Abstract
Context
Formalin-fixed paraffin-embedded (FFPE) tissue is a standard for specimen preservation, and as such FFPE tissue banks are an untapped resource of histologically-characterized specimens for retrospective biomarker investigation for pancreatic disease.
Objectives
We use liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) to compare FFPE specimens from three different diseases of the exocrine pancreas.
Design
We investigated the proteomic profile of FFPE pancreatic tissue from 9 archived specimens that were histologically classified as: autoimmune pancreatitis (n=3), chronic pancreatitis (n=3), and pancreatic cancer (n=3), using LC-MS/MS.
Setting
This is a proteomic analysis experiment of FFPE pancreatic tissue in an academic center.
Patients
FFPE tissue specimens were provided by Dana-Farber/Harvard Cancer Center (Boston, MA, USA).
Interventions
FFPE tissue specimens were collected via routine surgical resection procedures.
Main outcome measures
We compared proteins identified from chronic pancreatitis, autoimmune pancreatitis, and pancreatic cancer FFPE pancreatic tissue.
Results
We identified 386 non-redundant proteins from 9 specimens. Following our filtering criteria, 73, 29, and 53 proteins were identified exclusively in autoimmune pancreatitis, chronic pancreatitis, and pancreatic cancer specimens, respectively.
Conclusions
We report that differentially-expressed proteins can be identified among FFPE tissues specimens originating from individuals with different histological diagnoses. These proteins merit further confirmation with a greater number of specimens and orthogonal validation, such as immunohistochemistry. The mass spectrometry-based methodology used herein has the potential to enhance diagnostic biomarker and therapeutic target discovery, further advancing pancreatic research.
Keywords: Autoimmune Diseases, Biological Markers, Pancreas, Pancreatic Neoplasms, Pancreatitis, Chronic
INTRODUCTION
Diseases of the pancreas affect more than 1 million individuals in the United States annually, resulting in nearly $3 billion in direct and indirect medical costs [1]. Clarification of the biomolecular mechanisms of pancreatic diseases, such as pancreatitis and pancreatic cancer, is vital in identifying diagnostic biomarkers of early disease and developing targeted treatments to modify and/or retard disease progression.
Autoimmune pancreatitis is a benign idiopathic inflammatory disease of the pancreas resulting in masses and duct strictures. A recent study from Japan determined the prevalence of this rare disease to be 0.8/100,000 [2]. Another study estimated that 5–6% of patients classified previously as having idiopathic chronic pancreatitis have had diagnoses reclassified as autoimmune pancreatitis [3, 4]. Autoimmune pancreatitis may closely resemble pancreatic carcinoma, both clinically and radiographically, and thus the two pancreatic diseases can be difficult to differentiate [5, 6]. Current laboratory tests and biopsies by fine needle aspiration are limited in distinguishing the two diseases and cannot definitively rule out malignancy. In fact, over 10% of patients undergoing surgery for suspected pancreatic cancer, instead have autoimmune pancreatitis [7]. Therefore, improved diagnostic methods are needed to distinguish the two diseases. To that end, we propose a mass spectrometry-based proteomic study of formalin-fixed paraffin-embedded (FFPE) tissue for the discovery of biomarker candidates. We have applied previously similar techniques to differentiate late stage chronic pancreatitis from pancreatic cancer [8]; however, we now present the use of this technique in a more clinically relevant context that is the unmet need of distinguishing between autoimmune pancreatitis and pancreatic cancer.
Formalin fixing and paraffin embedding tissue is the standard technique for preserving specimens in hospital pathology departments. FFPE tissue banks are a rich resource for retrospective protein biomarker investigation. These types of specimens have historically been used to investigate the cellular localization of specific proteins via antibody-based immunohistochemistry [9]. Immunohistochemistry requires a priori knowledge of the proteins to be specifically targeted making it unsuitable for large-scale protein identification. State-of-the-art proteomics techniques offer an unbiased exploratory approach, which if applied to tissue repositories, may enhance our understanding of pancreatic disease pathogenesis and pathophysiology, potentially identifying novel targets for immunohistochemistry analysis.
Mass spectrometry-based proteomics is quickly becoming the model strategy for unbiased large-scale protein investigation [10]. Archived FFPE pancreatic tissue specimens offer a robust sample set with which to investigate altered biochemical pathways and uncover potential biomarkers of pancreatic disease. Analogous techniques have been applied previously to FFPE tissues from various organs [11]. The major objectives of this proteomic investigation are: 1) to identify proteins present in FFPE pancreatic tissue using liquid chromatography-coupled with tandem mass spectrometry (LC-MS/MS), and 2) to compare the proteomic profiles of autoimmune pancreatitis, chronic pancreatitis, and pancreatic cancer FFPE pancreas tissue specimens. To our knowledge, we report the first comparison of FFPE pancreas tissue from autoimmune pancreatitis, chronic pancreatitis, and pancreatic cancer.
METHODS
Study Design and Setting
Proteomic analysis of archived formalin-fixed paraffin-embedded (FFPE) pancreas tissue in an academic center.
MATERIALS
FFPE tissue specimens were provided by Dana-Farber/Harvard Cancer Center (Boston, MA, USA). Heptane (product #51750) was from Sigma-Aldrich (St. Louis, MO, USA). Pep-clean C18 spin columns (product #89870) were from Thermo Scientific (Waltham, MA, USA). Sequencing-grade modified trypsin (V5111) was obtained from Promega (Madison, WI, USA). Other reagents and solvents were purchased from Sigma-Aldrich (St. Louis, MO, USA) and Burdick & Jackson (Morristown, NJ, USA), respectively.
Formalin Fixing and Paraffin Embedded of Pancreatic Tissue
FFPE tissue specimens were fixed in 10% neutral-buffered formalin for 48 h (Dana-Farber/Harvard Cancer Center, Research Pathology Core Laboratory). Routine paraffin treatment was performed as follows: specimens were treated twice with 70% ethanol for 45 minutes, twice with 80% ethanol for 60 minutes, twice with 95% ethanol for 60 minutes, thrice with 100% ethanol for 60 minutes, twice with #83 xylene substitute for 60 minutes, and twice with paraffin for 60 minutes. Tissue sections of 5 µm thickness were cut from the FFPE whole-mount pancreatic tissue block, mounted on standard glass slides, and heated for 60 min at 60°C. Slides were stored at room temperature until use.
Experimental Workflow
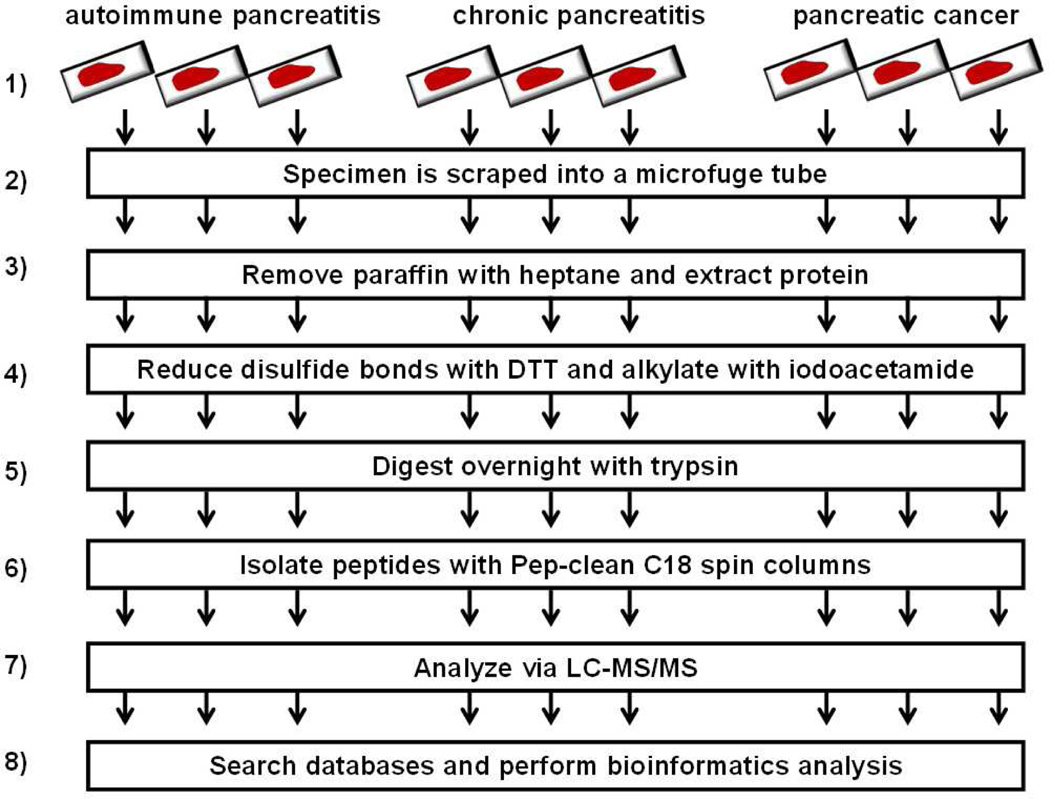
The experimental analysis is outline in Figure 1: 1) FFPE tissue specimens were obtained from the Dana-Farber/Harvard Cancer Center core facility; 2) a 1.5 cm x 1 cm x 5 µm slice of the FFPE tissue was scraped from the slide into a microcentrifuge tube; 3) paraffin was removed with heptane; 4) disulfide bonds were reduced with dithiotreitol (DTT) and alkylated with iodoacetamide; 5) sample was digested overnight with trypsin; 6) peptides were isolated with C18 spin columns; 7) LC-MS/MS analysis was performed; and 8) database searching and bioinformatics processing was performed.
Figure 1.
Experimental workflow. 1) FFPE tissue specimens were obtained; 2) a 1.5 cm 1.5 × 1 cm x 5 µm slice of the specimen was scraped into a microcentrifuge tube; 3) paraffin was removed with heptane; 4) disulfide bonds were reduced with dithiotreitol (DTT) and alkylated with iodoacetamide; 5) specimens were digested overnight with trypsin; 6) peptides were isolated; 7) peptides were analyzed via LC-MS/MS; and 8) bioinformatics analysis was performed.
Sample Preparation for Mass Spectrometry Analysis
The sample preparation protocol for mass spectrometry analysis was assembled from several sources [11, 12, 13, 14, 15]. After removing the excess paraffin from the tissue slice, the FFPE pancreatic tissue specimen from each glass slide was scraped, using a clean razor blade, into a 2 mL microcentrifuge tube. To remove the remaining paraffin, 0.5 mL of heptane was added to each sample, followed by vigorous vortexing for 10 seconds and incubation at room temperature for 1 h. Twenty-five microliters of methanol was then added to each tube, followed by vigorous vortexing for 10 seconds and centrifugation (20,000 g for 2 min at 4°C). Immediately following centrifugation, the upper (heptane) layer was discarded and the lower layer was allowed to evaporate. Proteins were then extracted by resuspending the dried material in 250 µL of 6 M guanidine-HCl/50 mM ammonium bicarbonate/20 mM DTT, pH 8.5, briefly sonicating, and incubating at 70°C for 1 h. After cooling to room temperature, iodoacetamide was added to a final concentration of 40 mM and the sample was incubated in the dark for 1 h. The alkylation reaction was quenched by adding 3 µL of 2 M DTT.
In preparation for tryptic digestion, the sample was diluted 1:6 with 50 mM ammonium bicarbonate (pH 8.1) to reduce the concentration of guanidine-HCl to 1 M. Each sample was incubated with 2.5 µg of trypsin. Following the incubation, the reaction was acidified with formic acid to a final concentration of 0.1% and evaporated via vacuum centrifugation. To remove substances which may interfere with mass spectrometry, peptides were isolated with C18 spin columns according to the manufacturer’s instructions. Samples were again vacuum centrifuged until dry and stored at −80°C until analysis. Immediately prior to analysis, the peptides were resuspended in sample loading buffer (5% formic acid, 5% acetonitrile, 90% water).
Mass Spectrometry
Mass spectrometry analysis was performed at the Proteomics Center at Children’s Hospital Boston, MA, USA. The resuspended peptides were fractionated using reversed-phase high pressure liquid chromatography (HPLC; Thermo Scientific, Waltham, MA, USA) and the gradient-eluted peptides were analyzed using an LTQ FT Ultra mass spectrometer (Thermo Scientific, Waltham, MA, USA). The liquid chromatography columns (15 cm x 100 µm inner diameter) were packed in-house with Magic C18 (5 µm, 100 Å; Michrom BioResources, Auburn, CA, USA), into PicoTips (New Objective, Woburn, MA, USA). Samples were analyzed with a 90 minute linear gradient (0–35% acetonitrile with 0.2% formic acid) and data were acquired in a datadependent manner, in which MS/MS fragmentation was performed on the six most intense peaks of every full MS scan.
ETHICS
The study was approved by our Institutional Review Committee. This protocol was approved by the Institutional Review Board at Brigham and Women’s Hospital and Children’s Hospital Boston (IRB # 2007-P-002480/1).
Written informed consent was obtained from each patient and the study protocol conforms to the ethical guidelines of the “World Medical Association (WMA) Declaration of Helsinki - Ethical Principles for Medical Research Involving Human Subjects” adopted by the 18th WMA General Assembly, Helsinki, Finland, June 1964 and amended by the 59th WMA General Assembly, Seoul, South Korea, October 2008.
STATISTICS
Bioinformatics and Data Analysis
Raw files were converted to mascot generic files (mgf) using MSconvert [16] for downstream database searching in ProteinPilot (v.4; AB SCIEX, Foster City, CA, USA). All data generated were searched against the UniProt human database (downloaded November 11, 2011) using the Paragon algorithm [17], which is integrated into the ProteinPilot search engine. Search parameters were set as follows: 1) sample type: identification; 2) Cys alkylation: iodoacetamide; 3) instrument: Orbitrap/FT (1–3 ppm) or LTQ FT Ultra mass spectrometer (Thermo Scientific, Waltham, MA, USA) MS/MS; 4) special factors: none; 5) identification focus: none; 6) database: UniProt human; and 7) search effort: thorough identification. A 1% false discovery rate (FDR) for protein identifications was determined using the Posterior Error Probability algorithm integrated into ProteinPilot. Relative protein quantification was performed using label-free spectral counting, which compared the number of identified tandem mass spectra for the same protein across multiple data sets [18, 19].
RESULTS
Proteomic Analysis of FFPE Tissue Identified Several Hundred Proteins Using Mass Spectrometry-Based Proteomic Techniques
FFPE tissue from nine histologically-classified archived specimens (autoimmune pancreatitis, n=3; chronic pancreatitis, n=3; and pancreatic cancer, n=3) were subjected to mass spectrometry-based proteomic analysis, as depicted in Figure 1. The analysis of each specimen, was performed using a single 1.5 cm x 1 cm x 5 µm slice of FFPE tissue. Mass spectrometric analysis of all 9 FFPE specimens identified a total of 386 non-redundant proteins, which we list in Supplementary Table 1. A nonredundant protein is one that has been counted only once, regardless the number of specimens in which it has been identified.
Non-Redundant Proteins Were Exclusive to Each Pancreatic Disease Cohort
We summarize the number of non-redundant proteins identified in this study in Table 1. Considering each specimen individually, we identified 131, 141, and 103 non-redundant proteins in each autoimmune pancreatitis (AIP) specimen; 188, 138, and 153 non-redundant proteins in each chronic pancreatitis (CP) specimen; and 150, 190, and 140 non-redundant proteins in each pancreatic cancer (PC) specimen. We then merged the three lists of proteins identified in each cohort to determine the number of non-redundant proteins identified within each cohort, as listed in Table 1. The numbers of non-redundant proteins identified in each cohort were: 211 in autoimmune pancreatitis, 265 in chronic pancreatitis, and 280 in pancreatic cancer.
Table 1.
Summary of the number of proteins identified from FFPE tissue specimens by cohort. A total of 386 non-redundant proteins were identified in the study.
| Cohort | Specimen | No. of non-redundant identified proteins |
||
|---|---|---|---|---|
| In each specimen |
Within each cohort |
Exclusive to each cohort |
||
| AIP | #1 | 131 | 211 | 29 |
| #2 | 141 | |||
| #3 | 103 | |||
| CP | #1 | 188 | 265 | 53 |
| #2 | 138 | |||
| #3 | 153 | |||
| PC | #1 | 150 | 280 | 73 |
| #2 | 190 | |||
| #3 | 140 | |||
AIP: autoimmune pancreatitis; CP: chronic pancreatitis; PC: pancreatic cancer
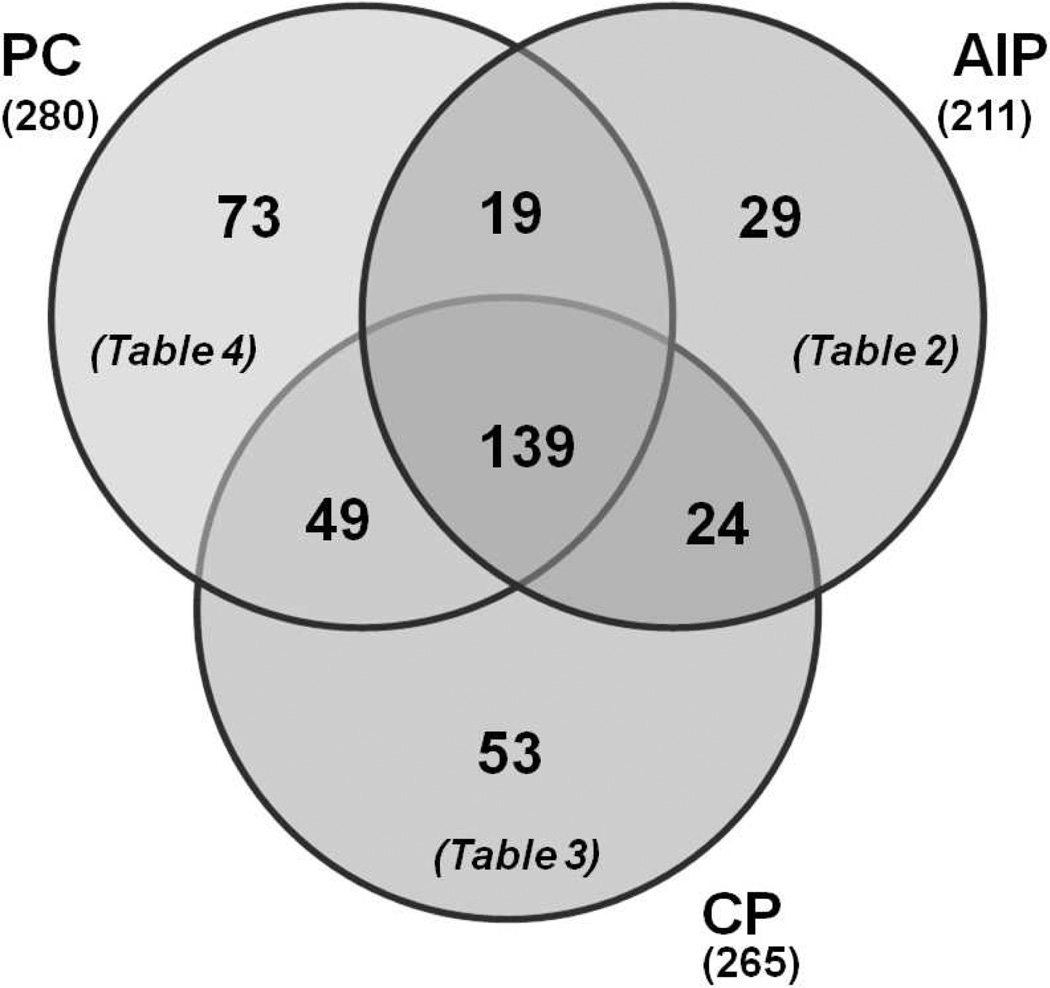
We then compared proteins among cohorts to identify those exclusive to each cohort and those common to the three cohorts (Figure 2). We identified 29 proteins exclusive to the autoimmune pancreatitis cohort (Table 2), 53 proteins exclusive to the chronic pancreatitis cohort (Table 3), and 73 proteins exclusive to the pancreatic cancer cohort (Table 4). Proteins identified exclusively in the autoimmune pancreatitis specimens include immunoglobulins and histocompatibility antigens. Epiplakin, protein disulfide-isomerase, and mucin 2 were among the proteins that were identified exclusively in the pancreatic cancer specimens. Hornerin and several ribosomal subunits were identified exclusively in the chronic pancreatitis specimens. Our analysis also identified 139 non-redundant proteins common to all three cohorts, 24 common to only autoimmune pancreatitis and chronic pancreatitis (Supplementary Table 2), 19 common to only autoimmune pancreatitis and pancreatic cancer (Supplementary Table 3) and 49 common to only pancreatic cancer and chronic pancreatitis (Supplementary Table 4).
Figure 2.
Distribution of proteins among the three cohorts. Venn diagrams showing unique and overlapping proteins among the three cohorts. Also indicated in the figure are references to the tables in which these sets of proteins are listed.
AIP: autoimmune pancreatitis; CP: chronic pancreatitis; PC: pancreatic cancer
Table 2.
Twenty-nine proteins identified exclusively in autoimmune pancreatitis and not in the other two cohorts, as illustrated in the Venn diagram in Figure 2.
| Name | UniProt ID | Spectral counts |
No. of specimens in which a protein was identified |
|||
|---|---|---|---|---|---|---|
| AIP#1 | AIP#2 | AIP#3 | Mean | |||
| 40S ribosomal protein S16 | P62249 | 0 | 20 | 0 | 7 | 1 |
| Actin-related protein 2/3 complex subunit 4 | P59998 | 22 | 0 | 0 | 7 | 1 |
| Adenylate kinase 2, mitochondrial | P54819 | 15 | 0 | 32 | 16 | 2 |
| Band 3 anion transport protein | P02730 | 0 | 32 | 0 | 11 | 1 |
| BCL2/adenovirus E1B 19 kDa protein-interacting protein 2 | Q12982 | 0 | 6 | 0 | 2 | 1 |
| Beta-1,4-galactosyltransferase 3 | O60512 | 27 | 0 | 0 | 9 | 1 |
| Clathrin light chain A | P09496 | 0 | 14 | 0 | 5 | 1 |
| Coronin-1A | P31146 | 15 | 0 | 0 | 5 | 1 |
| Coronin-1C | Q9ULV4 | 0 | 0 | 41 | 14 | 1 |
| Eosinophil peroxidase | P11678 | 99 | 0 | 0 | 33 | 1 |
| Heat shock protein beta-6 | O14558 | 0 | 0 | 35 | 12 | 1 |
| Histone H2A type 1 | P0C0S8 | 0 | 109 | 0 | 36 | 1 |
| Histone H2A type 1-J | Q99878 | 96 | 0 | 73 | 56 | 2 |
| Histone H2B type 1-C/E/F/G/I | P62807 | 156 | 0 | 0 | 52 | 1 |
| HLA class I histocompatibility antigen, A-30 alpha chain | P16188 | 27 | 0 | 0 | 9 | 1 |
| HLA class II histocompatibility antigen, DRB1-1 beta chain | P04229 | 32 | 0 | 0 | 11 | 1 |
| HLA class II histocompatibility antigen, DRB1-11 beta chain | P20039 | 20 | 0 | 0 | 7 | 1 |
| Ig gamma-1 chain C region | P01857 | 0 | 16 | 0 | 5 | 1 |
| Ig heavy chain V-III region BRO | P01766 | 0 | 12 | 0 | 4 | 1 |
| Ig heavy chain V-III region KOL | P01772 | 0 | 14 | 0 | 5 | 1 |
| Inner centromere protein | Q9NQS7 | 67 | 0 | 0 | 22 | 1 |
| Keratin, type I cuticular Ha3-I | O76009 | 20 | 0 | 0 | 7 | 1 |
| Keratin, type II cuticular Hb6 | O43790 | 42 | 0 | 0 | 14 | 1 |
| Laminin subunit alpha-5 | O15230 | 0 | 99 | 0 | 33 | 1 |
| Myosin-11 | P35749 | 0 | 0 | 185 | 62 | 1 |
| Synemin | O15061 | 0 | 0 | 147 | 49 | 1 |
| Tropomyosin beta chain | P07951 | 0 | 0 | 88 | 29 | 1 |
| Tubulin alpha-1B chain | P68363 | 0 | 0 | 32 | 11 | 1 |
| Ubiquitin-like modifier-activating enzyme 1 | P22314 | 45 | 0 | 0 | 15 | 1 |
AIP: autoimmune pancreatitis
Table 3.
Fifty-three proteins identified exclusively in chronic pancreatitis specimens and not in the other two cohorts, as illustrated in the Venn diagram in Figure 2.
| Name | UniProt ID | Spectral counts |
No. of specimens in which a protein was identified |
|||
|---|---|---|---|---|---|---|
| CP#1 | CP#2 | CP#3 | Mean | |||
| 28S ribosomal protein S29, mitochondrial | P51398 | 0 | 0 | 21 | 7 | 1 |
| 40S ribosomal protein S26 | P62854 | 0 | 0 | 18 | 6 | 1 |
| 60S ribosomal protein L10 | P27635 | 23 | 43 | 0 | 22 | 2 |
| 60S ribosomal protein L38 | P63173 | 0 | 0 | 11 | 4 | 1 |
| 60S ribosomal protein L5 | P46777 | 29 | 0 | 0 | 10 | 1 |
| 60S ribosomal protein L6 | Q02878 | 61 | 0 | 62 | 41 | 2 |
| 60S ribosomal protein L9 | P32969 | 21 | 43 | 14 | 26 | 3 |
| AF4/FMR2 family member 4 | Q9UHB7 | 0 | 0 | 80 | 27 | 1 |
| Annexin A6 | P08133 | 40 | 0 | 0 | 13 | 1 |
| Antigen peptide transporter 1 | Q03518 | 44 | 0 | 0 | 15 | 1 |
| Arf-GAP domain and FG repeats-containing protein 1 | P52594 | 11 | 0 | 16 | 9 | 2 |
| Beta-2-microglobulin | P61769 | 6 | 0 | 0 | 2 | 1 |
| BTB/POZ domain-containing protein KCTD12 | Q96CX2 | 15 | 0 | 0 | 5 | 1 |
| Cathepsin B | P07858 | 8 | 0 | 0 | 3 | 1 |
| Coagulation factor XIII A chain | P00488 | 25 | 0 | 0 | 8 | 1 |
| Complement C4-B | P0C0L5 | 0 | 0 | 80 | 27 | 1 |
| Cytosol aminopeptidase | P28838 | 27 | 0 | 0 | 9 | 1 |
| F-actin-capping protein subunit alpha-1 | P52907 | 32 | 0 | 0 | 11 | 1 |
| F-actin-capping protein subunit alpha-2 | P47755 | 23 | 0 | 0 | 8 | 1 |
| Glucosidase 2 subunit beta | P14314 | 48 | 0 | 0 | 16 | 1 |
| Heat shock-related 70 kDa protein 2 | P54652 | 0 | 0 | 48 | 16 | 1 |
| Histone H1.0 | P07305 | 0 | 0 | 43 | 14 | 1 |
| Histone H2A.J | Q9BTM1 | 0 | 89 | 0 | 30 | 1 |
| Histone H2B type 1-L | Q99880 | 0 | 0 | 160 | 53 | 1 |
| HLA class II histocompatibility antigen, DRB1-15 beta chain | P01911 | 21 | 0 | 0 | 7 | 1 |
| HLA class II histocompatibility antigen, DRB1-7 beta chain | P13761 | 23 | 0 | 0 | 8 | 1 |
| Hornerin | Q86YZ3 | 0 | 77 | 68 | 48 | 2 |
| Ig heavy chain V-I region EU | P01742 | 19 | 0 | 0 | 6 | 1 |
| Ig kappa chain V-I region HK101 | P01601 | 25 | 0 | 0 | 8 | 1 |
| Ig kappa chain V-III region POM | P01624 | 15 | 0 | 0 | 5 | 1 |
| Insulin | P01308 | 0 | 22 | 0 | 7 | 1 |
| Junction plakoglobin | P14923 | 0 | 43 | 0 | 14 | 1 |
| Lithostathine-1-beta | P48304 | 0 | 0 | 5 | 2 | 1 |
| Lysyl oxidase homolog 1 | Q08397 | 0 | 48 | 0 | 16 | 1 |
| Mimecan | P20774 | 0 | 36 | 0 | 12 | 1 |
| Moesin | P26038 | 29 | 0 | 0 | 10 | 1 |
| Neurosecretory protein VGF | O15240 | 0 | 31 | 0 | 10 | 1 |
| Non-secretory ribonuclease | P10153 | 0 | 0 | 7 | 2 | 1 |
| Pancreatic alpha-amylase | P04746 | 0 | 29 | 0 | 10 | 1 |
| Pancreatic triacylglycerol lipase | P16233 | 0 | 0 | 27 | 9 | 1 |
| Peroxiredoxin-5, mitochondrial | P30044 | 11 | 0 | 0 | 4 | 1 |
| Phosphoglycerate mutase 1 | P18669 | 13 | 0 | 0 | 4 | 1 |
| Probable ATP-dependent RNA helicase DDX17 | Q92841 | 0 | 0 | 55 | 18 | 1 |
| Protein canopy homolog 2 | Q9Y2B0 | 6 | 0 | 0 | 2 | 1 |
| Protein disulfide-isomerase A6 | Q15084 | 25 | 0 | 0 | 8 | 1 |
| Putative 40S ribosomal protein S26-like 1 | Q5JNZ5 | 11 | 0 | 0 | 4 | 1 |
| Putative 60S ribosomal protein L39-like 5 | Q59GN2 | 8 | 0 | 0 | 3 | 1 |
| Serum amyloid A protein | P02735 | 0 | 0 | 16 | 5 | 1 |
| Serum amyloid P-component | P02743 | 0 | 14 | 0 | 5 | 1 |
| Sulfatase-modifying factor 2 | Q8NBJ7 | 0 | 19 | 16 | 12 | 2 |
| Thioredoxin domain-containing protein 5 | Q8NBS9 | 23 | 0 | 0 | 8 | 1 |
| Tryptophan-tRNA ligase, cytoplasmic | P23381 | 13 | 0 | 0 | 4 | 1 |
| Tubulin alpha-1A chain | Q71U36 | 0 | 38 | 0 | 13 | 1 |
CP: chronic pancreatitis
Table 4.
Seventy-tree proteins identified exclusively in pancreatic cancer specimens and not in the other two cohorts, as illustrated in the Venn diagram in Figure 2.
| Name | UniProt ID | Spectral counts |
No. of specimens in which a protein was identified |
|||
|---|---|---|---|---|---|---|
| PC#1 | PC#2 | PC#3 | Mean | |||
| 14-3-3 protein epsilon | P62258 | 0 | 31 | 0 | 10 | 1 |
| 40S ribosomal protein S18 | P62269 | 0 | 27 | 0 | 9 | 1 |
| 40S ribosomal protein S24 | P62847 | 15 | 49 | 0 | 21 | 2 |
| 40S ribosomal protein S3 | P23396 | 0 | 27 | 0 | 9 | 1 |
| 40S ribosomal protein SA | P08865 | 20 | 25 | 0 | 15 | 2 |
| 60S acidic ribosomal protein P0 | P05388 | 0 | 40 | 0 | 13 | 1 |
| 60S acidic ribosomal protein P2 | P05387 | 0 | 4 | 0 | 1 | 1 |
| 60S ribosomal protein L37a | P61513 | 24 | 0 | 0 | 8 | 1 |
| Acyl-coenzyme A synthetase ACSM6, mitochondrial | Q6P461 | 18 | 0 | 0 | 6 | 1 |
| Aldehyde dehydrogenase family 16 member A1 | Q8IZ83 | 24 | 0 | 0 | 8 | 1 |
| Aldehyde dehydrogenase, mitochondrial | P05091 | 22 | 0 | 0 | 7 | 1 |
| Alpha-1-acid glycoprotein 1 | P02763 | 0 | 0 | 12 | 4 | 1 |
| Alpha-1-antichymotrypsin | P01011 | 0 | 0 | 16 | 5 | 1 |
| Anterior gradient protein 2 homolog | O95994 | 16 | 0 | 0 | 5 | 1 |
| Anterior gradient protein 3 homolog | Q8TD06 | 0 | 0 | 5 | 2 | 1 |
| Apolipoprotein E | P02649 | 0 | 0 | 28 | 9 | 1 |
| Azurocidin | P20160 | 27 | 29 | 0 | 19 | 2 |
| Band 4.1-like protein 2 | O43491 | 31 | 0 | 0 | 10 | 1 |
| Carcinoembryonic antigen-related cell adhesion molecule 5 | P06731 | 0 | 0 | 16 | 5 | 1 |
| Catalase | P04040 | 13 | 0 | 0 | 4 | 1 |
| Catenin alpha-2 | P26232 | 67 | 0 | 0 | 22 | 1 |
| Chymotrypsin-C | Q99895 | 0 | 13 | 0 | 4 | 1 |
| Complement C4-A | P0C0L4 | 0 | 0 | 56 | 19 | 1 |
| Complement component C9 | P02748 | 0 | 0 | 21 | 7 | 1 |
| Dolichyl-diphosphooligosaccharide-protein glycosyltransferase 1 | P04843 | 0 | 47 | 0 | 16 | 1 |
| ELAV-like protein 1 | Q15717 | 24 | 0 | 0 | 8 | 1 |
| Epiplakin | P58107 | 165 | 0 | 171 | 112 | 2 |
| Erythrocyte band 7 integral membrane protein | P27105 | 0 | 0 | 28 | 9 | 1 |
| Fatty acid synthase | P49327 | 45 | 0 | 0 | 15 | 1 |
| Fatty acid-binding protein, liver | P07148 | 13 | 0 | 0 | 4 | 1 |
| Filamin-B | O75369 | 51 | 0 | 0 | 17 | 1 |
| Heat shock protein HSP 90-beta | P08238 | 62 | 0 | 0 | 21 | 1 |
| Heterogeneous nuclear ribonucleoprotein Q | O60506 | 0 | 51 | 0 | 17 | 1 |
| Heterogeneous nuclear ribonucleoproteins C1/C2 | P07910 | 29 | 0 | 0 | 10 | 1 |
| Histone H2A type 2-A | Q6FI13 | 0 | 87 | 0 | 29 | 1 |
| Histone H2A.Z | P0C0S5 | 34 | 0 | 0 | 11 | 1 |
| Histone H2B type 1-O | P23527 | 140 | 0 | 0 | 47 | 1 |
| Histone H2B type 2-F | Q5QNW6 | 0 | 0 | 185 | 62 | 1 |
| HLA class I histocompatibility antigen, Cw-7 alpha chain | P10321 | 22 | 0 | 0 | 7 | 1 |
| HLA class II histocompatibility antigen, DRB1-9 beta chain | Q9TQE0 | 15 | 0 | 0 | 5 | 1 |
| IgGFc-binding protein | Q9Y6R7 | 42 | 0 | 0 | 14 | 1 |
| Importin subunit beta-1 | Q14974 | 22 | 0 | 0 | 7 | 1 |
| Integrin alpha-M | P11215 | 0 | 0 | 68 | 23 | 1 |
| Keratin, type I cuticular Ha6 | O76013 | 31 | 0 | 0 | 10 | 1 |
| Keratin, type I cytoskeletal 16 | P08779 | 53 | 0 | 0 | 18 | 1 |
| Keratin, type II cuticular Hb5 | P78386 | 33 | 0 | 0 | 11 | 1 |
| Keratin, type II cytoskeletal 7 | P08729 | 83 | 60 | 105 | 83 | 3 |
| Lactotransferrin | P02788 | 22 | 0 | 37 | 20 | 2 |
| L-lactate dehydrogenase A chain | P00338 | 25 | 0 | 30 | 18 | 2 |
| Mucin-2 | Q02817 | 34 | 0 | 0 | 11 | 1 |
| Mucin-5B | Q9HC84 | 0 | 0 | 91 | 30 | 1 |
| Myeloblastin | P24158 | 18 | 0 | 9 | 9 | 2 |
| Myosin-14 | Q7Z406 | 123 | 0 | 0 | 41 | 1 |
| Nicotinate phosphoribosyltransferase | Q6XQN6 | 7 | 0 | 0 | 2 | 1 |
| Outer dense fiber protein 3 | Q96PU9 | 0 | 0 | 14 | 5 | 1 |
| PDZ and LIM domain protein 1 | O00151 | 18 | 0 | 0 | 6 | 1 |
| Phenylalanine-tRNA ligase alpha chain | Q9Y285 | 0 | 22 | 0 | 7 | 1 |
| Protein disulfide-isomerase A3 | P30101 | 31 | 47 | 0 | 26 | 2 |
| Protein DJ-1 | Q99497 | 0 | 27 | 0 | 9 | 1 |
| Protein S100-A8 | P05109 | 0 | 0 | 9 | 3 | 1 |
| Proteolipid protein 2 | Q04941 | 0 | 4 | 0 | 1 | 1 |
| Putative trypsin-6 | Q8NHM4 | 0 | 0 | 47 | 16 | 1 |
| Retinal dehydrogenase 1 | P00352 | 0 | 31 | 0 | 10 | 1 |
| Solute carrier family 12 member 2 | P55011 | 51 | 0 | 0 | 17 | 1 |
| Spectrin beta chain, brain 1 | Q01082 | 71 | 0 | 0 | 24 | 1 |
| Staphylococcal nuclease domain-containing protein 1 | Q7KZF4 | 0 | 65 | 0 | 22 | 1 |
| Superoxide dismutase | P00441 | 9 | 0 | 0 | 3 | 1 |
| Translocon-associated protein subunit delta | P51571 | 0 | 9 | 0 | 3 | 1 |
| Transthyretin | P02766 | 0 | 0 | 9 | 3 | 1 |
| Tripeptidyl-peptidase 1 | O14773 | 11 | 0 | 19 | 10 | 2 |
| Trypsin-3 | P35030 | 0 | 63 | 0 | 21 | 1 |
| Tubulin beta-4B chain | P68371 | 45 | 25 | 0 | 23 | 2 |
| Tubulin-specific chaperone A | O75347 | 9 | 0 | 0 | 3 | 1 |
PC: pancreatic cancer
DISCUSSION
We have successfully applied LC-MS/MS analysis to FFPE tissue of autoimmune pancreatitis, chronic pancreatitis, and pancreatic cancer specimens. In total, we have identified 386 non-redundant proteins among all 9 specimens. By comparing these proteins, we have identified those that were found exclusively in autoimmune pancreatitis (29 proteins), chronic pancreatitis (53 proteins), and pancreatic cancer (73 proteins). A previously published study analyzed pancreatic cancer FFPE tissue using a Liquid Tissue (Expression Pathology, Rockville, MD, USA) workflow for a global proteomics analysis [20]. FFPE tissue processing techniques and mass spectrometry strategies analogous to those which we performed have been used previously to study tissues of other organ systems, including prostate [21], cochlea [14], liver [13, 22], glioblastoma [23], renal carcinoma [24], kidney glomeruli [25], mesenchymal tissue [12], and B-cell lymphoma cells [26], as well as our own recent study comparing pancreatic cancer and chronic pancreatitis [8]. We, however, present the first comparative LC-MS/MS analysis of FFPE pancreas tissue from individuals with autoimmune pancreatitis, chronic pancreatitis, and pancreatic cancer.
Our mass spectrometry-based analysis revealed several proteins as exclusive to specimens from particular cohorts and which may merit further investigation. Genetic and immunologic factors impact the pathogenesis of certain pancreatic diseases [27], and several proteins identified exclusively in a particular disease cohort may directly reflect the presence of that disease process. Histocompatibility antigen (HLA) DRB1-1 beta chain (HLA-DRB1 beta chain) may be correlated to autoimmune pancreatitis. Studies have demonstrated that HLA class II DRB1-1 beta chain allele (HLA-DRB1*0401) may have a protective role in autoimmune pancreatitis [28], while others have investigated allelic polymorphisms in the genes of the major histocompatibility complex (MHC) region, in particular HLA-DRB1*0401, suggest this allele is a susceptibility factor in patients with chronic pancreatitis [27]. Also identified exclusively in autoimmune pancreatitis, Ig gamma-1 chain (IGHG1) has been shown to down-regulate the cytotoxic activity of natural killer cells through inhibition of antibody-dependent cellular cytotoxicity [29].
Proteins identified exclusively in pancreatic cancer samples included: epiplakin, mucin2 (MUC2), and protein disulfide-isomerase A3. In a mouse model study, epiplakin was found in centroacinar cells and duct cells in the adult pancreas. Its presence has been observed in pancreatic intraepithelial neoplasia, previously identified as pancreatic ductal adenocarcinoma precursor lesions [30]. Mucins are large multifunctional glycoproteins that play an important role in the ductal structures within the pancreas. MUC2 has been shown to be expressed mainly in intraductal papillar mucinous neoplasms (IPMNs), but is occasionally also detected in aggressive pancreatic tumors [31]. Protein disulfide-isomerase A3 (ERp57) is a member of the protein disulfide-isomerase (PDI) family that is involved in the cleavage of disulfide bonds between cysteine residues. This enzyme has been implicated mainly in Alzheimer’s disease, but also has been associated with the pathogenesis of cancers, including pancreatic cancer [32]. Hornerin and several ribosomal subunits were identified exclusively in the chronic pancreatitis specimens; however, there is currently no other evidence that these proteins have a role in the development or progression of chronic pancreatitis. Following orthogonal validation, the proteins described above maybe potential diagnostic biomarkers of their respective diseases, as well as targets for directed investigations into disease pathogenesis and progression. Further biological and biochemical studies (e.g., immunohistochemistry using larger cohorts), must be performed to analyze further these selected proteins.
A comparison of frozen verses FFPE tissue would reveal further insight into the utility of FFPE specimens in pancreatic research. However, standardized conditions for frozen tissue are not well established. Such conditions include the initial freezing state (e.g., flash-freezing in liquid nitrogen or dry ice) or procedures for long-term storage (e.g., storage in liquid nitrogen or −80°C). In contrast, FFPE specimens are typically processed with standardized methodologies and are inherently stable at room temperature or below. Studies have also indicated that fresh-frozen tissue do not properly maintain cellular morphology [33, 34]. Moreover, recent studies of follicular lymphoma [11], renal carcinoma [24], ear canal [14], breast cancer [35], and liver [15], have identified similar numbers of proteins using either FFPE tissue or frozen tissue. However, no such comparison has been performed using pancreatic tissue. A systematic study of a statistically significant number of FFPE and frozen pancreatic tissue may provide further evidence as to the benefits of one method over the other for mass spectrometry-based proteomic analyses.
Diagnostic specificity and relatively standardized specimen preservation renders FFPE tissue analysis superior to other organ-specific biomarker discovery strategies that typically analyze body fluids, such as blood derivatives, urine, or proximal fluids [36, 37, 38, 39]. As such, recent efforts have attempted to further standardize collection, handling, and storage of various body fluid specimens [36, 37, 40, 41, 42, 43, 44, 45, 46]. Nevertheless, meaningful inter-laboratory data comparisons have historically been limited due to differences in specimen handling. Moreover, the availability of vast archives of patient samples with clearly defined clinical histories, diagnoses, and outcomes enhances the robustness and utility of FFPE-based investigations. The value of tissuebased analysis is not limited to biomarker discovery; thorough analysis may also identify therapeutic targets and improve understanding of the molecular basis of pathogenesis, pathophysiology and clinical course. In addition, miRNA and/or mRNA studies can also be performed on FFPE tissue [47, 48, 49, 50]. Data from these studies can be compared to proteomics-based FFPE analysis, providing insight into organ- or diseasespecific translational regulation. However, to achieve the maximum analytical depth, more efficient protein (or peptide) extraction protocols must be developed and potential specimen preparation artifacts, such as amino acid residue modifications, must be reduced or eliminated.
The identification of a greater number of proteins than presented herein would be possible by overcoming procedural limitations. For example, harsh FFPE tissue preparation conditions can limit protein identification. More specifically, irreversible formalin-induced intra- and intermolecular crosslinking in FFPE specimens often hinders the solubility of proteins, complicating the extraction of these proteins and, potentially, larger peptides [51]. During tissue fixation, formalin adds a methylene hydrate group to the side chain of certain amino acids resulting in a crosslinking methylene bridge (both inter- and intra-peptide) which will likely prevent peptide identification [52, 53]. Moreover, chemical modifications of side chain moieties of lysyl, arginyl, tyrosyl, histidyl, and seryl are common during FFPE preparation [33], and further limit protein identifications. In addition, in nucleic acid extraction from FFPE tissue, extensive periods of fixation significantly reduces RNA extraction [54] and typically results in DNA fragmentation [55], and by extension, such effects may also be prevalent in proteins. As FFPE sections generally consist of different cell types, sample heterogeneity may be an issue. If a more homogeneous sample is needed, laser-capture microdissection (LCM) and serial sectioning may be implemented for such analyses [25, 56, 57]. Cellular microdissection decreases sample complexity thereby increasing analytical depth. Improvements in sample fixation and analysis may overcome these obstacles, and greatly increase the depth of the proteomic analysis of wellpreserved FFPE specimens. In addition, future MSbased studies will achieve greater depth of quantitative proteomic analysis by exploiting the higher resolution, high mass accuracy and higher scanning speeds possible with emerging mass spectrometry platforms, such as the Orbitrap Elite [58] and the Q-Exactive [59] (Thermo Scientific, Waltham, MA, USA).
CONCLUSION
We have shown that differentially-expressed proteins can be identified among FFPE tissues specimens originating from individuals with autoimmune pancreatitis, chronic pancreatitis, and pancreatic cancer. With improvements in experimental methods, LC-MS/MS-based proteomic analysis of FFPE pancreatic tissue specimens may provide novel therapeutic targets and/or a means of high-throughput validation of current diagnostic biomarkers. With further investigation, this knowledge may result in methods enabling the discrimination between autoimmune pancreatitis and pancreatic cancer, which represents an unmet medical need. Given the plethora of FFPE specimen archives, large databases of disease-specific proteins may be assembled for validation via orthogonal methods, such as immunohistochemistry studies. The methodology described herein, in addition to the proteins which we have identified as differentially expressed, may offer a scaffold upon which to build further FFPE-based studies of pancreatic diseases. The work which we present demonstrates the potential of using LCMS/MS for the analysis of archived FFPE pancreatic tissue, particularly to differentiate autoimmune pancreatitis and pancreatic cancer, and provides a basis upon which biomarker studies can be developed further.
Supplementary Material
Acknowledgments
Funds were provided by the following NIH grants: 1 F32 DK085835-01A1 (J.A.P.), 1 R21 DK081703-01A2 (D.L.C.) and 5 P30 DK034854-24 (Harvard Digestive Diseases Center; D.L.C.). In addition, we would like to thank the Burrill family for their generous support through the Burrill Research Grant. We would also like to thank members of the Steen Lab at Children’s Hospital Boston, MA, USA, in particular John FK Sauld and Ali Ghoulidi, as well as Linda S Lee from the Center for Pancreatic Disease at Brigham and Women’s Hospital, Boston, MA, USA for their assistance.
Abbreviations
- AIP
autoimmune pancreatitis
- CP
chronic pancreatitis
- DTT
dithiotreitol
- FFPE
formalin-fixed paraffin-embedded
- LC-MS/MS
liquid chromatography coupled with tandem mass spectrometry
- PC
pancreatic cancer
Footnotes
Conflict of interest The authors declare no competing interests
References
- 1.NIH, Opportunities and Challenges in Digestive Diseases Research: Recommendations of the National Commission on Digestive Diseases. U.S. Department of Health and Human Services. National Institutes of Health. 2009:159–167. NIH publ. no. 08-6514.
- 2.Nishimori I, Tamakoshi A, Otsuki M. Prevalence of autoimmune pancreatitis in Japan from a nationwide survey in 2002. J Gastroenterol. 2007;42(Suppl 18):6–8. doi: 10.1007/s00535-007-2043-y. [DOI] [PubMed] [Google Scholar]
- 3.Pearson RK, Longnecker DS, Chari ST, Smyrk TC, Okazaki K, Frulloni L, Cavallini G. Controversies in clinical pancreatology: autoimmune pancreatitis: does it exist? Pancreas. 2003;27:1–13. doi: 10.1097/00006676-200307000-00001. [DOI] [PubMed] [Google Scholar]
- 4.Nishimori I, Tamakoshi A, Kawa S, Tanaka S, Takeuchi K, Kamisawa T, Saisho H, Hirano K, Okamura K, Yanagawa N, Otsuki M. Influence of steroid therapy on the course of diabetes mellitus in patients with autoimmune pancreatitis: findings from a nationwide survey in Japan. Pancreas. 2006;32:244–248. doi: 10.1097/01.mpa.0000202950.02988.07. [DOI] [PubMed] [Google Scholar]
- 5.Finkelberg DL, Sahani D, Deshpande V, Brugge WR. Autoimmune pancreatitis. The New England journal of medicine. 2006;355:2670–2676. doi: 10.1056/NEJMra061200. [DOI] [PubMed] [Google Scholar]
- 6.Pannala R, Chari ST. Autoimmune pancreatitis. Curr Opin Gastroenterol. 2008;24:591–596. doi: 10.1097/MOG.0b013e32830b10d2. [DOI] [PubMed] [Google Scholar]
- 7.Law R, Bronner M, Vogt D, Stevens T. Autoimmune pancreatitis: a mimic of pancreatic cancer. Cleveland Clinic journal of medicine. 2009;76:607–615. doi: 10.3949/ccjm.76a.09039. [DOI] [PubMed] [Google Scholar]
- 8.Paulo JA, Lee LS, Banks PA, Steen H, Conwell DL. Proteomic analysis of formalin-fixed paraffin-embedded pancreatic tissue using liquid chromatography tandem mass spectrometry. Pancreas. 2012;41:175–185. doi: 10.1097/MPA.0b013e318227a6b7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi SR, Cote RJ, Taylor CR. Antigen retrieval techniques: current perspectives. J Histochem Cytochem. 2001;49:931–937. doi: 10.1177/002215540104900801. [DOI] [PubMed] [Google Scholar]
- 10.Steen H, Mann M. The ABC’s (and XYZ’s) of peptide sequencing. Nature reviews. 2004;5:699–711. doi: 10.1038/nrm1468. [DOI] [PubMed] [Google Scholar]
- 11.Reimel BA, Pan S, May DH, Shaffer SA, Goodlett DR, McIntosh MW, Yerian LM, Bronner MP, Chen R, Brentnall TA. Proteomics on Fixed Tissue Specimens - A Review. Curr Proteomics. 2009;6:63–69. doi: 10.2174/157016409787847420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balgley BM, Guo T, Zhao K, Fang X, Tavassoli FA, Lee CS. Evaluation of archival time on shotgun proteomics of formalinfixed and paraffin-embedded tissues. Journal of proteome research. 2009;8:917–925. doi: 10.1021/pr800503u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang X, Feng S, Tian R, Ye M, Zou H. Development of efficient protein extraction methods for shotgun proteome analysis of formalin-fixed tissues. Journal of proteome research. 2007;6:1038–1047. doi: 10.1021/pr0605318. [DOI] [PubMed] [Google Scholar]
- 14.Palmer-Toy DE, Krastins B, Sarracino DA, Nadol JB, Jr, Merchant SN. Efficient method for the proteomic analysis of fixed and embedded tissues. Journal of proteome research. 2005;4:2404–2411. doi: 10.1021/pr050208p. [DOI] [PubMed] [Google Scholar]
- 15.Xu H, Yang L, Wang W, Shi SR, Liu C, Liu Y, Fang X, Taylor CR, Lee CS, Balgley BM. Antigen retrieval for proteomic characterization of formalin-fixed and paraffin-embedded tissues. Journal of proteome research. 2008;7:1098–1108. doi: 10.1021/pr7006768. [DOI] [PubMed] [Google Scholar]
- 16.Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24:2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shilov IV, Seymour SL, Patel AA, Loboda A, Tang WH, Keating SP, Hunter CL, Nuwaysir LM, Schaeffer DA. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics. 2007;6:1638–1655. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Dong MQ, Venable JD, Au N, Xu T, Park SK, Cociorva D, Johnson JR, Dillin A, Yates JR., 3rd Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science. 2007;317:660–663. doi: 10.1126/science.1139952. [DOI] [PubMed] [Google Scholar]
- 19.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 20.Cheung W, Darfler MM, Alvarez H, Hood BL, Conrads TP, Habbe N, Krizman DB, Mollenhauer J, Feldmann G, Maitra A. Application of a global proteomic approach to archival precursor lesions: deleted in malignant brain tumors 1 and tissue transglutaminase 2 are upregulated in pancreatic cancer precursors. Pancreatology : official journal of the International Association of Pancreatology. 2008;8:608–616. doi: 10.1159/000161012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hood BL, Darfler MM, Guiel TG, Furusato B, Lucas DA, Ringeisen BR, Sesterhenn IA, Conrads TP, Veenstra TD, Krizman DB. Proteomic analysis of formalin-fixed prostate cancer tissue. Mol Cell Proteomics. 2005;4:1741–1753. doi: 10.1074/mcp.M500102-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Ostasiewicz P, Zielinska DF, Mann M, Wisniewski JR. Proteome, Phosphoproteome, and N-Glycoproteome Are Quantitatively Preserved in Formalin-Fixed Paraffin-Embedded Tissue and Analyzable by High-Resolution Mass Spectrometry. J Proteome Res. 2010 doi: 10.1021/pr100234w. [DOI] [PubMed] [Google Scholar]
- 23.Guo T, Wang W, Rudnick PA, Song T, Li J, Zhuang Z, Weil RJ, DeVoe DL, Lee CS, Balgley BM. Proteome analysis of microdissected formalin-fixed and paraffin-embedded tissue specimens. J Histochem Cytochem. 2007;55:763–772. doi: 10.1369/jhc.7A7177.2007. [DOI] [PubMed] [Google Scholar]
- 24.Shi SR, Liu C, Balgley BM, Lee C, Taylor CR. Protein extraction from formalin-fixed, paraffin-embedded tissue sections: quality evaluation by mass spectrometry. J Histochem Cytochem. 2006;54:739–743. doi: 10.1369/jhc.5B6851.2006. [DOI] [PubMed] [Google Scholar]
- 25.Waanders LF, Chwalek K, Monetti M, Kumar C, Lammert E, Mann M. Quantitative proteomic analysis of single pancreatic islets. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18902–18907. doi: 10.1073/pnas.0908351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crockett DK, Lin Z, Vaughn CP, Lim MS, Elenitoba-Johnson KS. Identification of proteins from formalin-fixed paraffinembedded cells by LC-MS/MS. Lab Invest. 2005;85:1405–1415. doi: 10.1038/labinvest.3700343. [DOI] [PubMed] [Google Scholar]
- 27.Cavestro GM, Frulloni L, Neri TM, Seghini P, Nouvenne A, Zanetti A, Bovo P, Di Mario F, Okolicsanyi L, Cavallini G. Association of HLA-DRB1*0401 allele with chronic pancreatitis. Pancreas. 2003;26:388–391. doi: 10.1097/00006676-200305000-00013. [DOI] [PubMed] [Google Scholar]
- 28.Freitag TL, Cham C, Sung HH, Beilhack GF, Durinovic-Bello I, Patel SD, Bronson RT, Schuppan D, Sonderstrup G. Human risk allele HLA-DRB1*0405 predisposes class II transgenic Ab0 NOD mice to autoimmune pancreatitis. Gastroenterology. 2010;139:281–291. doi: 10.1053/j.gastro.2010.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X, Ni R, Chen J, Liu Z, Xiao M, Jiang F, Lu C. The presence of IGHG1 in human pancreatic carcinomas is associated with immune evasion mechanisms. Pancreas. 2011;40:753–761. doi: 10.1097/MPA.0b013e318213d51b. [DOI] [PubMed] [Google Scholar]
- 30.Yoshida T, Shiraki N, Baba H, Goto M, Fujiwara S, Kume K, Kume S. Expression patterns of epiplakin1 in pancreas, pancreatic cancer and regenerating pancreas. Genes Cells. 2008;13:667–678. doi: 10.1111/j.1365-2443.2008.01196.x. [DOI] [PubMed] [Google Scholar]
- 31.Levi E, Klimstra DS, Andea A, Basturk O, Adsay NV. MUC1 and MUC2 in pancreatic neoplasia. J Clin Pathol. 2004;57:456–462. doi: 10.1136/jcp.2003.013292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao Z, Liu H, Wang X, Li Z. Separation and identification of HSP-associated protein complexes from pancreatic cancer cell lines using 2D CN/SDS-PAGE coupled with mass spectrometry. J Biomed Biotechnol. 2011;2011:193052. doi: 10.1155/2011/193052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fox CH, Johnson FB, Whiting J, Roller PP. Formaldehyde fixation. J Histochem Cytochem. 1985;33:845–853. doi: 10.1177/33.8.3894502. [DOI] [PubMed] [Google Scholar]
- 34.McKenna RJ, Jr, Houck WV. New approaches to the minimally invasive treatment of lung cancer. Curr Opin Pulm Med. 2005;11:282–286. doi: 10.1097/01.mcp.0000166589.08880.44. [DOI] [PubMed] [Google Scholar]
- 35.Becker KF, Schott C, Hipp S, Metzger V, Porschewski P, Beck R, Nährig J, Becker I, Höfler H. Quantitative protein analysis from formalin-fixed tissues: implications for translational clinical research and nanoscale molecular diagnosis. The Journal of Pathology. 2007;211:370–378. doi: 10.1002/path.2107. [DOI] [PubMed] [Google Scholar]
- 36.Thongboonkerd V. Proteomics of human body fluids : principles, methods, and applications. Totowa, N.J: Humana Press; 2007. [Google Scholar]
- 37.Decramer S, Gonzalez de Peredo A, Breuil B, Mischak H, Monsarrat B, Bascands JL, Schanstra JP. Urine in clinical proteomics. Mol Cell Proteomics. 2008;7:1850–1862. doi: 10.1074/mcp.R800001-MCP200. [DOI] [PubMed] [Google Scholar]
- 38.Good DM, Thongboonkerd V, Novak J, Bascands JL, Schanstra JP, Coon JJ, Dominiczak A, Mischak H. Body fluid proteomics for biomarker discovery: lessons from the past hold the key to success in the future. J Proteome Res. 2007;6:4549–4555. doi: 10.1021/pr070529w. [DOI] [PubMed] [Google Scholar]
- 39.Paik YK, Kim H, Lee EY, Kwon MS, Cho SY. Overview and introduction to clinical proteomics. Methods Mol Biol. 2008;428:1–31. doi: 10.1007/978-1-59745-117-8_1. [DOI] [PubMed] [Google Scholar]
- 40.Barratt J, Topham P. Urine proteomics: the present and future of measuring urinary protein components in disease. Cmaj. 2007;177:361–368. doi: 10.1503/cmaj.061590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hortin GL, Sviridov D. Diagnostic potential for urinary proteomics. Pharmacogenomics. 2007;8:237–255. doi: 10.2217/14622416.8.3.237. [DOI] [PubMed] [Google Scholar]
- 42.Muller H, Brenner H. Urine markers as possible tools for prostate cancer screening: review of performance characteristics and practicality. Clinical chemistry. 2006;52:562–573. doi: 10.1373/clinchem.2005.062919. [DOI] [PubMed] [Google Scholar]
- 43.Munro NP, Cairns DA, Clarke P, Rogers M, Stanley AJ, Barrett JH, Harnden P, Thompson D, Eardley I, Banks RE, Knowles MA. Urinary biomarker profiling in transitional cell carcinoma. International journal of cancer. 2006;119:2642–2650. doi: 10.1002/ijc.22238. [DOI] [PubMed] [Google Scholar]
- 44.Thongboonkerd V. Urinary proteomics: towards biomarker discovery, diagnostics and prognostics. Molecular bioSystems. 2008;4:810–815. doi: 10.1039/b802534g. [DOI] [PubMed] [Google Scholar]
- 45.Paulo JA, Lee LS, Wu B, Repas K, Banks PA, Steen H, Conwell D. Cytokine protein microarray analysis of pancreatic fluid in tandem with the endoscopic pancreatic function test (ePFT) Pancreatology, Submitted. 2010 [Google Scholar]
- 46.Tammen H. Specimen collection and handling: standardization of blood sample collection. Methods Mol Biol. 2008;428:35–42. doi: 10.1007/978-1-59745-117-8_2. [DOI] [PubMed] [Google Scholar]
- 47.Nonn L, Vaishnav A, Gallagher L, Gann PH. mRNA and micro-RNA expression analysis in laser-capture microdissected prostate biopsies: valuable tool for risk assessment and prevention trials. Exp Mol Pathol. 2010;88:45–51. doi: 10.1016/j.yexmp.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ikenaga N, Ohuchida K, Mizumoto K, Yu J, Fujita H, Nakata K, Ueda J, Sato N, Nagai E, Tanaka M. S100A4 mRNA is a diagnostic and prognostic marker in pancreatic carcinoma. J Gastrointest Surg. 2009;13:1852–1858. doi: 10.1007/s11605-009-0978-4. [DOI] [PubMed] [Google Scholar]
- 49.Liu A, Tetzlaff MT, Vanbelle P, Elder D, Feldman M, Tobias JW, Sepulveda AR, Xu X. MicroRNA expression profiling outperforms mRNA expression profiling in formalin-fixed paraffin-embedded tissues. Int J Clin Exp Pathol. 2009;2:519–527. [PMC free article] [PubMed] [Google Scholar]
- 50.Gjerdrum LM, Abrahamsen HN, Villegas B, Sorensen BS, Schmidt H, Hamilton-Dutoit SJ. The influence of immunohistochemistry on mRNA recovery from microdissected frozen and formalin-fixed, paraffin-embedded sections. Diagn Mol Pathol. 2004;13:224–233. doi: 10.1097/01.pdm.0000134779.45353.d6. [DOI] [PubMed] [Google Scholar]
- 51.Ahram M, Flaig MJ, Gillespie JW, Duray PH, Linehan WM, Ornstein DK, Niu S, Zhao Y, Petricoin EF, 3rd, Emmert-Buck MR, Evaluation of ethanol-fixed. paraffin-embedded tissues for proteomic applications. Proteomics. 2003;3:413–421. doi: 10.1002/pmic.200390056. [DOI] [PubMed] [Google Scholar]
- 52.Rait VK, Xu L, O’Leary TJ, Mason JT. Modeling formalin fixation and antigen retrieval with bovine pancreatic RNase A II. Interrelationship of cross-linking, immunoreactivity, and heat treatment. Lab Invest. 2004;84:300–306. doi: 10.1038/labinvest.3700041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rait VK, O’Leary TJ, Mason JT. Modeling formalin fixation and antigen retrieval with bovine pancreatic ribonuclease A: Istructural and functional alterations. Lab Invest. 2004;84:292–299. doi: 10.1038/labinvest.3700045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Benchekroun M, DeGraw J, Gao J, Sun L, von Boguslawsky K, Leminen A, Andersson LC, Heiskala M. Impact of fixative on recovery of mRNA from paraffin-embedded tissue. Diagn Mol Pathol. 2004;13:116–125. doi: 10.1097/00019606-200406000-00008. [DOI] [PubMed] [Google Scholar]
- 55.Romero RL, Juston AC, Ballantyne J, Henry BE. The applicability of formalin-fixed and formalin fixed paraffin embedded tissues in forensic DNA analysis. J Forensic Sci. 1997;42:708–714. [PubMed] [Google Scholar]
- 56.Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA, Liotta LA. Science (New York, N.Y) 274: 1996. Laser capture microdissection; pp. 998–1001. [DOI] [PubMed] [Google Scholar]
- 57.Patel V, Hood BL, Molinolo AA, Lee NH, Conrads TP, Braisted JC, Krizman DB, Veenstra TD, Gutkind JS. Proteomic analysis of laser-captured paraffin-embedded tissues: a molecular portrait of head and neck cancer progression. Clin Cancer Res. 2008;14:1002–1014. doi: 10.1158/1078-0432.CCR-07-1497. [DOI] [PubMed] [Google Scholar]
- 58.Michalski A, Damoc E, Lange O, Denisov E, Nolting D, Muller M, Viner R, Schwartz J, Remes P, Belford M, Dunyach JJ, Cox J, Horning S, Mann M, Makarov A. Ultra High Resolution Linear Ion Trap Orbitrap Mass Spectrometer (Orbitrap Elite) Facilitates Top Down LC MS/MS and Versatile Peptide Fragmentation Modes. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.O111.013698. O111 013698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Michalski A, Damoc E, Hauschild JP, Lange O, Wieghaus A, Makarov A, Nagaraj N, Cox J, Mann M, Horning S. Mass spectrometry-based proteomics using Q Exactive, a highperformance benchtop quadrupole Orbitrap mass spectrometer, Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.011015. M111 011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.