 200. Our K-center approach does not allow any group to have more than 200 sequences.
After dividing the target sequences into N2 groups, each group is aligned, resulting in N2 pre-aligned groups. We have previously used a progressive method with the sum-of-pairs BLOSUM62 [29] scores to align sequences within each group. Such an approach does not perform as well as some modern alignment methods. In the later development of PROMALS3D, we used MAFFT (options: --maxiterate 1000 --localpair) to perform alignment within each group to obtain better alignment quality for each pre-aligned group.
2.3 Aligning pre-aligned groups with enhanced evolutionary and structural information
The core steps of the PROMALS3D method involve using advanced techniques to align the relatively divergent pre-aligned groups with additional information from sequence and structure databases. First, a representative sequence is selected from each pre-aligned group, giving rise to N2 representatives. Instead of using the longest sequence as the representative as in our original PROMALS method, we select the representative sequence that has the highest average similarity to other sequences in the same pre-aligned group.
Each representative sequence is subject to PSI-BLAST [30] iterations against the UniRef90 database [31] to retrieve sequence homologs. The sequence profile of PSI-BLAST searches is used to predict secondary structures by PSIPRED [32].
For each pair of representative sequences, we used a probabilistic model to obtain posterior profile-profile alignment probabilities for each position pair via the forward-backward algorithm. Strictly speaking, our probabilistic model for profile-profile comparison is not a hidden Markov model (HMM) as originally proposed [19], but a Conditional Random Field (CRF) [33], since we allowed observation-dependent transitions between hidden states. In our model, the transition probabilities depend on predicted secondary structures, which are used as a type of observations. Like that in HMMs, the forward-backward algorithm is applicable to CRFs to obtain posterior alignment probabilities, which serve as profile-derived alignment constraints.
PSI-BLAST profile is used to search a sequence database with known structures to retrieve homologs with 3D structures (homolog3Ds). Multiple homolog3Ds could be identified and used for one representative sequence, e.g., if it contains several distinct domains with known spatial structures. Structure-derived alignment constraints for two representative sequences are deduced from profile-based representative-to-homolog3D alignments and structure-based homolog3D-to-homolog3D alignments [23].
Profile-derived alignment constraints and structure-derived alignment constraints are combined for all pairs of representatives. These constraints are subject to consistency measure to derive consistency-based scoring function.
The N2 representatives are then progressively aligned by the consistency-based scoring function, with the aligning order following a UPGMA tree estimated for the representative sequences.
The pre-aligned groups are merged to the MSA of the N2 representatives to form an MSA of N1 target sequences. Finally, the clusters with highly similar sequences obtained at the initial clustering step are merged to the MSA of N1 target sequences to form the MSA of all input sequences.
3. PROMALS3D USAGE AND PRACTICAL ISSUES
PROMALS3D is available as a web server as well as a downloadable package at http://prodata.swmed.edu/PROMALS3D.
PROMALS3D web server allows input of both sequences and structures. The web server extracts sequences from input structures and combines them with input sequences to form the final input sequence set. The web server also prepares structural alignments for input structures and feeds them as structural constraints to the PROMALS3D program. On the other hand, the PROMALS3D downloadable package currently only takes sequences as input.
If only structures are input to the PROMALS3D web server, the final alignment is a consistency-based multiple structure alignment that integrates both structural information and homolog-derived sequence information.
Input sequences should be in FASTA format and should not have identical names. Certain characters in sequence names are changed to "_", including space, tab, and *?'`";&\|/{})(][$, but .(dot) and - are kept.
PROMALS3D web server [25] offers various options of customization of the final alignment output, such as displaying the alignment with sequences colored by predicted secondary structures and showing a consensus sequence and positional conservation indices [34].
The UPGMA tree built for target sequences is reported. Since it is based on a very crude measurement of evolutionary distances, it would not serve well for phylogenetic purposes.
The structure database is regularly updated in an automatic fashion. The structure database contains a non-redundant set of structures from the PDB database. The CD-HIT program is used to cluster sequences with 3D structures at the 70% identity level. Within each cluster, one representative structure is selected. X-ray structures are preferred over NMR or CryoEM structures. Among the X-ray structures, the one with the lowest RMSD is selected as the representative. The representative structures that are classified in the SCOP database are further split into structural domains according to SCOP domain definitions.
The PROMALS3D web server also offers an option to use PROMALS [19] to align within each pre-aligned group instead of MAFFT. This option is currently not available in the downloadable package. PROMALS uses sequence homologs and predicted secondary structures, and thus often produces better alignment results. This is helpful to achieve better overall alignment quality when pre-aligned groups themselves contain divergent sequences. However, database searching in PROMALS is more time-consuming compared to MAFFT.
Three options of structural alignments and their combinations are offered: DaliLite [35], FAST [36], and TM-align [37]. DaliLite gives slightly better results than FAST and TM-align [23]. Using combinations of them also provides slight improvement of alignment accuracy [23]. The default option of the PROMALS3D web server is the combination of FAST and TMalign. DaliLite is computationally intensive when the structures are large (e.g., with more than 500 residues).
In addition to input sequences, PROMALS3D also allows input of alignment constraints (user-defined constraints).
While PROMALS3D compares favorably to a number of other methods on an average basis [23], it does not mean it can outperform any method for all alignment cases. For regions with uncertainty, inspection of results produced by other methods could be helpful.
The advantage of PROMALS3D is the incorporation of information from homologous sequences and structures. However, mistakes may be introduced in the process. For example, PSI-BLAST may find non-homologous sequences (profile corrupt), and the PSI-BLAST alignment between the query and its hits may contain errors that could lead to inferior profile or wrong profile-profile alignment. The PSI-BLAST results of sequence and structure database searches are kept and can be accessed from the PROMALS3D web server.
Alignment mistakes could also be caused by wrong secondary structure predictions. While PSIPRED secondary structure prediction accuracy is on average about 70~80%, it is more difficult to obtain accurate predictions for beta-strands and in cases where few homologous sequences exist.
PROMALS3D method generally works best when sequences are of similar lengths and do not contain large non-homologous regions (e.g. inserted non-homologous domains).
Difficult cases that PROMALS3D may not perform well on include sequences with repeats, duplications or circular permutations, sequences with many disordered regions or low complexity regions, and sequences with predicted transmembrane segments.
Input datasets with many long sequences (e.g., >1000 amino acid residues) may cause memory crash. In these cases, reduction of the number of pre-aligned groups is recommended, which can be done by setting lower distance cutoff (-id_thr option) or setting lower maximum number of pre-aligned groups allowed (-max_group_number option).
ACKNOWLEDGEMENTS
The work is supported in part by the National Institutes of Health (GM094575 to NVG) and the Welch Foundation (I-1505 to NVG).
REFERENCES
- 1.Do CB, Katoh K. Protein multiple sequence alignment. In: Walker J, editor. Methods Mol Biol. 1st edn. Vol. 484. Totowa: Humana Press; 2008. pp. 379–413. [DOI] [PubMed] [Google Scholar]
- 2.Pei J. Multiple protein sequence alignment. Curr Opin Struct Biol. 2008;18(3):382–386. doi: 10.1016/j.sbi.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 3.Notredame C. Recent evolutions of multiple sequence alignment algorithms. PLoS Comput Biol. 2007;3(8):e123. doi: 10.1371/journal.pcbi.0030123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edgar RC, Batzoglou S. Multiple sequence alignment. Curr Opin Struct Biol. 2006;16(3):368–373. doi: 10.1016/j.sbi.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Wallace IM, Blackshields G, Higgins DG. Multiple sequence alignments. Curr Opin Struct Biol. 2005;15(3):261–266. doi: 10.1016/j.sbi.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 6.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 7.Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147(1):195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 8.Lipman DJ, Altschul SF, Kececioglu JD. A tool for multiple sequence alignment. Proc Natl Acad Sci U S A. 1989;86(12):4412–4415. doi: 10.1073/pnas.86.12.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L, Jiang T. On the complexity of multiple sequence alignment. J Comput Biol. 1994;1(4):337–348. doi: 10.1089/cmb.1994.1.337. [DOI] [PubMed] [Google Scholar]
- 10.Feng DF, Doolittle RF. Progressive sequence alignment as a prerequisite to correct phylogenetic trees. J Mol Evol. 1987;25(4):351–360. doi: 10.1007/BF02603120. [DOI] [PubMed] [Google Scholar]
- 11.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302(1):205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 15.Do CB, Mahabhashyam MS, Brudno M, Batzoglou S. ProbCons: Probabilistic consistency-based multiple sequence alignment. Genome Res. 2005;15(2):330–340. doi: 10.1101/gr.2821705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pei J, Grishin NV. MUMMALS: multiple sequence alignment improved by using hidden Markov models with local structural information. Nucleic Acids Res. 2006;34(16):4364–4374. doi: 10.1093/nar/gkl514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sadreyev R, Grishin N. COMPASS: a tool for comparison of multiple protein alignments with assessment of statistical significance. J Mol Biol. 2003;326(1):317–336. doi: 10.1016/s0022-2836(02)01371-2. [DOI] [PubMed] [Google Scholar]
- 18.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21(7):951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 19.Pei J, Grishin NV. PROMALS: towards accurate multiple sequence alignments of distantly related proteins. Bioinformatics. 2007;23(7):802–808. doi: 10.1093/bioinformatics/btm017. [DOI] [PubMed] [Google Scholar]
- 20.Deng X, Cheng J. MSACompro: protein multiple sequence alignment using predicted secondary structure, solvent accessibility, and residue-residue contacts. BMC Bioinformatics. 2011;12:472. doi: 10.1186/1471-2105-12-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chothia C, Lesk AM. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986;5(4):823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Armougom F, Moretti S, Poirot O, Audic S, Dumas P, Schaeli B, Keduas V, Notredame C. Expresso: automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee. Nucleic Acids Res. 2006;34(Web Server issue):W604–W608. doi: 10.1093/nar/gkl092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pei J, Kim BH, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36(7):2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou H, Zhou Y. SPEM: improving multiple sequence alignment with sequence profiles and predicted secondary structures. Bioinformatics. 2005;21(18):3615–3621. doi: 10.1093/bioinformatics/bti582. [DOI] [PubMed] [Google Scholar]
- 25.Pei J, Tang M, Grishin NV. PROMALS3D web server for accurate multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36(Web Server issue):W30–W34. doi: 10.1093/nar/gkn322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22(13):1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 27.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33(2):511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pei J, Sadreyev R, Grishin NV. PCMA: fast and accurate multiple sequence alignment based on profile consistency. Bioinformatics. 2003;19(3):427–428. doi: 10.1093/bioinformatics/btg008. [DOI] [PubMed] [Google Scholar]
- 29.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 1992;89(22):10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzek BE, Huang H, McGarvey P, Mazumder R, Wu CH. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics. 2007;23(10):1282–1288. doi: 10.1093/bioinformatics/btm098. [DOI] [PubMed] [Google Scholar]
- 32.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292(2):195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 33.Lafferty J, McCallum A, Pereira F. Conditional random fields: Probabilistic models for segmenting and labeling sequence data. Proc 18th International Conf on Machine Learning; 2001. pp. 282–289. [Google Scholar]
- 34.Pei J, Grishin NV. AL2CO: calculation of positional conservation in a protein sequence alignment. Bioinformatics. 2001;17(8):700–712. doi: 10.1093/bioinformatics/17.8.700. [DOI] [PubMed] [Google Scholar]
- 35.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16(6):566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 36.Zhu J, Weng Z. FAST: a novel protein structure alignment algorithm. Proteins. 2005;58(3):618–627. doi: 10.1002/prot.20331. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33(7):2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]