Abstract
Context
We have shown previously that trichloroacetic acid precipitation is an effective method of protein extraction from pancreatic fluid for downstream biomarker discovery, compared to other common extraction methods tested.
Objective
We aim to assess the utility of ultracentrifugation as an alternative method of protein extraction from pancreatic fluid.
Design
Proteins extracted from trichloroacetic acid- and ultracentrifugation-precipitated pancreatic fluid were identified using mass spectrometry techniques (in-gel tryptic digestion followed by liquid chromatography-tandem mass spectrometry; GeLC-MS/MS). Data were analyzed using Proteome Discoverer and Scaffold 3.
Setting
This is a proteomic analysis experiment of endoscopically collected fluid in an academic center.
Patients
The study population included adult patients referred to the Center for Pancreatic Disease at Brigham and Women’s Hospital, Boston, MA, USA for the evaluation of abdominal pain and gastrointestinal symptoms.
Interventions
Secretin-stimulated pancreatic fluid was collected as standard of care for the evaluation of abdominal pain and gastrointestinal symptoms.
Main outcome measures
We compared proteins identified via standard trichloroacetic acid precipitation and this alternative ultracentrifugation strategy.
Results
A subset of pancreatic fluid proteins was identified via the ultracentrifugation method. Of these proteins, similar numbers were obtained from fully tryptic or semi-tryptic database searching. Proteins identified in the ultracentrifugation-precipitated samples included previously identified biomarker candidates of chronic pancreatitis.
Conclusions
This alternative ultracentrifugation strategy requires less time and fewer handling procedures than standard trichloroacetic acid precipitation, at the expense of higher sample volume. As such, this method is well suited for targeted assays (i.e., dot blotting or targeted mass spectrometry) if the protein of interest is among those readily identified by ultracentrifugation-promoted precipitation.
Keywords: Biological Markers, Chromatography, Liquid, Pancreas, Pancreatic Function Tests, Pancreatic Juice, Tandem Mass Spectrometry
INTRODUCTION
Chronic pancreatitis is a disease characterized by chronic inflammation and progressive irreversible fibrosis resulting in the loss of exocrine and endocrine function. Patients often present with chronic abdominal pain with or without bouts of acute pancreatitis, and in advanced disease may develop fat and protein malabsorption, and diabetes. Disorders of the pancreas affect more than 1 million persons in the United States, costing nearly $3 billion annually [1, 2]. As such, the development of a practical diagnostic test for early chronic pancreatitis, prior to irreversible gland injury and dysfunction, would permit early intervention to potentially retard or ameliorate the disease.
Current clinical diagnosis of chronic pancreatitis relies on various radiologic imaging (e.g., CT and MRI), endoscopic imaging (e.g., EUS) and functional assessments (e.g., pancreas function testing and fecal elastase-1) [3]. Objective pathologic structural and functional findings are evident only in moderate to advanced disease, at which point the inflammatory and fibrotic changes are irreversible. Currently, pancreas function testing is the non-histological surrogate gold standard used to diagnose moderate-to-advanced chronic pancreatitis [4]. The degree of pancreatic dysfunction is determined by measuring specific concentrations of cellular secretory components in pancreatic fluid following hormone (secretin or cholecystokinin) stimulation [5]. The need persists for strategies and technologies designed to diagnose early chronic pancreatitis.
The proteomic analysis of pancreatic fluid offers exceptional promise for the discovery of diagnostic protein biomarkers for early chronic pancreatitis [6, 7]. We have demonstrated previously that pancreatic fluid can be readily collected via the secretin-stimulated endoscopic pancreatic function test (ePFT) in volumes of 2 to 8 mL, and as such is a suitable specimen for mass spectrometry-based proteomic investigations [7, 8, 9, 10, 11]. We have screened a series of precipitation strategies to optimize protein extraction from pancreatic fluid [11], and we have demonstrated that pancreatic fluid can be maintained at 4°C for several hours with minimal proteolysis. We determined that trichloroacetic acid (TCA) precipitation was able to: 1) concentrate proteins; 2) eliminate non-protein contaminants; and 3) quickly inactivate pancreatic proteases (which are active at physiological pH). Using this strategy, we assembled a preliminary list of potential chronic pancreatitis biomarkers meriting further investigation [8].
A recently published study of the bile proteome used differential ultracentrifugation to obtain distinct subsets of proteins [12, 13, 14]. This ultracentrifugation strategy allowed for the precipitation of a subset of biliary proteins by subjecting bile to high centrifugal force. Compared to TCA precipitation, ultracentrifugation requires fewer manual steps, thereby reducing the potential for sample loss. Furthermore, ultracentrifugation can be performed at 4°C, making it a potentially useful technique in the proteomic analysis of pancreatic fluid, as we have shown that a temperature at or below 4°C is an effective method of minimizing protease activity [11].
We aim to assess the utility of ultracentrifugation as an alternative to TCA precipitation for protein extraction prior to downstream in-gel tryptic digestion followed by liquid chromatography-tandem mass spectrometry (GeLC-MS/MS) proteomic analyses. We then compared standard database searching with tryptic specificity and semi-tryptic specificity to determine if non-tryptic peptide termini are more common in ultracentrifugation-precipitated samples compared to TCA-precipitated samples. We subsequently mined the proteomic data for evidence of exosomal marker proteins [15, 16].
The techniques outlined here can be used as a platform to perform larger biomarker validation and potentially discovery studies in chronic pancreatitis. This strategy may be particularly useful in targeting specific proteins previously determined to have a role in pancreatic disease.
METHODS
Study Population
The study population included adult patients referred to the Center for Pancreatic Disease at Brigham and Women’s Hospital, Boston, MA, USA for the evaluation of abdominal pain and gastrointestinal symptoms. To rule out pancreatic disease, all subjects underwent the following: 1) comprehensive review of history and physical examination; 2) review of radiologic and endoscopic data; and 3) upper endoscopy with ePFT followed by a gastric and duodenal mucosal biopsy. Pancreatic fluid samples from six patients were selected based upon availability. Samples were processed individually, but the data were pooled to reduce sample-to-sample variability [17, 18].
Materials
ChiRhoStim® synthetic human secretin was from ChiRhoClin (Burtonsville, MD, USA). SeeBluePlus2 Pre-Stained standard (LC5925), lithium dodecyl sulfate (LDS) Laemmli sample buffer (NP0008), NuPAGE 4–12% Bis-Tris polyacrylamide gels (NP0335), SimplyBlue Coomassie stain (LC0665), and 2-(N-morpholino)ethanesulfonic acid-sodium dodecyl sulfate (MES-SDS) electrophoresis buffer (NP002) were from Invitrogen (Carlsbad, CA, USA). Sequencing-grade modified trypsin (V5111) was obtained from Promega (Madison, WI, USA). Other reagents and solvents were from Sigma-Aldrich (St. Louis. MO, USA) and Burdick and Jackson (Morristown, NJ, USA), respectively.
Experimental Workflow
Figure 1 illustrates the general workflow for the overall analysis as follows: 1) ePFT sample collection; 2) centrifugation to remove particulates; 3) protein precipitation/ultracentrifugation; 4) pellet solubilized in LDS Laemmli loading buffer; 5) SDS-PAGE fractionation; 6) in-gel tryptic digestion; 7) LC-MS/MS peptide mass determination; and 8) bioinformatic data processing.
Figure 1.
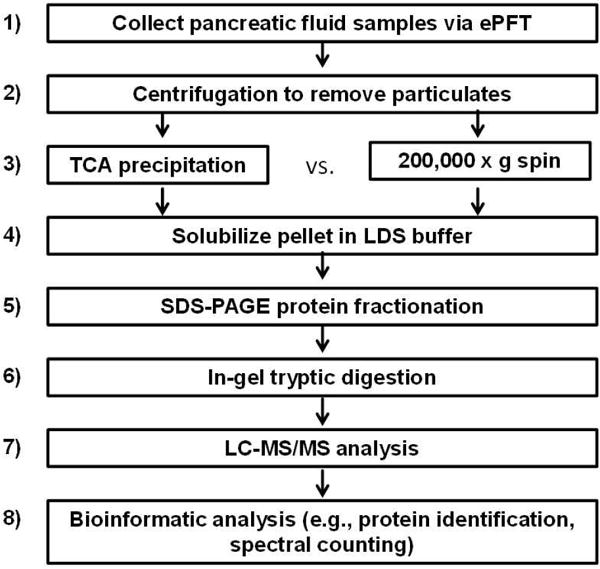
Experimental workflow. 1) Pancreatic fluid was collected via ePFT. 2) Samples were centrifuged to remove any particulates. 3) Pancreatic fluid was precipitated with TCA or subjected to ultracentrifugation at 200,000 g for 2 h. 4) The pellet was solubilized in LDS Laemmli loading buffer. 5) Proteins were fractionated by SDS-PAGE. 6) Proteins were in-gel typically digested. 7) Resulting peptides were subjected to LC-MS/MS analysis. 8) Proteins were identified via database searching using Proteome Discoverer (Thermo Scientific, Waltham, MA, USA) and Scaffold 3 (Proteome Software Inc., Portland, OR, USA) software.
Pancreatic Fluid Collection (ePFT Method)
The secretin-stimulated ePFT procedure was performed as described previously [19]. A peak pancreatic fluid bicarbonate concentration of 80 mEq/L is two standard deviations below the mean and considered the lower limit of normal [20, 21]. Duodenal aspirates were collected at 0, 5, 10, 15, 20, 30, 45 and 60 minutes after secretin stimulation. Only the 30-minute time point was used for the ensuing analysis, according to previously published methods [10].
Pancreatic Fluid Sample Preparation
Pancreatic fluid specimens for proteomic analysis were collected on ice, centrifuged at 4°C at 14,000 rpm for 15 minutes to remove cellular debris, aliquoted (500 μL), and stored at −80°C until analysis. Protein concentration was determined using the BioRAD (Hercules, CA, USA) protein assay according to the manufacturer’s instructions. We have omitted protease inhibitors as we demonstrated previously that at 4°C, little proteolysis in pancreatic fluid occurs activity [11], without the caveats associated with the addition of protease inhibitors in mass spectrometry experiments [22, 23, 24].
TCA Precipitation of Pancreatic Fluid
The proteins from 6 pancreatic fluid specimens (200 μL) were isolated by precipitation with the addition of 12.5% TCA, as described previously [10, 11]. This process limits protein degradation by instantaneously deactivating enzymes and removing salts that will interfere with the subsequent electrophoretic mobility-based fractionation by SDS-PAGE. The precipitated protein pellets were re-dissolved in 20 μL of reducing LDS Laemmli buffer [25] (with 10 mM dithiothreitol) for 1 h at 56°C and alkylated with 1% acrylamide at room temperature in the dark for 30 minutes for subsequent GeLC-MS/MS analysis.
Ultracentrifugation of Pancreatic Fluid
As an alternative to TCA precipitation, protein precipitation by ultracentrifugation was performed using pancreatic fluid from the same 6 patients used for TCA precipitation. One milliliter of pancreatic fluid (approximately 1 mg of protein) was deposited into a 13×51 mm Beckman (Brea, CA, USA) ultracentrifuge tube (#349622). The sample was subjected to ultracentrifugation at 200,000 g for 2 h at 4°C using a Beckmann TLA100.3 rotor on a Beckman TL-100 tabletop ultracentrifuge. Following centrifugation, the supernatant was aspirated completely, and a translucent pellet could be seen in the tube. The pellet was redissolved in 20 μL of reducing LDS Laemmli buffer [25] (with 10 mM dithiothreitol), incubated for 1 h at 56°C and alkylated with 1% acrylamide at room temperature for 30 minutes for subsequent GeLC-MS/MS analysis.
SDS-PAGE Fractionation and In-Gel Tryptic Digestion Followed by Liquid Chromatography-Tandem Mass Spectrometry (GeLC-MS/MS) of Pancreatic Fluid Specimens
In total, 6 gel lanes each of TCA- and ultracentrifugation-precipitated pancreatic fluid were subjected to SDS-PAGE and in-gel tryptic digestion followed by reversed-phase liquid chromatography in-line with a tandem mass spectrometer (GeLC-MS/MS). The proteins were fractionated using 4–12% NuPAGE pre-cast SDS-PAGE gels at 175 V for 45 minutes using MES-SDS buffer. Each gel lane was then divided into 7 sections and subject to in-gel tryptic digestion [26,27]. SDS-PAGE analysis revealed that the TCA precipitated samples were more concentrated than the ultracentrifugation-precipitated samples, as such we diluted the TCA precipitated samples five-fold in loading buffer (5% formic acid, 5% acetonitrile in water) prior to GeLC-MS/MS. The extracted peptides from each gel section were subjected to peptide fractionation using reversed-phase high performance liquid chromatography (HPLC; Thermo Scientific, Waltham, MA, USA) and the gradient-eluted peptides were analyzed by a hyphenated linear trap quadrupole-Fourier Transform ion cyclotron resonance (LTQ-FTICR) mass spectrometer (Thermo Scientific, Waltham, MA, USA). The liquid chromatography columns (15 cm × 100 μm internal diameter) were packed in-house (Magic C18, 5 μm, 100 Å beads; Michrom BioResources, Auburn, CA, USA) into PicoTips (New Objective, Woburn, MA, USA). Samples were analyzed with a 90 minute linear gradient (5–35% acetonitrile with 0.2% formic acid) and data were acquired in a data dependent manner, in which MS/MS fragmentation was performed using the 6 most intense peaks of every full MS scan.
Database Searching
All data generated were searched against the International Protein Index human database (IPI v3.69; EMBL-EBI, Hinxton, Cambridgeshire, United Kingdom; http://www.ebi.ac.uk/IPI/IPIhelp.html) using the Mascot search engine (v.2.204; Matrix Science, Boston, MA, USA) through Proteome Discoverer (v 1.3; Thermo Scientific, Waltham, MA, USA). Two miscleavages per peptide were allowed and mass tolerances of ±10 ppm for precursor ions and ±0.8 Da for fragment ions were used. Amino acid modifications were as follows: fixed: propionamide (Cys); variable: deamidation (Asn/Gln) and oxidation (Met).
DAVID Functional Annotation Bioinformatics Microarray Analysis
We used the Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Database (http://david.abcc.ncifcrf.gov/) interface [28,29] to classify proteins according to the Protein ANalysis THrough Evolutionary Relationships (PANTHER) protein class [30].
ETHICS
The study was approved by our Institutional Review Committee. This protocol was approved by the Institutional Review Board at Brigham and Women’s Hospital and Children’s Hospital Boston (IRB #2007-P-002480/1). The study population included adult patients referred to the Center for Pancreatic Disease at Brigham and Women’s Hospital, Boston, MA, USA for the evaluation of abdominal pain and gastrointestinal symptoms.
Both oral and written consent was talent and the study protocol conform to the ethical guidelines of the “World Medical Association (WMA) Declaration of Helsinki – Ethical Principles for Medical Research Involving Human Subjects” adopted by the 18th WMA General Assembly, Helsinki, Finland, June 1964 and amended by the 59th WMA General Assembly, Seoul, South Korea, October 2008.
STATISTICS
Scaffold (version Scaffold 3.30.07, Proteome Software Inc., Portland, OR, USA) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if greater than 95% probability as specified by the PeptideProphet™ algorithm [31] (Proteome Software, Inc., Portland, OR, USA). Only protein identifications with greater than 99% probability were accepted. Protein probabilities were assigned by the ProteinProphet™ algorithm (Proteome Software, Inc., Portland, OR, USA) [32]. The dataset was searched against the target database and a decoy database, determining the false discovery rate to be 0.1% at the peptide level; the latter featured the reversed amino acid sequences of all the entries in the IPI-human database (v3.69) [33, 34]. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Normalized spectral counts were determined from the Scaffold output.
RESULTS
Isolating Proteins from Pancreatic Fluid by Either TCA or Ultracentrifugation Precipitation Resulted in Different SDS-PAGE Banding Patterns
A total of 6 pancreatic fluid specimens underwent SDS-PAGE analysis (Figure 2). One aliquot of each specimen was subjected to TCA precipitation and the second aliquot to ultracentrifugation precipitation, for a total of 12 samples. The TCA-precipitated samples showed protein banding patterns similar to those seen in previous pancreatic fluid TCA precipitations [8, 10, 11, 35]. However, these protein banding patterns differed substantially from those observed in the ultracentrifugation-precipitated samples; the ultracentrifugation samples were far less concentrated and fewer stained bands were observed. Many of the highly abundant proteins observed in the TCA-precipitated samples were not observed in the ultracentrifugation-precipitated samples. It was likely that these proteins were very soluble and thus did not precipitate from solution. In a prior study using precipitation methods other than TCA, we noted smearing of the low molecular weight protein bands, indicating these proteins had undergone substantial proteolysis [11]. Such smearing was not observed in the ultracentrifugation-precipitated protein lanes, and as such we suspect that the ultracentrifugation samples had not undergone extensive proteolysis post-centrifugation. However, further data accessing protein degradation, such as the addition of labeled standards, are needed to support this claim. This finding is expected as the centrifugations were performed at 4°C, a temperature which we have shown does not result in noticeable proteolysis over the 2-hour precipitation period [11].
Figure 2.
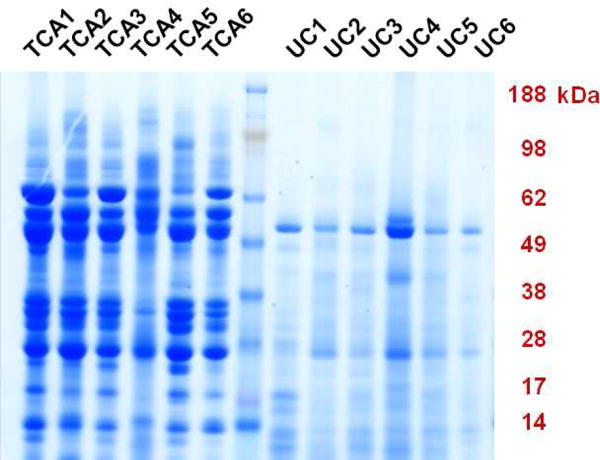
SDS-PAGE protein fractionation. SDS-PAGE images of trichloroacetic acid- (TCA) and ultracentrifugation (UC) precipitated protein samples. Subjects were coded 1 through 6; samples labeled with the same numerical designation following the TCA or UC notation were collected from the same subject.
Pancreatic Fluid Proteins Were Identified Via Ultracentrifugation Without TCA Precipitation
We identified a total of 440 non-redundant proteins using the standard TCA-precipitation followed by GeLC-MS/MS method, and a total of 196 proteins using the ultracentrifugation precipitation method (Supplementary Table). One hundred and eighty three of the 440 (approximately 43%) proteins identified using TCA precipitation were also identified using ultracentrifugation precipitation (Figure 3a). A total of 257 proteins were identified exclusively in the TCA-precipitated samples, while only 13 were exclusive to the ultracentrifugation-precipitated samples (Figure 3a). These 13 ultracentrifugation-specific proteins were listed in Table 1. Many of these proteins had less than five spectral counts; however, several with higher counts may merit further investigation, particularly cathepsin G, eosinophil peroxidase, ferritin heavy chain and mucin-1. Although limited, ultracentrifugation precipitation would aid assays designed to target these particular proteins relevant to pancreatic disease.
Figure 3.
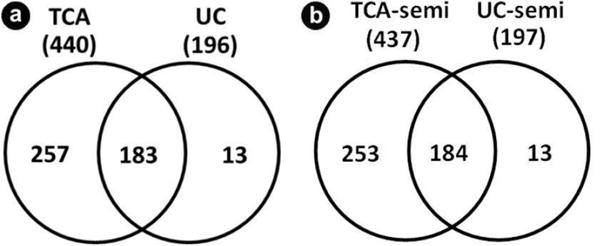
Comparison of proteins identified in the two precipitation methods. a. Venn diagram of non-redundant proteins comparing those identified (with fully tryptic specificity) by trichloroacetic acid (TCA) precipitation and ultracentrifugation (UC) precipitation. b. Semi-tryptic searching for both TCA (TCA-semi) and UC (UC-semi) precipitation.
Table 1.
Proteins identified exclusively in the ultracentrifugation-precipitated samples using fully tryptic specificity for database searching.
| Protein name | UniProt accession number | Average spectral counts
|
|
|---|---|---|---|
| Trichloroacetic acid (TCA) | Ultracentrifugation (UC) | ||
| Acid sphingomyelinase-like phosphodiesterase 3b | Q92485 | 0 | 4 |
| Alkaline phosphatase, tissue-nonspecific isozyme | P05186 | 0 | 2 |
| Calmodulin-like protein 3 | P27482 | 0 | 1 |
| Cathepsin G | P08311 | 0 | 6 |
| Complement component C9 | P02748 | 0 | 1 |
| C-reactive protein | P02741 | 0 | 2 |
| Ectonucleotidepyrophosphatase family member 7 | Q6UWV6 | 0 | 3 |
| Eosinophil peroxidase | P11678 | 0 | 9 |
| Ferritin heavy chain | P02794 | 0 | 6 |
| Histone H2B type 1-K | O60814 | 0 | 1 |
| Isoform 1 of Plakophilin-1 | Q13835-2 | 0 | 1 |
| Mucin-1 | P15941 | 0 | 5 |
| Pyruvate kinase isozymes M1/M2 | P14618 | 0 | 1 |
Database Searching for Semi-Tryptic Proteins Resulted in Few Additional Protein Identifications in Both TCA- and Ultracentrifugation-Precipitated Samples
We repeated the database search using semi-tryptic enzyme specificity, rather than the typical fully tryptic search parameter. As fully tryptic peptides were cleaved at the both peptide termini at either a lysine or arginine residue, semi-tryptic peptides have only one tryptic terminus as the other terminus was cleaved potentially by another protease. Semi-tryptic searches require longer database search times due to the increase in possible peptide matches, resulting in an enlarged search space. We compared protein identifications in TCA-precipitated samples to that in ultracentrifugation-precipitated samples searched using semi-tryptic specificity (Figure 3b). The result was similar to the comparisons of the precipitation methods using fully tryptic specificity (Figure 3a). Forty-two percent of the proteins that were identified using TCA precipitation were also identified using ultracentrifugation precipitation. In all, 13 proteins were identified in the ultracentrifugation-precipitated samples and not identified in the TCA-precipitated sample (Table 2). As in the prior comparison of fully tryptic proteins, many of the identified proteins had relatively low spectral counts (i.e., less than five). As in the fully tryptic comparison, the proteins eosinophil peroxidase, ferritin heavy chain and mucin-1 were also identified exclusively in the ultracentrifugation-precipitated samples when using the semi-tryptic database search. Interestingly, the spectral counts of these proteins were lower than those of proteins searched with full trypsin specificity, again potentially a result of increased search space, while retaining the 0.1% peptide false discovery rate.
Table 2.
Proteins identified exclusively in the ultracentrifugation-precipitated samples using semi-tryptic specificity for database searching.
| Protein name | UniProt accession number | Average spectral counts
|
|
|---|---|---|---|
| Trichloroacetic acid semi-tryptic | Ultracentrifugation semi-tryptic | ||
| Acid sphingomyelinase-like phosphodiesterase 3b | Q92485 | 0 | 4 |
| Alkaline phosphatase, tissue-nonspecific isozyme | P05186 | 0 | 2 |
| Annexin A11 | P50995 | 0 | 2 |
| Calmodulin-like protein 3 | P27482 | 0 | 1 |
| C-reactive protein | P02741 | 0 | 1 |
| Ectonucleotidepyrophosphatase family member 7 | Q6UWV6 | 0 | 3 |
| Eosinophil peroxidase | P11678 | 0 | 8 |
| Ferritin heavy chain | P02794 | 0 | 5 |
| Histone H2B type 1-K | O60814 | 0 | 2 |
| Isoform 1 of Plakophilin-1 | Q13835-2 | 0 | 1 |
| Mucin-1 | P15941 | 0 | 4 |
| WD repeat-containing protein 67 | Q96DN5 | 0 | 2 |
| Zinc finger protein 337 | Q9Y3M9 | 0 | 2 |
Gene Ontology Classification Revealed Mostly Subtle Differences in Classification Categories Between Proteins Isolated from TCA and Ultracentrifugation Precipitation
Using the PANTHER classification system, we performed gene ontology analysis comparing the proteins identified in the TCA precipitation samples (n=440) to those identified in the ultracentrifugation precipitation (n=196) samples. Only categories with 6 or more proteins were considered. We investigated the three domains of gene ontology classification: cellular compartment, biological process and molecular function. Analysis of cellular compartment (Figure 4a) revealed that cytoskeletal and intracellular proteins comprised a greater proportion of proteins in the TCA-precipitated samples compared to proteins in the ultracentrifugation-precipitated samples. Compared to the TCA-precipitated samples, a marginally greater proportion of proteins in the ultracentrifugation-precipitated samples were localized to the extracellular matrix, extracellular region, immunoglobulin complexes and protein complexes. Analysis of the biological processes (Figure 4b) did not reveal many differences in the protein classifications between the two precipitation techniques. Proteins involved in metabolic processes made up a slightly greater proportion of proteins identified in the TCA-precipitated samples. Similarly, proteins involved in the immune response and response to stimulus constituted a slightly greater proportion of proteins in the ultracentrifugation samples, reflecting a similarly greater proportion of immunoglobulin complex proteins identified by the cellular component classification. Comparison of the molecular function category (Figure 4c) also resulted in some slight differences. Compared to ultracentrifugation-precipitated proteins, the TCA-precipitated proteins were comprised of a marginally greater proportion of proteins with oxidoreductase activity and protein binding capability. This observation is consistent with the greater proportion of metabolic process proteins and intracellular proteins observed in the cellular compartment and biological process domains. In contrast, proportionally more peptidase (protease) activity-related proteins, as well as serine peptidase inhibitor (a protease inhibitor) activity-related proteins, were identified in the ultracentrifugation-precipitated sample. These classes of proteins are of particular interest in that several proteases and protease inhibitors have been identified as potential biomarkers of chronic pancreatitis (Table 3) [8, 9].
Figure 4.
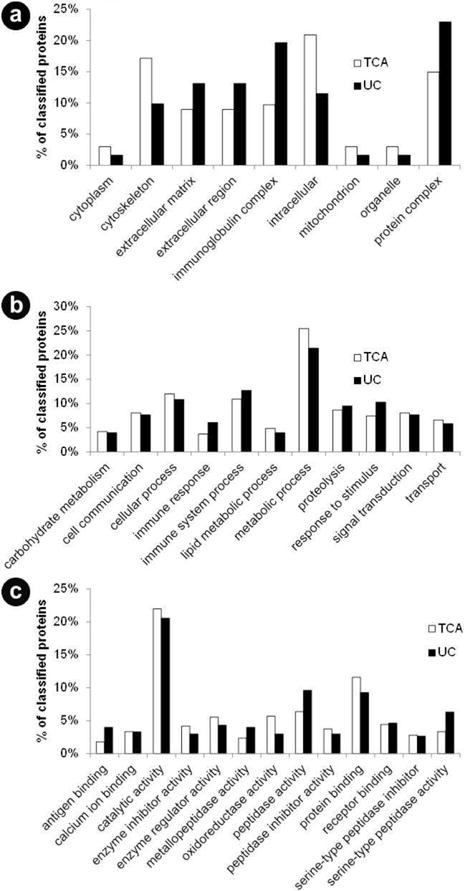
Gene ontology analysis. Using the Panther classification system, gene ontology analysis revealed differences in the cellular compartments (a.), biological processes (b.), and molecular functions of proteins identified in trichloroacetic acid-precipitated (TCA) samples compared to proteins in the ultracentrifugation-precipitated samples (UC) (c.). Note that the data is normalized for each sample as 440 proteins were considered for TCA and 196 for UC.
Table 3.
Previously-identified potential biomarkers of chronic pancreatitis which were also identified in the ultracentrifugation-precipitated samples.
| Protein name | UniProt accession number | Sum of spectral counts for sample type
|
|||
|---|---|---|---|---|---|
| Trichloroacetic acid | Trichloroacetic acid semi-tryptic | Ultracentrifugation | Ultracentrifugation semi-tryptic | ||
| Alpha-1-antichymotrypsin | P01011 | 76 | 70 | 3 | 3 |
| Alpha-1-antitrypsin | P01009 | 732 | 708 | 711 | 663 |
| Alpha-amylase 1 | P04745 | 1,001 | 1,032 | 2,145 | 1,944 |
| Alpha-amylase 2B | P19961 | 998 | 1,054 | 2,506 | 2,299 |
| Aminopeptidase N | P15144 | 306 | 291 | 161 | 186 |
| Bile salt-activated lipase | P19835 | 662 | 649 | 121 | 143 |
| Carboxypeptidase A1 | P15085 | 1,628 | 1,528 | 1,559 | 1,653 |
| Carboxypeptidase A2 | P48052 | 134 | 132 | 63 | 57 |
| Carboxypeptidase B | P15086 | 1,181 | 1,163 | 992 | 1,024 |
| Cathepsin G | P08311 | 2 | 3 | 26 | 35 |
| Chymotrypsin-C | Q99895 | 293 | 287 | 180 | 254 |
| Chymotrypsin-like elastase family member 2A | P08217 | 957 | 971 | 350 | 389 |
| Chymotrypsin-like elastase family member 2B | P08218 | 338 | 481 | 111 | 121 |
| Chymotrypsin-like elastase family member 3A | P09093 | 573 | 605 | 891 | 911 |
| Chymotrypsin-like elastase family member 3B | P08861 | 245 | 361 | 475 | 379 |
| Chymotrypsinogen B2 | Q6GPI1 | 819 | 837 | 127 | 83 |
| Colipase | P04118 | 70 | 93 | 33 | 32 |
| Complement C3 | P01024 | 642 | 605 | 76 | 89 |
| Gastric triacylglycerol lipase | P07098 | 91 | 104 | 30 | 32 |
| Haptoglobin | P00738 | 326 | 302 | 4 | 5 |
| Mucin-5AC (Fragments) | P98088 | 251 | 240 | 30 | 22 |
| Mucin-5B | Q9HC84 | 150 | 140 | 188 | 189 |
| Neprilysin | P08473 | 45 | 42 | 89 | 104 |
| Pancreatic alpha-amylase | P04746 | 1,137 | 1,188 | 2,798 | 2,640 |
| Pancreatic lipase-related protein 1 | P54315 | 51 | 49 | 49 | 24 |
| Pancreatic lipase-related protein 2 | P54317 | 191 | 192 | 194 | 194 |
| Pancreatic triacylglycerol lipase | P16233 | 1,485 | 1,640 | 1,243 | 1,136 |
We Found no Evidence that These Ultra-centrifugation-Precipitated Proteins Were of Exosomal Origin
We suspected that pancreatic fluid may be rich is exosomes, as these small membrane vesicles have been identified and extensively studied in urine [14, 36, 37, 38, 39, 40, 41]. However, our gene ontology analysis did not identify a significant number of membrane proteins in either the TCA- or ultracentrifugation-precipitated samples. We searched an online database, ExoCarta (http://www.exocarta.org/) [15, 16], which lists the top 27 exosomal marker proteins. Only two of these validated exosomal markers were identified in the pancreatic fluid samples: phosphoglycerate kinase 1 and actin, cytoplasmic 1. These proteins were identified in greater abundance in the TCA precipitated samples, and therefore provide no evidence that proteins exclusive to ultracentrifugation-precipitation were exosomal in origin. While we did not definitively identify exosomal marker proteins, it is inconclusive as to whether exosomes are present in pancreatic fluid as these marker proteins may have been below the detection limit of the instrument.
DISCUSSION
We have identified a subset of proteins in pancreatic fluid using ultracentrifugation-promoted protein precipitation in lieu of TCA precipitation. Using ultracentrifugation precipitation coupled with GeLC-MS/MS, we identified approximately 40% of the proteins identified using traditional TCA precipitation. Unlike many other previously-tested pancreatic fluid protein extraction methods [11], ultracentrifugation precipitation did not result in a high degree of proteolysis, typically observed as low molecular weight smears on SDS-PAGE images. Furthermore, ultracentrifugation-based precipitation requires only a single aspiration prior to SDS-PAGE, reducing handling procedures and potential for sample loss and as such is more amenable to robotic processing. The major caveat of this simplified procedure is that it requires at least 5 times the volume of pancreatic fluid per analysis compared to TCA precipitation: 200 μL needed for TCA precipitation versus 1 mL for the ultracentrifugation precipitation strategy described herein.
Secretin-stimulation and ePFT collection facilitates obtaining enough pancreatic fluid to perform the ultracentrifugation method, whereas more common collection techniques cannot safely collect such volumes [19, 20, 21, 42, 43, 44]. To date, several mass spectrometry-based proteomic investigations of pancreatic fluid have been performed using specimens collected surgically or via endoscopic retrograde cholangiopancreatography (ERCP) [45, 46, 47, 48, 49, 50, 51, 52, 53] which collect up to 10-fold lower volumes of pancreatic fluid than and at most twice the concentration of ePFT-collected fluid. From these experiments, the number of proteins identified varied from 22 to over 170 based on the mass spectrometry methodology utilized. More recently, the proteome of normal pancreatic fluid has been elucidated, which discovered over 270 proteins, again from ERCP-collected fluid [54]. The method of sample collection is a major caveat when comparing proteomic data our ePFT-collected fluid sampled to other published datasets.
In our prior studies using TCA protein precipitation, we identified several dozen potential protein biomarkers for chronic pancreatitis [8, 9]. Several of these proteins (including pancreatic enzymes (amylases, lipases, chymotrypsinogens, and carboxypeptidases), enzyme inhibitors (alpha-1-antitrypsin and alpha-1-antichymotrypsin), mucins, haptoglobin and neprilysin) were also identified in ultracentrifugation-precipitated pancreatic fluid (Table 3). These findings demonstrate that ultracentrifugation may be used as an alternative to TCA precipitation to evaluate certain biomarker candidates of chronic pancreatitis, as well as other pancreatic diseases in targeted assays. However, a major caveat is that incomplete precipitation due to ultracentrifugation may limit the number of proteins that are available for these targeted assays. Further analysis is warranted to ensure that reproducibility of the quantitative data is not hindered by different degrees of precipitation among samples which are treated similarly.
Interestingly, few additional proteins were identified by semi-tryptic peptide searching in both the TCA- and ultracentrifugation-precipitated samples. Semi-tryptic peptides appear when a peptide is cleaved up- or downstream of a canonical tryptic cleavage site. Such a phenomenon is typically a result of endogenous proteases, which are abundant in pancreatic fluid. The results presented here, and unpublished data, demonstrated that, generally, few additional proteins are identified in pancreatic fluid when semi-tryptic peptides are included in the database searches, suggesting that our standardized collection method is effective in limiting endogenous proteolysis [11]. Semi-tryptic searches, although easily implemented, are computationally intensive and can increase database search times several fold, and thus for the analysis herein are generally not computationally efficient.
SDS-PAGE banding patterns and mass spectrometric analysis of the ultracentrifugation-precipitated samples demonstrated fewer proteins compared to the TCA-precipitated samples. Soluble proteins typically remain in solution following ultracentrifugation at 200,000 g, leading us to postulate initially that the appearance of these proteins in the ultracentrifugation precipitate may be a result of the precipitation of exosomes or other small membrane vesicles that may be present in pancreatic fluid. Exosomes are small membrane vesicles (30 to 90 nm) originating from a variety of cell types [55], which fuse with the plasma membrane and empty their contents into another cell. Although not yet investigated in the context of pancreatic disease, exosomes have been implicated in the adaptive immune response and inflammatory response [56] in other diseases, and have been identified in urine [14, 36, 37, 38, 39, 40, 41]. While microvesicles in human body fluids such as blood [57, 58, 59] and urine [14, 36, 37, 38, 39, 40, 41] have been thoroughly investigated, to date, no investigation of such microvesicles in human pancreatic fluid has been published. Although we were unable to identify exosomal markers in our analysis, it remains possible that such proteins are indeed present, but at a level below the detection limit of our mass spectrometry-based strategy. The use of more concentrated protein samples, targeted mass spectrometry-based assays, or traditional techniques (such as western or dot blotting), along with the proper controls may result in a better assessment of the presence of exosomes in pancreatic fluid.
In addition to co-precipitation with microvesicles, protein aggregation may also contribute to ultracentrifugation-based precipitation of pancreatic fluid proteins. Secretin-stimulated, ePFT-collected pancreatic fluid typically has a protein concentration ranging from 1–2 mg/mL, a relatively high concentration which may promote protein aggregation and induce precipitation during high-speed centrifugation. Although it was our intention to enrich, rather than purify, the ultracentrifugation-precipitated proteins, we omitted washing the pellet to prevent pelleted protein loss. In addition, analysis of the post-ultracentrifugation supernatant may also reveal the degree of incomplete protein precipitation. If purer ultracentrifugation pellets are needed, potential carryover from the supernatant may be reduced by the washing with a non-detergent buffer. It may also be of interest to use differential centrifugation to define in more depth the proteome of pancreatic fluid. Further studies are necessary to determine the extent of these effects.
It may be expected that protein complexes will be precipitated using our ultracentrifugation strategy. As such, there is some evidence supporting the plausibility of protein complexes in the ultracentrifugation-precipitated pancreatic fluid. Zymogen granules are small secretory organelles in pancreatic acinar cells that contain inactivated digestive enzymes. These intercellular vesicles are typically isolated from acinar tissue and their ability to be isolated in pancreatic fluid has not yet been determined, although there presence may be possible due to cell lysis [60, 61, 62]. Studies have shown that pancreatic enzymes, such as lipase, trypsin, chymotrypsin, and carboxypeptidase are abundant, as well as inhibitors, such as alpha-1-antitrypsin are abundant in zymogen granules [60, 63]. Proteins with the highest spectral counts in the ultracentrifugation-precipitated samples included these pancreatic enzymes. Similarly, trypsin is known to form complexes with anti-1-antitrypisn, and both of these molecules have are among those with the highest spectral counts. Further investigation will be needed to assess the presence of such complexes, such as analysis via native gels or blue native-PAGE [64].
In summary, we have shown that ultracentrifugation may be a viable alternative to TCA precipitation for the proteomic analysis of pancreatic fluid. Approximately 40% of the proteins identified in TCA-precipitated pancreatic fluid were also identified in the ultracentrifugation-precipitated samples. The primary advantages of precipitation by ultracentrifugation include minimal sample processing and no incubation with strong acid. The benefits of ultracentrifugation precipitation over TCA precipitation come at the expense of sample volume, as ultracentrifugation requires at least five times the sample volume. Further studies are needed to understand better the mechanisms by which these pancreatic fluid proteins precipitate when subjected to ultracentrifugation. The ultra-centrifugation precipitation strategy outlined herein represents a method of studying the proteome of pancreatic fluid with minimal sample processing, which may be useful in the search for biomarkers of pancreatic disease.
Supplementary Material
Acknowledgments
Funds were provided by the following NIH grants: 1 F32 DK085835-01A1 (J.A.P.), 1 R21 DK081703-01A2 (D.L.C.) and5 P30 DK034854-24 (Harvard Digestive Diseases Center; D.L.C.). In addition, we would like to thank the Burrill family for their generous support through the Burrill Research Grant. We would also like to thank members of the Steen Laboratory at Children’s Hospital Boston and Harvard Medical School
Abbreviations
- ePFT
endoscopic pancreatic function test
- GeLC-MS/MS
in-gel tryptic digestion followed by liquid chromatography-tandem mass spectrometry
- LDS
lithium dodecyl sulfate
- MES
2-(N-morpholino)ethanesulfonic acid
- TCA
trichloroacetic acid
- UC
ultracentrifugation
Footnotes
Conflicts of interests The authors declare no competing interests
References
- 1.Otsuki M. Chronic pancreatitis in Japan: epidemiology, prognosis, diagnostic criteria, and future problems. J Gastroenterol. 2003;38:315–326. doi: 10.1007/s005350300058. [DOI] [PubMed] [Google Scholar]
- 2.Whitcomb DC, Yadav D, Adam S, Hawes RH, Brand RE, Anderson MA, Money ME, Banks PA, Bishop MD, Baillie J, Sherman S, DiSario J, Burton FR, Gardner TB, Amann ST, Gelrud A, Lo SK, DeMeo MT, Steinberg WM, Kochman ML, Etemad B, Forsmark CE, Elinoff B, Greer JB, O’Connell M, Lamb J, Barmada MM. Multicenter approach to recurrent acute and chronic pancreatitis in the United States: the North American Pancreatitis Study 2 (NAPS2) Pancreatology. 2008;8:520–531. doi: 10.1159/000152001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dominguez Munoz JE. Diagnosis of chronic pancreatitis: Functional testing. Best Pract Res Clin Gastroenterol. 2010;24:233–241. doi: 10.1016/j.bpg.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 4.Chowdhury R, Bhutani MS, Mishra G, Toskes PP, Forsmark CE. Comparative analysis of direct pancreatic function testing versus morphological assessment by endoscopic ultrasonography for the evaluation of chronic unexplained abdominal pain of presumed pancreatic origin. Pancreas. 2005;31:63–68. doi: 10.1097/01.mpa.0000164451.69265.80. [DOI] [PubMed] [Google Scholar]
- 5.DiMagno EP, Go VL, Summerskill WH. Relations between pancreatic enzyme ouputs and malabsorption in severe pancreatic insufficiency. N Engl J Med. 1973;288:813–815. doi: 10.1056/NEJM197304192881603. [DOI] [PubMed] [Google Scholar]
- 6.Paulo JA, Kadiyala V, Banks PA, Steen H, Conwell DL. Mass spectrometry-based proteomics for translational research: a technical overview. The Yale journal of biology and medicine. 2012;85:59–73. [PMC free article] [PubMed] [Google Scholar]
- 7.Paulo JA, Lee LS, Wu B, Banks PA, Steen H, Conwell DL. Mass spectrometry-based proteomics of endoscopically collected pancreatic fluid in chronic pancreatitis research. Proteomics Clin Appl. 2011;5:109–120. doi: 10.1002/prca.201000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paulo JA, Kadiyala V, Lee LS, Banks PA, Conwell DL, Steen H. Proteomic Analysis (GeLC-MS/MS) of ePFT-Collected Pancreatic Fluid in Chronic Pancreatitis. J Proteome Res. 2012:1897–1912. doi: 10.1021/pr2011022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paulo JA, Lee LS, Banks PA, Steen H, Conwell DL. Difference gel electrophoresis identifies differentially expressed proteins in endoscopically collected pancreatic fluid. Electrophoresis. 2011;32:1939–1951. doi: 10.1002/elps.201100203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paulo JA, Lee LS, Wu B, Repas K, Mortele KJ, Banks PA, Steen H, Conwell DL. Identification of pancreas-specific proteins in endoscopically (endoscopic pancreatic function test) collected pancreatic fluid with liquid chromatography–tandem mass spectrometry. Pancreas. 2010;39:889–896. doi: 10.1097/MPA.0b013e3181cf16f4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paulo JA, Lee LS, Wu B, Repas K, Banks PA, Conwell DL, Steen H. Optimized sample preparation of endoscopic collected pancreatic fluid for SDS-PAGE analysis. Electrophoresis. 2010;31:2377–2387. doi: 10.1002/elps.200900762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farina A, Dumonceau JM, Delhaye M, Frossard JL, Hadengue A, Hochstrasser DF, Lescuyer P. A step further in the analysis of human bile proteome. Journal of proteome research. 2011;10:2047–2063. doi: 10.1021/pr200011b. [DOI] [PubMed] [Google Scholar]
- 13.Farina A, Dumonceau JM, Frossard JL, Hadengue A, Hochstrasser DF, Lescuyer P. Proteomic analysis of human bile from malignant biliary stenosis induced by pancreatic cancer. J Proteome Res. 2009;8:159–169. doi: 10.1021/pr8004925. [DOI] [PubMed] [Google Scholar]
- 14.Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathivanan S, Simpson RJ. ExoCarta: A compendium of exosomal proteins and RNA. Proteomics. 2009;9:4997–5000. doi: 10.1002/pmic.200900351. [DOI] [PubMed] [Google Scholar]
- 16.Mathivanan S, Fahner CJ, Reid GE, Simpson RJ. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic acids research. 2012;40:D1241–1244. doi: 10.1093/nar/gkr828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diz AP, Truebano M, Skibinski DO. The consequences of sample pooling in proteomics: an empirical study. Electrophoresis. 2009;30:2967–2975. doi: 10.1002/elps.200900210. [DOI] [PubMed] [Google Scholar]
- 18.Karp NA, Lilley KS. Investigating sample pooling strategies for DIGE experiments to address biological variability. Proteomics. 2009;9:388–397. doi: 10.1002/pmic.200800485. [DOI] [PubMed] [Google Scholar]
- 19.Stevens T, Conwell D, Zuccaro G, Van Lente F, Khandwala F, Hanaway P, Vargo JJ, Dumot JA. Analysis of pancreatic elastase-1 concentrations in duodenal aspirates from healthy subjects and patients with chronic pancreatitis. Dig Dis Sci. 2004;49:1405–1411. doi: 10.1023/b:ddas.0000042238.80040.cc. [DOI] [PubMed] [Google Scholar]
- 20.Stevens T, Conwell DL, Zuccaro G, Van Lente F, Khandwala F, Purich E, Vargo JJ, Fein S, Dumot JA, Trolli P, O’Laughlin C. Electrolyte composition of endoscopically collected duodenal drainage fluid after synthetic porcine secretin stimulation in healthy subjects. Gastrointestinal endoscopy. 2004;60:351–355. doi: 10.1016/s0016-5107(04)01809-7. [DOI] [PubMed] [Google Scholar]
- 21.Wu B, Conwell DL. The endoscopic pancreatic function test. The American journal of gastroenterology. 2009;104:2381–2383. doi: 10.1038/ajg.2008.181. [DOI] [PubMed] [Google Scholar]
- 22.Finnie C, Svensson B. Proteolysis during the isoelectric focusing step of two-dimensional gel electrophoresis may be a common problem. Anal Biochem. 2002;311:182–186. doi: 10.1016/s0003-2697(02)00389-5. [DOI] [PubMed] [Google Scholar]
- 23.Marshall J, Kupchak P, Zhu W, Yantha J, Vrees T, Furesz S, Jacks K, Smith C, Kireeva I, Zhang R, Takahashi M, Stanton E, Jackowski G. Processing of serum proteins underlies the mass spectral fingerprinting of myocardial infarction. J Proteome Res. 2003;2:361–372. doi: 10.1021/pr030003l. [DOI] [PubMed] [Google Scholar]
- 24.Rai AJ, Gelfand CA, Haywood BC, Warunek DJ, Yi J, Schuchard MD, Mehigh RJ, Cockrill SL, Scott GB, Tammen H, Schulz-Knappe P, Speicher DW, Vitzthum F, Haab BB, Siest G, Chan DW. HUPO Plasma Proteome Project specimen collection and handling: towards the standardization of parameters for plasma proteome samples. Proteomics. 2005;5:3262–3277. doi: 10.1002/pmic.200401245. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Neubauer G, Mann M. Mapping of phosphorylation sites of gelisolated proteins by nanoelectrospray tandem mass spectrometry: potentials and limitations. Anal Chem. 1999;71:235–242. doi: 10.1021/ac9804902. [DOI] [PubMed] [Google Scholar]
- 27.Steen H, Kuster B, Fernandez M, Pandey A, Mann M. Detection of tyrosine phosphorylated peptides by precursor ion scanning quadrupole TOF mass spectrometry in positive ion mode. Analytical chemistry. 2001;73:1440–1448. doi: 10.1021/ac001318c. [DOI] [PubMed] [Google Scholar]
- 28.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome biology. 2003;4:P3. [PubMed] [Google Scholar]
- 29.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 30.Thomas PD, Kejariwal A, Campbell MJ, Mi H, Diemer K, Guo N, Ladunga I, Ulitsky-Lazareva B, Muruganujan A, Rabkin S, Vandergriff JA, Doremieux O. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic acids research. 2003;31:334–341. doi: 10.1093/nar/gkg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Analytical chemistry. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 32.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Analytical chemistry. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 33.Elias JE, Gibbons FD, King OD, Roth FP, Gygi SP. Intensity-based protein identification by machine learning from a library of tandem mass spectra. Nature biotechnology. 2004;22:214–219. doi: 10.1038/nbt930. [DOI] [PubMed] [Google Scholar]
- 34.Moore RE, Young MK, Lee TD. Method for screening peptide fragment ion mass spectra prior to database searching. J Am Soc Mass Spectrom. 2000;11:422–426. doi: 10.1016/S1044-0305(00)00097-0. [DOI] [PubMed] [Google Scholar]
- 35.Paulo J, Vaezzadeh A, Conwell D, Lee R, Steen H. Sample Handling of Body Fluids for Proteomics. In: Ivanov A, Lazarev A, editors. Sample Preparation in Biological Mass Spectrometry. Springer; New York, NY: 2011. [Google Scholar]
- 36.Chen M, Wang K, Zhang L, Li C, Yang Y. The discovery of putative urine markers for the specific detection of prostate tumor by integrative mining of public genomic profiles. PLoS One. 2011;6:e28552. doi: 10.1371/journal.pone.0028552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z, Hill S, Luther JM, Hachey DL, Schey KL. Proteomic analysis of urine exosomes by multidimensional protein identification technology (MudPIT) Proteomics. 2012;12:329–338. doi: 10.1002/pmic.201100477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Conde-Vancells J, Rodriguez-Suarez E, Gonzalez E, Berisa A, Gil D, Embade N, Valle M, Luka Z, Elortza F, Wagner C, Lu SC, Mato JM, Falcon-Perez M. Candidate biomarkers in exosome-like vesicles purified from rat and mouse urine samples. Proteomics Clin Appl. 2010;4:416–425. doi: 10.1002/prca.200900103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gonzales PA, Zhou H, Pisitkun T, Wang NS, Star RA, Knepper MA, Yuen PS. Isolation and purification of exosomes in urine. Methods in molecular biology (Clifton, N.J) 2010;641:89–99. doi: 10.1007/978-1-60761-711-2_6. [DOI] [PubMed] [Google Scholar]
- 40.Knepper MA, Pisitkun T. Exosomes in urine: who would have thought…? Kidney international. 2007;72:1043–1045. doi: 10.1038/sj.ki.5002510. [DOI] [PubMed] [Google Scholar]
- 41.Keller S, Rupp C, Stoeck A, Runz S, Fogel M, Lugert S, Hager HD, Abdel-Bakky MS, Gutwein P, Altevogt P. CD24 is a marker of exosomes secreted into urine and amniotic fluid. Kidney international. 2007;72:1095–1102. doi: 10.1038/sj.ki.5002486. [DOI] [PubMed] [Google Scholar]
- 42.Conwell DL, Zuccaro G, Jr, Vargo JJ, Morrow JB, Obuchowski N, Dumot JA, Trolli PA, Burton A, O’Laughlin C, Van Lente F. An endoscopic pancreatic function test with cholecystokinin-octapeptide for the diagnosis of chronic pancreatitis. Clin Gastroenterol Hepatol. 2003;1:189–194. doi: 10.1053/cgh.2003.50028. [DOI] [PubMed] [Google Scholar]
- 43.Conwell DL, Zuccaro G, Jr, Vargo JJ, Trolli PA, Vanlente F, Obuchowski N, Dumot JA, O’Laughlin C. An endoscopic pancreatic function test with synthetic porcine secretin for the evaluation of chronic abdominal pain and suspected chronic pancreatitis. Gastrointestinal endoscopy. 2003;57:37–40. doi: 10.1067/mge.2003.14. [DOI] [PubMed] [Google Scholar]
- 44.Conwell DL, Zuccaro G, Morrow JB, Van Lente F, O’Laughlin C, Vargo JJ, Dumot JA. Analysis of duodenal drainage fluid after cholecystokinin (CCK) stimulation in healthy volunteers. Pancreas. 2002;25:350–354. doi: 10.1097/00006676-200211000-00005. [DOI] [PubMed] [Google Scholar]
- 45.Chen R, Brentnall TA, Pan S, Cooke K, Moyes KW, Lane Z, Crispin DA, Goodlett DR, Aebersold R, Bronner MP. Quantitative proteomics analysis reveals that proteins differentially expressed in chronic pancreatitis are also frequently involved in pancreatic cancer. Mol Cell Proteomics. 2007;6:1331–1342. doi: 10.1074/mcp.M700072-MCP200. [DOI] [PubMed] [Google Scholar]
- 46.Chen R, Pan S, Cooke K, Moyes KW, Bronner MP, Goodlett DR, Aebersold R, Brentnall TA. Comparison of pancreas juice proteins from cancer versus pancreatitis using quantitative proteomic analysis. Pancreas. 2007;34:70–79. doi: 10.1097/01.mpa.0000240615.20474.fd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen R, Yi EC, Donohoe S, Pan S, Eng J, Cooke K, Crispin DA, Lane Z, Goodlett DR, Bronner MP, Aebersold R, Brentnall TA. Pancreatic cancer proteome: the proteins that underlie invasion, metastasis, and immunologic escape. Gastroenterology. 2005;129:1187–1197. doi: 10.1053/j.gastro.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 48.Cui Y, Tian M, Zong M, Teng M, Chen Y, Lu J, Jiang J, Liu X, Han J. Proteomic Analysis of Pancreatic Ductal Adenocarcinoma Compared with Normal Adjacent Pancreatic Tissue and Pancreatic Benign Cystadenoma. Pancreatology. 2008;9:89–98. doi: 10.1159/000178879. [DOI] [PubMed] [Google Scholar]
- 49.Gronborg M, Bunkenborg J, Kristiansen TZ, Jensen ON, Yeo CJ, Hruban RH, Maitra A, Goggins MG, Pandey A. Comprehensive proteomic analysis of human pancreatic juice. Journal of proteome research. 2004;3:1042–1055. doi: 10.1021/pr0499085. [DOI] [PubMed] [Google Scholar]
- 50.Gronborg M, Kristiansen TZ, Iwahori A, Chang R, Reddy R, Sato N, Molina H, Jensen ON, Hruban RH, Goggins MG, Maitra A, Pandey A. Biomarker discovery from pancreatic cancer secretome using a differential proteomic approach. Mol Cell Proteomics. 2006;5:157–171. doi: 10.1074/mcp.M500178-MCP200. [DOI] [PubMed] [Google Scholar]
- 51.Ke E, Patel BB, Liu T, Li XM, Haluszka O, Hoffman JP, Ehya H, Young NA, Watson JC, Weinberg DS, Nguyen MT, Cohen SJ, Meropol NJ, Litwin S, Tokar JL, Yeung AT. Proteomic Analyses of Pancreatic Cyst Fluids. Pancreas. 2009;38:33–42. doi: 10.1097/MPA.0b013e318193a08f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li C, Simeone DM, Brenner DE, Anderson MA, Shedden KA, Ruffin MT, Lubman DM. Pancreatic Cancer Serum Detection Using a Lectin/Glyco-Antibody Array Method. J Proteome Res. 2008;8:483–492. doi: 10.1021/pr8007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao Y, Lee WN, Lim S, Go VL, Xiao J, Cao R, Zhang H, Recker RR, Xiao GG. Quantitative proteomics: measuring protein synthesis using 15N amino acid labeling in pancreatic cancer cells. Analytical chemistry. 2009;81:764–771. doi: 10.1021/ac801905g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Doyle C, Yancey K, Pitt H, Wang M, Bemis K, Yip-Schneider M, Sherman S, Lillemoe K, Goggins M, Schmidt C. The proteome of normal pancreatic juice. Pancreas. 2012;41:186–194. doi: 10.1097/MPA.0b013e31822862f6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics. 2009;6:267–283. doi: 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- 56.Keller S, Sanderson MP, Stoeck A, Altevogt P. Exosomes: from biogenesis and secretion to biological function. Immunol Lett. 2006;107:102–108. doi: 10.1016/j.imlet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 57.Grant R, Ansa-Addo E, Stratton D, Antwi-Baffour S, Jorfi S, Kholia S, Krige L, Lange S, Inal J. A filtration-based protocol to isolate human plasma membrane-derived vesicles and exosomes from blood plasma. J Immunol Methods. 2011;371:143–151. doi: 10.1016/j.jim.2011.06.024. [DOI] [PubMed] [Google Scholar]
- 58.Li QL, Bu N, Yu YC, Hua W, Xin XY. Exvivo experiments of human ovarian cancer ascites-derived exosomes presented by dendritic cells derived from umbilical cord blood for immunotherapy treatment, Clinical medicine. Oncology. 2008;2:461–467. doi: 10.4137/cmo.s776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int Immunol. 2005;17:879–887. doi: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- 60.Rindler MJ. Isolation of zymogen granules from rat pancreas. Curr Protoc Cell Biol. 2006 doi: 10.1002/0471143030.cb0318s29. Chapter 3. Unit 3 18. [DOI] [PubMed] [Google Scholar]
- 61.De Lisle RC, Schulz I, Tyrakowski T, Haase W, Hopfer U. Isolation of stable pancreatic zymogen granules. Am J Physiol. 1984;246:G411–418. doi: 10.1152/ajpgi.1984.246.4.G411. [DOI] [PubMed] [Google Scholar]
- 62.Paquet MR, St-Jean P, Roberge M, Beaudoin AR. Isolation of zymogen granules from rat pancreas and characterization of their membrane proteins. European journal of cell biology. 1982;28:20–26. [PubMed] [Google Scholar]
- 63.Chen X, Walker AK, Strahler JR, Simon ES, Tomanicek-Volk SL, Nelson BB, Hurley MC, Ernst SA, Williams JA, Andrews PC. Organellar proteomics: analysis of pancreatic zymogen granule membranes. Mol Cell Proteomics. 2006;5:306–312. doi: 10.1074/mcp.M500172-MCP200. [DOI] [PubMed] [Google Scholar]
- 64.Swamy M, Siegers GM, Minguet S, Wollscheid B, Schamel WW. Blue native polyacrylamide gel electrophoresis (BN-PAGE) for the identification and analysis of multiprotein complexes. Sci STKE. 2006;2006:p14. doi: 10.1126/stke.3452006pl4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


